Abstract
Piwi-interacting RNAs (piRNAs) and CRISPR RNAs (crRNAs) are two recently discovered classes of small noncoding RNA that are found in animals and prokaryotes, respectively. Both of these novel RNA species function as components of adaptive immune systems that protect their hosts from foreign nucleic acids—piRNAs repress transposable elements in animal germlines, whereas crRNAs protect their bacterial hosts from phage and plasmids. The piRNA and CRISPR systems are nonhomologous but rather have independently evolved into logically similar defense mechanisms based on the specificity of targeting via nucleic acid base complementarity. Here we review what is known about the piRNA and CRISPR systems with a focus on comparing their evolutionary properties. In particular, we highlight the importance of several factors on the pattern of piRNA and CRISPR evolution, including the population genetic environment, the role of alternate defense systems and the mechanisms of acquisition of new piRNAs and CRISPRs.
Keywords: piRNA, CRISPR, co-evolution, transposable elements, phage, plasmids
INTRODUCION
In the last decade, a myriad of novel noncoding RNA species have been discovered, many of them very small in size (∼20–40 nt) [1]. Molecular pathways that involve small noncoding RNAs binding to an Argonaute protein are often referred to as RNAi-related pathways. There are several classes of Argonaute proteins, most famously the Ago-class Argonautes that bind to microRNAs and that are firmly established as fundamentally important regulators of gene expression in many areas of animal, plant and viral biology [2]. In this review, we will focus on small RNAs that bind to Piwi-class Argonaute proteins, called Piwi-interacting RNAs (piRNAs). Piwi proteins are only found in animals in which they are generally found highly expressed in the germline. Consistently, the best understood function of piRNAs is their role in defense against transposable elements in the germline.
There are also several classes of small noncoding RNAs that do not participate in RNAi-related pathways, and in this review, we will also discuss the CRISPR (clustered regularly interspaced short palindromic repeats) RNAs (crRNAs) that are found only in prokaryotes. Although there exists an archaeal Argonaute homolog [3], crRNAs do not bind to Argonaute proteins. Indeed, it has been suggested that the archaeal Argonaute may have a role in DNA rather than RNA modification [4] and that it may be involved in a completely separate prokaryotic defense mechanism from the CRISPR system [5]. Instead, the crRNAs bind to a different protein called Cas. Together, the prokaryotic CRISPR-Cas system functions as an adaptive defense mechanism against phage and plasmids.
Despite their lack of homology, there is a very clear logical similarity between the piRNA and CRISPR systems. In both systems, sequences from the invading nucleic acid are incorporated into specific loci in the host genome. When these sequences are transcribed and processed into small RNAs, the small RNAs can then guide repressive molecular complexes to invading nucleic acids in trans. Both the piRNA and CRISPR systems are thus clear examples of Lamarckian mechanisms in which environmental factors directly cause heritable genetic changes [6]. The similarity between piRNAs and CRISPRs has been observed many times—indeed CRISPRs were at one time speculated to be an RNAi-related system [7]—and there are already a number of recent reviews of piRNA [8, 9] and CRISPR biology [10–12]. However, there has been much less consideration of the evolutionary properties of these two fascinating molecular systems. Accordingly, our goal in this review is to survey the literature on piRNAs and CRISPRs with an emphasis on aspects of their biology most relevant to their evolution and to highlight factors which may cause the two systems to behave differently from an evolutionary standpoint.
OVERVIEW OF THE PIRNA SYSTEM
Although hints of the piRNA system were observed in Drosophila as early as 2001 [13], piRNAs were definitively discovered by several groups independently in 2006 by immunoprecipitating Piwi protein from mammalian testis and sequencing the bound small RNAs [14–17]. PiRNAs are ∼26–30 nt in mammals although their lengths can be slightly different in other animals. PiRNAs have essentially no known defining sequence characteristics beyond a very strong propensity for a 5′-uridine and a weaker bias toward an adenosine at position 10. PiRNAs are in general difficult to predict bioinformatically and must instead be defined biochemically. However, protocols for immunoprecipitating Piwi protein are still an active area of research [18] and there are no definitive sets of piRNA genes yet because the population of piRNAs is typically very large (in the hundreds of thousands) and complex. Caenorhabditis elegans piRNAs may be significantly different from mammalian and Drosophila piRNAs because they have a different length (21 nt), and there appears to be a conserved promoter motif upstream of many piRNAs [19], suggesting that each piRNA is a separate transcription unit, unlike piRNAs in mammals and Drosophila which are typically expressed in long polycistronic transcripts.
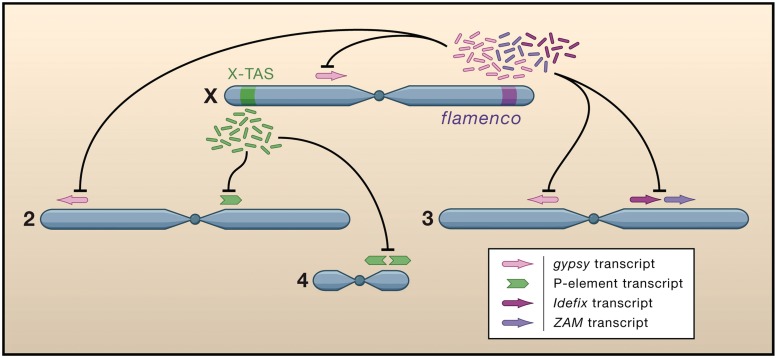
Unlike other small RNAs from the RNAi-related pathway, such as microRNAs and small interfering RNAs, which are produced from double-stranded intermediates by the Dicer enzyme, piRNAs are thought to be produced from long polycistronic RNA transcripts by a Dicer-independent mechanism in mammals and Drosophila. Note that unlike the CRISPR system described below, piRNA populations are very complex and piRNAs appear to be produced by quasi-random cleavage of the primary piRNA transcript [20]. That is, while piRNAs almost always start with a U and there are biases for particular sequences to be cleaved as piRNAs, there is a strong random component that determines which sequences of the primary transcript are processed into piRNAs (hence the term ‘quasi-random’). PiRNA 3′-end formation is poorly understood and is an object of active research [21]. However, piRNA 5′-end formation was addressed by several key papers [22, 23]. The authors studied master loci that control transposable element proliferation in Drosophila but were molecularly uncharacterized for many years because of the apparent lack of functional sequences at the loci, other than a jumble of transposable element insertions. These master loci were found to produce piRNAs that repress transposable elements in trans [22] (Figure 1). The authors proposed the Ping-Pong mechanism [22, 23] in which primary piRNAs cleave sense transposon transcripts and simultaneously produce secondary piRNAs from the sense transposons that then cleave antisense transposon transcripts. This mechanism thus depends on the transcription of both sense and anti-sense transposon transcripts. An alternate view is that piRNAs are in fact produced through a double-stranded intermediate [24] based on the recent reports of the existence of an RNA-dependent RNA polymerase in Drosophila [25]. However, the existence of a Drosophila RDRP remains controversial, and this view remains a minority interpretation at the present time.
Figure 1:
PiRNAs expressed from discrete loci in the Drosophila genome (X-TAS and Flamenco) repress transposable elements in trans (gypsy, P-element, Idefix, ZAM). Reprinted from ‘Mighty Piwis Defend Germline against Genome Intruders’, K. A. O’Donnell and J.D. Boeke, Cell 2007;129(1):37–44, with permission from Elsevier.
Since the Ping-Pong mechanism is a positive feedback loop, one question is how the Ping-Pong mechanism is started in the first place. In Drosophila, a partial answer is provided by the fact that piRNAs are deposited maternally into the embryo [26, 27]. PiRNAs can thus be inherited epigenetically across generations. A second answer comes from evidence in Drosophila, where primary piRNAs are produced in the somatic follicle cells and delivered to the germline to start the Ping-Pong cycle [28, 29]. A similar mechanism is found in Arabidopsis for a different class of small RNAs [30], suggesting that this may be a universal mechanism where transposons are activated outside of the germline to generate small RNAs, thus reducing the chance of deleterious transposon insertions in the germline. A third possibility is suggested by a related system of RNAi and heterochromatin formation in fission yeast, in which degradation products from random abundant transcripts are used to prime Argonaute proteins and start a positive feedback loop [31].
Aside from the piRNAs that are derived from repetitive elements and involved in the Ping-Pong mechanism, there are classes of piRNAs that are not repetitive. For example, the piRNA populations expressed at different stages of mammalian testis development are distinct and those found at the pachytene stage are depleted in repetitive sequences [32]. In addition, some piRNAs are found in genes and are assumed to repress their host transcripts [33]. Finally, there is some evidence that piRNAs are functional in the brain in rat [34]. The connection between neural expression of piRNAs and the expression of transposable elements in the mammalian brain [35] has been observed and is clearly intriguing, but there is currently no evidence to further connect these two aspects of neuroscience. In the rest of this review, we will focus on the repetitive piRNAs that are involved in the Ping-Pong mechanism and repress transposable elements because they are much better understood than the nonrepetitive piRNAs.
Overview of piRNA evolution
The piRNA system is known to be ancient as Piwi proteins, and the Ping-Pong signature are conserved in basal metazoans [36]. However, no Piwi homologs have been found outside animals so the piRNA system appears to be an animal-specific innovation. Between closely related species, the genomic locations of many piRNA clusters are conserved, but the sequences of the piRNAs themselves are not conserved between rat and mouse [37], C. elegans and C. briggsae [19] or Drosophila melanogaster and D. simulans [38]. Thus, the overall picture of piRNA evolution at the sequence level is one of very rapid evolution.
A recent study of human piRNAs by one of the authors suggested that there is strong negative selection at the sequence level for human piRNAs but only in the three African populations and not any of the eight non-African populations studied [39]. This observation is consistent with a recent report that African populations have much higher rates of transposon insertion than other populations [40]. A further intriguing observation from the analysis of human piRNAs and transposable elements is the depletion of piRNA matches in the reverse transcriptase region of human LINE-1 elements, though not mouse LINE-1 elements [39]. This observation suggests the possibility that at least one reverse transcriptase might be functional for the host and therefore protected from piRNA-mediated repression.
Beyond sequence divergence, it is also interesting to study the relationship of piRNA clusters and copy number changes, as an increase in copy number could potentially increase the level of gene expression of piRNAs. Assis and Kondrashov studied the evolution of piRNA clusters between mouse and rat and found a very high rate of piRNA cluster duplication, which they suggested is indicative of positive selection for higher expression level of piRNAs [37].
Although the piRNA system is not understood well enough for detailed mathematical modeling, there has been one attempt by Lu and Clark [41] at modeling piRNA-transposable element co-evolution using computer simulations. From their simulation, they suggested that retrotransposon insertions that are repressed by piRNAs can reach high frequencies or even be fixed in the population because their deleterious effect is attenuated by piRNA repression.
The idea that the piRNA pathway and transposable elements might co-evolve in a Red Queen-like scenario has been explored by a number of authors. In this scenario, alternating rounds of adaptation and counter-adaptation would lead to increased rates of positive selection. In a molecular evolution analysis examining species across the Drosophila genus, it was found that a higher transposable element abundance is positively correlated with greater codon bias in piRNA pathway genes but not an increased rate of amino acid substitution in these genes [42]. The authors suggested that these observations indicate that positive selection on piRNA pathway genes occurs mainly at the level of translation efficiency mediated by codon usage (although other explanations for codon bias are possible) as opposed to amino acid substitution [42]. Further, a resequencing study of a number of defense genes in D. melanogaster and D. simulans concluded that RNAi genes have the highest rate of adaptive evolution over all immune-system genes [43]. Subsequent studies also found recurrent adaptation across the twelve sequenced Drosophila genomes for a number of piRNA pathway genes, including SPN-E, AUB, KRIMP, SQU and ZUC [44], as well as Rhino [45]. Overall, these studies are consistent with elevated rates of evolution on piRNA pathway genes, consistent with its role in genome defense. While the molecular details of the Red Queen scenario for piRNAs and transposable elements are unclear, certain aspects of transposable element evolution, such as a higher global transposition rate, could select for certain features of piRNA-pathway genes, such as stronger binding affinity of the proteins for piRNAs.
PiRNAs and phenotypic capacitors
An interesting and somewhat contentious aspect of the role of the piRNA system in evolution is its role in canalization. Canalization, most famously associated with Waddington [46], refers to the buffering of genetic or environmental insults to ensure developmental robustness. In a seminal paper, Rutherford and Lindquist [47] suggested that Hsp90, a protein chaperone, is a phenotypic capacitor in Drosophila, meaning that it buffers genetic variation but when it is compromised, that variation is revealed in multiple mutant phenotypes, at least some of which could be adaptive in certain environments [48]. Similar results were subsequently demonstrated in Arabidopsis [49], suggesting that Hsp90 might play an evolutionarily conserved role as a phenotypic capacitor.
The connection between canalization and the piRNA system comes from a recent report that in Drosophila, Hsp90 regulates the piRNA pathway, which in turn regulates the insertion of transposons [50]. It was further suggested that Hsp90 interacts in a protein complex with Piwi protein and mediates canalization by epigenetic silencing of genetic variation and suppressing transposon insertion [51]. Thus, one potential mechanism by which the disruption of Hsp90 creates phenotypic variation is not by revealing previously cryptic variation as suggested by Rutherford and Lindquist but rather through de novo mutations generated by transposon insertions. For this to be true, a strong bias in the preference in genome position for transposition insertion dependent on genetic background is required, and while such a preference is known to exist, it is not clear if it is strong enough to fully explain the results of the Rutherford and Lindquist experiments. Also, the piRNA study [51] showed an effect on gene regulation separable from the effect on transposons. Conversely, imprecise transposon deletions could have a mutagenic effect and would necessarily be in the same place in the genome so more work needs to be done to define the exact role of piRNAs in canalization.
OVERVIEW OF CRISPRs
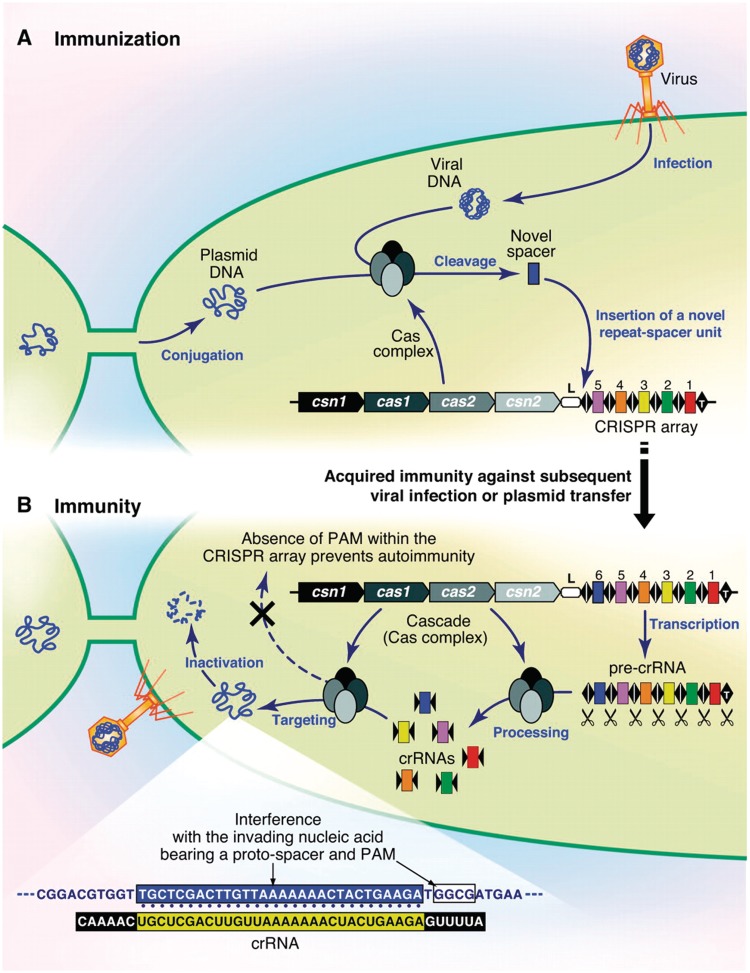
CRISPR loci were initially reported simply as arrays of DNA repeats in Escherichia coli [52] in 1987 and subsequently named ‘CRISPR’ in 2002 when it was observed that such arrays were common in prokaryotes [53]. In 2005, several groups found that CRISPR spacers often have similarity to foreign DNAs, especially phage, suggesting a role in cellular defense [54, 55]. Finally, molecular experiments in 2007 and 2008 showed that CRISPRs indeed confer immunity to phage [56] and plasmids [57] (Figure 2).
Figure 2:
(A) Sequences from viruses or plasmids are cleaved into novel spacers and inserted into discrete CRISPR array loci. (B) The CRISPR array is transcribed into a pre-crRNA which is processed into individual crRNAs. These small RNAs are bound by proteins from the Cas complex and used to guide the Cas proteins to target invading nucleic acids. PAM (protospacer-associated motif) distinguishes self from nonself to prevent autoimmunity. From ‘CRISPR/Cas, the Immune System of Bacteria and Archaea’, P. Horvath and R. Barrangou, Science 327:5962, 2010. Reprinted with permission from AAAS.
Databases of CRISPRs from sequenced prokaryotic genomes have been created [58] and current estimates indicate that nearly all archaea and about half of all bacteria contain CRISPRs. A prokaryotic cell can contain one or more CRISPR cassettes that are made up of alternating sequences of repeats and spacers. The spacers encode the functional RNA units that are often homologous to phage or plasmid sequences and can direct cleavage of those molecules in trans. The repeats may be recognized by a protein that processes the long RNA transcript into individual spacer units. The number of CRISPR loci per genome ranges from 1 to 15, and each varies in length from several to a few hundred spacers, up to a longest known locus containing 587 spacers. The repeat and spacer sizes are typically 21–48 bp for repeats and 26–72 bp for spacers. In general, the CRISPR repeats are not conserved at the sequence level beyond a few short conserved sequences such as GTTTg/c at the 5′-end and GAAAC at the 3′-end.
The mechanism of CRISPR-mediated defense seems to depend on the particular prokaryotic species, and crRNAs can direct cleavage of either DNA [59] or RNA [60]. However, the distinction between DNA and RNA targeting will not be important for the evolutionary perspective we adopt here. CrRNAs bind to Cas (CRISP-associated) proteins, which are typically encoded in the genome close to the CRISPR array. Several of the Cas proteins together form the Cascade complex. The classification of CRISPR-Cas systems has been revised a number of times, but the most recent study classifies them into three major types (Types I–III) [61]. The details of these different Cas systems are beyond the scope of this manuscript, and the interested reader is referred instead to other recent reviews on the subject [62]. The Cas proteins function in three distinct steps—to integrate nucleic acid fragments as new spacers, to cleave the precursor crRNA and finally to cleave the target.
Review of CRISPR evolution
One of the most interesting aspects of CRISPR loci is their linear organization—new CRIPSR spacers are always inserted at one end of the locus—which makes them a unique temporal record of past phage invasions [12]. Note that there are occasionally deletions of spacers so the linear ordering is only approximate. CRISPRs have also been well-studied in the context of metagenomics [63] where the simple presence of CRISPR cassettes is sufficient to link prokaryote species to their phage invaders.
CRISPRs can be located on plasmids and horizontally transferred between prokaryotes [64], and indeed CRISPRs are believed to have originated in thermophilic Archaea before spreading via horizontal transfer to other prokaryotes [65]. Conversely, CRISPRs can prevent horizontal gene transfer by repression of plasmids and thus may contribute to the formation of independent bacterial lineages, similar to other prokaryotic repressive mechanisms of plasmids [66].
One study that looked at the distribution of CRISPR cassettes in 290 strains of E. coli found that closely related strains generally had identical CRISPRs, whereas distantly related strains had completely different CRISPRs, suggesting rare but dramatic change in CRISPR spacer content over evolutionary time rather than small gradual changes [67]. Metagenomic studies of CRISPRs showed that there is a history of selective sweeps at CRISPR loci [68] and a history of polymorphism at old CRISPR spacers [69], consistent with their role in genome defense. Conversely, phage are known to escape CRISPR targeting by mutation and deletion of bases [59, 70] or shuffling of sequences [63]. Finally, a recent molecular evolution study of Cas gene evolution found patterns of relatively fast evolutionary change, consistent with a co-evolutionary arms-race between CRISPRs and phage [71].
Mathematical models
The elegance of the CRISPR system has attracted the attention of a number of modeling groups, who have attempted to design simple mathematical models of CRISPR evolution, generally based on simple ordinary differential equations. In one study by Bruce Levin, the population dynamics of CRISPRs in bacterial populations growing in a chemostat were modeled using standard chemostat models [72]. The biological significance of this model has not yet been shown, but one could perform the actual experiment of growing phage and bacteria together in a chemostat and tracking the dynamics of their population growth over time to directly test the predictions of the model. Among other simple mathematical models [73–75], Haerter et al. [73] suggested that phage and bacteria can coexist even when the phage are much more diverse than the capacity of the CRISPR system, while He and Deem [74] used their model to show that the 5′ most spacer is expected to be the most diverse. A recent study by Childs et al. [75] described an explicit eco-evolutionary model of CRISPR evolution that produced many insights, including that CRISPRs induce host and viral diversification, punctuated replacement of strains and the emergence of coalitions of dominant host strains.
In our own agent-based simulations of bacteria and phage evolving with CRISPR, we observe two evolutionary stable modes depending on the cost of resistance, which we can interpret as a combination of the energetic cost of expressing the CRISPR and Cas transcripts and proteins with the fitness cost of occasional errors in self versus nonself discrimination that result in cleavage of the host prokaryotic genome. In our simulations, we observed one mode where the individuals hold a large number of CRISPRs and one mode where each individual holds a small number of CRISPRs, but in either case the bacterial population as a whole has a high diversity of CRISPRs, thus conferring group resistance to the phage (M. S. Kumar and K. C. Chen, unpublished data). These results are qualitatively similar to previous simulation results for the restriction–modification system in bacteria [76].
DIFFERENCES BETWEEN PIRNA AND CRISPR EVOLUTION
The piRNA and CRISPR systems are clearly very similar in their overall molecular logic, and the presence of such an RNA-based mechanism in all three kingdoms of life suggests that RNA may be uniquely well-suited for genome defense against foreign nucleic acids. However, there are also significant differences between piRNAs and CRISPRs, which affect their evolution, as we discuss in this section.
Significance of the population genetic environment
While both the piRNA and CRISPR pathways are expected to show elevated rates of evolution consistent with the Red Queen dynamics often seen in host–pathogen interactions, the rate of evolution might still be very different between the two systems because of their different population genetic environments. Three important aspects of the population genetic environment that merit consideration are the effective population size, the generation time of the organisms involved and the mutation rate.
From population genetics theory, the effective population size for a transposable element family is the effective population size of the host species multiplied by the average number of active copies of the transposable element per haploid genome [77]. The second quantity—the average number of active copies of the transposable element—varies by species and transposable element family. As a concrete example, there are estimated to be 80–100 active LINE-1 elements in the human genome. In the piRNA-transposable element system, the transposable elements are embedded in the host genome and thus are constrained to be replicated in the same generation time as the host genome. The transposable elements also have roughly the same mutation rate as the host genome. Even if the transposable elements are biased to certain parts of the genome, the differences in the local mutation rate are relatively minor. The mutation rate is a significant consideration because there can be mutations in transposable elements that are countered by compensatory mutations in piRNAs. This is a different mechanism of host response to the mechanism of incorporation of new piRNA sequences, which we discuss below in the section, ‘Significance of the insertion mechanism of new CRISPRs/piRNAs’.
In contrast to the piRNA-transposable element system, in the CRISPR-phage system, the phage are autonomous and typically have a census population size much larger than their prokaryotic hosts. Although it is difficult to estimate effective population sizes for bacteria or phage, let alone average effective population sizes over all bacteria and all phage that engage in the CRISPR system, a reasonable estimate is that the effective population size of phage is significantly higher than prokaryotes based on the large difference in census population sizes. A higher effective population size would imply a higher effectiveness of natural selection for phage compared to prokaryotes. Overall, we expect similar phage and prokaryotic generation times since the phage lysis time should be correlated with the prokaryotic cell division time. However, in some systems, phage have a faster generation time than their host bacteria, since the phage can lyse cells and reproduce on a time scale faster than a bacterial cell division. Furthermore, in each generation, many phage can be produced whereas the bacterial population is only doubled. Finally, phage mutation rates are often significantly (10–100 times) higher than bacteria, both for phage with DNA genomes and those with RNA genomes [78].
In sum, despite the scarcity of precise measurements of the relevant population genetic parameters and generalizing over a very large phylogenetic range, we observe the following broad patterns. Both phage and transposable elements have higher effective population sizes than their host species. Both phage and transposable element also have similar generation time to their host species, though the phage generation time can be faster and rate of growth higher compared to transposable elements which are constrained to have exactly the same generation time as their host. Thus, the largest difference between the population genetic enviroments of the piRNA and CRISPR systems is the mutation rate since the phage can mutate at a much faster rate and evade CRISPR-mediated repression than transposable elements can evade piRNA-mediated repression. These observations suggest that CRISPRs would be a relatively ineffective defense mechanism against phage compared to the efficacy of piRNAs against transposable elements. However, CRISPRs are backed up by alternative defense mechanisms, and the insertion rate of new CRISPR spacers may be much higher as discussed in the next two sections.
Significance of alternative mechanisms of genomic defense
It is important to place the defense mechanisms we are discussing in the context of other defense mechanisms in the cell. Indeed, given the intense pressure placed on prokaryotes by fast evolving phage, it is unsurprising that prokaryotes have evolved multiple redundant defense mechanisms. Several such defense mechanisms have been discussed in the literature, including the restriction–modification system (RM) [79] and envelope resistance [80]. A restriction–modification system consists of a pair of enzymes that recognize the same short DNA sequence. The restriction enzyme cuts all unmethylated target sequences while a methylase acts to methylate all of the host target sequences. The RM system thus serves as a defense mechanism against invading phage. Since restriction enzymes target short (roughly 6–8 bp) sites, each enzyme can target many phage genomes. Each CRISPR spacer, however, is in principle constrained to target a specific phage because of the requirement for complementarity to the entire RNA sequence (though see below on the possibility of CRISPR seed sequence). On the other hand, it is much faster to evolve a new crRNA, which can be produced by a Lamarckian mechanism, than a new RM system, which requires classical Darwinian evolution by random mutation and selection. The RM and CRISPR systems thus have different properties that may allow them to work well together in genome defense. In fact, Abedon suggested that CRISPRs are subsidiary to other defense mechanisms [81] using an argument similar to the logic of the vertebrate immune response where an innate immune response provides the first line of defense and an adaptive immune system the second line of defense. In the prokaryotic context, RM systems might play the role of the innate immune response and CRISPRs the adaptive immune system. Experimental evolution results have also indicated that envelope resistance (i.e. a structural modification that prevents adsorption of any phage into the cell) often develops in response to phage in lab cultures [82].
In the case of piRNAs, our population genetic arguments above suggest that they might be more effective at repressing transposable elements than CRISPRs are at repressing phage. Nonetheless, other molecular mechanisms also play a significant role in the repression of transposable elements in the germline. One important mechanism is DNA methylation that prevents transcription of transposable elements in the germline. While little is understood about the evolutionary properties of DNA methylation or how DNA methyl marks are directed to specific loci in the genome, intriguingly, piRNAs are also implicated in the maintenance of DNA methylation in mammals [83, 84]. When this mechanism is more fully worked out at the molecular level, it may be possible to start understanding the interplay between piRNA-mediated regulation of transposable elements at the chromatin level versus the RNA level. On a broader scale, RNAi-related systems in general are known to be involved in genome defense [85] and may have even originated for that purpose.
Significance of the insertion mechanism for new CRISPRs/piRNAs
In the CRISPR system, the CAS proteins provide an active mechanism for inserting new phage sequences into CRISPR loci. In principle, this should allow very fast adaptation to novel phage attacks, in contrast to the piRNA system, as described below. An intriguing aspect of the CRISPR system is the linear arrangement of CRISPR spacers since the newest CRISPR RNAs are inserted at the 5′-end of the CRISPR cassette. For the evolutionist, this arrangement conveniently gives the temporal history of phage infections, with the caveat that occasional deletions of spacers make the history only approximate [12]. For the prokaryotic host, it is still not clear if there is any biological significance to this arrangement. One potential benefit to the host could come from RNA polymerase drop off: since the entire CRISPR cassette is transcribed as a long transcript from which individual spacers are cleaved, even if the polymerase falls off the elongating transcript, relevant CRISPR spacers that target currently active phage will still be expressed.
Unlike the CRISPR case, there is no analogous linear organization for piRNAs. Current evidence suggests that the transposons either jump randomly into piRNA loci or perhaps have a mild preference for inserting into the piRNA loci [86]. Selection for relevant piRNA insertions then occurs at the level of individual germ cells, and in this way adaptation to the invasion of the new transposable element can occur over the lifetime of an individual [86]. Nonetheless, because the mechanism for new piRNA insertion is close to random, it appears to be more inefficient than the mechanism for new CRISPR insertion. Thus, although the population genetic arguments above suggest that piRNAs may be more effective than CRISRPs at repressing their targets, this may be countered by their more inefficient acquisition mechanism.
Significance of the CRISPR/piRNA targeting mechanism
Independent of their role as transposon repressors, piRNAs appear to have a role in the control of endogenous gene expression. Such roles include the control of mRNA translation, direction of both euchromatic and heterochromatic histone modifications and control of higher order chromatin structures [87]. These nontransposon related roles are expected to apply different evolutionary pressure to some piRNAs, perhaps more similar to the evolutionary properties of microRNAs [88]. It is not clear yet whether CRISPRs regulate host–gene expression, but it is certainly conceivable that they have been co-opted by the cell for this purpose.
The mechanism of crRNA targeting is a matter of debate in the current literature. Initial experiments showed that even one mismatch was enough to prevent CRISPR-mediated silencing [56]. Since this result cannot be explained thermodynamically, one possibility is that there is another system that senses the mismatch and prevents silencing. However, later studies suggested a less stringent requirement for base pairing [89, 90], and more recently evidence for a 7-nt seed sequence in E. coli CRISPRs [91] was presented, reminiscent of microRNA seeds [92]. The existence of a seed sequence would be highly significant for the evolution of CRISPRs since it would drastically reduce the amount of sequence in the CRISPR spacer under selective constraint and allow for rapid evolution of new targets. In contrast, there is currently no evidence for a seed sequences in piRNAs and current evidence points to a requirement for nearly complete complementarity over the full length of the piRNA for targeting. Another interesting feature of piRNAs is that there are many redundant piRNA sequences, whereas the same does not appear to be true for CRISPRs. Redundancy would also serve to reduce the evolutionary constraint on individual piRNA sequences.
CONCLUSIONS
In this review, we have compared the evolution of two recently discovered RNA-based adaptive defense mechanisms: the piRNA system in animals and the CRISPR system in prokaryotes. In the process, we have reviewed the aspects of piRNA and CRISPR biology that are most relevant for understanding and modeling their evolution. Overall, the evolutionary logic of the two systems is strikingly similar despite their lack of homology, perhaps pointing to the fundamental importance of RNA-based mechanisms in genome defense. However, as discussed in this review, many aspects of their molecular biology confer different evolutionary properties to the two systems. Several of the most basic evolutionary properties that still remain to be elucidated are: (i) the rate of evolution of piRNAs and CRISPRs at the sequence level; (ii) the rate of evolution of piRNA and CRISPR-generating loci at the level of copy number variation; and (iii) the true amount of sequence in each piRNA and CRISPR that is under selective constraint—particularly the question of whether there is a seed sequence or not. Beyond these basic questions of molecular evolution are broader evolutionary questions such as the interplay of the piRNA and CRISPR systems with alternative defense mechanisms against foreign nucleic acids, such as DNA methylation in the case of piRNAs, or restriction–modification systems and envelope resistance in the case of CRISPRs. Once we can compare the different defense mechanisms, we can study the conditions under which the piRNA or CRISPR system might play important roles in evolution. For example, it has been suggested that the restriction–modification system is important for colonization of new habitats but not in stable communities [82].
There is still a long way to go in understanding the basic molecular biology of piRNAs and CRISPRs and detailed quantitative models of their evolution are not easy to formulate at the present time. Nonetheless, we hope that by highlighting a number of conceptual evolutionary issues, we can help frame future experimental and computational studies of these important genetic mechanisms.
Key Points.
PiRNAs and CRISPRs are RNA-based adaptive immune systems against transposable elements and phage/plasmids, respectively.
Consistent with their role in genome defense, the piRNA and CRISPR pathways show elevated rates of evolution.
Despite their broad similarity, several factors cause different patterns of evolution in piRNAs and CRISPRs. These include the population genetic environment, the presence of other genome defense systems and the mechanism of acquisition of new piRNAs/CRISPRs.
FUNDING
This work was partially funded by the National Institutes of Health (R00HG004515 to K.C.C.).
Acknowledgements
We thank Siobain Duffy, Jody Hey, Mark Siegal, Eduardo Sontag and Robert Trivers for helpful discussions and Marc Friedlander for detailed comments on the manuscript.
Biographies
M. Senthil Kumar undertook graduate studies in computational biology at Carnegie Mellon University and Rutgers University. He is at present a research intern in Computational and Mathematical Biology at the Genome Institute of Singapore.
Kevin C. Chen is an assistant professor in the Department of Genetics and BioMaPS Institute for Quantitative Biology at Rutgers University. He is a recipient of the NIH Pathway to Independence Award.
References
- 1.Ketting R. The many faces of RNAi. Dev Cell. 2011;20:148–61. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song J, Smith S, Hannon G, et al. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–7. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 4.Ruvkun G. Tiny RNA: Where do we come from? What are we? Where are we going? Trends Plant Sci. 2008;13:313–6. doi: 10.1016/j.tplants.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Makarova K, Wolf Y, van der Oost J, et al. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct. 2009;4:29. doi: 10.1186/1745-6150-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin E, Wolf Y. Is evolution Darwinian or/and Lamarckian? Biol Direct. 2009;4:42. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova K, Grishin N, Shabalina S, et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siomi M, Sato K, Pezic D, et al. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 9.Malone C, Hannon G. Molecular evolution of piRNA and transposon control pathways in Drosophila. Cold Spring Harb Symp Quant Biol. 2009;74:225–34. doi: 10.1101/sqb.2009.74.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deveau H, Garneau J, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–93. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 11.Marrafini L, Sontheimer E. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vale P, Little T. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc Biol Sci. 2010;277:2097–103. doi: 10.1098/rspb.2010.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravin A, Naumova N, Tulin A, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–27. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 14.Lau N, Seto A, Kim J, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–7. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 15.Girard A, Sachidanandam R, Hannon G, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;7099:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 16.Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 17.Grivna S, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci. 2006;103:13415–20. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirino Y, Vourekas A, Khandros E, et al. Immunoprecipitation of piRNPs and directional, next generation sequencing of piRNAs. Methods Mol Biol. 2011;725:281–93. doi: 10.1007/978-1-61779-046-1_18. [DOI] [PubMed] [Google Scholar]
- 19.Ruby J, Jan C, Player C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 20.Betel D, Sheridan R, Marks D, et al. Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput Biol. 2007;3:e222. doi: 10.1371/journal.pcbi.0030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaoka S, Izumi N, Katsuma S, et al. 3' end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015–22. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 22.Brennecke J, Aravin A, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 23.Gunawardane L, Saito K, Nishida K, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 24.Birchler J. Ubiquitous RNA-dependent RNA polymerase and gene silencing. Genome Biol. 2009;10:243. doi: 10.1186/gb-2009-10-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipardi C, Paterson B. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc Natl Acad Sci USA. 2009;106:15645–50. doi: 10.1073/pnas.0904984106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Brennecke J, Malone C, Aravin A, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–92. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blumenstiel J, Hartl D. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci. 2005;102:15965–70. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Vagin V, Lee S, et al. Collapse of germline piRNAs in the absence of argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–21. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone C, Brennecke J, Dus M, et al. Specialized piRNA pathways act in germline and somatic tissues of the drosophila ovary. Cell. 2009;137:522–35. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slotkin R, Vaughn M, Borges F, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–72. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–16. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravin A, Sachidanandam R, Girard A, et al. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–7. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 33.Robine N, Lau N, Balla S, et al. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–76. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee E, Banerjee S, Zhou J, et al. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–9. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muotri A, Marchetto M, Coufal N, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–6. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimson A, Srivastava M, Fahey B, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–7. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Assis R, Kondrashov A. Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. Proc Natl Acad Sci. 2009;106:7079–82. doi: 10.1073/pnas.0900523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malone C, Hannon G. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukic S, Chen K. Human piRNAs are under selection in Africans and repress transposable elements. Mol Biol Evol. 2011;28:3061–7. doi: 10.1093/molbev/msr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewing A, Kazazian H. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2010;21:985–90. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, Clark A. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 2010;20:212–27. doi: 10.1101/gr.095406.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castillo D, Mell J, Box K, et al. Molecular evolution under increasing transposable element burden in Drosophila: a speed limit on the evolutionary arms race. BMC Evol Biol. 2011;11:258. doi: 10.1186/1471-2148-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obbard D, Welch J, Kim K, et al. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 2009;5:e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolaczkowski B, Hupalo D, Kern A. Recurrent adaptation in RNA interference genes across the Drosophila phylogeny. Mol Biol Evol. 2010;28:1033–42. doi: 10.1093/molbev/msq284. [DOI] [PubMed] [Google Scholar]
- 45.Vermaak D, Henikoff S, Malik H. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waddington C. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–5. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 47.Rutherford S, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–42. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 48.Jarosz D, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–4. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Queitsch C, Sangster T, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–24. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 50.Specchia V, Piacentini L, Tritto P, et al. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–5. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 51.Gangaraju V, Yin H, Weiner M, et al. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 2011;2:153–8. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–33. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jansen R, Embden J, Gaastra W, et al. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 54.Mojica F, Diez-Villasenor C, Garcia-Martinez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 55.Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–61. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 56.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 57.Marrafini L, Sontheimer E. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garneau J, Dupuis M, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 60.Hale C, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Makarova K, Haft D, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 63.Andersson A, Banfield J. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–50. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 64.Godde J, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–29. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 65.Makarova K, Aravind L, Wolf Y, et al. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Budroni S, Siena E, Dunning Hotopp J, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci USA. 2011;108:4494–9. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Touchon M, Charpentier S, Clermont O, et al. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J Bacteriol. 2011;193:2460–7. doi: 10.1128/JB.01307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyson G, Banfield J. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–7. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 69.Heidelberg J, Nelson W, Schoenfeld T, et al. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS ONE. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deveau H, Barrangou R, Garneau J, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takeuchi N, Wolf Y, Makarova K, et al. Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol. 2012;194:1216–25. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levin B. Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR-mediated adaptive immunity in bacteria. PLoS Genet. 2010;6:e1001171. doi: 10.1371/journal.pgen.1001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haerter J, Trusina A, Sneppen K. Targeted bacterial immunity buffers phage diversity. J Virol. 2011;85:10554–60. doi: 10.1128/JVI.05222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He J, Deem M. Heterogeneous diversity of spacers within CRISPR (clustered regularly interspaced short palindromic repeats) Phys Rev Lett. 2010;105:128102. doi: 10.1103/PhysRevLett.105.128102. [DOI] [PubMed] [Google Scholar]
- 75.Childs L, Held N, Young M, et al. Multi-scale model of CRISPR-induced coevolutionary dynamics: diversification at the interface of Lamarck and Darwin. Evolution. 2012 doi: 10.1111/j.1558-5646.2012.01595.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pagie L, Hogeweg P. Individual- and population-based diversity in restriction-modification systems. Bull Math Biol. 2000;62:759–74. doi: 10.1006/bulm.2000.0177. [DOI] [PubMed] [Google Scholar]
- 77.Dolgin E, Charlesworth B, Cutter A. Population frequencies of transposable elements in selfing and outcrossing Caenorhabditis nematodes. Genet Res, Camb. 2008;90:317–29. doi: 10.1017/S0016672308009440. [DOI] [PubMed] [Google Scholar]
- 78.Duffy S, Shackleton L, Holmes E. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–76. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 79.Wilson G, Murray N. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 80.Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abedon S. Facilitation of CRISPR adaptation. Bacteriophage. 2011;1:179–81. doi: 10.4161/bact.1.3.16709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Korona R, Levin B. Phage-mediated selection and the evolution and maintenance of restriction-modification. Evolution. 1993;47:556–75. doi: 10.1111/j.1558-5646.1993.tb02113.x. [DOI] [PubMed] [Google Scholar]
- 83.Watanabe T, Tomizawa S, Mitsuya K, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–52. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aravin A, Sachidanandam R, Bourc'his D, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cerutti H, Casas-Mollano J. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khurana J, Wang J, Xu J, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–63. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–76. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 89.Garrett R. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem Soc Trans. 2011;39:51–7. doi: 10.1042/BST0390051. [DOI] [PubMed] [Google Scholar]
- 90.Manica A, Zebec Z, Teichmann D, et al. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol. 2011;80:481–91. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 91.Semenova E, Jore M, Datsenko K, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lai E. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]