Abstract
Nematodes parasitize an alarming number of people and agricultural animals globally and cause debilitating morbidity and mortality. Anthelmintics have been the primary tools used to control parasitic nematodes for the past several decades, but drug resistance is becoming a major obstacle. Xenobiotic detoxification pathways defend against drugs and other foreign chemicals in diverse organisms, and evidence is accumulating that they play a role in mediating resistance to anthelmintics in nematodes. Related anti-oxidation pathways may also provide filarial parasites protection against host free radical-mediated immune responses. Upstream regulatory pathways have received almost no attention in nematode parasites despite their potential to co-regulate multiple detoxification and anti-oxidation genes. The NRF2 transcription factor mediates inducible detoxification and anti-oxidation defenses in mammals and recent studies have demonstrated that it promotes multidrug resistance in some human tumors. Recent studies in the free-living model nematode Caenorhabditis elegans have defined the homologous transcription factor SKN-1 as a master regulator of detoxification and anti-oxidation genes. Despite similar functions, SKN-1 and NRF2 have important differences in structure and regulatory pathways. Protein alignment and phylogenetic analyses indicate that these differences are shared among many nematodes making SKN-1 a candidate for specifically targeting nematode detoxification and anti-oxidation.
Keywords: Xenobiotic detoxification, anti-oxidation, anthelminthic, SKN-1, NRF2, gene regulation, drug target, Caenorhabditis elegans
INTRODUCTION
Parasitic nematodes are a major cause of human mortality and morbidity in tropical and sub-tropical climates (Chan 1997; Mathers et al. 2007) and infect millions of people in the United States and Europe who live in poverty (Hotez 2009). The World Health Organization estimates that two billion people are infected with parasitic nematodes world-wide (http://www.who.int/wormcontrol/statistics/). Despite being common, helminth-induced diseases receive less than 1% of research funds globally and are regarded as neglected tropical diseases (Hotez et al. 2008). Parasitic nematodes also burden human health and nutrition by parasitizing livestock and cause an estimated $80 billion loss of worldwide crop production each year (Jasmer et al. 2003). Programs to control parasitic nematodes in humans and animals rely heavily on the widespread administration of three classes of helminth targeting drugs, or anthelmintics: benzimidazoles, which target the cytoskeleton via β-tubulin; levamisole and related compounds, which are nicotinic receptor agonists; and macrocyclic lactones, which have a high affinity for glutamate-gated chloride channels (Holden-Dye and Walker 2007). Unfortunately, resistance is a widespread and growing problem (Kaplan 2004; Kaplan and Vidyashankar 2011). Multidrug resistance is especially problematic, because it reduces the efficacy of many drugs and is poorly understood (Gilleard 2006; James et al. 2009).
Drug resistance is a fundamental response in populations of cells and organisms when repeatedly exposed to sub-lethal doses of a toxic compound (Persidis 1999; Kaplan 2004; Prasad and Kapoor 2004; Gilleard 2006; James and Davey 2008). Nematodes have large population sizes and high genetic diversity, which facilitates the evolution of resistance (Kaplan 2004). This potential for drug resistance is best exemplified in livestock that are routinely treated with anthelmintics; resistance to every anthelmintic class of drugs has been reported for parasites of every major livestock host (Kaplan 2004; Gilleard 2006; Kaplan and Vidyashankar 2011). Studies of human parasites have also suggested the potential for resistance to emerge (Geerts and Gryseels 2001; Awadzi et al. 2004; Osei-Atweneboana et al. 2007).
It has been over 25 years since a new class of broadly applicable anthelmintics has been adopted for widespread use (Prichard and Geary 2008). Two new classes of anthelmintics are being implemented for use in livestock (Kaminsky et al. 2008; Little et al. 2010), but resistance is likely to occur once new drugs are used widely. Therefore, new strategies, drug targets, and tools are urgently needed. Knowledge of the underlying molecular and genetic mechanisms is essential to the proper design and implementation of strategies to monitor the development of anthelmintic resistance and to maintain and extend the useful life of currently used drugs (Prichard 1994; Gilleard 2006; Lespine et al. 2008). Unfortunately, these processes are poorly understood and tools for their analysis are sparse for parasitic nematodes that have limited genetic tractability (Geerts and Gryseels 2001; von Samson-Himmelstjerna and Blackhall 2005; Gilleard 2006; Holden-Dye and Walker 2007; Mitreva et al. 2007; James and Davey 2008).
NEMATODE PARASITES OF MAJOR CONCERN
A full description of parasitic nematodes is beyond the scope of this review. Here, we briefly describe a few groups of concern based on existing resistance, medical or agricultural impact, and risk for resistance (listed in Table 1). Readers are directed to other reviews for details on livestock (Kaplan 2004; Kaplan and Vidyashankar 2011) and human (Albonico et al. 2008; Keiser and Utzinger 2008; Bockarie and Deb 2010) parasites.
Table 1.
Nematode parasites of major concern
| Group of parasites | Examples | Common name | Clade | Host | Disease symptoms |
|---|---|---|---|---|---|
| Gastrointestinal/livestock | Haemonchus contortus | barber pole worm | V | sheep and goat | anemia, diarrhea, lethargy, death |
| Teladorsagia circumcincta | brown stomach worm | V | sheep and goat | ||
| Cooperia spp. | V | cattle | |||
| Ostertagia spp. | V | cattle | |||
| cyathostomins | V | horse | |||
| Parascaris equorum | III | horse | |||
| Gastrointestinal/human | Ascaris lumbricoides | roundworm | III | human | anemia, malnutrition, developmental and cognitive delay, intestinal obstruction, increased risk of other infections |
| Ancylostoma duodenale | human hookworm | V | human | ||
| Necator americanus | human hookworm | V | human | ||
| Trichuris trichiura | whipworm | I | human | ||
| Filarial/human | Wuchereria bancrofti | III | human | lymphoedema, hydrocoeles, elephantiasis, immune suppression, blindness, and skin disease |
|
| Brugia malayi | filarial nematode | III | human | ||
| Brugia timori | III | human | |||
| Loa loa | eye worm | III | human | ||
| Onchocerca volvulus | III | human |
Symptoms are listed by group (Kaplan 2004; Albonico et al. 2008; Keiser and Utzinger 2008; Bockarie and Deb 2010; Kaplan and Vidyashankar 2011). "Clade" refers to the five major phylogenetic groups within nematodes (Blaxter 1998; Blaxter et al., 1998). See text for details.
Gastrointestinal parasites of livestock animals are common and diverse. Gastrointestinal nematodes attach to the stomach or intestinal mucosa of the host and feed on blood causing anemia, malnutrition, dehydration, lethargy, and sometimes death. Important species include Haemonchus contortus and Teladorsagia circumcincta in sheep and goat, Cooperia spp. and Ostertagia spp. in cattle, and cyathostomins and Parascaris equorum in horses (Kaplan and Vidyashankar 2011). Livestock farming has relied on anthelmintics for over 30 years. In the past 10–15 years, resistance has grown into a problem that threatens the viability of entire industries (Kaplan 2004; Kaplan and Vidyashankar 2011). Resistance to all three classes of anthelmintics has now been reported for parasites of all common livestock hosts. The widespread and rapid growth of resistance in multiple parasite and host livestock combinations suggests that resistance is an inevitable outcome of prolonged anthelmintic use (Kaplan and Vidyashankar 2011).
A group of human gastrointestinal parasites transmitted by the soil (soil-transmitted helminths) are often considered and treated together. They include roundworm (Ascaris lumbricoides), hookworms (Ancylostoma duodenale and Necator americanus), and whipworm (Trichuris trichiura), which in combination infect as many as two billion people in tropical and developing regions (Bethony et al. 2006; Albonico et al. 2008). Heavy infections cause a range of symptoms including anemia, malnutrition, diarrhea, colitis, and impaired growth and cognitive development and may also increase susceptibility to malaria, tuberculosis, and HIV (Bethony et al. 2006; Albonico et al. 2008). The socioeconomic benefits of de-worming children are recognized and global programs based on mass drug treatment with benzimidazole, levamisole, and nicotinic agonist class drugs are in place to reduce infection rates (Albonico et al. 2008).
There are eight nematode species known to live in the lymphatic vessels of humans, and these species are thought to infect 120 million people worldwide with a total of 1.2 billion at risk (Bockarie and Deb 2010; Taylor et al. 2010). The most common species are Wuchereria bancrofti, Brugia malayi, and Brugia timori. Another important species is Onchocerca volvulus, which is thought to infect 37 million people mostly in Africa; adults live in subcutaneous and deep tissues and larvae migrate to the skin and eyes (Taylor et al. 2010). The diseases caused by these species are classified as filariasis (referring to small, thin larvae) with symptoms including lymphoedema, hydrocoeles, elephantiasis, immune suppression, blindness, and skin disease (Taylor et al. 2010). Transmission of filarial nematodes is by biting insects. The World Health Organization initiated a global program to eliminate filarial diseases in 1999, and over two billion treatments with benzimidazole and macrocyclic lactone class drugs have been administered (Gustavsen et al. 2009; Hooper et al. 2009).
Although mass drug administration programs have been effective in interrupting or decreasing human nematode infections (Hooper et al. 2009), reports are beginning to emerge that at least support the potential for resistance (Geerts and Gryseels 2000; Geerts and Gryseels 2001; Albonico et al. 2004; Keiser and Utzinger 2008); resistance in humans is difficult to verify because controlled experiments (such as those conducted in livestock) cannot be performed (Kaplan 2004). The unfortunate history of resistance in ruminant gastrointestinal nematodes serves as a warning that widespread resistance in human parasites may be inevitable with prolonged drug selection (Geerts and Gryseels 2001).
XENOBIOTIC DETOXIFICATION PROMOTES MULTIDRUG RESISTANCE
Multidrug resistance is well studied in pathogenic microbes, fungi, and cancer cells and is often caused by the increased expression and activity of xenobiotic detoxification enzymes (Persidis 1999; Prasad et al. 2002; Moye-Rowley and Scott 2003; Prasad and Kapoor 2004; Sipos and Kuchler 2006; Lubelski et al. 2007). Despite the fundamental and conserved role of xenobiotic detoxification in drug resistance, these processes are poorly understood in nematodes. The limited knowledge of these processes in helminths has been reviewed in detail recently (Cvilink et al. 2009) and we only summarize it here briefly.
The general strategy for cellular detoxification is conserved. In animal cells, xenobiotic detoxification is modeled as occurring in three sequential and interdependent phases. In phase I, enzymes such as cytochrome P450s, short-chain dehydrogenases, and reductases uncover or insert reactive and hydrophilic groups onto xenobiotics leaving them more accessible for further processing (Iyanagi 2007; Cvilink et al. 2009). The genome of Caenorhabditis elegans, the free-living genetic model nematode, is predicted to contain 86 cytochrome P450s and 68 short-chain dehydrogenases (Lindblom and Dodd 2006; the genome of Pristionchus pacificus, a beetle-associated nematode, is predicted to contain 198 cytochrome P450s (Dieterich et al. 2008).
In phase II, conjugating enzymes catalyze the addition of glutathione, glucuronic acid, amino acids, and sulphates to xenobiotics or their phase I metabolites, resulting in less toxic and more water soluble and excretable products (O'Brien and Tew 1996; McLellan and Wolf 1999; Strange et al. 2001; Townsend and Tew 2003; Iyanagi 2007; Torres-Rivera and Landa 2008). Glutathione and glutathione s-tranferases (GSTs) also reduce electrophilic xenobiotics and oxygen free radicals generated by some xenobiotics and host immune systems (O'Brien and Tew 1996; Strange et al. 2001; Torres-Rivera and Landa 2008). Nematode GSTs have also been shown to possess prostaglandin isomerase activity and have been hypothesized to play a role in signaling of development and modification of prostaglandin-regulated mammalian immune responses (Perbandt et al. 2008; Joachim et al. 2011; Joachim and Ruttkowski 2011). Similar to phase I, there is a tremendous diversity of phase II genes with 72 and 139 predicted glucuronosyltransferases in C. elegans and P. pacificus, respectively (Lindblom and Dodd 2006; Dieterich et al. 2008); there are 48 and 54 predicted GSTs in C. elegans and P. pacificus, respectively. Increased activity and/or expression of phase I and/or II genes is associated with cambenzadole (related to benzimidazole) resistance in H. contortus (major parasite of ruminants, commonly drug resistant, and important model for experimental parasitology) (Kwalek et al. 1984) and ivermectin (a macrocyclic lactone) resistance in C. elegans (James et al. 2009) and H. contortus (Sotirchos et al. 2008).
A recent study analyzed the transcriptome of C. elegans four hours after exposure to the widely used benzimidole albendazole and found six phase I and ten phase II enzymes among the 42 genes up-regulated (Laing et al. 2010). This study also demonstrated that albendazole metabolites produced in C. elegans were similar to those in H. contortus and distinct from metabolites in mammals suggesting that these two nematodes share common biochemical properties of detoxification that are distinct from vertebrates.
In phase III, transmembrane transporters export drugs and their metabolites out of cells. The major family of xenobiotic transporters is the ATP-binding cassette (ABC) protein family, which includes three subgroups thought to be involved in drug resistance: ABCB (p-glycoprotein, PGP and multidrug resistant, MDR proteins), ABCC (multidrug resistance related protein, MRP), and ABCG (includes breast cancer resistance protein). ABC proteins are found in all cellular forms of life (Cvilink et al. 2009) and are diverse in nematodes with at least 60 family members in C. elegans (Zhao et al. 2007), 129 in P. pacificus (Dieterich et al. 2008), and 33 in B. malayi (filarial parasite of humans) (Ardelli et al. 2010). Binding of xenobiotics to these integral membrane proteins is followed by ATP binding and conformational changes that cause export of compounds; ATP hydrolysis allows regeneration of the initial state of the protein (Schinkel and Jonker 2003). Comprehensive information on xenobiotic transporter diversity and function is well reviewed (Kerboeuf et al. 2003; Sheps et al. 2004; Alvarez et al. 2006; Lindblom and Dodd 2006). ABC transporters have a well established role in promoting chemotherapeutic resistance in some cancer cells and have been an important target for reversing resistance in humans (Kerboeuf et al. 2003). As a result, this phase of detoxification has received the most attention in parasites (Alvarez et al. 2006; Lespine et al. 2008).
Isolates of ivermectin-selected H. contortus have been shown to have elevated expression of an ABCB gene, and verapamil, an inhibitor of PGP, increased ivermectin efficacy in these resistant strains (Xu et al. 1998). Selection for multidrug resistance over multiple generations also resulted in elevated expression of ABC transporter genes in C. elegans, and verapamil partially reversed resistance (James et al. 2009). Shorter-term exposures of B. malayi and H. contortus to ivermectin and/or its analogue moxidectin have also been shown to cause an increase in the expression of ABCB genes (Prichard and Roulet 2007; Stitt et al. 2011).
Interestingly, detoxification gene numbers vary greatly between three nematode species in which they have been surveyed (B. malayi, C. elegans, and P. pacificus, see above) (Lindblom and Dodd 2006; Dieterich et al. 2008; Ardelli et al. 2010). Brugia malayi resides in the controlled interstitial environments of human and insect host tissues and has the fewest number of detoxification genes, C. elegans feeds on bacteria on rotting plant tissues (Kiontke et al. 2011), and P. pacificus feeds on bacteria, fungi, and other nematodes that grow on beetle carcasses and has the highest number of detoxification genes (Herrmann et al. 2007). Thus, detoxification gene number may be highly flexible in nematodes and appears to evolve to match the chemical complexity of each species’ environment (Dieterich et al. 2008). It will be interesting to learn if gastrointestinal parasites have a high number of detoxification genes to match their chemically diverse environments.
Given the tremendous diversity of detoxification genes and their fundamental and interdependent functions, many more detoxification genes in all three phases are likely to contribute to anthelmintic resistance in nematodes. Lowering costs of next-generation DNA and RNA sequencing and a growing number of sequenced parasite genomes (Ghedin et al. 2007; Mitreva et al. 2011; Sommer and Streit 2011) will soon permit genome-wide identification of genes with altered expression in resistant strains and association of alleles with resistance. These approaches will provide a starting point for generating new hypotheses about mechanisms of drug resistance, but ultimately target-specific pharmacological or genetic modulation will be needed to test the functional importance of individual genes and pathways.
Targeting detoxification
Lespine et al. (2008) recently proposed using inhibitors of xenobiotic detoxification and transport in combination with currently available anthelmintics as a strategy to treat infections by multidrug resistant parasitic nematodes. Inhibitors of glutathione synthesis and ABC transporters such as buthionine sulfoxamine and verapamil (among others) are providing early evidence that this strategy might be effective (Xu et al. 1998; Molento and Prichard 1999; James and Davey 2008; Bartley et al. 2009; Stitt et al. 2011). However, currently available drugs that inhibit xenobiotic detoxification and transport have two important limitations for their use in vivo. The first limitation is that they only affect a single protein or class of proteins. The second major limitation of currently available detoxification targeting drugs is that they also inhibit homologous pathways in mammalian hosts. Inhibition of host detoxification mechanisms has the potential to be extremely harmful. For example, mice, cattle, and dogs with deficiencies in ABCB function suffer from neurotoxicity of ivermectin, which is normally very safe for mammals (Seaman et al. 1987; Schinkel et al. 1994; Roulet et al. 2003). Detoxification is also essential for normal cellular metabolism and redox homeostasis (Cvilink et al. 2009). Therefore, multidrug resistance reversal compounds would ideally target multiple detoxification proteins and would be specific for nematodes.
TRANSCRIPTION FACTORS AS MASTER REGULATORS OF DETOXIFICATION GENES
In mammals, Drosophila (fruitfly), and C. elegans, detoxification pathways are tightly regulated so that basal activity is low and exposure to toxic xenobiotics or oxidants simultaneously activates the expression of multiple genes through inducible transcription factors. The gene regulatory pathways upstream from detoxification effectors have been largely ignored in parasitic helminths despite their potential to control multiple genes in all phases of detoxification.
In mammals, two nuclear hormone receptors, pregnane X receptor (PXR, NR1I2) and constitutive androstane receptor (CAR, NR1I3), are activated by exogenous and endogenous ligands and control the expression of cytochrome P450 genes (Pascussi et al. 2008). Although the C. elegans genome is predicted to encode over 270 nuclear hormone receptors, only NHR-8 is known to play a role in tolerance of xenobiotics (Lindblom et al. 2001). The genes regulated by NHR-8 have not been identified and far more work is needed to understand the contribution of this and other nuclear hormone receptors to xenobiotic metabolism and drug resistance in nematodes. In mammals, cytochrome P450 genes are also regulated by the aryl hydrocarbon receptor (AHR). ahr-1 is the single AHR homolog in C. elegans, but this gene is not required for cytochrome P450 gene expression and instead plays a role in development (Huang et al. 2004; Aarnio et al. 2010).
SKN-1 regulates multiple xenobiotic detoxification and anti-oxidation genes
The cap-n-collar family (CNC) of basic leucine zipper transcription factors regulates the expression of xenobiotic detoxification genes in C. elegans, Drosophila, and mammals (An and Blackwell 2003; Kobayashi et al. 2008; Misra et al. 2011), and therefore, may perform this function in all animals. Included in this family is the mammalian nuclear eurythroid 2-related factor 2 (NRF2), which was originally characterized as mediating cellular defense responses to oxidants (Kobayashi and Yamamoto 2006). It is now clear that NRF2 also mediates cellular defenses to diverse xenobiotics by controlling the expression of genes in all three phases of detoxification (e.g., Thimmulappa et al. 2002; Hu et al. 2006). Recent studies have demonstrated that enhanced NRF2 activity mediates multidrug resistance in some cancer cells (Townsend and Tew 2003; Ohta et al. 2008; Shibata et al. 2008; Singh et al. 2008; Wang et al. 2008; Hayes and McMahon 2009; Hayes et al. 2010; Kensler and Wakabayashi 2010). Genetic silencing of NRF2 was able to reverse resistance by decreasing the expression of several detoxification genes (Singh et al. 2008). These results highlight the potential for transcriptional regulators to orchestrate drug resistance through multiple genes and for targeting of a single transcription factor to reverse resistance.
SKN-1 is the single CNC protein in C. elegans. Like NRF2, SKN-1 is activated by oxidants, electrophiles, and diverse xenobiotics and confers resistance by activating detoxification genes (An and Blackwell 2003; An et al. 2005; Inoue et al. 2005; Kell et al. 2007; Hasegawa et al. 2008; Kahn et al. 2008; Tullet et al. 2008; Choe et al. 2009; Przybysz et al. 2009). Genome-wide transcriptional profiling of C. elegans with and without skn-1(RNAi) has identified genes regulated by the transcription factor under basal conditions and during exposure to oxidative stress induced by hyperbaric hyperoxia, arsenite, and tert-butyl hydroxide (Oliveira et al. 2009; Park et al. 2009). Two hundred thirty three genes required SKN-1 for full expression under non-stressed conditions. Partially overlapping sets of 211, 118, and 64 genes required SKN-1 for increased expression with hyperbaric hyperoxia, arsenite, and organic peroxide, respectively. Sixty five of the SKN-1 regulated genes identified in these studies are predicted to encode proteins that function in detoxification and/or anti-oxidation (Table 2). Functional categories include eight short-chain dehydrogeneases, 20 glutathione s-transferases, 10 glucuronosyltransferases, two cytochrome P450s, and two ABC superfamily genes. Importantly, the true number of genes regulated by SKN-1 is likely to be considerably higher because the statistical requirements applied to transcriptome data are stringent and only four conditions have been tested. Our own microarray analysis of a C. elegans strain with constitutively active SKN-1 identifies another 44 detoxification genes under positive control of SKN-1 including six ABCB subfamily members (Choe et al., unpublished data).
Table 2.
Anti-oxidation and detoxification genes regulated by SKN-1
| sequence | name | description | function | control | hyperoxia | arsenite | tBOOH |
|---|---|---|---|---|---|---|---|
| Y54G11A.5 | ctl-2 | catalase 2, peroxisomal | antioxidation | X | |||
| F26E4.12 | Glutathione peroxidase | antioxidation | X | X | X | X | |
| R03G5.5 | Glutathione peroxidase | antioxidation | X | X | |||
| C15F1.7 | sod-1 | superoxide dismutase 1 | antioxidation | X | |||
| C35B1.5 | Thioredoxin, nucleoredoxin and related proteins | antioxidation | X | X | X | ||
| Y52E8A.3 | Thioredoxin, nucleoredoxin and related proteins | antioxidation | X | X | |||
| K12G11.4 | sodh-2 | Alcohol dehydrogenase, class V | phase I | X | |||
| C54D1.4 | alh-10 | Aldehyde dehydrogenase | phase I | X | |||
| C07D8.6 | Aldo/keto reductase family proteins | phase I | X | ||||
| T08H10.1 | Aldo/keto reductase family proteins | phase I | X | ||||
| K09A11.2 | cyp-14A1 | Cytochrome P450 CYP2 subfamily | phase I | X | X | ||
| K09A11.3 | cyp-14A2 | Cytochrome P450 family 14A2 | phase I | X | |||
| C46H11.2 | Flavin-containing monooxygenase | phase I | X | ||||
| F17A9.4 | NADH:flavin oxidoreductase | phase I | X | ||||
| F56D5.3 | NADH:flavin oxidoreductase | phase I | X | ||||
| C55A6.5 | sdz-8 | Predicted short chain-type dehydrogenase | phase I | X | X | ||
| C55A6.6 | Predicted short chain-type dehydrogenase | phase I | X | X | X | ||
| C55A6.7 | Predicted short chain-type dehydrogenase | phase I | X | ||||
| F20G2.1 | Predicted short chain-type dehydrogenase | phase I | X | X | |||
| F20G2.2 | Predicted short chain-type dehydrogenase | phase I | X | X | |||
| F55E10.6 | Predicted short chain-type dehydrogenase | phase I | X | ||||
| K10H10.3 | dhs-8 | Predicted short chain-type dehydrogenase | phase I | X | X | ||
| R08H2.1 | dhs-23 | Predicted short chain-type dehydrogenase | phase I | X | X | ||
| F25D1.5 | Reductases with broad range of substrate specificities | phase I | X | ||||
| B0222.9 | xanthine dehydrogenase | phase I | X | ||||
| F37B12.2 | gcs-1 | Gamma-glutamylcysteine synthetase | phase II | X | |||
| C53D5.5 | Gamma-glutamyltransferase | phase II | X | ||||
| E01A2.1 | Glutamate-cysteine ligase regulatory subunit | phase II | X | ||||
| Y34D9A.6 | glrx-10 | Glutaredoxin and related proteins | phase II | X | X | X | |
| C02D5.3 | gsto-2 | Glutathione S-transferase | phase II | X | X | X | |
| D1053.1 | gst-42 | Glutathione S-transferase | phase II | X | |||
| F11G11.1 | gst-8 | Glutathione S-transferase | phase II | X | X | X | |
| F11G11.2 | gst-7 | Glutathione S-transferase | phase II | X | X | X | |
| F11G11.3 | gst-6 | Glutathione S-transferase | phase II | X | X | ||
| F35E8.8 | gst-38 | Glutathione S-transferase | phase II | X | X | X | |
| F37B1.2 | gst-12 | Glutathione S-transferase | phase II | X | X | X | |
| F37B1.3 | gst-14 | Glutathione S-transferase | phase II | X | X | X | |
| F56A4.4 | Glutathione S-transferase | phase II | X | X | |||
| K08F4.7 | gst-4 | Glutathione S-transferase | phase II | X | X | X | X |
| R03D7.6 | gst-5 | Glutathione S-transferase | phase II | X | X | ||
| R07B1.4 | gst-36 | Glutathione S-transferase | phase II | X | |||
| R107.7 | gst-1 | Glutathione S-transferase | phase II | X | X | ||
| T26C5.1 | gst-13 | Glutathione S-transferase | phase II | X | X | X | |
| Y1H11.2 | gst-35 | Glutathione S-transferase | phase II | X | X | ||
| Y45G12C.2 | gst-10 | Glutathione S-transferase | phase II | X | X | X | |
| Y53F4B.33 | gst-39 | Glutathione S-transferase | phase II | X | X | ||
| Y53F4B.37 | gst-32 | Glutathione S-transferase | phase II | X | |||
| Y53G8B.1 | Glutathione S-transferase | phase II | X | ||||
| F35E8.11 | cdr-1 | glutathione S-transferase-like protein | phase II | X | X | ||
| M176.2 | Glutathione synthetase | phase II | X | ||||
| F25B4.8 | Glutathione-dependent formaldehyde-activating enzyme | phase II | X | ||||
| Y38F2AR.12 | Hydantoinase B/oxoprolinase | phase II | X | ||||
| C46F11.2 | Orthologous to human mitochondrial glutathione reductase | phase II | X | ||||
| AC3.7 | ugt-1 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| B0310.5 | ugt-46 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| C08F11.8 | ugt-22 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| C17G1.3 | ugt-23 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| C18C4.3 | ugt-48 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | X | ||
| C35A5.2 | ugt-33 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| F35H8.6 | ugt-58 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| K04A8.10 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | ||||
| Y39G10AR.6 | ugt-31 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| ZC443.6 | ugt-16 | UDP-glucuronosyl and UDP-glucosyl transferase | phase II | X | |||
| W09D6.6 | hmt-1 | Heavy metal exporter HMT1, ABC superfamily | phase III | X | |||
| T02D1.5 | pmp-4 | Peroxisomal long-chain acyl-CoA transporter, ABC superfamily | phase III | X |
WormBase sequence and gene (when availible) names are listed. Genes are from Park et al., 2009 (hyperoxia) and Olivera et al., 2009 (control, arsenic, and tBOOH) using the authors' statistical stringencies. Genes in bold were identified as upregulated by albendazole in a transcriptomic analysis (Laing et al., 2010) and genes in bold and underlined were identified as upregulated in a candidate gene study in ivermectin resistance stains (James and Davey 2008). tBOOH = tert-butyl hydroxide
Data are sparse, but two studies have provided evidence that SKN-1 target genes may play a role in anthelminthic resistance in C. elegans. gcs-1, a well-studied target of SKN-1 that encodes the rate-limiting enzyme for glutathione synthesis (gamma-glutamylcysteine synthetase, GCS), was four-fold over-expressed in multidrug resistant strains of C. elegans and pharmacological inhibition of GCS with buthionine sulfoxamine reversed resistance (James and Davey 2008). Five of the 16 detoxification genes upregulated by albendazole are known to be regulated by SKN-1 (Laing et al. 2010). Taken together, the data available to date indicate that SKN-1 is a master regulator of multiple detoxification and anti-oxidation genes, some of which have been shown to be responsive to long-term selection or short-term exposure to anthelmintics.
SKN-1 is essential for embryonic development
Interestingly, skn-1 was originally identified in forward genetic screens for C. elegans mutants with defective embryonic development (Bowerman et al. 1992). Loss-of-function skn-1 mutant embryos do not develop intestinal or pharyngeal cells, and thus do not survive. SKN-1 is maternally transcribed but zygotically translated to directly induce the expression of two paralogous GATA-factor-related zinc-finger proteins MED-1/2 in four-cell stage embryos to specify cell fates for development of the posterior pharynx and intestine. In skn-1 mutants (skin in excess), blastomeres that usually form the pharynx and intestine instead differentiate into excess body muscle and hypodermis (or informally skin). Therefore, targeting SKN-1 has the potential to inhibit development in embryos in addition to inhibiting xenobiotic detoxification and multidrug resistance in larvae and adults.
SKN-1 STRUCTURE AND REGULATION ARE DISTINCT FROM NRF2
As described above, inhibition of host detoxification pathways can have dire consequences. Comparisons between CNC regulation and function in mammals and helminths can identify potential modes of action for pharmacological inhibitors that are specific to parasites. Studies to date have identified three distinctions between regulation and function of SKN-1 in C. elegans and NRF2 in mammals: 1) DNA binding domains, 2) mechanisms of transcriptional activation, and 3) regulation of protein levels. We discuss these here and present evidence that many nematodes share these distinctions.
SKN-1 structure is unique in nematodes
Despite being in the basic region leucine zipper (bZIP) family of DNA binding transcriptional regulators and sharing a conserved core basic region with other CNCs, SKN-1 is unique because it lacks a leucine zipper (Blackwell et al. 1994). Other known bZIP proteins have leucine-rich α-helix regions that mediate protein dimerization and stabilize core basic amino acid regions so that they can bind to two adjacent major grooves in DNA. The interaction between SKN-1 and its target DNA bases [(A/T)(A/T)T(G/A)TCAT] was studied in detail with biochemical and crystal structure analyses (Blackwell et al. 1994; Carroll et al. 1997; Pal et al. 1997; Rupert et al. 1998; Kophengnavong et al. 1999). These studies found that SKN-1 forms a unique monomeric DNA binding motif that uses a second and novel basic (positive) amino acid region that stabilizes DNA binding by interacting with the negative phosphate backbone (Rupert et al. 1998) and contributes to target DNA sequence specificity (Kophengnavong et al. 1999). Relative to human NRF2, C. elegans SKN-1 is also missing a region at its carboxyl terminus named Neh3, which contributes transcriptional activity by interacting with another DNA binding protein named CHD6 (Nioi et al. 2005).
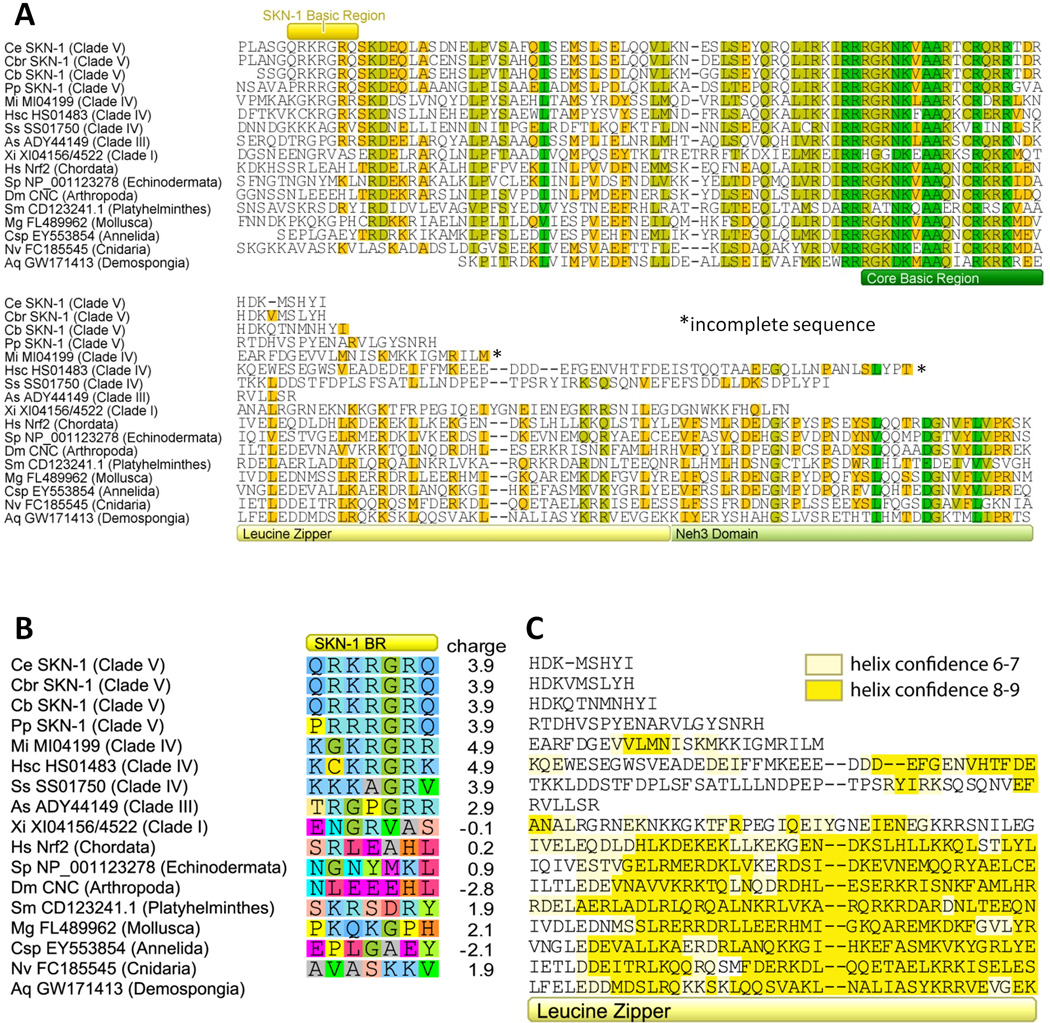
To determine if the novel SKN-1 basic region and loss of the leucine zipper and Neh3 features are shared by SKN-1 in other species, we performed BLAST searches using C. elegans SKN-1 and human NRF2 as the query protein sequences. Five major clades have been defined in the nematode phylum (I-V), with clades I and II the most basal and IV and V the most derived (Blaxter 1998; Blaxter et al. 1998). We compiled predicted SKN-1/NRF2 homologs from one species in clade I, one in clade III, three in clade IV, four in clade V, and eight other species representing diverse animal phyla. A protein alignment of the carboxyl-terminal regions of these predicted proteins is shown in Figure 1A (see the Supplemental Information for details of the alignment method). The core basic region, which interacts with the major grove in target DNA (Rupert et al. 1998), is found in all species. Conversely, nematode clades III, IV, and V share a unique upstream feature that corresponds to the second basic region originally identified in SKN-1 of C. elegans (Blackwell et al. 1994; Rupert et al. 1998). Figure 1B highlights amino acids that align with the C. elegans SKN-1 basic region by functional properties and lists the predicted charge for this region from each protein at pH 7.0. This short element carries a net charge of 2.9 to 4.9 at pH 7.0 in nematode clades III-V, but a charge of −2.8 to 2.1 in all other groups including Xiphinema index (plant pathogen vector) in nematode clade I. All nematode proteins from clades III-V also have a glycine at position 4 that contributes to DNA binding (Kophengnavong et al. 1999). As clades III-V form their own lineage within Nematoda (Blaxter 1998), this SKN-1 basic region most likely originated in their common ancestor.
Figure 1.
Alignment of carboxyl-terminal regions of SKN-1/NRF2 homologs from multiple nematodes and diverse animal phyla (A). Proteins are listed by species abbreviations, protein names or accession numbers, and phylum or nematode clade in parentheses. The alignment is color coded by degree of conservation. (B) Alignment of the SKN-1 basic region color coded by amino acid properties and listing the charge of the region. Species are Ce = C. elegans, Cb = C. briggsae, Cbr = C. brenneri, Pp = Pristionchus pacificus, Mi = Meloidogyne incognita, Hsc = Heterodera schachtii, Ss = Strongyloides stercoralis, As = Ascaris suum, Xi = Xiphinema index, Hs = Homo sapiens, Sp = Strongylocentrotus purpuratus, Dm = Drosophila melanogaster, Sm = Schistosoma mansoni, Mg = Mytilus galloprovincialis, Csp = Capitella sp., Nv = Nematostella vectensis, and Aq = Amphimedon queenslandica, Sequences are from WormBase (Ce, Cb, Cbr, and Pp), Nematode.net (Xi, Mi, Hsc, and Ss), or National Center for Biotechnology Information (all others). (C) Alignment of the leucine zipper color coded according to the confidence of an α-helix (“9” being highest). See Supplementary Information for details on the charge calculation and secondary structure predictions.
In turn, all predicted proteins from non-nematode species share a downstream leucine zipper and Neh3 domain (Figure 1A). Secondary structure prediction (PSIPRED) recovers an α-helix in human NRF2, which corresponds to its predicted leucine zipper (Moi et al. 1994), providing an important positive control for our other secondary structure predictions. A well-defined α-helix is also predicted in all proteins from other animal phyla, except Nematoda. The presence of this α-helix/leucine zipper includes the phyla Porifera (sponge) and Cnidaria (sea anemone) that are well accepted as basal within Metazoa (Halanych 2004; Ruiz-Trillo et al. 2008). Thus, this structural element extends at least as far back as to all animals. Conversely, the absence of this α-helix/leucine zipper includes clade I of nematodes, which is basally related to clades III-V (Blaxter 1998). Thus, this structural element was most likely lost within the nematode common ancestor.
The C-termini of all SKN-1 proteins from nematodes (except for Meloidogyne and Heterodera) are either known or inferred to be complete on the basis of an in-frame stop codon. Conversely, the completeness of the C-termini for Meloidogyne and Heterodera SKN-1 remain less clear as they lack stop codons. Nevertheless, this uncertainty about C-terminal completeness is regarded as of minor concern to our conclusion of an absent leucine zipper, because only the Heterodera sequence ends short of its full alignment to this domain (Figure 1A). The absence of Neh3 is less certain, because both of these sequences end before their full alignment to this domain.
These patterns suggest that the SKN-1 basic region could contribute to DNA binding in nematodes of clades III–V. Evolution of this region may have permitted, or compensated for, the loss or divergence of the leucine zipper. Loss of Neh3 by at least some nematodes suggests that SKN-1 may activate the transcription of target genes differently than in other animals including mammals (Nioi et al. 2005). Sequence data for SKN-1/NRF2 from more species within basal nematode clades and other Ecdysozoa (the superfamily that includes nematodes, arthropods, and several smaller phyla) are needed to refine the phylogenetic origin of these changes, and functional studies are needed to verify the role of the SKN-1 basic region in nematodes other than C. elegans.
SKN-1 regulation is distinct from NRF2 in mammals
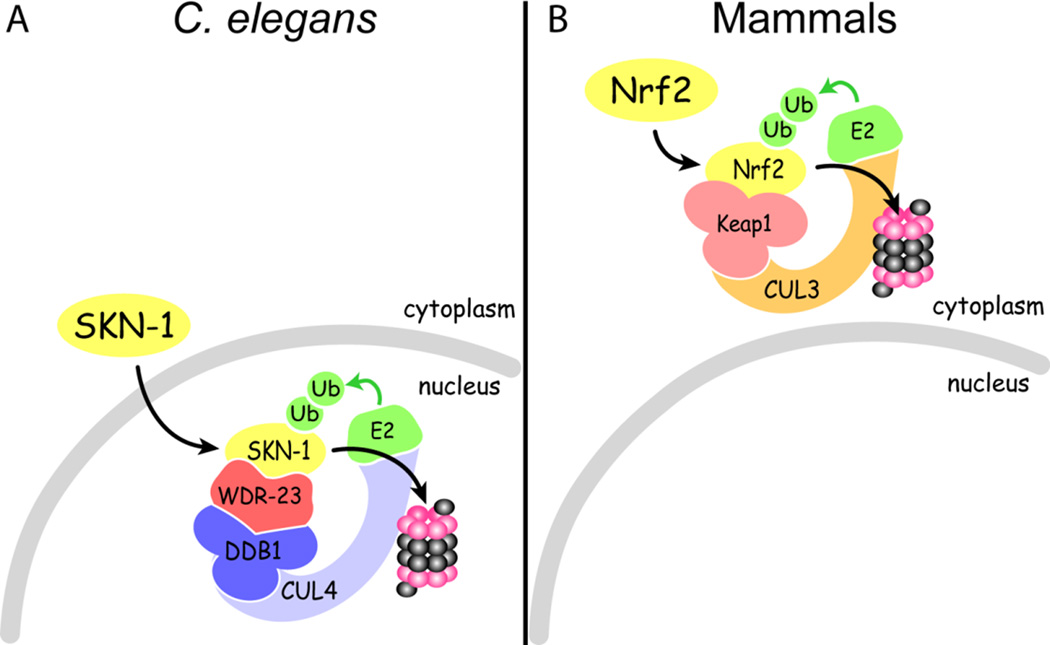
SKN-1 activity is low under basal conditions but is highly induced by exposure to xenobiotics and oxidants. To identify regulators of SKN-1, we recently performed a genome-wide RNAi screen for genes that repress transcription of the SKN-1 target gene gst-4. Proteasome subunits, a ubiquitin ligase, and a WD40 repeat protein named wdr-23 were among the genes identified (Choe et al. 2009). Subsequent genetic and biochemical studies support a model (Figure 2A) in which WDR-23 recruits SKN-1 to the CUL4/DDB1 ubiquitin ligase in the cell nucleus and targets the transcription factor for degradation by the proteasome. The principal role of WDR-23 in repressing SKN-1 was independently confirmed by a forward genetic screen (Hasegawa and Miwa 2010). Although the physiological function of NRF2 is very similar to SKN-1, it is regulated by a distinct ubiquitin ligase. In the cytoplasm of mammalian cells, NRF2 is recruited to the CUL3 ubiquitin ligase by a kelch repeat protein named KEAP1 (McMahon et al. 2003; McMahon et al. 2004; Kobayashi et al. 2006; Kobayashi et al. 2008; McMahon et al. 2010; Zhang 2010) (Figure 2B). A protein orthologous to KEAP1 (see our phylogenetic results below) also represses the Drosophila CNC indicating that this pathway is conserved (Sykiotis and Bohmann 2008). Although WD40 repeat and kelch repeat proteins both fold into β-propellers, they share no sequence homology (Chaudhuri et al. 2008; Hudson and Cooley 2008). These ubiquitin ligases directly repress their respective CNC proteins and exposure to oxidants or electrophiles is thought to activate the transcription factors by releasing them from repression (Choe et al. 2009; Giudice et al. 2010; Zhang 2010; Keum 2011).
Figure 2.
SKN-1 in C. elegans and NRF2 in mammals are regulated by distinct ubiquitin ligases. CUL3 and CUL4 are distinct ubiquitin ligases. WDR-23 and KEAP1 are distinct proteins that are thought to function in different cellular compartments (nucleus versus cytosol). Both pathways are thought to label their target with ubiquitin for proteasomal degradation.
Although the precise mechanisms of NRF2 activation by oxidants and electrophiles are still being defined and appear to vary by the chemical nature of the inducer, there are some well accepted models. In general, activation is thought to occur by direct modifications of KEAP1 or phosphorylation NRF2 that in turn promote stabilization of NRF2 (Kaspar et al. 2009). KEAP1 contains multiple redox-reactive cysteines that are directly modified by NRF2-activating electrophiles or endogenous stress-related chemicals (Holtzclaw et al. 2004; Kobayashi et al. 2009; Giudice et al. 2010; McMahon et al. 2010). Modification of KEAP1 stabilizes NRF2 either by preventing interaction (Dinkova-Kostova et al. 2002; Wakabayashi et al. 2004) or causing ubiquitinylation and degradation of KEAP1 (Eggler et al. 2005; Hong et al. 2005; Zhang et al. 2005).
A growing body of evidence indicates that multiple protein kinases also contribute to regulation of NRF2 abundance and activity (Giudice et al. 2010; Keum 2011). These include protein kinase C (Huang et al. 2002; Bloom and Jaiswal 2003), mitogen-activated protein kinases (MAPK) (Zipper and Mulcahy 2000; Buckley et al. 2003), PKR-like endoplasmic reticulum kinase (PERK) (Cullinan et al. 2004), and glycogen synthase kinase 3 (Salazar et al. 2006; Rojo et al. 2008; Rada et al. 2011). The molecular mechanisms linking all these kinases to NRF2 activation are still being defined, but some have been reported to modify interactions with KEAP1, modulate degradation by additional degradation pathways, or regulate nuclear export of the transcription factor (Huang et al. 2002; Bloom and Jaiswal 2003; Kaspar et al. 2009; Giudice et al. 2010; Keum 2011; Rada et al. 2011). Phosphorylation of a KEAP1 tyrosine residue has also been shown to play a role in NRF2 regulation (Jain et al. 2008).
Protein kinase pathways also regulate SKN-1 during stress. p38 MAPK (PMK-1) and extracellular regulated kinase (MPK-1) pathways directly phosphorylate SKN-1 at residues (S74 and S340) and promote nuclear accumulation and activation (Inoue et al. 2005; Okuyama et al. 2010). Four other kinases have been implicated in regulation of SKN-1 during stress, but it is not known if they phosphorylate the factor directly (Kell et al. 2007). GSK-3 and insulin-like receptor regulated kinases (SGK-1, AKT-1/2) inhibit SKN-1 nuclear accumulation via direct phosphorylation (An et al. 2005; Tullet et al. 2008). Using genetics, we provided evidence that WDR-23 acts downstream from at least three of these protein kinase pathways to regulate SKN-1, suggesting that phosphorylation may modify regulation by WDR-23 (Choe et al. 2009). Like KEAP1, WDR-23 has several cysteine residues (17). It is not yet known if electrophiles modify these cysteines and contribute to SKN-1 activation.
Given the importance of ubiquitin ligases to regulation of CNC proteins, it will be important to determine if WDR-23 or KEAP1 regulates SKN-1 in nematodes other than just C. elegans. WDR-23 is an evolutionarily ancient protein that is present in plants, fungi, and animals including probably all nematodes (Choe et al. 2009). The human and C. elegans WDR-23 orthologs interact with DDB1 via a specific ‘DWD’ domain (Angers et al. 2006; Jin et al. 2006; Lee and Zhou 2007; Choe et al. 2009) that is also conserved in plant WDR-23 orthologs (Lee et al. 2008) suggesting that the ubiquitin ligase function of the protein may be conserved in all eukaryotes. Alternatively, the CNC protein family is not present in plants and WDR23 does not appear to regulate or interact with NRF2 in human cells (Choe et al., unpublished observations) suggesting that WDR-23 has switched substrates during evolution.
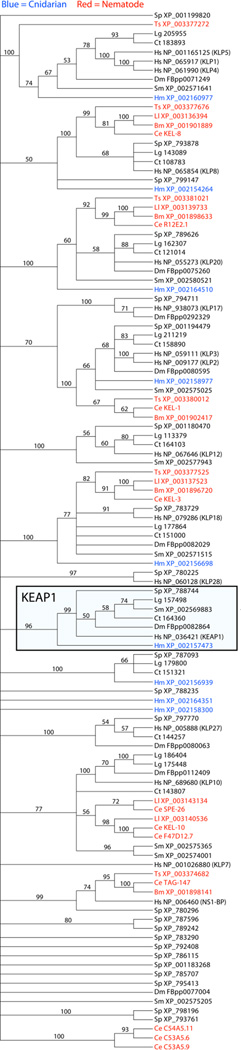
The kelch repeat protein family is found in bacteria, plants, fungi, and animals (Prag and Adams 2003). Surprisingly, a putative KEAP1 ortholog has not been described in C. elegans, raising the critical question of whether KEAP1 exists in nematodes and other related protostomes. Therefore, we performed maximum likelihood and Bayesian phylogenetic analyses of protein sequences found by BLAST searching with human KEAP1 in representatives of nematode clades I, III, and V and diverse animal phyla (Figures 3 and S1, see Supplementary Information for details of the analyses). The final alignment of kelch repeat proteins covers 988 amino acids in 114 different protein sequences from four nematodes [Trichenella spiralis (tissue-dwelling parasite of humans), Loa loa (filarial parasite of humans), B. malayi, and C. elegans)], six other invertebrate phyla, and human. The bootstrap maximum likelihood tree, which is more conservative than the Bayesian tree in terms of its group support, provides clear evidence for the recognition of a well-defined subfamily that includes human KEAP1 (Figure 3). Human KEAP1 first strongly groups (99% bootstrap score) with a single sequence from each of the major bilaterian phyla of Annelida, Echinodermata, Mollusca, and Platyhelminths and includes the previously described KEAP1-like protein from Drosophila (Sykiotis and Bohmann 2008). This group then joins (96% score) with a single sequence from Cnidaria. As Cnidaria is phylogenetically basal within Eumetazoa (Halanych 2004; Dunn et al. 2008), these strong bootstrap results allow for the recognition of a KEAP1 subfamily that spans all multicellular animals more recent than sponges and some other less-known groups. The Bayesian phylogeny provides further support for the same KEAP1 subfamily (100% posterior probability), but with better resolution of relationships within the subfamily (Figure S1).
Figure 3.
Majority-rule consensus tree summarizing the 1,000 phylogenies from bootstrap maximum likelihood analysis. Bootstrap scores are presented next to those internal branches with >50% support. Each sequence name consists of a two-letter abbreviation for its genus/species followed by its database accession number in FlyBase (Dm), Joint Genome Institute (Ct and Lg), WormBase (Ce), and National Center for Biotechnology Information (all others). The names for the human sequences also include specific kelch protein designations (in parentheses) as recognized in the literature. Nematode and hydra sequences are highlighted in red and blue, respectively. The box highlights those sequences that are inferred as orthologs of the KEAP1 subfamily according to these results. Abbreviations are Hm - Hydra magnipapillata (Cnidaria), Sm - Schistosoma mansoni (Platyhelminths), Ts - Trichinella spiralis (Nematoda, clade I), Bm - Brugia malayi (Nematoda, clade III), Ll - Loa loa (Nematoda, clade III), Ce – C. elegans (Nematoda, clade V), Lg - Lottia gigantean (Mollusca), Ct - Capitella teleta (Annelida), Dm - Drosophila melanogaster (Arthropoda), Sp - Strongylocentrotus purpuratus (Echinodermata), and Hs - Homo sapiens (Chordata).
Nematodes are well accepted as members of the Ecdyzoa protostome clade (Halanych 2004; Giribet et al. 2009). Therefore, it is surprising that there are no nematode sequences that join with Drosophila (arthropod) KEAP1. Indeed, no nematode sequence is found anywhere within the entire KEAP1 subfamily (Figures 3 and S1). Thus, our trees strongly agree that the Kelch repeat proteins of the four represented nematodes are all paralogs of KEAP1 and that none are orthologs. By definition, orthologs are more closely related to each other than are paralogs (Goodman et al. 1979). Thus, our successful recovery of more distantly related paralogs from the genomic databases of these nematodes serves as a critical positive control that these four species are indeed missing a KEAP1 ortholog (Hyndman et al. 2009). Given that our four species of nematodes belong to distantly related clades of Nematoda, I, III, and V (Blaxter et al. 1998), the lack of a KEAP1 ortholog may be phylum wide. If so, the absence of KEAP1 would be attributed to the loss and/or silencing of the gene in the nematode common ancestor or in an even older ancestor that nematodes share with other ecdyzoan phyla. Gene loss has been shown to be particularly high for nematodes (Mitreva et al. 2011). Further work is needed to determine if WDR-23 functionally compensates for the loss of KEAP1 in nematodes other than C. elegans.
SUMMARY
Drug resistance is a major obstacle to control of parasitic nematodes and mechanisms of resistance are poorly understood in this group of helminths. In diverse organisms, multidrug resistance is often associated with or caused by changes to xenobiotic detoxification mechanisms. Transcription factors such as SKN-1 regulate multiple detoxification genes and are promising but largely unexplored candidates for understanding and reversing resistance. SKN-1 is also essential for embryonic development. Despite very similar functions, many aspects of SKN-1 structure and regulation in C. elegans are distinct from NRF2 in mammals. Comparative analyses predict that SKN-1 structure and regulation could be similar among many nematodes, but unique relative to other animal phyla (see Table 3 for a summary) providing promising targets for specificity. Given that many parasitic nematodes have limited genetic tractability, the development of small molecule modulators of SKN-1 would provide useful tools for studying the function of this transcriptional pathway. Transgenic C. elegans reporter strains for SKN-1 activity have been developed and perform well in whole-animal high-throughput assays making screening for small-molecule inhibitors feasible (Leung et al. 2011).
Table 3.
Summary of evidence for the presence or absence of SKN-1/NRF2 domains, KEAP1, and WDR-23
| Taxononmic group | LeuZ | SKN-1 BR | Neh3 | KEAP1 | WDR-23 |
|---|---|---|---|---|---|
| Demospongia | + | ? | + | ? | + |
| Cnidaria | + | - | + | + | + |
| Platyhelminthes | + | - | + | + | + |
| Annelida | + | - | + | + | + |
| Mollusca | + | - | + | + | + |
| Nematoda Clade I | - | - | - | - | + |
| Nematoda Clade II | ? | ? | ? | ? | ? |
| Nematoda Clade III | - | + | - | - | + |
| Nematoda Clade IV | - | + | ? | ? | + |
| Nematoda Clade V | - | + | - | - | + |
| Arthropoda | + | - | + | + | + |
| Echinodermata | + | - | + | + | + |
| Chordata | + | - | + | + | + |
+ = present, - = absent , ? = not tested or tested but conclusion is not clear, LeuZ = leucine zipper, and BR = basic region.
Important questions concerning SKN-1 function and regulation include:
Are mutations in wdr-23 or skn-1 associated with multidrug resistance in parasitic nematodes?
What genes does SKN-1 regulate in parasitic species and are these associated with multidrug resistance?
Does WDR-23 regulate SKN-1 in parasitic nematodes?
Is SKN-1 activated by any of the common anthelmintics?
Does SKN-1 defend parasites from host-immune responses?
Is the function of SKN-1 in embryonic development conserved in other nematodes?
How do WDR-23 and SKN-1 interact in C. elegans and how conserved are the interaction motifs among other nematodes?
Supplementary Material
Acknowledgments
Writing of this paper was supported by NSF grant IOS-1120130 to KPC and NIH grant R21NS067678-01.
Footnotes
DECLARATION OF INTEREST
The authors report no declarations of interest.
REFERENCES
- Aarnio V, Storvik M, et al. Fatty acid composition and gene expression profiles are altered in aryl hydrocarbon receptor-1 mutant Caenorhabditis elegans. Comp Biochem Physiol C Toxicol Pharmacol. 2010;151(3):318–324. doi: 10.1016/j.cbpc.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Albonico M, Allen H, et al. Controlling soil-transmitted helminthiasis in pre-school-age children through preventive chemotherapy. PLoS Negl Trop Dis. 2008;2(3):e126. doi: 10.1371/journal.pntd.0000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albonico M, Engels D, et al. Monitoring drug efficacy and early detection of drug resistance in human soil-transmitted nematodes: a pressing public health agenda for helminth control. Int J Parasitol. 2004;34(11):1205–1210. doi: 10.1016/j.ijpara.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Alvarez AI, Merino G, et al. Role of ABC transporters in veterinary drug research and parasite resistance. Curr Drug Deliv. 2006;3(2):199–206. doi: 10.2174/156720106776359195. [DOI] [PubMed] [Google Scholar]
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17(15):1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Vranas K, et al. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. PNAS. 2005;102(45):16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Li T, et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Ardelli BF, Stitt LE, et al. Inventory and analysis of ATP-binding cassette (ABC) systems in Brugia malayi. Parasitol. 2010;137(8):1195–1212. doi: 10.1017/S0031182010000120. [DOI] [PubMed] [Google Scholar]
- Awadzi K, Boakye DA, et al. An investigation of persistent microfilaridermias despite multiple treatments with ivermectin, in two onchocerciasis-endemic foci in Ghana. Ann Trop Med Parasitol. 2004;98(3):231–249. doi: 10.1179/000349804225003253. [DOI] [PubMed] [Google Scholar]
- Bartley DJ, McAllister H, et al. P-glycoprotein interfering agents potentiate ivermectin susceptibility in ivermectin sensitive and resistant isolates of Teladorsagia circumcincta and Haemonchus contortus. Parasitol. 2009;136(9):1081–1088. doi: 10.1017/S0031182009990345. [DOI] [PubMed] [Google Scholar]
- Bethony J, Brooker S, et al. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet. 2006;367(9521):1521–1532. doi: 10.1016/S0140-6736(06)68653-4. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, et al. Formation of a monomeric DNA binding domain by SKN-1 bZIP and homeodomain elements. Science. 1994;266(5185):621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- Blaxter M. Caenorhabditis elegans is a nematode. Science. 1998;282(5396):2041–2046. doi: 10.1126/science.282.5396.2041. [DOI] [PubMed] [Google Scholar]
- Blaxter ML, De Ley P, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392(6671):71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem. 2003;278(45):44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- Bockarie MJ, Deb RM. Elimination of lymphatic filariasis: do we have the drugs to complete the job? Curr Opin Infect Dis. 2010;23(6):617–620. doi: 10.1097/QCO.0b013e32833fdee5. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, et al. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68(6):1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Buckley BJ, Marshall ZM, et al. Nitric oxide stimulates Nrf2 nuclear translocation in vascular endothelium. Biochem Biophys Res Commun. 2003;307(4):973–979. doi: 10.1016/s0006-291x(03)01308-1. [DOI] [PubMed] [Google Scholar]
- Carroll AS, Gilbert DE, et al. SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes Dev. 1997;11(17):2227–2238. doi: 10.1101/gad.11.17.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. The global burden of intestinal nematode infections - Fifty years on. Parasitol Today. 1997;13(11):438–443. doi: 10.1016/s0169-4758(97)01144-7. [DOI] [PubMed] [Google Scholar]
- Chaudhuri I, Soding J, et al. Evolution of the beta-propeller fold. Proteins. 2008;71(2):795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- Choe KP, Przybysz AJ, et al. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009;29(10):2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, et al. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvilink V, Lamka J, et al. Xenobiotic metabolizing enzymes and metabolism of anthelminthics in helminths. Drug Metab Rev. 2009;41(1):8–26. doi: 10.1080/03602530802602880. [DOI] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 2008;40(10):1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. PNAS. 2002;99(18):11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C, Hejnol A, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Eggler AL, Liu GW, et al. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. PNAS. 2005;102(29):10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts S, Gryseels B. Drug resistance in human helminths: current situation and lessons from livestock. Clin Microbiol Rev. 2000;13(2):207–222. doi: 10.1128/cmr.13.2.207-222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts S, Gryseels B. Anthelmintic resistance in human helminths: a review. Trop Med Int Health. 2001;6(11):915–921. doi: 10.1046/j.1365-3156.2001.00774.x. [DOI] [PubMed] [Google Scholar]
- Ghedin E, Wang S, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317(5845):1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS. Understanding anthelmintic resistance: The need for genomics and genetics. Int J Parasitol. 2006;36(12):1227–1239. doi: 10.1016/j.ijpara.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Giribet G, Dunn C, et al. In: Assembling the spiralian tree of life. Animal evolution: Genomes, fossils, and trees. Telford M, Littlewood D, et al., editors. Oxford, UK: Oxford Univ. Press; 2009. pp. 52–64. [Google Scholar]
- Giudice A, Arra C, et al. Review of molecular mechanisms involved in the activation of the Nrf2-ARE signaling pathway by chemopreventive agents. Methods Mol Biol. 2010;647:37–74. doi: 10.1007/978-1-60761-738-9_3. [DOI] [PubMed] [Google Scholar]
- Goodman M, Czelusniak J, et al. Fitting the gene lineage into its species lineage. A parsimony strategy illustrated by cladograms constructed from globin sequences. Syst Zool. 1979;28:132–163. [Google Scholar]
- Gustavsen KM, Bradley MH, et al. GlaxoSmithKline and Merck: private-sector collaboration for the elimination of lymphatic filariasis. Ann Trop Med Parasitol. 2009;103(Suppl 1):S11–S15. doi: 10.1179/000349809X12502035776478. [DOI] [PubMed] [Google Scholar]
- Halanych K. The new view of animal phylogeny. Ann Rev Ecol Evol Systematics. 2004;35:229–256. [Google Scholar]
- Hasegawa K, Miwa J. Genetic and cellular characterization of Caenorhabditis elegans mutants abnormal in the regulation of many phase II enzymes. PLoS ONE. 2010;5(6):e11194. doi: 10.1371/journal.pone.0011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Miwa S, et al. Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol Sci. 2008;101(2):215–225. doi: 10.1093/toxsci/kfm276. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34(4):176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M, et al. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13(11):1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- Herrmann M, Mayer WE, et al. The nematode Pristionchus pacificus (Nematoda: Diplogastridae) is associated with the oriental beetle Exomala orientalis (Coleoptera: Scarabaeidae) in Japan. Zoolog Sci. 2007;24(9):883–889. doi: 10.2108/zsj.24.883. [DOI] [PubMed] [Google Scholar]
- Holden-Dye L, Walker RJ. Anthelmintic drugs. WormBook. T. C. e. R. Community, WormBook. 2007 doi: 10.1895/wormbook.1.143.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzclaw WD, Dinkova-Kostova AT, et al. Protection against electrophile and oxidative stress by induction of phase 2 genes: the quest for the elusive sensor that responds to inducers. Adv Enz Reg. 2004;44(1):335–367. doi: 10.1016/j.advenzreg.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Hong F, Sekhar KR, et al. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280(36):31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- Hooper PJ, Bradley MH, et al. The global programme to eliminate lymphatic filariasis: health impact during its first 8 years (2000–2007) Ann Trop Med Parasitol. 2009;103(Suppl 1):S17–S21. doi: 10.1179/000349809X12502035776513. [DOI] [PubMed] [Google Scholar]
- Hotez P. Neglected diseases amid wealth in the United States and Europe. Health Aff (Millwood) 2009;28(6):1720–1725. doi: 10.1377/hlthaff.28.6.1720. [DOI] [PubMed] [Google Scholar]
- Hotez PJ, Brindley PJ, et al. Helminth infections: the great neglected tropical diseases. J Clin Invest. 2008;118(4):1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Xu C, et al. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006;79(20):1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Huang HC, Nguyen T, et al. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277(45):42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- Huang X, Powell-Coffman JA, et al. The AHR–1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Dev. 2004;131(4):819–828. doi: 10.1242/dev.00959. [DOI] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell Biochem. 2008;48:6–19. doi: 10.1007/978-0-387-09595-0_2. [DOI] [PubMed] [Google Scholar]
- Hyndman KA, Miyamoto MM, et al. Phylogeny, taxonomy, and evolution of the endothelin receptor gene family. Mol Phylogenet Evol. 2009;52(3):677–687. doi: 10.1016/j.ympev.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19(19):2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyanagi T. Molecular mechanism of phase I and phase II drug-metabolizing enzymes: Implications for detoxification. Int Rev Cyto. 2007;260:35–112. doi: 10.1016/S0074-7696(06)60002-8. [DOI] [PubMed] [Google Scholar]
- Jain AK, Mahajan S, et al. Phosphorylation and dephosphorylation of tyrosine 141 regulate stability and degradation of INrf2: a novel mechanism in Nrf2 activation. J Biol Chem. 2008;283(25):17712–17720. doi: 10.1074/jbc.M709854200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- James CE, Davey MW. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int J Parasitol. 2008;39(2):213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- James CE, Hudson AL, et al. Drug resistance mechanisms in helminths: is it survival of the fittest? Trends Parasitol. 2009;25(7):328–335. doi: 10.1016/j.pt.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Jasmer DP, Goverse A, et al. Parasitic nematode interactions with mammals and plants. Ann Rev Phyto. 2003;41(1):245–270. doi: 10.1146/annurev.phyto.41.052102.104023. [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, et al. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23(5):709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Joachim A, Lautscham E, et al. Oesophagostomum dentatum: effect of glutathione S-transferase (GST) inhibitors on GST activity and larval development. Exp Parasitol. 2011;127(4):762–767. doi: 10.1016/j.exppara.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Joachim A, Ruttkowski B. Prostaglandin D(2) synthesis in Oesophagostomum dentatum is mediated by cytosolic glutathione S-transferase. Exp Parasitol. 2011;127(2):604–606. doi: 10.1016/j.exppara.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Kahn NW, Rea SL, et al. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409(1):205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- Kaminsky R, Ducray P, et al. A new class of anthelmintics effective against drug-resistant nematodes. Nature. 2008;452(7184):176–180. doi: 10.1038/nature06722. [DOI] [PubMed] [Google Scholar]
- Kaplan RM. Drug resistance in nematodes of veterinary importance: a status report. Trend Parasitol. 2004;20(10):477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kaplan RM, Vidyashankar AN. An inconvenient truth: Global worming and anthelmintic resistance. Vet Parasitol. 2011;186(1–2):70–78. doi: 10.1016/j.vetpar.2011.11.048. [DOI] [PubMed] [Google Scholar]
- Kaspar JW, Niture SK, et al. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47(9):1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser J, Utzinger J. Efficacy of current drugs against soil-transmitted helminth infections: systematic review and meta-analysis. JAMA. 2008;299(16):1937–1948. doi: 10.1001/jama.299.16.1937. [DOI] [PubMed] [Google Scholar]
- Kell A, Ventura N, et al. Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Rad Biol Med. 2007;43(11):1560–1566. doi: 10.1016/j.freeradbiomed.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31(1):90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerboeuf D, Blackhall W, et al. P-glycoprotein in helminths: function and perspectives for anthelmintic treatment and reversal of resistance. Int J Antimicrobial Agents. 2003;22(3):332–346. doi: 10.1016/s0924-8579(03)00221-8. [DOI] [PubMed] [Google Scholar]
- Keum YS. Regulation of the Keap1/Nrf2 system by chemopreventive sulforaphane: implications of posttranslational modifications. Ann N Y Acad Sci. 2011;1229:184–189. doi: 10.1111/j.1749-6632.2011.06092.x. [DOI] [PubMed] [Google Scholar]
- Kiontke KC, Felix MA, et al. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11(1):339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kang M-I, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26(1):221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Li L, et al. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2008;29(2):493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Li L, et al. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29(2):493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enz Reg. 2006;46(1):113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Kophengnavong T, Carroll AS, et al. The SKN-1 amino-terminal arm is a DNA specificity segment. Mol Cell Biol. 1999;19(4):3039–3050. doi: 10.1128/mcb.19.4.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwalek J, Rew R, et al. Glutathione-S-transferase, a possible drug metabolizing enzyme, in Haemonchus contortus: comparative activity of cambendazole-resistant and susceptible strain. Int J Parasitol. 1984;14:173–175. doi: 10.1016/0020-7519(84)90045-6. [DOI] [PubMed] [Google Scholar]
- Laing ST, Ivens A, et al. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem J. 2010;432(3):505–514. doi: 10.1042/BJ20101346. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Terzaghi W, et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20(1):152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26(6):775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Lespine A, Alvinerie M, et al. ABC transporter modulation: a strategy to enhance the activity of macrocyclic lactone anthelmintics. Trend Parasitol. 2008;24(7):293–298. doi: 10.1016/j.pt.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Leung CK, Deonarine A, et al. High-throughput screening and biosensing with fluorescent C. elegans strains. J Vis Exp. 2011;(51):e2745. doi: 10.3791/2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J Exp Zool A Comp Exp Biol. 2006;305(9):720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom TH, Pierce GJ, et al. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Cur Biol. 2001;11(11):864–868. doi: 10.1016/s0960-9822(01)00236-6. [DOI] [PubMed] [Google Scholar]
- Little PR, Hodges A, et al. Field efficacy and safety of an oral formulation of the novel combination anthelmintic, derquantel-abamectin, in sheep in New Zealand. N Z Vet J. 2010;58(3):121–129. doi: 10.1080/00480169.2010.67513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelski J, Konings WN, et al. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007;71(3):463–476. doi: 10.1128/MMBR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers CD, Ezzati M, et al. Measuring the burden of neglected tropical diseases: The global burden of disease framework. PLoS Negl Trop Dis. 2007;1(2):e114. doi: 10.1371/journal.pntd.0000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan LI, Wolf CR. Glutathione and glutathione-dependent enzymes in cancer drug resistance. Drug Resist Updat. 1999;2(3):153–164. doi: 10.1054/drup.1999.0083. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, et al. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- McMahon M, Lamont DJ, et al. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. PNAS. 2010;107(44):18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Thomas N, et al. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive neh2 degron and the redoxinsensitive neh6 degron. J Biol Chem. 2004;279(30):31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- Misra JR, Horner MA, et al. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011;25(17):1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M, Jasmer DP, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43(3):228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M, Zarlenga DS, et al. Parasitic nematodes--From genomes to control. Vet Parasitol. 2007;148(1):31–42. doi: 10.1016/j.vetpar.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, et al. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. PNAS. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molento MB, Prichard RK. Effects of the multidrug-resistance-reversing agents verapamil and CL 347,099 on the efficacy of ivermectin or moxidectin against unselected and drug-selected strains of Haemonchus contortus in jirds (Meriones unguiculatus) Parasitol Res. 1999;85(12):1007–1011. doi: 10.1007/s004360050673. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley, Scott W. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Progress Nucleic Acid Res Mol Biol. 2003;73:251–279. doi: 10.1016/s0079-6603(03)01008-0. [DOI] [PubMed] [Google Scholar]
- Nioi P, Nguyen T, et al. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. 2005;25(24):10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien ML, Tew KD. Glutathione and related enzymes in multidrug resistance. Eur J Cancer. 1996;32A(6):967–978. doi: 10.1016/0959-8049(96)00051-2. [DOI] [PubMed] [Google Scholar]
- Ohta T, Iijima K, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68(5):1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Okuyama T, Inoue H, et al. The ERK-MAPK pathway regulates longevity through SKN-1 and insulin-like signaling in Caenorhabditis elegans. J Biol Chem. 2010;285(39):30274–30281. doi: 10.1074/jbc.M110.146274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RP, Abate JP, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8(5):524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Atweneboana MY, Eng JK, et al. Prevalence and intensity of Onchocerca volvulus infection and efficacy of ivermectin in endemic communities in Ghana: a twophase epidemiological study. Lancet. 2007;369(9578):2021–2129. doi: 10.1016/S0140-6736(07)60942-8. [DOI] [PubMed] [Google Scholar]
- Pal S, Lo MC, et al. Skn-1: evidence for a bipartite recognition helix in DNA binding. PNAS. 1997;94(11):5556–5561. doi: 10.1073/pnas.94.11.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-K, Tedesco PM, et al. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8(3):258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi J-M, Gerbal-Chaloin S, et al. The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Ann Rev Pharm Toxicol. 2008;48(1):1–32. doi: 10.1146/annurev.pharmtox.47.120505.105349. [DOI] [PubMed] [Google Scholar]
- Perbandt M, Hoppner J, et al. Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus. J Mol Biol. 2008;377(2):501–511. doi: 10.1016/j.jmb.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Persidis A. Cancer multidrug resistance. Nat Biotech. 1999;17(1):94–95. doi: 10.1038/5289. [DOI] [PubMed] [Google Scholar]
- Prag S, Adams JC. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Kapoor K. Multidrug resistance in yeast Candida. Int Rev Cyto. 2004;242:215–248. doi: 10.1016/S0074-7696(04)42005-1. [DOI] [PubMed] [Google Scholar]
- Prasad R, Lata Panwar S, et al. Drug resistance in yeasts - an emerging scenario. Adv Mico Physiol. 2002;46:155–201. doi: 10.1016/s0065-2911(02)46004-3. [DOI] [PubMed] [Google Scholar]
- Prichard R. Anthelmintic resistance. Vet Parasitol. 1994;54(1–3):259–268. doi: 10.1016/0304-4017(94)90094-9. [DOI] [PubMed] [Google Scholar]
- Prichard RK, Geary TG. Drug discovery: Fresh hope to can the worms. Nature. 2008;452(7184):157–158. doi: 10.1038/452157a. [DOI] [PubMed] [Google Scholar]
- Prichard RK, Roulet A. ABC transporters and beta-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitol. 2007;134((Pt 8)):1123–1132. doi: 10.1017/S0031182007000091. [DOI] [PubMed] [Google Scholar]
- Przybysz AJ, Choe KP, et al. Increased age reduces DAF-16 and SKN-1 signaling and the hormetic response of Caenorhabditis elegans to the xenobiotic juglone. Mech Age Dev. 2009;130(6):357–369. doi: 10.1016/j.mad.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada P, Rojo AI, et al. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31(6):1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo AI, Sagarra MR, et al. GSK-3beta down-regulates the transcription factor Nrf2 after oxidant damage: relevance to exposure of neuronal cells to oxidative stress. J Neurochem. 2008;105(1):192–202. doi: 10.1111/j.1471-4159.2007.05124.x. [DOI] [PubMed] [Google Scholar]
- Roulet A, Puel O, et al. MDR1-deficient genotype in Collie dogs hypersensitive to the P-glycoprotein substrate ivermectin. Eur J Pharmacol. 2003;460(2–3):85–91. doi: 10.1016/s0014-2999(02)02955-2. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, et al. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25(4):664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Rupert PB, Daughdrill GW, et al. A new DNA-binding motif in the Skn-1 binding domain-DNA complex. Nat Struct Biol. 1998;5(6):484–491. doi: 10.1038/nsb0698-484. [DOI] [PubMed] [Google Scholar]
- Salazar M, Rojo AI, et al. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem. 2006;281(21):14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003;55(1):3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- Schinkel AH, Smit JJ, et al. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77(4):491–502. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Seaman JT, Eagleson JS, et al. Avermectin B1 toxicity in a herd of Murray Grey cattle. Aust Vet J. 1987;64(9):284–285. doi: 10.1111/j.1751-0813.1987.tb15963.x. [DOI] [PubMed] [Google Scholar]
- Sheps JA, Ralph S, et al. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Gen Biol. 2004;5(3):R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135(4):1358–1368. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- Singh A, Boldin-Adamsky S, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68(19):7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos G, Kuchler K. Fungal ATP-binding cassette (ABC) transporters in drug resistance & detoxification. Curr Drug Targets. 2006;7(4):471–481. doi: 10.2174/138945006776359403. [DOI] [PubMed] [Google Scholar]
- Sommer RJ, Streit A. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annu Rev Genet. 2011;45:1–20. doi: 10.1146/annurev-genet-110410-132417. [DOI] [PubMed] [Google Scholar]
- Sotirchos IM, Hudson AL, et al. Thioredoxins of a parasitic nematode: comparison of the 16- and 12-kDA thioredoxins from Haemonchus contortus. Free Radic Biol Med. 2008;44(12):2026–2033. doi: 10.1016/j.freeradbiomed.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Stitt LE, Tompkins JB, et al. ABC transporters influence sensitivity of Brugia malayi to moxidectin and have potential roles in drug resistance. Exp Parasitol. 2011;129(2):137–144. doi: 10.1016/j.exppara.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Strange RC, Spiteri MA, et al. Glutathione-S-transferase family of enzymes. Mut Res. 2001;482(1–2):21–26. doi: 10.1016/s0027-5107(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14(1):76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Hoerauf A, et al. Lymphatic filariasis and onchocerciasis. Lancet. 2010;376(9747):1175–1185. doi: 10.1016/S0140-6736(10)60586-7. [DOI] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- Torres-Rivera A, Landa A. Glutathione transferases from parasites: A biochemical view. Acta Tropica. 2008;105(2):99–112. doi: 10.1016/j.actatropica.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22(47):7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JMA, Hertweck M, et al. Direct inhibition of the longevity-promoting factor SKN–1 by insulin-like signaling in C. elegans. Cell. 2008;132(6):1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G, Blackhall W. Will technology provide solutions for drug resistance in veterinary helminths? Vet Parasitol. 2005;132(3–4):223–239. doi: 10.1016/j.vetpar.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. PNAS. 2004;101(7):2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-J, Sun Z, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29(6):1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Molento M, et al. Ivermectin resistance in nematodes may be caused by alteration of P-glycoprotein homolog. Mol Biochem Parasitol. 1998;91(2):327–335. doi: 10.1016/s0166-6851(97)00215-6. [DOI] [PubMed] [Google Scholar]
- Zhang DD. The Nrf2-Keap1-ARE signaling pathway: The regulation and dual function of Nrf2 in cancer. Antioxid Redox Signal. 2010;13(11):1623–1626. doi: 10.1089/ars.2010.3301. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, et al. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280(34):30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Thomas JH, et al. Comparative genomics and adaptive selection of the ATP-binding- cassette gene family in Caenorhabditis species. Genetics. 2007;175(3):1407–1418. doi: 10.1534/genetics.106.066720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipper LM, Mulcahy RT. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem Biophys Res Commun. 2000;278(2):484–492. doi: 10.1006/bbrc.2000.3830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.