Abstract
The aim of this study was to investigate the molecular mechanisms underlying drought acclimation in coffee plants by the identification of candidate genes (CGs) using different approaches. The first approach used the data generated during the Brazilian Coffee expressed sequence tag (EST) project to select 13 CGs by an in silico analysis (electronic northern). The second approach was based on screening macroarrays spotted with plasmid DNA (coffee ESTs) with separate hybridizations using leaf cDNA probes from drought-tolerant and susceptible clones of Coffea canephora var. Conilon, grown under different water regimes. This allowed the isolation of seven additional CGs. The third approach used two-dimensional gel electrophoresis to identify proteins displaying differential accumulation in leaves of drought-tolerant and susceptible clones of C. canephora. Six of them were characterized by MALDI-TOF-MS/MS (matrix-assisted laser desorption-time of flight-tandem mass spectrometry) and the corresponding proteins were identified. Finally, additional CGs were selected from the literature, and quantitative real-time polymerase chain reaction (qPCR) was performed to analyse the expression of all identified CGs. Altogether, >40 genes presenting differential gene expression during drought acclimation were identified, some of them showing different expression profiles between drought-tolerant and susceptible clones. Based on the obtained results, it can be concluded that factors involved a complex network of responses probably involving the abscisic signalling pathway and nitric oxide are major molecular determinants that might explain the better efficiency in controlling stomata closure and transpiration displayed by drought-tolerant clones of C. canephora.
Keywords: Coffea canephora, differential expression, drought acclimation, proteomic, real-time PCR, candidate gene
Introduction
Coffee is one of the commodities most exchanged in international markets, with Brazil being the country with the largest production (Lashermes et al., 2008). It is cultivated in >80 countries and ∼25 million people depend on coffee for their livelihoods in Latin America, Africa, and Asia. The annual world production (134 million bags of 60 kg coffee beans in 2010, www.ico.org) is always subjected to regular oscillations mainly explained by the natural biennial cycle (larger production in one year and lesser production in the next year) and adverse effects of climatic conditions. Periods of drought and high temperature are key factors affecting coffee plant development and production (DaMatta and Ramalho, 2006). In the case of severe drought, flowering can be affected, leading to abortion of developing fruits, and under extreme conditions it may even cause plant death. As a consequence of global warming, coffee-growing geographical regions could also suffer delocalization (Assad et al., 2004), leading to important environmental, economic, and social problems. In such a context, the generation of drought-tolerant coffee varieties has now turned into one of the priorities of many coffee research institutes.
The coffee genus contains >100 species (Davis et al., 2006), with Coffea arabica and C. canephora being the two most cultivated species. Concerning drought tolerance, it is well known that genetic variability exists within the Coffea genus, as the Kouillou group appears to be more tolerant to drought than Robusta (currently defined as the SG2 group) types of C. canephora (Montagnon and Leroy, 1993; Montagnon, 2000). Under severe drought, Boyer (1965, 1969) showed that plants from the Kouillou group maintained stomatal opening, and consequently an active photosynthesis, while stomata of Robusta plants were completely closed under such conditions. The same author also reported more efficient root water absorption for the Kouillou plants that could explain its drought tolerance despite its maintenance of stomatal opening. Among the strategies displayed by coffee plants to cope with drought, leaf folding and inclination that reduce the leaf surface, water loss by transpiration, and exposure to high irradiance were commonly observed for Guinean and SG1 genotypes (Montagnon and Leroy, 1993). Leaf abscission is then reduced, favouring a rapid recovery of vegetation with the return of the rains. Such a trait can be considered as a selective advantage when compared with the dramatic leaf abscission that characterizes SG2 genotypes.
In addition to affecting coffee productivity, shape, and bean defects (Carr, 2001; Silva et al., 2005), large variations in rainfall and temperature also influence the biochemical composition (e.g. reducing sugars, proteins, and caffeine) of beans (Mazzafera, 2007) and consequently the final cup quality (Camargo et al., 1992). For example, bean caffeine content was shown to decrease with irrigation and temperature (Silva et al., 2005). In the same work, high protease and polyphenol oxidase activities were correlated with higher temperature and lower rainfall. Nevertheless, before analysing the interaction between quality and drought adaptation, the molecular control of drought tolerance in terms of physiological behaviour needs to be unravelled.
The identification and selection of C. canephora clones combining drought tolerance with other agronomic traits (e.g. efficient flowering, productivity, and vigour) is of particular interest to generate new varieties better adapted to climate changes. During the last decade, several drought-tolerant clones of C. canephora var. Conilon have been characterized as vigorous plants with high productivity throughout years under drought stress (Ferrão et al., 2000; Fonseca et al., 2004). Fingerprint analyses also revealed that these Conillon clones belong to the SG1 group of C. canephora (Lambot et al., 2008). Physiological analyses suggested that drought tolerance could be a direct consequence of better root development (Pinheiro et al., 2005). In another study, Lima et al. (2002) suggested that enhanced activity of antioxidant enzymes might also be involved in the drought tolerance mechanism. A key role of ascorbate peroxidase (APX) was postulated to allow clone 14 to cope with potential increases of H2O2 under drought conditions, as an increased (38%) activity of this enzyme was found for this clone upon drought stress (Pinheiro et al., 2004). When looking for activities of sucrose-metabolizing enzymes [e.g. acid invertase, sucrose synthase, and sucrose phosphate synthase (SPS)] as well as for contents of hexoses, sucrose, starch, and amino acids in leaves, Praxedes et al. (2006) observed a maintenance of SPS activity with the decrease of pre-dawn leaf water potential (Ψpd) for the drought-tolerant clone 120 but not for the drought-sensitive clones. According to DaMatta et al. (2003), the better crop yield of a drought-tolerant clone compared with a drought-sensitive clone is mainly associated with the maintenance of leaf area and tissue water potential that are consequences of reduced stomatal conductance (g s). Taken together, these observations suggest the existence of different biological mechanisms conferring drought tolerance in C. canephora, the molecular nature of which is still largely unknown.
Simkin et al. (2008) demonstrated that drought affected the leaf expression of several gene-encoding enzymes of the carotenoid biosynthesis pathway, therefore suggesting that changes in carotenoid content and activation of the xanthophyll cycle may play some role in drought tolerance, as also emphasized in previous studies (Davison et al., 2002; Du et al., 2010). Up-regulation of genes (CaGolS) encoding galactinol synthase upon drought was also observed (dos Santos et al., 2011). In that case, no concomitant accumulation of galactinol was observed, probably because this sugar was used as a substrate for the biosynthesis of high molecular weight polysaccharides. In a recent publication, differential gene expression from both subgenomes of C. arabica was studied through a coffee-specific microarray and showed preferential expression of C. canephora homologues at higher temperatures and expression of C. eugenioides homologue genes at lower temperatures (Bardil et al., 2011; Privat et al., 2011).
The recent advances in coffee genomics including expressed sequence tag (EST) sequencing projects (Lin et al., 2005; Poncet et al., 2006; Vieira et al., 2006; Vidal et al., 2010; Mondego et al., 2011), the construction of bacterial artificial chromosome (BAC) libraries (Noir et al., 2004; Leroy et al., 2005), and genetic maps (Crouzillat et al., 2004; Lefebvre-Pautigny et al., 2010; Leroy et al., 2011), as well as the improvement of coffee genetic transformation techniques (Alpizar et al., 2008; Ribas et al., 2011) now open the way to study the molecular and genetic determinism of drought tolerance and other important agronomic traits, leading to the identification of molecular markers that could be used to speed up coffee breeding programmes (Lashermes et al., 2008; De Kochko et al., 2010).
The search for candidate genes (CGs) for drought tolerance in coffee has now been initiated by an in silico analysis of the Brazilian Coffee Genome database (Vieira et al., 2006; Mondego et al., 2011). As part of this project, two EST libraries from leaves of plants of C. arabica var. Catuaí (considered as drought sensitive) and C. canephora clone 14 (drought tolerant) submitted to drought were generated. The plant materials of C. arabica and C. canephora were obtained from field and pot trials, respectively. More than 15 000 clones were sequenced and, after trimming and clustering, resulted in 10 924 reads grouped on 6141 unigenes (1779 contigs and 4362 singlets). The approaches used to identify CGs underlying drought acclimation (DA) in coffee consisted of an in silico analysis of ESTs generated from drought-exposed and untreated leaf cDNA libraries of C. canephora plants (Mondego et al., 2011), transcription profiling by macroarrays and quantitative real-time polymerase chain reaction (qPCR) analyses of drought-submitted and control tissues, as well as protein profiling by two-dimensional gel electrophoresis (2-DE) coupled with tryptic peptide identification by matrix-assisted laser desorption-time of flight-tandem mass spectrometry (MALDI-TOF-MS/MS). These integrated analyses resulted in the identification of >40 candidate drought-responsive genes. Results showing differential gene expression in leaves of sensitive versus tolerant clones of C. canephora cultivated in the greenhouse and submitted or not to water limitation are presented and discussed.
Materials and methods
Plant material
Clones of C. canephora Pierre var. Conilon tolerant (14 and 120) and susceptible (22) to drought were obtained as rooted stem cuttings from the Institute for Research and Rural Assistance (Incaper, Vitoria, Espirito Santo, Brazil) (Ferrão et al., 2000; Fonseca et al., 2004). These clones were grown in greenhouse (Federal University of Viçosa-UFV, Minas Gerais, Brazil) conditions [25 °C, 70% relative, average mid-day photosynthetic photon flux (PPF) of 900 μmol m−2 s−1] individually in pots of 12 litres of a mixture of soil, sand, and manure (3:1:1, v/v/v). After 6 months, plants of each clone were separated into two groups: the first one received regular irrigation (control) while watering was suspended for the second group until plants reached nearly –3.0 MPa (DA; e.g. clone 22, 6 d of DA; clones 14 and 120, 12 d of DA) (Marraccini et al., 2011). For both conditions, six plants (biological repetitions) were analysed. Fresh leaves (fully expanded leaves corresponding to the third pair in plagiotropic branches) were used for physiological and molecular analyses. In that case, leaves were collected in the daytime (at ∼10:00 h), immediately frozen in liquid nitrogen, and further stored at –80°C for protein and RNA extractions.
Physiological analyses
DA was evaluated by measuring Ψpd with a Scholander-type pressure chamber. Ψpd was regularly followed to reach near –3.0 MPa for drought-acclimated plants (Silva, 2007; Silva et al., 2010). The net CO2 assimilation rate (A), stomatal conductance to water vapour (g s), and internal to ambient CO2 concentration ratio (C i/C a) were measured between 10:00 h and noon under artificial and saturating PPF with a portable open-system infrared gas analyser (LCpro+, Analytical Development Co. Ltd, Hoddesdon, UK) under a relative humidity of ∼80%. The chlorophyll a fluorescence parameters were measured using a portable pulse amplitude modulation fluorometer (FMS2, Hansatech, King’s Lynn, Norfolk, UK). The maximum photochemical efficiency of PSII (F v/F m), the coefficient of photochemical quenching (q P), the quantum yield of photosystem II (PSII) electron transport (ΦPSII), the non-photochemical quenching (NPQ), and the fraction of PPF absorbed in PSII antennae and used neither in photochemistry nor dissipated thermally (P E) were measured as previously described (DaMatta et al., 2002; Lima et al., 2002; Pinheiro et al., 2004). For each parameter, values represent the mean ±SD of five replicates (Table 1).
Table 1.
Effects of the drought on leaf pre-dawn water potential (Ψpd in MPa), rate of decrease of Ψpd (RDPWP in MPa d−1 m−2), net CO2 assimilation rate (A in μmol m−2s−1), stomatal conductance (gs in mmol m−2 s−1), internal to ambient CO2 concentration ratio (Ci/Ca), maximum photochemical efficiency of PSII (Fv/Fm), quantum yield of PSII electron transport (ΦPSII), photochemical (qP) and Stern–Volmer non-photochemical (qN) quenching coefficients, and the fraction of PPF absorbed in PSII antennae and used neither in photochemistry nor dissipated thermally (PE) of clones 14, 22, and 120 of C. canephora
Different upper case letters denote significant differences among means of the two genotypes in irrigated conditions. Different lower case letters represent significant differences among means of the genotypes submitted to drought by the Newman–Keuls test at P ≤ 0.05 (clone effect). Means for plants submitted to drought marked with an asterisk differ from those of control plants by F-test at P ≤ 0.05 (treatment effect).
| Parameters | Drought-tolerant clone (14) | Drought-sensitive clone (22) | Drought-tolerant clone (120) | |||
| Control | Drought | Control | Drought | Control | Drought | |
| Ψpd | –0.02±0.01 A | –3.02±0.12 a* | –0.03±0.00 A | –3.01±0.11 a* | –0.03±0.01 A | –3.1±0.15 a* |
| RDPWP | – | 0.67±0.04 c | – | 1.01±0.04 a | – | 0.84±0.03 b |
| A | 9.40±0.34 A | 2.62±0.27 a* | 9.35±0.17 A | 0.95±0.23 b* | 9.62±0.61 A | 2.57±0.14 a* |
| g s | 60.00±5.00 B | 13.00±3.50 a* | 105.00±9.50 A | 5.00±3.00 a* | 72.00±7.00 B | 10.00±1.00 a* |
| C i/C a | 0.520±0.040 B | 0.380±0.040 b* | 0.670±0.040 A | 0.520±0.040 a* | 0.520±0.050 B | 0.320±0.050 b* |
| F v/F m | 0.840±0.011 A | 0.842±0.011 a | 0.831±0.011 A | 0.800±0.011 b* | 0.830±0.001 A | 0.850±0.004 a |
| ΦPSII | 0.455±0.049 A,B | 0.287±0.050 a,b* | 0.472±0.049 B | 0.210±0.050 b* | 0.577±0.048 A | 0.395±0.046 a* |
| q P | 0.713±0.051 A,B | 0.495±0.051 a,b* | 0.697±0.051 B | 0.362±0.051 b* | 0.815±0.033 A | 0.615±0.055 a* |
| q N | 0.665±0.056 A | 0.717±0.057 a | 0.642±0.056 A | 0.543±0.057 a | 0.560±0.067 A | 0.652±0.021 a |
| P E | 0.252±0.040 A | 0.425±0.040 a,b* | 0.242±0.039 A | 0.508±0.039 a* | 0.152±0.027 A | 0.332±0.048 b* |
In silico identification of candidate genes
For in silico expression analysis, contig and singlet frequencies across the libraries were obtained from the data set derived from the CAP3 assembly. A first set of CGs corresponded to genes exclusively expressed in the leaf cDNA library of drought-acclimated clone 14 (SH3) from C. canephora. Other CGs came from (i) the comparison of the cDNA library from leaves of drought-acclimated plants of clone 14 (SH3) with the leaf cDNA library (LF1) of the BP409 variety (Lin et al., 2005) of C. canephora and (ii) the comparison of the cDNA library from leaves of plants submitted to drought (SH2) with well-watered plants conditions (LV8 and LV9) for C. arabica var. Catuaí (Vieira et al., 2011). In all cases, CcUBQ10 transcripts were used as an internal control of expression. Fisher’s statistical test (Fisher, 1922) was performed, with a threshold of 4 indicating a significant variation in expression. Sequence homologies were detected by BLAST analyses (Altschul et al., 1990) against the GenBank and SGN (SOL Genomics Network; Mueller et al., 2005) databases.
RNA extraction
Samples were ground into a powder in liquid nitrogen and total RNAs were extracted as described previously (Marraccini et al., 2011). RNA quantification was performed using a NanoDrop™ 1000 Spectrophotometer (Waltham, MA, USA).
Northern blot experiment
A 15 μg aliquot of total RNA was fractionated on a 1.2% (w/v) agarose gel containing 2.2 M formaldehyde in MOPS buffer. The homogeneity of the amount of RNA deposited was checked according to the abundance of 26S and 18S rRNA on gels stained with ethidium bromide. Total RNA was transferred to Hybond N+ membranes which were hybridized at 65 °C in modified Church and Gilbert buffer (7% SDS, 10 mM EDTA, 0.5 M sodium phosphate pH 7.2). Probes corresponding to the CcCAB1, CcRBCS1, CcACBP1, CcELIP1, CcMET1, CcMPR1, CcUNK10, CcGRP1, and CcUBQ10 genes were amplified by conventional PCR using universal primers from a plasmid harbouring the EST sequences GT647358, GT649534, GT646045, GT647659, GT650680, GT648734, GT648004, GT650953, and GW488515, respectively (Vidal et al., 2010; Mondego et al., 2011), and labelled by random priming with [α-32P]dCTP (Amersham Biosciences, São Paulo, SP, Brazil) as previously described (Geromel et al., 2006). Washes were performed at 65 °C in 2× SSC (1× SSC=150 mM sodium chloride and 15 mM sodium citrate, pH 7.0)–0.1% SDS (2× 15 min) with a final stringent wash in 0.1× SSC, 0.1% SDS (2× 15 min). Membranes were exposed with BAS-MS 2340 IP support and data were acquired using a Fluorescent Image Analyzer FLA-3000 (Fujifilm Life Science, Woodbridge, CT, USA).
Real-time qPCR assays
To eliminate contaminant genomic DNA, samples were treated with RQ1 RNase-free DNase according to the manufacturer’s instructions (Promega, Madison, WI, USA) and RNA quality was verified by agarose gel electrophoresis and visual inspection of the rRNA bands upon ethidium bromide staining. Synthesis of the first-strand cDNA was done by treating 1 μg of total RNA with the ImProm-II™ Reverse Transcription System with oligo(dT15) according to the manufacturer’s recommendations (Promega). The absence of contaminating genomic DNA in the cDNA preparations was checked by conventional PCR using the SUS10/SUS11 primer pair that spans introns 5–9 of the CcSUS1 gene (AJ880768) encoding isoform 1 of the sucrose synthase (Leroy et al., 2005; Geromel et al., 2006). In that case, PCR was carried out with 1 μl of synthesized cDNA with a PTC-100 Thermocycler (MJ Research), using GoTaq DNA polymerase according to the supplier’s instructions (Promega) under the following conditions: initial denaturation 94 °C, 2 min; followed by 40 cycles of 94 °C, 30 s; 55 °C, 30 s; and 72 °C, 3 min, and a final extension step of 72 °C, 6 min. In such conditions, the amplification of a 667 bp fragment characterized the CcSUS1 cDNA and the absence of the corresponding genomic sequence is indicated by the lack of an amplicon at bp 1130 (data not shown).
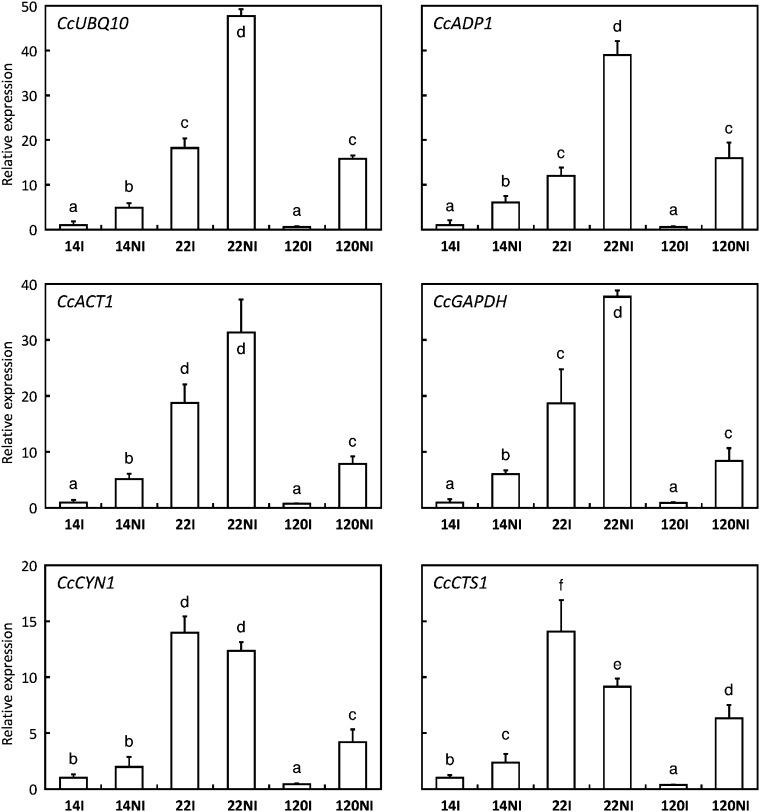
Real-time qPCR assays were carried out with the synthesized single-stranded cDNA described above and using the protocol recommended for the use of 7500 Fast Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA). cDNA preparations were diluted (1/25 to 1/100) and tested by qPCR using CG primer pairs (Table 3) designed employing the Primer Express software (Applied Biosystems) and preliminarily tested for their specificity and efficiency against a mix of cDNA or using cloned ESTs (data not shown). The qPCR was performed with 1 μl of diluted single-stranded cDNA and 0.2 μM (final concentration) of each primer in a final volume of 10 μl with SYBR green fluorochrome (SYBRGreen qPCR Mix-UDG/ROX, Invitrogen). The reaction mixture was incubated for 2 min at 50 °C (uracil DNA-glycosylase treatment), then for 5 min at 95 °C (inactivation of uracil DNA-glycosylase), followed by 40 amplification cycles of 3 s at 95 °C, 30 s at 60 °C. Data were analysed using the SDS 2.1 software (Applied Biosystems) to determine the cycle threshold (Ct) values. The specificity of the PCR products generated for each set of primers was verified by analysing the T m (dissociation) of amplified products. PCR efficiency (E) was estimated using absolute fluorescence data captured during the exponential phase of amplification of each reaction with the equation (1+E)=10(–1/slope) (Ramakers et al., 2003). Efficiency values were taken into account in all subsequent calculations. Gene expression levels were normalized to expression levels of CcUBQ10 or CcGAPDH as a constitutive reference (Barsalobres-Cavallari et al., 2009). For the CcUNK10 gene expression study, relative quantification was also normalized with CcADP1, CcACT1, CcCYN1, and CcCTS1 as alternative housekeeping genes (Fig. 4). Expression was expressed as relative quantification by applying the formula (1+E)−ΔΔCt, where ΔCt target=Ct target gene–Ct reference gene and ΔΔCt=ΔCt target–ΔCt internal calibrator, the internal calibrator always being the 14I sample with relative quantification equal to 1.
Table 3.
Candidate genes and corresponding primers used for qPCR experiments Coffee sequences were selected (E-value ≤1e−10) from public databases with the BLAST programs (Altschul et al., 1990). Gene names were assigned based on the best BLASTX hit obtained by comparing the selected coffee ESTs with public databases. GenBank (GB) accession numbers of coffee EST sequences are also given. Primers were designed using the Primer Express software (Applied Biosystems).
| Protein name | Gene name | GB numbers | Primer name | Primer sequence | bp | CG |
| Ubiquitin | CcUBQ10 | GW488515 | BUBI-F | 5′-AAGACAGCTTCAACAGAGTACAGCAT-3′ | 104 | R* |
| BUBI-R | 5′-GGCAGGACCTTGGCTGACTATA-3′ | |||||
| Glyceraldehyde-3-phosphate dehydrogenase | CcGAPDH | GW445811 | GAPDH-F | 5′-TTGAAGGGCGGTGCAAA-3′ | 59 | R* |
| GAPDH-R | 5′-AACATGGGTGCATCCTTGCT-3′ | |||||
| Cathepsin | CcCTS1 | GW442860 | CATHEP-F | 5′-CCCATTATCGTTCTGGTGTTTACAA-3′ | 80 | R |
| CATHEP-R | 5′-ACCCCATCCAATCAGCTTTACA-3′ | |||||
| Actin | CcACT1 | GW428479 | ACTIN-F | 5′-GGATTTCCAGGGCTGAATATGA-3′ | 80 | R |
| ACTIN-R | 5′-CGATACGATAGTAGCAGCTTAAAAGC-3′ | |||||
| ADP-ribosylation factor (ARF) | CcADP1 | GW450736 | ADP-F | 5′-ATGAATGCTGCTGAAATAACTGATAAG-3′ | 80 | R |
| ADP-R | 5′-GCACAGGTGCTTTGGATATACCA-3′ | |||||
| Cyclophilin | CcCYN1 | GW488988 | CYCLO-F | 5′-TCTTCATCTGCACCGACAAGA 3′5′-CACATCCATACCCTCCGTGAT-3′ | 80 | R |
| CYCLO-R | ||||||
| WRKY transcription factor | CcWRKY2 | GT647873 | 18089-F | 5′-CATGATCCCAAAGCAGTTATTACG-3′ | 67 | 1 |
| 18089-R | 5′-TTCTGGCAGTTGGTACATCATGAT-3′ | |||||
| Photosystem I subunit XI | CcPSAL1 | GT646615 | ND | 1 | ||
| Early light-induced protein (ELIP) | CcELIP1 | GT647659 | ** | 1 | ||
| Early light-induced protein (ELIP) | CcELIP2 | GT647647 | ** | 1 | ||
| Unknown protein 1 | CcUNK1 | DV689820 | 182052-F | 5′-TATAGTGTTTATGGTGTGGCTTTCAGT-3′ | 79 | 1 |
| 182052-R | 5′-GTACCACCGTAGGGAGACGTATG-3′ | |||||
| Catalase | CcCAT1 | GT649488 | 18297-F | 5′-TGACAGAACCTGCAGTAAAGACAGTATT-3′ | 146 | 2 |
| 18297-R | 5′-AACCAACCAAATGAACGAACAAT-3′ | |||||
| Catalase | CcCAT2 | GT648692 | SH3055G111-F | 5′-GATACCCAGCGACACCGTCTT-3′ | 70 | 5 |
| SH3055G111-R | 5′-GATGAGCACACTTGGGAGCAT-3′ | CcCAT1 | ||||
| Acyl-CoA-binding protein | CcACBP1 | GT646045 | 9158-F | 5′-AATACTACCAATGCAAGCAAGCTTA-3′ | 128 | 2 |
| 9158-R | 5′-CCTTCCATGCATCCCACTTT-3′ | |||||
| Chlorophyll a/b-binding protein | CcCAB1 | GT647358 | 18230-F | 5′-CTCTGAACTTCACCAGCTCTTCAA-3′ | 140 | 2 |
| 18230-R | 5′-GAGCTCGTCGTCCAATGAAGA-3′ | |||||
| Metallothionein like protein | CcMET1 | GT650680 | ** | 2 | ||
| GLB2-like non-symbiotic haemoglobin | CcNSH1 | GT649198 | RBCS3'-F | 5′-CTAAGGGATACGGATGAAATTCCA-3′ | 102 | 2 |
| RBCS3'-R | 5′-CTCTCGCAGCTGCACTACTGATT-3′ | |||||
| Glycine-rich protein | CcGRP1 | GT650953 | GC182-F | 5′-AAGCCAGCTTCTAGCTATGTCATGA-3′ | 100 | 2 |
| GC182-R | 5′-GATCCAGCACTTATTTGAGATCACA-3′ | |||||
| Mannose 6-phosphate reductase | CcMPR1 | GT648734 | LPSH3069F05-F | 5′-AATCAGCAATTACAGCGTTTTGC-3′ | 100 | 2,3 |
| LPSH3069F05-R | 5′-AGTGACACAGATGCCGTGCTT-3′ | |||||
| Aldose-phosphate reductase | CcAPR1 | GT649483 | LPSH3060G02-F | 5′-TGAAGCCAGCTGTGAATCAACT-3′ | 100 | 5 |
| LPSH3060G02-R | 5′-GTGTGGGCAGTGACACAGATG-3′ | CcMPR1 | ||||
| Unknown protein 10 | CcUNK10 | GT648004 | D18240-F | 5′-TAGCCTTGTTCTTTTAGGGAGTCTTATC-3′ | 134 | 2 |
| D18240-R | 5′-AGAGCTTCGTCCAGGAAGAAGA-3′ | |||||
| TNF-associated factor (TRAF) | CcTRAF1 | GT652792 | LP-12677-FLP-12677-R | 5′-GACTCTGGGCACATCGTGATAG-3′5′-TGCGAAGGGAAAGAATGGAA-3′ | 100 | 3 |
| Prephenate dehydrogenase | CcPDH1 | GT655248 | LP175502-F | 5′-GCAGGGACATGGACGAATTT-3′ | 100 | 3 |
| LP175502-R | 5′-TGAAACGGCATGGACTTGAC-3′ | |||||
| Unknown protein 8 | CcUNK8 | DV695331 | LP18100-F | 5′-CTCGCGTGGCCGAGATT-3′ | 100 | 3 |
| LP18100-R | 5′-CCCTCACATTTCCACGTGAAT-3′ | |||||
| Dehydrin | CcDH3 | DV689895 | 18390-F | 5′-TTAATAGCAGCTTTCCAGTGTGTCA-3′ | 100 | 3 |
| 18390-F | 5′-GATCCCCCAAAAAGCAGAAAA-3′ | |||||
| EDR1-like MAPKK kinase | CcEDR1 | GW457961 | LPSH3054B02-F | 5′-TGTCAAATTGATGAAAAGCGAAGA-3′ | 100 | 3 |
| LPSH3054B02-R | 5′-AAGGAAAAGTAGGAAATCAGCCAAA-3′ | |||||
| EDR1-like MAPKK kinase | CcEDR2 | DV681462 | EDR13-FEDR13-R | 5′ CGGCATAAGAGCGAGTGGAA 3′5′ ATGCAATCGCTGGTGTAGAAAA 3′ | 70 | 6 |
| Small heat shock protein (sHSP) | CcHSP1 | GW447897 | LPSH3056B04-F | 5′-GGTAGGACGCCATGGGAGAT-3′ | 100 | 3, 4 |
| LPSH3056B04-R | 5′-CCTCAACCCACACCTTCACAT-3′ | |||||
| Carbonic anhydrase | CcCA1 | GT008701 | HRAM1-F | 5′-AAGGCCATTGTCGGACTTCA-3′ | 100 | 4 |
| HRAM1-R | 5′-TTGTTTGCAACTCTGCAGTGATT-3′ | |||||
| Type-2C protein phosphatase (PP2C) | CcPP2C | GW435032 | HRAM2-F | 5′-CGAAGAAATCAGGCGTATCAGAG-3′ | 101 | 4 |
| HRAM2-R | 5′-TAAACCGCACGTCACCAAAAG-3′ | |||||
| Oxygen-evolving complex PSII (psbO) | CcPSBO | GT647517 | HRAM12-F | 5′-TGAATTTCTCGTGCCATCATACA-3′ | 101 | 4 |
| HRAM12-R | 5′-CTCCAGCAGGCAATGCAACT-3′ | |||||
| Oxygen-evolving complex PSII (psbP) | CcPSBP | GW488995 | HRAM7-F | 5′-AGCTGTCGATTCCCTCAAAATG-3′ | 100 | 4 |
| HRAM7-R | 5′-GAGACACTGCTGTTGCTATCGAA-3′ | |||||
| Oxygen-evolving complex PSII (psbQ) | CcPSBQ | GT645658 | HRAM36-F | 5′-CAGGGCAATCAAGGTTGGA-3′ | 102 | 4 |
| HRAM36-R | 5′-CGGTCCTTGAGTGGCAAATC-3′ | |||||
| Rubisco small chain (RbcS) | CcRBCS1 | GT649534 | 18244-F | 5′-CACCAACTGGAAAGTTGAAGAA-3′ | 169 | 6 |
| 18244-R | 5′-TATCCCGGTGACCTGTGGTATT-3′ | |||||
| Subtilisin-like serine protease | CcSDD1 | GT659840 | SDD1-F | 5′-GAGCCCCGATTGATCTTCTG-3′ | 101 | 6 |
| SDD1-R | 5′-ACTCAGCCCCAAAAGGGTTAA−3′ | |||||
| Glucosyltransferase arbutin synthase | CcGAS1 | GT721173 | ARBU09-F | 5′-AAAGCGTTGCAGGAGCAAGA-3′ | 80 | 6 |
| ARBU09-R | 5′-CTGTCCCCTGAGCCCATCT-3′ | |||||
| Abscisic acid receptor | CcPYL3 | GT720590 | PYL31-F | 5′-CGGTGACGACTGTCCATGAG-3′ | 80 | 6 |
| PYL31-R | 5′-CGGCACGTCAACGATATACG-3′ | |||||
| Abscisic acid receptor | CcPYL7 | DV704550 | PYL72-F | 5′-GAGAAGCACATTCTTGGGATCAA-3′ | 80 | 6 |
| PYL72-R | 5′-GGATGCACGGTAAGGATGGA-3′ | |||||
| Amidase (IAA synthesis) | CcAMI1 | GT651356 | HD141612-F | 5′-AGATGTCTGCCACAGTGTGAAGA-3′ | 80 | 6 |
| HD141612-R | 5′-GGAGTGCAAGGATGCCAAAA-3′ | |||||
| RD29-like protein | CcRD29 | GT660256 | RD29-F | 5′-TGATGATCAAGATCCCCAACAC-3′ | 100 | 6 |
| RD29-R | 5′-CTTCGCTTTCGCCTTCACTT-3′ | |||||
| AP2/ERF DREB-like | CcDREB1 | GW463524 | DREBA09-F | 5′-CAATGCCTGCAAAGCCAATTA-3′ | 80 | 6 |
| DREBA09-R | 5′-TTTTCCTGCCTGCACGTTTC-3′ | |||||
| NAC-RD26-like | CcRD26 | GT003652 | NACRD26-F | 5′-TTTGGCCCTGCGCTCTAGT-3′ | 98 | 6 |
| NACRD26-R | 5′-AAGCGGGTCAGTTTCTCGAA-3′ | |||||
| MYB-type 2 transcription factor | CcMYB1 | GT689406 | MYB61-F | 5′-CCCGGCAATCTTCCAGCTA−3′ | 100 | 6 |
| MYB61-R | 5′-TCAAGCGTGGCAACTTCACT−3′ | |||||
| Caffeoyl-CoA 3-O methyltransferase | CcCCoAOMT1 | GT715419 | CCOAOMT-F | 5′-GACACACCAGCCCTTTCGAT-3′ | 100 | 6 |
| CCOAOMT-R | 5′-CAGCTCTCGCTCTTCCAGAAG-3′ | |||||
| Clp protease ATP-binding subunit | CcCLP1 | GT651512 | CLP1-F | 5′-CCACCCCAGTAGGACCAGAAA-3′ | 80 | 6 |
| CLP1-R | 5′-CATTCGTCGTGCTCGTGTTG-3′ | |||||
| Ascorbate peroxidase | CcAPX1 | GT697455 | ASCPER1-F | 5′-GACCTGAACAATGCCCAGAAG-3′ | 70 | 6 |
| ASCPER1-R | 5′-CGTAAATGAGCAGCAGGTGATG-3′ | |||||
| Ascorbate peroxidase | CcAPX2 | EE193467 | ASCPER6-F | 5′-AGACCGTGTCTCAAACCGACTAC-3′ | 80 | 5 CcAPX1 |
| ASCPER6-R | 5′ GTTGATCTGTTGGCCCAAAGA 3′ | |||||
| Leucine zipper (ABA signalling) | CcABI5 | GT645781 | ABI52-F | 5′-GCCGCTGCAGCCTCTATTT-3′ | 99 | 6 |
| ABI52-R | 5′-AGCTGAGCGTTGTTCCCTATCT-3′ | |||||
| Leucine zipper (ABA signalling) | CcAREB1 | EE198828 | AREB11-F | 5′-TGTTGTAAGGGAGGATGCTCAA-3′ | 100 | 6 |
| AREB11-R | 5′-TCCAAACCCAAAAGCCTGAT-3′ | |||||
| Homeobox leucine zipper hypothetical protein | CcHDZ1 | GT724869 | HD68272-F | 5′-GACGTTGACGGACGAGAACA−3′ | 100 | 6 |
| HD68272-R | 5′-TGTAGCCGCTGGCAACTG−3′ | |||||
| Homeobox leucine zipper hypothetical protein | CcHDZ2 | GW447493 | HD111162-F | 5′-AGTGCCAGGGAAAAGAATTCC-3′ | 100 | 6 |
| HD111162-R | 5′-TCCGGCTGTCTCTTTCTCATG-3′ |
R, reference genes tested, with CcUBQ10 and CcGAPDH being used as reference genes in qPCR experiments (*, **. gene expression tested by northern blot but not by qPCR); ND, gene expression not determined.
The size of the amplicon in bp is also indicated. The selected candidate genes (CGs) corresponded to (1) genes exclusively overexpressed in the leaf SH3 cDNA library of drought-tolerant clone 14 of C. canephora var. Conilon subjected to drought (Fig. 1A); (2) genes mainly overexpressed in the SH3 cDNA library (Fig. 1B); (3) genes identified by the macroarray experiment (Fig. 2); (4) genes encoding proteins showing differential accumulation by 2-DE (Fig. 3); (5) genes homologous to other CGs; and (6) genes selected from the literature. CGs are ordered as presented in the text.
Fig. 4.
Relative expression of the CcUNK10 gene standardized with different reference genes. Transcript abundances were analysed by qPCR in leaves of drought-tolerant (14 and 120) and drought-susceptible (22) clones of C. canephora grown with (I) or without (NI) irrigation and strandardized independently with the CcUBQ10 (ubiquitin), CcADP1 (ADP ribosylation factor), CcACT1 (actin), CcCYN1 (cyclophilin), CcCTS1 (cathepsin), and CcGAPDH (glyceraldehyde-3-phosphate dehydrogenase) genes as references. Values are the mean of at least three technical repetitions ±SD. The significance of expression level differences between the treatments was evaluated using the pairwise Wilcoxon rank test (non-parametric test). Treatments sharing the same letter are not significantly different.
Screening of macroarrays
The inserts of 3388 clones of a unigene set based on clustering ESTs from drought-acclimated cDNA libraries from C. arabica var. Catuaí (SH2) and C. canephora clone 14 (SH3) (Vieira et al., 2006; Mondego et al., 2011) were PCR amplified with the universal M13 reverse and forward primers. The concentration and quality of PCR products were checked on agarose gels. Aliquots of 50 μl each (±250 ng μl−1) were transferred to 384 plates. An equal volume of dimethylsulphoxide (DMSO) was added to each well. Spotting of the PCR products on nylon Hybond-N+ membranes (GE Healthcare) was performed using a Q-Bot (Genetix Inc.). Each PCR product was spotted in duplicate, in a 3×2 format, totalling 6912 spots including 68 controls. After spotting, nylon membranes were treated with denaturing and neutralizing solutions, according to the manufacturer. DNA was fixed onto membranes by UV cross-linking. Membranes were further hybridized independently with probes corresponding to single-stranded cDNAs obtained by reverse transcription of 30 μg of total RNA extracted from leaves of C. canephora clones 22 and 14 grown with (I) or without (NI) irrigation. Labelling was performed by random priming with [α-33P]dCTP. After overnight hybridization, nylon membranes were washed and revealed as described for northern blot experiments. Differentially expressed genes were identified using the ArrayGauge software (Fuji).
Proteome analysis by 2-DE
Proteins were extracted from leaves of clones 22 and 14 of C. canephora by a modified phenol/SDS method (Ramos et al., 2007) followed by 2-DE. The first dimension (isoelectric focusing) was carried out using 13 cm IPG strips (pH 3–10 or pH 4–7) in an IPGphor system (GE Healthcare), and the samples (500–1000 μg of proteins) were loaded and separated at 20 °C for 12 h. The second dimension was in an SDS–PAGE (11%) using the Hoefer SE600 Ruby system (GE Healthcare) with a 15 mA gel−1 for 45 min and a 30 mA gel−1 for 180 min at 12 °C. Gels were stained with Coomassie Blue G-250 and R-350, digitalized using an UMAX Image Scanner, and analysed with ImageMaster 2D Platinum 6.0 software (GE Healthcare). Protein spots differentially expressed were removed manually from gels and analysed by mass spectrometry using a MALDI-TOF/TOF (Auto-flex, Bruker) mass spectrometer.
Protein identification by MS
The proteins were identified by PMF (peptide mass fingerprinting) using PiumsGUI2.2, and MS/MS ion search using the software X!Tandem. The obtained sequences were screened against the SOL Genomics Network and other coffee sequences available in public databases. The packages Trans-Proteomic Pipeline (TPP) and Scaffold were used to analyse protein data. Results were further verified by de novo sequencing data using the FlexAnalysis software (Bruker Daltonics).
Results
Physiological characteristics of C. canephora clones
A leaf Ψpd close to zero confirmed the untreated condition of irrigated plants (Table 1). In addition, values of A, F v/F m of PSII, ΦPSII, q N, and P E were equal for clone 22 (drought-sensitive) and the drought-tolerant clones 14 and 120 under irrigation. The main differences between these clones concerned the g s values that were ∼2-fold higher for clone 22 than for clones 14 and 120. Under irrigation, the q P of drought tolerance was similar for all the clones.
After suspension of watering, clone 22 showed a faster decline [rate of decrease in the pre-dawn leaf water potential (RDPWP)] of Ψpd than the drought-tolerant clones to reach drought conditions (–3.0 MPa). Regarding the drought-tolerant clones, it is worth noting that the decrease of RDPWP was faster for clone 120 than for clone 14, but both reached –3.0 MPa 12 d after suspension of watering. The delay observed for clone 120 compared with clone 14 in initiation of its reduction of Ψpd explains the differences observed between these two drought-tolerant clones (Marraccini et al., 2011). When established, DA led to decreases in both g s and A, 80% and 73% for clone 14; 72% and 86% for clone 120; and 95% and 90% for clone 22, respectively. Independently of the clones, the g s reduction was greater than the decline of A, and was accompanied by a decrease in the C i/C a ratios. Altogether, these data clearly indicate that the inhibition of photosynthesis by drought tolerance is probably related to stomatal closure.
Regarding the maximum photochemical efficiency of PSII (evaluated by the F v/F m ratio), similar values were observed under irrigated (control) and non-irrigated conditions for the drought-tolerant clones 14 and 120. Even if a slight decrease in the F v/F m ratio was observed for the drought-sensitive clone under drought conditions, F v/F m values were close to that considered as an optimum (∼0.80), therefore indicating that no photoinhibitory damage occurred for clone 22. For all the clones, the reduction of A under DA was not accompanied by an increase of q N. However, all clones showed a reduction of q P, demonstrating that PSII excitation pressure increased. This indicates that a fraction of closed PSII reaction centresled to a decrease in ΦPSII observed during DA.
Selection of candidate genes
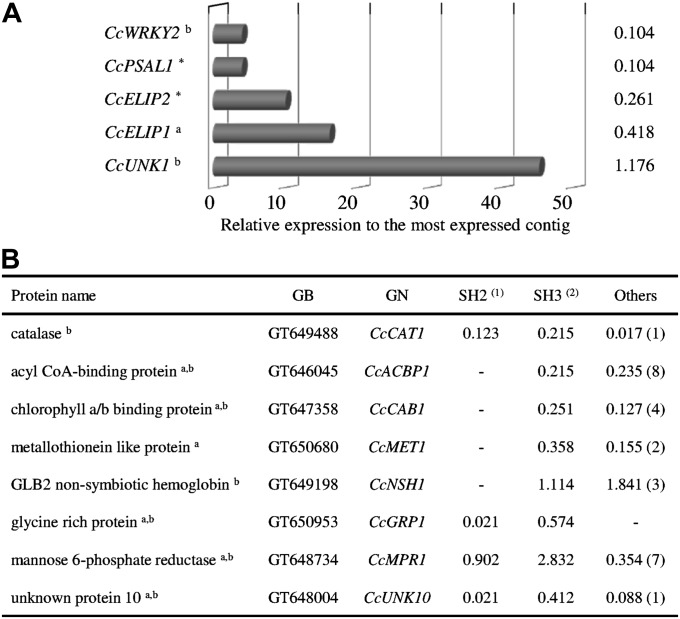
Based on the Brazilian Coffee Genome database, a first set of five CGs exclusively expressed in the leaf cDNA library (SH3) of drought-tolerant clone 14 of C. canephora subjected to drought was selected (Fig. 1A). It contained the genes encoding a WRKY transcription factor (CcWRKY2) and a psaL subunit of PSI (CcPSAL1); two putative early light-induced proteins (CcELIP1 and CcELIP2) that shared >99% amino acid identity, and a ‘no hit’ protein (CcUNK1; with UNK for unknown) were also identified.
Fig. 1.
In silico analysis of coffee ESTs overexpressed under drought conditions. (A) ESTs exclusively overexpressed in the leaves of drought-tolerant clone 14 of C. canephora var. Conilon subjected to drought (SH3 cDNA library). The numbers indicate the percentage of reads in the library (n total=5743 reads). Results of TBLASTX against the non-redundant protein sequence database (NR; E-value cut-off of 1e−10) were as follows: CcWRKY2 encoding a WRKY transcription factor (Ricinus communis, XP002529048, E-value 1e−115), CcPSAL1 (GT646615) encoding the photosystem I subunit XI (Nicotiana attenuata, AAO85557, E-value 2e−103), CcELIP2 (GT647647) encoding isoform 2 of a putative early light-induced protein (Ricinus communis, XP002517068, E-value 3e−59), and CcELIP1 encoding isoform 1 of a putative early light-induced protein (Camellia sinensis, ACB20694, E-value 3e−63). No hits were found for the CcUNK1 (DV689820) gene. (B) ESTs from C. canephora mainly overexpressed in the SH3 cDNA library (results of Fisher’s test). Values indicate the percentage of ESTs in the corresponding cDNA libraries. Numbers of other cDNA libraries containing reads are indicated in parentheses. Total reads in SH2 [(1) 5053] and SH3 [(2) 5743]. GenBank accession numbers (GB) and gene names (GN) of coffee ESTs are also given. a, Gene expression analysed by northern blot; b, gene expression analysed by qPCR; *, gene expression not determined.
A second set of CGs corresponded to reads overexpressed in leaf cDNA libraries from drought-acclimated plants of C. canephora (SH3) but also present in other libraries. This was the case for genes coding for a catalase (CcCAT1), an acyl-CoA-binding protein (CcACBP1), chlorophyll a/b- (CAB) binding protein (CcCAB1), a metallothionein-like protein (CcMET1), a GLB2-like non-symbiotic haemoglobin (CcNSH1), a glycine-rich protein (CcGRP1), a mannose 6-phosphate reductase (CcMPR1: EC 1.1.1.224), and an unknown protein (CcUNK10), all of them presenting higher in silico expression in the SH3 library of C. canephora than in the SH2 library of C. arabica var. Catuaí (Fig. 1B).
Identification of CG genes by the screening of macroarrays
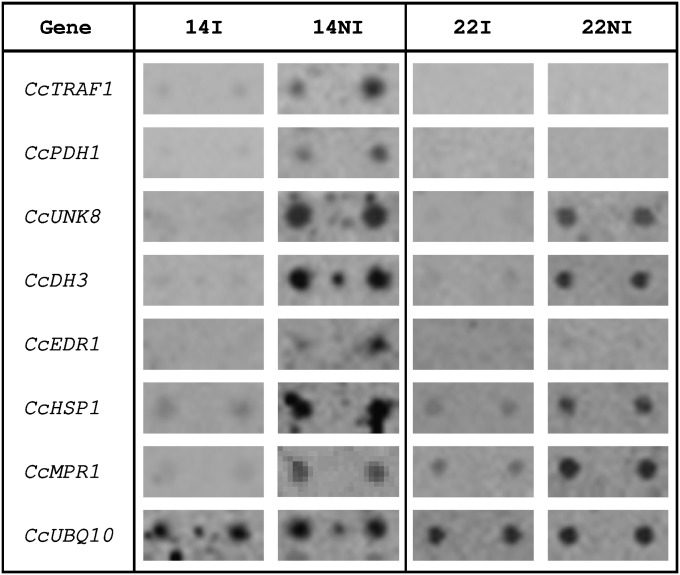
Another set of CGs was selected by screening macroarray membranes containing ESTs representing coffee unigenes expressed in SH2 and SH3 cDNA libraries, with cDNA probes corresponding to RNA extracted from leaves of C. canephora clones 22 and 14 subjected or not to drought conditions. This allowed the selection of 28 CGs that showed differential expression between the two clones of C. canephora and the water treatments. Hybridization results, presented for some of them (Fig. 2), concerned genes coding for a TRAF-like [tumour necrosis factor receptor (TNFR)-associated factor] protein (CcTRAF1), a pre-phenate dehydrogenase (CcPDH1: EC 1.3.1.12), an unknown protein 8 (CcUNK8), a dehydrin identical to the CcDH3 protein (Hinninger et al., 2006), an EDR1 (enhanced disease resistance)-like mitogen-activated protein kinase kinase (MAPKK) kinase protein (CcEDR1) previously reported to function as a negative regulator of disease resistance and ethylene-induced senescence (Tang et al., 2005), a small heat shock protein (CcHSP1), and the previously identified CcMPR1. The expression of these genes was further tested by qPCR.
Fig. 2.
Identification of candidate genes by the screening of macroarrays (reverse northern). Membranes were hybridized with cDNA probes representing RNA extracted from leaves of C. canephora clones 22 and 14 grown with (I) or without (NI) irrigation. Hybridization results are shown for the CcTRAF1, CcPDH1, CcUNK8, CcDH3, CcEDR1, CcHSP1, and CcMPR1 genes, and also for the constitutive CcUBQ10 gene. For all these genes, expression was analysed by qPCR.
Protein expression pattern in response to drought
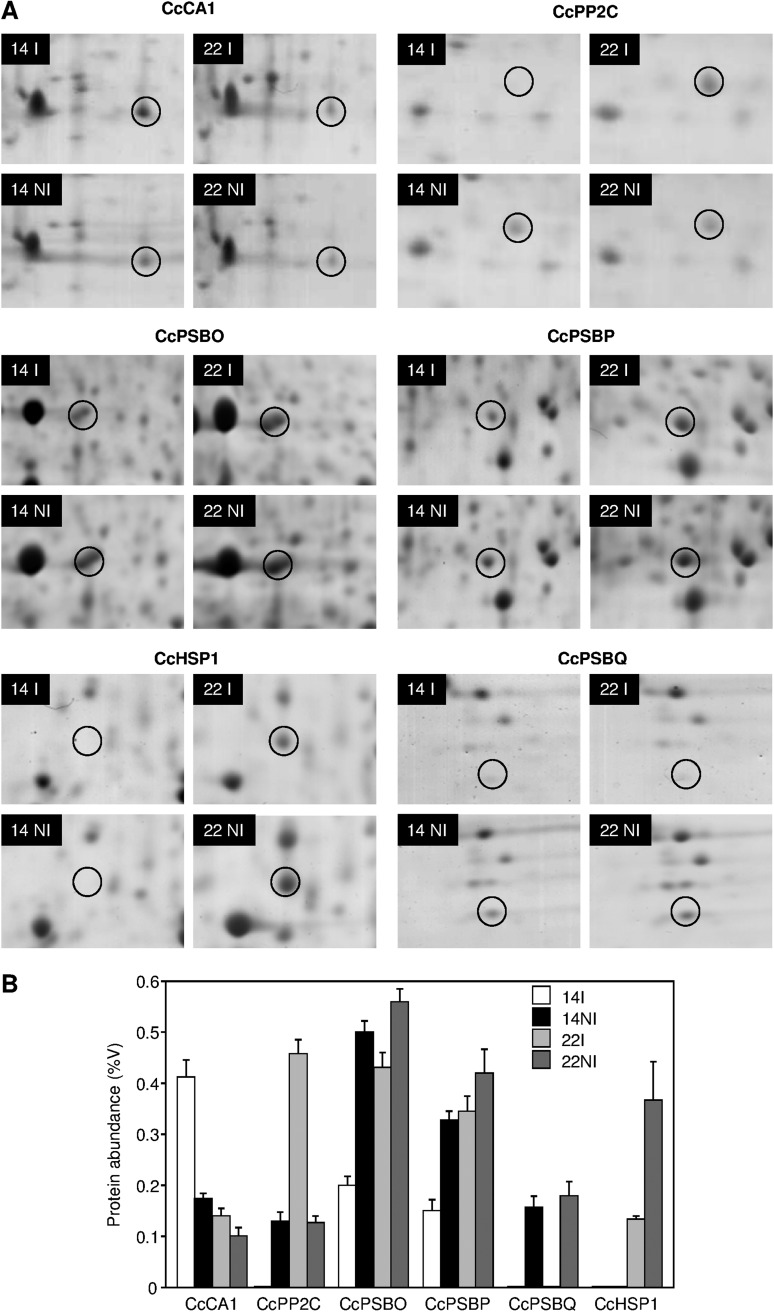
Finally, a 2-DE approach was also used to search for other CGs. The 2-DE of soluble proteins extracted from leaves of C. canephora clones 14 and 22 (under both I and NI conditions) usually led to the identification of 700–1000 well resolved spots (data not shown). Comparisons between these gels permitted selection of ∼40 intense proteins showing differential accumulation between coffee clones and with drought conditions applied to the plants. Some of these proteins were further sequenced by MALDI-TOF-MS/MS (Table 2). Among them, a chloroplastic carbonic anhydrase-like protein (CcCA1: EC 4.2.1.1) was identified in higher amounts in clone 14 than in clone 22 under untreated (I) conditions and decreased with drought (NI) for both clones (Fig. 3). For a type-2C protein phosphatase (CcPP2C: EC 3.1.3.16), a decrease with drought was observed in leaves of clone 22 while the opposite was observed in clone 14. 2-DE profiles also showed differential accumulation of extrinsic proteins of PSII involved in the oxygen-evolving complex (OEC), such as CcPSBO and CcPSBP proteins, with a higher amount in clone 22 than in clone 14 and increased accumulation under drought for both clones. For CcPSBQ, an increased amount of protein was found for both clones upon drought. A small heat shock protein (CcHSP1, described before) accumulated with DA in leaves of the drought-susceptible clone 22 but not in those of the drought-tolerant clone 14. For all these proteins, corresponding coffee SGN (Mueller et al., 2005) contig or EST (Lin et al., 2005; Vieira et al., 2006) sequences with high similarity were identified and used to design specific primers to analyse the effects of drought on gene expression by qPCR (Table 3).
Table 2.
Identification of leaf coffee proteins presenting differential accumulation during drought acclimation
Protein names and corresponding peptide sequences obtained by MALDI-TOF-MS/MS are indicated and validated statistically by Scaffold Score expressed as a percentage (%). The observed isoelectric point (pI) and molecular weight (MW) were calculated from the 2-DE gels by ImageMaster Platinum 6.0 Software. Homology searches were done with the TBLASTN program (Altschul et al., 1990) using an E-value cut-off of 1e−10. GenBank (GB) and SGN (http://solgenomics.net, Mueller et al., 2005) accession numbers of ESTs and contigs containing the sequenced peptides are indicated.
| Protein name | Gene name | Sequences | Scaffold Score (%) | Accession nos | pI/MW |
| Carbonic anhydrase | CcCA1 | EKYETNPALFGQLAK | 91 | GT008701 | 6.85/23 |
| YMIVACADSR | 85 | ||||
| VNPSLILSLQPGDAFIVR | 80 | ||||
| SVANLVPPYDQLK | 90 | ||||
| HSEFGAAIEFAVLHLK | 70 | ||||
| VENIVVIGHSACGGIK | 60 | ||||
| GLMSIPEDGTTSTDFIEDWVK | 90 | ||||
| VKTEHGSKPLPEQIVLAE | 95 | ||||
| Type-2C protein phosphatase (PP2C) | CcPP2C | KTSTSLMALSSPQLR | 90 | GW435032 | 5.26/22 |
| EEMEDDVIIVR | 95 | SGN-U631279 | |||
| ALEEAFESADMK | 85 | ||||
| EAGGWISNGR | 85 | ||||
| Oxygen-evolving complex PSII (psbO) | CcPSBO | KDGIDYAAVTVQLPGGERV | 90 | GT647517 | 5.46/33 |
| RLTYDEIQSK | 74 | SGN-U629535 | |||
| Oxygen-evolving complex PSII (psbP) | CcPSBP | WNPSKEVEFPGQVLR | 90 | GW488995 | 5.76/29 |
| YEDNFDSNSSVSVIIIPCDKK | 50 | ||||
| SITDYGPPEEFLSK | 95 | ||||
| VDYLLGK | 52 | ||||
| TEAEGGFEPNTVATANILEQETPIV | 55 | ||||
| TADGDEGGKHQLIR | 90 | ||||
| Oxygen-evolving complex PSII (psbQ) | CcPSBQ | RFFIQALPPAGAAARA | 90 | GT645658 | 10.01/15 |
| DLDLPLKDR | 74 | SGN-U637442 | |||
| LKAELLR | 62 | ||||
| STPEAEKYYAATVSTLNDVLSK | 95 | ||||
| YDLNTVISAKPK | 90 | ||||
| Small heat shock protein (sHSP) | CcHSP1 | SAPVAPIGVLDRFPTAR | 95 | GW447897 | 5.87/22 |
| TVQQMMETMER | 91 | ||||
| NEDGEEEEKNEWSAK | 90 | ||||
| LMEDPFAYSGGWPSPLAPDTGGYSR | 60 | ||||
| GRTPWEIK | 60 | ||||
| ALPENAQF | 70 |
Fig. 3.
Protein responses in coffee leaf exposed to drought. (A) Plants were irrigated (I) or not irrigated (NI); leaf soluble proteins were extracted and separated by two-dimensional gel electrophoresis (2-DE), stained by Coomassie Brillant Blue, and scanned. The results shown are representative of at least three repetitions. Spots corresponding to CcCA1 (carbonic anhydrase) and CcPP2C (type-2C protein phosphatase) proteins were analysed using strips with an immobilized linear pH gradient of 3–10. Spots corresponding to CcPSBO (PSII oxygen evolving complex protein, OEC), CcPSBP (OEC), and CcHSP1 (small heat shock protein) proteins were analysed using strips with an immobilized linear pH gradient of 4–7, and CcPSBQ (OEC) with pH gradient 6–11. Proteins were further characterized by MALDI-TOF-MS/MS (see Table 2). (B) Normalized protein abundance of the coffee leaves extracted from clones 14 and 22 of C. canephora subjected (NI) or not subjected (I) to drought. Protein abundance was deduced from the 2-DE using ImageMaster Platinum 6.0 software and is expressed as a percentage of volume (%V) which was calculated from the gel images as the volume of a specific spot divided by the sum of the volume of all other spots present in the gel multiplied by 100. Volume percentage averages were calculated from 2-DE gels loaded with three biological replicates.
Expression profiles of candidate genes
As described before, CGs were selected by either electronic northern, screening of macroarrays, or after the comparison of 2-DE gels. This list was also increased by adding several other CGs as yet not reported in the literature [e.g. Responsive to Dehydration (RD)-type genes) to respond upon drought conditions. Expression profiles of CGs were further checked in leaves of C. canephora clones 14, 22, and 120 by classical northern blot and qPCR experiments. In some cases, expression profiles were also assessed by both methods. These analyses led to the identification of differentially expressed genes under irrigated and non-irrigated conditions. These results are detailed below in separated sections.
Analysis of CcUNK10 gene expression standardized with different housekeeping genes
Even thought it codes for an unknown function, the study of CcUNK10 is of a particular interest because this gene was preferentially expressed in the drought-sensitive clone 22, mainly under untreated condition. but was also highly induced by drought. Because the accuracy of qPCR always requires the use of a reference gene to normalize expression levels, several housekeeping genes were tested to standardize CcUNK10 expression (Fig. 4). It is worth noting that CcUNK10 expression profiles were similar when using the CcUBQ10 (ubiquitin), CcADP1 (ADP ribosylation factor), CcACT1 (actin), and CcGAPDH (glyceraldehyde-3-phosphate dehydrogenase) genes as references to standardize the results of relative quantification. In these cases, the CcUNK10 expression levels were always conserved [22NI>22I=120NI≥14NI>14I=120I] but differed from those that used CcCYN1 (cyclophilin) and CcCTS1 (cathepsin) as references. Therefore, qPCR analyses of all identified CGs were performed using CcUBQ10 or CcGAPDH.
Genes with reduced expression under drought acclimation
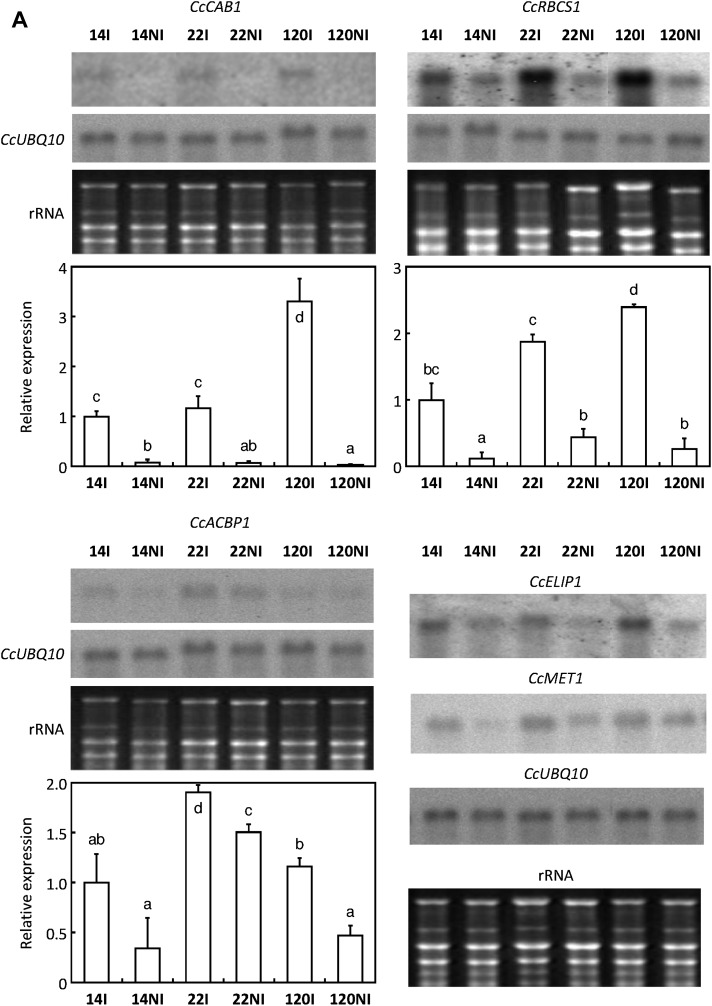
Expression of genes coding for CAB-binding protein (CcCAB1), the small subunit (CcRBCS1) of ribulose 1,5-bisphosphate carboxylase (Marraccini et al., 2003), and an acyl-CoA-binding protein (CcACBP1: EC 3.4.17.3) was studied by northern blot and qPCR experiments (Fig, 5A). For all clones of C. canephora tested, expression of CcCAB1 and CcRBCS1 genes decreased greatly with drought. For both genes, expression levels were higher under irrigated conditions for clone 120 as compared with clones 14 and 22. For CcACBP1, expression upon drought and irrigated conditions was higher in clone 22 than in both tolerant clones 14 and 120. The decrease under the NI condition was higher in the tolerant clones. For all these genes, expression profiles obtained by northern blot and qPCR were identical. Northern blot experiments also showed that drought caused the down-regulation of CcELIP1 and CcMET1 gene expression for all C. canephora clones tested (Fig, 5A).
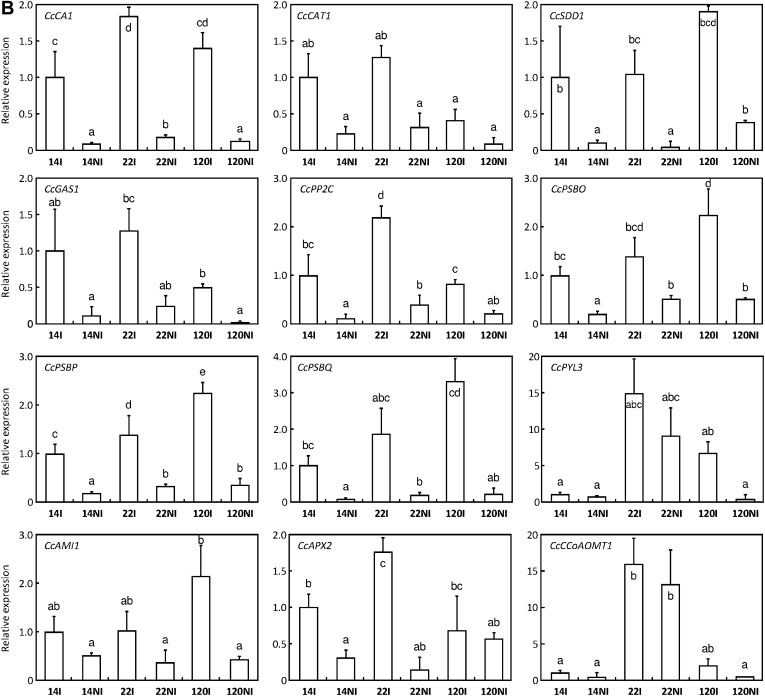
QPCR experiments also demonstrated that expression of the genes CcCA1, CcCAT1, CcSDD1 (coding for a subtilisin-like serine protease), CcGAS1 (coding for a glucosyltransferase arbutin synthase, EC 2.4.1.218), and CcPP2C declined with drought, except for clone 120 in the case of CcCAT1 where this decline was not statistically different (Fig. 5B). In the same way, expression of CcPSBO, CcPSBP, and CcPSBQ genes encoding OEC proteins of PSII was also greatly reduced under DA (Fig. 5B). Down-regulated expression of CcAMI1 (coding for an amidase involved in indole acetic acid synthesis), CcCCoAOMT1 coding for the caffeoyl-coenzyme A 3-O methyltransferase, (Lepelley et al., 2007) and CcAPX2 (coding for an ascorbate peroxidase) was also observed upon drought (Fig. 5B). Interestingly, for CcPYL3 [coding for an abscisic acid (ABA) receptor], down-regulation was clearly observed only for clone 120 (Fig. 5B).
Fig. 5.
Expression profiles of genes down-regulated during drought acclimation. Gene expression was analysed in leaves of clones 14, 22, and 120 of C. canephora subjected (NI) or not (I) to drought. (A) Abundances of specific transcripts were analysed by northern blot and qPCR for CcCAB1, CcRBCS1, and CcACBP1. Transcript abundances of CcELIP1 and CcMET1 genes were monitored only by northern blot. For this analysis, total RNA stained with ethidium bromide was used to monitor the equal loading of the samples. Constitutive expression of the CcUBQ10 gene is also shown. (B) For other genes, transcript abundances were analysed by qPCR. The expression of the CcUBQ10 gene was used as a reference to measure the relative quantification that corresponds to the mean of three technical repetitions ±SD. The gene names are indicated in the histograms. Results are expressed using 14I as an internal calibrator. In each case, values of relative quantification correspond to the mean of at least three technical repetitions ±SD. Significance of expression level differences between the treatments was evaluated using the pairwise Wilcoxon rank test (non-parametric test). Treatments sharing the same letter are not significantly different.
Genes with increased expression under drought acclimation
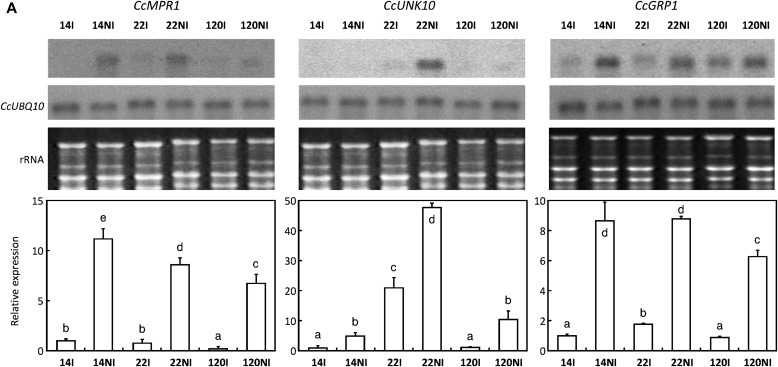
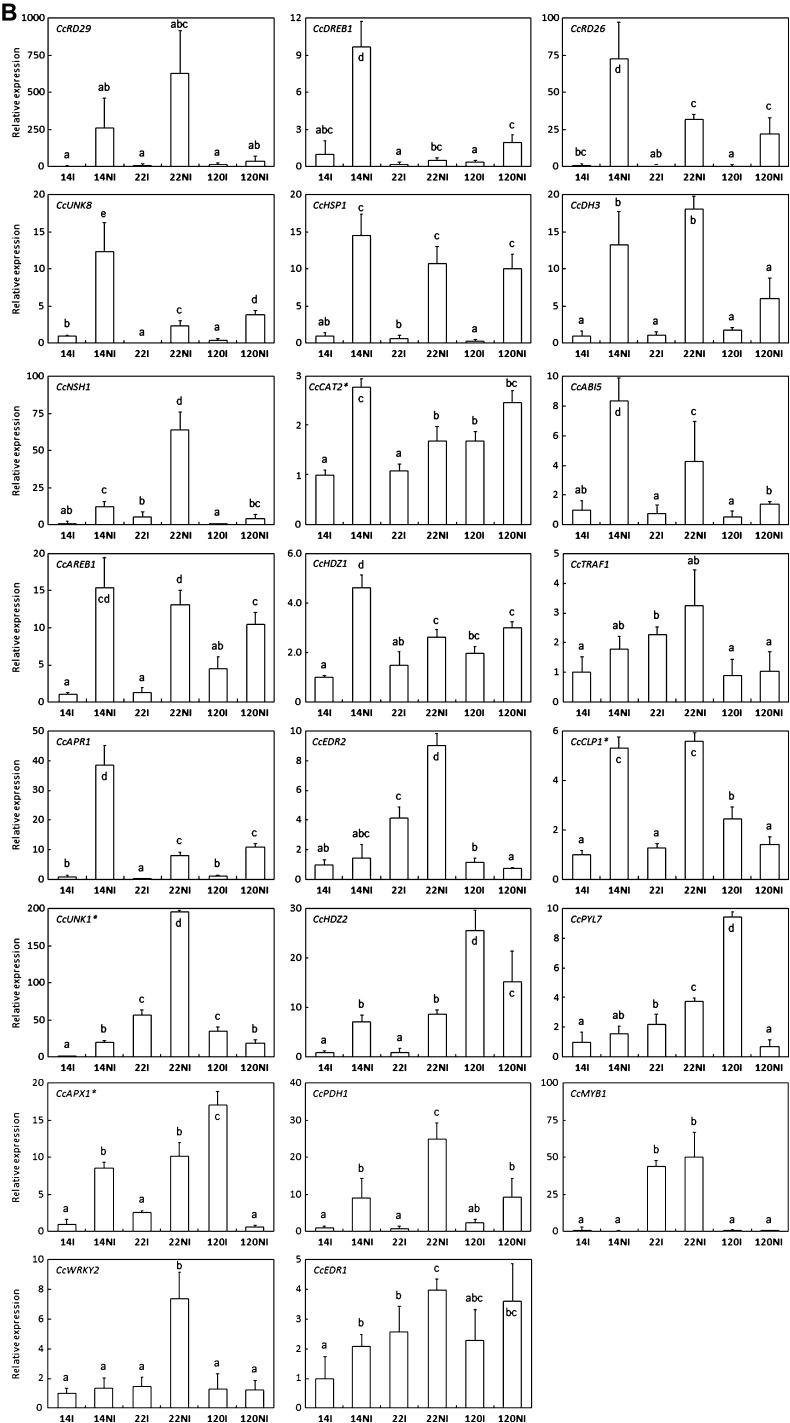
As demonstrated by northern blot and qPCR analyses, drought increased the expression of CcMPR1, CcUNK10, and CcGRP1 genes (Fig. 6A). On the other hand, water limitation also increased the expression of CcRD29, CcDREB1, and CcRD26 genes coding for RD29-like protein, and the AP2/ERF DREB-like and NAC-RD26-like transcription factors, respectively (Fig. 6B). It is worth noting that higher levels of induction upon drought were observed for CcDREB1 and CcRD26 in the drought-tolerant clone 14 as compared with the other clones tested under the same conditions. A similar differential pattern was also observed for the regulatory genes CcABI5, CcAREB1, and CcHDZ1 (Fig. 6B). Furthermore, the CcHSP1, CcDH3, CcAPR1, and CcCAT2, as well as the CcUNK8 expression profile followed a similar pattern (Fig. 6B). In summary, the results show that for all these genes mentioned above, clone 14 displays a clear stronger induction upon DA (12 d of DA for both clones) than clone 120, suggesting that different mechanisms may account for their drought tolerance phenotype.
Fig. 6.
Expression profiles of genes up-regulated during drought acclimation. Gene expression was analysed in leaves of clones 14, 22, and 120 of C. canephora subjected (NI) or not (I) to drought. (A) Abundances of specific transcripts were analysed by northern blot and qPCR for CcMPR1, CcUNK10, and CcGRP1. For northern blot analyses, total RNA stained with ethidium bromide was used to monitor the equal loading of the samples. Constitutive expression of the CcUBQ10 gene is also shown. (B) For other genes, transcript abundances were analysed by qPCR. The expression of the CcUBQ10 gene was used as a reference to measure the relative quantification, except for the genes marked with an asterisk that used the expression of the CcGAPDH gene as reference. The gene names are indicated in the histograms. Results are expressed using 14I as an internal calibrator. In each case, values of relative quantification correspond to the mean of at least three technical repetitions ±SD. Significance of expression level differences between the treatments was evaluated using the pairwise Wilcoxon rank test (non-parametric test). Treatments sharing the same letter are not significantly different.
In addition, the expression patterns displayed by other regulatory genes such as CcERD2, CcHDZ2, and the ABA receptor-encoding gene CcPYL7, whereby induction upon DA was observed for clone 14 and down-regulation was observed for clone 120, also corroborates this hypothesis (Fig. 6B). Other genes such as CcCLP1, CcUNK1, and CcAPX1 also follow the same pattern, but in the case of CcTRAF1, clone 120 displayed unchanged levels upon drought (Fig. 6B).
The expression of CcNSH1, CcPDH1, and CcWRKY2 was also increased by drought, at higher levels in the drought-sensitive clone 22 than in both tolerant clones 14 and 120 (Fig. 6B). Similarly, higher expression levels of the CcMYB1 gene coding for a helix–turn–helix MYB-type 2 transcription factor was observed for clone 22 as compared with both tolerant clones, but in this case induction upon DA was not statistically different. Finally, transcript accumulation upon DA was also observed for CcEDR1 in all clones tested (Fig. 6B).
Discussion
The analyses of physiological parameters performed under irrigation showed higher values of g s and C i/C a for the drought-sensitive clone 22 compared with the drought-tolerant clones 14 and 120. For the latter, the results presented here were identical to those previously reported for the same clone by Pinheiro et al. (2004). Taken together, these analyses suggested that the drought-sensitive clone 22 had a lesser efficiency in controlling stomatal closure and transpiration than the drought-tolerant clones 14 and 120, a situation that could explain its faster decline of Ψpd and greater reduction of A under drought. Because the stomatal closure reduced transpiration more than photosynthesis, the drought-tolerant clones 14 and 120 should have higher water use efficiency than the drought-sensitive clone 22 as previously observed by carbon isotope discrimination for other clones of C. canephora (Lima et al., 2002; DaMatta et al., 2003). The data presented here also suggested that the photosynthetic inhibition was mainly due to stomatal closure. However, non-stomatal limitation of photosynthesis, possibly associated with the decrease of Rubisco regeneration and carboxylation, could also occur (Kanechi et al., 1996; DaMatta et al., 1997, 2002). The reduction of ΦPSII observed under DA, for the drought-tolerant clones 14 and 120, was not accompanied by variations of F v/F m, probably being associated with the inhibition of PSII. Such a reduction should reflect a mechanism of photoprotection under DA, by adjusting the rate of electron transport to the rate reducing power, for example through photorespiration.
In order to study the molecular changes that occurred during DA, an integrated analysis was performed to identify proteins and CGs presenting differential expression profiles under drought stress, using different approaches. For all the CGs identified, a qPCR analysis was used to ascertain the expression pattern upon DA. The accuracy of qPCR experiments requires a reference gene to normalize expression levels, such as CcUBQ10 that was used as a reference gene in coffee (C. arabica L. cv. Laurina) to standardize expression levels of a large set of genes during fruit development (Salmona et al., 2008; Joët et al., 2009). Recently, Cruz et al. (2009) recommended using CcUBQ10 and CcGAPDH genes as internal references because of their stable expression in drought-acclimated leaves of C. arabica cv. Catuaí vermelho. In addition, the use of CcGAPDH as a reference gene was also recommended to realize accurate and reliable normalization of tissue/organ-specific gene expression in C. arabica (Barsalobres-Cavallari et al., 2009). In the present work, expression levels of the CcUNK10 gene in leaves of C. canephora were identical using either CcUBQ10 or CcGAPDH as reference genes, and the former was chosen to standardize most of the qPCR experiments performed.
The results presented here indicated that a number of genes were down-regulated and others up-regulated under drought. Furthermore, some genes displayed a clear differential pattern of expression between the clones tested. Based on the biological functions of these genes, the results demonstrate that different categories of the signalling pathways of drought response in coffee have been identified; and these are discussed below, focusing mainly on the responses displayed by the drought-tolerant clones.
Core components and modulators of ABA signalling
Results of qPCR assays of two coffee homologues of the recently identified PIR/PYL/RCAR type of ABA receptors (Ma et al., 2009; Cutler et al., 2010; Hubbard et al., 2010) indicated that CcPYL3 and CcPYL7 expression was higher in clone 120 than in clone 14 under irrigated conditions and its expression decreased upon DA. On the other hand, CcPYL7 expression was induced by drought only in clone 14, suggesting a probable involvement of the ABA signalling pathway in coffee responses to drought. This result is in agreement with data presented by Sun et al. (2011), where expression of some PYL- encoding genes from tomato was down-regulated by DA and some others genes showed an increase in expression.
In coffee, Silva (2007) showed that severe drought (Ψpd ∼ –3.0 MPa) effectively led to an increase of ABA quantified in leaves of both drought-tolerant (clone 120) and sensitive (clone 109A) clones of C. canephora var. Conilon. According to the newly developed model for early ABA signalling, a core mechanism is based on a three-component system made up of three protein classes: PIR/PYL/RCARs, protein phosphatase 2Cs (PP2Cs), and SNF1-related protein kinase 2s (SnRK2s) (Hauser et al., 2011). Data presented here for CcPP2C indicated a decreased expression of this gene upon DA and lower expression for both drought-tolerant clones under irrigated conditions as compared with clone 22. This result also supports a probable active ABA signalling pathway in coffee in response to drought and is in agreement with the newly established model for ABA signalling, in which the PYR/RCARs act as ABA receptors, the PP2Cs act as negative regulators of the pathway, and SnRK2s act as positive regulators of downstream signalling (Ma et al., 2009; Park et al., 2009). In that way, a double-negative regulatory pathway is established, whereby ABA-bound PYR/RCARs inhibit PP2C activity and PP2Cs inactivate SnRK2s (Park et al., 2009; Umezawa et al., 2009; Vlad et al., 2009). Thus, in the absence of ABA, the PP2Cs are active and repress SnRK2 activity and downstream signalling. In the presence of ABA, PYR/RCARs interact with PP2Cs and inhibit phosphatase activity, allowing SnRK2 activation and phosphorylation of target proteins. Furthermore, in addition to the lower gene expression, the protein content of CcPP2C was lower for clone 14 under irrigated conditions as compared with clone 22, pointing to a potentially more active ABA signalling pathway in clone 14. Based on these results, and considering that coffee PP2Cs function as a negative regulator of ABA signalling, the down-regulation of this gene under drought would probably sensitize coffee plants to ABA and its downstream responses.
Secondary messengers
A clear difference in the expression pattern of CcNSH1, encoding a non-symbiotic haemoglobin, was observed between the clones studied. Under irrigated conditions clone 22 showed a higher expression of CcNSH1 (>5-fold) than both drought-tolerant clones, and a 7-fold increased expression was also observed for clone 22 upon DA as compared with the drought-tolerant clones 14 and 120 under the same conditions. Non-symbiotic haemoglobins have been shown to be involved in scavenging of nitric oxide (NO) molecules, which play a key role in oxygen sensing/balancing in plants and animals (Dordas et al., 2003; Perazzolli et al., 2006). Therefore, the results suggest that NO levels might be elevated in the drought-tolerant clones which could be due to the lower scavenging capacity of CcNSH1 observed for these genotypes. In recent years, NO has emerged as an important endogenous plant signalling molecule that mediates many developmental and physiological processes (Neill et al., 2008). In leaves, NO mediates ABA-induced stomatal closure (Desikan et al., 2002, 2004; Garcia-Mata and Lamattina, 2002), modulates the rate of leaf expansion (Leshem and Haramaty, 1996), and may affect metabolic processes such as photosynthesis (Hill and Bennett, 1970) and respiration (Zottini et al., 2002).
Furthermore, ABA also stimulates the generation of H2O2 in guard cells, and pharmacological and genetic data demonstrate that NO accumulation in these cells is dependent on such production. Recent data have extended this model to maize mesophyll cells where the induction of antioxidant defences by drought and ABA required the generation of H2O2 and NO in addition to the activation of a MAPK. The present results demonstrate that there is an increased expression of CcEDR1 (similar to the ENHANCED DISEASE RESISTANCE 1 from Arabidopsis) upon drought for all clones tested. In contrast, CcERD2 displays a decrease in expression upon DA in clone 120. EDR1 encodes a CTR1-like kinase which was previously reported to function as a negative regulator of disease resistance and ethylene-induced senescence. Tang et al. (2005) reported that the edr1 mutant displays enhanced stress responses and spontaneous necrotic lesions under drought conditions in the absence of pathogen, suggesting that EDR1 is also involved in stress response signalling and cell death regulation. MAPKs are abundant in all eukaryotes and phosphorylate a wide range of target proteins, including other kinases and/or transcription factors (Colcombet and Hirt, 2008). Diverse stimuli, including pathogens, abiotic stress, H2O2, ABA, and phytohormones, activate MAPKs. A role for MAPKs in ABA signalling has already been demonstrated (Colcombet and Hirt, 2008) as well as a negative regulation of MAPK pathways by PP2Cs (Schweighofer et al., 2004).
Published data suggest that drought and salinity induce NO generation which activates cellular processes that afford some protection against the oxidative stress associated with these conditions. Thus, the data suggest an emerging model of stress responses in which ABA has several functions, including the rapid induction of stomatal closure to reduce transpiration water loss and the activation of antioxidant defences to combat oxidative stress, processes involving NO as a key signalling intermediate (Neill et al., 2008).
Transcription factors
Differential patterns of expression between clones 22 and the drought-tolerant clones 14 and 120 were also observed for genes involved in transcriptional regulation. The qPCR results presented here demonstrated that water deficit induced differential effects between the two drought-tolerant clones on the expression of regulatory genes such as CcABI5, CcAREB1, CcHDZ1, CcHDZ2, CcDREB1, and CcRD26. Upon drought, clone 14 displayed a higher increase in mRNA levels for CcABI5, CcAREB1, CcHDZ1, CcHDZ2, CcDREB1, and CcRD26 as compared with clones 120, indicating that different mechanisms may account for their drought tolerance phenotype. In agreement with this hypothesis, in the case of CcHDZ2, clone 120 even showed a decrease in expression upon DA. Those genes comprise components of the ABA-dependent (CcABI5 and CcAREB1) and ABA-independent (CcHDZ1, CcHDZ2, CcDREB1, and CcRD26) signalling pathways. In plants, it is well known that two pathways of signal transduction exist in the control of stress-related genes: one dependent on ABA and another independent of this phytohormone (Yamaguchi-Shinozaki and Shinozaki, 2005). In this scheme, it was demonstrated that transcription factors belonging to the ERF/AP2 family (e.g. DREB), but also NAC/HD-ZIP transcription factors, controlled the expression of the RD29A gene, encoding a late embryogenesis abundant (LEA)-like hydrophilic protein, co-operatively or separately in an ABA-independent manner (Qiang et al., 2000). The very high expression of CcRD29 observed in the leaves of clones 14 and 22 upon drought is noteworthy.
The results obtained by qPCR for CcMYB, encoding a MYB protein displaying high similarity to MYB4 from Arabidopsis, shows a very different pattern of expression between the coffee clones tested. The expression of CcMYB1 is almost absent in the drought-tolerant clones 14 and 120, and a 50-fold difference in expression is observed when compared with the results obtained for clone 22. In Arabidopsis, a group of MYB transcription factors have been reported to regulate the biosynthesis of secondary metabolites (Stracke et al., 2001), and MYB4 acts as a transcription repressor inhibiting expression of the cinnamate 4-hydroxylase (C4H) gene, which encodes a key enzyme in hydroxycinnamate ester biosynthesis. In a myb4 knockout mutant, hydroxycinnamate esters accumulate, resulting in tolerance of the mutant plant to UV-B (Jin et al., 2000). In that sense, due to the absence of negative regulation performed by CcMYB1, the drought-tolerant clones might have a higher content of secondary metabolites such as the antioxidant chlorogenic acid, a hydroxycinnamic acid synthesized by the same pathway. A higher content of antioxidant would certainly contribute to drought tolerance. However, a pattern of expression similar to that of CcMYB1 (e.g. higher expression in clone 22 and almost null expression for the drought-tolerant clones 14 and 120) was observed for CcCCoAOMT1, the first enzyme in the phenylpropanoid pathway. Further analysis of the metabolite content of these clones is needed to clarify this point.
From the set of transcription factor genes tested, only CcWRKY2 had unchanged expression upon drought for both drought-tolerant clones tested. However, a significant induction of CcWRKY2 was observed for the drought-sensitive clone 22 under drought. Recently, the Arabidopsis WRKY2 transcription factor has been shown to mediate seed germination and post-germination arrest of development by ABA (Jiang and Yu, 2009). Analysis of the WRKY2 expression level in ABA-insensitive and ABA-deficient mutants further indicated that ABA-induced WRKY2 accumulation requires ABI5, ABI3, ABA2, and ABA3. Furthemore, the authors demonstrated that the ABA hypersensitivity of the wrky2 mutants is attributable to elevated mRNA levels of ABI5, ABI3, and ABI5-induced Em1 and Em6 in the mutants (Jiang and Yu, 2009). The results presented here also show a similar pattern of expression as, upon drought, both tolerant clones displayed negligible expression of CcWRKY2. On the other hand, increased expression of CcABI5 was observed for both clones under the same drought condition.
Photosynthesis-related genes
As previously reported in Arabidopsis (Seki et al., 2002), a clear down-regulated expression of CcCAB1, CcPSBO, CcPSBP, CcPSBQ, and CcRBCS1 was observed in leaves of C. canephora clones under drought. Drought also repressed the expression of CcCA1 coding for the carbonic anhydrase (CA) supplying CO2 for Rubisco (Badger and Price, 1994). In barley, CA activity increased in response to elevated ABA concentration, and in wheat CA activity increased during the first period of drought in flag leaves of drought-resistant genotypes but was shown to decrease at the last stage of vegetation (Popova et al., 1996; Guliyev et al., 2008). Northern blot analysis shows that CcELIP1 was also down-regulated by drought, for all clones evaluated. The early light-induced proteins (ELIPs) belong to the multigenic family of light-harvesting complexes, which bind chlorophyll and absorb solar energy in green plants. Reports indicate that ELIPs fulfil a photoprotective function that could involve either the binding of chlorophylls released during turnover of pigment-binding proteins or the stabilization of the proper assembly of those proteins during high light stress (Hutin et al., 2002). Taken together, these results clearly demonstrated that leaf expression of several photosynthetic genes was greatly affected by DA in C. canephora.
Furthermore, results of a 2-DE experiment indicated identical responses for CcCA1-encoded protein, with a reduction under DA in leaf protein extracts for both clones 14 and 22 of C. canephora. Other differential responses between gene expression and protein accumulation were also clearly observed for CcPSBP, CcPSBQ, and CcPSBO, with reduced gene expression under DA and a higher amount of corresponding proteins under drought than under the irrigated condition. This situation, similar to that observed for the Rubisco protein (Marraccini et al., 2011), could be explained by (i) the action of chaperones that could prevent the protein degradation under DA; (ii) the up-regulation of other gene alleles to compensate the down-regulation of those measured during DA; and (iii) the protein translation that occurred from mRNA transcribed during the night. In that case, nocturnal accumulation of mRNAs could participate in maintaining the high daytime amount of the corresponding protein even under a sharp reduction in gene expression. Then, this should favour a quick recovery of photosynthetic capacity under favourable environmental conditions and help coffee plants to cope with drought.
Drought-responsive structural genes
Genes involved in DA such as that encoding a small heat shock protein (CcHSP1), a CcDH3 coding for a coffee dehydrin, and CcCLP1, coding for an ATP-dependent calpain protease, also displayed differential responses upon drought and between the drought-tolerant clones. Heat shock proteins, dehydrins, and ATP-dependent calpain proteases have been previously reported to be involved in protein folding and protein turnover upon DA (Close et al., 1993; Zagdanska and Wishnewski, 1998; Demirevska et al., 2008). The results of protein analysis by a 2-DE experiment also confirmed the up-regulation of CcHSP1, particularly for clone 22. For clone 14, the same explanation given above for photosynthesis-related genes could apply to explain the differences between gene expression and protein accumulation. The importance of narrowing down the right protein isoform or allele is further supported by the results obtained here, whereby upon drought, the catalase-encoding gene CcCAT1 is down-regulated and CcCAT2 (encoding another catalase) is up-regulated. Similar results were obtained for the ascorbate peroxidase genes CcAPX2 (down-regulated) versus CcAPX1 (up-regulated). Furthermore, in the case of CcAPX1, a clear differential response was observed between the drought-tolerant clones 14 and 120, as it is induced upon DA in clone 14 and down-regulated in clone 120. The involvement of another APX in drought responses in clone 120 has still to be investigated. In the case of the oxidoreductase genes CcAPR1 and CcMPR1, up-regulation upon drought was observed for all clones tested. However, higher induction for clone 14 was observed compared with clone 120. Therefore, the results obtained for CcCAT2, CcAPX1, and CcMPR1 indicate a stronger induction of the coffee antioxidant and osmoprotection systems upon DA in clone 14 as compared with the other clones.
Pinheiro et al. (2004) reported that clone 14 displayed increased activity (38%) of APX, an enzyme which has high affinity for H2O2, as a result of drought (Smirnoff, 1995; Polle, 2001; Mittler, 2002). The authors concluded that APX might have a key role in allowing clone 14 to cope with potential increases in H2O2 under drought conditions. In the same vein, the present qPCR results for CcAPX1 show a clear induction of this gene upon drought. In addition, a clear difference in expression between clones 14 and 120 was observed, as CcAPX1 decreases upon DA, suggesting a role for CcAPX1 in the drought tolerance phenotype in clone 14. However, clone 22 displayed a similar pattern of induction of CcAPX1 as clone 14 and, therefore, further studies aiming at characterizing other APX-encoding genes from coffee and their expression upon DA might be needed.
The results presented here also identified DA-inducible expression of the CcPDH1 gene coding for a prephenate dehydrogenase protein and down-regulation of the CcGAS1 gene encoding arbutin synthase (Hefner et al., 2002) with water limitation in C. canephora. The first enzyme catalyses the aromatization of prephenate to form tyrosine precursors and subsequently phenylalanine, and might affect the synthesis of several secondary metabolites (Warpeha et al., 2006; Urano et al., 2009), and CcGAS1 is involved the synthesis of arbutin (4-hydroxyphenyl-β-D-glucopyranoside), a phenolic glucoside compound that accumulates in leaves of some drought- and cold-tolerant plant species such as Myrothamnus flabellifolia where it could represent 25% of leaf dry weight (Bianchi et al., 1993; Hincha et al., 1999). As previously mentioned for CcCCoAOMT1, to address the role of arbutin and other phenolic compounds to prevent cell damage under abiotic stresses in coffee, further analysis of the metabolite content of these clones should be performed.
Down-regulated expression of the CcSDD1 gene encoding a subtilisin-like serine protease (SDD: stomatal density and distribution) involved in stomata development was also observed under drought. In Arabidopsis, sdd mutants showed increased stomata number, indicating that this gene is a negative regulator of guard cell formation (Von Groll et al., 2002). Compared with the CcUBQ10 expression level, the expression level of CcSDD1 reported here in C. canephora leaves was low. This is expected considering that expression of this gene is mainly observed in stomata precursor cells. For this reason, the analysis of CcSDD1 expression in meristem cells of stressed and unstressed coffee plants should be of particular interest.
As already reported before in other plants (Dubos et al., 2003; Le Provost et al., 2003; Rodriguez et al., 2006; Wang et al., 2009), stress-inducible expression of the CcGRP1 gene coding for a glycine-rich protein was also observed. Currently, the role of GRPs in abiotic stress in plants is still unknown, but these proteins were localized in the cell wall of many higher plants where they may play a role in its maintenance, reinforcement, and repair during the dehydration–rehydration process (Harrak et al., 1999; Ringli et al., 2001; Le Provost et al., 2003; Mousavi and Hotta, 2005). Some drought-inducible genes were also found whose functions in relation to drought are still poorly understood. Results have identified three novel genes (CcUNK1, CcUNK8, and CcUNK10) highly induced upon drought. The functions of these genes are not yet known, and further research is needed to characterize their function.
In many cases, ABA was shown to control the expression of several stress-responsive genes (Seki et al., 2002) such as HSP- and dehydrin-encoding genes, among several others. Due to the oxidative burst caused by drought and the protective role of HSPs, dehydrins, catalases, APXs, and oxidoreductases, the overexpression of their corresponding genes reported here is in accordance with responses usually observed in other plant species submitted to water limitations (Shinozaki and Yamaguchi-Shinozaki, 1997).
The leaf ABA contents of clones 14 and 22 analysed in this study are unknown but should increase under drought, as already shown for clone 120 of C. canephora var. Conilon (Silva, 2007). The results found for CGs differentially expressed upon DA and the comparison between the responses of drought-tolerant genotypes, presented here, lead to the first insight on the molecular players involved in drought responses in coffee. The identified CGs belong to different classes of genes, such as ABA perception and signalling, transcriptional regulation, and drought-responsive structural genes. Photosynthesis-related genes have also been identified. Based on the obtained results, it can be concluded that a complex network of responses probably involving the ABA signalling pathway and NO might be the major molecular determinants to explain the better efficiency in controlling stomatal closure and transpiration displayed by the drought-tolerant clone 14 of C. canephora.
Some of the identified CGs have recently been mapped (Leroy et al., 2011), and for several of these genes work is now in progress to analyse their corresponding genomic copy and promoter regions to verify if single nucleotide polymorphisms (SNPs) and/or insertion/deletions (INDELs) could be related to the differential expression profiles observed for these genes in drought-tolerant and sensitive clones of C. canephora. Such an approach will be facilitated with the forthcoming sequencing of the whole C. canephora genome (De Kochko et al., 2010).
Acknowledgments
This work was carried out under the project of scientific cooperation Embrapa-Cirad ‘Genetic determinism of drought tolerance in coffee’. The authors acknowledge financial support from the Brazilian Coffee R&D Consortium, FINEP, INCT-Café (CNPq/FAPEMIG), and Fundação Araucária (Brazil). The authors would also like to thank Drs Aymbiré Francisco Almeida da Fonseca, and Romário Gava Ferrão from the INCAPER Institute for providing plant materials, as well as Dr Marcelo E. Loureiro from UFV for the experimental design of the physiological analyses.
Glossary
Abbreviations
- 2-DE
immobilized pH gradient 2-dimensional gel electrophoresis
- CG
candidate gene
- DA
drought acclimation
- qPCR
quantitative polymerase chain reaction
- SD
standard deviation
References
- Alpizar E, Dechamp E, Lapeyre-Montes F, Guilhaumon C, Bertrand B, Jourdan C, Lashermes P, Etienne H. Agrobacterium rhizogenes-transformed roots of coffee (Coffea arabica): conditions for long-term proliferation, and morphological and molecular characterization. Annals of Botany. 2008;101:929–940. doi: 10.1093/aob/mcn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Assad ED, Pinto HS, Zullo J, Jr, Ávila AMH. Impacto das mudanças climáticas no zoneamento agroclimático do café no Brasil. Pesquisa Agropecuária Brasileira. 2004;39:1057–1064. [Google Scholar]
- Badger MR, Price GD. The role of carbonic anhydrase in photosynthesis. Annual Review of Plant Physiology and Plant Molecular Biology. 1994;45:369–392. [Google Scholar]
- Bardil A, Almeida JD, Combes MC, Lashermes P, Bertrand B. Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytologist. 2011;192:760–774. doi: 10.1111/j.1469-8137.2011.03833.x. [DOI] [PubMed] [Google Scholar]
- Barsalobres-Cavallari CF, Severino FE, Maluf MP, Maia IG. Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Molecular Biology. 2009;10:1. doi: 10.1186/1471-2199-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi G, Gamba A, Limiroli R, Pozzi N, Elster R, Salamini F, Bartels D. The unusual sugar composition in leaves of the resurrection plant. Myrothamnus flabellifolia. Physiologia Plantarum. 1993;87:223–226. [Google Scholar]
- Boyer J. Comportement hydrique de deux grands groupes de Coffea canephora de Côte d’Ivoire. Café Cacao Thé. 1965;9:263–282. [Google Scholar]
- Boyer J. Etude expérimentale des effets du régime d’humidité du sol sur la croissance végétative, la floraison et la fructification des caféiers Robusta. Café Cacao Thé. 1969;13:187–200. [Google Scholar]
- Camargo AP, Santinato R, Cortez JG. Aptidão climática para qualidade da bebida nas principais regiões cafeeiras de Arábica no Brasil. In: Anais do 18° Congresso Brasileiro de Pesquisas Cafeeiras, Araxá, Minas Gerais. Brasil. 1992:70–74. [Google Scholar]
- Carr MKV. The water relations and irrigation requirements of coffee. Experimental Agriculture. 2001;37:1–36. [Google Scholar]
- Colcombet J, Heribert H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochemical Journal. 2008;413:217–226. doi: 10.1042/BJ20080625. [DOI] [PubMed] [Google Scholar]
- Close TJ, Fenton RD, Moonan F. A view of plant dehydrins using antibodies specific to the carboxy terminal peptide. Plant Molecular Biology. 1993;23:279–286. doi: 10.1007/BF00029004. [DOI] [PubMed] [Google Scholar]
- Crouzillat D, Rigoreau M, Bellanger L, Priyono P, Mawardi S, Syahrudi McCarthy J, Tanksley S, Zaenudin I, Pétiard V. A Robusta consensus genetic map using RFLP and microsatellite markers for the detection of QTL. In: 20th International Conference on Coffee Science, Bangalore-India, CDrom, B202. 2004 [Google Scholar]
- Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros LMG, Romano E, Sá MFG Vaslin M, Alves-Ferreira M. Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Molecular Breeding. 2009;23:607–616. [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams S. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- DaMatta FM, Chaves ARM, Pinheiro HA, Ducatti C, Loureiro ME. Drought tolerance of two field-grown clones of Coffea canephora. Plant Science. 2003;164:111–117. [Google Scholar]
- DaMatta FM, Loos RA, Silva EA, Loureiro ME. Limitations to photosynthesis in Coffea canephora as a result of nitrogen and water availability. Journal of Plant Physiology. 2002;159:975–981. [Google Scholar]
- DaMatta FM, Maestri M, Barros RS. Photosynthetic performance of two coffee species under drought. Photosynthetica. 1997;34:257–264. [Google Scholar]
- DaMatta FM, Ramalho JC. Impact of drought and temperature stress on coffee physiology and production: a review. Brazilian Journal of Plant Physiology. 2006;18:55–81. [Google Scholar]
- Davis AP, Govaerts R, Bridson DM, Stoffelen P. An annotated taxonomic conspectus of the genus Coffea (Rubiaceae) Biological Journal of the Linnean Society. 2006;152:465–512. [Google Scholar]
- Davison PA, Hunter CN, Horton P. Overexpression of betacarotene hydroxylase enhances stress tolerance in Arabidopsis. Nature. 2002;418:203–206. doi: 10.1038/nature00861. [DOI] [PubMed] [Google Scholar]
- De Kochko A, Akaffou S, Andrade AC, et al. Advances in Coffea genomics. Advances in Botanical Research. 2010;53:23–53. [Google Scholar]
- Demirevska K, Simova-Stoilova L, Vassileva V, Vaseva I, Grigorova B, Feller U. Drought-induced leaf protein alterations in sensitive and tolerant wheat varieties. General and Applied Plant Physiology. 2008;34:79–102. [Google Scholar]
- Desikan R, Cheung M-K, Bright J, Henson D, Hancock JT, Neill SJ. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. Journal of Experimental Botany. 2004;55:205–212. doi: 10.1093/jxb/erh033. [DOI] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancock J, Neill S. Proceedings of the National Academy of Sciences. USA 99; 2002. A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana ; pp. 16314–16318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos TB, Budzinski IGF, Marur CJ, Petkowicz CLO, Pereira LFP, Vieira LGE. Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiology and Biochemistry. 2011;49:441–448. doi: 10.1016/j.plaphy.2011.01.023. [DOI] [PubMed] [Google Scholar]
- Dordas C, Rivoal J, Hill RD. Plant haemoglobins, nitric oxide and hypoxic stress. Annals of Botany. 2003;91:173–178. doi: 10.1093/aob/mcf115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wang N, Cui F, Li X, Xiao J, Xiong L. Characterization of the β-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiology. 2010;154:1304–1318. doi: 10.1104/pp.110.163741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Le Provost G, Pot D, Salin F, Lalane C, Madur D, Frigerio J-M, Plomion C. Identification and characterization of water-deficit responsive genes in hydroponically grown maritime pine (Pinus pinaster) seedlings. Tree Physiology. 2003;23:169–179. doi: 10.1093/treephys/23.3.169. [DOI] [PubMed] [Google Scholar]
- Ferrão RG, Fonseca AFA, Silveira JSM, Ferrão MAG, Bragança SM. EMCAPA 8141—Robustão Capixaba, variedade clonal de café conilon tolerante à seca, desenvolvida para o estado do Espírito Santo. Revista Ceres. 2000;273:555–560. [Google Scholar]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. Journal of the Royal Statistical Society. 1922;85:87–94. [Google Scholar]
- Fonseca AFA, Ferrão MAG, Ferrão RG, Verdin Filho AC, Volpi PS, Zucateli F. Conilon Vitória—Incaper 8142: improved Coffea canephora var. Kouillou clone cultivar for the State Espírito Santo. Crop Breeding and Applied Biotechnology. 2004;4:1–3. [Google Scholar]
- Garcia-Mata C, Lamattina L. Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiology. 2002;128:790–792. doi: 10.1104/pp.011020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geromel C, Ferreira LP, Guerreiro SMC, Cavalari AA, Pot D, Pereira LFP, Leroy T, Vieira LGE, Mazzafera P, Marraccini P. Biochemical and genomic analysis of sucrose metabolism during coffee (Coffea arabica) fruit development. Journal of Experimental Botany. 2006;57:3243–3258. doi: 10.1093/jxb/erl084. [DOI] [PubMed] [Google Scholar]
- Guliyev N, Bayramov S, Babayev H. Effect of water deficit on Rubisco and carbonic anhydrase activities in different wheat genotypes. In: Allen JF, Grantt E, Golbeck JH, Osmond R, editors. Photosynthesis, Energy from the Sun. 14th International Congress on Photosynthesis. Glasgow: Springer; 2008. pp. 1465–1468. [Google Scholar]
- Harrak H, Chamberland H, Plante M, Bellemare G, Lafontaine JG, Tabaeizadeh Z. A proline-, threonine-, and glycine-rich protein down-regulated by drought is localized in the cell wall of xylem elements. Plant Physiology. 1999;121:557–564. doi: 10.1104/pp.121.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology. 2011;21:346–355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefner T, Arend J, Warzecha H, Siems K, Stöckigt J. Arbutin synthase, a novel member of the NRD1β glycosyltransferase family, is a unique multifunctional enzyme converting various natural products and xenobiotics. Bioorganic and Medicinal Chemistry Letters. 2002;10:1731–1741. doi: 10.1016/s0968-0896(02)00029-9. [DOI] [PubMed] [Google Scholar]
- Hill AC, Bennet JH. Inhibition of apparent photosynthesis by nitrogen oxides. Atmospheric Environment. 1970;4:341–348. [Google Scholar]
- Hincha DK, Oliver AE, Crowe JH. Lipid composition determines the effects of arbutin on the stability of membranes. Biophysical Journal. 1999;77:2024–2034. doi: 10.1016/S0006-3495(99)77043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinniger C, Caillet V, Michoux F, Ben Amor M, Tanksley S, Lin C, McCarthy J. Isolation and characterization of cDNA encoding three dehydrins expressed during Coffea canephora (Robusta) grain development. Annals of Botany. 2006;97:755–765. doi: 10.1093/aob/mcl032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes and Development. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin C, Havaux M, Carde JP, Kloppstech K, Meierhoff K, Hoffman N, Nussaume L. Double mutation cpSRP43-/cpSRP54- is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light harvesting chlorophyll proteins. The Plant Journal. 2002;29:531–543. doi: 10.1046/j.0960-7412.2001.01211.x. [DOI] [PubMed] [Google Scholar]
- Jiang W, Yu D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biology. 2009;9:96–110. doi: 10.1186/1471-2229-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO Journal. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët T, Laffargue A, Salmona J, Doulbeau S, Descroix F, Bertrand B, de Kochko A, Dussert S. Metabolic pathways in tropical dicotyledonous albuminous seeds: Coffea arabica as a case study. New Phytologist. 2009;182:146–162. doi: 10.1111/j.1469-8137.2008.02742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanechi M, Uchida NU, Yasuda T, Yamaguchi T. Non-stomatal inhibition associated with inactivation of Rubisco in dehydrated coffee leaves under unshaded and shaded conditions. Plant and Cell Physiology. 1996;37:455–460. [Google Scholar]
- Lambot C, Crouzillat D, Fonseca AFA, Leloup V, Haiduc A, Broun P, Pétiard V, Ferrão MAG, Ferrão RG. Evaluation of conilons for genetic diversity, cup quality and biochemical composition. Proceedings of the 22th International Scientific Colloquium on Coffee. 2008 Campinas, SP, Brazil, A128. [Google Scholar]
- Lashermes P, Andrade AC, Etienne H. Genomics of coffee, one of the world’s largest traded commodities. In: Moore H, Ming R, editors. Genomics of tropical crop plants. Berlin: Springer; 2008. pp. 203–226. [Google Scholar]
- Le Provost G, Paiva J, Pot D, Brach J, Plomion C. Seasonal variation in transcript accumulation in wood forming tissues of maritime pine (Pinus pinaster Ait.) with emphasis on a cell wall glycine rich protein. Planta. 2003;217:820–830. doi: 10.1007/s00425-003-1051-2. [DOI] [PubMed] [Google Scholar]
- Leshem YY, Haramathy E. The characterization and contrasting effects of nitric oxide free radical in vegetative stress and senescence of Pisum sativum foliage. Journal of Plant Physiology. 1996;148:258–263. [Google Scholar]
- Lefebvre-Pautigny F, Wu FN, et al. High resolution synteny maps allowing direct comparisons between the coffee and tomato genomes. Tree Genetics and Genomes. 2010;6:565–577. [Google Scholar]
- Lepelley M, Cheminade G, Tremillon N, Simkin A, Caillet V, McCarthy J. Chlorogenic acid synthesis in coffee: an analysis of CGA content and real-time RT-PCR expression of HCT, HQT. C3H1 and CCoAOMT1 genes during grain development in C. canephora. Plant Science. 2007;172:978–996. [Google Scholar]
- Leroy T, De Bellis F, Legnate H, et al. Improving quality of African robustas: QTLs for yield- and quality-related traits in Coffea canephora. Tree Genetics and Genomes. 2011;7:781–798. [Google Scholar]
- Leroy T, Marraccini P, Dufour M, et al. Construction and characterization of a Coffea canephora BAC library to study the organization of sucrose biosynthesis genes. Theoretical and Applied Genetics. 2005;111:1032–1041. doi: 10.1007/s00122-005-0018-z. [DOI] [PubMed] [Google Scholar]
- Lima ALS, DaMatta FM, Pinheiro HA, Totola MR, Loureiro ME. Photochemical responses and oxidative stress in two clones of Coffea canephora under water stress. Environmental and Experimental Botany. 2002;47:239–247. [Google Scholar]
- Lin C, Mueller LA, McCarthy J, Crouzillat D, Pétiard V, Tanksley SD. Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts. Theoretical and Applied Genetics. 2005;112:114–130. doi: 10.1007/s00122-005-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Marraccini P, Courjault C, Caillet V, Lausanne F, Lepage B, Rogers WJ, Tessereau S, Deshayes A. Rubisco small subunit of Coffea arabica: cDNA sequence, gene cloning and promoter analysis in transgenic tobacco plants. Plant Physiology and Biochemistry. 2003;41:17–25. [Google Scholar]
- Marraccini P, Freire LP, Alves GSC, et al. RBCS1 expression in coffee: Coffea orthologs, Coffea arabica homeologs, and expression variability between genotypes and under drought stress. BMC Plant Biology. 2011;11:85. doi: 10.1186/1471-2229-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzafera P. Estresse hìdrico e conseqüencias na composição das sementes de café e qualidade da bebida. In: Garcia Salva TJ, Guerreiro Filho O, Thomaziello RA, Fazuoli LC, editors. Cafés de qualidade: aspectos tecnológicos, científicos e comerciais. IAC: Campinas, SP, Brazil; 2007. pp. 73–90. [Google Scholar]
- Mittler R. Oxidative stress, antioxidants, and stress tolerance. Trends in Plant Science. 2002;9:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mondego JMC, Vidal RO, Carazzolle MF, et al. An EST-based analysis identifies new genes and reveals distinctive gene expression features of Coffea arabica and. Coffea canephora. BMC Plant Biology. 2011;11:30. doi: 10.1186/1471-2229-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnon C. PhD Thesis. Montpellier University, France; 2000. Optimisation des gains génétiques dans le schéma de sélection récurrente réciproque de Coffea canephora Pierre. [Google Scholar]
- Montagnon C, Leroy T. Réaction à la sécheresse de jeunes caféiers Coffea canephora de Côte-d’Ivoire appartenant à différents groupes génétiques. Café Cacao Thé. 1993;37:179–190. [Google Scholar]
- Mousavi A, Hotta Y. Glycine-rich proteins: a class of novel proteins. Applied Biochemistry and Biotechnology. 2005;120:169–174. doi: 10.1385/abab:120:3:169. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, et al. The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiology. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, Morris P, Ribeiro D, Wilson I. Nitric oxide, stomatal closure, and abiotic stress. Journal of Experimental Botany. 2008;59:165–176. doi: 10.1093/jxb/erm293. [DOI] [PubMed] [Google Scholar]
- Noir S, Patheyron S, Combes MC, Lashermes P, Chalhoub B. Construction and characterization of a BAC library for genome analysis of the allotetraploid coffee species (Coffea arabica L.) Theoretical and Applied Genetics. 2004;109:225–230. doi: 10.1007/s00122-004-1604-1. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazzolli M, Romero-Puertas MC, Delledonne M. Modulation of nitric oxide bioactivity by plant haemoglobins. Journal of Experimental Botany. 2006;57:479–488. doi: 10.1093/jxb/erj051. [DOI] [PubMed] [Google Scholar]
- Pinheiro HA, DaMatta FM, Chaves ARM, Fontes EPB, Loureiro ME. Drought tolerance in relation to protection against oxidative stress in clones of Coffea canephora subjected to long-term drought. Plant Science. 2004;167:1307–1314. [Google Scholar]
- Pinheiro HA, DaMatta FM, Chaves ARM, Loureiro ME, Ducatti C. Drought tolerance is associated with rooting depth and stomatal control of water use in clones of. Coffea canephora. Annals of Botany. 2005;96:101–108. doi: 10.1093/aob/mci154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle A. 2001. Dissection of the superoxide dismutase–ascorbate–glutathione pathway by metabolic modeling: computer analysis as a step towards flux analysis. Plant Physiology. 126:445–462. doi: 10.1104/pp.126.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet V, Rondeau M, Tranchand C, Cayrel A, Hamon S, de Kochko A, Hamon P. SSR mining in coffee tree EST databases: potential use of EST-SSRs as marker across Coffea genus. Molecular and General Genetics. 2006;276:436–449. doi: 10.1007/s00438-006-0153-5. [DOI] [PubMed] [Google Scholar]
- Popova LP, Tsonev TD, Lazova GN, Stoinova ZG. Drought and ABA-induced changes in photosynthesis of barley plants. Physiologia Plantarum. 1996;96:623–629. [Google Scholar]
- Praxedes SC, DaMatta FM, Loureiro ME, Ferrão MAG, Cordeiro AT. Effects of long-term soil drought on photosynthesis and carbohydrate metabolism in mature robusta coffee (Coffea canephora Pierre var. kouillou) leaves. Environmental and Experimental Botany. 2006;56:263–273. [Google Scholar]
- Privat I, Bardil A, Bombarely Gomez A, et al. The ‘PUCE CAFE’ project: the first 15K coffee microarray, a new tool for discovering candidate genes correlated to agronomic and quality traits. BMC Genomics. 2011;12:5. doi: 10.1186/1471-2164-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Nanming Z, Yamaguchi-Shinozaki K, Shinozaki K. Regulatory role of DREB transcription factors in plant drought, salt and cold tolerance. Chinese Science Bulletin. 2000;45:970–975. [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Ramos HJO, Seixas DF, Pereira LFP, et al. V Simpósio de pesquisa dos cafés do Brasil, Águas de Lindóia, SP: Embrapa Café, CD-rom; 2007. Análise proteômica do estresse hídrico em cafeeiro. [Google Scholar]
- Ribas AF, Dechamp E, Champion A, Bertrand B, Combes MC, Verdeil JL, Lapeyre F, Lashermes P, Etienne H. Agrobacterium-mediated genetic transformation of Coffea arabica (L.) is greatly enhanced by using established embryogenic callus cultures. BMC Plant Biology. 2011;11:92. doi: 10.1186/1471-2229-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C, Keller B, Ryser U. Glycine-rich proteins as structural components of plant cell walls. Cellular and Molecular Life Science. 2001;58:1430–1441. doi: 10.1007/PL00000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Canales E, Borroto CJ, Carmona E, López J, Pujol M, Borrás-Hidalgo O. Identification of genes induced upon water-deficit stress in a drought-tolerant rice cultivar. Journal of Plant Physiology. 2006;163:577–584. doi: 10.1016/j.jplph.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Salmona J, Dussert S, Descroix F, de Kochko A, Bertrand B, Joet T. Deciphering transcriptional networks that govern Coffea arabica seed development using combined cDNA array and real-time RT-PCR approaches. Plant Molecular Biology. 2008;66:105–124. doi: 10.1007/s11103-007-9256-6. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiology. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva EA, Mazzafera P, Brunini O, Sakai E, Arruda FB, Mattoso LHC, Carvalho CRL, Pires RCM. The influence of water management and environmental conditions on the chemical composition and beverage quality of coffee beans. Brazilian Journal of Plant Physiology. 2005;17:229–238. [Google Scholar]
- Silva VA. PhD Thesis. Federal University of Viçosa, Brazil; 2007. Caracterização fisiológica da tolerância à seca em Coffea canephora: contribuição relativa do sistema radicular e da parte aérea. 68. [Google Scholar]
- Silva VA, Antunes WC, Guimarães BLS, Paiva RMC, Silva VF, Ferrão MAG, DaMatta FM, Loureiro ME. Physiological response of Conilon coffee clone sensitive to drought grafted onto tolerant rootstock. Pesquisa Agropecuária Brasileira. 2010;45:457–464. [Google Scholar]
- Simkin AJ, Moreau H, Kuntz M, Pagny G, Lin C, Tanksley S, McCarthy J. An investigation of carotenoid biosynthesis in Coffea canephora and Coffea arabica. Journal of Plant Physiology. 2008;165:1087–1106. doi: 10.1016/j.jplph.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Tang D, Christiansen KM, Innes RW. Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiology. 2005;138:1018–1026. doi: 10.1104/pp.105.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N. Antioxidant systems and plant response to the environment. In: Smirnoff N, editor. Environment and plant metabolism—flexibility and acclimation. Oxford: BIOS Scientific Publishers; 1995. pp. 217–243. [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in. Arabidopsis thaliana. Current Opinion in Plant Biology. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. Journal of Experimental Botany. 2011;62:5659–5669. doi: 10.1093/jxb/err252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, Kobayashi M, Koshiba T, Kamiya Y, Shinozaki K. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana . The Plant Journal. 2006;46:171–182. doi: 10.1111/j.1365-313X.2006.02683.x. [DOI] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. The Plant Journal. 2009;57:1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- Vidal RO, Mondego JMC, Pot D, Ambrósio AB, Andrade AC, Pereira LFP, Colombo CA, Vieira LGE, Carazzolle MF, Pereira GAG. A high-throughput data mining of single nucleotide polymorphisms in Coffea species expressed sequence tags suggests differential homeologous gene expression in the allotetraploid. Coffea arabica. Plant Physiology. 2010;154:1053–1066. doi: 10.1104/pp.110.162438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis . The Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira LGE, Andrade AC, et al. Brazilian coffee genome project: an EST-based genomic resource. Brazilian Journal of Plant Physiology. 2006;18:95–108. [Google Scholar]
- Von Groll U, Berger D, Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. The Plant Cell. 2002;14:1527–1539. doi: 10.1105/tpc.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shang H, Liu Y, Zheng M, Wu R, Phillips J, Bartels D, Deng X. A role for a cell wall localized glycine-rich protein in dehydration and rehydration of the resurrection plant. Boea hygrometrica. Plant Biology. 2009;11:837–848. doi: 10.1111/j.1438-8677.2008.00187.x. [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Lateef SS, Lapik Y, Anderson M, Lee B-S, Kaufman LS. G-protein-coupled receptor 1, G-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis . Plant Physiology. 2006;140:844–855. doi: 10.1104/pp.105.071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Zagdańska B, Wiśniewski K. ATP-dependent proteolysis contributes to the acclimation-induced drought resistance in spring wheat. Acta Physiologiae Plantarum. 1998;20:55–58. [Google Scholar]
- Zottini M, Formentin E, Scattolin M, Carimi F, Lo Schiavo F, Terzi M. Nitric oxide affects plant mitochondrial functionality. in vivo. FEBS Letters. 2002;515:75–78. doi: 10.1016/s0014-5793(02)02438-9. [DOI] [PubMed] [Google Scholar]