Abstract
Considerable support exists for roles of metabolism in modulating the carcinogenic properties of chemicals. In particular, many of these compounds are procarcinogens that require activation to electrophilic forms to exert genotoxic effects. We systematically analyzed the existing literature on metabolism of carcinogens by human enzymes, which has been developed largely in the past 25 years. The metabolism and especially bioactivation of carcinogens are dominated by cytochrome P450 enzymes (66% of bioactivations). Within this group, six P450s—1A1, 1A2, 1B1, 2A6, 2E1, and 3A4—accounted for 77% of the P450 activation reactions. The roles of these P450s can be compared with those estimated for drug metabolism and should be considered in issues involving enzyme induction, chemoprevention, molecular epidemiology, inter-individual variations, and risk assessment.
INTRODUCTION
Knowledge that chemicals can cause cancer goes back to at least 1761 with the report by Hill1 that the use of tobacco snuff was related to oral cancer in humans. More than 100 years ago, Rehn2 reported an association of bladder cancer with occupations in the so-called aniline dye factories. Experimental studies in animals showed that chemicals cause cancer, beginning with reports on coal tar in rabbits by Yamagiwa.3 Classic studies with the polycyclic aromatic hydrocarbon benzo[a]pyrene followed.4 Fieser5 and others had suggested that metabolism (of carcinogens1) plays a role in cancer, and extensive animal studies by James and Elizabeth Miller6-8 validated the concept (Figure 1). Incorporation of the capability for metabolism led to the success of the bacterial Ames test9 for mutagenicity and testing for carcinogenic potential.
Figure 1.
General paradigm of metabolism of chemical carcinogens.
The roles of individual enzymes in carcinogen metabolism has been studied extensively, and roles of many human P450s in carcinogen activation have been characterized.10 Much of this work was first done in medium-throughput screens, e.g. bacterial genotoxicity, and then extended with more detailed studies of reaction products and DNA adducts.10-13 Studies with P450s led the way but similar approaches have been used with other enzymes known to have roles in the metabolism of xenobiotic chemicals.14,15 Research in this area has been important in several applied disciplines and approaches. These include molecular epidemiology, which is an attempt to relate risk from carcinogens to the enzymes present in an individual.16-18 Another area of interest is chemoprevention, where a major strategy involves either inhibiting enzymes that activate carcinogens or inducing enzymes that inactive them.19,20
Several efforts have been made to delineate the levels of expression of individual enzymes, especially P450s, in humans.21 Another approach is to analyze the fractions of the enzymes involved in reactions. Such analyses have been reported for drugs, for all “xenobiotic-metabolizing enzymes”22,23 and for P450s.22-25 The analyses are rather consistent with each other and generally accepted, both with marketed drugs and new chemical entities (drug candidates). Of particular note are the findings that i) ~75% of enzymatic reaction with drugs are catalyzed by P450s,23 ii) ~90% of the P450 reactions can be accounted for by a set of five P450s: 1A2, 2C9, 2C19, 2D6, and 3A4,23-25 and iii) the largest fraction of the P450 reactions are catalyzed by P450 3A enzymes, particularly P450 3A4.23-25
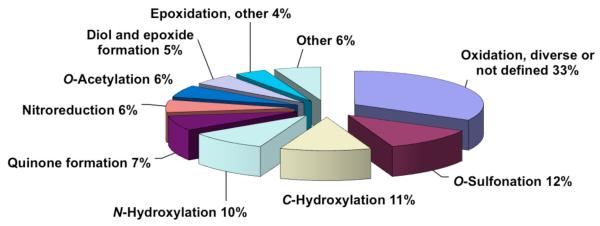
To our knowledge, there has not been a similar effort to categorize all of the the human enzymes involved in the metabolism of carcinogens. We thought that this would be a useful exercise in the light of continuing scientific interest in chemical carcinogenesis, cancer chemoprevention, and molecular epidemiology of cancer. We report our analysis of the literature in parts—general chemicals (environmental/industrial),Footnote 1 drugs, and natural/physiological compounds—as well as an overall analysis of all literature carcinogens for which information about metabolism is available. The results show a dominant role for P450s, especially the three Family 1 P450 enzymes (1A1, 1A2, 1B1) and P450s 2A6, 2E1, and 3A4 (Figure 2). The aldo-keto reductase (AKR) enzymes are also highly represented.26
Figure 2.
Enzyme contributions to activation of carcinogens (from Table 1). A: Fractions of activation reactions attributed to groups of enzymes. The analysis is based on 713 reactions. B: Fractions of P450 activation reactions attributed to individual human P450 enzymes (from a total of 473 reactions considered). See text for discussion.
ASSIGNMENTS OF ROLES OF ENZYMES
Most of the literature on the roles of human enzymes in carcinogen metabolism has been developed in the last 25 years, e.g. a review by one of us in 198827 had only a very limited discussion of this aspect. The present literature analysis is a continuation of the work done by one of us (S. Rendic) on literature searches on the metabolism of drugs and other chemicals catalyzed by human P450s, for more than 15 years ending in February 2012. Extensive key-word literature searches were done using the PubMed database, accessing the MEDLINE database of references and abstracts. In the latter stages, the existing literature on metabolism of carcinogens and the original papers was systematically analyzed, extracting those data contributing in a “significant way” to the activation and/or detoxication of general chemicals, drugs, and physiological compounds. (This is a qualitative evaluation, and the reader is referred to a more comprehensive list in Supporting Information Table S1.)
The results are presented in several tables (Tables 1-4), including activation reactions with all chemicals (Table 1) (exclusive of “weak” reactions), followed by the activation of physiological/natural compounds (Table 2) and drugs (Table 3). Detoxication reactions are presented in Table 4. For convenience, PMID numbers of references are included in the tables to facilitate searches and retrievals. In considering the results for all types of metabolism, it is clear that P450s are dominant. The three activation tables (Tables 1-3) contain only data for what are deemed “significant activation”. All activation data (potent and weak) are presented as a single table in the Supporting Information (Table S1).
Table 1. Data on individual enzymes and chemicals, but not including weak bioactivation (see Supporting Information for inclusion of weak activation data).
Note: the term “diol” is used in stead of “dihydrodiol” for convenience with the PAHs.
| enzyme | category | subcategory | compound | reaction | remarks | references | PubMed ID |
|---|---|---|---|---|---|---|---|
| AKR1A1 | chemical | PAH, metabolite | (±)-benzo[a]pyrene-7,8- dihydrodiol |
oxidation, o-quinone formation, preferential for (−)-7R,8R-oxidation |
activation | 26, 28-33 | 16411658, 11306097, 9973208, 11535067, 15720144, 17295519, 18788756 |
| AKR1A1 | chemical | PAH, metabolite | 12-methylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation |
activation | 28 | 11306097 |
| AKR1A1 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | oxidation, o-quinone formation (medium Km, high activity, high efficiency) |
activation | 28, 30, 34 | 11306097, 11535067, 16946553 |
| AKR1A1 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone form. |
activation | 28,30 | 11306097, 11535067 |
| AKR1A1 | chemical | PAH, metabolite | 7-methylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation, preferential for (−)3S,4S-oxidation |
activation | 28, 30 | 11306097, 11535067 |
| AKR1A1 | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol | oxidation, o-quinone formation, preferential for (−)3R,4R-oxidation |
activation | 28, 30 | 11306097, 11535067 |
| AKR1A1 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation, o-quinone formation |
activation | 28 | 11306097 |
| AKR1B1 | chemical | PAH | (+)-benz[a]anthracene- 3S,4S-diol |
oxidation, o-quinoneform, stereospecific for (+)-7S-,8S |
activation | 33 | 18788756 |
| AKR1B1 | chemical | PAH, metabolite | (+)-benzo[a]pyrene-7S,8S- diol |
oxidation, o-quinone formation, stereospecific for (+)-7S-,8S |
activation | 33 | 18788756 |
| AKR1B1 | chemical | PAH, metabolite | (+)-S,S-benzo[g]chrysene- 11,12-diol |
oxidation, o-quinone formation, stereospecific for (+)-7S-,8S |
activation | 33 | 18788756 |
| AKR1B10 | chemical | PAH, metabolite | (−)-R,R- and (+)-S,S- benzo[g]chrysene-11,12-diol |
oxidation, o-quinone formation |
activation | 33 | 18788756 |
| AKR1B10 | chemical | PAH, metabolite | (+)-benz[a]anthracene-3S,4S- diol |
oxidation, o-quinone formation, stereospecific for (+)-7S-,8S |
activation | 33 | 18788756 |
| AKR1B10 | chemical | PAH, metabolite | (+)-benzo[a]pyrene-7S,8S- diol |
oxidation, o-quinone formation, stereospecific for (+)-7S-,8S |
activation | 33 | 18788756 |
| AKR1B10 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation |
activation | 33 | 18788756 |
| AKR1C1 | chemical | PAH, metabolite | (+,−)- and (−)- benzo[a]pyrene-7,8-diol |
oxidation, o-quinone formation |
activation |
29, 33, 35, 36 |
9973208, 18788756, 11978787, 11060293 |
| AKR1C1 | chemical | PAH, metabolite | 5-methylchrysene-7,8-diol | oxidation, o-quinone formation |
activation | 35, 36 | 11978787, 11060293 |
| AKR1C1 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation, minor enzyme |
activation | 35, 36 | 11978787, 11060293 |
| AKR1C1 | chemical | PAH, metabolite | 7-methylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation, minor enzyme |
activation | 35 | 11978787 |
| AKR1C1 | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C1 | chemical | aromatic hydrocarbon | benzene diol | oxidation, o-quinone formation |
activation | 36, 37 | 11060293, 15026176 |
| AKR1C1 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | oxidation, o-quinone formation |
activation | 35, 36 | 11978787, 11060293 |
| AKR1C1 | chemical | PAH, metabolite | naphthalene 1,2-diol | oxidation, o-quinone formation, major enzyme |
activation | 35, 36 | 11978787, 11060293 |
| AKR1C2 | chemical | PAH, metabolite | (±)- and (−)-benzo[a]pyrene- 7,8-diol |
oxidation, o-quinone formation |
activation | 29, 33, 35 | 9973208, 18788756, 11978787 |
| AKR1C2 | chemical | PAH, metabolite | 5-methylchrysene-7,8-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C2 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C2 | chemical | PAH, metabolite | 7-methylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C2 | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C2 | chemical | PAH, metabolite | benzene diol | oxidation, o-quinone formation |
activation | 37 | 15026176 |
| AKR1C2 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol |
oxidation, o-quinone form. |
activation | 35 | 11978787 |
| AKR1C2 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C2 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C2 | chemical | PAH, metabolite | naphthalene 1,2-diol | oxidation, o-quinone formation, major enzyme |
activation | 35 | 11978787 |
| AKR1C3 | chemical | PAH, metabolite | (+,−)- and (−)- benzo[a]pyrene-7,8- dihydrodiol |
oxidation, o-quinone formation |
activation | 29, 33, 35 | 9973208, 18788756, 11978787 |
| AKR1C3 | chemical | PAH, metabolite | 5-methylchrysene-7,8-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C3 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C3 | chemical | PAH, metabolite | 7-methylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C3 | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C3 | chemical | aromatic hydrocarbon, metabolite |
benzene diol | oxidation, o-quinone formation |
activation | 37 | 15026176 |
| AKR1C3 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol |
oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C3 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C3 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C3 | chemical | PAH, metabolite | naphthalene 1,2-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C4 | chemical | PAH, metabolite | (±)- and (−)-benzo[a]pyrene- 7,8-diol |
oxidation, o-quinone formation |
activation | 29, 35 | 9973208, 11978787 |
| AKR1C4 | chemical | PAH, metabolite | (±)-benzo[a]pyrene-7,8-diol | oxidation, o-quinone formation |
activation | 33 | 18788756 |
| AKR1C4 | chemical | PAH, metabolite | 5-methylchrysene-7,8-diol | oxidation, o-quinone formation, major enzyme |
activation | 35 | 11978787 |
| AKR1C4 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation, major enzyme |
activation | 35 | 11978787 |
| AKR1C4 | chemical | PAH, metabolite | 7-methylbenz[a]anthracene- 3,4-diol |
oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C4 | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C4 | chemical | aromatic hydrocarbon, metabolite |
benzene diol | oxidation, o-quinone formation |
activation | 37 | 15026176 |
| AKR1C4 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol |
oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C4 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | oxidation, o-quinone formation, major enzyme |
activation | 35 | 11978787 |
| AKR1C4 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| AKR1C4 | chemical | PAH, metabolite | naphthalene 1,2-diol | oxidation, o-quinone formation |
activation | 35 | 11978787 |
| COX-1 | chemical | PAH, metabolite | (±)- and (+)-benzo[a]pyrene- 7,8-diol |
oxidation | activation | 38 | 11159734 |
| COX-1 | chemical | heterocyclic amine | 2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (MeIQx) |
oxidation | activation | 38 | 11159734 |
| COX-1 | chemical | arylamine | 4,4′-methylene bis(2- chloroaniline) (MOCA) |
oxidation | potent activation |
38 | 11159734 |
| COX-1 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl | oxidation | activation | 38 | 11159734 |
| COX-1 | chemical | arylamine | benzidine | oxidation | activation | 38 | 11159734 |
| COX-1 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor, DNA binding |
ellipticine | oxidation | activation | 39, 40 | 16936898, 21753906 |
| COX-2 | chemical | PAH, metabolite | (±)- and (+)-benzo[a]pyrene- 7,8-diol |
oxidation | activation | 38 | 11159734 |
| COX-2 | chemical | heterocyclic amine | 2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (MeIQx) |
oxidation | activation | 38 | 11159734 |
| COX-2 | chemical | arylamine | 4,4′-methylene bis(2- chloroaniline) (MOCA) |
oxidation | potent activation |
38 | 11159734 |
| COX-2 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl | oxidation | activation | 38 | 11159734 |
| COX-2 | chemical | arylamine | benzidine | oxidation | activation | 38 | 11159734 |
| COX-2 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor, and DNA binding |
ellipticine | oxidation | activation | 39, 40 | 16936898, 21753906 |
| CYP1A1 | chemical | PAH, metabolite | (±)-, (−)-, and (+)- benzo[a]pyrene-7,8-diol |
trans-(anti)-7,8- dihydroxy-9,10-epoxy- 7,8,9,10-tetrahydroformation (trans-diol epoxide formation); oxidation |
potent activation |
13, 31, 32, 34, 41-52 |
2509067, 8674051, 7955101, 11502724, 11238186, 8043197, 7581497, 11952781, 15720144, 10426814, 8293790, 16946553, 16885195, 17295519, 17525473, 21028851 |
| CYP1A1 | chemical | nitroarene | 1,8-dinitropyrene | nitroreduction | potent activation |
53 | 11113705 |
| CYP1A1 | chemical | arylamine, metabolite of 1-nitropyrene |
1-aminopyrene | oxidation | activation | 54 | 11525925 |
| CYP1A1 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4- (diallylamino)-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2H- benzotriazole (PBTA-8) |
oxidation | activation | 55 | 18562244 |
| CYP1A1 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4- (diethylamino)-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2H- benzotriazole (PBTA-7) |
oxidation | activation | 55 | 18562244 |
| CYP1A1 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4-amino- 5-methoxyphenyl]-5-amino- 7-bromo-4-chloro-2H- benzotriazole (PBTA-4) |
oxidation | activation | 55 | 18562244 |
| CYP1A1 | chemical | benzotriazole | 2-[2-(acetylamino)-4-amino- 5-methoxyphenyl]-5-amino- 7-bromo-4-chloro-2H- benzotriazole (PBTA-4) |
oxidation | activation | 56 | 21786339 |
| CYP1A1 | chemical | arylamine, metabolite | 2-acetylaminofluorene (2- AAF) |
N-hydroxylation, oxidation |
activation | 41, 43, 57 | 8674051, 11502724, 15279838 |
| CYP1A1 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N-hydroxylation, oxidation |
activation |
41, 43, 57- 61 |
8674051, 11502724, 15279838, 9111224, 9855011, 21081470, 1377247 |
| CYP1A1 | chemical | heterocyclic amine | 2-amino-3,4- dimethylimidazo[4,5- f]quinoline (MeIQ) |
N-hydroxylation, oxidation |
potent activation |
41, 43, 49, 56, 62, 63 |
8674051, 11502724, 10426814, 21786339, 11473383, 8200084 |
| CYP1A1 | chemical | heterocyclic amine | 2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (MeIQx) |
N-hydroxylation, oxidation |
activation |
41, 43, 49, 58, 62-64 |
8674051, 11502724, 10426814, 9111224, 11473383, 8200084, 17627018 |
| CYP1A1 | chemical | heterocyclic amine | 2-amino-3- methylimidazo[4,5- f]quinoline (IQ) |
N-hydroxylation, oxidation |
potent activation |
41, 43, 49, 58, 61, 65 |
8674051, 11502724, 10426814, 9111224, 11377247, 9918136 |
| CYP1A1 | chemical | heterocyclic amine | 2-amino-6- methyldipyrido[1,2-a:3′,2′-d]- imidazole (Glu-P-1) |
N-hydroxylation, oxidation |
activation | 41, 43, 61 | 8674051, 11502724, 11377247 |
| CYP1A1 | chemical | arylamine | 2-aminoanthracene |
N-hydroxylation, oxidation (high activity) |
potent activation |
41, 43, 61, 66 |
8674051, 11502724, 11377247, 9685642 |
| CYP1A1 | chemical | arylamine | 2-aminofluorene |
N-hydroxylation, oxidation |
activation |
41, 43, 49, 61 |
8674051, 11502724, 10426814, 11377247 |
| CYP1A1 | chemical | nitroarene | 2-nitronaphthalene | nitroreduction | activation | 67 | 10521697 |
| CYP1A1 | chemical | nitroarene | 2-nitropyrene | 2-aminopyrene formation (nitroreduction) |
activation | 41 | 8674051 |
| CYP1A1 | chemical | nitrosamine | 3-(n-nitrosomethylamino) propiona ldehyde |
oxidation | activation | 68 | 15725615 |
| CYP1A1 | chemical | nitrosamine | 3-(N- nitrosomethylamino) propioni trile |
oxidation (at high concentrations) |
activation | 68, 69 | 15725615, 16720019 |
| CYP1A1 | chemical | nitroarene | 3,6-dinitrobenzo[e]pyrene | nitroreduction | activation | 70 | 19393727 |
| CYP1A1 | chemical | heterocyclic amine | 3-Amino-1,4-dimethyl-5H- pyrido[4,3-b]indole (Trp-P-1) |
N-hydroxylation, oxidation |
potent activation |
41, 43, 49, 61, 66, 71 |
8674051, 11502724, 10426814, 11377247, 9685642, 9721189 |
| CYP1A1 | chemical | heterocyclic amine | 3-amino-1-methyl-5H- pyrido[4,3-b]indole (Trp-P-2) |
N-hydroxylation, oxidation |
activation |
41, 43, 49, 61-63, 66 |
8674051, 11502724, 10426814, 11377247, 11473383, 8200084, 9685642 |
| CYP1A1 | chemical | arylamine, metabolite | 3-aminobenzanthrone | N-hydroxylation | activation | 72, 73 | 15885895, 16601755 |
| CYP1A1 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene (3-MeO- AAB) |
oxidation | potent activation |
41, 43, 49, 66 |
8674051, 11502724, 10426814, 9685642 |
| CYP1A1 | chemical | PAH | 3-methylcholanthrene (3MC) | oxidation | activation | 74 | 11360624 |
| CYP1A1 | chemical | PAH, metabolite | 3-methylcholanthrene-11,12- diol, 3MC-11,12-diol |
oxidation | activation | 43 | 11502724 |
| CYP1A1 | natural compound |
indole, alkylating, pulmonary toxin; in higher concentrations in mammalian digestive tract and coal tar |
3-methylindole, skatole | epoxidation (3- methyloxindole formation); dehydrogenation (desaturation, 3- methyleneindolenine formation), low Km, medium activity, high efficiency |
activation | 75-78 | 8558432, 11408359, 12563100, 20795680 |
| CYP1A1 | chemical | nitroarene | 3-nitrobenzanthrone | nitroreduction | activation | 73, 79, 80 | 16601755, 12740904, 12782579 |
| CYP1A1 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl |
N-hydroxylation, oxidation |
activation | 41, 58, 66 | 8674051, 9111224, 9685642 |
| CYP1A1 | chemical | PAH, metabolite | 5,6-dimethylchrysene-1,2- diol |
oxidation | activation |
34, 43, 49, 81 |
16946553, 11502724, 10426814, 14720319 |
| CYP1A1 | chemical | PAH | 5-methylchrysene | 1,2-dihydrodiol formation (medium Km, high activity, high efficiency), oxidation |
activation | 43, 81-83 | 11502724, 14720319, 8542586, 18992797 |
| CYP1A1 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | oxidation | potent activation |
34, 41, 43, 49, 81, 83 |
16946553, 8674051, 11502724, 10426814, 14720319, 18992797 |
| CYP1A1 | chemical | arylamine | 6-aminochrysene | oxidation (high activity) | potent activation |
41, 43, 66 | 8674051, 11502724, 9685642 |
| CYP1A1 | chemical | arylamine, metabolite | 6-aminochrysene-1,2-diol | diol epoxide formation, oxidation |
activation | 41, 84, 85 | 8674051, 8118930, 8330339 |
| CYP1A1 | chemical | PAH | 6-methylchrysene | 1,2-dihydrodiol formation | activation | 82 | 8542586 |
| CYP1A1 | chemical | nitroarene | 6-nitrochrysene | oxidation | activation | 43 | 11502724 |
| CYP1A1 | chemical | PAH | 7,12- dimethylbenz[a]anthracene |
oxidation (low Km, high activity and efficiency) |
potent activation |
43, 49, 81, 86, 87 |
11502724, 10426814, 14720319, 12584184, 20507880 |
| CYP1A1 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
3,4-dihydrodiol-1,2- epoxide formation (medium Km, high activity, high efficiency), oxidation |
potent activation |
34, 43, 49, 66, 74, 81 |
16946553, 11502724, 10426814, 9685642, 11360624, 14720319 |
| CYP1A1 | chemical |
N-heterocyclic aromatic hydrocarbon |
7H-dibenzo[c,g]carbazole | oxidation | potent activation |
88-91 | 10984687, 12034315, 15534862, 21809388 |
| CYP1A1 | chemical | PAH, aza-aromatic | 7-methylbenz[c]acridine | 3,4-dihydrodiol formation | activation | 92 | 7866988 |
| CYP1A1 | chemical | PAH, aza-aromatic | 7-methylbenz[c]acridine | oxidation | potent activation |
92 | 7866988 |
| CYP1A1 | chemical | PAH, metabolite | 9-hydroxybenzo[a]pyrene | oxidation | activation | 43 | 11502724 |
| CYP1A1 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 | epoxidation 8,9-, oxidation |
activation |
41, 57, 61, 93, 94 |
8674051, 15279838, 11377247, 7923587, 8200084 |
| CYP1A1 | chemical | heterocyclic amine | aminomethylphenylnorharma n |
N-hydroxylation | activation | 95 | 17067997 |
| CYP1A1 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid I | nitroreduction | potent activation |
96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A1 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid II | nitroreduction | activation | 96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A1 | chemical | PAH | benz[a]anthracene | oxidation | activation | 43, 81, 100 | 11502724, 14720319 |
| CYP1A1 | chemical | PAH, metabolite | benz[a]anthracene-1,2-diol | oxidation | activation |
34, 43, 74, 101 |
16946553, 11502724, 11360624, 11377097 |
| CYP1A1 | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol | oxidation | activation | 34, 43, 81 | 16946553, 11502724, 14720319 |
| CYP1A1 | chemical | PAH, metabolite | benz[a]anthracene-5,6-diol | oxidation | activation | 34, 43 | 16946553, 11502724 |
| CYP1A1 | chemical | diphenylmethanol, metabolite |
benzhydrol | oxidation | activation | 102 | 12160905 |
| CYP1A1 | chemical | PAH | benzo[a]perylene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | benzo[a]pyrene |
trans-7,8-dihydroxy-9,10- epoxy-7,8,9,10- tetrahydro- formation (low activity, medium activity, or high activity, high efficiency); 1,6-,3,6-, 6,12-dione (quinone formation, low activity); oxidation |
activation |
41, 43, 50- 52, 57, 81, 93, 94, 104-108 |
8674051, 11502724, 16885195, 17525473, 21028851, 15279838 14720319, 7923587, 8200084, 9806168, 11513247, 8037457, 1486866, 19501186 |
| CYP1A1 | chemical | PAH, metabolite | benzo[b]fluoroanthene-9,10- diol |
oxidation | potent activation |
34, 41, 43, 49, 66, 81 |
16946553, 8674051, 11502724, 10426814, 9685642, 14720319 |
| CYP1A1 | chemical | PAH | benzo[c]phenanthrene | dihydrodiol 3,4-, 1,2- epoxide formation (major enzyme); oxidation |
activation | 109 | 11409939 |
| CYP1A1 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol |
dihydrodiol 3,4-, 1,2- epoxide formation; oxidation |
activation | 43, 49 | 11502724, 10426814 |
| CYP1A1 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | oxidation | activation |
34, 43, 49, 81 |
16946553, 11502724, 10426814, 14720319 |
| CYP1A1 | chemical | aromatic ketone, diphenyl ketone |
benzophenone | oxidation | activation | 102 | 12160905 |
| CYP1A1 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation | potent activation |
43, 49, 81 | 11502724, 10426814, 14720319 |
| CYP1A1 | chemical | PAH | cyclopenta[c,d]pyrene | oxidation | activation | 110 | 7923587 |
| CYP1A1 | Drug | imidazole; anticancer, alkylating |
dacarbazine |
N-demethylation (major extrahepatic enzyme) |
activation | 111 | 10473105 |
| CYP1A1 | chemical | PAH, metabolite, aza- aromatic |
dibenz[a,h]acridine | 10,11-diol formation | potent activation |
112 | 15144224 |
| CYP1A1 | chemical | PAH | dibenz[a,h]anthracene | oxidation | activation | 43 | 11502724 |
| CYP1A1 | chemical | PAH, aza-aromatic | dibenz[a,j]acridine | 3,4-dihydrodiol formation | activation | 92 | 7866988 |
| CYP1A1 | chemical | PAH | dibenzo[a,e]fluoranthene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | dibenzo[a,e]pyrene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | dibenzo[a,f]fluoranthene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | dibenzo[a,h]pyrene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | dibenzo[a,k]fluoranthene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | dibenzo[a,l]pyrene | (−)-syn- and (−)-anti- 11,12-dihydrodiol-13,14- epoxide formation (medium Km, high activity, high efficiency); oxidation |
potent activation |
43, 81, 103, 113-119 |
11502724, 14720319, 10613181, 9625737, 10207125, 10493514, 10506751, 8968059, 16581046, 17509623 |
| CYP1A1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
11,12-dihydrodiol-13,14- epoxide formation (medium Km, high activity, high efficiency); oxidation |
potent activation |
34, 43, 49, 71, 81, 113-115, 118-120 |
16946553, 11502724, 10426814, 9721189, 14720319, 9625737, 10207125, 10493514, 16581046, 17509623, 16485905 |
| CYP1A1 | chemical | PAH | dibenzo[b,k]fluoranthene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | physiologi -cal compound |
estrogen | 17β−estradiol | C2-hydroxylation (major reaction, medium Km, high activity, high efficiency), major metabolite and major extrahepatic enzyme; C4- hydroxylation (minor reaction, medium Km, medium efficiency, low activity), oxidation, 3,4- quinone formation (lower activity); oxidation, 2,3- quinone formation; C16α- hydroxylation (high Km, low activity) |
potent activation |
71, 106, 121-130 |
9721189, 8037457, 7826886, 9625734, 9054608, 9667077, 8930523, 11555828, 12865317, 15784278, 16112414, 17570247 |
| CYP1A1 | physiologi cal compound |
estrogen | estrone | C2-hydroxylation (major reaction, medium Km, low activity), oxidation, 2,3- quinone formation; C4- hydroxylation (medium Km, low activity, or medium activity); C16α- hydroxylation (minor reaction, very low activity) |
activation |
49, 127, 130, 131 |
10426814, 12865317, 17570247, 15805301 |
| CYP1A1 | chemical | PAH, metabolite | fluoranthene-2,3-diol | oxidation | activation | 34, 43 | 16946553, 11502724 |
| CYP1A1 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | oxidation (at high concentrations) |
activation | 87 | 20507880 |
| CYP1A1 | chemical | PAH | naphtho[1,2-k]fluoranthene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | naphtho[2,1-a]pyrene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical | PAH | naphtho[2,3-a]pyrene | oxidation | activation | 103 | 10613181 |
| CYP1A1 | chemical |
N-heterocyclic aromatic hydrocarbon, dibenzocarbazole |
N- methyldibenzo[c,g]carbazole |
oxidation | potent activation |
88-91 | 10984687, 12034315, 15534862, 21809388 |
| CYP1A1 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanabasine | oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP1A1 | chemical | nitrosamine |
N-nitrosodibutylamine (N, N- dibutylnitrosamine) |
oxidation | activation | 134 | 11600130 |
| CYP1A1 | chemical | nitrosamine | N-nitrosodiethylamine (N, N-diethylnitrosamine) | oxidation | activation | 132-134 | 11774366, 12214673, 11600130 |
| CYP1A1 | chemical | nitrosamine |
N-nitrosodi-n-propylamine (N-nitrosodipropylamine) |
oxidation | activation | 134 | 11600130 |
| CYP1A1 | chemical | nitrosamine | N-nitrosoethylbutylamine | oxidation (major enzyme) | activation | 134 | 11600130 |
| CYP1A1 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP1A1 | chemical | nitrosamine | N-nitrosomethylethylamine | oxidation | activation | 134 | 11600130 |
| CYP1A1 | chemical | nitrosamine | N-nitrosomethylpropylamine | oxidation | activation | 69, 134 | 16720019, 11600130 |
| CYP1A1 | chemical | nitrosamine | N-nitrosomorpholine | oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP1A1 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP1A1 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OH- tetrahydrofuran formation); oxidation |
activation | 132, 133 | 11774366, 12214673 |
| CYP1A1 | chemical | azoarylamine | o-aminoazotoluene | oxidation | activation | 41, 66 | 8674051, 9685642 |
| CYP1A1 | chemical | diphenylketone, metabolite |
p-benzoylphenol,4- hydroxybenzophenone |
oxidation | activation | 102 | 12160905 |
| CYP1A1 | chemical | PAH | phenanthrene | oxidation to 1,2- (major reaction), 9,10-, and 3,4- dihydrodiols (minor reactions) and phenols, at high concentration |
activation | 46, 135 | 7581497, 19766613 |
| CYP1A1 | chemical | aza-aromatic | Sudan I | oxidation, major enzyme | activation | 136, 137 | 12384524, 17159775 |
| CYP1A2 | chemical | PAH, metabolite | (±)-, (−)-, and (+)- benzo[a]pyrene-7,8- dihydrodiol |
trans-(anti)-7,8- dihydroxy-9,10-epoxy- 7,8,9,10-tetrahydro- formation; oxidation |
activation |
13, 34, 41- 43, 120, 138, 139 |
2509067, 16946553, 8674051, 7955101, 1502724, 16485905, 9014198, 2803520 |
| CYP1A2 | chemical | triazole | 1-aminobenzotriazole (1- ABT) |
oxidation | activation | 140 | 17584015 |
| CYP1A2 | chemical | arylamine, metabolite of 1-nitropyrene |
1-aminopyrene | oxidation | activation |
54, 141- 144 |
11525925, 15728263, 15843388, 17158518, 9860501 |
| CYP1A2 | chemical | PAH, aza-aromatic | 1-azabenzo[a]pyrene | oxidation | potent activation |
145 | 14729370 |
| CYP1A2 | chemical | arylamine, metabolite | 2-acetylaminofluorene (2- AAF) |
N-hydroxylation (major enzyme), oxidation |
potent activation |
12, 41, 57, 107, 139, 146, 147 |
2655891, 8674051, 15279838, 1486866, 2803520, 11375903, 15450435 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N-hydroxylation, oxidation (high activity, major enzyme, major reaction) |
potent activation |
57-61, 63, 107, 146, 148-156, 157 |
15279838, 9111224, 9855011, 21081470, 11377247, 8200083, 1486866, 11375903, 8082563, 1913651, 9705755, 11013410, 10503887, 12351158, 14744142, 14725854, 15073045, 16167840 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-3,4,8- trimethylimidazo[4,5- ]quinoxaline (DiMeIQx) |
N-hydroxylation | potent activation |
107 | 1486866 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-3,4- dimethylimidazo[4,5- f]quinolone (MeIQ) |
N-hydroxylation; oxidation (major enzyme) |
potent activation |
12, 41, 56, 61-63, 71, 94, 107, 120, 150, 151, 154, 155, 158 |
2655891, 8674051, 21786339, 11377247, 11473383, 8200083, 9721189, 8200084, 1486866, 16485905, 9705755, 11013410, 14744142, 14725854, 10861951 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (MeIQx) |
N-hydroxylation (major enzyme, high activity) |
potent activation |
12, 41, 58, 61-63, 94, 107, 151- 155 |
2655891, 8674051, 9111224, 11377247, 11473383, 8200083, 8200084, 1486866, 11013410, 10503887, 12351158, 14744142, 14725854 |
| CYP1A2 | chemical | arylamine, heterocyclic |
2-amino-3-methyl-9H- pyrido[2,3-b]indole (MeAαC) |
N-hydroxylation, oxidation |
potent activation |
159 | 14729582 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-3- methylimidazo[4,5- f]quinolone (IQ) |
N-hydroxylation, oxidation (high activity, major enzyme) |
potent activation |
12, 41, 58, 61, 65, 71, 141-144, 151, 154, 155, 160- 163 |
2655891, 8674051, 9111224, 11377247, 9918136, 9721189, 15728263, 15843388, 17158518, 9860501, 11013410, 14744142, 14725854, 1486866, 2813353, 9675256, 10023085, 15089095 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-6- methyldipyrido[1,2-a:3′,2′-d]- imidazole (Glu-P-1) |
N-hydroxylation, oxidation (high activity, major enzyme) |
potent activation |
12, 41, 61, 107, 139, 154, 155, 160 |
2655891, 8674051, 11377247, 1486866, 2803520, 14744142, 14725854, 2813353 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-α-carboline | oxidation | activation | 149, 164 | 1913651, 8801053 |
| CYP1A2 | chemical | arylamine | 2-aminoanthracene |
N-hydroxylation, oxidation (major enzyme) |
potent activation |
12, 41, 61, 107, 139, 141-144, 162, 165 |
2655891, 8674051, 11377247, 1486866, 2803520, 15728263, 15843388, 17158518, 9860501, 10023085, 9477228 |
| CYP1A2 | chemical | heterocyclic amine | 2-aminodipyrido[1,2-a:3,2′- d]-imidazole (Glu-P-2) |
oxidation | activation |
12, 107, 139 |
2655891, 1486866, 2803520 |
| CYP1A2 | chemical | arylamine | 2-aminofluorene (2-AF) |
N-hydroxylation, oxidation (major enzyme, high activity) |
potent activation |
12, 41, 61, 107, 139, 166-168 |
2655891, 8674051, 11377247, 1486866, 2803520, 15840428, 16372832, 10727902 |
| CYP1A2 | chemical | arylamine | 2-naphthylamine (β- naphthylamine) |
N-hydroxylation, oxidation |
activation |
12, 61, 107, 147, 160 |
2655891, 11377247, 1486866, 15450435, 2813353 |
| CYP1A2 | chemical | nitroarene | 2-nitrofluoranthene | nitroreduction | activation | 53 | 11113705 |
| CYP1A2 | chemical | nitrosamine | 3-(N- nitrosomethylamino) propiona ldehyde |
oxidation | activation | 68 | 15725615 |
| CYP1A2 | chemical | nitroarene | 3,6-dinitrobenzo[e]pyrene | nitroreduction | activation | 70 | 19393727 |
| CYP1A2 | chemical | nitroarene | 3-acetylaminobenzanthrone |
N-hydroxylation (concentration dependent) |
activation | 79 | 12740904 |
| CYP1A2 | chemical | heterocyclic amine | 3-amino-1,4-dimethyl-5H- pyrido[4,3-b]indole (Trp-P-1) |
N-hydroxylation; oxidation (major enzyme) |
activation |
12, 41, 61, 139, 151 |
2655891, 8674051, 11377247, 2803520, 11013410 |
| CYP1A2 | chemical | heterocyclic amine | 3-amino-1-methyl-5H- pyrido[4,3-b]indole (Trp-P-2) |
N-hydroxylation, oxidation (major enzyme) |
activation |
12, 41, 61, 62, 94, 107, 151, 160 |
2655891, 8674051, 11377247, 11473383, 8200084, 1486866, 11013410, 2813353 |
| CYP1A2 | chemical | arylamine, metabolite | 3-aminobenzanthrone |
N-hydroxylation (major enzyme, concentration dependent) |
activation | 72, 73 | 15885895, 16601755 |
| CYP1A2 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene |
oxidation | activation | 41 | 8674051 |
| CYP1A2 | chemical | arylamine | 3′-methyl-4- dimethylaminazobenzene |
oxidation | potent activation |
169 | 10720750 |
| CYP1A2 | chemical | nitroarene | 3-nitrobenzanthrone | nitroreduction | activation | 73, 79, 80 | 16601755, 12740904, 12782579 |
| CYP1A2 | chemical | nitroarene | 3-nitrofluoranthene | nitroreduction | activation | 53 | 11113705 |
| CYP1A2 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl |
N-hydroxylation, oxidation |
activation |
12, 41, 58, 107, 147, 160, 170, 171 |
2655891, 8674051, 9111224, 1486866, 15450435, 2813353, 9163700, 16988941 |
| CYP1A2 | natural compound |
furanoterpene produced by sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation (major enzyme) | activation | 172, 173 | 1651809, 15892579 |
| CYP1A2 | chemical | nitroarene | 4-nitropyrene | 4-aminopyrene formation (nitroreduction) |
activation | 174 | 10197616 |
| CYP1A2 | chemical | PAH, metabolite | 5,6-dimethylchrysene-1,2- diol |
oxidation | activation | 34, 43, 81 | 16946553, 11502724, 14720319 |
| CYP1A2 | chemical |
N-heterocyclic aromatic hydrocarbon, |
5,9- dimethyldibenzo[c,g]carbazol e |
oxidation | activation | 88-91 | 10984687, 12034315, 15534862, 21809388 |
| CYP1A2 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | oxidation | activation |
31, 34, 43, 81 |
16946553, 8674051, 11502724, 14720319 |
| CYP1A2 | chemical | arylamine | 6-aminochrysene | oxidation | activation |
41, 66, 84, 85, 141- 144 |
8674051, 9685642, 8118930, 8330339, 15728263, 15843388, 17158518, 9860501 |
| CYP1A2 | chemical | arylamine, metabolite | 6-aminochrysene-1,2-diol | diol epoxide formation; oxidation |
activation | 41, 84, 85 | 8674051, 8118930, 8330339 |
| CYP1A2 | chemical | PAH | 7,12- dimethylbenz[a]anthracene |
oxidation | activation | 43, 81, 86 | 11502724, 14720319, 12584184 |
| CYP1A2 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
oxidation | activation | 34, 43, 81 | 16946553, 11502724, 14720319 |
| CYP1A2 | chemical |
N-heterocyclic aromatic hydrocarbon |
7H-dibenzo[c,g]carbazole | oxidation | activation | 88-90 | 10984687, 12034315, 15534862 |
| CYP1A2 | chemical | PAH, aza-aromatic | 7-methylbenz[c]acridine | 3,4-dihydrodiol formation | activation | 92 | 7866988 |
| CYP1A2 | chemical | PAH, aza-aromatic | 7-methylbenz[c]acridine | oxidation | activation | 92 | 7866988 |
| CYP1A2 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation (both exo- 8,9- and endo-8,9-), oxidation |
activation |
11, 12, 41, 42, 57, 61, 93, 94, 162, 175-182 |
2492107, 2509067, 8674051, 7955101, 15279838, 11377247, 7923587, 8200084, 10023085, 2162057, 766804, 8261428, 12079611, 1902334, 11782366, 16385575, 16608170 |
| CYP1A2 | chemical | heterocyclic amine | aminomethylphenylnorharma n |
N-hydroxylation | activation | 95 | 17067997 |
| CYP1A2 | chemical | arylamine, heterocyclic |
aminophenylharman | N-hydroxylation | activation | 95 | 17067997 |
| CYP1A2 | chemical | arylamine, heterocyclic |
aminophenylnorharman | N-hydroxylation | activation | 95 | 17067997 |
| CYP1A2 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid I | nitroreduction | potent activation |
96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A2 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid II | nitroreduction | potent activation |
96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A2 | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol | oxidation | activation | 34, 43, 81 | 16946553, 11502724, 14720319 |
| CYP1A2 | chemical | diphenylmethanol, metabolite |
benzhydrol | oxidation | activation | 102 | 12160905 |
| CYP1A2 | chemical | PAH, metabolite | benzo[b]fluoroanthene-9,10- diol |
oxidation | activation |
34, 41, 43, 81 |
16946553, 8674051, 11502724, 14720319 |
| CYP1A2 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol |
oxidation | activation | 43 | 11502724 |
| CYP1A2 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | oxidation | activation | 34, 43, 81 | 16946553, 11502724, 14720319 |
| CYP1A2 | chemical | aromatic ketone, diphenyl ketone |
benzophenone | oxidation | activation | 102 | 12160905 |
| CYP1A2 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation | activation | 34, 41, 81 | 16946553, 11502724, 14720319 |
| CYP1A2 | drug | imidazole; anticancer, alkylating |
dacarbazine |
N-demethylation (major enzyme) |
potent activation |
111 | 10473105 |
| CYP1A2 | natural compound |
bicyclic monoterpene | Δ3-carene | epoxidation (high Km, medium activity) |
activation | 183 | 16379671 |
| CYP1A2 | chemical | PAH | dibenz[a,h]anthracene | 3,4-dihydrodiol formation | activation | 184 | 8638931 |
| CYP1A2 | chemical | PAH, aza-aromatic | dibenz[a,j]acridine | 3,4-dihydrodiol formation | activation | 92 | 7866988 |
| CYP1A2 | chemical | PAH | dibenzo[a,l]pyrene | (−)-anti-11,12- dihydrodiol-13,14- epoxide formation, oxidation |
activation | 81 | 14720319 |
| CYP1A2 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
oxidation | activation | 34, 71, 81 | 16946553, 9721189, 14720319 |
| CYP1A2 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C12- and C13- (low activity) |
activation |
39, 40, 185-189 |
16936898, 21753906, 11755121, 12123750, 15548707, 17197724, 21683692 |
| CYP1A2 | physiologi cal compound |
estrogen | 17β-estradiol | C2-hydroxylation (major reaction, medium Km, medium activity, medium efficiency), major metabolite and major enzyme in liver; C4- hydroxylation (minor reaction); C16α- hydroxylation (major enzyme, high Km, no activity, or low activity) |
activation |
71, 106, 122-124, 126-129, 190-194 |
9721189, 8037457, 9625734, 9054608, 9667077, 11555828, 12865317, 15784278, 16112414, 1449532, 9635876, 11454902, 11741520, 14703066 |
| CYP1A2 | natural compound |
alkenylbenzene; occurs in a variety of foods including essential oils of tarragon, sweet basil, sweet fennel, anis |
estragole | C1′-hydroxylation (major enzyme, medium Km, low activity) |
potent activation |
195-197 | 17407329, 15914212, 21459083 |
| CYP1A2 | physiologi cal compound |
estrogen | estrone | C2-hydroxylation (medium Km, high activity, major metabolite); C4- hydroxylation (medium Km, medium activity, very low activity); C16α- hydroxylation (minor reaction, very low activity) |
activation |
49, 122, 123, 127, 128, 191, 192, 198 |
10426814, 9625734, 9054608, 12865317, 15784278, 9635876, 11454902, 16537715 |
| CYP1A2 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | oxidation (at high concentration) |
activation | 87 | 20507880 |
| CYP1A2 | natural compound |
phenylpropene; from Rhizoma acorigraminei |
methyleugenol | C1′-hydroxylation (medium Km, major enzyme) |
activation | 196, 199 | 15914212, 16411663 |
| CYP1A2 | chemical | arylamine, metabolite |
N-acetyl-N-hydroxy-3- aminobenzanthrone |
oxidation, at higher concentrations |
activation | 79 | 12740904 |
| CYP1A2 | chemical | PAH | naphthalene | oxidation | activation | 200 | 11356140 |
| CYP1A2 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-3- aminobenzanthrone |
reduction to amine | activation | 79 | 12740904 |
| CYP1A2 | chemical |
N-heterocyclic aromatic hydrocarbon |
N- methyldibenzo[c,g]carbazole |
oxidation | activation | 88-91 | 10984687, 12034315, 15534862, 21809388 |
| CYP1A2 | chemical | nitrosamine | N-nitrosoethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP1A2 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP1A2 | chemical | nitrosamine | N-nitrosomethylethylamine | oxidation | activation | 134 | 11600130 |
| CYP1A2 | chemical | nitrosamine | N-nitrosomethylpropylamine | oxidation | activation | 69, 134 | 16720019, 11600130 |
| CYP1A2 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OHtetrahydrofuran formation) |
activation | 132, 133 | 11774366, 12214673 |
| CYP1A2 | chemical | azoarylamine | o-aminoazotoluene | oxidation | activation | 41, 66 | 8674051, 9685642 |
| CYP1A2 | chemical | o-methoxyaniline |
o-anisidine (2- methoxyaniline) |
N-hydroxylation (major enzyme with recombinant model), oxidation |
activation | 147, 201 | 15450435, 15828049 |
| CYP1A2 | chemical | diphenylketone, metabolite |
p-benzoylphenol (4- hydroxybenzophenone) |
oxidation | activation | 102 | 12160905 |
| CYP1A2 | chemical | PAH | phenanthrene | oxidation to 1,2- (major reaction), 3,4-, and 9,10- dihydrodiols and phenols |
activation | 46, 135 | 7581497, 19766613 |
| CYP1A2 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation | 41 | 8674051 |
| CYP1A2 | chemical | aromatic hydrocarbon, alkyl benzene |
styrene (vinyl benzene) | oxidation, 7,8-oxide formation |
activation | 202-207 | 9253143, 7696548, 11407535, 12616646, 12834847, 18266326 |
| CYP1B1 | chemical | PAH, metabolite | (±)-, (−)-, and (+)- benzo[a]pyrene-7,8- dihydrodiol |
trans-(anti)-7,8- dihydroxy-9,10-epoxy- 7,8,9,10-tetrahydro- formation, trans-diol epoxide form (low Km, high activity, high efficiency); oxidation |
potent activation |
26, 31, 32, 34, 41, 43, 49, 52, 120, 208-210 |
16411658, 15720144, 17295519, 16946553, 8674051, 11502724, 10426814, 21028851, 16485905, 12628515, 12807732, 16551781 |
| CYP1B1 | chemical | nitroarene | 1,8-dinitropyrene | nitroreduction | potent activation |
53 | 11113705 |
| CYP1B1 | chemical | arylamine, metabolite of 1-nitropyrene |
1-aminopyrene | oxidation | potent activation |
54 | 11525925 |
| CYP1B1 | chemical | PAH | 2,3-dihydroxy-2,3- dihydrofluoranthene |
oxidation | activation | 66 | 9685642 |
| CYP1B1 | chemical | heterocyclic amine | 2-amino-3,4- dimethylimidazo[4,5- f]quinolone (MeIQ) |
N-hydroxylation, oxidation |
activation |
41, 49, 61, 100, 210, 211 |
8674051, 10426814, 11377247, 11502724, 16551781, 11719446 |
| CYP1B1 | chemical | heterocyclic amine | 2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (MeIQx) |
N- hydroxylation, oxidation |
activation | 41, 49, 61 | 8674051 10426814, 11377247 |
| CYP1B1 | chemical | heterocyclic amine | 2-amino-3- methylimidazo[4,5- f]quinolone (IQ) |
N-hydroxylation, oxidation |
activation |
41, 49, 61, 65, 100 |
8674051, 10426814, 11377247, 9918136, 11502724 |
| CYP1B1 | chemical | arylamine | 2-aminoanthracene |
N-hydroxylation, oxidation (high activity) |
potent activation |
41, 61, 100, 210 |
8674051, 11377247, 11502724, 16551781 |
| CYP1B1 | chemical | arylamine | 2-aminofluorene (2-AF) |
N-hydroxylation, oxidation |
potent activation |
41, 49, 61, 100, 210 |
8674051, 10426814, 11377247, 11502724, 16551781 |
| CYP1B1 | chemical | nitroarene | 2-nitrofluoranthene | nitroreduction | potent activation |
53 | 11113705 |
| CYP1B1 | chemical | nitroarene | 2-nitropyrene | 2-aminopyrene formation (nitroreduction) |
potent activation |
41, 100 | 8674051, 11502724 |
| CYP1B1 | chemical | nitrosamine | 3-(N- nitrosomethylamino) propiona ldehyde |
oxidation | activation | 68 | 15725615 |
| CYP1B1 | chemical | heterocyclic amine | 3-amino-1,4-dimethyl-5H-pyrido[4,3-b]indole (Trp-P-1) |
N-hydroxylation, oxidation |
potent activation |
41, 49, 61, 66, 71, 100, 210 |
8674051, 10426814, 11377247, 9685642, 9721189, 11502724, 16551781 |
| CYP1B1 | chemical | heterocyclic amine | 3-amino-1-methyl-5H- pyrido[4,3-b]indole (Trp-P-2) |
N-hydroxylation; oxidation |
activation |
41, 49, 61, 62, 66, 71, 100 |
8674051, 10426814, 11377247, 11473383, 9685642, 11502724 |
| CYP1B1 | chemical | arylamine, metabolite | 3-aminobenzanthrone | N-hydroxylation | activation | 212 | 15310241 |
| CYP1B1 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene |
oxidation | potent activation |
41, 49, 66, 100, 210 |
8674051, 10426814, 9685642, 11502724, 16551781 |
| CYP1B1 | chemical | nitroarene | 3-nitrofluoranthene | nitroreduction | activation | 53 | 11113705 |
| CYP1B1 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
oxidation | activation |
132, 133, 213-217 |
11774366, 12214673, 1312898, 7595636, 8806763, 10803680, 9106248 |
| CYP1B1 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl |
N-hydroxylation; oxidation |
activation | 41, 218 | 8674051, 19274671 |
| CYP1B1 | chemical | PAH, metabolite | 5,6-dimethylchrysene-1,2- diol |
oxidation | activation |
34, 41, 43, 49, 66, 81, 210 |
16946553, 8674051, 11502724, 10426814, 9685642, 14720319, 16551781 |
| CYP1B1 | chemical | PAH | 5-methylchrysene | oxidation | activation | 83, 210 | 18992797 16551781 |
| CYP1B1 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | oxidation (medium Km, high activity, high efficiency) |
potent activation |
34, 41, 43, 49, 66, 81, 83, 120, 210 |
16946553, 8674051, 11502724, 10426814, 9685642, 14720319, 18992797, 16485905, 16551781 |
| CYP1B1 | chemical | arylamine | 6-aminochrysene | oxidation | potent activation |
41, 43 | 8674051, 11502724 |
| CYP1B1 | chemical | arylamine, metabolite | 6-aminochrysene-1,2-diol | diolepoxide formation, oxidation |
potent activation |
41, 84, 85 | 8674051, 8118930, 8330339 |
| CYP1B1 | chemical | nitroarene | 6-nitrochrysene | nitroreduction; 5,6- quinone formation |
activation | 41, 43, 219 | 8674051, 11502724, 8481905 |
| CYP1B1 | chemical | PAH | 7,12- dimethylbenz[a]anthracene |
oxidation (low Km, high activity and efficiency) |
activation |
43, 49, 81, 86, 210 |
11502724, 10426814, 14720319, 12584184, 16551781 |
| CYP1B1 | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
3,4-dihydrodiol-1,2- epoxide formation (medium Km, high activity, high efficiency); oxidation |
potent activation |
34, 41, 43, 49, 66, 81 |
16946553, 8674051, 11502724, 10426814, 9685642, 14720319 |
| CYP1B1 | chemical | PAH, metabolite | 9-hydroxybenzo[a]pyrene | oxidation | activation | 43 | 11502724 |
| CYP1B1 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | oxidation | activation |
61, 210, 217 |
11377247, 16551781, 9106248 |
| CYP1B1 | chemical | diphenylmethanol, metabolite |
benzhydrol | oxidation | activation | 102 | 12160905 |
| CYP1B1 | chemical | PAH | benzo[a]pyrene |
trans-7,8-dihydroxy-9,10- epoxy-7,8,9,10- tetrahydro- formation (medium Km, high activity, high efficiency); 1,6-,3,6-dione (quinone form., low activity); oxidation (major enzyme) |
activation |
34, 41, 43, 49, 52, 61, 81, 104, 208, 210, 220-222 |
16946553, 8674051, 11502724, 10426814, 21028851, 11377247, 14720319, 9806168, 12628515, 16551781, 10409402, 11465393, 15958554 |
| CYP1B1 | chemical | PAH, metabolite | benzo[b]fluoroanthene-9,10- diol |
oxidation | activation |
34, 43, 49, 50, 81 |
16946553, 8674051, 11502724, 10426814, 14720319 |
| CYP1B1 | chemical | PAH | benzo[c]phenanthrene | dihydrodiol 3,4-, 1,2- epoxide formation (major enzyme); oxidation |
activation |
43, 81, 109, 217, 223 |
11502724, 14720319, 11409939, 9168260, 21781864 |
| CYP1B1 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol |
dihydrodiol 3,4-, 1,2- epoxide formation |
activation | 43, 49 | 11502724, 10426814 |
| CYP1B1 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | oxidation | potent activation |
34, 41, 43, 49, 66, 81 |
16946553, 8674051, 11502724, 10426814, 9685642, 14720319 |
| CYP1B1 | chemical | aromatic ketone, diphenyl ketone |
benzophenone | oxidation | activation | 102 | 12160905 |
| CYP1B1 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation | potent activation |
34, 41, 43, 49, 66, 81, 210 |
16946553, 8674051, 11502724, 10426814, 9685642, 14720319, 16551781 |
| CYP1B1 | chemical | PAH | cyclopenta[c,d]pyrene | oxidation | activation | 217 | 9106248 |
| CYP1B1 | chemical | PAH, aza-aromatic | dibenz[a,h]acridine | 10,11-diol formation | activation | 111 | 15144224 |
| CYP1B1 | chemical | PAH | dibenzo[a,l]pyrene | (−)-anti-11,12- dihydrodiol-13,14- epoxide formation (medium Km, high activity, high efficiency); oxidation |
potent activation |
43, 81, 113-119, 208, 210, 224 |
11502724, 14720319, 9625737, 10207125, 10493514, 10506751, 8968059, 16581046, 17509623, 12628515, 16551781, 17623886 |
| CYP1B1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
11,12-dihydrodiol-13,14- epoxide formation (medium Km, high activity, high efficiency) |
potent activation |
34, 41, 43, 49, 71, 81, 113-115, 118, 120, 208, 210 |
16946553, 8674051, 11502724, 10426814, 9721189, 14720319, 9625737, 10207125, 10493514, 16581046, 17509623, 16485905, 12628515, 16551781 |
| CYP1B1 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor and DNA binding |
ellipticine | oxidation | activation |
39, 185- 188 |
16936898, 11755121, 12123750, 15548707, 17197724 |
| CYP1B1 | physiologi cal compound |
estrogen | 17β-estradiol | C4-hydroxylation (major enzyme, medium Km, medium activity, medium and low efficiency), oxidation, 3,4-quinone formation; C2- hydroxylation (low activity, minor reaction), oxidation, 2,3-quinone formation; C16α- hydroxylation (minor enzyme, medium and high Km, low activity) |
potent activation |
49, 71, 121-130, 194, 211, 221, 225- 228 |
10426814, 9721189, 7826886, 9625734, 9054608, 9667077, 8930523, 11555828, 12865317, 15784278, 16112414, 17570247, 14703066, 11719446, 11465393, 8790407, 7568105, 10862525, 10910054 |
| CYP1B1 | physiologi cal compound |
estrogen | estrone | C4-hydroxylation (low Km, major reaction); C2- hydroxylation (low activity, minor reaction), oxidation, 2,3-quinone formation; |
potent activation |
49, 127, 128, 130, 198 |
10426814, 12865317, 15784278, 17570247, 16537715 |
| CYP1B1 | chemical | nitrosamine |
N-nitrosodiethylamine (N,N- diethylnitrosamine) |
oxidation | activation | 132-134 | 11774366, 12214673, 11600130 |
| CYP1B1 | chemical | nitrosamine | N-nitrosoethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP1B1 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP1B1 | chemical | nitrosamine | N-nitrosomethylpropylamine | oxidation | activation | 69, 134 | 16720019, 11600130 |
| CYP1B1 | chemical | nitrosamine | N-nitrosomorpholine | oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP1B1 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OHtetrahydrofuran formation), oxidation |
activation | 132, 133 | 11774366, 12214673 |
| CYP1B1 | chemical | diphenylketone, metabolite |
p-benzoylphenol,4- hydroxybenzophenone |
oxidation | activation | 102 | 12160905 |
| CYP1B1 | chemical | PAH | phenanthrene | oxidation to 9,10- (major reaction), and 1,2- and 3,4- (minor reaction) dihydrodiols and phenols |
activation | 135 | 19766613 |
| CYP1B1 | chemical | nitroarene | 1-nitropyrene | 1-aminopyrene form. (nitroreduction), at low concentrations, epoxidation C4,5-, at high concentration |
potent activation |
53, 54, 229 | 11113705, 11525925, 15310239 |
| CYP2A6 | chemical | haloalkane | 1, 2-dibromoethane (ethylene dibromide) |
oxidation to 2- bromoacetaldehyde |
activation | 230 | 8870687 |
| CYP2A6 | chemical | diene | 1,3-butadiene | butadiene monoxide (epoxybutene) formation (high activity) |
activation | 231-233 | 7586124, 8901879, 9016811 |
| CYP2A6 | chemical | nitrile, herbicide | 2,6-dichlorobenzonitrile (dichlobenil) |
epoxidation, C2,3- | activation | 234, 235 | 8649351, 8863822 |
| CYP2A6 | chemical | arylamine | 2,6-dimethylaniline |
N-hydroxylation (at higher concentrations), major enzyme |
activation | 236 | 11409937 |
| CYP2A6 | chemical | nitrosamine | 3-(N- nitrosomethylamino) propiona ldehyde |
oxidation | activation | 68 | 15725615 |
| CYP2A6 | chemical | nitrosamine | 3-(N- nitrosomethylamino) propioni trile |
oxidation (at high concentrations, major enzyme) |
potent activation |
68, 69 | 15725615, 16720019 |
| CYP2A6 | chemical | arylamine, metabolite | 3-aminobenzanthrone | N-hydroxylation | activation | 212 | 15310241 |
| CYP2A6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
Cα-hydroxylationmethylene (lactol/acid formation or Cα-methyl (diol/acid formation) |
activation |
170, 237, 238 |
9163700, 12975327, 21473878 |
| CYP2A6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
Cα-hydroxylationmethylene (keto aldehyde and keto alcohol formation), high Km, low activity, oxidation |
activation |
66, 132- 134, 141- 143, 213- 216, 238- 246 |
9685642, 11774366, 12214673, 11600130, 15728263, 15843388, 17158518, 1312898, 7595636, 8806763, 10803680, 11016631, 15333516, 17671098, 21473878, 10837014, 11080669, 12920169, 1423839 |
| CYP2A6 | chemical | arylamine | 4,4′-methylene bis(2- chloroaniline) (MOCA) |
oxidation, N- hydroxylation |
activation |
65, 106, 248, 249 |
9685642, 1486866, 1944238, 1740010 |
| CYP2A6 | chemical | arylamine | 6-aminochrysene | oxidation | activation | 248 | 1944238 |
| CYP2A6 | chemical | diphenylmethanol, metabolite |
benzhydrol | oxidation (major enzyme) | activation | 102 | 12160905 |
| CYP2A6 | chemical | aromatic ketone, diphenyl ketone |
benzophenone | oxidation (major enzyme) | activation | 102 | 12160905 |
| CYP2A6 | chemical | aliphatic epoxide, metabolite |
butadiene monoxide (1,2- epoxy-3-butene) |
diepoxybutane meso- (major) and (±)- formation (at high concentrations) |
activation | 231-233 | 7586124, 8901879, 9016811 |
| CYP2A6 | chemical | haloalkane | chloroform (trichloromethane) |
dehalogenation, reductive (at high concentrations); oxidation (major enzyme at high concentrations) |
activation | 250 | 12584152 |
| CYP2A6 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (minor enzyme, high Km); oxidation |
activation |
101, 108, 251-254 |
11377097, 19501186, 8242617, 9010702, 10348794, 10692561 |
| CYP2A6 | natural compound |
alkenylbenzene; occurs in a variety of foods including essential oils of tarragon, sweet basil, sweet fennel, anis |
estragole | C1′-hydroxylation (major enzyme, medium Km, medium activity) |
potent activation |
195, 197 | 17407329, 21459083 |
| CYP2A6 | chemical | phosphoramide | hexamethylphosphoramide | oxidation, formaldehyde production |
activation | 235, 255 | 8863822, 9007030 |
| CYP2A6 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation (minor reaction, high Km), oxidation (at high concentration) |
activation |
87, 247, 253 |
20507880, 8242617, 10348794 |
| CYP2A6 | natural compound |
furanocoumarin; anti- psoriatic, photosensitizer, found in several species of plants |
methoxalen (8- methoxypsoralen, xanthotoxin) |
epoxidation (furanoepoxide formation) and hydrolysis |
activation | 140 | 17584015 |
| CYP2A6 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanabasine | oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP2A6 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanatabine | oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP2A6 | chemical | nitrosamine |
N-nitrosodibutylamine (N, N- dibutylnitrosamine) |
oxidation | activation | 244, 134 | 10837014, 11600130 |
| CYP2A6 | chemical | nitrosamine |
N-nitrosodiethylamine (N,N- diethylnitrosamine) |
oxidation | potent activation |
106, 132- 134, 141- 144, 245- 247 |
1486866, 11774366, 12214673, 11600130, 15728263, 15843388, 17158518, 9860501, 10837014, 11080669, 1423839 |
| CYP2A6 | chemical | nitrosamine |
N-nitrosodi-n-propylamine (N-nitrosodipropylamine) |
oxidation | potent activation |
134, 141- 143, 244 |
11600130, 15728263, 15843388, 17158518, 10837014 |
| CYP2A6 | chemical | nitrosamine | N-nitrosoethylbutylamine | oxidation (major enzyme) | potent activation |
134, 244 | 11600130, 10837014 |
| CYP2A6 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation (major enzyme) | potent activation |
134, 244 | 11600130, 10837014 |
| CYP2A6 | chemical | nitrosamine | N-nitrosomethylethylamine | oxidation | potent activation |
134, 243 | 11600130, 10837014 |
| CYP2A6 | chemical | nitrosamine | N-nitrosomethylphenylamine | oxidation | activation |
134, 243, 245 |
11600130, 10837014, 11080669 |
| CYP2A6 | chemical | nitrosamine | N-nitrosomethylpropylamine | oxidation | activation |
68, 134, 244 |
16720019, 11600130, 10837014 |
| CYP2A6 | chemical | nitrosamine | N-nitrosomorpholine | oxidation (major enzyme) | activation | 132, 133 | 11774366, 12214673 |
| CYP2A6 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N-nitrosonornicotine, NNN) | hydroxylation C5′- (lactol formation, medium Km, high to medium activity), oxidation (major enzyme) |
activation |
132-134, 244, 256- 258 |
11774366, 12214673, 11600130, 15651850, 10837014, 9276639, 7646564 |
| CYP2A6 | chemical | nitrosamine | N-nitrosopiperidine | Cα-hydroxylation (2-OH- tetrahydropyran and 2- OH-5- methyltetrahydropyran formation), major enzyme; oxidation |
activation |
132, 133, 258 |
11774366, 12214673, 15651850 |
| CYP2A6 | chemical | nitrosamine | N-nitrosopyrrolidine, | Cα-hydroxylation (2-OH- tetrahydrofuran formation); oxidation (major enzyme) |
activation |
132-134, 244, 258 |
11774366, 12214673, 11600130, 15651850, 10837014 |
| CYP2A6 | chemical | diphenylketone, metabolite |
p-benzoylphenol (4- hydroxybenzophenone) |
oxidation (major enzyme) | activation | 102 | 12160905 |
| CYP2A6 | natural compound |
Methylenedioxypheny l (benzodioxole) |
safrole | C1′-hydroxylation (major enzyme at low concentrations), medium Km, medium activity |
activation |
195, 196, 259, 260 |
17407329, 15914212, 15377158, 15310247 |
| CYP2A13 | chemical | nitrosamine | 3-(N- nitrosomethylamino) propiona ldehyde |
oxidation (major enzyme) | potent activation |
68 | 15725615 |
| CYP2A13 | natural compound |
indole, alkylating, pulmonary toxin; in higher concentrations in mammalian digestive tract and coal tar |
3-methylindole (skatole) | dehydrogenation (desaturation, 3- methyleneindolenine form., low Km, medium activity, high efficiency), epoxidation (3- methyloxindole formation) |
potent activation |
77, 261 | 20795680, 19608696 |
| CYP2A13 | chemical | nitrosamine | 3-N-nitrosoguvacoline | oxidation (major enzyme) | activation | 68, 69 | 15725615, 16720019 |
| CYP2A13 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
Cα-hydroxylation-methyl (keto alcohol formation), major enzyme, medium Km, medium activity, or high activity |
potent activation |
237-240, 262-266 |
11016631, 12975327, 15333516, 15528319, 15962925, 12130698, 16917071, 17671098, 19074523, 21473878 |
| CYP2A13 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl |
N-hydroxylation; oxidation |
activation | 58, 171 | 9111224, 16988941 |
| CYP2A13 | chemical | difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation 8,9-, oxidation |
potent activation |
181, 265 | 16385575, 16917071 |
| CYP2A13 | natural compound |
furanocoumarin; anti- psoriatic, photosensitizer, found in bergamot essential oil, in other citrus essential oils, and in grapefruit juice |
bergapten, 5- methoxypsoralen |
epoxidation and hydrolysis to dihydrodiol |
activation | 267 | 20798279 |
| CYP2A13 | chemical | nitrosamine | N-nitrosomethylpropylamine | oxidation | potent activation |
69, 134 | 16720019, 11600130 |
| CYP2A13 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
hydroxylation C5′- (lactol formation, medium Km, high activity) and C2′- (keto alcohol formation, medium Km, low activity) |
activation | 258, 266 | 19074523, 15651850 |
| CYP2A13 | chemical | nitrosamine | N-nitrosopiperidine | Cα-hydroxylation (2-OH- tetrahydropyran and 2- OH-5-methyl tetrahydrofuran formation) |
activation | 258 | 15651850 |
| CYP2A13 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OH- tetrahydrofuran formation) |
activation | 258 | 15651850 |
| CYP2A13 | chemical | aromatic hydrocarbon, alkyl benzene |
styrene (vinyl benzene) | oxidation, 7,8-oxide formation |
activation | 207 | 18266326 |
| CYP2B6 | chemical | haloalkane | 1, 2-dibromoethane (ethylene dibromide) |
oxidation to 2- bromoacetaldehyde |
activation | 230 | 8870687 |
| CYP2B6 | chemical | haloalkane | 2, 2-dichloro-1,1,1- trifluoroethane (HCFC-123) |
oxidation | activation | 268, 269 | 11684364, 11684365 |
| CYP2B6 | chemical | arylamine, metabolite | 3-aminobenzanthrone | N-hydroxylation | activation | 212 | 15310241 |
| CYP2B6 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene |
oxidation | activation | 270 | 7905383 |
| CYP2B6 | chemical | nitroarene | 3-nitrobenzanthrone | nitroreduction | potent activation |
80 | 12782579 |
| CYP2B6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
hydroxylation, alphamethyl (keto alcohol form.), major reaction and Cα-methylene (keto aldehyde form.), minor reaction |
activation |
74, 213- 216, 252, 271, 272 |
11360624, 1312898, 7595636, 8806763, 10803680, 12920169, 9280407, 16174803 |
| CYP2B6 | chemical | unsaturated | 4-vinyl-1-cyclohexene (S)- and (R)- |
epoxidation 7,8- (major reaction, stereoselective for (R)-); epoxidation 1,2- |
activation | 100, 273 | 11502734, 11159809 |
| CYP2B6 | chemical | arylamine | 6-aminochrysene | oxidation | activation | 85, 270 | 8330339, 7905383 |
| CYP2B6 | chemical | herbicide, chloroacetamide |
alachlor | oxidation | activation | 274 | 11133395 |
| CYP2B6 | chemical | herbicide, chloroacetamide |
butachlor | oxidation | activation | 274 | 11133395 |
| CYP2B6 | chemical | haloalkane | chloroform (trichloromethane) |
oxidation (at high conc.) | activation | 250 | 12584152 |
| CYP2B6 | drug | azaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (major enzyme, major reaction, high Km, high activity); oxidation |
potent activation |
74, 101, 108, 251- 254, 271, 275 |
11360624, 11377097, 19501186, 8242617, 9010702, 10348794, 10692561, 9280407, 15919850 |
| CYP2B6 | chemical | polycyclic aromatic hydrocarbon (PAH) |
dibenz[a,h]anthracene | 3,4-dihydrodiol form. | activation | 184 | 8638931 |
| CYP2B6 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (S)- (high Km, major enzyme), oxidation (at high concentrations) |
activation |
87, 259, 261, 262, 275-278 |
20507880, 8242617, 10348794, 10692561, 15919850, 10534317, 15821045, 16854777 |
| CYP2B6 | chemical | herbicide, chloroacetamide |
metolachlor | oxidation | activation | 274 | 11133395 |
| CYP2B6 | chemical | aziridine |
N, N’, N”-triethylene thiophosphoramide (thioTEPA) |
desulfuration, TEPA formation (major enzyme) |
activation | 140, 279 | 17584015, 12107550, |
| CYP2B6 | chemical | nitrosamine | N-nitrosomorpholine | oxidation | activation | 74 | 11360624 |
| CYP2B6 | chemical | o-methoxyaniline |
o-anisidine (2- methoxyaniline) |
N-hydroxylation | activation | 201 | 15828049 |
| CYP2B6 | chemical | aromatic hydrocarbon, alkyl benzene |
styrene (vinyl benzene) | oxidation (major enzyme in liver microsomes at high concentration) |
activation |
202-205, 2780 |
9253143, 7696548, 11407535, 12616646, 16125881 |
| CYP2C8 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP2C8 | chemical | haloalkane | chloroform (trichloromethane) |
dehalogenation reductive (at high concentrations) |
activation | 258 | 12584152 |
| CYP2C8 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation (minor enzyme, high Km), oxidation (at high concentrations) |
activation |
87, 259, 261 |
20507880, 8242617, 10348794 |
| CYP2C8 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP2C8 | chemical | nitrosamine | N-nitrosomethylpropylamine | oxidation | activation | 134 | 11600130 |
| CYP2C8 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OH- tetrahydrofuran formation); oxidation |
activation | 132, 133 | 11774366, 12214673 |
| CYP2C8 | chemical | aromatic hydrocarbons, alkyl benzene |
styrene (vinyl benzene) | oxidation (major enzyme in liver microsomes at high concentrations) |
activation |
202-205, 280 |
9253143, 7696548, 11407535, 12616646, 16125881 |
| CYP2C9 | chemical | diene | 1,3-butadiene | butadiene monoxide (epoxybutene) formation (high activity) |
activation | 231-233 | 7586124, 8901879, 9016811 |
| CYP2C9 | chemical | triazole | 1-aminobenzotriazole (1- ABT) |
oxidation | activation | 140 | 17584015 |
| CYP2C9 | chemical | PAH | 7,12- dimethylbenz[a]anthracene |
oxidation | activation | 43, 81, 87 | 11502724, 14720319, 20507880 |
| CYP2C9 | chemical | PAH | benzo[c]phenanthrene | oxidation | activation | 81 | 14720319 |
| CYP2C9 | chemical | aliphatic epoxide, metabolite |
butadiene monoxide (1, 2- epoxy-3-butene) |
diepoxybutane meso- and (±)- formation |
activation | 231-233 | 7586124, 8901879, 9016811 |
| CYP2C9 | chemical | haloalkane | chloroform (trichloromethane) |
dehalogenation reductive (at high concentrations) |
activation | 250 | 12584152 |
| CYP2C9 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (low Km, low activity, major enzyme at low concentrations); oxidation |
activation |
101, 108, 251-254 |
11377097, 19501186, 8242617, 9010702, 10348794, 10692561 |
| CYP2C9 | chemical | PAH | dibenz[a,h]anthracene | 3,4-dihydrodiol formation | activation | 184 | 8638931 |
| CYP2C9 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation (low Km), oxidation (at high concentration) |
activation | 87, 259 | 20507880, 8242617 |
| CYP2C9 | natural compound |
phenylpropene, from Rhizoma acorigraminei |
methyleugenol | C1′-hydroxylation (medium activity, high Km), at high concentration |
activation | 196, 199 | 15914212, 16411663 |
| CYP2C19 | chemical | triazole | 1-aminobenzotriazole (1- ABT) |
oxidation | activation | 140 | 17584015 |
| CYP2C19 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation (major enzyme) | activation | 173 | 15892579 |
| CYP2C19 | chemical | haloalkane | chloroform (trichloromethane) |
dehalogenation reductive (at high concentration); oxidation (at high concentration) |
activation | 250 | 12584152 |
| CYP2C19 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (low Km, low activity); oxidation |
activation |
108, 251- 254 |
19501186, 8242617, 9010702, 10348794, 10692561 |
| CYP2C19 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (S)- (minor reaction, high Km), oxidation (at high concentration) |
activation |
87, 259, 253, 276 |
20507880, 8242617, 10348794, 10534317 |
| CYP2C19 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP2C19 | chemical | nitrosamine | N-nitrosomethylethylamine | oxidation | activation | 134 | 11600130 |
| CYP2C19 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OH- tetrahydrofuran formation) |
activation | 132, 133 | 11774366, 12214673 |
| CYP2D6 | chemical | triazole | 1-aminobenzotriazole (1- ABT) |
oxidation | activation | 140 | 17584015 |
| CYP2D6 | chemical | nitrosamine | 3-(N- nitrosomethylamino) propiona ldehyde |
oxidation | activation | 68 | 15725615 |
| CYP2D6 | chemical | nitroarene | 3-nitrobenzanthrone | nitroreduction | potent activation |
80 | 12782579 |
| CYP2D6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
Cα- hydroxylation,methylene |
activation | 170 | 9163700 |
| CYP2D6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
Cα-hydroxylation, methyl (keto alcohol formation), high Km, medium activity, or high activity, major reaction and Cα- methylene (keto aldehyde formation), high Km, low activity, minor reaction |
activation |
170, 213- 216, 281 |
9163700, 1312898, 7595636, 8806763, 10803680, 8485585 |
| CYP2D6 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 173 | 15892579 |
| CYP2D6 | chemical | PAH | 7,12- dimethylbenz[a]anthracene |
oxidation | activation | 87 | 20507880 |
| CYP2D6 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | oxidation | activation | 61 | 11377247 |
| CYP2D6 | chemical | haloalkane | chloroform (trichloromethane) |
dehalogenation, reductive (at high concentration) |
activation | 250 | 12584152 |
| CYP2D6 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor and DNA binding |
ellipticine | oxidation N2-; hydroxylation, C13- (low activity) |
activation |
39, 40, 187, 188 |
16936898, 21753906, 15548707, 17197724 |
| CYP2D6 | drug | oxazaphosporine; anticancer, nitrogen mustard |
alkylating ifosfamide |
oxidation (at high concentration) |
activation | 87 | 20507880 |
| CYP2E1 | chemical | haloalkane | 1, 2-dibromoethane (ethylene dibromide) |
oxidation to 2- bromoacetaldehyde |
activation |
107, 230, 180 |
1486866, 8870687, 1664256 |
| CYP2E1 | chemical | haloalkane | 1, 2-dichloroethane (ethylene dichloride) |
oxidation | activation | 107, 282 | 1486866, 1664256 |
| CYP2E1 | chemical | haloalkane | 1, 2-dichloropropane (propylene dichloride) |
oxidation | activation | 107, 282 | 1486866, 1664256 |
| CYP2E1 | chemical | hydrazine | 1, 2-dimethylhydrazine | oxidation | activation | 283 | 15576447 |
| CYP2E1 | chemical | haloalkane | 1,1,2-trichloroethane | oxidation | activation | 284 | 8671747 |
| CYP2E1 | chemical | haloalkene | 1,1,3-trichloropropene | oxidation | activation | 284 | 8671747 |
| CYP2E1 | chemical | haloalkene | 1,1-dichloroethylene (vinylidene chloride) |
epoxidation | activation | 285 | 15319346 |
| CYP2E1 | chemical | diene | 1,3-butadiene | butadiene monoxide (epoxybutene) (S)- and (R)- formation (high activity, major enzyme) |
potent activation |
231-233, 286 |
7586124, 8901879, 9016811, 9635416 |
| CYP2E1 | chemical | halobenzene | 1,4-dichlorobenzene | oxidation | activation | 287 | 9817075 |
| CYP2E1 | chemical | triazole | 1-aminobenzotriazole (1- ABT) |
oxidation | activation | 140 | 17584015 |
| CYP2E1 | chemical | haloalkane | 2, 2-dichloro-1,1,1- trifluoroethane (HCFC-123) |
oxidation | activation |
268, 269, 288, 289 |
11684364, 11684365, 7975716, 8199305 |
| CYP2E1 | chemical | halobenzene | 2,3-dichlorobutane | oxidation | activation | 284 | 8671747 |
| CYP2E1 | chemical | nitrile, herbicide | 2,6-dichlorobenzonitrile (dichlobenil) |
epoxidation, C2,3- | activation | 234,235 | 8649351, 8863822 |
| CYP2E1 | chemical | arylamine | 2,6-dimethylaniline |
N-hydroxylation (at higher concentration) |
activation | 236 | 11409937 |
| CYP2E1 | chemical | arylamine | 2-aminoanthracene | N-hydroxylation | activation | 61 | 11377247 |
| CYP2E1 | chemical | arylamine | 2-aminofluorene (2-AF) | N-hydroxylation | activation | 61 | 11377247 |
| CYP2E1 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 173 | 15892579 |
| CYP2E1 | chemical | unsaturated | 4-vinyl-1-cyclohexene, (S)- and (R)- |
epoxidation 7,8-, stereoselective for (S)-; epoxidation 1,2-, stereoselective for (R)- |
activation | 100, 273 | 11502734, 11159809 |
| CYP2E1 | chemical | acrylic amide | acrylamide | epoxidation to glycidamide |
activation | 290, 291 | 19190172, 20209648 |
| CYP2E1 | chemical | aliphatic nitrile | acrylonitrile (vinyl cyanide, cyanoethylene) |
oxidation (2- cyanoethylene oxide formation) |
activation |
107, 282, 290 |
1486866, 1664256, 8117926 |
| CYP2E1 | chemical | oxidoazanium | azoxymethane | oxidation | activation | 283 | 15576447 |
| CYP2E1 | chemical | aromatic hydrocarbon | benzene | hydroxylation, aromatic (via benzene oxide, muconic acid, and benzoquinone formation, major enzyme at low concentrations) |
activation |
107, 110, 282, 293- 295 |
1486866, 7923572, 1664256, 10207612, 11083083, 15122651 |
| CYP2E1 | chemical | aliphatic epoxide, metabolite |
butadiene monoxide (1, 2- epoxy-3-butene) |
diepoxybutane meso- (major) and (±)- formation, high activity, major enzyme |
potent activation |
231-233, 296 |
7586124, 8901879, 9016811, 17298833 |
| CYP2E1 | chemical | haloalkane | carbon tetrachloride | dechlorination reductive (at low concentrations), oxidative stress induction |
potent activation |
106, 297- 300 |
1486866, 8571359, 10731522, 8471158, 12235922 |
| CYP2E1 | chemical | haloalkane | chloroform (trichloromethane) |
dehalogenation reductive (at high concentration, major enzyme); oxidation (major enzyme at low concentration) |
potent activation |
66, 107, 252, 282, 299, 301 |
9685642, 1486866, 12584152, 1664256, 8471158, 15129551 |
| CYP2E1 | chemical | haloalkene | chloroprene | epoxidation | activation | 302 | 11397396 |
| CYP2E1 | drug | platinum-containing; anticancer |
cisplatin | oxidation | activation | 303, 304 | 16251482, 17761302 |
| CYP2E1 | drug | imidazole; anticancer, alkylating |
dacarbazine | N-demethylation | activation | 111 | 10473105 |
| CYP2E1 | chemical | haloalkene dichloromethane (methylene chloride) |
dehalogenation | oxidative | activation |
107, 282, 284, 297 |
1486866, 1664256, 8671747, 8571359 |
| CYP2E1 | chemical | organic solvents, alcohol |
ethanol | oxidation, reactive oxygen species production |
activation | 305-309 | 7687464, 16052683, 16878272, 16356668, 21146245 |
| CYP2E1 | natural compound |
carbamic acid derivative; fermentation by- product |
ethyl carbamate (urethane) | oxidation to vinyl carbamate epoxide |
activation |
107, 282, 310-312 |
1486866, 1664256, 9344892, 9150748, 11181492 |
| CYP2E1 | chemical | furan | furan | oxidation, cis-2-butene- 1,4-dial formation |
activation | 313 | 16006568 |
| CYP2E1 | chemical | organic solvents, alkylformamide |
N, N-dimethylformamide(DMF) |
N-demethylation (high activity) |
activation | 314, 315 | 8477011, 11684354 |
| CYP2E1 | chemical | organic solvents, alkylformamide |
N-methylformamide | oxidation (methylisocyanate formation) |
activation | 316 | 1538706 |
| CYP2E1 | chemical | nitrosamine | N-nitrosodiethanolamine | oxidation | activation | 317 | 18616954 |
| CYP2E1 | chemical | nitrosamine |
N-nitrosodiethylamine (N,N- diethylnitrosamine,) |
oxidation (major enzyme) | potent activation |
107, 132- 134, 141- 144, 244, 318, 319 |
1486866, 11774366, 12214673, 11600130, 15728263, 15843388, 17158518, 9860501, 10837014, 11733072, 14669323 |
| CYP2E1 | chemical | nitrosamine |
N-nitrosodimethylamine (N,N-dimethylnitrosamine, DMN) |
oxidation (major enzyme) | activation |
107, 134, 170, 244, 246, 317, 318, 320, 321, 322 |
1486866, 11600130, 9163700, 10837014, 1423839, 18616954, 11733072, 8692217, 10366544, 15668106 |
| CYP2E1 | chemical | nitrosamine |
N-nitrosodi-n-propylamine (N-nitrosodipropylamine) |
Cα-hydroxylation and N- depropylation (major enzyme); oxidation |
activation |
134, 141- 143, 244, 318, 323- 325 |
11600130, 15728263, 15843388, 17158518, 10837014, 11733072, 9247615, 8824531, 10910959 |
| CYP2E1 | chemical | nitrosamine | N-nitrosoethylbutylamine | oxidation | activation | 134, 244 | 11600130, 10837014 |
| CYP2E1 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation | potent activation |
107, 134, 244 |
1486866, 11600130, 10837014, |
| CYP2E1 | chemical | nitrosamine | N-nitrosomethylethylamine | oxidation | potent activation |
134, 244 | 11600130, 10837014 |
| CYP2E1 | chemical | nitrosamine | N-nitrosomethylpropylamine | oxidation | potent activation |
134, 244 | 11600130, 10837014 |
| CYP2E1 | chemical | nitrosamine | N-nitrosomorpholine | oxidation | activation |
132, 133, 326 |
11774366, 12214673, 10461547 |
| CYP2E1 | chemical | nitrosamine |
N-nitroso-N- methylbenzylamine |
oxidation | activation | 107 | 1486866 |
| CYP2E1 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
hydroxylation C5′- (lactol formation, low activity) |
activation |
134, 244, 247, 256, 257 |
11600130, 10837014, 1423839, 9276639, 7646564 |
| CYP2E1 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OH- tetrahydrofuran formation); oxidation |
activation |
132-134, 244, 327 |
11774366, 12214673, 11600130, 10837014, 17457417 |
| CYP2E1 | chemical | o-methoxyaniline |
o-anisidine (2- methoxyaniline) |
N-hydroxylation (major enzyme in microsomal model) |
potent activation |
201, 328 | 15828049, 21217841 |
| CYP2E1 | chemical | aromatic hydrocarbons, alkyl benzene |
styrene (vinyl benzene) | oxidation, medium Km, major enzyme in liver at low concentration, stereoselective |
activation |
66, 107, 202-205, 280, 282, 329 |
9685642, 1486866, 9253143, 7696548, 11407535, 12616646, 16125881, 1664256, 16872732 |
| CYP2E1 | chemical | haloalkene | tetrachloroethylene | oxidation | activation | 284 | 8671747 |
| CYP2E1 | chemical | haloalkene | trichloroethylene (TCE) | oxidation to trichloroethylene oxide and chloral hydrate form ation (major enzyme) |
activation |
107, 282, 330-333 |
1486866, 1664256, 9070354, 10807551, 11304134, 15987776 |
| CYP2E1 | chemical | vinyl halide | vinyl bromide (bromoethylene) |
oxidation | activation |
66, 107, 282 |
9685642, 1486866, 1664256 |
| CYP2E1 | chemical | carbamic acid derivative, metabolite |
vinyl carbamate | epoxide formation | activation |
66, 107, 282, 312 |
9685642, 1486866, 1664256, 11181492 |
| CYP2E1 | chemical | vinyl halide | vinyl chloride (chloroethylene) |
oxidation | potent activation |
66, 107, 282, 299 |
9685642, 1486866, 1664256, 8471158 |
| CYP2E1 | chemical | cyclohexane derivative | vinylcyclohexane | oxidation (epoxidation) | activation | 329 | 16872732 |
| CYP2F1 | natural compound |
indole, alkylating, pulmonary toxin; present in higher concentrations in mammalian digestive tract and coal tar |
3-methylindole, skatole | dehydrogenation (desaturation, 3- methyleneindolenine form., low Km, medium activity, high efficiency), major enzyme |
potent activation |
75-78, 334, 335 |
8558432, 11408359, 12563100, 20795680, 10383923, 17962375 |
| CYP2F1 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
hydroxylation, α-methyl (keto alcohol formation) |
activation | 213-216 | 1312898, 7595636, 8806763, 10803680 |
| CYP2F1 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 172 | 1651809 |
| CYP2F1 | chemical | PAH | naphthalene | oxidation | activation | 334 | 10383923 |
| CYP2F1 | chemical | aromatic hydrocarbon, alkyl benzene |
styrene (vinyl benzene) | oxidation (major enzyme in lung microsomes) |
activation |
202-205, 278 |
9253143, 7696548, 11407535, 12616646, 16125881 |
| CYP2W1 | chemical | PAH, metabolite | (±)-benzo[a]pyrene-7,8-diol | oxidation | activation | 210 | 16551781 |
| CYP2W1 | chemical | arylamine | 2-aminoanthracene | oxidation | activation | 210 | 16551781 |
| CYP2W1 | chemical | arylamine | 2-aminofluorene (2-AF) | oxidation | activation | 210 | 16551781 |
| CYP2W1 | chemical | heterocyclic amine | 3-amino-1,4-dimethyl-5H- pyrido[4,3-b]indole (Trp-P-1) |
oxidation | activation | 210 | 16551781 |
| CYP2W1 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene |
oxidation | activation | 210 | 16551781 |
| CYP2W1 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | oxidation | activation | 210 | 16551781 |
| CYP2W1 | Natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | oxidation | activation | 210 | 16551781 |
| CYP2W1 | chemical | PAH, metabolite | chrysene-1,2-diol | oxidation | activation | 210 | 16551781 |
| CYP2W1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
oxidation | activation | 210 | 16551781 |
| CYP2W1 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation | 210 | 16551781 |
| CYP3A4 | chemical | nitroarene | 1, 2-dihydro-1,2-dihydroxy- 6-nitrochrysene |
oxidation | activation | 219 | 8481905 |
| CYP3A4 | chemical | nitroarene | 6-nitrochrysene | oxidation | activation | 12 | 2655891 |
| CYP3A4 | chemical | nitroarene | 1,6-dinitropyrene | nitroreduction | activation | 174 | 10197616 |
| CYP3A4 | chemical | triazole | 1-aminobenzotriazole (1- ABT) |
oxidation | activation | 140 | 17584015 |
| CYP3A4 | chemical | arylamine, metabolite of 1-nitropyrene |
1-aminopyrene | oxidation | activation | 54 | 11525925 |
| CYP3A4 | chemical | nitroarene | 1-nitropyrene | epoxidation C4,5-, minor reaction |
activation |
54, 107, 174 |
11525925, 1486866, 10197616 |
| CYP3A4 | chemical | arylamine | 2-aminofluorene | oxidation | activation | 61, 336 | 9328287, 11377247 |
| CYP3A4 | chemical | nitroarene | 3,6-dinitrobenzo[e]pyrene | niroreduction and O- acetylation |
activation | 70 | 19393727 |
| CYP3A4 | chemical | heterocyclic amine | 3-amino-1,4-dimethyl-5H- pyrido[4,3-b]indole (Trp-P-1) |
N-hydroxylation | potent activation |
12, 61 | 2655891, 11377247 |
| CYP3A4 | chemical | heterocyclic amine | 3-amino-1-methyl-5H- pyrido[4,3-b]indole (Trp-P-2) |
N-hydroxylation | potent activation |
61 | 11377247 |
| CYP3A4 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene |
oxidation | activation | 336 | 9328287 |
| CYP3A4 | chemical | arylamine | 4,4′-methylene bis(2- chloroaniline) (MOCA) |
oxidation, N- (major enzyme) |
activation |
66, 107, 249 |
9685642, 1486866, 1740010 |
| CYP3A4 | Natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | epoxidation; oxidation, minor enzyme |
activation |
140, 172, 173 |
17584015, 1651809, 15892579 |
| CYP3A4 | chemical | nitroarene | 4-nitropyrene | 4-aminopyrene formation (nitroreduction), major enzyme |
activation | 174 | 10197616 |
| CYP3A4 | chemical |
N-heterocyclic aromatic hydrocarbon, dibenzocarbazole |
5,9- dimethyldibenzo[c,g]carbazol e |
oxidation | activation | 337 | 21798277 |
| CYP3A4 | chemical | arylamine | 6-aminochrysene | oxidation (high activity) | potent activation |
12, 84, 85, 107, 336, 338 |
2655891, 8118930, 8330339, 1486866, 9328287, 9493761 |
| CYP3A4 | chemical | arylamine, metabolite | 6-aminochrysene-1,2-diol | Diol epoxide formation; oxidation |
activation | 84, 85 | 8118930, 8330339 |
| CYP3A4 | chemical |
N-heterocyclic aromatic hydrocarbon, dibenzocarbazole |
7H-dibenzo[c,g]carbazole | oxidation | activation | 337 | 21798277 |
| CYP3A4 | chemical | PAH, aza-aromatic | 7-methylbenz[c]acridine | oxidation. | potent activation |
92 | 7866988 |
| CYP3A4 | chemical | PAH, metabolite | benzo[a]pyrene 7,8-diol | oxidation | activation | 12 | 2655891 |
| CYP3A4 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation exo-C8,9- (major activating enzyme), oxidation |
activation |
11, 12, 42, 57, 61, 175-180, 182, 336, 339-341 |
2492107, 2655891, 7955101, 15279838, 11377247, 2162057, 7766804, 8261428, 12079611, 1902334, 11782366, 16608170, 9328287, 1643250, 7545582, 7850790 |
| CYP3A4 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin G1 (AFG1) | oxidation | activation |
11, 12, 107, 148, 341- 343 |
2492107, 2655891, 1486866, 8082563, 7850790, 352361, 12849689 |
| CYP3A4 | chemical | herbicide, chloroacetamide |
alachlor | oxidation | activation | 274 | 11133395 |
| CYP3A4 | chemical | herbicide, chloroacetamide |
butachlor | oxidation | activation | 274 | 11133395 |
| CYP3A4 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation; oxidation |
activation |
108, 251- 254, 275, 344 |
19501186, 8242617, 9010702, 10348794, 10692561, 9923542, 15919850 |
| CYP3A4 | chemical | PAH), aza-aromatic | dibenz[a,j]acridine | 3,4-dihydrodiol formation | potent activation |
92 | 7866988 |
| CYP3A4 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C13- (major enzyme) and C12- (minor reaction); oxidation N2- (major enzyme |
potent activation |
39, 40, 287-290, 345 |
16936898, 21753906, 11755121, 12123750, 15548707, 17197724, 20576524 |
| CYP3A4 | physiologi cal compound |
estrogen | 17β-estradiol | C2-hydroxylation (major reaction, major enzyme, medium Km, medium efficiency, high activity), major metabolite and major enzyme in liver; oxidation, 2,3-quinone formation (lower activity); C4- hydroxylation (minor reaction, major enzyme, medium Km, medium activity, medium efficiency); oxidation, 3,4-quinone formation; C16α-hydroxylation (high Km, low activity) |
activation |
107, 122- 124, 126- 130, 190- 193, 346, 347 |
1486866, 9625734, 9054608, 9667077, 11555828, 12865317, 15784278, 16112414, 17570247, 1449532, 9635876, 11454902, 11741520, 10821664, 12124305 |
| CYP3A4 | physiologi cal compound |
estrogen | estrone | C2-hydroxylation (high Km, major metabolite, low activity); oxidation, 2,3- quinone formation; C4- hydroxylation (high Km, low activity, major enzyme); C16α- hydroxylation (high Km, low activity, major enzyme) |
activation |
48, 122- 124, 127, 128, 130, 191, 192 |
10426814, 9625734, 9054608, 9667077, 12865317, 15784278, 17570247, 9635876, 11454902 |
| CYP3A4 | drug | estradiol derivative; estrogen, contraceptive |
17α-ethynylestradiol (ethi- nylestradiol 17α-) |
oxygenation (2- hydroxylation, 17α- inactivation) |
activation | 140, 348 | 17584015, 17251390 |
| CYP3A4 | drug | antimitotic, epipodophyllotoxin, topoisomerase II inhibitor |
etoposide (VP-16) |
O-demethylation (catechol formation), high Km, high activity, major enzyme |
activation | 349-351 | 8114683, 9456308, 17168690 |
| CYP3A4 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (R)- (high Km, high activity), oxidation at high concentrations |
potent activation |
87, 251, 254, 275- 277, 344, 352-355 |
20507880, 8242617, 10692561, 15919850, 10534317, 15821045, 9923542, 8161344, 10101149, 10348794, 16854777 |
| CYP3A4 | chemical | herbicide, chloroacetamide |
metolachlor | oxidation | activation | 274 | 11133395 |
| CYP3A4 | natural compound |
alkaloid, pyrrolizidine, genotoxic |
monocrotaline | dehydrogenation | activation | 356 | 15649625 |
| CYP3A4 | chemical | aziridine |
N, N’, N”-triethylene thiophosphoramide (thioTEPA) |
desulfuration, TEPA formation (minor enzyme) |
activation | 279 | 12107550 |
| CYP3A4 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanabasine | oxidation (major enzyme) | activation | 132, 133 | 11774366, 12214673 |
| CYP3A4 | chemical | nitrosamine |
N-nitrosodibutylamine (N,N- dibutylnitrosamine) |
oxidation | activation | 134 | 11600130 |
| CYP3A4 | chemical | nitrosamine | N-nitrosodiethylamine (N,N-diethylnitrosamine) | oxidation | activation | 134, 255 | 11600130, 1423839 |
| CYP3A4 | chemical | nitrosamine | N-nitrosoethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP3A4 | chemical | nitrosamine | N-nitrosomethylbutylamine | oxidation | activation | 134 | 11600130 |
| CYP3A4 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N-nitrosonornicotine, NNN) | hydroxylation C2′- (keto alcohol formation); oxidation |
activation |
132, 133, 264, 265 |
11774366, 12214673, 9276639, 7646564 |
| CYP3A4 | chemical | nitrosamine | N-nitrosopiperidine | Cα-hydroxylation (2-OH- tetrahydropyran and 2- OH-5- methyltetrahydrofuran formation); oxidation |
activation | 132, 133 | 11774366, 12214673 |
| CYP3A4 | chemical | nitrosamine | N-nitrosopyrrolidine | Cα-hydroxylation (2-OH- tetrahydrofuran formation); oxidation |
activation | 132, 133 | 11774366, 12214673 |
| CYP3A4 | natural compound |
pyrrolizidine alkaloid from Senecio retrorsus |
retrorsine | dehydrogenation | activation | 356 | 15649625 |
| CYP3A4 | natural compound |
pyrrolizidine alkaloid, food contaminant (meat, milk, and honey) |
riddelline | dehydrogenation | activation | 356 | 15649625 |
| CYP3A4 | natural compound |
pyrrolizidine alkaloid, genotoxic |
senecionine | dehydrogenation | activation | 107, 357 | 1486866, 2009596 |
| CYP3A4 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation |
11, 12, 66, 107, 336, 341 |
2492107, 2655891, 9685642, 1486866, 9328287, 7850790 |
| CYP3A4 | drug | triphenylethyleneamin e; antiestrogen, estrogen receptor modulator |
tamoxifen | Cα-hydroxylation (major enzyme), catechol formation, oxidation, at high concentrations |
activation |
87, 354, 358-364 |
20507880, 10348797, 12018981, 12971802, 14678348, 15159443, 16533026, 12124303, 12419838 |
| CYP3A4 | chemical | organophosphate | tris(2,3- dibromopropyl)phosphate |
oxidation | activation | 12, 107 | 2655891, 1486866 |
| CYP3A5 | chemical | PAH, aza-aromatic | 7-methylbenz[c]acridine | oxidation | potent activation |
92 | 7866988 |
| CYP3A5 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation exo-8,9- (major reaction); oxidation |
activation | 182, 365 | 16608170, 7893152 |
| CYP3A5 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (very low activity) activation to cytotoxic metabolites |
activation | 253 | 10348794 |
| CYP3A5 | chemical | PAH, aza-aromatic | dibenz[a,j]acridine | 3,4-dihydrodiol formation | activation | 92 | 7866988 |
| CYP3A5 | physiologi cal compound |
estrogen | 17β-estradiol | C2-hydroxylation; C4- hydroxylation (major reaction); C16α- hydroxylation (low activity) |
activation |
127, 128, 191, 192, 347 |
12865317, 15784278, 9635876, 11454902, 12124305 |
| CYP3A5 | drug | estradiol derivative; estrogen, contraceptive |
17α-ethynylestradiol (ethinylestradiol, 17α-) |
oxygenation (2- hydroxylation, 17α- mechanism-based inactivation) |
activation | 140, 348 | 17584015, 17251390 |
| CYP3A5 | drug | antimitotic, epipodophyllotoxin, topoisomerase II inhibitor |
etoposide (VP-16) |
O-demethylation (catechol formation), medium Km, high activity, minor enzyme |
activation | 349 | 8114683 |
| CYP3A5 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation, stereoselective for (R)- |
activation |
253, 276, 277, 353, 355 |
10348794, 10534317, 15821045, 10101149, 16854777 |
| CYP3A5 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanabasine | oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP3A5 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP3A7 | chemical | arylamine | 2-aminofluorene (2-AF) | oxidation | activation | 336 | 9328287 |
| CYP3A7 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene |
oxidation | activation | 336 | 9328287 |
| CYP3A7 | chemical | arylamine | 6-aminochrysene | oxidation | activation | 336, 338 | 9328287, 9493761 |
| CYP3A7 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation exo-C8,9-; oxidation |
activation |
107, 182, 336, 338, 341, 366 |
1486866, 16608170, 9328287, 9493761, 7850790, 9044840 |
| CYP3A7 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin G1 (AFG1) | oxidation | activation | 341 | 7850790 |
| CYP3A7 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation | activation | 253, 275 | 10348794, 15919850 |
| CYP3A7 | physiologi cal compound |
estrogen | 17β-estradiol | C2-hydroxylation (medium Km, low activity, major reaction); C4- hydroxylation (low activity, high Km); C16α- hydroxylation (very low activity, high Km) |
activation |
127, 128, 347 |
12865317, 15784278, 12124305, |
| CYP3A7 | physiologi cal compound |
estrogen | estrone | C2-hydroxylation (medium Km, medium activity); C4- hydroxylation (low activity, medium Km); C16α-hydroxylation (medium Km, medium activity) |
activation | 127, 128 | 12865317, 15784278 |
| CYP3A7 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (R)- (high Km, medium (S-) and high (R-) activity, minor enzyme and reaction) |
activation | 253, 273 | 10348794, 15919850 |
| CYP3A7 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation |
336, 338, 341 |
9328287, 9493761, 7850790 |
| CYP4B1 | chemical | arylamine | 2-aminofluorene (2-AF) |
N-hydroxylation; oxidation |
activation | 367, 368 | 11396967, 11062028 |
| CYP4B1 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 172 | 1651809 |
| CYP4B1 | chemical | aromatic hydrocarbon | benzene | oxidation (at high conc., minor reaction) |
activation | 294 | 11083083 |
|
CYP19A1 (aromatase) |
physiologi cal compound |
estrogen | 17β-estradiol | C2-hydroxylation (medium Km) |
activation | 125, 369 | 8930523, 8476762 |
|
CYP19A1 (aromatase) |
physiologi cal compound |
estrogen | estrone | C2-hydroxylation (medium Km) |
activation | 125, 369 | 8930523, 8476762 |
|
Epoxide hydrolase, EH |
chemical | PAH, metabolite | benzo[a]pyrene-7,8-oxide | hydrolysis to benzo[a]pyrene -7,8-diol |
activation | 51, 220 | 21028851, 10409402 |
| FMO1 | chemical | thiocarbamide | thiourea |
S-oxidation (medium Km, high activity) |
activation | 370, 371 | 10901713, 14976351 |
| FMO2 | chemical | thiocarbamide | 1-phenylthiourea | S-oxidation | activation | 372 | 15144220 |
| FMO2 | chemical | thiocarbamide | α-naphthylthiourea | S-oxidation | activation | 372 | 15144220 |
| FMO2 | chemical | thiocarbamide | ethylenethiourea | S-oxidation | activation | 372 | 15144220 |
| FMO2 | chemical | thiocarbamide | thiourea |
S-oxidation (medium Km, high activity) |
activation | 372, 373 | 15144220, 11744609 |
| FMO3 | chemical | thiocarbamide | thiourea |
S-oxidation (medium Km, high activity) |
activation | 374 | 12093470 |
| GST | chemical | haloalkane | 1, 2-dibromoethane (ethylene dibromide) |
GSH conjugation | activation | 375-378 | 8330352, 16948056, 12542971, 15554237 |
| GST | chemical | haloalkane | 1, 2-dichloroethane, ethylene dichloride |
GSH conjugation | activation | 375, 378 | 8330352, 15554237 |
| GST | chemical | haloalkene | chlorotrifluoroethene | GSH conjugation | activation | 378 | 15554237 |
| GST | chemical | haloalkane | dichloromethane, methylene chloride |
GSH conjugation | activation |
284, 377, 378 |
8671747, 12542971, 15554237 |
| GST | chemical | haloalkene | hexachlorobutadiene | GSH conjugation | activation | 378 | 15554237 |
| GST | chemical | haloalkene | tetrafluoroethene | GSH conjugation | activation | 378 | 15554237 |
| GST | chemical | haloalkene | trichloroethene | GSH conjugation | activation | 378 | 15554237 |
| GST T1-1 | chemical | haloalkane | 1, 2-dibromoethane (ethylene dibromide) |
GSH conjugation | activation | 378 | 8565128 |
| GST T1-1 | chemical | haloalkane | dibromomethane (methylene dibromide) |
GSH conjugation | activation | 378 | 8565128 |
| GST T1-1 | chemical | haloalkane | diepoxybutane (butadiene diepoxide) |
GSH conjugation | activation | 379, 380 | 8565128, 222181695 |
| GST A1-1 | chemical | haloalkane | diepoxybutane, butadiene diepoxide |
GSH conjugation | activation | 380 | 222181695 |
| GST A3-3 | chemical | haloalkane | diepoxybutane (butadiene diepoxide) |
GSH conjugation | activation | 380 | 222181695 |
| GST M1-1 | chemical | haloalkane | diepoxybutane (butadiene diepoxide) |
GSH conjugation | activation | 380 | 222181695 |
| GST P1-1 | chemical | haloalkane | diepoxybutane (butadiene diepoxide) |
GSH conjugation | activation | 380 | 222181695 |
|
Lactoperoxi dase (LPO) |
chemical | arylamine, metabolite | 3-aminobenzanthrone | N-oxidation | activation | 72, 73 | 15885895, 16601755 |
|
Lactoperoxi dase (LPO) |
drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
Ellipticine | oxidation | activation | 39, 40 | 16936898, 21753906 |
|
Myeloperoxi dase (MPO) |
chemical | arylamine, metabolite | 3-aminobenzanthrone | N-oxidation | activation | 72, 73 | 15885895, 16601755 |
|
Myeloperoxi dase (MPO) |
drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | oxidation | activation | 39, 40 | 16936898, 21753906 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
1,2-dibromoethane (ethylene dibromide) |
conjugation | activation | 381, 382 | 12151404, 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
dibromomethane (methylene dibromide) |
conjugation | activation | 382, 383 | 15257623, 15206895 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
bromomethyl acetate | conjugation | activation | 383 | 15206895 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
dichloromethane (methylene dichloride) |
conjugation | activation | 382 | 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
bromochloromethane | conjugation | activation | 382 | 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
1,2-diiodoethane (ethylene diiodide) |
conjugation | activation | 382 | 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
1,2-bromochloroethane | conjugation | activation | 382 | 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
1,3-dibromopropane | conjugation | activation | 382 | 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
1,4-dibromobutane | conjugation | activation | 382 | 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
1,3-diiodopropane | conjugation | activation | 382 | 15257623 |
|
O6- alkylguanine DNA alkyl transferase (MGMT) |
chemical | bifunctional electrophile |
diepoxybutane (butadiene diepoxide) |
conjugation | activation | 382 | 15257623 |
|
NADPH- cytochrome P450 reductase (POR) |
chemical | nitroarene | 1,8-dinitropyrene | nitroreduction | activation | 53, 384 | 11113705, 15606153 |
|
NADPH- cytochrome P450 reductase (POR) |
chemical | nitroarene | 1-nitro-6-nitrosopyrene | reduction | activation | 384 | 15606153 |
|
NADPH- cytochrome P450 reductase (POR) |
chemical | nitroarene | 1-nitro-8-nitrosopyrene | reduction | activation | 384 | 15606153 |
|
NADPH- cytochrome P450 reductase (POR) |
chemical | nitroarene | 3,6-dinitrobenzo[e]pyrene | niroreduction and O- acetylation |
activation | 70 | 19393727 |
|
NADPH- cytochrome P450 reductase (POR) |
chemical | nitroarene | 3-nitrobenzanthrone | nitroreduction | activation | 80 | 12782579 |
|
NADPH- cytochrome P450 reductase (POR) |
chemical | quinone | anthraquinone | reduction | activation | 385 | 11697035 |
|
NADPH- cytochrome P450 reductase (POR) |
drug | dihydroxyanthraquino ne; laxative |
danthron | reduction | activation | 383 | 11697035 |
|
Xanthine oxidoreducta se (XOR) |
chemical | nitroarene | 2-nitroanisole | nitroreduction to hydroxylamine |
activation | 328 | 21217841 |
|
Xanthine oxidoreducta se (XOR) |
chemical | nitroarene | 3-nitrobenzanthrone | nitroreduction to hydroxylamine |
activation | 79 | 12740904 |
| NAT1 | chemical | nitroarene | 1,8-dinitropyrene |
O-acetylaton after nitroreduction |
activation | 386 | 10357791 |
| NAT1 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4- (diallylamino)-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2H-benzotriazole (PBTA-8) |
O-acetylation | activation | 55 | 18562244 |
| NAT1 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4- (diethylamino)-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2H- benzotriazole (PBTA-7) |
O-acetylation | activation | 55 | 18562244 |
| NAT1 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4-[bis(2- hydroxyethyl)amino]-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2H- benzotriazole (PBTA-6) |
O-acetylation | activation | 55 | 18562244 |
| NAT1 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4-amino- 5-methoxyphenyl]-5-amino- 7-bromo-4-chloro-2H- benzotriazole (PBTA-4) |
O-acetylation | activation | 55 | 18562244 |
| NAT1 | chemical | arylamine, metabolite | 2-acetylaminofluorene (2- AAF) |
O-acetylation after N-hydroxylation | potent activation |
147 | 15450435 |
| NAT1 | chemical | heterocyclic amine | 2-amino-6- methyldipyrido[1,2-a:3′,2′-d]- imidazole (Glu-P-1) |
O-acetylation after N- hydroxylation |
activation | 386 | 10357791 |
| NAT1 | chemical | arylamine | 2-aminofluorene (2-AF) |
O-acetylaton after N- hydroxylation |
activation | 386, 387 | 10357791, 1617672 |
| NAT1 | chemical | arylamine | 2-naphthylamine |
O-acetylation after N- hydroxylation |
activation | 147 | 15450435 |
| NAT1 | chemical | nitroaromatic | 2-nitrofluorene |
O-acetylaton after nitroreduction |
activation | 386 | 10357791 |
| NAT1 | chemical | nitroarene | 3-acetylaminobenzanthrone |
O-acetylation after N- hydroxylation, at higher conc. |
potent activation |
79 | 12740904 |
| NAT1 | chemical | arylamine, metabolite | 3-aminobenzanthrone |
O-acetylation after N- hydroxylation, at higher conccentrations |
potent activation |
79 | 12740904 |
| NAT1 | chemical | nitroarene | 3-nitrobenzanthrone |
O-acetylation after nitro- reduction to hydroxylamine, at higher concentrations |
potent activation |
73, 79, 388 | 16601755, 12740904, 12419844 |
| NAT1 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl (4-ABP) |
O-acetylation after N- hydroxylation |
potent activation |
147, 389 | 15450435, 8353847 |
| NAT1 | chemical | arylamine, heterocyclic |
aminophenylnorharman |
O-acetylation after N- hydroxylation |
activation | 95 | 17067997 |
| NAT1 | chemical | arylamine | benzidine |
O-acetylation after N- hydroxylation |
activation | 147, 387 | 15450435, 1617672 |
| NAT1 | chemical | arylamine, metabolite |
N-acetyl-N-hydroxy-3- aminobenzanthrone |
O-acetylation at higher concentrations |
potent activation |
79, 387 | 12740904, 12419844 |
| NAT1 | chemical | arylamine, tobacco smoke compound, metabolite |
N-hydroxy-4-aminobiphenyl (N-OH-4-ABP) |
O-acetylation | activation | 389 | 8353847 |
| NAT1 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-2-aminofluorene (N-OH-2-AF) |
O-acetylation | activation | 389 | 8353847 |
| NAT1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy- aminomethylphenylnorharma n |
O-acetylation | activation | 95 | 17067997 |
| NAT1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy- aminophenylnorharman |
O-acetylation | activation | 95 | 17067997 |
| NAT1 | chemical | nitroarene | nitrofen |
O-acetylation after nitroreduction |
activation | 390 | 14754874 |
| NAT1 | chemical | o-methoxyaniline |
o-anisidine, 2- methoxyaniline |
O-acetylation after N- hydroxylation |
activation | 147 | 15450435 |
| NAT2 | chemical | nitroarene | 1,8-dinitropyrene |
O-acetylaton after nitroreduction |
potent activation |
386 | 10357791 |
| NAT2 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4- (diallylamino)-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2Hbenzotriazole (PBTA-8) |
oxidation | activation | 55 | 18562244 |
| NAT2 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4- (diethylamino)-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2Hbenzotriazole (PBTA-7) |
oxidation | activation | 55 | 18562244 |
| NAT2 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4-[bis(2- hydroxyethyl)amino]-5- methoxyphenyl]-5-amino-7- bromo-4-chloro-2H- benzotriazole (PBTA-6) |
oxidation | activation | 55 | 18562244 |
| NAT2 | chemical | 2-phenylbenzotriazole | 2-[2-(acetylamino)-4-amino- 5-methoxyphenyl]-5-amino- 7-bromo-4-chloro-2H- benzotriazole (PBTA-4) |
oxidation | activation | 55 | 18562244 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
2-amino-3,4- dimethylimidazo[4,5- f]quinolone (MeIQ) |
O-acetylation after N- hydroxylation |
activation | 386, 387 | 10357791, 1617672 |
| NAT2 | chemical | heterocyclic amine | 2-amino-6- methyldipyrido[1,2-a:3′,2′-d]- imidazole (Glu-P-1) |
O-acetylation after N- hydroxylation |
activation | 386 | 10357791 |
| NAT2 | chemical | nitroarene | 2-nitrobenzanthrone |
O-acetylation after reduction to hydroxylamine |
activation | 391 | 17483118 |
| NAT2 | chemical | nitroarene | 3-acetylaminobenzanthrone |
O-acetylation after Nhydroxylation, at higher concentration |
potent activation |
79 | 12740904 |
| NAT2 | chemical | arylamine, metabolite | 3-aminobenzanthrone |
O-acetylation after Nhydroxylation, at higher concentration |
potent activation |
79 | 12740904 |
| NAT2 | chemical | nitroarene | 3-nitrobenzanthrone |
O-acetylation after nitro- reduction to hydroxylamine, at higher concentration |
potent activation |
73, 79, 388, 391 |
16601755, 12740904, 12419844, 17483118 |
| NAT2 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl (4-ABP) |
O-acetylation after N- hydroxylation |
activation | 147, 389 | 15450435, 8353847 |
| NAT2 | chemical | arylamine | 6-aminochrysene |
O-acetylation after N- hydroxylation |
activation | 386 | 10357791 |
| NAT2 | chemical | arylamine, heterocyclic |
aminomethylphenylnorharma n |
O-acetylation after N-hydroxylation | activation | 95 | 17067997 |
| NAT2 | chemical | arylamine, heterocyclic |
aminophenylharman | O-acetylation after N-hydroxylation | activation | 95 | 17067997 |
| NAT2 | chemical | arylamine, heterocyclic |
aminophenylnorharman |
O-acetylation after N- hydroxylation |
activation | 95 | 17067997 |
| NAT2 | chemical | arylamine, metabolite |
N-acetyl-N-hydroxy-3- aminobenzanthrone |
O-acetylation at higher conc. |
potent activation |
79, 388 | 12740904, 12419844 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methylimidazo[4,5- f]quinolone (N-hydroxyisoIQ) |
O-acetylation | activation | 392 | 7697826 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-3,4- dimethylimidazo[4,5- f]quinolone (N-hydroxy- MeIQ) |
O-acetylation | activation | 392 | 7697826 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (N-hydroxy- MeIQx) |
O-acetylation | activation | 392 | 7697826 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-3- methylimidazo[4,5- f]quinolone (N-hydroxy-IQ) |
O-acetylation | potent activation |
392, 393 | 7697826, 12067565 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-6- methyldipyrido[1,2-a:3′,2′-d]- imidazole (N-hydroxy-Glu-P- 1) |
O-acetylation | activation | 392 | 7697826 |
| NAT2 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-2- aminobenzanthrone |
O-acetylation | activation | 391 | 17483118 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2- aminodipyrido[1,2-a:3,2′-d]- imidazole (N-hydroxy-Glu-P- 2) |
O-acetylation | ativation | 392 | 7697826 |
| NAT2 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-3- aminobenzanthrone |
O-acetylation | activation | 391 | 17483118 |
| NAT2 | chemical | arylamine, tobacco smoke compound, metabolite |
N-hydroxy-4-aminobiphenyl (N-OH-4-ABP) |
O-acetylation | activation | 389 | 8353847 |
| NAT2 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxyaminofluorene (N-OH-2-AF) |
O-acetylation | activation | 389 | 8353847 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N- hydroxyaminomethylphenyln orharman |
O-acetylation | activation | 95 | 17067997 |
| NAT2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N- hydroxyaminophenylnorharm an |
O-acetylation | activation | 95 | 17067997 |
| NAT2 | chemical | o-methoxyaniline |
o-anisidine, 2- methoxyaniline |
O-acetylation after N- hydroxylation |
activation | 147 | 15450435 |
|
Prostaglandi n H synthase (PHS, COX) |
chemical | arylamine, metabolite | 3-aminobenzanthrone | N-oxidation | activation | 72, 73 | 15885895, 16601755 |
|
Quinone oxidoreducta se (NQO1) |
chemical | nitroarene | 3-nitrobenzanthrone | nitroreduction (major enzyme) |
potent activation |
73, 80 | 16601755, 12782579 |
| SULT1A1 | chemical | PAH, metabolite | (−)-1-hydroxyethylpyrene | O-sulfonation | activation | 169, 394 | 10720750, 11535246 |
| SULT1A1 | chemical | PAH, metabolite | 1-(1-pyrenyl)ethanol | O-sulfonation | activation | 395 | 9141497 |
| SULT1A1 | chemical | PAH, metabolite | 1-hydroxymethylpyrene | O-sulfonation | potent activation |
169, 390, 396, 397 |
10720750, 14754874, 10657971, 17936463 |
| SULT1A1 | natural compound |
1-methoxy-3- indolylmethyl glucosinolate breakdown product, in many Brassica vegetables |
1-methoxy-3-indolylmethyl- alcohol |
O-sulfonation | activation | 398 | 20846518 |
| SULT1A1 | chemical | benzylic alcohol, nitroatromatic |
2,4-dinitrobenzylalcohol | O-sulfonation | activation |
169, 394, 399 |
10720750, 11535246, 11154739 |
| SULT1A1 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
O-sulfonation after oxidation |
activation | 400 | 22006426 |
| SULT1A1 | chemical | heterocyclic amine | 2-amino-6- methyldipyrido[1,2-a:3′,2′-d]- imidazole (Glu-P-1) |
O-sulfonation after N- hydroxylation |
potent activation |
401 | 22072630 |
| SULT1A1 | chemical | benzylic alcohol | 2-aminobenzylalcohol | O-sulfonation | activation | 3979 | 11154739 |
| SULT1A1 | chemical | arylamine, heterocyclic |
2-hydroxylamino-3-methyl- 9H-pyrido[2,3-b]indole (N- OH-MeAαC) |
O-sulfonation | potent activation |
160 | 14729582 |
| SULT1A1 | chemical | hydroxylamine, heterocyclic |
2-hydroxylamino-5- phenylpyridine |
O-sulfonation | activation | 394 | 11535246 |
| SULT1A1 | chemical | nitroarene | 2-nitrobenzanthrone |
O-sulfonation after reduction to hydroxylamine |
activation | 391 | 17483118 |
| SULT1A1 | chemical | nitroarene | 3,9-dinitrofluoranthene | O-sulfonation | potent activation |
401 | 22072630 |
| SULT1A1 | chemical | nitroarene | 3-acetylaminobenzanthrone |
O-sulfonation after N- hydroxylation, at higher concentrations |
potent activation |
79 | 12740904 |
| SULT1A1 | chemical | arylamine, metabolite | 3-aminobenzanthrone |
O-sulfonation after N- hydroxylation, at higher concentrations |
potent activation |
79 | 12740904 |
| SULT1A1 | chemical | azoaromatic amine | 3-methoxy-4- aminoazobenzene |
O-sulfonation | potent activation |
401 | 22072630 |
| SULT1A1 | chemical | nitroarene | 3-nitrobenzanthrone |
O-sulfonation after nitroreduction to hydroxylamine |
potent activation |
73, 79, 364, 391, 401 |
16601755, 12740904, 12419844, 17483118, 22072630 |
| SULT1A1 | drug | triphenylethyleneamin e; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen | O-sulfonation | activation | 402, 403 | 12034366, 21537383 |
| SULT1A1 | chemical | furaldehyde derivative | 5-hydroxymethylfurfural | O-sulfonation | activation |
322, 397, 400 |
15668106, 17936463, 22006426 |
| SULT1A1 | chemical | nitroaromatic | 5-nitroacenaphthene | O-sulfonation | potent activation |
401 | 22072630 |
| SULT1A1 | chemical | PAH, metabolite | 7-hydroxy-7,8,9,10- tetrahydrobenzo[a]pyrene |
O-sulfonation | activation | 395 | 9141497 |
| SULT1A1 | natural compound |
nephrotoxin, Aristolochia fangchi compound, nitroarene |
aristolochic acids I and II |
O-sulfonation after nitroreduction to hydroxylamine |
potent activation |
404 | 16161050 |
| SULT1A1 | chemical | furan | furan |
O-sulfonation after oxidation |
activation | 322 | 15668106 |
| SULT1A1 | chemical | furan derivative | furfuryl alcohol | O-sulfonation | activation | 405 | 21729924 |
| SULT1A1 | chemical | arylamine, metabolite |
N-acetyl-N-hydroxy-3- aminobenzanthrone |
O-sulfonation, at higher concentrations |
potent activation |
79, 364 | 12740904, 12419844 |
| SULT1A1 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2-acetylamino-3- methyl-5-phenylpyridine |
O-sulfonation | activation | 399 | 11154739 |
| SULT1A1 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH-2-AAF) |
O-sulfonation | activation | 169, 406 | 10720750, 11535243 |
| SULT1A1 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2-acetylaminol-5- phenylpyridine |
O-sulfonation | activation | 399 | 11154739 |
| SULT1A1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-hydroxy-PhIP) |
O-sulfonation | potent activation |
393, 399 | 12067565, 11154739 |
| SULT1A1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-3- methylimidazo[4,5- f]quinolone (N-hydroxy-IQ) |
O-sulfonation | potent activation |
407 | 16708048 |
| SULT1A1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-6- methyldipyrido[1,2-a:3′,2′-d]- imidazole (N-hydroxy-Glu-P- 1) |
O-sulfonation | activation | 408 | 7834621 |
| SULT1A1 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-2- aminobenzanthrone |
O-sulfonation | activation | 401 | 17483118 |
| SULT1A1 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-3- aminobenzanthrone |
O-sulfonation | activation | 391 | 17483118 |
| SULT1A1 | chemical | arylamine, tobacco smoke compound, metabolite |
N-hydroxy-4-aminobiphenyl (N-OH-4-ABP) |
O-sulfonation | activation | 409 | 7859374 |
| SULT1A1 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-2-aminofluorene (N-OH-4-AF) |
O-sulfonation | activation | 408 | 7834621 |
| SULT1A1 | chemical | nitroarene | nitrofen |
O-sulfonation after nitroreduction to hydroxylamine |
potent activation |
322, 390 | 15668106, 14754874 |
| SULT1A1 | chemical | nitrosamine |
N-nitrosodimethylamine (N, N-dimethylnitrosamine, DMN) |
O-sulfonation after oxidation |
activation | 322 | 15668106 |
| SULT1A1 | chemical | heterocyclic amine |
N-OH-4,4′-methylenebis(2- chloroaniline) (N-OHMOCA) |
O-sulfonation | activation | 408 | 7834621 |
| SULT1A2 | chemical | PAH, metabolite | (−)-1-hydroxyethylpyrene | O-sulfonation | activation | 394 | 11535246 |
| SULT1A2 | chemical | PAH, metabolite | 1-hydroxymethylpyrene | O-sulfonation | activation | 394, 410 | 11535246, 12464797 |
| SULT1A2 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
O-sulfonation after oxidation |
activation | 400 | 22006426 |
| SULT1A2 | chemical | arylamine, heterocyclic |
2-hydroxylamino-3-methyl- 9H-pyrido[2,3-b]indole (N- OH-MeAαC) |
O-sulfonation | activation | 159 | 14729582 |
| SULT1A2 | chemical | hydroxylamine, heterocyclic |
2-hydroxylamino-5- phenylpyridine |
O-sulfonation | potent activation |
394, 410 | 11535246, 12464797 |
| SULT1A2 | chemical | nitroarene | 3-acetylaminobenzanthrone |
O-sulfonation after N- hydroxylation, at higher concentrations |
potent activation |
79 | 12740904 |
| SULT1A2 | chemical | arylamine, metabolite | 3-aminobenzanthrone |
O-sulfonation after N- hydroxylation, at higher concentrations |
potent activation |
79 | 12740904 |
| SULT1A2 | chemical | nitroarene | 3-nitrobenzanthrone |
O-sulfonation after nitroreduction to hydroxylamine |
activation | 73, 79 | 16601755, 12740904 |
| SULT1A2 | chemical | furaldehyde derivative | 5-hydroxymethylfurfural | O-sulfonation | activation | 400 | 22006426 |
| SULT1A2 | chemical | arylamine, metabolite |
N-acetyl-N-hydroxy-3- aminobenzanthrone |
O-sulfonation, at higher concentrations |
potent activation |
79, 388 | 12740904, 12419844 |
| SULT1A2 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2-acetylamino-3- methyl-5-phenylpyridine |
O-sulfonation | activation | 399 | 11154739 |
| SULT1A2 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
O-sulfonation | potent activation |
169, 394, 410 |
10720750, 11535246, 12464797 |
| SULT1A2 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2-acetylaminol-5- phenylpyridine |
O-sulfonation | activation | 399 | 11154739 |
| SULT1A2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
O-sulfonation | potent activation |
393, 399 | 12067565, 11154739 |
| SULT1A2 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-2-aminofluorene (N-OH-2-AF) |
O-sulfonation | potent activation |
401 | 22072630 |
| SULT1A3 | natural compound |
alkenylbenzene; occurs in a variety of foods including essential oils of tarragon, sweet basil, sweet fennel, anis, metabolite |
1′-hydroxyestragole | O-sulfonation | activation | 197, 401 | 21459083, 22072630 |
| SULT1A3 | chemical | PAH, metabolite | 1-hydroxymethylpyrene | O-sulfonation | activation |
169, 394, 396 |
10720750, 11535246, 10657971 |
| SULT1A3 | chemical | nitroalkane | 2-nitropropane |
O-sulfonation, propane 2- nitronate formation |
activation | 169, 396 | 1072075, 10657971 |
| SULT1A3 | natural compound |
alkenylbenzene; occurs in a variety of foods including essential oils of tarragon, sweet basil, sweet fennel, anis |
estragole |
O-sulfonation after C1′- hydroxylation |
activation | 401 | 22072630 |
| SULT1A3 | chemical | nitroarene | nitrofen |
O-sulfonation after nitroreduction to hydroxylamine |
activation | 390 | 14754874 |
| SULT1B1 | chemical | PAH, aldehyde | 1-formylpyrene | O-sulfonation | activation | 399 | 11154739 |
| SULT1B1 | chemical | PAH, metabolite | 1-hydroxymethylpyrene | O-sulfonation | activation | 394 | 11535246 |
| SULT1B1 | chemical | arylamine, heterocyclic |
2-hydroxylamino-3-methyl- 9H-pyrido[2,3-b]indole (N- OH-MeAαC) |
O-sulfonation | activation | 159 | 14729582 |
| SULT1B1 | chemical | hydroxylamine, heterocyclic |
2-hydroxylamino-5- phenylpyridine |
O-sulfonation | activation | 394 | 11535246 |
| SULT1B1 | chemical | PAH, metabolite | 4- hydroxycyclopenta[d,e.f]chry sene |
O-sulfonation | potent activation |
169, 394 | 10720750, 11535246 |
| SULT1B1 | chemical | PAH, metabolite | 6- hydroxymethylbenzo[a]pyren e |
O-sulfonation | potent activation |
394 | 11535246 |
| SULT1B1 | natural compound |
nephrotoxin, Aristolochia fangchi compound, nitroarene |
aristolochic acids I and II |
O-sulfonation after nitroreduction to hydroxylamine |
activation | 394 | 16161050 |
| SULT1C1 | chemical | PAH, metabolite | (−)-1-hydroxyethylpyrene | O-sulfonation | activation | 392 | 11535246 |
| SULT1C1 | chemical | arylamine, heterocyclic |
2-hydroxylamino-3-methyl- 9H-pyrido[2,3-b]indole (N- OH-MeAαC) |
O-sulfonation | activation | 169 | 14729582 |
| SULT1C1 | chemical | nitroarene | nitrofen |
O-sulfonation after nitroreduction to hydroxylamine |
activation | 390 | 14754874 |
| SULT1C2 | chemical | PAH, metabolite | (−)-1-hydroxyethylpyrene | O-sulfonation | activation | 394 | 11535246 |
| SULT1C2 | chemical | PAH, metabolite | 1-hydroxymethylpyrene | O-sulfonation | activation | 394 | 11535246 |
| SULT1C2 | chemical | furan derivative | 2,5-(bishydroxymethyl)furan | O-sulfonation | activation | 411 | 21825114 |
| SULT1C2 | chemical | furan derivative | 5-methylfurfural | O-sulfonation | activation | 411 | 21825114 |
| SULT1C2 | chemical | furan derivative | furfuryl alcohol | O-sulfonation | activation | 411 | 21825114 |
| SULT1C3 | chemical | PAH, metabolite | (±)-, (+)-, and (−)-1- hydroxyethylpyrene |
O-sulfonation | activation | 397 | 17936463 |
| SULT1C3 | chemical | methylenedioxyphenyl , benzodioxole, metabolite |
1′-hydroxysafrole |
O-sulfonation, at high conc. |
activation | 397 | 17936463 |
| SULT1C3 | chemical | PAH, metabolite | 6- hydroxymethylanthanthrene |
O-sulfonation | potent activation |
397 | 17936463 |
| SULT1C3 | chemical | PAH, metabolite | 6- hydroxymethylbenzo[a]pyren e |
O-sulfonation | potent activation |
397 | 17936463 |
| SULT1E1 | chemical | PAH, metabolite | (±)-, (+)-, and (−)-1- hydroxyethylpyrene |
O-sulfonation | potent activation |
169, 394, 397 |
10720750, 11535246, 17936463 |
| SULT1E1 | chemical | PAH, metabolite | 1-(1-pyrenyl)ethanol | O-sulfonation | activation | 395 | 9141497 |
| SULT1E1 | chemical | PAH, metabolite | 10-hydroxy-7,8,9,10- tetrahydrobenzo[a]pyrene |
O-sulfonation | activation | 395 | 9141497 |
| SULT1E1 | chemical | PAH, metabolite | 1-hydroxymethylpyrene | O-sulfonation | potent activation |
394, 396, 397 |
11535246, 10657971, 17936463 |
| SULT1E1 | chemical | PAH, metabolite | 4- hydroxycyclopenta[def]chrys ene |
O-sulfonation | activation | 169, 394 | 10720750, 11535246 |
| SULT2A1 | chemical | PAH, metabolite | (±)-, (+)-, and (−)-1- hydroxyethylpyrene |
O-sulfonation | activation |
169, 394, 397 |
10720750, 11535246, 17936463 |
| SULT2A1 | chemical | PAH, metabolite | 1-(1-pyrenyl)ethanol, I-HEP | O-sulfonation | activation | 395 | 9141497 |
| SULT2A1 | chemical | PAH, metabolite | 1-hydroxy-3- methylcholanthrene |
O-sulfonation | activation | 395 | 9141497 |
| SULT2A1 | chemical | PAH, metabolite | 1-hydroxymethylpyrene | O-sulfonation | potent activation |
169, 394, 396 |
10720750, 11535246, 10657971 |
| SULT2A1 | chemical | PAH, metabolite | 2-hydroxymethylpyrene | O-sulfonation | activation | 395 | 9141497 |
| SULT2A1 | drug | pregnane, antiandrogen, metabolite |
3α-hydroxycyproterone acetate |
O-sulfonation | activation | 394 | 11535246 |
| SULT2A1 | chemical | PAH, metabolite | 6- hydroxymethylbenzo[a]pyren e |
O-sulfonation | potent activation |
169, 394 | 10720750, 11535246 |
| SULT2A1 | chemical | PAH, metabolite | 7-hydroxy-12- methylbenz[a]anthracene |
O-sulfonation | activation | 394 | 9141497 |
| SULT2A1 | drug | triphenylethyleneamin e; antiestrogen, estrogen receptor modulator, metabolite |
α-hydroxytamoxifen | O-sulfonation | activation | 412, 413 | 9855017, 15371299 |
| SULT2A1 | drug | thioxanthenone; schistosomicide |
hycanthone | O-sulfonation | potent activation |
394, 395 | 11535246, 9141497 |
| SULT2E1 | chemical | PAH | 1-acetylpyrene | O-sulfonation | potent activation |
399 | 11154739 |
| SULT2E1 | drug | thioxanthenone; schistosomicide |
hycanthone | O-sulfonation | activation | 394 | 11535246 |
Table 4. Detoxication Reactions.
| enzyme | category | subcategory | compound | reaction | references | PubMed ID |
|---|---|---|---|---|---|---|
| AKR1B1 | natural compound |
carbonyl, unsaturated | 4-hydroxynonenal | reduction | 414 | 21329684 |
| AKR1B1 | natural compound |
carbonyl, unsaturated | acrolein | reduction (low activity) |
414 | 21329684 |
| AKR1B1 | natural compound |
carbonyl, unsaturated | crotonaldehyde | reduction (low activity) |
414 | 21329684 |
| AKR1B1 | natural compound |
GSH conjugate, metabolite |
GS-2-hexenal | reduction (high activity) |
414 | 21329684 |
| AKR1B1 | natural compound |
carbonyl, unsaturated | GS-4-hydroxynonanal | reduction (high activity) |
414 | 21329684 |
| AKR1B1 | natural compound |
carbonyl, unsaturated | GS-butanal | reduction (high activity) |
414 | 21329684 |
| AKR1B1 | natural compound |
carbonyl, unsaturated | GS-propanal | reduction | 414 | 21329684 |
| AKR1B1 | natural compound |
GSH conjugate, metabolite |
GS-trans-2-hexenal | reduction | 414 | 21329684 |
| AKR1B1 | natural compound |
carbonyl, unsaturated | trans-2,4-hexedinal | reduction | 414 | 21329684 |
| AKR1B1 | natural compound |
carbonyl, unsaturated | trans-2-hexenal | reduction | 414 | 21329684 |
| AKR1B10 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
reduction | 415 | 16381663 |
| AKR1B10 | natural compound |
carbonyl, unsaturated | 4-hydroxynonenal | reduction (high activity) |
414, 416, 417 |
21329684 19013440, 19563777, |
| AKR1B10 | natural compound |
carbonyl, unsaturated | 4-methylpentanal | reduction | 416 | 19013440 |
| AKR1B10 | natural compound |
carbonyl | 4-oxonon-2-enal | reduction | 416 | 19013440 |
| AKR1B10 | natural compound |
carbonyl | acrolein | reduction | 414, 417 | 21329684 19563777 |
| AKR1B10 | natural compound |
carbonyl | crotonaldehyde | reduction (high activity) |
414, 417 | 21329684 19563777 |
| AKR1B10 | natural compound |
GSH conjugate, metabolite |
GS-2-hexenal | reduction | 414 | 21329684 |
| AKR1B10 | natural compound |
GSH conjugate, metabolite |
GS-acrolein | reduction | 417 | 19563777 |
| AKR1B10 | natural compound |
carbonyl, unsaturated | GS-butanal | reduction | 414 | 21329684 |
| AKR1B10 | natural compound |
GSH conjugate, metabolite |
GS-crotonaldehyde | reduction | 417 | 19563777 |
| AKR1B10 | natural compound |
carbonyl, unsaturated | GS-propanal | reduction (low activity) |
414 | 21329684 |
| AKR1B10 | natural compound |
GSH conjugate, metabolite |
GS-trans-2, 4-hexadienal | reduction | 417 | 19563777 |
| AKR1B10 | natural compound |
GSH conjugate, metabolite |
GS-trans-2-hexenal | reduction (high activity) |
414, 417 | 21329684 19563777 |
| AKR1B10 | natural compound |
carbonyl, unsaturated | trans-2, 4-hexadienal | reduction | 417 | 19563777 |
| AKR1B10 | natural compound |
carbonyl, unsaturated | trans-2,4-hexedinal | reduction | 414 | 21329684 |
| AKR1B10 | natural compound |
carbonyl, unsaturated | trans-2-hexenal | reduction (high activity) |
414, 417 | 21329684 19563777 |
| AKR1C1 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
reduction | 418, 419 | 11037109, 11306090 |
| AKR1C2 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
reduction | 418, 419 | 11037109, 11306090 |
| AKR1C4 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
reduction | 418, 419 | 11037109, 11306090 |
| AKR7A2 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 dialdehyde | reduction | 420, 421 | 10383892, 17537398 |
| AKR7A3 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products, metabolite |
aflatoxin B1 dialdehyde | reduction | 420-422 | 10383892, 17537398, 18416522 |
| CYP1A1 | chemical | nitroarene | 2-nitroanisole | demethylation, O- hydroxylation, C2-, C2-C5-, and C2- C6- |
423, 424 | 15144223, 17159769 |
| CYP1A1 | natural compound |
nephrotoxin, Aristolochia fangchi compound, nitroarene |
aristolochic acid I | hydroxylation, C8- (major enzyme) |
99 | 22086975 |
| CYP1A1 | chemical | PAH | benzo[a]pyrene | hydroxylation, 3- (low Km, or medium Km, medium activity, or high activity) |
44, 46, 48, 104-106, 425-427 |
11238186, 7581497, 8293790, 9806168, 11513247, 8037457, 7766605, 9152602, 7703357 |
| CYP1A1 | chemical | PAH | benzo[a]pyrene | hydroxylation 3- (low activity) |
44, 138 | 11238186 9014198 |
| CYP1A1 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor, and DNA binding |
ellipticine | hydroxylation, C7- and C9- (major enzyme, low activity) |
39, 187, 189 | 16936898, 15548707, 21683692 |
| CYP1A1 | chemical | aza-aromatic | Sudan I | hydroxylation, C4′- (high activity), hydroxylation, C6- (low activity); oxidation, major enzyme |
136, 137 | 12384524, 17159775 |
| CYP1A2 | chemical | heterocyclic amine | 2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (MeIQx) |
C8-oxidation (carboxylic acid form.) |
428, 429 | 10220313, 11258970 |
| CYP1A2 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | hydroxylation (AFM1 formation, major enzyme and reaction); hydroxylation, C3α- (AFQ1 formation, low activity); demethylation, O- (AFP1 formation, very low activity) |
176-178 | 7766804, 8261428, 12079611 |
| CYP1A2 | natural compound |
nephrotoxin, Aristolochia fangchi compound, nitroarene |
aristolochic acid I | hydroxylation, C8- (major enzyme) |
99 | 22086975 |
| CYP1A2 | chemical | PAH | benzo[a]pyrene | 3- hydroxylation (low activity) |
46, 104, 106, 425- 427, 430 |
7581497, 9806168, 8037457, 7766605, 9152602, 7703357, 1551116 |
| CYP1A2 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C7- and C9- (major enzyme in liver, low activity) |
39, 187, 189 | 16936898, 15548707, 21683692 |
| CYP1A2 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | C7- hydroxylation (major enzyme) |
187 | 15548707 |
| CYP1B1 | chemical | PAH | benzo[a]pyrene | 3-hydroxylation |
104, 222, 426 |
9806168, 15958554, 9152602 |
| CYP1B1 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C7- (very low activity) and C9- |
187 | 15548707 |
| CYP2A6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
oxidation, N- | 265 | 12975327 |
| CYP2A6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
oxidation, N- | 265 | 12975327 |
| CYP2A6 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | hydroxylation (AFM1 formation, low activity); hydroxylation, C3α- (AFQ1 formation, low activity); demethylation, O- (AFP1 formation, major reaction), at high substrate concentrations |
181 | 16385575 |
| CYP2A13 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
oxidation, N- | 265 | 12975327 |
| CYP2A13 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | hydroxylation (AFM1 formation, low activity); hydroxylation, C3α- (AFQ1 formation, low activity); demethylation, O- (AFP1 formation, very low activity) |
181 | 16385575 |
| CYP2B6 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
oxidation, N- | 265 | 12975327 |
| CYP2C19 | chemical | PAH | benzo[a]pyrene | hydroxylation 3- |
138, 425, 427 |
9014198, 7766605, 7703357, |
| CYP2C8 | chemical | PAH | benzo[a]pyrene | hydroxylation 3- |
425, 427, 430 |
7766605, 7703357, 1551116 |
| CYP2C9 | chemical | PAH | benzo[a]pyrene | hydroxylation 3- (major enzyme) |
138, 425, 427, 430 |
9014198, 7766605, 7703357, 1551116 |
| CYP2D6 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C7- (low activity) and C9- |
187 | 15548707 |
| CYP2E1 | chemical | nitroarene | 2-nitroanisole | demethylation, O- hydroxylation, C2-, C2-C5-, and C2- C6- (major enzyme) |
423, 424 | 15144223, 17159769 |
| CYP2E1 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanone (NNK) |
oxidation, N- | 265 | 12975327 |
| CYP2E1 | chemical | PAH | benzo[a]pyrene | hydroxylation 3- (low activity) |
425, 427 | 7766605, 7703357 |
| CYP3A4 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | hydroxylation, C3α- (AFQ1 form.), medium Km, medium activity, major reaction, major enzyme |
176-178, 339 |
7766804, 8261428, 12079611, 1643250 |
| CYP3A4 | chemical | PAH | benzo[a]pyrene | hydroxylation 3- (major enzyme) |
138, 425- 427, 430 |
9014198, 7766605, 9152602, 7703357, 1551116 |
| CYP3A4 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C9- (very low activity) |
187 | 15548707 |
| CYP3A4 | chemical | aza-aromatic | Sudan I | hydroxylation, C4′- and C6 (low activity) |
136 | 12384524 |
| CYP3A5 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | hydroxylation, C3α- (AFQ1 form.) |
182, 365 | 16608170, 7893152 |
| CYP3A7 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | hydroxylation, C3α - (AFQ1 formation) |
182 | 16608170 |
|
Epoxide hydrolase, EH |
chemical | haloalkene, metabolite | chloroprene epoxide (1- chloroethenyl oxirane) |
hydrolysis | 431 | 14565770 |
|
Epoxide hydrolase, EH |
natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products, metabolite |
aflatoxin B1-8,9-epoxide | hydrolysis, aflatoxin-8,9- dihydrodiol formation |
432, 433 | 8781383, 9115980 |
|
Epoxide hydrolase, EH |
chemical | PAH, metabolite | benzo[a]pyrene-7,8-oxide | hydrolysis to benzo[a]pyrene- 7,8-diol |
52, 220 | 21028851, 10409402 |
| GST | physiological compound |
estrogen, metabolite | 4-hdroxyestrone-o-quinone | covalent binding | 434 | 18588320 |
| GST | chemical | acrylic amide | acrylamide | GSH conjugation | 435 | 19904761 |
| GSTM | chemical | aromatic hydrocarbon, alkyl benzene, metabolite |
styrene 7,8-oxide | GSH conjugation | 436, 437 | 3224538, 3692493 |
| GSTA1 | chemical | PAH | dibenzo[a,l]pyrene | GSH conjugation after oxidation |
119 | 17509623 |
| GSTA1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
GSH conjugation after oxidation |
119, 438 | 17509623, 9855012 |
| GSTA1-1 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-dihydrodiol 9,10-epoxide |
GSH conjugation (high activity) |
439-441 | 8706254, 11849043, 12067250 |
| GSTA1-1 | drug | oxazaphosporine; anticancer, mitrogen mustard, alkylating, metabolite |
4-hydroxycyclophosphamide | GSH conjugation | 442 | 7954469 |
| GSTA1-1 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products, metabolite |
aflatoxin B1-8,9-epoxide | GSH conjugation | 375, 443 | 8330352, 9675258 |
| GSTA1-1 | chemical | PAH, metabolite | benzo[c]chrysene-9,10-diol 11,12-epoxide |
GSH conjugation | 439 | 8706254 |
| GSTA1-1 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol 1,2-epoxide |
GSH conjugation | 439 | 8706254 |
| GSTA1-1 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol 13,14-epoxide |
GSH conjugation | 439 | 8706254 |
| GSTA1-1 | chemical | PAH, metabolite | chrysene-trans-1,2- dihydrodiol-3,4-epoxide |
GSH conjugation | 439 | 8706254 |
| GSTA1-1 | chemical | PAH, metabolite | dibenz[a,h]anthracene-3,4- dihydrodiol 1,2-epoxide |
GSH conjugation | 439 | 8706254 |
| GSTA1-1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol 13,14-epoxide |
GSH conjugation (high activity) |
441 | 12067250 |
| GSTA1-1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-acetoxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-acetoxy-PhIP) |
reduction in reaction with GST (major enzyme) |
406, 444 | 11535243, 8069858 |
| GSTA1-1 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH-2-AAF) |
GSH conjugation | 375 | 8330352 |
| GSTA1-1 | drug | phosphoramide mustard ; anticancer, alkylating, metabolite of cyclophosphamide |
phosphoramide mustard | GSH conjugation | 442 | 7954469 |
| GSTA1-2 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating, metabolite |
4-hydroxycyclophosphamide | GSH conjugation | 442 | 7954469 |
| GSTA1-2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-acetoxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-acetoxy-PhIP) |
reduction in reaction with GST (low activity) |
444 | 8069858 |
| GSTA2-2 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products, metabolite |
aflatoxin B1-8,9-epoxide | GSH conjugation | 420 | 10383892 |
| GSTA2-2 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-acetoxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-acetoxy-PhIP) |
Reduction in reaction with GST (low activity) |
406 | 11535243 |
| GSTA2-2 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol 13,14-epoxide |
GSH conjugation (low activity) |
441 | 12067250 |
| GSTA3-3 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol 13,14-epoxide |
GSH conjugation | 441 | 12067250 |
| GSTM1 | chemical | PAH, metabolite | phenanthrene-9,10-epoxide | GSH conjugation | 445 | 16978029 |
| GSTM1-1 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products, metabolite |
aflatoxin B1-8,9-epoxide | GSH conjugation, major enzyme |
432, 443, 433 |
8781383, 9675258, 9115980 |
| GSTM1-1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-acetoxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-acetoxy-PhIP) |
reduction in reaction with GST (low activity) |
404 | 11535243 |
| GSTM1-1 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol 9,10-epoxide |
GSH conjugation | 440, 446 | 11849043, 9403173 |
| GSTM1-1 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
GSH conjugation after oxidation |
51, 447 | 17525473, 12507920 |
| GSTM1-1 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating, metabolite |
4-hydroxycyclophosphamide | GSH conjugation | 442 | 7954469 |
| GSTM1-1 | chemical | PAH | benzo[a]pyrene | GSH conjugation after oxidation |
51 | 17525473 |
| GSTM1-1 | chemical | PAH, metabolite | benzo[c]chrysene-9,10-diol 11,12-epoxide |
GSH conjugation | 446 | 9403173 |
| GSTM1-1 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol 13,14-epoxide |
GSH conjugation | 446 | 9403173 |
| GSTM1-1 | chemical | PAH, metabolite | chrysene-trans-1,2-diol-3,4- epoxide |
GSH conjugation | 446 | 9403173 |
| GSTM1-1 | chemical | PAH | dibenz[a,h]anthracene-3,4- diol-1,2-epoxide |
GSH conjugation | 446 | 9403173 |
| GSTM1-1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol 13,14-epoxide |
GSH conjugation | 446 | 9403173 |
| GSTP1-1 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol 9,10-epoxide |
GSH conjugation |
438, 440, 446, 448, 449 |
9855012, 11849043 9403173, 9299520, 9525277 |
| GSTP1-1 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
GSH conjugation after oxidation |
50, 450 | 16885195, 10344744 |
| GSTP1-1 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating, metabolite |
4-hydroxycyclophosphamide | GSH conjugation | 442 | 7954469 |
| GSTP1-1 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating, metabolite |
4-hydroxyifosfamide | GSH conjugation | 451 | 8555414 |
| GSTP1-1 | chemical | quinoline | 4-nitroquinoline 1-oxide (NQO) |
GSH conjugation | 452, 453 | 11108662, 15766272 |
| GSTP1-1 | chemical | PAH | 5-methylchrysene | GSH conjugation after oxidation |
83 | 18992797 |
| GSTP1-1 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | GSH conjugation after oxidation |
83 | 18992797 |
| GSTP1-1 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol 3,4-epoxide |
GSH conjugation | 454 | 9771942 |
| GSTP1-1 | chemical | PAH, metabolite | 6-methylchrysene-1,2-diol 3,4-epoxide |
GSH-conjugation | 454 | 9771942 |
| GSTP1-1 | chemical | PAH | benzo[a]pyrene | GSH conjugation after oxidation |
50 | 16885195 |
| GSTP1-1 | chemical | PAH, metabolite | benzo[c]chrysene-9,10-diol 11,12-epoxide |
GSH conjugation | 446, 455 | 9403173, 9827546 |
| GSTP1-1 | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol-1,2-epoxide |
GSH conjugation |
438, 446, 455, 456 |
9855012, 9403173, 9827546, 9850062 |
| GSTP1-1 | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol- 13,14-epoxide |
GSH conjugation |
438, 446, 455, 456 |
9855012, 9403173, 9827546, 9850062 |
| GSTP1-1 | chemical | PAH, metabolite | chrysene-trans-1,2- dihydrodiol-3,4-epoxide |
GSH conjugation |
446, 448, 449 |
9403173, 9299520, 9525277 |
| GSTP1-1 | chemical | PAH, metabolite | dibenz[a,h]anthracene-3,4- diol-1,2-epoxide |
GSH-conjugation | 446, 449 | 9403173, 9525277 |
| GSTP1-1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol-13,14-epoxide |
GSH conjugation |
438, 446, 455, 457 |
9855012, 9403173, 9827546, 9687571 |
| GSTP1-1 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating, metabolite |
ifosfamide mustard | GSH conjugation | 451 | 8555414 |
| GSTP1-1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-acetoxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-acetoxy-PhIP) |
reduction in reaction with GST (low activity) |
406, 444 | 11535243, 8069858 |
| GSTP1-1 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
GSH conjugation | 375 | 8330352 |
| GSTP1-1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
reduction in reaction with GST |
458 | 11196146 |
| GSTT1-1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-acetoxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-acetoxy-PhIP) |
reduction in reaction with GST (low activity) |
406 | 11535243 |
| GSTT2-1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-acetoxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-acetoxy-PhIP) |
reduction in reaction with GST (low activity) |
406 | 11535243 |
|
NAD(P)H- dependent quinone oxidoredu ctase, NQO1 |
physiological compound |
estrogen, metabolite | 4-hydroxyestrone-o-quinone | reduction (low activity) |
434 | 18588320 |
|
NADH cytochrom e b5 reductase, b5R; cytochrom e b5, CYB5 |
chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
reduction | 459 | 17040106 |
|
NADH cytochrom e b5 reductase, b5R; cytochrom e b5, CYB5 |
chemical | arylamine, tobacco smoke compound, metabolite |
N-hydroxy-4-aminobiphenyl (N-OH-4-ABP) |
reduction (low activity) |
459, 460 | 17040106, 21447608 |
| NAT1 | chemical | heterocyclic amine | 2-amino-3- methylimidazo[4,5- f]quinolone (IQ) |
N-acetylation (low activity) |
389 | 8353847 |
| NAT1 | chemical | arylamine | 2-aminofluorene (2-AF) |
N-acetylation (low activity) |
389 | 8353847 |
| NAT1 | chemical | PAH | 2-naphthylamine |
N-acetylation (low activity) |
389 | 8353847 |
| NAT1 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl (4-ABP) |
N-acetylation (low activity) |
389 | 8353847 |
| NAT1 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminobiphenyl (N-acetoxy-4-ABP) |
N-acetylation (low activity) |
389 | 8353847 |
| NAT1 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH-2-AAF) |
N-acetylation (low activity) |
389 | 8353847 |
| NAT1 | chemical | arylamine, tobacco smoke compound, metabolite |
N-hydroxy-4-aminobiphenyl (N-OH-4-ABP) |
N-acetylation (low activity) |
389 | 8353847 |
| NAT1 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-2-aminofluorene (N-OH-2-AF) |
N-acetylation (low activity) |
389 | 8353847 |
| NAT2 | chemical | heterocyclic amine | 2-amino-3- methylimidazo[4,5- f]quinolone (IQ) |
N-acetylation | 389 | 8353847 |
| NAT2 | chemical | arylamine | 2-aminofluorene (2-AF) | N-acetylation | 389 | 8353847 |
| NAT2 | chemical | arylamine | 2-naphthylamine | N-acetylation | 389 | 8353847 |
| NAT2 | chemical | arylamine, tobacco smoke compound |
4-aminobiphenyl (4-ABP) |
N-acetylation | 389 | 8353847 |
| NAT2 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminobiphenyl (N-acetoxy-4-ABP) |
N-acetylation | 389 | 8353847 |
| NAT2 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH-2-AAF) |
N-acetylation | 389 | 8353847 |
| NAT2 | chemical | arylamine, tobacco smoke compound, metabolite |
N-hydroxy-4-aminobiphenyl (N-OH-4-ABP) |
N-acetylation | 389 | 8353847 |
| NAT2 | chemical | hydroxylamine, arylamine, metabolite |
N-hydroxy-2-aminofluorene (N-OH-2-AF) |
N-acetylation | 389 | 8353847 |
| S-COMT | physiological compound |
estrogen, metabolite | 2-hydroxyestradiol |
O-methylation, 2- OH and 3-OH |
461, 462 | 11606384, 12360102 |
| S-COMT | physiological compound |
estrogen, metabolite | 2-hydroxyestrone |
O-methylation, 2- OH and 3-OH |
462 | 12360102 |
| S-COMT | physiological compound |
estrogen, metabolite | 4-hydroxyestradiol |
O-methylation, 4- OH |
462 | 12360102 |
| S-COMT | physiological compound |
estrogen, metabolite | 4-hydroxyestrone |
O-methylation, 4- OH |
462 | 12360102 |
| S-COMT | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | 12-methylbenz[a]anthracene- 3,4-diol |
O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | 5-methylchrysene-7,8-diol | O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | 7,12- dimethylbenz[a]anthracene- 3,4-diol |
O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | 7-methylbenz[a]anthracene- 3,4-diol |
O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | benz[a]anthracene-3,4-diol |
O-methylation (low activity) |
463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | benzo[c]phenanthrene-3,4- diol |
O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | benzo[g]chrysene-11,12-diol | O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | chrysene-1,2-diol | O-methylation | 463 | 21622560 |
| S-COMT | chemical | PAH, metabolite | chrysene-3,4-diol |
O-methylation (low activity) |
463 | 21622560 |
| SULT | chemical | heterocyclic amine | 2-amino-3- methylimidazo[4,5- f]quinolone (IQ) |
N-sulfamate formation (low activity) |
464 | 7744696 |
| SULT | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (N-OH-MeIQx) |
N-sulfation after N- hydroxylation |
429, 465 | 11258970 8844796 |
| UGT | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-3,8- dimethylimidazo[4,5- f]quinoxaline (N-OH-MeIQx) |
N-glucuronidation | 429, 465 | 11258970 8844796 |
| UGT1A1 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
O-glucuronidation | 466 | 11929814 |
| UGT1A1 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N2- (major metabolite) and N3-glucuronidation |
467, 468 | 11408353, 17638922 |
| UGT1A1 | physiological compound |
estrogen, metabolite | 2-hydroxyestradiol | O-glucuronidation | 469 | 15117964 |
| UGT1A1 | physiological compound |
estrogen, metabolite | 2-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A1 | physiological compound |
estrogen, metabolite | 4-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A1 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
O-glucuronidation | 470 | 21780761 |
| UGT1A1 | physiological compound |
estrogen | 17β-estradiol | O-glucuronidation | 469 | 15117964 |
| UGT1A1 | physiological compound |
estrogen | estrone | O-glucuronidation | 469 | 15117964 |
| UGT1A1 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N2- (major metabolite) and N3- glucuronidation, major enzyme |
146, 467, 468, 471, 472 |
11375903, 11408353, 17638922, 15310245, 15708579 |
| UGT1A1 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen |
O-glucuronidation (low activity) |
402, 473, 474 |
12034366, 16480962, 17664247 |
| UGT1A3 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N2- and N3- glucuronidation |
475 | 10357796 |
| UGT1A3 | physiological compound |
estrogen, metabolite | 2-hydroxyestradiol |
O-glucuronidation (low activity) |
469 | 15117964 |
| UGT1A3 | physiological compound |
estrogen, metabolite | 2-hydroxy-estrone |
O-glucuronidation (low activity) |
469 | 15117964 |
| UGT1A3 | physiological compound |
estrogen, metabolite | 4-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A3 | physiological compound |
estrogen | 17β-estradiol | O-glucuronidation | 469 | 15117964 |
| UGT1A3 | physiological compound |
estrogen | estrone | O-glucuronidation | 469 | 15117964 |
| UGT1A3 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
N-glucuronidation (low activity) |
146 | 11375903 |
| UGT1A3 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N2- and N3- glucuronidation (major metabolite) |
146, 467, 468, 471 |
11375903, 11408353, 17638922, 15310245 |
| UGT1A4 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N2- and N3- glucuronidation (major metabolite), low activity |
467, 468 | 11408353, 17638922 |
| UGT1A4 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
N-glucuronidation | 476-478 | 14871856, 14709623, 18238858 |
| UGT1A4 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen | N-glucuronidation |
278, 402, 473, 474, 479 |
16884532, 12034366, 16480962, 17664247, 15135306 |
| UGT1A4 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
O-glucuronidation | 470 | 21780761 |
| UGT1A4 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N2- (major metabolite) and N3-glucuronidation |
467, 468, 471 |
11408353, 17638922, 15310245 |
| UGT1A4 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanabasine | O-glucuronidation | 478 | 18238858 |
| UGT1A4 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator |
tamoxifen | N-glucuronidation | 278 | 16884532 |
| UGT1A6 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen |
O-glucuronidation (low activity) |
473 | 16480962 |
| UGT1A6 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH-2AAF) |
N-glucuronidation (low activity) |
146 | 11375903 |
| UGT1A6 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N-glucuronidation (low activity) |
146 | 11375903 |
| UGT1A7 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
O-glucuronidation | 466 | 11929814 |
| UGT1A7 | physiological compound |
estrogen, metabolite | 2-hydroxyestradiol | O-glucuronidation | 469 | 15117964 |
| UGT1A7 | physiological compound |
estrogen, metabolite | 2-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A7 | physiological compound |
estrogen, metabolite | 4-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A7 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen |
O-glucuronidation (low activity) |
473, 474 | 16480962, 17664247 |
| UGT1A7 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
O-glucuronidation | 470 | 21780761 |
| UGT1A7 | physiological compound |
estrogen | 17β-estradiol |
O-glucuronidation (low activity) |
469 | 15117964 |
| UGT1A7 | physiological compound |
estrogen | estrone |
O-glucuronidation (low activity) |
469 | 15117964 |
| UGT1A7 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
N-glucuronidation (high activity) |
146 | 11375903 |
| UGT1A7 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N2- and N3- glucuronidation (low activity) |
146, 467, 468, 471 |
11375903, 11408353, 17638922, 15310245 |
| UGT1A8 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N2- and N3- glucuronidation |
466 | 10357796 |
| UGT1A8 | physiological compound |
estrogen, metabolite | 2-hydroxyestradiol | O-glucuronidation | 469 | 15117964 |
| UGT1A8 | physiological compound |
estrogen, metabolite | 2-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A8 | physiological compound |
estrogen, metabolite | 4-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A8 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen |
O-glucuronidation (low activity) |
473, 474 | 16480962, 17664247 |
| UGT1A8 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
O-glucuronidation | 470 | 21780761 |
| UGT1A8 | physiological compound |
estrogen | 17β-estradiol | O-glucuronidation | 469 | 15117964 |
| UGT1A8 | physiological compound |
estrogen | estrone |
O-glucuronidation (low activity) |
469 | 15117964 |
| UGT1A8 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
N-glucuronidation (low activity) |
146 | 11375903 |
| UGT1A8 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N2- (major metabolite) and N3-glucuronidation |
146, 467, 468, 471 |
11375903, 11408353, 17638922, 15310245 |
| UGT1A9 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
O-glucuronidation | 466 | 11929814 |
| UGT1A9 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N2- (major metabolite) and N3- glucuronidation, low activity |
467, 468, 475 |
11408353, 17638922, 10357796 |
| UGT1A9 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
O-glucuronidation | 477, 480 | 14709623, 11038164 |
| UGT1A9 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen |
O-glucuronidation (low activity) |
402, 473, 474 |
12034366, 16480962, 17664247 |
| UGT1A9 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
O-glucuronidation (high activity) |
470 | 21780761 |
| UGT1A9 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
N-glucuronidation (high activity) |
146 | 11375903 |
| UGT1A9 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N2- and N3- glucuronidation (major metabolite) |
146, 467, 468, 471, 475 |
11375903, 11408353, 17638922, 15310245, 10357796 |
| UGT1A10 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
O-glucuronidation (major enzyme) |
466, 481 | 11929814, 16510539 |
| UGT1A10 | chemical | heterocyclic amine | 2-amino-1-methyl-6- phenylimidazo[4,5- b]pyridine (PhIP) |
N2- and N3- glucuronidation |
468, 475 | 17638922, 10357796 |
| UGT1A10 | physiological compound |
estrogen, metabolite | 2-hydroxyestradiol |
O-glucuronidation (high activity) |
469 | 15117964 |
| UGT1A10 | physiological compound |
estrogen, metabolite | 2-hydroxyestrone | O-glucuronidation | 469 | 15117964 |
| UGT1A10 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
O-glucuronidation | 482 | 20007297 |
| UGT1A10 | physiological compound |
estrogen, metabolite | 4-hydroxyestrone |
O-glucuronidation (high activity) |
469 | 15117964 |
| UGT1A10 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen |
O-glucuronidation (low activity) |
473, 474 | 16480962, 17664247 |
| UGT1A10 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
O-glucuronidation | 468 | 21780761 |
| UGT1A10 | physiological compound |
estrogen | 17β-estradiol |
O-glucuronidation (high activity) |
467 | 15117964 |
| UGT1A10 | physiological compound |
estrogen | estrone |
O-glucuronidation (high activity) |
467 | 15117964 |
| UGT1A10 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
N-glucuronidation (low activity) |
146 | 11375903 |
| UGT1A10 | chemical | hydroxylamine, heterocyclic amine, metabolite |
N-hydroxy-2-amino-1- methyl-6-phenylimidazo[4,5- b]pyridine (N-OH-PhIP) |
N2- (major metabolite) and N3- glucuronidation, high activity |
146, 465, 466, 469 |
11375903, 11408353, 17638922, 15310245 |
| UGT2A1 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
O-glucuronidation | 483 | 21164388 |
| UGT2A1 | chemical | PAH, metabolite | 5-methylchrysene-1,2-diol | O-glucuronidation | 481 | 21164388 |
| UGT2B10 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
O-glucuronidation | 478 | 18238858 |
| UGT2B10 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanabasine | O-glucuronidation | 478 | 18238858 |
| UGT2B10 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanatabine | O-glucuronidation | 478 | 18238858 |
| UGT2B10 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanatabine | O-glucuronidation | 468, 478 | 17638922, 18238858 |
| UGT2B10 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
O-glucuronidation | 478 | 18238858 |
| UGT2B10 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
O-glucuronidation | 468, 478 | 17638922, 18238858 |
| UGT2B15 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen | O-glucuronidation |
402, 473, 474 |
12034366, 16480962, 17664247 |
| UGT2B17 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
O-glucuronidation | 484 | 17416778 |
| UGT2B17 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen | O-glucuronidation | 474 | 17664247 |
| UGT2B7 | chemical | PAH, metabolite | (+)- and (−)-benzo[a]pyrene- 7,8-diol |
O-glucuronidation | 466 | 11929814 |
| UGT2B7 | chemical | tobacco-specific nitrosamine |
4-(methylnitrosamino)-1-(3- pyridyl)-1-butanol (NNAL) |
O-glucuronidation |
476, 477, 480 |
14871856, 14709623, 11038164 |
| UGT2B7 | chemical | PAH, metabolite | dibenzo[a,l]pyrene-11,12- diol |
O-glucuronidation | 470 | 21780761 |
| UGT2B7 | chemical | hydroxamic acid, heterocyclic amine, metabolite |
N-hydroxy-2- acetylaminofluorene (N-OH- 2-AAF) |
N-glucuronidation | 146 | 11375903 |
| UGT2B7 | drug | triphenylethyleneamine ; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen | O-glucuronidation | 474 | 17664247 |
Table 2. Bioactivation of Natural Compounds.
| enzyme | category | subcategory | compound | reaction | remarks | references | PubMed ID |
|---|---|---|---|---|---|---|---|
| CYP1A1 | natural compound |
indole, alkylating, pulmonary toxin; higher concentrations in mammalian digestive tract and coal tar |
3-methylindole (skatole) epoxidation (3- methyloxindole formation); dehydrogenation (desaturation, 3- methyleneindolenine form), low Km, medium |
activity, high efficiency | activation | 75-78 | 8558432, 11408359, 12563100, 20795680 |
| CYP1A1 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation 8,9-; oxidation |
activation |
41, 57, 61, 93, 94 |
8674051, 15279838, 11377247, 7923587, 8200084 |
| CYP1A1 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid I | nitroreduction | potent activation |
96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A1 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid II | nitroreduction | activation | 96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A1 | physiological compound |
estrogen | 17β−estradiol | C2-hydroxylation (major reaction, medium Km, high activity, high efficiency), major metabolite and major extrahepatic enzyme; C4-hydroxylation (minor reaction, medium Km, medium efficiency, low activity), oxidation, 3,4- quinone formation (lower activity); oxidation, 2,3-quinone formation; C16α- hydroxylation (high Km, low activity) |
potent activation |
71, 106, 121-130 |
9721189, 8037457, 7826886, 9625734, 9054608, 9667077, 8930523, 11555828, 12865317, 15784278, 16112414, 17570247 |
| CYP1A1 | physiological compound |
estrogen | estrone | C2-hydroxylation (major reaction, medium Km, low activity), oxidation, 2,3-quinone formation; C4-hydroxylation (medium Km, low activity, or medium activity); C16α- hydroxylation (minor reaction, very low activity) |
activation |
49, 127, 130, 131 |
10426814, 12865317, 17570247, 15805301 |
| CYP1A1 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP1A2 | natural compound |
furanoterpene produced by sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation (major enzyme) |
activation | 172, 173 | 1651809, 15892579 |
| CYP1A2 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation (both exo- 8,9- and endo-8,9-); oxidation |
activation |
11, 12, 13, 41, 42, 57, 61, 93, 94, 162, 175- 182 |
2492107 2655891, 2509067, 8674051, 7955101, 15279838, 11377247, 7923587, 8200084, 10023085, 2162057 7766804, 8261428, 12079611, 1902334, 11782366, 16385575, 16608170 |
| CYP1A2 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid I | nitroreduction | potent activation |
96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A2 | natural compound |
phenanthroic acid derivative; nephrotoxin, found in the Aristolochiaceae family of plants |
aristolochic acid II | nitroreduction | potent activation |
96-99 | 11511187, 15386410, 16125300, 22086975 |
| CYP1A2 | natural compound |
bicyclic monoterpene | Δ3-carene | epoxidation (high Km, medium activity) |
activation | 183 | 16379671 |
| CYP1A2 | physiological compound |
estrogen | 17β-estradiol | C2-hydroxylation (major reaction, medium Km, medium activity, medium efficiency), major metabolite and major enzyme in liver; C4-hydroxylation (minor reaction); C16α- hydroxylation (major enzyme, high Km, no activity, or low activity) |
activation |
71, 106, 122-124, 126-129, 190-194 |
9721189, 8037457, 9625734, 9054608, 9667077, 11555828, 12865317, 15784278, 16112414, 1449532, 9635876, 11454902, 11741520, 14703066 |
| CYP1A2 | natural compound |
alkenylbenzene; occurs in a variety of foods including essential oils of tarragon, sweet basil, sweet fennel, anis |
estragole | C1′-hydroxylation (major enzyme, medium Km, low activity) |
potent activation |
195-197 | 17407329, 15914212, 21459083 |
| CYP1A2 | physiological compound |
estrogen | estrone | C2-hydroxylation (medium Km, high activity, major metabolite); C4- hydroxylation (medium Km, medium activity, very low activity); C16α-hydroxylation (minor reaction, very low activity) |
activation |
49, 122, 123, 127, 128, 191, 192, 198 |
10426814, 9625734, 9054608, 12865317, 15784278, 9635876, 11454902, 16537715 |
| CYP1A2 | natural compound |
phenylpropene, from Rhizoma acorigraminei |
methyleugenol | C1′-hydroxylation (medium Km, major enzyme) |
activation | 196, 199 | 15914212, 16411663 |
| CYP1A2 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation | 41 | 8674051 |
| CYP1B1 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | oxidation | activation |
61, 210, 217 |
11377247, 16551781, 9106248 |
| CYP1B1 | physiological compound |
estrogen | 17β-estradiol | C4-hydroxylation (major enzyme, medium Km, medium activity, medium and low efficiency), oxidation, 3,4-quinone formation; C2-hydroxylation (low activity, minor reaction), oxidation, 2,3-quinone formation; C16α- hydroxylation (minor enzyme, medium and high Km, low activity) |
potent activation |
49, 71, 121-130, 194, 211, 221, 225- 228 |
10426814, 9721189, 7826886, 9625734, 9054608, 9667077, 8930523, 11555828, 12865317, 15784278, 16112414, 17570247, 14703066, 11719446, 11465393, 8790407, 7568105, 10862525, 10910054 |
| CYP1B1 | physiological compound |
estrogen | estrone | C4-hydroxylation (low Km, major reaction); C2- hydroxylation (low activity, minor reaction), oxidation, 2,3-quinone formation; |
potent activation |
49, 127, 128, 130, 198 |
10426814, 12865317, 15784278, 17570247, 16537715 |
| CYP2A6 | natural compound |
alkenylbenzene; occurs in a variety of foods |
including essential oils of tarragon, sweet basil, sweet fennel, anis estragole |
C1′-hydroxylation (major enzyme, medium Km, medium activity) |
potent activation |
195, 197 | 17407329, 21459083 |
| CYP2A6 | natural compound |
furanocoumarin; anti- psoriatic, photosensitizer, found in several species of plants |
methoxalen (8- methoxypsoralen, xanthotoxin) |
epoxidation (furanoepoxide formation) and hydrolysis |
activation | 140 | 17584015 |
| CYP2A6 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
hydroxylation C5′- (lactol formation, medium Km, high to medium activity); oxidation (major enzyme) |
activation |
132-134, 244, 256- 258 |
11774366, 12214673, 11600130, 15651850, 10837014, 9276639, 7646564 |
| CYP2A6 | natural compound |
methylenedioxypheny, benzodioxole |
safrole | C1′-hydroxylation (major enzyme at low concentrations), medium Km, medium activity |
activation |
195, 196, 259, 260 |
17407329, 15914212, 15377158, 15310247 |
| CYP2A13 | natural compound |
indole, alkylating, pulmonary toxin; in higher concentrations in mammalian digestive tract and coal tar |
3-methylindole, skatole | dehydrogenation (desaturation, 3- methyleneindolenine formation, low Km, medium activity, high efficiency); epoxidation (3-methyloxindole formation) |
potent activation |
77, 261 | 20795680, 19608696 |
| CYP2A13 | chemical | difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation 8,9-; oxidation |
potent activation |
181, 265 | 16385575, 16917071 |
| CYP2A13 | natural compound |
furanocoumarin; anti- psoriatic, photosensitiser, found in bergamot essential oil, in other citrus essential oils, and in grapefruit juice |
bergapten (5- methoxypsoralen) |
epoxidation and hydrolysis to dihydrodiol |
activation | 267 | 20798279 |
| CYP2A13 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
hydroxylation C5′- (lactol formation, medium Km, high activity) and C2′- (keto alcohol formation, medium Km, low activity) |
activation | 258, 266 | 19074523, 15651850 |
| CYP2C9 | natural compound |
phenylpropene; from Rhizoma acorigraminei |
methyleugenol | C1′-hydroxylation (medium activity, high Km), at high concentration |
activation | 196, 199 | 15914212, 16411663 |
| CYP2C19 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation (major enzyme) |
activation | 173 | 15892579 |
| CYP2D6 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 173 | 15892579 |
| CYP2D6 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | oxidation | activation | 61 | 11377247 |
| CYP2E1 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusariumsolani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 173 | 15892579 |
| CYP2E1 | natural compound |
carbamic acid derivative; fermentation by-product |
ethyl carbamate (urethane) | oxidation to vinyl carbamate epoxide |
activation |
107, 282, 310-312 |
1486866, 1664256, 9344892, 9150748, 11181492 |
| CYP2E1 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
hydroxylation C5′- (lactol formation, low activity) |
activation |
134, 244, 247, 256, 265 |
11600130, 10837014, 1423839, 9276639, 7646564 |
| CYP2F1 | natural compound |
indole, alkylating, pulmonary toxin; present in higher concentrations |
in mammalian digestive tract and coal tar |
3-methylindole, skatole dehydrogenation (desaturation, 3- methyleneindolenine form., low Km, medium activity, high efficiency), major enzyme |
potent activation |
75-79, 334, 335 |
8558432, 11408359, 12563100, 20795680, 10383923, 17962375 |
| CYP2F1 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 172 | 1651809 |
| CYP2W1 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | oxidation | activation | 210 | 16551781 |
| CYP2W1 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation | 210 | 16551781 |
| CYP3A4 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | epoxidation; oxidation, minor enzyme |
activation |
140, 172, 173 |
17584015, 1651809, 15892579 |
| CYP3A4 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation exo-C8,9- (major activating enzyme), oxidation |
activation |
11, 12, 41, 57, 61, 175-180, 182, 336, 339-341 |
2492107, 2655891, 7955101, 15279838, 11377247, 2162057, 7766804, 8261428, 12079611, 1902334, 11782366, 16608170, 9328287, 1643250, 7545582, 7850790 |
| CYP3A4 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin G1 (AFG1) | oxidation | activation |
11, 12, 107, 341-343 |
2492107, 2655891, 1486866, 8082563, 7850790, 352361, 12849689 |
| CYP3A4 | physiological compound |
estrogen | 17β-estradiol | C2-hydroxylation (major reaction, major enzyme, medium Km, medium efficiency, high activity), major metabolite and major enzyme in liver; oxidation, 2,3-quinone formation (lower activity); C4- hydroxylation (minor reaction, major enzyme, medium Km, medium activity, medium efficiency); oxidation, 3,4-quinone formation; C16α-hydroxylation (high Km, low activity) |
activation |
107, 122- 124, 126- 130, 190- 193, 346, 347 |
1486866, 9625734, 9054608, 9667077, 11555828, 12865317, 15784278, 16112414, 17570247, 1449532, 9635876, 11454902, 11741520, 10821664, 12124305 |
| CYP3A4 | physiological compound |
estrogen | estrone | C2-hydroxylation (high Km, major metabolite, low activity); oxidation, 2,3-quinone formation; C4-hydroxylation (high Km, low activity, major enzyme); C16α- hydroxylation (high Km, low activity, major enzyme) |
activation |
49, 122- 124, 127, 128, 130, 191, 192 |
10426814, 9625734, 9054608, 9667077, 12865317, 15784278, 17570247, 9635876, 11454902 |
| CYP3A4 | natural compound |
alkaloid, pyrrolizidine, genotoxic |
monocrotaline | dehydrogenation | activation | 356 | 15649625 |
| CYP3A4 | natural compound |
nitrosamine, tobacco- specific |
N’-nitrosonornicotine (N- nitrosonornicotine, NNN) |
hydroxylation C2′- (keto alcohol formation); oxidation |
activation |
132, 133, 256, 257 |
11774366, 12214673, 9276639, 7646564 |
| CYP3A4 | natural compound |
pyrrolizidine alkaloid from Senecio retrorsus |
retrorsine | dehydrogenation | activation | 356 | 15649625 |
| CYP3A4 | natural compound |
pyrrolizidine alkaloid, food contaminant (meat, milk, and honey) |
riddelliine | dehydrogenation | activation | 356 | 15649625 |
| CYP3A4 | natural compound |
pyrrolizidine alkaloid, genotoxic |
senecionine | dehydrogenation | activation | 107, 357 | 1486866, 2009596 |
| CYP3A4 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation |
11, 12, 66, 107, 336, 341 |
2492107, 2655891, 9685642, 1486866, 9328287, 7850790 |
| CYP3A5 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation exo-8,9- (major reaction); oxidation |
activation | 182, 365 | 16608170, 7893152 |
| CYP3A5 | physiological compound |
estrogen | 17β-estradiol | C2-hydroxylation; C4- hydroxylation (major reaction); C16α- hydroxylation (low activity) |
activation |
127, 128, 191, 192, 347 |
12865317, 15784278, 9635876, 11454902, 12124305 |
| CYP3A5 | chemical | tobacco-specific nitrosamine |
N’-nitrosoanabasine | oxidation | activation | 132, 133 | 11774366, 12214673 |
| CYP3A7 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin B1 (AFB1) | epoxidation exo-C8,9-; oxidation |
activation |
107, 182, 336, 338, 341, 366 |
1486866, 16608170, 9328287, 9493761, 7850790, 9044840 |
| CYP3A7 | natural compound |
difuranocoumarin; mycotoxin, produced by Aspergillus species on food products |
aflatoxin G1 (AFG1) | oxidation | activation | 341 | 7850790 |
| CYP3A7 | physiological compound |
estrogen | 17β-estradiol | C2-hydroxylation (medium Km, low activity, major reaction); C4-hydroxylation (low activity, high Km); C16α-hydroxylation (very low activity, high Km) |
activation |
127, 128, 347 |
12865317, 15784278, 12124305, |
| CYP3A7 | physiological compound |
estrogen | estrone | C2-hydroxylation (medium Km, medium activity); C4- hydroxylation (low activity, medium Km); C16α-hydroxylation (medium Km, medium activity) |
activation | 127, 128 | 12865317, 15784278 |
| CYP3A7 | natural compound |
furanoxanthone; mycotoxin, produced by Aspergillus species |
sterigmatocystin | oxidation | activation |
336, 338, 341 |
9328287, 9493761, 7850790 |
| CYP4B1 | natural compound |
furanoterpene produced in sweet potatoes infected with Fusarium solani; pulmonary toxin, alkylating |
4-ipomeanol | oxidation | activation | 172 | 1651809 |
|
CYP19A1 (aromatase) |
physiological compound |
estrogen | estradiol 17β- | C2-hydroxylation (medium Km) |
activation | 125, 369 | 8930523, 8476762 |
|
CYP19A1 (aromatase) |
physiological compound |
estrogen | estrone | C2-hydroxylation (medium Km) |
activation | 125, 369 | 8930523, 8476762 |
| SULT1A1 | natural compound |
1-methoxy-3- indolylmethyl glucosinolate breakdown product, in many Brassica vegetables |
1-methoxy-3- indolylmethyl-alcohol |
O-sulfonation | activation | 398 | 20846518 |
| SULT1A1 | natural compound |
nephrotoxin, Aristolochia fangchi compound, nitroarene |
aristolochic acids I and II |
O-sulfonation after nitroreduction to hydroxylamine |
potent activation |
404 | 16161050 |
| SULT1A3 | natural compound |
alkenylbenzene; occurs in a variety of foods including essential oils of tarragon, sweet basil, sweet fennel, anis, metabolite |
1′-hydroxyestragole | O-sulfonation | activation | 197, 401 | 21459083, 22072630 |
| SULT1A3 | natural compound |
alkenylbenzene; occurs in a variety of foods including essential oils of tarragon, sweet basil, sweet fennel, anis |
estragole |
O-sulfonation after C1′- hydroxylation |
activation | 401 | 22072630 |
| SULT1B1 | natural compound |
nephrotoxin, Aristolochia fangchi compound, nitroarene |
aristolochic acids I and II |
O-sulfonation after nitroreduction to hydroxylamine |
activation | 394 | 16161050 |
Table 3. Activation of Drugs.
| enzyme | category | subcategory | compound | reaction | remarks | references | PubMed ID |
|---|---|---|---|---|---|---|---|
| COX-1 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor, and DNA binding |
ellipticine | oxidation | activation | 39, 40 | 16936898, 21753906 |
| CYP1A1 | drug | imidazole; anticancer, alkylating | dacarbazine |
N-demethylation (major extrahepatic enzyme) |
activation | 111 | 10473105 |
| CYP1A1 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | oxidation (at high conc.) | activation | 87 | 20507880 |
| CYP1A2 | drug | imidazole; anticancer, alkylating | dacarbazine |
N-demethylation (major enzyme) |
potent activation |
111 | 10473105 |
| CYP1A2 | drug | pyrido-carbazole; antineoplastic, alkaloid, apocyanaceae plant compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C12- and C13- (low activity) |
activation |
39, 40, 185- 189 |
16936898, 21753906, 11755121, 12123750, 15548707, 17197724, 21683692 |
| CYP1A2 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | oxidation (at high concentration) |
activation | 87 | 20507880 |
| CYP1B1 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor and DNA binding |
ellipticine | oxidation | activation | 39, 185-188 | 16936898, 11755121, 12123750, 15548707, 17197724 |
| CYP2A6 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (minor enzyme, high Km); oxidation |
activation |
101, 108, 251-254 |
11377097, 19501186, 8242617, 9010702, 10348794, 10692561 |
| CYP2A6 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation (minor reaction, high Km); oxidation (at high concentration) |
activation | 87, 251, 253 | 20507880, 8242617, 10348794 |
| CYP2B6 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (major enzyme, major reaction, high Km, high activity); oxidation |
potent activation |
74, 101, 108, 251-254, 271, 275 |
11360624, 11377097, 19501186, 8242617, 9010702, 10348794, 10692561, 9280407, 15919850 |
| CYP2B6 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (S)-(high Km, major enzyme); oxidation (at high concentrations) |
activation |
87, 251-254, 275-277, 355 |
20507880, 8242617, 10348794, 10692561, 15919850, 10534317, 15821045, 16854777 |
| CYP2C8 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation (minor enzyme, high Km), oxidation (at high concentrations) |
activation | 87, 251, 253 | 20507880, 8242617, 10348794 |
| CYP2C9 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (low Km, low activity, major enzyme at low concentration); oxidation |
activation |
101, 108, 251-254 |
11377097, 19501186, 8242617, 9010702, 10348794, 10692561 |
| CYP2C9 | Drug | Oxazaphosporine; Anticancer, Nitrogen mustard alkylating |
Ifosfamide | C4-hydroxylation (low Km), Oxidation (at high concentrations) |
activation | 87, 251 | 8242617, 20507880 |
| CYP2C9 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation (low Km), oxidation (at high concentration) |
activation | 86, 251 | 20507880, 8242617 |
| CYP2C19 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (S)- (minor reaction, high Km), oxidation (at high concentration) |
activation |
87, 251, 253, 276 |
20507880, 8242617, 10348794, 10534317 |
| CYP2D6 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plant compound, topoisomerase II inhibitor and DNA binding |
ellipticine | oxidation N2-; hydroxylation, C13-(low activity) |
activation |
39, 40, 187, 188 |
16936898, 21753906, 15548707, 17197724 |
| CYP2D6 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | oxidation (at high concentration) |
activation | 87 | 20507880 |
| CYP2E1 | drug | platinum-containing; anticancer | cisplatin | oxidation | activation | 303, 304 | 16251482, 17761302 |
| CYP2E1 | drug | imidazole; anticancer, alkylating | dacarbazine | N-demethylation | activation | 111 | 10473105 |
| CYP3A4 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation; oxidation | activation |
108, 251-254, 275, 344 |
19501186, 8242617, 9010702, 10348794, 10692561, 9923542, 15919850 |
| CYP3A4 | drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | hydroxylation, C13- (major enzyme) and C12- (minor reaction); oxidation N2- (major enzyme) |
potent activation |
39, 40, 287- 290, 345 |
16936898, 21753906, 11755121, 12123750, 15548707, 17197724, 20576524 |
| CYP3A4 | drug | estradiol derivative; estrogen, contraceptive |
17α-ethynylestradiol (ethinylestradiol 17α-) |
oxygenation (2- hydroxylation, 17α- inactivation) |
activation | 140, 348 | 17584015, 17251390 |
| CYP3A4 | drug | antimitotic, epipodophyllotoxin, topoisomerase II inhibitor |
etoposide (VP-16) |
O-demethylation (catechol formation), high Km, high activity, major enzyme |
activation | 349-351 | 8114683, 9456308, 17168690 |
| CYP3A4 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (R)- (high Km, high activity); oxidation at high concentration |
potent activation |
87, 251, 275- 277, 344, 352-355 |
20507880, 8242617, 10692561, 15919850, 10534317, 15821045, 9923542, 8161344, 10101149, 10348794, 16854777 |
| CYP3A4 | drug | triphenylethyleneamine; antiestrogen, estrogen receptor modulator |
tamoxifen | Cα-hydroxylation (major enzyme); catechol formation; oxidation, at high concentration |
activation |
87, 354, 358- 364 |
20507880, 10348797, 12018981, 12971802, 14678348, 15159443, 16533026, 12124303, 12419838 |
| CYP3A5 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation (very low activity) activation to cytotoxic metabolites |
activation | 253 | 10348794 |
| CYP3A5 | drug | estradiol derivative; estrogen, contraceptive |
17α-ethynylestradiol (ethinylestradiol, 17α-) |
oxygenation (2- hydroxylation, 17α- mechanism-based inactivation) |
activation | 140, 348 | 17584015, 17251390 |
| CYP3A5 | drug | antimitotic, epipodophyllotoxin, topoisomerase II inhibitor |
etoposide (VP-16) |
O-demethylation (catechol formation), medium Km, high activity, minor enzyme |
activation | 349 | 8114683 |
| CYP3A5 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation, stereoselective for (R)- |
activation |
253, 276, 277, 353, 355 |
10348794, 10534317, 15821045, 10101149, 16854777 |
| CYP3A7 | drug | oxazaphosporine; anticancer, nitrogen mustard, alkylating |
cyclophosphamide | C4-hydroxylation | activation | 253, 275 | 10348794, 15919850 |
| CYP3A7 | drug | oxazaphosporine; anticancer, nitrogen mustard alkylating |
ifosfamide | C4-hydroxylation stereoselective for (R)- (high Km, medium (S-) and high (R-) activity, minor enzyme and reaction) |
activation | 253, 275 | 10348794, 15919850 |
|
Lactoperox idase (LPO) |
drug | pyrido-carbazole; antineoplastic, alkaloid, Apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
Ellipticine | oxidation | activation | 39, 40 | 16936898, 21753906 |
|
Myelopero xidase (MPO) |
drug | pyrido-carbazole; antineoplastic, alkaloid, apocyanaceae plants compound, topoisomerase II inhibitor and DNA binding |
ellipticine | oxidation | activation | 39, 40 | 16936898, 21753906 |
|
NADPH- cytochrom e P450 reductase (POR) |
drug | dihydroxyanthraquinone, laxative | danthron | reduction | activation | 385 | 11697035 |
| SULT1A1 | drug | triphenylethyleneamine; antiestrogen, estrogen receptor modulator, metabolite |
4-hydroxytamoxifen | O-sulfonation | activation | 402, 403 | 12034366, 21537383 |
| SULT2A1 | drug | pregnane, antiandrogen, metabolite | 3α- hydroxycyproterone acetate |
O-sulfonation | activation | 394 | 11535246 |
| SULT2A1 | drug | triphenylethyleneamine; antiestrogen, estrogen receptor modulator, metabolite |
α-hydroxytamoxifen | O-sulfonation | activation | 412, 413 | 9855017, 15371299 |
| SULT2A1 | drug | thioxanthenone, schistosomicide | hycanthone | O-sulfonation | potent activation |
394, 395 | 11535246, 9141497 |
| SULT2E1 | drug | thioxanthenone, schistosomicide | hycanthone | O-sulfonation | activation | 394 | 11535246 |
The classification of compounds into Tables 2 and 3 is somewhat arbitrary. Most of the data on activation of carcinogenic chemicals is with “chemicals” and only a limited amount with drugs and physiological compounds/natural products. We selected some compounds found in nature that are known to be carcinogens under some conditions, e.g. estrogens. Natural products are included (although one could also consider the PAHs to be natural, too). The set of drugs is mainly those used to treat cancer by DNA alkylation, topoisomerase poisons, etc. These compounds are often tumorigenic themselves and have been included. The term “chemicals” is used to describe these components that do not fit well into the natural product or drug classification. Even here there is room for change, e.g. many of the nitrosamines can be formed from secondary amines in vivo.
Another point that should be made is that we use the term carcinogen broadly, including some compounds that are “cancer suspects” and might have caused cancer at very high doses in experimental animal models. Inclusion in the tables here does not necessarily carry an endorsement as a human carcinogen for any regulatory purposes. Some of the compounds cited here are used effectively as drugs, and some are physiological compounds known to be important in normal homeostasis, e.g. estrogens.
The analysis of greatest interest is the activation of chemical carcinogens, and the results are summarized in Figure 2 as well as in Table 1, the main point of this review. The most striking aspect is the dominant role of the P450 enzymes. Interestingly, beyond these the AKR enzymes have a role that exceeds that of any other enzyme group, driven by their reported roles in PAH activation to quinones.485
Of the P450s, six P450s—1A1, 1A2, 1B1, 2A6, 2E1, and 3A4—account for 77% of the reported activations (Figure 2B). The Family 1 enzymes are prominent in the activation of PAHs and heterocyclic aromatic amines, plus arylamines and a variety of other compounds. The values for P450s 2A6 and 2E1 are driven by their roles in the metabolism of N-nitrosamines and a variety of low Mr commodity chemicals, including several vinyl monomers.282
INTERPRETATIONS OF THE ASSIGNMENT RESULTS
Activation vs. Detoxication
We have presented separate tables for activation (Tables 1-3) and detoxication of carcinogens (Table 4). However, distinguishing a role for an enzyme in this regard may be difficult. E.g. there is the classic case of the action of epoxide hydrolase in benzo[a]pyrene activation (Figure 3).486 Another example has a single enzyme forming two different products from the same substrate, the P450 3A4-catalyzed oxidation of AFB1 to AFQ1 (3-hydroxylation, detoxication) and AFB1 8,9-exo-epoxide (activation) (Figure 4).176 The activation of the anticancer drug ellipticine is catalyzed by cyclooxygenases, peroxidases, and P450 enzymes (Tables 1, 2). Hydroxylations at positions C12- and C13, as well as N2-oxidation, are associated to the activation of ellipticine to toxic metabolites. These reactions are catalyzed mainly by the P450 1A1, 1A2, 1B1, and 3A4 enzymes. The same P450 enzymes catalyze detoxication of ellipticine by hydroxylation at the C7- and C9-positions, and the major enzymes considered are P450s 1A1 and 1A2 (Table 4). It has been reported that the balance of activation vs. deactivation of this drug is dependent on cytochrome b5, in that cytochrome b5 enhances production of 12-hydroxy and 13-hydroxyellipticine and thus changes the product ratio in a favor of increased formation of covalent ellipticine-DNA adducts. The effect of cytochrome b5 might be even more pronounced in vivo, in that it has been reported that ellipticine increases levels of cytochrome b5 in rat liver.39,187,189
Figure 3.
Diol-epoxide pathway of benzo[a]pyrene activation.486
Figure 4.
Oxidation of AFB1 by P450 3A4.176
Another issue involves tissue selectivity. As a case in point, GSH transferases catalyze the conjugation of bifunctional electrophiles (e.g., ethylene dibromide, Figure 5) with GSH to form half-mustards and then episulfonium ions.377 These reactive species can react with DNA and are considered to be involved in chemical carcinogenesis. However, if such reactions occur in erythrocytes there is no DNA and this might be considered a detoxication.
Figure 5.
Conjugation of ethylene dibromide with GSH.377
Selectivity of activation vs. detoxication reactions is observed in the acetylation of N-hydroxy heterocyclic and aromatic amines (Table 1 vs. Table 4). The vast majority of heterocyclic amines do not undergo detoxication by N-acetylation because most heterocyclic amines are poor substrates for NATs. These compounds do undergo bioactivation by NATs via O-acetylation, following P450-mediated N-oxidation of the exocyclic amine groups. In contrast to heterocyclic amines, many aromatic amines do undergo detoxication by N-acetylation with NATs.
As mentioned before, chemical carcinogens are activated by a number of enzymes, of which the major ones are P450, SULT, AKR, and NAT (Figure 2 and Table 2), while the data on activation of natural/physiological compounds show that primarily P450 (92%, of which the major ones are 1A1, 1A2, 1B1, and Subfamily 3A) and SULT (8%) enzymes participate in their activation (Table 3). Similarly to general chemicals, drugs are activated by P450s (76%), SULT (12%), LPO (7%), and COX (5%) enzymes. Major P450 enzymes assigned are 3A4 (20%) and 1A2 (11%), and participation of others is low (Table 2).
When considering activation vs. detoxication reactions for a specific compound and/or reaction, attention should be given to the experimental conditions applied and the properties of the compounds and metabolites formed. For instance, 2-nitroanisole is (under oxidative conditions) detoxicated by C2-, C5-, and C6-hydroxylations catalyzed by P450 2E1, 1A1, and 2B6 enzymes (2E1 being the major one). However, under anaerobic conditions activation by nitroreduction prevails due to catalysis by XOR. In addition, 2-nitroanisole and its metabolite 2-nitrophenol induce P450 1A2 and NQO1 (in rats), thus providing the possibility to influence their own detoxication and/or activation pathways.423,424
Our approach here has been to list enzymes under both activation and detoxication in cases that are deemed to be duplicative.
Influence of the Diversities of Different Chemical Classes
In making the assignments shown in Figure 2, the number of compounds available can be considered a contributor to the reported results. For instance, it is known that P450s 1A1 and 1B1 (and AKR enzymes) can oxidize many PAHs.220,485,486 Many of these are available, given the long-standing interest in individual PAHs,4 and have been tested with P450s.41,109,229 Likewise, many arylamines and heterocyclic aromatic amines are known and have been tested with P450 1A2.12 Further, many N-nitrosamines (and vinyl monomers) have been tested with P450s 2A6 and 2E1.247,282 It is possible that, in the future, the availability of a large number of analogs in another class of carcinogens might lead to more testing and shift the balance of the results in Figure 2.
Another point to be made here is that our classification includes compounds shown to be pro-mutagens, to bind covalently to DNA, etc. We do not have evidence that all of the compounds listed in our tables actually cause tumors in experimental animals or humans, although we believe that there is a likelihood that they do at some dose (readers are referred to the National Toxicology Program, International Agency on Cancer Research, and other sources for in vivo cancer results and classifications of human carcinogens).
Types of Reactions
Another analysis involves the type of reactions involved in bioactivation reactions (Figure 6). As seen there, 11 reactions account for 94% of the total, each representing 5-12%. The O-acetylation and O-sulfonation conjugation collectively account for 18%. Nitroreductions (6%) plus other reductions (0.3%) constitute 6.3%. Most of the other reactions are oxidations, together accounting for ~ 73%. Of these, N-hydroxylation (10%) and C-hydroxylation (11%) are the most prominent.
Figure 6.
Analysis of types of activation reactions (data of Table 1, total of 799 reactions). See text for discussion.
The results support the general view that there are many ways to activate procarcinogens (Figures 1, 3-5). As with the classification by enzymes (vide infra) there are caveats about representation based on the number of compounds experimentally available.
We have not analyzed the entries in Table 1 in the context of the chemical nature of the substrate, but this is rather obvious from the nature of Figure 6. Epoxidations involve olefins and aryl rings, nitro reductions involve nitro groups, N-hydroxylations involve arylamines and heterocyclic amines (O-acetylation involves the products), and O-sulfonation involves hydroxylarylamines and benzylic allylic alcohols. Cα-Hydroxylation is prominent for N-nitrosamines. Thus, a single group of chemicals does not dominate.
Weaker Activations
Several enzymes and their reactions have been included in the analyses, although the evidence for significance of their roles in rather weak. There was not a logical reason to delete these from our analysis nor a means of setting a strict benchmark for strong vs. weak roles because of the diversity of assays used. Further, “weak activation” (included in Table 1) might become “activation” or “potent activation” either following ingestion of certain enzyme inducers and/or the expression of a more active variant of the enzyme, as exemplified in several animal models.27
In this regard, we had reported a major role for P450 2C9 in benzo[a]pyrene 3-hydroxylation in human liver.430 Although this reaction has been studied for many years and is the basis of the classic “AHH” activity,487 it is generally not considered to be a bioactivation process, especially in liver. P450 enzymes such as those in the P450 2D6 and Subfamily 2C have been tested for several activities and do have low levels of activity (Table 1), but there is little if any evidence that these drug-metabolizing enzymes have major roles in chemical carcinogenesis in humans.
Similar points can also be raised about the activation of AFB1. Although a number of forms of human P450 have some capability of activating AFB1 (Table 1),139 the evidence is very limited that most of these have relevance. The established target of AFB1 in the liver, and enzymes that are predominantly expressed in other tissues are not very relevant. The existing literature clearly shows roles of primarily two P450s, 3A4 and 1A2.432,443,488 P450 3A4 forms the highly mutagenic exo-8,9-poxide; P450 1A2 forms a roughly equimolar mixture of the dangerous exo-plus the endo-epoxide, the latter of which is essentially non-genotoxic.176,432,489 The situation is complicated in that both of these enzymes also catalyze AFB1 detoxication reactions, 3α-hydroxylation in the case of P450 3A4 (AFQ1) and 9a-hydroxylation in the case of P450 1A2 (AFM1).176
The information presented in this review is relevant in the context of translational studies. A case in point involves P450 2D6 and lung cancer. The interest began even before the characterization of P450 2D6, with a report that individuals with lung cancer showed a low representation of phenotypically poor metabolizers of debrisoquine, subsequently confirmed as a P450 2D6 prototypic substrate.490 These results led to the consideration of the hypothesis that P450 2D6 is involved in the bioactivation of a major carcinogen leading to lung cancer. The level of P450 2D6 in lung tissue is low,491 but it is conceivable that systemic exposure to an entity produced in the liver could be involved. However, attempts to identify a major role of P450 2D6 in the activation of carcinogens have been resoundingly negative.492,493 Further, studies on the genotoxicity of crude cigarette smoke condensates and liver microsomes showed a role for P450 1A2 but not P450 2D6, based on the use of inhibitors etc.149
An alternate hypothesis, given the lack of evidence for a role of P450 2D6 in bioactivation of carcinogens, is that the CYP2D6 gene is linked to the expression of an oncogene. However, no evidence for this hypothesis exists and the known major genetic defect regulating P450 2D6 is aberrant RNA splicing, which is not likely to involve co-regulation of nearby genes.494 Additional epidemiology studies have generally not confirmed a major effect of P450 2D6 expression related to any kind of lung cancer.495,496 In our opinion, resources could be used more effectively if sound experimental studies preceded expensive epidemiological studies with marginal bases of biological causality.
Analysis of Enzymes Involved in Detoxication
Analysis of the data on detoxication is presented in Figure 7. GST and UGT reactions account for > 50% of the reactions, which is not surprising. Two other transferases, NAT and COMT, are also prominent. The fraction attributed to AKR reactions is surprisingly high. Also surprising is the low fraction attributed to epoxide hydrolase, which seem surprising in light of the notoriety of epoxides in toxicology and drug metabolism circles.497 However, epoxide hydrolase is rather ineffective in hydrolyzing some of the most reactive epoxides498 and AFB1 8,9-epoxides.433 However, the roles of epoxide hydrolase and sulfotransferases in detoxication may be underestimated because of the nature of the reactions that have been reported to date with the human enzymes. We suspect that there is more literature using animal epoxide hydrolases that has not been re-done with the human enzymes, and the overall picture (Figure 7) might be misleading. The results of Figure 7 can be contrasted with those in Figures 2 and 6, when the SULT enzymes figure in many bioactivation reactions. However, judging sulfotransferases to be primary bioactivation enzymes, as opposed to detoxication, may not be a proper conclusion. P450s are involved in ~ 14% of the detoxication reactions with carcinogens (Figure 7), but this estimate may not be accurate. As with all of the enzymes of interest here, the assays for bioactivation (e.g. Ames test, umu assays, covalent binding) are often easier to set up than those that would accurately measure detoxication, and the literature may be misleading as to the relative importance of the detoxication enzymes.
Figure 7.
Analysis of detoxication reactions. A: Enzymes involved in detoxication. B: Reactions involved in detoxication. Data are from Table 4 (total of 281 reactions). See text for discussion.
Potential Roles of “Orphan” Enzymes
The analysis of enzymes and P450s (Figure 2) is based on existing knowledge (of the enzymes, as well as the carcinogens), and the pattern might change with time. With the (human) P450s, ~ 1/4 can still be considered “orphans,” in the sense that limited information is available about their catalytic activities and their roles in physiological processes.499 Further, only limited information is available about roles in carcinogen metabolism. An exception in this regard is P450 2W1, which has been shown (like P450 1B1) to activate several classes of carcinogens.210 Of interest is the reported expression of P450 2W1 only in tumor tissue.500 Another orphan P450, P450 4F11, was found not to have appreciable activity towards any carcinogens tested.501
The role of P450 2S1 in the activation of carcinogens is controversial, as well as almost all other potential substrates. Following the initial discovery of human P450 2S1,502 it was reported that the enzyme would oxidize naphthalene.503 However, this report was not confirmed and no evidence for a role of P450 2S1 in the activation of any carcinogens was seen.210 Further, the only substrate reported and independently confirmed for P450 2S1 is the drug candidate 1,4-bis{[2-dimethylamino-N-oxide)ethyl]amino}-5,8-dihydroxyanthracene-9,10-dione (AQ4N).504,505 Some carcinogens can be activated by P450 2S1 in the presence of oxygen surrogates (hydroperoxides)506,507 but the significance of these reactions is unknown, in that P450 2S1 has been demonstrated to be rapidly reduced by NADPH-P450 reductase in the usual manner.505 Nevertheless, expression of P450 2S1 in a mammalian cell line did lead to the formation of products of benzo[a]pyrene, indicating some mechanism of function.506,507
A number of the other P450 orphans have been expressed508 but apparently roles in carcinogen metabolism have not been investigated. This same statement can be applied to other enzymes under consideration, regarding recently discovered gene products.
We should point out that our analyses are based largely on studies done with the “wild type,” or most abundant genetic variants, in the human population. A treatment of all of the implicaitons of variations is beyond the scope of this review and indeed the catalytic efficiencies of only a subset of the variants of these enzymes has been determined with any substrates (e.g., http://www.cypalleles.ki.se/) and few with carcinogens. However, if a racial group exists in which the frequency of a fuctional polymorphism is high, then the balance of enzyme involvement (e.g., Figure 2B) could be shifted.
COMPARISONS OF PATTERNS FOR THE METABOLISM OF DRUGS AND CARCINOGENS
We25,509-512 and others22-24 have presented compilations of the roles of P450s and other enzymes involved in the metabolism of drugs. Comprehensive and up-to-date information on the metabolism of chemicals (including drugs and physiological compounds) in humans and animal models is available in a form of Web searchable absorption-distribution-metabolism-excretion (ADME) database (http://jp.fujitsu.com/group/kyushu/en/services/admedatabase/). The generally accepted view is that, for the drugs that undergo metabolism, almost 75% involve P450 reactions.23-25 Five P450s—1A2, 2C9, 2C19, 2D6, and 3A4—are involved in ~ 90% of these P450 reactions.23-25 This situation, i.e. a large segment of drug metabolism being controlled by a few enzymes, has been useful in being able to rapidly define metabolism issues in drug development.
In an analysis made previously with a total of 7906 entries (metabolic reactions catalyzed by P450s with different compounds as substrates),513 2065 entries are related to P450 Subfamily 3A enzymes, i.e. 26% of the total. Making similar analysis for clinically significant drugs,513 P450 3A4 and 3A5 enzymes participated in 34% of the total P450-catalyzed metabolic reactions, less than the ~50% presented by others.23,24 The contribution of the P450 Subfamily 3A enzymes may be overestimated, or the differences may reflect the time period sampled or the possible differences between sets of drugs published in the open literature vs. proprietary drugs and those in development used in the analyses.
The results shown in Figure 2 can be compared to drug metabolism. The first point is that a similar fraction of the total bioactivation (68%) is due to P450 enzymes (Figure 2A). The five P450s involved in drug metabolism are “replaced” with six—1A1, 1A2, 1B1, 2A6, 2E1, and 3A4—that collectively account for 77% of the P450-mediated reactions.
The numbers resulting from the present analysis of updated earlier data513 on all compounds fit better to the data we report in the activation of the carcinogens by human P450s, and can be used for comparisons. E.g., participation of P450s 1A1, 1A2 and 2E1 in carcinogen activation reactions is 20, 17, and 11%, respectively, and their participation in total metabolic reactions presented here is 7, 10, and 5%, respectively. The participation of P450 1B1 in activation is 11%, and in total metabolism reactions is 3%. However, the participation of P450 3A4 in activation reactions is 10% and its participation in all metabolic reactions is 20%.
The participation of P450 2C9 in activation reactions is ~2% and in total metabolism reactions is 9%; the participation of P450 2C19 in activation reactions is 1% and in total metabolism reactions is 8%. Finally, the apparent participation of P450 2D6 in activation reactons is ~2% and in total metabolism reactions is 10%. We make the general conclusion that the contribution of “toxicologically significant” enzymes (e.g. P450s 1A1, 1A2, 1B1, 2A6, 2E1) is greater in activation reactions and less in considerations of total metabolism. The opposite is the case for generally-detoxicating enzymes, e.g. P450s 2D6, 3A4 (with exceptions), and the 2C Subfamily.
APPLICATIONS
The information presented here is intended to provide a summary to readers who are interested in the literature on human enzymes involved in chemical carcinogenesis reported to date. Further, the analyses (Figure 2) have uses in themselves, despite the stated caveats, in evaluating the individual enzymes.
The analysis provides some guidance in translational applications. For instance, the information presented in Figure 2 can direct the efforts of those in the field of molecular epidemiology of cancer as to which genes and single nucleotide polymorphisms might be most profitable to study. Likewise, some guidance is provided to those in the field of chemoprevention as to which enzymes and reactions might be most useful to inhibit or induce. Such considerations also apply in general issues of risk assessment, in evaluating issues such as inducibility and inter-individual variation.
The analysis (Figure 2) is also useful in basic research. For one thing, it was a matter of curiosity for us—after studying the area for so long513—to know which P450s are most prominent in chemical carcinogenesis. Thus, justification is clearly provided for studying the six P450s cited in Figure 2B (1A1, 1A2, 1B1, 2A6, 2E1, 3A4). Of the six, only P450 1A1 has not been reported at the level of a crystal structure. However, none of these P450 structures has been solved with a carcinogen bound. An overall hope is to use our knowledge of structure-activity relationships to predict which new chemicals will be activated and whether products will be toxic or carcinogenic. In addition, there are numerous basic questions regarding how P450s, including those cited here, such as: What is the molecular basis of catalytic selectivity? What factors determine rates of particular P450 reactions? What is the molecular basis of the cooperativity seen in P450s, including some of those described here, e.g. P450 3A4 and also P450 1A2 (with pyrene).514
It should be emphasized again that almost all of the work cited here was done in the past 25 years.27 In dealing with an effort of this scope, we might have overlooked some useful papers in the field and apologize to those authors in advance. An Excel file of an expansion of Table 1 is included for the ease of those readers who wish to search it or use it to develop or update their own databases.
Supplementary Material
ACKNOWLEDGEMENT
We thank L. M. Folkman and particulary K. Trisler for assistance in preparation of the manuscript. We also acknowledge the pioneering efforts of Dr. Frederick J. DiCarlo and his contributions to the area of xenobiotic metabolism. This review is dedicated to the memory of two eminent cancer researchers who passed away recently, Drs. Fred F. Kadlubar and Donald M. Jerina. Both played major roles in the work summarized here and are missed in this field.
FUNDING SOURCES
This study was supported in part by National Institutes of Health grants R37 CA090426 and R01 ES010546 (F.P.G.).
ABBREVIATIONS
- AF
aflatoxin
- AGT or MGMT (used in tables)
O6-alkylguanine DNA-alkyltransferase
- AKR
aldo-keto reductase
- COX (used in tables) or PTGS
cyclooxygenase (prostaglandin synthase)
- P450 or CYP (used in tables)
cytochrome P450
- FMO
microsomal flavin-containing monoxygenase
- GST
glutathione (GSH) transferase
- HAA
heterocyclic arylamine
- NAT
N-acetyltransferase
- NPR or POR (used in tables)
NADPH-P450 reductase
- NQO
NADPH-quinone reductase
- PAH
polycyclic aromatic hydrocarbon
- SULT
sulfotransferase
- UGT
UDP glucuronosyl transferase
- XOR
xanthine oxidoreductase
Footnotes
Footnote 1: For convenience we will use the term “carcinogens” to refer to both carcinogens that act directly (e.g., modifying DNA) and to procarcinogens (i.e., those that require metabolism to be converted to act on biological targets). In the tables the term “chemicals” is used for those chemicals that are not drugs or natural products.
REFERENCES
- (1).Hill J, Baldwin R. In: Cautions Against the Immoderate Use of Snuff. 2nd ed Jackson J, editor. London: 1761. [Google Scholar]
- (2).Rehn L. Über Blasentumoren bei Fuchsinarbeitern. Archiv. Clin. Chirgurie. 1895;50:588–600. [Google Scholar]
- (3).Yamagiwa K, Ichikawa K. Experimentelle Studie über die Pathogenese der Epithelialgeschwulste. Mitt. Med. Fak. Tokio. 1915;15:295–344. [Google Scholar]
- (4).Cook JW, Hewett CL, Hieger I. The isolation of a cancer-producing hydrocarbon from coal tar. Parts I, II, and III. J. Chem. Soc. 1933:394–405. [Google Scholar]
- (5).Fieser LF. Carcinogenic activity, structure and chemical reactivity of polynuclear hydrocarbons. Am. J. Cancer. 1938;34:37–124. [Google Scholar]
- (6).Miller EC, Miller JA. The presence and significance of bound amino azodyes in the livers of rats fed p-dimethylaminoazobenzene. Cancer Res. 1947;7:468–480. [Google Scholar]
- (7).Miller EC. Studies on the formation of protein-bound derivatives of 3,4-benzpyrene in the epidermal fraction of mouse skin. Cancer Res. 1951;11:100–108. [PubMed] [Google Scholar]
- (8).Miller JA. Carcinogenesis by chemicals: an overview. G. H. A. Clowes Memorial Lecture. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- (9).Ames BN, Durston WE, Yamasaki E, Lee FD. Carcinogens are mutagens: A simple test system combining liver homogenates for activation and bacteria for detection. Proc. Natl. Acad. Sci. U. S. A. 1973;70:2281–2285. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Guengerich FP, Shimada T. Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem. Res. Toxicol. 1991;4:391–407. doi: 10.1021/tx00022a001. [DOI] [PubMed] [Google Scholar]
- (11).Shimada T, Guengerich FP. Evidence for cytochrome P-450NF, the nifedipine oxidase, being the principal enzyme involved in the bioactivation of aflatoxins in human liver. Proc. Natl. Acad. Sci. U. S. A. 1989;86:462–465. doi: 10.1073/pnas.86.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Shimada T, Iwasaki M, Martin MV, Guengerich FP. Human liver microsomal cytochrome P-450 enzymes involved in the bioactivation of procarcinogens detected by umu gene response in Salmonella typhimurium TA1535/pSK1002. Cancer Res. 1989;49:3218–3228. [PubMed] [Google Scholar]
- (13).Shimada T, Martin MV, Pruess-Schwartz D, Marnett LJ, Guengerich FP. Roles of individual human cytochrome P450 enzymes in the bioactivation of benzo[a]pyrene, 7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene, and othe dihydrodiol derivatves of polycyclic aromatic hydrocarbons. Cancer Res. 1989;49:6304–6312. [PubMed] [Google Scholar]
- (14).Thier R, Pemble SE, Taylor JB, Humphreys WG, Persmark M, Ketterer B, Guengerich FP. Expression of mammalian glutathione S-transferase 5-5 in Salmonella typhimurium TA1535 leads to base-pair mutations upon exposure to dihalomethanes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8576–8580. doi: 10.1073/pnas.90.18.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Glatt H, Bartsch I, Christoph S, Coughtrie MWH, Falany CN, Hagen M, Landsiedel R, Pabel U, Phillips DH, Seidel A, Yamazoe Y. Sulfotransferase-mediated activation of mutagens studied using heterologous expression systems. Chem.-Biol. Interact. 1998;109:195–219. doi: 10.1016/s0009-2797(97)00133-6. [DOI] [PubMed] [Google Scholar]
- (16).Kellerman G, Luyten-Kellerman M, Shaw CR. Genetic variation of aryl hydrocarbon hydroxylase in human lymphocytes. Am. J. Human Genet. 1973;25:327–331. [PMC free article] [PubMed] [Google Scholar]
- (17).Kellerman G, Shaw CR, Luyten-Kellerman M. Aryl hydrocarbon hydroxylase inducibility and bronchogenic carcinoma. New Engl. J. Med. 1973;298:934–937. doi: 10.1056/NEJM197311012891802. [DOI] [PubMed] [Google Scholar]
- (18).Butler MA, Lang NP, Young JF, Caporaso NE, Vineis P, Hayes RB, Teitel CH, Massengill JP, Lawsen MF, Kadlubar FF. Determination of CYP1A2 and NAT2 phenotypes in human populations by analysis of caffeine urinary metabolites. Pharmacogenetics. 1992;2:116–127. doi: 10.1097/00008571-199206000-00003. [DOI] [PubMed] [Google Scholar]
- (19).Pantuck EJ, Pantuck CB, Garland WA, Min BH, Wattenberg LW, Anderson KE, Kappas A, Conney AH. Stimulatory effect of brussels sprouts and cabbage on human drug metabolism. Clin. Pharmacol. Therapeut. 1979;25:88–95. doi: 10.1002/cpt197925188. [DOI] [PubMed] [Google Scholar]
- (20).Wattenberg LW. Inhibition of neoplasia by minor dietary constituents. Cancer Res. Suppl. 1983;43:2448–2453. [PubMed] [Google Scholar]
- (21).Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P450 enzymes involved in the oxidation of drugs, carcinogens, and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Therapeut. 1994;270:414–423. [PubMed] [Google Scholar]
- (22).Evans WE, Relling MV. Pharmacogenomics: translating function genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- (23).Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE. Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab. Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- (24).Wienkers LC, Heath TG. Predicting in vivo drug interactions from in vitro drug discovery data. Nat. Rev. Drug Discov. 2005;4:825–833. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- (25).Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochhemistry. 3rd ed Kluwer Academic-Plenum Press; New York: 2005. pp. 377–530. [Google Scholar]
- (26).Jiang H, Vudathala DK, Blair IA, Penning TM. Competing roles of aldo-keto reductase 1A1 and cytochrome P4501B1 in benzo[a]pyrene-7,8-diol activation in human bronchoalveolar H358 cells role of AKRs in P4501B1 induction. Chem. Res. Toxicol. 2006;19:68–78. doi: 10.1021/tx0502488. [DOI] [PubMed] [Google Scholar]
- (27).Guengerich FP. Roles of cytochrome P-450 enzymes in chemical carcinogenesis and cancer chemotherapy. Cancer Res. 1988;48:2946–2954. [PubMed] [Google Scholar]
- (28).Palackal NT, Burczynski ME, Harvey RG, Penning TM. Metabolic activation of polycyclic aromatic hydrocarbon trans-dihydrodiols by ubiquitously expressed aldehyde reductase (AKR1A1) Chem.-Biol. Interact. 2001;130-132:815–824. doi: 10.1016/s0009-2797(00)00237-4. [DOI] [PubMed] [Google Scholar]
- (29).Burczynski ME, Lin HK, Penning TM. Isoform-specific induction of a human aldo-keto reductase by polycyclic aromatic hydrocarbons (PAHs), electrophiles, and oxidative stress implications for the alternative pathway of PAH activation catalyzed by human dihydrodiol dehydrogenase. Cancer Res. 1999;59:607–614. [PubMed] [Google Scholar]
- (30).Palackal NT, Burczynski ME, Harvey RG, Penning TM. The ubiquitous aldehyde reductase (AKR1A1) oxidizes proximate carcinogen trans-dihydrodiols to o-quinones. Potential role in polycyclic aromatic hydrocarbon activation. Biochemistry. 2001;40:10901–10910. doi: 10.1021/bi010872t. [DOI] [PubMed] [Google Scholar]
- (31).Jiang H, Shen YM, Quinn AM, Penning TM. Competing roles of cytochrome P450 1A1/1B1 and aldo-keto reductase 1A1 in the metabolic activation of (±)-7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene in human bronchoalveolar cell extracts. Chem. Res. Toxicol. 2005;18:365–374. doi: 10.1021/tx0497245. [DOI] [PubMed] [Google Scholar]
- (32).Ruan Q, Gelhaus SL, Penning TM, Harvey RG, Blair IA. Aldo-keto reductase- and cytochrome P450-dependent formation of benzo[a]pyrene-derived DNA adducts in human bronchoalveolar cells. Chem. Res. Toxicol. 2007;20:424–431. doi: 10.1021/tx060180b. [DOI] [PubMed] [Google Scholar]
- (33).Quinn AM, Harvey RG, Penning TM. Oxidation of PAH trans-dihydrodiols by human aldo-keto reductase AKR1B10. Chem. Res. Toxicol. 2008;21:2207–2215. doi: 10.1021/tx8002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab. Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- (35).Palackal NT, Lee SH, Harvey RG, Blair IA, Penning TM. Activation of polycyclic aromatic hydrocarbon trans-dihydrodiol proximate carcinogens by human aldo-keto reductase (AKR1C) enzymes and their functional overexpression in human lung carcinoma (A549) cells. J. Biol. Chem. 2002;277:24799–24808. doi: 10.1074/jbc.M112424200. [DOI] [PubMed] [Google Scholar]
- (36).Burczynski ME, Sridhar GR, Palackal NT, Penning TM. The reactive oxygen species- and Michael acceptor-inducible human aldo-keto reductase AKR1C1 reduces the α,β-unsaturated aldehyde 4-hydroxy-2-nonenal to 1,4-dihydroxy-2-nonene. J. Biol. Chem. 2001;276:2890–2897. doi: 10.1074/jbc.M006655200. [DOI] [PubMed] [Google Scholar]
- (37).Penning TM, Jin Y, Steckelbroeck S, Lanisnik Rizner T, Lewis M. Structure-function of human 3α-hydroxysteroid dehydrogenase genes and proteins. Mol. Cell Endocrinol. 2004;215:63–72. doi: 10.1016/j.mce.2003.11.006. [DOI] [PubMed] [Google Scholar]
- (38).Wiese FW, Thompson PA, Kadlubar FF. Carcinogen substrate specificity of human COX-1 and COX-2. Carcinogenesis. 2001;22:5–10. doi: 10.1093/carcin/22.1.5. [DOI] [PubMed] [Google Scholar]
- (39).Stiborová M, Rupertová M, Schmeiser HH, Frei E. Molecular mechanisms of antineoplastic action of an anticancer drug ellipticine. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2006;150:13–23. doi: 10.5507/bp.2006.002. [DOI] [PubMed] [Google Scholar]
- (40).Stiborová M, Paljaková J, Martinkova E, Borek-Dohalská L, Eckschlager T, Kizek R, Frei E. Ellipticine cytotoxicity to cancer cell lines—a comparative study. Interdiscip. Toxicol. 2011;4:98–105. doi: 10.2478/v10102-011-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR. Activation of chemically diverse procarcinogens by human cytochrome P450 1B1. Cancer Res. 1996;56:2979–2984. [PubMed] [Google Scholar]
- (42).Shimada T, Gillam EM, Sandhu P, Guo Z, Tukey RH, Guengerich FP. Activation of procarcinogens by human cytochrome P450 enzymes expressed in Escherichia coli. Simplified bacterial systems for genotoxicity assays. Carcinogenesis. 1994;15:2523–2529. doi: 10.1093/carcin/15.11.2523. [DOI] [PubMed] [Google Scholar]
- (43).Shimada T, Oda Y, Gillam EMJ, Guengerich FP, Inoue K. Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab. Dispos. 2001;29:1176–1182. [PubMed] [Google Scholar]
- (44).Schwarz D, Kisselev P, Cascorbi I, Schunck WH, Roots I. Differential metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8-dihydrodiol by human CYP1A1 variants. Carcinogenesis. 2001;22:453–459. doi: 10.1093/carcin/22.3.453. [DOI] [PubMed] [Google Scholar]
- (45).Shou M, Korzekwa KR, Crespi CL, Gonzalez FJ, Gelboin HV. The role of 12 cDNA-expressed human, rodent, and rabbit cytochromes P450 in the metabolism of benzo[a]pyrene and benzo[a]pyrene trans-7,8-dihydrodiol. Mol. Carcinog. 1994;10:159–168. doi: 10.1002/mc.2940100307. [DOI] [PubMed] [Google Scholar]
- (46).Doehmer J, Holtkamp D, Soballa V, Raab G, Schmalix W, Seidel A, Greim H, Jacob J. Cytochrome P450 mediated reactions studied in genetically engineered V79 Chinese hamster cells. Pharmacogenetics. 1995;5:91–96. doi: 10.1097/00008571-199512001-00008. [DOI] [PubMed] [Google Scholar]
- (47).Kisselev P, Schwarz D, Platt KL, Schunck WH, Roots I. Epoxidation of benzo[a]pyrene-7,8-dihydrodiol by human CYP1A1 in reconstituted membranes. Eur. J. Biochem. 2002;269:1799–1805. doi: 10.1046/j.1432-1033.2002.02848.x. [DOI] [PubMed] [Google Scholar]
- (48).Schmalix WA, Maser H, Kiefer F, Reen R, Wiebel FJ, Gonzalez F, Seidel A, Glatt H, Greim H, Doehmer J. Stable expression of human cytochrome P450 1A1 cDNA in V79 Chinese hamster cells and metabolic activation of benzo[a]pyrene. Eur. J. Pharmacol. 1993;248:251–261. doi: 10.1016/0926-6917(93)90052-r. [DOI] [PubMed] [Google Scholar]
- (49).Shimada T, Watanabe J, Kawajiri K, Sutter TR, Guengerich FP, Gillam EM, Inoue K. Catalytic properties of polymorphic human cytochrome P450 1B1 variants. Carcinogenesis. 1999;20:1607–1614. doi: 10.1093/carcin/20.8.1607. [DOI] [PubMed] [Google Scholar]
- (50).Kushman ME, Kabler SL, Fleming MH, Ravoori S, Gupta RC, Doehmer J, Morrow CS, Townsend AJ. Expression of human glutathione S-transferase P1 confers resistance to benzo[a]pyrene or benzo[a]pyrene 7,8-dihydrodiol mutagenesis, macromolecular alkylation and formation of stable N2-Gua-BPDE adducts in stably tranfected V79MZ cells co-expressing hCYP1A1. Carcinogenesis. 2007;28:207–214. doi: 10.1093/carcin/bgl125. [DOI] [PubMed] [Google Scholar]
- (51).Kushman ME, Kabler SL, Ahmad S, Doehmer J, Morrow CS, Townsend AJ. Protective efficacy of hGSTM1-1 against B[a]P and (+)- or (−)-B[a]P-7,8-dihydrodiol cytotoxicity, mutagenicity, and macromolecular adducts in V79 cells coexpressing hCYP1A1. Toxicol. Sci. 2007;99:51–57. doi: 10.1093/toxsci/kfm133. [DOI] [PubMed] [Google Scholar]
- (52).Gelhaus SL, Harvey RG, Penning TM, Blair IA. Regulation of benzo[a]pyrene-mediated DNA- and glutathione-adduct formation by 2,3,7,8-tetrachlorodibenzo-p-dioxin in human lung cells. Chem. Res. Toxicol. 2011;24:89–98. doi: 10.1021/tx100297z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Yamazaki H, Hatanaka N, Kizu R, Hayakawa K, Shimada N, Guengerich FP, Nakajima M, Yokoi T. Bioactivation of diesel exhaust particle extracts and their major nitrated polycyclic aromatic hydrocarbon components, 1-nitropyrene and dinitropyrenes, by human cytochromes P450 1A1, 1A2, and 1B1. Mut. Res. 2000;472:129–38. doi: 10.1016/s1383-5718(00)00138-8. [DOI] [PubMed] [Google Scholar]
- (54).Hatanaka N, Yamazaki H, Oda Y, Guengerich FP, Nakajima M, Yokoi T. Metabolic activation of carcinogenic 1-nitropyrene by human cytochrome P450 1B1 in Salmonella typhimurium strain expressing an O-acetyltransferase in SOS/umu assay. Mut. Res. 2001;497:223–233. doi: 10.1016/s1383-5718(01)00254-6. [DOI] [PubMed] [Google Scholar]
- (55).Oda Y, Watanabe T, Terao Y, Nukaya H, Wakabayashi K. Genotoxic activation of 2-phenylbenzotriazole-type compounds by human cytochrome P4501A1 and N-acetyltransferase expressed in Salmonella typhimurium umu strains. Mut. Res. 2008;654:52–57. doi: 10.1016/j.mrgentox.2008.04.013. [DOI] [PubMed] [Google Scholar]
- (56).Marumoto S, Oda Y, Miyazawa M. Antigenotoxic activity of naturally occurring furanocoumarins. Environ. Mol. Mutagen. 2011;52:646–57. doi: 10.1002/em.20665. [DOI] [PubMed] [Google Scholar]
- (57).Yamazaki Y, Fujita K, Nakayama K, Suzuki A, Nakamura K, Yamazaki H, Kamataki T. Establishment of ten strains of genetically engineered Salmonella typhimurium TA1538 each co-expressing a form of human cytochrome P450 with NADPH-cytochrome P450 reductase sensitive to various promutagens. Mut. Res. 2004;562:151–62. doi: 10.1016/j.mrgentox.2004.06.003. [DOI] [PubMed] [Google Scholar]
- (58).Hammons GJ, Milton D, Stepps K, Guengerich FP, Tukey RH, Kadlubar FF. Metabolism of carcinogenic heterocyclic and aromatic amines by recombinant cytochrome P450 enzymes. Carcinogenesis. 1997;18:851–854. doi: 10.1093/carcin/18.4.851. [DOI] [PubMed] [Google Scholar]
- (59).Crofts FG, Sutter TR, Strickland PT. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human cytochrome P4501A1 and P4501B1. Carcinogenesis. 1998;19:1969–1973. doi: 10.1093/carcin/19.11.1969. [DOI] [PubMed] [Google Scholar]
- (60).Cheung C, Loy S, Li GX, Liu AB, Yang CS. Rapid induction of colon carcinogenesis in CYP1A-humanized mice by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and dextran sodium sulfate. Carcinogenesis. 2011;32:233–239. doi: 10.1093/carcin/bgq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Oda Y, Aryal P, Terashita T, Gillam EM, Guengerich FP, Shimada T. Metabolic activation of heterocyclic amines and other procarcinogens in Salmonella typhimurium umu tester strains expressing human cytochrome P4501A1, 1A2, 1B1, 2C9, 2D6, 2E1, and 3A4 and human NADPH-P450 reductase and bacterial O-acetyltransferase. Mut. Res. 2001;492:81–90. doi: 10.1016/s1383-5718(01)00154-1. [DOI] [PubMed] [Google Scholar]
- (62).Josephy PD, Batty SM, Boverhof DR. Recombinant human P450 forms 1A1, 1A2, and 1B1 catalyze the bioactivation of heterocyclic amine mutagens in Escherichia coli lacZ strains. Environ. Mol. Mutagen. 2001;38:12–8. doi: 10.1002/em.1045. [DOI] [PubMed] [Google Scholar]
- (63).Edwards RJ, Murray BP, Murray S, Schulz T, Neubert D, Gant TW, Thorgeirsson SS, Boobis AR, Davies DS. Contribution of CYP1A1 and CYP1A2 to the activation of heterocyclic amines in monkeys and human. Carcinogenesis. 1994;15:829–836. doi: 10.1093/carcin/15.5.829. [DOI] [PubMed] [Google Scholar]
- (64).Bendaly J, Zhao S, Neale JR, Metry KJ, Doll MA, States JC, Pierce WM, Jr., Hein DW. 2-Amino-3,8-dimethylimidazo-[4,5-f]quinoxaline-induced DNA adduct formation and mutagenesis in DNA repair-deficient Chinese hamster ovary cells expressing human cytochrome P4501A1 and rapid or slow acetylator N-acetyltransferase 2. Cancer Epidemiol. Biomarkers Prev. 2007;16:1503–1509. doi: 10.1158/1055-9965.EPI-07-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Williams JA, Stone EM, Millar BC, Gusterson BA, Grover PL, Phillips DH. Determination of the enzymes responsible for activation of the heterocyclic amine 2-amino-3-methylimidazo (4,5-f) quinoline in the human breast. Pharmacogenetics. 1999;8:519–528. doi: 10.1097/00008571-199812000-00009. [DOI] [PubMed] [Google Scholar]
- (66).Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mut. Res. 1998;400:201–213. doi: 10.1016/s0027-5107(98)00037-2. [DOI] [PubMed] [Google Scholar]
- (67).Sasaki JC, Arey J, Eastmond DA, Parks KK, Phousongphouang PT, Grosovsky AJ. Evidence for oxidative metabolism in the genotoxicity of the atmospheric reaction product 2-nitronaphthalene in human lymphoblastoid cell lines. Mut. Res. 1999;445:113–25. doi: 10.1016/s1383-5718(99)00118-7. [DOI] [PubMed] [Google Scholar]
- (68).Miyazaki M, Sugawara E, Yoshimura T, Yamazaki H, Kamataki T. Mutagenic activation of betel quid-specific N-nitrosamines catalyzed by human cytochrome P450 co-expressed with NADPH-cytochrome P450 reductase in Salmonella typhimurium YG7108. Mut. Res. 2005;581:165–71. doi: 10.1016/j.mrgentox.2004.12.002. [DOI] [PubMed] [Google Scholar]
- (69).Aiub CA, Mazzei JL, Pinto LF, Felzenszwalb I. Evaluation of nitroreductase and acetyltransferase participation in N-nitrosodiethylamine genotoxicity. Chem.-Biol. Interact. 2006;161:146–154. doi: 10.1016/j.cbi.2006.03.012. [DOI] [PubMed] [Google Scholar]
- (70).Oda Y, Hirayama T, Watanabe T. Genotoxic activation of the environmental pollutant 3,6-dinitrobenzo[e]pyrene in Salmonella typhimurium umu strains expressing human cytochrome P450 and N-acetyltransferase. Toxicol. Lett. 2009;188:258–262. doi: 10.1016/j.toxlet.2009.04.010. [DOI] [PubMed] [Google Scholar]
- (71).Shimada T, Wunsch RM, Hanna IH, Sutter TR, Guengerich FP, Gillam EM. Recombinant human cytochrome P450 1B1 expression. Escherichia coli. Arch. Biochem. Biophys. 1998;357:111–120. doi: 10.1006/abbi.1998.0808. [DOI] [PubMed] [Google Scholar]
- (72).Arlt VM, Henderson CJ, Wolf CR, Schmeiser HH, Phillips DH, Stiborová M. Bioactivation of 3-aminobenzanthrone, a human metabolite of the environmental pollutant 3-nitrobenzanthrone: evidence for DNA adduct formation mediated by cytochrome P450 enzymes and peroxidases. Cancer Lett. 2006;234:220–231. doi: 10.1016/j.canlet.2005.03.035. [DOI] [PubMed] [Google Scholar]
- (73).Stiborová M, Arlt VM, Henderson CJ, Wolf CR, Frei E, Schmeiser HH, Phillips DH. Molecular mechanism of genotoxicity of the environmental pollutant 3-nitrobenzanthrone. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2005;149:191–197. doi: 10.5507/bp.2005.025. [DOI] [PubMed] [Google Scholar]
- (74).Wu J, Dong H, Cai Z, Yu Y. Stable expression of human cytochrome CYP2B6 and CYP1A1 in Chinese hamster CHL cells. Their use in micronucleus assays. Chin. Med. Sci. J. 1997;12:148–155. [PubMed] [Google Scholar]
- (75).Thornton-Manning J, Appleton JL, Gonzalez FJ, Yost GS. Metabolism of 3-methylindole by vaccinia-expressed P450 enzymes. Correlation of 3-methyleneindolenine formation and protein-binding. J. Pharmacol. Exp. Ther. 1999;276:21–29. [PubMed] [Google Scholar]
- (76).Lanza DL, Yost GS. Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome P450 enzymes. Drug Metab. Dispos. 2001;29:950–953. [PubMed] [Google Scholar]
- (77).Nichols WK, Mehta R, Skordos K, Mace K, Pfeifer AM, Carr BA, Minko T, Burchiel SW, Yost GS. 3-Methylindole-induced toxicity to human bronchial epithelial cell lines. Toxicol. Sci. 2003;71:229–236. doi: 10.1093/toxsci/71.2.229. [DOI] [PubMed] [Google Scholar]
- (78).Weems JM, Lamb JG, D’Agostino J, Ding X, Yost GS. Potent mutagenicity of 3-methylindole requires pulmonary cytochrome P450-mediated bioactivation. A comparison to the prototype cigarette smoke mutagens B(a)P and NNK. Chem. Res. Toxicol. 2010;23:1682–1690. doi: 10.1021/tx100147z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Arlt VM, Glatt H, Muckel E, Pabel U, Sorg BL, Seidel A, Frank H, Schmeiser HH, Phillips DH. Activation of 3-nitrobenzanthrone and its metabolites by human acetyltransferases, sulfotransferases and cytochrome P450 expressed in Chinese hamster V79 cells. Int. J. Cancer. 2003;105:583–592. doi: 10.1002/ijc.11143. [DOI] [PubMed] [Google Scholar]
- (80).Arlt VM, Stiborová M, Hewer A, Schmeiser HH, Phillips DH. Human enzymes involved in the metabolic activation of the environmental contaminant 3-nitrobenzanthrone. Evidence for reductive activation by human NADPH cytochrome P450 reductase. Cancer Res. 2003;63:2752–2761. [PubMed] [Google Scholar]
- (81).Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Koehl W, Amin S, Staretz ME, Ueng Y-F, Yamazaki H, Tateishi T, Guengerich FP, Hecht SS. Metabolism of 5-methylchrysene and 6-methylchrysene by human hepatic and pulmonary cytochrome P450 enzymes. Cancer Res. 1996;56:316–324. [PubMed] [Google Scholar]
- (83).Ahmad S, Kabler SL, Rudd L, Amin S, Doehmer J, Morrow CS, Townsend AJ. Cytotoxicity and mutagenicity of 5-methylchrysene and its 1,2-dihydrodiol in V79MZ cells modified to express human CYP1A1 or CYP1B1, in the presence or absence of human GSTP1 coexpression. Toxicol. Lett. 2008;183:99–104. doi: 10.1016/j.toxlet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Yamazaki H, Mimura M, Oda Y, Gonzalez FJ, El-Bayoumy K, Chae HY, Guengerich FP, Shimada T. Activation of trans-1,2-dihydro-1,2-dihydroxy-6-aminochrysene to genotoxic metabolites by rat and human cytochromes P450. Carcinogenesis. 1994;15:465–470. doi: 10.1093/carcin/15.3.465. [DOI] [PubMed] [Google Scholar]
- (85).Yamazaki H, Mimura M, Oda Y, Inui Y, Shiraga T, Iwasaki K, Guengerich FP, Shimada T. Roles of different forms of cytochrome P450 in the activation of the promutagen 6-aminochrysene to genotoxic metabolites in human liver microsomes. Carcinogenesis. 1993;14:1271–1278. doi: 10.1093/carcin/14.7.1271. [DOI] [PubMed] [Google Scholar]
- (86).Buters J, Quintanilla-Martinez L, Schober W, Soballa VJ, Hintermair J, Wolff T, Gonzalez FJ, Greim H. CYP1B1 determines susceptibility to low doses of 7,12-dimethylbenz[a]anthracene-induced ovarian cancers in mice correlation of CYP1B1-mediated DNA adducts with carcinogenicity. Carcinogenesis. 2003;24:327–334. doi: 10.1093/carcin/24.2.327. [DOI] [PubMed] [Google Scholar]
- (87).Hashizume T, Yoshitomi S, Asahi S, Uematsu R, Matsumura S, Chatani F, Oda H. Advantages of human hepatocyte-derived transformants expressing a series of human cytochrome P450 isoforms for genotoxicity examination. Toxicol. Sci. 2010;116:488–497. doi: 10.1093/toxsci/kfq154. [DOI] [PubMed] [Google Scholar]
- (88).Gabelova A, Bacova G, Ruzekova L, Farkasova T. Role of cytochrome P4501A1 in biotransformation of a tissue specific sarcomagen N-methyldibenzo[c,g]carbazole. Mut. Res. 2000;469:259–269. doi: 10.1016/s1383-5718(00)00087-5. [DOI] [PubMed] [Google Scholar]
- (89).Gabelova A, Farkasova T, Bacova G, Robichova S. Mutagenicity of 7H-dibenzo[c,g]carbazole and its tissue specific derivatives in genetically engineered Chinese hamster V79 cell lines stably expressing cytochrome P450. Mut. Res. 2002;517:135–45. doi: 10.1016/s1383-5718(02)00055-4. [DOI] [PubMed] [Google Scholar]
- (90).Gabelova A, Binkova B, Valovicova Z, Sram RJ. DNA adduct formation by 7H-dibenzo[c,g]carbazole and its tissue- and organ-specific derivatives in Chinese hamster V79 cell lines stably expressing cytochrome P450 enzymes. Environ. Mol. Mutagen. 2004;44:448–458. doi: 10.1002/em.20073. [DOI] [PubMed] [Google Scholar]
- (91).Gábelová A, Valovičová Z, Mesárosová M, Trilecová L, Hrubá E, Marvanová S, Krčmár P, Milcová A, Schmuczerová J, Vondráček J, Machala M, Topinka J. Genotoxicity of 7H-dibenzo[c,g]carbazole and its tissue-specific derivatives in human hepatoma HepG2 cells is related to CYP1A1/1A2 expression. Environ. Mol. Mutagen. 2011;52:636–645. doi: 10.1002/em.20664. [DOI] [PubMed] [Google Scholar]
- (92).Roberts-Thompson SJ, McManus ME, Tukey RH, Gonzalez FJ, Holder GM. Metabolism of polycyclic aza-aromatic carcinogens catalyzed by four expressed human cytochromes P450. Cancer Res. 1995;55:1052–1059. [PubMed] [Google Scholar]
- (93).Penman BW, Chen L, Gelboin HV, Gonzalez FJ, Crespi CL. Development of a human lymphoblastoid cell line constitutively expressing human CYP1A1 cDNA substrate specificity with model substrates and promutagens. Carcinogenesis. 1994;15:1931–1937. doi: 10.1093/carcin/15.9.1931. [DOI] [PubMed] [Google Scholar]
- (94).Sengstag C, Eugster HP, Würgler FE. High promutagen activating capacity of yeast microsomes containing human cytochrome P-450 1A and human NADPH-cytochrome P-450 reductase. Carcinogenesis. 1994;15:837–843. doi: 10.1093/carcin/15.5.837. [DOI] [PubMed] [Google Scholar]
- (95).Oda Y, Totsuka Y, Wakabayashi K, Guengerich FP, Shimada T. Activation of aminophenylnorharman, aminomethylphenylnorharman and aminophenylharman to genotoxic metabolites by human N-acetyltransferases and cytochrome P450 enzymes expressed in Salmonella typhimurium umu tester strains. Mutagenesis. 2006;21:411–416. doi: 10.1093/mutage/gel047. [DOI] [PubMed] [Google Scholar]
- (96).Stiborová M, Frei E, Wiessler M, Schmeiser HH. Human enzymes involved in the metabolic activation of carcinogenic aristolochic acids. Evidence for reductive activation by cytochromes P450 1A1 and 1A2. Chem. Res. Toxicol. 2001;14:1128–1137. doi: 10.1021/tx010059z. [DOI] [PubMed] [Google Scholar]
- (97).Stiborová M, Frei E, Hodek P, Wiessler M, Schmeiser HH. Human hepatic and renal microsomes, cytochromes P450 1A1/2, NADPH-cytochrome P450 reductase and prostaglandin H synthase mediate the formation of aristolochic acid-DNA adducts found in patients with urothelial cancer. Int. J. Cancer. 2005;113:189–197. doi: 10.1002/ijc.20564. [DOI] [PubMed] [Google Scholar]
- (98).Stiborová M, Sopko B, Hodek P, Frei E, Schmeiser HH, Hudecek J. The binding of aristolochic acid I to the active site of human cytochromes P450 1A1 and 1A2 explains their potential to reductively activate this human carcinogen. Cancer Lett. 2005;229:193–204. doi: 10.1016/j.canlet.2005.06.038. [DOI] [PubMed] [Google Scholar]
- (99).Stiborová M, Levová K, Bárta F, Shi Z, Frei E, Schmeiser HH, Nebert DW, Phillips DH, Arlt VM. Bioactivation versus detoxication of the urothelial carcinogen aristolochic acid I by human cytochrome P450 1A1 and 1A2. Toxicol. Sci. 2012;125:345–358. doi: 10.1093/toxsci/kfr306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Fontaine SM, Hoyer PB, Halpert JR, Sipes IG. Role of induction of specific hepatic cytochrome P450 isoforms in epoxidation of 4-vinylcyclohexene. Drug Metab. Dispos. 2001;29:1236–1242. [PubMed] [Google Scholar]
- (101).Yoshitomi S, Ikemoto K, Takahashi J, Miki H, Namba M, Asahi S. Establishment of the transformants expressing human cytochrome P450 subtypes in HepG2, and their applications on drug metabolism and toxicology. Toxicol. In Vitro. 2001;15:245–256. doi: 10.1016/s0887-2333(01)00011-x. [DOI] [PubMed] [Google Scholar]
- (102).Takemoto K, Yamazaki H, Nakajima M, Yokoi T. Genotoxic activation of benzophenone and its two metabolites by human cytochrome P450s in SOS/umu assay. Mut. Res. 2002;519:199–204. doi: 10.1016/s1383-5718(02)00141-9. [DOI] [PubMed] [Google Scholar]
- (103).Durant JL, Lafleur AL, Busby WF, Jr., Donhoffner LL, Penman BW, Crespi CL. Mutagenicity of C24H14 PAH in human cells expressing CYP1A1. Mut. Res. 1999;446:1–14. doi: 10.1016/s1383-5718(99)00135-7. [DOI] [PubMed] [Google Scholar]
- (104).Kim JH, Stansbury KH, Walker NJ, Trush MA, Strickland PT, Sutter TR. Metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8-diol by human cytochrome P450 1B1. Carcinogenesis. 1998;19:1847–1853. doi: 10.1093/carcin/19.10.1847. [DOI] [PubMed] [Google Scholar]
- (105).Schwarz D, Kisselev P, Honeck H, Cascorbi I, Schunck WH, Roots I. Co-expression of human cytochrome P4501A1 (CYP1A1) variants and human NADPH-cytochrome P450 reductase in the baculovirus/insect cell system. Xenobiotica. 2001;31:345–356. doi: 10.1080/00498250110055947. [DOI] [PubMed] [Google Scholar]
- (106).Guo Z, Gillam EM, Ohmori S, Tukey RH, Guengerich FP. Expression of modified human cytochrome P450 1A1 in Escherichia coli: Effects of 5′ substitution, stabilization, purification, spectral characterization, and catalytic properties. Arch. Biochem. Biophys. 1994;312:436–446. doi: 10.1006/abbi.1994.1330. [DOI] [PubMed] [Google Scholar]
- (107).Guengerich FP, Shimada T, Raney KD, Yun CH, Meyer DJ, Ketterer B, Harris TM, Groopman JD, Kadlubar FF. Elucidation of catalytic specificities of human cytochrome P450 and glutathione S-transferase enzymes and relevance to molecular epidemiology. Environ. Health Perspect. 1992;98:75–80. doi: 10.1289/ehp.929875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Hashizume T, Yoshitomi S, Asahi S, Matsumura S, Chatani F, Oda H. In vitro micronucleus test in HepG2 transformants expressing a series of human cytochrome P450 isoforms with chemicals requiring metabolic activation. Mut. Res. 2009;677:1–7. doi: 10.1016/j.mrgentox.2009.03.009. [DOI] [PubMed] [Google Scholar]
- (109).Baum M, Amin S, Guengerich FP, Hecht SS, Kohl W, Eisenbrand G. Metabolic activation of benzo[c]phenanthrene by cytochrome P450 enzymes in human liver and lung. Chem. Res. Toxicol. 2001;14:686–693. doi: 10.1021/tx000240s. [DOI] [PubMed] [Google Scholar]
- (110).Seaton MJ, Schlosser PM, Bond JA, Medinsky MA. Benzene metabolism by human liver microsomes in relation to cytochrome P4502E1. Carcinogenesis. 1994;15:1799–1806. doi: 10.1093/carcin/15.9.1799. [DOI] [PubMed] [Google Scholar]
- (111).Reid JM, Kuffel MJ, Miller JK, Rios R, Ames MM. Metabolic activation of dacarbazine by human cytochromes P450: The role of CYP1A1, CYP1A2, and CYP2E1. Clin. Cancer Res. 1999;5:2192–2197. [PubMed] [Google Scholar]
- (112).Yuan ZX, Kumar S, Sikka HC. Comparative metabolism of the aza polynuclear aromatic hydrocarbon dibenz[a,h]acridine by recombinant human and rat cytochrome P450s. Chem. Res. Toxicol. 17:672–678. doi: 10.1021/tx049979i. [DOI] [PubMed] [Google Scholar]
- (113).Luch A, Coffing SL, Tang YM, Schneider A, Soballa V, Greim H, Jefcoate CR, Seidel A, Greenlee WF, Baird WM, Doehmer J. Stable expression of human cytochrome P450 1B1 in V79 Chinese hamster cells and metabolically catalyzed DNA adduct formation of dibenzo[a,l]pyrene. Chem. Res. Toxicol. 1998;11:686–695. doi: 10.1021/tx970236p. [DOI] [PubMed] [Google Scholar]
- (114).Luch A, Schober W, Soballa VJ, Raab G, Greim H, Jacob J, Doehmer J, Seidel A. Metabolic activation of dibenzo[a,l]pyrene by human cytochrome P450 1A1 and P450 1B1 expressed in V79 Chinese hamster cells. Chem. Res. Toxicol. 1999;12:353–364. doi: 10.1021/tx980240g. [DOI] [PubMed] [Google Scholar]
- (115).Luch A, Kishiyama S, Seidel A, Doehmer J, Greim H, Baird WM. The K-region trans-8,9-diol does not significantly contribute as an intermediate in the metabolic activation of dibenzo[a,l]pyrene to DNA-binding metabolites by human cytochrome P450 1A1 or 1B1. Cancer Res. 1999;59:4603–4609. [PubMed] [Google Scholar]
- (116).King LC, Adams L, Allison J, Kohan MJ, Nelson G, Desai D, Amin S, Ross JA. A quantitative comparison of dibenzo[a,l]pyrene-DNA adduct formation by recombinant human cytochrome P450 microsomes. Mol. Carcinog. 1999;26:74–82. [PubMed] [Google Scholar]
- (117).Shou M, Krausz KW, Gonzalez FJ, Gelboin HV. Metabolic activation of the potent carcinogen dibenzo[a,l]pyrene by human recombinant cytochromes P450, lung and liver microsomes. Carcinogenesis. 1996;17:2429–2433. doi: 10.1093/carcin/17.11.2429. [DOI] [PubMed] [Google Scholar]
- (118).Schober W, Luch A, Soballa VJ, Raab G, Stegeman JJ, Doehmer J, Jacob J, Seidel A. On the species-specific biotransformation of dibenzo[a,l]pyrene. Chem.-Biol. Interact. 2006;161:37–48. doi: 10.1016/j.cbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- (119).Kushman ME, Kabler SL, Ahmad S, Doehmer J, Morrow CS, Townsend AJ. Cytotoxicity and mutagenicity of dibenzo[a,l]pyrene and (±)-dibenzo[a,l]pyrene-11,12-dihydrodiol in V79MZ cells co-expressing either hCYP1A1 or hCYP1B1 together with human glutathione-S-transferase A1. Mut. Res. 2007;624:80–87. doi: 10.1016/j.mrfmmm.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Shimada T, Guengerich FP. Inhibition of human cytochrome P450 A1-, 1A2-, and 1B1-mediated activation of procarcinogens to genotoxic metabolites by polycyclic aromatic hydrocarbons. Chem. Res. Toxicol. 2006;19:288–294. doi: 10.1021/tx050291v. [DOI] [PubMed] [Google Scholar]
- (121).Spink DC, Hayes CL, Young NR, Christou M, Sutter TR, Jefcoate CR, Gierthy JF. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on estrogen metabolism in MCF-7 breast cancer cells: evidence for induction of a novel 17β-estradiol 4-hydroxylase. J. Steroid Biochem. Mol Biol. 1994;51:251–258. doi: 10.1016/0960-0760(94)90037-x. [DOI] [PubMed] [Google Scholar]
- (122).Yamazaki H, Shaw PM, Guengerich FP, Shimada T. Roles of cytochromes P450 1A2 and 3A4 in the oxidation of estradiol and estrone in human liver microsomes. Chem. Res. Toxicol. 1998;11:659–665. doi: 10.1021/tx970217f. [DOI] [PubMed] [Google Scholar]
- (123).Shou M, Korzekwa KR, Brooks EN, Krausz KW, Gonzalez FJ, Gelboin HV. Role of human hepatic cytochrome P450 1A2 and 3A4 in the metabolic activation of estrone. Carcinogenesis. 1997;18:207–214. doi: 10.1093/carcin/18.1.207. [DOI] [PubMed] [Google Scholar]
- (124).Niwa T, Yabusaki Y, Honma K, Matsuo N, Tatsuta K, Ishibashi F, Katagiri M. Contribution of human hepatic cytochrome P450 isoforms to regioselective hydroxylation of steroid hormones. Xenobiotica. 1998;6:539–547. doi: 10.1080/004982598239290. [DOI] [PubMed] [Google Scholar]
- (125).Okubo K, Jinbo M, Toma Y, Shimizu Y, Yanaihara T. Aromatase and estrogen 2-hydroxylase activities of human placental microsomes in pregnancy-induced hypertension. Endocr. J. 1996;43:363–368. doi: 10.1507/endocrj.43.363. [DOI] [PubMed] [Google Scholar]
- (126).Badawi AF, Cavalieri EL, Rogan EG. Role of human cytochrome P450 1A1, 1A2, 1B1, and 3A4 in the 2-, 4-, and 16α-hydroxylation of 17β-estradiol. Metabolism. 2001;50:1001–1003. doi: 10.1053/meta.2001.25592. [DOI] [PubMed] [Google Scholar]
- (127).Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT. Characterization of the oxidative metabolites of 17β-estradiol and estrone formed by 15 selectively expressed human cytochrome P450 isoforms. Endocrinology. 2003;144:3382–3398. doi: 10.1210/en.2003-0192. [DOI] [PubMed] [Google Scholar]
- (128).Zhu BT, Lee AJ. NADPH-dependent metabolism of 17β-estradiol and estrone to polar and nonpolar metabolites by human tissues and cytochrome P450 isoforms. Steroids. 2005;70:225–244. doi: 10.1016/j.steroids.2005.01.002. [DOI] [PubMed] [Google Scholar]
- (129).Tsuchiya Y, Nakajima M, Yokoi T. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 2005;227:115–124. doi: 10.1016/j.canlet.2004.10.007. [DOI] [PubMed] [Google Scholar]
- (130).Zhang Y, Gaikwad NW, Olson K, Zahid M, Cavalieri EL, Rogan EG. Cytochrome P450 isoforms catalyze formation of catechol estrogen quinones that react with DNA. Metabolism. 2007;56:887–894. doi: 10.1016/j.metabol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- (131).Kisselev P, Schunck WH, Roots I, Schwarz D. Association of CYP1A1 polymorphisms with differential metabolic activation of 17β-estradiol and estrone. Cancer Res. 2005;65:2972–2978. doi: 10.1158/0008-5472.CAN-04-3543. [DOI] [PubMed] [Google Scholar]
- (132).Fujita K, Kamataki T. Predicting the mutagenicity of tobacco-related N-nitrosamines in humans using 11 strains of Salmonella typhimurium YG7108, each coexpressing a form of human cytochrome P450 along with NADPH-cytochrome P450 reductase. Environ. Mol. Mutagen. 2001;38:339–346. doi: 10.1002/em.10036. [DOI] [PubMed] [Google Scholar]
- (133).Kamataki T, Fujita K, Nakayama K, Miyamoto M, Ariyoshi N. Role of human cytochrome P450 (CYP) in the metabolic activation of nitrosamine derivatives: application of genetically engineered Salmonella expressing human CYP. Drug Metab. Rev. 2002;34:667–676. doi: 10.1081/dmr-120005668. [DOI] [PubMed] [Google Scholar]
- (134).Fujita K, Kamataki T. Role of human cytochrome P450 (CYP) in the metabolic activation of N-alkylnitrosamines: Application of genetically engineered Salmonella typhimurium YG7108 expressing each form of CYP together with human NADPH-cytochrome P450 reductase. Mut. Res. 2001;483:35–41. doi: 10.1016/s0027-5107(01)00223-8. [DOI] [PubMed] [Google Scholar]
- (135).Schober W, Pusch G, Oeder S, Reindl H, Behrendt H, Buters JT. Metabolic activation of phenanthrene by human and mouse cytochromes P450 and pharmacokinetics in CYP1A2 knockout mice. Chem.-Biol. Interact. 2010;183:57–66. doi: 10.1016/j.cbi.2009.09.008. [DOI] [PubMed] [Google Scholar]
- (136).Stiborová M, Martinek V, Rydlova H, Hodek P, Frei E. Sudan I is a potential carcinogen for humans. Evidence for its metabolic activation and detoxication by human recombinant cytochrome P450 1A1 and liver microsomes. Cancer Res. 2002;62:5678–5684. [PubMed] [Google Scholar]
- (137).Stiborová M, Martinek V, Schmeiser HH, Frei E. Modulation of CYP1A1-mediated oxidation of carcinogenic azo dye Sudan I and its binding to DNA by cytochrome b5. Neuro Endocrinol. Lett. 2006;27(Suppl.2):35–39. [PubMed] [Google Scholar]
- (138).Gautier JC, Leceour S, Cosme J, Perret A, Urban P, Beaune P, Pompon D. Contribution of human cytochrome P450 to benzo[a]pyrene and benzo[a]pyrene-7,8-dihydrodiol metabolism, as predicted from heterologous expression in yeast. Pharmacogenetics. 1996;6:489–499. doi: 10.1097/00008571-199612000-00002. [DOI] [PubMed] [Google Scholar]
- (139).Aoyama T, Gonzalez FJ, Gelboin HV. Human cDNA-expressed cytochrome P450 IA2 mutagen activation and substrate specificity. Mol. Carcinog. 1989;2:192–198. doi: 10.1002/mc.2940020405. [DOI] [PubMed] [Google Scholar]
- (140).Kalgutkar AS, Obach RS, Maurer TS. Mechanism-based inactivation of cytochrome P450 enzymes: Chemical mechanisms, structure-activity relationships and relationship to clinical drug-drug interactions and idiosyncratic adverse drug reactions. Curr. Drug Metab. 2007;8:407–447. doi: 10.2174/138920007780866807. [DOI] [PubMed] [Google Scholar]
- (141).Duarte MP, Palma BB, Gilep AA, LaiRes A, Oliveira JS, Usanov SA, Rueff J, Kranendonk M. The stimulatory role of human cytochrome b5 in the bioactivation activities of human CYP1A2, 2A6 and 2E1. A new cell expression system to study cytochrome P450 mediated biotransformation. Mutagenesis. 2005;20:93–100. doi: 10.1093/mutage/gei012. [DOI] [PubMed] [Google Scholar]
- (142).Duarte MP, Palma BB, LaiRes A, Oliveira JS, Rueff J, Kranendonk M. Escherichia coli BTC, a human cytochrome P450 competent tester strain with a high sensitivity towards alkylating agents involvement of alkyltransferases in the repair of DNA damage induced by aromatic amines. Mutagenesis. 2005;20:199–208. doi: 10.1093/mutage/gei028. [DOI] [PubMed] [Google Scholar]
- (143).Duarte MP, Palma BB, Gilep AA, LaiRes A, Oliveira JS, Usanov SA, Rueff J, Kranendonk M. The stimulatory role of human cytochrome b5 in the bioactivation activities of human CYP1A2, 2A6 and 2E1. A new cell expression system to study cytochrome P450-mediated biotransformation (a corrigendum report on Duarte et al. (2005) Mutagenesis 20, 93-100) Mutagenesis. 2007;22:75–81. doi: 10.1093/mutage/gei012. [DOI] [PubMed] [Google Scholar]
- (144).Loeppky RN, Fuchs A, Janzowski C, Humberd C, Goelzer P, Schneider H, Eisenbrand G. Probing the mechanism of the carcinogenic activation of N-nitrosodiethanolamine with deuterium isotope effects. In vivo induction of DNA single-strand breaks and related in vitro assays. Chem. Res. Toxicol. 1998;11:1556–1566. doi: 10.1021/tx9801716. [DOI] [PubMed] [Google Scholar]
- (145).Yamada K, Suzuki T, Hakura A, Mizutani T, Saeki K. Metabolic activation of 10-aza-substituted benzo[a]pyrene by cytochrome P450 1A2 in human liver microsomes. Mut. Res. 2004;557:159–165. doi: 10.1016/j.mrgentox.2003.10.007. [DOI] [PubMed] [Google Scholar]
- (146).Yueh MF, Nguyen N, Famourzadeh M, Strassburg CP, Oda Y, Guengerich FP, Tukey RH. The contribution of UDP-glucuronosyltransferase 1A9 on CYP1A2-mediated genotoxicity by aromatic and heterocyclic amines. Carcinogenesis. 2001;22:943–950. doi: 10.1093/carcin/22.6.943. [DOI] [PubMed] [Google Scholar]
- (147).Oda Y. Analysis of the involvement of human N-acetyltransferase 1 in the genotoxic activation of bladder carcinogenic arylamines using a SOS/umu assay system. Mut. Res. 2004;554:399–406. doi: 10.1016/j.mrfmmm.2004.06.033. [DOI] [PubMed] [Google Scholar]
- (148).Gonzalez FJ, Gelboin HV. Role of human cytochromes P450 in the metabolic activation of chemical carcinogenesis and toxins. Drug Metab. Rev. 1994;26:165–183. doi: 10.3109/03602539409029789. [DOI] [PubMed] [Google Scholar]
- (149).Shimada T, Guengerich FP. Activation of amino-α-carboline, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, and a copper phthalocyanine cellulose extract of cigarette smoke condensate by cytochrome P-450 enzymes in rat and human liver microsomes. Cancer Res. 1991;51:5284–5291. [PubMed] [Google Scholar]
- (150).Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem. Res. Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- (151).Aryal P, Terashita T, Guengerich FP, Shimada T, Oda Y. Use of genetically engineered Salmonella typhimurium OY1002/1A2 strain coexpressing human cytochrome P450 1A2 and NADPH-cytochrome P450 reductase and bacterial O-acetyltransferase in SOS/umu assay. Environ. Mol. Mutagen. 2000;36:121–126. doi: 10.1002/1098-2280(2000)36:2<121::aid-em6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- (152).Turesky RJ, Constable A, Fay LB, Guengerich FP. Interspecies differences between rat and human P450 1A2 in the metabolism of heterocyclic aromatic amines. Cancer Lett. 1999;143:109–112. doi: 10.1016/s0304-3835(99)00137-8. [DOI] [PubMed] [Google Scholar]
- (153).Turesky R, Guengerich FP, Guillouzo A, Langouët S. Metabolism of heterocyclic aromatic amines by human hepatocytes and cytochrome P4501A2. Mut. Res. 2002;506-507:187–95. doi: 10.1016/s0027-5107(02)00165-3. [DOI] [PubMed] [Google Scholar]
- (154).Kim D, Guengerich FP. Selection of human cytochrome P450 1A2 mutants with enhanced catalytic activity for heterocyclic amine N-hydroxylation. Biochemistry. 2004;43:981–938. doi: 10.1021/bi035593f. [DOI] [PubMed] [Google Scholar]
- (155).Zhou H, Josephy PD, Kim D, Guengerich FP. Functional characterization of four allelic variants of human cytochrome P450 1A2. Arch. Biochem. Biophys. 2004;422:23–30. doi: 10.1016/j.abb.2003.11.019. [DOI] [PubMed] [Google Scholar]
- (156).Walters DG, Young PJ, Agus C, Knize MG, Boobis AR, Gooderham NJ, Lake BG. Cruciferous vegetable consumption alters the metabolism of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Carcinogenesis. 2004;25:1659–1669. doi: 10.1093/carcin/bgh164. [DOI] [PubMed] [Google Scholar]
- (157).Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, Dalton TP, Nebert DW, Idle JR, Gonzalez FJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem. Res. Toxicol. 2005;18:1471–1478. doi: 10.1021/tx050136g. [DOI] [PubMed] [Google Scholar]
- (158).Josephy PD, Bibeau KL, Evans DH. Activation of MeIQ (2-amino-3,4-dimethylimidazo-[4,5-f]quinoline) by sequence variants of recombinant human cytochrome P450 1A2. Environ. Mol. Mutagen. 2000;35:328–335. [PubMed] [Google Scholar]
- (159).Glatt H, Pabel U, Meinl W, Frederiksen H, Frandsen H, Muckel E. Bioactivation of the heterocyclic aromatic amine 2-amino-3-methyl-9H-pyrido [2,3-b]indole (MeAαC) in recombinant test systems expressing human xenobiotic-metabolizing enzymes. Carcinogenesis. 2004;25:801–807. doi: 10.1093/carcin/bgh077. [DOI] [PubMed] [Google Scholar]
- (160).Butler MA, Iwasaki M, Guengerich FP, Kadlubar FF. Human cytochrome P-450PA (P-4501A2), the phenacetin O-deethylase, is primarily responsible for the hepatic 3-demethylation of caffeine and N-oxidation of carcinogenic arylamines. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7696–7700. doi: 10.1073/pnas.86.20.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Barcelo S, Mace K, Pfeifer AM, Chipman JK. Production of DNA strand breaks by N-nitrosodimethylamine and 2-amino-3-methylimidazo[4,5-f]quinoline in THLE cells expressing human CYP isoenzymes and inhibition by sulforaphane. Mut. Res. 1998;402:111–120. doi: 10.1016/s0027-5107(97)00288-1. [DOI] [PubMed] [Google Scholar]
- (162).Kranendonk M, Fisher CW, Roda R, Carreira F, Theisen P, LaiRes A, Rueff J, Vermeulen NP, Estabrook RW. Escherichia coli MTC, a NADPH cytochrome P450 reductase competent mutagenicity tester strain for the expression of human cytochrome P450. Comparison of three types of expression systems. Mut. Res. 1999;439:287–300. doi: 10.1016/s1383-5718(98)00193-4. [DOI] [PubMed] [Google Scholar]
- (163).Kim D, Kadlubar FF, Teitel CH, Guengerich FP. Formation and reduction of aryl and heterocyclic nitroso compounds and significance in the flux of hydroxylamines. Chem. Res. Toxicol. 2004;17:529–536. doi: 10.1021/tx034267y. [DOI] [PubMed] [Google Scholar]
- (164).Raza H, King RS, Squires RB, Guengerich FP, Miller DW, Freeman JP, Lang NP, Kadlubar FF. Metabolism of 2-amino-α-carboline, a food-borne heterocyclic amine mutagen and carcinogen, by human and rodent liver microsomes and by human cytochrome P4501A2. Drug Metab. Dispos. 1996;24:395–400. [PubMed] [Google Scholar]
- (165).Josephy PD, Evans DH, Parikh A, Guengerich FP. Metabolic activation of aromatic amine mutagens by simultaneous expression of human cytochrome P450 1A2, NADPH-cytochrome P450 reductase, and N-acetyltransferase in Escherichia coli. Chem. Res. Toxicol. 1998;11:70–74. doi: 10.1021/tx970171q. [DOI] [PubMed] [Google Scholar]
- (166).Stanley LA, Skare JA, Doyle E, Powrie R, D’Angelo D, Elcombe CR. Lack of evidence for metabolism of p-phenylenediamine by human hepatic cytochrome P450 enzymes. Toxicology. 2005;210:147–157. doi: 10.1016/j.tox.2005.01.019. [DOI] [PubMed] [Google Scholar]
- (167).Scheuenpflug J, Krebsfanger N, Doehmer J. Heterologous co-expression of human cytochrome P450 1A2 and polymorphic forms of N-acetyltransferase 2 for studies on aromatic amines in V79 Chinese hamster cells. Altern. Lab. Anim. 2005;33:561–577. doi: 10.1177/026119290503300609. [DOI] [PubMed] [Google Scholar]
- (168).Kappers WA, van Och FM, de Groene EM, Horbach GJ. Comparison of three different in vitro mutation assays used for the investigation of cytochrome P450-mediated mutagenicity of nitro-polycyclic aromatic hydrocarbons. Mut. Res. 2000;466:143–159. doi: 10.1016/s1383-5718(00)00015-2. [DOI] [PubMed] [Google Scholar]
- (169).Glatt H, Engelke CE, Pabel U, Teubner W, Jones AL, Coughtrie MW, Andrae U, Falany CN, Meinl W. Sulfotransferases: Genetics and role in toxicology. Toxicol. Lett. 2000;112-113:341–348. doi: 10.1016/s0378-4274(99)00214-3. [DOI] [PubMed] [Google Scholar]
- (170).Anderson KE, Hammons GJ, Kadlubar FF, Potter JD, Kaderlik KR, Ilett KF, Minchin RF, Teitel CH, Chou HC, Martin MV, Guengerich FP, Barone GW, Lang NP, Peterson LA. Metabolic activation of aromatic amines by human pancreas. Carcinogenesis. 1997;18:1085–1092. doi: 10.1093/carcin/18.5.1085. [DOI] [PubMed] [Google Scholar]
- (171).Nakajima M, Itoh M, Sakai H, Fukami T, Katoh M, Yamazaki H, Kadlubar FF, Imaoka S, Funae Y, Yokoi T. CYP2A13 expressed in human bladder metabolically activates 4-aminobiphenyl. Int. J. Cancer. 2006;119:2520–2526. doi: 10.1002/ijc.22136. [DOI] [PubMed] [Google Scholar]
- (172).Czerwinski M, McLemore TL, Philpot RM, Nhamburo PT, Korzekwa K, Gelboin HV, Gonzalez FJ. Metabolic activation of 4-ipomeanol by complementary DNA-expressed human cytochromes P-450. Evidence for species-specific metabolism. Cancer Res. 1991;51:4636–4638. [PubMed] [Google Scholar]
- (173).Baer BR, Rettie AE, Henne KR. Bioactivation of 4-ipomeanol by CYP4B1. Adduct characterization and evidence for an enedial intermediate. Chem. Res. Toxicol. 2005;18:855–864. doi: 10.1021/tx0496993. [DOI] [PubMed] [Google Scholar]
- (174).Chae Y-H, Thomas T, Guengerich FP, Fu PP, El-Bayoumy K. Comparative metabolism of 1-, 2-, and 4-nitropyrene by human hepatic and pulmonary microsomes. Cancer Res. 1999;59:1473–1480. [PubMed] [Google Scholar]
- (175).Aoyama T, Yamano S, Guzelian PS, Gelboin HV, Gonzalez FJ. 5 of 12 forms of vaccinia virus-expressed human hepatic cytochrome-P450 metabolically activate aflatoxin B1. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4790–4793. doi: 10.1073/pnas.87.12.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Ueng Y-F, Shimada T, Yamazaki H, Guengerich FP. Oxidation of aflatoxin B1 by bacterial recombinant human cytochrome P450 enzymes. Chem. Res. Toxicol. 1995;8:218–225. doi: 10.1021/tx00044a006. [DOI] [PubMed] [Google Scholar]
- (177).Gallagher EP, Wienkers LC, Stapleton PL, Kunze KL, Eaton DL. Role of human microsomal and human complementary DNA-expressed cytochromes P4501A2 and P4503A4 in the bioactivation of aflatoxin B1. Cancer Res. 1994;54:101–108. [PubMed] [Google Scholar]
- (178).Van Vleet TR, Klein PJ, Coulombe RA., Jr. Metabolism and cytotoxicity of aflatoxin B1 in cytochrome P-450-expressing human lung cells. J. Toxicol. Environ. Health A. 2002;65:853–867. doi: 10.1080/00984100290071216. [DOI] [PubMed] [Google Scholar]
- (179).Ramsdell HS, Parkinson A, Eddy AC, Eaton DL. Bioactivation of aflatoxin B1 by human liver microsomes: Role of cytochrome P450 IIIA enzymes. Toxicol. Appl. Pharmacol. 1991;108:436–447. doi: 10.1016/0041-008x(91)90090-2. [DOI] [PubMed] [Google Scholar]
- (180).Van Vleet TR, Mace K, Coulombe RA., Jr. Comparative aflatoxin B(1) activation and cytotoxicity in human bronchial cells expressing cytochromes P450 1A2 and 3A4. Cancer Res. 2002;62:105–112. [PubMed] [Google Scholar]
- (181).He XY, Tang L, Wang SL, Cai QS, Wang JS, Hong JY. Efficient activation of aflatoxin B1 by cytochrome P450 2A13, an enzyme predominantly expressed in human respiratory tract. Int. J. Cancer. 2006;118:2665–2671. doi: 10.1002/ijc.21665. [DOI] [PubMed] [Google Scholar]
- (182).Kamdem LK, Meineke I, Godtel-Armbrust U, Brockmoller J, Wojnowski L. Dominant contribution of P450 3A4 to the hepatic carcinogenic activation of aflatoxin B1. Chem. Res. Toxicol. 2006;19:577–86. doi: 10.1021/tx050358e. [DOI] [PubMed] [Google Scholar]
- (183).Duisken M, Benz D, Peiffer TH, Blomeke B, Hollendert J. Metabolism of 3-carene by human cytochrome P450 enzymes. Identification and characterization of two new metabolites. Curr. Drug Metab. 2005;6:593–601. doi: 10.2174/138920005774832614. [DOI] [PubMed] [Google Scholar]
- (184).Shou MG, Krausz KW, Gonzalez FJ, Gelboin HV. Metabolic activation of the potent carcinogen dibenzo[a,h]anthracene by cDNA-expresed human cytochromes P450. Arch. Biochem. Biophys. 1996;328:201–207. doi: 10.1006/abbi.1996.0161. [DOI] [PubMed] [Google Scholar]
- (185).Stiborová M, Bieler CA, Wiessler M, Frei E. The anticancer agent ellipticine on activation by cytochrome P450 forms covalent DNA adducts. Biochem. Pharmacol. 2001;62:1675–1684. doi: 10.1016/s0006-2952(01)00806-1. [DOI] [PubMed] [Google Scholar]
- (186).Frei E, Bieler CA, Arlt VM, Wiessler M, Stiborová M. Covalent binding of the anticancer drug ellipticine to DNA in V79 cells transfected with human cytochrome P450 enzymes. Biochem. Pharmacol. 2002;64:289–295. doi: 10.1016/s0006-2952(02)01072-9. [DOI] [PubMed] [Google Scholar]
- (187).Stiborová M, Sejbal J, Borek-Dohalska L, Aimova D, Poljakova J, Forsterova K, Rupertova M, Wiesner J, Hudecek J, Wiessler M, Frei E. The anticancer drug ellipticine forms covalent DNA adducts, mediated by human cytochromes P450, through metabolism to 13-hydroxyellipticine and ellipticine N2-oxide. Cancer Res. 2004;64:8374–8380. doi: 10.1158/0008-5472.CAN-04-2202. [DOI] [PubMed] [Google Scholar]
- (188).Stiborová M, Borek-Dohalska L, Aimova D, Kotrbova V, Kukackova K, Janouchova K, Rupertova M, Ryslava H, Hudecek J, Frei E. Oxidation pattern of the anticancer drug ellipticine by hepatic microsomes— similarity between human and rat systems. Gen. Physiol, Biophys. 2006;25:245–261. [PubMed] [Google Scholar]
- (189).Kotrbová V, Mrázová B, Moserová M, Martínek V, Hodek P, Hudeček J, Frei E, Stiborová M. Cytochrome b5 shifts oxidation of the anticancer drug ellipticine by cytochromes P450 1A1 and 1A2 from its detoxication to activation, thereby modulating its pharmacological efficacy. Biochem. Pharmacol. 82:669–680. doi: 10.1016/j.bcp.2011.06.003. [DOI] [PubMed] [Google Scholar]
- (190).Kerlan V, Dreano Y, Bercovici JP, Beaune PH, Floch HH, Berthou F. Nature of cytochromes P450 involved in the 2-/4-hydroxylations of estradiol in human liver microsomes. Biochem. Pharmacol. 1992;44:1745–1756. doi: 10.1016/0006-2952(92)90068-t. [DOI] [PubMed] [Google Scholar]
- (191).Huang Z, Guengerich FP, Kaminsky LS. 16α-Hydroxylation of estrone by human cytochrome P4503A4/5. Carcinogenesis. 1998;19:867–872. doi: 10.1093/carcin/19.5.867. [DOI] [PubMed] [Google Scholar]
- (192).Lee AJ, Kosh JW, Conney AH, Zhu BT. Characterization of the NADPH-dependent metabolism of 17β-estradiol to multiple metabolites by human liver microsomes and selectively expressed human cytochrome P450 3A4 and 3A5. J. Pharmacol. Exp. Ther. 2001;298:420–432. [PubMed] [Google Scholar]
- (193).Cheng ZN, Shu Y, Liu ZQ, Wang LS, Ou-Yang DS, Zhou HH. Role of cytochrome P450 in estradiol metabolism in vitro. Acta Pharmacol. Sin. 2001;22:148–154. [PubMed] [Google Scholar]
- (194).Modugno F, Knoll C, Kanbour-Shakir A, Romkes M. A potential role for the estrogen-metabolizing cytochrome P450 enzymes in human breast carcinogenesis. Breast Cancer Res. Treat. 2003;82:191–197. doi: 10.1023/B:BREA.0000004376.21491.44. [DOI] [PubMed] [Google Scholar]
- (195).Jeurissen SM, Punt A, Boersma MG, Bogaards JJ, Fiamegos YC, Schilter B, van Bladeren PJ, Cnubben NH, Rietjens IMMC. Human cytochrome P450 enzyme specificity for the bioactivation of estragole and related alkenylbenzenes. Chem. Res. Toxicol. 2007;20:798–806. doi: 10.1021/tx700012d. [DOI] [PubMed] [Google Scholar]
- (196).Rietgens IMMC, Boersma MG, van der Woude H, Jeurissen SM, Schutte MF, Alink GM. Flavonoids and alkenylbenzenes: mechanisms of mutagenic action and carcinogenic risk. Mut. Res. 2002;574:124–138. doi: 10.1016/j.mrfmmm.2005.01.028. [DOI] [PubMed] [Google Scholar]
- (197).Chen XW, Serag ES, Sneed KB, Zhou SF. Herbal bioactivation, molecular targets and the toxicity relevance. Chem.-Biol. Interact. 2011;192:161–176. doi: 10.1016/j.cbi.2011.03.016. [DOI] [PubMed] [Google Scholar]
- (198).Cribb AE, Knight MJ, Dryer D, Guernsey J, Hender K, Tesch M, Saleh TM. Role of polymorphic human cytochrome P450 enzymes in estrone oxidation. Cancer Epidemiol. Biomarkers Prev. 2006;15:551–558. doi: 10.1158/1055-9965.EPI-05-0801. [DOI] [PubMed] [Google Scholar]
- (199).Jeurissen SM, Bogaards JJ, Boersma MG, ter Horst JP, Awad HM, Fiamegos YC, van Beek TA, Alink GM, Sudholter EJ, Cnubben NH, Rietjens IM. Human cytochrome P450 enzymes of importance for the bioactivation of methyleugenol to the proximate carcinogen 1′-hydroxymethyleugenol. Chem. Res. Toxicol. 2006;19:111–116. doi: 10.1021/tx050267h. [DOI] [PubMed] [Google Scholar]
- (200).Urban P, Jobert AS, Laine R, Pompon D. Cytochrome P450 (CYP) mutants and substrate-specificity alterations: segment-directed mutagenesis applied to human CYP1A1. Biochem. Soc. Trans. 2001;29:128–135. doi: 10.1042/0300-5127:0290128. [DOI] [PubMed] [Google Scholar]
- (201).Stiborová M, Miksanova M, Sulc M, Rydlova H, Schmeiser HH, Frei E. Identification of a genotoxic mechanism for the carcinogenicity of the environmental pollutant and suspected human carcinogen o-anisidine. Int. J. Cancer. 2005;116:667–678. doi: 10.1002/ijc.21122. [DOI] [PubMed] [Google Scholar]
- (202).Kim H, Wang RS, Elovaara E, Raunio H, Pelkonen O, Aoyama T, Vainio H, Nakajima T. Cytochrome P450 isozymes responsible for the metabolism of toluene and styrene in human liver microsomes. Xenobiotica. 1997;27:657–665. doi: 10.1080/004982597240253. [DOI] [PubMed] [Google Scholar]
- (203).Nakajima T, Elovaara E, Gonzalez FJ, Gelboin HV, Raunio H, Pelkonen O, Vainio H, Aoyama T. Styrene metabolism by cDNA-expressed human hepatic and pulmonary cytochromes P450. Chem. Res. Toxicol. 1994;7:891–896. doi: 10.1021/tx00042a026. [DOI] [PubMed] [Google Scholar]
- (204).Wenker MA, Kezic S, Monster AC, De Wolff FA. Metabolism of styrene in the human liver in vitro. Interindividual variation and enantioselectivity. Xenobiotica. 2001;31:61–72. doi: 10.1080/00498250010031638. [DOI] [PubMed] [Google Scholar]
- (205).Lewis DF, Sams C, Loizou GD. A quantitative structure-activity relationship analysis on a series of alkyl benzenes metabolized by human cytochrome P450 2E1. J. Biochem. Mol. Toxicol. 2003;17:47–52. doi: 10.1002/jbt.10055. [DOI] [PubMed] [Google Scholar]
- (206).Estavillo C, Lu Z, Jansson I, Schenkman JB, Rusling JF. Epoxidation of styrene by human cyt P450 1A2 by thin film electrolysis and peroxide activation compared to solution reactions. Biophys. Chem. 2003;104:291–296. doi: 10.1016/s0301-4622(02)00383-6. [DOI] [PubMed] [Google Scholar]
- (207).Fukami T, Katoh M, Yamazaki H, Yokoi T, Nakajima M. Human cytochrome P450 2A13 efficiently metabolizes chemicals in air pollutants: naphthalene, styrene, and toluene. Chem. Res. Toxicol. 2008;21:720–725. doi: 10.1021/tx700325f. [DOI] [PubMed] [Google Scholar]
- (208).Guengerich FP, Chun Y-J, Kim D, Gillam EMJ, Shimada T. Cytochrome P450 1B1: a target for inhibition in anticarcinogenesis strategies. Mut. Res. 2003;523-524:173–182. doi: 10.1016/s0027-5107(02)00333-0. [DOI] [PubMed] [Google Scholar]
- (209).Mammen JS, Pittman GS, Li Y., Abou-, Zahr F, Bejjani BA, Bell DA, Strickland PT, Sutter TR. Single amino acid mutations, but not common polymorphisms, decrease the activity of CYP1B1 against (−)-benzo[a]pyrene-7R-trans-7,8-dihydrodiol. Carcinogenesis. 2003;24:1247–1255. doi: 10.1093/carcin/bgg088. [DOI] [PubMed] [Google Scholar]
- (210).Wu Z-L, Sohl CD, Shimada T, Guengerich FP. Recombinant enzymes over-expressed in bacteria show broad catalytic specificity of human cytochrome P450 2W1 and limited activity of human cytochrome P450 2S1. Mol. Pharmacol. 2006;69:2007–2014. doi: 10.1124/mol.106.023648. [DOI] [PubMed] [Google Scholar]
- (211).Chun Y-J, Kim S, Kim D, Lee SK, Guengerich FP. A new selective and potent inhibitor of human cytochrome P450 1B1 and its application to antimutagenesis. Cancer Res. 2001;61:8164–8170. [PubMed] [Google Scholar]
- (212).Arlt VM, Hewer A, Sorg BL, Schmeiser HH, Phillips DH, Stiborová M. 3-Aminobenzanthrone, a human metabolite of the environmental pollutant 3-nitrobenzanthrone, forms DNA adducts after metabolic activation by human and rat liver microsomes. Evidence for activation by cytochrome P450 1A1 and P450 1A2. Chem. Res. Toxicol. 2004;17:1092–1101. doi: 10.1021/tx049912v. [DOI] [PubMed] [Google Scholar]
- (213).Smith TJ, Guo Z, Gonzalez FJ, Guengerich FP, Stoner GD, Yang CS. Metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in human lung and liver microsomes and cytochromes P-450 expressed in hepatoma cells. Cancer Res. 1992;52:1757–1763. [PubMed] [Google Scholar]
- (214).Smith TJ, Stoner GD, Yang CS. Activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in human lung microsomes by cytochromes P450, lipoxygenase, and hydroperoxides. Cancer Res. 1995;55:5566–5573. [PubMed] [Google Scholar]
- (215).Patten CJ, Smith TJ, Murphy SE, Wang MH, Lee J, Tynes RE, Koch P, Yang CS. Kinetic analysis of the activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone by heterologously expressed human P450 enzymes and the effect of P450-specific chemical inhibitors on this activation in human liver microsomes. Arch. Biochem. Biophys. 1996;333:127–138. doi: 10.1006/abbi.1996.0373. [DOI] [PubMed] [Google Scholar]
- (216).Abdel-Rahman SZ, Salama SA, Au WW, Hamada FA. Role of polymorphic CYP2E1 and CYP2D6 genes in NNK-induced chromosome aberrations in cultured human lymphocytes. Pharmacogenetics. 10:239–249. doi: 10.1097/00008571-200004000-00005. [DOI] [PubMed] [Google Scholar]
- (217).Crespi CL, Penman BW, Steimel DT, Smith T, Yang CS, Sutter TR. Development of a human lymphoblastoid cell line constitutively expressing human CYP1B1 cDNA. Substrate specificity with model substrates and promutagens. Mutagenesis. 1997;12:83–9. doi: 10.1093/mutage/12.2.83. [DOI] [PubMed] [Google Scholar]
- (218).Ketelslegers HB, Godschalk RW, Eskens BJ, Dallinga JW, Gottschalk RW, van Schooten FJ, van Delft JH, Kleinjans JC. Potential role of cytochrome P450-1B1 in the metabolic activation of 4-aminobiphenyl in humans. Mol. Carcinog. 2009;48:685–691. doi: 10.1002/mc.20530. [DOI] [PubMed] [Google Scholar]
- (219).Chae YH, Yun C-H, Guengerich FP, Kadlubar FF, El-Bayoumy K. Roles of human hepatic and pulmonary cytochrome P450 enzymes in the metabolism of the environmental carcinogen 6-nitrochrysene. Cancer Res. 1993;53:2028–2034. [PubMed] [Google Scholar]
- (220).Shimada T, Gillam EM, Oda Y, Tsumura F, Sutter TR, Guengerich FP, Inoue K. Metabolism of benzo[a]pyrene to trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene by recombinant human cytochrome P450 1B1 and purified liver epoxide hydrolase. Chem. Res. Toxicol. 1999;12:623–629. doi: 10.1021/tx990028s. [DOI] [PubMed] [Google Scholar]
- (221).Shimada T, Watanabe J, Inoue K, Guengerich FP, Gillam EMJ. Specificity of 17β-oestradiol and benzo[a]pyrene oxidation by polymorphic human cytochrome P4501B1 variants substituted at residues 48, 119 and 432. Xenobiotica. 2001;31:163–176. doi: 10.1080/00498250110043490. [DOI] [PubMed] [Google Scholar]
- (222).Aklillu E, Ovrebo S, Botnen IV, Otter C, Ingelman-Sundberg M. Characterization of common CYP1B1 variants with different capacity for benzo[a]pyrene-7,8-dihydrodiol epoxide formation from benzo[a]pyrene. Cancer Res. 2005;65:5105–5111. doi: 10.1158/0008-5472.CAN-05-0113. [DOI] [PubMed] [Google Scholar]
- (223).Einolf HJ, Story WT, Marcus CB, Larsen MC, Jefcoate CR, Greenlee WF, Yagi H, Jerina DM, Amin S, Park SS, Gelboin HV, Baird WM. Role of cytochrome P450 enzyme induction in the metabolic activation of benzo[c]phenentrene in human cell lines and mouse epidermis. Chem. Res. Toxicol. 1997;10:609–617. doi: 10.1021/tx960174n. [DOI] [PubMed] [Google Scholar]
- (224).Mahadevan B, Luch A, Atkin J, Haynes M, Nguyen T, Baird WM. Inhibition of human cytochrome P450 1B1 further clarifies its role in the activation of dibenzo[a,l]pyrene in cells in culture. J. Biochem. Mol. Toxicol. 2007;21:101–109. doi: 10.1002/jbt.20168. [DOI] [PubMed] [Google Scholar]
- (225).Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17β-Estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Liehr JG, Ricci MJ, Jefcoate CR, Hannigan EV, Hokanson JA, Zhu BT. 4-Hydroxylation of estradiol by human uterine myometrium and myoma microsomes: Implications for the mechanism of uterine tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9220–9224. doi: 10.1073/pnas.92.20.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Li DN, Seidel A, Pritchard MP, Wolf CR, Friedberg T. Polymorphisms in P450 CYP1B1 affect the conversion of estradiol to the potentially carcinogenic metabolite 4-hydroxyestradiol. Pharmacogenetics. 2000;10:343–353. doi: 10.1097/00008571-200006000-00008. [DOI] [PubMed] [Google Scholar]
- (228).Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF. Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: Association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res. 2000;60:3440–3444. [PubMed] [Google Scholar]
- (229).Sun Y-W, Guengerich FP, Sharma AK, Boyiri T, Amin S, El-Bayoumy K. Human cytochrome P450s 1A1 and 1B1 catalyze ring oxidation but not nitroreduction of environmental pollutants mono-nitropyrene isomers in primary cultures of human breast cells and cultured MCF-10A and MCF-7 cell lines. Chem. Res. Toxicol. 2004;17:1077–1085. doi: 10.1021/tx049889d. [DOI] [PubMed] [Google Scholar]
- (230).Wormhoudt LW, Ploemen JH, de Waziers I, Commandeur JN, Beaune PH, van Bladeren PJ, Vermeulen NPE. Inter-individual variability in the oxidation of 1,2-dibromoethane. Use of heterologously expressed human cytochrome P450 and human liver microsomes. Chem.-Biol. Interact. 1996;101:175–192. doi: 10.1016/0009-2797(96)03723-4. [DOI] [PubMed] [Google Scholar]
- (231).Seaton MJ, Follansbee MH, Bond JA. Oxidation of 1,2-epoxy-3-butene to 1,2 3,4-diepoxybutane by cDNA-expressed human cytochromes P450 2E1 and 3A4 and human, mouse and rat liver microsomes. Carcinogenesis. 1995;16:2287–2293. doi: 10.1093/carcin/16.10.2287. [DOI] [PubMed] [Google Scholar]
- (232).Elfarra AA, Krause RJ, Selzer RR. Biochemistry of 1,3-butadiene metabolism and its relevance to 1,3-butadiene-induced carcinogenicity. Toxicology. 1996;113:23–30. doi: 10.1016/0300-483x(96)03423-3. [DOI] [PubMed] [Google Scholar]
- (233).Krause RJ, Elfarra AA. Oxidation of butadiene monoxide to meso- and (±)-diepoxybutane by cDNA-expressed human cytochrome P450s and by mouse, rat, and human liver microsomes. Evidence for preferential hydration of meso-diepoxybutane in rat and human liver microsomes. Arch. Biochem. Biophys. 1997;337:176–184. doi: 10.1006/abbi.1996.9781. [DOI] [PubMed] [Google Scholar]
- (234).Ding X, Spink DC, Bhama JK, Sheng JJ, Vaz AD, Coon MJ. Metabolic activation of 2,6-dichlorobenzonitrile, an olfactory-specific toxicant, by rat, rabbit, and human cytochromes P450. Mol. Pharmacol. 1996;49:1113–1121. [PubMed] [Google Scholar]
- (235).Liu C, Zhuo X, Gonzalez FJ, Ding X. Baculovirus-mediated expression and characterization of rat CYP2A3 and human CYP2A6: Role in metabolic activation of nasal toxicants. Mol. Pharmacol. 1996;50:781–788. [PubMed] [Google Scholar]
- (236).Gan J, Skipper PL, Tannenbaum SR. Oxidation of 2,6-dimethylaniline by recombinant human cytochrome P450s and human liver microsomes. Chem. Res. Toxicol. 2001;14:672–677. doi: 10.1021/tx000181i. [DOI] [PubMed] [Google Scholar]
- (237).Jalas JR, Ding X, Murphy SE. Comparative metabolism of the tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol by rat cytochrome P450 2A3 and human cytochrome P450 2A13. Drug Metab. Dispos. 2003;31:1199–1202. doi: 10.1124/dmd.31.10.1199. [DOI] [PubMed] [Google Scholar]
- (238).Chiang HC, Wang CY, Lee HL, Tsou TC. Metabolic effects of CYP2A6 and CYP2A13 on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK)-induced gene mutation—a mammalian cell-based mutagenesis approach. Toxicol. Appl. Pharmacol. 2011;253:145–152. doi: 10.1016/j.taap.2011.03.022. [DOI] [PubMed] [Google Scholar]
- (239).Su T, Bao Z, Zhang QY, Smith TJ, Hong JY, Ding X. Human cytochrome P450 CYP2A13: Predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 2000;60:5074–5079. [PubMed] [Google Scholar]
- (240).He XY, Shen J, Ding X, Lu AYH, Hong JY. Identification of critical amino acid residues of human CYP2A13 for the metabolic activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco-specific carcinogen. Drug Metab. Dispos. 2004;32:1516–1521. doi: 10.1124/dmd.104.001370. [DOI] [PubMed] [Google Scholar]
- (241).Seidel A, Soballa VJ, Raab G, Greim H, Grimmer G, Jacob J, Doehmer J. Regio- and stereoselectivity of metabolism of benzo[c]phenanthrene mediated by genetically engineered V79 Chinese hamster cells expressing rat and human cytochromes P450. Environ. Toxicol. Pharmacol. 1998;5:179–196. doi: 10.1016/s1382-6689(97)10073-4. [DOI] [PubMed] [Google Scholar]
- (242).Zhang X, D’Agostino J, Wu H, Zhang QY, von Weymarn L, Murphy SE, Ding X. CYP2A13: Variable expression and role in human lung microsomal metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. J. Pharmacol. Exp. Ther. 2007;323:570–578. doi: 10.1124/jpet.107.127068. [DOI] [PubMed] [Google Scholar]
- (243).Wong HL, Murphy SE, Hecht SS. Cytochrome P450 2A-catalyzed metabolic activation of structurally similar carcinogenic N’-nitrosonornicotine, N-nitrosopiperidine, and N-nitrosopyrrolidine. Chem. Res. Toxicol. 2005;18:61–69. doi: 10.1021/tx0497696. [DOI] [PubMed] [Google Scholar]
- (244).Kushida H, Fujita K, Suzuki A, Yamada M, Endo T, Nohmi T, Kamataki T. Metabolic activation of N-alkylnitrosamines in genetically engineered Salmonella typhimurium expressing CYP2E1 or CYP2A6 together with human NADPH-cytochrome P450 reductase. Carcinogenesis. 2000;21:1227–1232. [PubMed] [Google Scholar]
- (245).Kushida H, Fujita K, Suzuki A, Yamada M, Nohmi T, Kamataki T. Development of a Salmonella tester strain sensitive to promutagenic N-nitrosamines. Expression of recombinant CYP2A6 and human NADPH-cytochrome P450 reductase in S. typhimurium YG7108. Mut. Res. 2000;471:135–143. doi: 10.1016/s1383-5718(00)00117-0. [DOI] [PubMed] [Google Scholar]
- (246).Smith GB, Bend JR, Bedard LL, Reid KR, Petsikas D, Massey TE. Biotransformation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in peripheral human lung microsomes. Drug Metab. Dispos. 2003;31:1134–1141. doi: 10.1124/dmd.31.9.1134. [DOI] [PubMed] [Google Scholar]
- (247).Yamazaki H, Inui Y, Yun C-H, Mimura M, Guengerich FP, Shimada T. Cytochrome P450 2E1 and 2A6 enzymes as major catalysts for metabolic activation of N-nitrosodialkylamines and tobacco-related nitrosamines in human liver microsomes. Carcinogenesis. 1992;13:1789–1794. doi: 10.1093/carcin/13.10.1789. [DOI] [PubMed] [Google Scholar]
- (248).Yun CH, Shimada T, Guengerich FP. Purification and characterization of human liver microsomal cytochrome P450 2A6. Mol. Pharmacol. 1994;40:679–685. [PubMed] [Google Scholar]
- (249).Yun CH, Shimada T, Guengerich FP. Contributions of human liver cytochrome P450 enzymes to the N-oxidation of 4,4′-methylene-bis(2-chloroaniline) Carcinogenesis. 1992;13:217–222. doi: 10.1093/carcin/13.2.217. [DOI] [PubMed] [Google Scholar]
- (250).Gemma S, Vittozzi L, Testai E. Metabolism of chloroform in the human liver and identification of the competent P450s. Drug Metab. Dispos. 2003;31:266–274. doi: 10.1124/dmd.31.3.266. [DOI] [PubMed] [Google Scholar]
- (251).Chang TKH, Weber GF, Crespi CL, Waxman DJ. Differential activation of cyclophosphamide and iphosphamide by cytochromes P-450 2B and 3A in human liver microsomes. Cancer Res. 1993;53:5629–5637. [PubMed] [Google Scholar]
- (252).Bohnenstengel F, Hofmann U, Eichelbaum M, Kroemer HK. Characterization of the cytochrome P450 involved in side chain oxidation of cyclophosphamide in humans. Eur. J. Clin. Pharmacol. 1996;51:297–301. doi: 10.1007/s002280050201. [DOI] [PubMed] [Google Scholar]
- (253).Roy P, Yu LJ, Crespi CL, Waxman DJ. Development of a substrate-activity based approach to identify the major human liver P-450 catalysts of cyclophosphamide and ifosfamide activation based on cDNA-expressed activities and liver microsomal P-450 profiles. Drug Metab. Dispos. 1999;27:655–666. [PubMed] [Google Scholar]
- (254).Huang Z, Roy P, Waxman DJ. Role of human liver microsomal CYP3A4 and CYP2B6 in catalyzing N-dechloroethylation of cyclophosphamide and ifosfamide. Biochem. Pharmacol. 2000;59:961–972. doi: 10.1016/s0006-2952(99)00410-4. [DOI] [PubMed] [Google Scholar]
- (255).Thornton-Manning JR, Nikula KJ, Hotchkiss JA, Avila KJ, Rohrbacher KD, Ding X, Dahl AR. Nasal cytochrome P450 2A: Identification, regional localization, and metabolic activity toward hexamethylphosphoramide, a known nasal carcinogen. Toxicol. Appl. Pharmacol. 1997;142:22–30. doi: 10.1006/taap.1996.7975. [DOI] [PubMed] [Google Scholar]
- (256).Patten CJ, Smith TJ, Friesen MJ, Tynes RE, Yang CS, Murphy SE. Evidence for cytochrome P450 2A6 and 3A4 as major catalysts for N’-nitrosonornicotine α-hydroxylation by human liver microsomes. Carcinogenesis. 1997;18:1623–1630. doi: 10.1093/carcin/18.8.1623. [DOI] [PubMed] [Google Scholar]
- (257).Berkman CE, Park SB, Wrighton SA, Cashman JR. In vitro-in vivo correlations of human (S)-nicotine metabolism. Biochem. Pharmacol. 1995;50:565–570. doi: 10.1016/0006-2952(95)00168-y. [DOI] [PubMed] [Google Scholar]
- (258).Wong HL, Murphy SE, Hecht SS. Cytochrome P450 2A-catalyzed metabolic activation of structurally similar carcinogenic nitrosamines: N’-nitrosonornicotine enantiomers, N-nitrosopiperidine, and N-nitrosopyrrolidine. Chem. Res. Toxicol. 2005;18:61–69. doi: 10.1021/tx0497696. [DOI] [PubMed] [Google Scholar]
- (259).Jeurissen SM, Bogaards JJ, Awad HM, Boersma MG, Brand W, Fiamegos YC, van Beek TA, Alink GM, Sudholter EJ, Cnubben NH, Rietjens IMCM. Human cytochrome P450 enzyme specificity for bioactivation of safrole to the proximate carcinogen 1′-hydroxysafrole. Chem. Res. Toxicol. 2004;17:1245–1250. doi: 10.1021/tx040001v. [DOI] [PubMed] [Google Scholar]
- (260).Ueng Y-F, Hsieh CH, Don MJ, Chi CW, Ho LK. Identification of the main human cytochrome P450 enzymes involved in safrole 1′-hydroxylation. Chem. Res. Toxicol. 2004;17:1151–1156. doi: 10.1021/tx030055p. [DOI] [PubMed] [Google Scholar]
- (261).D’Agostino J, Zhuo X, Shadid M, Morgan DG, Zhang X, Humphreys WG, Shu YZ, Yost GS, Ding X. The pneumotoxin 3-methylindole is a substrate and a mechanism-based inactivator of CYP2A13, a human cytochrome P450 enzyme preferentially expressed in the respiratory tract. Drug Metab. Dispos. 2009;37:2018–2027. doi: 10.1124/dmd.109.027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (262).Bao Z, He XY, Ding X, Prabhu S, Hong JY. Metabolism of nicotine and cotinine by human cytochrome P450 2A13. Drug Metab. Dispos. 2005;33:258–261. doi: 10.1124/dmd.104.002105. [DOI] [PubMed] [Google Scholar]
- (263).Wong HL, Zhang X, Zhang QY, Gu J, Ding X, Hecht SS, Murphy SE. Metabolic activation of the tobacco carcinogen 4-(methylnitrosamino)-(3-pyridyl)-1-butanone by cytochrome P450 2A13 in human fetal nasal microsomes. Chem. Res. Toxicol. 2005;18:913–918. doi: 10.1021/tx0500777. [DOI] [PubMed] [Google Scholar]
- (264).Zhang X, Su T, Zhang QY, Gu J, Caggana M, Li H, Ding X. Genetic polymorphisms of the human CYP2A13 gene: Identification of single-nucleotide polymorphisms and functional characterization of an Arg257Cys variant. J. Pharmacol. Exp. Ther. 2002;302:416–423. doi: 10.1124/jpet.302.2.416. [DOI] [PubMed] [Google Scholar]
- (265).Wang SL, He XY, Shen J, Wang JS, Hong JY. The missense genetic polymorphisms of human CYP2A13. Functional significance in carcinogen activation and identification of a null allelic variant. Toxicol. Sci. 2006;94:38–45. doi: 10.1093/toxsci/kfl081. [DOI] [PubMed] [Google Scholar]
- (266).Schlicht KE, Berg JZ, Murphy SE. Effect of CYP2A13 active site mutation N297A on metabolism of coumarin and tobacco-specific nitrosamines. Drug Metab. Dispos. 2009;37:665–671. doi: 10.1124/dmd.108.025072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (267).Goto T, Moriuchi H, Fu X, Ikegawa T, Matsubara T, Chang G, Uno T, Morigaki K, Isshiki K, Imaishi H. The effects of single nucleotide polymorphisms in CYP2A13 on metabolism of 5-methoxypsoralen. Drug Metab. Dispos. 2010;38:2110–2116. doi: 10.1124/dmd.110.034553. [DOI] [PubMed] [Google Scholar]
- (268).White IN, De Matteis F. The role of CYP forms in the metabolism and metabolic activation of HCFCs and other halocarbons. Toxicol. Lett. 2001;124:121–128. doi: 10.1016/s0378-4274(00)00288-5. [DOI] [PubMed] [Google Scholar]
- (269).White IN, Razvi N, Gibbs AH, Davies AM, Manno M, Zaccaro C, Matteis FD, Pahler A, Dekant W. Neoantigen formation and clastogenic action of hydrochlorofluorocarbons-123 and perchloroethylene in human MCL-5 cells. Toxicol. Lett. 2001;124:129–138. doi: 10.1016/s0378-4274(00)00281-2. [DOI] [PubMed] [Google Scholar]
- (270).Mimura M, Baba T, Yamazaki H, Ohmori S, Inui Y, Gonzalez FJ, Guengerich FP, Shimada T. Characterization of cytochrome P-450 2B6 in human liver microsomes. Drug Metab. Dispos. 1993;21:1048–1056. [PubMed] [Google Scholar]
- (271).Code EL, Crespi CL, Penman BW, Gonzalez FJ, Chang TK, Waxman DJ. Human cytochrome P4502B6. Interindividual hepatic expression, substrate specificity, and role in procarcinogen activation. Drug Metab. Dispos. 25:985–993. [PubMed] [Google Scholar]
- (272).Dicke KE, Skrlin SM, Murphy SE. Nicotine and 4-(methylnitrosoamino)-1-(3-pyridyl)-butanone metabolism by cytochrome P450 2B6. Drug Metab. Dispos. 2005;33:1760–1764. doi: 10.1124/dmd.105.006718. [DOI] [PubMed] [Google Scholar]
- (273).Fontaine SM, Mash EA, Hoyer PB, Sipes IG. Stereochemical aspects of vinylcyclohexene bioactivation in rodent hepatic microsomes and purified human cytochrome P450 enzyme systems. Drug Metab. Dispos. 2001;29:179–184. [PubMed] [Google Scholar]
- (274).Coleman S, Linderman R, Hodgson E, Rose RL. Comparative metabolism of chloroacetamide herbicides and selected metabolites in human and rat liver microsomes. Environ. Health Perspect. 2000;108:1151–1157. doi: 10.1289/ehp.001081151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (275).Chen CS, Jounaidi Y, Waxman DJ. Enantioselective metabolism and cytotoxicity of R-ifosfamide and S-ifosfamide by tumor cell-expressed cytochromes P450. Drug Metab. Dispos. 2005;33:1261–1267. doi: 10.1124/dmd.105.004788. [DOI] [PubMed] [Google Scholar]
- (276).Roy P, Tretyakov O, Wright J, Waxman DJ. Stereoselective metabolism of ifosfamide by human P-450s 3A4 and 2B6. Favorable metabolic properties of R-enantiomer. Drug Metab. Dispos. 1999;27:1309–1318. [PubMed] [Google Scholar]
- (277).McCune JS, Risler LJ, Phillips BR, Thummel KE, Blough D, Shen DD. Contribution of CYP3A5 to hepatic and renal ifosfamide N-dechloroethylation. Drug Metab. Dispos. 2005;33:1074–1081. doi: 10.1124/dmd.104.002279. [DOI] [PubMed] [Google Scholar]
- (278).Sun D, Chen G, Dellinger RW, Duncan K, Fang JL, Lazarus P. Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res. 2006;8:R50. doi: 10.1186/bcr1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (279).Jacobson PA, Green K, Birnbaum A, Remmel RP. Cytochrome P450 isozymes 3A4 and 2B6 are involved in the in vitro human metabolism of thiotepa to TEPA. Cancer Chemother. Pharmacol. 2002;49:461–467. doi: 10.1007/s00280-002-0453-3. [DOI] [PubMed] [Google Scholar]
- (280).Ma M, Umemura T, Mori Y, Gong Y, Saijo Y, Sata F, Kawai T, Kishi R. Influence of genetic polymorphisms of styrene-metabolizing enzymes and smoking habits on levels of urinary metabolites after occupational exposure to styrene. Toxicol. Lett. 2005;160:84–91. doi: 10.1016/j.toxlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- (281).Penman BW, Reece J, Smith T, Yang CS, Gelboin HV, Gonzalez FJ, Crespi CL. Characterization of a human cell line expressing high levels of cDNA-derived CYP2D6. Pharmacogenetics. 1993;3:28–39. doi: 10.1097/00008571-199302000-00003. [DOI] [PubMed] [Google Scholar]
- (282).Guengerich FP, Kim D-H, Iwasaki M. Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem. Res. Toxicol. 1991;4:168–179. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- (283).Cheung C, Yu AM, Ward JM, Krausz KW, Akiyama TE, Feigenbaum L, Gonzalez FJ. The cyp2e1-humanized transgenic mouse: Role of cyp2e1 in acetaminophen hepatotoxicity. Drug Metab. Dispos. 2005;33:449–457. doi: 10.1124/dmd.104.002402. [DOI] [PubMed] [Google Scholar]
- (284).Doherty AT, Ellard S, Parry EM, Parry JM. An investigation into the activation and deactivation of chlorinated hydrocarbons to genotoxins in metabolically competent human cells. Mutagenesis. 1996;11:247–274. doi: 10.1093/mutage/11.3.247. [DOI] [PubMed] [Google Scholar]
- (285).Simmonds AC, Reilly CA, Baldwin RM, Ghanayem BI, Lanza DL, Yost GS, Collins KS, Forkert PG. Bioactivation of 1,1-dichloroethylene to its epoxide by CYP2E1 and CYP2F enzymes. Drug Metab. Dispos. 2004;32:1032–1039. [PubMed] [Google Scholar]
- (286).Nieusma JL, Claffey DJ, Koop DR, Chen W, Peter RM, Nelson SD, Ruth JA, Ross D. Oxidation of 1,3-butadiene to (R)- and (S)-butadiene monoxide by purified recombinant cytochrome P450 2E1 from rabbit, rat and human. Toxicol. Lett. 1998;95:123–129. doi: 10.1016/s0378-4274(98)00026-5. [DOI] [PubMed] [Google Scholar]
- (287).Nedelcheva V, Gut I, Soucek P, Frantík E. Cytochrome P450 catalyzed oxidation of monochlorobenzene, 1,2- and 1,4-dichlorobenzene in rat, mouse, and human liver microsomes. Chem.-Biol. Interact. 1998;115:53–70. doi: 10.1016/s0009-2797(98)00058-1. [DOI] [PubMed] [Google Scholar]
- (288).Herbst J, Köster U, Kerssebaum R, Dekant W. Role of P4502E1 in the metabolism of 1,1,2,2,-tetrafluoro-1-(2,2,2-trifluoroethoxy)ethane. Xenobiotica. 1994;24:507–516. doi: 10.3109/00498259409043253. [DOI] [PubMed] [Google Scholar]
- (289).Urban G, Speerschnaider P, Dekant W. Metabolism of the chlorofluorocarbon substitute 1,1-dichloro-2,2,2-trifluoroethane by rat and human liver microsomes. The role of cytochrome P450 2E1. Chem. Res. Toxicol. 1994;7:170–176. doi: 10.1021/tx00038a009. [DOI] [PubMed] [Google Scholar]
- (290).Doroshyenko O, Fuhr U, Kunz D, Frank D, Kinzig M, Jetter A, Reith Y, Lazar A, Taubert D, Kirchheiner J, Baum M, Eisenbrand G, Berger FI, Bertow D, Berkessel A, Sörgel F, Schömig E, Tomalik-Scharte D. In vivo role of cytochrome P450 2E1 and glutathione-S-transferase activity for acrylamide toxicokinetics in humans. Cancer Epidemiol. Biomarkers Prev. 2009;18:433–443. doi: 10.1158/1055-9965.EPI-08-0832. [DOI] [PubMed] [Google Scholar]
- (291).Koyama N, Yasui M, Oda Y, Suzuki S, Satoh T, Suzuki T, Matsuda T, Masuda S, Kinae N, Honma M. Genotoxicity of acrylamide in vitro. Acrylamide is not metabolically activated in standard in vitro systems. Environ. Mol. Mutagen. 2011;52:11–19. doi: 10.1002/em.20560. [DOI] [PubMed] [Google Scholar]
- (292).Kedderis GL, Batra R, Koop DR. Epoxidation of acrylonitrile by rat and human cytochromes P450. Chem. Res. Toxicol. 1993;6:866–871. doi: 10.1021/tx00036a017. [DOI] [PubMed] [Google Scholar]
- (293).Nedelcheva V, Gut I, Soucek P, Tichavska B, Tynkova L, Mraz J, Guengerich FP, Ingelman-Sundberg M. Metabolism of benezene in human liver microsomes. Individual variations in relation to CYP2E1 expression. Arch. Toxicol. 1999;73:33–40. doi: 10.1007/s002040050583. [DOI] [PubMed] [Google Scholar]
- (294).Powley MW, Carlson GP. Cytochromes P450 involved with benzene metabolism in hepatic and pulmonary microsomes. J. Biochem. Mol. Toxicol. 2000;14:303–309. doi: 10.1002/1099-0461(2000)14:6<303::AID-JBT2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- (295).Sheets PL, Yost GS, Carlson GP. Benzene metabolism in human lung cell lines BEAS-2B and A549 and cells overexpressing CYP2F1. J. Biochem. Mol. Toxicol. 2004;18:92–99. doi: 10.1002/jbt.20010. [DOI] [PubMed] [Google Scholar]
- (296).Boysen G, Scarlett CO, Temple B, Combs TP, Brooks NL, Borchers CH, Swenberg JA. Identification of covalent modifications in P450 2E1 by 1,2-epoxy-3-butene in vitro. Chem.-Biol. Interact. 2007;166:170–175. doi: 10.1016/j.cbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- (297).Raucy JL. Risk assessment toxicity from chemical exposure resulting from enhanced expression of CYP2E1. Toxicology. 1995;105:217–224. doi: 10.1016/0300-483x(95)03216-3. [DOI] [PubMed] [Google Scholar]
- (298).Zangar RC, Benson JM, Burnett VL, Springer DL. Cytochrome P450 2E1 is the primary enzyme responsible for low-dose carbon tetrachloride metabolism in human liver microsomes. Chem.-Biol. Interact. 2000;125:233–243. doi: 10.1016/s0009-2797(00)00149-6. [DOI] [PubMed] [Google Scholar]
- (299).Raucy JL, Krane JC, Lasker JM. Bioactivation of halogenated hydrocarbons by cytochrome P4502E1. Crit. Rev. Toxicol. 1993;23:1–20. doi: 10.3109/10408449309104072. [DOI] [PubMed] [Google Scholar]
- (300).Takahashi S, Takahashi T, Mizobuchi S, Matsumi M, Morita K, Miyazaki M, Namba M, Akagi R, Hirakawa M. Increased cytotoxicity of carbon tetrachloride in a human hepatoma cell line overexpressing cytochrome P450 2E1. J. Int. Med. Res. 2002;30:400–405. doi: 10.1177/147323000203000406. [DOI] [PubMed] [Google Scholar]
- (301).Lipscomb JC, Barton HA, Tornero-Velez R, Evans MV, Alcasey S, Snawder JE, Laskey J. The metabolic rate constants and specific activity of human and rat hepatic cytochrome P-450 2E1 toward toluene and chloroform. J. Toxicol. Environ. Health A. 2004;67:537–53. doi: 10.1080/15287390490425588. [DOI] [PubMed] [Google Scholar]
- (302).Himmelstein MW, Carpenter SC, Hinderliter PM, Snow TA, Valentine R. The metabolism of β-chloroprene: preliminary in-vitro studies using liver microsomes. Chem.-Biol. Interact. 2001;135-136:267–84. doi: 10.1016/s0009-2797(01)00214-9. [DOI] [PubMed] [Google Scholar]
- (303).Lu Y, Cederbaum AI. Cisplatin-induced hepatotoxicity is enhanced by elevated expression of cytochrome P450 2E1. Toxicol. Sci. 2006;89:515–523. doi: 10.1093/toxsci/kfj031. [DOI] [PubMed] [Google Scholar]
- (304).Lu Y, Cederbaum A. The mode of cisplatin-induced cell death in CYP2E1-overexpressing HepG2 cells: Modulation by ERK, ROS, glutathione, and thioredoxin. Free Radic. Biol. Med. 2007;43:1061–1075. doi: 10.1016/j.freeradbiomed.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (305).Dai Y, Rashba-Step J, Cederbaum AI. Stable expression of human cytochrome P4502E1 in HepG2 cells. Characterization of catalytic activities and production of reactive oxygen intermediates. Biochemistry. 1993;32:6928–6937. doi: 10.1021/bi00078a017. [DOI] [PubMed] [Google Scholar]
- (306).Liu LG, Yan H, Yao P, Zhang W, Zou LJ, Song FF, Li K, Sun XF. CYP2E1-dependent hepatotoxicity and oxidative damage after ethanol administration in human primary hepatocytes. World J. Gastroenterol. 2005;11:4530–4535. doi: 10.3748/wjg.v11.i29.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (307).Cederbaum AI. CYP2E1. Biochemical and toxicological aspects and role in alcohol-induced liver injury. Mt. Sinai J. Med. 2006;73:657–672. [PubMed] [Google Scholar]
- (308).Shimada M, Liu L, Nussler N, Jonas S, Langrehr JM, Ogawa T, Kaminishi M, Neuhaus P, Nussler AK. Human hepatocytes are protected from ethanol-induced cytotoxicity by DADS via CYP2E1 inhibition. Toxicol. Lett. 2006;163:242–249. doi: 10.1016/j.toxlet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- (309).Magne L, Blanc E, Legrand B, Lucas D, Barouki R, Rouach H, Garlatti M. ATF4 and the integrated stress response are induced by ethanol and cytochrome P450 2E1 in human hepatocytes. J. Hepatol. 2011;54:729–737. doi: 10.1016/j.jhep.2010.07.023. [DOI] [PubMed] [Google Scholar]
- (310).Forkert PG, Lee RP. Metabolism of ethyl carbamate by pulmonary cytochrome P450 and carboxylesterase isozymes. Involvement of CYP 2E1 and hydrolase A. Toxicol. Appl. Pharmacol. 1997;146:245–254. doi: 10.1006/taap.1997.8233. [DOI] [PubMed] [Google Scholar]
- (311).Hubner P, Groux PM, Weibel B, Sengstag C, Horlbeck J, Leong-Morgenthaler PM, Luthy J. Genotoxicity of ethyl carbamate (urethane) in Salmonella, yeast and human lymphoblastoid cells. Mutat. Res. 1997;390:11–19. doi: 10.1016/s0165-1218(96)00160-7. [DOI] [PubMed] [Google Scholar]
- (312).Forkert PG, Lee RP, Reid K. Involvement of CYP2E1 and carboxylesterase enzymes in vinyl carbamate metabolism in human lung microsomes. Drug Metab. Dispos. 2001;29:258–263. [PubMed] [Google Scholar]
- (313).Peterson LA, Cummings ME, Vu CC, Matter BA. Glutathione trapping to measure microsomal oxidation of furan to cis-2-butene-1,4-dial. Drug Metab. Dispos. 2005;33:1453–1458. doi: 10.1124/dmd.105.004432. [DOI] [PubMed] [Google Scholar]
- (314).Mraz J, Jheeta P, Gescher A, Hyland R, Thumel K, Threadgill MD. Investigation of the mechanistic basis of N,N-dimethylformamide toxicity. Metabolism of N,N-dimethylformamide and its deuterated isotopomers by cytochrome P450 2E1. Chem. Res. Toxicol. 1993;6:197–207. doi: 10.1021/tx00032a009. [DOI] [PubMed] [Google Scholar]
- (315).Amato G, Grasso E, Longo V, Gervasi PG. Oxidation of N,N-dimethylformamide and N,N-diethylformamide by human liver microsomes and human recombinant P450s. Toxicol. Lett. 2001;124:11–19. doi: 10.1016/s0378-4274(01)00324-1. [DOI] [PubMed] [Google Scholar]
- (316).Hyland R, Gescher A, Thumel K, Schiller C, Jheeta P, Mynett K, Smith AW, Mraz J. Metabolic oxidation and toxification of N-methylformamide catalyzed by the cytochrome P450 isoenzyme CYP2E1. Mol. Pharmacol. 1992;41:259–266. [PubMed] [Google Scholar]
- (317).Liu Y, Glatt H. Mutagenicity of N-nitrosodiethanolamine in a V79-derived cell line expressing two human biotransformation enzymes. Mut. Res. 2008;643:64–69. doi: 10.1016/j.mrfmmm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- (318).Cooper MT, Porter TD. Cytochrome b(5) coexpression increases the CYP2E1-dependent mutagenicity of dialkylnitrosamines in methyltransferase-deficient strains of Salmonella typhimurium. Mut. Res. 2001;484:61–68. doi: 10.1016/s0027-5107(01)00236-6. [DOI] [PubMed] [Google Scholar]
- (319).Zhuge J, Luo Y, Yu YN. Heterologous expression of human cytochrome P450 2E1 in HepG2 cell line. World J. Gastroenterol. 2003;9:2732–2736. doi: 10.3748/wjg.v9.i12.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (320).Nakagawa T, Sawada M, Gonzalez FJ, Yokoi T, Kamataki T. Stable expression of human CYP2E1 in Chinese hamster cells: High sensitivity to N,N-dimethylnitrosamine in cytotoxicity testing. Mutat. Res. 1996;354:181–186. doi: 10.1016/s0165-1161(96)90015-1. [DOI] [PubMed] [Google Scholar]
- (321).Lin HL, Parsels LA, Maybaum J, Hollenberg PF. N-Nitrosodimethylamine-mediated cytotoxicity in a cell line expressing P450 2E1. Evidence for apoptotic cell death. Toxicol. Appl. Pharmacol. 1999;157:117–124. doi: 10.1006/taap.1999.8651. [DOI] [PubMed] [Google Scholar]
- (322).Glatt H, Schneider H, Liu Y. V79-hCYP2E1-hSULT1A1, a cell line for the sensitive detection of genotoxic effects induced by carbohydrate pyrolysis products and other food-borne chemicals. Mut. Res. 2005;580:41–52. doi: 10.1016/j.mrgentox.2004.11.005. [DOI] [PubMed] [Google Scholar]
- (323).Bellec G, Dreano Y, Bail JP, Menez JF, Berthou F. Cytochrome P450 hydroxylation of carbon atoms of the alkyl chain of symmetrical N-nitrosodialkylamines by human liver microsomes. Mutat. Res. 1997;377:199–209. doi: 10.1016/s0027-5107(97)00073-0. [DOI] [PubMed] [Google Scholar]
- (324).Bellec G, Dreano Y, Lozach P, Menez JF, Berthou F. Cytochrome P450 metabolic dealkylation of nine N-nitrosodialkylamines by human liver microsomes. Carcinogenesis. 1996;17:2029–2034. doi: 10.1093/carcin/17.9.2029. [DOI] [PubMed] [Google Scholar]
- (325).Teiber JF, Hollenberg PF. Identification of the human liver microsomal cytochrome P450s involved in the metabolism of N-nitrosodi-n-propylamine. Carcinogenesis. 2000;21:1559–1566. [PubMed] [Google Scholar]
- (326).Ekins S, Wrighton SA. The role of CYP2B6 in human xenobiotic metabolism. Drug Metab. Rev. 1999;31:719–754. doi: 10.1081/dmr-100101942. [DOI] [PubMed] [Google Scholar]
- (327).Krishnan S, Hvastkovs EG, Bajrami B, Jansson I, Schenkman JB, Rusling JF. Genotoxicity screening for N-nitroso compounds. Electrochemical and electrochemiluminescent detection of human enzyme-generated DNA damage from N-nitrosopyrrolidine. Chem. Commun (Camb) 2007;7:1713–1715. doi: 10.1039/b703012f. [DOI] [PubMed] [Google Scholar]
- (328).Stiborová M, Naiman K, Martínková M, Martínek V, Svobodová M, Schmeiser HH, Frei E. Genotoxic mechanisms for the carcinogenicity of the environmental pollutants and carcinogens o-anisidine and 2-nitroanisole follow from adducts generated by their metabolite N-(2-methoxyphenyl)-hydroxylamine with deoxyguanosine in DNA. Interdiscip. Toxicol. 2009;2:24–27. doi: 10.2478/v10102-009-0004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (329).Chung JK, Yuan W, Liu G, Zheng J. Investigation of bioactivation and toxicity of styrene in CYP2E1 transgenic cells. Toxicology. 226:99–106. doi: 10.1016/j.tox.2006.06.001. [DOI] [PubMed] [Google Scholar]
- (330).Lipscomb JC, Garrett CM, Snawder JE. Cytochrome P450-dependent metabolism of trichloroethylene. Interindividual differences in humans. Toxicol. Appl. Pharmacol. 1997;142:311–318. doi: 10.1006/taap.1996.8040. [DOI] [PubMed] [Google Scholar]
- (331).Lash LH, Fisher JW, Lipscomb JC, Parker JC. Metabolism of trichloroethylene. Environ. Health Perspect. 2000;108(Suppl 2):177–200. doi: 10.1289/ehp.00108s2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (332).Cai H, Guengerich FP. Reaction of trichloroethylene and trichloroethylene oxide with cytochrome P450 enzymes. Inactivation and sites of modification. Chem. Res. Toxicol. 2001;14:451–458. doi: 10.1021/tx0002280. [DOI] [PubMed] [Google Scholar]
- (333).Forkert PG, Baldwin RM, Millen B, Lash LH, Putt DA, Shultz MA, Collins KS. Pulmonary bioactivation of trichloroethylene to chloral hydrate relative contributions of CYP2E1, CYP2F, and CYP2B1. Drug Metab. Dispos. 2005;33:1429–1437. doi: 10.1124/dmd.105.005074. [DOI] [PubMed] [Google Scholar]
- (334).Lanza DL, Code E, Crespi CL, Gonzalez FJ, Yost GS. Specific dehydrogenation of 3-methylindole and epoxidation of naphthalene by recombinant human CYP2F1 expressed in lymphoblastoid cells. Drug Metab. Dispos. 1999;27:798–803. [PubMed] [Google Scholar]
- (335).Kartha JS, Yost GS. Mechanism-based inactivation of lung-selective cytochrome P450 CYP2F enzymes. Drug Metab. Dispos. 2008;36:155–162. doi: 10.1124/dmd.107.017897. [DOI] [PubMed] [Google Scholar]
- (336).Gillam EM, Wunsch RM, Ueng Y-F, Shimada T, Reilly PE, Kamataki T, Guengerich FP. Expression of cytochrome P450 3A7 in Escherichia coli. Effects of 5′ modification and catalytic characterization of recombinant enzyme expressed in bicistronic format with NADPH-cytochrome P450 reductase. Arch. Biochem. Biophys. 1997;346:81–90. doi: 10.1006/abbi.1997.0286. [DOI] [PubMed] [Google Scholar]
- (337).Mesárosová M, Valovičová Z, Srančíková A, Krajčovičová Z, Milcová A, Sokolová R, Schmuczerová J, Topinka J, Gábelová A. The role of human cytochrome P4503A4 in biotransformation of tissue-specific derivatives of 7H-dibenzo[c,g]carbazole. Toxicol. Appl. Pharmacol. 2011;255:307–315. doi: 10.1016/j.taap.2011.06.027. [DOI] [PubMed] [Google Scholar]
- (338).Hakkola J, Pelkonen O, Pasanen M, Raunio H. Xenobiotic-metabolizing cytochrome P450 enzymes in the human feto-placental unit. Role in intrauterine toxicity. Crit. Rev. Toxicol. 1998;28:35–72. doi: 10.1080/10408449891344173. [DOI] [PubMed] [Google Scholar]
- (339).Raney KD, Shimada T, Kim DH, Groopman JD, Harris TM, Guengerich FP. Oxidation of aflatoxins and sterigmatocystin by human liver microsomes: significance of aflatoxin Q1 as a detoxication product of aflatoxin B1. Chem. Res. Toxicol. 1992;5:202–210. doi: 10.1021/tx00026a009. [DOI] [PubMed] [Google Scholar]
- (340).De Groene EM, Seinen W, Horbach GJ. A NIH/3T3 cell line stably expressing human cytochrome P450-3A4 used in combination with a lacZ’ shuttle vector to study mutagenicity. Eur. J. Pharmacol. 1995;293:47–53. doi: 10.1016/0926-6917(95)90017-9. [DOI] [PubMed] [Google Scholar]
- (341).Hashimoto H, Nakagawa T, Yokoi T, Sawada M, Itoh S, Kamataki T. Fetus-specific CYP3A7 and adult-specific CYP3A4 expressed in Chinese hamster CHL cells have similar capacity to activate carcinogenic mycotoxins. Cancer Res. 1995;55:787–791. [PubMed] [Google Scholar]
- (342).Buening MK, Fortner JG, Kappas A, Conney AH. 7,8-Benzoflavone stimulates the metabolic activation of aflatoxin B1 to mutagens by human liver. Biochem. Biophys. Res. Commun. 1978;82:348–355. doi: 10.1016/0006-291x(78)90616-2. [DOI] [PubMed] [Google Scholar]
- (343).Sabater Vilar M, Kuilman-Wahls ME, Fink-Gremmels J. Inhibition of aflatoxin B1 mutagenicity by cyclopiazonic acid in the presence of human liver preparations. Toxicol. Lett. 2003;143:291–299. doi: 10.1016/s0378-4274(03)00196-6. [DOI] [PubMed] [Google Scholar]
- (344).Philip PA, Ali-Sadat S, Doehmer J, Kocarek T, Akhtar A, Lu H, Chan KK. Use of V79 cells with stably transfected cytochrome P450 cDNAs in studying the metabolism and effects of cytotoxic drugs. Cancer Chemother. Pharmacol. 1999;43:59–67. doi: 10.1007/s002800050863. [DOI] [PubMed] [Google Scholar]
- (345).Stiborová M, Rupertová M, Frei E. Cytochrome P450-and peroxidase-mediated oxidation of anticancer alkaloid ellipticine dictates its anti-tumor efficiency. Biochim. Biophys. Acta. 2011;1814:175–185. doi: 10.1016/j.bbapap.2010.05.016. [DOI] [PubMed] [Google Scholar]
- (346).Hosea NA, Miller GP, Guengerich FP. Elucidation of distinct ligand binding sites for cytochrome P450 3A4. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- (347).Williams JA, Ring BJ, Cantrell VE, Jones DR, Eckstein J, Ruterbories K, Hamman MA, Hall SD, Wrighton SA. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab. Dispos. 2002;30:883–891. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- (348).Lin HL, Hollenberg PF. The inactivation of cytochrome P450 A5 by 17α-ethynylestradiol is cytochrome b5-dependent: Metabolic activation of the ethynyl moiety leads to the formation of glutathione conjugates, a heme adduct, and covalent binding to the apoprotein. J. Pharmacol. Exp. Ther. 2007;321:276–287. doi: 10.1124/jpet.106.117861. [DOI] [PubMed] [Google Scholar]
- (349).Relling MV, Nemec J, Schuetz EG, Schuetz JD, Gonzalez FJ, Korzekwa KR. O-Demethylation of epipodophyllotoxins is catalyzed by human cytochrome P450. Mol. Pharmacol. 1994;45:352–358. [PubMed] [Google Scholar]
- (350).Zhao XJ, Kawashiro T, Ishizaki T. Mutual inhibition between quinine and etoposide by human liver microsomes. Evidence for cytochrome P450 3A4 involvement in their major metabolic pathways. Drug Metab. Dispos. 1998;26:188–191. [PubMed] [Google Scholar]
- (351).Zheng N, Pang S, Oe T, Felix CA, Wehrli S, Blair IA. Characterization of an etoposide-glutathione conjugate derived from metabolic activation by human cytochrome P450. Curr. Drug Metab. 2006;7:897–911. doi: 10.2174/138920006779010638. [DOI] [PubMed] [Google Scholar]
- (352).Walker D, Flinois J-P, Monkman SC, Beloc C, Boddy AV, Cholerton S, Daly AK, Lind MJ, Pearson ADJ, Beaune PH, Idle JR. Identification of the major human hepatic cytochrome P450 involved in activation and N-dechloroethylation of ifosfamide. Biochem. Pharmacol. 1994;47:1157–1163. doi: 10.1016/0006-2952(94)90387-5. [DOI] [PubMed] [Google Scholar]
- (353).Granvil CP, Madan A, Sharkawi M, Parkinson A, Wainer IW. Role of CYP2B6 and CYP3A4 in the in vitro N-dechloroethylation of (R)- and (S)-ifosfamide in human liver microsomes. Drug Metab. Dispos. 1999;27:533–541. [PubMed] [Google Scholar]
- (354).Dehal SS, Kupfer D. Cytochrome P-450 3A and 2D6 catalyze ortho hydroxylation of 4-hydroxytamoxifen and 3-hydroxytamoxifen (droloxifene) yielding tamoxifen catechol: Involvement of catechols in covalent binding to hepatic proteins. Drug Metab. Dispos. 1999;27:681–688. [PubMed] [Google Scholar]
- (355).Lu H, Wang JJ, Chan KK, Philip PA. Stereoselectivity in metabolism of ifosfamide by CYP3A4 and CYP2B6. Xenobiotica. 2006;36:367–385. doi: 10.1080/00498250600598486. [DOI] [PubMed] [Google Scholar]
- (356).Wang YP, Yan J, Fu PP, Chou MW. Human liver microsomal reduction of pyrrolizidine alkaloid N-oxides to form the corresponding carcinogenic parent alkaloid. Toxicol. Lett. 2005;155:411–420. doi: 10.1016/j.toxlet.2004.11.010. [DOI] [PubMed] [Google Scholar]
- (357).Miranda CL, Reed RL, Guengerich FP, Buhler DR. Role of cytochrome P450IIIA4 in the metabolism of the pyrrolizidine alkaloid senecionine in human liver. Carcinogenesis. 1991;12:515–519. doi: 10.1093/carcin/12.3.515. [DOI] [PubMed] [Google Scholar]
- (358).Notley LM, De Wolf CJ, Wunsch RM, Lancaster RG, Gillam EM. Bioactivation of tamoxifen by recombinant human cytochrome P450 enzymes. Chem. Res. Toxicol. 2002;15:614–622. doi: 10.1021/tx0100439. [DOI] [PubMed] [Google Scholar]
- (359).Kim SY, Suzuki N, Santosh Laxmi YR, Rieger R, Shibutani S. α-Hydroxylation of tamoxifen and toremifene by human and rat cytochrome P450 3A subfamily enzymes. Chem. Res. Toxicol. 2003;16:1138–1144. doi: 10.1021/tx0300131. [DOI] [PubMed] [Google Scholar]
- (360).Coller JK, Krebsfaenger N, Klein K, Wolbold R, Nussler A, Neuhaus P, Zanger UM, Eichelbaum M, Murdter TE. Large interindividual variability in the in vitro formation of tamoxifen metabolites related to the development of genotoxicity. Br. J. Clin. Pharmacol. 2004;57:105–11. doi: 10.1046/j.1365-2125.2003.01970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (361).Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J. Pharmacol. Exp. Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- (362).Notley LM, Crewe KH, Taylor PJ, Lennard MS, Gillam EMJ. Characterization of the human cytochrome P450 forms involved in metabolism of tamoxifen to its α-hydroxy and α,4-dihydroxy derivatives. Chem. Res. Toxicol. 2005;18:1611–1618. doi: 10.1021/tx050140s. [DOI] [PubMed] [Google Scholar]
- (363).Crewe HK, Notley LM, Wunsch RM, Lennard MS, Gillam EMJ. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes. Formation of the 4-hydroxy, 4′-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab. Dispos. 2002;30:869–874. doi: 10.1124/dmd.30.8.869. [DOI] [PubMed] [Google Scholar]
- (364).Boocock DJ, Brown K, Gibbs AH, Sanchez E, Turteltaub KW, White IN. Identification of human CYP forms involved in the activation of tamoxifen and irreversible binding to DNA. Carcinogenesis. 2002;23:1897–1901. doi: 10.1093/carcin/23.11.1897. [DOI] [PubMed] [Google Scholar]
- (365).Gillam EMJ, Guo Z, Ueng Y-F, Yamazaki H, Cock I, Reilly PEB, Hooper WD, Guengerich FP. Expression of cytochrome P450 3A5 in Escherichia coli. Effects of 5′ modification, purification, spectral characterization, reconstitution conditions, and catalytic activities. Arch. Biochem. Biophys. 1995;317:374–384. doi: 10.1006/abbi.1995.1177. [DOI] [PubMed] [Google Scholar]
- (366).Li Y, Yokoi T, Katsuki M, Wang J-S, Groopman JD, Kamataki T. In vivo activation of aflatoxin B1 in C57BL/6N mice carrying a human fetus-specific CYP3A7 gene. Cancer Res. 1997;57:641–645. [PubMed] [Google Scholar]
- (367).Imaoka S, Hayashi K, Hiroi T, Yabusaki Y, Kamataki T, Funae Y. A transgenic mouse expressing human CYP4B1 in the liver. Biochem. Biophys Res. Commun. 2001;284:757–762. doi: 10.1006/bbrc.2001.5055. [DOI] [PubMed] [Google Scholar]
- (368).Imaoka S, Yoneda Y, Sugimoto T, Hiroi T, Yamamoto K, Nakatani T, Funae Y. CYP4B1 is a possible risk factor for bladder cancer in humans. Biochem. Biophys. Res. Commun. 2000;277:776–780. doi: 10.1006/bbrc.2000.3740. [DOI] [PubMed] [Google Scholar]
- (369).Osawa Y, Higashiyama T, Shimizu Y, Yarborough C. Multiple functions of aromatase and the active site structure. Aromatase is the placental estrogen 2-hydroxylase. J. Steroid Biochem. Mol Biol. 1993;44:469–480. doi: 10.1016/0960-0760(93)90252-r. [DOI] [PubMed] [Google Scholar]
- (370).Kim YM, Ziegler DM. Size limits of thiocarbamides accepted as substrates by human flavin-containing monooxygenase 1. Drug Metab. Dispos. 2000;28:1003–1006. [PubMed] [Google Scholar]
- (371).Furnes B, Schlenk D. Evaluation of xenobiotic N- and S-oxidation by variant flavin-containing monooxygenase 1 (FMO1) enzymes. Toxicol. Sci. 2004;78:196–203. doi: 10.1093/toxsci/kfh079. [DOI] [PubMed] [Google Scholar]
- (372).Henderson MC, Krueger SK, Stevens JF, Williams DE. Human flavin-containing monooxygenase form 2. S-Oxygenation sulfenic acid formation from thioureas and oxidation of glutathione. Chem. Res. Toxicol. 2004;17:633–640. doi: 10.1021/tx034253s. [DOI] [PubMed] [Google Scholar]
- (373).Krueger SK, Martin SR, Yueh MF, Pereira CB, Williams DE. Identification of active flavin-containing monooxygenase isoform 2 in human lung and characterization of expressed protein. Drug Metab. Dispos. 2002;30:34–41. doi: 10.1124/dmd.30.1.34. [DOI] [PubMed] [Google Scholar]
- (374).Smith PB, Crespi C. Thiourea toxicity in mouse C3H/10T1/2 cells expressing human flavin-dependent monooxygenase 3. Biochem. Pharmacol. 2002;63:1941–1948. doi: 10.1016/s0006-2952(02)00978-4. [DOI] [PubMed] [Google Scholar]
- (375).Simula TP, Glancey MJ, Wolf CR. Human glutathione S-transferase-expressing Salmonella typhimurium tester strains to study the activation/detoxification of mutagenic compounds: studies with halogenated compounds, aromatic amines and aflatoxin B1. Carcinogenesis. 1993;14:1371–1376. doi: 10.1093/carcin/14.7.1371. [DOI] [PubMed] [Google Scholar]
- (376).Josephy PD, Taylor PL, Vervaet G, Mannervik B. Screening and characterization of variant Theta-class glutathione transferases catalyzing the activation of ethylene dibromide to a mutagen. Environ. Mol. Mutagen. 2006;47:657–665. doi: 10.1002/em.20252. [DOI] [PubMed] [Google Scholar]
- (377).Guengerich FP. Activation of dihaloalkanes by thiol-dependent mechanisms. J. Biochem. Molec. Biol. 2003;36:20–27. doi: 10.5483/bmbrep.2003.36.1.020. [DOI] [PubMed] [Google Scholar]
- (378).Anders MW. Glutathione-dependent bioactivation of haloalkanes and haloalkenes. Drug Metab. Rev. 2004;36:583–594. doi: 10.1081/dmr-200033451. [DOI] [PubMed] [Google Scholar]
- (379).Thier R, Pemble S, Kramer H, Taylor JB, Guengerich FP, Ketterer B. Human glutathione S-transferase T1-1 enhances mutagenicity of 1,2-dibromoethane, dibromomethane, and 1,2,3,4-diepoxybutane in Salmonella typhimurium. Carcinogenesis. 1996;17:163–166. doi: 10.1093/carcin/17.1.163. [DOI] [PubMed] [Google Scholar]
- (380).Cho S-H, Guengerich FP. Conjugation of butadiene diepoxide with glutathione yields DNA adducts in vitro and in vivo. Chem. Res. Toxicol. 2012;25 doi: 10.1021/tx200471x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (381).Liu L, Pegg AE, Williams KM, Guengerich FP. Paradoxical enhancement of the toxicity of 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2002;277:37920–37928. doi: 10.1074/jbc.M205548200. [DOI] [PubMed] [Google Scholar]
- (382).Valadez JG, Liu L, Loktionova NA, Pegg AE, Guengerich FP. Activation of bis-electrophiles to mutagenic conjugates by human O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- (383).Liu L, Williams KM, Guengerich FP, Pegg AE. O6-Alkylguanine-DNA alkyltransferase has opposing effects in modulating the genotoxicity of dibromomethane and bromomethyl acetate. Chem. Res. Toxicol. 2004;17:742–752. doi: 10.1021/tx049958o. [DOI] [PubMed] [Google Scholar]
- (384).Murata M, Ohnishi S, Seike K, Fukuhara K, Miyata N, Kawanishi S. Oxidaative DNA damage induced by carcinogenic dinitropyrenes in the presence of P450 reductase. Chem. Res. Toxciol. 2004;17:1750–1756. doi: 10.1021/tx0497550. [DOI] [PubMed] [Google Scholar]
- (385).Ohkuma Y, Hiraku Y, Kawanishi S. Sequence-specific DNA damage induced by carcinogenic danthron and anthraquinone in the presence of Cu(II), cytochrome P450 reductase and NADPH. Free Radic. Res. 2001;34:595–604. doi: 10.1080/10715760100300491. [DOI] [PubMed] [Google Scholar]
- (386).Oda Y, Yamazaki H, Shimada T. Role of human N-acetyltransferases, NAT1 or NAT2, in genotoxicity of nitroarenes and aromatic amines in Salmonella typhimurium NM6001 and NM6002. Carcinogenesis. 1999;20:1079–1083. doi: 10.1093/carcin/20.6.1079. [DOI] [PubMed] [Google Scholar]
- (387).Grant DM, Josephy PD, Lord HL, Morrison LD. Salmonella typhimurium strains expressing human arylamine N-acetyltransferases: Metabolism and mutagenic activation of aromatic amines. Cancer Res. 1992;52:3961–3964. [PubMed] [Google Scholar]
- (388).Arlt VM, Glatt H, Muckel E, Pabel U, Sorg BL, Schmeiser HH, Phillips DH. Metabolic activation of the environmental contaminant 3-nitrobenzanthrone by human acetyltransferases and sulfotransferase. Carcinogenesis. 2002;23:1937–1945. doi: 10.1093/carcin/23.11.1937. [DOI] [PubMed] [Google Scholar]
- (389).Hein DW, Doll MA, Rustan TD, Gray K, Feng Y, Ferguson RJ, Grant DM. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633–1638. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- (390).Glatt H, Meinl W. Use of genetically manipulated Salmonella typhimurium strains to evaluate the role of sulfotransferases and acetyltransferases in nitrofen mutagenicity. Carcinogenesis. 2004;25:779–786. doi: 10.1093/carcin/bgh070. [DOI] [PubMed] [Google Scholar]
- (391).Arlt VM, Glatt H, Gamboa da Costa G, Reynisson J, Takamura-Enya T, Phillips DH. Mutagenicity and DNA adduct formation by the urban air pollutant 2-nitrobenzanthrone. Toxicol. Sci. 2007;98:445–457. doi: 10.1093/toxsci/kfm103. [DOI] [PubMed] [Google Scholar]
- (392).Wild D, Feser W, Michel S, Lord HL, Josephy PD. Metabolic activation of heterocyclic aromatic amines catalyzed by human arylamine N-acetyltransferase isozymes (NAT1 and NAT2) expressed in Salmonella typhimurium. Carcinogenesis. 1995;16:643–648. doi: 10.1093/carcin/16.3.643. [DOI] [PubMed] [Google Scholar]
- (393).Muckel E, Frandsen H, Glatt HR. Heterologous expression of human N-acetyltransferases 1 and 2 and sulfotransferase 1A1 in Salmonella typhimurium for mutagenicity testing of heterocyclic amines. Food Chem. Toxicol. 2002;40:1063–1068. doi: 10.1016/s0278-6915(02)00032-7. [DOI] [PubMed] [Google Scholar]
- (394).Glatt H, Boeing H, Engelke CE, Ma L, Kuhlow A, Pabel U, Pomplun D, Teubner W, Meinl W. Human cytosolic sulphotransferases: Genetics, characteristics, toxicological aspects. Mut. Res. 2001;482:27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- (395).Glatt H. Sulfation and sulfotransferases. Bioactivation of mutagens via sulfation. FASEB J. 1997;11:314–321. doi: 10.1096/fasebj.11.5.9141497. [DOI] [PubMed] [Google Scholar]
- (396).Kreis P, Brandner S, Coughtrie MW, Pabel U, Meinl W, Glatt H, Andrae U. Human phenol sulfotransferases hP-PST and hM-PST activate propane 2-nitronate to a genotoxicant. Carcinogenesis. 2000;21:295–299. doi: 10.1093/carcin/21.2.295. [DOI] [PubMed] [Google Scholar]
- (397).Meinl W, Donath C, Schneider H, Sommer Y, Glatt H. SULT1C3, an orphan sequence of the human genome, encodes an enzyme activating various promutagens. Food Chem. Toxicol. 2008;46:1249–1256. doi: 10.1016/j.fct.2007.08.040. [DOI] [PubMed] [Google Scholar]
- (398).Glatt H, Baasanjav-Gerber C, Schumacher F, Monien BH, Schreiner M, Frank H, Seidel A, Engst W. 1-Methoxy-3-indolylmethyl glucosinolate. A potent genotoxicant in bacterial and mammalian cells. Mechanisms of bioactivation. Chem.-Biol. Interact. 2011;192:81–86. doi: 10.1016/j.cbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- (399).Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem.-Biol. Interact. 2000;129:141–170. doi: 10.1016/s0009-2797(00)00202-7. [DOI] [PubMed] [Google Scholar]
- (400).Svendsen C, Meinl W, Glatt H, Alexander J, Knutsen HK, Hjertholm H, Rasmussen T, Husøy T. Intestinal carcinogenesis of two food processing contaminants, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 5-hydroxymethylfurfural, in transgenic FVB min mice expressing human sulfotransferases. Mol. Carcinog. 2011 doi: 10.1002/mc.20869. doi 10.1002/mc.20869. [DOI] [PubMed] [Google Scholar]
- (401).Oda Y, Zhang Y, Buchinger S, Reifferscheid G, Yang M. Roles of human sulfotransferases in genotoxicity of carcinogens using genetically engineered umu test strains. Environ. Mol. Mutagen. 2012;253:152–164. doi: 10.1002/em.20696. [DOI] [PubMed] [Google Scholar]
- (402).Nishiyama T, Ogura K, Nakano H, Ohnuma T, Kaku T, Hiratsuka A, Muro K, Watabe T. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem. Pharmacol. 2002;63:1817–1830. doi: 10.1016/s0006-2952(02)00994-2. [DOI] [PubMed] [Google Scholar]
- (403).Mercer KE, Apostolov EO, da Costa GG, Yu X, Lang P, Roberts DW, Davis W, Basnakian AG, Kadlubar FF, Kadlubar SA. Expression of sulfotransferase isoform 1A1 (SULT1A1) in breast cancer cells significantly increases 4-hydroxytamoxifen-induced apoptosis. Int. J Mol. Epidemiol. Genet. 2010;1:92–103. [PMC free article] [PubMed] [Google Scholar]
- (404).Meinl W, Pabel U, Osterloh-Quiroz M, Hengstler J-G, Glatt H. Human sulphotransferases are involved in the activation of aristolochic acids and are expressed in renal target tissue. Int. J. Cancer. 2006;118:1090–1097. doi: 10.1002/ijc.21480. [DOI] [PubMed] [Google Scholar]
- (405).Monien BH, Herrmann K, Florian S, Glatt H. Metabolic activation of furfuryl alcohol: formation of 2-methylfuranyl DNA adducts in Salmonella typhimurium strains expressing human sulfotransferase 1A1 and in FVB/N mice. Carcinogenesis. 2011;32:1533–1539. doi: 10.1093/carcin/bgr126. [DOI] [PubMed] [Google Scholar]
- (406).Coles B, Nowell SA, MacLeod SL, Sweeney C, Lang NP, Kadlubar FF. The role of human glutathione S-transferases (hGSTs) in the detoxification of the food-derived carcinogen metabolite N-acetoxy-PhIP, and the effect of a polymorphism in hGSTA1 on colorectal cancer risk. Mut. Res. 2001;482:3–10. doi: 10.1016/s0027-5107(01)00187-7. [DOI] [PubMed] [Google Scholar]
- (407).Al-Buheissi SZ, Patel HR, Meinl W, Hewer A, Bryan RL, Glatt H, Miller RA, Phillips DH. N-Acetyltransferase and sulfotransferase activity in human prostates: Potential for carcinogen activation. Pharmacogenet. Genomics. 2006;16:391–399. doi: 10.1097/01.fpc.0000204998.22301.09. [DOI] [PubMed] [Google Scholar]
- (408).Chou HC, Lang NP, Kadlubar FF. Metabolic activation of N-hydroxy arylamines and N-hydroxy heterocyclic amines by human sulfotransferase(s) Cancer Res. 1995;55:525–529. [PubMed] [Google Scholar]
- (409).Chou HC, Lang NP, Kadluber FF. Metabolic activation of the N-hydroxy derivative of the carcinogen 4-aminobiphenyl by human tissue sulfotransferases. Carcinogenesis. 1995;16:413–417. doi: 10.1093/carcin/16.2.413. [DOI] [PubMed] [Google Scholar]
- (410).Meinl W, Meerman JH, Glatt H. Differential activation of promutagens by alloenzymes of human sulfotransferase 1A2 expressed in Salmonella typhimurium. Pharmacogenetics. 2002;12:677–689. doi: 10.1097/00008571-200212000-00002. [DOI] [PubMed] [Google Scholar]
- (411).Glatt H, Schneider H, Murkovic M, Monien BH, Meinl W. Hydroxymethyl-substituted furans: Mutagenicity in Salmonella typhimurium strains engineered for expression of various human and rodent sulphotransferases. Mutagenesis. 2012;27:41–48. doi: 10.1093/mutage/ger054. [DOI] [PubMed] [Google Scholar]
- (412).Shibutani S, Shaw PM, Suzuki N, Dasaradhi L, Duffel MW, Terashima I. Sulfation of α-hydroxytamoxifen catalyzed by human hydroxysteroid sulfotransferase results in tamoxifen-DNA adducts. Carcinogenesis. 1998;19:2007–2011. doi: 10.1093/carcin/19.11.2007. [DOI] [PubMed] [Google Scholar]
- (413).Apak TI, Duffel MW. Interactions of the stereoisomers of alpha-hydroxytamoxifen with human hydroxysteroid sulfotransferase SULT2A1 and rat hydroxysteroid sulfotransferase STa. Drug Metab. Dispos. 2004;32:1501–1508. doi: 10.1124/dmd.104.000919. [DOI] [PubMed] [Google Scholar]
- (414).Shen Y, Zhong L, Johnson S, Cao D. Human aldo-keto reductases B1 and 1B10: a comparative study on their enzyme activity toward electrophilic carbonyl compounds. Chem.-Biol. Interact. 2011;191:192–198. doi: 10.1016/j.cbi.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (415).Martin HJ, Breyer-Pfaff U, Wsol V, Venz S, Block S, Maser E. Purification and characterization of AKR1B10 from human liver: role in carbonyl reduction of xenobiotics. Drug Metab. Dispos. 2006;34:464–470. doi: 10.1124/dmd.105.007971. [DOI] [PubMed] [Google Scholar]
- (416).Martin HJ, Maser E. Role of human aldo-keto-reductase AKR1B10 in the protection against toxic aldehydes. Chem.-Biol. Interactions. 2009;178:145–150. doi: 10.1016/j.cbi.2008.10.021. [DOI] [PubMed] [Google Scholar]
- (417).Zhong L, Liu Z, Yan R, Johnson S, Zhao Y, Fang X, Cao D. Aldo-keto reductase family 1B10 protein detoxifies dietary and lipid-derived α, β-unsaturated carbonyls at physiological levels. Biochem. Biophys. Res. Commun. 2009;387:245–250. doi: 10.1016/j.bbrc.2009.06.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (418).Atalla A, Breyer-Pfaff U, Maser E. Purification and characterization of oxidoreductases-catalyzing carbonyl reduction of the tobacco-specific nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human liver cytosol. Xenobiotica. 2000;30:755–769. doi: 10.1080/00498250050119826. [DOI] [PubMed] [Google Scholar]
- (419).Atalla A, Maser E. Characterization of enzymes participating in carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human placenta. Chem.-Biol. Interact. 2001;130-132:737–748. doi: 10.1016/s0009-2797(00)00304-5. [DOI] [PubMed] [Google Scholar]
- (420).Knight LP, Primiano T, Groopman JD, Kensler TW, Sutter TR. B1-metabolizing member of the aldo-keto reductase superfamily, AKR7A3. Carcinogenesis. 1999;20:1215–1223. doi: 10.1093/carcin/20.7.1215. [DOI] [PubMed] [Google Scholar]
- (421).Penning TM, Drury JE. Human aldo-keto reductases: Function, gene regulation, and single nucleotide polymorphisms. Arch. Biochem. Biophys. 2007;464:241–250. doi: 10.1016/j.abb.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (422).Bodreddigari S, Jones LK, Egner PA, Groopman JD, Sutter CH, Roebuck BD, Guengerich FP, Kensler TW, Sutter TR. Protection against aflatoxin B1-induced cytotoxicity by expression of the cloned aflatoxin B1-aldehyde reductases rat AKR7A1 and human AKR7A3. Chem. Res. Toxicol. 2008;21:1134–1142. doi: 10.1021/tx7004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (423).Miksanova M, Sulc M, Rydlova H, Schmeiser HH, Frei E, Stiborová M. Enzymes involved in the metabolism of the carcinogen 2-nitroanisole: evidence for its oxidative detoxication by human cytochromes P450. Chem. Res. Toxicol. 2004;17:663–671. doi: 10.1021/tx0499721. [DOI] [PubMed] [Google Scholar]
- (424).Dracinska H, Miksanova M, Svobodova M, Smrcek S, Frei E, Schmeiser HH, Stiborová M. Oxidative detoxication of carcinogenic 2-nitroanisole by human, rat and rabbit cytochrome P450. Neuro. Endocrinol. Lett. 2006;27(Suppl. 2):9–13. [PubMed] [Google Scholar]
- (425).Jones JP, Shou M, Korzekwa KR. Stereospecific activation of the procarcinogen benzo[a]pyrene: a probe for the active sites of the cytochrome P450 superfamily. Biochemistry. 1995;34:6956–6961. doi: 10.1021/bi00021a007. [DOI] [PubMed] [Google Scholar]
- (426).Shimada T, Gillam EM, Sutter TR, Strickland PT, Guengerich FP, Yamazaki H. Oxidation of xenobiotics by recombinant human cytochrome P450 1B1. Drug Metab. Dispos. 1997;25:617–622. [PubMed] [Google Scholar]
- (427).Bauer E, Guo Z, Ueng Y-F, Bell LC, Guengerich FP. Oxidation of benzo[a]pyrene by recombinant human cytochrome P450 enzymes. Chem. Res. Toxicol. 1995;8:136–142. doi: 10.1021/tx00043a018. [DOI] [PubMed] [Google Scholar]
- (428).Parikh A, Josephy PD, Guengerich FP. Selection and characterization of human cytochrome P450 1A2 mutants with altered catalytic properties. Biochemistry. 1999;38:5283–5289. doi: 10.1021/bi990142+. [DOI] [PubMed] [Google Scholar]
- (429).Langouët S, Welti DH, Kerriguy N, Fay LB, Huynh-Ba T, Markovic J, Guengerich FP, Guillouzo A, Turesky RJ. Metabolism of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline-8-carboxylic acid is a major detoxification pathway catalyzed by cytochrome P450 1A2. Chem. Res. Toxicol. 2001;14:211–221. doi: 10.1021/tx000176e. [DOI] [PubMed] [Google Scholar]
- (430).Yun C-H, Shimada T, Guengerich FP. Roles of human liver cytochrome P-450 2C and 3A enzymes in the 3-hydroxylation of benzo(a)pyrene. Cancer Res. 1992;52:1868–1874. [PubMed] [Google Scholar]
- (431).Munter T, Cottrell L, Golding BT, Watson WP. Detoxication pathways involving glutathione and epoxide hydrolase in the in vitro metabolism of chloroprene. Chem. Res. Toxicol. 2003;16:1287–1297. doi: 10.1021/tx034107m. [DOI] [PubMed] [Google Scholar]
- (432).Guengerich FP, Johnson WW, Ueng Y-F, Yamazaki H, Shimada T. Involvement of cytochrome P450s, glutathione S-transferase, and epoxide hydrolase in the metabolism of aflatoxin B1 and relevance to risk of human liver cancer. Environ. Health Perspect. 1996;104(Suppl. 3):557–562. doi: 10.1289/ehp.96104s3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (433).Johnson WW, Ueng Y-F, Yamazaki H, Shimada T, Guengerich FP. Role of microsomal epoxide hydrolase in the hydrolysis of aflatoxin B1 8,9-epoxide. Chem. Res. Toxicol. 1997;10:672–676. doi: 10.1021/tx960209j. [DOI] [PubMed] [Google Scholar]
- (434).Chandrasena RE, Edirisinghe PD, Bolton JL, Thatcher GR. Problematic detoxification of estrogen quinones by NAD(P)H-dependent quinone oxidoreductase and glutathione-S-transferase. Chem. Res. Toxicol. 2008;21:1324–1329. doi: 10.1021/tx8000797. [DOI] [PubMed] [Google Scholar]
- (435).Pernice R, Hauder J, Koehler P, Vitaglione P, Fogliano V, Somoza V. Effect of sulforaphane on glutathione-adduct formation and on glutathione S-transferase-dependent detoxification of acrylamide in Caco-2 cells. Mol Nutr Food Res. 2009;53:1540–1550. doi: 10.1002/mnfr.200900447. [DOI] [PubMed] [Google Scholar]
- (436).Pacifici GM, Guthenberg C, Warholm M, Mannervik B, Rane A. Conjugation of styrene oxide by the basic and acidic forms of glutathione transferase in the human fetal liver. Dev. Pharmacol. Ther. 1988;11:243–251. doi: 10.1159/000457695. [DOI] [PubMed] [Google Scholar]
- (437).Pacifici GM, Warholm M, Guthenberg C, Mannervik B, Rane A. Detoxification of styrene oxide by human liver glutathione transferase. Hum. Toxicol. 1987;6:483–489. doi: 10.1177/096032718700600606. [DOI] [PubMed] [Google Scholar]
- (438).Seidel A, Friedberg T, Löllmann B, Schwierzok A, Funk M, Frank H, Holler R, Oesch F, Glatt H. Detoxification of optically active bay- and fjord-region polycyclic aromatic hydrocarbon dihydrodiol epoxides by human glutathione transferase P1-1 expressed in Chinese hamster V79 cells. Carcinogenesis. 1998;19:1975–1981. doi: 10.1093/carcin/19.11.1975. [DOI] [PubMed] [Google Scholar]
- (439).Jernstrom B, Funk M, Frank H, Mannervik B, Seidel A. Glutathione S-transferase A1-1-catalysed conjugation of bay and fjord region diol epoxides or polycyclic aromatic hydrocarbons with glutathione. Carcinogenesis. 1996;17:1491–1498. doi: 10.1093/carcin/17.7.1491. [DOI] [PubMed] [Google Scholar]
- (440).Sundberg K, Dreij K, Seidel A, Jernström B. Glutathione conjugation and DNA adduct formation of dibenzo[a,l]pyrene and benzo[a]pyrene diol epoxides in V79 cells stably expressing different human glutathione transferases. Chem. Res. Toxicol. 2002;15:170–179. doi: 10.1021/tx015546t. [DOI] [PubMed] [Google Scholar]
- (441).Dreij K, Sundberg K, Johansson AS, Nordling E, Seidel A, Persson B, Mannervik B, Jernström B. Catalytic activities of human alpha class glutathione transferases toward carcinogenic dibenzo[a,l]pyrene diol epoxides. Chem. Res. Toxicol. 2002;15:825–831. doi: 10.1021/tx025519i. [DOI] [PubMed] [Google Scholar]
- (442).Dirven HA, van Ommen B, van Bladeren PJ. Involvement of human glutathione S-transferase isoenzymes in the conjugation of cyclophosphamide metabolites with glutathione. Cancer Res. 1994;54:6215–6220. [PubMed] [Google Scholar]
- (443).Guengerich FP, Johnson WW, Shimada T, Ueng YF, Yamazaki H, Langouët S. Activation and detoxication of aflatoxin B1. Mut. Res. 1998;402:121–128. doi: 10.1016/s0027-5107(97)00289-3. [DOI] [PubMed] [Google Scholar]
- (444).Lin D, Meyer DJ, Ketterer B, Lang NP, Kadlubar FF. Effects of human and rat glutathione S-transferases on the covalent DNA binding of the N-acetoxy derivatives of heterocyclic amine carcinogens in vitro: a possible mechanism of organ specificity in their carcinogenesis. Cancer Res. 1994;54:4920–4926. [PubMed] [Google Scholar]
- (445).Upadhyaya P, Rao P, Hochalter JB, Li ZZ, Villalta PW, Hecht SS. Quantitation of N-acetyl-S-(9,10-dihydro-9-hydroxy-10-phenanthryl)-L-cysteine in human urine: Comparison with glutathione-S-transferase genotypes in smokers. Chem. Res. Toxicol. 2006;19:1234–1240. doi: 10.1021/tx060096w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (446).Sundberg K, Widersten M, Seidel A, Mannervik B, Jernström B. Glutathione conjugation of bay- and fjord-region diol epoxides of polycyclic aromatic hydrocarbons by glutathione transferases M1-1 and P1-1. Chem. Res. Toxicol. 1997;10:1221–1227. doi: 10.1021/tx970099w. [DOI] [PubMed] [Google Scholar]
- (447).Alexandrov K, Cascorbi I, Rojas M, Bouvier G, Kriek E, Bartsch H. CYP1A1 and GSTM1 genotypes affect benzo[a]pyrene DNA adducts in smokers’ lung: Comparison with aromatic/hydrophobic adduct formation. Carcinogenesis. 2002;23:1969–1977. doi: 10.1093/carcin/23.12.1969. [DOI] [PubMed] [Google Scholar]
- (448).Hu X, Xia H, Srivastava SK, Herzog C, Awasthi YC, Ji X, Zimniak P, Singh SV. Activity of four allelic forms of glutathione S-transferase hGSTP1-1 for diol epoxides of polycyclic aromatic hydrocarbons. Biochem. Biophys. Res. Commun. 1997;238:397–402. doi: 10.1006/bbrc.1997.7311. [DOI] [PubMed] [Google Scholar]
- (449).Sundberg K, Johansson AS, Stenberg G, Widersten M, Seidel A, Mannervik B, Jernström B. Differences in the catalytic efficiencies of allelic variants of glutathione transferase P1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis. 1998;19:433–436. doi: 10.1093/carcin/19.3.433. [DOI] [PubMed] [Google Scholar]
- (450).Hu X, Herzog C, Zimniak P, Singh SV. Differential protection against benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide-induced DNA damage in HepG2 cells stably transfected with allelic variants of pi class human glutathione S-transferase. Cancer Res. 1999;59:2358–2362. [PubMed] [Google Scholar]
- (451).Dirven HA, Megens L, Oudshoorn MJ, Dingemanse MA, van Ommen B, van Bladeren PJ. Glutathione conjugation of the cytostatic drug ifosfamide and the role of human glutathione S-transferases. Chem. Res. Toxicol. 1995;8:979–986. doi: 10.1021/tx00049a012. [DOI] [PubMed] [Google Scholar]
- (452).Morrow CS, Smitherman PK, Townsend AJ. Role of multidrug-resistance protein 2 in glutathione S-transferase P1-1-mediated resistance to 4-nitroquinoline 1-oxide toxicities in HepG2 cells. Mol. Carcinog. 2000;29:170–178. doi: 10.1002/1098-2744(200011)29:3<170::aid-mc6>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- (453).Peklak-Scott C, Townsend AJ, Morrow CS. Dynamics of glutathione conjugation and conjugate efflux in detoxification of the carcinogen, 4-nitroquinoline 1-oxide: contributions of glutathione, glutathione S-transferase, and MRP1. Biochemistry. 2005;44:4426–4433. doi: 10.1021/bi047810y. [DOI] [PubMed] [Google Scholar]
- (454).Hu X, Pal A, Krzeminski J, Amin S, Awasthi YC, Zimniak P, Singh SV. Specificities of human glutathione S-transferase isozymes toward anti-diol epoxides of methylchrysenes. Carcinogenesis. 1998;19:1685–1689. doi: 10.1093/carcin/19.9.1685. [DOI] [PubMed] [Google Scholar]
- (455).Sundberg K, Seidel A, Mannervik B, Jernström B. Detoxication of carcinogenic fjord-region diol epoxides of polycyclic aromatic hydrocarbons by glutathione transferase P1-1 variants and glutathione. FEBS Lett. 1998;438:206–210. doi: 10.1016/s0014-5793(98)01291-5. [DOI] [PubMed] [Google Scholar]
- (456).Hu X, Xia H, Srivastava SK, Pal A, Awasthi YC, Zimniak P, Singh SV. Catalytic efficiencies of allelic variants of human glutathione S-transferase P1-1 toward carcinogenic anti-diol epoxides of benzo[c]phenanthrene and benzo[g]chrysene. 1998;s:5340–5343. [PubMed] [Google Scholar]
- (457).Fields WR, Morrow CS, Doss AJ, Sundberg K, Jernström B, Townsend AJ. Overexpression of stably transfected human glutathione S-transferase P1-1 protects against DNA damage by benzo[a]pyrene diol-epoxide in human T47D cells. Mol. Pharmacol. 1998;54:298–304. doi: 10.1124/mol.54.2.298. [DOI] [PubMed] [Google Scholar]
- (458).Nelson CP, Kidd LC, Sauvageot J, Isaacs WB, De Marzo AM, Groopman JD, Nelson WG, Kensler TW. Protection against 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine cytotoxicity and DNA adduct formation in human prostate by glutathione S-transferase P1. Cancer Res. 2001;61:103–109. [PubMed] [Google Scholar]
- (459).Kurian JR, Chin NA, Longlais BJ, Hayes KL, Trepanier LA. Reductive detoxification of arylhydroxylamine carcinogens by human NADH cytochrome b5 reductase and cytochrome b5. Chem. Res. Toxicol. 2006;19:1366–1373. doi: 10.1021/tx060106t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (460).Rhoads K, Sacco JC, Drescher N, Wong A, Trepanier LA. Individual variability in the detoxification of carcinogenic arylhydroxylamines in human breast. Toxicol. Sci. 2011;121:245–256. doi: 10.1093/toxsci/kfr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (461).Lavigne JA, Goodman JE, Fonong T, Odwin S, He P, Roberts DW, Yager JD. The effects of catechol-O-methyltransferase inhibition on estrogen metabolite and oxidative DNA damage levels in estradiol-treated MCF-7 cells. Cancer Res. 2001;61:7488–7494. [PubMed] [Google Scholar]
- (462).Goodman JE, Jensen LT, He P, Yager JD. Characterization of human soluble high and low activity catechol-O-methyltransferase catalyzed catechol estrogen methylation. Pharmacogenetics. 2002;12:517–28. doi: 10.1097/00008571-200210000-00003. [DOI] [PubMed] [Google Scholar]
- (463).Zhang L, Jin Y, Chen M, Huang M, Harvey RG, Blair IA, Penning TM. Detoxication of structurally diverse polycyclic aromatic hydrocarbon (PAH) o-quinones by human recombinant catechol-O-methyltransferase (COMT) via O-methylation of PAH catechols. J. Biol. Chem. 2011;286:25644–25654. doi: 10.1074/jbc.M111.240739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (464).Ozawa S, Nagata K, Yamazoe Y, Kato R. Formation of 2-amino-3-methylimidazo[4,5-f]quinoline- and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline-sulfamates by cDNA-expressed mammalian phenol sulfotransferases. Jpn. J. Cancer Res. 1995;86:264–269. doi: 10.1111/j.1349-7006.1995.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (465).Turesky RJ, Fay LB, Welti DH. Metabolism of heterocyclic aromatic amines and strategies of human biomonitoring. In: Adamson RH, Gustafsson J-Å, Ito N, Nagao M, Wakabayashi K, Yamazoe Y, editors. Heterocyclic Amines and Human Cancer. Vol. 23. Princeton Sci. Publ.; Princeton, NJ: 1995. pp. 59–68. Proc. 23rd Int. Princess Takamatsu Cancer Sympos., Tokyo, 10-12 November, 1992. [PubMed] [Google Scholar]
- (466).Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, Lazarus P. Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res. 2002;62:1978–1986. [PubMed] [Google Scholar]
- (467).Malfatti MA, Felton JS. N-Glucuronidation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and N-hydroxy-PhIP by specific human UDP-glucuronosyltransferases. Carcinogenesis. 2001;22:1087–1093. doi: 10.1093/carcin/22.7.1087. [DOI] [PubMed] [Google Scholar]
- (468).Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis. 2007;28:2412–2418. doi: 10.1093/carcin/bgm164. [DOI] [PubMed] [Google Scholar]
- (469).Basu NK, Kubota S, Meselhy MR, Ciotti M, Chowdhury B, Hartori M, Owens IS. Gastrointestinally distributed UDP-glucuronosyltransferase 1A10, which metabolizes estrogens and nonsteroidal anti-inflammatory drugs, depends upon phosphorylation. J. Biol. Chem. 2004;279:28320–28329. doi: 10.1074/jbc.M401396200. [DOI] [PubMed] [Google Scholar]
- (470).Olson KC, Sun D, Chen G, Sharma AK, Amin S, Ropson IJ, Spratt TE, Lazarus P. Characterization of dibenzo[a,l]pyrene-trans-11,12-diol (dibenzo[def,p]chrysene) glucuronidation by UDP-glucuronosyltransferases. Chem. Res. Toxicol. 2011;24:1549–1559. doi: 10.1021/tx200178v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (471).Malfatti MA, Felton JS. Human UDP-glucuronosyltransferase A1 is the primary enzyme responsible for the N-glucuronidation of N-hydroxy-PhIP in vitro. Chem. Res. Toxicol. 2004;17:1137–1144. doi: 10.1021/tx049898m. [DOI] [PubMed] [Google Scholar]
- (472).Malfatti MA, Wu RW, Felton JS. The effect of UDP-glucuronosyltransferase 1A1 expression on the mutagenicity and metabolism of the cooked-food carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in CHO cells. Mut. Res. 2005;570:205–214. doi: 10.1016/j.mrfmmm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- (473).Ogura K, Ishikawa Y, Kaku T, Nishiyama T, Ohnuma T, Muro K, Hiratsuka A. Quaternary ammonium-linked glucuronidation of trans-4-hydroxytamoxifen, an active metabolite of tamoxifen, by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem. Pharmacol. 2006;71:1358–1369. doi: 10.1016/j.bcp.2006.01.004. [DOI] [PubMed] [Google Scholar]
- (474).Sun D, Sharma AK, Dellinger RW, Blevins-Primeau AS, Balliet RM, Chen G, Boyiri T, Amin S, Lazarus P. Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab. Dispos. 2007;35:2006–2014. doi: 10.1124/dmd.107.017145. [DOI] [PubMed] [Google Scholar]
- (475).Nowell SA, Massengill JS, Williams S, Radominska-Pandya A, Tephly TR, Cheng Z, Strassburg CP, Tukey RH, MacLeod SL, Lang NP, Kadlubar FF. Glucuronidation of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human microsomal UDP-glucuronosyltransferases: Identification of specific UGT1A family isoforms involved. Carcinogenesis. 1999;20:1107–1114. doi: 10.1093/carcin/20.6.1107. [DOI] [PubMed] [Google Scholar]
- (476).Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–1196. doi: 10.1158/0008-5472.can-03-3219. [DOI] [PubMed] [Google Scholar]
- (477).Wiener D, Doerge DR, Fang JL, Upadhyaya P, Lazarus P. Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab. Dispos. 2004;32:72–79. doi: 10.1124/dmd.32.1.72. [DOI] [PubMed] [Google Scholar]
- (478).Chen G, Dellinger RW, Sun D, Spratt TE, Lazarus P. Glucuronidation of tobacco-specific nitrosamines by UGT2B10. Drug Metab Dispos. 2008;36:824–830. doi: 10.1124/dmd.107.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (479).Kaku T, Ogura K, Nishiyama T, Ohnuma T, Muro K, Hiratsuka A. Quaternary ammonium-linked glucuronidation of tamoxifen by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem. Pharmacol. 2004;67:2093–2102. doi: 10.1016/j.bcp.2004.02.014. [DOI] [PubMed] [Google Scholar]
- (480).Ren Q, Murphy SE, Zheng Z, Lazarus P. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab. Dispos. 2000;28:1352–1360. [PubMed] [Google Scholar]
- (481).Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab. Dispos. 2006;34:943–949. doi: 10.1124/dmd.105.009100. [DOI] [PubMed] [Google Scholar]
- (482).Balliet RM, Chen G, Dellinger RW, Lazarus P. UDP-glucuronosyltransferase 1A10: activity against the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol, and a potential role for a novel UGT1A10 promoter deletion polymorphism in cancer susceptibility. Drug Metab. Dispos. 2010;38:484–90. doi: 10.1124/dmd.109.030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (483).Bushey RT, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. Characterization of UDP-glucuronosyltransferase 2A1 (UGT2A1) variants and their potential role in tobacco carcinogenesis. Pharmacogenet. Genomics. 2011;21:55–65. doi: 10.1097/FPC.0b013e328341db05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (484).Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, Chase GA, Richie J, Lazarus P. The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:823–828. doi: 10.1158/1055-9965.EPI-06-0823. [DOI] [PubMed] [Google Scholar]
- (485).Penning TM. Aldo-keto reductases and formation of polycyclic aromatic hydrocarbon o-quinones. Methods Enzymol. 2004;378:31–67. doi: 10.1016/S0076-6879(04)78003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (486).Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes memorial lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- (487).Nebert DW, Gelboin HV. Substrate-inducible microsomal arylhydroxylase in mammalian cell culture: assay and properties of induced enzyme. J. Biol. Chem. 1968;243:6242–6249. [PubMed] [Google Scholar]
- (488).Guengerich FP, Johnson WW. Kinetics of hydrolysis and reaction of aflatoxin B1 exo-8,9-epoxide and relevance to toxicity and detoxication. Drug Metab. Rev. 1999;31:141–158. doi: 10.1081/dmr-100101911. [DOI] [PubMed] [Google Scholar]
- (489).Iyer R, Coles B, Raney KD, Thier R, Guengerich FP, Harris TM. DNA adduction by the potent carcinogen aflatoxin B1: mechanistic studies. J. Am. Chem. Soc. 1994;116:1603–1609. [Google Scholar]
- (490).Distlerath LM, Reilly PEB, Martin MV, Davis GG, Wilkinson GR, Guengerich FP. Purification and characterization of the human liver cytochromes P-450 involved in debrisoquine 4-hydroxylation and phenacetin O-deethylation, two prototypes for genetic polymorphism in oxidative drug metabolism. J. Biol. Chem. 1985;260:9057–9067. [PubMed] [Google Scholar]
- (491).Lo Guidice JM, Marez D, Sabbagh N, Legrand-Andreoletti M, Spire C, Alcaïde E, Lafitte JJ, Broly F. Evidence for CYP2D6 expression in human lung. Biochem. Biophys. Res. Commun. 1997;241:79–85. doi: 10.1006/bbrc.1997.7775. [DOI] [PubMed] [Google Scholar]
- (492).Wolff T, Distlerath LM, Worthington MT, Groopman JD, Hammons GJ, Kadlubar FF, Prough RA, Martin MV, Guengerich FP. Substrate specificity of human liver cytochrome P-450 debrisoquine 4-hydroxylase probed using immunochemical inhibition and chemical modeling. Cancer Res. 1985;45:2116–2122. [PubMed] [Google Scholar]
- (493).Crespi CL, Penman BW, Gelboin HV, Gonzalez FJ. A tobacco smoke-derived nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, is activated by multiple human cytochrome P450s including the polymorphic human cytochrome P4502D6. Carcinogenesis. 1991;12:1197–1201. doi: 10.1093/carcin/12.7.1197. [DOI] [PubMed] [Google Scholar]
- (494).Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, Gelboin HV, Hardwick JP, Meyer UA. Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature. 1988;331:442–446. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- (495).Christensen PM, Gøtzsche PC, Brøsen K. The sparteine/debrisoquine (CYP2D6) oxidation polymorphism and the risk of lung cancer: a meta-analysis. Eur. J. Clin. Pharmacol. 1997;51:389–393. doi: 10.1007/s002280050219. [DOI] [PubMed] [Google Scholar]
- (496).Rostami-Hodjegan A, Lennard MS, Woods HF, Tucker GT. Meta-analysis of studies of the CYP2D6 polymorphism in relation to lung cancer and Parkinson’s disease. Pharmacogenetics. 1998;8:227–238. doi: 10.1097/00008571-199806000-00005. [DOI] [PubMed] [Google Scholar]
- (497).Guengerich FP. Cytochrome P450 oxidation in the generation of reactive electrophiles: Epoxidations and related reactions. Arch. Biochem. Biophys. 2003;409:59–71. doi: 10.1016/s0003-9861(02)00415-0. [DOI] [PubMed] [Google Scholar]
- (498).Gozukara EM, Belvedere G, Robinson RC, Deutsch J, Coon MJ, Guengerich FP, Gelboin HV. The effect of epoxide hydratase on benzo[a]pyrene diol epoxide hydrolysis and binding to DNA and mixed-function oxidase proteins. Mol. Pharmacol. 1981;19:153–161. [PubMed] [Google Scholar]
- (499).Guengerich FP, Tang Z, Salamanca-Pinzon SG, Cheng Q. Characterizing proteins of unknown function: Orphan cytochrome P450s as a paradigm. Mol. Interventions. 2010:153–163. doi: 10.1124/mi.10.3.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (500).Karlgren M, Gomez A, Stark K, Svard J, Rodriguez-Antona C, Oliw E, Bernal ML, Ramón y Cajal S, Johansson I, Ingelman-Sundberg M. Tumor-specific expression of the novel cytochrome P450 enzyme, CYP2W1. Biochem. Biophys. Res. Commun. 2006;341:451–458. doi: 10.1016/j.bbrc.2005.12.200. [DOI] [PubMed] [Google Scholar]
- (501).Tang Z, Salamanca-Pinzon SG, Wu Z-L, Xiao Y, Guengerich FP. Human cytochrome P450 4F11: heterologous expression in bacteria, purification, and characterization of catalytic function. Arch. Biochem. Biophys. 2010;494:86–93. doi: 10.1016/j.abb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (502).Rylander T, Neve EPA, Ingelman-Sundberg M, Oscarson M. Identification and tissue distribution of the novel human cytochrome P450 2S1 (CYP2S1) Biochem. Biophys. Res. Commun. 2001;281:529–535. doi: 10.1006/bbrc.2001.4390. [DOI] [PubMed] [Google Scholar]
- (503).Karlgren M, Miura S, Ingelman-Sundberg M. Novel extrahepatic cytochrome P450s. Toxicol. Appl. Pharmacol. 2005;207(Suppl. 1):57–61. doi: 10.1016/j.taap.2004.12.022. [DOI] [PubMed] [Google Scholar]
- (504).Nishida CR, Lee M, Ortiz de Montellano PR. Efficient hypoxic activation of the anticancer agent AQ4N by CYP2S1 and CYP2W1. Mol. Pharmacol. 2010;78:497–502. doi: 10.1124/mol.110.065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (505).Xiao Y, Shinko R, Guengerich FP. Cytochrome P450 2S1 is reduced by NADPH-cytrochrome P450 reductase. Drug Metab. Dispos. 2011;39:944–946. doi: 10.1124/dmd.111.039321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (506).Bui PH, Hsu EL, Hankinson O. Fatty acid hydroperoxides support cytochrome P450 2S1-mediated bioactivation of benzo[a]pyrene-7,8-dihydrodiol. Mol. Pharmacol. 2009;76:1044–1052. doi: 10.1124/mol.109.057760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (507).Bui PH, Hankinson O. Functional characterization of human cytochrome P450 2S1 using a synthetic gene-expressed protein. Escherichia coli. Mol. Pharmacol. 2009;76:1031–1043. doi: 10.1124/mol.109.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (508).Guengerich FP, Cheng Q. Orphans in the human cytochrome P450 family: Approaches to discovering function and relevance to pharmacology. Pharmacol. Rev. 2011;63:684–699. doi: 10.1124/pr.110.003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (509).Rendic S, Di Carlo FJ. Human cytochrome P450 enzymes: a status report summarizing their reacitons, substrates, inducers, and inhibitors. Drug Metab. Rev. 1997;29:413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- (510).Rendic S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002;34:83–448. doi: 10.1081/dmr-120001392. [DOI] [PubMed] [Google Scholar]
- (511).Guengerich FP, Rendic S. Update information on drug metabolism systems-2009, Part I. Drug Metab. Rev. 2010;11:1–3. doi: 10.2174/138920010791110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (512).Rendic S, Guengerich FP. Summary of information on human cytochrome P450 enzyme and transporters: “Non-chemical” effectors influencing expression and activity. Curr. Drug Metab. 2010;11:4–84. doi: 10.2174/138920010791110917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (513).Guengerich FP. Separation and purification of multiple forms of microsomal cytochrome P-450. Activities of different forms of cytochrome P-450 towards several compounds of environmental interest. J. Biol. Chem. 1977;252:3970–3979. [PubMed] [Google Scholar]
- (514).Sohl CD, Isin EM, Eoff RL, Marsch GA, Stec DF, Guengerich FP. Cooperativity in oxidation reactions catalyzed by cytochrome P450 1A2. Highly cooperative pyrene hydroxylation and multiphasic kinetics of ligand binding. J. Biol. Chem. 2008;283:7293–7308. doi: 10.1074/jbc.M709783200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.