Abstract
Fragile X syndrome is the leading cause of inherited mental impairment and is associated with expansions of CGG repeats within the FMR1 gene. To detect expanded CGG repeats, we developed a dual-mode single-molecule fluorescence assay that allows acquisition of two parallel, independent measures of repeat number based on (1) the number of Cy3-labeled probes bound to the repeat region and (2) the physical length of the electric field-linearized repeat region, obtained from the relative position of a single Cy5 dye near the end of the repeat region. Using target strands derived from cell-line DNA with defined numbers of CGG repeats, we show that this assay can rapidly and simultaneously measure the repeats of a collection of individual sample strands within a single field of view. With a low occurrence of false positives, the assay differentiated normal CGG repeat lengths (CGGN, N = 23) and expanded CGG repeat lengths (CGGN, N = 118), representing a premutation disease state. Further, mixtures of these DNAs gave results that correlated with their relative populations. This strategy may be useful for identifying heterozygosity or for screening collections of individuals, and it is readily adaptable for screening other repeat disorders.
Keywords: Fragile X syndrome, Single molecule, Total internal reflection fluorescence microscopy (TIRFM), Heterozygosity, Dual mode, Photobleaching, Diagnostic assay
Introduction
Microsatellites are regions of DNA in which simple sequences of one to six nucleotides are repeated multiple times. Trinucleotide repeat sequences form a class of microsatellites that commonly impact phenotype and are linked to more than 30 genetic diseases. The lengths of these repeat sequences can increase over time and over generations, leading to more severe disease symptoms and earlier onset [1–3].
Fragile X syndrome is the leading cause of inherited mental impairment and is associated with the expansion of CGG repeats within the FMR1 gene. CGG expansions >200 repeats disrupt synthesis of the Fragile X mental retardation protein (FMRP), which is required for neural development during embryogenesis. The phenotype for Fragile X syndrome includes cognitive dysfunction, hypersensitivity, speech delay, and physical abnormalities [4]. Unaffected individuals have CGG repeat lengths between 6 and 54 repeats, with 45–54 repeats termed the gray zone. Premutation carriers (55–200 CGG), both male and female, may develop Fragile X tremor ataxia syndrome (FXTAS), despite earlier normal cognitive functioning [5]. Female premutation carriers are also at risk for premature ovarian failure [6]. Expansion to the full mutation from the gray-zone allele can occur in two generations [7, 8]. There is thus a strong desire to develop rapid, robust assays for the lengths of these expansions to identify people that have premutations; i.e., expansions that are almost long enough to result in a disease state but do not give a strong phenotype.
Methods to diagnose Fragile X syndrome are generally based on determining the CGG repeat length. Southern blot analysis can be used to measure the repeat number accurately for full-length mutations and large premutations, but this approach is time intensive. Recently developed PCR methods [9–14] have improved the accuracy and range of measured CGG repeat lengths, and capillary gel electrophoresis (CAGE) is increasingly used to quantify the PCR products [8–10]. Still, a truly high-throughput assay would be highly beneficial for screening and for identifying heterozygous individuals with one allele in the premutation or full-mutation range.
Several single-molecule strategies have been developed to extract sequence information from individual molecules, using either fluorescence- or electrochemical-based methods. The fluorescence-based approaches include polymerase-directed single-nucleotide incorporation [15–17], barcoding [18–20], and length measurements [21, 22]. Non-optical methods include protein nanopores [23] and solid-state devices [24, 25]. An advantage of single-molecule approaches is their scalability to larger platforms, allowing for highly parallel sample processing while greatly reducing the amount of material needed. By acquiring signals from individual molecules, single-molecule methods can detect small sub-populations, an ability that would be advantageous in large-population screening.
Here, we present a novel single-molecule fluorescence (SMF) assay that can screen mixtures of normal and pre-mutation levels of CGG repeats. Using wide-field total internal reflection fluorescence microscopy (TIRFM), we collect data simultaneously from scores of individual molecules. This dual-mode SMF assay acquires two independent measures of the CGG repeat number from individual CGG repeat sequences. By binding sequence-specific probes to ssDNA derived from the repeat region, fluorescence intensity readouts of the number of probes bound can be obtained. In parallel, the sub-diffraction limit physical lengths of the repeat region, now in duplex form by annealing of the probe, can be directly measured by super-resolution imaging [26–30]. The distance between a single Cy5 at the untethered end of the target and the centroid of fluorescence of the bound probes is determined by separately imaging the dyes and determining their relative positions.
The acquisition of two independent measurements of the CGG repeat number for each sample reduces the overall likelihood of false positives for premutations. If the false positives reported by each method are largely uncorrelated, the expected probability of false positives reported by this dual-mode approach would approach the product of the false-positive rates associated with each method, yielding a substantially lower overall rate. Indeed, coupling the length measurement to the intensity measurement decreased the overall false-positive rate by fourfold, the same within error as the product of the false-positive rates of each method. In addition, this SMF assay facilitates the identification of multiple populations such as would be present in samples from heterozygous individuals or from collections of individuals. Although the strict accuracy of the CGG repeat number is less than achieved with other methods (e.g., Southern blot, CAGE), this method can rapidly differentiate normal and premutation molecules with a low probability of false positives, making it applicable to rapidly screen populations that include both homozygotes and heterozygotes.
Materials and Methods
Preparation of CGG Repeats and DNA Rulers
Synthetic CGG target strands containing 5, 10, and 15 CGG repeats were purchased from Integrated DNA Technologies (Coralville, IA). DNA targets containing larger numbers of repeats were prepared as PCR products from cell-line DNA (23, 56, 118, and 340 repeats) with a master mix from Asuragen containing GC-rich AMP buffer, FMR1 primers, and GC-rich polymerase mix [14]. The forward primer was 5′-phosphorylated to facilitate digestion of the complementary strand by lambda exonuclease in a subsequent step. Samples were amplified with an initial heat denaturation step of 98°C for 5 min. This step was followed by 25 cycles of 97°C for 35 s, 62°C for 35 s, and 72°C for 4 min, with a final of extension at 72°C for 10 min. The single-stranded PCR products were prepared by lambda exonuclease digestion to facilitate hybridization of the fluorescently labeled probes to the target sequence.
DNA rulers of defined lengths were designed to calibrate the electrokinetic stretching method. First, deletion mutants with varying lengths (23–500 bp) between two constant primer regions were constructed from a pMAL plasmid with a Quikchange site-directed mutagenesis kit (Agilent, Santa Clara, CA). The rulers were then generated from the deletion mutants by primer extension with Pfu Hotstart DNA polymerase (Agilent, Santa Clara, CA) using dye-labeled constant region primers (forward primer: 5′-biotin-GGTGA(Cy3)AATCATGCCGAACATCCCG and reverse primer: 5′-GATGGCGCGAATGT(Cy5)CATCAGA ACG). The sequences of the mutants were confirmed by sequencing, and the DNA ruler lengths were confirmed by gel electrophoresis on 3% agarose gels with a 100-bp DNA ladder (New England Biolabs, Ipswich, MA).
Target/Probe Annealing
The CGG repeat-containing ssDNAs were incubated with Cy3-labeled [5′-Cy3-CCGCCGCCGCCGCCG; (CCG)5] repeat probes and a Cy5-labeled probe (5′-Cy5-CATCTT CTCTTCAGCCCTGCTAGCGCCGGGAGC) complementary to a flanking region (Integrated DNA Technologies, Coralville, IA). The samples were heated to 95°C and then cycled 100 times above and below the Tm of the repeat probe to optimize binding in 100 mM NaCl, 25 mM Tris–Cl, pH 8.0.
Flow Chamber Preparation
Quartz slides (G. Finkenbeiner Inc., Waltham, MA) were coated with methoxy poly (ethylene glycol) succinimidyl valerate (mPEG-SVA) to minimize nonspecific adsorption of the labeled samples and doped with 1–2% biotinylated mPEG (Laysan Bio Inc., Arab, AL). Silver-based electrodes were deposited on the quartz surface by using a conductive microtip pen (Chemtronics, Kennesaw, GA). The slide was incubated with 0.1 mg/mL streptavidin (Invitrogen, Carlsbad, CA). The biotinylated target–probe complexes were added to the sample chamber and immobilized to the surface through the biotin–streptavidin linkage. The unbound complexes were washed out during buffer exchange. For length measurements, a constant electric field (~104 V/m) was applied across the sample chamber to linearize and orient the target strands.
SMF Microscopy
The slide was mounted on an inverted microscope (Olympus IX-71) with a 60× water immersion objective. The samples were separately excited by prism-type total internal reflection of a 532 nm laser (Crystalaser, Reno, NV) and a 632 nm laser (CVI Melles Griot, Albuquerque, NM). The images were acquired with a cooled I-Penta-MAX IIC CCD (Princeton Instruments, Trenton, NJ). The image capture frequency was 2–10 Hz. A deoxygenating imaging buffer, used to slow photobleaching, consisted of 0.5× TBE (50 mM Tris, 41 mM boric acid, 0.5 mM EDTA), 0.8 mM Trolox, 12% glucose, 0.1 mg/mL glucose oxidase (Sigma), and 0.04 mg/mL catalase (Roche).
Fluorescence Intensity Measurements and Dye Counting
The molecules were initially excited at 532 nm to measure the total intensity of the Cy3-labeled probes bound to the targets. The molecules were imaged for ~60 s to observe a sufficient number of discrete losses in intensity due to the photobleaching of individual Cy3 dyes. The photobleaching events were identified by a two-tailed sliding t test. The magnitude of the decrease in intensity due to photobleaching was calculated from the difference between the intensities before and after the photobleaching event. The average intensity of the measured photobleaching events was used to calculate the number of probes bound to each target from the total intensity for each molecule relative to that of each photobleaching step.
Sub-Diffraction Length Measurements
The physical length of the CGG target sequence was directly determined by measuring the distance between the centroid of the Cy3-labeled CGG repeat probes, before applying the electric field, and the Cy5-labeled probe attached to the non-CGG flanking region after application of the electric field [29]. Because the distance between the dyes is less than the diffraction limit, each dye was separately imaged. The positions of the dyes were found by a nonlinear least squares fit to a two-dimensional Gaussian function with a background,
| (1) |
where A is the height, (x0, y0) is the centroid of the point source, σy and σx are the widths of the point-spread function, and z is the background. The distance between the two dyes could then be determined from the relative positions of the two dyes,
| (2) |
where the pixel size of the magnified image is 95.4 nm. The measured distances were converted to the number of repeats by using the standard value of helical rise per base pair (0.34 nm/bp).
Dual-Mode Analysis
Because of the inherent heterogeneities in the measurements, criteria were designed to identify the molecules for which the repeat numbers from the intensity and length measurements were correlated. A molecule was considered to have agreeing values for the two measurements if either of the two following two conditions were satisfied: (1) the difference between the two measurements was <30 repeats or (2) the fractional difference between the two measurements was <0.4. These two conditions were implemented to minimize biasing in the identification of similar values for longer or shorter repeats. The analysis was performed without referencing the actual repeat length. The distributions of the measured CGG repeat values were fit with two Gaussian functions to determine the average values and the relative fractions of the repeat populations.
Results and Discussion
To determine the CGG repeat numbers of the sample DNA strands, we developed a dual-mode assay to simultaneously measure two independent values for each target strand that could be correlated to the CGG repeat number: (1) the number of bound probe oligonucleotides, which is derived from the fluorescence intensity of target-bound and dye-labeled probes and (2) the direct physical length of the repeat region.
Determination of CGG Repeat Length Based on Fluorescence Intensity
To validate the strategy of using a fluorescence readout to count the number of dyes annealed to CGG repeat sequences, we tested DNA target strands spanning the normal, gray-zone, and premutation disease states (5–118 repeats). For repeat lengths of 5, 10, and 15 CGG repeats, we used synthetic DNA targets strands, and for longer CGG repeat sequences (23, 56, 96, and 118 repeats), we used PCR followed by lambda exonuclease digestion to generate ssDNA. Figure 1 depicts the probe binding strategy for the targeted molecules. The DNA target strands were incubated with Cy3-(CCG)5 probes designed to bind to the repeat region. The targets were also annealed with a Cy5-labeled strand complementary to a constant (non-CGG repeating sequence) region flanking the repeat sequence as a control for identifying overlapping, unresolved molecules. The probe-bound targets were immobilized onto streptavidin-treated PEG-coated slides through biotin–streptavidin linkages. Using wide-field TIRFM, time traces for scores of molecules were simultaneously collected for each field of view by direct excitation with a 532 nm laser. Figure 2a shows representative three-dimensional screen-shots for immobilized probe-bound targets with CGG repeat lengths of 10, 15, 56, and 118. The four images have the same intensity scale. The average intensities of the probe-annealed targets increased dramatically with the higher CGG repeat numbers, indicating that more probes were bound to the longer targets. However, visual inspection of the individual intensities for (CGG)56 and (CGG)118 reveals significant variation, suggesting populations of targets with different numbers of bound probes.
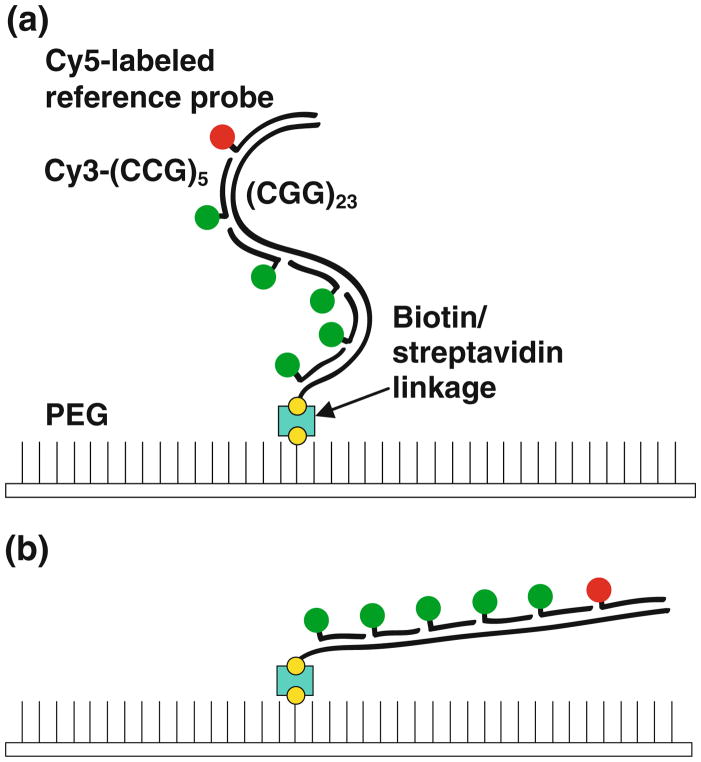
Fig. 1.
A schematic of the dye-labeling strategy for the dual-mode SMF assay. a In the absence of the electric field, the target molecules are randomly oriented with respect to the tethered end. The target strand is bound with Cy3-(CCG)5 probes targeted to the repeat region (Cy3 shown schematically in green) and a Cy5-labeled probe targeted to a flanking non-repeat region (red). The biotin linkages on the PEG surface and the target strand are shown in yellow. The streptavidin molecule is indicated in cyan. b Upon application of the electric field, the molecules become oriented and extended in the direction of the field, increasing the distance of the Cy5 probe from the tethered end (Color figure online)
Fig. 2.
Intensity-based probe-counting measurements of the varying CGG repeat lengths. a Intensity distributions and screenshots for four CGG repeat lengths (10, 15, 56, and 118). The dashed line in the histograms indicates the expected number of (GCC)5-Cy3 probes bound to the target strands. b Representative time traces for the fluorescence intensity of immobilized (CGG)5, (CGG)10, (CGG)15, and (CGG)23 molecules that exhibit the expected number of stepwise decreases in intensity from photobleaching of Cy3 dyes. c The relationship between the target CGG repeat length and the average number of probes bound. The line indicates the expected number of probes bound for optimal probe binding to the target
To accurately assess the number of probes bound to the target strands as a measure of the CGG repeat length and to minimize the effect of variations in dye emission due to nonuniform illumination, dye orientation, or conformation on the total intensity, we developed a calibration procedure based on dye photobleaching. For the probe-bound target (CGG)5, the time traces predominantly exhibited a complete loss of fluorescence in a single step, which reflects the irreversible photobleaching of a single Cy3 dye (Fig. 2b). As the CGG repeat length increased, a greater number of photo-bleaching events were observed. In the time trace shown for (CGG)23, four discrete decreases in intensity occurred, with each corresponding to the photobleaching of an individual Cy3 dye. Ideally, the total number of observed photobleaching events could be used as a metric for the number of probes bound to the target [e.g., the four events in Fig. 2b for (CGG)23 correspond to four probes bound to the target]. However, for longer targets, this strategy was not practical because the solution conditions required to slow photobleaching enough to ensure that all individual photobleaching steps are resolved would lead to the requirement for very long observation times to achieve complete photobleaching of all targets in a given field of view. Therefore, the magnitudes of the stepwise decreases in intensity due to the photobleaching of the individual Cy3 dyes per molecule were collected to determine the average intensity for a dye-labeled probe bound to the target. The number of bound probes per target was then calculated as the ratio of the initial intensity and the average photobleaching step size.
The histograms in Fig. 2a show the distribution for the number of probes bound to individual target molecules as determined from the dye-counting procedure. The expected number of probes for each repeat size is derived from the maximum number of (CCG)5 probes that could fully pair with each target size. As expected, the histograms for (CGG)10 and (CGG)15 show well-defined Gaussian-like distributions centered at ~2 and 3, respectively. For these targets with numbers of CGG repeats in the “normal” range, the targets can be discriminated in terms of their CGG repeat number by the detected number of probes bound to the individual target molecules. The results indicate that the probes are likely fully base paired to the target or are occupying enough of the CGG repeats to make additional probe binding unlikely.
On the other hand, (CGG)56 and (CGG)118 show significant heterogeneity, with only a small fraction of the molecules exhibiting optimal probe binding and most of the molecules displaying less than optimal probe binding. We first considered the possibility that the degree of heterogeneity was actually uniform for the DNAs of different lengths but appeared less significant for the shorter DNAs because a significant fraction of target molecules did not bind any fluorescent probes and were therefore not included in the analysis. Such a result would be expected if the principal sources of heterogeneity were incomplete fluorescent labeling of the oligonucleotide probe or incomplete binding of the probe (in a manner that did not depend on the length of the repeating sequence). Considering the average signal from the (CGG)118 target, each stretch of five repeats would have a 64% chance of having a labeled probe bound if this were the sole source of heterogeneity. We then performed Monte Carlo simulations to test whether this model would reproduce the observed data for the shorter DNA targets. However, the simulations did not adequately reproduce the data, giving much greater representation of molecules with low signal and an overall shifting of the peak, relative to the data (not shown). Thus, we conclude that there is more heterogeneity for the targets with larger numbers of repeats. This increase may reflect the ability of longer CGG repeats to form more stable intramolecular hairpin structures [31–33], which would reduce the number of available sites for probe binding [34]. It is also likely that some heterogeneity arises from incomplete PCR (see below), producing a sub-population that has optimal probe binding but gives reduced signal because of the reduced repeat length; i.e., these molecules represent true negatives. Agarose gels of the PCR products indicated the absence of a distinct band associated with any length heterogeneities (data not shown), but this result does not rule out a broad distribution of molecules with different lengths.
In summary, the dye-counting strategy accurately reports the number of bound probes at low CGG repeat numbers, suggesting optimal base pairing between the probes and the targets. However, the measured repeat lengths deviate from the expected length for the higher CGG repeat numbers, starting at the gray-zone length (CGG)56. The fractional occupancy falls to approximately two-thirds the expected number for (CGG)118. For all of the targets, a sub-population of the targets is optimally probe bound with the expected number of probes, but the heterogeneity in the fractional occupancy makes it difficult to assess accurately CGG repeat numbers beyond “normal” state lengths.
Importantly, the false-positive rate for the (CGG)23 targets, defined as the fraction of “normal” repeat length strands that were identified as corresponding to premutation lengths, was 7.2% (15 of 206 molecules). This highlights the sensitivity of this method to accurately report the presence of “normal” CGG repeat lengths with a modest level of false positives.
Direct Physical Length Measurement of CGG Repeats
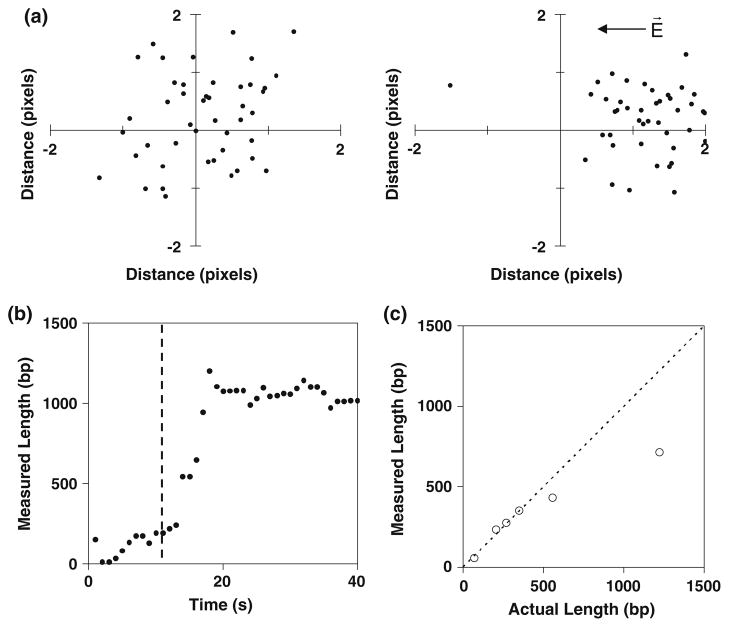
As an orthogonal approach to determine the CGG repeat number, we set out to use electrokinetic stretching and super-resolution imaging to determine the physical length of the CGG repeats by measuring the physical length of the repeat region. As controls, Cy3-, Cy5-labeled biotinylated DNA duplexes with a defined number of base pairs were constructed by primer extension so that the Cy3 dye was located proximal to the biotinylated end and the Cy5 dye was located near the untethered end. For the duplexes examined, the lengths were well below the diffraction limit. To achieve super-resolution imaging, the Cy3 and Cy5 dyes were separately imaged. The distance between the Cy3 and Cy5 dyes was then determined from their positions defined by point-spread functions (see Eqs. 1, 2). An electric field was applied to linearize single immobilized molecules, as shown in Fig. 1b [20, 21, 35, 36]. For a 350-bp duplex in the absence of the electric field, the positions of the Cy5 dyes were randomly oriented with respect to the Cy3 dyes. In the presence of the electric field, the Cy5 dyes became oriented toward the cathode, and the distance from the Cy3 dyes increased, indicating that the immobilized DNA was elongated and oriented in response to the electric field (Fig. 3a). Figure 3b shows a representative time trace that tracks the distance of the untethered Cy5-labeled end of the duplex relative to the tethered Cy3-labeled end for a 1,200-bp duplex. When an electric field (~104 V/m) was applied, the distance between the two dyes increased, approaching the fully extended length. The physical lengths were measured for duplexes ranging from 67 to 1,200 bp. The average measured lengths of the molecules increased with the number of base pairs (Fig. 3c). Similar to the intensity method, the measured values deviated from the actual length with increasing length. The shortening of the average length relative to the expected length may result from incomplete elongation by the electric field, increased formation of intramolecular contacts for the longer targets, and length heterogeneities due to incomplete PCR of the repeat region.
Fig. 3.
Length-based measurements for different CGG repeat lengths. a For a 350-bp duplex DNA in the absence of the field, the Cy5-labeled untethered ends of the molecules are randomly oriented with respect to the Cy3 dyes near the biotinylated end. For each molecule, the position of the Cy3 was placed at the origin. In the presence of the field, the molecules become oriented toward the cathode. The units of the axes are pixels (1 pixel = 95.4 nm). The arrow indicates the direction of the electric field. b A representative time trace for the electrokinetic stretching of a 1,200-bp duplex. The vertical line indicates the onset time of the electric field. c The averaged measured lengths (bp) of the immobilized strands increase with the actual length
If the false positives reported by each method were uncorrelated, the expected probability of false positives reported by utilizing two measurements for each target strand would be expressed as the product of the false-positive rates associated with each method. The false-positive rate measured for (CGG)23 by the length measurement was ~24% (11 of 45 molecules). Given that the intensity measurement has a false-positive rate of 7.2%, the dual-mode method could yield an overall false-positive rate of 1.8%, a significant improvement in accuracy.
Dual-Mode SMF assay
For each molecule, two measures of the CGG repeat numbers were acquired, one based on the number of probes bound to the target and another from the length of the target. The total intensities of the probe-bound targets and photobleaching step size of the Cy3-labeled target-bound probes were used to calculate the number of probes bound to each target. For the physical length measurement, the position of the centroid of the Cy3 fluorescence was determined by a point-spread function and defined as the location of the tethered end of the immobilized molecule. The centroid of the total Cy3 fluorescence can function as the location of the tethered end of the target strand in the absence of the field because the tethered molecule will be freely rotating about the tether such that the time-averaged position of the dyes would be centered at the tether location [37]. An electric field was then applied across the sample chamber, and the Cy5 dye was directly excited with the 632 nm laser to acquire the position of the Cy5 dye located at the untethered end. The physical length of the repeat sequence was then calculated from the differences in the coordinates of the centroid corresponding to the Cy3 dyes and the Cy5 dye (Eq. 2). The molecules with correlated repeat numbers from the intensity and length measurements were identified according to the criteria described in “Materials and Methods”.
Mixtures of Different CGG Repeat Numbers
Four mixtures with different relative amounts of (CGG)23 and (CGG)118 were examined using the dual-mode SMF assay. Approximately 200 molecules were measured for each mixture, with the repeat numbers for each molecule determined by the intensity and length measurements. Based on the matching criteria and in agreement with the error rates from the separate measurement modes, more than half of the molecules had correlating CGG repeat lengths from the two measurements. The average values of the CGG repeat lengths from these correlated molecules were similar to the expected CGG repeat number. The homogeneous (CGG)23 samples gave a well-defined unimodal distribution with averages of 18 and 19 repeats for the intensity and length measurements, respectively (Fig. 4a). The 70% confidence intervals were 11–24 for the intensity measurement and 11–26 from the length measurement. Based on the false-positive rates of the dye-counting method and the length measurements, the dual-mode approach was expected to produce 1.8% false positives. Indeed, the false-positive frequency was 2.1%, indicating that most of the false positives from the individual assays are uncorrelated.
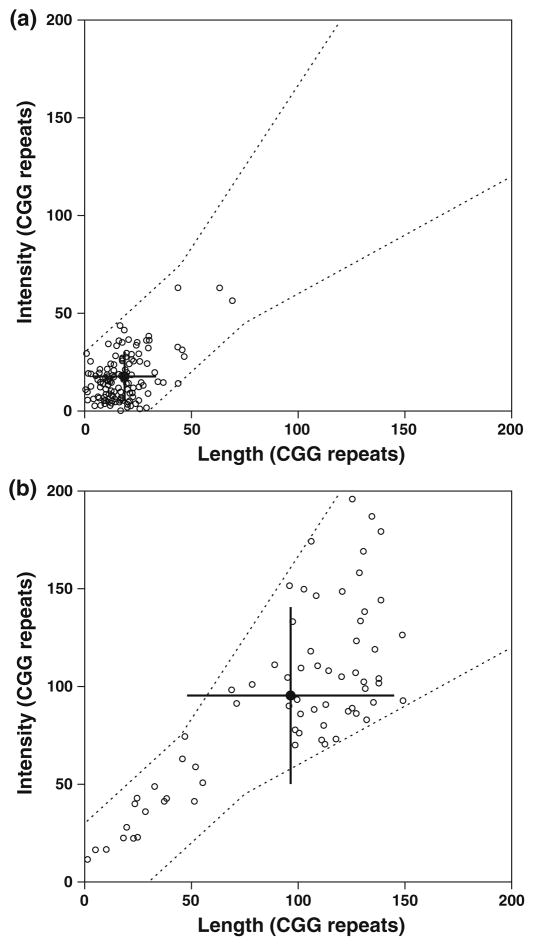
Fig. 4.
Scatter plots showing the distribution of the measured CGG repeat sizes for a (CGG)23 and b (CGG)118, in which the length-derived measurements correspond to the x-axis and the dye-counting measurements correspond to the y-axis. The filled symbol indicates the average of the two measurements. The error bars show the standard deviations. The averages of the correlated molecules are similar to the expected values. For (CGG)23, 151 of the 206 (73.3%) analyzed molecules had correlated length and dye-count measurements, as indicated by the boundaries defined by the dashed lines. Data that gave uncorrelated measurements, falling outside of this boundary, were rejected from further analysis and are therefore not included in the plots. For (CGG)118, 93 of 231 (40.2%) of the analyzed molecules had correlated length and dye-count measurements. These values are consistent with the expected fraction of correlated molecules between the two measurements for (CGG)23 and (CGG)118 with 71.6 and 42.9%, respectively. The true-negative values by the dye-counting and length measurements were 92.8 and 77.2%, respectively. The true-positive values by the dye-counting and length measurements were 54.6 and 78.0%
The repeat values for the (CGG)118 samples were higher and more broadly distributed than the (CGG)23 samples (Fig. 4b). The average repeat numbers were significantly larger, 98 by the intensity method and 97 by the length method. The 70% confidence intervals from intensity and length were 72–122 and 73–120, respectively. The (CGG)118 values appeared to have a bimodal distribution, and ~76% of the molecules were within the expected range. The combined average (average of both methods for each molecule) of the major population was 101 repeats.
As expected, the experiments with a 3:1 mixture of (CGG)23 and (CGG)118 strands, 3(CGG)23·1(CGG)118, showed the presence of a population with a larger repeat length that was not present in the homogeneous (CGG)23 samples (Fig. 5a). The major population represents 84% of the molecules with an average repeat number of 20. The smaller population has an average repeat number of 90. As shown in Fig. 5b, the experiments with a 1:3 mixture of (CGG)23 and (CGG)118, 1(CGG)23·3(CGG)118, gave an increase in the fraction of the longer repeat number population, compared to 3(CGG)23·1(CGG)118. Based on the Gaussian fits, the larger population represents 60% of the molecules with an average repeat number of 104. The smaller population has an average repeat number of 40, somewhat larger than the actual length.
Fig. 5.
Scatter plots of the correlated dual-mode measurements for the heterogeneous mixtures a 3(CGG)23·1(CGG)118 and b 1(CGG)23·3(CGG)118. For the 3(CGG)23·1(CGG)118 mixture, 103 of 186 (55.4%) molecules had correlated measurements of the CGG repeat size. For the 1(CGG)23·3(CGG)118 mixture, 98 of 223 (43.9%) molecules had correlated measurements of the CGG repeat size. Data that gave uncorrelated measurements were rejected from further analysis and are not shown in the plots
Figure 6 shows a plot of the fractions of (CGG)23 and (CGG)118 target molecules that gave values indicating the expected length ranges in the mixtures. Both repeat numbers are linear over the relative fractions. The y-intercept for (CGG)118 underscores the presence of a persistent level of heterogeneity associated with the longer repeat length. Based on the detected negative population for the dye-counting (45%) and length (22%) measurements for (CGG)118, the expected percentage of false negatives for this premutation length target DNA was 10%; i.e., we would expect that 10% of the targets would coincidentally give false measurements by both methods that correspond to smaller numbers of repeats, within the normal range. However, a larger fraction, 24% of the molecules, corresponded to normal lengths. Thus, in contrast to the situation with the uncorrelated false positives, a fraction of the false negatives are correlated. These molecules may possess secondary structure, which could reduce the number of binding sites and the length of the DNA. It is also possible that some reflect truncated PCR synthesis and a sub-population of true negatives, i.e., molecules that are included in the sample of 118 repeat molecules but that actually possess fewer repeats. With the possibility of true negatives, the fraction of shorter products of 24% is likely to represent an upper limit on the false negative rate of the assay. Further study would be needed to establish the origins of this population.
Fig. 6.
The fraction of target (CGG)23 and (CGG)118 molecules that gave measurements corresponding the correct length range, plotted against the expected fraction for the homogenous and heterogeneous mixtures of (CGG)23 and (CGG)118. Only target molecules that gave correlated values from the dual measurements were considered in this analysis. The filled squares correspond to (CGG)23, and the filled circles correspond to (CGG)118. The average values and the relative fractions are determined from bimodal Gaussian fits. Linear fits to the data are shown. The deviations between the expected and the actual fractions for the varying ratios of the two lengths are quantitatively accounted for by the presence of a fraction of the (CGG)118 samples (~24%) that appear to possess a smaller number of repeats (see text)
Conclusions
The results demonstrate the feasibility and advantages of using a dual-mode SMF assay to measure the number of CGG repeats in DNA samples. This assay can discriminate between a “normal” CGG repeat length, (CGG)23, and a length corresponding to the premutation disease state, (CGG)118 and has potential applications in large-scale molecular genetic testing. Such testing has been advised for individuals who have a family history of Fragile X syndrome or undiagnosed cognitive disabilities as well as for females who are known carriers of Fragile X syndrome or have a family history of premature ovarian insufficiency [38]. DNA corresponding to the repeat region can be PCR amplified using probes targeted to a flanking region [39], as was done here, with the single-molecule approach offering the advantage of increased sensitivity relative to electrophoretic methods. This increased sensitivity could allow the pooling of samples from many individuals for rapid screening, reducing the time and cost per sample. Ultimately, it may be possible to perform such a single-molecule assay using unamplified genomic DNA, with the repeat region extracted using known restriction enzymes [39] and immobilized by hybridization to a tagged oligonucleotide. Such a procedure could greatly increase the throughput, relative to PCR-based methods, either with pooled samples from multiple individuals or with an addressable array format for individual, rapid screening of a large number of individuals. Finally, this dual-mode SMF assay may also be useful for screening of other trinucleotide repeat disorders.
Contributor Information
Brian Cannon, Department of Chemistry and Biochemistry and the Institute for Cellular and Molecular Biology, The University of Texas, 1 University Station A4800, Austin, TX 78712, USA.
Cynthia Pan, Department of Chemistry and Biochemistry and the Institute for Cellular and Molecular Biology, The University of Texas, 1 University Station A4800, Austin, TX 78712, USA.
Liangjing Chen, Asuragen Inc., Austin, TX 78744, USA.
Andrew G. Hadd, Asuragen Inc., Austin, TX 78744, USA
Rick Russell, Email: rick_russell@mail.utexas.edu, Department of Chemistry and Biochemistry and the Institute for Cellular and Molecular Biology, The University of Texas, 1 University Station A4800, Austin, TX 78712, USA.
References
- 1.Brouwer JR, Willemsen R, Oostra BA. Micro-satellite repeat instability and neurological disease. Bioessays. 2009;1:71–83. doi: 10.1002/bies.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nature Reviews Molecular Cell Biology. 2010;3:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 3.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nature Reviews Genetics. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile X syndrome. Annual Review of Genomics and Human Genetics. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 5.Hagerman RJ, Hull CE, Safanda JF, Carpenter I, Staley LW, O’Connor RA, et al. High functioning fragile X males: Demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. American Journal of Medical Genetics. 1994;4:298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- 6.Coffey SM, Cook K, Tartaglia N, Tassone F, Nguyen DV, Pan R, et al. Expanded clinical phenotype of women with the FMR1 premutation. American Journal of Medical Genetics A. 2008;146A:1009–1016. doi: 10.1002/ajmg.a.32060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terracciano A, Pomponi MG, Marino GM, Chiurazzi P, Rinaldi MM, Dobosz M, et al. Expansion to full mutation of a FMR1 intermediate allele over two generations. European Journal of Human Genetics. 2004;12:333–336. doi: 10.1038/sj.ejhg.5201154. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Carvajal I, Lopez Posadas B, Pan R, Raske C, Hagerman PJ, Tassone F. Expansion of an FMR1 grey-zone allele to a full mutation in two generations. Journal of Molecular Diagnostics. 2009;11:306–310. doi: 10.2353/jmoldx.2009.080174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Hadd A, Sah S, Filipovic-Sadic S, Krosting J, Sekinger E, et al. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. Journal of Molecular Diagnostics. 2010;12:589–600. doi: 10.2353/jmoldx.2010.090227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyon E, Laver T, Yu P, Jama M, Young K, Zoccoli M, et al. A simple, high-throughput assay for fragile X expanded alleles using triple repeat primed PCR and capillary electrophoresis. Journal of Molecular Diagnostics. 2010;12:505–511. doi: 10.2353/jmoldx.2010.090229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hantash FM, Goos DG, Tsao D, Quan F, Buller-Burckle A, Peng M, et al. Qualitative assessment of FMR1 (CGG)n triplet repeat status in normal, intermediate, premutation, full mutation, and mosaic carriers in both sexes: Implications for fragile X syndrome carrier and newborn screening. Genetics Medicine. 2010;12:162–173. doi: 10.1097/GIM.0b013e3181d0d40e. [DOI] [PubMed] [Google Scholar]
- 12.Khaniani MS, Kalitsis P, Burgess T, Slater HR. An improved diagnostic PCR assay for identification of cryptic heterozygosity for CGG triplet repeat alleles in the fragile X gene (FMR1) Molecular Cytogenetics. 2008;1:5. doi: 10.1186/1755-8166-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todorov T, Todorova A, Georgieva B, Mitev V. A unified rapid PCR method for detection of normal and expanded trinucleotide alleles of CAG repeats in huntington chorea and CGG repeats in fragile X syndrome. Molecular Biotechnology. 2010;2:150–154. doi: 10.1007/s12033-010-9260-y. [DOI] [PubMed] [Google Scholar]
- 14.Filipovic-Sadic S, Sah S, Chen L, Krosting J, Sekinger E, Zhang W, et al. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clinical Chemistry. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nature Biotechnology. 2009;27:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 17.Schlapak R, Kinns H, Wechselberger C, Hesse J, Howorka S. Sizing trinucleotide repeat sequences by single-molecule analysis of fluorescence brightness. ChemPhysChem. 2007;8:1618–1621. doi: 10.1002/cphc.200700163. [DOI] [PubMed] [Google Scholar]
- 18.Jo K, Dhingra DM, Odijk T, de Pablo JJ, Graham MD, Runnheim R, et al. A single-molecule barcoding system using nanoslits for DNA analysis. Proceedings of the National Academy of Sciences USA. 2007;8:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao M, Gordon MP, Phong A, Ha C, Chan TF, Cai D, et al. Determination of haplotypes from single DNA molecules: A method for single-molecule barcoding. Human Mutation. 2007;9:913–921. doi: 10.1002/humu.20528. [DOI] [PubMed] [Google Scholar]
- 20.Burton RE, White EJ, Foss TR, Phillips KM, Meltzer RH, Kojanian N, et al. A microfluidic chip-compatible bioassay based on single-molecule detection with high sensitivity and multiplexing. Lab on a Chip. 2010;10:843–851. doi: 10.1039/b922106a. [DOI] [PubMed] [Google Scholar]
- 21.Chan EY, Goncalves NM, Haeusler RA, Hatch AJ, Larson JW, Maletta AM, et al. DNA mapping using microfluidic stretching and single-molecule detection of fluorescent site-specific tags. Genome Research. 2004;14:137–1146. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Protozanova E, Zhang M, White EJ, Mollova ET, Broeck DT, Fridrikh SV, et al. Fast high-resolution mapping of long fragments of genomic DNA based on single-molecule detection. Analytical Biochemistry. 2010;1:83–90. doi: 10.1016/j.ab.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 23.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proceedings of the National Academy of Sciences USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine PM, Gong P, Levicky R, Shepard KL. Real-time, multiplexed electrochemical DNA detection using an active complementary metal-oxide-semiconductor biosensor array with integrated sensor electronics. Biosensor Bioelectron-ics. 2009;7:1995–2001. doi: 10.1016/j.bios.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 26.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 27.Qu X, Wu D, Mets L, Scherer NF. Nanometer-localized multiple single-molecule fluorescence microscopy. Proceedings of the National Academy of Sciences USA. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon MP, Ha T, Selvin PR. Single-molecule high-resolution imaging with photobleaching. Proceedings of the National Academy of Sciences USA. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proceedings of the National Academy of Sciences USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annual Reviews in Biochemistry. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadel Y, Weisman-Shomer P, Fry M. The fragile X syndrome single strand d(CGG)n nucleotide repeats readily fold back to form unimolecular hairpin structures. Journal of Biological Chemistry. 1995;270:28970–28977. doi: 10.1074/jbc.270.48.28970. [DOI] [PubMed] [Google Scholar]
- 32.Paiva AM, Sheardy RD. Influence of sequence context and length on the structure and stability of triplet repeat DNA oligomers. Biochemistry. 2004;43:14218–14227. doi: 10.1021/bi0494368. [DOI] [PubMed] [Google Scholar]
- 33.Jarem DA, Huckaby LV, Delaney S. AGG interruptions in (CGG)(n) DNA repeat tracts modulate the structure and thermodynamics of non-B conformations in vitro. Biochemistry. 2010;49:6826–6837. doi: 10.1021/bi1007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Völker J, Klump HH, Breslauer KJ. DNA energy landscapes via calorimetric detection of microstate ensembles of metastable macrostates and triplet repeat diseases. Proceedings of the National Academy of Sciences USA. 2008;105:18326–18330. doi: 10.1073/pnas.0810376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maier B, Seifert U, Rädler JO. Elastic response of DNA to external electric fields in two dimensions. Europhysics Letters. 2002;60:622–628. [Google Scholar]
- 36.Randall GC, Schultz KM, Doyle PS. Methods to electrophoretically stretch DNA: Microcontractions, gels, and hybrid gel-microcontraction devices. Lab on a Chip. 2006;6:516–525. doi: 10.1039/b515326c. [DOI] [PubMed] [Google Scholar]
- 37.Nelson PC, Zurla Z, Brogioli D, Beausang JF, Finzi L, Dunlap D. Tethered particle motion as a diagnostic of DNA tether length. Journal of Physical Chemistry B. 2006;110:17260–17267. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 38.American College of Obstetricians and Gynecologists. Carrier screening for fragile X syndrome. Committee Opinion No. 469. Obstetrics and Gynecology. 2010;116:1008–1010. doi: 10.1097/AOG.0b013e3181fae884. [DOI] [PubMed] [Google Scholar]
- 39.Spector EB, Kronquist KE. ACMG standards and guidelines for clinical genetics laboratories. 2005. Fragile X: Technical standards and guidelines. [Google Scholar]