Abstract
Background
Elucidating the selective and neutral forces underlying molecular evolution is fundamental to understanding the genetic basis of adaptation. Plants have evolved a suite of adaptive responses to cope with variable environmental conditions, but relatively little is known about which genes are involved in such responses. Here we studied molecular evolution on a genome-wide scale in two species of Cardamine with distinct habitat preferences: C. resedifolia, found at high altitudes, and C. impatiens, found at low altitudes. Our analyses focussed on genes that are involved in stress responses to two factors that differentiate the high- and low-altitude habitats, namely temperature and irradiation.
Results
High-throughput sequencing was used to obtain gene sequences from C. resedifolia and C. impatiens. Using the available A. thaliana gene sequences and annotation, we identified nearly 3,000 triplets of putative orthologues, including genes involved in cold response, photosynthesis or in general stress responses. By comparing estimated rates of molecular substitution, codon usage, and gene expression in these species with those of Arabidopsis, we were able to evaluate the role of positive and relaxed selection in driving the evolution of Cardamine genes. Our analyses revealed a statistically significant higher rate of molecular substitution in C. resedifolia than in C. impatiens, compatible with more efficient positive selection in the former. Conversely, the genome-wide level of selective pressure is compatible with more relaxed selection in C. impatiens. Moreover, levels of selective pressure were heterogeneous between functional classes and between species, with cold responsive genes evolving particularly fast in C. resedifolia, but not in C. impatiens.
Conclusions
Overall, our comparative genomic analyses revealed that differences in effective population size might contribute to the differences in the rate of protein evolution and in the levels of selective pressure between the C. impatiens and C. resedifolia lineages. The within-species analyses also revealed evolutionary patterns associated with habitat preference of two Cardamine species. We conclude that the selective pressures associated with the habitats typical of C. resedifolia may have caused the rapid evolution of genes involved in cold response.
Keywords: Molecular evolution, 454 next generation sequencing, adaptive traits, habitat preference, Cardamine
Background
Organisms adapt to different habitats through natural selection, which favors the fixation of alleles that increase the fitness of the individual that bears them. However, it is quite difficult to identify the locus/loci targeted by selection. One reason is that the number of loci involved in a particular adaptation and their phenotypic effects vary depending on the genetic architecture underlying the adaptive trait [1]. Another reason is that often the sequence and/or information about the gene(s) involved in the adaptive response is unavailable.
Two main approaches are used to identify genetic signatures of adaptive evolution and link them to phenotypic traits. The first is the candidate gene approach, whereby signatures of positive selection (e.g. the over-representation of specific polymorphisms) are identified using a population genetics framework in those genes previously known to be involved in the phenotypic trait of interest (e.g. [2-7]). The advantage of this approach is that the consequences of molecular variation on phenotypes can be inferred and their adaptive significance evaluated [8]. On the other hand, this approach assumes a comprehensive knowledge of the function of such genes and may neglect other genes relevant to the phenotypic trait. The second is the genome-wide approach, where the pattern of molecular evolution of hundreds to thousands of genes scattered across the genome are analyzed simultaneously (e.g. [9-14]). As high throughput technologies are becoming cheaper and more accessible, this approach is increasingly gaining attention from the evolutionary biology research community. An important advantage of this method is that it does not rely on prior information about gene sequence, and therefore it is well suited for studying non-model species. The possibility of performing genome scale studies in non-model organisms is indeed a powerful approach to address the genetic basis of specific adaptations, which can only be obtained by choosing the appropriate organism and ecological context. However, the typically poor knowledge about gene function in non-model organisms often prevents a comprehensive understanding of the adaptive significance of eventual signatures of positive selection. A useful compromise is to use the genome-wide approach in species closely related to well-studied model organisms, so that gene function can be inferred by comparative analyses. In this way it is possible to exploit what is known about the ecology and the life history of the species, and thus the approach is particularly suited to identifying genes involved in species-specific and habitat-specific adaptations (e.g. [11,14]).
Environmental variation is a key factor driving adaptive evolution and determining the ecological niche of a species. Plants and other sessile organisms are particularly affected by circadian and seasonal oscillations in abiotic factors. For example, sudden drops in temperature, high levels of solar irradiation and limited access to water are common sources of environmental stress, especially at high altitude in mountainous regions [15]. These stressors affect the evolution of species living in these habitats, and their capacity to adapt to these stressors ultimately determines their distribution [16-19]. To cope with drops in temperature, plants have developed a series of physiological adaptations that rely on the up- and down-regulation of cold responsive genes triggered by cold exposure (e.g. [20,21]; reviewed in [22]). Similarly, cold temperatures and high irradiation are not favorable to efficient photosynthesis, and consequently, a suite of photoprotective strategies are required for survival and reproduction at high altitudes (e.g. [23-27]). A fairly detailed understanding of the relevant regulatory pathways and gene function in the model species Arabidopsis thaliana now exists; however, little is known about their adaptive role, particularly in relation to the diverse habitats present along an altitudinal gradient.
In order to assess the adaptive role of cold responsive genes, as well of the genes involved in photosynthesis, we compared patterns of gene evolution in congeneric species living at different altitudes. We performed molecular evolution analyses in two closely related Brassicaceae species adapted to non-overlapping altitudinal ranges: Cardamine resedifolia, a perennial species usually growing under conditions of high irradiation and severe temperature oscillations between 1,500 and 3,500 meters above sea level; and C. impatiens, an annual nemoral species normally growing between 300 and 1,500 meters above sea level [28]. The distinct habitats associated with high and low altitudes make C. resedifolia and C. impatiens ideal species for studying the adaptive significance of the genes involved in altitude-related stress responses. It should also be noted that the two species may differ in their outcrossing rates, even though conclusive evidence is still lacking: C. impatiens is mainly selfing [29], although some populations have been reported as partially outcrossing [30]; C. resedifolia is also a predominantly selfing species, but the precise outcrossing rate is unknown [31].
The two Cardamine species described above are closely related to A. thaliana [32,33], making it possible to apply the extensive knowledge about the gene functions of this model organism to our study. Gene sequences of both C. resedifolia and C. impatiens were obtained by high-throughput sequencing technology and subsequently identified and partitioned into functional classes based on gene similarity to the A. thaliana orthologues. Then, we used complementary approaches, such as analyses of non-synonymous and synonymous substitutions and of their ratio, of levels of selective pressure, codon usage, and gene expression, to quantify the difference in adaptive evolution between functional classes and between species. This allowed us to infer the adaptive significance of genes involved in adaptation to cold stress and photosynthesis at high altitude and put them into an ecological context.
Results
We obtained sequences representing the C. resedifolia and C. impatiens transcriptomes using high-throughput sequencing. Genes and their putative function were identified by comparison of the sequences obtained here to the available annotated A. thaliana gene sequences. To minimize the chance of mistaking a paralogue for an orthologue, we considered as triplets of putative orthologues only those consisting of reciprocal best hits, i.e. those where the three sequences were consistently found as best hit matches of one another. Eventually, we obtained 2,922 triplets of (partial) nuclear genes, with a mean (± standard error) length for the A. thaliana orthologues of 594.0 ± 5.8 base pairs (bp), corresponding to a mean coverage of 46.3 ± 0.5% of the full-length gene sequence. In C. impatiens, the mean sequence length was 592.3 ± 5.8 bp, while in C. resedifolia the mean sequence length was 592.2 ± 5.8 bp.
We partitioned these genes according to their putative function, and then focused our analyses on those functional classes that are associated with the adaptive response to high altitude (Additional File 1). In particular, according to the annotation in A. thaliana, 56 genes were involved in cold acclimation (CGO), 67 were involved in UV-B and high irradiation response (PGO), 332 were involved in general stress responses (SGO). To these three classes we added a manually compiled class that included 55 genes functionally characterized as cold responsive genes (CRG).
Rate of molecular substitution in Cardamine
The analysis of the rates of nucleotide substitution revealed faster molecular evolution along the C. resedifolia lineage than along the C. impatiens lineage (Table 1). Because the rates of sequence evolution were highly correlated with the length of the A. thaliana orthologous gene (P < 0.0005 for all correlations; see Additional File 2; here and henceforth gene length is referred to the coding portion of the gene), we corrected these measures accordingly by using the residuals of such correlations (Additional File 3). While the rate of synonymous substitution, dS, was similar between lineages (Wilcoxon rank sum test, two-sided P = 0.1830), the rate of non-synonymous substitution, dN, was larger in the C. resedifolia lineage than in the C. impatiens lineage (P = 0.0001). Conversely, the ratio ω = dN/dS, was significantly larger in the C. impatiens than in the C. resedifolia lineage (P = 3 × 10-9).
Table 1.
Rate of molecular substitution in Cardamine.
| C. impatiens | C. resedifolia | ||||
|---|---|---|---|---|---|
| n a | mean (SE)b | n a | mean (SE)b | P c | |
| dN | 2913 | 0.0073 (0.0002) | 2913 | 0.0083 (0.0002) | 0.0001 |
| dS | 2913 | 0.0592 (0.0007) | 2913 | 0.0665 (0.0010) | 0.1830 |
| dN/dS | 2847 | 0.1763 (0.0058) | 2875 | 0.1632 (0.0044) | 3 × 10-9 |
a Number of genes.
b Mean and standard error.
c Wilcoxon test probability after correcting for gene length (partial correlation)
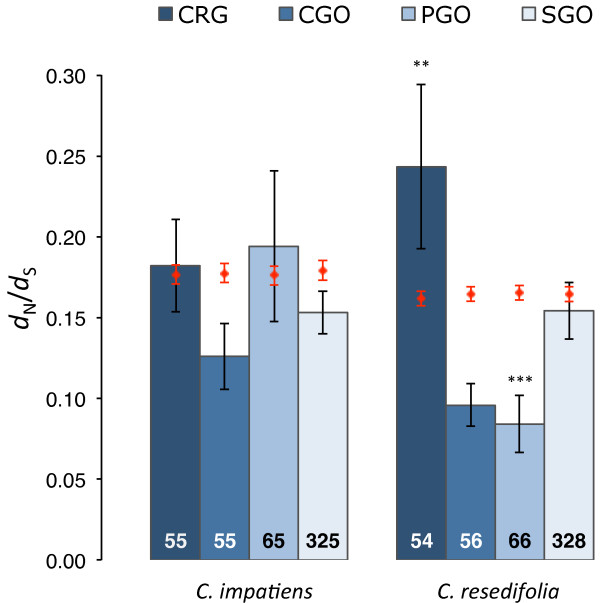
For each functional class, both the dN and the dS rates, as well their ratio ω, were not significantly different between the two lineages after correcting for multiple testing (corrected P > 0.025 for all comparisons-step-down Holm-Bonferroni method [34] applied to 8 tests for either dN, dS, and ω; Figure 1; Additional File 3). Selective constraints varied however across functional classes within the same lineage: dN was significantly lower in PGO than in other genes in the C. resedifolia lineage (corrected P = 3 × 10-5), and was significantly higher in CRG than in other genes in both Cardamine lineages (corrected P < 0.015 for both comparisons). The statistically significant differences were not due to random effects resulting from the small sample size of the functional classes relative to the full dataset. In fact, a bootstrap approach revealed that at most 0.2% of the randomized datasets produced Wilcoxon tests P values lower than those observed (10,000 replicates). Since there was a significant correlation between dN and the length of the A. thaliana orthologue (Spearman's ρ > -0.067, P < 0.0001, for both lineages), we also re-analyzed the comparisons using the residuals of the correlation. The within-lineage differences were still statistically significant, thus excluding the possibility that such differences were a result of length heterogeneity across our gene sets (Additional File 3). Similarly, the mean values of the residuals for dN and dS still did not differ between lineages, with the exception of the PGO genes, where the dN residuals were lower in C. resedifolia than in C. impatiens (corrected P = 0.0048).
Figure 1.
Levels of selective pressure in C. impatiens and C. resedifolia orthologous genes. Mean values of ω are reported for genes functionally characterized as cold responsive (CRG), and for genes annotated as involved in cold response (CGO), photosynthesis (PGO) and general stress responses (SGO). Text in bars denotes the number of genes; error bars denote the standard error of the mean. For each functional class, the mean residual values of the correlation between levels of selective pressure and A. thaliana gene length were compared to the mean estimated for the genes not in such functional class (identified by red dots) using a Wilcoxon test: * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
The analysis of the levels of selective pressure, ω, provided compelling evidence that different selective forces are dominating the evolution of the four functional categories in the two Cardamine lineages (Figure 1; Additional File 3). Again, as a result of the correlation between substitution rates and gene length, we report the results of the residuals of such correlations. In the C. resedifolia lineage, the mean value of ω was significantly lower in PGO than in other genes (P < 0.0005), consistent with intense purifying selection. This is in sharp contrast with what observed in CRG, where the mean value of ω was significantly higher than in other genes (P = 0.0057), indicating either relaxed or positive selection. Instead, in the C. impatiens lineage, the levels of selective pressure in genes of the four functional classes were similar to those at the genome-wide level (P > 0.05, for all comparisons).
In C. resedifolia, selective pressures were different between genes involved in cold response classified as CGO and those classified as CRG. The two datasets have 12 genes in common (Additional File 1), whose mean ω did not differ from that of the remaining genes (P = 0.3649). However, the genes unique to CGO and CRG had a mean ω in line with what was observed in the complete datasets. That is, genes unique to the CGO category had a mean ω significantly lower than the remaining genes (P = 0.0056), confirming strong purifying selection in CGO. Conversely, genes exclusive to the CRG category had a mean ω significantly higher than the other genes (P = 0.0070), indicating either relaxed or positive selection in these genes.
The rate of evolution and the occurrence of positive selection were further investigated by analyzing the pattern of molecular substitution of each single gene. The most sensitive test for this purpose is one that employs branch-site models, which aim to detect positive selection affecting a few sites along particular lineages. When C. impatiens was used as the foreground lineage (test BSCi) the test identified one outlier with FDR < 0.20. This number increased substantially when the C. resedifolia lineage was used instead (test BSCi, FDR < 0.20), with seven genes showing evidence for positive selection along this lineage (Table 2). Two genes among these eight outliers were identified as being involved in general stress responses (not more than expected by chance; Fisher exact test, two-tailed P = 0.228).
Table 2.
Fast evolving genes identified by the branch, site, and branch-site codon substitution models at a FDR threshold of 0.20.
| Gene | Function | LRTa |
|---|---|---|
| AT1G07890b | Ascorbate peroxidase; response to salt and heat stress | BSCr |
| AT1G14610 | Aminoacyl-tRNA ligase | BSCr |
| AT1G21680 | Unknown protein | BSCr |
| AT1G49750 | Leucine-rich repeat family protein | S21, S87 |
| AT1G54040b | Enzyme regulator; response to jasmonic acid stimulus, leaf senescence | BSCr |
| AT1G71040b | Copper ion binding, oxidoreductase; response to phosphate starvation | B, BCi |
| AT2G25840 | Aminoacyl-tRNA ligase | BCr |
| AT2G31610b | 40S ribosomal protein; response to salt stress | S21, S87 |
| AT3G06130 | Metal ion binding | BSCr |
| AT3G52910 | Growth regulating factor, transcription activator | BSCr |
| AT4G17520 | Putative nuclear RNA-binding protein | S21, S87, BSCi |
| AT5G06980 | Unknown protein | S21, S87 |
| AT5G20900 | Jasmonate-ZIM-domain protein | BSCr |
| AT5G26830 | Threonyl-tRNA synthetase and ligase | B, BCi |
| AT5G62680 | Transport family protein | BCi |
a Likelihood Ratio Tests (LRT) in which the alternative model explained the pattern of codon substitution better than the null model, and with a false discovery rate lower than 0.20. Patterns of codon substitution were tested using branch models (B tests), site models (S tests), and branch-site models (BS tests). B tests compared models M0 and M0'. S tests compared model M2a to M1a (S21) and model M8 to M7 (S87). BS tests were based on the branch-site model A, test 2. Subscripts in the B and BS tests indicate whether the C. impatiens (Ci) or the C. resedifolia (Cr) lineages were used as foreground branches (see main text for details).
b gene of the SGO functional class.
To increase the chance of discovering interesting candidate genes, we also employed additional maximum likelihood ratio tests based on branch or site codon substitution models [35]. These tests identified several genes whose pattern was better explained by the occurrence of non-neutral evolution (Additional Files 4 and 5). Branch models identified three genes (with FDR < 0.20) with a pattern of nucleotide substitution that fit a model allowing two ω rates, one for the focal C. impatiens branch and a second for the rest of the phylogenetic tree (BCi test). A single gene was detected if the C. resedifolia branch was used as the focal branch instead (BCr test; Table 2). Using site models, the fit of the data under a neutral model was compared to that under a model of positive selection. In the first comparison, the neutral model admitted one class of sites with 0 < ω < 1, while the selection model allowed the presence of a second class of sites with ω > 1 (S21 test). The second, more powerful (less robust), approach compared more realistic models where (nearly) neutrally evolving sites were partitioned in ten classes with ω values drawn from a beta distribution (S78 test). The two likelihood ratio tests consistently identified four genes (with FDR < 0.20) showing evidence for the action of positive selection (Table 2).
As expected, the tests based on either the branch, site or branch-site models resulted in the detection of sets of outliers with little overlap. Among the three genes detected by more than one test, two (AT1G71040 and AT5G26830) were detected by both the B and the BCi tests, and one (AT4G17520) was detected by the S21, S87 and BSCi tests.
To exclude the possibility that substitution rate patterns were the result of orthology mis-assignment, effectively inflating interspecific divergence, we compared homologous sequences from A. thaliana, Cardamine and other plant species within a phylogenetic framework. Specifically, we searched for putative paralogues of the gene triplets, for each of the 15 candidate genes (Table 2). In six such cases we verified the presence of putative paralogues using BLAST searches (Additional File 6). The A. thaliana and the two putative Cardamine orthologues identified by the reciprocal best-hit approach always clustered together in the tree, supporting their orthology and the results of the likelihood ratio tests.
Expression breadth, expression level and rate of molecular substitution
The breadth of expression of a gene, namely whether it has tissue-specific expression or is expressed broadly over more tissues, has been shown to be correlated with several patterns of molecular evolution (e.g. [36-40]).
To evaluate such effects in Cardamine, we correlated the levels of selective pressure, ω, to the breadth of expression measured across tissues, across developmental time points, and along stress treatments. For these analyses we used the expression patterns of A. thaliana orthologues as proxies for expression in Cardamine (see Materials and Methods), assuming that orthologous genes will maintain similar expression patterns [41-45]. These analyses found strong evidence that, in both Cardamine lineages, genes evolved at different rates and under different selective regimes depending on their breadth of expression (Table 3; Additional File 7). The ratio ω decreased significantly with the increase in the temporal breadth of expression, i.e. the number of flower and leaf developmental stages at which a gene was expressed (P < 10-8, for both comparisons). The same statistically significant trend held when correlating ω with the spatial breadth of expression (P < 10-8), i.e. the number of tissues/organs where the gene is expressed and the associated organ specificity index τ [46]. Because gene length was significantly correlated with both ω (ρ > 0.112, P < 10-7, for both Cardamine lineages) and expression breadth (ρ < -0.069, P < 0.0005, for all measures), we repeated our analyses using the residuals of such correlations (Additional File 7). The partial correlations coefficients were still statistically highly significant (P < 10-7, for all comparisons) and indicated that the correlations between ω and breadth of expression were independent from gene length. Thus, a broad breadth of expression imposes selective constraints limiting the possible action of positive selection. Conversely, genes expressed at one developmental time point or in specific organs evolve faster, as a result of either relaxed or positive selection.
Table 3.
Correlation between breadth of expression and level of selective pressure, dN/dS.
| Breadth typea | C. impatiens | C. resedifolia | ||
|---|---|---|---|---|
| Flower development | -0.156 | ****^ | -0.156 | ****^ |
| Leaf development | -0.115 | ****^ | -0.145 | ****^ |
| Organs | -0.131 | ****^ | -0.135 | ****^ |
| Organ specificity τ | 0.125 | ****^ | 0.113 | ****^ |
| UV-B stress | 0.061 | **§ | 0.036 | NS |
| Salt stress | 0.057 | **§ | 0.070 | ***§ |
| Osmotic stress | 0.039 | * | 0.057 | **§ |
| Drought stress | 0.050 | * | 0.032 | NS |
| Cold stress | 0.043 | * | 0.052 | **§ |
* Spearman's P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, NS = non significant.
^ partial correlations are significant after correcting for the length of the A. thaliana orthologue.
§ significant after Holm-Bonferroni multiple test correction (only for stress responses, n = 10)
a Breadth of expression can be either spatial (i.e., number of tissues in which a gene is expressed) or temporal (when a gene is expressed, during either development or stress exposure). Note that the organ-specificity index τ is inversely correlated with the number (breadth) of organs at which a gene is expressed.
When ω was compared to the duration (i.e., the persistence) of the stress response, we obtained mixed results that depended on the stress type (Table 3). In general, the ratio ω was positively correlated to the persistence of gene expression during stress responses (Additional File 7). After a Holm-Bonferroni correction for multiple testing, ω was significantly correlated to the duration of the responses to salt and UV-B in the C. impatiens lineage (ρ > 0.057, corrected P < 0.025, for both correlations); and to the duration of the responses to salt, osmotic and cold stress in the C. resedifolia lineage (ρ > 0.051, corrected P < 0.05, for all three correlations).
In addition to the breadth of expression, the level of gene expression (in A. thaliana) was also an important determinant of ω (Table 4; Additional File 8). For instance, there was a statistically highly significant negative correlation between the mean gene expression and ω in both Cardamine lineages (ρ < 0.248, P < 10-30). This correlation also remained highly significant when the mean expression level was controlled for the effects of gene length (P < 10-30; correlation between gene length and mean expression level, ρ = -0.344, P < 10-15). Thus, we conclude that selective constraints may limit the evolution of proteins encoded by highly expressed genes in Cardamine.
Table 4.
Correlation between level of selective pressure dN/dS and level of gene expression
| C. impatiens | C. resedifolia | ||||||
|---|---|---|---|---|---|---|---|
| Gene FCa | n b | mean (SE)c | ρd | P d | ρd | P d | |
| Mean Expression | |||||||
| All genes | 2922 | 8.72 (0.03) | -- | -0.249 | **** | -0.297 | **** |
| Cold response (CRG) | 55 | 8.49 (0.21) | NS | -0.363 | ** | -0.310 | * |
| Cold (CGO) | 56 | 10.16 (0.22) | **** | -0.199 | NS | -0.251 | NS |
| Photosynthesis (PGO) | 67 | 10.29 (0.21) | **** | -0.203 | NS | -0.365 | ** |
| Stress (SGO) | 332 | 9.69 (0.09) | **** | -0.221 | **** | -0.209 | *** |
| Maximum Expression | |||||||
| All genes | 2922 | 11.02 (0.03) | -- | -0.185 | **** | -0.246 | **** |
| Cold response (CRG) | 55 | 12.51 (0.20) | **** | -0.126 | NS | -0.190 | NS |
| Cold (CGO) | 56 | 13.06 (0.24) | **** | -0.184 | NS | -0.097 | NS |
| Photosynthesis (PGO) | 67 | 12.97 (0.21) | **** | -0.198 | NS | -0.367 | ** |
| Stress (SGO) | 332 | 12.26 (0.10) | **** | -0.083 | NS | -0.100 | NS |
* Spearman correlation P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, NS = non significant.
a Gene functional class.
b Number of genes.
c Mean/maximum expression was estimated for all genes and genes included in each functional class and tested (Wilcoxon test) against mean for genes not in that functional class.
d Spearman's correlation between level of gene expression and ω estimated along the two Cardamine lineages.
The relationship between gene expression and rate of evolution indicates that the different selective pressures we observed in genes involved in stress response may be correlated to their expression level. For instance, genes of the CGO and PGO functional classes had a calculated expression level higher than other genes (P < 10-8, for all comparisons), and also had lower ω along the C. resedifolia lineage than the genome-wide mean (see Figure 1). To investigate the association between gene expression and ω, we then calculated the residuals of their correlation and compared them across functional classes (Additional File 9). The corrected ω were no longer statistically significant for PGO and CGO genes, suggesting that the levels of selective pressure in these genes were highly associated with their high expression levels. Interestingly, removing the effect of expression level produced a statistically significant difference in ω between SGO genes and the other genes (P < 0.005, for both Cardamine lineages). Such differences suggests heterogeneity in the association between rates of molecular evolution and expression levels in SGO genes, likely due to the diverse nature of the stress responses to which these genes are responding. Results did not change if we analyzed the correlation between maximum, rather than mean, expression level and ω (Additional Files 8 and 9), further indicating a strong association between the expression pattern of a gene and its rate of molecular evolution.
Codon usage in Cardamine genes
Synonymous codon usage can be under weak selection and lead to non-random use of the codons coding for the same aminoacid. Due to Hill-Robertson interference, such bias is expected to correlate with the occurrence of positive selection, so that it will be lower in genes that experience (recurrent) episodes of positive selection [47-49]. Consistent with this prediction, in both Cardamine lineages there was a statistically significant negative correlation between ω and codon bias, measured as Fop (frequency of the optimal codon (both ρ < -0.135, P < 10-10). This correlation also held when controlling for the effects of other determinants of codon usage, such as expression levels, gene length and GC content (e.g. as reported in [50]). For instance, in both Cardamine lineages, Fop was significantly correlated with expression level (ρ > 0.325 and P < 10-50), gene length (ρ < -0.240, P < 10-39), and GC content at the third codon position (ρ > 0.490, P < 10-100; note that most preferred codons end in either G or C, see Additional File 10). However, partial correlation coefficients (which control for dependence of these variables) between codon usage and ω remained highly significant (-0.042 < ρ < -0.121, 0.02 <P < 10-10).
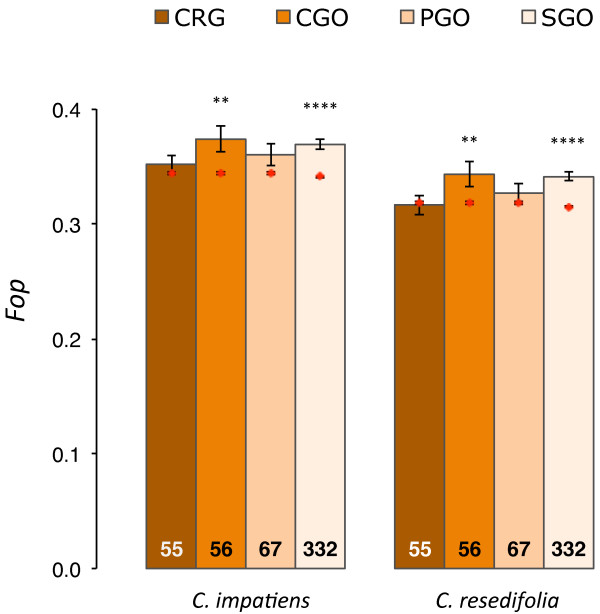
We then analyzed codon usage for the genes in each of the four functional classes. Genes involved in stress responses generally had larger codon bias compared to other genes (Figure 2; Additional File 11). The difference was statistically significant for CGO and SGO genes in both Cardamine lineages (P < 0.01, for both comparisons), while it was not significantly different for CRG and PGO genes (P > 0.05, for all comparisons). However, such differences were no longer significant when controlling for gene expression level (Additional File 11), suggesting that the expression pattern influenced codon usage more than the distinct selective pressures to which they are subject. Thus, codon usage bias does not support the frequent action of positive selection in genes of the functional classes under consideration.
Figure 2.
Codon usage bias in C. impatiens and C. resedifolia orthologous genes. Mean values of the frequency of the optimal codon (Fop) are reported for genes functionally characterized as cold responsive genes (CRG), and for genes annotated as involved either in cold response (CGO), photosynthesis (PGO) and general stress responses (SGO). Text in bars denotes the number of genes; error bars denote the standard error of the mean. The mean values of each functional class were compared to the mean estimated for the genes not in that functional class (identified by red dots) using a Wilcoxon test: * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Discussion
Our analyses of protein-coding sequences highlighted lineage- and gene-specific evolutionary patterns in two species of Cardamine characterized by distinct habitat preferences, life-histories and, possibly, breeding systems. C. resedifolia and C. impatiens are not sister species [51]; hence, the substitution rates estimated in the present study apply to the lineages leading to the two species after the split from their common ancestor, rather than to these specific taxa as such. However, C. resedifolia, C. impatiens, and the clades they belong to, are characterized by divergent phenotypic and life-history traits that have profoundly shaped their evolutionary histories; for instance, the clade comprising C. resedifolia (group A in Figure 1 of Carlsen et al. [51]) is basal to the Cardamine radiation and includes only perennial species living in high altitude alpine habitats with well-developed petals indicating the existence of a mixed mating system. In contrast, C. impatiens is, to our knowledge, the only annual selfing species of its group (group C in Figure 1 of Carlsen et al. [51]), which includes species that are mainly found at moderate elevations. C. impatiens is further characterized by having very reduced or no petals, in agreement with a predominantly selfing mating system [29,30]. Thus, the observed patterns of molecular evolution reflect the differences in life history traits and habitats between the two lineages.
The effects of such differences on the rate of molecular evolution are quite complex. For instance, the rate of non-synonymous substitution was significantly higher for the C. resedifolia lineage than for that of C. impatiens. Previous studies have reported a higher substitution rate in annual plants compared to perennial plants [52-54] (but see [55]). However, our observations are in contrast with this prediction, since the annual C. impatiens had lower substitution rate than the perennial C. resedifolia. Moreover, since the rate of synonymous substitution was similar between lineages, there is no support for the hypothesis that mutation rate is higher along the C. resedifolia lineage than along that of C. impatiens.
The interspecific differences in non-synonymous substitution rates could also be the result of differences in effective population size [56,57]. For instance, low effective population size can affect the substitution rate by allowing slightly deleterious mutations to escape purifying selection and reach fixation (reviewed in [56]). Alternatively, high effective population size can increase the efficiency of positive selection by facilitating the fixation of favorable alleles in genes undergoing adaptive evolution [58]. Thus, a high dispersion in the distribution of dN across the genome is expected to be associated with a large effective population size, especially when there is recombination (e.g. [49,59]). The analysis of non-synonymous substitution fixation rate revealed that the variance in dN was significantly larger in C. resedifolia than in C. impatiens (6.6 × 10-15 vs. 9.2 × 10-15; F test, P < 10-15, also when correcting for gene length). This difference in protein evolution heterogeneity is thus consistent with more efficient positive selection and higher effective population size in C. resedifolia. Both of these factors, in turn, would explain why dN is higher in this lineage than in C. impatiens. Preliminary analyses of recombination levels and polymorphism suggest indeed that selfing rates are lower in C. resedifolia, and the effective population size is slightly larger than in C. impatiens (Ometto and Varotto, unpublished results).
Intriguingly, mean ω was larger in C. impatiens than in C. resedifolia. The fact that in C. impatiens ω was considerably lower than one is compatible with either moderately positive or relaxed selection. Three lines of evidence suggest a reduced efficiency of purifying selection in C. impatiens, rather than pervasive positive selection at a genome-wide level. Firstly, the analyses on dN suggest that positive selection was more efficient along the C. resedifolia lineage than the C. impatiens lineage. Secondly, only one gene (using a FDR threshold of 0.20; Table 2; Additional File 5) exhibited strong evidence for site-specific positive selection along the C. impatiens branch. By comparison, seven genes exhibited strong evidence for site-specific positive selection along the C. resedifolia branch. Finally, the analysis of codon usage failed to detect differences in the efficiency of positive selection between the two lineages. However, the contrast between rates of non-synonymous substitution and of ω warrants some caution in drawing firm conclusions. The most likely explanation for the opposite patterns observed in dN and dN/dS lies on the high heterogeneity in substitution rate among genes. For instance the correlations and the coefficients of determination in the regressions between dS and ω (ρ = -0.314, P < 10-15, R2 = 0.052 in C. resedifolia; ρ = -0.292, P < 10-15, R2 = 0.037 in C. impatiens) and between dN and ω (ρ = 0.880, P < 10-15, R2 = 0.326 in C. resedifolia; ρ = 0.879, P < 10-15, R2 = 0.305 in C. impatiens), suggest that substitution rates are relatively weak predictors of the actual selective pressure experienced by a gene. The statistically significant correlation between synonymous substitution rate and ω is rather unusual: previous studies established that the correlation is caused by a combination of (i) adjacent substitutions [60] and (ii) of a bias on substitution rate estimation due to either low divergence and/or statistical methods [60,61]. Appropriate analyses will be necessary to unravel the effects of such factors in our system. The complex dynamics of substitution rates are also evident in the statistically significant correlation to gene length. The influence of gene length on sequence evolution had been previously reported for Populus tremula [38], stressing the importance of accounting for all diverse determinants of levels of gene and protein evolution. Additional studies, including analyses on intraspecific polymorphism, will be certainly necessary to disentangle the neutral and selective forces that have shaped such patterns [62,63].
The molecular evolution analyses in stress-related genes also revealed important lineage-specific patterns that may be associated with the distinct life-history traits and habitats of C. resedifolia and C. impatiens. For instance, similar selective regimes affected the evolution of genes involved in stress response in the C. impatiens lineage. Conversely, there was a correlation between the type of stress response and the rate of molecular evolution along the C. resedifolia lineage; for example, genes involved in photosynthesis evolved slower than other genes, consistent with selective constraints that limited the accumulation of non-synonymous substitution. This makes sense given their involvement in a process that is particularly relevant in the high altitude habitat of C. resedifolia. Strangely, in C. resedifolia selection acted in opposite directions in the two functional classes associated with cold responses. That is, compared to the genome-wide levels of selective pressure, genes that were identified as involved in cold response based on functional studies (i.e., CRG genes) displayed significantly lower levels of selective pressure, while cold responsive (CGO) genes were under more selective constraints. Since there was little overlap between the two classes (Additional File 1), the discrepancy in the levels of selective pressure between CGO and CRG genes may have captured different selective pressures acting along the cold response pathway. However, the difference may also stem from the approaches used to define the two cold-response functional classes. Specifically, the gene ontology annotations of the CGO genes were taken from the TAIR database [64], which is a less accurate source of functional information than the direct assays used to define CRG genes. For instance, the TAIR annotations may be based solely on sequence or structural similarity, and a gene may have been annotated as cold responsive because it is indirectly up- or down-regulated in plants exposed to low temperatures (e.g. more than 3,000 genes were reported as cold responsive in A. thaliana [20]). However, without a comprehensive knowledge on the response pathway it is difficult to evaluate their role in cold response in Cardamine, especially considering that their expression patterns may differ from those of A. thaliana.
The levels of selective pressure were also associated with the duration and pattern of gene expression. For instance, in C. impatiens and C. resedifolia, ω was lower for genes that were expressed only briefly during specific stress responses than for genes that were up-regulated over a longer period of time (as inferred by A. thaliana expression patterns). Most importantly, the correlation existed only for specific stress responses in each species, suggesting habitat-specific selective pressures. In particular, in C. resedifolia, ω was significantly affected by the duration of the responses to osmotic, salt and cold stress. The responses to these three stresses involve partially overlapping pathways [65-67], as cold stress causes membrane leakage and, as a consequence, the activation of the physiological responses also observed in high salt and osmotic stresses [68]. These results support the adaptive relevance of these genes in response to the severe temperature changes that this species experiences in alpine habitats. On the other hand, in C. impatiens ω was significantly correlated with the extent of up-regulation in those genes involved in UV-B stress response. C. impatiens is a nemoral species that is likely to be particularly sensitive to the effect of exposure to UV-B light. Therefore, it is reasonable to expect that this species has evolved response mechanisms based on gene up-regulation to cope with discontinuous UV stress. It will be extremely interesting to experimentally measure the expression of the Cardamine genes in the functional classes considered, as species-specific adaptive variations in expression levels cannot be excluded.
While unequal selective pressures across the stress-related functional classes were well-documented here, we detected positive selection in only a few single genes. In plants, previous studies have found evidence of genome-wide positive selection in some species (e.g. [69-71]), but in most cases there was little indication of widespread adaptive evolution (e.g. [13,72-75]). Other studies have identified positive selection of some genes involved in stress response (e.g. cold-hardiness in conifers [5]; drought stress in wild tomatoes [7]). The most likely explanation for the low rate of adaptive evolution is that most plants have low effective population size [13,76], which ultimately diminishes the efficiency of positive selection. Estimation of the effective population size of C. resedifolia and C. impatiens will be useful to verify whether this parameter can explain the scarce evidence for positive selection in our dataset.
Four methodological issues may have further resulted in an underestimation of the genes targets of positive selection. The first is that the power of codon substitution models is fairly conservative compared to that of models incorporating polymorphism data (i.e., McDonald-Kreitman test [77]). The second is that our reciprocal best-hit approach was prone to miss genes with high sequence divergence caused by adaptive evolution, as it was intended to avoid an overestimation of divergence by comparing paralogues. In particular, this approach may have overlooked duplicated genes, which are common in A. thaliana [78,79] and in other plants (e.g. cottonwood [80], grapevine [81]), and which can undergo sub- or neo-functionalization driven by positive selection [82]. For instance, our dataset included only ~11% of all A. thaliana genes, and we analyzed between 17% and 48% of the genes annotated as involved in the stress responses considered in the present study. An examination of our dataset revealed that many of the well-characterized transcription factors involved in cold response (e.g. CBF genes, ICE1) were absent. This indicates either that we are missing many of the genes upstream of the stress response pathways, or that in Cardamine these transcription factors may be differently regulated than in Arabidopsis thaliana or do not participate to the cold response. However, it is unlikely that this pathway is missing, since cold response is well-conserved across distantly related taxa (e.g. Arabidopsis [83]; Citrus [84]; Solanum [85]; Poaceae [86]). A third issue is related to the use of partial genes, which may have reduced the power of the likelihood ratio tests as a result of an insufficient number of informative sites. Finally, genes expressed at a low level, including the aforementioned transcription factors, may be missing from our dataset because of insufficient coverage or normalization. Because genes expressed at a low level are those evolving faster, this bias in gene representation could contribute to the relatively low number of rapidly evolving genes identified in this study. Deeper sequencing efforts could undoubtedly improve the situation by increasing both coverage and average transcriptome length.
One C. impatiens gene and seven C. resedifolia genes showed signatures of positive selection under the branch-sites model of codon substitution. The C. impatiens gene orthologue of AT4G17520 has not been functionally characterized in A. thaliana, although it is known that the protein encoded by this genes displays homology to the hyaluronan/mRNA binding protein family, a group of proteins binding both specific RNA and a high-molecular-mass polysaccharide extremely abundant in the connective tissue and extracellular matrix of animals [87]. Specific studies will be necessary to understand its role in adaptive processes in the C. impatiens lineage. As for the C. resedifolia candidate genes, no functional information is available for the orthologue of AT1G21680, which codes for a protein of unknown function with homology to TolB, a protein involved in outer membrane stability and uptake of biomolecules in E. coli [88]. Instead, AT3G06130 is known to code for a protein putatively involved in metal ion binding, and is similar to proteins of the heavy metal transport/detoxification superfamily. Heavy metal hyperaccumulation has been documented in several Brassicaceae species [89] and C. resedifolia has been reported to accumulate large quantities of nickel from the nickel-rich debris of the glacial till, where this species usually grows [90]. Therefore, it is possible that the signature of positive selection identified in this orthologue is related to high contents of heavy metals in soil experienced by C. resedifolia. A third gene, orthologue of AT1G14610, is a valine-tRNA synthetase. Given the multiple metabolic pathways in which valine is involved (e.g. glucosinolate [91] and pantothenate biosynthesis [92], conjugation to plant hormones [93,94]), it would be highly speculative to associate this gene to adaptive processes in the C. resedifolia lineage.
Based on functional evidence from Arabidopsis and other plant species, two other genes identified as putative targets of positive selection in C. resedifolia may play major roles in the response against bacterial pathogens and insect herbivory. The first gene, AT5G20900, is part of the Jasmonate-ZIM-domain protein family [95,96], whose members are involved in response to wounding and herbivory [97]. The other candidate is the orthologue of AT1G54040, which codes for the Arabidopsis epithiospecifier protein (ESP). ESP catalyzes the formation of simpler nitriles and epithionitriles from glucosinolates, thus modulating the release of isothiocyanate, a metabolite involved in herbivory and pathogen defense in Brassicaceae [98]. In two closely related Boechera species (Brassicaceae), the level of glucosinolates is negatively correlated with elevation preferences and growth rates, but positively correlated with drought tolerance [99]. However, the response to herbivory as a function of elevation is not uniform across plants, with some species experiencing more (e.g. [100]) and other less (e.g. [101-103]) damage with increase in elevation. Interestingly, a positive correlation with levels of insect herbivory was found for C. cordifolia exposed to full sunlight, possibly resulting from moderate water stress associated with a different insect guild compared to shadowy environments [104]. These observations provide the framework to experimentally test whether the signature of positive selection identified in AT5G20900 and AT1G54040 orthologues might be related to the higher exposition to sun, lower water availability or slower growth rate characterizing C. resedifolia as compared to C. impatiens. Interestingly, the orthologues of AT1G07890 and AT3G52910, may also be related to the light regimes characterizing C. resedifolia and C. impatiens habitats. AT1G07890 codes for a cytosolic ascorbate peroxidase (APX1) that scavenges hydrogen peroxide in plant cells [105], thus reducing the accumulation of reactive oxygen species (ROS) that cause cellular damage through protein oxidation [106]. The product of AT3G52910 is a transcriptional activator of the growth regulating factor family. Genes from this family have been demonstrated to be involved in the regulation of cell expansion and division in leaves, cotyledons and petals [107,108]. Strikingly, the product of AT3G52910 is one of the proteins oxidized in apx1 mutant plants in response to moderate light stress [106], indicating that it could also be part of the signaling cascade activated by ROS stress.
The site codon substitution models also identified putative targets of positive selection that deserve further characterization. In particular, the orthologue of AT2G31610 codes for a ribosomal protein (RPS3A) that is involved in response to salt and genotoxic stress in A. thaliana [109]. This gene is part of the ribosomal protein S3 family, which includes three paralogues (AT2G31610, AT3G53870 and AT5G35530) with similar sequences and function. Gene families can undergo relaxed selection immediately following duplication [110]; however, this does not seem to be the case for AT2G31610, as the phylogenetic tree clearly indicates that the duplication occurred before the split between the Arabidopsis and Cardamine lineages (Additional File 6).
Additional studies will be necessary to corroborate these findings and link the evolutionary pattern of each gene to its phenotypic effects. In particular, the use of intraspecific variation and functional analyses in the model species A. thaliana will be crucial to ascertaining whether positive selection or relaxed selection accelerated the evolution of these genes and their relevance in adaptive processes in Cardamine.
Conclusions
Overall, our results highlight the importance of employing complementary approaches to studying the genetic bases of adaptation in non-model species. Our use of comparative genomics on congeneric species identified evolutionary patterns that aid the understanding of the extrinsic and intrinsic factors driving plant adaptation. In the case of Cardamine, intrinsic factors (the breeding system, demography) most likely contribute to the different levels of selective pressure in C. resedifolia compared to C. impatiens lineages. In addition, extrinsic factors (stress responses associated with habitats preferences) seem to be the primary drivers of heterogeneity in the levels of selective pressure observed among genes in C. resedifolia.
Methods
Plant material
Cardamine resedifolia and C. impatiens seeds were collected in Trentino-Alto Adige (south-eastern Alps, Italy) from the wild populations found at the localities 'Solda' (46°30'52"N, 10°33'36"E; 2660 m above mean sea level) and 'Spormaggiore' (46°13'56"N, 11°03'30"E; 420 m AMSL), respectively. After three days of cold stratification in the dark, seedlings were potted in commercial soil GS90L and grown for 15 days in a controlled environment chamber under long day photoperiod (16-h light, 25°C; 8-h dark, 23°C), with illumination of 120 μmol m-2 sec-1 from cool white lights.
Sampling and RNA extraction
We enriched our mRNA library with transcripts from cold responsive genes by exposing plants to cold stress before sampling. In particular, based on the activation and kinetics of the cold responsive pathway of the close relative Arabidopsis thaliana (e.g. [111]), plants at the six-leaves stage were transferred to a growth chamber at 4°C with 35 μmol m-2 sec-1 continuous light for cold acclimation. Whole aerial parts from nine plants were harvested, pooled and plunged into liquid nitrogen immediately before, and at 15', 30', 1 h, 2 h, 3 h, 4 h, 6 h, 8 h, 12 h, 24 h, and 7 days after cold treatment (12 samples per species).
Total RNA was extracted separately from each sample using the Spectrum™ Plant Total RNA Kit (SIGMA, MO, USA), quantified in a spectrophotometer and quality controlled using electrophoretic separation in agarose gel. Approximately 1.7 μg of total RNA from each sample was pooled per species, and mRNA isolation was carried out with 20 μg of pooled RNA with Ambion Poly(A)Purist™ Kit (Life Technologies, CA, USA). mRNA quality/quantity was assessed with the RNA 6000 bioanalyzer chip (Agilent, CA, USA).
cDNA synthesis, normalization and high throughput sequencing
Double-stranded cDNA was synthesized using the SMART cDNA library construction kit (Clontech, USA). A modified oligo-dT primer (AAGCAGTGGTATCAACGCAGAGTGGCCGAGGCGGCC(T)20VN*) was used for first-strand synthesis. After second strand synthesis, double-strand cDNA was purified with QIAquick PCR purification kit (Qiagen, Germany).
To enrich for rare transcripts, 2.0 μg of cDNA from each species were normalized using the Trimmer-Direct Kit (Evrogen, Russia) according to the manufacturer's instructions. 500 ng of normalized cDNA library for each species were used for 454 library preparation and simultaneously sequenced in one run on a GS FLX titanium instrument (Roche-454, USA) according to manufacturer's instructions.
The sequencing data are deposited in the EMBL ENA SRA under the accession number ERA032352.
Reads assembly and orthologous gene set identification
The sequencing run produced 396,602 reads for the C. impatiens sample, with mean ± standard error (SE) length of 295.7 ± 0.2 base pairs (bp) (median = 306 bp). The run also produced 442,030 reads for the C. resedifolia sample, with mean length of 296.0 ± 0.2 bp (median = 306 bp). Reads were assembled using the GS de novo assembler software version 2.3 (Roche) using a minimum overlap of 40 bp and an identity of 100%. These stringent parameters were chosen to reduce the probability of co-aligning possible paralogous genes, and to maximize the probability of aligning reads from the same allele (although our samples consisted of inbred plants, we cannot exclude the possibility of heterozygosity in our samples). As an additional quality-control step, we only considered contigs longer than 250 bp. The assembly of the C. resedifolia reads resulted in 10,456 contigs with a mean length of 702.3 ± 3.6 bp, each covered at a mean depth of 29.9 ± 0.3 reads. For C. impatiens, the assembly resulted in 9,484 contigs with a mean length of 664.9 ± 3.7 bp, each covered at a mean depth of 29.9 ± 0.4 reads.
The identification of the orthologous sequences was done using the C. impatiens contigs, the C. resedifolia contigs and Arabidopsis thaliana coding sequences (TAIR9 release [64]). First, we formatted all three datasets as BLAST databases, using the dustmasker sequence filtering application for the two Cardamine datasets. Then we searched orthologues by running a total of six pairwise BLASTn using the BLAST+ tools suite [112]. The best-hit search was optimized using the parameters "-best_hit_overhang 0.18 -softmasking F". To reduce spurious matches, best hits were retained for the next step only if 1) at least 70% of their aligned sequences matched the respective queries; and 2) if either the query or the best hit sequences aligned for at least 60% of their length in the pairwise alignment. Finally, to reduce the chance of mistaking a paralogue for an orthologue, we identified as triplets of putative orthologues only those consisting of reciprocal best hits (RBH) [113], i.e. those where the three sequences were consistently found as best hit matches of one another. This approach resulted in a total of 2,922 triplets of putative orthologues, each triplet corresponding to a single nuclear gene. For comparison, when RBH were identified between C. resedifolia and C. impatiens only, we obtained 4,624 pairs of putative orthologues.
Sequence alignments
After aligning the sequences of each triplet with Clustal W 2.0 [114], we extracted the portion of the alignments containing all three orthologous sequences and trimmed partial codons at the 5' and 3' ends (based on the A. thaliana sequence).
A potential caveat is the presence of a highly variable region present in those genes that are associated with the chloroplast. This region codes for an N-terminal transit peptide that will eventually be cleaved after targeting [115,116] and is enriched in hydroxylated residues and deficient in acidic ones [117]. Since several aminoacids can fulfill such biochemical requirements, functionally equivalent transit peptides can accumulate non-synonymous substitutions particularly fast, and may bias genome-wide and gene-specific estimates of aminoacid substitutions. Therefore, we first identified eventual transit peptides in the chloroplast-targeting A. thaliana genes present in our dataset using the ChloroP program [118], and subsequently removed them from the alignments.
The 2,922 A. thaliana aligned sequences of our final dataset had a mean ± SE length of 594.0 ± 5.8 bp (median = 531 bp; mode = 369 bp). In C. impatiens, the mean length was 592.3 ± 5.8 bp (median = 528 bp; mode = 384 bp). In C. resedifolia, the mean length was 592.2 ± 5.8 bp (median = 528 bp; mode = 384 bp).
Gene annotation and ontology
Gene ontology (GO) and gene annotation were based on the A. thaliana genome annotation [119] available at TAIR (retrieved December 2009 [64]).
GO was used to discriminate genes involved in cold acclimation (CGO), in photosynthesis (as a proxy for UV-B and high irradiation response) (PGO), and those broadly involved in stress resistance (SGO; Additional File 12). In addition, we compiled a list of cold responsive genes (CRG; Additional File 13) that satisfied one of the following two conditions. The first condition was that these genes were involved in cold resistance based on known functional assays; these genes include transcription factors (ICE1 [120], CBF1, CBF2 and CBF3 [121-123], ZAT12 [21], HOS1 [124], and ESK1 [125]), and other genes active at different points along the cold acclimation response pathways (reviewed in [22]). The second condition was that the genes had to be reported as cold responsive (up-regulated in any pathway) at least twice, either in genome-wide expression studies [20,21,126-128], in their annotation description, or in the aforementioned functional studies. Such cross-validation was useful to prevent the inclusion of false-positives, since many of the ~3,000 genes identified as cold responsive in genome-wide studies are probably not primarily nor directly involved in cold resistance.
Among the 2,922 genes analyzed in the present study, 56 were included in the CGO functional class (representing 25% of all A. thaliana genes with such annotations), 67 in the PGO class (48%), 332 in the SGO class (17%), and 55 in the CRG class (18% of the genes included in our list). Note that some genes are present in more than one functional class, since some genes are involved in several stress-related pathways (Additional File 1). As a result of this non-independence, it was not possible to make direct statistical comparisons across functional classes.
Analysis of the rate of molecular and protein evolution
A typical signature of positive selection is a high rate of non-synonymous substitution, dN (leading to aminoacid changes), compared to synonymous substitution, dS. This because synonymous substitutions accumulate nearly neutrally, while non-synonymous substitutions are subject to selective pressures of varying degree and sign. In general, the ratio ω = dN/dS measures the levels of selective pressure operating in a protein coding gene: the value is less than 1 if the gene is under purifying selection, equal to 1 if the gene is evolving neutrally, and greater than one if positive selection has accelerated the fixation of aminoacid changes.
For each gene, we used the program PAML 4.4 [35] to test different models of substitution rates (e.g. [129,130]). We used seven likelihood ratio tests for identifying candidates with distinct evolutionary histories, for instance genes whose substitution rates varied among lineages and/or among coding sites. For this reason, it is assumed that the outlier gene sets are, at most, only partially overlapping. For sake of completeness, we provide the results of all analyses, but we focus in particular on the branch-site test of positive selection, the most appropriate for detecting candidate genes under positive selection in either the C. resedifolia or the C. impatiens lineage (see below). First we compared the models that assume one or more substitution rates across the phylogeny. We estimated dS and dN between pairs of species and over all branches of the phylogenetic tree using the "one-ratio" branch model (M0), which assumes a constant dN/dS ratio, ω, across the phylogeny (with model = 0 and NSsites = 0; A. thaliana was used as the outgroup to C. impatiens and C. resedifolia). The likelihood of this model was compared to that of the ''free-ratio'' model (M0' [131]), which allows ω to vary among branches of the tree (model = 1 and NSsites = 0). Each comparison, i.e. twice the likelihood difference (2Δλ), was tested using a χ2 test with 3 degrees of freedom (which corresponds to the number of branches minus one). In a second approach, the model M0 was compared to branch models where we assumed two ω, the first for either the C. resedifolia or C. impatiens branch, and the second for the other branches (model = 2 and NSsites = 0). In this case the likelihood ratio test was performed using 1 degree of freedom.
The occurrence of positive selection was tested by comparing the likelihoods of (nearly) neutral models to those of models that allow for the occurrence of positive selection. In a first approach we compared the likelihood of a model (M1a) that assumes two sets of sites with neutral (ω = 1) or nearly neutral evolution (0 < ω < 1), to a model (M2a) with an additional class of sites with ω > 1 (M1a: model = 0 and NSsites = 1; M2a: model = 0 and NSsites = 2). In a second more realistic approach, we compared the likelihood of a model (M7) where ten site classes have ω values drawn from a β distribution (model = 0 and NSsites = 7) to a model (M8) that incorporates an additional class of sites under positive selection (model = 0 and NSsites = 8). In this case each comparison was tested using a χ2 test with 2 degrees of freedom.
Finally, we used the branch-site test of positive selection to detect positive selection affecting a few sites along particular lineages (i.e. in the foreground branches). In this test (branch-site model A, test 2 [132]), ω can vary both among sites in the protein and across branches on the tree (model = 2 NSsites = 2). We set as foreground branches either the C. resedifolia or the C. impatiens lineages and allowed two ω along the branches. The null model fixes ω2 to one (fix_omega = 1, omega = 1), while the positive selection model allows ω2 to be larger than one (fix_omega = 0, omega = 1). The likelihood ratio test had one degree of freedom.
To account for multiple testing, for each likelihood ratio test, we estimated the false discovery rate (FDR) using the qvalue package [133] implemented in R [134]. Only genes with a FDR lower than 0.20 were discussed further in the main text. This threshold was chosen to allow between-tests comparisons and to account for the different power of the likelihood ratio tests (e.g. the S21 test is less powerful than the S87 test). Moreover, the fact that we used only partial genes in our analyses, and that the phylogeny included only three species, may have considerably reduced the power of these tests (see PAML documentation [35]).
Rates of synonymous and non-synonymous substitution used in the analyses were those estimated by the ''free-ratio'' model along the C. resedifolia and the C. impatiens branches.
Breadth and levels of expression
Since gene expression data are not yet available for C. impatiens and C. resedifolia, here we assumed that data for the closely related species A. thaliana can serve as a proxy for expression levels in all three species. This is reasonable, given that a recent study suggests quite similar patterns of expression between A. thaliana and the far more divergent species Silene latifolia [135].
For most of the genes in our list we could calculate the breadth of expression based on their expression levels collected across the developmental gradient in several organs of A. thaliana [136]. The expression levels were originally inferred from the intensity of each gene's hybridization onto an Affymetrix microarray [136]. For our analyses, log2 transformed gene expression intensities (which were) were back-transformed by calculating the power of two of their values. The spatial breadth of expression was estimated as the number of organs (minimum 1, maximum 14) where the gene expression value was larger than 75 (arbitrarily chosen as threshold to reduce false positives, see [40]; values ranged from 0 to 66,760, with a median of 460). For organs represented by more developmental stages, we considered a combined value that equaled 1 only when the gene was expressed in at least one developmental stage. In a second approach, we used the organ specificity index τ [46], which also includes the information on the level of expression. This index has a value of 0 when the gene is expressed equally across organs, while it approaches 1 when the gene has organ-specific expression. We also estimated the temporal breadth of expression in leaves and flowers as the number of developmental time points in which the gene was expressed at a level larger than 75.
In addition, we calculated the breadth of expression as a function of the kinetics of a gene in the presence of an abiotic stress. Our rationale for choosing this estimate is that genes expressed briefly during a stress response may be subject to different selective pressures compared to those expressed continuously. For our purpose, we used the AtGenExpress dataset [128], which contains time series of expression data collected from plants subjected to one of the following stress treatments: UV-B radiations, drought, salt, cold and osmosis. For every stress treatment, we assigned to each gene a value corresponding to the number of time-points at which its expression in treated plants was at least 3 times higher than in control plants [128].
Finally, we estimated the mean and maximum levels of expression of the genes from data reported in [136]. Because stress responses are typically not constitutive, using maximum expression levels reduces the possibility to overlook transient peaks of expression.
Codon usage analysis
We estimated codon bias using the frequency of optimal codons (Fop [137], where stronger synonymous codon usage bias is identified by larger Fop values. This index was calculated using the program CodonW (version 1.4.2 [138]). First we inferred the preferred codon usage for each species using the correspondence analysis of relative synonymous codon usage approach as implemented in CodonW. Briefly, putative optimal (preferred) codons are identified as those that are significantly overrepresented in genes with high codon bias compared to those with low bias [139]. To avoid a bias due to the heterogeneous composition of our gene dataset (enriched in stress related genes, see above), we inferred the preferred codons only from genes annotated as ribosomal (n = 77), which, being highly expressed, should be enriched in optimal codons. Similarly, we discarded 11 (partial) genes shorter than 100 codons [140,141]. We then let CodonW to automatically identify codon usage on the 50% highest- and lowest-biased genes (see Additional File 10 for a list of putative optimal codons in Cardamine). This percentage was chosen because in A. thaliana it maximized the agreement between our estimate and what is reported in the literature [139]. Indeed, optimal codons identified by using higher or lower percentages of the ribosomal genes, or the 5% highest- and lowest-biased genes among all those sequenced, had a lower agreement (data not shown). Therefore, we can assume that the preferred codon usage patterns identified for the two Cardamine species are also very close to the real ones.
Authors' contributions
LO contributed to develop the design of the study, performed the bioinformatics and statistical analyses, and drafted the manuscript. ML carried out all molecular work related to the 454 sequencing, and participated in manuscript drafting. LB contributed to the sequence evolution analyses. CV conceived and coordinated the study, participated in data analysis, and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Genes in functional classes. Venn-diagrams showing the numbers of genes in the functional classes considered in this study.
Correlation between substitution rate and gene length. Plot showing the correlation between the length of the A. thaliana orthogous gene and the substitution rate in C. impatiens and C. resedifolia genes.
Rate of synonymous and non-synonymous substitution in Cardamine genes. I. Mean substitution rate in C. resedifolia and C. impatiens genes included in the four functional classes considered in this study. Statistical comparisons within and between lineages are also reported, including those calculated for values corrected for gene length.
Number of genes identified by likelihood ratio tests that compared branch, site and branch-site codon substitution models. Number of genes identified (at decreasing probabilities thresholds) as putative targets of positive selection by likelihood ratio tests based on branch models (B tests, Table S4.1), site models (S tests, Table S4.2), and branch-site models (BS tests, Table S4.3).
Description of the top ten genes identified by likelihood ratio tests that compared various codon substitution models. Top ten genes identified as putative targets of positive selection by likelihood ratio tests based on branch models (B tests, Tables S5.1-S5.3), site models (S tests, Tables S5.4-S5.5), and branch-site models (BS tests, Tables S5.6-S5.7).
Phylogenetic trees of candidate genes. Phylogenetic trees for the five genes that were putative targets for positive selection (at FDR < 0.20) according to likelihood ratio tests based on the site and branch-site codon substitution models implemented in PAML.
Correlation between the temporal breadth of expression and levels of selection. Spearman's correlations between levels of selection and both the spatial and temporal breadth of expression.
Correlation between rate of molecular evolution and level of gene expression. Mean and maximum expression levels, and Spearman's correlations between such expression levels and levels of selection for the genes included in the four functional classes considered in this study.
Rate of synonymous and non-synonymous substitution in Cardamine genes. II. Mean substitution rate in C. resedifolia and C. impatiens genes included in the four functional classes considered in this study. Comparisons of the values corrected for expression levels.
Optimal codons in Cardamine. Putative optimal codons identified in C. resedifolia and C. impatiens.
Codon usage bias in Cardamine genes. Mean codon usage bias, measured as Fop, in the four functional classes considered in this study. Statistical results of the comparisons between functional classes and species are also reported.
Definition of the Gene Ontology (GO) terms associated with the functional classes analyzed in this study. GO terms and associated function used to identify genes in our dataset that were putatively involved in cold response, photosynthesis, and general stress response.
Cold responsive genes. List of genes identified as involved in cold response in previous studies.
Contributor Information
Lino Ometto, Email: lino.ometto@fmach.it.
Mingai Li, Email: mingai.li@fmach.it.
Luisa Bresadola, Email: luisa.bresadola@fmach.it.
Claudio Varotto, Email: claudio.varotto@fmach.it.
Acknowledgements
We thank F. Prosser for indication of the Cardamine populations used in this study and H.C. Hauffe for critical reading of the manuscript. We wish to thank four anonymous referees, whose insightful comments significantly improved the manuscript. This work was funded by the Autonomous Province of Trento (Italy), under the ACE-SAP project (regulation number 23, June 12th 2008, of the Servizio Università e Ricerca Scientifica).
References
- Orr HA. The genetic theory of adaptation: a brief history. Nat Rev Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Hoekstra HE, D'Agostino SL. The genetic basis of adaptive melanism in pocket mice. Proc Natl Acad Sci USA. 2003;100:5268–5273. doi: 10.1073/pnas.0431157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Sabatino SJ, Hoffmann FG, Gering EJ, Moriyama H, Ferrand N, Monteiro B, Nachman MW. The molecular basis of high-altitude adaptation in deer mice. PLoS Genet. 2007;3:e45. doi: 10.1371/journal.pgen.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Ungerer MC. Relaxed selection on the CBF/DREB1 regulatory genes and reduced freezing tolerance in the southern range of Arabidopsis thaliana. Mol Biol Evol. 2008;25:2547–2555. doi: 10.1093/molbev/msn196. [DOI] [PubMed] [Google Scholar]
- Eckert AJ, Wegrzyn JL, Pande B, Jermstad KD, Lee JM, Liechty JD, Tearse BR, Krutovsky KV, Neale DB. Multilocus patterns of nucleotide diversity and divergence reveal positive selection at candidate genes related to cold hardiness in coastal Douglas Fir (Pseudotsuga menziesii var. menziesii) Genetics. 2009;183:289–298. doi: 10.1534/genetics.109.103895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol Ecol. 2010;19:760–774. doi: 10.1111/j.1365-294X.2009.04508.x. [DOI] [PubMed] [Google Scholar]
- Xia H, Camus-Kulandaivelu L, Stephan W, Tellier A, Zhang Z. Nucleotide diversity patterns of local adaptation at drought-related candidate genes in wild tomatoes. Mol Ecol. 2010;19:4144–4154. doi: 10.1111/j.1365-294X.2010.04762.x. [DOI] [PubMed] [Google Scholar]
- Storz JF, Wheat CW. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010;64:2489–2509. doi: 10.1111/j.1558-5646.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, Sninsky JJ, Adams MD, Cargill M. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen K, Reusch TBH. Genome scans detect consistent divergent selection among subtidal vs. intertidal populations of the marine angiosperm Zostera marina. Mol Ecol. 2007;16:5156–5167. doi: 10.1111/j.1365-294X.2007.03577.x. [DOI] [PubMed] [Google Scholar]
- Namroud M-C, Beaulieu J, Juge N, Laroche J, Bousquet J. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol Ecol. 2008;17:3599–3613. doi: 10.1111/j.1365-294X.2008.03840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann TI, Song B-H, Windsor AJ, Mitchell-Olds T, Dixon CJ, Kapralov MV, Filatov DA, Eyre-Walker A. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27:1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C. Alpine plant life. Functional plant ecology of high mountain ecosystems. 2. Berlin, Germany: Springer; 2003. [Google Scholar]
- Zhen Y, Ungerer MC. Clinal variation in freezing tolerance among natural accessions of Arabidopsis thaliana. New Phytol. 2008;177:419–427. doi: 10.1111/j.1469-8137.2007.02262.x. [DOI] [PubMed] [Google Scholar]
- Alexander JM, Kueffer C, Daehler CC, Edwards PJ, Pauchard A, Seipel T. MIREN Consortium. Assembly of nonnative floras along elevational gradients explained by directional ecological filtering. Proc Natl Acad Sci USA. 2011;108:656–661. doi: 10.1073/pnas.1013136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins SM, Dobrowski SZ, Greenberg JA, Abatzoglou JT, Mynsberge AR. Changes in climatic water balance drive downhill shifts in plant species' optimum elevations. Science. 2011;331:324–327. doi: 10.1126/science.1199040. [DOI] [PubMed] [Google Scholar]
- Montesinos-Navarro A, Wig J, Pico FX, Tonsor SJ. Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol. 2011;189:282–294. doi: 10.1111/j.1469-8137.2010.03479.x. [DOI] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005;1:e26. doi: 10.1371/journal.pgen.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Ruelland E, Vaultier M-N, Zachowski A, Hurry V. Cold signalling and cold acclimation in plants. Adv Bot Res. 2009;49:35–150. [Google Scholar]
- Streb P, Shang W, Feierabend J, Bligny R. Divergent strategies of photoprotection in high-mountain plants. Planta. 1998;207:313–324. doi: 10.1007/s004250050488. [DOI] [Google Scholar]
- Germino M, Smith W. High resistance to low-temperature photoinhibition in two alpine, snowbank species. Physiol Plantarum. 2000;110:89–95. doi: 10.1034/j.1399-3054.2000.110112.x. [DOI] [Google Scholar]
- Frohnmeyer H, Staiger D. Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 2003;133:1420–1428. doi: 10.1104/pp.103.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb P, Aubert S, Gout E, Bligny R. Cold- and light-induced changes of metabolite and antioxidant levels in two high mountain plant species Soldanella alpina and Ranunculus glacialis and a lowland species Pisum sativum. Physiol Plantarum. 2003;118:96–104. doi: 10.1034/j.1399-3054.2003.00099.x. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Fujii N, Setoguchi H. Molecular evolution of phytochromes in Cardamine nipponica (Brassicaceae) suggests the involvement of PHYE in local adaptation. Genetics. 2009;182:603–614. doi: 10.1534/genetics.109.102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeschimann D, Lauber K, Moser DM, Theurillat J-P. Flora Alpina. Bern, Switzerland: Haupt Verlag; 2004. [Google Scholar]
- Kimata M. Comparative Studies on the Reproductive Systems of Cardamine flexuosa, C. impatiens, C. scutata, and C. lyrata, Cruciferae. Bot Mag Tokyo. 1983;96:299–312. doi: 10.1007/BF02488175. [DOI] [Google Scholar]
- Kucera J, Lihova J, Marhold K. Taxonomy and phylogeography of Cardamine impatiens and C. pectinata (Brassicaceae) Bot J Linn Soc. 2006;152:169–195. doi: 10.1111/j.1095-8339.2006.00559.x. [DOI] [Google Scholar]
- Lihova J, Carlsen T, Brochmann C, Marhold K. Contrasting phylogeographies inferred for the two alpine sister species Cardamine resedifolia and C. alpina (Brassicaceae) J Biogeogr. 2009;36:104–120. doi: 10.1111/j.1365-2699.2008.01998.x. [DOI] [Google Scholar]
- Bailey CD, Koch MA, Mayer M, Mummenhoff K, O'Kane SL, Warwick SI, Windham MD, Al-Shehbaz IA. Toward a global phylogeny of the Brassicaceae. Mol Biol Evol. 2006;23:2142–2160. doi: 10.1093/molbev/msl087. [DOI] [PubMed] [Google Scholar]
- Couvreur TLP, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae) Mol Biol Evol. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand J Statist. 1979;6:65–70. [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Gehring C, Hurst LD. Gametophytic selection in Arabidopsis thaliana supports the selective model of intron length reduction. PLoS Genet. 2005;1:e13. doi: 10.1371/journal.pgen.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Gene expression and protein length influence codon usage and rates of sequence evolution in Populus tremula. Mol Biol Evol. 2007;24:836–844. doi: 10.1093/molbev/msl212. [DOI] [PubMed] [Google Scholar]
- Colinas J, Schmidler SC, Bohrer G, Iordanov B, Benfey PN. Intergenic and genic sequence lengths have opposite relationships with respect to gene expression. PLoS ONE. 2008;3:e3670. doi: 10.1371/journal.pone.0003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camiolo S, Rau D, Porceddu A. Mutational biases and selective forces shaping the structure of Arabidopsis genes. PLoS ONE. 2009;4:e6356. doi: 10.1371/journal.pone.0006356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan ET, Quon GT, Chua G, Babak T, Trochesset M, Zirngibl RA, Aubin J, Ratcliffe MJH, Wilde A, Brudno M, Morris QD, Hughes TR. Conservation of core gene expression in vertebrate tissues. J Biol. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng-Bradley X, Rung J, Parkinson H, Brazma A. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 2010;11:R124. doi: 10.1186/gb-2010-11-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanderbali AS, Yoo M-J, Zahn LM, Brockington SF, Wall PK, Gitzendanner MA, Albert VA, Leebens-Mack J, Altman NS, Ma H, Depamphilis CW, Soltis DE, Soltis PS. Conservation and canalization of gene expression during angiosperm diversification accompany the origin and evolution of the flower. Proc Natl Acad Sci USA. 2010;107:22570–22575. doi: 10.1073/pnas.1013395108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small R, Cronn R, Wendel J. Use of nuclear genes for phylogeny reconstruction in plants. Aust Syst Bot. 2004;17:145–170. doi: 10.1071/SB03015. [DOI] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. doi: 10.1017/S0016672300010156. [DOI] [PubMed] [Google Scholar]
- Kliman RM, Hey J. Hill-Robertson interference in Drosophila melanogaster: reply to Marais, Mouchiroud and Duret. Genet Res. 2003;81:89–90. doi: 10.1017/S0016672302006067. [DOI] [PubMed] [Google Scholar]
- Betancourt AJ, Presgraves DC. Linkage limits the power of natural selection in Drosophila. Proc Natl Acad Sci USA. 2002;99:13616–13620. doi: 10.1073/pnas.212277199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Mouchiroud D, Duret L. Neutral effect of recombination on base composition in Drosophila. Genet Res. 2003;81:79–87. doi: 10.1017/S0016672302006079. [DOI] [PubMed] [Google Scholar]
- Carlsen T, Bleeker W, Hurka H, Elven R, Brochmann C. Biogeography and phylogeny of Cardamine (Brassicaceae) Ann Mo Bot Gard. 2009;96:215–236. doi: 10.3417/2007047. [DOI] [Google Scholar]
- Andreasen K, Baldwin BG. Unequal evolutionary rates between annual and perennial lineages of checker mallows (Sidalcea, Malvaceae): evidence from 18S-26S rDNA internal and external transcribed spacers. Mol Biol Evol. 2001;18:936–944. doi: 10.1093/oxfordjournals.molbev.a003894. [DOI] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Müller K, Albach DC. Evolutionary rates in Veronica L. (Plantaginaceae): disentangling the influence of life history and breeding system. J Mol Evol. 2010;70:44–56. doi: 10.1007/s00239-009-9307-5. [DOI] [PubMed] [Google Scholar]
- Whittle C-A, Johnston MO. Broad-scale analysis contradicts the theory that generation time affects molecular evolutionary rates in plants. J Mol Evol. 2003;56:223–233. doi: 10.1007/s00239-002-2395-0. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr Opin Genet Dev. 2001;11:685–690. doi: 10.1016/S0959-437X(00)00254-9. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Welch JJ, Bromham L. Watching the clock: studying variation in rates of molecular evolution between species. Trends Ecol Evol. 2010;25:495–503. doi: 10.1016/j.tree.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009. [DOI] [PubMed]
- Presgraves DC. Recombination enhances protein adaptation in Drosophila melanogaster. Curr Biol. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. [DOI] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. The positive correlation between dN/dS and dS in mammals is due to runs of adjacent substitutions. Mol Biol Evol. 2011;28:1371–1380. doi: 10.1093/molbev/msq320. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Z, Vang S, Yu J, Wong GK-S, Wang J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J Mol Evol. 2009;68:414–423. doi: 10.1007/s00239-009-9222-9. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JBW, Künstner A, Nam K, Jakobsson M, Ellegren H. Nonlinear dynamics of nonsynonymous (dN) and synonymous (dS) substitution rates affects inference of selection. Genome Biol Evol. 2009;1:308–319. doi: 10.1093/gbe/evp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Information Resource. http://www.arabidopsis.org
- Kreps J, Wu Y, Chang H, Zhu T, Wang X, Harper J. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 2007;8:125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Tominaga Y, Nakagawara C, Shigematsu S, Minami A, Kawamura Y. Responses of the plasma membrane to low temperatures. Physiol Plantarum. 2006;126:81–89. doi: 10.1111/j.1399-3054.2005.00594.x. [DOI] [Google Scholar]
- Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol Biol Evol. 2010;27:650–660. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 2010;27:1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Scotti-Saintagne C, Scotti I, Lai Z, Rieseberg LH. Genomic patterns of adaptive divergence between chromosomally differentiated sunflower species. Mol Biol Evol. 2009;26:1341–1355. doi: 10.1093/molbev/msp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- Foxe JP, Dar V-U-N, Zheng H, Nordborg M, Gaut BS, Wright SI. Selection on amino acid substitutions in Arabidopsis. Mol Biol Evol. 2008;25:1375–1383. doi: 10.1093/molbev/msn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin MT, Casa AM, Sun H, Murray SC, Paterson AH, Aquadro CF, Kresovich S. Challenges of detecting directional selection after a bottleneck: lessons from Sorghum bicolor. Genetics. 2006;173:953–964. doi: 10.1534/genetics.105.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Liberles DA. A systematic search for positive selection in higher plants (Embryophytes) BMC Plant Biol. 2006;6:12. doi: 10.1186/1471-2229-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasburg JL, Kane NC, Raduski AR, Bonin A, Michelmore R, Rieseberg LH. Effective population size is positively correlated with levels of adaptive divergence among annual sunflowers. Mol Biol Evol. 2011;28:1569–1580. doi: 10.1093/molbev/msq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Maere S, de Bodt S, Raes J, Casneuf T, van Montagu M, Kuiper M, van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon A-F, Weissenbach J, Quétier F, Wincker P. French-Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 2001;125:89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champ KI, Febres VJ, Moore GA. The role of CBF transcriptional activators in two Citrus species (Poncirus and Citrus) with contrasting levels of freezing tolerance. Physiol Plantarum. 2007;129:529–541. doi: 10.1111/j.1399-3054.2006.00826.x. [DOI] [Google Scholar]
- Pennycooke JC, Cheng H, Roberts SM, Yang Q, Rhee SY, Stockinger EJ. The low temperature-responsive, Solanum CBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Mol Biol. 2008;67:483–497. doi: 10.1007/s11103-008-9333-5. [DOI] [PubMed] [Google Scholar]
- Tondelli A, Francia E, Barabaschi D, Pasquariello M, Pecchioni N. Inside the CBF locus in Poaceae. Plant Sci. 2011;180:39–45. doi: 10.1016/j.plantsci.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Huang L, Grammatikakis N, Yoneda M, Banerjee SD, Toole BP. Molecular characterization of a novel intracellular hyaluronan-binding protein. J Biol Chem. 2000;275:29829–29839. doi: 10.1074/jbc.M002737200. [DOI] [PubMed] [Google Scholar]
- Lazzaroni JC, Germon P, Ray MC, Vianney A. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett. 1999;177:191–197. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C, Schat H. Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 2009;181:759–776. doi: 10.1111/j.1469-8137.2008.02748.x. [DOI] [PubMed] [Google Scholar]
- Vergnano Gambi O, Gabbrielli R, Pancaro L. Nickel, chromium and cobalt in plants from italian serpentine areas. Acta Oecol-Oec Plant. 1982;3:291–306. [Google Scholar]
- Mikkelsen MD, Petersen BL, Olsen CE, Halkier BA. Biosynthesis and metabolic engineering of glucosinolates. Amino Acids. 2002;22:279–295. doi: 10.1007/s007260200014. [DOI] [PubMed] [Google Scholar]
- Jones C, Dancer J, Smith A, Abell C. Evidence for the pathway to pantothenate in plants. Can J Chem. 1994;72:261–263. doi: 10.1139/v94-039. [DOI] [Google Scholar]
- Ludwig-Müller J. Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot. 2011;62:1757–1773. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- Wang L, Allmann S, Wu J, Baldwin IT. Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 2008;146:904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol. 2008;11:486–494. doi: 10.1016/j.pbi.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Manuel Franco-Zorrilla J, Pauwels L, Witters E, Isabel Puga M, Paz-Ares J, Goossens A, Reymond P, De Jaeger G, Solano R. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J. The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell. 2001;13:2793–2807. doi: 10.1105/tpc.010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen R, Steffes L, Wolf J, Brown P, Matzner S, Siemens DH. Evolution of drought tolerance and defense: dependence of tradeoffs on mechanism, environment and defense switching. Oikos. 2008;117:231–244. doi: 10.1111/j.2007.0030-1299.16111.x. [DOI] [Google Scholar]
- Zehnder CB, Stodola KW, Joyce BL, Egetter D, Cooper RJ, Hunter MD. Elevational and seasonal variation in the foliar quality and arthropod community of Acer pensylvanicum. Environ Entomol. 2009;38:1161–1167. doi: 10.1603/022.038.0424. [DOI] [PubMed] [Google Scholar]
- Reynolds B, Crossley D. Spatial variation in herbivory by forest canopy arthropods along an elevation gradient. Environ Entomol. 1997;26:1232–1239. [Google Scholar]
- Suzuki S. Leaf phenology, seasonal changes in leaf quality and herbivory pattern of Sanguisorba tenuifolia at different altitudes. Oecologia. 1998;117:169–176. doi: 10.1007/s004420050645. [DOI] [PubMed] [Google Scholar]
- Hengxiao G, McMillin J, Wagner M, Zhou J, Zhou Z, Xu X. Altitudinal variation in foliar chemistry and anatomy of yunnan pine, Pinus yunnanensis, and pine sawfly (Hym., Diprionidae) performance. J Appl Entomol. 1999;123:465–471. doi: 10.1046/j.1439-0418.1999.00395.x. [DOI] [Google Scholar]
- Louda S, Rodman J. Insect herbivory as a major factor in the shade distribution of a native crucifer (Cardamine cordifolia A. Gray, bittercress) J Ecol. 1996;84:229–237. doi: 10.2307/2261358. [DOI] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R. Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J. 2003;34:185–201. doi: 10.1046/j.1365-313x.2003.01715.x. [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I-P, Haehnel U, Altschmied L, Schubert I, Puchta H. The transcriptional response of Arabidopsis to genotoxic stress-a high-density colony array study (HDCA) Plant J. 2003;35:771–786. doi: 10.1046/j.1365-313X.2003.01847.x. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SG, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Robinson C, Ellis RJ. Transport of proteins into chloroplasts. Partial purification of a chloroplast protease involved in the processing of important precursor polypeptides. Eur J Biochem. 1984;142:337–342. doi: 10.1111/j.1432-1033.1984.tb08291.x. [DOI] [PubMed] [Google Scholar]
- Soll J, Tien R. Protein translocation into and across the chloroplastic envelope membranes. Plant Mol Biol. 1998;38:191–207. doi: 10.1023/A:1006034020192. [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, Moseyko N, Yoo D, Xu I, Zoeckler B, Montoya M, Miller N, Weems D, Rhee SY. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004;135:745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee B-H, Hong X, Agarwal M, Zhu J-K. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001;127:910–917. doi: 10.1104/pp.010548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Pérez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–470. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Gene Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Browse J. eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Lee B-H, Henderson DA, Zhu J-K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen A. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Storey J. A direct approach to false discovery rates. J Roy Stat Soc B. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Qiu S, Bergero R, Zeng K, Charlesworth D. Patterns of codon usage bias in Silene latifolia. Mol Biol Evol. 2011;28:771–780. doi: 10.1093/molbev/msq251. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- CodonW. http://codonw.sourceforge.net/
- Chiapello H, Lisacek F, Caboche M, Hénaut A. Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene. 1998;209:GC1–GC38. doi: 10.1016/S0378-1119(97)00671-9. [DOI] [PubMed] [Google Scholar]
- Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol. 2002;19:1390–1394. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- Wright SI, Yau CBK, Looseley M, Meyers BC. Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol Biol Evol. 2004;21:1719–1726. doi: 10.1093/molbev/msh191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes in functional classes. Venn-diagrams showing the numbers of genes in the functional classes considered in this study.
Correlation between substitution rate and gene length. Plot showing the correlation between the length of the A. thaliana orthogous gene and the substitution rate in C. impatiens and C. resedifolia genes.
Rate of synonymous and non-synonymous substitution in Cardamine genes. I. Mean substitution rate in C. resedifolia and C. impatiens genes included in the four functional classes considered in this study. Statistical comparisons within and between lineages are also reported, including those calculated for values corrected for gene length.
Number of genes identified by likelihood ratio tests that compared branch, site and branch-site codon substitution models. Number of genes identified (at decreasing probabilities thresholds) as putative targets of positive selection by likelihood ratio tests based on branch models (B tests, Table S4.1), site models (S tests, Table S4.2), and branch-site models (BS tests, Table S4.3).
Description of the top ten genes identified by likelihood ratio tests that compared various codon substitution models. Top ten genes identified as putative targets of positive selection by likelihood ratio tests based on branch models (B tests, Tables S5.1-S5.3), site models (S tests, Tables S5.4-S5.5), and branch-site models (BS tests, Tables S5.6-S5.7).
Phylogenetic trees of candidate genes. Phylogenetic trees for the five genes that were putative targets for positive selection (at FDR < 0.20) according to likelihood ratio tests based on the site and branch-site codon substitution models implemented in PAML.
Correlation between the temporal breadth of expression and levels of selection. Spearman's correlations between levels of selection and both the spatial and temporal breadth of expression.
Correlation between rate of molecular evolution and level of gene expression. Mean and maximum expression levels, and Spearman's correlations between such expression levels and levels of selection for the genes included in the four functional classes considered in this study.
Rate of synonymous and non-synonymous substitution in Cardamine genes. II. Mean substitution rate in C. resedifolia and C. impatiens genes included in the four functional classes considered in this study. Comparisons of the values corrected for expression levels.
Optimal codons in Cardamine. Putative optimal codons identified in C. resedifolia and C. impatiens.
Codon usage bias in Cardamine genes. Mean codon usage bias, measured as Fop, in the four functional classes considered in this study. Statistical results of the comparisons between functional classes and species are also reported.
Definition of the Gene Ontology (GO) terms associated with the functional classes analyzed in this study. GO terms and associated function used to identify genes in our dataset that were putatively involved in cold response, photosynthesis, and general stress response.
Cold responsive genes. List of genes identified as involved in cold response in previous studies.