Abstract
The use of CD133 as a marker for identifying and isolating colon cancer stem cells (CSCs) is controversial. In particular, it is not clear whether the pattern of expression is influenced by the tumor microenvironment. We analysed the expression of CD133 in the SW620 colon cancer cell line, which produces subpopulations with different levels of CD133. CD133neg and CD133hi SW620 cells were isolated and compared in hanging drop spheroid cultures. The expression of CD133 was examined under different culture conditions and following nutrient withdrawal. The results showed that SW620 cells express the CD133 antigen at variable levels. CD133neg and CD133hi SW620 cells can be separated by fluorescence-activated cell sorting and, when grown as adherent cells in liquid culture, are serially passaged without changes to the CD133 levels. However, the CD133 antigen is re-expressed when the CD133neg cells are deprived of nutrients or grown as non-adherent cells in spheroid cultures. Our observations indicate that the presence of the CD133 antigen does not identify a stem cell population of SW620 colorectal cancer cells. The expression of CD133 is dynamic and is modulated during certain cell culture conditions, suggesting that changes in expression may be mediated by the energy supply available to the cells.
Keywords: CD133, colorectal cancer, cancer stem cells, microenvironment
Introduction
Cancer stem cells (CSCs) are defined as the subset of tumor cells that have an increased ability to self-renew and generate the diverse cells that comprise the tumor (1,2). CSCs are considered to be responsible for tumor initiation, metastasis and resistance to treatments (3). Cell surface markers which allow the isolation of CSCs are important in the investigation of the biological behavior of CSCs and the development of targeted therapies.
CD133, also termed Prominin-1, is a transmembrane pentaspan protein that was first described as a surface antigen specific for human hematopoietic stem and progenitor cells (4,5) and subsequently recognized as a stem cell marker for other normal tissues. Notably, CD133 has been reported to be a marker of putative CSCs in brain tumors, hepatocellular carcinoma, melanoma and prostate, colon, lung, ovarian and pancreatic cancers (6–16). This has led to CD133 being referred to as ‘the molecule of the moment’ (17) in stem cell and cancer research. However, the significance of CD133 as a marker for identifying and isolating colon CSCs is controversial (18–21). CD133 expression, as assessed by antibody staining, appears to be restricted to a small population of colon cancer cells which form tumors that recapitulate the morphology of the original tumor in immunodeficient mice (9–11). By contrast, studies monitoring CD133 expression using a reporter gene or polyclonal antibodies have shown that CD133 is not restricted to CSCs in primary colon cancer (18–20). Moreover, CD133neg and CD133hi metastatic colon cancer cells are able to initiate tumors (20). Results of a recent study have shown that CD133 is expressed by CSCs and differentiated tumor cells, but the AC133 epitope is lost following CSC differentiation (21). In view of these contrasting reports, the relevance of CD133 expression to colorectal tumorigenesis should be considered as a work in progress.
In this study, we analyzed the expression of CD133 in the SW620 colon cancer cell line to study the correlation between the levels of CD133 expression and the biology of the cells. We detected a variation in CD133 expression in different environments, including culture conditions and nutrient withdrawal. Contrary to the current paradigm, our data indicate that the expression of the CD133 antigen is not static, but is regulated by the microenvironment of the cells; this plasticity may explain the high expression of CD133 in tumors.
Materials and methods
Cell lines and cell culture
Human SW620 colon cancer cells (22) (ATCC, Manassas, VA, USA) were routinely passaged in RPMI-1640 medium (Gibco, Carlsbad, CA, USA)/10% fetal bovine serum (FBS, Gibco) with the following additives: hydrocortisone (1 μg/ml), thioglycerol (0.01 μg/ml), insulin (0.025 U/ml), penicillin G (60 mg/l) and streptomycin (100 mg/l). The cells were cultured at 37°C in a humidified atmosphere containing 10% CO2.
Flow cytometry
Single-cell suspensions were obtained by trypsinization of the cells in adherent cultures or of the spheroids grown in hanging drops. Cell preparations were stained with antibodies against human CD133 (AC133, 1:40; Miltenyi Biotec, Gladbach, Germany) followed by Alexa488 anti-mouse IgG, or with APC-conjugated anti-human CD133/1 and CD133/2 antibodies (Miltenyi Biotec). The cells were analysed on a FACSCalibur flow cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA). Dead cells, cell debris, doublets and aggregates were excluded from the analysis using forward and side scatter and pulse-width gating. The cells were prepared for sorting by staining with human CD133/1 (1:10, APC conjugated; Miltenyi Biotec) and 1 μg/ml propidium iodide (PI) to exclude dead cells during sorting. Cell sorting was performed using a FACSAria (Becton-Dickinson). Matched isotype primary antibodies were used as controls.
Spheroid culture in hanging drops
Cells were prepared as single cell suspensions. The cells were counted and resuspended in medium (RPMI-1640 with 20% FBS and antibiotics) to a concentration of 1.6×104 cells/ml in a sterile basin. An 8-channel pipette was used to make rows of 30 μl drops (500 cells/drop) on the up-turned inner surface of the lid of a tissue culture dish. The lid was then inverted and placed on top of a culture dish containing 10 ml of PBS. The drops were incubated at 37°C and 10% CO2 for 6 days. The resulting spheroids were photographed in phase contrast using a Nikon microscope with a ×10 lens.
Immunofluorescence and confocal microscopy
The sorted cells were seeded on glass cover slips, fixed in 4% paraformaldehyde and blocked in PBS with 0.2% BSA for 30 min at room temperature prior to incubation with mouse anti-human CD133/1 antibody (1:40; Miltenyi Biotec) followed by goat anti-mouse Alexa488 (Invitrogen, Carlsbad, CA, USA). The nuclei of the cells were stained by incubating with DAPI (1 μg/ml) for 5 min. The coverslips were mounted on microscope slides. Confocal microscopy was performed using a Nikon digital eclipse C1si confocal microscope with EN-C1 software.
Immunohistochemistry
The spheroids were collected and frozen individually in OCT compound (ProSciTech, Thuringowa, Queensland, Australia). Frozen sections of the spheroids were fixed in acetone (−20°C, 10 min) and rehydrated in PBS. Endogenous peroxidases were inactivated by immersing the sections in 0.3% hydrogen peroxide for 20 min. The sections were incubated overnight at 4°C in a humidified chamber with mouse anti-human monoclonal CD133/2 antibody (1:40; Miltenyi Biotec) followed by biotinylated secondary antibody (VECTASTAIN ABC kit, Vector Laboratories, Burlingame, CA, USA) for 30 min at room temperature. Each section was further incubated in VECTASTAIN ABC reagent for 30 min at room temperature. The sections were developed using DAB (Vector Laboratories) as the substrate and then counterstained with hematoxylin. Negative controls were stained in parallel substituting an isotype control with the primary antibody.
Western blot analysis
Whole protein lysates of cultured cells and spheroids were used in these experiments. A total of 50 μg of protein was used per lane. The primary antibodies were anti-β-tubulin mouse mAb (1:1000) and anti-CD133 (1:200 mouse-anti-CD133/1 at W6B3C1; Miltenyi Biotec). The secondary antibody was Odyssey anti-mouse 800 (1:10000). Reactive bands were visualized using an Odyssey infrared photometer (Li-Cor, Lincoln, NE, USA) according to the manufacturer’s instructions.
Quantitative real time-PCR (qRT-PCR)
Total RNA was prepared from the cultured cells and spheroids using the RNeasy extraction kit (GE Healthcare, Piscataway, NJ, USA) and reverse transcribed using high-capacity cDNA reverse transcription kits (Applied Biosystems, Mulgrave, Victoria, Australia) according to the manufacturer’s instructions. qRT-PCR was performed using a 7300 Fast Real-Time PCR system (Applied Biosystems) using SYBR-Green PCR Master mix (Applied Biosystems). The human-specific intron-spanning primer pairs for CD133 were provided by Qiagen (catalog number: QT00075586; Hilden, Germany). The following primer pair was used for GAPDH: forward, CAATGACCCCTTCATTGACC; reverse, TGATGACAAGCTTCCCGTTC. The cycle conditions were as follows: 1 cycle at 50°C for 2 min, followed by 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 sec and 60°C for 1 min. The specificity of the PCR products was tested using dissociation curves. The relative values of the transcripts were calculated using the 2-ΔΔCt method, where ΔCt is equal to the difference in threshold cycles of the target and reference.
Induction of CD133 by nutrient withdrawal
CD133neg and CD133hi cells were plated in complete medium at 1×106 cells/well. After one day, the medium was changed to DMEM, 5% FBS (control), DMEM with no serum or glucose-free DMEM with 5% FBS. The cells were harvested 4 days later and CD133 expression was monitored by fluorescence-activated cell sorting (FACS). The median and peak fluorescence channels were determined using the Stat program in CellQuest.
Statistical analysis
Results are presented as the mean ± SD for three repeated individual experiments for each group. Statistical analyses were performed using SPSS software (version 10.0; SPSS, Inc., Chicago, IL, USA). Correlations between the sample groups and molecular variables were calculated using the paired t-test. P<0.05 was considered to indicate a statistically significant result.
Results
CD133 expression in the SW620 colon cancer cell line
SW620 cells express two distinct levels of CD133 as assessed by FACS. CD133neg cells accounted for 70% and CD133hi cells for 30% of the SW620 cells (Fig. 1). The two subpopulations were always present, irrespective of the batch of cells or passage number; however, the relative percentage of each subpopulation varied occasionally between cultures.
Figure 1.

CD133 expression in SW620 cells. SW620 cells were analysed by FACS using CD133/1 antibody and Alexa488 anti-mouse IgG. Solid purple, CD133 reactivity of SW620 cells; green overlay, isotype control; FACS, fluorescence-activated cell sorting.
CD133neg and CD133hi expression remains stable following sorting
To investigate the stability of CD133 expression in the negative and positive subsets, SW620 cells were sorted into separate CD133neg and CD133hi pools. The two subpopulations were grown separately and analysed for CD133 expression 8 days after sorting by flow cytometry (Fig. 2A) and immunofluorecence microscopy (Fig. 2B). The two methods showed that CD133 expression in CD133neg and CD133hi SW620 cells is stable during normal passage in culture.
Figure 2.

CD133 expression in cultured CD133neg and CD133hi SW620 cells. (A) CD133 expression for CD133neg and CD133hi SW620 cells 8 days after sorting. Solid purple, CD133 reactivity of CD133neg cells; green overlay, isotype control; red overlay, CD133 reactivity of CD133hi cells. (B) Immunofluorescence staining for CD133 of CD133neg and CD133hi SW620 cells 8 days after sorting. CD133, green; DAPI, blue; magnification, ×600.
CD133 expression in SW620 cells is induced by anchorage-independent growth or nutrient withdrawal
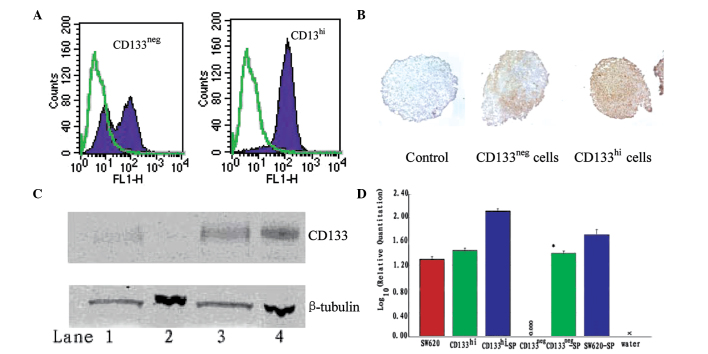
The expression of CD133 was stable in selected subpopulations under standard culture conditions (Fig. 2). To test whether this observation was also true under anchorage-independent conditions, we collected SW620 spheroids and monitored the CD133 expression by FACS analysis. Spheroids (30 per cell line) were collected, pooled and trypsinized to yield single cell suspensions. The cells were then stained with CD133 antibody for FACS analysis. There was significant CD133 re-expression in CD133neg cells grown as spheroids, with >50% of the cells converting to CD133hi; CD133 expression in the CD133hi spheroids remained stable (Fig. 3A). Control cells grown as monolayers and tested in parallel remained true to the original phenotype (data not shown), demonstrating that the re-expression of CD133 was induced by anchorage-independent growth. The re-expression of CD133 in the spheroids was confirmed by immunohistochemical staining and western blotting. A strong, patchy expression of the CD133 antigen was detected in the CD133neg cell spheroids (Fig. 3B), consistent with the distribution detected in primary colon tumors (9); the marked increase in CD133 protein expression was confirmed by immunoblotting (Fig. 3C). At the mRNA level, the level of the CD133 gene expression in the spheroids of CD133neg cells was significantly increased compared with that of cells from the monolayer cultures; a smaller but consistent increase in CD133 gene expression was also detected in CD133hi and parental SW620 cells grown as spheroids (Fig. 3D).
Figure 3.
CD133 expression in CD133neg and CD133hi SW620 cell spheroids. (A) Within each cell line, spheroids were pooled and disrupted into single cell suspensions. CD133 reactivity was assessed by FACS. Solid purple, CD133; green overlay, negative control. (B) Immunohistochemical staining of CD133 in spheroids. Brown, CD133; blue, nuclear stain; magnification ×200. (C) CD133 protein expression in spheroids compared with cell monolayers by western blot analysis. Upper panel, CD133 reactivity; lower panel, β-tubulin reactivity. Lane 1, CD133neg spheroids; lane 2, CD133neg cell monolayers; lane 3, CD133hi spheroids; lane 4, CD133hi monolayers. (D) Expression of CD133 by qRT-PCR, standardized to CD133 expression of CD133neg monolayer culture cells. *P<0.05 compared with CD133neg monolayer culture cells. SP, spheroid; FACS, fluorescence-activated cell sorting; qRT-PCR, quantitative real time-PCR.
The re-expression of CD133 in spheroids, where cells are packed tightly in a small volume of medium, raises the possibility that nutrient or oxygen deficiency regulates CD133 antigen expression. To test this hypothesis, monolayer cultures of CD133neg cells were exposed to low oxygen, low serum, low glucose or low glucose plus low serum. After 5 days, the CD133 expression of the cells under each culture condition was monitored by FACS analysis. Cells cultured without FBS and without glucose had low viability following 5 days in culture and were not analysed further, but all the other samples had high viability. A reduction in oxygen level to 2% did not affect CD133 antigen expression (data not shown). CD133neg cells grown in medium without glucose showed a marked shift in CD133 expression, whereas FBS withdrawal did not have a significant effect (Fig. 4). Neither glucose nor serum withdrawal had an effect on CD133 expression in CD133hi SW620 cells (Fig. 4).
Figure 4.
Effects of glucose and serum withdrawal on CD133 expression. CD133neg and CD133hi cells were plated in 6-well plates with normal medium. After one day the medium was changed to DMEM plus 5% FBS (Control), DMEM with no serum (No serum) or glucose-free DMEM with 5% FBS (No glucose). The cells were harvested five days later and CD133 expression was monitored by FACS. Solid purple, CD133 reactivity of CD133neg cells; green overlay, negative control; red overlay, CD133 reactivity of CD133hi cells. Median and peak fluorescence channels were determined using the Stat program in CellQuest. FBS, fetal bovine serum; FACS, fluorescence-activated cell sorting.
Discussion
CD133 expression characterizes immature cells in the lower part of the intestinal crypt and is suggested to be a hallmark of colorectal cancer stem cells (CRCS) in primary tumors (9–11). In the present study, we investigated the role of CD133 in a colon cancer cell line, SW620, in which CD133neg and CD133hi cells are present. CD133neg and CD133hi subpopulations were obtained by FACS sorting and used for functional studies. Following serial passages, the expression of CD133 (detected with the AC133 epitope antibody) was stable in the two subsets. Notably, the difference in the CD133 mRNA level between the CD133hi and CD133neg cells was confirmed at the RNA level by qRT-PCR (Fig. 3D). Doubts have been shed on the reliability of AC133 as an accurate measure of CD133 expression, with reported AC133 epitope loss upon CSC differentiation (21) and cell cycle-dependent variation of the CD133 epitope (23). Our results have shown that CD133 antibody staining of the AC133 epitope is correlated with CD133 mRNA expression in SW620 cells under different growth conditions and that CD133 expression is stable during the monolayer culture of SW620.
As a model for CD133 expression during the tumorigenesis of colon cancer cells, CD133neg and CD133hi cells were grown in spheroid cultures using the hanging drop method. This method is based on the ability of cells to aggregate and form homogeneous spheroids without the need for polymer scaffolds such as matrigel, polyglycolic acid or microporous supports (24). The spheroids are in vitro 3D tissue structures that mimic the in vivo tissue organization and microenvironment of tumors (25,26). We detected the significant re-expression of CD133 in CD133neg cells at the protein and mRNA level during spheroid formation. CD133 re-expression in CD133neg cells only occurred when the cells were grown as spheroid and not when passaged as adherent cells. There are two possible explanations for this phenomenon: a switch from cell/matrix to cell/cell interactions or reduced nutrient availability resulting from high local cell densities. Since there are relative nutrient and oxygen supply insufficiencies in the microenvironment of primary human tumors and xenograft cells (27), and considering that CD133 is a marker of bioenergetic stress in human glioma (28), we postulated that the CD133 re-expression observed in our study was correlated with the energy metabolism of the cells. Nutrient withdrawal markedly induced CD133 expression in CD133neg SW620 monolayers cultured without glucose. Our findings suggest that CD133 re-expression in the tumorigenesis of CD133neg SW620 cells may be mediated by the microenvironment of the tumor cells.
CD133 appears to have no obvious functional role in driving tumorigenesis, invasion and metastasis (29,30); however, CD133 is expressed in tumors, particularly at the invasive front or in conditions that mimic invasion (31). Our present findings support these results and complement them by showing the upregulation of CD133 in SW620 cells under the conditions of glucose deprivation.
In conclusion, our data indicate a plasticity of CD133 antigen expression, which makes it unsuitable as a marker of either colon stem cells or colon CSCs. It is likely that CD133 expression is maintained or induced during tumor formation and serves as a marker of cells undergoing metabolic stress responses. The potential role of CD133 in protecting cells from bioenergetic stress, and its possible link to the maintenance of an immature phenotype, requires further investigation.
Acknowledgements
We thank Francesca Walker, Huihua Zhang and Tony Burgess (Ludwig Institute, Melbourne) for assistance with all experiments.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells - perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 3.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 4.Yin AH, Miraglia S, Zanjani ED, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 5.Miraglia S, Godfrey W, Yin AH, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013–5021. [PubMed] [Google Scholar]
- 6.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 7.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 9.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 10.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 11.Todaro M, Alea MP, Di Stefano AB, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 13.Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Monzani E, Facchetti F, Galmozzi E, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43:935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 15.Curley MD, Therrien VA, Cummings CL, et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27:2875–2883. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 16.Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008;214:3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 18.Shmelkov SV, Butler JM, Hooper AT, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horst D, Kriegl L, Engel J, Kirchner T, Jung A. CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br J Cancer. 2008;99:1285–1289. doi: 10.1038/sj.bjc.6604664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima M, Ishii G, Atsumi N, Fujii S, Saito N, Ochiai A. Immunohistochemical detection of CD133 expression in colorectal cancer: A clinicopathological study. Cancer Sci. 2008;99:1578–1583. doi: 10.1111/j.1349-7006.2008.00849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemper K, Sprick MR, de Bree M, et al. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010;70:719–729. doi: 10.1158/0008-5472.CAN-09-1820. [DOI] [PubMed] [Google Scholar]
- 22.Leibovitz A, Stinson JC, McCombs WB, III, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 23.Jaksch M, Múnera J, Bajpai R, Terskikh A, Oshima RG. Cell cycle-dependent variation of a CD133 epitope in human embryonic stem cell, colon cancer, and melanoma cell lines. Cancer Res. 2008;68:7882–7886. doi: 10.1158/0008-5472.CAN-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotech Bioeng. 2003;83:173–180. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]
- 25.Dertinger H, Hulser DF. Intercellular communication in spheroids. Recent Results Cancer Res. 1984;95:67–83. doi: 10.1007/978-3-642-82340-4_4. [DOI] [PubMed] [Google Scholar]
- 26.Sutherland RM. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science. 1988;240:177–184. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 27.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 28.Griguer CE, Oliva CR, Gobin E, Marcorelles P, Benos DJ, Lancaster JR, Gillespie GY. CD133 is a marker of bioenergetic stress in human glioma. PLoS One. 2008;3:e3655. doi: 10.1371/journal.pone.0003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du L, Wang H, He L, Zhang J, Ni B, Wang X, Jin H, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 30.Horst D, Scheel SK, Liebmann S, Neumann J, Maatz S, Kirchner T, Jung A. The cancer stem cell marker CD133 has high prognostic impact but unknown functional relevance for the metastasis of human colon cancer. J Pathol. 2009;219:427–434. doi: 10.1002/path.2597. [DOI] [PubMed] [Google Scholar]
- 31.Kirkland SC. Type I collagen inhibits differentiation and promotes a stem cell-like phenotype in human colorectal carcinoma cells. Br J Cancer. 2009;101:320–326. doi: 10.1038/sj.bjc.6605143. [DOI] [PMC free article] [PubMed] [Google Scholar]