Abstract
Botulinum neurotoxins (BoNTs, serotypes A–G) are the most deadly substances known. Here, we investigated how BoNT/E, a serotype that causes human botulism, translocates into the cytosol of neurons. Analogous to BoNT/B, BoNT/E required binding of the coreceptor, GT1b, to undergo significant secondary structural changes and transform into a hydrophobic protein at low pH. These data indicate that both serotypes act as coincidence detectors for both GT1b and low pH, to undergo translocation. However, BoNT/E translocated much more rapidly than BoNT/B. Also, BoNT/E required only GT1b, and not low pH, to oligomerize, whereas BoNT/B required both. In further contrast to the case of BoNT/B, low pH alone altered the secondary structure of BoNT/E to some degree and resulted in its premature inactivation. Hence, comparison of two BoNT serotypes revealed that these agents exhibit both convergent and divergent responses to receptor interactions, and pH, in the translocation pathway.
Botulinum neurotoxins (BoNTs, serotypes A–G) are the most potent toxins known.1 They target presynaptic nerve terminals with high specificity, causing a severe disease termed botulism. These toxins are produced as 150 kDa proteins by bacteria belonging to the genus Clostridium. Each toxin consists of a heavy chain (HC, 100 kDa) and a light chain (LC, 50 kDa). The HC binds to receptors on the neuronal surface and mediates toxin uptake. Recent studies indicate that the BoNTs utilize synaptic vesicle (SV) recycling as their major endocytic pathway,2−14 with the exception of BoNT/C.15 The HC also mediates the translocation of the LC from the SV/endosomal lumen into the cytosol.16 The LC then acts as a specific protease that cleaves SNARE (soluble NSF attachment protein receptor) proteins, thereby inhibiting neurotransmitter release. Such blockade results in flaccid paralysis at the neuromuscular junction and can result in respiratory failure and death.1 Because of their extreme potency (lethal dose ranging from 0.1 to 1 ng/kg), BoNTs are listed as Category A bioterrorism agents by the Centers for Disease Control and Prevention (CDC).17,18 Paradoxically, BoNTs are also widely used to treat a variety of diseases and are also used cosmetically to treat wrinkles.19
The mechanism by which BoNTs translocate their LCs from the SV/endosome lumen into the cytosol remains the least understood step in intoxication.16 Previous studies indicate that low pH in the endosomal lumen is required for the translocation step: neutralizing the pH with chloroquine, ammonium chloride, or methylamine hydrochloride or blocking endosomal acidification by specific vacuolar H+-ATPase inhibitors antagonized the action of all of the BoNTs.20−23 Electrophysiological studies revealed that addition of BoNTs to the low-pH, oxidizing side of a membrane resulted in single-channel formation in vitro; the channel activity was restricted to the HC.16,24−26 These data led to the hypothesis that low endosomal pH triggers the HC to form a membrane channel that mediates LC translocation. Because BoNTs are synthesized as soluble proteins, transforming them into membrane channels must involve major structural rearrangements; however, low pH alone failed to trigger any structural changes in BoNT/B, and this serotype was structurally stable from pH 7 to 4.27
Recent studies have shed new light on this apparent paradox.28 It was found that BoNT/B is a coincidence detector: binding to the coreceptor molecule, the ganglioside GT1b, allows the toxin to sense low pH and transform into oligomeric hydrophobic membrane-associated channels. This form of coincidence detection, i.e., the simultaneous requirement for both receptor binding and low pH, ensures that translocation occurs at the right time and place. GT1b-dependent oligomerization of BoNT/B in solution requires a pH of ≤5; at pH 5, the BoNT/B HC self-assembles into membrane-bound double- and triple-doughnut structures that resemble other protein translocation complexes.28 However, whether the concept of coincidence detection applies to any other BoNT serotype has yet to be determined: the underlying mechanisms that mediate their conversion into membrane channels remain elusive, and whether receptor molecules play a role in shaping the behavior of these toxins for translocation has not been addressed.
To begin to address these questions, we studied how BoNT/E senses low pH to transform into a hydrophobic protein. In contrast to the case for BoNT/B, which has a slow onset (e.g., 27% of subjects exhibit symptoms in ≤1 day), BoNT/E causes human botulism rapidly and has the shortest onset time (97% of subjects exhibit symptoms in ≤1 day).29−31 In this study, we have compared these two serotypes with the goal of gaining novel insights into the means by which translocation, one of the major steps that underlies intoxication by BoNTs, is triggered. The ganglioside GT1b serves as a coreceptor (in conjunction with protein receptors) for both BoNT/B and -E.13,32−36 We found that while both toxins act as coincidence detectors for receptor (GT1b) and low pH, they also exhibit striking differences with regard to how they respond to each of these signals. We speculate that these differences might underlie the distinct kinetics of translocation of these two toxin serotypes to affect the time course of intoxication.
Experimental Procedures
Toxins and Antibodies
BoNT/E and -B and rabbit polyclonal anti-BoNT/E and anti-BoNT/B antibodies were prepared as described previously.6,28,37 Monoclonal antibodies against SNAP-25 (71.1), syntaxin (HPC-1), and synaptobrevin II (syb, 69.1) were kindly provided by R. Jahn (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany).
Concanamycin A (con A) Assay
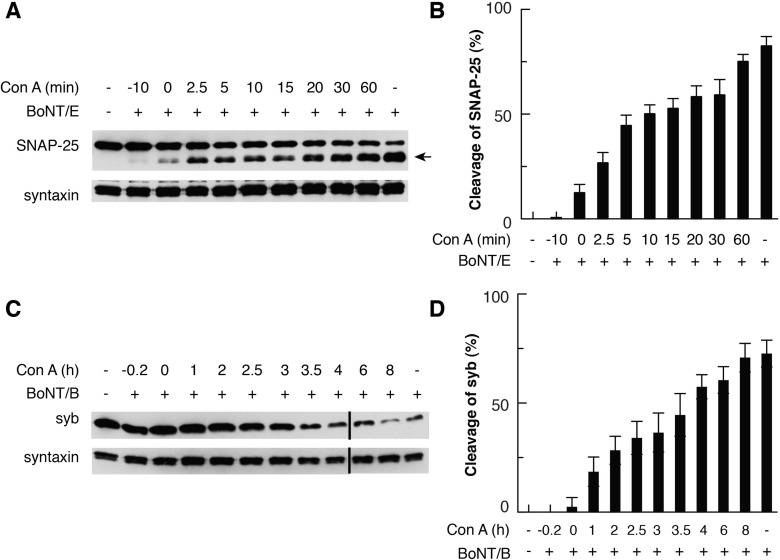
All procedures involving animals were conducted according to National Institutes of Health guidelines, as approved by the Animal Care and Use Committee of the University of Wisconsin. Cultures of rat embryonic (E17-18) hippocampal neurons were prepared as previously described.28 Experiments were performed on neurons that were 15–20 days old. Neurons were treated with BoNT/E or -B (10 nM) in high-K+ buffer [85 mM NaCl, 60 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5.5 mM glucose, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 10 mM sodium 2-(N-morpholino)ethanesulfonate (NaMES) (pH 7.4)] for 5 min. Toxins were washed off using low-K+ buffer [140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5.5 mM glucose, 10 mM HEPES, and 10 mM NaMES (pH 7.4)]. Where appropriate, con A (0.05 μM, Sigma-Aldrich) was added to the culture media before, with, or after toxin exposure at the indicated time. Neurons were incubated for 12 h, and cells were lysed as described previously.28 Cleavage of SNAP-25 by BoNT/E, or syb by BoNT/B, was monitored via immunoblot analysis. Syntaxin served as an internal loading control. Cleavage of SNAP-25 or syb was quantified using data obtained from at least eight independent trials.
Blue Native-PAGE (BN-PAGE) Assay
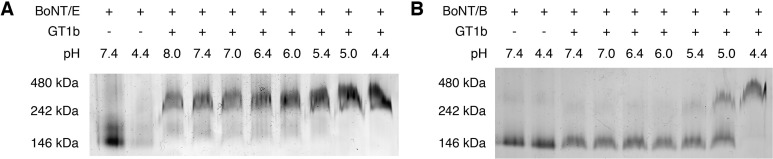
BN-PAGE assays were performed as previously described,28 using the Native-PAGE Novex Bis-Tris gel system (Invitrogen). BoNT/E or -B (100 nM) was incubated in the absence or presence of ganglioside GT1b (10 μM) at the indicated pH and 37 °C for 2 h. Samples were analyzed by BN-PAGE followed by either silver staining (Figure 2) or immunoblot using anti-BoNT/E antibodies (Figure S1 of the Supporting Information).
Figure 2.
GT1b allows oligomerization of BoNT/E at neutral pH. (A) BN-PAGE analysis of BoNT/E at the indicated pH in the absence and presence of GT1b. (B) BN-PAGE analysis of BoNT/B is included as a control. Representative gels, from three independent trials, are shown.
Circular Dichroism (CD) Spectroscopy
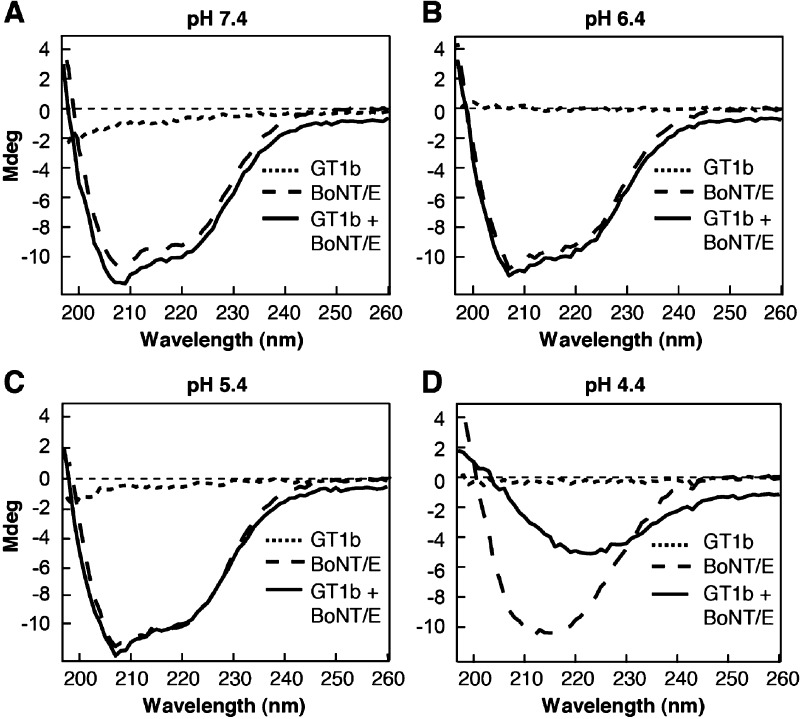
CD experiments were performed using an Aviv model 202SF spectrometer as described previously.28 Far-ultraviolet (UV) CD spectra (from 197 to 260 nm) of BoNT/E (0.67 μM) were obtained at the indicated pH and 37 °C using 1 mm path length quartz cuvettes. GT1b (30 μM) was then directly added to samples, and spectra were measured again. All spectra were corrected by subtracting spectra of the buffer alone. Helical content was estimated on the basis of the molar ellipticity value at 222 nm as described previously.28
Triton X-114 Partitioning Assay
Triton X-114 partitioning assays were performed as described previously.28,38 BoNT/E or -B (0.045 mg/mL, 300 nM) was incubated with or without GT1b (90 μM) at the indicated pH for 1 h at 37 °C before Triton X-114 partitioning assays were performed. The buffer consisted of 10 mM HEPES, 10 mM NaMES, 10 mM sodium acetate, and 150 mM NaCl, with the pH adjusted to the desired values. Triton X-114 was added to samples to a final concentration of 1% (v/v), and samples were vortexed for 15 min at 4 °C and centrifuged at 3000 g for 5 min to remove insoluble material. The supernatants were collected, incubated at 30 °C for 5 min, laid on top of a sucrose cushion [containing 6% (w/v) sucrose and 0.06% Triton X-114 in the above buffer, at the indicated pH], and centrifuged at 830 g for 3 min at room temperature. The detergent phase and aqueous phase were separated, and the aqueous phase was re-extracted with Triton X-114 as detailed above. The detergent phases were combined, and all samples were precipitated with 4 volumes of acetone overnight at −30 °C. The pellets were dissolved in SDS sample buffer and subjected to immunoblot analysis using anti-BoNT/E or anti-BoNT/B antibodies. The fraction of each protein in the detergent phase was quantified using data obtained from three independent trials.
Off-Pathway Assay
Off-pathway assays were performed as described previously.28 Briefly, BoNT/E (concentration as indicated) was incubated in high-K+ buffer at pH 7.4 or 4.4 for 2 h at 37 °C. Where appropriate, the pH of samples containing BoNT/E at pH 4.4 was reversed to 7.4, and BoNT/E was incubated at 37 °C for the indicated time. Toxins were then incubated with cultured rat hippocampal neurons for 5 min. Neurons were washed and incubated in neuronal media at 37 °C for 20 h. Off-pathway assays of BoNT/B (10 nM) were also performed as controls. Cells were harvested, and cleavage of SNAP-25 by BoNT/E, or cleavage of syb by BoNT/B, was monitored via immunoblot analysis using anti-SNAP-25 or anti-syb antibodies. The cleavage of SNAP-25 was quantified using data obtained from at least three independent trials.
Results
Time Courses for Blocking the Action of BoNT/E and -B in Neurons by con A
Translocation is a key step during BoNT intoxication,16 and the kinetics of this step can potentially impact the time course for the onset of botulism in host organisms. Current evidence indicates that BoNT/E translocates faster than BoNT/A in neurons,23,39 but we still lack a general understanding of the translocation rates for other BoNT serotypes. Specifically, BoNT/B is also a common cause of human botulism1 and is used clinically,19 but the time course for translocation of this serotype remains completely unknown.
To estimate the rate of BoNT/B translocation, we used con A, a specific inhibitor of vacuolar H+-ATPase,23 in rat hippocampal neurons. BoNT LC translocation requires low endosomal pH;1,16 after toxins are internalized into SVs/endosomes via endocytosis, blockade of acidification at different time points by con A traps the remaining LCs within the SVs/endosomes, thereby preventing SNARE cleavage by LCs in the cytosol. Thus, assaying for SNARE cleavage under these conditions provides a straightforward way to estimate BoNT translocation rates.23,39 Experiments utilizing BoNT/E were performed as controls.
Without con A, treating neurons with 10 nM BoNT/E or -B in high-K+ buffer (to stimulate SV recycling) for 5 min resulted in approximately 75–80% substrate cleavage during a subsequent 12 h incubation. Substrate cleavage was monitored by immunoblot analysis using antibodies against SNAP-25 (cleaved by BoNT/E) or syb (cleaved by BoNT/B) (Figure 1). Cleavage was completely blocked when con A was added to cells 10 min before toxin treatment. When con A was applied 10 min after BoNT/E, ∼50% cleavage of SNAP-25 was observed; BoNT/E became completely insensitive to con A when con A was added 1 h after toxin internalization (Figure 1A,B). In accordance with previous studies, these data suggest that BoNT/E completes translocation within 1 h of toxin internalization.23,39 In contrast, application of con A 1 h after BoNT/B addition severely hindered BoNT/B translocation, as less than ∼25% cleavage of syb was observed, and BoNT/B did not become completely insensitive to con A until con A was added more than 6 h after toxin treatment (Figure 1C,D). These results are unlikely due to slow cell surface binding and internalization of BoNT/B into SV/endosomes; as for BoNT/E,23,36 the majority of BoNT/B was already internalized into SVs/endosomes during the 5 min high-K+ buffer stimulation, and remaining toxin molecules on the cell surface after high-K+ buffer treatment were removed by extensive washing.4,10 As con A affects only translocation, our data suggest that BoNT/B translocates much more slowly than BoNT/E.
Figure 1.
Time courses for blocking the action of BoNT/E and -B in neurons by con A. Immunoblot analysis of SNAP-25 cleavage by BoNT/E (A, quantified in panel B) or syb cleavage by BoNT/B (C, quantified in panel D) in cultured rat hippocampal neurons. con A was added to neurons at the indicated time, either before, during, or after toxin exposure. SNARE cleavage was assayed 12 h after toxin exposure. The arrow indicates the cleaved form of SNAP-25. Syntaxin served as an internal loading control. The black line indicates lanes, from the same blot, that were juxtaposed. Data are means ± SEM (n = 8 in panel B; n = 9 in panel D).
Binding to GT1b Allows BoNT/E To Form Oligomers at Neutral pH
We then investigated the prerequisite factors for BoNT/E and -B translocation, which might underlie different translocation rates. As noted above, previous studies revealed that BoNT/B is a coincidence detector for low pH and the coreceptor GT1b; both are simultaneously required for BoNT/B to transform into an oligomeric membrane-bound channel-like structure.28 Moreover, low pH and GT1b also drive the oligomerization of BoNT/B, and this oligomerization step might contribute to efficient translocation, as suggested by studies of other bacterial toxins.40,41 To address whether BoNT/E also responds to low pH and GT1b by forming oligomers, we subjected this serotype to BN-PAGE under a variety of conditions. Oligomerization of BoNT/B was monitored as a control. In the absence of GT1b, BoNT/E, similar to BoNT/B, remained monomeric at pH 7.4 and 4.4 (Figure 2A and Figure S1 of the Supporting Information; note that the toxin did not stain well with silver staining at pH 4.4, but immunoblot analysis revealed that the toxin remained monomeric at this pH value). In the presence of GT1b, BoNT/B assembled into oligomers when the pH was <5.4 (Figure 2B; see also ref (28)). Surprisingly, binding of GT1b allowed BoNT/E to efficiently form oligomers at all pH values examined, even at pH 8 (Figure 2A). These data indicate that while the oligomerization of BoNT/B can occur only in acidic SVs/endosomes after internalization, BoNT/E may already undergo oligomerization upon binding to GT1b on the cell surface, potentially contributing to fast translocation.
BoNT/E Changes Conformation and Becomes Hydrophobic in the Presence of GT1b at Low pH
BoNTs have the remarkable ability to transform from soluble proteins into membrane-bound channels that mediate translocation.16,28 We reiterate that for BoNT/B, both oligomerization and transformation into a membrane protein require the presence of GT1b and low pH. However, this question has not been addressed for BoNT/E or any other serotype. We therefore conducted CD spectroscopy measurements, to probe structural changes in BoNT/E, at different pH values in the presence and absence of GT1b. Far-UV CD spectra were collected from 197 to 260 nm (Figure 3), and the helical content was estimated on the basis of the molar ellipticity at 222 nm.42 BoNT/E alone at pH 7.4 had a helical content of ∼27.5% (Figure 3A), consistent with a value of ∼26% as determined via crystallography.43 Lowering the pH from 7.4 to 5.4 did not significantly affect the shape of the spectra (Figure 3B,C), and the helical contents of BoNT/E at pH 6.4 and 5.4 were ∼27.3 and ∼29.9%, respectively. However, at pH 4.4, the peak of the BoNT/E spectrum was slightly shifted to the right (Figure 3D), but the helical content remained ∼27.6%. At pH ≥5.4, addition of GT1b did not significantly change the spectra of BoNT/E (Figure 3A–C); the helical content was ∼28.1% at pH 7.4, ∼28.9% at pH 6.4, and ∼28.8% at pH 5.4. However, as for BoNT/B,28 addition of GT1b to BoNT/E samples at pH 4.4 resulted in a major change in the shape of the spectrum (Figure 3D) that was associated with a reduction in helical content to ∼11.7%.
Figure 3.
GT1b triggers major conformational changes in BoNT/E at low pH. Far-UV CD spectra of BoNT/E in the absence (dashed lines) and presence (solid lines) of GT1b at pH 7.4 (A), 6.4 (B), 5.4 (C), and 4.4 (D). CD spectra of GT1b alone were included as controls (dotted lines).
To further test whether the observed structural changes in BoNT/E triggered by GT1b at low pH were associated with its conversion into a hydrophobic protein, we performed Triton X-114 partitioning assays. The detergent Triton X-114 remains homogeneous in solution at low temperature but partitions into aqueous and detergent phases at room temperature. This provides a convenient way to distinguish integral or lipid-anchored proteins (partitioning in the detergent phase) from soluble or peripheral proteins (partitioning in the aqueous phase).28,38 Parallel experiments using BoNT/B were included as controls (Figure 4A, bottom panel, and Figure 4B; see also ref (28)). Similar to BoNT/B, BoNT/E partitioned largely in the aqueous phase at both pH 7.4 and 4.4 in the absence of GT1b (Figure 4A, top panel, and Figure 4B). Binding to GT1b at pH 7.4 did not cause a significant increase in the level of partitioning of BoNT/E into the detergent phase. However, when the pH was lowered to 4.4, in the presence of GT1b, ∼80% of BoNT/E partitioned into the detergent phase. This was not due to tighter binding of BoNT/E at lower pH, because BoNT/E binds GT1b much less avidly at acidic pH than neutral pH.44 Overall, the data reported in this section indicate that BoNT/E requires a change in pH similar to that of BoNT/B to undergo the GT1b-dependent conversion into a hydrophobic protein.
Figure 4.
BoNT/E becomes hydrophobic at low pH only in the presence of GT1b. (A) Triton X-114 partitioning assays were conducted at the indicated pH in the absence or presence of GT1b, and BoNT/E in the total input (T), aqueous (A), and detergent (D) phases was detected by immunoblot analysis. (B) Partitioning of BoNT/E was quantified. Triton X-114 partitioning assays using BoNT/B were performed as controls.28 Data are means ± SEM (n = 3).
BoNT/E Enters an Off Pathway at Low pH
As shown in the preceding section, in the absence of GT1b, BoNT/E exhibited a slight right shift in its CD spectra when the pH was lowered from 7.4 to 4.4 (Figure 3D). This behavior is clearly different from that of BoNT/B, which is structurally stable from pH 7.4 to 4.4.27,28 It appears that low pH may be able to alter the structure of isolated BoNT/E in solution to some degree, even in the absence of GT1b.
To assess the functional impact of the low-pH-induced structural changes in BoNT/E, we conducted an “off-pathway” assay, again using cultured neurons as the target.28 This assay was devised to determine whether incubating the toxin, free in solution, at low pH, would result in premature conformational changes that result in unfolding and inactivation (i.e., entry into an “off pathway”). Such pathways exist for some bacterial toxins, including anthrax toxin,45 but not for BoNT/B (Figure 5A; see also ref (28)). In sharp contrast to BoNT/B, preincubation of BoNT/E at pH 4.4 for 2 h abolished its ability to enter neurons and cleave SNAP-25 (Figure 5A,B). We also reversed the preincubation pH back to 7.4, after a 2 h incubation at pH 4.4, before applying toxins to neurons and found that the cleavage of SNAP-25 was only partially recovered (∼40%) (Figure 5C,D), so the off pathway appears to be largely irreversible. These data indicate that unlike that of BoNT/B, low pH alone disrupts the structure of BoNT/E, leading it to enter an off pathway.
Figure 5.
Low-pH pretreatment inactivates BoNT/E. (A and B) BoNT/E was incubated at the indicated pH for 2 h at 37 °C, before being added to rat hippocampal neurons. Neurons were lysed after 20 h, and cleavage of SNAP-25 by BoNT/E was monitored by immunoblot analysis (A, quantified in panel B). Cleavage of syb by BoNT/B, under the same conditions, was measured as a control. Syntaxin served as an internal loading control. (C and D) BoNT/E was incubated at pH 4.4 for 2 h, before the pH of the samples was reversed back to 7.4 for the indicated period of time. BoNT/E was then incubated with neurons, and cleavage of SNAP-25 was monitored by immunoblot analysis (C, quantified in panel D). The effect of BoNT/E, which had not been pretreated with low-pH buffer, was analyzed as a control. Arrows indicate the cleaved form of SNAP-25. Data are means ± SEM from at least three independent trials.
Discussion
Many pathogenic bacteria produce protein toxins that alter the function of host cells. Among these toxins, BoNTs are the most deadly.1 BoNTs act by targeting presynaptic nerve terminals; however, the mechanism by which these agents translocate into the cytosol of host cells to exert their effects remains elusive.16 This study is focused on the translocation of BoNT/E, the serotype with the fastest onset for causing human botulism.29−31
Recent studies have provided new insights into the factors the trigger translocation of BoNT/B and revealed that this serotype requires both binding to the coreceptor molecule GT1b and low pH to transform into an oligomeric membrane protein.28 In this study, we found that oligomerization of BoNT/E is dependent only on GT1b and does not require low pH. In addition, oligomerization of BoNT/E was not associated with significant changes in secondary structure (Figures 3 and 4)28 and was not blocked by toosendanin (data not shown), a BoNT translocation inhibitor that hinders BoNT/B oligomerization.28 These findings indicate that while BoNT/B can only oligomerize after internalization and acidification of the organelle that mediated uptake, BoNT/E might undergo oligomerization on the cell surface, possibly contributing to its fast translocation. This is reminiscent of other bacterial toxins, such as anthrax toxin, whereby oligomerization occurs upon receptor binding on the cell surface, and the soluble oligomer is further converted into a membrane channel by low endosomal pH.41,46
Structural studies revealed that the domain arrangement of BoNT/E at neutral pH is different from that of BoNT/A and -B.43,47−49 In BoNT/E, the receptor-binding domain is on the same side of the translocation domain of the HC as the LC, and all three domains have mutual interfaces. This is different from the case for BoNT/A and -B, in which the LC and receptor-binding domain are on opposite sides of the translocation domain. The unique domain arrangement within BoNT/E has been proposed to contribute to the rapid kinetics of translocation of this serotype.43 Thus, it will be interesting to determine whether the unique domain arrangement with BoNT/E underlies its ability to oligomerize in a pH-independent manner upon binding GT1b.
In addition, it has been hypothesized that BoNT/E might have a less stringent requirement at low pH, and that this might also contribute to fast translocation.39 However, we found that BoNT/E has a pH dependence for transforming into a hydrophobic protein, in the presence of GT1b, similar to that of BoNT/B, a serotype that translocates with relatively slow kinetics. Hence, the rapid translocation of the BoNT/E LC does not appear to be due to a greater sensitivity of this serotype to low pH.
For bacterial toxins that require low pH for translocation, such as diphtheria toxin,50 histidine residues within the translocation domain serve as pH sensors: protonation of histidines at low pH triggers conformational changes that result in channel formation.51−53 While histidines serve as common low-pH sensors for a number of processes involving protein–membrane interactions51,54,55 and/or molten globule formation,56,57 BoNT/E has only one histidine within the translocation domain, and there are no histidines in the translocation domain of BoNT/B. These findings indicate that either BoNTs use different sensors within the translocation domain or pH sensing is mediated by other regions of these proteins. Moreover, recent studies revealed that deletion of the receptor-binding domain of BoNT/A alone (or together with the LC) disrupted low-pH sensing activity and the mutant (composed of the translocation domain and the LC or the translocation domain alone) was constitutively active for channel formation, even at neutral pH.26,58 These findings, in conjunction with our observations that BoNT/B and -E require binding to GT1b to transform into hydrophobic proteins at low pH, indicate that the receptor-binding domain somehow plays a crucial role in sensing low pH for translocation. In this regard, it is notable that the receptor binding domains of both serotypes contain histidines: two in BoNT/B and five in BoNT/E.
In summary, this work suggests that BoNT/E, similar to BoNT/B, is a coincidence detector for the coreceptor GT1b and low pH for translocation, and that the receptor-binding domain within these toxins may play a crucial role in sensing the low pH for translocation. Nonetheless, BoNT/E exhibited some striking differences from BoNT/B. (1) It translocates much more rapidly than BoNT/B in cultured hippocampal neurons. (2) Low pH alone disrupts the structure of BoNT/E to some degree and drives the toxin into an off pathway. (3) GT1b triggered BoNT/E oligomerization even at neutral pH. Therefore, though these toxins operate in similar ways, comparison of two serotypes has revealed marked differences in their responses to the factors that trigger translocation, raising the possibility that these differences contribute to the distinct translocation rates of different serotypes.
We note that motor neurons exhibit somewhat different expression patterns regarding protein receptors for BoNTs, and gangliosides, as compared to the cultured central nervous system neurons used for our studies, and it has been proposed that these differences may explain the higher apparent sensitivities of motor neuron terminals to the toxins.59 This view is supported by the finding that gangliosides serve as critical cofactors for toxin translocation (the current study and see also ref (28)). Future studies, monitoring BoNT translocation directly at the terminals of different classes of neurons, especially at the neuromuscular junction, will shed light on this issue.
Acknowledgments
We thank A. Figueroa Bernier for helping to prepare rat hippocampal neurons.
Glossary
Abbreviations
- BN-PAGE
blue native-PAGE
- BoNT
botulinum neurotoxin
- BoNT/B
botulinum neurotoxin type B
- BoNT/C
botulinum neurotoxin type C
- BoNT/E
botulinum neurotoxin type E
- CD
circular dichroism
- CDC
Centers for Disease Control and Prevention
- con A
concanamycin A
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HC
heavy chain
- LC
light chain
- NaMES
sodium 2-(N-morpholino)ethanesulfonate
- SEM
standard error of the mean
- SNARE
soluble NSF attachment protein receptor
- SV
synaptic vesicle
- syb
synaptobrevin II
- UV
ultraviolet.
Supporting Information Available
BN-PAGE followed by silver staining or immunoblot analysis of BoNT/E (Figure S1). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by a Grant AI057744 from the National Institutes of Health to E.R.C. E.R.C. is an Investigator of the Howard Hughes Medical Institute.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Schiavo G.; Matteoli M.; Montecucco C. (2000) Neurotoxins affecting neuroexocytosis. Physiol. Rev. 80, 717–766. [DOI] [PubMed] [Google Scholar]
- Nishiki T.; Kamata Y.; Nemoto Y.; Omori A.; Ito T.; Takahashi M.; Kozaki S. (1994) Identification of protein receptor for Clostridium botulinum type B neurotoxin in rat brain synaptosomes. J. Biol. Chem. 269, 10498–10503. [PubMed] [Google Scholar]
- Dong M.; Yeh F.; Tepp W. H.; Dean C.; Johnson E. A.; Janz R.; Chapman E. R. (2006) SV2 is the protein receptor for botulinum neurotoxin A. Science 312, 592–596. [DOI] [PubMed] [Google Scholar]
- Dong M.; Richards D. A.; Goodnough M. C.; Tepp W. H.; Johnson E. A.; Chapman E. R. (2003) Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J. Cell Biol. 162, 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiki T.; Tokuyama Y.; Kamata Y.; Nemoto Y.; Yoshida A.; Sekiguchi M.; Takahashi M.; Kozaki S. (1996) Binding of botulinum type B neurotoxin to Chinese hamster ovary cells transfected with rat synaptotagmin II cDNA. Neurosci. Lett. 208, 105–108. [DOI] [PubMed] [Google Scholar]
- Dong M.; Liu H.; Tepp W. H.; Johnson E. A.; Janz R.; Chapman E. R. (2008) Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol. Biol. Cell 19, 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel A.; Karnath T.; Henke T.; Bigalke H.; Binz T. (2004) Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J. Biol. Chem. 279, 30865–30870. [DOI] [PubMed] [Google Scholar]
- Mahrhold S.; Rummel A.; Bigalke H.; Davletov B.; Binz T. (2006) The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 580, 2011–2014. [DOI] [PubMed] [Google Scholar]
- Rummel A.; Eichner T.; Weil T.; Karnath T.; Gutcaits A.; Mahrhold S.; Sandhoff K.; Proia R. L.; Acharya K. R.; Bigalke H.; Binz T. (2007) Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double-receptor concept. Proc. Natl. Acad. Sci. U.S.A. 104, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.; Tepp W. H.; Liu H.; Johnson E. A.; Chapman E. R. (2007) Mechanism of botulinum neurotoxin B and G entry into hippocampal neurons. J. Cell Biol. 179, 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.; Tepp W. H.; Johnson E. A.; Dong M. (2011) Botulinum neurotoxin D uses synaptic vesicle protein SV2 and gangliosides as receptors. PLoS Pathog. 7, e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.; Chen C.; Barbieri J. T.; Kim J. J.; Baldwin M. R. (2009) Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry 48, 5631–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel A.; Hafner K.; Mahrhold S.; Darashchonak N.; Holt M.; Jahn R.; Beermann S.; Karnath T.; Bigalke H.; Binz T. (2009) Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J. Neurochem. 110, 1942–1954. [DOI] [PubMed] [Google Scholar]
- Yeh F. L.; Dong M.; Yao J.; Tepp W. H.; Lin G.; Johnson E. A.; Chapman E. R. (2010) SV2 mediates entry of tetanus neurotoxin into central neurons. PLoS Pathog. 6, e1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.; Berntsson R. P.; Tepp W. H.; Pitkin R. M.; Johnson E. A.; Stenmark P.; Dong M. (2012) Botulinum neurotoxin D-C uses synaptotagmin I/II as receptors and human synaptotagmin II is not an effective receptor for type B, D-C, and G toxins. J. Cell Sci. DOI: 10.1242/jcs.103564 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montal M. (2010) Botulinum neurotoxin: A marvel of protein design. Annu. Rev. Biochem. 79, 591–617. [DOI] [PubMed] [Google Scholar]
- Arnon S. S.; Schechter R.; Inglesby T. V.; Henderson D. A.; Bartlett J. G.; Ascher M. S.; Eitzen E.; Fine A. D.; Hauer J.; Layton M.; Lillibridge S.; Osterholm M. T.; O’Toole T.; Parker G.; Perl T. M.; Russell P. K.; Swerdlow D. L.; Tonat K. (2001) Botulinum toxin as a biological weapon: Medical and public health management. JAMA, J. Am. Med. Assoc. 285, 1059–1070. [DOI] [PubMed] [Google Scholar]
- Rotz L. D.; Khan A. S.; Lillibridge S. R.; Ostroff S. M.; Hughes J. M. (2002) Public health assessment of potential biological terrorism agents. Emerging Infect. Dis. 8, 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco C.; Molgo J. (2005) Botulinal neurotoxins: Revival of an old killer. Curr. Opin. Pharmacol. 5, 274–279. [DOI] [PubMed] [Google Scholar]
- Simpson L. L. (1983) Ammonium chloride and methylamine hydrochloride antagonize clostridial neurotoxins. J. Pharmacol. Exp. Ther. 225, 546–552. [PubMed] [Google Scholar]
- Simpson L. L. (1982) The interaction between aminoquinolines and presynaptically acting neurotoxins. J. Pharmacol. Exp. Ther. 222, 43–48. [PubMed] [Google Scholar]
- Simpson L. L.; Coffield J. A.; Bakry N. (1994) Inhibition of vacuolar adenosine triphosphatase antagonizes the effects of clostridial neurotoxins but not phospholipase A2 neurotoxins. J. Pharmacol. Exp. Ther. 269, 256–262. [PubMed] [Google Scholar]
- Keller J. E.; Cai F.; Neale E. A. (2004) Uptake of botulinum neurotoxin into cultured neurons. Biochemistry 43, 526–532. [DOI] [PubMed] [Google Scholar]
- Sheridan R. E. (1998) Gating and permeability of ion channels produced by botulinum toxin types A and E in PC12 cell membranes. Toxicon 36, 703–717. [DOI] [PubMed] [Google Scholar]
- Hoch D. H. (1985) Botulinum, tetanus, and diphtheria toxin channels in planar lipid bilayers. Doctoral Dissertation, Albert Einstein College of Medicine, Bronx, NY.
- Fischer A.; Sambashivan S.; Brunger A. T.; Montal M. (2012) Beltless translocation domain of botulinum neurotoxin A embodies a minimum ion-conductive channel. J. Biol. Chem. 287, 1657–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy S.; Kumaran D.; Keller J.; Swaminathan S. (2004) Role of metals in the biological activity of Clostridium botulinum neurotoxins. Biochemistry 43, 2209–2216. [DOI] [PubMed] [Google Scholar]
- Sun S.; Suresh S.; Liu H.; Tepp W. H.; Johnson E. A.; Edwardson J. M.; Chapman E. R. (2011) Receptor binding enables botulinum neurotoxin B to sense low pH for translocation channel assembly. Cell Host Microbe 10, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff B. A.; Griffin P. M.; McCroskey L. M.; Smart J. F.; Wainwright R. B.; Bryant R. G.; Hutwagner L. C.; Hatheway C. L. (1992) Clinical and laboratory comparison of botulism from toxin types A, B, and E in the United States, 1975–1988. J. Infect. Dis. 166, 1281–1286. [DOI] [PubMed] [Google Scholar]
- Simpson L. L. (1980) Kinetic studies on the interaction between botulinum toxin type A and the cholinergic neuromuscular junction. J. Pharmacol. Exp. Ther. 212, 16–21. [PubMed] [Google Scholar]
- Simpson L. L.; Dasgupta B. R. (1983) Botulinum neurotoxin type E: Studies on mechanism of action and on structure-activity relationships. J. Pharmacol. Exp. Ther. 224, 135–140. [PubMed] [Google Scholar]
- Kozaki S.; Kamata Y.; Watarai S.; Nishiki T.; Mochida S. (1998) Ganglioside GT1b as a complementary receptor component for Clostridium botulinum neurotoxins. Microb. Pathog. 25, 91–99. [DOI] [PubMed] [Google Scholar]
- Nishiki T.; Tokuyama Y.; Kamata Y.; Nemoto Y.; Yoshida A.; Sato K.; Sekiguchi M.; Takahashi M.; Kozaki S. (1996) The high-affinity binding of Clostridium botulinum type B neurotoxin to synaptotagmin II associated with gangliosides GT1b/GD1a. FEBS Lett. 378, 253–257. [DOI] [PubMed] [Google Scholar]
- Rummel A.; Eichner T.; Weil T.; Karnath T.; Gutcaits A.; Mahrhold S.; Sandhoff K.; Proia R. L.; Acharya K. R.; Bigalke H.; Binz T. (2007) Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double-receptor concept. Proc. Natl. Acad. Sci. U.S.A. 104, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata Y.; Kozaki S.; Sakaguchi G.; Iwamori M.; Nagai Y. (1986) Evidence for direct binding of Clostridium botulinum type E derivative toxin and its fragments to gangliosides and free fatty acids. Biochem. Biophys. Res. Commun. 140, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Dong M.; Liu H.; Tepp W. H.; Johnson E. A.; Janz R.; Chapman E. R. (2008) Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol. Biol. Cell 19, 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. J.; Siegel L. S. (1986) Purification of type E botulinum neurotoxin by high-performance ion exchange chromatography. Anal. Biochem. 156, 213–219. [DOI] [PubMed] [Google Scholar]
- Bordier C. (1981) Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256, 1604–1607. [PubMed] [Google Scholar]
- Wang J.; Meng J.; Lawrence G. W.; Zurawski T. H.; Sasse A.; Bodeker M. O.; Gilmore M. A.; Fernandez-Salas E.; Francis J.; Steward L. E.; Aoki K. R.; Dolly J. O. (2008) Novel chimeras of botulinum neurotoxins A and E unveil contributions from the binding, translocation, and protease domains to their functional characteristics. J. Biol. Chem. 283, 16993–17002. [DOI] [PubMed] [Google Scholar]
- Menestrina G.; Schiavo G.; Montecucco C. (1994) Molecular mechanisms of action of bacterial protein toxins. Mol. Aspects Med. 15, 79–193. [DOI] [PubMed] [Google Scholar]
- Young J. A.; Collier R. J. (2007) Anthrax toxin: Receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 76, 243–265. [DOI] [PubMed] [Google Scholar]
- Chen Y. H.; Yang J. T.; Martinez H. M. (1972) Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry 11, 4120–4131. [DOI] [PubMed] [Google Scholar]
- Kumaran D.; Eswaramoorthy S.; Furey W.; Navaza J.; Sax M.; Swaminathan S. (2009) Domain organization in Clostridium botulinum neurotoxin type E is unique: Its implication in faster translocation. J. Mol. Biol. 386, 233–245. [DOI] [PubMed] [Google Scholar]
- Kamata Y.; Kozaki S.; Sakaguchi G. (1988) Effects of pH on the binding of Clostridium botulinum type E derivative toxin to gangliosides and phospholipids. FEMS Microbiol. Lett. 55, 71–76. [Google Scholar]
- Sun J.; Vernier G.; Wigelsworth D. J.; Collier R. J. (2007) Insertion of anthrax protective antigen into liposomal membranes: Effects of a receptor. J. Biol. Chem. 282, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Miller C. J.; Elliott J. L.; Collier R. J. (1999) Anthrax protective antigen: Prepore-to-pore conversion. Biochemistry 38, 10432–10441. [DOI] [PubMed] [Google Scholar]
- Fischer A.; Montal M. (2007) Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proc. Natl. Acad. Sci. U.S.A. 104, 10447–10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S.; Eswaramoorthy S. (2000) Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat. Struct. Biol. 7, 693–699. [DOI] [PubMed] [Google Scholar]
- Lacy D. B.; Tepp W.; Cohen A. C.; DasGupta B. R.; Stevens R. C. (1998) Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat. Struct. Biol. 5, 898–902. [DOI] [PubMed] [Google Scholar]
- Blewitt M. G.; Chung L. A.; London E. (1985) Effect of pH on the conformation of diphtheria toxin and its implications for membrane penetration. Biochemistry 24, 5458–5464. [DOI] [PubMed] [Google Scholar]
- Perier A.; Chassaing A.; Raffestin S.; Pichard S.; Masella M.; Menez A.; Forge V.; Chenal A.; Gillet D. (2007) Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. J. Biol. Chem. 282, 24239–24245. [DOI] [PubMed] [Google Scholar]
- Kyrychenko A.; Posokhov Y. O.; Rodnin M. V.; Ladokhin A. S. (2009) Kinetic intermediate reveals staggered pH-dependent transitions along the membrane insertion pathway of the diphtheria toxin T-domain. Biochemistry 48, 7584–7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnin M. V.; Kyrychenko A.; Kienker P.; Sharma O.; Vargas-Uribe M.; Collier R. J.; Finkelstein A.; Ladokhin A. S. (2011) Replacement of C-terminal histidines uncouples membrane insertion and translocation in diphtheria toxin T-domain. Biophys. J. 101, L41–L43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason A. J.; Gasnier C.; Kichler A.; Prevost G.; Aunis D.; Metz-Boutigue M. H.; Bechinger B. (2006) Enhanced membrane disruption and antibiotic action against pathogenic bacteria by designed histidine-rich peptides at acidic pH. Antimicrob. Agents Chemother. 50, 3305–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann T.; Mueller D. S.; Mark A. E.; Young P. R.; Kobe B. (2006) The role of histidine residues in low-pH-mediated viral membrane fusion. Structure 14, 1481–1487. [DOI] [PubMed] [Google Scholar]
- Jamin M.; Antalik M.; Loh S. N.; Bolen D. W.; Baldwin R. L. (2000) The unfolding enthalpy of the pH 4 molten globule of apomyoglobin measured by isothermal titration calorimetry. Protein Sci. 9, 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.; Song J. (2005) Molecular mechanism underlying the thermal stability and pH-induced unfolding of CHABII. J. Mol. Biol. 348, 205–218. [DOI] [PubMed] [Google Scholar]
- Fischer A.; Mushrush D. J.; Lacy D. B.; Montal M. (2008) Botulinum neurotoxin devoid of receptor binding domain translocates active protease. PLoS Pathog. 4, e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verderio C.; Rossetto O.; Grumelli C.; Frassoni C.; Montecucco C.; Matteoli M. (2006) Entering neurons: Botulinum toxins and synaptic vesicle recycling. EMBO Rep. 7, 995–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.