Abstract
Human embryonic stem cells provide an alternative to using human embryos for studying developmentally regulated gene expression. The co-expression of high levels of embryonic ε and fetal γ globin by the hESC-derived erythroblasts allows the interrogation of ε globin regulation at the transcriptional and epigenetic level which could only be attained previously by studying cell lines or transgenic mice. In this study, we compared the histone modifications across the β globin locus of the undifferentiated hESCs and hESC-, FL-, and mobilized PB CD34+ cells-derived erythroblasts, which have distinct globin expression patterns corresponding to their developmental stages. We demonstrated that the histone codes employed by the β globin locus are conserved throughout development. Furthermore, in spite of the close proximity of the ε globin promoter, as compared to the β or γ globin promoter, with the LCR, a chromatin loop was also formed between the LCR and the active ε globin promoter, similar to the loop that forms between the β or γ globin promoters and the LCR, in contrary to the previously proposed tracking mechanism.
Introduction
The human β globin locus, located on chromosome 11 and spanning roughly 100 kb, encodes five functional globin genes: ε, Gγ, Aγ, δ, and β, that are arranged in the order according to their developmental stage-specific expression which involves two hemoglobin switches, in response to the differential local oxygen pressure during development (reviewed in [1]). Embryonic ε globin gene is predominantly expressed by the transiently circulated, large, mostly nucleated primitive erythroblasts originating from the yolk sac. The first hemoglobin switch entails the emergence of small, enucleated definitive erythroid cells from the fetal liver (FL) at approximately 6 to 8 weeks of gestation expressing fetal Gγ and Aγ globin genes with their ε globin gene silenced. The second hemoglobin switch occurs around birth when the bone marrow becomes the major hematopoietic tissue and generates enucleated erythroid cells expressing mainly adult β globin. A regulatory domain named the locus control region (LCR) is located upstream of the ε globin. The LCR, characterized by 5 DNase I hypersensitive sites (HSs) each containing multiple transcriptional factor binding motifs, has been shown to participate in the regulation of hemoglobin expression.
The temporal and tissue-specific expression characteristics of the β globin locus have made it an attractive experimental model for studying the regulation of mammalian gene expression. An enormous amount of work has been done to elucidate the regulation of fetal to adult globin switching (reviewed in [2]), partly due to the fact that a better understanding of the silencing and reactivation of fetal globin may lead to therapeutic opportunities for the treatment of hemoglobinopathies such as beta thalassemia. In comparison, the regulation of embryonic ε globin expression has been studied less extensively. The difficulties in obtaining a sufficient number of primary cells expressing embryonic ε globin have largely limited the related research to employing K562 cells [3, 4], a human leukemia cell line that express both embryonic and fetal but not adult hemoglobins, and transgenic mice carrying human β globin locus [5, 6]. Recently, the establishment of methods allowing generation of a large quantity of embryonic globin-expressing erythroblasts from human embryonic stem cells (hESCs) lead to studies comparing the epigenetic landscapes of β globin locus of hESC-derived erythroid cells, FL, and bone marrow erythroblasts. It has been found that complex developmental patterns of histone modifications as well as the formation of extended DNA hypomethylation domains are associated with the human β-locus globin switch [7, 8]. In this study, we provide further evidence that a looping mechanism is associated with the expression of ε globin in these hESC-derived erythroblasts, just as it is associated with the expression of fetal γ globin in FL cells and the expression of adult β globin in adult peripheral blood (PB) CD34+ cells derived-erythroid cells.
Materials and Methods
hESC culture
hESC line H1 (NIH code WA01) was cultured as previously described [9]. Briefly, H1 was propagated on irradiated murine embryonic fibroblasts (MEFs) on gelatin coated tissue culture plates (BD falcon, San Jose, CA) in ES medium consisted of Dulbecco modified Eagle medium/F12 medium supplemented with 15% knock-out serum replacement, 1 mM sodium pyruvate (all 3 from Invitrogen, Carlsbad, CA), 0.1 mM β-mercaptoethanol (Sigma, St Louis, MO), 0.1 mM minimum essential media (MEM) nonessential amino acids (Mediatech, Herndon, VA), 1 × penicillin/streptomycin (Mediatech), and 2 ng/mL basic fibroblast growth factor (bFGF) (Peprotech, Rocky Hill, NJ). For MEF-free cultures, hESCs were transferred to tissue culture plates coated with matrigel (BD Biosciences, San Jose, CA) diluted 1:50 with PBS (Thermo Fisher Scientific, Waltham, MA) and cultured with MEF-conditioned ES medium (MEF-CM) for 3 generations. MEF-CM was prepared by incubating 50 mL of ES medium per 6 × 106 irradiated MEFs at 37°C, 5% CO2, in a humidified incubator for 3 days. bFGF was added to a final concentration of 4 ng/mL. These MEF-free hESCs were used for chromatin immunoprecipitation (ChIP) assays.
Erythroid differentiation
hESCs were induced to undergo hematopoietic differentiation via embryoid body (EB) formation [10]. Briefly, hESCs cultured with MEFs were harvested by gentle scraping and seeded onto ultra-low attachment plates (Corning, Acton, MA) in EB medium consisting of Iscove MEM (IMDM) (Mediatech), 15% ES-qualified FBS (Invitrogen), 10% protein-free hybridoma medium II (PFHM-II) (Invitrogen), 300 μg/mL iron-saturated human transferrin (Sigma), 50 μg/mL ascorbic acid (Sigma), 0.1 mM β-mercaptoethanol, 1 × penicillin/streptomycin, and 2 mM L-glutamine (Mediatech). After 7 days, EBs were dissociated into single cells and cultured in an erythroid-inducing medium composed of Stemline hematopoietic stem cell growth and expansion medium (Sigma) supplemented with 2 mML-glutamine, 1× penicillin/streptomycin, 0.1 mM MEM nonessential amino acids, 0.1 mM β-mercaptoethanol, 200 μg/ml iron-saturated transferrin,10 μM ethanolamine, 10 μg/ml insulin (all from Sigma), 6 U/ml erythropoietin, 50 ng/ml stem cell factor (both from Amgen, Thousand Oakes, CA), 20 ng/ml interleukin-3, 20 ng/ml interleukin-6, 40 ng/ml insulin-like growth factor-1, 10 ng/ml vascular endothelial growth factor (all from Peprotech, Rocky Hill, NJ), 5% protein free hybridoma medium, 1× insulin-transferrin-selenium (both from Invitrogen), and 1× EX-CYTE (Millipore, Billerica, MA). for 14 days. The purity of cells was confirmed using flow cytometry by staining cells with phycoerythrin (PE) conjugated anti-glycophorin-A (Gly-A) antibody (DAKO, Glostrup, Denmark). When the erythroid population comprised less than 93% of total cells, they were subjected to further enrichment using the MACS system per manufacturer’s instruction (Miltenyi Biotech, Auburn, CA).
CD34+ cells from healthy volunteer-donor mobilized PB and dissociated FL cells (50–100-day gestation) were cultured in the same erythroid-inducing medium to generate adult- and fetal-type erythroid cells. FL cells were obtained from the fetal tissue repository (University of Washington Birth Defects Research Laboratory) and PB-derived CD34+ cells were obtained from German Red Cross Blood Service, or from the NIH repository (Fred Hutchinson Cancer Research Center) with permission of the University of Washington Institutional Review Board. Cell purity was also confirmed and further enriched if necessary as described above. β-locus globin expression patterns were determined by RNase protection assay as previously described [11] and by RNA-sequencing.
Phenotypic analysis of erythroblasts
The expression of erythroid marker glycophorin-A and leukocyte common antigen CD45 was examined using flow cytometry analysis. Antibodies used were glycophorin-A-PE (DAKO) and allophycocyanin (APC) conjugated CD45 (BD Biosciences). Cells were washed once with PBS-0.5% BSA and then incubated with antibodies for 30 minutes at 4°C, washed, resuspended in PBS-0.5% BSA containing 100 ng/mL propidium iodide (Sigma) for dead cell exclusion. Cells were acquired using FACScalibur (BD Biosciences), and results were analyzed with Cell Quest Pro (BD Biosciences). Gatings were set according to isotype controls.
Chromatin Immunoprecipitation (ChIP) assays
Two ChIP protocols were used. For RNA polymerase II (Pol II), transcription factor IID (TFIID), TFIIB, and monomethylated H3 lysine 27 (H3K27me1), ChIP assays were performed as described by Yin et al [12]. For trimethylated H3 lysine 4 (H3K4me3) and trimethylated H3 lysine 27 (H3K27me3), ChIP assays were performed as described by Kimura et al [13] as part of ChIP sequencing procedure. For both procedures, chromatin was fragmented to roughly 200-500 bp in size by sonication with a microtip (Fisher Sonic Dismembrator 500) (Thermo Fisher Scientific). Antibodies used were: Pol II (SC-899), TFIID (SC-273X), TFIIB (SC-225), (all three were from Santa Cruz, CA), AcH3 (Upstate/Millipore, 06-599, Billerica, MA), H3K27me1 (Upstate/Millipore, 07-448), H3K27me3 (Upstate/Millipore, 07-449), and H3K4me3 (Cell signaling, 9751, Danvers, MA). For PolII, TFIID, TFIIB, and H3K27me1, the relative enrichment of fragments was determined by quantitative real-time polymerase chain reactions (PCR) using FastStart SYBR Green Master (Roche Applied Science, Indianapolis, IN) in the MJ Research DNA Engine Opticon 2 (Bio-rad, Hercules, CA) and analyzed with MJ Opticon Monitor Analysis Software Version 2.02. Human genomic DNA (Bioline, Taunton, MA) was used to generate standard curves. The PCR conditions were: 10 min at 95 °C, followed by 40 cycles of 40 s at 95 °C, 1 min at 60 °C, and 1 min at 72 °C, and then 5 min at 72 °C, followed by melting curve analysis. Primer sequences are provided in Supplementary Table 1. One way analysis of variance (ANOVA) followed by Tukey’s Post Hoc Test was performed using GraphPad InState version 3.10 (GraphPad Software, Inc. La Jolla, CA) to determine whether the differences between means of each group were statistically significant. When 2 bars are labeled with the same letter, it indicates that these 2 sets of data are not statistically significantly different. For H3K4me3, H3K27me2, and AcH3, ChIP sequencing was performed. The libraries were constructed according to the Illumina (San Diego, CA) protocol. Briefly, purified ChIP DNA was end-repaired using End-It DNA end-repair kit (Epicentre, Madison, WI.) and adenine was added to the 3′ ends using Klenow fragment (3′-5′ exo-) (New England Biolabs (NEB), Ipswich, MA). Adapters (Illumina) were ligated to the DNA fragments using T4 DNA ligase (NEB) and the products were PCR amplified (Phusion, NEB) and size-selected on 2% agarose gels. The libraries were sequenced on Genome Analyzer IIX or Hi-Seq 2000 platforms (both from Illumina) by the High-througput Genomics Center (University of Washington, Seattle, WA). Regions of local enrichment of short-read sequence tags mapped to the genome were identified using HotSpot algorithm (http://www.uwencode.org/proj/hotspot-ptih/). One percent false discovery rate thresholds (FDR 0.01) were computed for each cell type by applying the HotSpot algorithm to an equivalent number of random uniquely mapping 36mers. Data alignment was performed using Bowtie aligner (version 0.12.7) (http://bowtie-bio.sourceforge.net/index.shtml). The raw data have been deposited to NCBI Gene Expression Omnibus with accession number GSE35375.
Chromatin Conformation Capture (3C) assays
The higher chromatin structure of the erythroblasts of the three developmental stages was analyzed using 3C assays essentially as previously described [14]. Briefly, cross-linked DNA was prepared from the nuclei of erythroblasts and subjected to digestion with restriction enzyme HindIII (New England Biolabs, Ipswich, MA). Afterward, DNA fragments were ligated with T4 DNA ligase (New England Biolabs). The DNA was concentrated by ethanol precipitation and subjected to RNase A treatment and further purification by phenol/chlorophorm extraction and ethanol precipitation. Control template was similarly prepared from mixed equimolar amounts of BAC Clone CTD2596M16 containing human β-globin locus and BAC Clone RP11-687D2 containing human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene. The ligation products were quantitated by real time PCR using Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) or Universal Probe Master (Roche Applied Science) with probes (5′ FAM/3′ BHQ-1, Biosearch Technologies, Novato, CA) specific to ε, γ, β globin or GAPDH fragments. The ligation frequencies between the globin site pairs were normalized and expressed as the percentage of the interaction observed within GAPDH gene. Primer and probes sequences are provided in Supplementary Table 2.
RNA-sequencing
Total RNA was extracted from erythroblasts using TRIzol® Reagent (Invitrogen, Carlsbad, CA) per manufacturer’s instruction. 18S and 28S rRNAs were depleted from total RNA using Ribosomal RNA Depletion Kit (Applied Biosystems). cDNA library was constructed using SOLiD Whole Transcriptome Analysis Kit (Applied Biosystems). Massively parallel ligation sequencing was carried out to 50 bases using Applied Biosystems SOLiD System according to manufacturer’s instruction. Corona_lite_plus (version 4.2.1) from Applied Biosystems were used to map sequence reads to human reference sequence (hg18) from University of California – Santa Cruz. Ribosome RNA reads filtering was executed at the beginning of reads mapping with full length of 50bp. The remaining reads were mapped to the reference with the length of 45bp and 40bp respectively. Five mismatches were allowed at each mapping step. Reads mapped to unique loci and a fraction of those mapped to multiple loci (2-10) were used for further analysis. To calculate gene expression intensity, the read counts were normalized to reads per kilobase of exon model per million mapped reads (RPKM). Spots where RPKM were less than 0.01 for genes in all three libraries were removed from analysis.
Results
The three erythroid populations represent three distinct developmental stages with unique globin expression profiles
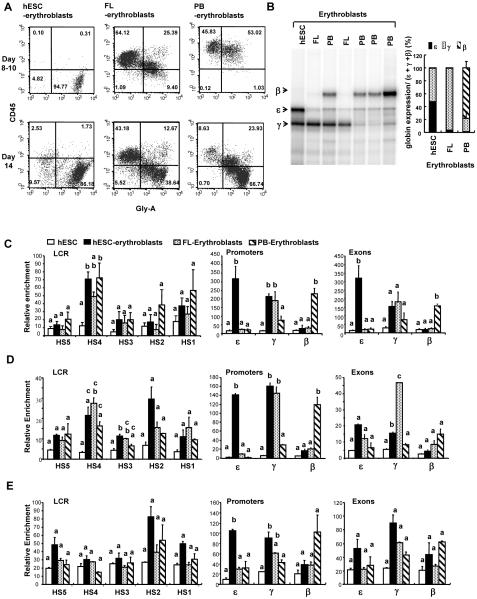
To study the epigenetic regulation of hemoglobin switching in different developmental stages, we first obtained nucleated erythroblasts by differentiating hESCs, FL, and adult PB CD34+ cells in the erythroid inducing medium. The expression of surface marker CD45 and glycophorin-A was examined between day 8 and day 14 after cells were directed to undergo erythroid differentiation (Figure 1A). Interestingly, while the immature erythroid cells from FL and PB CD34+ cells expressed leukocyte common antigen CD45, the immature erythroid cells from hESCs lacked such expression. With further maturation, both FL- and PB-derived erythroblasts lost CD45 expression. RNase protection assays revealed that hESC-derived erythroblasts expressed high levels of embryonic ε and fetal γ globin with little adult β globin whereas FL-derived erythroblasts mainly expressed fetal γ globin and PB-derived erythroblasts mainly expressed adult β globin, although occasionally had elevated fetal γ globin expression (Figure 1B). The respective globin expression patterns of these three types of erythroblasts were also confirmed using RNA-sequencing (Supplementary Table 3). These results demonstrated that the three erythroid populations under investigation here possessed unique globin expression patterns associated with embryonic/early fetal, fetal, and adult developmental stages.
Figure 1. The unique globin expression patterns and transcriptional machinery recruitment to the β globin locus of three developmentally distinct erythroid populations.
(A) The CD45 and glycophorin-A expression by immature and mature erythroblasts derived from hESC, FL, or PB-CD34+ cells. Erythroblasts were generated from dissociated day-7 embryoid bodies prepared from hESCs, from FL mononuclear cells, or from mobilized PB CD34+ cells. Cells suspensions were collected between day 8 and day 14, stained with CD45-APC and glycophorin-A-PE, and enumerated using flow cytometry. Gatings were set based on isotype controls. (B) The β locus globins expression by the three erythroid populations. RNA was prepared from day-14 erythroblasts and RNase protection assay was carried out to determine the levels of ε, γ, and β globin transcripts. Occasionally, elevated levels of γ globin expression by PB CD34+ cells-derived erythroblasts were detected. A non-specific band slightly higher than the band for ε globin was detected in all samples. Results from RNase protection were also confirmed using RNA-sequencing (Supplementary Table 3). (C) Recruitment of RNA polymerase II to the β globin locus in undifferentiated hESCs and three erythroid populations. (D) Recruitment of TFIIB to the β globin locus. (E) Recruitment of TFIID to the β globin locus. For (C), (D), and (E), glycophorin-A+ FL erythroblasts were enriched using MACS prior to preparing chromatin for ChIP assay. The relative degree of enrichment for each target sequence was determined by first quantifying against a standard curve generated using purified human genomic DNA and then normalizing to the level of human GAPDH promoter present in each individual precipitation. Data shown are mean ± standard error of mean (SEM). One-way ANOVA was performed followed by Tukey’s Post Hoc Test to determine whether any two means of a specific target are statistically significantly different (p<0.05). When two bars are labeled with the same letter, it denotes that these means are not statistically significantly different.
ChIP assays were then performed with chromatin obtained from undifferentiated hESCs as well as the three different erythroid populations to interrogate whether the distinct globin expression patterns were a reflection of the recruitment of transcriptional complexes to the specific globin regions (Figures 1C). While little RNA Pol II was found to be associated with the entire β globin locus of undifferentiated hESCs, elevated levels of RNA Pol II were found to be associated with the LCR, especially the DNaseI HS4 of the three erythroid populations (Figure 1C). Much higher levels of RNA Pol II were detected at the globin promoters and exons in accordance with the unique globin expression pattern of the specific erythroid population. Specifically, RNA Pol II was associated with both embryonic ε and γ globin promoters and exons of hESC-derived erythroblasts; with only γ globin promoters and exons of FL-derived erythroblasts; and mainly with β globin promoter and exon of PB-derived erythroblasts. The recruitment of transcription factor IIB (TFIIB) and TFIID, which are part of the RNA Pol II preinitiation complex, showed similar patterns with elevated LCR recruitment in erythroid cells as compared to undifferentiated hESCs, and globin gene-specific recruitment that was consistent with the globins expressed by the individual erythroid populations (Figures 1D, 1E). Together, these data show that the unique globin patterns displayed by these three developmentally distinct erythroid populations were a result of targeted recruitment of RNA polymerase II and its preinitiation complex to the specific globin genes, and not a result of failed elongation by non-expressed genes.
Epigenetic markings are conserved in the three erythroid populations in accordance with their unique globin expression profiles
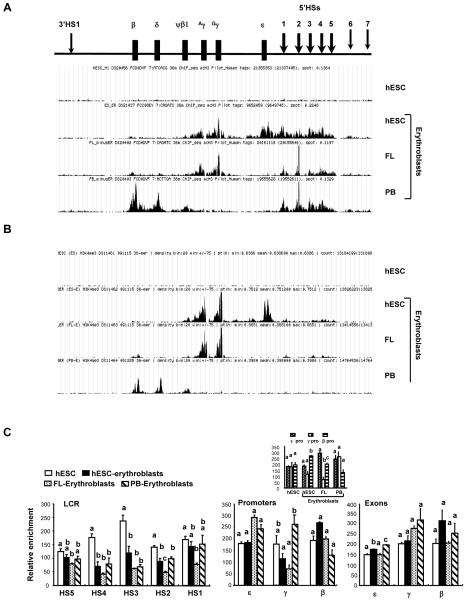
Covalent modifications of the N-terminal tails of histones alter the association between histones and genomic DNA and lead to changes in chromatin structure and in the accessibility of the gene to transcriptional machineries. To study whether the same histone codes are employed by the β globin locus throughout development, we performed ChIP assays to study the epigenetic landscapes of β globin locus in undifferentiated hESCs and the three erythroid populations (Figure 2). Histone H3 hyperacetylation (AcH3) was observed in the LCR of the three erythroid populations as opposed to the undifferentiated hESCs (Figure 2A). Hyperacetylated histone H3 was also found in the active globin promoters and exons, confirming that while the LCR has acquired an open chromatin conformation in erythroid cells of any developmental stage, only the temporally active globin genes have acquired such structure. The developmentally silenced globin genes are situated within a closed, hypoacetylated domain. Furthermore, histone H3 lysine 4 trimethylation (H3K4me3), another mark for an active chromatin domain, was studied (Figure 2B). Consistent with a closed chromatin conformation, no H3K4me3 peaks were detected in the β globin locus of undifferentiated hESCs. Interestingly, while active chromatin mark AcH3 encompassed both the LCR and active globin genes of erythroid cells (Figure 2B), only chromatins of active globin regions, but not the LCR, were marked by high levels of H3K4me3 (Figure 2B). This pattern was consistent in all three erythroid populations studied. Finally, the distribution of histone H3 lysine 27 monomethylation (H3K27me1) and trimethylation (H3K27me3) was examined in the four cell populations. We were unable to detect specific H3K27me3 enrichment across the β globin locus of the four cell types with ChIP sequencing although its enrichment was detected at other loci (data not shown). H3K27me1 was decreased in the HSs 2, 3, and 4 of the LCR of erythroid cells in comparison to undifferentiated hESCs (Figure 2C). Such differences among the cell types were not evident in the promoter or exon regions. However, when ε, γ, and β promoters within individual cell types were compared, we found that the active globin promoters, as opposed to dormant globin promoters, had the lowest level of H3K27me1 in the erythroid cells (Figure 2C insert). Our data contrast the recent findings by Kim and Kim that the β-globin locus is marked by H3K27me3 in non-erythroid 293FT cells and by H3K27me1 in K562 erythroid cells [15]. The reason for these disparities is not clear. However, our data do agree with other groups who find that H3K27me1 is a repressive chromatin mark associated with the formation of heterochromatin [16, 17], that H3K27me1 removal may be a general prerequisite for the initiation of high-level transcription [18], and that H3K27me1 mark is depleted at the β globin region as compared to ε and γ globin regions in adult CD36+ erythroid precursors [19]. Overall, our studies show that the β globin locus employs similar epigenetic markings throughout development to form a chromatin structure that allows the selective expression of developmentally regulated globins.
Figure 2. Epigenetic landscapes of the β globin locus of undifferentiated hESCs and erythroblasts derived from hESC, FL, and PB.
Quantification of (A) histone H3 acetylation (AcH3), (B) H3K4 trimethylation (H3K4me3) and (C) H3K27 monomethylation (H3K27me1) distribution across the β globin locus using ChIP assays. (A) and (B) were quantified by high throughput sequencing using Illumina Hi-seq 2000 (A), or Genome Analyzer IIX (B). Regions of local enrichment of short-read sequence tags mapped to the genome were identified using HotSpot algorithm. Data alignment was performed using Bowtie aligner. (C) was quantified by real time PCR. Normalization and statistical analyses were performed as described in Figure 1. An additional panel for the H3K27me1 enrichment in the promoter regions is provided to assist side-by-side comparison of the relative distribution of H3K27me1 across the ε, γ, and β globin promoters in each specific cell type.
Physical proximity of the LCR and globin promoters changes according to the developmental stage of the erythroid cells
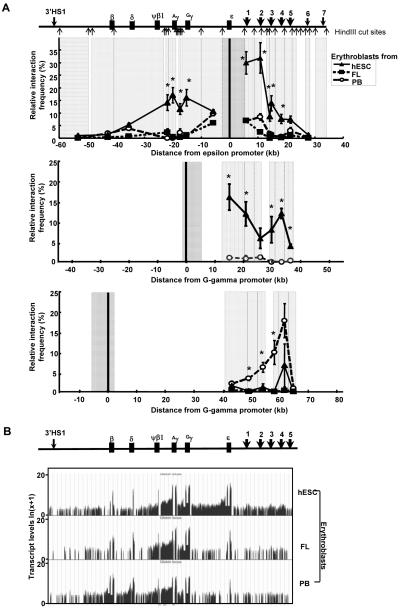
It has been shown in the transgenic mice that an active chromatin hub forms between the developmentally active globin genes and the HSs of LCR with the inactive globin genes looped out [20]. Here we interrogated the dynamic long-range interactions across the β globin locus in the three developmentally distinct erythroid populations using 3C assays (Figure 3A). In hESC-derived erythroblasts, the interaction frequencies between the ε globin promoter and the HSs 1-4 of the LCR were found to be significantly higher than the frequencies observed in the FL- and PB-derived erythroblasts, suggesting the formation of a chromatin loop between the active ε promoter and the LCR in the hESC-derived erythroblasts. Furthermore, in the FL- and PB-derived erythroblasts, the interaction frequencies between ε globin promoter and the γ globin genes decreased as the distance from the ε globin promoter increased (Figure 3A, top panel). In contrast, high levels of interaction between ε globin promoter and both γ globin genes (Gγ and Aγ) were observed in the hESC-derived erythroblasts. No such increased interaction was found between ε globin promoter and the δ and β globin genes in the hESC-derived erythroblasts as compared to the FL- and PB-derived erythroblasts. When the interaction frequencies between Gγ promoter and the LCR was characterized, significantly higher levels of interactions were found in the hESC-derived erythroblasts than those in the PB-derived erythroblasts (Figure 3A, middle panel). Similar levels of interaction frequencies between Gγ promoter and the LCR in the FL-derived erythroblasts as compared to the hESC-derived erythroblasts were also observed in a separate experiment (Supplementary Figure 1). On the contrary, only PB-derived erythroblasts showed significant interactions between the β globin promoter and the LCR (Figure 3A, bottom panel), consistent with the finding that these cells were the only studied cell type that expressed high levels of β globin. Together, these data suggest that in hESC-derived erythroblasts where high levels of ε and γ globins were expressed with little adult δ or β globin genes, the active ε, Gγ, Aγ globin genes and the LCR were spatially organized into a physically closely associated hub while the inactive δ and β globin genes were situated outside the hub.
Figure 3. Long range interaction between globin genes and the LCR.
(A) Physical proximity between embryonic ε, fetal γ, or adult β globin promoters and the other genetic elements of the β globin locus as revealed by chromatin conformation capture studies. HindIII cut sites relative to various genetic elements are shown. Anchor fragments encompassing the promoters investigated (thick black lines) are shown in dark gray. Fragments analyzed are shown in light gray. There was insufficient amount of FL templates for analyzing the interactions between Gγ promoter and the HSs of the LCR (middle panel) but data from an independent experiment are included in Supplementary Figure 1. Data shown are mean ± standard error of mean (SEM). One-way ANOVA followed by Tukey’s Post Hoc Test was performed to analyze whether the means differ statistically significantly in the experiments where ε or β promoters were used as the anchor. For the experiment using fragment enclosing Gγ promoter as the anchor, Student’s t test was performed. * denotes that the frequencies found in hESC-derived erythroblasts are significantly different from both the FL- and PB-derived erythroblasts (top panel), or from the PB-derived erythroblasts (middle panel) * in the bottom panel indicates that the interaction frequencies found in the PB-derived erythroblasts are significantly different from those found in both FL- and hESC-derived erythroblasts. (B) Levels of transcripts across the β globin locus determined by RNA-sequencing. Data shown are genome browser view of RPKM normalized read counts mapped to the β globin locus. These read counts were further ln(x+1) transformed to enable viewing of low abundance transcripts. The untransformed RPKM normalized read counts are included in Supplementary Table 3.
It has also been proposed that a tracking or a facilitated tracking and transcription mechanism is responsible for the expression of embryonic ε globin in K526 cells [3, 4]. We nextstudied both the genic and intergenic transcriptions across the beta globin locus in the three erythroblasts populations using RNA-sequencing to obtain absolute quantifications of transcripts (Figure 3B, Supplementary Table 3). Our data confirm that intergenic transcription is a wide-spread phenomenon in β globin locus of erythroid cells, as previously described by Miles et al. [21] and not necessarily specific to the intergenic region leading up to ε globin promoter. While RNA PolII did appear to track between the LCR and the ε globin promoter, given the low abundance of HS1-ε transcripts relative to other intergenic transcripts, such as Aγ - Ψβ1 and Ψβ1- δ, whether this tracking is responsible for the ε globin activation in hESC-derived erythroblasts remains to be determined.
Discussion
An extensive body of work has shown that the modifications of histones, the fundamental constituent of nucleosomes and chromatin fibers, alter gene expression by affecting overall chromatin structure and by modulating the binding of effector molecules (reviewed in [22]). The vast number of possible modifications and the crosstalk between modifications give the higher organisms tight control and fine tune capability required for spatially and temporally regulated gene expression. Chromatin profilings in ESCs, which possess both indefinite self-renewal capability and differentiation potential, have revealed bivalency characterized by the co-existence of repressive chromatin mark (H3K27me3) and active chromatin marks including histone acetylation and H3K4me3 at the promoters of the genes critical for developmental decisions [23-25]. In addition, certain localized epigenetic markings for the later formation of an active tissue-specific chromatin domain in differentiated cells, including VpreB1 and λ5 genes for B cells [26], and the LCR for erythroid cells [27] are found to be already established in the murine ESCs. Furthermore, epigenetic pre-marking and recruitment of transcriptional factors at the enhancers of silent tissue-specific genes including liver-specific Alb1, macrophage/dendritic cell-specific Il12b, and thymocyte-specific Ptcra genes at the ESCs stage appear to be important for the transcriptional activation of these genes in differentiated cells [28, 29]. In contrast to these findings, we did not find evidence of either bivalency or active epigenetic marks in the entire β globin locus of hESCs. The recent finding that places hESC and mESC in different developmental stages [30] may have implications on why active LCR is already formed in the ESCs of murine but not in those of human origin. The heterogeneity of the ESCs population, as demonstrated by the disappearance of bivalency marks when ESCs are subfractionated [31], may also provide an alternative explanation. Nevertheless, the active LCR is formed in the hESC-derived erythroblasts similar to that in the FL-derived and PB-derived erythroblasts. The activation of the LCR may have occurred at the stage of multipotent hematopoietic stem cells during hESC differentiation as CD34+CD38− cells have been shown to express embryonic ε and fetal γ globin mRNA [32] and the acquisition of chromatin signatures for LCR activation has been noted in the multipotent human hematopoietic stem cells [19].
The characteristic co-expression of high levels of embryonic ε and γ globins [9] makes hESC-derived erythroblasts an ideal model system for studying the transcriptional and epigenetic regulation of embryonic ε globin expression, which previously has been confined to utilizing transgenic mice carrying human β globin locus, and K562 cells. Thus far, the studies conducted analyzing the epigenetic landscapes of hESC-derived erythroblasts are limited. It was found that while β globin locus genes contain no CpG islands, domains of DNA hypomethylation spanning thousands of base pairs within domains of acetylated histones are established around the most highly expressed genes during each developmental stage when comparing hESC-derived erythroblasts to uncultured FL and bone marrow cells [7, 8]. Consistent with these findings, our study also showed that the LCR is hyperacetylated in erythroblasts of all developmental stages, but such hyperacetylation is confined to only the actively expressed globin genes of each specific developmental stage. Furthermore, we provided evidence that H3K4me3 co-localized with AcH3 at the actively transcribed globin genes and H3K27me1 was selectively depleted from the actively transcribed globin promoters. Together with the results by Lathrop et al. [8], and Hsu et al. [7], it was revealed that throughout development, human primary erythroblasts employ the same histone codes including hypomethylation, histone acetylation, H3K4me2, H3K4me3, and selective depletion of H3K27me1 and H3K9me2 to ensure proper temporal expression of specific globin genes. The role of various methyltransferases (reviewed in [33]) and polycomb group (reviewed in [34]) in the establishment and maintenance of these domains and globin gene regulation requires further studies.
Higher-order chromatin structures, such as chromatin looping, have been implicated in participating in the activation and repression of genes (reviewed in [35, 36]). The long-range physical interactions between the HSs of the LCR and the developmentally active globin genes have been demonstrated in the endogenous mouse β locus, as well as the human β globin locus in transgenic mice [20, 37, 38] with the potential involvement of BCL11A and Sox6 [39], which may participate in a large transcriptional interactome [40]. The long-range interactions between LCR and the active β locus globin genes, to our best knowledge, have not been shown in the primary human erythroid cells. While there is little doubt that a looping mechanism would contribute to the long-range activation of γ and β globin genes in primary FL- and PB-derived erythroblasts, respectively, by the LCR, a different mechanism might be involved in the activation of embryonic ε globin, given the close proximity between ε globin and the LCR. Indeed, a tracking mechanism has been proposed to be responsible for the transfer of RNA polymerase II to the ε globin promoter, based on the continuity of histone modifications, including AcH3, AcH4, and H3K4me2 and the RNA polymerase II distribution from the LCR to the ε globin promoter, and the presence of intergenic transcripts between HS2 and ε promoter, in the ε globin-expressing K562 cells [3]. While we also detected intergenic transcripts, confirming RNA tracking between the LCR and the ε globin gene, and wide distribution of acetylated H3 in the LCR and ε globin gene, H3K4me3 mapping clearly showed that the LCR had relatively low H3K4me3 marking as compared to the ε globin gene. Therefore, in the ε globin-expressing hESC-derived erythroblasts, the LCR and ε globin gene were not situated within a continuous chromatin domain. Most importantly, the increased cross-linking frequencies between the ε globin promoter and the HSs of the LCR in the hESC-derived erythroblasts as compared to FL- and PB-derived erythroblasts, revealed by chromatin conformation capture, suggest a physical proximity between the LCR and the ε globin promoter closer than otherwise would have been expected based solely on the distance, base-pair-wise, between these two genetic elements. Although our results are consistent with a looping mechanism contributing to the activation of ε globin promoter, we can not rule out a facilitated tracking and transcription mechanism that leads to the eventual formation of a looped active chromatin hub, as proposed by Zhu et al. [4]. However, as the levels of HS1-ε intergenic transcripts detected were not proportional to the levels of ε globin expressed, how much the RNA polymerase II tracking between the LCR and the ε globin contributes to the eventual ε globin activation remains unclear. Finally, as we have previously demonstrated that hESC-derived erythroblasts co-express embryonic ε and fetal γ globins at the single cell level, a previously proposed flip-flop mechanism [41] may be at play here. The increased cross-linking frequencies between the LCR, ε globin and both Gγ and Aγ globin genes in these cells support that ε, Gγ, and Aγ globin promoters are held at close physical proximity to the LCR, which can potentially facilitate the co-expression of these globin genes.
Overall, our studies provide a comprehensive comparison of chromatin modifications of the β globin locus of undifferentiated hESCs and three erythroblast populations of distinct developmental stages. We found that the locus is inactive and without epigenetic pre-marking in the hESCs. Once activated in the erythroblasts, the locus employs the same histone codes throughout development to mark actively expressed genes and utilizes the looping mechanism to facilitate transcription.
Supplementary Material
Acknowledgments
The authors express their gratitude towards Drs. Theresa Canfield, Erika Giste, Richard Sandstrom, and R. Scott Hansen for assistance with high throughput sequencing. This research was supported by the National Institute of Health grants DK077864 (K-H.C), HL46557 (T.P.) and 1RC2HG005654 (J.A.S.).
Footnotes
Author Disclosure Statement:
The authors declare no potential conflicts of interest.
References
- 1.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Experimental hematology. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilber A, Nienhuis AW, Persons DA. Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood. 2011;117:3945–3953. doi: 10.1182/blood-2010-11-316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim A, Dean A. Developmental stage differences in chromatin subdomains of the beta-globin locus. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7028–7033. doi: 10.1073/pnas.0307985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic acids research. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupon JW, Wang SZ, Gnanapragasam M, Labropoulos S, Ginder GD. MBD2 contributes to developmental silencing of the human epsilon-globin gene. Blood cells, molecules & diseases. 2011;46:212–219. doi: 10.1016/j.bcmd.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamura E, Matsuzaki H, Campbell AD, Engel JD, Fukamizu A, Tanimoto K. All of the human beta-type globin genes compete for LCR enhancer activity in embryonic erythroid cells of yeast artificial chromosome transgenic mice. Faseb J. 2009;23:4335–4343. doi: 10.1096/fj.09-137778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu M, Richardson CA, Olivier E, Qiu C, Bouhassira EE, Lowrey CH, Fiering S. Complex developmental patterns of histone modifications associated with the human beta-globin switch in primary cells. Experimental hematology. 2009;37:799–806. e794. doi: 10.1016/j.exphem.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lathrop MJ, Hsu M, Richardson CA, Olivier EN, Qiu C, Bouhassira EE, Fiering S, Lowrey CH. Developmentally regulated extended domains of DNA hypomethylation encompass highly transcribed genes of the human beta-globin locus. Exp Hematol. 2009;37:807–813. e802. doi: 10.1016/j.exphem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang KH, Nelson AM, Cao H, Wang L, Nakamoto B, Ware CB, Papayannopoulou T. Definitive-like erythroid cells derived from human embryonic stem cells coexpress high levels of embryonic and fetal globins with little or no adult globin. Blood. 2006;108:1515–1523. doi: 10.1182/blood-2005-11-011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang KH, Nelson AM, Fields PA, Hesson JL, Ulyanova T, Cao H, Nakamoto B, Ware CB, Papayannopoulou T. Diverse hematopoietic potentials of five human embryonic stem cell lines. Experimental cell research. 2008;314:2930–2940. doi: 10.1016/j.yexcr.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navas PA, Peterson KR, Li Q, Skarpidi E, Rohde A, Shaw SE, Clegg CH, Asano H, Stamatoyannopoulos G. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Molecular and cellular biology. 1998;18:4188–4196. doi: 10.1128/mcb.18.7.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin W, Barkess G, Fang X, Xiang P, Cao H, Stamatoyannopoulos G, Li Q. Histone acetylation at the human beta-globin locus changes with developmental age. Blood. 2007;110:4101–4107. doi: 10.1182/blood-2007-05-091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell structure and function. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 14.Fang X, Xiang P, Yin W, Stamatoyannopoulos G, Li Q. Cooperativeness of the higher chromatin structure of the beta-globin locus revealed by the deletion mutations of DNase I hypersensitive site 3 of the LCR. Journal of molecular biology. 2007;365:31–37. doi: 10.1016/j.jmb.2006.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YW, Kim A. Characterization of histone H3K27 modifications in the beta-globin locus. Biochemical and biophysical research communications. 2011;405:210–215. doi: 10.1016/j.bbrc.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nature structural & molecular biology. 2009;16:763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob Y, Stroud H, Leblanc C, Feng S, Zhuo L, Caro E, Hassel C, Gutierrez C, Michaels SD, Jacobsen SE. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature. 2010;466:987–991. doi: 10.1038/nature09290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Molecular and cellular biology. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell stem cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nature genetics. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 21.Miles J, Mitchell JA, Chakalova L, Goyenechea B, Osborne CS, O’Neill L, Tanimoto K, Engel JD, Fraser P. Intergenic transcription, cell-cycle and the developmentally regulated epigenetic profile of the human beta-globin locus. PloS one. 2007;2:e630. doi: 10.1371/journal.pone.0000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell stem cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 26.Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Molecular and cellular biology. 2005;25:1804–1820. doi: 10.1128/MCB.25.5.1804-1820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levings PP, Zhou Z, Vieira KF, Crusselle-Davis VJ, Bungert J. Recruitment of transcription complexes to the beta-globin locus control region and transcription of hypersensitive site 3 prior to erythroid differentiation of murine embryonic stem cells. The FEBS journal. 2006;273:746–755. doi: 10.1111/j.1742-4658.2005.05107.x. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes & development. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 31.Hong SH, Rampalli S, Lee JB, McNicol J, Collins T, Draper JS, Bhatia M. Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell stem cell. 2011;9:24–36. doi: 10.1016/j.stem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Lu SJ, Li F, Vida L, Honig GR. CD34+CD38- hematopoietic precursors derived from human embryonic stem cells exhibit an embryonic gene expression pattern. Blood. 2004;103:4134–4141. doi: 10.1182/blood-2003-10-3575. [DOI] [PubMed] [Google Scholar]
- 33.Hosey AM, Chaturvedi CP, Brand M. Crosstalk between histone modifications maintains the developmental pattern of gene expression on a tissue-specific locus. Epigenetics. 2010;5:273–281. doi: 10.4161/epi.5.4.11522. [DOI] [PubMed] [Google Scholar]
- 34.Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the Polycomb perspective. Developmental cell. 2010;19:651–661. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochimica et biophysica acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng W, Blobel GA. Do chromatin loops provide epigenetic gene expression states? Current opinion in genetics & development. 2010;20:548–554. doi: 10.1016/j.gde.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nature genetics. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 38.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Molecular cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes & development. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gribnau J, de Boer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. The EMBO journal. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.