Abstract
Nuclear pore complexes (NPCs) perforate the double-layered nuclear envelope and form the main gateway for molecular exchange between nucleus and cytoplasm of the eukaryotic cell. Because NPCs are extraordinarily complex and large, thus challenging to investigate on a molecular level, they are still rather poorly understood, despite their pivotal role in cellular homeostasis. To decipher the NPC structure at high resolution, the prerequisite to fully understand its function, a tailored approach is necessary that feeds from complimentary data, obtained at largely different spatial resolutions. The problem is further complicated by the dynamic nature of the NPC, manifested in flexible regions and dynamic components. Here we summarize the current state of these structural efforts, describe the breakthroughs of recent years, point out the existing disputes in the field, and give an outlook of what we should expect to happen in the near future.
Introduction
Compartmentalization is the decisive difference between eukaryotic and prokaryotic cells. The generation of diverse membrane-enclosed organelles allowed eukaryotes to develop a much higher degree of regulation and the optimization of biological processes in specific reaction compartments[1]. Gene transcription into mature mRNA occurs in the nucleus, enclosed within the double-layered nuclear envelope (NE), while protein translation exclusively takes place in the cytoplasm. The regulatory advantage of this setup generates the need for enormous transport capacities to ferry cargo between nucleo- and cytoplasm across the NE. Nuclear pore complexes (NPCs) are large protein assemblies that are embedded in circular openings in the NE, where inner and outer nuclear membranes are fused [2] NPCs constitute the principal gateway across the NE. They have an estimated mass of 40-60 MDa, and are composed of about 30 different proteins, collectively called nucleoporins or nups [3,4]. Because of an apparent 8fold rotational symmetry of the NPC, nups are estimated to occur in multiples of 8 copies, perhaps totaling 456 proteins [5]. In the simplest approximation, the NPC can be likened to a torus of about 100 nm diameter, with a central aqueous hole of about 50 nm across that serves as the main transport gate. Ions, metabolites, and cargo smaller than ∼40 kDa - ignoring a few notable exceptions - freely diffuse through the NPC. For larger cargo, specific nuclear transport receptors (NTRs) facilitate the transport across the NPC. The central cavity of the NPC is lined with proteins that contain hydrophilic fiber-like extensions interspersed with easily recognizable hydrophobic phenylalanine-glycine dipeptide repeats, hence they are collectively called FG-Nups [2]. These extensions emanate into and crowd the aqueous transport channel thereby forming a barrier and a specific milieu that large cargo cannot penetrate on its own. Large import cargo typically contains a nuclear localization signal (NLS), a peptide motif that is recognized by importing NTRs [6],[7]. NTRs have affinity for FG-Nups and thus an NTR-cargo complex can partition into the transport channel. Release on the nuclear side is achieved by the GTP-bound G protein Ran, which competes with cargo for NTR binding. Ran is kept GTP-bound in the nucleus due to the chromatin bound Ran-specific guanine nucleotide exchange factor RCC1, while the Ran GTPase activating protein RanGAP is localized to the cytoplasmic side of the NPC. Nuclear protein export starts with the recognition of nuclear export signals (NESs) on the cargo protein by NTRs that from a ternary complex with RanGTP [8],[9]. The assembled exporting NTR-RanGTP-cargo complex can now partition into the central transport channel, while cargo-release at the cytoplasmic side is achieved by RanGTP hydrolysis.
Although some of the basic principles that govern nucleo-cytoplasmic transport are now quite well understood, many specific questions remain. To name a few: 1) whether or not the FG-Nup network is uniform or whether it offers distinct routes for specific substrates is not clear; 2) the mechanism of membrane-bound cargo crossing the NE is poorly understood; 3) the molecular details of NPC involvement in gene regulation and other emerging functions are still largely obscure. For these and many more reasons it is essential to arrive at an understanding of the NPC architecture at the atomic level. Only when we understand the precise location of all nucleoporins within the NPC proper will it be possibleto probe all functions of this formidable machine and really arrive at a detailed understanding.
Other large protein assemblies, like the photosystems I and II, the ribosome, the proteasome, the RNA polymerases, as well as the clathrin and the COPII vesicle coats are already described in atomic or near-atomic detail, and they all profited from combined structural approaches integrating data of different size scales. For the NPC, due its size and complexity, this task is even more formidable but the results of recent years obtained in a variety of laboratories using different approaches are encouraging and clearly show that the field is on the right track to solve the problem. Here we summarize and discuss the structural data that is available for the NPC, the integration of it and how it will lead to an unambiguous description of the structure in the future.
What constitutes the NPC?
With any large assembly structure the question is how one defines its boundaries. For the NPC, it is typically defined biochemically, from fractionating eukaryotic cells and analyzing the composition of the most highly enriched NPC fraction[10,11]. This approach led to the description of about 30 proteins as nucleoporins, since they were abundantly found in this fraction, largely overlapped when different organisms were analyzed, and generally had no other function already assigned. However, the average dwell time of most nups at the NPC has been analyzed by fluorescence microscopy and found to vary by several orders of magnitude [12]. This means, only a subset of nups is stably integrated into the NPC, while others are rather dynamically associated. This behavior contributed to the fact that the nup stoichiometry is not known with high accuracy [11],[10]. Thus, the boundaries of the NPC are somewhat arbitrarily defined. This is further complicated by the fact that NPC has not yet been reconstituted from purified components, a method that likely would reveal the minimal set of proteins essential for building the NPC. The modular nature of the NPC allows to classify nucleoporins into categories: ∼15 scaffold nucleoporins form the stable NPC core, ∼10 nucleoporins contain phenylalanine-glycine (FG)-extensions, are typically more dynamic and form the main transport barrier, and 3-4 nucleoporins are membrane-embedded and presumably anchor the NPC in the NE [3]. A large body of data points to the conclusion that the set of ∼15 scaffold nucleoporins assembles into the basic NPC structure, to which the FG-nups get recruited subsequently. Thus, in a stepwise approach, the scaffold structure is what needs to be established first before we can arrive at a complete description of the NPC.
Electronmicroscopic Analysis of the Overall Structure of the NPC
Due to its size, complexity, and dynamic nature the NPC cannot be isolated in intact form from the cell, and reconstitution from its purified components has not been established either. These circumstances so far exclude the synergistic application of the two standard structure determination methods for large assemblies, X-ray crystallography and single-particle cryo-electronmicroscopy (cryo-EM), in a way similar to what was achieved for the clathrin and COPII vesicle coats. The best resolved structures of intact NPC are obtained through scanning and transmission electron microscopy, as well as cryo-electron tomography (cryo-ET) (Figure 1A). These pictures all coherently reveal the 8-fold rotational symmetry of the NPC scaffold around the central transport channel that can be seen in all preparations from highly diverged species. These preparations also reveal the overall dimensions of the NPC. Major discrepancies are visible when comparing tomographic reconstructions of the NPC from Dictyostelium discoideum, Xenopus laevis, and Homo sapiens, which roughly match in overall diameter and the width of the central channel, but differ by almost factor two in height [13-15]. Whether these differences are due to preparation, composition, or a combination thereof still awaits clarification. The cryo-ET reconstructions reach a resolution of around 6 nm, enough to appreciate that the main NPC scaffold ring is not uniformly electron-dense, but rather has a sponge-like appearance of tubular connections interspersed with cavities [16]. Various averaging methods have been employed, suggesting that the NPC scaffold itself may exhibit dynamic behavior in addition to the well established flexibility of the FG-rich extensions, which cannot be resolved by EM techniques.
Figure 1. Structural aspects of the NPC and scaffold models.
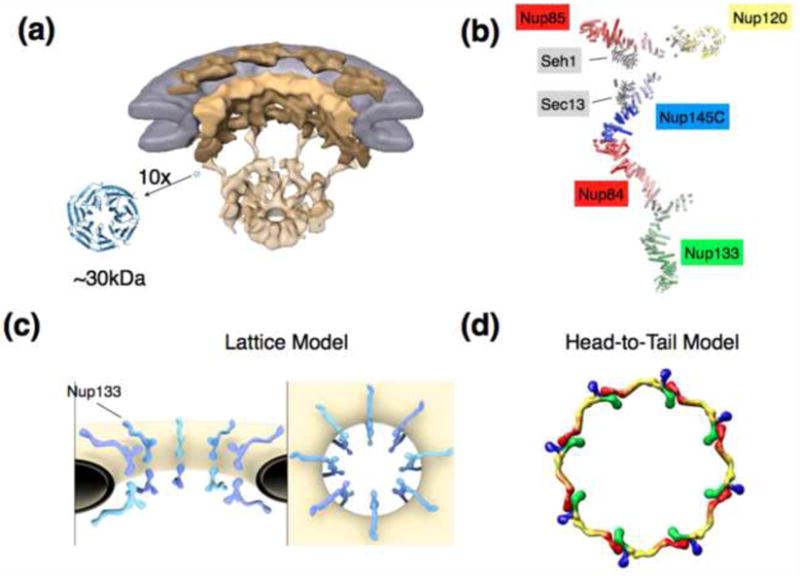
A) Overall architecture of the NPC determined by cryo-electron tomography and shown in a cut-away view. The nuclear envelope is colored in grey, scaffolding components of the NPC depicted in shades of yellow [16]. For scale, a canonical β-propeller in 10fold magnification is shown. β-Propellers, together with various helical units, make up a large portion of the stable NPC scaffold. Fitting of crystal structures into the cryo-ET structures is unreliable due to the resolution gap.
B) Composite structure of the heptameric 575 kDa Y-complex, a major scaffolding component of the NPC. The composite is generated based on individual crystal structures [18-22], modeling [31], interaction data, and EM pictures [17,39].
C) Arrangement of the Y-complexes within the NPC as suggested by the lattice model [18,20]. Two Y-complexes are related by a 2-fold symmetry axis and span the width of the NPC, with Nup133 pointing outward. Likely 8 such pairs, consistent with the 8fold rotational symmetry of the NPC, are positioned around the central transport axis of the NPC. Whether or not the two Y complexes directly bind or not is still unresolved. Lateral connection between Y-complexes most likely occurs via other components of the NPC, but this is also not determined yet.
D) Alternative model for the Y-complex arrangement within the pore. 8 Y-complexes are directly connected in head-to-tail fashion and form a ring [5,24,44,46]. Some authors argue for an arrangement with 2 stacked rings [5], others for a configuration with 4 stacked rings per NPC [24,44]. Color coding: Nup133 (red), Nup84/Nup145C/Sec13 (yellow), Nup85/Seh1 (blue), Nup120 (green).
Crystallographic Analysis of Individual Nups and Their Subcomplexes
Over the past decade many crystal structures of nucleoporins have been determined. The first such structures comprised only domains of individual nups, but over time larger structures including ternary complexes of close to 200 kDa were successfully tackled. At this stage, the collection of available crystal structures comprises about 30% of the main scaffold. Most of the ∼15 structural nucleoporins are organized into two large subcomplexes, the Y-complex with seven universally conserved members, and the Nic96 complex with five members. The Y-complex, composed of Seh1, Sec13, Nup84, Nup85, Nup120, Nup133, and Nup145C is the better understood assembly, largely due to its extraordinary in vitro stability [17]. The available crystal structures already cover about 90% of the 575 kDa complex (Figure 1B)[18-25]. The branched Y-structure is entirely built from β-propellers and elongated helical domains. Some of the helical domains are clearly evolutionary related and likely diverged from a common ancestor, despite very low sequence homology. The most prominent helical topology is the characteristic fold-back arrangement of the ∼25 helices in the (ancestral coatomer element 1) ACE1 module, specifically found in Nup84, Nup85, Nup145C of the Y-complex, as well as Nic96 and, curiously, Sec31 and Sec16 of the COPII vesicle coating system [20,26]. This discovery experimentally established the evolutionarily relationship between the NPC and COPII as two curved-membrane-stabilizing assemblies[20,27-29]. Moreover, Nup133 is clearly related to Nup157/170 of the Nic96 complex[22], while Nup120 with its more intricate helical arrangement seems to have evolved rather independently. Distinctions between the β-propellers incorporated into the NPC scaffold is often limited to modest modulations of surface features. Notably, the multi-functional β-propeller proteins Sec13 and Seh1, both fulfilling additional tasks outside of the NPC and also promiscuously binding to other proteins, are an exception, in that they build seven-bladed ß-propeller structures by acquiring one blade in trans from their respective binding partners to close and stabilize their open 6-bladed propeller. This peculiar protein-protein interaction theme might be specific to Sec13 and Seh1, at least it has so far neither been observed nor predicted for other proteins.
The structural characterization of the Nic96 complex is more fragmentary, so far only individual protein domains of three components have been reported [22,30,31]. Cell biological and biochemical data now start to establish the direct protein contacts between the members of the Nic96 complex, i.e. Nup53, Nic96, Nup157, Nup188, and Nup192 [32,33],[34-36]. Structural data, protein predictions, and evolutionary considerations suggest that the Y- and the Nic96 complexes are partially related, but there are also substantial differences.
While the collection of available structures is very informative it still is only a collection of mosaic blocks that need to be fitted together properly in order to determine the whole picture. Ideally, overlapping complexes of all scaffold components will be available eventually, which at that time will allow for the unambiguous description of the NPC. However, we are not there yet and thus the relative spatial arrangement of the Nup subcomplexes is vigorously debated in the field.
Bridging the Resolution Gap
Crystal structures can be fitted approximately into EM maps of sufficient, subnanometer resolution [37]. At 6-7 Å resolution secondary structure elements start to become visible, at which point docking of crystal structures is quite accurate[38]. So far though, the available EM maps of the NPC are far from this resolution, with the tomographic reconstructions extending to around ∼ 6 nm and the best random conicaltilt reconstruction of the assembled Y-complex limited to 3.5 nm [39]. Because of this resolution gap, data from the different imaging techniques cannot yet be used synergistically in ways we have seen with other systems in the past, most notably the elegant combined cryo-EM – X-ray crystallography approaches used to arrive at a pseudo-atomic resolution of the assembled clathrin and COPII vesicle coats [40-42]. Regardless, Kampmann and Blobel attempted to fit crystal structures into their 3.5 nm Y-complex structure and the result is at least consistent with previously published data that established the arrangement of the seven conserved nups in the Y-complex [17,39].
A big challenge en route toward a molecular description of the NPC scaffold is to figure out the relative orientation of the subcomplexes with respect to each other, as well as to determine their stoichiometry accurately (Figure 1C, D). Interaction data that confidently places subcomplexes in their proper neighborhood within the NPC is not published, presumably because these inter-subcomplex interactions are not very stable in isolation and thus are difficult to tract experimentally. Short of such data, we suggested a Y-complex arrangement rooted in the common ancestry of ACE1 proteins within the NPC and the outer coat of COPII. This lattice model postulates that the Nup84-Nup145C unit of the Y complex forms an edge element aligned along the positive curvature of the pore membrane, in analogy to its cousin, the Sec31-Sec31 edge element in COPII, for which experimental data exists [18] (Figure 1C, Table 1). Further, the lattice model suggests that Nup133 is positioned facing outward from the NPC, based on in vivo fluorescence data [43]. Two Y complexes come close in head-to-head fashion in the equatorial plane, but whether or not they bind directly or are bridged by other nup(s) is a matter of speculation. The Blobel and Hoelz laboratories suggested a diametrically opposed model, in which 8 Y-complexes are arranged equatorially in head-to-tail fashion (Figure 1D). Four such rings are suggested to stack on top of one another [44]. This model is essentially rooted in crystal packing contacts observed in crystallized fragments of the Y-complex, which were interpreted as physiologically relevant. Steric incompatibility between the initial model and subsequent structural studies, however, is a conflicting issue that is difficult to reconcile[18,20,23,25,44].
Table 1. Characteristic features of current NPC models.
| Model | Y-cplx orientation (long axis rel. to central 8fold) | Number of Y-rings | Relative position of Nic96-cplx | Y-cplxs per ring | Position ofneighboringY-cplx | Y-Y contact | Refs |
|---|---|---|---|---|---|---|---|
| Lattice Model | parallel | 2 | unclear, possibly intercalating laterally | Likely 8 | Head-to-Head | Direct or indirect | [18,20,29] |
| Fence-pole Model | perpendicular | 4 | forms aconcentric inner shell | 8 | Head-to-Tail | Direct, via Nup120-Nup133 | [24,44] |
| Computation al Model | perpendicular | 2 | 2 rings sandwiched between Y complex rings | 8 | Head-to-Tail | Direct | [5] |
Excitingly, super-resolution microscopy might remedy the situation in the foreseeable future. It allows for a spatial resolution that should enable the distinction of individual subcomplex within an intact NPC after fluorescently labeling them. In a pilot study the relative position of several spatially separated nucleoporins were already positioned with an accuracy that should allow for the distinction of individual subcomplexes [45]. Another novel approach is to figure out the relative orientation of specific nucleoporins within the NPC by recording fluorescence-anisotropy. In this method a nucleoporin is tagged with a rigidly-tethered GFP molecule, whose fluorescence is measured in an angle-dependent way relative to the orientation of the NE-embedded NPC [46]. Anisotropy reports that the nucleoporin adopts a specific orientation relative to the NPC, but due to the indirect measurement one can envision severe artifacts. This method was used to support the idea, that 8 Y-complexes form a ring in head-to-tail orientation with the long axis oriented parallel to the plane of the NE (Figure 1D). In order to be fully convincing, matching data using other nucleoporins needs to be measured as well.
The Computational Approach
To integrate positional and interaction data from a plethora of low resolution techniques a computational approach was developed with the goal to arrive at the architecture of the NPC entirely without the use of crystallographic data [5,47]. The results of these studies are indeed impressive and led to a first description of the NPC with an attempt to position all nucleoporin molecules within an energy-minimized assembly. The resolution of this arrangement is somewhat arbitrarily defined but does not exceed ∼5 nm. Thus, the protein positions can at best be described as tentative. Whether this computational approach can be further improved and be used to also integrate crystallographic data yet awaits proof. To properly integrate data of vastly different resolution and quality appears to be a formidable challenge.
Outlook
The problem of solving the structure of the NPC has a come a long way from being just a figment of the imagination to the situation that we face today. Electron-microscopic techniques have established the overall dimensions of the NPC from various eukaryotes, crystallographic approaches have resulted in an impressive array of partial structures that have steadily increased in size and complexity over the past few years. For the published structural analyses of large (1 MDa or more) protein assemblies in general, specifically tailored approaches had to be taken in each. ‘One-size fits all’ approaches do not work for problems of this complexity. It is obvious that a complete crystallographic analysis of the NPC seems out of reach for many reasons. However, every additional piece of structural data at near-atomic resolution that becomes available will limit further the contrasting models that are currently entertained for the NPC structure. Reproducible and accurate interaction data might be the key to soon come close to an undisputable assembly structure of the NPC as a first step. Single-particle EM reconstruction, not yet successfully employed, could proof crucial to bridge the resolution gap between X-ray crystallography and cryo-tomography. Once a convincing NPC structure at high resolution is established it will be interesting to see how it compares to the ancestrally-related COPII vesicle coat, whether the modular character of the NPC is indeed used to build NPCs of different composition, and whether new conclusions about the FG-repeat extensions and their involvement in facilitated transport will and can be drawn.
Acknowledgments
We would like to thank Ohad Medalia and Martin Kampmann for providing figures. This work was supported by NIH Grant GM077537.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavalier-Smith T. Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol Direct. 2010;5:7. doi: 10.1186/1745-6150-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran E, Wente S. Dynamic Nuclear Pore Complexes: Life on the Edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 3.Brohawn SG, Partridge JR, Whittle JRR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onischenko E, Weis K. Nuclear pore complex-a coat specifically tailored for the nuclear envelope. Curr Opin Cell Biol. 2011;23:293–301. doi: 10.1016/j.ceb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 6.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 7.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Güttler T, Görlich D. Ran-dependent nuclear export mediators: a structural perspective. EMBO J. 2011;30:3457–3474. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol. 2010;20:247–252. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 13.Elad N, Maimon T, Frenkiel-Krispin D, Lim RYH, Medalia O. Structural analysis of the nuclear pore complex by integrated approaches. Curr Opin Struct Biol. 2009;19:226–232. doi: 10.1016/j.sbi.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Maimon T, Medalia O. Perspective on the metazoan nuclear pore complex. Nucleus. 2010;1:383–386. doi: 10.4161/nucl.1.5.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 17.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Brohawn SG, Schwartz TU. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol. 2009;16:1173–1177. doi: 10.1038/nsmb.1713. Structural evidence that ACE1 proteins not only are shared between COPII coats and the NPC as building blocks, but that they also assemble in related fashion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leksa NC, Brohawn SG, Schwartz TU. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure. 2009;17:1082–1091. doi: 10.1016/j.str.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whittle JRR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. Journal of Biological Chemistry. 2009;284:28442–28452. doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proceedings of the National Academy of Sciences. 2009;106:14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy V, Hsia KC, Debler EW, Kampmann M, Davenport AM, Blobel G, Hoelz A. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proceedings of the National Academy of Sciences. 2009;106:17693–17698. doi: 10.1073/pnas.0909373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittle JRR, Schwartz TU. Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol. 2010;190:347–361. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 28.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brohawn SG, Schwartz TU. A lattice model of the nuclear pore complex. Commun Integr Biol. 2009;2:205–207. doi: 10.4161/cib.2.3.7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Jeudy S, Schwartz TU. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem. 2007;282:34904–34912. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- 32.Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009;185:475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. Substantive study on the molecular interactions within the Nic96 complex. Use of proteins from a thermotolerant yeast holds promise for yielding more stable proteins for in vitro studies. [DOI] [PubMed] [Google Scholar]
- 35.Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachdev R, Sieverding C, Flötenmeyer M, Antonin W. The C-terminal domain of Nup93 is essential for assembly of the structural backbone of nuclear pore complexes. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen JZ, Fürst J, Chapman MS, Grigorieff N. Low-resolution structure refinement in electron microscopy. J Struct Biol. 2003;144:144–151. doi: 10.1016/j.jsb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat Protoc. 2031;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 41.Stagg SM, LaPointe P, Razvi A, Gürkan C, Potter CS, Carragher B, Balch WE. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 43.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MGL, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. First report using super resolution microscopy to study the spatial distribution of nucleoporins with the NPC. Even better resolved mapping attempts using related technology are expected to gather more insight into assembly and stoichiometry of nucleoporins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Kampmann M, Atkinson CE, Mattheyses AL, Simon SM. Mapping the orientation of nuclear pore proteins in living cells with polarized fluorescence microscopy. Nat Struct Mol Biol. 2011;18:643–649. doi: 10.1038/nsmb.2056. Technically interesting and innovative approach to map the orientation of nucleoporins within the nuclear pore complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]


