Abstract
A single stranded DNA fragment was modified with alkylating derivatives of oligonucleotides complementary to a certain nucleotide sequences in the fragment. The derivatives carried aromatic 2-chloroethylamino groups at their 3'- or 5'-terminal nucleotide residues. Some of the derivatives carried both alkylating group and intercalating phenazine group which stabilized complementary complexes. It was found that these oligonucleotide derivatives modify the DNA fragment in a specific way near the target complementary nucleotide sequences, and the DNA fragment can be cleaved at the alkylated nucleotides positions. Alkylating derivatives carrying phenazine groups were found to be the most efficient in reaction with the DNA fragment.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asseline U., Delarue M., Lancelot G., Toulmé F., Thuong N. T., Montenay-Garestier T., Hélène C. Nucleic acid-binding molecules with high affinity and base sequence specificity: intercalating agents covalently linked to oligodeoxynucleotides. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3297–3301. doi: 10.1073/pnas.81.11.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikova A. M., Zarytova V. F., Grineva N. I. Synthesis of ribonucleosides and diribonucleoside phosphates containing 2-chloroethylamine and nitrogen mustard residues. Tetrahedron Lett. 1967 Sep;37:3557–3562. doi: 10.1016/s0040-4039(01)89794-x. [DOI] [PubMed] [Google Scholar]
- Chu B. C., Orgel L. E. Nonenzymatic sequence-specific cleavage of single-stranded DNA. Proc Natl Acad Sci U S A. 1985 Feb;82(4):963–967. doi: 10.1073/pnas.82.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer G. B., Dervan P. B. Sequence-specific cleavage of single-stranded DNA: oligodeoxynucleotide-EDTA X Fe(II). Proc Natl Acad Sci U S A. 1985 Feb;82(4):968–972. doi: 10.1073/pnas.82.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
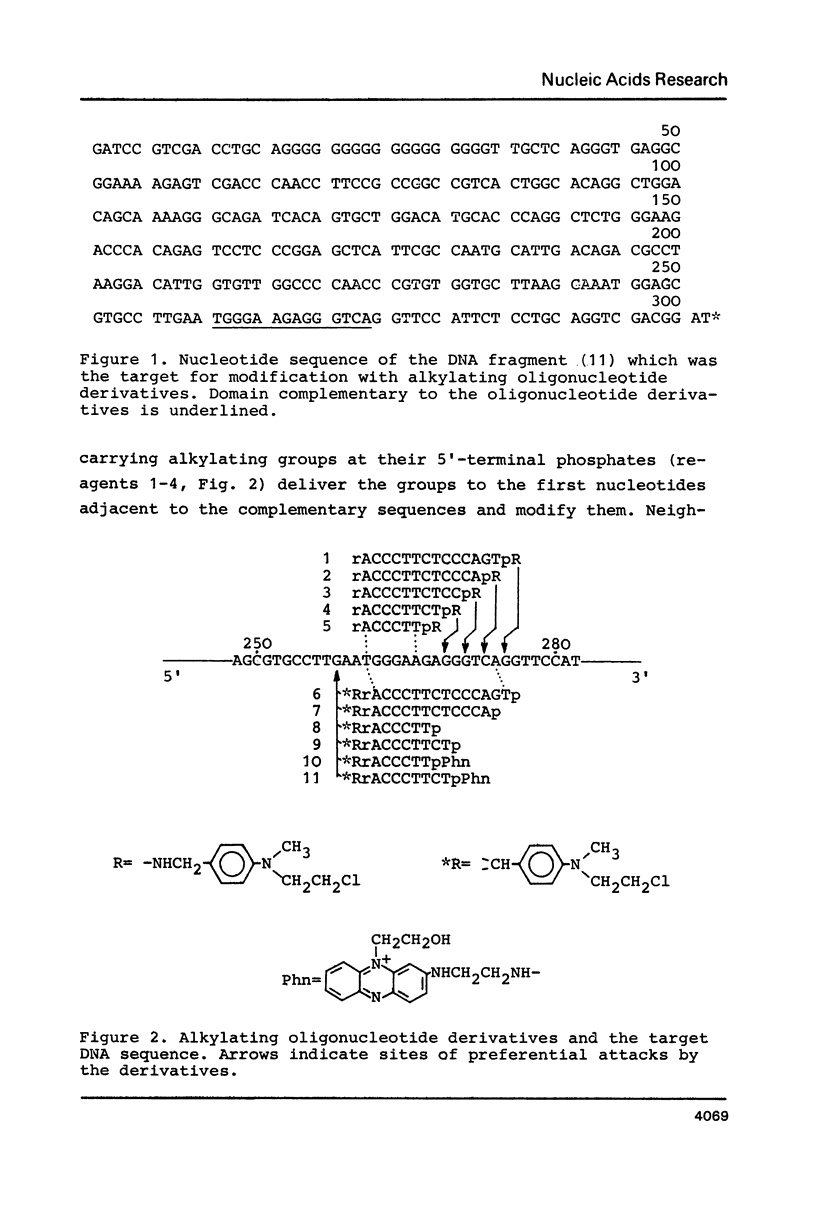
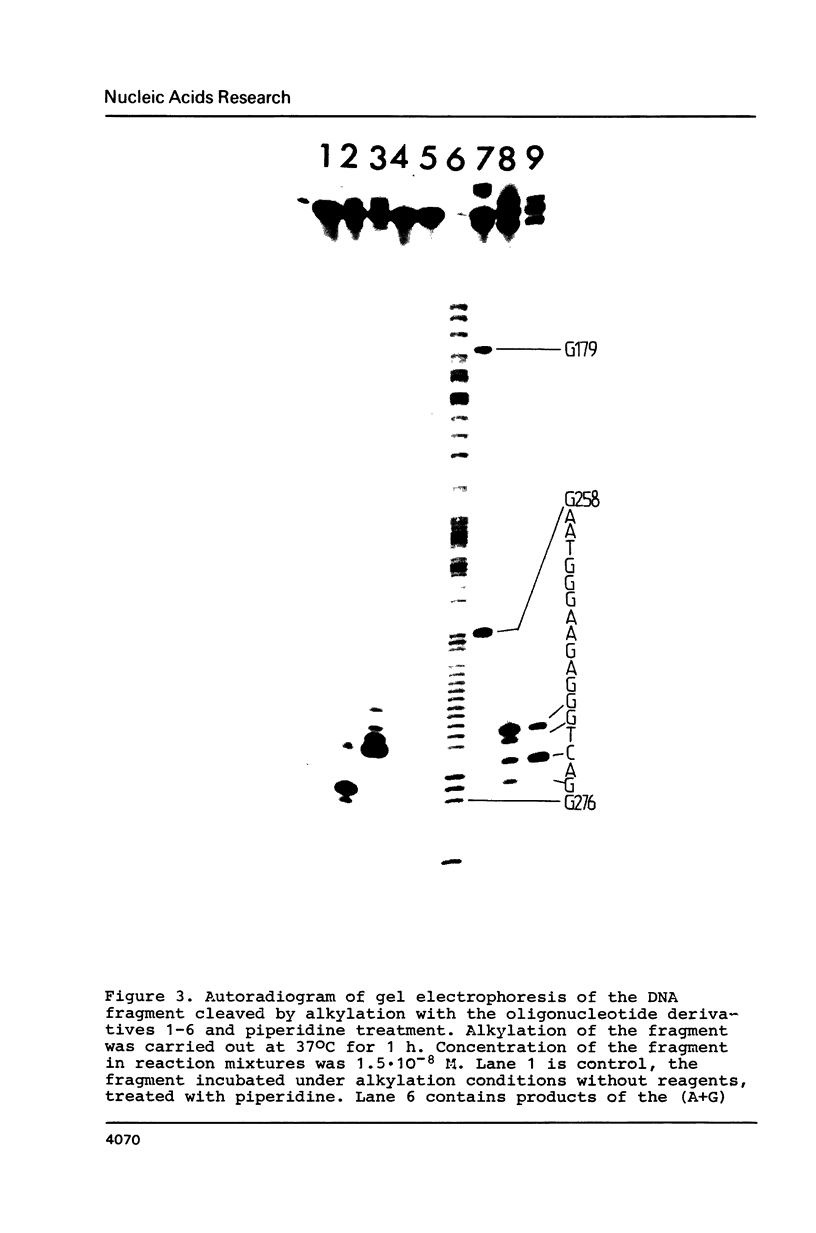
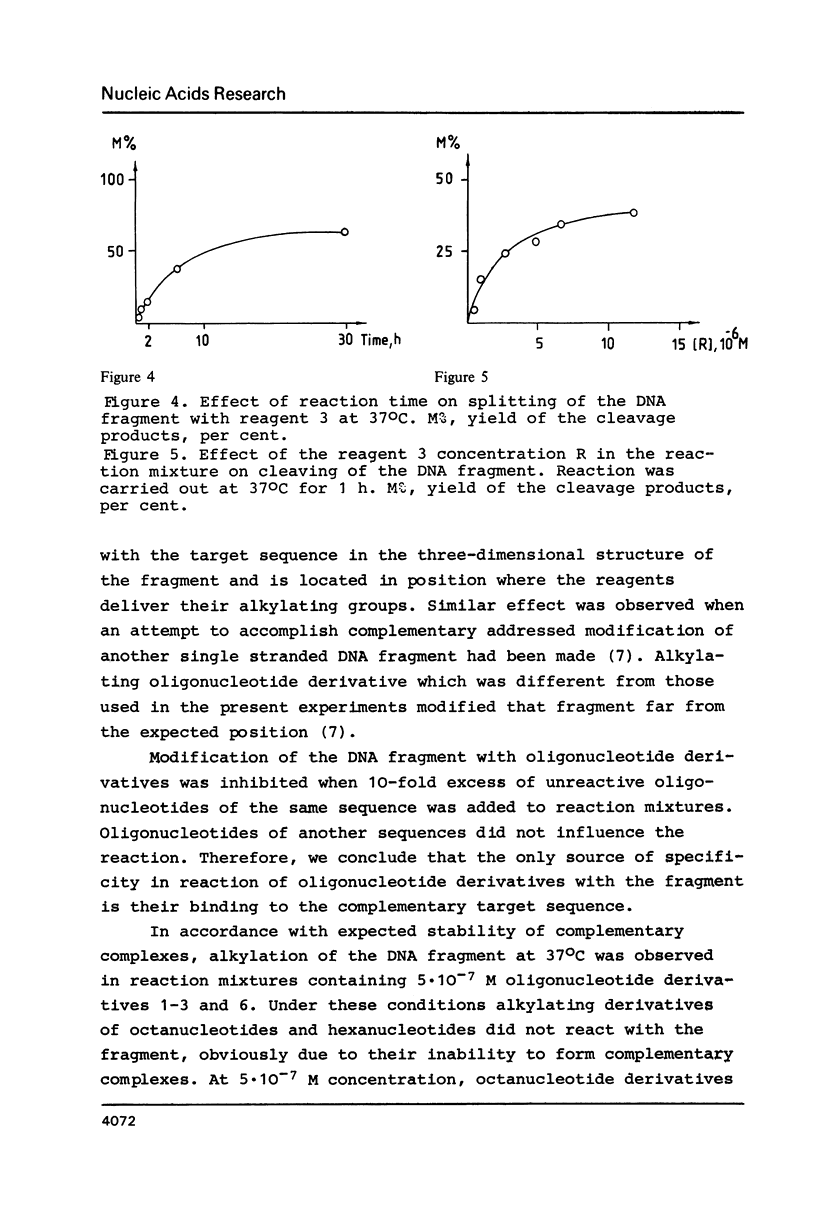
- Grachev M. A., Oshevskii S. I. Issledovanie "adresovannoi" khimicheskoi modifikatsii fragmenta odnonitevoi DNK. Dokl Akad Nauk SSSR. 1983;272(5):1259–1262. [PubMed] [Google Scholar]
- Grineva N. I., Karpova G. G. Complementarily addressed modification of rRNA with p-(chloroethylmethylamino)benzylidene hexanucleotides. FEBS Lett. 1973 Jun 1;32(2):351–355. doi: 10.1016/0014-5793(73)80871-3. [DOI] [PubMed] [Google Scholar]
- Grineva N. I., Karpova G. G., Kuznetsova L. M., Venkstern T. V., Bayev A. A. Complementary addressed modification of yeast tRNA Val 1 with alkylating derivative of d(pC-G)-A. The positions of the alkylated nucleotides and the course of the alkylation in the complex. Nucleic Acids Res. 1977;4(5):1609–1631. doi: 10.1093/nar/4.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorre D. G., Vlassov V. V. Complementary-addressed (sequence-specific) modification of nucleic acids. Prog Nucleic Acid Res Mol Biol. 1985;32:291–320. doi: 10.1016/s0079-6603(08)60352-9. [DOI] [PubMed] [Google Scholar]
- Kolchanov N. A., Solov'ev V. V., Zharkikh A. A. Vysokaia nasyshchennost' priamymi povtorami v genakh RNK-polimeraz po dannym kontekstnogo analiza. Dokl Akad Nauk SSSR. 1983;273(3):741–744. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Patton J. R., Chae C. B. A method for isolation of a large amount of a single-stranded DNA fragment. Anal Biochem. 1982 Oct;126(1):231–234. doi: 10.1016/0003-2697(82)90134-8. [DOI] [PubMed] [Google Scholar]
- Summerton J. Intracellular inactivation of specific nucleotide sequences: a general approach to the treatment of viral diseases and virally-mediated cancers. J Theor Biol. 1979 May 7;78(1):77–99. doi: 10.1016/0022-5193(79)90327-8. [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Gaidamakov S. A., Gorn V. V., Grachev S. A. Sequence-specific chemical modification of a 365-nucleotide-long DNA fragment with an alkylating oligonucleotide derivative. FEBS Lett. 1985 Mar 25;182(2):415–418. doi: 10.1016/0014-5793(85)80345-8. [DOI] [PubMed] [Google Scholar]