Abstract
Resistance to the experimental human cytomegalovirus (CMV) UL97 kinase inhibitor maribavir has been mapped to UL97 mutations at codons 353, 397, 409 and 411, in the kinase ATP-binding region, and to mutations in the UL27 gene. We studied the maribavir susceptibility phenotypes of additional UL97 mutations observed in vitro and in clinical trials, and the effect of simultaneous mutation in both UL97 and UL27. In vitro selection under maribavir identified a new locus of UL97 mutation within the conserved kinase p-loop (L337M), which conferred low grade maribavir resistance (3.5-fold increased EC50) without ganciclovir cross-resistance. During maribavir Phase III CMV prevention clinical trials, 3 previously unknown UL97 sequence variants were detected in plasma samples after 27 to 98 days of drug exposure (I324V, S334G and S386L). These variants did not confer any drug resistance despite proximity to mutations that confer maribavir resistance. The UL27 resistance mutation R233S, when added to strains containing UL97 mutations L337M or V353A, doubled their maribavir EC50s. These results expand the range of UL97 maribavir-resistance mutations into another part of the kinase ATP-binding region, but offer no genotypic evidence that development of drug resistance affected the outcomes of Phase III maribavir clinical trials after drug exposure of up to 14 weeks. There is a potential for increased maribavir resistance in UL27–UL97 double mutants.
Keywords: cytomegalovirus, maribavir, UL97, kinase, UL27, antiviral drug resistance
1. Introduction
The human cytomegalovirus (CMV) UL97 kinase is prominently involved in antiviral therapy for this leading opportunistic viral pathogen. By enabling the initial phosphorylation of ganciclovir that is necessary for its antiviral activity, the UL97 kinase provides selective drug activation in virus-infected cells, and UL97 mutation is the dominant pathway for ganciclovir resistance (Chou, 2008). UL97 also plays major facilitating roles during normal viral replication (Marschall et al., 2011), as illustrated by the severe growth defect of CMV strains lacking a functioning UL97 gene product (Prichard, 2009). Thus, UL97 is itself an antiviral drug target.
The UL97 kinase inhibitor maribavir (MBV) has undergone considerable antiviral drug development. Favorable preclinical studies showing good in vitro anti-CMV potency (Biron et al., 2002) and early clinical studies indicating a significant in vivo antiviral effect without dose-limiting toxicity (Lalezari et al., 2002; Winston et al., 2008) were followed by Phase III clinical trials that showed a surprising lack of efficacy of low-dose MBV in preventing CMV viremia or disease after stem cell transplantation (Marty et al., 2011); a parallel trial in liver transplantation was halted. Case reports of MBV at higher doses as salvage therapy of CMV disease refractory to existing therapy suggest potential therapeutic utility of the drug (Avery et al., 2010) while providing a cautionary instance of MBV resistance (Strasfeld et al., 2010).
In vitro studies showed that UL97 mutations at codons 353, 397, 409 and 411 in the ATP binding region confer moderate to high-level MBV resistance without ganciclovir cross-resistance (Biron et al., 2002; Chou and Marousek, 2008; Chou et al., 2007). Subsequently, some of the same mutations T409M and H411Y/N were identified in vivo (Strasfeld et al., 2010). Diverse mutations in the CMV gene UL27 can also confer low-grade MBV resistance (Chou, 2009; Komazin et al., 2003). Since further treatment trials are planned for MBV, continued study of MBV resistance is needed.
2. Materials and Methods
2.1 Viral strains and clones
CMV laboratory strains and their bacterial artificial chromosome (BAC) clones, modified from reference strain AD169 to incorporate a secreted alkaline phosphatase (SEAP) reporter gene for rapid viral quantitation, have been used extensively for recombinant phenotyping (Chou, 2009, 2010; Chou and Marousek, 2008).
2.2 In vitro viral propagation under MBV
Ten new in vitro propagation experiments of the error-prone exonuclease mutant T2294 (UL54 D413A) under increasing concentrations of MBV were performed as previously described (Chou and Marousek, 2008). Extracts of infected cell cultures were sampled approximately every 5 passages for the emergence of mutations in UL97 and UL27.
2.3 Clinical study specimens
Studies 300 (ClinicalTrials.gov identifier NCT00411645) (Marty et al., 2011) and 301 (ClinicalTrials.gov identifier NCT00497796) were Phase III, multi-center trials in allogeneic hematopoietic cell transplantation (HCT) (study 300) or liver transplantation (study 301) where subjects were randomized to receive prophylactic MBV 100 mg orally twice daily versus placebo (study 300) or oral ganciclovir (study 301) for 12–14 weeks after transplantation, or until the detection of CMV viremia. Plasma samples were collected weekly while on study drug and periodically after discontinuation. CMV plasma viral loads were monitored using the commercial COBAS Amplicor PCR-based assay (Roche).
2.4 Genotypic analysis
Plasma samples with CMV loads >2000 copies/mL were selected from subjects who received MBV in studies 300 and 301, regardless of duration of drug exposure. The latest sample was analyzed from subjects with multiple CMV-positive samples. Nested PCR was performed on plasma DNA extracts using Taq polymerase to amplify UL97 nucleotides 761 to 1504, and UL27 nucleotides −69 to 1640. Automated fluorescent dyedeoxy sequencing data (Big Dye version 3.1, Applied Biosystems) were compared to CMV reference strain AD169 (Genbank X17403) for UL97 codons 301–460 and UL27 codons 1–450. DNA extracts from in vitro selection experiments were similarly amplified and sequenced, except without the need for nested PCR, and sequencing included all 608 codons of UL27.
2.5 Recombinant phenotyping
This was accomplished by mutagenesis of a baseline BAC clone of a drug-susceptible CMV strain, followed by viral yield reduction assay for drug susceptibility (Chou, 2010; Hakki et al., 2011). UL97 mutations were introduced by cotransfection of a transfer vector with BAC BA12 (for strain T3138), or in all other cases by recombination of a transfer vector and BAC BA9, as previously described (Chou, 2010). The UL27 mutation R233S known to confer low-grade MBV resistance (Chou et al., 2004) was introduced into BAC clones by “en passant” mutagenesis using long primers specific for this mutation, as described (Hakki et al., 2011). Other UL27 mutations were introduced by recombination of a transfer vector with BAC BA101 followed by Flp recombinase induction, as described (Hakki et al., 2011). BAC DNA was transfected into human foreskin fibroblast (HFF) cultures and the resulting live virus strains were validated for intended sequences in the genes being mutagenized (Chou, 2010; Hakki et al., 2011). Yield reduction assays for MBV susceptibility were performed in human embryonic lung (HEL) fibroblast cultures to determine the drug concentration required to reduce supernatant SEAP production by 50% (EC50) (Chou, 2009; Chou and Marousek, 2008; Hakki et al., 2011). Viral growth curves and ganciclovir susceptibility were determined in HFF cultures (Chou, 2010; Chou et al., 2007).
3. Results
3.1 In vitro selected UL97 and UL27 mutations
Table 1 shows the outcome of 10 new in vitro MBV selection experiments. All except one evolved previously reported mutations (Chou and Marousek, 2008), typically beginning with UL97 mutations H411Y/N, followed by mutations such as V353A that combined to confer high-level MBV resistance (Chou and Marousek, 2008). In experiment M58, UL97 mutation L337M and UL27 mutation D534Y were first detected at passage 7, each as an approximately equal mixture with wild type sequence. By passage 10, wild type sequence was no longer detected at these codons, and by passage 20, UL97 mutation H411L had completely replaced its wild type counterpart. The newly discovered mutation L337M is located in the UL97 ATP-binding kinase subdomain I (codons 337–345) (Hanks et al., 1988; Lurain et al., 2001). UL27 mutations D534Y and R448P (experiment M76) are also new.
Table 1.
UL97 and UL27 mutations selected in vitro under MBV
| Expt. No.a | UL97 mutationsb | UL27 mutationsb | MBV concn (μM)c |
|---|---|---|---|
| M53 | H411Y (6) | 0.5 | |
| M54 | H411N/Y (6) | 0.5 | |
| M57 | H411N (6) | 0.5 | |
| M58 | L337M (7) H411L (16) | D534Y (7) | 5 |
| M71 | H411L (5) V353A (15) | 15 | |
| M72 | H411Y (10) V353A (15) T409M (18) | 5 | |
| M73 | H411N (5) H411L (10) V353A (15) | 25 | |
| M74 | H411Y (5) V353A (10) | 10 | |
| M75 | T409M (5) H411Y (15) V353A (15) | 15 | |
| M76 | H411Y (5) V353A (10) | R448P (10) | 5 |
Each numbered experiment is an independent serial propagation of virus under drug
Passage number at first detection of mutation shown in parentheses.
Newly identified mutations shown in bold
MBV concentration at the highest culture passage number listed
3.2 Genotypic analysis of clinical trial specimens
Among a total of 598 subjects who received MBV, CMV genotypic data were obtained on 60 and 50 plasma samples respectively in studies 300 and 301. The samples had a median viral load of 6765 copies/mL in study 300, and 5685 copies/mL in study 301. The duration of MBV exposure prior to sampling ranged from 2 to 85 days (median 24.5 days) in study 300 and 43 to 106 days (median 95 days) in study 301. Sequence data were analyzed for all samples at UL97 codons 301–460 and UL27 codons 1–450, which include the locations of all known MBV resistance mutations when the study was conducted (Chou, 2009; Chou and Marousek, 2008). The UL97 sequence was well conserved; only 6 samples showed any amino acid variation from the AD169 reference sequence. There were 2 instances of Q449K, a known baseline sequence polymorphism (Lurain et al., 2001) one instance of M460I, a known ganciclovir-resistance mutation (Chou, 2008) that increases MBV susceptibility (Shannon-Lowe and Emery, 2010), and 3 previously unknown sequence variants as shown in Table 2. Because of the prophylactic design of the clinical trials, there were no pre-therapy viral sequences for comparison. I324V and S334G are located just upstream of subdomain I (Lurain et al., 2001) while S386L is near the mutation L397R known to confer high-grade MBV resistance (Biron et al., 2002; Chou and Marousek, 2008).
Table 2.
Uncharacterized UL97 sequence variants detected in Phase III MBV trials
| Subject No. | Study | UL97 changea | UL27 changea | MBV exposure (days) | Days off MBVb | CMV loadc |
|---|---|---|---|---|---|---|
| 15424 | 300 | I324V | P8S R30C | 27 | 2 | 11200 |
| 3013506 | 301 | S334G | 98 | 42 | 15200 | |
| 3018005 | 301 | S386L | 98 | 29 | 2200 |
Sequence changes from reference CMV strain AD169 not known to be baseline interstrain sequence polymorphisms
Number of days from end of MBV prophylaxis to sample collection
Plasma viral load of specimen used for genotyping, CMV genome copies/mL
In UL27, all samples except one showed some amino acid variation from the AD169 reference sequence, reflecting considerable sequence polymorphism in this gene (Chou et al., 2004; Hantz et al., 2009). None contained a UL27 mutation known to confer MBV resistance (Chou, 2009; Chou et al., 2004; Komazin et al., 2003). However, among MBV recipients, uncharacterized UL27 amino acid changes were found in 15 of 60 (25%) samples in study 300 and 13 of 50 (26%) samples in study 301 (Table 3). UL27 sequences were also determined for a sample of placebo recipients in study 300 and 8 of 15 (53%) cases showed uncharacterized UL27 changes. Among the 3 specimens with unknown UL97 changes (Table 2), only one had unknown UL27 changes, which were distant from known MBV resistance mutations (Chou, 2009; Chou et al., 2004; Komazin et al., 2003).
Table 3.
Uncharacterized UL27 sequence variants found in MBV recipients
| Study 300 | Study 301 | Number of cases per varianta |
|---|---|---|
| P8S, P16Sb, R30C, R46H, R57H, A138V, E165K, A258T, A267E, A272V, S312F, T342K, Q424H | P3S, P9S, P12Sc, P16L, A20Vc, R64H, P143S, A242V, A267T, A278T, N300Sc | 1 |
| G291del(GNHD), H334R, A387V | G291del(GNHD) | 2 |
| R246H | R246Hb | 4 |
Total across both studies. Some individuals had more than one variant
Associated clinical CMV isolate tested maribavir susceptible by plaque reduction assay
Also found in placebo recipient in Study 300. Other variants from placebo recipients: P12S, P79S, G237S, R238H, N296S, G299ins(NHGG), A311V
3.3 Recombinant phenotyping of new UL97 and UL27 mutations
Two independently constructed UL97 L337M mutant viruses showed the same low grade MBV resistance (3.4- to 3.5-fold increased EC50), less than the 9- to >200-fold increased EC50 reported for other UL97 MBV-resistance mutations (Table 4) (Chou, 2008). The L337M mutation did not affect ganciclovir susceptibility (EC50 = 1.17 ± 0.33 μM in 8 replicates, vs. 1.14 ± 0.40 μM for control strain T3261 (Chou, 2010)). The mutant grew normally in cell culture (Fig. 1), similar to other MBV-resistant UL97 mutants (Chou et al., 2007). UL97 variants I324V, S334G and S386L conferred no MBV resistance. UL27 mutants R448P and D534Y had slightly reduced MBV susceptibilities (~50% increases in EC50; p<0.001 for both mutations vs. wild type control, by unpaired Student t test).
Table 4.
Genotypes and phenotypes of recombinant CMV strains
| BACa | Virusb | UL97 genotype
|
UL27 genotype
|
Maribavir susceptibility
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mutation | Otherc | Mutation | Otherc | EC50, μMd | Sde | Ratiof | Ng | ||
| T2211 | H587Y | 0.11 | 0.03 | 71 | |||||
| BA29 | T3261 | Frt | 0.12 | 0.04 | 56 | ||||
| BA93 | T3386 | H587Y | Frt | 0.10 | 0.02 | 85 | |||
| BA194 | T3735 | Frt | R233S | 0.20 | 0.03 | 1.8 | 7 | ||
| BA148 | T3555 | H587Y | R448P | Frt | 0.17 | 0.04 | 1.5 | 23 | |
| BA147 | T3554 | H587Y | D534Y | Frt | 0.15 | 0.03 | 1.4 | 16 | |
| BA118 | T3433 | I324V | Frt | 0.07 | 0.01 | 0.6 | 9 | ||
| BA119 | T3434 | S334G | Frt | 0.12 | 0.03 | 1.1 | 11 | ||
| BA195 | T3667 | S334G | Frt | R233S | 0.22 | 0.06 | 2.0 | 14 | |
| T3138 | L337M | 0.37 | 0.08 | 3.4 | 21 | ||||
| BA15 | T3178 | L337M | Frt | 0.39 | 0.09 | 3.5 | 39 | ||
| BA194 | T3666 | L337M | Frt | R233S | 0.79 | 0.23 | 7.2 | 9 | |
| BA202 | T3679 | V353A | Frt | 1.27 | 0.37 | 12 | 8 | ||
| BA196 | T3668 | V353A | Frt | R233S | 2.94 | 0.51 | 27 | 13 | |
| BA110 | T3424 | S386L | Frt | 0.10 | 0.03 | 0.9 | 11 | ||
Bacterial artificial chromosome clone name
Recombinant virus strain
Other UL97 genetic differences from strain AD169, unrelated to drug resistance
Frt = 34-bp Flp recombinase recognition site upstream of gene
By SEAP yield reduction assay, mean value of the number of assays shown
Items associated with decreased MBV susceptibility shown in bold
Standard deviation of the number of assays shown
EC50 compared with median EC50 of baseline strains
Number of replicates (performed on at least 4 separate dates)
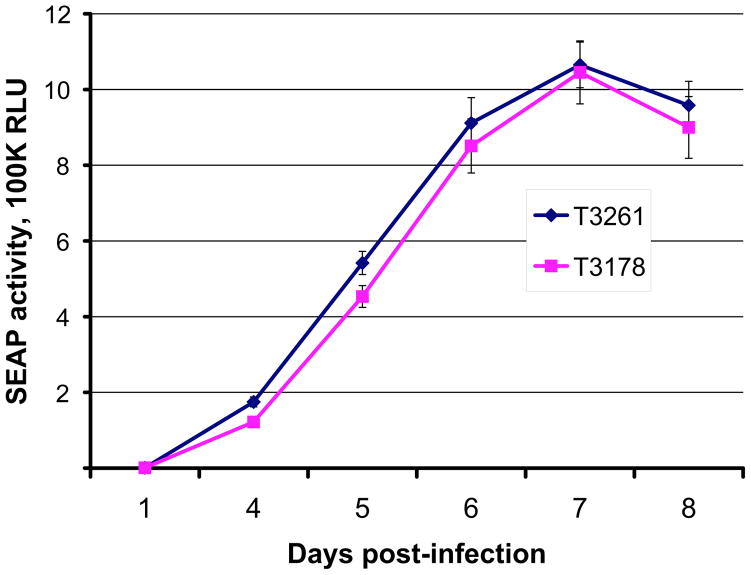
Figure 1. Growth curves of UL97 L337M mutant (T3178) and wild type CMV (T3261).
Growth was assayed in HFF cultures by supernatant SEAP activity (shown as 105 RLU, relative light units) at days 4 to 8 following equivalent low multiplicity infection as calibrated by SEAP activity at 24 hours post-infection (Chou et al., 2007). Each data point is the mean ± standard deviation of 4 replicates. Results shown are representative of 4 independent experiments.
3.4 MBV susceptibility of UL27–UL97 double mutants
While the clinical relevance of UL97 and UL27 mutations that individually confer low-grade MBV resistance in vitro remains unclear, mutations occurring in both UL27 and UL97 might combine to result in a greater overall degree of resistance. Because this had not been tested by recombinant phenotyping, CMV strains containing the well-characterized UL27 mutation R233S (Chou et al., 2004; Hakki et al., 2011) and the UL97 mutations L337M or V353A were constructed (Table 4). The introduction of UL27 mutation R233S into strains containing UL97 mutations L337M or V353A resulted in an approximately 2-fold increase in MBV EC50 over that of the UL97 single mutants, consistent with the effect of R233S alone (Table 4). Thus, the combination of two low-grade resistance mutations in UL27 and UL97 had a multiplier effect, resulting in a level of MBV resistance over baseline comparable to the level of ganciclovir resistance commonly observed in clinical isolates (Chou, 2008).
4. Discussion
New in vitro data expand the range of CMV UL97 structural domains involved in MBV resistance to include the conserved kinase p-loop, but nearby UL97 sequence variants detected in Phase III clinical trials did not confer MBV resistance.
Based on previous structure models of MBV binding to UL97 (Chou and Marousek, 2008) and biochemical evidence of MBV as an ATP-competitive UL97 kinase inhibitor (Shannon-Lowe and Emery, 2010), it was anticipated that MBV resistance mutations might occur in the ATP-binding p-loop (subdomain I, codons 337–345), which contains the 3 characteristic glycine resides at positions 338, 340 and 343 (Hanks et al., 1988). We now show that the p-loop mutation L337M is uncommonly selected in vitro to confer low-grade MBV resistance. The L337 residue is well conserved among diverse kinases (Hanks et al., 1988) but is evidently tolerant of mutation without significant impact on CMV replication (Fig. 1).
Despite proximity to known MBV resistance mutations, the unknown UL97 sequence variants I324V, S334G and S386L encountered in the clinical trials did not affect MBV susceptibility when assessed by recombinant phenotyping. These variants may represent unrecognized baseline polymorphism, given that UL97 codons below 400 are not routinely sequenced in the current diagnostic assay for ganciclovir resistance.
In contrast to the case of MBV resistance that developed within 10 weeks of drug exposure in an attempt to treat a starting plasma viral load of 1.8 million copies/mL, and associated with emergence of UL97 mutations T409M, H411Y and H411N (Strasfeld et al., 2010), no UL97 mutations known to confer MBV resistance were found in the Phase III CMV prevention clinical trials, including in study 301 where 50 samples represented a median duration of drug exposure of 14 weeks. The median viral load of ~6000 copies/mL, being orders of magnitude less than in the case report, is the most likely explanation for the difference in outcomes. In ganciclovir and valganciclovir CMV prevention trials involving similar patient populations, genotypic evidence of resistance was uncommon (0–3% of subjects), with no clear disease association when detected (Boivin et al., 2005; Chou et al., 2010).
Diverse mutations in the poorly understood CMV gene UL27 that slightly decrease MBV susceptibility continue to be discovered in vitro, including R448P and D534Y (Table 1). These mutations appear to be an adaptation to loss of UL97 kinase activity (Chou, 2009), possibly related to the proposed function of UL27 in downregulating the cellular protein Tip60 early in the viral infection cycle (Reitsma et al., 2011) and/or nucleolar localization of UL27 (Hakki et al., 2011). The clinical significance of UL27 mutations that confer low-level resistance (2–3 fold) remains uncertain. By permitting continued viral replication in the presence of MBV, such mutations may facilitate the emergence of additional mutations that, either singly or in combination with the pre-existing mutation, result in greater resistance to MBV. As shown here by construction of UL27–UL97 double mutants, two low-level resistance mutations together can have a multiplier effect on the MBV EC50.
Approximately 25% of the specimens from MBV recipients in the clinical trials contained uncharacterized UL27 amino acid variants, but so did an even larger fraction of specimens from placebo recipients, presumably reflecting the incompletely explored sequence polymorphism of this gene. The same fraction of samples showing such variants in studies 300 and 301 despite the much longer median MBV exposure duration in study 301 also suggests that most of these changes are unrecognized baseline sequence polymorphisms. A considerable recombinant phenotyping effort would be required to determine the effect of each of the new UL27 variants on MBV susceptibility, including possible effects in combination with changes in UL97, but some of the uncharacterized UL27 variants found in MBV recipients were also noted in placebo recipients or in connection with clinical isolates that tested susceptible (Table 3).
This study was focused on UL97 because of the consistent selection of localized UL97 mutations after MBV exposure in vitro. Without detection of any UL97 mutations similar to those that confer MBV resistance in vitro, it is implausible that drug resistance was an important factor in the MBV studies 300 and 301 failing to achieve their intended endpoints. Long experience with ganciclovir has shown that in vitro selected mutations were a reliable guide to the UL97 codon ranges later found to be mutated in resistant clinical isolates (Chou, 2008). However, genotypic resistance testing in the Phase III trials was limited to viral genes and codon ranges containing established MBV resistance mutations. It is possible that mutations elsewhere in the CMV genome can affect MBV susceptibility. This is a continuing area of investigation that will be guided by further in vitro selection studies and by genotypic data from future MBV recipients.
In the context of current results and the favorable toxicity profile of MBV, it would appear reasonable to explore further the clinical use of the drug. There is a need to optimize dosing and consider study endpoints (Marty and Boeckh, 2011), drug combinations and host cell conditions that may affect antiviral utility (Chou, 2008). A new Phase II dose ranging trial has recently been registered in Europe to compare MBV and valganciclovir for treatment of CMV infections in transplant recipients without end-organ disease (EudraCT number 2010-024247-32).
5. Conclusions
5.1
The CMV UL97 kinase region known to be involved in maribavir resistance is now expanded to include the ATP-binding p-loop (Hanks subdomain I), as exemplified by mutation L337M.
5.2
Previously unknown UL97 sequence variants I324V, S334G and S386L, encountered in Phase III maribavir clinical trials after 4 to 14 weeks of drug exposure, were shown to have no effect on maribavir susceptibility in vitro.
5.3
Mutations in UL27 and UL97 that individually conferred low-level maribavir resistance combined to confer a moderate level of maribavir resistance similar to that seen in typical ganciclovir-resistant clinical isolates.
Highlights.
CMV UL97 kinase ATP-binding p-loop is newly implicated in maribavir resistance
UL97 variants found in maribavir clinical trials conferred no maribavir resistance
UL27 and UL97 mutations combined to increase the level of maribavir resistance
Acknowledgments
We thank Gail Marousek, Alwin Borgmann, Victor Wong, Coyne Drummond and Ben Houser for technical assistance; Juli Miller and Walter Tatarowicz (ViroPharma) for assembling information on the clinical trial specimens. Genotyping of clinical study specimens was supported by ViroPharma in a Department of Veterans Affairs Collaborative Research and Development Agreement; all other aspects of this research were supported by NIH grant RO1-39938 and Department of Veterans Affairs research funds. MH was supported by a faculty development grant from the Sunlin and Priscilla Chou Foundation.
Footnotes
Conflicts of Interest: Stephen Villano is an employee of ViroPharma Incorporated. No other authors have any conflicts to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avery RK, Marty FM, Strasfeld L, Lee I, Arrieta A, Chou S, Tatarowicz W, Villano S. Oral maribavir for treatment of refractory or resistant cytomegalovirus infections in transplant recipients. Transpl Infect Dis. 2010;12:489–496. doi: 10.1111/j.1399-3062.2010.00550.x. [DOI] [PubMed] [Google Scholar]
- Biron KK, Harvey RJ, Chamberlain SC, Good SS, Smith AA, 3rd, Davis MG, Talarico CL, Miller WH, Ferris R, Dornsife RE, Stanat SC, Drach JC, Townsend LB, Koszalka GW. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob Agents Chemother. 2002;46:2365–2372. doi: 10.1128/AAC.46.8.2365-2372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin G, Goyette N, Gilbert C, Humar A, Covington E. Clinical impact of ganciclovir-resistant cytomegalovirus infections in solid organ transplant patients. Transpl Infect Dis. 2005;7:166–170. doi: 10.1111/j.1399-3062.2005.00112.x. [DOI] [PubMed] [Google Scholar]
- Chou S. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev Med Virol. 2008;18:233–246. doi: 10.1002/rmv.574. [DOI] [PubMed] [Google Scholar]
- Chou S. Diverse cytomegalovirus UL27 mutations adapt to loss of viral UL97 kinase activity under maribavir. Antimicrob Agents Chemother. 2009;53:81–85. doi: 10.1128/AAC.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Recombinant phenotyping of cytomegalovirus UL97 kinase sequence variants for ganciclovir resistance. Antimicrob Agents Chemother. 2010;54:2371–2378. doi: 10.1128/AAC.00186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Marousek G, Boivin G, Goyette N, Farhan M, Ives JA, Elston R. Recombinant phenotyping of cytomegalovirus sequence variants detected after 200 or 100 days of valganciclovir prophylaxis. Transplantation. 2010;90:1409–1413. doi: 10.1097/TP.0b013e3181fdd9d2. [DOI] [PubMed] [Google Scholar]
- Chou S, Marousek GI. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J Virol. 2008;82:246–253. doi: 10.1128/JVI.01787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Marousek GI, Senters AE, Davis MG, Biron KK. Mutations in the human cytomegalovirus UL27 gene that confer resistance to maribavir. J Virol. 2004;78:7124–7130. doi: 10.1128/JVI.78.13.7124-7130.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Van Wechel LC, Marousek GI. Cytomegalovirus UL97 kinase mutations that confer maribavir resistance. J Infect Dis. 2007;196:91–94. doi: 10.1086/518514. [DOI] [PubMed] [Google Scholar]
- Hakki M, Drummond C, Houser B, Marousek G, Chou S. Resistance to maribavir is associated with the exclusion of pUL27 from nucleoli during human cytomegalovirus infection. Antiviral Res. 2011;92:313–318. doi: 10.1016/j.antiviral.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hantz S, Couvreux A, Champier G, Trapes L, Cotin S, Denis F, Bouaziz S, Alain S. Conserved domains and structure prediction of human cytomegalovirus UL27 protein. Antivir Ther. 2009;14:663–672. [PubMed] [Google Scholar]
- Komazin G, Ptak RG, Emmer BT, Townsend LB, Drach JC. Resistance of human cytomegalovirus to the benzimidazole L-ribonucleoside maribavir maps to UL27. J Virol. 2003;77:11499–11506. doi: 10.1128/JVI.77.21.11499-11506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalezari JP, Aberg JA, Wang LH, Wire MB, Miner R, Snowden W, Talarico CL, Shaw S, Jacobson MA, Drew WL. Phase I dose escalation trial evaluating the pharmacokinetics, anti-human cytomegalovirus (HCMV) activity, and safety of 1263W94 in human immunodeficiency virus-infected men with asymptomatic HCMV shedding. Antimicrob Agents Chemother. 2002;46:2969–2976. doi: 10.1128/AAC.46.9.2969-2976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurain NS, Weinberg A, Crumpacker CS, Chou S. Sequencing of cytomegalovirus UL97 gene for genotypic antiviral resistance testing. Antimicrob Agents Chemother. 2001;45:2775–2780. doi: 10.1128/AAC.45.10.2775-2780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M, Feichtinger S, Milbradt J. Regulatory roles of protein kinases in cytomegalovirus replication. Adv Virus Res. 2011;80:69–101. doi: 10.1016/B978-0-12-385987-7.00004-X. [DOI] [PubMed] [Google Scholar]
- Marty FM, Ljungman P, Papanicolaou GA, Winston DJ, Chemaly RF, Strasfeld L, Young JA, Rodriguez T, Maertens J, Schmitt M, Einsele H, Ferrant A, Lipton JH, Villano SA, Chen H, Boeckh M. Maribavir prophylaxis for prevention of cytomegalovirus disease in recipients of allogeneic stem-cell transplants: a phase 3, double-blind, placebo-controlled, randomised trial. Lancet Infect Dis. 2011;11:284–292. doi: 10.1016/S1473-3099(11)70024-X. [DOI] [PubMed] [Google Scholar]
- Marty FM, Boeckh M. Maribavir and human cytomegalovirus – what happened in the clinical trials and why might the drug have failed? Curr Opin Virol. 2011;1:555–562. doi: 10.1016/j.coviro.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Prichard MN. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev Med Virol. 2009;19:215–229. doi: 10.1002/rmv.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma JM, Savaryn JP, Faust K, Sato H, Halligan BD, Terhune SS. Antiviral inhibition targeting the HCMV kinase pUL97 requires pUL27-dependent degradation of Tip60 acetyltransferase and cell-cycle arrest. Cell Host Microbe. 2011;9:103–114. doi: 10.1016/j.chom.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Shannon-Lowe CD, Emery VC. The effects of maribavir on the autophosphorylation of ganciclovir resistant mutants of the cytomegalovirus UL97 protein. Herpesviridae. 2010;1:4. doi: 10.1186/2042-4280-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasfeld L, Lee I, Villano S, Chou S. Virologic characterization of multi-drug-resistant cytomegalovirus infection in two transplant recipients treated with maribavir. J Infect Dis. 2010;202:104–108. doi: 10.1086/653122. [DOI] [PubMed] [Google Scholar]
- Winston DJ, Young JA, Pullarkat V, Papanicolaou GA, Vij R, Vance E, Alangaden GJ, Chemaly RF, Petersen F, Chao N, Klein J, Sprague K, Villano SA, Boeckh M. Maribavir prophylaxis for prevention of cytomegalovirus infection in allogeneic stem-cell transplant recipients: a multicenter, randomized, double-blind, placebo-controlled, dose-ranging study. Blood. 2008;111:5403–5410. doi: 10.1182/blood-2007-11-121558. [DOI] [PMC free article] [PubMed] [Google Scholar]