SUMMARY
Nucleotide excision repair (NER) has long been known to remove DNA lesions induced by chemical carcinogens, and the molecular mechanism has been partially elucidated. Here we demonstrate that in S.pombe a DNA recognition protein, alkyltransferase-like 1 (Atl1), can play a pivotal role in selecting a specific NER pathway dependent on the nature of the DNA modification. The relative ease of dissociation of Atl1 from DNA containing small O6-alkylguanines allows accurate completion of global genome repair (GGR), whereas strong Atl1 binding to bulky O6-alkylguanines blocks GGR, stalls the transcription machinery and diverts the damage to transcription-coupled repair. Our findings redraw the initial stages of the NER process in those organisms that express an alkyltransferase-like gene and raise the question of whether or not O6-alkylguanine lesions that are poor substrates for the alkyltransferase proteins in higher eukaryotes might, by analogy, signal such lesions for repair by NER.
INTRODUCTION
Alkylating agents are a diverse class of genotoxins that elicit a wide range of adverse biological effects in living organisms. Alkylation of the O6-position of guanine in DNA, if not repaired before DNA replication, can give rise to transition mutations and recombination events in surviving cells, or can be cytotoxic. In the case of O6-methylguanine (O6-MeG), toxicity is due to the action of the post-replication mismatch repair (MMR) system. O6-alkylguanines (O6-alkG) in DNA have long been known to be substrates for O6-alkylguanine-DNA alkyltransferases (AGTs), proteins that transfer the alkyl group to a cysteine residue in the highly conserved active site PCHRV/I motif. AGTs are thereby able to protect cells and organisms against the biological effects of alkylating agents (Margison et al., 2002).
The alkyltransferase-like (ATL) proteins are a recently-discovered family that exists in prokaryotes and lower eukaryotes (Margison et al., 2003). They have amino acid sequence motifs that resemble those found in AGTs, but the cysteine residue in the binding site motif is replaced, usually by tryptophan. We previously demonstrated that ATL proteins from E. coli (eATL) and S. pombe (Atl1) lack the ability to carry out repair by alkyl group transfer or removal, and that they did not display the glycosylase or endonuclease activity that is characteristic of the base excision repair (BER) pathway (Pearson et al., 2005; Pearson et al., 2006). However, deletion of the Atl1-encoding gene in S. pombe increased its sensitivity to the toxic effects of a number of alkylating agents (Margison et al., 2007; Pearson et al., 2006) and the mutagenic effects of the methylating agent, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (Tubbs et al., 2009). Thus Atl1 is involved in protecting S. pombe against the biological effects of alkylating agents.
Crystal structures of Atl1 bound to oligodeoxyribonucleotide (ODN) containing O6-MeG or O6-(4-oxo-4-(3-pyridyl)butyl)guanine (O6-PobG) have shown that the alkylated guanine is rotated into a binding pocket and that there is considerable DNA bending (45°). Previous genetic analyses showed that atl1 is epistatic with rad13 (human XPG) and swi10 (human ERCC1) (Tubbs et al., 2009). Together, these results suggest that Atl1 binding to O6-alkylguanines results in structural changes in DNA that presents these lesions to the nucleotide excision repair (NER) pathway. However, prior to the current study, it remained unknown if the global genome repair (GGR) or transcription coupled repair (TCR) branches of NER were engaged by Atl1.
In the present report, we have carried out epistasis and cell cycle analysis of the interactions between atl1 and GGR and TCR genes. We show that Atl1 is vital for initiating the processing of damage produced by simple alkylating agents principally via the GGR pathway. In contrast, Atl1 exacerbates the toxicity of lesions generated by bulky alkylating agents in NER-defective cells. This occurs more extensively in cells defective in TCR, probably as a direct or indirect consequence of stalling at replication forks. The crystal structures and interaction kinetics of Atl1 with a range of O6-alkylguanines of different complexity suggest that different affinities of Atl1 for simple and bulky DNA lesions determine which pathway is engaged in lesion processing.
RESULTS
NER pathway selection by Atl1 is alkylating agent-dependent
The processing of DNA lesions by NER involves damage recognition, repair complex assembly and damage removal. Broadly, in humans, damage in transcribed genes can stall transcription complexes resulting in CSB-mediated TCR while damage elsewhere in the genome is recognised by the DDB1–DDB2 and XPC-hHR23b complexes that initiate GGR. Both pathways subsequently involve the transcription complex TFIIH, XPA and RPA, the elimination of the damage by the endonucleases XPF and XPG and the restoration of the DNA by polymerase and ligase.
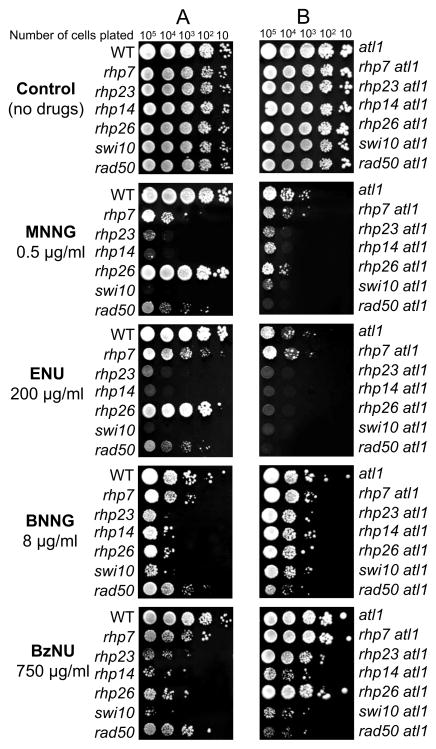
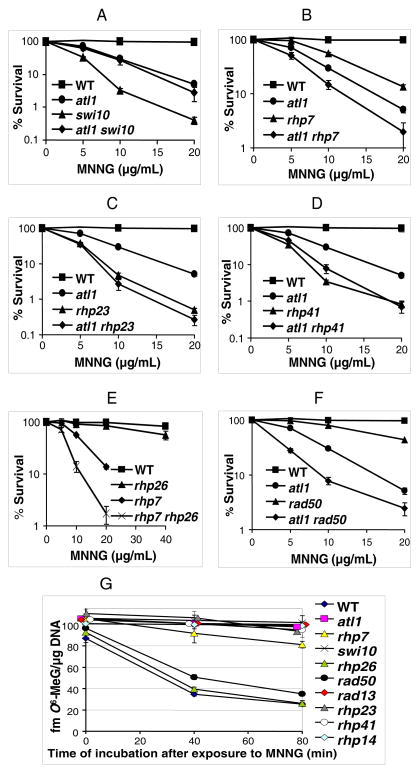
In S.pombe, standard “spot” and clonogenic assays showed that deletion of atl1 or deletions of the GGR genes, rhp23, rhp14, swi10, rhp41, and rhp7 (human HR23B, XPA, ERCC1, XPC and DDB2) sensitised cells to MNNG, whereas deletions of the TCR gene, rhp26 (human CSB) or the homologous recombination (HR) gene rad50 (human Rad50), did not (Figure 1A, 2). We also demonstrated that atl1 is epistatic with rhp23, rhp41 and swi10 but additional deletion of atl1 increased MNNG toxicity in the rhp7 and rhp14 deletants (Figure 1B, 2A–D). The very high sensitivity of the rhp7 rhp26 double deletant strain (Figure 2E) strain suggests that TCR can partially compensate for GGR deficiency when cells are exposed to MNNG. The atl1 rad50 double deletant was also extremely sensitive (Figure 2F), indicating that HR is a backup pathway in the absence of Atl1.
Figure 1. Agar plate (“spot”) assays for atl1-, NER- and HR - deficient mutants after treatment with MNNG, ENU, BNNG and BzNU.
(A) Sensitivity of WT strain (GM1) and single deletants of rhp7 (GM56), rhp23 (GM50), rhp14 (GM25), rhp26 (GM51), swi10 (OL455) and rad50 (GM11)
(B) Sensitivity of atl1 single (GM3) and the double deletants: atl1 rhp7 (GM61), atl1 rhp23 (GM100), atl1 rhp14 (GM32), atl1 rhp26 (GM60), atl1 swi10 (GM48) and atl1 rad50 (GM+15). See also Figure S1.
Figure 2. Survival of WT and deletant S. pombe strains and persistence of O6-MeG in DNA after exposure to MNNG.
(A–F) MNNG-sensitivity measured by clonogenic survival assays for WT (GM1) and deletants: atl1 (GM3)
(A) swi10 (OL455), atl1 swi10 (GM48)
(B) rhp7 (GM56), atl1 rhp7 (GM61)
(C) rhp23 (GM50), atl1 rhp23 (GM100)
(D) rhp41 (TM4), atl1 rhp41 (GM46)
(E) rhp26 (GM51), rhp7 (GM56), rhp7 rhp26 (GM121)
(F) rad50 (GM11), atl1 rad50 (GM15)
(G) The amount of O6-MeG in DNA at various times after 10 min exposure to MNNG (10 μg/mL).
Values are the mean of triplicate or quadruplicate treatments and error bars represent standard error of the mean.
The atl1 and swi10 deletants were not sensitive to another commonly used methylating agent methyl methanesulfonate (MMS, Figure S1), which generates relatively low levels of O6-MeG in DNA (Beranek, 1990; Singer and Grunberger, 1983). This agrees with the suggestion that O6-MeG is the principal lethal lesion induced by MNNG. To confirm this, we determined O6-MeG levels in DNA extracted from various strains following a short exposure to MNNG. In WT, rhp26Δ and rad50Δ cells there was a rapid decrease in O6-MeG levels with time (Figure 2G) with more than 50% repair (~1200 O6-MeG residues removed) within 40 min. In contrast, there was little or no removal of O6-MeG in the swi10, rad13, rhp23, rhp14 and rhp41 deletants up to 80 min after treatment. There was also some repair of O6-MeG in the rhp7 deletant, but to a much lesser extent than in the WT strain (Figure 2G). O6-MeG therefore persisted in the DNA of those deletants that displayed increased sensitivity to MNNG but was removed from the DNA of the resistant strains.
We then examined sensitivity of WT strain and atl1, rhp7, rhp23, rhp14, rhp26, swi10 and rad50 deletants to the ethylating agent N-ethyl-N-nitrosourea (ENU) and two agents that introduce bulkier lesions into DNA i.e. N-butyl-N′-nitro-N-nitrosoguanidine (BNNG) and N-benzyl-N-nitrosourea (BzNU). Using spot assays, we were intrigued to find that while deletion of atl1 increased sensitivity to ENU, it had no effect on sensitivity to BNNG or BzNU (Figure 1B). Moreover, while deletion of the TCR gene rhp26 had no effect on sensitivity to ENU it extensively increased sensitivity to BNNG and BzNU. Deletion of the GGR genes rhp23, rhp14 and swi10 significantly increased the toxicity to all alkylating agents whereas deletion of rhp7 had only marginal effects on sensitivity in all cases (Figure 1A).
Our earlier investigations had suggested that Atl1 would bind to a range of O6-alkylguanines in DNA (Pearson et al., 2006), so the lack of any effect of atl1 deletion on sensitivity to the bulkier alkylating agents was unexpected. Investigating the basis of this in double deletants, we found that deletion of atl1 complemented the sensitivity of all NER-deficient strains, but particularly the rhp26 deletant, to BNNG and BzNU (Figure 1B). The effect of atl1 deletion on the sensitivity to BNNG and BzNU was only observed in the rad50Δ strain, indicating that the HR pathway compensates for the absence of Atl1.
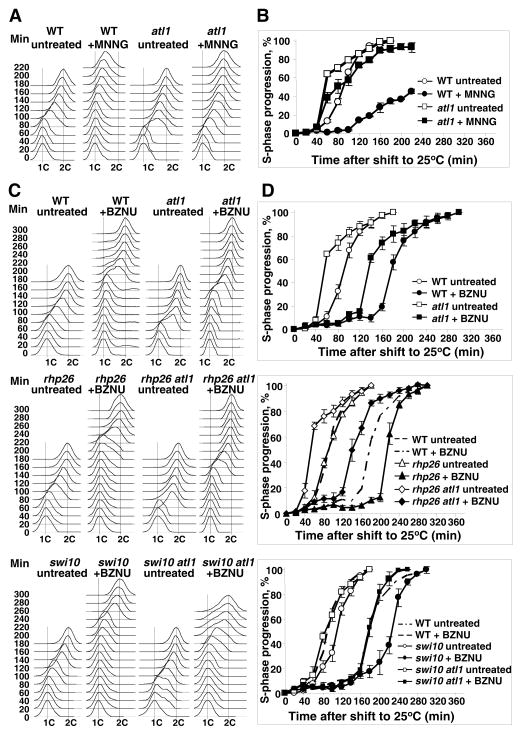
Atl1 binding delays chromosome replication after high dose MNNG or low dose BzNU
To examine if Atl1 binding to O6-alkylguanine residues might result in a DNA replication block, we used flow cytometry to measure S-phase progression after release of G1-arrested cdc10-M17 following brief exposure to MNNG or BzNU. The shift of the histogram peaks over time (Figure 3A and C) was used to quantitate progression through S-phase (Figure 3B and D). At low doses of MNNG, there was no indication of delayed progression in WT or atl1, swi10 or rhp26 deletants (data not shown). However, after a very high dose of MNNG there was an additional delay of ~60 min in WT cells. This delay was essentially attributable to Atl1 since the progression profile after MNNG- treatment of the atl1 deletant overlapped with that of the untreated WT (Figure 3B).
Figure 3. Flow cytometric quantitation of S-phase progression after MNNG and BzNU treatment.
Cdc10-M17 cells grown at 25°C and synchronized in G1 by shifting to 37°C for 4.5 hours. Before release from arrest, cells were treated with BNNG or MNNG (10 min), centrifuged and resuspended in YEL. Samples were taken every 20 min and nuclear DNA content was measured by flow cytometry.
(A) Overlays of S-phase flow cytometry histogram stacks and
(B) S-phase progression after treatment with high concentrations of MNNG leading to ~0.5% cell survival. S-phase progression (%) was measured by the relative shifting of the mean of S-phase peaks from unreplicated 1C toward fully replicated 2C values preset as 0% and 100% respectively.
(C) Overlays of S-phase flow cytometry histogram stacks and
(D) S-phase progression comparing WT (SP413), atl1 (GM62), rhp26 (GM106), rhp26 atl1 (GM107), swi10 (GM108), swi10 atl1 (GM109) strains after treatment with BzNU (0.5 mg/mL). Values are the mean of triplicate experiments and error bars represent SEM. The values of cell survival were as follows: wild-type (27%), atl1 (32.7%), rhp26 (13%), rhp26 atl1 (17.5%), swi10 (8.8%), swi10 atl1 (25.8%). See also Figure S3.
To investigate G1-arrest by BzNU, we used doses that resulted in ~27% cell survival: higher doses caused complete suppression of replication (data not shown). We observed a substantial (~80 min) delay after releasing the replication block in WT cells treated with BzNU. Deletion of atl1 shortened this delay by ~40 min. These data suggest that the delay in replication onset is, at least in part, Atl1-mediated. Deletions of the rhp26 and swi10 genes had little or no impact on the delay of replication onset in untreated cells, but caused a very substantial delay in BzNU treated cells. Again, deletion of the atl1 gene shortened the S-phase delay in both of these deletants but the extent of the shortening was much more substantial in the rhp26 deletant strain (Figure 3C).
O6-methylguanine is less toxic than O6-benzylguanine
To compare the relative toxic effects of O6-MeG and O6-BnG in chromosomal DNA, we treated S. pombe wild-type cells with two concentrations each of MNNG and BzNU which resulted in ~60% and ~30% survival. We found ~6030 and ~8030 O6-MeGs and ~185 and ~206 O6-BnGs per cell respectively (Table S3), suggesting that O6-BnG is approximately 20 times more toxic than O6-MeG: on average one O6-BnG per 72000 bases will kill ~60% of cells.
Structural analysis of Atl1 bound to O6-alkylguanine-containing ODN duplexes
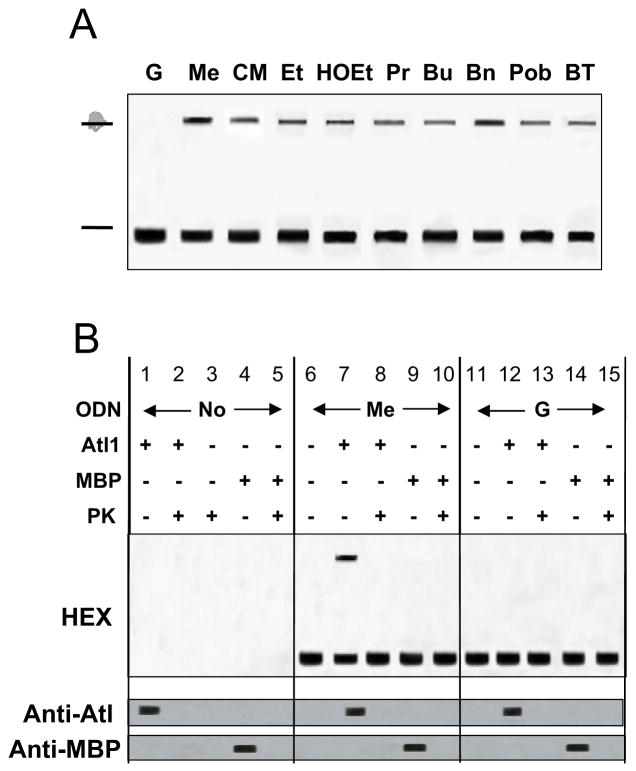
To determine if the above observations might reflect differences in Atl1-O6-alkylguanine interactions, we initially used electrophoretic mobility shift assays (EMSA) to assess binding to ODN duplexes containing a range of O6-alkylguanines. No shift was seen with the control (G-containing) ODN, but ODN containing single O6-methyl (Me), carboxymethyl (CM), -ethyl (Et), -hydroxyethyl (HOEt), -propyl (Pr), -butyl (Bu) -benzyl (Bn), -pyridyloxobutyl (Pob) or -4-bromothenyl (BT) guanine residues were shifted (Figure 4A). These ODN shifts were ablated by proteinase K digestion of the Atl1-ODN complex and no shifts were seen with the non-DNA binding protein maltose-binding protein (MBP) (Figure 4B and 4S).
Figure 4. Qualitative assessment of the binding of Atl1 to ODN by EMSA.
(A) Atl1 binds specifically to ODNs containing O6-alkylguanines.
ODNs were annealed by mixing complement (5′-[HEX]-GGGCCAGCACGGAGCTGCAGTTC) with the “lesion”-containing ODN (5′-GAACTXCAGCTCCGTGCTGGCCC, where X is guanine (G) or the modified bases: O6-methyl (Me), -carboxymethyl (CM), -ethyl (Et), -hydroxyethyl (HOEt), -n-propyl (Pr), -butyl (Bu) -benzyl (Bn), -pyridyloxobutyl (Pob) and -4-bromothenyl (BT). Atl1 (5 μM) was incubated with duplex ODN (1 μM) in total volume of 10 μL of reaction buffer then electrophoresed. Gels were scanned using a Pharos laser scanner (BioRad).
(B) Additional EMSA controls for O6-MeG-containing ODN which include incubation with the non-DNA binding protein MBP and incubation of Atl1, MBP or the ODN incubation mixes with proteinase K (PK). The upper gel photograph was obtained a Pharos laser scanner and the gel was electroblotted to nitrocellulose. The Atl1 (centre gel photograph) and MBP proteins (lower gel photograph) and their degradation by PK were visualized by western blot with rabbit anti-Atl1 polyclonal antiserum or anti-MBP antibodies followed by incubation with horseradish peroxidase-labeled anti-rabbit antibodies and detection by enhanced chemiluminescence. Data for all other ODNs are shown in Figure S4.
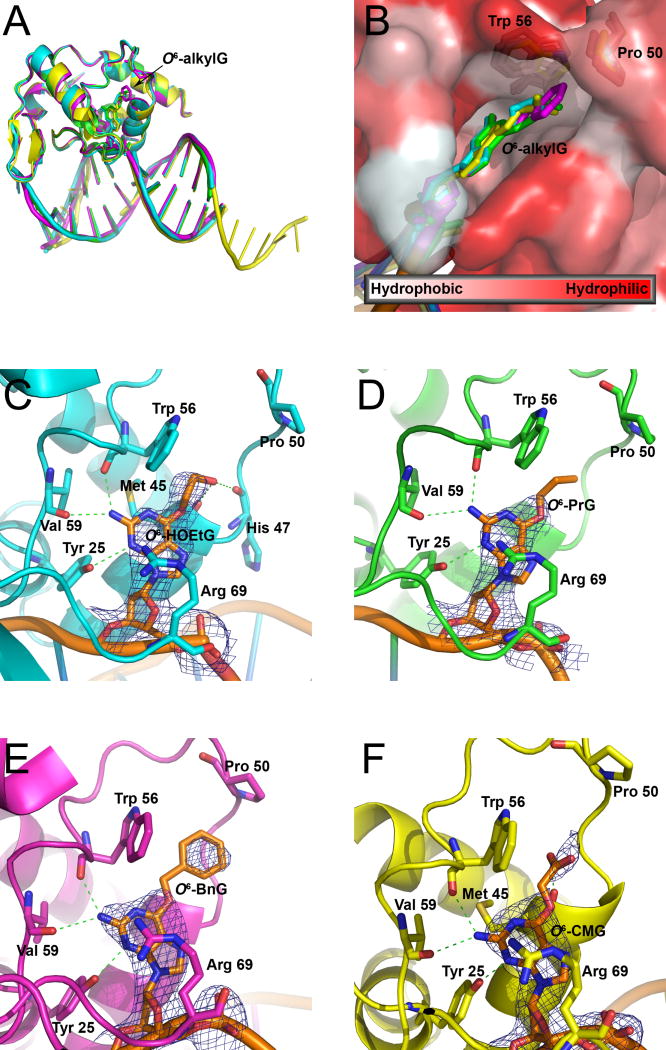
We then solved crystal structures for Atl1 in complex with duplex ODNs containing O6-HOEtG, O6-PrG, O6-BnG and O6-CMG (Figure 5C–F, Table 1). All complexes crystallized in space group P6122 or P41212 with very similar overall structures (Figure 5A). For each lesion, the damaged base is rotated out of the DNA helix and into the Atl1 binding pocket, with Arg 39 donating a hydrogen bond to the orphaned cytosine to stabilize the extrahelical DNA conformation. Also, in all cases, the DNA is bent ~45°. These observations are consistent with previous crystal structures of Atl1 in complex with ODN duplexes of the same sequence containing O6-MeG or O6-PobG (6). O6-HOEtG, O6-PrG, O6-BnG and O6-CMG adducts fit within the predominantly hydrophobic Atl1 binding pocket, which is capped by Pro 50, without significant rearrangement of side chains for surrounding amino acid residues. The predominant interactions between the hydroxyethyl, propyl, and benzyl side chains of the adducts and the amino acid side chains in the interior of the Atl1 binding pocket are hydrophobic in nature (Figure 5B). Additional hydrogen bonds with neighbouring carbonyl oxygen atoms stabilize the HOEtG and CMG adducts. Unable to make similar interactions, the propyl and benzyl groups show increased mobility as signified by higher B factors and reduced electron density for these adducts. The higher mobility of propyl and benzyl groups may facilitate their alignment with the hydrophobic interior of the binding pocket and contribute to the high affinity of Atl1 for these alkyl groups. Together, these results indicate that a variety of lesions can be accommodated in the Atl1 binding pocket. Larger groups make stronger hydrophobic interactions than smaller groups or groups that can establish weaker hydrogen bonding interactions, suggesting the larger lesions may bind Atl1 more tightly. Since this might influence the downstream interactions that determine how the lesion is processed, we undertook quantitative assessment of Atl1 binding using lesion-containing ODN duplexes.
Figure 5. Crystal structures for Atl1 in complex with ODN containing O6-HOEtG (cyan) O6-PrG (green), O6-BnG (magenta), or O6-CMG (yellow).
(A) Structural overlay of Atl1:ODN complexes showing similar DNA bend.
(B) Molecular surface of Atl1 lesion-binding pocket colored by hydrophobicity (white=most hydrophobic; red=least hydrophobic) and overlaid with Atl1:ODN complex structures. Trp 56 and Pro 50 are shown for reference.
(C–F) Binding of O6-HOEtG (C), O6-PrG (D), O6-BnG (E), and O6-CMG (F) within the Atl1 binding pocket. 2Fo-Fc omit electron density (blue) is shown for each O6-alkylG adduct (orange). Hydrogen bonds are shown as dashed green lines. The accompanying crystallographic data are listed in Table 1.
Table 1.
X-ray Crystallography Data Collection and Refinement Statistics
| O6-HOEtG | O6-PrG | O6-BnG | O6-CMG | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6122 | P6122 | P6122 | P41212 |
| Cell dimensions | ||||
| a, b, c (Å) | 59.8, 59.8, 232.7 | 59.5, 59.5, 236.9 | 59.9, 59.9, 237.1 | 85.2, 85.2, 150.7 |
| αβγ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 50.0-3.10 (3.21-3.10) | 50.0-3.05 (3.16-3.05) | 50.0-2.84 (2.94-2.84) | 50.0-2.85 (2.95-2.85) |
| Rsym | 6.5 (51.6) | 5.0 (34.9) | 9.0 (45.9) | 6.2 (49.9) |
| I/σI | 49.0 (2.2) | 59.5 (2.7) | 22.8 (2.1) | 31.2 (1.9) |
| Completeness (%) | 97.1 (77.5) | 97.6 (82.3) | 93.0 (61.1) | 95.6 (77.0) |
| Redundancy | 14.0 | 15.2 | 10.1 | 6.7 |
| Refinement | ||||
| Resolution (Å) | 38.7-3.10 | 34.8-3.05 | 39.0-2.84 | 43.3 - 2.85 |
| No. reflections | 4488 | 4797 | 5915 | 11727 |
| Rwork/Rfree | 22.5/28.3 | 25.0/29.5 | 17.3/25.3 | 22.0/28.2 |
| No. atoms | ||||
| Protein | 891 | 898 | 891 | 1782 |
| Ligand/ion | 530 | 530 | 534 | 1076 |
| Water | 4 | 1 | 0 | 0 |
| B-factors | ||||
| Protein | 147.0 | 139.5 | 105.9 | 122.6 |
| Ligand/ion | 137.1 | 126.2 | 113.6 | 126.6 |
| Water | 98.8 | 88.79 | N/A | N/A |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.010 | 0.010 | 0.008 | 0.011 |
| Bond angles (°) | 1.6 | 1.6 | 1.3 | 2.0 |
One crystal was used for each dataset. Values in parentheses are for highest-resolution shell.
The affinity of Atl1 for O6-alkylguanines increases with the size of the alkyl group
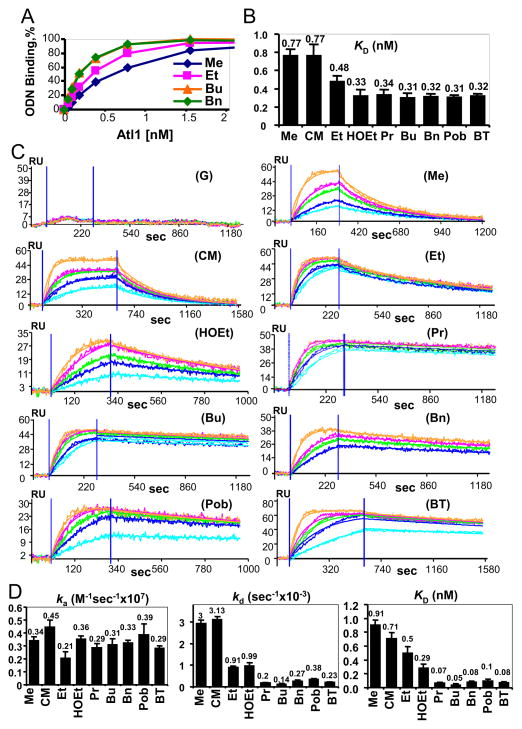
Binding of Atl1 to control or modified duplex ODN that had been immobilized via a 5′-terminal biotin to SA-coated microtitre plates was quantified by ELISA. Typical Atl1 concentration-dependence curves and extrapolated KD values are shown in Figure 6A and B respectively. No Atl1 binding was seen using control (G-containing) duplex ODN (data not shown) and its affinity for duplex ODNs containing O6-MeG or O6-CMG was similar but less than for the other duplexes, the most potent of which contained O6-BuG, O6-BnG, O6-PobG and O6-BTG: binding to O6-EtG showed an intermediate value.
Figure 6. Quantitative assessment of the binding of Atl1 to ODN by ELISA and SPR.
(A) Direct ELISA: effect of incubation of increasing concentrations of Atl1 with ODN duplexes bound via 5′-terminal biotin to SA-coated plates; typical binding curves.
(B) IC50 values extrapolated from the direct ELISA curves.
(C) SPR sensograms for the indicated ODN.
(D) SPR-determined interaction constants ka (association ‘on’ rate), kd (dissociation rate) and KD (dissociation constant). Values are means of at least triplicate determinations and error bars indicate standard error of the mean.
We next used surface plasmon resonance (SPR) to quantify the binding of Atl1 to, and dissociation from, control or modified duplex ODN. Sensorgrams (Figure 6C) indicated similarities in association (“on”) rates (ka values 0.21–0.45 M−1sec−1 × 107), but substantial differences in dissociation (“off”) rates (kd values 0.14 – 3.13 sec−1 × 10−3 (Figure 6D). The resulting KD values ranged over more than one order of magnitude (0.05–0.91 nM; Figure 6D) and clearly demonstrated that the affinities of Atl1 for bulky O6-alkylguanines (0.05–0.1 nM) were substantially higher than for the simpler lesions (0.5–0.91 nM).
DISCUSSION
The NER pathways have not been extensively investigated in S. pombe, but by analogy with the systems in S. cerevisiae and higher eukaryotes, are likely to be differentiated by the lesion recognition step. In GGR, DNA damage is detected throughout the genome of S. cerevisiae by the RAD7-RAD16-Ecl1-Cullin3 (NEF4) complex sharing certain functional similarities with human DDB1-DDB2-Roc1-cullin4 complex (Fuss and Tainer, 2011; Prakash and Prakash, 2000; Reed, 2011). The equivalent complex in S. pombe is rhp7-rhp16-based (Lombaerts et al., 1999). In S. cerevisiae, the initial step is followed by the recruitment of the RAD4-RAD23-Rad33 complex (Rhp41-Rhp23-Cdc31 in S. pombe and XPC-hHR23b-centrin-2 in H. sapiens, (den Dulk et al., 2008; Lombaerts et al., 2000; Marti et al., 2003; Nouspikel, 2009; Paoletti et al., 2003). In TCR, damage in the transcribed strand stalls the transcription complex and CSB (RAD26 in S. cerevisiae and Rhp26 in S. pombe) initiates the repair process (Fousteri and Mullenders, 2008; Yasuhira et al., 1999). Subsequently the two pathways merge with the recruitment of transcription factor II H (TFIIH) and various other factors that eventually result in the excision of the lesion within a short fragment (23–27 nucleotides) of DNA (reviewed in (Fousteri and Mullenders, 2008; Fuss and Tainer, 2011; Nouspikel, 2009)).
Atl1 protects against the toxic effects of MNNG
The present results indicate that in S.pombe, protection against the lethal effects of MNNG requires the GGR-specific genes rhp7, rhp23 and rhp41, but not the TCR-specific gene rhp26, the BER gene apn2 (Figure S1) or the HR gene rad50. We also show that the lesion predominantly responsible for killing by MNNG is O6-MeG.
Atl1 binds strongly to DNA containing O6-MeG in vitro, and might thus be expected to fulfil the role of the Rhp7 complex (see above). However, the rhp7 deletant was moderately sensitive to MNNG and impaired in its ability to remove O6-MeG. Thus Rhp7 seems to be at least partially involved in these processes. In human cells, what seems to be important for the subsequent step in NER is that opposite the damaged bases there are unpaired bases to which the XPC-hHR23b-centrin-2 complex binds (Min and Pavletich, 2007; Nouspikel, 2009). Our crystal structures show that Atl1 binding to O6-MeG generates a single unpaired C in the opposite strand, and while this might be anticipated to be capable of recruiting the Rhp41 complex, there would then be no obvious role for the Rhp7 complex. It therefore seems reasonable to propose a model (Figure 7) in which, for the majority of O6-MeG lesions, the Rhp7 complex facilitates Rhp41 binding via an interaction with the Atl1- lesion complex, possibly involving chromatin remodelling, or displacement of Atl1, and hence generating a more extensive DNA melting region for Rhp41 complex binding.
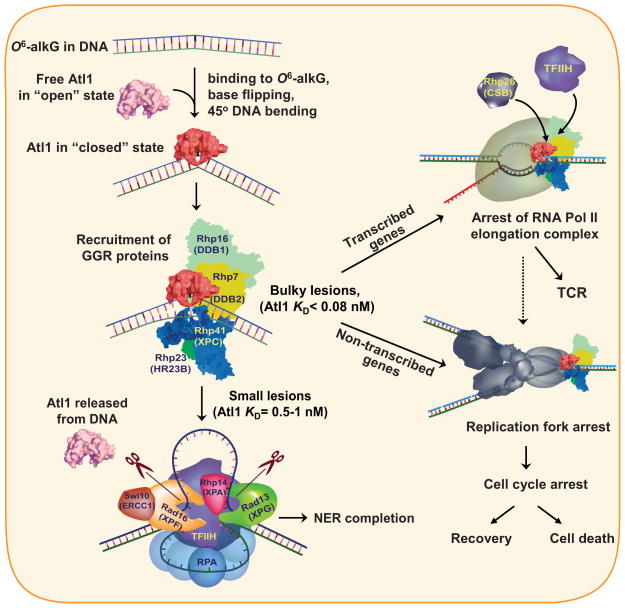
Figure 7. Schematic representation of the role of Atl1 in repair of O6-alkylguanines.
Atl1 binding to small O6-alkylguanine modifications activates GGR whereas bulky O6-alkylguanines engage both GGR and TCR, block chromosomal replication and invoke cell cycle arrest or cell death.
The toxicity of O6-MeG is likely to be a consequence of replication of O6-MeG-containing DNA giving rise to O6-MeG:T mispairs. These are recognised by the post replication MMR system, which results in a reiterative cycle of resynthesis and degradation of the T-containing strand (Hickman and Samson, 2004). After a further round of replication the gapped duplex results in DNA double-strand breaks (DSBs) that can be processed by recombination repair or give rise to lethality. That the atl1 rad50 double deletant was extremely sensitive to MNNG toxicity (Figure 1 and 2F) confirms that the toxic effects of MNNG can be rescued by the HR machinery.
After high doses of MNNG, the abundance of Rhp7/Rhp16 and Rhp41/Rhp23 complexes may not be sufficient to initiate GGR at all DNA lesions and consequently some Atl1-O6-MeG complexes may persist long enough to block DNA replication, either directly or by stalling RNA polymerase II. Indeed, after exposure of WT cells to high doses of MNNG, we observed substantial delay in S-phase progression and degradation of the RNA polymerase II large subunit, Rpb1 that was completely dependent on Atl1 (Figure 3A and B, S2). This indicates that Atl1-O6-MeG complexes can stall RNA polymerase II: the stalled transcription complex probably then stalls DNA replication.
Processing of O6-alkylguanines generated by higher alkylating agents
As we had seen with MNNG, deletion of atl1 increased the sensitivity of S. pombe to ENU but we were surprised to observe no impact on the sensitivity to BNNG or BzNU which generate the bulkier lesions, O6-BuG and O6-BnG in DNA. Furthermore, deletion of the TCR gene rhp26, which had no effect on sensitivity to MNNG or ENU, vastly increased sensitivity to BNNG and BzNU, while deletion of swi10, rhp7, rhp23 and rhp14 increased sensitivity to all of these alkylating agents (Figure 1A). For these bulkier lesions, the increased sensitivity in NER deletants was complemented by deletion of atl1 and to a greater extent in the rhp26 deletant. In addition, BzNU pulse treatment of G1-arrested WT cells resulted in a profound delay in DNA replication onset that was substantially shortened in the atl1-deletant. There were also increases in the length of the S-phase delay in rhp26 and swi10 deletants and again these were reduced by the additional deletion of atl1, the most extensive effect occurring in the rhp26 deletant. These observations indicate that in the NER deletants, Atl1 binding to the bulkier O6-alkylguanines leads to both transcription and replication blockage as shown in our model (Figure 7) and seen after very high doses of MNNG.
So why is there no phenotypic effect of atl1 deletion? Given that the delay of replication onset was also observed in the atl1 deletant, and all alt1-NER double deletants have shown increased sensitivity to the bulky agents, it is likely that, in the absence of Atl1, bulky O6-alkylguanines can be processed by TCR or GGR, but less effectively, causing replication arrest, but without increased killing. In UV-irradiated S. pombe cells, bulky cyclobutane pyrimidine dimers (CPD) initiate TCR when they occupy the translocation site within the RNA pol II core subunit (Brueckner et al., 2007). Thus in the absence of Atl1, bulky O6-alkylguanines may mimic the effect of CPD. That rad50 deletion significantly increased sensitivity of atl1 deficient cells to BNNG and BzNU (Figure 1B) indicates that HR is involved in recovery from replication fork arrest and explains why the atl1 deletant is not sensitive to bulky agents. The ability of BzNU to block replication explains why O6-BnG appears to be ~20 times more toxic than O6-MeG. Another consequence of this is that WT, atl1 and other NER deletant cells progressing through S-phase become substantially more sensitive to killing by BZNU, but not MNNG (Figure S3).
In E.coli the ATL protein strongly enhances the repair of O6-HOEt-, -1-hydroxypropyl- and -2-hydroxypropyl- guanine by NER (Mazon et al., 2009), and protects these adducts, but not O6-MeG, against MMR-mediated toxicity (Mazon et al., 2010). This could also be explained by a high affinity of eATL for bulky O6-alkylguanines and indicates functional similarity between the S.pombe and E.coli proteins.
Atl1 complexes with DNA containing a variety of O6-alkylguanines share common structures but different affinities
We sought to establish a biochemical basis for the effects of Atl1 by determination of its crystal structures and kinetic interaction characteristics with ODN containing different O6-alkylguanines.
The crystal structures show, in all cases, that the alkylated base is flipped out of the helix and the DNA phosphodiester backbone undergoes a 45° bend. This remarkable similarity of the three-dimensional structures for all of the Atl1-DNA complexes suggests that neither the shape nor the extent of DNA helix distortion introduced by Atl1 can adequately explain ability of Atl1 to shuttle repair via different repair pathways for small and bulky O6-alkylguanines. However, the Atl1 binding pocket can indeed accommodate a very wide variety of lesions, allowing numerous possibilities for interaction of the alkyl groups with amino acid residues in and around the binding pocket. Thus within the binding pocket, longer and bulkier groups make more hydrophobic interactions than smaller groups, or groups that can form weaker hydrogen bonds, potentially allowing tighter binding of the larger lesions.
Given the above observations, we used ELISA to determine equilibrium binding constants and SPR to measure the association and dissociation rates of the Atl1-DNA complexes and the corresponding dissociation constants. The ELISA results revealed similar and relatively low Atl1 affinity to ODN containing O6-MeG and O6-CMG and much higher affinities to substrates containing the other lesions, with O6-EtG being intermediate. In SPR measurements, we found that the association (‘on’) rates for all ODN were very similar, but the rates of dissociation (“off” rates) for those containing O6-MeG and O6-CMG were more than one order of magnitude higher than those of ODN containing Pr, Bu, Bn, BT and Pob lesions. Overall, the dissociation constants (KD) for this ODN series decreased in the order Me>CM>Et>HOEt>(Pr, Bu, Bn, BT, Pob) implying that the bulkiness of alkyl modification indeed determines the strength of Atl1 binding. We conclude that Atl1 forms more stable complexes with bulky O6-alkylguanine residues: these results are consistent with the crystal structures and explain the phenotypes we have described.
Model for the role of Atl1 in processing small and bulky O6-alkylguanine lesions in DNA
Because Atl1 forms very similar structures with both small and bulky O6-alkylguanines it is reasonable to suggest that any subsequent interactions with the downstream protein complexes rhp7/rhp16 and rhp41/rhp23, will also be identical, up to the point where Atl1 displacement from DNA takes place (Figure 7). Relatively facile dissociation of Atl1 from DNA containing small O6-alkylguanines (KD=0.5–0.91 nM) promotes accurate completion of GGR, whereas strong Atl1 binding to bulky O6-alkylguanines (KD<0.1 nM) blocks GGR. When bulky O6-alkylguanines are present in transcribed regions, Atl1, in complex with GGR damage recognition proteins, stalls the transcription machinery and diverts the damage to TCR, by activating Rhp26 and recruiting TFIIH and other downstream NER factors. In non-transcribed regions of the genome or in transcribed regions under circumstances where any of the NER factors are limited, the Atl1 complex will ultimately stall DNA replication complexes, leading to cell cycle arrest and/or cell death.
In conclusion, we provide evidence that the same DNA damage sensing protein can direct similar DNA lesions down different repair pathways according to the affinity for its substrates. For the tightly binding lesions, this might be considered a new branch of NER that fuses GGR to TCR.
Given the mechanistic similarities between ATL proteins and the mainly eukaryotic AGT proteins, these findings raise the question of whether or not O6-alkylguanines that are poor substrates for the alkyl transfer function of AGTs might, by analogy, signal such lesions for repair by NER. Another possibility is that eukaryotes may possess a functional homologue of ATL that would provide an alternative mechanism for processing O6-alkylguanines in DNA, a pathway that would have far-reaching clinical significance.
EXPERIMENTAL PROCEDURES
ODN syntheses
ODNs containing O6-methylguanine (O6-MeG) were obtained commercially (DNA Technology). The 23-mer sequence used was based on previous studies of the action of MGMT and Atl1 (Pearson et al., 2005) and was not based on the sequence of any known gene. ODNs containing, O6- carboxymethyl (CM), -ethyl (Et), -hydroxyethyl (HOEt), -n-propyl (Pr), -n-butyl (Bu) -benzyl (Bn), -pyridyloxobutyl (Pob) and -4-bromothenyl (BT) guanine residues were synthesized by reaction of the appropriate alcohol during post-DNA synthesis chemistry as described (Millington et al., 2012; Shibata et al., 2006). All O6-alkylguanine containing ODNs were purified by reversed phase HPLC (Millington et al., 2012; Shibata et al., 2006); confirmatory analysis was done by mass spectrometry (Shibata et al., 2006). The sequences of the various ODN used in these studies, and the positions of the modified bases (indicated by X) are given in Table S2. To produce duplexes, the ODN were annealed to complements with biotin or HEX at the 5′-end.
Atl1 purification
For EMSA, ELISA and some SPR studies, Atl1 was expressed as an MBP-fusion protein from the pMAL-2c expression vector (New England Biolabs) construct as described by Pearson et al., (2005, 2006) with minor modifications. MBP-Atl1 fusion protein (40 mg) was then cleaved with 0.1%w/v of Factor Xa (1 mg/mL, NEB); at room temperature for 2 hours. The efficiency of the reaction was assessed by resolving the cleavage products on a 15% SDS-polyacrylamide gel. The cleavage reaction was then applied to a Superdex200™ prep grade column (16 mm × 60 mm; GE Heathcare) that was pre-equilibrated with 50 mM Tris-HCl (pH 8.3), 100 mM NaCl. The column was eluted at a flow rate of 0.8 mL/min and 1.6 mL fractions were collected. Protein elution was monitored by absorption at 215 nm using a flow cell. Pooled Atl1 containing fractions were further purified through amylose columns to remove remaining uncleaved MBP-fusion protein.
X-ray crystallography
For the XRC studies, C-terminally hexahistidine-tagged Atl1 was expressed and purified as described previously (Tubbs et al., 2009). Single-strand ODN of sequence 5′-GCCATGXCTAGTA-3′, where X=O6-HOEtG, O6-PrG, O6-BnG or O6-CMG, were annealed with equimolar complementary ODN of sequence 5′-CTACTAGCCATGG-3′ to produce 13-mer double-strand DNA with a 5′-overhang on either end. This ODN was then mixed with purified Atl1 at an ODN:protein molar ratio of 1.5:1.
Crystallization and X-ray Diffraction Data Collection
Crystals were grown by the hanging drop vapor diffusion method mixing 1 μL of protein-DNA complex with 1 μL well solution.
Diffraction data for the Atl1:O6-BnG-ODN complex were collected at SSRL beamline 11-1 at a wavelength of 0.97945 on a MAR325 detector. Diffraction data for the Atl1:O6-HOEtG-ODN, Atl1:O6-PrG-ODN, and Atl1:O6-CMG-ODN complexes were collected at ALS beamline 12.3.1 at a wavelength of 1.1158 Å on an ADSC Q315 detector. Diffraction data were processed with HKL2000 (Otwinowski and Minor, 1997). Structures were solved by molecular replacement with Phaser (McCoy et al., 2007), using the Atl1 structure (pdb 3GVA) as a search model for Atl1–DNA complexes.
Crystallographic Structure Refinement and Analysis
Crystallographic refinement was done with Python-based Hierarchical ENvironment for Integrated Xtallography (PHENIX) (Adams et al., 2010). Coot was used for manual model building into 2Fo − Fc and composite omit and simulated annealing omit 2Fo − Fc and Fo − Fc electron density maps (Emsley and Cowtan, 2004). DNA was built into the model after two rounds of refinement. Water molecules were added to regions of greater than 3σ in a Fo − Fc difference map after two rounds of refinement. Structural superimpositions were done with Sequoia (Bruns et al., 1999). Structure figures were made with PyMol (http://www.pymol.org).
EMSA methodology
ODNs were annealed by mixing equimolar amounts of O6-alkylguanine-containing ODN and complement that was fluorescently labeled at the 5′-end with HEX, in 500 mM NaCl, heating in a dry-block at 80°C for 5 min, then allowing to cool to room temperature. Aliquots of the ODN were incubated with Atl1 (5 uL of 1pmole/uL) in total volume of 20 μL of buffer I (50 mM Tris-HCl, 1 mM EDTA, 3 mM dithiothreitol, pH 8.3) for 1 hour at 37°C and then subjected to non-denaturing gel electrophoresis (15% acrylamide) in TBE buffer (89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA) at 100V for 2 hours. Gels were scanned using the HEX settings on a Pharos laser scanner (BioRad). Additional EMSA controls included incubation with the non-DNA binding protein MBP (5 uL of 1pmole/uL in 20 μL buffer I) and incubation of Atl1, MBP or the ODN incubation mixes with amounts of proteinase K (Sigma; 2 ul of a 10 ug/mL solution in water) that had been shown to degrade the amounts of the individual proteins used. The presence of the Atl1 and MBP proteins and their degradation by PK was shown by electrophoretic transfer of the bands to nitrocellulose membranes (GE healthcare) at 100V for 1 hour and probing with anti-Atl1 rabbit polyclonal antiserum that we had generated to purified recombinant Atl1 or anti-MBP antibodies (New England Biolab) followed by incubation with horseradish peroxidase-labeled anti-rabbit antibodies (DakoCytomation) and detection by enhanced chemiluminescence (GE Healthcare).
Direct ELISA methodology
O6-alkylguanine-containing ODNs were annealed to the biotinylated complement, immobilized on streptavidin (SA) coated 96-well plates and incubated with incrementally increasing concentrations of Atl1. Following sequential 1-hour room temperature incubations with anti-Atl1 antibodies, goat anti rabbit HRP was added and binding quantified by measurement of chemiluminescence on a TECAN GENios plate-reader. Binding was expressed as % of ODN bound. Binding curves were plotted and KD values i.e. the concentration of Atl1 (nM) at which 50% of the target ODN is bound, determined.
Yeast strains, media and standard genetic methods
The genotypes of S. pombe strains used in this study are listed in Table S1. The majority of strains used in this study originated from GM1 (Pearson et al., 2006). The disruptions of the rhp7, rhp26 and rad23 genes in GM1 strain were done by one step PCR-based gene targeting (Bahler et al., 1998; Longtine et al., 1998).
The standard media and general methods were as described (Gutz et al., 1974; Moreno et al., 1991). Yeast extract agar (YEA) contained 0.5% Difco yeast extract, 3% glucose, 1.8% agar; yeast extract liquid (YEL) - 0.5% Difco yeast extract and 3% glucose. Adenine, leucine, uracil, histidine, lysine (100 mg/l each) were added to YEA/YEL where required.
Agar plate (“spot”) assays
YEA plates were prepared by adding the appropriate volume of stock solutions of different alkylating agents (100 mg/mL in dry DMSO) to cooled molten agar. Serial 1 in 10 dilutions of S. pombe cultures containing 5 × 107 cells/mL were spotted on YEA plates (5 μl per spot). Photographs were taken after 4–5 days of incubation at 30°C for cdc10+ strains and 25°C for cdc10-M17 mutants.
Determination of MNNG sensitivity by clonogenic survival assay
To determine MNNG sensitivity, yeast cells were streaked to single colonies on YEA plates and grown for 4–5 days. Suspensions of cells derived from a single colony (107 cell/mL) were exposed to different concentrations of MNNG for 10 min in YEL then diluted 100-fold in distilled water to stop the reaction. Serial cell dilutions were then plated in YEA medium to attain growth of approximately 200 colonies per plate. Cell titer was estimated by plating the original cell suspension without MNNG treatment. After 7 days of growth at 30°C, cell survival (%) was calculated as ratio between the number of colonies and the number of cells plated.
SPR analyses
The interaction of Atl1 with ODN containing O6-Me, O6-CM, O6-Et, O6-HOEt, O6-Pr, O6-Bu, O6-Bn, O6-Pob and O6- BT guanine and guanine-containing control ODN (Table S2) were analysed using Bio-Rad ProteOn “XPR36” SPR system and neutravidin-coated NLC sensor chips. Proteon Manager software was used to fit the binding curves and calculate the binding constants and data were presented in the form of “on” (ka) and “off” (kd) rates and dissociation constants (KD).
Analysis of S-phase progression by FACS analysis
Cdc10-M17 cells were synchronized in G1-S by incubation at 37°C for 4 h (Kim and Huberman, 2001); then treated with MNNG or BZNU for 10 min and shifted to 25°C. At 20 min intervals, aliquots of cells (100 μl) were taken for flow cytometry analysis. Samples for FACS were prepared as described previously (Doll et al., 2005; Kim and Huberman, 2001) with minor modifications. S-phase progression was assessed using FlowJo software as described (Willis and Rhind, 2009). The DNA peak position for the zero time sample was used to set the 1C value (pre-replication) and the 2C value (post replication) was set from the location of the DNA peak in the late time points for the untreated control samples. For each time point, S-phase progression was quantitated using the equation: % progression = (C − A)/(B − A), where A = 1C, B = 2C, and C = mean values for S-phase peaks.
Determination of O6-MeG and O6-BnG levels in S. pombe cells after treatment with MNNG and BzNU
S. pombe cells were grown in YEL to an OD600 of 0.5 and treated with 120 and 240 μg/mL MNNG for 10 minutes at 30°C. Treatment with 1 mg/mL BzNU for 60 and 90 min was in TE, pH 8.1 to increase BZNU hydrolysis. Cells were then centrifuged (800 × g for 2 minutes) and resuspended in the same volume of TE, pH 8.1. After treatment with MNNG and BzNU, cells were harvested by centrifugation as above and snap frozen in dry ice. DNA was isolated by phenol extraction and ethanol precipitation. O6-MeG and O6-BnG in DNA were quantified using a modification of the standard MGMT activity assay procedure as described previously (Watson and Margison, 2000; Watson et al., 2009).
Supplementary Material
Highlights.
Atl1 binds to and forms similar structures with a wide range of O6-alkylguanines
Relatively weak Atl1 binding to simple lesions initiates GGR
Strong binding to more complex lesions activates TCR and stalls DNA replication
Acknowledgments
This work was supported by Cancer Research UK, National Institutes of Health grant CA097209 (JAT), the Skaggs Institute for Chemical Biology (JLT), EPSRC (CLM), BBSRC (OJW, DMW), CHEMORES (GPM), and the Royal Thai Government (PS).
X-ray diffraction technologies at the SIBYLS beamline 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory, are supported by the U.S. Department of Energy (DOE) program Integrated Diffraction Analysis Technologies (IDAT). Diffraction data collection at the Stanford Synchrotron Radiation Light source is supported by DOE and NIH.
We acknowledge the help of Dr. Jonathan Popplewell of Bio-Rad Laboratories in the analysis of SPR data and Grant Guenther and Matthew Kroeger for purifying the Atl1 protein used for crystallographic studies.
Footnotes
The authors declare that there are no conflicts of interest.
Accession Numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4ENJ (Atl1:O6-HOEtG), 4ENK (Atl1:O6-PrG), 4ENM (Atl1:O6-BnG), and 4ENN (Atl1:O6-CMG).
Further details for all these analyses are in the Supplementary Methods.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- Bruns CM, Hubatsch I, Ridderstrom M, Mannervik B, Tainer JA. Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J Mol Biol. 1999;288:427–439. doi: 10.1006/jmbi.1999.2697. [DOI] [PubMed] [Google Scholar]
- den Dulk B, van Eijk P, de Ruijter M, Brandsma JA, Brouwer J. The NER protein Rad33 shows functional homology to human Centrin2 and is involved in modification of Rad4. DNA Repair (Amst) 2008;7:858–868. doi: 10.1016/j.dnarep.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Doll E, Molnar M, Hiraoka Y, Kohli J. Characterization of rec15, an early meiotic recombination gene in Schizosaccharomyces pombe. Curr Genet. 2005;48:323–333. doi: 10.1007/s00294-005-0024-3. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. In: Handbook of genetics. King RC, editor. Vol. 1. NY: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- Hickman MJ, Samson LD. Apoptotic Signaling in Response to a Single Type of DNA Lesion, O6-Methylguanine. Molecular cell. 2004;14:105–116. doi: 10.1016/s1097-2765(04)00162-5. [DOI] [PubMed] [Google Scholar]
- Kim SM, Huberman JA. Regulation of replication timing in fission yeast. Embo J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaerts M, Goeloe JI, den Dulk H, Brandsma JA, Brouwer J. Identification and characterization of the rhp23(+) DNA repair gene in Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2000;268:210–215. doi: 10.1006/bbrc.2000.2100. [DOI] [PubMed] [Google Scholar]
- Lombaerts M, Peltola PH, Visse R, den Dulk H, Brandsma JA, Brouwer J. Characterization of the rhp7(+) and rhp16(+) genes in Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:3410–3416. doi: 10.1093/nar/27.17.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Margison GP, Butt A, Pearson SJ, Wharton S, Watson AJ, Marriott A, Caetano CM, Hollins JJ, Rukazenkova N, Begum G, Santibanez-Koref MF. Alkyltransferase-like proteins. DNA Repair (Amst) 2007;6:1222–1228. doi: 10.1016/j.dnarep.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Margison GP, Povey AC, Kaina B, Santibanez Koref MF. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2003;24:625–635. doi: 10.1093/carcin/bgg005. [DOI] [PubMed] [Google Scholar]
- Margison GP, Santibanez Koref MF, Povey AC. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis. 2002;17:483–487. doi: 10.1093/mutage/17.6.483. [DOI] [PubMed] [Google Scholar]
- Marti TM, Kunz C, Fleck O. Repair of damaged and mismatched DNA by the XPC homologues Rhp41 and Rhp42 of fission yeast. Genetics. 2003;164:457–467. doi: 10.1093/genetics/164.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G, Philippin G, Cadet J, Gasparutto D, Fuchs RP. The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O(6)-alkylguanine adducts in E. coli. DNA Repair (Amst) 2009;8:697–703. doi: 10.1016/j.dnarep.2009.01.022. [DOI] [PubMed] [Google Scholar]
- Mazon G, Philippin G, Cadet J, Gasparutto D, Modesti M, Fuchs RP. Alkyltransferase-like protein (eATL) prevents mismatch repair-mediated toxicity induced by O6-alkylguanine adducts in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:18050–18055. doi: 10.1073/pnas.1008635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millington CL, Watson AJ, Marriott AS, Margison GP, Povey AC, Williams DM. Convenient and efficient syntheses of oligodeoxyribonucleotides containing O6-(carboxymethyl)guanine and O6-(4-oxo-4-(3-pyridyl)butyl)guanine. Nucleosides Nucleotides Nucleic Acids. 2012;31:328–338. doi: 10.1080/15257770.2012.656784. [DOI] [PubMed] [Google Scholar]
- Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nouspikel T. DNA repair in mammalian cells: Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Bordes N, Haddad R, Schwartz CL, Chang F, Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol Biol Cell. 2003;14:2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson SJ, Ferguson J, Santibanez-Koref M, Margison GP. Inhibition of O6-methylguanine-DNA methyltransferase by an alkyltransferase-like protein from Escherichia coli. Nucleic Acids Res. 2005;33:3837–3844. doi: 10.1093/nar/gki696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson SJ, Wharton S, Watson AJ, Begum G, Butt A, Glynn N, Williams DM, Shibata T, Santibanez-Koref MF, Margison GP. A novel DNA damage recognition protein in Schizosaccharomyces pombe. Nucleic Acids Res. 2006;34:2347–2354. doi: 10.1093/nar/gkl270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- Reed SH. Nucleotide excision repair in chromatin: Damage removal at the drop of a HAT. DNA Repair (Amst) 2011;10:734–742. doi: 10.1016/j.dnarep.2011.04.029. [DOI] [PubMed] [Google Scholar]
- Shibata T, Glynn N, McMurry TB, McElhinney RS, Margison GP, Williams DM. Novel synthesis of O6-alkylguanine containing oligodeoxyribonucleotides as substrates for the human DNA repair protein, O6-methylguanine DNA methyltransferase (MGMT) Nucleic Acids Res. 2006;34:1884–1891. doi: 10.1093/nar/gkl117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. New York: Plenum Press; 1983. [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AJ, Margison GP. O6-alkylguanine-DNA alkyltransferase assay. Methods Mol Biol. 2000;152:49–61. doi: 10.1385/1-59259-068-3:49. [DOI] [PubMed] [Google Scholar]
- Watson AJ, Middleton MR, McGown G, Thorncroft M, Ranson M, Hersey P, McArthur G, Davis ID, Thomson D, Beith J, et al. O(6)-methylguanine-DNA methyltransferase depletion and DNA damage in patients with melanoma treated with temozolomide alone or with lomeguatrib. Br J Cancer. 2009;100:1250–1256. doi: 10.1038/sj.bjc.6605015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis N, Rhind N. Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint. Mol Biol Cell. 2009;20:819–833. doi: 10.1091/mbc.E08-08-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhira S, Morimyo M, Yasui A. Transcription dependence and the roles of two excision repair pathways for UV damage in fission yeast Schizosaccharomyces pombe. J Biol Chem. 1999;274:26822–26827. doi: 10.1074/jbc.274.38.26822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.