Arabidopsis FANCM, a homolog of human FANCM, is involved in the suppression of somatic homologous recombination. Here, it is shown that At-FANCM also plays a key role in meiosis, as it ensures the controlled formation of genetic exchange by suppressing erroneous interactions between parental genomes and by controlling meiotic crossover formation between homologs.
Abstract
The human hereditary disease Fanconi anemia leads to severe symptoms, including developmental defects and breakdown of the hematopoietic system. It is caused by single mutations in the FANC genes, one of which encodes the DNA translocase FANCM (for Fanconi anemia complementation group M), which is required for the repair of DNA interstrand cross-links to ensure replication progression. We identified a homolog of FANCM in Arabidopsis thaliana that is not directly involved in the repair of DNA lesions but suppresses spontaneous somatic homologous recombination via a RecQ helicase (At-RECQ4A)–independent pathway. In addition, it is required for double-strand break–induced homologous recombination. The fertility of At-fancm mutant plants is compromised. Evidence suggests that during meiosis At-FANCM acts as antirecombinase to suppress ectopic recombination-dependent chromosome interactions, but this activity is antagonized by the ZMM pathway to enable the formation of interference-sensitive crossovers and chromosome synapsis. Surprisingly, mutation of At-FANCM overcomes the sterility phenotype of an At-MutS homolog4 mutant by apparently rescuing a proportion of crossover-designated recombination intermediates via a route that is likely At-MMS and UV sensitive81 dependent. However, this is insufficient to ensure the formation of an obligate crossover. Thus, At-FANCM is not only a safeguard for genome stability in somatic cells but is an important factor in the control of meiotic crossover formation.
INTRODUCTION
The human hereditary disease Fanconi anemia (FA) was first described by Guido Fanconi in 1927 (Fanconi, 1927). FA patients suffer from a wide range of symptoms, including developmental defects, increased cancer incidence, and breakdown of the hematopoietic system (Neveling et al., 2009).
To date, mutations in at least 15 different genes have been associated with FA. Despite the diversity of clinical symptoms and the number of genes implicated in the disease, there are a limited number of cellular phenotypes. These include a disturbance of cell cycle progression, apoptosis, spontaneous chromosome breakage, and radial chromosomes, which are indicative of an underlying defect in genome stability. For diagnostic purposes, the elevated sensitivity of FA cells to genotoxins like mitomycin C (MMC) and cisplatin [cis-diamminedichloroplatinum(II)], which cause DNA interstrand cross-links (ICLs) is used. It seems that FA is a result of problems in repairing ICLs, which in turn disrupts transcription, replication, and mitosis (de Winter and Joenje, 2009).
Most FA proteins interact in a core complex and all 15 are needed for ICL repair. At a stalled replication fork (e.g., caused by an ICL), the FA core complex monoubiquitinates a heterodimer of FANCD2/FANCI (for Fanconi anemia complementation group D2/Fanconi anemia complementation group I), which is then recruited to the lesion, where it activates downstream FA proteins and other repair and recombination factors. The protein responsible for guiding the FA core complex to DNA lesions is FANCM (for Fanconi anemia complementation group M), which has a strong preference to bind branched DNA structures in vitro (e.g., Holliday junctions [HJs] and replication forks) (Gari et al., 2008a).
Human FANCM consists of an N-terminal bipartite SF2 helicase domain (composed of a DEXDc and a HELICc domain), which has been shown to have ATPase and double-stranded DNA translocase activity in vitro but no helicase activity on a number of DNA substrates (Meetei et al., 2005; Gari et al., 2008a, 2008b). FANCM is able to branch migrate HJs and replication forks in vitro, which may enable it to remodel replication forks to produce so-called chicken-foot structures, a proposed intermediate in many repair and recombination reactions at stalled replication forks. The FANCM C terminus contains an inactive endonuclease domain that is similar to those found in XPF family proteins, such as XPF and MUS81. This domain is rendered inactive in all animal FANCM homologs by mutations in several functionally essential amino acids (Meetei et al., 2005). FANCM also functions independently of the FA core complex, where it helps to regulate cell cycle checkpoints in response to DNA lesions in S-phase (Collis et al., 2008; Luke-Glaser et al., 2010; Schwab et al., 2010).
The only FA homolog conserved in the budding yeast Saccharomyces cerevisiae is Mph1 (for Mutator phenotype1), an ortholog of FANCM. In contrast with FANCM, sensitivity to ICL-inducing agents has not been described for mph1 mutant cells. The connection to DNA repair at stalled replication forks has been conserved, though, since mph1 mutants are sensitive to treatment with genotoxins that lead to replication stress (e.g., methyl methanesulfonate [MMS]) (Scheller et al., 2000). In vitro activities also differ from animal FANCM; Mph1 has a true helicase activity and can unwind double-stranded DNA in a 3′ to 5′ direction (Prakash et al., 2005). Sc-MPH1 is epistatic to a number of budding yeast homologous recombination (HR) genes, and the somatic HR rate is increased in mph1 mutant cells (Schürer et al., 2004). Recently, it was shown that Mph1 participates in D-loop formation in HR, together with the recombinase Rad54 (for radiation sensitive54) (Panico et al., 2010; Ede et al., 2011). In double mutants of MPH1 and SGS1 (for slow growth suppressor1), the sole budding yeast RecQ helicase homolog, the HR rate is higher than the rate of either single mutant, indicating two separate pathways with Mph1 and Sgs1 functioning in either one to suppress somatic HR (Schürer et al., 2004). Homozygous diploids of the mph1 mutant exhibit a slight sporulation defect, but spore survival is not different from the wild type (Scheller et al., 2000). This suggests that Mph1 has little or no meiotic function in budding yeast. The association of Mph1 with HR and translesion synthesis places the protein at repair and recombination processes at stalled replication forks, similar to animal FANCM. Similar functions in HR at double-strand breaks (DSBs) and stalled replication forks have also been reported for the Schizosaccharomyces pombe ortholog, Fml1 (for FANCM-like protein1) (Sun et al., 2008).
Key aspects of meiotic recombination are conserved between animals, yeast, and plants (Osman et al., 2011). Following the formation of DSBs by Spo11 (for sporulation11) (Keeney et al., 1997; Hartung and Puchta, 2000; Grelon et al., 2001; Stacey et al., 2006; Hartung et al., 2007b), recombination is initiated between homologous chromosomes by the recombinases Rad51 (for radiation sensitive51) and Dmc1 (for disrupted meiotic cDNA1) (Klimyuk and Jones, 1997; Li et al., 2004; Shinohara and Shinohara, 2004; Seeliger et al., 2012). These proteins facilitate the invasion of single-stranded ends of a DSB into homologous sequences, creating a D-loop structure. The invading strand is then elongated by DNA polymerases. In general, most DSBs are processed to noncrossover (NCO) products with a small proportion becoming crossovers (COs) (Sanchez-Moran et al., 2007). Studies in budding yeast suggest that NCOs form via the synthesis-dependent strand-annealing pathway (Allers and Lichten, 2001). Here, the invading strand is elongated and removed from the D-loop to reanneal to the second end of the DSB; remaining gaps are closed and nicks are sealed. The majority of COs, on average ∼85%, arise via the formation of a joint-molecule intermediate, the double-Holliday junction (dHJ) (Szostak et al., 1983). dHJ formation is dependent on the ZMM (Zip1, Zip2, Zip3, Zip4, Msh4 (for MutS homolog4), Msh5, and Mer3) proteins, first identified in budding yeast and also found in Arabidopsis thaliana (Börner et al., 2004; Osman et al., 2011). The dHJs are then resolved into COs by a yet to be determined resolvase. A characteristic of these COs is that they exhibit interference, whereby the position of one CO decreases the probability of a second in an adjacent chromosomal region (Jones, 1984). A proportion of COs arises via a ZMM-independent process that appears to involve the structure-specific endonuclease At-MUS81 (for MMS and UV sensitive81) (Hartung et al., 2006; Geuting et al., 2009). These COs are interference insensitive, exhibit a random numerical distribution, and amount to ∼15% of the total in the wild type (Higgins et al., 2008a). Recently, At-TOP3α (for Topoisomerase3α) and At-RMI1 (for RecQ-mediated genome instability1) have been shown to be essential for meiosis in Arabidopsis. It is proposed they act by dissolution of dHJs and account for some NCO products (Chelysheva et al., 2008; Hartung et al., 2008).
Here, we report the analysis of a FANCM/Mph1 homolog in the model plant Arabidopsis. We show that contrary to the phenotype of its fungal and animal orthologs, At-FANCM has no direct role in DNA repair, including the repair of ICLs, but is important for the choice of HR pathways in somatic cells. Surprisingly, At-fancm mutant plants are compromised in their fertility. Our analysis indicates that At-FANCM is required for ordered meiotic CO formation and normal chromosome synapsis. Evidence suggests that it acts as an antirecombinase, but this activity is antagonized by the ZMM pathway to enable the formation of interference-sensitive COs.
RESULTS
Identification of Plant FANCM Homologs
We identified a locus (At1g35530, named At-FANCM) in the model plant Arabidopsis whose predicted protein sequence had an amino acid identity of 25.2 and 22.1% compared with Hs-FANCM and Sc-Mph1, respectively.
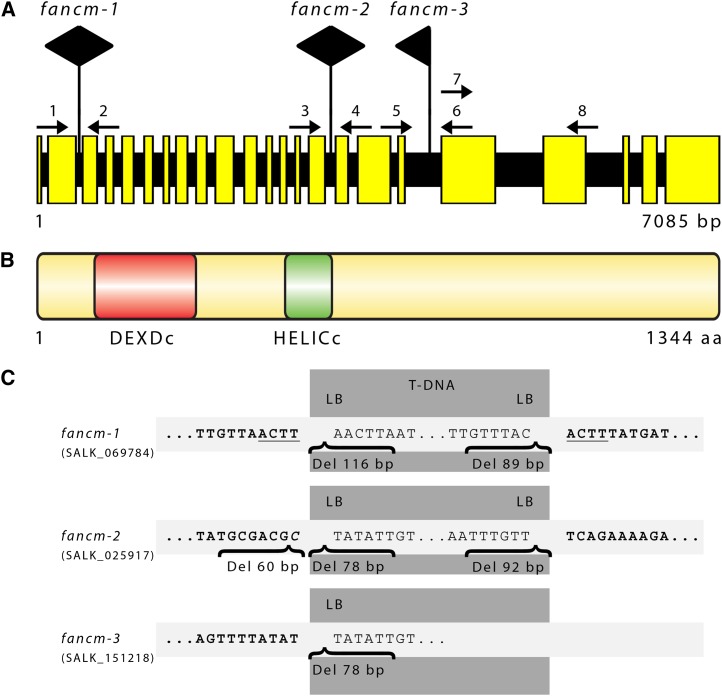
We analyzed the At-FANCM cDNA by sequencing of PCR fragments amplified from Arabidopsis seedling mRNA. Contrary to in silico predictions, the At-FANCM open reading frame (ORF) has a length of 4035 bp (submitted to GenBank; accession number JQ278026). Comparison of the sequenced ORF and the most recent prediction (AT1G35530.2, accession number sequence 6530298622 [The Arabidopsis Information Resource] or NM_001198212 [National Center for Biotechnology Information]) showed that the splice junctions of introns 3, 15, and 20 were incorrectly annotated, and intron 22 was not recognized. The At-FANCM genomic locus has a length of 7085 bp and is composed of 23 exons and 22 introns (Figure 1A). At-FANCM comprises 1344 amino acids, and it contains an N-terminal helicase domain consisting of DEXDc and HELICc domains, similar to its animal and fungal homologs (Figure 1B; see Supplemental Figure 1A and Supplemental Data Set 1 online). The C-terminal XPF endonuclease-like domain that is conserved in animal FANCM homologs is missing in At-FANCM and other plant and fungal homologs analyzed (see Supplemental Figure 1A online).
Figure 1.
Gene and Protein Structure of At-FANCM.
(A) At-FANCM is split into 23 exons and 22 introns with a length of 7085 bp from start to stop codon. T-DNA insertions of mutant lines fancm-1, fancm-2, and fancm-3 were detected in introns 2, 15, and 18, respectively. Large black arrows point in the direction of left border sequences. Small numbered arrows refer to the primers used in genotyping. Primers 7 and 8 were used to assess presence of At-FANCM genomic sequence downstream of the insertion of line fancm-3.
(B) The At-FANCM protein has a length of 1344 amino acids (aa) and contains a bipartite helicase domain composed of a DEXDc and a HELICc domain at its N terminus.
(C) Detailed analysis of the T-DNA insertion loci. Genomic sequences are shown in bold type, and sequence duplications are underlined. Insertion of foreign sequences is in italic, deletions are named “Del,” and their respective length is given.
Analysis of microarray expression data of the probe set identifier 262036_at using the Arabidopsis eFP browser showed elevated expression of At-FANCM in shoot apical meristem development, seed stages 9 and 10, but also in flower stages 9 to 11, as well as in stamens in stages 12 and 15 (Winter et al., 2007). Furthermore, in a coexpressed gene network constructed by ATTED-II (Obayashi et al., 2009), the meiotic genes At-SPO11-1, At-SYN3/At-RAD21.2 (for radiation sensitive21.1), and At-PRD1 (for putative recombination initiation defect1) (among others) were directly connected with At-FANCM, indicating a high degree of coexpression.
We obtained three predicted At-FANCM T-DNA insertion lines from the SALK collection (Alonso et al., 2003), named fancm-1 (SALK_069784), fancm-2 (SALK_025917), and fancm-3 (SALK_151218), and characterized them in detail (Figure 1C). The insertion sites of all three mutant lines were verified by sequencing of the gene/T-DNA junctions at both ends, except for fancm-3, where the downstream junction could not be amplified, but amplification of an adjacent region indicated that no large rearrangements occurred due to the T-DNA insertion. The insertions of fancm-1, fancm-2, and fancm-3 reside in introns 2, 15, and 18, respectively. Real-time quantitative PCR expression analysis of the fancm mutant lines tested transcription in regions of the At-FANCM ORF 5′, 3′, and across the T-DNA insertions. Across each T-DNA insertion, only minor levels of expression could be found in mutant lines. 5′ of the T-DNA insertions, fancm-2 and fancm-3 were expressed at similar levels as Columbia-0 (Col-0), while in fancm-1, expression was reduced to a third of Col-0. 3′ of the T-DNA insertions, expression of both fancm-1 and fancm-3 was much lower than Col-0, at 0.05 and 0.12 times, respectively. At this 3′ region, expression of fancm-2 was ∼2 times higher than Col-0, although with a high variance (see Supplemental Figure 2 online). Expression of gene fragments might lead to translation into protein fragments, which might possess residual activity. In either case, expression of a full-length FANCM protein can be excluded because of the inserted T-DNA.
Since insertion of T-DNAs into the Arabidopsis genome might lead to reciprocal chromosome translocations with a probability of 20% (Clark and Krysan, 2010), we tested all three fancm mutant lines by backcrossing homozygous plants with their wild-type Col-0 and assessing pollen viability in the respective heterozygous F1 progeny. If a translocation occurred, 50% of pollen should be inviable (Curtis et al., 2009), which we tested by esterase activity staining with fluorescein diacetate (Heslop-Harrison and Heslop-Harrison, 1970). None of the three fancm mutant lines showed pollen viability in the range of 50%. In fact, the pollen viability of heterozygous fancm-1, fancm-2, and fancm-3 was at ∼80%.
At-FANCM Does Not Appear to Be Implicated in Somatic DNA Repair
Similar to other members of the FA pathway, animal FANCM has been shown to be involved in the repair of ICLs. Animal fancm mutant cells display sensitivity to cross-linking agents (e.g., MMC and cisplatin). By contrast, ICL repair does not appear to be a major role for budding yeast Mph1, since its mutants are more sensitive to genotoxins like the methylating agent MMS. We therefore tested At-FANCM for a potential role in a number of DNA repair pathways. One-week-old plantlets were grown in the presence of different concentrations of genotoxins for a further 2 weeks. Impaired DNA repair in the mutant would result in slower growth and a lower fresh weight of the mutant plantlets compared with the wild type. Surprisingly, the T-DNA insertion lines fancm-1, fancm-2, and fancm-3 did not show any difference in sensitivity to MMC, cisplatin, or MMS compared with wild-type plantlets (see Supplemental Figure 3 online). Contrary to its animal and fungal homologs, At-FANCM does not seem to have a direct function in the repair of these types of DNA damage. Furthermore, we also could not detect increased sensitivity to bleomycin, camptothecin, hydroxyurea, or raltitrexed (see Supplemental Figure 3 online).
At-FANCM Defines an RECQ4A-Independent Pathway for the Suppression of Spontaneous HR
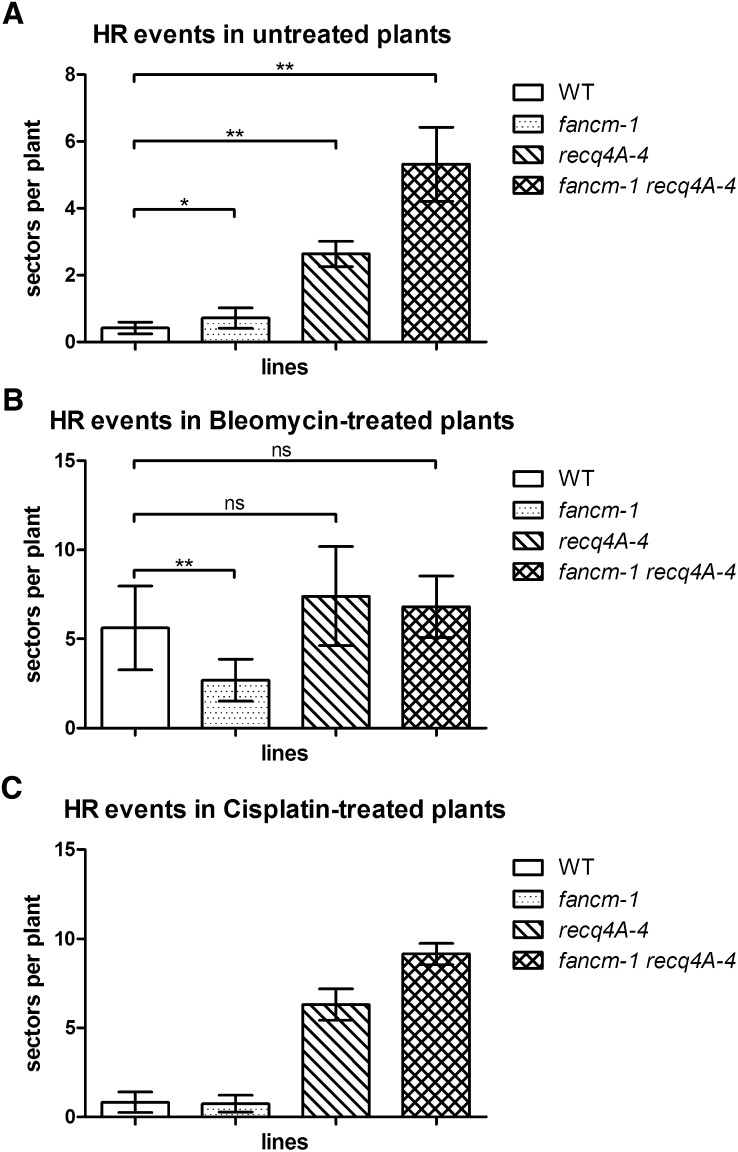
Previously, we have shown that the RecQ family helicase At-RECQ4A, a homolog of human BLM (for Bloom syndrome helicase) and yeast Sgs1 helicases, suppresses spontaneous HR events in somatic cells together with its partners At-RMI1 and At-TOP3A (Hartung et al., 2007a, 2008). To perform a similar analysis, we crossed fancm-1 mutant plants with the HR reporter line IC9 (Molinier et al., 2004). Interestingly, At-FANCM also seems to suppress spontaneous HR events because fancm-1 plants showed a slightly elevated rate of HR compared with the wild type (Figure 2A; 0.715 versus 0.414, P = 0.026, n = 6). The elevation was not as high as in recq4A-4 plants. To define whether or not At-FANCM is epistatic to At-RECQ4A, we produced the double mutant fancm-1 recq4A-4, which displayed an increase in HR above both single mutants (Figure 2A; recq4A-4 versus fancm-1 recq4A-4: 2.633 versus 5.313, P = 0.0022, n = 6). Thus, At-FANCM is not epistatic to At-RECQ4A with respect to CO suppression; both proteins seem to act in independent pathways.
Figure 2.
Somatic HR Frequencies.
Recombination frequencies in the IC9 recombination reporter background.
(A) Spontaneous HR frequencies in untreated plants were slightly but significantly higher in fancm-1 compared with the wild type (WT) (mean 0.715 versus 0.414 sectors per plant). In recq4A-4, the HR frequency was higher still with 2.633 spp. With 5.313 spp, the double mutant fancm-1 recq4A-4 had a HR frequency significantly higher than either single mutant, indicating two parallel pathways of HR suppression.
(B) Following induction of DSBs by 5 µg/mL bleomycin, the mean HR frequency of wild-type plants was at 5.62 spp and that of recq4A-4 was not significantly different from the wild type with 7.398 spp. In fancm-1, there was a significant reduction in HR frequency to 2.685 spp compared with the wild type, while the double mutant had 6.8 spp and was not different from the wild type or recq4A-4.
(C) Treatment with cisplatin induces DNA ICLs. Induction of HR with 3 µM cisplatin increased the HR rate of wild-type plants to 0.82 spp (compared with 0.414 spp uninduced). Similar to the uninduced assay, the fancm-1 recq4A double mutant exhibits a synergistic increase in HR.
At-FANCM Is Required for DSB-Induced HR in Somatic Cells
After the induction of DSBs by treatment with bleomycin, the HR rate in fancm-1 was significantly lower than in the wild type (Figure 2B; 2.685 versus 5.62, P = 0.0047, n = 6). This is contrary to the results for fancm-1 without DSB induction and shows that At-FANCM promotes HR events following a DSB. This also indicates that spontaneous HR events detected with the assay system used here do not necessarily occur to repair DSBs. As published earlier (Hartung et al., 2007a), the HR rate in recq4A-4 was not different from the wild type after DSB induction by bleomycin. The HR rate of the fancm-1 recq4A-4 double mutant also was not different from recq4A-4 or the wild type in this experiment (Figure 2B).
To test whether DNA damaging agents that are not capable of directly inducing DSBs have a similar effect on HR, we used the genotoxic agent cisplatin that reacts with bases in DNA to form covalent cross-links. These in turn, if unrepaired, can lead to stalled replication forks and subsequent repair processes that include nucleotide excision repair and HR (De Silva et al., 2000; Räschle et al., 2008). Exposure of wild-type plants to 3 µM cisplatin increased the HR rate by approximately twofold (Figure 2C). Interestingly, in the fancm-1 recq4A-4 double mutant cisplatin treatment led to a HR rate that was higher than that of the respective single mutants and similar to the results in the uninduced assay (Figure 2C). At-FANCM and At-RECQ4A therefore function in two separate pathways to suppress HR to repair DNA cross-link lesions. Furthermore, this result might indicate that the nature of spontaneous HR events as quantified in the uninduced assays is due to naturally occurring DNA cross-links than DSBs.
Reduced Fertility in fancm Mutants Is Due to a Defect in Meiosis
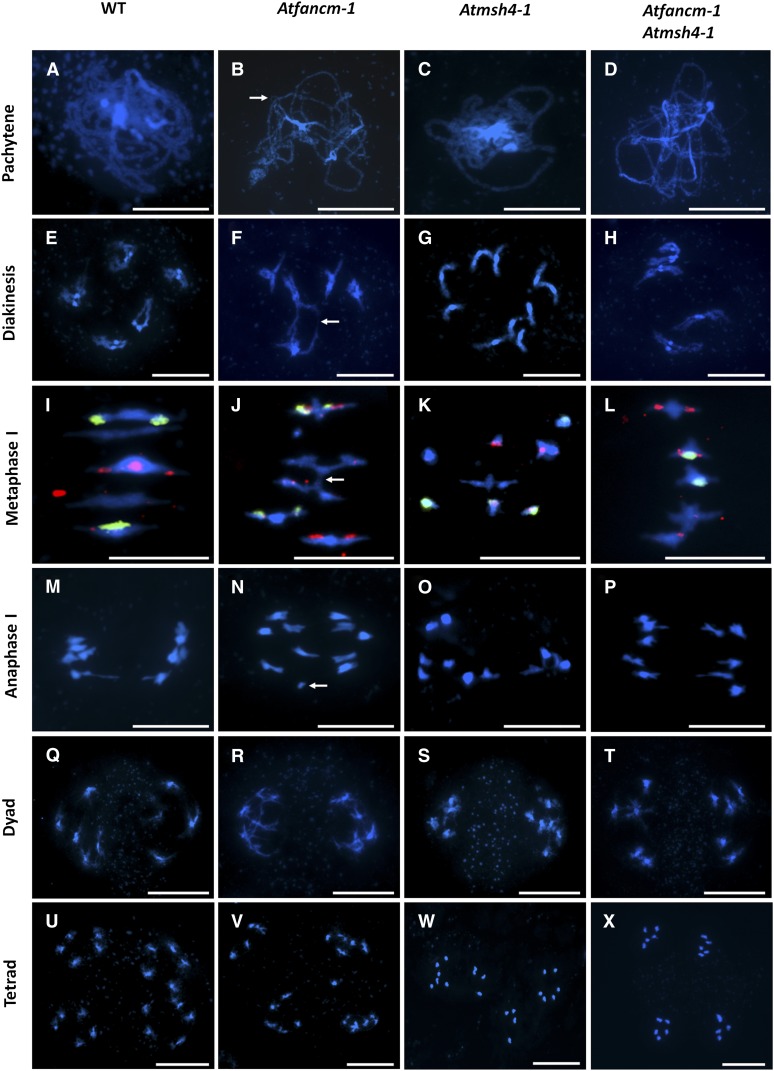
Arabidopsis produces ∼50 seeds per silique in the wild type. During the growth of fancm mutant plants, we noticed a reduction in the seed set by all three mutant lines. The mean seed set in fancm-1 was 41.36 (n = 10) compared with 51.48 (n = 10) in the wild type. To investigate the basis of the fertility defect, we analyzed 4′,6-diamidino-2-phenylindole (DAPI)-stained chromosome spread preparations from fancm-1 pollen mother cells undergoing meiosis using fluorescence microscopy. Early meiotic stages from G2 through leptotene to zygotene appeared normal in the mutant. In wild-type pachytene nuclei, the homologous chromosomes were fully synapsed, held in close apposition along their entire length by the synaptonemal complex (SC), which forms during zygotene and reaches completion at the onset of pachytene (Figure 3A). However, in fancm-1, we routinely observed meiocytes that based on the degree of chromosome condensation were at pachytene but where bivalent formation was incomplete, which could indicate a synapsis defect (Figure 3B). Evidence of chromosomal breaks and interlocks, where synapsis of two chromosomes is topologically hindered by other DNA strands, was also apparent (Figure 3B, arrow). During diakinesis and metaphase I in both wild-type and fancm-1, homologous chromosomes were visible as five bivalents linked by chiasmata (Figures 3E, 3F, 3I, and 3J). Interbivalent connections were also prevalent in the mutant, whether these all arose through recombination or were due to unresolved interlocks or chromatin stickiness that is sometimes observed was difficult to discern (Figures 3F and 3J, arrow). Following the first division at anaphase I, chromosome fragments were sometimes observed in the mutant indicative of a defect in recombinational repair (Figure 3N, arrow). Evidence of fragmentation was also visible at the tetrad stage following the second meiotic division (Figure 3V). A survey of 50 meiocytes at metaphase I revealed that the frequency of interchromosome connections per nucleus for fancm-1 was 0.52, compared with 0.11 in the wild type.
Figure 3.
Representative Meiotic Stages from Pollen Mother Cells.
DAPI-stained chromatin spreads of wild-type (WT) ([A], [E], [I], [M], [Q], and [U]), fancm-1 ([B], [F], [J], [N], [R], and [V]), msh4-1 ([C], [G], [K], [O], [S], and [W]), and fancm-1 msh4-1 ([D], [H], [L], [P], [T], and [X]) meiocytes. Metaphase I stages are additionally labeled with 45S (green) and 5S (red) fluorescence in situ hybridization probes to distinguish chromosomes. In pachytene ([A] to [D]), incomplete synapsis, chromatin breaks, and chromosomal interlocks ([B], arrow) are visible in fancm-1 as well as the double mutant. In diakinesis ([E] to [H]), bivalents form in all lines except msh4-1. In fancm-1 and the double mutant, however, connections between bivalents are often detected. Such chromatin bridges are also visible in metaphase I ([I] to [L]) nuclei of fancm-1 ([J], arrow) and fancm-1 msh4-1, leading to unequal distribution of chromosomes in anaphase I ([M] to [P], arrow), which is also visible at the tetrad stage ([U] to [X]). Bars = 10 µm.
Loss of At-FANCM Compromises the Formation of SC and Late Recombination Intermediates
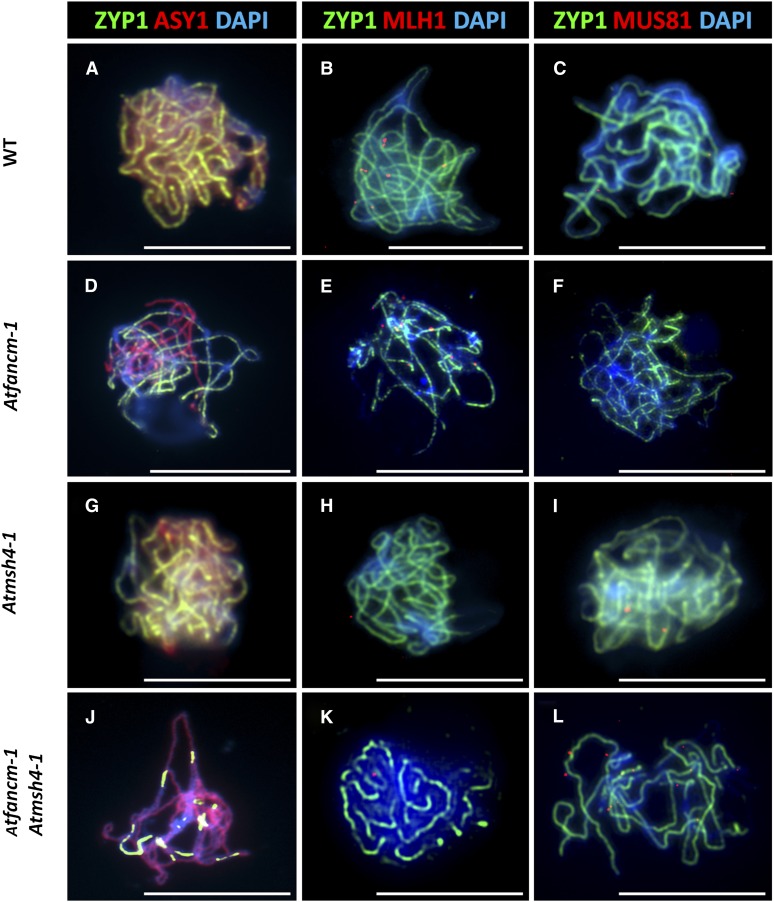
To further analyze the meiotic phenotype of fancm-1, immunolocalization studies were conducted. Localization of the axial element protein At-ASY1 (for asynaptic1) at leptotene in fancm-1 was indistinguishable from the wild type, suggesting chromosome axis formation is normal in the mutant (Figures 4A and 4D). SC formation can be monitored by immunostaining of the transverse filament protein At-ZYP1 (Higgins et al., 2005). Here, a striking difference between the wild type and fancm-1 meiocytes was observed. In wild-type meiocytes at pachytene, At-ZYP1 forms a continuous signal along the synapsed chromosomes, which is accompanied by a marked depletion in the ASY1 staining (Figure 3A). In corresponding fancm-1 nuclei, the ZYP1 signal was generally incomplete, with ASY1 remaining on the unsynapsed regions (Figure 3D). This could indicate that fancm-1 meiocytes fail to complete synapsis possibly due to the observed chromosomal interlocks and interchromosome connections that might slow prophase I progression. This was supported by the fact that wild-type buds of ∼600 µm had completed meiosis and contained only tetrads and pollen, whereas at this stage, fancm-1 buds contained cells at stages ranging from zygotene through to tetrads and pollen.
Figure 4.
Dual Immunolocalization of Meiotic Proteins ZYP1, ASY1, MLH1, and MUS81 in Pachytene Stage Male Meiocytes.
Representative cells of the wild type (WT) ([A] to [C]), fancm-1 ([D] to [F]), msh4-1 ([G] to [I]), and fancm-1 msh4-1 ([J] to [L]) are shown. SC axial element protein ZYP1 is stained green in all cells ([A] to [L]). Detection of SC lateral element ASY1 ([A], [D], [G], and [J]; red) shows incomplete synapsis in fancm-1 and fancm-1 msh4-1 ([D] and [J]). Staining of MLH1 ([B], [E], [H], and [K]) or MUS81 ([C], [F], [I], and [L]) in red shows a reduction of MLH1 foci in msh4-1 and fancm-1 msh4-1 and an increase of MUS81 foci in Atfancm-1 msh4-1. Chromatin is also stained with DAPI. Bars = 10 µm.
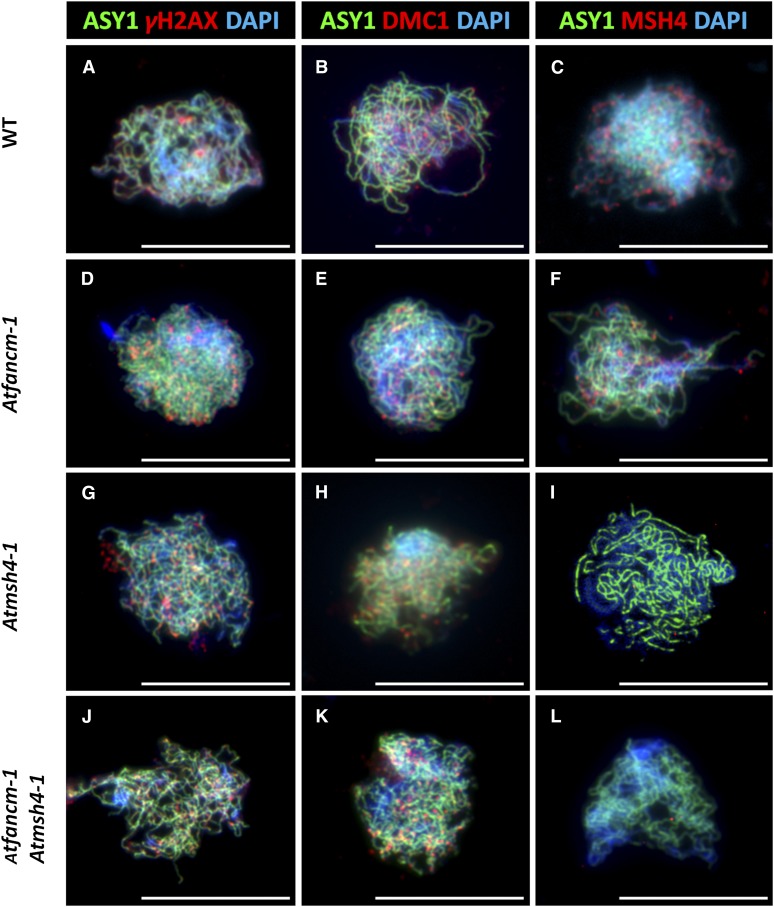
To try to establish the cause of synaptic defect, we monitored recombination in fancm-1. Although the direct detection of DSBs is thus far not possible in Arabidopsis, DSB formation can be inferred from the number of γH2AX foci that form in early leptotene. γH2AX is the phosphorylated form of the DSB repair specific histone variant H2AX that accumulates at the sites of DSBs (Rogakou et al., 1999). Similarly, the number of foci corresponding to the strand-exchange proteins At-DMC1 and At-RAD51 also reflects the number of recombination initiation events (Ferdous et al., 2012). Our analysis revealed that the number of γH2AX, DMC1, and RAD51 foci in fancm-1 and wild-type meiocytes at leptotene was not significantly different (Figures 5A, 5B, 5D, and 5E; mean number of foci fancm-1 versus the wild type [n = 5]; γH2AX 142.2 versus 160.8, P = 0.24; DMC1 148.2 versus 141.0, P = 0.44; RAD51 146.8 versus 143.8, P = 0.79). This suggests that DSB formation and the early stages in recombination are unaffected in fancm-1. This is consistent with the observation that ASY1 localization in the mutant is normal, since defects in axis morphogenesis can affect DSB formation (Schwacha and Kleckner, 1997; Xu et al., 1997; Ferdous et al., 2012). We next investigated the localization of the MutS homolog At-MSH4, which based on biochemical studies of the human protein, is thought to act in a complex with At-MSH5 to stabilize progenitor HJs (Snowden et al., 2004). There was no apparent difference in the number of MSH4 foci that associated with the chromosome axes at leptotene in fancm-1 compared with the wild type (Figures 5C and F; 154.6 versus 140.0, P = 0.48, n = 5). In both cases, this was followed by a gradual decrease in the number of foci through to pachytene (data not shown). We then analyzed the localization of At-MLH1 in fancm-1 and wild-type meiocytes at pachytene. The MutL homolog At-MLH1 is a component of late recombination nodules, and the number of MLH1 foci corresponds to the number of interference-sensitive COs (Marcon and Moens, 2003; Jackson et al., 2006; Chelysheva et al., 2010). In the wild type, the mean number of MLH1 foci per cell was 9.8 with a range of 8 to 12 (Figure 4B). In fancm-1, there was a slight reduction in the mean number of MLH1 foci to 9.1 with a range of 6 to 12 (Figure 4E). This suggested that a small proportion of interfering COs are lost in the mutant.
Figure 5.
Dual Immunolocalization of Meiotic Proteins ASY1, γH2AX, DMC1, and MSH4 in Pollen Mother Cells.
Representative cells of the wild type (WT) ([A] to [C]), fancm-1 ([D] to [F]), msh4-1 ([G] to [I]), and fancm-1 msh4-1 ([J] to [L]) are shown. SC lateral element protein ASY1 is stained green in all cells ([A] to [L]). Numbers of DSB marker γH2AX ([A], [D], [G], and [J]; red) and recombinase DMC1 ([B], [E], [H], and [K]; red) are similar in all lines. MSH4 protein foci ([C], [F], [I], and [L]; red) can be detected in the wild type and fancm-1 in similar quantities but not in msh4-1 or the double mutant. Chromatin is also stained with DAPI. Bars = 10 µm.
Analysis of At-FANCM Function within Meiotic Pathways
To analyze the basis of the defects in synapsis and meiotic recombination observed in fancm-1 in more detail, the mutant was crossed to several recombination pathway mutants.
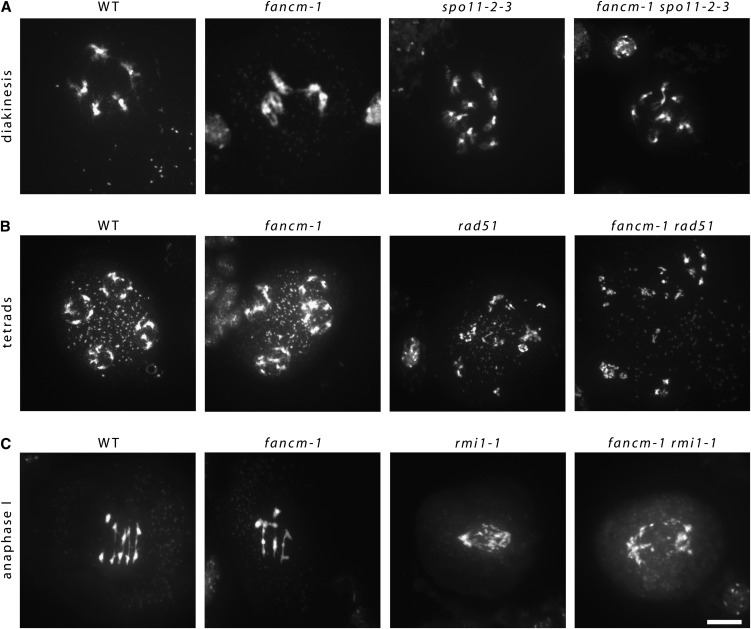
To determine if the chromosome fragments observed in fancm-1 arose in meiotic S-phase rather than from a meiotic recombination defect, we constructed an fancm-1 spo11-2-3 double mutant. The spo11-2-3 mutant displays intact univalents in meiosis I that are randomly distributed due to the lack of DSBs to initiate meiotic recombination and is therefore virtually sterile (Figure 6A) (Hartung et al., 2007b). The fancm-1 spo11-2-3 double mutant was also practically sterile, while the number of seeds per silique in fancm-1 was lower than the wild type but higher than either spo11-2-3 or the double mutant. Cytogenetic analysis of fancm-1 spo11-2-3 meiocytes revealed univalents in meiosis I with no evidence of chromosome interactions or fragmentation (Figure 6A). Thus, the fact that spo11-2-3 is able to suppress the fancm-1 phenotype indicates that At-FANCM has a meiotic function following DSB formation.
Figure 6.
Epistasis Analysis of Meiosis Phenotypes.
Chromatin spreads of double mutant fancm-1 spo11-2-3, fancm-1 rad51, and fancm-1 rmi1-1 as well as the respective single mutant and wild-type (WT) meiocytes. Shown are informative meiotic stages.
(A) and (B) Diakinesis meiocytes of the wild type and fancm-1 are paired into five bivalents (A), while there are 10 univalents visible in corresponding meiocytes of spo11-2-3 as well as the double mutant line fancm-1 spo11-2-3 (B). In rad51 mutant meiocytes, SPO11-induced DSBs cannot be repaired, which results in a random segregation of chromatin fragments seen in tetrads. Similarly, in the double mutant fancm-1 rad51, chromatin fragmentation is detectable, but not in fancm-1 or wild-type meiocytes.
(C) In the wild type and fancm-1 anaphase I, homologous chromosomes are pulled toward opposite poles. In rmi1-1, there are unresolved recombination intermediates between homologous chromosomes that result in chromosome bridges and fragmentation. Similar defects can be found in the double mutant fancm-1 rmi1-1. Bar = 10 µm.
Studies have shown that loss of At-MSH4, At-MSH5, or At-MLH3, the functional partner of At-MLH1, slows meiotic progression in Arabidopsis, but SC formation is otherwise normal (Higgins et al., 2004, 2008b; Jackson et al., 2006). By contrast, mutations affecting the strand-exchange proteins At-RAD51 and At-DMC1 prevent SC formation (Sanchez-Moran et al., 2007). Since substantial SC formation occurs in fancm-1 and localization of DMC1 and RAD51 appeared normal, it seems likely that At-FANCM functions downstream of the strand-exchange proteins. Consistent with this, analysis of an fancm-1 rad51 double mutant revealed that rad51 suppressed the fancm-1 phenotype since extensive chromosome fragmentation was observed (Figure 6B). We next analyzed a fancm-1 msh4-1 double mutant. Although SC formation is normal, albeit delayed, in a msh4-1 mutant, loss of the gene results in a strong reduction in fertility due to a dramatic reduction in CO formation (Higgins et al., 2004, 2008b; Lu et al., 2008). Surprisingly, fancm-1 msh4-1 homozygous plants produced considerably more seeds per silique than msh4-1 (19.88 versus 4.0, respectively), although this was not as high as fancm-1 (41.36). Thus, mutation of At-FANCM partially rescued the fertility defect of msh4-1. Cytogenetical analysis of chromosome spread preparations from fancm-1 msh4-1 meiocytes showed that the increased fertility relative to msh4-1 was associated with an increase in chiasmata, but as in fancm-1, SC formation appeared incomplete (Figure 3D). This suggested that loss of At-FANCM suppresses the msh4-1 recombination phenotype.
Topoisomerase 3α and RMI1 are essential during meiosis in Arabidopsis, where they are thought to dissolve a subset of dHJs into NCO products (Chelysheva et al., 2008; Hartung et al., 2008). In the At-rmi1-1 mutant, extensive chromosome fragmentation occurs at the metaphase I to anaphase I transition when the unrepaired recombination intermediates that have accumulated break under division spindle tension. This leads to meiotic arrest and complete sterility. The fancm-1 rmi1-1 double mutant was also sterile. Cytogenetic analysis of fancm-1 rmi1-1 meiocytes showed that the meiotic phenotype of the double mutant was indistinguishable from that of rmi1-1 in anaphase I (Figure 6C). Furthermore, we did not observe any meiosis II meiocytes in the fancm-1 rmi1-1 double mutant, as was already reported for rmi1 single mutants (Chelysheva et al., 2008; Hartung et al., 2008). Our results support these earlier studies that suggest At-RMI1 is important for removing a subset of dHJs during pachytene. They are also consistent with the immunolocalization studies that reveal that the number of At-MLH1 foci, which are thought to mark CO sites, is only slightly reduced in fancm-1 (see below).
Analysis of CO/Chiasma Formation in fancm-1 msh4-1 Double Mutants
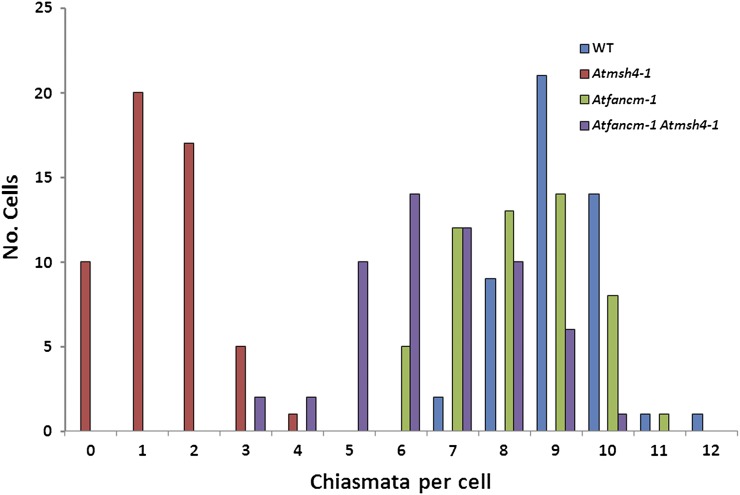
To investigate the nature of COs in fancm-1 msh4-1, we conducted a thorough analysis of chiasma formation in the various mutant lines. Chiasma formation was determined in metaphase I meiocytes following fluorescence in situ hybridization with 45S and 5S rDNA probes to allow identification of the individual bivalents (Figures 3I to 3L) (Sanchez-Moran et al., 2002). In the wild type, the mean chiasma frequency was 9.13 (Figure 7; ±0.14 se; n = 50). The number of chiasmata per cell fell in a range from 7 to 12 with over 90% in the 8 to 10 class. In the initial cytogenetical analysis of fancm-1, we noted that although five bivalents were present at metaphase I, the number of chiasmata associated with these appeared reduced in comparison to the wild type. Analysis of 50 nuclei confirmed a modest yet significant reduction in the mean chiasma frequency to 8.19 (Figure 7; ±0.18 se; P < 0.001). This figure was consistent with the number of MLH1 foci observed at pachytene (see above). Although up to 11 chiasmata were observed in a single fancm-1 nucleus, there was a general shift to a lower and broader range than that observed in the wild type with as few as six chiasmata observed in five of the nuclei in the analysis. Nevertheless, univalents were not observed in the sample surveyed, suggesting that despite the reduction in chiasmata, the obligate CO is maintained (Jones, 1984; Jones and Franklin, 2006). Consistent with this and in common with the wild type, the chiasma distribution in fancm-1 deviated significantly from a Poisson distribution [X(11)2 = 34.7, P > 0.001], suggesting that CO interference still operates in this mutant line. Analysis of msh4-1 revealed a mean chiasma frequency of 1.38 (Figure 7), which is very similar to that previously recorded for this mutant (Higgins et al., 2004). The mean chiasma frequency for the fancm-1 msh4-1 mutant was 6.6 (Figure 7; ±0.21 se). Moreover, the chiasma frequency for msh4-1 in this study and previously (Higgins et al., 2004, 2008a, 2008b) fits a Poisson distribution, whereas the data for the fancm-1 msh4-1 double mutant does not [X(10)2 = 17.6, P = 0.07]. This indicates that chiasma distribution in fancm-1 msh4-1 is not numerically random and is overdistributed around the mean, similar to the wild type. Chiasmata frequency for the double mutant was significantly lower than the fancm-1 mutant (P < 0.001). Inspection of the number of chiasmata in individual nuclei revealed a substantial overlap in the range compared with the single mutant. However, in contrast with fancm-1, where the cells with only six chiasmata all contained five bivalents, in the double mutant univalents were detected in 28.6% of the cells with this number of chiasmata. Unsurprisingly, this increased to 70% in the fancm-1 msh4-1 cells with only five chiasmata.
Figure 7.
Distribution of Chiasmata Numbers per Cell in the Wild Type, msh4-1, fancm-1, and fancm-1 msh4-1.
Chiasmata numbers were counted in metaphase I cells stained with DAPI as well as 5S and 45S fluorescence in situ hybridization probes in the wild type (WT; blue), msh4-1 (red), fancm-1 (green), and the double mutant fancm-1 msh4-1 (purple). The number of chiasmata was slightly reduced in fancm-1 compared with the wild type. The strongly reduced chiasmata count of msh4-1 could be partially rescued by further mutation of At-FANCM, as can be seen in the respective double mutant. Interestingly, in fancm-1, univalents were never observed, although as few as six chiasmata were counted in some cells, indicating that an obligate CO per chromosome is maintained.
In Arabidopsis zmm mutants, a small proportion of COs remain, some of which are dependent on At-MUS81 (Higgins et al., 2008a). Immunolocalization of MUS81 in wild-type plants revealed the presence of around 150 foci on chromosome spreads at leptotene, suggesting that the protein is associated with the majority if not all recombination initiation events. The number of MUS81 foci reduced through zygotene such that by mid-late pachytene the mean number was 1.6 (n = 18; Figure 4C). A similar set of events occurs in fancm-1 such that by pachytene the mean number of foci was 1.7 (n = 17; Figure 4F), consistent with the hypothesis that the MUS81 foci observed in pachytene correspond to the sites of some or all of the ZMM-independent COs. Since the number of COs in fancm-1 msh4-1 was significantly increased relative to msh4-1, we determined if there was a coordinate effect on the number of MUS81 foci. This proved to be the case, as we observed an increase in the mean number of MUS81 foci at pachytene to 7.9 (n = 16) per cell in the double mutant (Figure 4L). Immunolocalization with an anti-At-MLH1 antibody revealed that the mean number of MLH1 foci in the double mutant was not substantially different from that in msh4-1 (Figures 4H and 4K; 1.6 versus 0.9, P = 0.054). This suggests that the low number of COs dependent on MUS81in wild-type plants must be due to a restriction on that pathway that is lifted in the absence of fancm mutant plants. Therefore, At-FANCM seems to suppress recombination intermediates that would lead into the MUS81-dependent pathway. That there is no concomitant decrease of MLH1 foci in fancm might indicate a more complex situation than a simple switch between two CO-producing pathways that is regulated by FANCM.
DISCUSSION
How Did ICL Repair Evolve?
In vertebrates, the FA proteins are essential for the recognition and repair of DNA ICLs. The loss of any one FA protein renders the cell hypersensitive to ICL inducing agents. To date, in budding yeast, only one homolog of 15 FA proteins, Mph1, appears conserved, but it does not share the ICL repair function with the human homolog. Here, we report that the Arabidopsis homolog of human FANCM and budding yeast Mph1, At-FANCM, has no direct function in the repair of DNA ICLs. Moreover, it is also not involved in the repair of other kinds of lesions like alkylation damage by MMS, as is the case with Mph1. Hence, it seems that the sole FA homolog that is conserved in the eukaryotic kingdoms of plants, fungi, and animals has functionally diverged.
The whole FA protein complement seems to be present only in vertebrates, since in other animals and plants, just a subset of FA homologs is conserved: FANCM, FANCL (for Fanconi anemia complementation group L), FANCD2, FANCD1 (for Fanconi anemia complementation group D1), FANCJ (for Fanconi anemia complementation group J), and FANCO (for Fanconi anemia complementation group O) (reviewed in Patel and Joenje, 2007; Knoll and Puchta, 2011). Interestingly, direct evidence of enzymatic function in animals has been reported only for the conserved subset of FA proteins. Our analysis of the plant FANCM homolog indicates that the specialized ICL repair function of the FA proteins was an evolutionarily recent acquisition within the animal lineage, perhaps linked with the origin of the full FA protein complement. This argument is supported by observations of ICL repair pathways outside the animal kingdom that are unrelated to FA homologs. Our recent description of three independent pathways to repair cisplatin-induced lesions in Arabidopsis, dependent on At-RECQ4A, At-MUS81, and At-RAD5A (for radiation hypersensitive5A), respectively, affirms this hypothesis (Mannuss et al., 2010). In the light of these results, it will be interesting to explore the function of further nonanimal FA homologs to determine if they too have a role other than ICL repair. Another interesting question arises from the observation that the core complex proteins are not widely conserved. How do the few ancestrally conserved FA homologs, most importantly FANCM on the one hand and FANCD2/FANCI on the other hand, work together without protein interactions via the core complex to link them? Additionally, the absence of the C-terminal XPF family endonuclease domain in FANCM homologs outside of vertebrates poses the question if this domain strongly modulates the observed differences in function between these homologs.
Antagonistic Functions of At-FANCM in Somatic HR
Although At-FANCM does not seem to be directly involved in repair of ICLs, we show here that it functions in the regulation of spontaneous as well as bleomycin-induced HR in somatic cells and also HR following DNA cross-link damage. Loss of At-FANCM leads to an increase of spontaneous HR events, indicating a suppressive function. This is a phenotype similar to that of an mph1 mutant in yeast, which also displays a slightly elevated HR rate (Schürer et al., 2004). Also similar to results in budding yeast, the spontaneous HR rate is strongly increased in mutants of At-RECQ4A, the Arabidopsis homolog of the RecQ helicase Sc-SGS1 (Gangloff et al., 1994; Hartung et al., 2007a). In both yeast and Arabidopsis, double mutants of Sc-MPH1 and Sc-SGS1 or At-FANCM and At-RECQ4A, respectively, display an increase in their HR rate above that of either single mutant. At-FANCM and At-RECQ4A therefore seem to function in two parallel pathways that suppress HR events. In yeast mph1 mutants, there has been no report on the HR rate after induction of DSBs. Here, we show that At-fancm mutant plants have a lower than wild-type HR rate after treatment with the DSB-inducing agent bleomycin. Contrary to spontaneous HR events, At-FANCM seems to promote HR in DSB repair. This raises a question as to the role of spontaneous HR events, if it is not DSB repair. We addressed this question by studying HR rates following induction with cisplatin. Contrary to the bleomycin results, induction of HR by the cross-linking agent cisplatin leads to phenotypes similar to these in the uninduced assays, with At-FANCM and At-RECQ4A acting in two distinct HR-suppressing pathways. This indicates naturally occurring DNA cross-linking reactions (e.g., during cell metabolism) as a source of HR reactions in uninduced assays. It was recently shown that DNA adduct and cross-link–forming aldehydes, either endogenously produced or exogenously applied, can lead to genotoxic lesions that are repaired via the FA pathway in chicken DT40 cells as well as mice (Langevin et al., 2011; Rosado et al., 2011). Plant metabolism is a ready source of a large number of aldehydes that could lead to adducts and cross-links in plant nuclear DNA that need to be repaired via HR (Marnett, 1999; Hirayama et al., 2004; Wei et al., 2009). Furthermore, animal FA proteins as well as yeast Mph1 have been linked with repair and recombination events at stalled replication forks, especially ICL repair at forks in the FA case. In plant somatic cells, conservative HR is a minor DSB repair pathway, with nonhomologous end joining and single-strand annealing providing the majority of repair events (Siebert and Puchta, 2002; Puchta, 2005). However, in S-phase, at stalled replication forks in particular, HR has to occur since DNA breaks will lead to a single broken DSB end that cannot be repaired by nonhomologous end joining. Thus, structural differences of the recombination intermediates might be the basis of the phenotypic difference of fancm HR rates: While At-FANCM promotes HR in DSB repair, it suppresses HR following DNA cross-links and/or stalled replication forks.
At-FANCM Functions Downstream of Meiotic DSBs
Aside from BRCA2 (for breast cancer susceptibility gene2) (FANCD1), no clear data have been presented yet on a role of other FA proteins in germ cells or meiosis, although there are indications that there might still be undiscovered functions. In FA patients as well as FA model organisms (including FANCM), gonadal development is usually compromised; it is thought to be caused by endocrine disorders, though (Trivin et al., 2007; Bakker et al., 2009). Similarly, diploid mph1 mutant yeast cells have been shown to exhibit a sporulation defect, while spore survival is not different from the wild type (Scheller et al., 2000). The authors of that study concluded that budding yeast Mph1 could not have an important role in meiosis. Nevertheless, in this study, we have shown that At-FANCM is involved in meiotic CO formation in Arabidopsis.
Since animal FANCM and yeast Mph1 function to repair damaged replication forks, one might anticipate that the meiotic phenotype of the At-fancm-1 mutant could be due to unrepaired damage during meiotic S-phase. However, based on the fertility and meiotic phenotypes of the double mutants, this seems unlikely. Double mutants of At-fancm with either At-spo11-2 or At-rad51 show that the fancm phenotype is hypostatic to the respective second mutant. This clearly indicates that At-FANCM functions after At-SPO11-1/SPO11-2–dependent induction of DSBs and single-strand invasion of the donor strand by single-stranded DNA-RAD51 (and single-stranded DNA-DMC1) filaments. The extensive SC formation observed in the fancm-1 mutant also supports this conclusion.
At-FANCM Is Required for Coupling Meiotic Recombination and Synapsis
Meiotic CO formation is a highly controlled process that ensures at least one, obligate CO per bivalent. Multiple COs on the same chromosome are for the most part spaced apart due to the imposition of CO interference, the basis of which remains to be determined (Jones, 1984; Jones and Franklin, 2006; Berchowitz and Copenhaver, 2010). CO designation occurs at an early stage in the recombination pathway, with formation dependent on the ZMM proteins and normally coupled to SC formation (Börner et al., 2004).
In budding yeast, the pro-CO activity of the ZMMs has been shown to antagonize the anti-CO activity of the Sgs1 helicase. The role of Sgs1 in the formation of COs initially emerged from studies in the BR strain of budding yeast where a sgs1 mutant was reported to exhibit a modest increase of 1.2- to 1.4-fold in crossing over in allelic intervals (Rockmill et al., 2003). Studies in the SK1 strain did not detect a statistically significant effect on allelic COs over several genetic intervals that were analyzed, but an increase in ectopic crossing was observed, leading the authors to conclude that loss of Sgs1 has, at most, a modest effect on CO formation (Jessop et al., 2006). Molecular analysis of recombination intermediates from the sgs1 mutant using two-dimensional gel analysis revealed the formation of aberrant multichromatid joint molecules that can also be visualized by electron microscopy of DNA isolated from the mutant (Oh et al., 2007). The strong CO defect observed in zmm mutants is suppressed by sgs1 (Jessop et al., 2006; Oh et al., 2007). Physical analysis of recombination intermediates from a msh5 mutant revealed that the formation of interhomolog dHJs was delayed relative to the wild type and did not accumulate to the same level. These defects were suppressed by the sgs1 mutation. Thus, it was concluded Sgs1 possesses a strong anticrossover activity that prevents aberrant recombination and is antagonized by pro-crossover factors, such as Msh4/Msh5 and Mlh1/Mlh3 (Jessop et al., 2006; Oh et al., 2007).
Unlike budding yeast, it is not possible to conduct a direct physical analysis of joint molecules formed during meiosis in Arabidopsis. Nevertheless, in many respects, our data suggest that the role of At-FANCM during meiosis may be quite similar to that of Sc-Sgs1. In particular, the detection of aberrant At-FANCM–dependent ectopic connections, together with the phenotypes of the At-msh4 mutant and fancm-1 msh4 double mutant, support this view. It is suggested that the ZMM proteins are components of late recombination nodules that protect CO-designated intermediates from the antirecombination activity of Sgs1 and other helicases (Börner et al., 2004; Fung et al., 2004; Jessop et al., 2006). In support of this, loss of Sgs1 in yeast does not affect the formation of NCOs, which are thought to diverge at an earlier stage in the recombination pathway (Allers and Lichten, 2001; Börner et al., 2004). Data presented here are consistent with a similar role for At-FANCM. In common with budding yeast, most COs in wild-type Arabidopsis are interference sensitive. As a result, their numerical distribution is nonrandom and does not fit a Poisson distribution (Higgins et al., 2004, 2008b). In the fancm-1 msh4-1 double mutant, chiasmata/COs are restored to ∼70% wild-type level. Immunolocalization studies suggest that unlike the wild type, they do not appear to be dependent on At-MLH1, but arise via the activity of At-MUS81. In the wild type, interference insensitive COs, some of which require MUS81, account for ∼15% of the total and exhibit a Poisson distribution (Higgins et al., 2008a). By contrast, the numerical distribution of the COs in fancm-1 msh4-1 deviates from a Poisson distribution. An attractive explanation for this phenomenon is that they arise from intermediates that are CO designated, but in the absence of a functioning ZMM pathway, CO imposition is absent such that some are not repaired as COs. As a result, an obligate CO is not assured, and although five bivalents are often present, a proportion of meiocytes contain univalents at metaphase.
One slight difference between At-fancm-1 and Sc-sgs1 is the effect that loss of the proteins has on allelic COs. In Sc-sgs1, any effect is marginal and subject to strain variability (see above) (Jessop et al., 2006). Loss of At-FANCM resulted in a significant, albeit small, reduction in the mean number of chiasmata relative to the wild type (8.19 versus 9.13). Poisson distribution analysis revealed that COs in the fancm mutant were not randomly distributed, suggesting they were interference sensitive. Cells containing either six or seven chiasmata were more frequent in fancm-1 compared with the wild type (17 versus 2); nevertheless, univalents were not observed in these cells. This would suggest a tendency to maintain an obligate CO on each bivalent and given an overall reduction in CO number, an apparent increase in CO interference. If so, the underlying reason remains unclear; nevertheless, it may be a symptom of disrupting the normal coupling between recombination and SC formation in fancm-1. The link between meiotic recombination and synapsis has previously been observed in other mutants. For example, recombination pathway mutants, such as At-msh4 and At-msh5, exhibit both a reduction in COs and a delay in SC formation (Higgins et al., 2004, 2008b; Jackson et al., 2006). Loss of the SC component At-ZYP1 results in a synaptic failure coupled with a modest overall reduction in COs and a tendency for ectopic COs (Higgins et al., 2005). Although COs are formed at approaching wild-type levels in fancm-1, our study reveals a recombination defect that results in interchromosomal connections, interlocks, and fragmentation. These are accompanied by a failure to form complete SCs. Recent studies in Sordaria macrospora have also reported that synapsis is prevented by stalled recombination events associated with interlocked regions (Storlazzi et al., 2010). Thus, our studies are a further illustration of the close coupling that occurs between meiotic recombination and chromosome synapsis.
Functional Divergence of Helicases in Meiosis
That At-FANCM and Sc-Sgs1 may perform related functions sheds light on a puzzle arising from previous studies of the RecQ helicase At-RECQ4A (Hartung et al., 2007a; Higgins et al., 2011). In common with Sc-Sgs1, At-RECQ4A is related to the human BLM helicase (Hartung et al., 2007a). Analyses of At-RECQ4A in vegetative cells revealed functional similarity to Sc-Sgs1 (Hartung et al., 2006, 2007a, 2008). Surprisingly, however, this did not extend to their meiotic role. It appears that loss of At-RECQ4A results in a relatively mild meiotic defect due to the formation of chromatin bridges between the telomeres of nonhomologous chromosomes (Higgins et al., 2011). This functional divergence is intriguing but may be quite common among the helicases. It will be of the utmost interest to elucidate whether the human FANCM homolog has a role in meiotic recombination as well.
In Caenorhabditis elegans, the RTEL-1 helicase functions as a meiotic antirecombinase, such that an increase in COs occurs in its absence. Evidence indicates that Ce-RTEL-1 may promote synthesis-dependent strand annealing by disassembling D-loop intermediates (Youds et al., 2010). Interestingly, budding yeast lacks an RTEL-1 homolog but one exists in Arabidopsis (Knoll and Puchta, 2011). Hence, it will be of interest to determine if the plant protein is also required during meiosis.
METHODS
Primers Used in Quantitative PCR Expression Analysis
Primer pairs flanking each T-DNA insertion in lines fancm-1, fancm-2, and fancm-3 as well as the corresponding regions in wild-type Col-0 were used in quantitative PCR reactions to assess expression levels across the insertion. Further regions 5′ and 3′ of all insertions were amplified in all lines (see Supplemental Figure 2 online): region 5′, FANCM-9 (5′-TCGTCATCCCATTTCACTC-3′) and FANCM-10 (5′-GCTGCCTCAGGATCAATC-3′); region T-DNA fancm-1, FANCM-9 and FANCM-11 (5′-GCAAAGCCACCAATGTATTC-3′); region T-DNA fancm-2, FANCM-12 (5′-CAACGGATGGGAAGAACTG-3′) and FANCM-13 (5′-GCAATGTCTGGAAGTGAGG-3′); region T-DNA fancm-3, FANCM-14 (5′-TGTTGGAGAAATTGTGTTATC-3′) and FANCM-15 (5′-CTCTCCTGCCAATTCGTTA-3′); region 3′, FANCM-16 (5′-GATGTCGGCTGATGAGAAC-3′) and FANCM-8 (5′-GTAATGGTGACTGGCTGAG-3′). Reference genes used in this study were At3g18780 (ACT2) and At4g34270 (Czechowski et al., 2005), amplified with primer pairs ACT2-fw (5′-ATTCAGATGCCCAGAAGTCTTGTTC-3′) and ACT2-rev (5′-GCAAGTGCTGTGATTTCTTTGCTCA-3′) or At4g34270-F1 (5′-AGATGAACTGGCTGACAATG-3′) and At4g34270-R1 (5′-TGTTGCTTCTCTCCAACAGT-3′), respectively. Expression levels of fancm lines were calculated relative to Col-0 in each region, respectively, after normalization to the geometric mean of the reference genes.
Primers Used in Amplification of At-FANCM cDNA Fragments
Starting from an oligo(dT)18 reverse-transcribed cDNA pool of 2-week-old Arabidopsis thaliana seedlings, overlapping fragments of At-FANCM were amplified and sequenced. The primer pairs for the fragments were as follows: FANCM-17 (5′-CCGCCATTCTCTGTGTCTC-3′) and FANCM-2 (5′-CCTCAATCTGCTGCATCAC-3′); FANCM-1 (5′-GGATCTAGGGTTCCAATAG-3′) and FANCM-18 (5′-AGCATATGCGTCTCTGCAG-3′); FANCM-19 (5′-GCTAATGTATCTCCTCTGAG-3′) and FANCM-20 (5′-CTCCAATCCTCAGGAATAG-3′); FANCM-21 (5′-AAGCACCTTAGAGACAACAG-3′) and FANCM-22 (5′-CTTAAAGGGTTCAACGAATTG-3′). Primers FANCM-2, FANCM-1, FANCM-19, and FANCM-21 were used for sequencing of the fragments.
Although we were not able to establish the 5′ and 3′ ends of the cDNA by rapid amplification of cDNA ends, we were able to amplify PCR fragments with primers located 150 bp upstream of the start and 108 bp downstream of the stop codon, respectively. Primers located a short distance further outside did not amplify a fragment, indicating that we were able to characterize the ORF almost completely.
Primers Used for PCR-Based Genotyping of T-DNA Insertion Lines
Two primer pairs were used to genotype each T-DNA insertion line. One primer pair was located up- and downstream of the insertion site to detect wild-type loci, while the other primer pair contained one genomic and one T-DNA primer to detect the T-DNA. In fancm-1, the wild-type PCR was done with primers FANCM-1 and FANCM-2, while the T-DNA PCR was done with primers FANCM-1 and LBd1 (5′-TCGGAACCACCATCAAACAG-3′). In fancm-2, the wild-type PCR was done with primers FANCM-3 (5′-GTCGCAACATCTATTGGTG-3′) and FANCM-4 (5′-TAACCCTCAGACCTGTATC-3′), while the T-DNA PCR was done with primers FANCM-4 and LBd1. In fancm-3, the wild-type PCR was done with primers FANCM-5 (5′-GGAAGTCAACACATCACAG-3′) and FANCM-6 (5′-GTCCTGTTCTCGTAGTGG-3′), while the T-DNA PCR was done with primers FANCM-5 and LBd1. To assess the integrity of the At-FANCM locus 3′ of the fancm-3 T-DNA insertion, a fragment was amplified with primers FANCM-7 (5′-TAACGAATTGGCAGGAGAG-3′) and FANCM-8.
Plant Handling and Growth Conditions
Plants were grown in a greenhouse under constant 22°C with a light phase of 16 h and a dark phase of 8 h, except for genotoxin and HR assays. For crosses, excess flower buds and already formed siliques were removed from prospective mother plants. Then, sepals, petals, and stamens were dissected from flower buds. The remaining gynoecium was pollinated by application of mature stamens from the father plant to the stigma. Seeds from crosses were propagated through F1 and F2 generations and genotyped by PCR with primers for the respective loci. F3 seeds were usually used for experiments.
Seeds of T-DNA insertion lines were obtained from the SALK, GABI, or SAIL collections (Sessions et al., 2002; Alonso et al., 2003; Rosso et al., 2003).
For assays, seeds were surface sterilized in 4% NaOCl and stratified at 4°C overnight. Germination occurred on plates containing solid germination medium (GM). Plantlets were grown in CU-36L4 growth chambers (Percival Scientific) with a light phase of 16 h at 22°C and a dark phase of 8 h at 20°C. Genotoxins used in these assays were bleomycin sulfate, MMC (both Duchefa Biochemie), cisplatin, hydroxyurea, MMS (Sigma-Aldrich Chemie), and raltitrexed (AK Scientific).
RNA Extraction and cDNA Cloning
Two-week-old seedlings of Arabidopsis were frozen in liquid nitrogen. Total RNA was then extracted using the RNeasy plant mini kit (Qiagen). A cDNA pool was constructed with an oligo(dT)18 primer and the RevertAid First-Strand cDNA synthesis kit (Fermentas). Sequencing of the fragments was performed by GATC Biotech. Final assembly of the cDNA sequence was done in SeqMan 5.03 (DNASTAR).
Somatic Genotoxin Sensitivity and HR Assays
Both assays were performed as recently described (Hartung et al., 2007a). For the sensitivity assay, 1-week-old plantlets were transferred to six-well plates containing 5 mL liquid GM medium each, supplemented with genotoxins of different concentrations. Growth continued for 13 d, after which the plantlets were dried with paper towels and fresh weight was established with an analytical balance.
For the HR assay, 50 1-week-old plantlets containing the desired mutation as well as the IC9 HR reporter construct (Molinier et al., 2004) were transferred to halved Petri dishes containing 10 mL liquid GM medium, one-half supplemented with genotoxins. Growth continued for 8 d, followed by the GUS staining reaction for 2 d at 37°C and an extraction of plant pigments in 70% ethanol at 65°C overnight. Blue sectors were scored using a binocular microscope.
Cytological Methods
Preparations of Arabidopsis male meiocytes were performed as described (Armstrong et al., 2009). The primary antibodies used in this study were anti-ASY1 (Armstrong et al., 2002), anti-ZYP1 (Higgins et al., 2005), anti-RAD51 (Mercier et al., 2003), and anti-MUS81 (Higgins et al., 2008a). In the case of simultaneous staining of proteins with primary antibodies from the same species, the first primary antibody was covered with labeled Fab fragments before application of the second primary antibody (Seeliger et al., 2012). Analysis of chiasmata and statistical analysis was performed as described (Sanchez-Moran et al., 2002; Higgins et al., 2004).
Statistical Methods
For statistical analysis of HR data, two-tailed P values of pairwise comparisons of the data acquired in the different plant lines were performed with the Mann-Whitney test with a confidence interval of 95%. The reported P values are either exact values or Gaussian approximations.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JQ278026. The Arabidopsis Genome Initiative locus identifier of At-FANCM is At1g35530.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis of FANCM Homologs.
Supplemental Figure 2. Expression Analysis of the At-FANCM Locus in fancm Mutant Lines.
Supplemental Figure 3. Genotoxin Sensitivity Assays.
Supplemental Data Set 1. Sequence Alignment Used to Produce Supplemental Figure 1B.
Supplementary Material
Acknowledgments
We thank Nancy Kleckner for insightful discussions and Mandy Meier and Sabrina Wagner for technical assistance. This work was funded by the European Research Council Advanced Grant “COMREC” and Deutsche Forschungsgemeinshaft Grant Pu 137-11 to H.P. Work in the Franklin laboratory is funded by Biotechnology and Biological Science Research Council.
AUTHOR CONTRIBUTIONS
A.K., J.D.H., F.C.H.F., and H.P. designed the research. A.K., J.D.H., K.S., S.J.R., N.J.D., M.B., and S.S. performed research. A.K., J.D.H., F.C.H.F., and H.P. analyzed data. A.K., J.D.H., F.C.H.F., and H.P. wrote the article.
Glossary
- FA
Fanconi anemia
- MMC
mitomycin C
- ICL
interstrand cross-link
- HR
homologous recombination
- MMS
methyl methanesulfonate
- HJ
Holliday junctions
- NCO
noncrossover
- CO
crossovers
- DSB
double-strand break
- dHJ
double-Holliday junction
- ORF
open reading frame
- DAPI
4′,6-diamidino-2-phenylindole
- SC
synaptonemal complex
- GM
germination medium
References
- Allers T., Lichten M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Sanchez-Moran E., Franklin F.C. (2009). Cytological analysis of Arabidopsis thaliana meiotic chromosomes. In Methods in Molecular Biology: Meiosis: Cytological Methods, Vol. 2, S. Keeney, ed (New York: Humana Press; Springer), pp. 131–145. [DOI] [PubMed]
- Bakker S.T., van de Vrugt H.J., Rooimans M.A., Oostra A.B., Steltenpool J., Delzenne-Goette E., van der Wal A., van der Valk M., Joenje H., te Riele H., de Winter J.P. (2009). Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum. Mol. Genet. 18: 3484–3495 [DOI] [PubMed] [Google Scholar]
- Berchowitz L.E., Copenhaver G.P. (2010). Genetic interference: Don’t stand so close to me. Curr. Genomics 11: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner G.V., Kleckner N., Hunter N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Grandont L., Vrielynck N., le Guin S., Mercier R., Grelon M. (2010). An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129: 143–153 [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Vezon D., Belcram K., Gendrot G., Grelon M. (2008). The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet. 4: e1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.A., Krysan P.J. (2010). Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 64: 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis S.J., Ciccia A., Deans A.J., Horejsí Z., Martin J.S., Maslen S.L., Skehel J.M., Elledge S.J., West S.C., Boulton S.J. (2008). FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell 32: 313–324 [DOI] [PubMed] [Google Scholar]
- Curtis M.J., Belcram K., Bollmann S.R., Tominey C.M., Hoffman P.D., Mercier R., Hays J.B. (2009). Reciprocal chromosome translocation associated with TDNA-insertion mutation in Arabidopsis: Genetic and cytological analyses of consequences for gametophyte development and for construction of doubly mutant lines. Planta 229: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva I.U., McHugh P.J., Clingen P.H., Hartley J.A. (2000). Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20: 7980–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter J.P., Joenje H. (2009). The genetic and molecular basis of Fanconi anemia. Mutat. Res. 668: 11–19 [DOI] [PubMed] [Google Scholar]
- Ede C., Rudolph C.J., Lehmann S., Schürer K.A., Kramer W. (2011). Budding yeast Mph1 promotes sister chromatid interactions by a mechanism involving strand invasion. DNA Repair (Amst.) 10: 45–55 [DOI] [PubMed] [Google Scholar]
- Fanconi G. (1927). Familiäre infantile perniziosaartige Anämie (perniziöses Blutbild und Konstitution). In Jahrbuch für Kinderheilkunde (Berlin: Karger), pp. 257–280.
- Ferdous M., Higgins J.D., Osman K., Lambing C., Roitinger E., Mechtler K., Armstrong S.J., Perry R., Pradillo M., Cuñado N., Franklin F.C.H. (2012). Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8: e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung J.C., Rockmill B., Odell M., Roeder G.S. (2004). Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802 [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. (1994). The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., Décaillet C., Delannoy M., Wu L., Constantinou A. (2008b). Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. USA 105: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., Décaillet C., Stasiak A.Z., Stasiak A., Constantinou A. (2008a). The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- Geuting V., Kobbe D., Hartung F., Dürr J., Focke M., Puchta H. (2009). Two distinct MUS81-EME1 complexes from Arabidopsis process Holliday junctions. Plant Physiol. 150: 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M., Vezon D., Gendrot G., Pelletier G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Puchta H. (2000). Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 28: 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H. (2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34: 4438–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Knoll A., Wurz-Wildersinn R., Puchta H. (2008). Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007a). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Wurz-Wildersinn R., Fuchs J., Schubert I., Suer S., Puchta H. (2007b). The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J., Heslop-Harrison Y. (1970). Evaluation of pollen viability by enzymatically induced fluorescence; intracellular hydrolysis of fluorescein diacetate. Stain Technol. 45: 115–120 [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Armstrong S.J., Franklin F.C., Jones G.H. (2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Buckling E.F., Franklin F.C., Jones G.H. (2008a). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Ferdous M., Osman K., Franklin F.C. (2011). The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 65: 492–502 [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Vignard J., Mercier R., Pugh A.G., Franklin F.C., Jones G.H. (2008b). AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55: 28–39 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Fujishige N., Kunii T., Nishimura N., Iuchi S., Shinozaki K. (2004). A novel ethanol-hypersensitive mutant of Arabidopsis. Plant Cell Physiol. 45: 703–711 [DOI] [PubMed] [Google Scholar]
- Jackson N., Sanchez-Moran E., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C. (2006). Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 25: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Rockmill B., Roeder G.S., Lichten M. (2006). Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.H. (1984). The control of chiasma distribution. Symp. Soc. Exp. Biol. 38: 293–320 [PubMed] [Google Scholar]
- Jones G.H., Franklin F.C. (2006). Meiotic crossing-over: Obligation and interference. Cell 126: 246–248 [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Klimyuk V.I., Jones J.D. (1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11: 1–14 [DOI] [PubMed] [Google Scholar]
- Knoll A., Puchta H. (2011). The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 62: 1565–1579 [DOI] [PubMed] [Google Scholar]
- Langevin F., Crossan G.P., Rosado I.V., Arends M.J., Patel K.J. (2011). Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475: 53–58 [DOI] [PubMed] [Google Scholar]
- Li W., Chen C., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Liu X., An L., Zhang W., Sun J., Pei H., Meng H., Fan Y., Zhang C. (2008). The Arabidopsis MutS homolog AtMSH5 is required for normal meiosis. Cell Res. 18: 589–599 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S., Luke B., Grossi S., Constantinou A. (2010). FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 29: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannuss A., Dukowic-Schulze S., Suer S., Hartung F., Pacher M., Puchta H. (2010). RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E., Moens P. (2003). MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165: 2283–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett L.J. (1999). Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 424: 83–95 [DOI] [PubMed] [Google Scholar]
- Meetei A.R., et al. (2005). A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37: 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Armstrong S.J., Horlow C., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C. (2003). The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K., Endt D., Hoehn H., Schindler D. (2009). Genotype-phenotype correlations in Fanconi anemia. Mutat. Res. 668: 73–91 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37(Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.D., Lao J.P., Hwang P.Y., Taylor A.F., Smith G.R., Hunter N. (2007). BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K., Higgins J.D., Sanchez-Moran E., Armstrong S.J., Franklin F.C. (2011). Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 190: 523–544 [DOI] [PubMed] [Google Scholar]
- Panico E.R., Ede C., Schildmann M., Schürer K.A., Kramer W. (2010). Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast 27: 11–27 [DOI] [PubMed] [Google Scholar]
- Patel K.J., Joenje H. (2007). Fanconi anemia and DNA replication repair. DNA Repair (Amst.) 6: 885–890 [DOI] [PubMed] [Google Scholar]
- Prakash R., Krejci L., Van Komen S., Anke Schürer K., Kramer W., Sung P. (2005). Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 280: 7854–7860 [DOI] [PubMed] [Google Scholar]
- Puchta H. (2005). The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Räschle M., Knipscheer P., Enoiu M., Angelov T., Sun J., Griffith J.D., Ellenberger T.E., Schärer O.D., Walter J.C. (2008). Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134: 969–980 Erratum. Cell 137: 972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Fung J.C., Branda S.S., Roeder G.S. (2003). The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13: 1954–1962 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon C., Redon C., Bonner W.M. (1999). Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado I.V., Langevin F., Crossan G.P., Takata M., Patel K.J. (2011). Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat. Struct. Mol. Biol. 18: 1432–1434 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Armstrong S.J., Santos J.L., Franklin F.C., Jones G.H. (2002). Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E., Santos J.L., Jones G.H., Franklin F.C. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21: 2220–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J., Schürer A., Rudolph C., Hettwer S., Kramer W. (2000). MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürer K.A., Rudolph C., Ulrich H.D., Kramer W. (2004). Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics 166: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R.A., Blackford A.N., Niedzwiedz W. (2010). ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 29: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. (1997). Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135 [DOI] [PubMed] [Google Scholar]
- Seeliger K., Dukowic-Schulze S., Wurz-Wildersinn R., Pacher M., Puchta H. (2012). BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 193: 364–375 [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Shinohara M. (2004). Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet. Genome Res. 107: 201–207 [DOI] [PubMed] [Google Scholar]
- Siebert R., Puchta H. (2002). Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T., Acharya S., Butz C., Berardini M., Fishel R. (2004). hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Stacey N.J., Kuromori T., Azumi Y., Roberts G., Breuer C., Wada T., Maxwell A., Roberts K., Sugimoto-Shirasu K. (2006). Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 48: 206–216 [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Gargano S., Ruprich-Robert G., Falque M., David M., Kleckner N., Zickler D. (2010). Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Nandi S., Osman F., Ahn J.S., Jakovleska J., Lorenz A., Whitby M.C. (2008). The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell 32: 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver T.L., Rothstein R.J., Stahl F.W. (1983). The double-strand-break repair model for recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- Trivin C., Gluckman E., Leblanc T., Cousin M.N., Soulier J., Brauner R. (2007). Factors and markers of growth hormone secretion and gonadal function in Fanconi anemia. Growth Horm. IGF Res. 17: 122–129 [DOI] [PubMed] [Google Scholar]
- Wei Y., Lin M., Oliver D.J., Schnable P.S. (2009). The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem. 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Weiner B.M., Kleckner N. (1997). Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11: 106–118 [DOI] [PubMed] [Google Scholar]
- Youds J.L., Mets D.G., McIlwraith M.J., Martin J.S., Ward J.D., ONeil N.J., Rose A.M., West S.C., Meyer B.J., Boulton S.J. (2010). RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.