Abstract
Telomerase, an enzyme responsible for the maintenance of linear chromosome ends, is precisely regulated during plant development. In animals, involvement of the epigenetic state of the telomerase reverse transcriptase (TERT) gene in the complex regulation of telomerase activity has been reported. To reveal whether epigenetic mechanisms participate in the regulation of plant telomerase, the relationship between telomerase activity in tissues of Arabidopsis thaliana and DNA methylation and histone modifications in the A. thaliana TERT (AtTERT) upstream region was studied. As expected, a gradual decrease of telomerase activity during leaf maturation was observed. A different pattern with a more progressive loss of telomerase activity and AtTERT transcription during leaf development was revealed in MET1 gene-knockout mutants. Analysis of DNA methylation in the AtTERT upstream region showed low levels of methylated cytosines without notable differences between telomerase-positive and telomerase-negative wild-type tissues. Surprisingly, a high level of CG methylation was found in the AtTERT coding region, although this type of methylation is a characteristic attribute of constitutively expressed genes. Analysis of chromatin modifications in the AtTERT upstream region and in exon 5 showed increased loading of the H3K27me3 mark in the telomerase-negative mature leaf compared to telomerase-positive seedlings, whereas H3K4me3, H3K9Ac, and H3K9me2 were approximately at the same level. Consistently, the chromatin structure of the AtTERT gene was maintained. These results are discussed in the context of the general involvement of epigenetic mechanisms in the regulation of gene expression and with respect to similar studies performed in animal models.
Keywords: Arabidopsis thaliana, developmental regulation, DNA methylation, histone modifications, telomerase
Introduction
Telomerase is a ribonucleoprotein enzyme complex responsible for the synthesis of telomeres, specialized nucleoprotein structures at the ends of linear eukaryotic chromosomes. Telomerase consists of a catalytic subunit, telomerase reverse transcriptase (TERT), and a telomerase RNA (TR) subunit which serves as a template for the elongation of the telomere motif. Telomerase activity is strictly regulated during plant development. Analysis of model plants including Arabidopsis (Fitzgerald et al., 1996), Silene latifolia ( Riha et al., 1998), tobacco (Fajkus et al., 1998), barley (Heller et al., 1996), soybean (Fitzgerald et al., 1996), and tomato (Broun et al., 1992) has revealed active telomerase in organs and tissues containing dividing meristem cells (seedlings, root tips, blossoms, floral buds) and in cell cultures (Fajkus et al., 1996). On the other hand, telomerase activity was abolished in organs formed by terminally differentiated cells, e.g. stems or mature leaves. In this respect, the pattern of telomerase activity in plants resembles that in humans, but with a notable difference: telomerase down-regulation in terminally differentiated plant cells is reversible and highly dynamic, as is their differentiation status itself, and reflects the totipotent character of plant cells (Fajkus et al., 1998). Nevertheless, the general pattern of telomerase activity is different even among mammalian model species; while in most human adult somatic tissues telomerase expression and activity are undetectable or very low, mouse somatic tissue cells express a detectable amount of the TERT mRNA (Horikawa et al., 2005). This indicates that developmental regulation of telomerase is not driven by simple and generally valid mechanisms.
In plants, the molecular mechanisms of telomerase regulation at both the cellular and organism levels are far from being elucidated. These processes include regulation of telomerase transcription (Fitzgerald et al., 1996; Oguchi et al., 1999), alternative splicing of TERT gene transcripts (Heller-Uszynska et al., 2002; Rossignol et al., 2007), and post-translational modifications of telomerase (Oguchi et al., 2004). A recent description of the Arabidopsis TR subunit suggested a possible involvement of two variant RNA subunits in formation of telomerase nucleoprotein complexes, yielding telomerases of different activity (Cifuentes-Rojas et al., 2011). Moreover, strong regulatory elements downstream of the transcription start site were identified in our previous study (Fojtova et al., 2011), demonstrating an enormous complexity of the plant telomerase regulation process.
The involvement of the chromatin state and epigenetic mechanisms in regulation of the TERT gene were demonstrated in animal models (reviewed in (Zhu et al., 2011)). Hyperacetylated and H3K4-methylated histones were associated with human TERT (hTERT) expression in telomerase-positive cells, while H3K9 and H3K20 methylation marked histones in telomerase-negative cells. Although the sequence of the hTERT locus including the promoter region meets parameters for the CpG islands, no unambiguous correlation between promoter methylation and activity exists. Association of hTERT promoter methylation with the loss of its activity is evidenced by the demethylation-induced increase of hTERT transcription in immortalized fibroblasts (Devereux et al., 1999) and binding of methyl-CpG-binding domain protein 2 (MBD 2) to the hypermethylated hTERT promoter in HeLa cells (Chatagnon et al., 2009). On the other hand, demethylation in tumour cell lines with high telomerase activity was correlated with a significant reduction of hTERT transcription (Guilleret and Benhattar, 2003). It is supposed that in this case methylation prevents the transcriptional repressors from binding, but a small methylation-free region near the transcription start site is able to ensure hTERT transcription. Nevertheless, in most normal somatic cells with a basal level of telomerase activity the hTERT promoter is hypomethylated (Dessain et al., 2000). As regards chromatin structure, it was shown that cell differentiation was associated with the loss of DNaseI-hypersensitive sites in the human and mouse TERT promoters and their upstream regions (Wang et al., 2009), showing that changes of chromatin structure leading to its more condensed state are connected with hTERT transcriptional silencing.
In this work, telomerase activity during Arabidopsis thaliana development was correlated with the A. thaliana TERT (AtTERT) epigenetic pattern. While DNA methylation did not seem to be involved in the gradual attenuation of telomerase transcription during leaf maturation, the repressive chromatin modification signal – trimethylation of lysine 27 in histone H3 (H3K27me3) – was installed in the AtTERT upstream and gene body regions in telomerase-negative tissue. Nevertheless, no significant change in the general chromatin structure accompanied the H3K27me3 loading, and developmentally silenced AtTERT maintained the euchromatin-specific modifications.
Materials and methods
Plant material
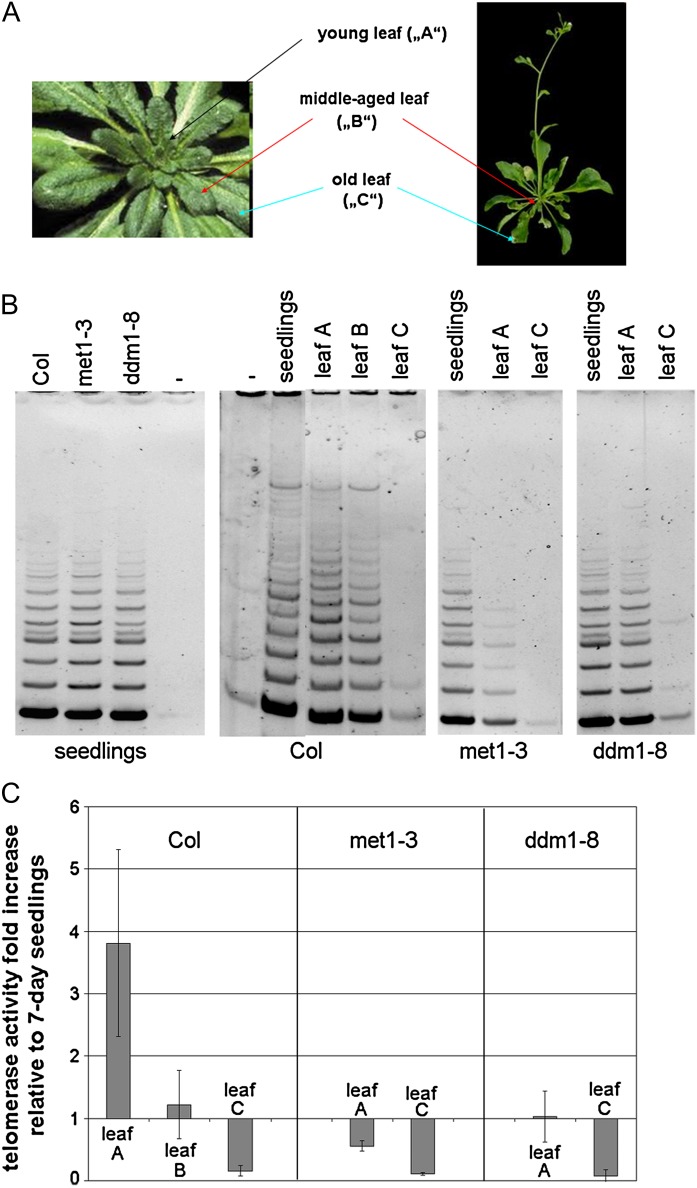
A. thaliana seedlings of the Columbia-0 ecotype and a ddm1 (At5g66750) mutant (ddm1-8 strain, SALK000590) were purchased from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003), and seedlings of the mutant plant with a T-DNA insertion in the MET1 gene (At5g49160, met1-3 strain; Saze et al., 2003) were kindly provided by Dr Ales Pecinka (GMI, Vienna, Austria). Primers for genotyping are described in Supplementary Table S1. Seeds were placed on half-strength Murashige–Skoog (Duchefa Biochemie, Haarlem, The Netherlands) agar plates and grown under cycles of 8 h light (illumination 100 μmol m−2 s−1), at 21 °C and 16 h dark at 19 °C. After 7 days, seedlings were collected for analyses. Plants were grown in soil from 2 week-old seedlings under the same light/dark conditions favouring leaf growth. Leaves were harvested from 6–8 week-old plants as depicted in Fig. 1A.
Fig. 1.
Telomerase activity dynamics in Arabidopsis wild-type and methylation mutant tissues. (A) Strategy for collection of leaves at different developmental stages. (B) In vitro telomerase activity assays. Telomerase activity was determined in extracts from 7 day seedlings and from leaves collected from the Columbia wild-type (Col), met1-3, and ddm1-8 plants using the TS21 and TELPR primers (Supplementary Table S1). Lanes show: –, negative controls (no protein extract in the reaction); leaf A, young leaf; leaf B, middle-aged leaf; leaf C, mature leaf. (C) Quantitative analysis of telomerase activity in leaves. Analysis was based on SYBR Green I fluorescence detection and was performed using at least two biological replicates (three technical replicates for each). The ΔCt method (Pfaffl, 2004) was used to calculate relative telomerase activity. Analyses were done for two biological replicates in three technical replicates; error bars show SD.
Analysis of telomerase activity (TRAP assay)
Telomerase extracts from Arabidopsis tissues were prepared as described (Fitzgerald et al., 1996; Sykorova et al., 2003). Telomerase activity was analysed according to the protocol in (Fajkus et al., 1998). First, 1 μl of 10 μM TS21 substrate primer (Supplementary Table S1) was mixed with 1 μl of telomerase extract (protein concentration 50 ng μl−1). Primer elongation proceeded in 25 μl of the reaction buffer at 26 °C for 45 min. After extension, telomerase was heat-inactivated and samples were cooled to 80 °C. Then, 1 μl of 10 μM TELPR reverse primer (Supplementary Table S1) and 2 units of DyNAzymeII DNA polymerase (Finnzymes, Espoo, Finland) were added to start PCR amplification of telomerase extension products (35 cycles of 95 °C/30 s, 65 °C/30 s, 72 °C/30 s) followed by a final extension (72 °C/5 min). Products of TRAP reactions were analysed by electrophoresis on a 12.5% polyacrylamide gel in 0.5×TBE buffer; the gel was stained with GelStar Nucleic Acid Gel Stain (LONZA, Basel, Switzerland) and signals were visualized using the LAS-3000 system (FujiFilm, Tokyo, Japan). Telomerase activity and processivity were deduced from the intensity and extension of the TRAP products ladder, respectively.
The quantitative version of the TRAP assay was performed as described in Herbert et al. (2006) using FastStart SYBR Green Master (Roche, Basel, Switzerland) and TS21 and TELPR primers. Samples were analysed in triplicates in a 20 μl reaction mix. Ct values were determined using Rotorgene6000 (Qiagen, Hilden, Germany) software and relative telomerase activity was calculated by the ΔCt method (Pfaffl, 2004).
RNA isolation and RT-PCR analysis
Total RNA was isolated from Arabidopsis tissues using the RNeasy Plant Mini Kit (Qiagen) followed by DNase I treatment (TURBO DNA-free; Applied Biosystems/Ambion, Foster City, CA, USA) according to the manufacturer's instructions. The quality and quantity of RNA was checked by electrophoresis on 1% (w/v) agarose gels and by absorbance measurements (NanoPhotometr IMPLEN). cDNA was prepared by reverse transcription of 1 μg of RNA using M-MuLV reverse transcriptase (New England Biolabs, Hitchin, Herts, UK) and Random Nonamers (Sigma-Aldrich, St Louis, MO, USA). Quantification of the AtTERT transcript relative to the ubiquitin reference transcript was done using FastStart SYBR Green Master (Roche) on the Rotorgene6000 (Qiagen). One μl of five-times-diluted cDNA was added to the 20 μl reaction mix; the final concentration of each forward and reverse primer was 0.25 μM (Supplementary Table S1). Reactions were done in triplicates; the PCR programme consisted of 15 min of initial denaturation at 94 °C followed by 40 cycles of 30 s at 94 °C, 30 s at 56 °C, and 30 s at 72 °C. Analyses were performed for at least two biological replicates in three technical replicates. Transcription in the respective tissue was calculated as the fold increase/decrease relative to wild-type 7 day seedlings (ΔΔCt method (Pfaffl, 2004)).
DNA isolation and analysis of DNA methylation
Total genomic DNA was isolated from 1 g of 7 day seedlings and 6–8 week-old leaves by the cetyltrimethylammonium bromide method as described in Kovarik et al. (2000).
Bisulphite conversion of genomic DNA was done by the EpiTect Bisulfite Kit (Qiagen) in which non-methylated cytosines are converted to uracils and amplified as thymines in the subsequent PCR, while 5-methylcytosines are resistant in this reaction (Clark et al., 1994). Sequences of primers for amplification of the AtTERT upstream region (284 bp fragment) and AtTERT exon 5 (476 bp fragment) are listed in Supplementary Table S1. PCR was done using DyNAzymeII DNA polymerase in a programme consisting of initial denaturation (2 min) and 40 cycles of 30 s at 94 °C, 30 s at 56 °C and 40 s at 72 °C followed by a final extension (72 °C/5 min). PCR products were cloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA) and sequenced (Macrogene, Seoul, South Korea). Methylation of cytosines located in the respective sequence context was analysed by CyMATE software (Hetzl et al., 2007) in the 224 bp AtTERT upstream region (close to the ATG site) where the sequencing signals were convincingly seen, and in the AtTERT exon 5.
Analysis of histone modifications by chromatin immunoprecipitation
Histone modifications by chromatin immunoprecipitation (ChIP) were analysed using the EpiQuik™ Plant Chip Kit (Epigentek, Farmingdale, NY, USA). Chromatin was crosslinked for 15 min in 1% formaldehyde (Sigma-Aldrich), fragmented by sonication (Bioruptor; Diagenode, Liège, Belgium) to an average fragment length of 500 bp, and immunoprecipitated by antibodies against H3K9me2 (Abcam, Cambridge, UK), H3K4me3 (Abcam), H3K9Ac (Abcam), or H3K27me3 (Millipore, Billerica, MA, USA). A total of 20 ng of purified DNA from the immunoprecipitated fractions was subjected to PCR using primers for the AtTERT upstream region and the exon 5 (Supplementary Table S1) and DyNAzymeII DNA polymerase with the same PCR programme as described for analysis of DNA methylation. Quantitative PCR was performed as described for RT-PCR analysis. Results were evaluated statistically using the two-tailed Student's t test; a P value of <0.05 was considered as statistically significant.
Results
Gradual decrease of telomerase activity during leaf maturation
Telomerase activity was determined by the TRAP assay in A. thaliana 7 day seedlings and leaves of different ages (Fig. 1A). A gradual decrease of telomerase activity during leaf maturation was observed (Fig. 1B, 1C). Telomerase activity comparable to or even higher than that in 7 day seedlings was observed in young leaves (leaf A, Fig. 1A). In a so-called middle-aged leaf (leaf B), telomerase activity was around the level in seedlings and was absent or very low in mature leaves (leaf C).
The dynamics of telomerase activity during plant development are believed to be correlated with TERT promoter activity, i.e. with the level of the transcript for the telomerase protein subunit. A good correlation between AtTERT transcription and telomerase activity was observed (Supplementary Fig. S1).
Telomerase dynamics is affected in leaves of the met1-3 mutant
Numerous studies have shown that DNA methylation plays an important role in the modulation of promoter activity. Although the precise range of the AtTERT promoter has not been characterized yet, a minimal telomerase promoter has recently been identified using a collection of T-DNA insertion lines as a sequence 271 bp upstream of the ATG signal (Fojtova et al., 2011). To investigate cytosine methylation in this putative promoter, primers delimiting the region from position –284 to the ATG codon (Supplementary Table S1) were designed to amplify sodium bisulphite-modified DNA templates. The number of methylated cytosines is very low in this region and there is no notable difference in cytosine methylation between telomerase-positive (7 day seedlings) and -negative (mature leaves) tissues (Supplementary Fig. S2). Based on these results, DNA methylation in the putative promoter region is not a dominant factor in the regulation of AtTERT transcription.
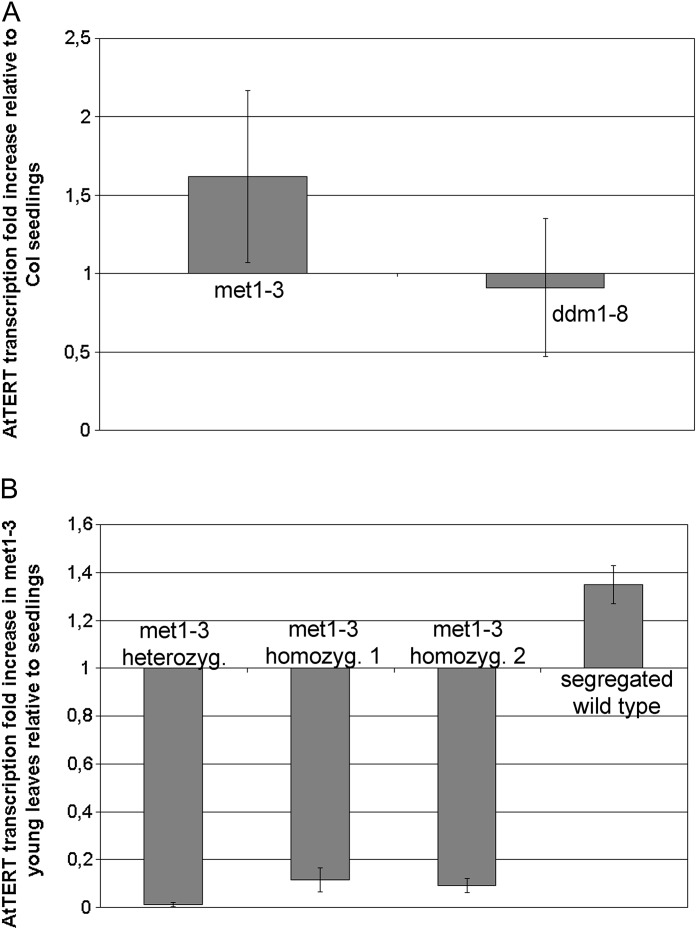
The above results were complemented by an analysis of mutant plants defective in pathways crucial for the maintenance of cytosine methylation. Analysis of telomerase activity in seedlings of met1-3 and ddm1-8 mutants revealed patterns fully comparable to the wild type (Fig. 1B, left panel), and no significant changes in AtTERT transcription were detected (Fig. 2A).
Fig. 2.
Analysis of AtTERT transcription in methylation mutants. (A) AtTERT transcription in 7 day seedlings from methylation mutants. Analysis was done using four biological replicates of seedlings from four ddm1-8 homozygous plants and from four met1-3 heterozygous plants. Amplification of a 110 bp fragment of the AtTERT exon 1 was expressed relative to the ubiquitin endogenous control. The ΔΔCt method (Pfaffl, 2004) was used to calculate AtTERT transcription. Error bars show SD. No significant change of the AtTERT transcript level as compared to wild-type seedlings was observed. Col, Columbia wild-type. (B) AtTERT transcription in young leaves from met1-3 plants (one heterozygous and two homozygous representatives; homozygous plants were selected with extremely low frequency and did not grow up to the reproductive stage, as previously reported by Saze et al., 2003) and from segregated wild-type plants. AtTERT transcription in met1-3 young leaves was significantly lower compared to wild-type samples (Supplementary Fig. S1).
The ddm1-8 mutant plants showed telomerase activities in young (leaf A) and old (leaf C) leaves to be more or less comparable to those in leaves of the corresponding developmental stage in wild-type plants (Fig. 1B), although – according to the results of quantitative assays (Fig. 1C) – telomerase activity in the ddm1-8 young leaf is lower than in wild-type Columbia tissues, approaching the value observed in the middle-aged wild-type leaves (leaf B). Surprisingly, in repeated analyses met1-3 mutants revealed considerably lower telomerase activity in young leaves (Fig. 1B, 1C). The amounts of AtTERT transcript in young leaves of one plant heterozygous for a T-DNA insertion in the MET1 gene and of two plants homozygous for this insertion were close to the detection limit (Fig. 2B), i.e. significantly lower than in the corresponding wild-type samples, in which AtTERT transcription was even higher as compared to that in 7 day seedlings (Supplementary Fig. S1). Based on these analyses, it is possible to hypothesize that a complex pattern of phenotypic defects connected to the loss of MET1 function (Mathieu et al., 2007) encompasses disruption of telomerase developmental regulation, leading to an early loss of telomerase expression and activity in the mutant leaves. But in contrast to observations suggesting that the phenotypic consequences of CG methylation erasure are not simply overcome by the reintroduction of the function of both MET1 alleles, AtTERT transcription (Fig. 2B) and telomerase activity (not shown) are fully reverted in plants segregated from the mutant background.
AtTERT is CG-methylated in the gene body region
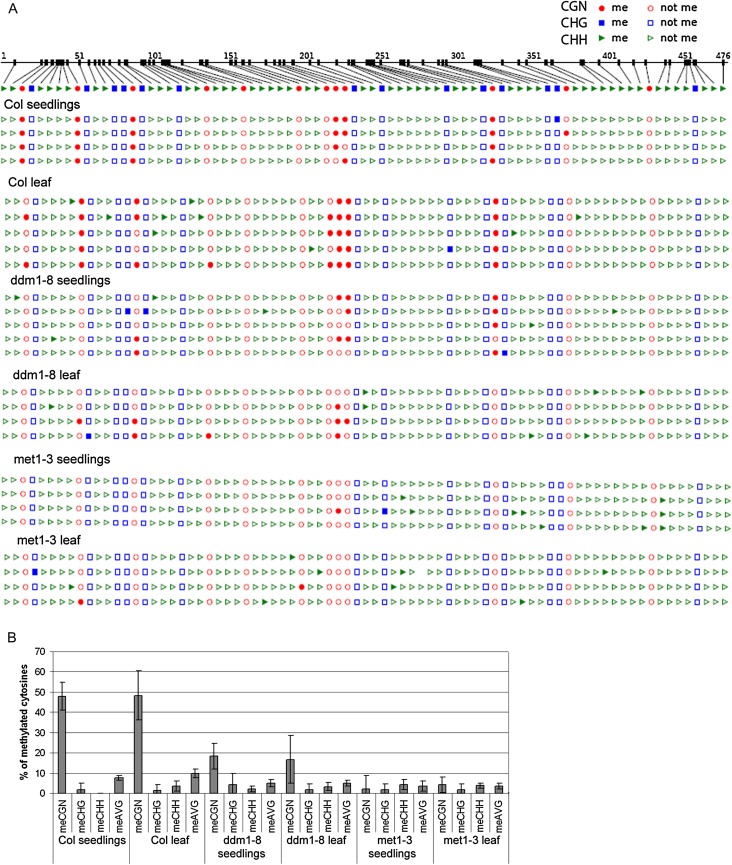
Although the AtTERT putative promoter region is not methylated in either of the tissues tested, methylation in exon 5 in Arabidopsis was detected by high-throughput methylation analysis (http://signal.salk.edu/cgi-bin/methylome?GENE=At5g16850) (Zhang et al., 2006). In agreement with this report, a relatively high level of CG methylation was detected in 7 day seedlings and mature leaves of wild-type plants using a primer set delimiting a 476 bp region of the AtTERT fifth exon (Fig. 3). CHG and CHH methylations were low and close to the average levels reported for the whole A. thaliana genome (6.7 and 1.7%, respectively). A significant decrease of methylation in CG doublets was observed in met1-3 and ddm1-8 tissues, which was more pronounced in met1-3 mutants where CG methylation dropped to a level comparable to those of CHG and CHH. As in the wild type, the amount of methylated cytosines was comparable in different tissues of both mutant plants (Fig. 3).
Fig. 3.
Analysis of DNA methylation in AtTERT exon 5 by bisulphite genomic sequencing. (A) Distribution of methylated cytosines along the 476 bp region of exon 5. Seven day seedlings and mature leaves (leaf C, Fig. 1A) from wild-type and methylation mutants were subjected to analysis. Col, Columbia wild-type; CG methylation, red circles; CHG methylation, blue squares; methylation of cytosines in a non-symmetrical sequence context, green triangles; filled symbols, methylated cytosine; empty symbols, non-methylated cytosine. Twelve cytosines in CG, 14 cytosines in CHG, and 53 cytosines in CHH were evaluated. (B) Graphical representation of the methylated cytosine content in the respective sequence context in tissues of wild-type and methylation mutant plants. Note the comparable level of methylated cytosines in tissues of the same genotype. In all met1-3 and ddm1-8 clones, the level of methylated cytosines in a CG sequence context was decreased significantly, and this drop was more pronounced in the met1-3 mutant background. meAVG, average cytosine methylation.
AtTERT gene silencing is accompanied by increased loading of the H3K27me3 epigenetic mark, but the region maintains its euchromatic nature
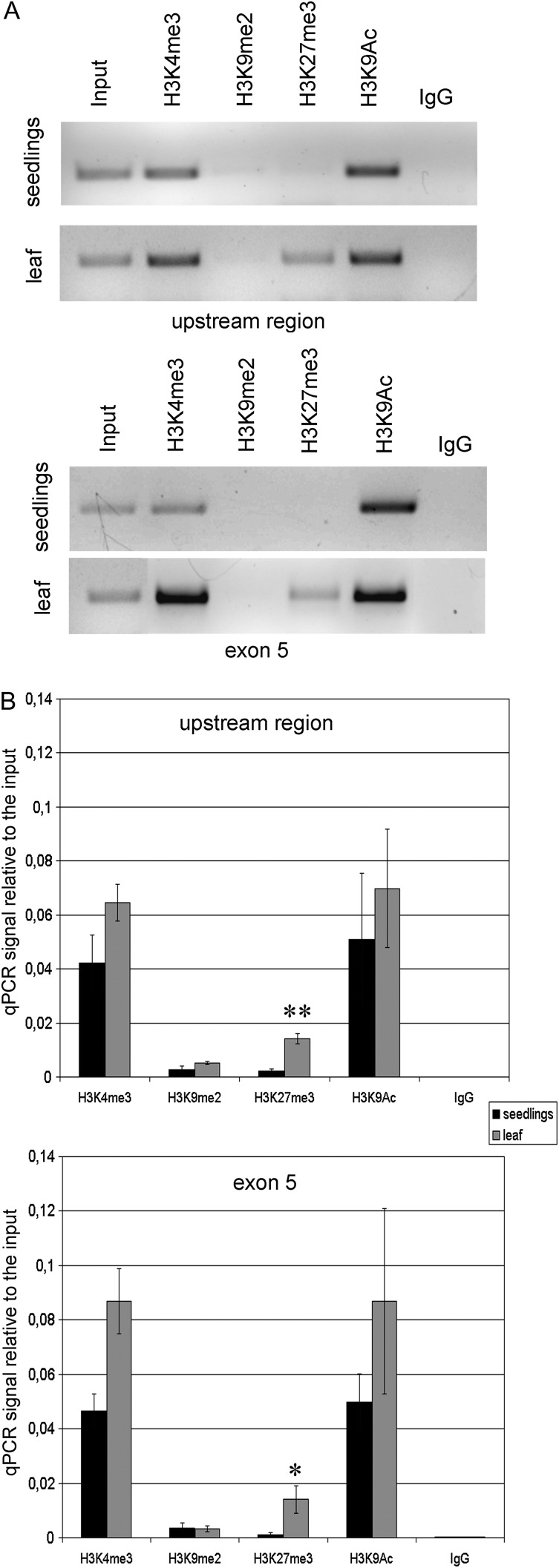
Modifications of histone amino acid residues represent crucial determinants of chromatin structure and activity of the corresponding DNA regions. We analysed the distribution of four selected chromatin epigenetic marks in the AtTERT upstream region and in exon 5 of telomerase-positive (7 day seedlings) and telomerase-negative (mature leaf) tissues. Crosslinked and sonicated chromatin was immunoprecipitated using antibodies against H3K4me3 (a euchromatin-specific epigenetic mark), H3K9me2 (heterochromatin, mainly constitutive), H3K27me3 (heterochromatin in developmentally silenced regions), and H3K9Ac (euchromatin). Primers for PCR covered the region from –336 bp to the ATG position and 476 bp region of exon 5 (Supplementary Table S1).
AtTERT upstream and exon 5 regions are clearly associated with the euchromatin-specific histone modifications H3K4me3 and H3K9Ac in both telomerase-positive and telomerase-negative tissues, while levels of H3K9me2 are low (Fig. 4). These results suggest that the general chromatin environment in the AtTERT gene is not markedly altered during plant development. Correspondingly, the pattern of micrococcal nuclease digestion of AtTERT chromatin is comparable in nuclei isolated from 7 day seedlings and from mature leaves (Supplementary Fig. S3). In agreement with previously published results obtained from analysis of 10 day seedlings (Turck et al., 2007; Zhang et al., 2007; Roudier et al., 2011), low signals for the H3K27me3 fraction were detected in 7 day seedlings. In the mature leaf, the intensity of this signal increased significantly in both analysed regions (Fig. 4). A similar pattern of distribution of chromatin marks was observed in 7 day seedlings and mature leaves of met1-3 mutant plants (Supplementary Fig. S4).
Fig. 4.
Analysis of histone modifications in the AtTERT upstream region and in exon 5 by ChIP. DNAs from immunoprecipitated fractions of chromatin were purified and a 336 bp region upstream of the ATG signal and a 476 bp region of the fifth exon were amplified using classical (A) or quantitative (B) PCR (qPCR). (A) A representative example of PCR amplification of the AtTERT upstream region and of exon 5 in immunoprecipitated fractions. Signals of euchromatin-specific marks (H3K4me2, H3K9Ac) were strong in both tissues analysed; signals for the modification typical for constitutive heterochromatin (H3K9me2) were below the detection limit. Note the distinct H3K27me3 band in the leaf samples. (B) Two biological replicates of wild-type seedlings and mature leaves were immunoprecipitated and subjected to quantitative PCR. Signal from the immunoprecipitated fractions was expressed relative to that from the total input chromatin. The amount of the H3K27me3 mark increased in the telomerase-negative tissue (leaf) in both regions analysed (P < 0.01 in the AtTERT upstream region; P < 0.05 in the exon 5).
Discussion
Epigenetic modifications of promoter sequences are strong determinants of their transcriptional potency. While promoter-associated DNA methylation is generally considered as a silencing mark, the pattern of histone modifications is more complex and displays both organism- and locus-specific features (Fransz et al., 2006; Hon et al., 2009).
A detailed methylation map of the A. thaliana genome was obtained using high-throughput sequencing approaches (Zhang et al., 2006), and the low level of methylated cytosines observed in the AtTERT upstream region (Supplementary Fig. S2) is in accordance with this map. Using mammalian models, convincing data showing involvement of epigenetic mechanisms in telomerase developmental regulation have been reported. While the function of TERT promoter methylation was rather ambiguous, as an increase of promoter activity was observed to accompany both hypo- and hypermethylated states, the role of native chromatin environment including histone modifications for tight hTERT gene regulation was clearly demonstrated (reviewed in Zhu et al., 2011). In our study, no correlation was observed between AtTERT transcription and methylation of the putative minimal promoter. Nevertheless, based on the conclusions of Vaughn et al. (2007), methylation of promoters in Arabidopsis is a relatively rare event and in this respect DNA methylation is not broadly involved in the regulation of gene expression.
Interestingly, along the AtTERT gene methylation in the CG sequence context was detected in the fifth exon (Fig. 3). This type of methylation, termed gene body methylation, has been found in both plant and mammalian genomes (Lorincz et al., 2004; Cokus et al., 2008; Lunerova-Bedrichova et al., 2008). In Arabidopsis, genes methylated in transcribed regions are generally constitutively expressed and display lower tissue specificity compared to genes with methylated promoters (Zhang et al., 2006). In a more detailed study (Aceituno et al., 2008) gene body methylation was negatively correlated with gene responsiveness, i.e. capacity to change expression under developmental and environmental stimuli. The protein subunit of telomerase does not fit this general rule, because its expression changes significantly during plant development and the function of the gene body methylation in the AtTERT locus remains enigmatic. A connection between AtTERT gene methylation and alternative splicing of the AtTERT transcript (Rossignol et al., 2007) may be a promising possibility. Unfortunately, testing of this hypothesis is methodically rather difficult due to the very low level of the alternatively spliced AtTERT transcript which reduces the reproducibility of quantitative RT-PCR assays (M. Fojtová, unpublished results).
Interesting findings arise from the analysis of the AtTERT transcription in met1-3 mutant leaves. Low levels of AtTERT transcripts and telomerase activity were found in developing leaves of met1-3 mutants (Fig. 1B, 1C, 2B), while in wild-type young leaves both values were even higher as compared to seedlings (Supplementary Fig. S1). This observation might indicate impaired developmental regulation of the AtTERT gene in met1 mutants. We should however be careful about drawing such conclusions, and take into consideration possible moderate variations in the leaf developmental stage (although in all cases, leaves of approximately the same age were collected for analyses), and the very small size (due to their retarded growth) of met1-3 mutants leaves considered as young leaves. Further comprehensive research including analysis of other crucial mutations in related pathways is necessary to clarify this topic definitely.
The recently published map of the main epigenetic states in Arabidopsis chromatin revealed distinct combinations of 12 chromatin marks defining active genes, repressed genes, silent repeat elements, and intergenic regions (Roudier et al., 2011). According to the present data, AtTERT chromatin was associated with the histone marks H3K4me3, H3K4me2, H3K36me3, and H3K27me1 in 10-day seedlings. Except for the H3K27me1 modification which is prevalent in silent transposable elements, the other modifications are convincingly linked with transcriptionally active genes, in accordance with our result demonstrating enrichment of AtTERT chromatin in H3K4me3 and H3K9Ac marks in 7 day seedlings. In telomerase-negative samples, increased H3K27me3 loading in the AtTERT upstream and exon 5 regions was observed (Fig. 4) while signals for the other modifications (H3K4me3, H3KAc, and H3K9me2) were more or less comparable in both telomerase-positive and telomerase-negative tissues. The simultaneous presence of H3K4me3 and H3K27me3 in this locus seems to be rather contradictory with the data of Roudier et al. (2011) and Ha et al. (2011), which show low association of these marks. Because the levels of H3K4me3 and H3K9Ac remained reproducibly high in both regions analysed and based on results of micrococcal nuclease digestion (Supplementary Fig. S3), one can speculate that – despite the increased H3K27me3 loading – the AtTERT chromatin maintained the euchromatin state in telomerase-negative tissue. Moreover, silencing of FLC gene transcription during the plant transition to flowering – representing a typical example of developmental gene regulation – is accompanied, besides distinctive H3K27me3 loading, by a significant decrease of H3K4me3 and H3KAc and even an increase of H3K9me2 (see Bastow et al., 2004; reviewed by Deal and Henikoff, 2011). To verify the necessity of H3K27me3 for the AtTERT silencing, analysis of telomerase dynamics in plants with loss of function of terminal flower 2 (TFL2)/like heterochromatin protein 1 (LHP1) might be informative. TFL2/LHP1 is essential for the establishment of the H3K27me3 repressive modification at developmentally regulated genes (Turck et al., 2007) and loss of its function leads to a broad range of developmental defects (Gaudin et al., 2001).
In recent reports, independence of H3K27me3 and DNA methylation (Zhang et al., 2007) and even mutual exclusivity of these modifications (Weinhofer et al., 2010) were demonstrated. In this context, essentially the same patterns of histone modifications, including increased H3K27me3 loading in telomerase-negative tissue in the methylation-free upstream region and in exon 5 with an increased level of methylated cytosines, are very interesting. Since histone modifications are comparable in mature leaves of the met1-3 mutant (Supplementary Fig. S4) and of the wild type (Fig. 4), it seems that H3K27me3 loading is in no way affected by cytosine methylation in exon 5.
Taken together, our analysis of the epigenetic states of the TERT gene in telomerase-positive and telomerase-negative Arabidopsis tissues reveals differential levels of H3K27me3 modification. Nevertheless, in contrast to the situation in mammalian cells where chromatin surrounding the active TERT gene is associated with euchromatin-specific histone modifications (hyperacetylation and H3K4 methylation), while chromatin of the silenced hTERT gene is marked by H3K9 methylation and H4K20 methylation (i.e. modifications typical for silenced and even heterochromatic regions; Wang et al., 2009), such notable changes of native chromatin environment are not associated with TERT gene silencing in Arabidopsis. Although immediate promoter status generally results from a complex interplay of many cellular factors, the observed differences between animal and plant cells in the mechanisms involved in developmental regulation of TERT may reflect a unique attribute of plants – their totipotency – which accords with a reversible and dynamic character of telomerase silencing (Fajkus et al., 1998).
Supplementary material
Supplementary material is available at JXB online.
Supplementary Table S1. Sequences of primers used in genotyping the mutant lines, telomerase activity assay, quantitative analysis of transcription, analysis of methylation by bisulphite genomic sequencing (BGS), and analysis of chromatin modifications (ChIP).
Supplementary Fig. S1. AtTERT transcription in wild-type leaves.
Supplementary Fig. S2. Analysis of DNA methylation in AtTERT upstream region by bisulphite genomic sequencing.
Supplementary Fig. S3. Micrococcal nuclease digestion of nuclei isolated from Arabidopsis seedlings and leaves.
Supplementary Fig. S4. Analysis of histone modifications in the met1-3 seedlings and mature leaves.
Acknowledgments
This work was supported by the Czech Science Foundation (P501/11/0596), the Grant Agency of the AS CR (IAA500040801), and the project 'CEITEC – Central European Institute of Technology' (CZ.1.05/1.1.00/02.0068) from the European Regional Development Fund.
Glossary
Abbreviations
- AtTERT
A. thaliana TERT
- ChIP
chromatin immunoprecipitation
- hTERT
human TERT
- TERT
telomerase reverse transcriptase
- TR subunit
telomerase RNA subunit
References
- Aceituno FF, Moseyko N, Rhee SY, Gutierrez RA. The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana . BMC Genomics. 2008;9:438. doi: 10.1186/1471-2164-9-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Broun P, Ganal MW, Tanksley SD. Telomeric arrays display high levels of heritable polymorphism among closely related plant varieties. Proceedings of the National Academy of Sciences, USA. 1992;89:1354–1357. doi: 10.1073/pnas.89.4.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatagnon A, Bougel S, Perriaud L, Lachuer J, Benhattar J, Dante R. Specific association between the methyl-CpG-binding domain protein 2 and the hypermethylated region of the human telomerase reverse transcriptase promoter in cancer cells. Carcinogenesis. 2009;30:28–34. doi: 10.1093/carcin/bgn240. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proceedings of the National Academy of Sciences, USA. 2011;108:73–78. doi: 10.1073/pnas.1013021107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Research. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. Histone variants and modifications in plant gene regulation. Current Opinion in Plant Biology. 2011;14:116–122. doi: 10.1016/j.pbi.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessain SK, Yu H, Reddel RR, Beijersbergen RL, Weinberg RA. Methylation of the human telomerase gene CpG island. Cancer Research. 2000;60:537–541. [PubMed] [Google Scholar]
- Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Research. 1999;59:6087–6090. [PubMed] [Google Scholar]
- Fajkus J, Kovarik A, Kralovics R. Telomerase activity in plant cells. FEBS Letters. 1996;391:307–309. doi: 10.1016/0014-5793(96)00757-0. [DOI] [PubMed] [Google Scholar]
- Fajkus J, Fulneckova J, Hulanova M, Berkova K, Riha K, Matyasek R. Plant cells express telomerase activity upon transfer to callus culture, without extensively changing telomere lengths. Molecular and General Genetics. 1998;260:470–474. doi: 10.1007/s004380050918. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE. Characterization and developmental patterns of telomerase expression in plants. Proceedings of the National Academy of Sciences, USA. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fojtova M, Peska V, Dobsakova Z, Mozgova I, Fajkus J, Sykorova E. Molecular analysis of the T-DNA insertion mutants identified putative regulatory elements in the AtTERT gene. Journal of Experimental Botany. 2011;62:5531–5545. doi: 10.1093/jxb/err235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, ten Hoopen R, Tessadori F. Composition and formation of heterochromatin in Arabidopsis thaliana . Chromosome Research. 2006;14:71–82. doi: 10.1007/s10577-005-1022-5. [DOI] [PubMed] [Google Scholar]
- Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- Guilleret I, Benhattar J. Demethylation of the human telomerase catalytic subunit (hTERT) gene promoter reduced hTERT expression and telomerase activity and shortened telomeres. Experimental Cell Research. 2003;289:326–334. doi: 10.1016/s0014-4827(03)00281-7. [DOI] [PubMed] [Google Scholar]
- Ha M, Ng DW, Li WH, Chen ZJ. Coordinated histone modifications are associated with gene expression variation within and between species. Genome Research. 2011;21:590–598. doi: 10.1101/gr.116467.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller K, Kilian A, Piatyszek MA, Kleinhofs A. Telomerase activity in plant extracts. Molecular and General Genetics. 1996;252:342–345. doi: 10.1007/BF02173780. [DOI] [PubMed] [Google Scholar]
- Heller-Uszynska K, Schnippenkoetter W, Kilian A. Cloning and characterization of rice (Oryza sativa L) telomerase reverse transcriptase, which reveals complex splicing patterns. Plant Journal. 2002;31:75–86. doi: 10.1046/j.1365-313x.2001.01337.x. [DOI] [PubMed] [Google Scholar]
- Herbert BS, Hochreiter AE, Wright WE, Shay JW. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nature Protocols. 2006;1:1583–1590. doi: 10.1038/nprot.2006.239. [DOI] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. The Plant Journal. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Computational Biology. 2009;5 doi: 10.1371/journal.pcbi.1000566. e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa I, Chiang YJ, Patterson T, Feigenbaum L, Leem SH, Michishita E, Larionov V, Hodes RJ, Barrett JC. Differential cis-regulation of human versus mouse TERT gene expression in vivo: identification of a human-specific repressive element. Proceedings of the National Academy of Sciences, USA. 2005;102:18437–18442. doi: 10.1073/pnas.0508964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Van Houdt H, Holy A, Depicker A. Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. FEBS Letters. 2000;467:47–51. doi: 10.1016/s0014-5793(00)01077-2. [DOI] [PubMed] [Google Scholar]
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nature Structural and Molecular Biology. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- Lunerova-Bedrichova J, Bleys A, Fojtova M, Khaitova L, Depicker A, Kovarik A. Trans-generation inheritance of methylation patterns in a tobacco transgene following a post-transcriptional silencing event. Plant Journal. 2008;54:1049–1062. doi: 10.1111/j.1365-313X.2008.03475.x. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Oguchi K, Liu H, Tamura K, Takahashi H. Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana . FEBS Letters. 1999;457:465–469. doi: 10.1016/s0014-5793(99)01083-2. [DOI] [PubMed] [Google Scholar]
- Oguchi K, Tamura K, Takahashi H. Characterization of Oryza sativa telomerase reverse transcriptase and possible role of its phosphorylation in the control of telomerase activity. Gene. 2004;342:57–66. doi: 10.1016/j.gene.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. Quantification strategies in real-time PCR. In: S.A. Bustin., editor. A-Z of Quantitative PCR. International University Line, La Jolla, CA; 2004. pp. 87–112. [Google Scholar]
- Riha K, Fajkus J, Siroky J, Vyskot B. Developmental control of telomere lengths and telomerase activity in plants. The Plant Cell. 1998;10:1691–1698. doi: 10.1105/tpc.10.10.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P, Collier S, Bush M, Shaw P, Doonan JH. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. Journal of Cellular Sciences. 2007;120:3678–3687. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO Journal. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nature Genetics. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Sykorova E, Lim KY, Chase MW, Knapp S, Leitch IJ, Leitch AR, Fajkus J. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): first evidence from eudicots. Plant Journal. 2003;34:283–291. doi: 10.1046/j.1365-313x.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genetics. 2007;3 doi: 10.1371/journal.pgen.0030086. e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhao Y, Hu C, Zhu J. Differential repression of human and mouse TERT genes during cell differentiation. Nucleic Acids Research. 2009;37:2618–2629. doi: 10.1093/nar/gkp125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I, Hehenberger E, Roszak P, Hennig L, Kohler C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genetics. 2010;6 doi: 10.1371/journal.pgen.1001152. e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzic M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, Colot V, Doerge RW, Martienssen RA. Epigenetic natural variation in Arabidopsis thaliana . PLoS Biology. 2007;5 doi: 10.1371/journal.pbio.0050174. e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biology. 2007;5 doi: 10.1371/journal.pbio.0050129. e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhao Y, Wang S. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell. 2011;1:22–32. doi: 10.1007/s13238-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.