Abstract
The mitogen-activated protein kinase kinases (the MAPK/ERK kinases; MKKs or MEKs) and their downstream substrates, the extracellular-regulated kinases have been intensively studied for their roles in development and disease. Until recently, it had been assumed any mutation affecting their function would have lethal consequences. However, the identification of MEK1 and MEK2 mutations in developmental syndromes as well as chemotherapy-resistant tumors, and the discovery of genomic variants in MEK1 and MEK2 have led to the realization the extent of genomic variation associated with MEKs is much greater than had been appreciated. In this review, we will discuss these recent advances, relating them to what is currently understood about the structure and function of MEKs, and describe how they change our understanding of the role of MEKs in development and disease.
Keywords: MEK, MAPK, ERK, cardio-facial cutaneous syndrome, cancer, SNP
INTRODUCTION
The mitogen-activated protein kinase kinases (the MAPK/ERK kinases; MKKs or MEKs) and their downstream substrates, the extracellular signal-regulated kinases (ERKs, also known as mitogen-activated protein kinase or MAPK) are involved in diverse biologic processes. These multipurpose kinases have been reported to have an essential role in fundamental cellular activities, including cell survival, proliferation, motility and differentiation. They are critical for angiogenesis [1] and immune response [2] and play important roles in development [3], where they are involved in meiosis [4], gastrulation [5], cell fate determination [6–10], organogenesis and limb patterning [11]. They are frequent targets of bacterial pathogens, including Bacillus anthracis (anthrax; [12]), Yersinia pestis (plague; [13]) and Shigella (Shigellosis; [14]). Due to these studies, it was previously assumed that germline mutations in MEKs or ERKs would be lethal. However, the startling observations of Rodriguez-Viciana et al. [15] that germline mutations in MEKs are associated with cardio-facio-cutaneous (CFC) syndrome abruptly changed that paradigm [16]. Since then, new findings have led to the growing awareness that the extent of genomic variation associated with MEKs is much greater than had been appreciated. In this review, we will discuss these recent advances, relating them to what is currently understood about the structure and function of MEKs, and describe how they change our understanding of the role of MEKs in development and disease.
MEK STRUCTURE AND FUNCTION
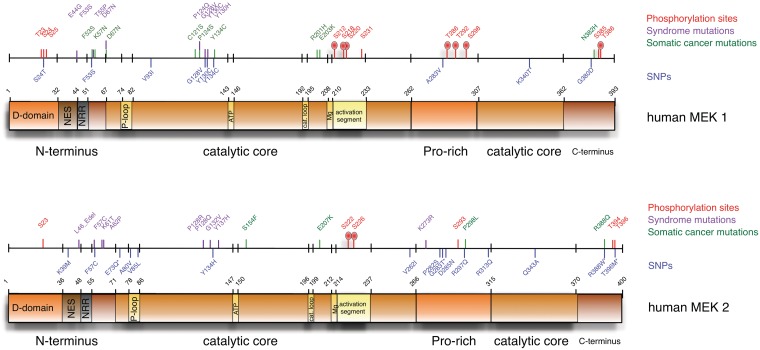
A genomic approach to biology is only meaningful when interpreted in a physiologic, functional context. Fortunately, we know a lot about the structure and function of both MEKs (reviewed by [17]). MEK1 and MEK2 are dual specificity kinases that phosphorylate ERK1/2 at residues T202/185 and Y204/187 (reviewed by [18]). The functional domains of human MEK1 and MEK2 are diagrammed in Figure 1. Both MEK1 and MEK2 contain a central catalytic domain flanked on either side by short amino-terminal and carboxy-terminal regions and containing a proline-rich insert. These kinases are highly homologous, sharing 80% overall similarity, with 90% amino acid identity in their kinase domains. However, their sequences diverge in the flanking regions and the proline-rich insert. Each of these regions incorporates specific elements that distinguish these kinases, influencing interactions with other proteins and modifying kinase activity.
Figure 1:
A linear model of MEK 1 and MEK 2 showing locations of identified functional regions and mutations. Linear models of MEK 1 and MEK 2 are used to indicate the locations of functional domains in both kinases. Above each is a scale with hash marks at the locations of predicted phosphorylation sites (red), experimentally identified phosphorylation sites (red hash with oval), syndrome-associated mutations (purple), somatic cancer mutations (green), and SNPs (blue). Numbers indicate amino acid position starting from the first Met residue.
The amino-terminal flanking regions of MEK1 and MEK2 (amino acids 1–67 and 1–71, respectively; Figure 1) share 58% of the sequence identity. However, their sequences are less similar in the first 32 or 36 residues (MEK1 or MEK2, respectively) than they are in the remaining 28 or 24 residues (22% homology vs. 82%). This is notable because this least similar region mediates interaction with their ERK substrates. The ERK docking site or D-domain encompasses a short stretch of basic and hydrophobic amino acids located within the first 10 residues [19]. The positively charged D-domain is essential for MEK binding to a complementary acidic common docking domain in the carboxy-termini of ERK1 and ERK2 [20, 21]. Deletion of the D-domain or mutation of either hydrophobic or basic residues reduced ERK phosphorylation [22, 23]. Similarly, proteolysis of the D-domain by anthrax lethal factor, the principle virulence factor secreted by B. anthracis, inhibits MEK association with and activation of ERK [12, 24, 25]. Genomic variants within this region may alter MEK:substrate interaction.
Between the D-domain and the catalytic core lies a nuclear export sequence (NES; amino acids 33–44 and 37–48; Figure 1). The NES is critical for MEK function and subcellular localization. MEKs normally reside in the cytoplasm. However, lysine to alanine substitution in the NES alters MEK steady-state cellular distribution to the cytoplasm and nucleus. This suggests MEKs translocate to the nucleus upon activation and then are rapidly exported to the cytoplasm in an NES-dependent manner [26, 27]. Genomic variants within this region may influence MEK subcellular localization.
Immediately following the NES lies a negative regulatory region (NRR; Figure 1). Deletion of residues 44–51 of MEK1 or 48–55 of MEK2 causes a 60- or 9-fold elevation in basal kinase activity, respectively [28]. These residues repress MEK activity by forcing an inactive conformation that disrupts the ATP-binding site [29]. Genomic variants within this region may alter MEK catalytic activity.
The proline-rich domain is located within the carboxy-terminal portion of the central conserved catalytic domain (amino acids 262–307 in MEK1 and 266–315 in MEK2; Figure 1) and is believed to mediate specific protein–protein interactions important for the regulation of the MEKs [30–32]. Residues at the end of this segment have been reported to mediate binding with the bacterial virulence factors B. anthracis LF [24] and Y. pestis YopJ [33]. The proline-rich domains of MEK1 and MEK2 show 60% homology. Genomic variants within this region may influence MEK activity through altered protein complex formation.
The structure of the catalytic kinase core is broadly similar to that of other kinases and may be divided into a small amino-terminal lobe and a larger carboxy terminal lobe (Figure 1). Located at the interface between these lobes are conserved regions playing important roles in ATP binding and hydrolysis, substrate recognition and phosphate transfer (reviewed by [17]). Genomic variants within any of these regions may decrease the catalytic activity of MEK. Critical sites of this catalytic core include the glycine-rich P-loop (amino acids 74–82 of MEK1 or 78–86 of MEK2), the Mg2+-positioning loop (amino acids 208–210 of MEK1 or 212–214 of MEK2), the ATP-binding site (amino acids 143–146 of MEK1 or 147–150 of MEK2), and the catalytic loop (amino acids 192–195 of MEK1 or 196–199 of MEK2).
The carboxy-terminal regions of MEK1 (amino acids 362–393) and MEK2 (amino acids 370–400) share 69% homology. Little is known of the function of this domain. However, Brunet et al. [34] reported that ERK phosphorylation of MEK1 at T386 is a component of a negative feedback loop regulating MEK1 inactivation.
Additional insight into MEK structure and function comes from phosphorylation studies. Major phosphorylation sites at S218 and S220 of MEK1 or S222 and S226 of MEK2 are located within the activation segment (amino acids 208–233 of MEK1 or 212–237 of MEK2; Figure 1, red hash marks with ovals). Phosphorylation at these sites causes a re-alignment of the loop that coordinates Mg2+ -positioning that is essential for ATP-binding and catalysis. Paradoxically, Gopalbhai et al. [35] has reported phosphorylation at S212 in the activation loop reduces MEK1 activity. These reports indicate genomic variants within the activation loop may enhance or suppress the catalytic activity of MEK.
Phosphorylation outside the activation segment also influences MEK activity. PAK1-mediated phosphorylation of S298 in the proline-rich region is reported to induce MEK1 autophosphorylation at S218 and S222 [36, 37]. Elsewhere within the proline-rich domain MEK1 but not MEK2 has been reported to be phosphorylated on T286 and T292 and inactivated by cdc2 [38]. In addition, Eblen et al. [39] have shown that ERK2-mediated phosphorylation of MEK1 at T292 blocks PAK-dependent phosphorylation of S298. ERK-mediated phosphorylation of T292 has also been reported by Brunet et al. [34]. In a recent study, Catalanotti et al. [40] reported alanine substitution on MEK1 at T292 enhanced both MEK1 and MEK2 activation and ERK phosphorylation. Conversely, mutation of the same residue to aspartic acid dampened MEK1 and MEK2 activation and ERK phosphorylation. How this occurs is not known but the ability of MEK1 to modulate MEK2 activity may be related to the ability of these two kinases to form heterodimers because an N87G mutation in MEK1 blocks formation of heterodimers and prolongs growth factor stimulated activation of MEK2. These studies indicate genomic variants within the proline-rich region may enhance or suppress the MEK activity. Other phosphorylation sites have been identified in kinomic studies [41–44] but the physiologic role of these is unknown (Figure 1, red hash marks lacking ovals).
MUTATIONAL ANALYSIS OF MEK IN DEVELOPMENT AND DISEASE
Initial insight into the central role of MEK in cell proliferation and tumorigenesis has come from the in vitro analysis of engineered mutations to MEK. As mentioned earlier, deletion of residues 44–51 of MEK1 or 48–55 of MEK2 causes a 60- or 9-fold elevation in basal kinase activity, respectively [28]. Site-directed mutagenesis switching serine phosphorylation targets in the activation segment to either aspartate or glutamate mimics phosphorylation by introduction of negatively charged side-chains and elevates MEK1 activity 300-fold above basal levels. This effect is doubled when the NRR is also deleted [28]. Constitutively active MEK1 mutants have been shown to enhance cell proliferation and differentiation, and to promote transformation of NIH3T3 cells, as evidenced by foci formation, growth in soft agar, and growth of tumors in nude mice [28, 45, 46]. Conversely, dominant negative MEK1 mutants prevent fibroblast proliferation [47], and can revert mos-, raf-, ras- and src-transformed 3T3 cells [46, 48].
Due to structural similarity, MEK1 and MEK2 are thought to be functionally redundant. However, genetic dissection of MEK1 and MEK2 using mouse models has suggested nonoverlapping functions. MEK1 null mice are embryonic lethal at E10.5 with placental vascularization defects [49], whereas MEK2 knockout mice are viable and fertile with no evidence of growth defects or anatomical alterations [50]. These models indicate that MEK1 can compensate for MEK2, but not vice versa. Interestingly, MEK1/2 double knockout in both murine and human epidermis result in epithelial hypoproliferation and inhibition of Raf-induced hyperproliferation, while neither MEK1 nor MEK2 knockout alone has any effect on epidermal development [51], suggesting that MEK1 and MEK2 are functionally redundant in the epidermis and that neither alone is sufficient for epidermal development.
Transgenic mouse models utilizing constitutively active or dominant negative MEK have also been very useful in delineating the roles of MEKs in development and disease. Multiple groups have shown that epidermal-specific expression of wild-type [52] or constitutively active MEK1 [53, 54] results in epidermal hyperproliferation, suppression of epidermal differentiation, and papilloma formation. Interestingly, overexpression of a kinase dead MEK1 mutant in the epidermis also had the ability to induce epidermal hyperplasia, indicating that MEK1 kinase activity is not necessary to increase epidermal proliferation [52]. Furthermore, MEK2 was unable to alter the proliferation and differentiation profile of epidermal cells in this mouse model [52]. This supports the hypothesis that MEK1 and MEK2 have nonoverlapping functions.
Other models that indicate the proliferative activity of MEK1 in vivo include cardiac- and chondrocyte-specific expression of a constitutively active MEK1 mutant, which resulted in cardiac hypertrophy and inhibition of chondrocyte differentiation, respectively [55, 56].
MEK-ASSOCIATED MUTATIONS IN DISEASE
CFC and related syndromes
CFC syndrome is a rare, autosomal and presumably dominant syndrome characterized by distinctive facial features, cardiac anomalies, hair and skin abnormalities, postnatal growth deficiency and hypotonia [57]. In landmark publications, Rodriguez-Viciana et al. [15] and Niihori et al. [58] showed that genetic mutations in genes participating with MEK signaling, including KRAS, BRAF, MEK1 and MEK2 are associated with CFC syndrome. Rodriguez-Viciana’s group screened a panel of blood samples from CFC patients for germline mutations in this pathway and found 11 different missense mutations in BRAF (9 of which had not been previously identified) in 18/23 patients. Likewise, Niihori’s group screened for mutations in 43 samples and found germline KRAS and BRAF mutations in 3 and 19 patients, respectively. No mutations were detected in the remaining 24 patient samples. Rodriguez-Viciana’s group also found that three of their remaining five patients harbored novel mutations in MEK1 (F53S, Y130C) and MEK2 (F57C) (Figure 1, purple hash marks). This is particularly noteworthy because it was the first identified instance of naturally occurring mutations in these genes. Their observations also offer interesting insight into the biochemistry of MEK 1 and MEK2 because none of these residues had previously been demonstrated to influence MEK biologic activities.
Since these initial reports, additional mutations in MEK1 and MEK2 resulting in increased kinase activity have been identified in 20–25% of CFC [59–63] (Figure 1, purple hash marks). CFC MEK mutations occur in two main clusters. The first cluster lies in and around the MEK NRR. In addition, two different deletions in the same area are documented in MEK2 in patients suffering from CFC. The most frequently documented mutation lies in the second cluster in the kinase domain encoding an amino acid change residue 130 from tyrosine to histidine, cytosine or asparagine [59, 60, 63–66]. The corresponding Y137H mutation was also found in MEK2 [62]. Hao et al. [33] have reported that the Y130H mutation constitutively activates MEK1. Along with amino acid 130, MEK1 mutations have been published at residues 128 and 124 and the corresponding residues 128 and 132 in MEK2. In MEK2, a third mutation has also been characterized in the proline-rich domain, causing a lysine to arginine switch at residue 273.
Although not all mutations have been examined, MEK1 and MEK2 mutations appear to result in a moderate increase of MEK activity [15, 60], but CFC mutant MEK kinase activity is not as robust as constitutively active MEK [15]. These assays were performed in the presence of BRAF. In fact, with the exception of F57C, BRAF phosphorylation is required for CFC MEK activation [67]. And while most of the MEK mutations identified cause constitutive activation of the MEK pathway, one mutation in an upstream activator (G596V BRAF) was deficient in its ability to activate MEK [15]. This seems paradoxical because it suggests that a similar phenotype may be caused by both activating and inactivating MEK signaling. A similar situation exists for the related autosomal dominant Noonan and LEOPARD syndromes, which are caused by gain-of-function mutations and inactivating mutations in the protein tyrosine phosphatase PTP11 (SHP2), leading to constitutive [68, 69] or impaired ERK activation [70], respectively. If the developmental processes underlying these syndromes require a transient activation of ERK, then the dampened or sustained elevation of ERK activity may be sufficient to disrupt it. Alternatively, loss of ERK signaling may trigger activation of a compensatory mechanism, such as an alternative RAF signaling pathway that mimics sustained activation of this pathway and elicits a similar phenotypic result.
Cancer
Approximately one-third of all cancer cases have aberrant MAPK pathway signaling [71, 72], making the members of this pathway attractive targets for therapy. RAS mutants are frequent in human cancer, with ∼20–25% of all tumors containing activating mutations in a RAS gene [73, 74]. Mutations in BRAF occur at a high frequency in a variety of tumor types, predominantly melanomas (50–70%), thyroid (30–50%), ovarian (30%) and colorectal cancer (CRC) (5–20%) [75, 76]. The predominant BRAF mutation is V600E, accounting for more than 90% of BRAF mutations identified [76]. The high frequency of BRAF mutations in human cancer has led to an intense focus on the development of therapeutic strategies to target BRAF. Although current strategies utilizing selective targeting of BRAF V600E have shown encouraging results in the clinic, therapeutic resistance is a continuing issue with response to this therapy. Although MEK mutations are rare in human cancer, MEK inhibitors have been developed as a therapeutic strategy to combat BRAF inhibitor resistance by targeting downstream effectors. To date, these MEK inhibitors have shown poor efficacy and activity in the clinic. However, with the emergence of resistance to BRAF therapy, and a higher than previously thought frequency of somatic MEK mutations (Figure 1, green hash marks; discussed below), these inhibitors are finding renewed clinical use.
Melanoma
More than 80% of the melanomas harbor either a NRAS or BRAF mutation, resulting in uncontrolled MAPK pathway activation in this cancer type. The BRAF V600E activating mutation accounts for the majority of BRAF mutations in melanoma. Interestingly, MEK mutations have been identified in melanomas exhibiting resistance to BRAF inhibitors, suggesting a possible mechanism for acquired resistance (discussed in more detail in the next section). Somatic mutations in MEK have not been investigated until recently, however. Analysis of melanoma tumor samples identified a low incidence of MEK1 mutations (3–8%) [77, 78]. The first identified melanoma-associated MEK1 mutation, K57N (Figure 1, green hash mark), had been previously reported in a lung adenocarcinoma and shown to be constitutively active (discussed in more detail below) [79]. This mutation is located between the nuclear export signal and catalytic domain of MEK1 [77]. Exome sequencing has since identified additional MEK1 and MEK2 somatic mutations in melanoma [78]. Of 127 melanoma samples analyzed, MEK1 mutations were identified in 7, while only one sample had a MEK2 mutation. Identified MEK1 mutations included P124S (in four samples), E203K (in one sample), F53L (in one sample) and N382H (in one sample), while the lone MEK2 mutation was S154F (Figure 1, green hash marks). It is interesting to note that these mutated residues overlap with those implicated in CFC [15]. Surprisingly, these somatic MEK mutations did not correlate with BRAF or NRAS mutation status, suggesting the requirement of additional modulation of the MAPK pathway, possibly in response to therapeutic resistance mechanisms.
Furthermore, analysis of melanoma metastases identified MEK mutations in samples that harbored BRAF mutations other than V600E (E203K and E207K for MEK1 and MEK2, respectively). Functional analysis indicated that E203K, P124S and F53L were constitutively active mutations for MEK1 [78].
Non-small cell lung cancer
Approximately 30% of the non-small cell lung cancer (NSCLC) cases have KRAS mutations, whereas only 1–3% harbor BRAF mutations [80]. Furthermore, BRAF mutations predominantly occur in adenocarcinomas, and are typically not V600E. Initial analysis of human lung cancer cell lines identified only one mutation, in MEK2, which led to an amino acid change (P298L; Figure 1, green hash mark) [81], suggesting MEK mutations in lung cancer are a rare event. Mutational analysis of primary NSCLC tumors identified a novel MEK1 somatic mutation (in 2 of 207 samples), K57N (Figure 1, green hash mark) [79]. This mutation, located in a nonkinase region of MEK1, was shown to confer constitutive activation of the downstream effector ERK1/2 [79].
An additional cohort of 280 NSCLC patients also identified K57N in one of the samples [82]. This MEK1 mutation was found to be exclusive of mutations of other components of the MAPK signaling pathway within both cohorts [79, 82].
Colorectal cancer
In CRC, RAS is mutated in ∼36% of cases, while BRAF mutations can be found in 9–11% of CRC [83]. A screen of 93 tumor samples and 22 cell lines identified a low rate of MEK1 (1%) and MEK2 (2%) somatic mutations [84]. As seen for somatic mutations in melanoma, the mutations identified in these CRC samples (R201H, E203K, Y134C, for MEK1; R388Q for MEK2) also lie in the same region of the protein as CFC germline mutations (Figure 1, compare purple hash marks to green hash marks). A separate mutational analysis also identified a low rate of MEK1 mutations in CRC samples (2.2%), the MEK1 mutation (D67N) having been previously characterized in ovarian cancer cells (discussed in the following section) [77].
Ovarian cancer
In low grade serous ovarian carcinomas, BRAF mutations are present in 28–37% of cases [85]. Mutational analysis of 15 ovarian cancer cell lines identified a MEK1 mutation in one of these lines, ES-2, while no mutations were found in MEK2 [86]. The novel MEK1 mutation, D67N (Figure 1, green hash mark), was demonstrated to be constitutively active based on increased ERK phosphorylation following transient transfection of 293T cells [86].
Cancer types that lack somatic MEK mutations
There are many tumor types where MEK mutations have yet to be identified. These types include thyroid cancer [77], breast cancer [84, 87], gliomas [88] and testicular cancer [89]. Interestingly, the majority of these cancer types has a low rate of RAS/BRAF mutations [76, 89–91]. The exception to this group is thyroid cancer, where up to 70% of the papillary thyroid cancer cases have BRAF V600E [92, 93]. This may seem surprising, as one would expect a low frequency of MEK1 mutations in cancers that have a high prevalence of RAS/BRAF mutations, due to the redundant nature of MAPK signaling activation from RAS/BRAF and MEK mutations. However, it appears that cancer types with frequent RAS/BRAF mutations also harbor a low frequency of MEK mutations as well (melanoma, colon and lung), while those that have rare RAS/BRAF also appear to not contain MEK mutations (breast, glioma and testicular). It is unclear whether MEK1 and MEK2 mutations occur at such as low rate that more samples require screening, or if they simply do not occur in these cancer types.
A mechanism for BRAF inhibitor resistance
Due to the high frequency of BRAF V600E mutations in multiple cancer types, selective BRAF inhibitors have been developed as a therapeutic strategy for patients with cancers harboring the V600E mutation. These inhibitors, vemurafenib and dabrefenib, showed promising results in phase 1 trials, but the responses were short-lived due to the development of resistance to these therapies [94, 95]. Although the mechanism of resistance to BRAF inhibitors is still unclear, many potential mechanisms have been suggested. These include activation of receptor tyrosine kinases such as insulin-like growth factor receptor 1 and platelet-derived growth factor receptor (reviewed in [94]) and epidermal growth factor receptor (EGFR) [96], NRAS up-regulation [97], and elevated CRAF levels [98].
A more recent alternative has been the acquisition of MEK1 mutations as a mechanism for BRAF inhibitor resistance. Mutagenesis screens of BRAF V600E containing cell lines identified constitutively active MEK1 mutations that conferred resistance to either MEK or BRAF inhibitors [99, 100]. More importantly, a screen of tumors from relapsed patients identified MEK1 mutations that conferred resistance to BRAF inhibition, as well as to MEK inhibitors [99]. Furthermore, an activating MEK1 C121S somatic mutation was identified in the melanoma sample that was resistant to the BRAF inhibitor PLX4032 [100]. This mutation was not found in the pretreated tumor, suggesting that acquisition of this MEK1 activating mutation is a possible mechanism for BRAF inhibitor resistance.
Genomic variation in MEK1 and MEK2
In the preceding discussion, we described genomic variants of MEK1 and MEK2 that were identified because they changed MEK activity, creating a developmental syndrome or providing a selective growth advantage. However, the human genome is riddled with single nucleotide polymorphisms that may or may not alter protein activity. Neither MEK is an exception in this regard. Publicly available data in dbSNP [101] and Ensembl lists 29 identified synonymous single nucleotide polymorphisms present in MEK1, as well as 10 nonsynonymous SNPs. In addition to this, more than 40 intronic SNPs have been identified for this gene. Similarly, for MEK2 the dbSNP and Ensembl databases list 36 identified synonymous single-nucleotide polymorphisms as well as 19 nonsynonymous SNPs. Remarkably, more than 207 intronic SNPs have been identified for this gene. Notably, many of these nonsynonymous SNPs map to functional regions of MEK1 and MEK2, indicating they may have phenotypic consequences (Figure 1, blue hash marks). These consequences may be severe as seen in CFC and related developmental syndromes. Alternatively, it is possible these polymorphisms have subtle effects that influence other traits linked to MEK activity.
SUMMARY
In this review article, we have discussed recent advances in the genomic study of MEK1 and MEK2, relating them to what is currently understood about the structure and function of MEKs, and describing how they have changed our understanding of the role of MEKs in development and disease. One of the more intriguing discoveries arising from sequencing studies such as the 1000 Genomes Project Consortium [102] and the Cancer Genome Atlas is the remarkable extent of genetic variation seen across the genome. This has led to a growing awareness that the extent of genomic variation associated with MEKs is much greater than initially had been appreciated. These studies are in their infancy so we may expect to add to the list of genomic variants in the next few years. The more complicated task that lies ahead is to decipher how this variation impacts cell and body function. Diseases such as CFC illustrate how these SNPs can influence development in profound and unexpected ways. Fortunately, these diseases are rare. The functional consequences of the mutations may be elucidated using protein or cell-based studies but transgenic or knock-in mouse models must be developed to guage their phenotypic consequences.
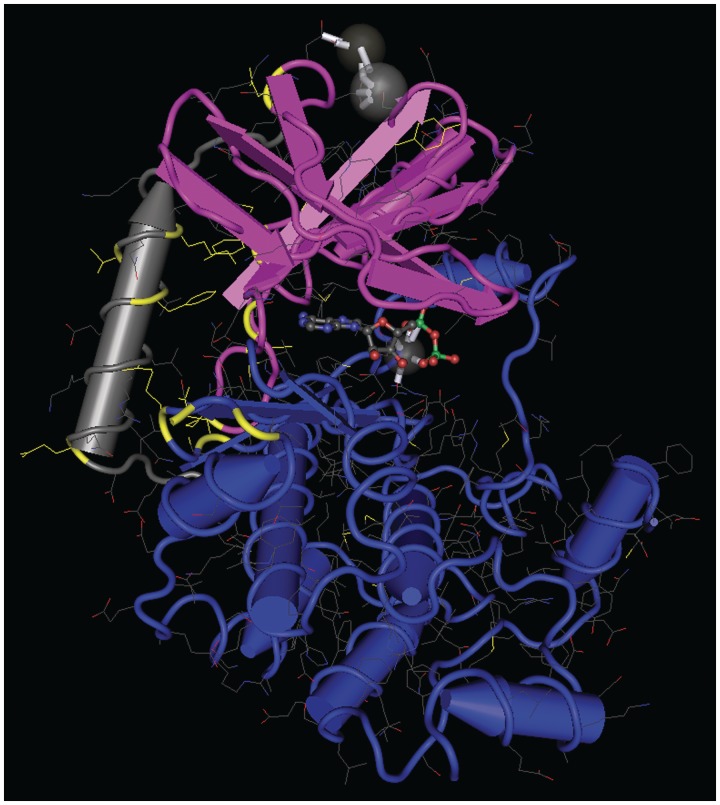
It is curious that the genomic variants cluster in functional regions in which we previously have identified mutations contributing to developmental syndromes or cancer progression (Figure 1). This is not coincidental. Although these clusters of mutations are separated in a linear representation of MEK1, when mapped on to a three-dimensional (3D) model of MEK1 they are brought closer together, forming a mutational hot-spot that is proximal to the NRR and the ATP-binding site and thus is well positioned to influence catalytic activity (Figure 2). We know the mutations associated with developmental syndromes cause a modest activation of MEK activity compared with activating mutations in the activation segment. This may be the key to why they are tolerated during development. Their hyperactivity may be tampered down by compensatory mechanisms that prevent unrestrained proliferation or neoplasia. The same explanation may be applied to non-syndromic germline SNPs. But why are related mutations observed in cancer and why are they more commonly observed in MEK1 whereas germline variants are more common in MEK2? To explain this, we propose MEK1 and MEK2 are differentially regulated. If MEK1 is subject to a lesser degree of feedback regulation it may be less tolerant of syndromic mutations during development and better capable of driving proliferation in cancer. In contrast, the ability to compensate for MEK2 activation would be an advantage during development but a disadvantage when trying to drive tumorigenesis or evade therapies targeting upstream activators. For example, it is possible that while the effects of MEK2 mutations are mitigated by negative feedback through heterodimerization with MEK1, activating MEK1 mutations cannot be repressed in a similar manner. Alternatively, other studies support differential roles of MEK1 and MEK2. For example, Scholl et al. [103] recently demonstrated that MEK1 but not MEK2 is required for DMBA/TPA-induced benign epidermal papilloma formation. On the other hand, Voisin et al. [104] have shown that shRNA-mediated MEK2 knockdown has much stronger inhibitory effect on colon cancer cell proliferation than MEK1 knockdown does, leading them to conclude that MEK2 is more important for colon cancer cell proliferation than MEK1. Similarly, our lab has shown MEK2 is sufficient for melanoma cell proliferation, but MEK1 is not [105]. Whatever the answer may be, it is clear that new insight into MEK genomics has changed our understanding of the role of MEKs in development and disease.
Figure 2:
A 3D model of MEK 1 showing a mutational hot spot. A 3D X-ray model of residues 35–382 of MEK 1 in a binary complex with ADP and Mg2+ [29] rendered in worms with Cn3D software was used to map the locations of syndromic and cancer-related mutations as well as germline variants. The model is oriented with the small amino-terminal catalytic lobe colored pink at the top and the large catalytic lobe in blue at the bottom. ADP and Mg2+ are shown in the cleft between the two lobes. The amino terminus is represented on the left in grey. Mutated residues are indicated in yellow.
Key Points.
The mitogen-activated protein kinase kinases and their downstream substrates, the extracellular signal-regulated kinases, play central roles in diverse biologic processes.
Functionally, both MEK 1 and MEK2 may be divided into several regulatory regions incorporating specific elements that distinguish these kinases and influence their interactions with other proteins.
Initial insight into the central role of MEK in cell proliferation and tumorigenesis has come from the in vitro analysis of engineered mutations to MEK.
Although germline MEK mutations are associated with the developmental syndrome CFC syndrome, somatic MEK mutations have been identified in a limited number of tumors as well as in chemotherapy-resistant tumors.
The unequal distribution of syndrome and cancer-associated mutations indicates MEK 1 and MEK 2 are differentially regulated.
FUNDING
We acknowledge financial support from the National Institutes of Health/National Cancer Institute (RC2CA148149) and the Dwight Reed Memorial Foundation.
Biographies
Jennifer Bromberg-White, PhD, is a research scientist in the Laboratory of Cancer and Developmental Cell Biology at the Van Andel Research Institute. Her research interests include delineating the roles of MKKs during developmental and pathological angiogenesis in the retina.
Nicholas Andersen, PhD, is a post-doctoral fellow in the Laboratory of Cancer and Developmental Cell Biology at the Van Andel Research Institute. His research interests include determining the involvement of MKK pathways signaling in initiating and driving tumor growth and angiogenesis.
Nicholas Duesbery, PhD, heads the Laboratory of Cancer and Developmental Cell Biology and is the director of the Center for Cancer and Cell Biology at the Van Andel Research Institute. His research and that of the lab focuses on defining the roles of MKK signaling in endothelial function during development and disease.
References
- 1.Depeille PE, Ding Y, Bromberg-White JL, et al. MKK signaling and vascularization. Oncogene. 2007;26:1290–6. doi: 10.1038/sj.onc.1210198. [DOI] [PubMed] [Google Scholar]
- 2.Mansfield PJ, Shayman JA, Boxer LA. Regulation of polymorphonuclear leukocyte phagocytosis by myosin light chain kinase after activation of mitogen-activated protein kinase. Blood. 2000;95:2407–12. [PubMed] [Google Scholar]
- 3.Widmann C, Gibson S, Jarpe MB, et al. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–80. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 4.Haccard O, Sarcevic B, Lewellyn A, et al. Induction of metaphase arrest in cleaving Xenopus embryos by MAP kinase. Science. 1993;262:1262–5. doi: 10.1126/science.8235656. [DOI] [PubMed] [Google Scholar]
- 5.Gotoh Y, Masuyama N, Suzuki A, et al. Involvement of the MAP kinase cascade in Xenopus mesoderm induction. EMBO J. 1995;14:2491–8. doi: 10.1002/j.1460-2075.1995.tb07246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornfeld K, Guan KL, Horvitz HR. The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev. 1995;9:756–68. doi: 10.1101/gad.9.6.756. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y, Han M, Guan KL. MEK-2, a Caenorhabditis elegans MAP kinase kinase, functions in Ras-mediated vulval induction and other developmental events. Genes Dev. 1995;9:742–55. doi: 10.1101/gad.9.6.742. [DOI] [PubMed] [Google Scholar]
- 8.Hsu JC, Perrimon N. A temperature-sensitive MEK mutation demonstrates the conservation of the signaling pathways activated by receptor tyrosine kinases. Genes Dev. 1994;8:2176–87. doi: 10.1101/gad.8.18.2176. [DOI] [PubMed] [Google Scholar]
- 9.Umbhauer M, Marshall CJ, Mason CS, et al. Mesoderm induction in Xenopus caused by activation of MAP kinase. Nature. 1995;376:58–62. doi: 10.1038/376058a0. [DOI] [PubMed] [Google Scholar]
- 10.Curran KL, Grainger RM. Expression of activated MAP kinase in Xenopus laevis embryos: evaluating the roles of FGF and other signaling pathways in early induction and patterning. Dev Biol. 2000;228:41–56. doi: 10.1006/dbio.2000.9917. [DOI] [PubMed] [Google Scholar]
- 11.Corson LB, Yamanaka Y, Lai KM, et al. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–37. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- 12.Duesbery NS, Webb CP, Leppla SH, et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–7. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee S, Keitany G, Li Y, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–4. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Xu H, Zhou Y, et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–3. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Viciana P, Tetsu O, Tidyman WE, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–90. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 16.Duesbery N, Vande Woude G. BRAF and MEK mutations make a late entrance. Sci STKE. 2006;2006:pe15. doi: 10.1126/stke.3282006pe15. [DOI] [PubMed] [Google Scholar]
- 17.Roskoski R., Jr MEK1/2 dual-specificity protein kinases: structure and regulation. Biochem Biophys Res Commun. 2012;417:5–10. doi: 10.1016/j.bbrc.2011.11.145. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Gibson TB, Robinson F, et al. MAP kinases. Chem Rev. 2001;101:2449–76. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 19.Xu B, Wilsbacher JL, Collisson T, et al. The N-terminal ERK-binding site of MEK1 is required for efficient feedback phosphorylation by ERK2 in vitro and ERK activation in vivo. J Biol Chem. 1999;274:34029–35. doi: 10.1074/jbc.274.48.34029. [DOI] [PubMed] [Google Scholar]
- 20.Yang SH, Whitmarsh AJ, Davis RJ, et al. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998;17:1740–9. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubinfeld H, Hanoch T, Seger R. Identification of a cytoplasmic-retention sequence in ERK2. J Biol Chem. 1999;274:30349–52. doi: 10.1074/jbc.274.43.30349. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901–8. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu B, Stippec S, Robinson FL, et al. Hydrophobic as well as charged residues in both MEK1 and ERK2 are important for their proper docking. J Biol Chem. 2001;276:26509–15. doi: 10.1074/jbc.M102769200. [DOI] [PubMed] [Google Scholar]
- 24.Chopra AP, Boone SA, Liang X, et al. Anthrax lethal factor proteolysis and inactivation of MAPK kinase. J Biol Chem. 2003;278:9402–6. doi: 10.1074/jbc.M211262200. [DOI] [PubMed] [Google Scholar]
- 25.Bardwell AJ, Abdollahi M, Bardwell L. Anthrax lethal factor-cleavage products of MAPK (mitogen-activated protein kinase) kinases exhibit reduced binding to their cognate MAPKs. Biochem J. 2004;378:569–77. doi: 10.1042/BJ20031382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaaro H, Rubinfeld H, Hanoch T, et al. Nuclear translocation of mitogen-activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc Natl Acad Sci USA. 1997;94:3742–7. doi: 10.1073/pnas.94.8.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda M, Gotoh I, Adachi M, et al. A novel regulatory mechanism in the mitogen-activated protein (MAP) kinase cascade. Role of nuclear export signal of MAP kinase kinase. J Biol Chem. 1997;272:32642–8. doi: 10.1074/jbc.272.51.32642. [DOI] [PubMed] [Google Scholar]
- 28.Mansour SJ, Candia JM, Gloor KK, et al. Constitutively active mitogen-activated protein kinase kinase 1 (MAPKK1) and MAPKK2 mediate similar transcriptional and morphological responses. Cell Growth Differ. 1996;7:243–50. [PubMed] [Google Scholar]
- 29.Fischmann TO, Smith CK, Mayhood TW, et al. Crystal structures of MEK1 binary and ternary complexes with nucleotides and inhibitors. Biochemistry. 2009;48:2661–74. doi: 10.1021/bi801898e. [DOI] [PubMed] [Google Scholar]
- 30.Catling AD, Schaeffer HJ, Reuter CW, et al. A proline-rich sequence unique to MEK1 and MEK2 is required for raf binding and regulates MEK function. Mol Cell Biol. 1995;15:5214–25. doi: 10.1128/mcb.15.10.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaeffer HJ, Catling AD, Eblen ST, et al. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–71. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 32.Jelinek T, Catling AD, Reuter CW, et al. RAS and RAF-1 form a signalling complex with MEK-1 but not MEK-2. Mol Cell Biol. 1994;14:8212–8. doi: 10.1128/mcb.14.12.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao YH, Wang Y, Burdette D, et al. Structural requirements for Yersinia YopJ inhibition of MAP kinase pathways. PLoS One. 2008;3:e1375. doi: 10.1371/journal.pone.0001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunet A, Pages G, Pouyssegur J. Growth factor-stimulated MAP kinase induces rapid retrophosphorylation and inhibition of MAP kinase kinase (MEK1) FEBS Lett. 1994;346:299–303. doi: 10.1016/0014-5793(94)00475-7. [DOI] [PubMed] [Google Scholar]
- 35.Gopalbhai K, Jansen G, Beauregard G, et al. Negative regulation of MAPKK by phosphorylation of a conserved serine residue equivalent to Ser212 of MEK1. J Biol Chem. 2003;278:8118–25. doi: 10.1074/jbc.M211870200. [DOI] [PubMed] [Google Scholar]
- 36.Slack-Davis JK, Eblen ST, Zecevic M, et al. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J Cell Biol. 2003;162:281–91. doi: 10.1083/jcb.200212141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park ER, Eblen ST, Catling AD. MEK1 activation by PAK: a novel mechanism. Cell Signal. 2007;19:1488–96. doi: 10.1016/j.cellsig.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossomando AJ, Dent P, Sturgill TW, et al. Mitogen-activated protein kinase kinase 1 (MKK1) is negatively regulated by threonine phosphorylation. Mol Cell Biol. 1994;14:1594–602. doi: 10.1128/mcb.14.3.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eblen ST, Slack-Davis JK, Tarcsafalvi A, et al. Mitogen-activated protein kinase feedback phosphorylation regulates MEK1 complex formation and activation during cellular adhesion. Mol Cell Biol. 2004;24:2308–17. doi: 10.1128/MCB.24.6.2308-2317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Catalanotti F, Reyes G, Jesenberger V, et al. A Mek1-Mek2 heterodimer determines the strength and duration of the Erk signal. Nat Struct Mol Biol. 2009;16:294–303. doi: 10.1038/nsmb.1564. [DOI] [PubMed] [Google Scholar]
- 41.Resing KA, Mansour SJ, Hermann AS, et al. Determination of v-Mos-catalyzed phosphorylation sites and autophosphorylation sites on MAP kinase kinase by ESI/MS. Biochemistry. 1995;34:2610–20. doi: 10.1021/bi00008a027. [DOI] [PubMed] [Google Scholar]
- 42.Beausoleil SA, Jedrychowski M, Schwartz D, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daub H, Olsen JV, Bairlein M, et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–48. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Oppermann FS, Gnad F, Olsen JV, et al. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8:1751–64. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansour SJ, Matten WT, Hermann AS, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–70. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 46.Cowley S, Paterson H, Kemp P, et al. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–52. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 47.Pages G, Lenormand P, L’Allemain G, et al. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci USA. 1993;90:8319–23. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okazaki K, Sagata N. MAP kinase activation is essential for oncogenic transformation of NIH3T3 cells by Mos. Oncogene. 1995;10:1149–57. [PubMed] [Google Scholar]
- 49.Giroux S, Tremblay M, Bernard D, et al. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr Biol. 1999;9:369–72. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 50.Belanger LF, Roy S, Tremblay M, et al. Mek2 is dispensable for mouse growth and development. Mol Cell Biol. 2003;23:4778–87. doi: 10.1128/MCB.23.14.4778-4787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scholl FA, Dumesic PA, Barragan DI, et al. Mek1/2 MAPK kinases are essential for mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell. 2007;12:615–29. doi: 10.1016/j.devcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Scholl FA, Dumesic PA, Khavari PA. Mek1 alters epidermal growth and differentiation. Cancer Res. 2004;64:6035–40. doi: 10.1158/0008-5472.CAN-04-0017. [DOI] [PubMed] [Google Scholar]
- 53.Hobbs RM, Silva-Vargas V, Groves R, et al. Expression of activated MEK1 in differentiating epidermal cells is sufficient to generate hyperproliferative and inflammatory skin lesions. J Invest Dermatol. 2004;123:503–15. doi: 10.1111/j.0022-202X.2004.23225.x. [DOI] [PubMed] [Google Scholar]
- 54.Arwert EN, Lal R, Quist S, et al. Tumor formation initiated by nondividing epidermal cells via an inflammatory infiltrate. Proc Natl Acad Sci USA. 2010;107:19903–8. doi: 10.1073/pnas.1007404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bueno OF, De Windt LJ, Tymitz KM, et al. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 2000;19:6341–50. doi: 10.1093/emboj/19.23.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murakami S, Balmes G, McKinney S, et al. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18:290–305. doi: 10.1101/gad.1179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds JF, Neri G, Herrmann JP, et al. New multiple congenital anomalies/mental retardation syndrome with cardio-facio-cutaneous involvement—the CFC syndrome. Am J Med Genet. 1986;25:413–27. doi: 10.1002/ajmg.1320250303. [DOI] [PubMed] [Google Scholar]
- 58.Niihori T, Aoki Y, Narumi Y, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–6. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 59.Nava C, Hanna N, Michot C, et al. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: genotype-phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–71. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Viciana P, Rauen KA. Biochemical characterization of novel germline BRAF and MEK mutations in cardio-facio-cutaneous syndrome. Methods Enzymol. 2008;438:277–89. doi: 10.1016/S0076-6879(07)38019-1. [DOI] [PubMed] [Google Scholar]
- 61.Allanson JE, Anneren G, Aoki Y, et al. Cardio-facio-cutaneous syndrome: does genotype predict phenotype? Am J Med Genet C Semin Med Genet. 2011;157:129–35. doi: 10.1002/ajmg.c.30295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulz AL, Albrecht B, Arici C, et al. Mutation and phenotypic spectrum in patients with cardio-facio-cutaneous and Costello syndrome. Clin Genet. 2008;73:62–70. doi: 10.1111/j.1399-0004.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- 63.Narumi Y, Aoki Y, Niihori T, et al. Molecular and clinical characterization of cardio-facio-cutaneous (CFC) syndrome: overlapping clinical manifestations with Costello syndrome. Am J Med Genet A. 2007;143A:799–807. doi: 10.1002/ajmg.a.31658. [DOI] [PubMed] [Google Scholar]
- 64.Dentici ML, Sarkozy A, Pantaleoni F, et al. Spectrum of MEK1 and MEK2 gene mutations in cardio-facio-cutaneous syndrome and genotype-phenotype correlations. Eur J Hum Genet. 2009;17:733–40. doi: 10.1038/ejhg.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoon G, Rosenberg J, Blaser S, et al. Neurological complications of cardio-facio-cutaneous syndrome. Dev Med Child Neurol. 2007;49:894–9. doi: 10.1111/j.1469-8749.2007.00894.x. [DOI] [PubMed] [Google Scholar]
- 66.Gripp KW, Lin AE, Nicholson L, et al. Further delineation of the phenotype resulting from BRAF or MEK1 germline mutations helps differentiate cardio-facio-cutaneous syndrome from Costello syndrome. Am J Med Genet A. 2007;143A:1472–80. doi: 10.1002/ajmg.a.31815. [DOI] [PubMed] [Google Scholar]
- 67.Senawong T, Phuchareon J, Ohara O, et al. Germline mutations of MEK in cardio-facio-cutaneous syndrome are sensitive to MEK and RAF inhibition: implications for therapeutic options. Hum Mol Genet. 2008;17:419–30. doi: 10.1093/hmg/ddm319. [DOI] [PubMed] [Google Scholar]
- 68.Araki T, Mohi MG, Ismat FA, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–57. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 69.Fragale A, Tartaglia M, Wu J, et al. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum Mutat. 2004;23:267–77. doi: 10.1002/humu.20005. [DOI] [PubMed] [Google Scholar]
- 70.Kontaridis MI, Swanson KD, David FS, et al. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–92. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 71.Martin GS. Cell signaling and cancer. Cancer Cell. 2003;4:167–74. doi: 10.1016/s1535-6108(03)00216-2. [DOI] [PubMed] [Google Scholar]
- 72.Hoshino R, Chatani Y, Yamori T, et al. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 1999;18:813–22. doi: 10.1038/sj.onc.1202367. [DOI] [PubMed] [Google Scholar]
- 73.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 74.Kiaris H, Spandidos D. Mutations of ras genes in human tumors (review. Int J Oncol. 1995;7:413–21. [PubMed] [Google Scholar]
- 75.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–85. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 76.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 77.Murugan AK, Dong J, Xie J, et al. MEK1 mutations, but not ERK2 mutations, occur in melanomas and colon carcinomas, but none in thyroid carcinomas. Cell Cycle. 2009;8:2122–4. doi: 10.4161/cc.8.13.8710. [DOI] [PubMed] [Google Scholar]
- 78.Nikolaev SI, Rimoldi D, Iseli C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2011;44:133–9. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 79.Marks JL, Gong Y, Chitale D, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–8. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitsudomi T, Viallet J, Mulshine JL, et al. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene. 1991;6:1353–62. [PubMed] [Google Scholar]
- 81.Bansal A, Ramirez RD, Minna JD. Mutation analysis of the coding sequences of MEK-1 and MEK-2 genes in human lung cancer cell lines. Oncogene. 1997;14:1231–4. doi: 10.1038/sj.onc.1200947. [DOI] [PubMed] [Google Scholar]
- 82.Sasaki H, Hikosaka Y, Kawano O, et al. MEK1 and AKT2 mutations in Japanese lung cancer. J Thorac Oncol. 2010;5:597–600. doi: 10.1097/JTO.0b013e3181d35236. [DOI] [PubMed] [Google Scholar]
- 83.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–7. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 84.Bentivegna S, Zheng J, Namsaraev E, et al. Rapid identification of somatic mutations in colorectal and breast cancer tissues using mismatch repair detection (MRD) Hum Mutat. 2008;29:441–50. doi: 10.1002/humu.20672. [DOI] [PubMed] [Google Scholar]
- 85.Singer G, Oldt R, III, Cohen Y, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–6. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 86.Estep AL, Palmer C, McCormick F, et al. Mutation analysis of BRAF, MEK1 and MEK2 in 15 ovarian cancer cell lines: implications for therapy. PLoS One. 2007;2:e1279. doi: 10.1371/journal.pone.0001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stephens P, Edkins S, Davies H, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–2. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 88.Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–91. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bignell G, Smith R, Hunter C, et al. Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer. 2006;45:42–6. doi: 10.1002/gcc.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Basto D, Trovisco V, Lopes JM, et al. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathol. 2005;109:207–10. doi: 10.1007/s00401-004-0936-x. [DOI] [PubMed] [Google Scholar]
- 91.Jeuken J, van den Broecke C, Gijsen S, et al. RAS/RAF pathway activation in gliomas: the result of copy number gains rather than activating mutations. Acta Neuropathol. 2007;114:121–33. doi: 10.1007/s00401-007-0239-0. [DOI] [PubMed] [Google Scholar]
- 92.Cohen Y, Xing M, Mambo E, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–7. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 93.Rusinek D, Szpak-Ulczok S, Jarzab B. Gene expression profile of human thyroid cancer in relation to its mutational status. J Mol Endocrinol. 2011;47:R91–103. doi: 10.1530/JME-11-0023. [DOI] [PubMed] [Google Scholar]
- 94.Villanueva J, Vultur A, Herlyn M. Resistance to BRAF inhibitors: unraveling mechanisms and future treatment options. Cancer Res. 2011;71:7137–40. doi: 10.1158/0008-5472.CAN-11-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alcala AM, Flaherty KT. BRAF inhibitors for the treatment of metastatic melanoma: clinical trials and mechanisms of resistance. Clin Cancer Res. 2012;18:33–9. doi: 10.1158/1078-0432.CCR-11-0997. [DOI] [PubMed] [Google Scholar]
- 96.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 97.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Montagut C, Sharma SV, Shioda T, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–61. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Emery CM, Vijayendran KG, Zipser MC, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Altshuler D, Durbin RM, Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scholl FA, Dumesic PA, Barragan DI, et al. Selective role for Mek1 but not Mek2 in the induction of epidermal neoplasia. Cancer Res. 2009;69:3772–8. doi: 10.1158/0008-5472.CAN-08-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Voisin L, Julien C, Duhamel S, et al. Activation of MEK1 or MEK2 isoform is sufficient to fully transform intestinal epithelial cells and induce the formation of metastatic tumors. BMC Cancer. 2008;8:337. doi: 10.1186/1471-2407-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee CS, Dykema KJ, Hawkins DM, et al. MEK2 is sufficient but not necessary for proliferation and anchorage-independent growth of SK-MEL-28 melanoma cells. PLoS One. 2011;6:e17165. doi: 10.1371/journal.pone.0017165. [DOI] [PMC free article] [PubMed] [Google Scholar]