Abstract
Overexpression of Arabidopsis Reversion-To-ethylene Sensitivity1 (RTE1) results in whole-plant ethylene insensitivity dependent on the ethylene receptor gene Ethylene Response1 (ETR1). However, overexpression of the tomato RTE1 homologue Green Ripe (GR) delays fruit ripening but does not confer whole-plant ethylene insensitivity. It was decided to investigate whether aspects of ethylene-induced growth and development of the monocotyledonous model plant rice could be modulated by rice RTE1 homologues (OsRTH genes). Results from a cross-species complementation test in Arabidopsis showed that OsRTH1 overexpression complemented the rte1-2 loss-of-function mutation and conferred whole-plant ethylene insensitivity in an ETR1-dependent manner. In contrast, OsRTH2 and OsRTH3 overexpression did not complement rte1-2 or confer ethylene insensitivity. In rice, OsRTH1 overexpression substantially prevented ethylene-induced alterations in growth and development, including leaf senescence, seedling leaf elongation and development, coleoptile elongation or curvature, and adventitious root development. Results of subcellular localizations of OsRTHs, each fused with the green fluorescent protein, in onion epidermal cells suggested that the three OsRTHs were predominantly localized to the Golgi. OsRTH1 may be an RTE1 orthologue of rice and modulate rice ethylene responses. The possible roles of auxins and gibberellins in the ethylene-induced alterations in growth were evaluated and the biological significance of ethylene in the early stage of rice seedling growth is discussed.
Keywords: Arabidopsis, coleoptile curvature, ethylene, OsRTH1, rice, RTE1
Introduction
Ethylene is a gaseous plant hormone, and Arabidopsis has been used as a eudicotyledonous model plant for study of ethylene signal transduction. Major components of the signalling pathway have been identified (Guzman and Ecker, 1990; Chao et al., 1997; Hua and Meyerowitz, 1998; Johnson and Ecker, 1998; Alonso et al., 1999; Wang et al., 2002, 2003; Guo and Ecker, 2003; Resnick et al., 2008). Ethylene signalling components were identified and functionally demonstrated in tomato, which indicated conserved signalling machinery across dicot species (Wilkinson et al., 1995; Tieman and Klee, 1999; Tieman et al., 2000, 2001; Adams-Phillips et al., 2004; Zhong et al., 2008).
At the top of the ethylene signal transduction hierarchy is a small family of ethylene receptors. In the absence of ethylene, ethylene receptors constitutively suppress responses that are ethylene inducible. Ethylene binding prevents the suppression of ethylene receptors, and responses can proceed (Klee, 2004). Arabidopsis has five ethylene receptors: ethylene response1 (ETR1), ETR2, ethylene response sensor1 (ERS1), ERS2, and ethylene insensitivity4 (EIN4) (Bleecker and Kende, 2000; Hall et al., 2007). A suppressor screen of the dominant ethylene-insensitive etr1-2 mutation revealed Reversion-to-Ethylene Sensitivity 1 (RTE1) as a positive regulator of the ethylene receptor gene ETR1. Arabidopsis RTE1 is predominantly localized to the Golgi and can physically associate with ETR1 at the endoplasmic reticulum (ER) (Resnick et al., 2006; Zhou et al., 2007; Dong et al., 2008, 2010). RTE1 overexpression can promote ETR1 receptor signalling and results in whole-plant ethylene insensitivity throughout development (Zhou et al., 2007). Green Ripe (GR) is an RTE1 homologue of tomato, and elevated GR expression delays fruit ripening but does not confer whole-plant ethylene insensitivity (Barry and Giovannoni, 2006). The ethylene signalling machinery may be highly conserved in higher plants but may be differentially regulated in different plant species.
Current knowledge of the role of ethylene in plants comes mainly from studies of dicotyledonous plant species. Rice is a monocot and a major crop in Asia; two major subspecies (japonica and indica), which probably diverged >0.44 million years ago, are widely cultivated (Ma and Bennetzen, 2004). The roles of ethylene in rice were primarily revealed in submergence responses of indica cultivars. Upon flooding, submerged deep-water rice plants produce ethylene, which induces biosynthesis of gibberellins (GAs) and degradation of abscisic acid (ABA), thus favouring internodal elongation so that submerged shoots can grow above the water surface to obtain oxygen. Accompanying internodal elongation is ethylene-induced cell death, which facilitates the emergence of adventitious roots from the node of stem sections (Metraux and Kende, 1983; Raskin and Kende, 1984; Satler and Kende, 1985; Saika et al., 2007; Fukao and Bailey-Serres, 2008a). Two quantitative trait loci (QTLs) responsible for the internodal elongation of deep-water rice have been cloned: these are SNORKEL genes (SK1 and SK2) encoding proteins of the ethylene response factor (ERF) family (Hattori et al., 2009). Of note, SK1 and SK2 are ethylene inducible but their functions in internodal elongation depend on GAs. Another type of survival strategy for rice is avoiding energy consumption. Submergence 1A (Sub1A), encoding an ERF protein, is present in a few indica cultivars and confers tolerance to complete submergence by restricting GA responses. Although Sub1A expression is ethylene inducible, it can function independently of ethylene actions (Fukao et al., 2006; Xu et al., 2006; Fukao and Bailey-Serres, 2008b ).
A recent study showed the rice ethylene receptor homologue OsETR2 with a role in promoting flowering of the japonica cultivar Zhonghua 11 (ZH11) (Wuriyanghan et al., 2009). Two aspects of ethylene-induced growth alteration, seedling height and primary root elongation, were modulated to different degrees in japonica cultivars overexpressing OsETR2, OsEIN2 (Ethylene-Insensitive 2), and OsEIN3 by chemical treatments that replaced ethylene or eliminated ethylene production (Jun et al., 2004; Mao et al., 2006; Wuriyanghan et al., 2009). Other aspects of ethylene response that alter rice seedling growth and development have yet to be studied. Of note, primary root growth and plant height can be modulated by auxins and GAs. Given that ethylene modulates Arabidopsis primary root growth and apical hook formation by modulating the function of DELLA protein (a GA response repressor) and auxin biosynthesis and polar transport, whether ethylene-induced alterations in rice growth are affected by the corresponding plant hormones needs to be investigated (Achard et al., 2003; Stepanova et al., 2005).
It was decided to investigate whether any OsRTH genes are functionally conserved in ethylene signalling in rice and whether OsRTH overexpression may address the functional significance of ethylene-dependent alterations in growth. The ethylene antagonist 1-methylcyclopropene (1-MCP) was used to evaluate ethylene responses modulated by OsRTH1 overexpression in rice. OsRTH1 overexpression conferred ethylene insensitivity to an extent similar to or even greater than that with 1-MCP treatment. The possible synergistic effects of ethylene with other plant hormones, auxins and GAs, on the ethylene-induced alterations in growth were evaluated. OsRTH1 may be functionally conserved in ethylene signalling, and the biological significance of ethylene in rice seedling growth is discussed.
Materials and methods
Plant materials and gas treatments
Rice (Oryza sativa L. ssp. japonica cv. Zhonghua 11, designated ZH11) was used throughout this study. Ethylene concentrations were measured by gas chromatography (GC) using a flame ionization detector (FID). 1-MCP (Rohm & Haas China, Beijing) was released in water as per the manufacturer’s instructions, and the concentration was determined by GC/FID. Unless specified, 5 μl l−1 1-MCP and 100 μl l−1 ethylene were applied to rice. Growth conditions and ethylene treatment for Arabidopsis were as described (Xie et al., 2006). For analysis of the triple-response phenotype in Arabidopsis seedlings, 20 μl l−1 ethylene was applied.
Laser scanning confocol microscopy (LSCM)
For subcellular localization study of fluorescence protein fusions, corresponding transgenes were delivered into onion epidermal cells by particle bombardment as described (Zhou et al., 2007). LSCM involved the Olympus FluoView FV1000 and FV10-ASW1.7 Viewer for data acquisition at the Core Facility Center of the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences.
Clones and transgenes
OsRTH1, OsRTH2, and OsRTH3 were cloned by PCR. For green fluorescent protein (GFP) fusion, corresponding clones were released by BamHI and ligated with GFP on pRTL2. The primers were OsRTH1-F (5′-CCGAATTCATGGCACCAAACAAAATTTCC TC-3′) and OsRTH1-R (5′-CCGGATCC TCAGCACACTAGATCCTTCATG-3′); OsRTH2-F (5′-ATGGATCCATGGAGGTTGAAGCTGCTTG-3′) and OsRTH2-R (5′-ATGGATCCTCAGCAGACCAAGCCCTTGA-3′); and OsRTH3-F (5′-ATGGATCCATGGAAACCGACAGAAGCCA-3′); and OsRTH3-R (5′-ATGGATCCCTACAACTCTACAAGGCTCT-3′).
Real-time quantitative reverse transcription-PCR (qRT-PCR)
qRT-PCR involved use of the StepOne real-time PCR system (Applied Biosystems). Total RNA was isolated by use of TaKaRa RNAiso Plus. cDNA was synthesized from mRNA by use of the PrimeScript RT reagent Kit and TaKaRa SYBR Premix Ex Taq. Ubiquitin and actin were used as the internal calibrator for Arabidopsis and rice, respectively. Melting curve analysis for each primer set suggested no non-specific priming (data not shown). Each measurement was repeated three times with three independent biological samples (n=3×3). The ubiquitin primers for qRT-PCR were as described by Zhang and Wen (2010) or the following: Sub 1C-F (5′-CTGCTCCGACGACCTGAT-3′) and Sub 1C-R (5′-TTAGGCGAGTCGCATGTCAA-3′); ADH2-F (5′-CCCATCCCTGGATTCAGGT-3′) and ADH2-R (5′-CACGAGGTAGGTGCTGATTGA-3′); SC129-F (5′-TGACGGTGTACGGTCCGAT-3′) and SC129-R (5′-TCGGCGTACTGGTCACAGAT-3′); OsActin-F(5′-GAAGATCACTGCCTTGCTCC-3′) and OsActin-R (5′-CGATAACAGCTCCTCTTGGC-3′); and OsRTH1-F (5′-ACTCATTTGTGGCAAACTGCTT-3') and OsRTH1-R (5′-ATCCTTCATGCAGTATACAGCA-3′).
Leaf senescence test and rice seedling growth
The rice leaf senescence test was as described (Kao and Yang, 1983) and involved 100 μl l−1ethylene. Chlorophyll content was measured 4 d after the treatment (Zhang and Wen, 2010). Rice seedlings were grown in an environmentally controlled phytotron (28±1 °C, 12 h/12 h day and night, 50–70% humidity, and average illumination at 482 μmol m−2 s−1).
Results
Sequence and gene structure of evolutionarily representative plant RTHs
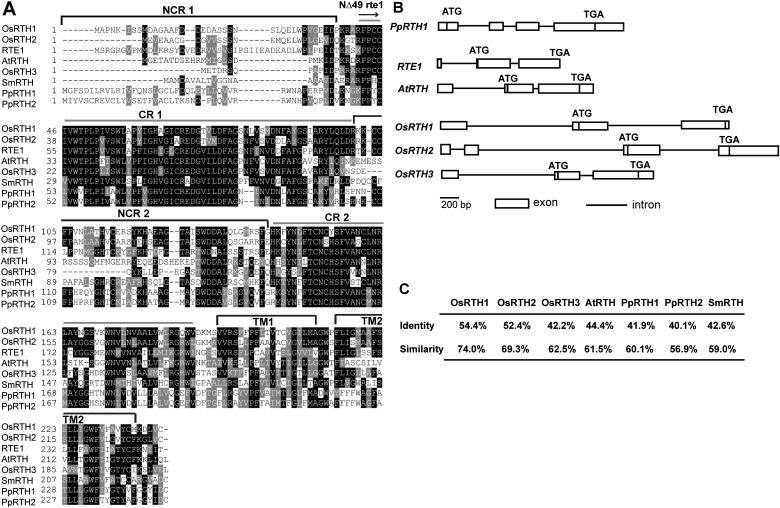
RTHs from other evolutionarily representative plant species could be retrieved from phytozome (http://www.phytozome.net/); their sequences are not shown in this study. The RTH sequence and gene structure of evolutionarily representative plant species, including Physcomitrella patens, Selaginella moellendorffii, Arabidopsis, and rice were compared (Banks et al., 2011).
In addition to the putative, C-terminal transmembrane domains, these RTHs have two conserved regions (CR1 and CR2) and two non-conserved regions (NCR1 and NCR2). NCR1 may not have a role in RTE1 function because the NCR1-lacking NΔ49rte1 isoform is still functional in ethylene signalling (Zhou et al., 2007) (Fig. 1A). The structures of AtRTH, OsRTH1 (Os01g0711600), and OsRTH3 (Os03G0799500) are similar to that of RTE1. They have three exons and two introns. The putative start codon is located at the beginning of exon 2 and the stop codon is located at the end of exon 3. PpRTH1 and OsRTH2 (Os05g0539800) are structurally distinct from RTE1 and have three introns and four exons (Fig. 1B). Because of the lack of genomic sequence information, the gene structures for the moss PpRTH2 and lycophyte SmRTH could not be compared. On pairwise sequence alignment of RTHs and RTE1 (http://www.ebi.ac.uk/Tools/psa/emboss_needle/), RTE1 had the highest sequence identity with and similarity to OsRTH1, and the lowest with AtRTH, OsRTH3, and RTHs of lower plants (Fig. 1C).
Fig. 1.
RTH sequence and gene structure. (A) Sequence alignment of evolutionarily representative plant RTHs. NCR, non-conserved region; CR, conserved region; TM, transmembrane domain; NΔ49rte1, the rte1 isoform lacking the N-terminus; the arrow indicates the start of NΔ49 rte1. (B) RTH gene structure. Rectangle indicate the exons and lines indicate the introns. (C) Sequence identity and similarity of plant RTHs compared with RTE1. Os, rice (Oryza sativa); At, Arabidopsis; Sm, Selaginella; Pp, Physcomitrella.
OsRTH1 and RTE1 had the highest sequence identity and similarity in gene structure. Although OsRTH2 and RTE1 showed high sequence homology, their gene structures differed. OsRTH3 and AtRTH were similar to RTE1 in gene structure but had relatively poor protein sequence identity and similarity. These results agree with a previous phylogenetic analysis showing OsRTH3 and AtRTH in the same clade, and RTE1, OsRTH1, and OsRTH2 in the same clade (Barry and Giovannoni, 2006). Thus, OsRTH1 probably has a role in ethylene signalling in rice.
Cross-species complementation test of OsRTHs in Arabidopsis
RTE1 overexpression leads to ethylene insensitivity in Arabidopsis, which depends on ETR1 (Resnick et al., 2006; Zhou et al., 2007). Cross-species complementation testing was used to determine whether any OsRTH genes are functionally conserved in ethylene signalling in Arabidopsis.
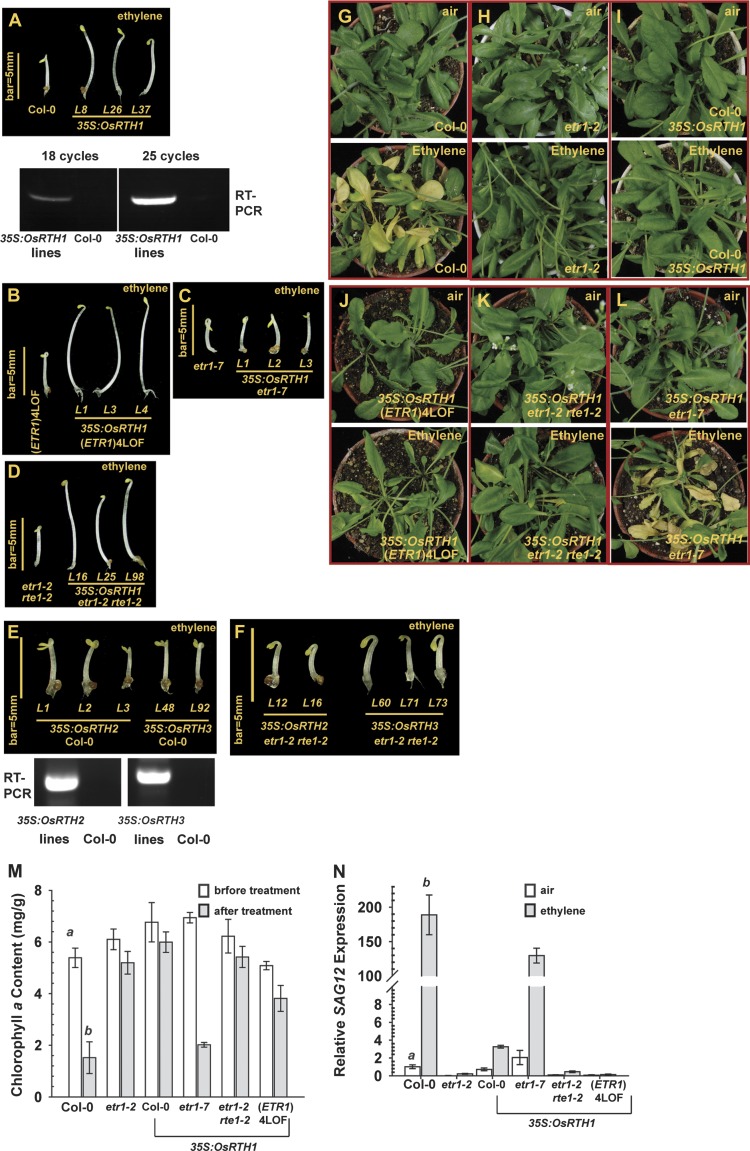
Ethylene treatment promotes the apical hook curvature and inhibits the hypocotyl and primary root growth of Arabidopsis etiolated seedlings, namely the seedling triple-response phenotype. With ethylene treatment, the seedling triple-response phenotype was prevented in wild-type (Col-0) seedlings expressing 35S:OsRTH1. Overexpression of the transgene was confirmed by RT-PCR (Fig. 2A). ETR1 is the only wild-type ethylene receptor gene in the receptor quadruple mutant ers1-2 etr2 ein4 ers2, designated (ETR1)4LOF (LOF indicating loss of function), which shows the seedling triple-response phenotype with ethylene treatment (Liu et al., 2010; Liu and Wen, 2012). 35S:OsRTH1 expression substantially rescued the hypocotyl growth inhibition of (ETR1)4LOF (Fig. 2B). As expected, OsRTH1 overexpression did not rescue the growth inhibition of ethylene-treated ETR1-defective etr1-7, and both etr1-7 and the transformation lines showed a seedling triple-response phenotype (Fig. 2C). Ethylene insensitivity conferred by etr1-2 depends on RTE1 (Resnick et al., 2006). With ethylene, etr1-2 rte1-2 seedlings showed a short seedling hypocotyl, and 35S:OsRTH1 expression rescued the growth inhibition (Fig. 2D).
Fig. 2.
Functional analyses of plant RTHs in Arabidopsis. The seedling triple-response phenotype of ethylene-grown wild type (Col-0) and 35S:OsRTH1 transformation lines (A), (ETR1)4LOF and a mutant expressing 35S:OsRTH1 (B), etr1-7 and a mutant expressing 35S:OsRTH1 (C), and etr1-2 rte1-2 and a mutant expressing 35S:OsRTH1 (D). (E) The seedling triple-response phenotype of the wild type (Col-0) expressing 35S:OsRTH2 and 35S:OsRTH3. (F) Seedling triple-response phenotype of etr1-2 rte1-2 expressing 35S:OsRTH2 and OsRTH3. Leaf senescence phenotype of the wild type (Col-0) (G) and ethylene-insensitive etr1-2 (H); phenotype of wild type (Col-0), (ETR1)4LOF (J), etr1-2 rte1-2 (K), and etr1-7 (L) expressing 35S:OsRTH1. Air and ethylene indicate the phenotype of the same plants before and after the treatment, respectively. Chlorophyll a measurement (M) and SAG12 expression (N) of the wild type (Col-0), etr1-2, and 35S:OsRTH1 transformants in the corresponding mutation background as indicated. Error bars indicate the standard error (SE) for the means of five measurements. RT-PCR, analysis of the mRNA level of corresponding transgenes at the translational level. a (air) and b (ethylene) indicate a statistically significant difference (α=0.01) between the wild type and mutant or transformation lines.
Unlike OsRTH1, neither OsRTH2 nor OsRTH3 overexpression prevented the ethylene-induced growth inhibition in wild-type (Col-0) seedlings. Overexpression of these transgenes was confirmed by RT-PCR (Fig. 2E). Neither transgene could complement the rte1-2 loss-of-function mutation, and the etr1-2 rte1-2 transformation mutant, which expressed 35S:OsRTH2 or 35S:OsRTH3, showed the ethylene-induced seedling triple-response phenotype (Fig. 2F).
The effect of OsRTH1 overexpression on other aspects of the ethylene response was examined. Wild-type (Col-0) but not ethylene-insensitive etr1-2 rosettes showed the leaf senescence phenotype with ethylene treatment (20 μl l−1) (Fig. 2G, H). The expression of 35S:OsRTH1 prevented the ethylene-induced leaf senescence phenotype in the wild type, (ETR1)4LOF, and etr1-2 rte1-2, but not in etr1-7 (Fig. 2I–L).
The degrees of leaf senescence were quantified by measuring the chlorophyll a content. The chlorophyll a content was greatly decreased with ethylene treatment in wild-type (Col-0) leaves and slightly decreased in etr1-2 leaves. With the 35S:OsRTH1 transgene, the chlorophyll a content was slightly reduced in wild-type, etr1-2 rte1-2, and (ETR1)4LOF leaves, and greatly reduced in etr1-7 leaves (Fig. 2M). Senescence Associated Gene12 (SAG12) expression is specifically associated with leaf senescence progression (Noh and Amasino, 1999; Grbić, 2003). Progression of leaf senescence was quantified by measuring SAG12 expression. With ethylene treatment for 48 h, the SAG12 level in wild-type leaves was substantially increased, up to 190-fold. SAG12 expression in etr1-2 leaves was extremely low and not induced. Wild-type plants expressing 35S:OsRTH1 showed relatively minor SAG12 induction, <4-fold. The SAG12 level was highly induced, up to 130-fold, in etr1-7 expressing 35S:OsRTH1. etr1-2 rte1-2 and (ETR1)4LOF expressing 35S:OsRTH1 did not show SAG12 induction (Fig. 2N).
Thus, OsRTH1 may be functionally conserved in regulating ETR1 receptor signalling when heterogeneously expressed in Arabidopsis. The other two OsRTH genes examined were unable to affect ethylene responses at the transcriptional level.
GFP-fused OsRTHs are associated with the Golgi
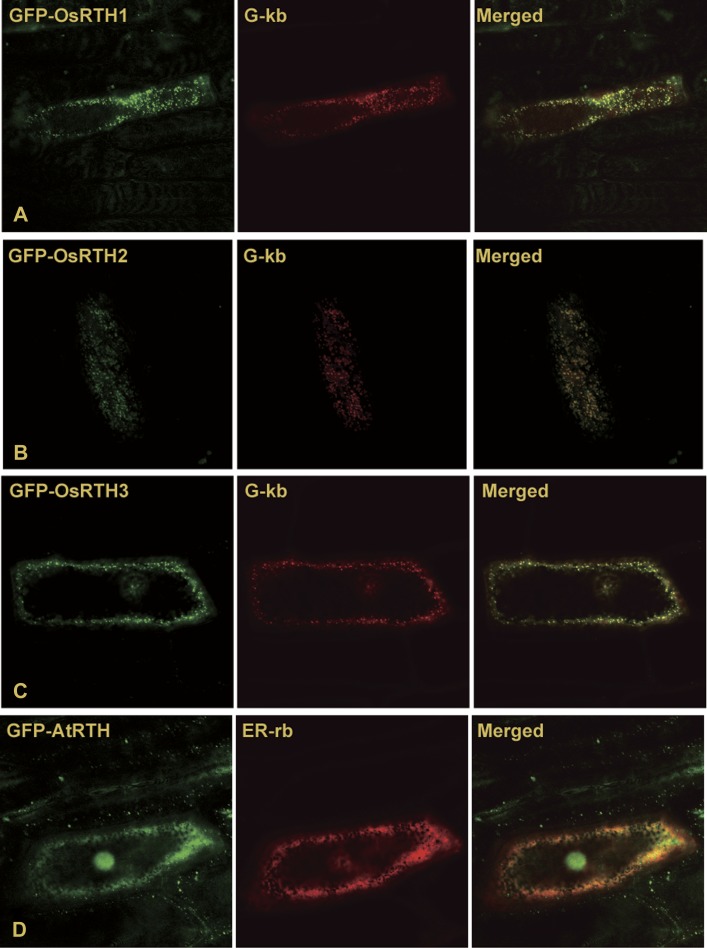
GFP-tagged RTE1 is predominantly localized to the Golgi apparatus when ectopically expressed in onion epidermal cells and in Arabidopsis (Zhou et al., 2007). Therefore, ectopically expressed RTE1 and its orthologues can correctly localize to the Golgi apparatus in cells of dicots (Arabidopsis) and monocots (onion). The subcellular localization of OsRTHs in onion epidermal cells was evaluated.
GFP was individually fused to OsRTH1, OsRTH2, aOsRTH3, and AtRTH. The resulting clones were co-expressed with a fluorescence protein-fused organelle marker in onion epidermal cells. GFP–OsRTHs co-localized with the Golgi-mCherry marker G-rk (Nelson et al., 2007) (Fig. 3A–C). AtRTH is the only RTE1 homologue in Arabidopsis and is believed not to be functional in ethylene signalling (Rivarola et al., 2009). The subcellular localization of AtRTH was evaluated to address whether the RTE1 homologue may localize in compartments distinct from the Golgi apparatus. GFP–AtRTH co-localized with the ER-mCherry marker ER-rb (Nelson et al., 2007) and in the nucleus (Fig. 3D). The subcellular localization of GFP–OsRTHs in rice cells was examined, but the fluorescence of GFP–OsRTHs was found to be extremely poor in the resulting transformation rice lines and it was not possile to determine their localizations (data not shown).
Fig. 3.
Subcellular localizations of GFP–RTHs in onion epidermal cells. Subcellular localizations of GFP–OsRTH1 (A), GFP–RTH2 (B), GFP–RTH3 (C), and GFP–AtRTH (E) determined by laser scanning confocol miscroscopy in onion epidermal cells co-expressing G-kb, the Golgi marker, and ER-rb, the ER marker.
The data suggest that AtRTH1 localizes to the ER and within the nucleus. The possibility that AtRTH may localize to other organelles if expressed in Arabidopsis cells cannot be excluded. Nevertheless, this possibility does not affect AtRTH being less likely to be associated with the Golgi apparatus. In contrast, OsRTHs may all localize predominantly in the Golgi apparatus.
OsRTH1 overexpression prevents ethylene-induced rice leaf senescence
Complementation testing revealed that OsRTH1 is an RTE1 orthologue. Elevated RTE1 expression results in a hypermorph that causes ethylene insensitivity. Next experiments were carried out to examine whether elevated OsRTH1 expression may confer ethylene insensitivity in rice. OsRTH1 overexpression and treatment with the ethylene antagonist 1-MCP were compared in terms of degree of ethylene response.
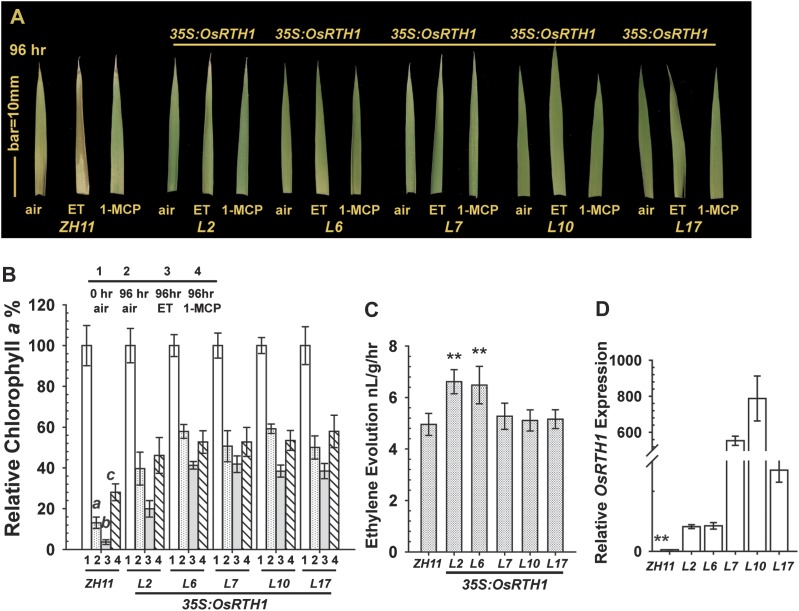
Ethylene can promote rice leaf senescence (Kao and Yang, 1983). Here it was found that detached leaf fragments of wild-type rice (ZH11) showed the leaf senescence phenotype, and the chlorophyll a content decreased to 13% after 4 d (96 h) in air. 1-MCP treatment attenuated the leaf senescence phenotype, and the chlorophyll a content was ∼28%. ZH11 expressing 35S:OsRTH1 did not show the leaf senescence phenotype, and the chlorophyll a content was relatively high (39–59%), regardless of 1-MCP treatment (Fig. 4A, B). With 96 h ethylene treatment, leaf fragments of ZH11 were completely yellow, and the chlorophyll a content was only 3.6%. 35S:OsRTH1 transformation lines did not show the leaf senescence phenotype, and the chlorophyll a content was still high after ethylene treatment, with the chlorophyll a content lower in the transformation line L2 than in the other lines (19.4% versus ∼40%) (Fig. 4A, B).
Fig. 4.
Rice leaf senescence test. (A) Senescence phenotypes of rice leaf in air, ethylene (ET), and 1-MCP. (B) Chlorophyll a content (%) relative to that before treatment (0 h). Data are the mean ±SD of five biological repeats. (C) Ethylene evolution of ZH11 and transformation rice lines. (D) Relative OsRTH1 expression in ZH11 and transformation rice lines (L). Data are the mean ±SD of each measurement. Ethylene, 100 μl l−1; 1-MCP, 5 μl l−1. a, b, and c, statistically significant difference (Fisher’s LSD, α=0.01) between the wild type (ZH11) and transformation rice lines for air (a), ethylene (ET, b), and 1-MCP (c) treatments. **Significant difference (Fisher’s LSD, α=0.01) among ZH11 and transformation rice lines.
The ethylene evolution in ZH11 and 35S:OsRTH1 lines was examined to evaluate whether the degree of leaf senescence in transformation lines was affected by ethylene production. The ethylene evolution of ZH11, L7, L10, and L17 was similar to but slightly lower than that of L2 and L6 (Fig. 4C). To confirm that prevention of the ethylene-induced senescence phenotype resulted from OsRTH1 overexpression, the mRNA level of OsRTH1 in each transformation rice line was compared with that in ZH11. OsRTH1 was overexpressed in each transformation line, and OsRTH1 expression was higher in lines L7 and L10 than in L2, L6, and L17 (Fig. 4D). Thus, the OsRTH1 level in transformation lines was sufficient to suppress the ethylene-induced leaf senescence to a great extent.
The inhibition of rice leaf senescence progression was stronger with OsRTH1 overexpression than with 1-MCP treatment. The ethylene evolution of air-grown 35S:OsRTH1 lines was not reduced and the delay in leaf senescence was not due to alterations in endogenous ethylene production.
OsRTH1 overexpression attenuates the expression of ethylene-inducible genes
Because elevated OsRTH1 overexpression efficiently prevented ethylene-induced leaf senescence, whether OsRTH1 overexpression could repress the expression of various ethylene-inducible genes was next examined.
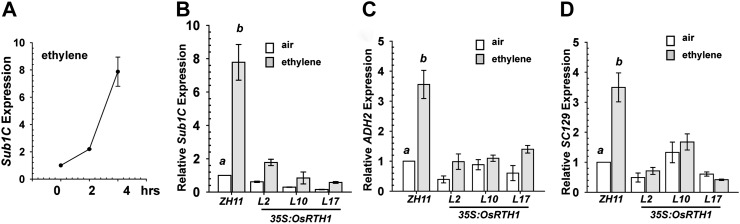
At the submergence locus, Sub1C is ethylene inducible in japonica and indica cultivars (Fukao et al., 2006). A 4 h ethylene treatment was sufficient to elevate Sub1C expression to nearly 8-fold in the wild-type rice (ZH11) (Fig. 5A, B). In contrast, Sub1C expression was highly attenuated in 35S:OsRTH1 lines with ethylene treatment (Fig. 5B). ADH2 encodes alcohol dehydrogenase2, and its expression can be induced by ethylene (Fukao et al., 2006). With ethylene treatment, ADH2 expression was ∼3-fold higher in ZH11 than in 35S:OsRTH1 lines (Fig. 5C). SC129 (AK104680), possibly encoding a glutathione S-transferase, is ethylene inducible (Jun et al., 2004). It was found SC129 expression in ZH11 was induced 3.5-fold by ethylene, with its induction weak in 35S:OsRTH1 lines (Fig. 5D).
Fig. 5.
Gene expression analyses. (A) Kinetics of Sub1C induction by ethylene treatment. Expression of Sub1C (B), ADH2 (C), and SC129 (D) of ZH11 and 35S:OsRTH1 transformation lines in air (white bars) and ethylene (grey bars). Data are the mean ±SE of three independent measurements with three repeats (n=3×3). a (air) and b (ethylene): significant difference (Fisher’s LSD, α=0.01) between ZH11 and transformation rice lines.
OsRTH1 overexpression prevents ethylene-induced alterations in coleoptile growth and development
Coleoptiles protect rice seedlings and are closed in darkness; ethylene promotes the coleoptile elongation of etiolated rice seedlings (Ku et al., 1970; Satler and Kende, 1985). Experiments were carried out to examine whether OsRTH1 overexpression can prevent ethylene-induced coleoptile elongation.
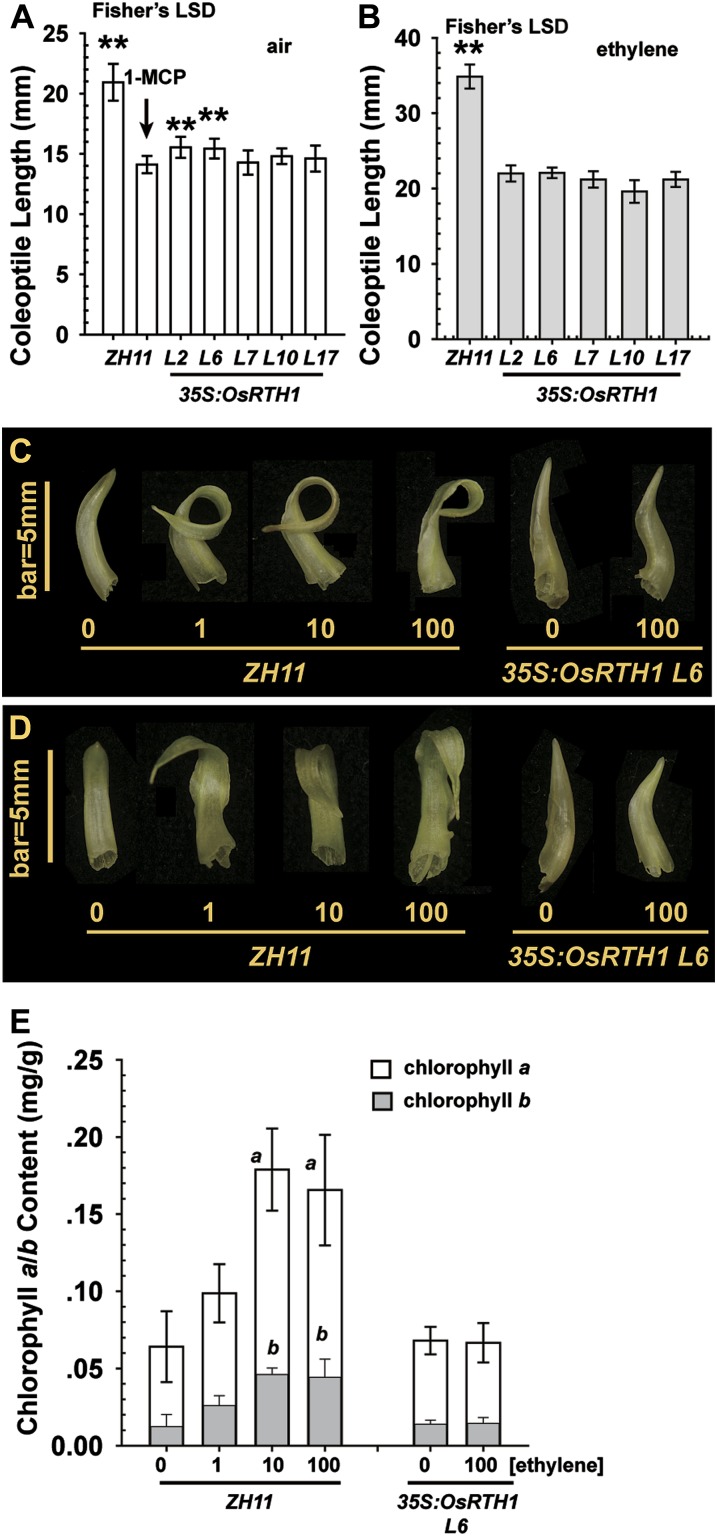
Etiolated rice seedlings with a coleoptile length of 2 mm were subjected to air, 1-MCP, or ethylene treatment in the dark for 5 d, and coleoptile length was measured. In air, the coleoptile length was 5–6 mm longer in ZH11 than in the transformation lines. As expected, 1-MCP had a similar effect to the 35S:OsRTH1 transgene in preventing coleoptile elongation, and coleoptile length was similar in 1-MCP-treated ZH11 and the transformation lines (Fig. 6A). Ethylene treatment substantially promoted coleoptile elongation, and the length was much longer, by 12–15 mm, in ZH11 than in the transformation lines (Fig. 6B). Therefore, OsRTH1 overexpression effectively prevented ethylene-induced coleoptile elongation in dark-grown rice seedlings.
Fig. 6.
Seedling coleoptile growth. Measurement of the coleoptile length of etiolated rice seedlings grown in air (A) and ethylene (B). The coleoptile phenotype of light-grown ZH11 and 35S:OsRTH1 (line L6) seedlings in air and ethylene, viewed from the side (C) and back (D). (E) Chlorophyll content of coleoptiles of light-grown ZH11 and 35S:OsRTH1 line L6. Numbers in (C), (D), and on the x-axis (E) indicate the ethylene concentration (μl l−1). Data are the mean ±SD for (A) and (B), n ≥ 15, and mean ± SE for (E) of 3–5 measurements. **Significant difference (Fisher’s LSD, α=0.01) between 1-MCP-treated ZH11 and air-grown ZH11 and transformation rice lines (A), or ethylene-treated ZH11 and transformation rice lines (B). a (chlorophyll a) and b (chlorophyll b) indicate identical chlorophyll contents between ZH11 treated with 10 μl l−1 and 100 μl l−1 ethylene (Student’s t-test, P > 0.05).
In addition, experiments were conducted to determine whether ethylene had any effect on the growth and development of light-grown seedling coleoptiles. Etiolated rice seedlings with a coleoptile length of 2–3 mm were transferred to growth under light for 4 d with various ethylene concentrations. Ethylene-treated ZH11 seedlings showed exaggerated coleoptile curvature, with minimal curvature of air-grown ZH11. An amount of 1 μl l−1 ethylene was sufficient to produce the curvature phenotype. In contrast, seedlings of the 35S:OsRTH1 transformation line L6 showed a relatively straight coleoptile, regardless of ethylene treatment (Fig. 6C). Consistent with the phenotype of the 35S:OsRTH1 transformation line L6, the other OsRTH1 overexpression lines produced a short and relatively straight coleoptile when grown under light, regardless of ethylene treatment (Supplementary Fig. S1 available at JXB online). Therefore, ethylene promoted the coleoptile curvature of light-grown seedlings.
The abaxial side of the coleoptile of ethylene-grown ZH11 was greener than that of air-grown ZH11. Ethylene treatment had little effect on the greening of the abaxial side of transformation coleoptiles (Fig. 6D). Therefore, the chlorophyll content was measured in ZH11 and 35S:OsRTH1 transformation line L6 coleoptiles. Ethylene treatment increased the chlorophyll content in ZH11 but not in 35S:OsRTH1 line L6 coleoptiles. The chlorophyll a/b content in ZH11 coleoptiles was identical with 10 μl l−1 and 100 μl l−1 ethylene treatment (Student’s t-test; P > 0.01). The chlorophyll content was identical in coleoptiles of air- and ethylene-grown 35S:OsRTH1 L6 and air-grown ZH11 (F test; P=0.3515 and F=1.1947 < Fcrit=4.459) (Fig. 6E). Thus, ethylene could produce coleoptile curvature and greening in light-grown seedlings. The degree of greening was associated with the chlorophyll content. Elevated OsRTH1 expression antagonized ethylene effects on coleoptile growth and development.
OsRTH1 overexpression inhibits ethylene-induced leaf elongation and development
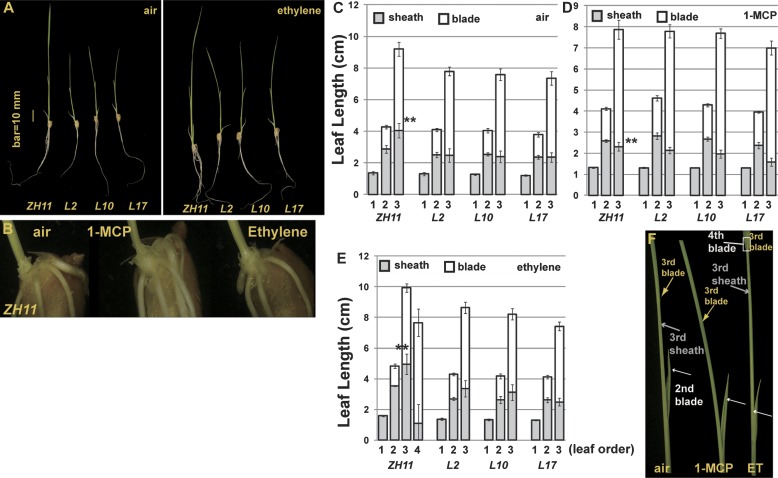
1-Aminocyclopropane-1-carboxylic acid (ACC) is the immediate ethylene biosynthesis precursor. ACC-treated rice seedlings are taller than untreated seedlings (Jun et al., 2004; Wuriyanghan et al., 2009). Here the effect of ethylene on rice seedling growth was evaluated. Germinating rice seedlings with a coleoptile of 2–3 mm were cultured hydroponically for 7 d in an environmentally controlled phytotron. ZH11 was taller than 35S:OsRTH1 transformation lines regardless of ethylene treatment (Fig. 7A). Plant height can be determined by the internode and leaf length. Experimnents were conducted to determine whether the internodes elongated with ethylene treatment. By peeling off outer leaves, we showed that the last leaf was attached to the grain and found no sign of internodal elongation regardless of ethylene or 1-MCP treatment (Fig. 7B).
Fig. 7.
Ethylene promotes the leaf growth of ZH11. (A) Seedling phenotype of ZH11 and 35S:OsRTH1 transformation lines in air and ethylene. (B) The internode is not visible in rice seedlings with the outer leaves removed. Length of individual leaves of rice seedlings grown in air (C), 1-MCP (D), and ethylene (E); the x-axis indicates the leaf order. (F) Shoot phenotype of ZH11 rice seedlings grown in air, 1-MCP, and ethylene (ET). The second blade is indicated by a white arrow; the third sheath by a grey arrow; the third blade by a yellow arrow; and the fourth leaf by a white box. **Significant difference (Fisher’s LSD, α=0.01) comparing the third-leaf sheath length between ZH11 and transformation rice lines.
The next aim was to examine whether leaf elongation resulted in taller seedlings. The rice leaf is composed of a leaf sheath and leaf blade, except that the first leaf does not have a blade. The first and second leaves of ZH11 and transformation lines were similar in length; however, the sheath of the third leaf was longer, by ∼1 cm (or 10%), in ZH11 than in the 35S:OsRTH1 transformation lines (Fig. 7C). 1-MCP treatment substantially inhibited growth of the third leaf of ZH11, and the length of each leaf was similar to that of air-grown 35S:OsRTH1 lines. As expected, 1-MCP did not affect the leaf length of 35S:OsRTH1 lines (Fig. 7C, D). Air-grown ZH11 had three leaves at day 7. Of note, with ethylene treatment, ZH11 seedlings had four leaves, whereas 35S:OsRTH1 seedlings had three. The third leaf sheath of ethylene-treated ZH11 was ∼2 cm (or 25%) longer than that of 35S:OsRTH1 lines (Fig. 7E, 7F). Therefore, ethylene promoted the elongation of the third sheath and the development of the fourth leaf at the stage examined. The third sheath elongation was responsible for the tall ethylene-treated ZH11.
1-MCP treatment and OsRTH1 overexpression had the same inhibitory effect on leaf growth and elongation promoted by ethylene. Ethylene may promote the leaf development so that ethylene-grown ZH11 seedlings had one more leaf at the stage examined.
Ethylene and GAs have different effects on seedling leaf elongation
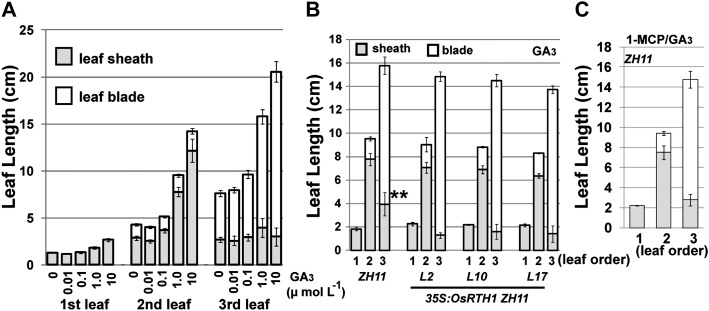
Ethylene can induce endogenous GA biosynthesis in indica deep-water cultivars (Metraux and Kende, 1983; Raskin and Kende, 1984; Saika et al., 2007; Jackson, 2008). Whether ethylene-induced leaf elongation of ZH11, a japonica cultivar, could be due to elevated GAs was next examined.
When supplemented in hydroponic culture, GA3 had little effect on leaf growth at concentrations <0.1 μmol l−1 (Fig. 8A). GA3 of 1.0 μmol−1 and 10 μmol l−1 substantially promoted the elongation of the second sheath and third blade (Fig. 8A). With GA3 at 1.0 μmol l−1, 35S:OsRTH1 transformation lines and 1-MCP-treated ZH11 showed a leaf growth pattern similar to that of GA3-treated ZH11, except that the third sheath was shorter, by ∼2–3 cm, for OsRTH1 overexpression lines and 1-MCP-treated ZH11 than ZH11 in the presence of GA3 treatment (Fig. 8B, C).
Fig. 8.
Effect of gibberellins on rice seedling growth. (A) Leaf length of ZH11 seedlings grown with GA3. (B) Leaf length of ZH11 and 35S:OsRTH1 transformation lines with 1 μM GA3. The x-axis indicates leaf order. (C) Leaf length of ZH11 seedlings with GA3 and 1-MCP. The x-axis indicates leaf order. ZH11, wild type; L2, L10, and L17 are three independent transformation lines. At least 15 seedlings were scored for each measurement (n ≥ 15). Data are the mean ±SD for each measurement. Ethylene: 100 μl l−1. **Significant difference (Fisher’s LSD, α=0.01) comparing the third-leaf sheath length between ZH11 and transformation lines in (B).
Thus, high GA3 treatment differentially promoted rice leaf elongation. The effects of GA3 and ethylene on leaf growth patterns were distinct. GAs increased the effects of 1-MCP treatment or OsRTH1 overexpression on inhibition of elongation of the third sheath. Thus, ethylene-induced leaf growth may not be primarily due to the effects of GAs.
OsRTH1 overexpression prevents ethylene-induced adventitious root growth
The rice root system mainly consists of a primary root and numerous adventitious roots. The adventitious root, also called the crown root, plays a major role in nutrition and water uptake (Inukai et al., 2005; Osmont et al., 2007; Rebouillat et al., 2009). The internodal adventitious root initiation, induced by ethylene, is coupled with the internodal elongation of indica deep-water cultivars (Mergemann and Sauter, 2000). The effects of ethylene and OsRTH1 overexpression on the adventitious root growth of ZH11 (a japonica cultivar) were thus examined.
Rice seedlings with a coleoptile of 2–3 mm were hydroponically grown for 7 d. Without exogenous ethylene, ZH11 showed approximately two more adventitious roots than did the transformation lines. Ethylene treatment increased the number of adventitious roots of ZH11 but not 35S:OsRTH1 lines by about two. 1-MCP treatment reduced the number of adventitious roots of ZH11 by two and had little effect on 35S:OsRTH1 lines. 1-MCP-treated ZH11 and 35S:OsRTH1 lines were similar in adventitious root number (Fig. 9A).
Fig. 9.
Adventitious root growth of ZH11 and 35S:OsRTH1 lines. Adventitious root number of hydroponically grown (A) and wet tissue-grown (B) ZH11 and 35S:OsRTH1 lines. ZH11, wild type; L2, L10, and L17 are three independent transformation lines. At least 15 seedlings were scored for each measurement (n ≥ 15). NAA, 0.1 μM; NPA, 1 μM; ethylene, 100 μl l−1; 1-MCP, 5 μl l−1.
Auxins are important to rice adventitious root growth (Yamamoto et al., 2007). The roles of auxins in ethylene-induced adventitious root growth were evaluated. Auxin [1-naphthaleneacetic acid (NAA)]-treated ZH11 had about one more adventitious root than untreated ZH11. The auxin transporter inhibitor 3-nitropropionic acid (NPA) severely inhibited adventitious root growth, and treated rice seedlings had only one adventitious root. ZH11 seedlings with ethylene treatment alone and those with ethylene and auxin treatment had the same number of adventitious roots. Auxin (NAA) alone or ethylene and auxin had minor effects on adventitious root growth of 35S:OSRTH1 lines. With NAA treatment, there were 3–4 fewer adventitious roots in 35S:OsRTH1 lines than in ZH11. The prevention of endogenous ethylene perception of ZH11 by 1-MCP reduced the adventitious root number, regardless of auxin treatment. The adventitious root number of 1-MCP/NAA-treated ZH11 was similar to that of NAA-treated 35S:OsRTH1 lines (Fig. 9A).
Ethylene and 1-MCP are gases, and their efficacy on root growth may be altered to an unknown extent because of the physical barrier of the aqueous solution. The effects of ethylene on the adventitious root growth of rice seedlings grown on wet tissues to eliminate the aqueous barrier were evaluated. Air-grown ZH11 seedlings had 2–3 more adventitious roots than did the 35S:OsRTH1 transformation lines. Ethylene treatment increased the number of adventitious roots by two for ZH11 but not for 35S:OsRTH1 lines. 1-MCP treatment reduced the number of adventitious roots by 2.5 for ZH11 but not for 35S:OsRTH1 lines. ZH11 and the 35S:OsRTH1 lines were similar in adventitious root number with 1-MCP treatment. Auxin (NAA) treatment increased the number of adventitious roots by 1.5 for ZH11 and by 1–2 for the 35S:OsRTH1 lines. Preventing endogenous ethylene perception by 1-MCP reduced the adventitious root number of ZH11 and NAA-treated ZH11 by 2 and 2.5, respectively. The adventitious root number of ZH11 was identical with 1-MCP and 1-MCP/NAA treatments (Student’s t- test and P=0.104). ZH11 and the 35S:OsRTH1 lines were identical in adventitious root number in the presence of NAA and 1-MCP (F test; F=0.949 < Fcrit 2.769, and P=0.423) (Fig. 9B).
Therefore, auxins were essential for basal-level adventitious root growth, and the effects of an excessive amount of exogenous auxins on root growth were marginal. In indica deep-water rice, ethylene-induced adventitious root growth is coupled with internodal elongation. Here it was shown that adventitious root growth induced by ethylene was independent of internodal elongation in the japonica cultivar ZH11. 1-MCP treatment and 35S:OsRTH1 had the same effects on inhibition of ethylene-induced adventitious root growth. Auxin-induced adventitious root growth was substantially attenuated by 1-MCP treatment or 35S:OsRTH1 expression, which implies the important roles of ethylene in auxin-induced adventitious root growth.
Discussion
In this study, OsRTH1 overexpression suppressed various aspects of ethylene response in Arabidopsis and rice, and complemented Arabidopsis rte1-2 mutation. Thus, the ethylene signalling machinery is highly conserved across higher plant species and favours OsRTH1 as an orthologue of Arabidopsis RTE1, although OsRTH1 functions were not inferred from phenotypes of loss-of-function mutants. Of note, rte1 loss-of-function mutants do not exhibit a discernible phenotype (Zhou et al., 2007; Resnick et al., 2008); rice mutants defective in OsRTH1 may not display a discernible phenotype, and the mutants may not be the best for inferring OsRTH1 functions.
Three OsRTH genes are identified in the rice genome, and OsRTH2 and OsRTH3 were unable to confer ethylene insensitivity when overexpressed. The rice genome has undergone gene loss after whole-genome duplication to form a stabilized diploid (Ma and Bennetzen, 2004, 2006; Salse et al., 2008). Truly redundant genes are evolutionarily unstable, and subfunctionalization or neofunctionalization stabilizes duplicated loci in the genome (Nowak et al., 1997). The localization of GFP–AtRTH to the ER and nucleus may indicate functional divergence of AtRTH, which does not have a role in ethylene signalling (Rivarola et al., 2009). In contrast, the present study suggested that the three OsRTHs, like RTE1, predominantly associated with the Golgi apparatus. The absence of non-Golgi OsRTHs may indicate loss of the non-Golgi OsRTHs during the rice genome diploidization, with Golgi-associated OsRTHs retained. Among these OsRTHs, only OsRTH1 acquired functions in ethylene signalling. Although it was not possible to demonstrate whether OsRTH2 and OsRTH3 are expressed at the translational level, analyses of gene structure and sequence identity and a previous phylogenetic analysis do not favour the two OsRTHs being functionally conserved in ethylene signaling (Barry and Giovannoni, 2006). Although within the same clade as RTE1 and OsRTH1, OsRTH2 may have not acquired functions in ethylene signalling. Alternatively, OsRTH2 could have lost its functions in ethylene signalling after mutation accumulation during evolution. AtRTH and OsRTH3 are in the same clade (Barry and Giovannoni, 2006), in agreement with results showing that OsRTH3 overexpression failed to confer ethylene insensitivity and complemented rte1-2 mutation.
Previous studies of ethylene effects on rice growth and development involved ACC or ethephon, chemicals that replace ethylene gas; two aspects of ethylene-induced growth alterations were differentially alleviated in japonica rice expressing OsETR2, OsEIN2, or OsEIN3 (Jun et al., 2004; Mao et al., 2006; Wuriyanghan et al., 2009). The growth alterations by ACC or ethephon included an increase in seedling height and a minor elongation in the primary root. Other aspects of growth and development that are modulated by ethylene must be investigated. Auxins and GAs may also affect root growth in Arabidopsis (Achard et al., 2003; Stepanova et al., 2005); whether the ethylene-induced alterations in rice seedling growth result from synergistic actions of these hormones is unknown.
Treatment with ethylene, rather than a chemical replacement, was used to evaluate the effects on aspects of rice growth and development, and 1-MCP treatment was used to evaluate the effects of OsRTH1 overexpression on the prevention of ethylene-induced alterations in growth. Ethylene promoted various aspects of rice growth and development, including coleoptile elongation of etiolated rice seedlings, coleoptile curvature and greening of light-grown rice seedlings, and the growth and development of rice seedling leaves and adventitious roots. Although GAs may promote leaf elongation, the leaf growth patterns of ethylene- and GA-treated rice seedlings were distinct, which indicates differential regulation of rice leaf growth by the two hormones. Auxins play major roles in adventitious root development; however, the effects of an excess amount of auxins were marginal. In contrast, the increase in adventitious root number caused by ethylene indicated a collaboration of auxins and ethylene in adventitious root development. Unlike in deep-water rice, in the japonica cultivar ZH11, adventitious growth promoted by ethylene was not associated with internodal elongation.
The ethylene-dependent alterations in growth of rice seedlings may reveal the biological significance of ethylene in rice seedling growth and development. Etiolated rice seedlings show coleoptile elongation induced by ethylene (Ku et al., 1970; Satler and Kende, 1985). Conceivably, when seedlings are germinated in the dark environment of soil, leaves are enclosed within a coleoptile. With ethylene production, the coleoptiles elongate and penetrate through the soil while protecting the shoots. Once the seedlings emerge from the soil and perceive light, ethylene promotes the coleoptile curvature, which facilitates exposure of seedling leaves to light. Meanwhile, ethylene-induced coleoptile greening may provide energy, from photosynthesis, for seedling growth. Once seedlings grow under light, ethylene promotes the growth and development of shoots and adventitious roots. The leaf elongation and development, promoted by ethylene, may facilitate early-stage rice seedling growth, and the increase in adventitious root number may help rice seedlings stand in the soft, muddy field and facilitate nutrition uptake. This study suggests that ethylene plays important roles in coordinating various environmental (darkness and light) and internal cues during early-stage rice seedling growth and development.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Coleoptile phenotype of light-grown 35S:OsRTH1 transformation rice lines grown in air and ethylene.
Acknowledgments
We thank our colleagues Z.H. He and H.X. Lin for rice cultivation at Experimental Stations, and Rohm & Haas China (Beijing) for 1-MCP. The organelle markers ER-rb and G-rk (Nelson et al., 2007) were from the Arabidopsis Biological Resource Center (CD3-960 and CD3-967). The rice transformation was carried out by Ms S.P. Xu. This work was supported by the Ministry of Science and Technology (2012AA10A302 and 2011CB100700), National Natural Sciences Foundation of China (31123006, 31070249, and 30430080), and Chinese Academy of Sciences (KSCX2-EW-J-12).
References
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. The Plant Cell. 2003;15:2816–2825. doi: 10.1105/tpc.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Phillips L, Barry C, Kannan P, Leclercq J, Bouzayen M, Giovannoni J. Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Molecular Biology. 2004;54:387–404. doi: 10.1023/B:PLAN.0000036371.30528.26. [DOI] [PubMed] [Google Scholar]
- Alonso J, Hirayama T, Roman G, Nourizadeh S, Ecker J. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ. Ripening in the tomato green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proceedings of the National Academy of Sciences, USA. 2006;103:7923–7928. doi: 10.1073/pnas.0602319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annual Review of Cell and Developmental Biology. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker J. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Dong C-H, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C. Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. Journal of Biological Chemistry. 2010;285:40706–40713. doi: 10.1074/jbc.M110.146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C-H, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. The Plant Journal. 2008;53:275–286. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. Ethylene—a key regulator of submergence responses in rice. Plant Science. 2008a;175:43–51. [Google Scholar]
- Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proceedings of the National Academy of Sciences, USA. 2008b;105:16814–16819. doi: 10.1073/pnas.0807821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. The Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V. SAG2 and SAG12 protein expression in senescing Arabidopsis plants. Physiologia Plantarum. 2003;119:263–269. [Google Scholar]
- Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. The Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B, Shakeel S, Schaller G. Ethylene receptors: ethylene perception and signal transduction. Journal of Plant Growth Regulation. 2007;26:118–130. [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. . Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. The Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB. Ethylene-promoted elongation: an adaptation to submergence stress. Annals of Botany. 2008;101:229–248. doi: 10.1093/aob/mcm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annual Review of Genetics. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Jun S-H, Han M-J, Lee S, Seo YS, Kim WT, An G. OsEIN2 is a positive component in ethylene signaling in rice. Plant and Cell Physiology. 2004;45:281–289. doi: 10.1093/pcp/pch033. [DOI] [PubMed] [Google Scholar]
- Kao CH, Yang SF. Role of ethylene in the senescence of detached rice leaves. Plant Physiology. 1983;73:881–885. doi: 10.1104/pp.73.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ. Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiology. 2004;135:660–667. doi: 10.1104/pp.104.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HS, Suge H, Rappaport L, Pratt HK. Stimulation of rice coleoptile growth by ethylene. Planta. 1970;90:333–339. doi: 10.1007/BF00386385. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wen C- K. Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiology. 2012;158:1193–1207. doi: 10.1104/pp.111.187757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Xu C, Wen C- K. Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in. Arabidopsis. BMC Plant Biology. 2010;10:60. doi: 10.1186/1471-2229-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proceedings of the National Academy of Sciences, USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proceedings of the National Academy of Sciences, USA. 2006;103:383–388. doi: 10.1073/pnas.0509810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Wang S, Jia Q, Wu P. OsEIL1, a rice hsomolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Molecular Biology. 2006;61:141–152. doi: 10.1007/s11103-005-6184-1. [DOI] [PubMed] [Google Scholar]
- Mergemann H, Sauter M. Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiology. 2000;124:609–614. doi: 10.1104/pp.124.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metraux J-P, Kende H. The role of ethylene in the growth response of submerged deep water rice. Plant Physiology. 1983;72:441–446. doi: 10.1104/pp.72.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Noh Y-S, Amasino RM. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Molecular Biology. 1999;41:181–194. doi: 10.1023/a:1006342412688. [DOI] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. Hidden branches: developments in root system architecture. Annual Review of Plant Biology. 2007;58:93–113. doi: 10.1146/annurev.arplant.58.032806.104006. [DOI] [PubMed] [Google Scholar]
- Raskin I, Kende H. Role of gibberellin in the growth response of submerged deep water rice. Plant Physiology. 1984;76:947–950. doi: 10.1104/pp.76.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouillat J, Dievart A, Verdeil J, Escoute J, Giese G, Breitler J, Gantet P, Espeout S, Guiderdoni E, Périn C. Molecular genetics of rice root development. Rice. 2009;2:15–34. [Google Scholar]
- Resnick JS, Rivarola M, Chang C. Involvement of RTE1 in conformational changes promoting ETR1 ethylene receptor signaling in Arabidopsis. The Plant Journal. 2008;56:423–431. doi: 10.1111/j.1365-313X.2008.03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen C-K, Shockey JA, Chang C. REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivarola M, McClellan CA, Resnick JS, Chang C. ETR1-specific mutations distinguish ETR1 from other Arabidopsis ethylene receptors as revealed by genetic interaction with RTE1. Plant Physiology. 2009;150:547–551. doi: 10.1104/pp.109.138461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, et al. Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8'-hydroxylase in rice. Plant and Cell Physiology. 2007;48:287–298. doi: 10.1093/pcp/pcm003. [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. The Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satler SO, Kende H. Ethylene and the growth of rice seedlings. Plant Physiology. 1985;79:194–198. doi: 10.1104/pp.79.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. The Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ. Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. The Plant Journal. 2001;26:47–58. doi: 10.1046/j.1365-313x.2001.01006.x. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Klee HJ. Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiology. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proceedings of the National Academy of Sciences, USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. The Plant Cell. 2002;14:S131–151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proceedings of the National Academy of Sciences, USA. 2003;100:352–357. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Wuriyanghan H, Zhang B, Cao W-H, et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. The Plant Cell. 2009;21:1473–1494. doi: 10.1105/tpc.108.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Liu Q, Wen CK. Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. 2006;142:492–508. doi: 10.1104/pp.106.082628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiology. 2007;143:1362–1371. doi: 10.1104/pp.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wen C- K. Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiology and Biochemistry. 2010;48:45–53. doi: 10.1016/j.plaphy.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Grierson D. Tomato ethylene receptor–CTR interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. Journal of Experimental Botany. 2008;59:965–972. doi: 10.1093/jxb/ern021. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liu Q, Xie F, Wen C- K. RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiology. 2007;145:75–86. doi: 10.1104/pp.107.104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.