Abstract
The bioactivation of both endogenous and equine estrogens to electrophilic quinoid metabolites has been postulated as a contributing factor in carcinogenic initiation and/or promotion in hormone sensitive tissues. Bearing structural resemblance to estrogens, extensive studies have shown that many selective estrogen receptor modulators (SERMs) are subject to similar bioactivation pathways. Lasofoxifene (LAS), a third generation SERM which has completed Phase III clinical trials for the prevention and treatment of osteoporosis, is currently approved in the European Union for this indication. Previously, Prakash et al. (Drug Metab. Dispos. 2008, 36, 1218-26) reported that similar to estradiol, two catechol regioisomers of LAS are formed as primary oxidative metabolites, accounting for roughly half of total LAS metabolism. However, the potential for further oxidation of these catechols to electrophilic o-quinones has not been reported. In the present study, LAS was synthesized and its oxidative metabolism investigated in vitro under various conditions. Incubation of LAS with tyrosinase, human liver microsomes, or rat liver microsomes in the presence of GSH as a trapping reagent resulted in formation of two mono-GSH and two di-GSH catechol conjugates which were characterized by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Similar conjugates were also detected in incubations with P450 3A4, P450 2D6, and P450 1B1 supersomes. Interestingly, these conjugates were also detected as major metabolites when compared to competing detoxification pathways such as glucuronidation and methylation. The 7-hydroxylasofoxifene (7-OHLAS) catechol regioisomer was also synthesized and oxidized either chemically or enzymatically to an o-quinone that was shown to form depurinating adducts with DNA. Collectively, these data show that analogous to estrogens, LAS is oxidized to catechols and o-quinones which could potentially contribute to in vivo toxicity for this SERM.
Keywords: lasofoxifene, 7-hydroxylasofoxifene, estradiol, cytochrome P450, o-quinone, GSH conjugate, depurinating adduct
Introduction
Selective estrogen receptor modulators (SERMs) are a class of compounds that act as estrogens in certain tissues while antagonizing the effects of estrogen in others, depending on cellular context.1 They have found clinical use in the treatment (tamoxifen) and prevention of breast cancer (tamoxifen, toremefine, raloxifene), postmenopausal osteoporosis (raloxifene), and infertility (clomifene).2 Lasofoxifene (LAS, Scheme 1) is a third generation SERM which has completed Phase III clinical trials for the prevention and treatment of postmenopausal osteoporosis.3 Displaying dramatically increased in vivo potency and oral bioavailability over earlier generation SERMs, LAS has been shown significantly to improve bone mineral density (BMD) and biomarkers for bone turnover in postmenopausal women.4-6 Results of the PEARL (Postmenopausal Evaluation and Risk-Reduction with Lasofoxifene) trial recently showed that women taking LAS were at a reduced risk for both vertebral and non-vertebral fracture, coronary heart disease, and stroke.7-10 Further findings of the same trial also demonstrated that LAS not only displayed superior skeletal efficacy compared to raloxifene, but also showed a dose-dependent reduction in risk for ER-positive invasive breast cancer while lacking endometrial stimulation, suggesting a potential utility in chemoprevention.5, 11-13 Although it has not received FDA approval in the US,14 LAS was approved by the European Commission for the treatment of osteoporosis in postmenopausal women at increased risk for fracture in March, 2009.15
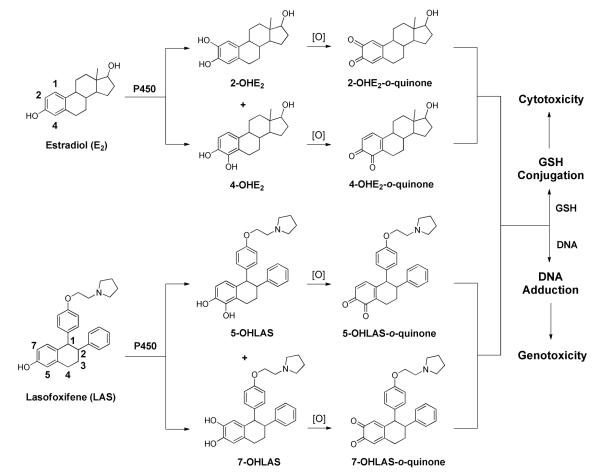
Scheme 1.
Proposed bioactivation of lasofoxifene compared to estradiol.
Conversely, while LAS and other earlier generation SERMs appear to exhibit favorable tissue selectivity, a growing body of evidence also demonstrates that analogous to estrogens, SERMs are susceptible to oxidative bioactivation.16-24 It is well established that E2 is metabolized to both 2-hydroxy and 4-hydroxy catechols by P450 isozymes, and that these catechols may further oxidize to electrophilic o-quinones and quinone methides.25, 26 The formation of such quinoids has been hypothesized to contribute to the initiation and/or promotion of certain estrogen dependent cancers through a variety of mechanisms, including DNA adduction, protein modification, and reactive oxygen species (ROS) generation.25-27 Similarly, the triphenylethylene SERM tamoxifen is metabolized to reactive carbocation,16 o-quinone17, 18 and quinone methide intermediates.19 The detection of DNA adducts in women undergoing tamoxifen therapy lends further support to a genotoxic pathway resulting from tamoxifen bioactivation.28 It has also been reported that raloxifene is a time-dependent inhibitor of P450 3A4 in vitro via di-quinone methide formation.20, 29 Moreover, studies by our lab and by others have established similar bioactivation pathways for other triphenylethylene (toremifene, droloxifene),17, 21 benzothiophene [desmethyl arzoxifene (DMA)],22, 23 benzopyran (acolbifene),22 and naphthol (LY2066948)24 based SERMs.
Unlike the first-generation triphenylethylene and second-generation benzothiophene SERMs, LAS possesses a tetralin core structurally similar to the fused A and B rings of 17β-estradiol (E2; Scheme 1), and like E2, binds selectively to ERα with high affinity (IC50 = 1.5 nM).30 The structural resemblance to E2 led us to hypothesize that LAS would form similar quinoid metabolites (Scheme 1). It has been previously reported that 5-hydroxylasofoxifene (5-OHLAS) and 7-hydroxylasofoxifene (75OHLAS) catechols are formed as primary oxidative metabolites of LAS;31 however, the potential for further oxidation of these catechols to electrophilic quinoids has not been investigated. In the current study, we have found that LAS can be oxidized both chemically and enzymatically to o-quinones which form conjugates with GSH. Furthermore, several depurinating DNA adducts were detected upon incubation of synthesized 7-OHLAS-o-quinone with deoxynucleosides or DNA. Taken together, these data suggest that the formation of reactive quinoids from catechol LAS metabolites may represent a plausible mechanism of potential toxicity for this SERM.
Materials and Methods
Caution
Lasofoxifene-o-quinone and estrogen-o-quinone were handled in accordance with the NIH Guidelines for the Laboratory Use of Chemical Carcinogens.32
Chemicals
Racemic LAS and 75OHLAS were synthesized as described previously.33 E2 was purchased from Steraloids Inc. (Newport, RI). Rat liver microsomes were prepared from the livers of female Sprague-Dawley rats, as previously described.34 Human liver microsomes (pooled from 15 individuals) were purchased from In Vitro Technologies (Baltimore, MD). Human cytochrome P450 supersomes were purchased from BD Biosciences (Woburn, MA). All other chemicals were purchased from Aldrich Chemical (Milwaukee, WI), Fisher Scientific (Itasca, IL), or Sigma (St. Louis, MO) unless stated otherwise.
Instrumentation
NMR spectra were obtained using either a Bruker Avance 400 MHz or Bruker DPX 400 MHz spectrometer. UV spectra were obtained using a Hewlett-Packard 8452A photodiode array UV/Vis spectrometer (Palo Alto, CA). LC5MS/MS analyses were performed using either of two instrument configurations: (1) An Agilent 6310 ion trap mass spectrometer (Agilent Technologies, Santa Clara, CA) coupled to an Agilent 1100 HPLC (Palo Alto, CA), or (2) an API 3000 triple quadrupole mass spectrometer (Applied Biosystem, Foster City, CA) coupled to an Agilent 1200 HPLC (Palo Alto, CA).
LC-MS/MS Methodology
Two methods were used for all LC-MS/MS analyses. Analysis of depurinating adducts from CT-DNA incubations was completed using the API 3000 triple quadrupole instrument. Samples were separated using a Phenomenex Kinetex C18 column (3 × 100 mm, 2.6 μm) and ADV-FFKIT filter (Analytical, Prompton Plains, NJ, USA) at a flow rate of 0.3 mL/min. The mobile phase was composed of solvent A (water containing 10% methanol and 0.1% formic acid) and solvent B (methanol containing 0.1% formic acid), beginning with 30% B, increasing to 98% B over 10 min, holding at 98% B for 10 min, and returning to 30% B over 2 min. The system was then allowed to equilibrate for 8 min before subsequent sample injections. Ions were detected in positive mode with electrospray ionization and multiple reaction monitoring carried out at 350 °C. Collision energies were optimized to 67 volts for 7-OHAS-Ade adducts, and 63 volts for 7-OHAS-Gua adducts. Fragmentations for the collision-induced dissociation of m/z 563→136, and m/z 579→152 were monitored for Ade and Gua adducts, respectively.
All other LC-MS/MS analyses were completed using the Agilent 6310 ion trap instrument equipped with an electrospray ionization source and measuring UV absorbance at 270 nm. Samples were separated using an Agilent Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm) at a flow rate of 1.0 mL/min. The mobile phase was composed of solvent A (water containing 10% methanol and 0.1% formic acid) and solvent B (acetonitrile containing 0.1% formic acid), beginning with 5% B, increasing to 60% B over 40 min, 90% B over 5 min, and then returning to 5% B over 3 min. The system was then allowed to equilibrate for 10 min before subsequent sample injections. Ions were detected in positive mode using collision-induced dissociation (CID) ionization with a resolving power of 5000 FWHM and mass accuracy of 0.1 amu.
Incubations with Tyrosinase
Solutions containing LAS or E2 (30 μm), tyrosinase (0.1 mg/mL), and GSH (1 mM) in 50 mM phosphate buffer (pH 7.4, 0.5 mL total volume) were incubated at 37 °C for 30 min. Reactions were terminated by chilling in an ice bath followed by the addition of perchloric acid (25 μL). Samples were then centrifuged (10,640g for 10 min at 4 °C), and supernatants were filtered and immediately analyzed by LC-MS/MS. Controls were performed by omission of tyrosinase or GSH.
Incubations with Rat/Human Liver Microsomes
Solutions containing LAS or E2 (30 μm), rat or human liver microsomes (1 nmol P450/mL), GSH (1 mM), and a NADPH-generating system (1 mM NADP+, 5 mM MgCl2, 5 mM isocitric acid, 0.2 unit/mL isocitrate dehydrogenase) in phosphate buffer (pH 7.4, 50 mM, 0.5 mL total volume) were incubated for 30 min at 37 °C. Reactions were quenched by chilling in ice followed by addition of perchloric acid (25 μL). Incubation mixtures were centrifuged (10,000 × g for 10 min at 4 °C), and supernatants were filtered and analyzed by LC-MS/MS. For control incubations, either NADP+ or GSH was omitted.
Incubations with P450 3A4, P450 2D6, or P450 1B1 Supersomes
Solutions containing LAS or E2 (30 μm), supersomes (P450 3A4, P450 2D6, or P450 1B1, 10 pmol/mL), GSH (1 mM), and a NADPH-generating system (1 mM NADP+, 5 mM MgCl2, 5 mM isocitric acid, 0.2 unit/mL isocitrate dehydrogenase) in phosphate buffer (pH 7.4, 50 mM, 0.5 mL total volume) were incubated for 30 min at 37 °C. Reactions were then chilled in ice, treated with perchloric acid (25 μL), and centrifuged (10,000 × g for 10 min at 4 °C). Supernatants were filtered and immediately analyzed by LC-MS/MS. For control experiments, either NADP+ or GSH was omitted.
Competition of Catechol LAS Glucuronidation with Catechol LAS Oxidation and Glutathione Conjugation
Solutions containing LAS (30 μm), rat liver microsomes (1 nmol P450/mL), a NADPH-generating system (1 mM NADP+, 5 mM MgCl2, 5 mM isocitric acid, 0.2 unit/mL isocitrate dehydrogenase), GSH (1 mM), uridine diphosphate glucuronic acid (UDPGA, 1 mM) and alamethicin (10 Rg/mg protein) were incubated for 30 min at 37 °C in 50 mM phosphate buffer (pH 7.4, 1 mL total volume). After chilling in ice, perchloric acid (50 μL) was added, and proteins were removed by centrifugation (10,000 × g for 10 min at 4 °C). Aliquots of supernatant were then analyzed by LC-MS/MS. For control experiments, either GSH or UDPGA was omitted.
Competition of Catechol LAS Methylation with Catechol LAS Oxidation and Glutathione Conjugation
Solutions containing LAS (30 μm), rat liver microsomes (1 nmol P450/mL), and catechol5O5methyltransferase (COMT, 1 mM) were incubated for 30 min at 37 °C in 50 mM phosphate buffer (pH 7.4, 1 mL total volume). Incubations were initiated by the addition of a solution containing magnesium chloride (1 mM), S-adenosyl methionine (SAM, 0.3 mM), NADP+ (1 mM), MgCl2 (5 mM), isocitric acid (5 mM), isocitrate dehydrogenase (0.2 unit/mL), and GSH (1 mM). Reactions were quenched by chilling in ice followed by addition of perchloric acid (50 μL). Proteins were removed by centrifugation (10,000 × g for 10 min at 4 °C), and aliquots (100 μL) of supernatant were analyzed by LC-MS/MS. For control experiments, either GSH or COMT was omitted.
LAS-o-Quinone Decomposition Kinetics
To a solution of LAS (500 μm) in anhydrous methanol (200 μL) was added 25iodoxybenzoic acid (0.84 mg, 30 equiv.) at room temperature. After stirring for 1 min, a yellow color developed and the reaction mixture was filtered. The LAS-o-quinone solution (100 μL) was immediately added to 50 mM phosphate buffer (0.9 mL, pH 7.4) at 37 °C. Disappearance of o-quinone was then monitored by measuring the decrease in UV absorbance at 378 nm. The half5life was subsequently determined by measuring the pseudo5first5order rate of decay of the absorbance signal at 378 nm according to the equation t1/2 = ln(2)/k.
Reaction of 7-OHLAS-o-Quinone with Deoxynucleosides
Solutions containing 7-OHLAS (30 μm), tyrosinase (0.1 mg/mL), and each of the four deoxynucleosides (dG, dA, dT, or dC, 300 μm) were incubated for 30 min at 37 °C in 50 mM phosphate buffer (pH 7.4, 0.5 mL total volume). Reactions were quenched by chilling in ice followed by addition of cold ethanol (1 mL). Protein was removed by centrifugation (10,000 × g for 10 min at 4 °C), the supernatant was concentrated to a volume of 0.5 mL, and aliquots of supernatant were analyzed by LC-MS/MS. For control experiments, either tyrosinase or deoxynucleoside was omitted.
Reaction of 7-OHLAS-o-Quinone with Calf Thymus DNA
To a solution of 7-OHLAS (2.53 mg, 5.89 μmol) in acetonitrile (500 μL) and dimethylformamide (100 μL) was added activated MnO2 (3.34 mg, 38.4 μmol, 6.5 equiv.) at 0 °C. The solution was stirred at 0 °C for 15 min and then filtered directly into a solution of CT-DNA (1 mg/mL in 50 mM phosphate buffer, pH 7.0, 5 mL total volume). The resulting mixture was incubated for 10 hr at 37 °C. Following incubation, cold ethanol (10 mL) was added and the solution was stored at −20 °C for 1 hr. Precipitated DNA was removed by centrifugation (3330 g for 15 min at 4 °C) and the supernatant was concentrated to a volume of 0.5 mL, loaded onto a PrepSep C18 solid-phase extraction cartridge, washed with 5% methanol in water (1 mL), and eluted with 6 mL of a solution of methanol/acetonitrile/water/formic acid (8:1:1:0.1, v/v). The eluate was evaporated to dryness under a stream of nitrogen, and the residue was reconstituted in 100 μL methanol containing 0.1% formic acid. Aliquots of the resulting solution were analyzed by LC-MS/MS.
Results
Incubations with Tyrosinase
Two mono-GSH and two di-GSH conjugates were detected in incubations of LAS with tyrosinase. The mono-GSH conjugates were arbitrarily assigned as OHLAS-SG1 and OHLAS-SG2 (Figure 1A), and were identified based upon detection of protonated molecular ions of m/z 735 [M+H]+. CID of molecular ions m/z 73-produced characteristic fragments of m/z 717, 642, 606, and 462 corresponding to loss of water, loss of water plus glycine residue, loss of γ-glutamyl group, and cleavage of alkyl thioether bond, respectively (Figure 1C). Typical fragmentations were also observed for di-GSH conjugates, which were arbitrarily assigned as OHLAS5diSG1 and OHLAS5diSG2 (Figure 1A) and identified based upon detection of protonated molecular ions of m/z 1040 [M+H]+. Fragment ions of m/z 1022, 893, 782, and 638 were observed, corresponding to loss of water, loss of γ-glutamyl group plus water, loss of two γ-glutamyl groups, and loss of one γ-glutamyl group coupled with alkyl thioether bond cleavage, respectively (Figure 1D).35 Similarly, four GSH conjugates (2-OHE2-diSG, 2-OHE2-SG1, 2-OHE2-SG2, and 4-OHE2-SG) were detected when E2 was incubated with tyrosinase in the presence of GSH (Figure 1B) as reported previously.36
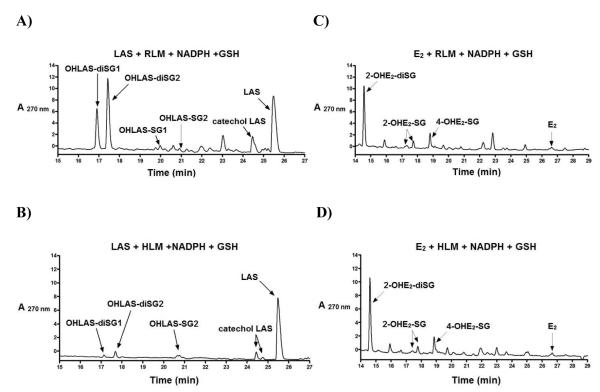
Figure 1.
Representative LC chromatograms of 30 RM LAS (A) and 30 RM E2 (B) incubated with tyrosinase (0.1 mg/mL) and GSH (1 mM) in 50 mM phosphate buffer (pH 7.4) for 30 min at 37 °C; Mass spectrometric analyses of OHLAS-SG1, OHLAS-SG2 (C), and OHLAS-diSG1, OHLAS-diSG2 (D).
Incubations with Rat/Human Liver Microsomes
All GSH conjugates of catechol LAS identified in tyrosinase incubations (OHLAS-SG1, OHLAS-SG2, OHLAS5diSG1, and OHLAS-diSG2) were also detected in rat liver microsomal incubations (Figure 2A). Significantly less metabolism was observed in human liver microsomal incubations, although three of the four conjugates (OHLAS-SG1, OHLAS-SG2, and OHLAS5diSG2) were detected (Figure 2B). All previously reported GSH conjugates of the E2 catechols (2-OHE2-diSG, 2-OHE2-SG1, 2-OHE2-SG2, and 4-OHE2-SG) seen in tyrosinase incubations, were also detected in both >rat and human liver microsomal incubations in similar relative amounts (Figures 2C, 2D).
Figure 2.
Representative LC chromatograms of 30 RM LAS (A, B) or 30 RM E2 (C, D) incubated with rat or human liver microsomes (1 nmol P450/mL) and GSH (1 mM) in the presence of a NADPH-generating system (1 mM NADP+, 5 mM MgCl2, 5 mM isocitric acid, 0.2 unit/mL isocitrate dehydrogenase) in 50 mM phosphate buffer (pH 7.4) for 30 min at 37 °C.
Incubations with P450 3A4, P450 2D6, or P450 1B1 Supersomes
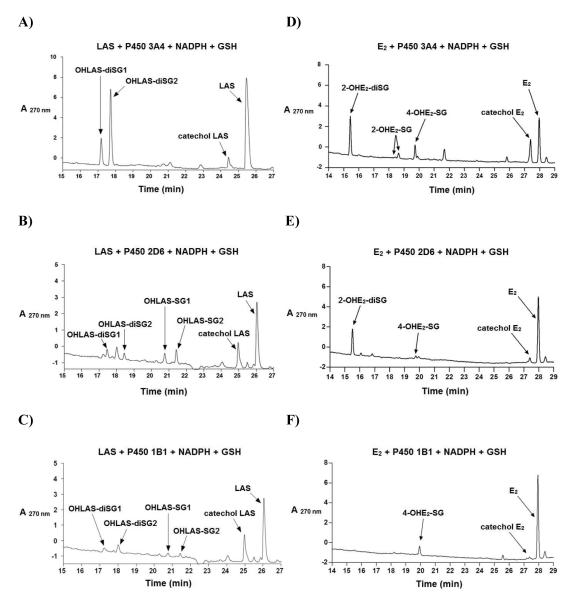
Incubations with P450 3A4 supersomes generated only di-GSH conjugates (OHLAS-diSG1, and OHLAS-diSG2) as major metabolites, whereas all four conjugates (OHLAS-SG1, OHLAS-SG2, OHLAS-diSG1, and OHLAS-diSG2) were detected in experiments with P450 1B1 and P450 2D6 supersomes (Figures 3A-C). By comparison, 2-OHE2-diSG was the major metabolite seen in incubations with E2 and P450 3A4 or P450 2D6 supersomes, while 4-OHE2-SG was the sole metabolite detected in experiments with P450 1B1 (Figures 3D-F), in accordance with previous studies.37
Figure 3.
Representative LC chromatograms of 30 ZM LAS (A, B, C) or 30 ZM E2 (D, E, F) incubated with P450 3A4, P450 2D6, or P450 1B1 (10 pmol/mL) supersomes, along with GSH (1 mM) and a NADPH-generating system (1 mM NADP+, 5 mM MgCl2, 5 mM isocitric acid, 0.2 unit/mL isocitrate dehydrogenase) in 50 mM phosphate buffer (pH 7.4) for 30 min at 37 °C.
Competition of Catechol LAS Glucuronidation or Methylation with Catechol LAS Oxidation and Glutathione Conjugation
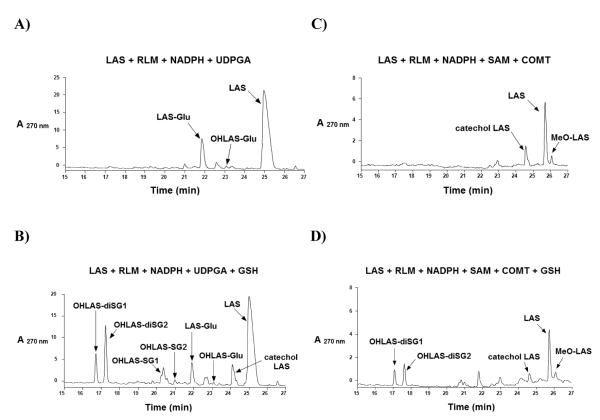
Because glucuronide and methyl ether metabolites of LAS catechols were previously reported whereas GSH conjugates were not observed,31 we investigated the competition between these detoxification pathways. Incubations of LAS with rat liver microsomes and UDPGA in the presence of a NADPH-generating system yielded the expected glucuronide conjugate of the parent compound (LAS-Glu) as a major product, as well as glucuronidated catechol (OHLAS-Glu) as a minor metabolite (Figure 4A). LAS-Glu was identified based upon detection of a molecular ion [M+H]+ at m/z 590 and a fragment ion of m/z 414 corresponding to glycosidic bond cleavage. OHLAS-Glu was detected as [M+H]+ at m/z 606 and gave similar fragmentation to m/z 430 as previously reported.31 Interestingly, with the inclusion of GSH as a trapping reagent in the above incubations, all four previously identified GSH conjugates (OHLAS-SG1, OHLAS-SG2, OHLAS-diSG1, and OHLAS-diSG2) were detected in addition to the glucuronide metabolites (Figure 4B).
Figure 4.
Representative LC chromatograms of 30 RM LAS, rat liver microsomes (1 nmol P450/mL), and a NADPH-generating system (1 mM NADP+, 5 mM MgCl2, 5 mM isocitric acid, 0.2 unit/mL isocitrate dehydrogenase), incubated in 50 mM phosphate buffer (pH 7.4) for 30 min at 37 °C along with either of the following: (A) UDPGA (1mM) and alamethicin (10 Rg/mg protein); (B) GSH (1 mM), UDPGA (1mM), and alamethicin (10 Rg/mg protein); (C) COMT (1mM) and SAM (0.3 mM); or (D) GSH (1 mM), COMT (1mM) and SAM (0.3 mM).
Similarly, incubations of LAS with rat liver microsomes, COMT and appropriate cofactors (see Materials and Methods) generated both catechol LAS and detectable amounts of methylated LAS catechol (MeO-LAS, Figure 4C). MeO-LAS was identified based upon detection of [M+H]+ at m/z 444 and fragment ion m/z 253, corresponding to loss of phenoxyethyl-pyrrolidine side chain, as previously reported.31 Again, inclusion of GSH as a trapping reagent in these incubations resulted in GSH conjugate detection (OHLAS-diSG1 and OHLAS-diSG2, Figure 4D).
LAS-o-Quinone Decomposition Kinetics
Similar to other SERM and estrogen o-quinones,38, 39 a strong absorbance at 378 nm was observed in the UV spectrum of the chemically generated LAS-o-quinone mixture which was absent from the spectrum of LAS itself. At physiological pH and temperature, the pseudo-first-order rate of decay of this signal was monitored and the half-life was calculated to be 55 ± 4 min according to the equation t1/2 = ln(2)/k.
Reaction of 7-OHLAS-o-Quinone with Deoxynucleosides or CT-DNA
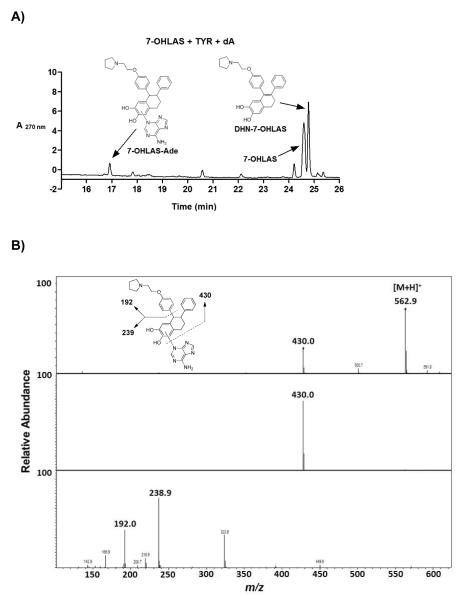
Incubation of 7-OHLAS and tyrosinase along with either of four deoxynucleosides (dG, dA, dT, or dC) resulted in detection of one depurinating adenine adduct (7-OHLAS-Ade, Figure 5A). This adduct was identified based upon detection of a molecular ion [M+H]+ at m/z 563. Fragment ions of m/z 430 and m/z 239 were also detected, corresponding to loss of adenine and subsequent loss of phenoxyethyl-pyrrolidine side chain, respectively (Figure 5B). Although the absolute structure of 7-OHLAS-Ade was not determined due to low yield, A-ring substitution by the N3 nitrogen of adenine was deemed most probable, as similar adduction has been observed for the case of estradiol.25, 40, 41
Figure 5.
Representative LC chromatogram of 30 RM 7-OHLAS, tyrosinase (0.1 mg/mL), and dA (300 μm) incubated in 50 mM phosphate buffer (pH 7.4) for 30 min at 37 °C (A) and MS-MS fragmentation of 7-OHLAS-Ade (B).
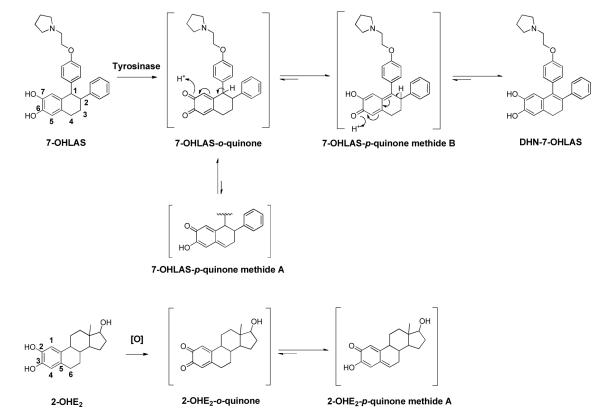
A less polar oxidative metabolite of 7-OHLAS was also detected at m/z 428 as a major product in tyrosinase incubations (Figure 5A). Fragment ions of m/z 330 and m/z 237 corresponding to loss of vinylpyrrolidine and loss of phenoxyethyl-pyrrolidine side chain, respectively, suggested a loss of two mass units from the tetralin ring (data not shown). This metabolite was tentatively assigned as the 1-2 unsaturated dihydronaphthyl analog of 7-OHLAS (DHN-7-OHLAS). Formation of DHN-7-OHLAS could occur through isomerization of 7-OHLAS-o-quinone to a p-Quinone methide followed by tautomerization to DHN-7-OHLAS (Scheme 2). This metabolite was only observed in the absence of GSH since GSH would trap the o-quinones prior to the tautomerization reaction. Similar pathways have previously been observed for catechol estrogens.36, 42
Scheme 2.
Proposed mechanism for formation of DHN-7-OHLAS via tyrosinase oxidation; Formation of 2-OHE2-p-Quinone methide A.
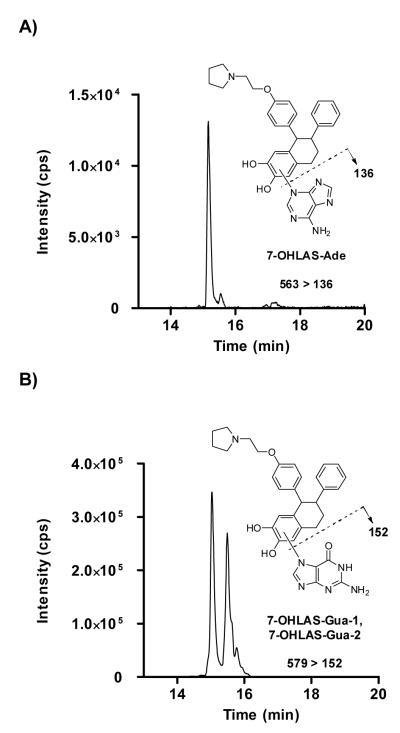
Incubation of chemically-oxidized 7-OHLAS-o-quinone with DNA resulted in detection of 7-OHLAS-Ade as well as two depurinating guanine adducts, arbitrarily assigned as 7-OHLAS-Gua-1 and 7-OHLAS-Gua-2 (Figure 6). While absolute structures of 7-OHLAS-Gua-1 and 7-OHLAS-Gua-2 were not determined due to low yield, A-ring substitution by the N7 nitrogen of guanine at the 5 and 8 positions of LAS was likely, again based upon similar adduction observed for depurinating guanine adducts of E2.25, 40, 41 Adenine and guanine adducts were detected with multiple reaction monitoring and collision-induced dissociation for the fragmentation pathways of m/z 563→136, and m/z 579→152, respectively (Figure 6).
Figure 6.
Detection of 7-OHLAS-Ade by multiple reaction monitoring (MRM) at m/z 563 > 136 (A) and 7-OHLAS-Gua51 and 7-OHLAS-Gua-2 by MRM at m/z 579 > 152 (B).
Discussion
The formation of reactive metabolites has been implicated in the toxicity of both estrogens and SERMs. E2 for example, is metabolized to both 2-hydroxy and 4-hydroxy catechols by P450 1A1 and P450 1B1, respectively.25 These catechols may be further oxidized to o-quinones which are capable of covalently modifying DNA and cellular proteins and may also generate reactive oxygen species (ROS) through redox cycling.25 Furthermore, extensive studies have shown that triphenylethylene, benzothiophene, and benzopyran SERMs are metabolized to a variety of quinoids, and for the case of tamoxifen, bioactivation has been associated with carcinogenesis.28
Previous reports have shown that analogous to E2, LAS is extensively metabolized to 5-OHLAS and 7-OHLAS catechols, mainly by P450 3A4 and P450 2D6, respectively.31 While conjugation of catechol LAS (O-methylation, glucuronidation, or sulfation) comprises a major route for detoxification and drug clearance,31 the potential for further oxidation of catechol LAS to o-quinones and subsequent conjugation with cellular nucleophiles has not been thoroughly scrutinized. Indeed for the case of E2, catechol oxidation to o-quinones followed by GSH conjugation is observed even in the presence of competing O-methylation.43
In the present study, LAS and 7-OHLAS were synthesized and oxidized to o-quinones both chemically and enzymatically. At physiological temperature and pH, LAS-o-quinone was found to have a half-life of approximately 55 min. This is comparable to half-lives observed for o-quinones formed from other SERMs, but substantially longer-lived than what is seen for estradiol-derived o-quinones (Table 1). Previous work has demonstrated that catechols which form long-lived o-quinones display increased cytotoxicity compared to those that are shorter-lived,38, 39 suggesting that LAS-o-quinones may have the potential to be more cytotoxic than those derived from E2.
Table 1.
Relative Reactivities of Estrogen and SERM o-Quinones
| Estrogen/SERM o-Quinone | Half-life (min) | Reference |
|---|---|---|
| 2-OHE2 a | 0.70 | (36) |
| 4-OHE2 a | 12 | (36) |
| 4-OHENb, c | 138 | (38) |
| Tamoxifen a | 80 | (50) |
| Raloxifene b | 69 | (39) |
| LY2066948 b | 234 | (24) |
| Lasofoxifene b | 55 | Present Study |
The rate of disappearance of the o-quinone was determined by monitoring the rate of apparent disappearance of the corresponding GSH conjugates by HPLC.
The disappearance of the quinoid species was followed by monitoring the decrease in UV absorbance at the appropriate wavelength in phosphate buffer (pH 7.4, 37 °C).
4-hydroxyequilenin.
Incubation of LAS with tyrosinase, a monooxygenase that efficiently oxidizes phenols to catechols and o-quinones,44, 45 yielded LAS-o-quinone which was trapped by GSH to produce two mono-GSH and two di-GSH catechol conjugates (Figure 1A). These conjugates were also observed in both rat and human liver microsomal incubations, although overall metabolism was notably attenuated in the case of the latter (Figures 2A, 2B). As P450 2D6 and P450 3A4 have previously been identified as the P450s primarily responsible for LAS catechol formation in vivo,31 and P450 1B1 is one of the key enzymes responsible for the metabolism of estrogens to genotoxic o-quinones extra-hepatically,37, 46, 47 we investigated LAS-o-quinone formation catalyzed by these isozymes. All LAS GSH conjugates seen in tyrosinase incubations were also detected in experiments with P450 2D6 (Figure 3B). Metabolic turnover with P450 3A4 appeared markedly enhanced, producing only di-GSH conjugates with OHLAS-diSG2 being the major metabolite (Figure 3A), suggesting LAS may be a better substrate for P450 3A4 than P450 2D6. Similar to P450 2D6, all four GSH conjugates were detected in experiments with P450 1B1 (Figure 3C). It is therefore plausible that LAS-o-quinones are capable of being generated in target tissues for estrogens and SERMs such as the breast, ovaries, and uterus where P450 1B1 is primarily expressed.46
After confirming that LAS catechols may be readily oxidized to their corresponding o-quinones in incubations with tyrosinase, microsomes, or supersomes, we next sought to determine whether similar oxidation would take place in the presence of competing detoxification pathways. Prakash, et al. reported previously that neither catechol nor glutathione conjugates of LAS were detected in humans or preclinical species, and concluded that this was likely due to rapid conjugative metabolism (O-methylation, glucuronidation, or sulfation) followed by urinary excretion.31 However, results of the present study suggest that LAS-o-quinone formation and subsequent thiol conjugation could represent a possible competing pathway, as GSH conjugates were observed alongside glucuronidated or O-methylated metabolites in incubations containing GSH in the presence of glucuronidating or methylating systems (Figure 4). Although LAS GSH conjugates have not been previously detected in vivo, our data clearly demonstrate that LAS catechols may further oxidize to thiophilic o-quinones. It is therefore not unreasonable to propose that a relatively long-lived LAS-o-quinone may be selective enough to react with thiol moieties of cellular proteins. Protein labeling has previously been observed for quinoids of both estrogens and SERMs,20, 25 and identification of protein targets for LAS-o-quinones will be a subject of further studies.
Finally, we aimed to investigate whether LAS-o-quinones could potentially form adducts with DNA, as such modification has been reported for quinoids of SERMs and estrogens.28, 48 In particular, depurinating adducts of estrogen o-quinones have been shown to generate apurinic sites on DNA that are prone to improper repair, resulting in mutations that are critical for initiation of breast, prostate, and other cancers.49 Incubation of 7-OHLAS with tyrosinase and deoxynucleosides resulted in detection of depurinating 7-OHLAS-Ade (Figure 5). Furthermore, 7-OHLAS-Ade was also detected along with depurinating 7-OHLAS-Gua-1 and 7-OHLAS-Gua-2 in incubations with CT-DNA (Figure 6). In contrast with what is observed for structurally similar 2-OHE2,41 stable DNA adducts were not detected in tyrosinase incubations with 7-OHLAS and deoxynucleosides. We speculate that the presence of an aromatic ring at the 1-position of the tetralin core of LAS (Scheme 1) is likely responsible for this observed difference in reactivity. 2-OHE2-o-quinone readily isomerizes to a p-Quinone methide A (Scheme 2) that reacts at C56 with the exocyclic amino group of purine nucleosides via 1,6-Michael addition to form stable adducts.41 Isomerization of 7-OHLAS-o-quinone to a structurally analogous 7-OHLAS-p-Quinone methide A would be disfavored over formation of a 7-OHLAS-p-Quinone methide B, as the latter is stabilized by extended pi-electron conjugation (Scheme 2). In support of this, DHN-7-OHLAS was detected as a major metabolite in incubations of 7-OHLAS with tyrosinase and deoxynucleosides (Figure 5A). In the absence of a strong nucleophile like GSH, it is likely that reaction of deoxynucleosides with 7-OHLAS-o-quinone to yield depurinating adducts competes with isomerization of 7-OHLAS-o-quinone to a 7-OHLAS-p-Quinone methide B which then tautomerizes to DHN-7-OHLAS. As the latter pathway is more energetically favored, formation of DHN-7-OHLAS predominates over formation of depurinating adducts (Figure 5A). Of note, 7-OHLAS was also observed to be an antiestrogen of sub-micromolar potency in Ishikawa/ECC-1 endometrial cancer cells, as measured by inhibition of E2-induced alkaline phosphatase induction (data not shown). This finding demonstrates that 7-OHLAS is also transported to the nucleus; thereby increasing the chance for interaction with DNA and potentially amplifying adduct formation. Taken together, these data may suggest that like E2, generation of apurinic sites by LAS-o-quinones is feasible and could conceivably result in similar carcinogenic initiation.
In conclusion, our data show that similar to endogenous estrogens, LAS catechols are further oxidized to electrophilic o-quinones which react with both GSH and DNA, and which could potentially cause toxicity in vivo. Moreover, while o-quinone formation from other SERMs such as tamoxifen, raloxifene and LY2066948 constitutes a relatively minor metabolic pathway,24, 39, 50 in this study LAS-o-quinones were observed as major metabolites, similar to what is observed for structurally similar estradiol. The results of this investigation therefore, will be useful in the design and development of novel ER ligands which maintain efficacy as SERMs while minimizing the potential problems associated with the formation of reactive intermediates.
Acknowledgment
We thank Atieh Hajirahimkhan and Ping Yao for antiestrogenic determination of 7-OHLAS in Ishikawa/ECC-1 cells.
Funding Support
This work was supported by NIH Grant CA79870.
Abbreviations
- 2-OHE2
2-hydroxyestradiol
- 4-OHE2
4-hydroxyestradiol
- 4-OHEN
4-hydroxyequilenin
- 5-OHLAS
5-hydroxylasofoxifene
- 7-OHLAS
7-hydroxylasofoxifene
- CID
collision-induced dissociation
- COMT
catechol-O-methyltransferase
- CT-DNA
calf thymus DNA
- DHN-7-OHLAS
1,2-dihydronaphthyl-7-hydroxylasofoxifene
- DMA
desmethyl arzoxifene
- E2
17β-estradiol
- ER
estrogen receptor
- FDA
Food and Drug Administration
- FWHM
full width half maximum
- HRT
hormone-replacement therapy
- LAS
lasofoxifene
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- SAM
S-adenosyl methionine
- SERM
selective estrogen receptor modulator
- UDPGA
uridine diphosphate glucuronic acid
References
- 1.Cho CH, Nuttall ME. Therapeutic potential of oestrogen receptor ligands in development for osteoporosis. Expert Opin. Emerg. Drugs. 2001;6:137–154. doi: 10.1517/14728214.6.1.137. [DOI] [PubMed] [Google Scholar]
- 2.Shelly W, Draper MW, Krishnan V, Wong M, Jaffe RB. Selective estrogen receptor modulators: an update on recent clinical findings. Obstet. Gynecol. Surv. 2008;63:163–181. doi: 10.1097/OGX.0b013e31816400d7. [DOI] [PubMed] [Google Scholar]
- 3.Gennari L, Merlotti D, De Paola V, Nuti R. Lasofoxifene: Evidence of its therapeutic value in osteoporosis. Core Evid. 2010;4:113–129. doi: 10.2147/ce.s6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson GM, Naunton M, Tichelaar LK, Gennari L. Lasofoxifene: selective estrogen receptor modulator for the prevention and treatment of postmenopausal osteoporosis. Ann. Pharmacother. 2011;45:499–509. doi: 10.1345/aph.1P604. [DOI] [PubMed] [Google Scholar]
- 5.Gennari L, Merlotti D, Stolakis K, Nuti R. Lasofoxifene, from the preclinical drug discovery to the treatment of postmenopausal osteoporosis. Expert Opin. Drug Discovery. 2011;6:205–217. doi: 10.1517/17460441.2011.547188. [DOI] [PubMed] [Google Scholar]
- 6.Obiorah I, Jordan VC. Progress in endocrine approaches to the treatment and prevention of breast cancer. Maturitas. 2011;70:315–321. doi: 10.1016/j.maturitas.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings SR, Ensrud K, Delmas PD, LaCroix AZ, Vukicevic S, Reid DM, Goldstein S, Sriram U, Lee A, Thompson J, Armstrong RA, Thompson DD, Powles T, Zanchetta J, Kendler D, Neven P, Eastell R. Lasofoxifene in postmenopausal women with osteoporosis. N. Engl. J. Med. 2010;362:686–696. doi: 10.1056/NEJMoa0808692. [DOI] [PubMed] [Google Scholar]
- 8.Ensrud K, LaCroix A, Thompson JR, Thompson DD, Eastell R, Reid DM, Vukicevic S, Cauley J, Barrett-Connor E, Armstrong R, Welty F, Cummings S. Lasofoxifene and cardiovascular events in postmenopausal women with osteoporosis: Five-year results from the Postmenopausal Evaluation and Risk Reduction with Lasofoxifene (PEARL) trial. Circulation. 2010;122:1716–1724. doi: 10.1161/CIRCULATIONAHA.109.924571. [DOI] [PubMed] [Google Scholar]
- 9.Lacroix AZ, Powles T, Osborne CK, Wolter K, Thompson JR, Thompson DD, Allred DC, Armstrong R, Cummings SR, Eastell R, Ensrud KE, Goss P, Lee A, Neven P, Reid DM, Curto M, Vukicevic S. Breast cancer incidence in the randomized PEARL trial of lasofoxifene in postmenopausal osteoporotic women. J. Natl. Cancer Inst. 2010;102:1706–1715. doi: 10.1093/jnci/djq415. [DOI] [PubMed] [Google Scholar]
- 10.Palacios S. Efficacy and safety of bazedoxifene, a novel selective estrogen receptor modulator for the prevention and treatment of postmenopausal osteoporosis. Curr. Med. Res. Opin. 2010;26:1553–1563. doi: 10.1185/03007991003795873. [DOI] [PubMed] [Google Scholar]
- 11.Cuzick J, DeCensi A, Arun B, Brown PH, Castiglione M, Dunn B, Forbes JF, Glaus A, Howell A, von Minckwitz G, Vogel V, Zwierzina H. Preventative therapy for breast cancer: a consensus statement. Lancet Oncol. 2011;12:496–503. doi: 10.1016/S1470-2045(11)70030-4. [DOI] [PubMed] [Google Scholar]
- 12.Gennari L, Merlotti D, Nuti R. Selective estrogen receptor modulator (SERM) for the treatment of osteoporosis in postmenopausal women: focus on lasofoxifene. Clin. Interv. Aging. 2010;5:19–29. doi: 10.2147/cia.s6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Villiers TJ. Selective estrogen receptor modulators in the treatment of osteoporosis: a review of the clinical evidence. Clin. Invest. 2011;1:719–724. [Google Scholar]
- 14.Pickar JH, MacNeil T, Ohleth K. SERMs: progress and future perspectives. Maturitas. 2010;67:129–138. doi: 10.1016/j.maturitas.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Lewiecki EM. Lasofoxifene for the prevention and treatment of postmenopausal osteoporosis. Ther. Clin. Risk Manag. 2009;5:817–827. doi: 10.2147/tcrm.s5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma M, Shubert DE, Lewis J, McGarrigle BP, Bofinger DP, Olson JR. Biotransformation of tamoxifen in a human endometrial explant culture model. Chem. Biol. Interact. 2003;146:237–249. doi: 10.1016/j.cbi.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Dowers TS, Qin ZH, Thatcher GR, Bolton JL. Bioactivation of selective estrogen receptor modulators (SERMs) Chem. Res. Toxicol. 2006;19:1125–1137. doi: 10.1021/tx060126v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan VC. Metabolites of tamoxifen in animals and man: identification, pharmacology, and significance. Breast Cancer Res. Treat. 1982;2:123–138. doi: 10.1007/BF01806449. [DOI] [PubMed] [Google Scholar]
- 19.Potter GA, McCague R, Jarman M. A mechanistic hypothesis for DNA adduct formation by tamoxifen following hepatic oxidative metabolism. Carcinogenesis. 1994;15:439–442. doi: 10.1093/carcin/15.3.439. [DOI] [PubMed] [Google Scholar]
- 20.Chen Q. Cytochrome P450 3A4-mediated bioactivation of raloxifene: irreversible enzyme inhibition and thiol adduct formation. Chem. Res. Toxicol. 2002;15:907–914. doi: 10.1021/tx0200109. [DOI] [PubMed] [Google Scholar]
- 21.Fan PW, Zhang F, Bolton JL. 4-Hydroxylated metabolites of the antiestrogens tamoxifen and toremifene are metabolized to unusually stable quinone methides. Chem. Res. Toxicol. 2000;13:45–52. doi: 10.1021/tx990144v. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Liu J, van Breemen RB, Thatcher GR, Bolton JL. Bioactivation of the selective estrogen receptor modulator desmethylated arzoxifene to quinoids: 4′-fluoro substitution prevents quinoid formation. Chem. Res. Toxicol. 2005;18:162–173. doi: 10.1021/tx049776u. [DOI] [PubMed] [Google Scholar]
- 23.Qin Z, Kastrati I, Ashgodom RT, Lantvit DD, Overk CR, Choi Y, van Breemen RB, Bolton JL, Thatcher GR. Structural modulation of oxidative metabolism in design of improved benzothiophene selective estrogen receptor modulators. Drug Metab. Dispos. 2009;37:161–169. doi: 10.1124/dmd.108.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gherezghiher TB, Michalsen B, Chandrasena EP, Qin Z, Sohn J, Thatcher GRJ, Bolton JL. The naphthol selective estrogen receptor modulator (SERM), LY2066948, is oxidized to an o-quinone analogous to the naphthol equine estrogen, equilenin. Chemico-Biological Interactions. 2012;196:1–10. doi: 10.1016/j.cbi.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolton JL, Thatcher GR. Potential mechanisms of estrogen quinone carcinogenesis. Chem. Res. Toxicol. 2008;21:93–101. doi: 10.1021/tx700191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavalieri E, Chakravarti D, Guttenplan J, Hart E, Ingle J, Jankowiak R, Muti P, Rogan E, Russo J, Santen R, Sutter T. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. Biochim. Biophys. Acta. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Cavalieri E, Frenkel K, Liehr JG, Rogan E, Roy D. Estrogens as endogenous genotoxic agents--DNA adducts and mutations. J. Natl .Cancer Inst. Monogr. 2000;27:75–93. doi: 10.1093/oxfordjournals.jncimonographs.a024247. [DOI] [PubMed] [Google Scholar]
- 28.Shibutani S, Ravindernath A, Suzuki N, Terashima I, Sugarman SM, Grollman AP, Pearl ML. Identification of tamoxifen-DNA adducts in the endometrium of women treated with tamoxifen. Carcinogenesis. 2000;21:1461–1467. [PubMed] [Google Scholar]
- 29.Moore CD, Reilly CA, Yost GS. CYP3A4-Mediated oxygenation versus dehydrogenation of raloxifene. Biochemistry. 2010;49:4466–4475. doi: 10.1021/bi902213r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke HZ, Paralkar VM, Grasser WA, Crawford DT, Qi H, Simmons HA, Pirie CM, Chidsey-Frink KL, Owen TA, Smock SL, Chen HK, Jee WS, Cameron KO, Rosati RL, Brown TA, Dasilva-Jardine P, Thompson DD. Effects of CP-336,156, a new, nonsteroidal estrogen agonist/antagonist, on bone, serum cholesterol, uterus and body composition in rat models. Endocrinology. 1998;139:2068–2076. doi: 10.1210/endo.139.4.5902. [DOI] [PubMed] [Google Scholar]
- 31.Prakash C, Johnson KA, Gardner MJ. Disposition of lasofoxifene, a next-generation selective estrogen receptor modulator, in healthy male subjects. Drug Metab. Dispos. 2008;36:1218–1226. doi: 10.1124/dmd.108.020404. [DOI] [PubMed] [Google Scholar]
- 32.NIH . NIH guidelines for the laboratory use of chemical carcinogens. NIH; Washington, DC: 1981. pp. 2381–2385. U.G.P.O. [Google Scholar]
- 33.Day WW, Johnson KA, Prakash CA, Eggler JF. Preparation of estrogen agonist/antagonist metabolites. 2001. PCT Int. Appl. WO 2001077093.
- 34.Thompson JA, Malkinson AM, Wand MD, Mastovich SL, Mead EW, Schullek KM, Laudenschlager WG. Oxidative metabolism of butylated hydroxytoluene by hepatic and pulmonary microsomes from rats and mice. Drug Metab. Dispos. 1987;15:833–840. [PubMed] [Google Scholar]
- 35.Dieckhaus CM, Fernandez-Metzler CL, King R, Krolikowski PH, Baillie TA. Negative ion tandem mass spectrometry for the detection of glutathione conjugates. Chem. Res. Toxicol. 2005;18:630–638. doi: 10.1021/tx049741u. [DOI] [PubMed] [Google Scholar]
- 36.Iverson SL, Shen L, Anlar N, Bolton JL. Bioactivation of estrone and its catechol metabolites to quinoid-glutathione conjugates in rat liver microsomes. Chem. Res. Toxicol. 1996;9:492–499. doi: 10.1021/tx950178c. [DOI] [PubMed] [Google Scholar]
- 37.Spink DC, Zhang F, Hussain MM, Katz BH, Liu X, Hilker DR, Bolton JL. Metabolism of equilenin in MCF-7 and MDA-MB-231 human breast cancer cells. Chem. Res. Toxicol. 2001;14:572–581. doi: 10.1021/tx000219r. [DOI] [PubMed] [Google Scholar]
- 38.Shen L, Pisha E, Huang Z, Pezzuto JM, Krol E, Alam Z, van Breemen RB, Bolton JL. Bioreductive activation of catechol estrogen-ortho-quinones: aromatization of the B ring in 4-hydroxyequilenin markedly alters quinoid formation and reactivity. Carcinogenesis. 1997;18:1093–1101. doi: 10.1093/carcin/18.5.1093. [DOI] [PubMed] [Google Scholar]
- 39.Yu L, Liu H, Li W, Zhang F, Luckie C, van Breemen RB, Thatcher GR, Bolton JL. Oxidation of raloxifene to quinoids: potential toxic pathways via a diquinone methide and o-quinones. Chemical Research in Toxicology. 2004;17:879–888. doi: 10.1021/tx0342722. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Gaikwad NW, K. O, Zahid M, Cavalieri E, Rogan E. Cytochrome P450 isoforms catalyze formation of catechol estrogen quinones that react with DNA. Metabolism. 2007;56:887–894. doi: 10.1016/j.metabol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating adducts: implications for tumor-initiating activity. Chem. Res. Toxicol. 2006;19:164–172. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 42.Bolton JL, Shen L. p-Quinone methides are the major decomposition products of catechol estrogen o-quinones. Carcinogenesis. 1996;17:925–929. doi: 10.1093/carcin/17.5.925. [DOI] [PubMed] [Google Scholar]
- 43.Butterworth M, Lau SS, Monks TJ. 17 beta-estradiol metabolism by hamster hepatic microsomes: comparison of catechol estrogen O-methylation with catechol estrogen oxidation and glutathione conjugation. Chem. Res. Toxicol. 1996;9:793–799. doi: 10.1021/tx9501952. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez-Ferrer A, Rodriguez-Lopez JN, Garcia-Canovas F, Garcia-Carmona F. Tyrosinase: a comprehensive review of its mechanism. Biochim. Biophys. Acta. 1995;1247:1–11. doi: 10.1016/0167-4838(94)00204-t. [DOI] [PubMed] [Google Scholar]
- 45.Pezzella A, Lista L, Napolitano A, d’Ischia M. Tyrosinase-catalyzed oxidation of 17beta-estradiol: structure elucidation of the products formed beyond catechol estrogen quinones. Chem. Res. Toxicol. 2005;18:1413–1419. doi: 10.1021/tx050060o. [DOI] [PubMed] [Google Scholar]
- 46.Yager JD. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 47.Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT. Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology. 2003;144:3382–3398. doi: 10.1210/en.2003-0192. [DOI] [PubMed] [Google Scholar]
- 48.Bolton JL, Yu L, Thatcher GR. Quinoids formed from estrogens and antiestrogens. Methods Enzymol. 2004;378:110–123. doi: 10.1016/S0076-6879(04)78006-4. [DOI] [PubMed] [Google Scholar]
- 49.Zahid M, Gaikwad NW, Rogan EG, Cavalieri EL. Inhibition of depurinating estrogen-DNA adduct formation by natural compounds. Chem. Res. Toxicol. 2007;20:1947–1953. doi: 10.1021/tx700269s. [DOI] [PubMed] [Google Scholar]
- 50.Zhang F, Fan PW, Liu X, Shen L, van Breemen RB, Bolton JL. Synthesis and reactivity of a potential carcinogenic metabolite of tamoxifen: 3,4-dihydroxytamoxifen-o-quinone. Chem. Res. Toxicol. 2000;13:53–62. doi: 10.1021/tx990145n. [DOI] [PubMed] [Google Scholar]