This study describes a conditional RNA interference system that enabled the knockdown of essential mitotic genes along with high-resolution live microscopy. Loss-of-function analyses of augmin and γ-TuRC constitute a proof of principle and provide insight into acentrosomal microtubule formation during plant mitosis.
Abstract
Mitosis is a fundamental process of eukaryotic cell proliferation. However, the molecular mechanisms underlying mitosis remain poorly understood in plants partly because of the lack of an appropriate model cell system in which loss-of-function analyses can be easily combined with high-resolution microscopy. Here, we developed an inducible RNA interference (RNAi) system and three-dimensional time-lapse confocal microscopy in the moss Physcomitrella patens that allowed in-depth phenotype characterization of the moss genes essential for cell division. We applied this technique to two microtubule regulators, augmin and γ-tubulin complexes, whose mitotic roles remain obscure in plant cells. Live imaging of caulonemal cells showed that they proceed through mitosis with continual generation and self-organization of acentrosomal microtubules. We demonstrated that augmin plays an important role in γ-tubulin localization and microtubule generation from prometaphase to cytokinesis. Most evidently, microtubule formation in phragmoplasts was severely compromised after RNAi knockdown of an augmin subunit, leading to incomplete expansion of phragmoplasts and cytokinesis failure. Knockdown of the γ-tubulin complex affected microtubule formation throughout mitosis. We conclude that postanaphase microtubule generation is predominantly stimulated by the augmin/γ-tubulin machinery in moss and further propose that this RNAi system serves as a powerful tool to dissect the molecular mechanisms underlying mitosis in land plants.
INTRODUCTION
Mitotic cell division is a classic example that emphasizes differences between the plant and animal kingdoms (Inoué and Sato, 1967). Although animals use centrosomes as dominant microtubule (MT) nucleation centers, land plants lack these structures (Murata et al., 2007; Marshall, 2009). Extended central spindle MTs are characteristic structures observed after sister chromatid separation in animals, which ensure cytokinesis. Land plants have seemingly different structures called phragmoplasts, which are assembled at the center of the segregating chromosomes during telophase and ensure cell plate formation (Lambert and Bajer, 1972; Otegui et al., 2005). However, the extent to which plant mitosis is different from that in animals is unclear (Lloyd and Chan, 2006).
A key mitotic event in every eukaryote is the formation of spindle MTs, and several molecular players have been identified in animal cells. At the centrosome, a potent MT nucleator, the γ-tubulin ring complex (γ-TuRC), is recruited and activated by the centrosomin family proteins (Drosophila melanogaster centrosomin, mammalian CDK5RAP2) and the polo-like kinase (Megraw et al., 2001; Goshima et al., 2007; Dobbelaere et al., 2008; Choi et al., 2010). In addition, at least two acentrosomal mechanisms are present in animal somatic cells that induce γ-TuRC–dependent spindle MT generation independent of centrosomes. The first is the chromatin-dependent mechanism in which chromosome-associated RCC1 (for regulator of chromosome condensation1) or Aurora B kinase induces MT generation around chromosomes by modulating the activities of several MT binding proteins, including TPX2 (for target protein for Xklp2), HURP (hepatoma upregulated protein), and kinesin-13s (Walczak and Heald, 2008). The other, which was more recently proposed, is the augmin-mediated mechanism in which existing spindle MTs recruit the eight-subunit protein complex augmin that further recruits γ-TuRC for new MT generation (Goshima et al., 2008; Goshima and Kimura, 2010). Therefore, the latter mechanism would ensure MT amplification that is analogous to the MT-dependent MT nucleation found in plant and yeast cells during interphase (Janson et al., 2005; Murata et al., 2005). A recent experiment suggested that all three of these mechanisms also contribute to postanaphase MT formation in human HeLa cells (Uehara and Goshima, 2010). However, unlike the centrosomin family proteins, the role of augmin in MT generation has not yet been firmly established. For example, biochemical evidence that augmin activates γ-TuRC for MT nucleation is still missing. Furthermore, a null mutant of augmin showed defective chromosome alignment and spindle morphology but did not show defective spindle MT assembly per se during female meiosis in Drosophila, in which centrosomes are naturally absent (Meireles et al., 2009).
In plant cells, the mechanism of spindle/phragmoplast MT generation is less understood. Land plants not only lack centrosomes but also lack the centrosomin family members or some chromatin pathway factors that are critical in animals (e.g., HURP). Augmin genes are present in the plant genome, and a mutant has been recently characterized in Arabidopsis thaliana (Ho et al., 2011). This mutant showed spindle and phragmoplast MT organization defects in microspores, but similar to Drosophila meiosis, it was not evident whether MT generation was impaired (Ho et al., 2011).
We reasoned that the mechanism of acentrosomal mitosis in plants or animal oocytes has been less completely explored partly because cell biological analyses, such as high-resolution microscopy combined with gene perturbation, are more difficult to apply. For example, although the tobacco (Nicotiana tabacum) bright yellow-2 cell line has been an excellent system for filming mitosis (e.g., Granger and Cyr, 2000; Kumagai et al., 2001; Hayashi et al., 2007; Kurihara et al., 2008; Smertenko et al., 2011), the current unavailability of full-genome sequences makes it more challenging to perform systematic gene knockdown analyses using RNA interference (RNAi) or gene targeting. The genetic approach has been predominantly undertaken in Arabidopsis, and a number of mitotic genes have been identified accordingly (e.g., Whittington et al., 2001; Ambrose et al., 2005; Bannigan et al., 2007). However, phenotypic characterization, particularly in living cells, is technically not so easy as in animal cell lines.
We began this study by searching for a potent plant cell type, which led to the idea that the moss Physcomitrella patens might be an ideal model system for studying mitosis. P. patens has 27 chromosomes, and its draft genome sequences (∼500 Mbp) are available (Rensing et al., 2008). It spends most of its life cycle in a haploid state, and several key molecular techniques, such as homologous recombination or RNAi, can be applied (Bezanilla et al., 2003, 2005; Cove et al., 2006; Prigge and Bezanilla, 2010). Caulonemal cells in protonemata, which are filamentous structures that appear after spore germination, experience less autofluorescence-related interference derived from chloroplasts; therefore, they are suitable for the live imaging of fluorescent protein-tagged markers (Hiwatashi et al., 2008; Vidali et al., 2009a, 2009b). To date, RNAi analyses of essential genes have been based on transient transformation with a plasmid from which double-stranded RNAs (dsRNAs) are constitutively produced. This has led to the identification of actin regulators of tip growth of chloronemal cells, another protonemata cell type, that appear immediately after protoplast transformation (Vidali et al., 2007, 2009b, 2010; Augustine et al., 2008). However, mitosis is an infrequent event in the chloronemal cell cycle (>24 h) (Cove, 2005), and RNAi and mitotic imaging have not yet been successfully combined using this approach. Moreover, if dsRNAs are constitutively induced (Harries et al., 2005), RNAi transgenic lines cannot be selected for the genes that are essential for moss viability. For the same reason, the gene disruption approach cannot be undertaken for essential genes in haploid moss.
In this study, we developed an inducible RNAi system in P. patens, in which target knockdown was confirmed for all nine tested genes. Using this system, we showed that the augmin protein complex is critical for the generation of MTs from prometaphase to cytokinesis. Interestingly, augmin played a dominant role in the formation of phragmoplast MTs.
RESULTS
Development of an Inducible RNAi System
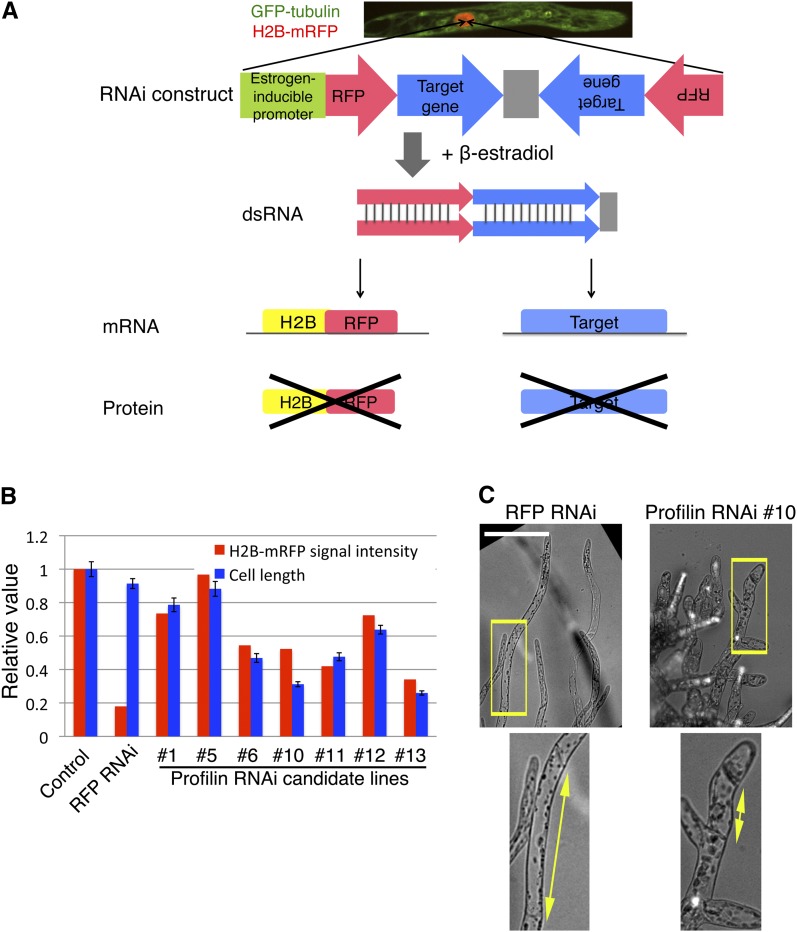
By testing a few possible promoter systems, we successfully developed an inducible RNAi system by introducing the XVE estrogen-inducible system that had been originally established in Arabidopsis (Zuo et al., 2000). In this system, β-estradiol would stimulate dsRNA formation for both the target gene and the RFP (red fluorescent protein) gene, resulting in the coknockdown of the RFP fusion protein and the target protein (Figure 1A; see Supplemental Figure 1A online). This construct was integrated with various copy numbers in moss cells that constitutively expressed green fluorescent protein (GFP)-tubulin and histone-RFP. To deduce the RNAi efficiency of each transgenic line, RFP intensities were quantified using a custom-made computer algorithm (see Supplemental Figure 1B online). Because histone is a very stable protein (Djondjurov et al., 1983), the histone-RFP signal would not completely disappear during several days after RNAi, allowing us to monitor chromosome behavior while RNAi is effective on the target protein (this was indeed the case; see below).
Figure 1.
Inducible RNAi System in the Moss P. patens.
(A) Scheme of the inducible RNAi system. The RNAi elements in the inducible RNAi vector construct were integrated into the genome of a transgenic line coexpressing GFP-tubulin and histone-RFP. Transcription followed by formation of dsRNA targeting histone-RFP, and a target gene was induced upon addition of β-estradiol to the medium. Supplemental Figure 1A online shows a more detailed map of this construct.
(B) Selection of inducible profilin RNAi lines. After selection of seven stable transformants (RNAi candidate lines), cells were treated with β-estradiol for 5 d, followed by measurement of histone-RFP intensity and cell length (the second cell from the apical tip was selected for length measurement) (error bars indicate sd; n = 24 to 36).
(C) Appearance of shorter cells after inducible profilin RNAi (line #10). Enlarged images are displayed at the bottom. Bar = 100 µm.
We first tested the validity of this RNAi system by targeting FtsZ (filamentous temperature–sensitive Z) and profilin genes, which are two of the several genes whose knockdowns are known to cause severe cellular defects in P. patens protonemata (Strepp et al., 1998; Vidali et al., 2007). Using the standard polyethylene glycol–mediated transformation followed by drug resistance–based selection, we obtained 12 and 14 RNAi candidate lines for these two genes and identified five and four lines for which RFP signal was reduced (summarized in Table 1). Interestingly, the mean RFP intensity and the phenotype appearance were correlated in 12 of 12 FtsZ RNAi lines (giant chloroplasts due to division failure) and seven of seven profilin RNAi lines (small cell size due to impaired tip growth) (Figures 1B and 1C; see Supplemental Figures 1C to 1E online). Furthermore, we confirmed a 77 or 90% reduction in FtsZ mRNA level using quantitative real-time PCR (qRT-PCR) (see Supplemental Figure 1F online). We concluded that RNAi worked efficiently for these two genes.
Table 1. Summary of RNAi Transgenic Line Selection.
| Gene | Paralogs (Identity) | RNAi Construct | RFP Intensity Measurement | RFP Signal Reduceda | Imaging | Phenotypes Detectedb |
|---|---|---|---|---|---|---|
| (Vector) | – | – | 11 | 3 | 3 | 0 |
| FtsZ | 2 | 1st | 12 | 5 | 5 | 5 |
| Profilin | 3 | 1st | 14 | 4 | 4 | 4 |
| XMAP215 | 2 (92%) | 1st | 17 | 10 | 6 | 3c |
| γ-Tubulin | 2 (99%) | 1st | 18 | 6 | 2 | 1d |
| 2nd | 23 | 6 | 6 | 4 | ||
| GCP4 | 1 | 1st | 21 | 7 | 2 | 2 |
| 2nd | 25 | 8 | 5 | 3 | ||
| 3rd | 42 | 11 | 10 | 3 | ||
| Mis12 | 1 | 1st | 19 | 8 | 6 | 0e |
| Aug3 | 1 | 1st | 32 | 11 | 6 | 4 |
| 2nd | 23 | 4 | 4 | 3 |
More than 60% reduction compared to the control line was selected in most cases.
We first observed the phenotype of RFP-reduced lines at day 3 or 4 after RNAi induction. When mitotic phenotypes were not observed, imaging was performed at day 7 for some RFP-reduced lines. If more than one line produced mitotic phenotypes at day 7, we did not further pursue the day 7 phenotype assessment for the rest of the lines. Therefore, more lines than listed here might actually produce phenotypes when cultured for a longer duration.
The cell growth phenotype was detected also for another RNAi construct (see Supplemental Table 3 online) that was transiently transformed into protoplasts.
Although only one line was obtained, the rescue experiment confirmed that the phenotype was derived from γ-tubulin knockdown.
This indicates either that Mis12 is nonessential for chromosome segregation in caulonemal cells or that residual Mis12 after RNAi suffice its mitotic function.
We next extended the RNAi targets to five gene families that might produce specific cellular defects (see Supplemental Figure 2A online). XMAP215 (for Xenopus microtubule-associated protein 215kD [Dis1/MOR1]) is an MT polymerase (Whittington et al., 2001; Kinoshita et al., 2002; Brouhard et al., 2008), whereas γ-tubulin and γ-tubulin complex protein4 (GCP4) are subunits of γ-TuRC, a MT nucleator (Figure 2A; see Supplemental Table 1 online) (Moritz and Agard, 2001; Kong et al., 2010). Mis12 (for Minichromosome instability12) is a conserved kinetochore protein (Goshima et al., 2003). Augmin is a recently identified eight-subunit protein complex that binds to MTs and whose knockdown induces various mitotic defects in animal cells (Figure 2A) (Goshima et al., 2008; Wu et al., 2008; Lawo et al., 2009; Uehara et al., 2009; Tsai et al., 2011). Gel filtration chromatography, immunoprecipitation followed by mass spectrometry, and BLAST homology search suggested that P. patens contains an augmin complex with at least five conserved subunits (see Supplemental Figures 2B and 2C online), implying that this multisubunit complex is conserved in plants and animals. We selected the Aug3 subunit of augmin (also called Dgt3 in fly and hDgt3 or HAUS3 in humans; see Supplemental Table 1 online), which is a core subunit and relatively well conserved among species. P. patens has two copies of γ-tubulin and XMAP215, which might function redundantly (amino acid sequences are >92% identical). Therefore, we designed RNA sequences that were highly conserved between the paralogs, which might lead to codepletion (see Supplemental Figure 2A online). To exclude the possibility that a phenotype was derived from the off-target effects of an RNAi construct, a nonoverlapping RNA that targeted a different region of a given gene was also designed for XMAP215, GCP4, and Aug3 (see Supplemental Figure 2A online).
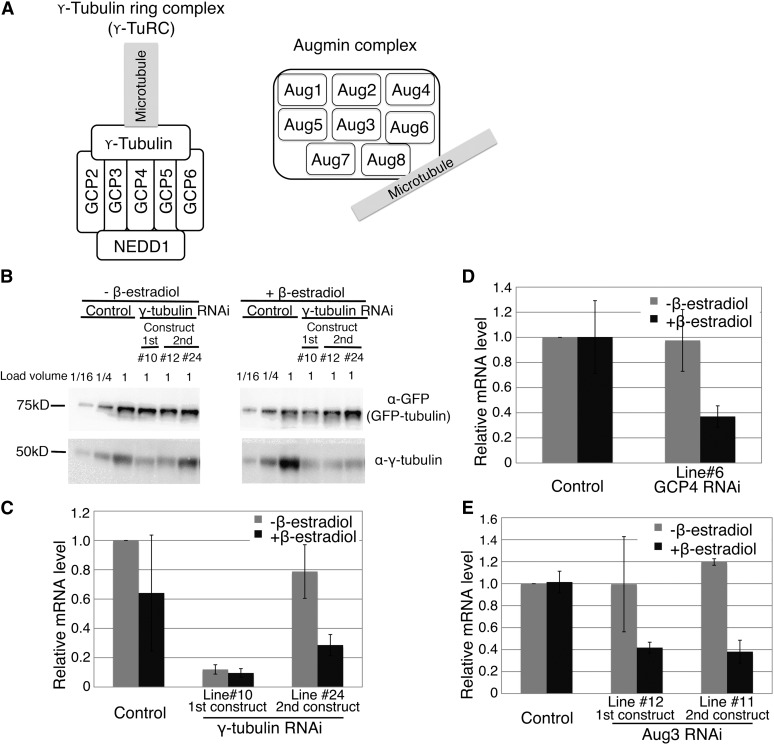
Figure 2.
Confirmation of RNAi Knockdown of γ-TuRC and Augmin Genes.
(A) Schematics of augmin and γ-tubulin ring complexes. It has been shown that the human Hice1/Dgt4/HAUS8 subunit of augmin (depicted here as Aug8) directly binds to MTs (Wu et al., 2008; Tsai et al., 2011), whereas γ-tubulin associates with minus ends of MTs (Moritz and Agard, 2001).
(B) Immunoblotting of γ-tubulin and control GFP-tubulin proteins in the cell extracts of the whole protonemata. Knockdown of the targeted proteins was detected in the RNAi lines. In one case, β-estradiol–dependent RNAi knockdown was clearly observed (line #24), whereas in the other case, further knockdown after β-estradiol addition was not so clear (e.g., line #10). See Supplemental Figures 2D and 2E online for other genes.
(C) to (E) qRT-PCR analysis to quantify mRNA levels of γ-tubulin, GCP4, and Aug3 with or without RNAi induction. Error bars indicate sd of two to three independent experiments.
We obtained four to 11 RNAi candidate lines in which RFP signal was reduced (Table 1). For each gene, a reduction in target protein or mRNA amount was confirmed by immunoblotting (Figure 2B; see Supplemental Figures 2D and 2E online) or qRT-PCR (Figures 2C to 2E), respectively, in the entire moss protonemata. It was likely that RNAi was also working in dividing caulonemal cells, which were used for observation of mitosis (this was confirmed for γ-tubulin by immunostaining; see below). Consistent with the results for FtsZ and profilin, we found a good correlation between RFP signal reduction and the appearance of a specific phenotype for nine of 11 GCP4 RNAi lines (see Supplemental Figure 2F online). Protein/mRNA reduction in the absence of β-estradiol was also observed in some lines (e.g., the first γ-tubulin RNAi construct, clone #10; Figures 2B and 2C); however, mitotic phenotypes were detected only after β-estradiol was added to these lines (see below), suggesting that these lines exist in a leaky RNAi condition (i.e., certain amounts of dsRNAs are expressed without β-estradiol). Further reduction of the protein content by β-estradiol treatment in actively dividing caulonemal cells, which was not clearly detected by bulk immunoblotting/RT-PCR, induced severe mitotic defects.
In summary, we tested nine genes, seven of which had not been characterized in P. patens, and obtained at least three independent RNAi transgenic lines for each. For all the genes, protein/mRNA knockdown was confirmed either directly (using immunoblotting or qRT-PCR for eight genes) or indirectly (appearance of the expected phenotype for the profilin gene). Noticeably, the RNAi line selection process can be completed in 6 weeks (from plasmid transformation to RFP/phenotype observation), which is shorter than the generation time of Arabidopsis (∼12 weeks); moreover, simultaneous handling of multiple genes is possible using a simple transformation protocol. These results are promising for future expansion of this system to other intracellular processes or to dozens of uncharacterized genes, like those forming a large protein family, such as kinesins.
Augmin Supports Spindle MT Amplification during Pre-Anaphase
In this study, we used the inducible RNAi system to critically evaluate the role of augmin during mitosis. We first performed long-term (∼15 h) imaging at 3-min intervals using a low-magnification lens to observe multiple control and Aug3 knockdown cells. The cell cycle of caulonemal apical cells was 5 to 6 h under our experimental conditions. During control cell division, we observed condensed pre-anaphase chromosomes for a fairly constant duration (9.5 ± 1.7 min; n = 103; we encountered only one case in which mitosis took >15 min) followed by chromosome segregation and phragmoplast formation (Figures 3A to 3C, Table 2; see Supplemental Movie 1 online). The mitotic duration was much shorter than other acentrosomal systems (Kurihara et al., 2008; Kitajima et al., 2011; Petry et al., 2011).
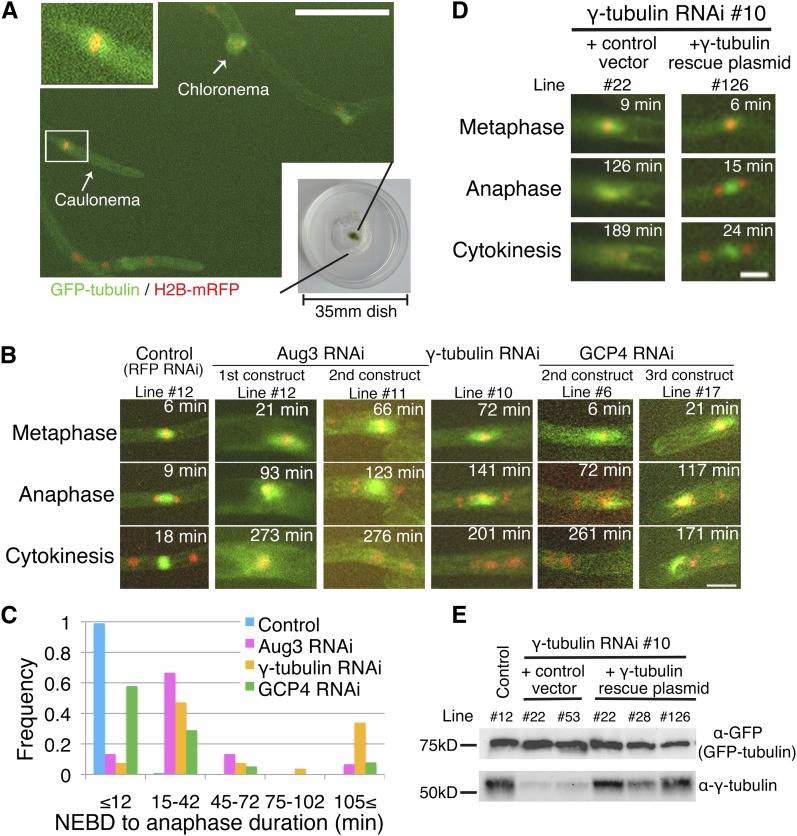
Figure 3.
Mitotic Phenotypes after γ-TuRC or Augmin Knockdown: Long-Term Imaging.
(A) Protonemata expressing GFP-tubulin (green) and histone-RFP (red) imaged using a ×10 objective lens. Insets show magnified images of metaphase spindles of a caulonemal cell. Bar = 100 µm. Also see Supplemental Movie 1 online.
(B) Mitotic defects observed after RNAi of Aug3 (an augmin subunit), γ-tubulin, and GCP4 (a γ-TuRC subunit). The control line shown here is the stable transformant of the pGG626 plasmid that knocks down only RFP. Note that although histone-RFP levels were reduced in these lines, we applied fluorescent light for enough duration to detect the remaining signals. Also see Supplemental Movie 2 online.
(C) Mitotic delay was observed after Aug3 (n = 15), γ-tubulin (n = 53), or GCP4 (n = 38) RNAi knockdown (control; n = 103). The duration was calculated based on time-lapse imaging in which images were acquired every 3 min.
(D) and (E) The γ-tubulin phenotype was rescued by expressing the RNAi-insensitive γ-tubulin gene. A few independent lines were selected and tested for phenotype appearance and protein expression. Bars = 20 µm.
Table 2. Parameters of Mitosis of Control and RNAi-Treated Cells.
| Cell Line | NEBD-Anaphase (min) (n)a | Metaphase Spindle Length (µm) (n) |
|---|---|---|
| Control | 9.5 ± 1.7 (103)b | 12.3 ± 1.0 (8)b |
| 12.1 ± 1.5 (11)c | ||
| γ-Tubulin RNAi (1st construct, line #10) | 69 ± 55 (53) | 16.3 ± 3.1 (5) |
| GCP4 RNAi (2nd construct, line #6) | 27 ± 41 (38) | 17.0 ± 2.4 (13) |
| Aug3 RNAi (1st construct, line #12) | 46 ± 61 (15) | 18.5 ± 4.0 (14) |
| γ-Tubulin RNAi (#10) + control vector | 56 ± 23 (seven lines, each >13 cells) | n.d. |
| γ-Tubulin RNAi (#10) + γ-Tubulin rescue plasmid | 17 ± 5 (five lines, each more than nine cells) | n.d. |
| γ-Tubulin RNAi, day 3 (2nd construct, line #24) | 11 ± 2.1 (19) | n.d. |
| γ-Tubulin RNAi, day 5 (2nd construct, line #24) | 26 ± 15 (31) | n.d. |
Mean ± sd is presented. n.d., not determined.
Based on the data obtained using long-term imaging with a 3-min interval. When 10- or 30-s interval imaging was performed with spinning-disk confocal microscopy, NEBD-to-anaphase duration in control cells was 10.6 ± 1.0 min (n = 10).
Data obtained in the GFP-tubulin/histone-RFP line with the control RNAi vector.
Data obtained in the GFP-tubulin/histone-RFP line.
By contrast, cells depleted of Aug3 showed prolonged mitosis (Figures 3B and 3C, Table 2; see Supplemental Movie 2 online). Bipolar spindle structures formed after nuclear envelope breakdown (NEBD) but were sometimes not maintained (5/66 cells) (Figure 3B, Line #12). In other cells, an anaphase-like event was later observed (Figure 3B, Line #11; 123 min) in which chromosomes that were attached to the spindle MTs were segregated followed by formation of a phragmoplast-like structure. However, phragmoplast expansion appeared to be incomplete in some cases, and decondensed chromosomes moved and became organized side by side, indicating incomplete cytokinesis (276 min). These mitotic defects were observed for two nonoverlapping RNAi constructs. These results suggested that augmin is required for proper spindle and phragmoplast formation.
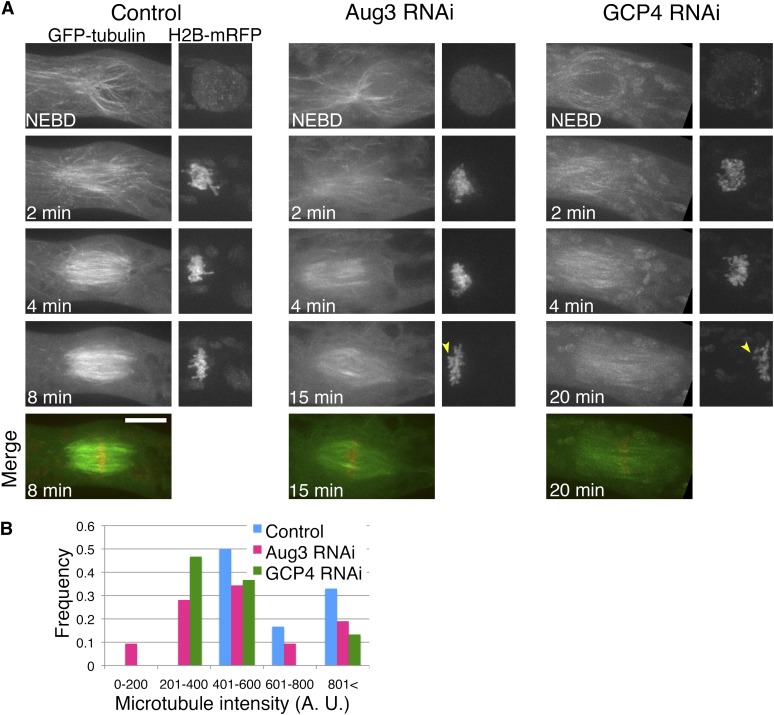
Next, living cells were imaged using three-dimensional spinning-disk confocal microscopy (Figure 4A; see Supplemental Movie 3 online). In both control and RNAi-treated cells, MTs gradually accumulated along the nuclear envelope (NE) during prophase, followed by NEBD. These MTs somewhat resemble the polar caps observed during prophase in some seed plant cells (Lloyd and Chan, 2006). However, the distribution of MTs around the NE was asymmetrical, with more MTs found on the apical side (see Supplemental Figures 3A and 3B online), as previously shown by immunostaining of MTs (Doonan et al., 1985). This finding contrasts with the random distribution seen in other acentrosomal animal systems (Mahoney et al., 2006; Schuh and Ellenberg, 2007), but it may be conserved in moss cells (Schmiedel and Schnepf, 1980). After NEBD occurred in the control cells, the spindle shape was initially biased; in the first ∼3 min, spindle MTs were more abundant on the tip-proximal side (Figure 4A, 2 min; see Supplemental Figure 3C online). Chromosome masses were initially surrounded by a fishing net–like MT network. MT distribution gradually became symmetrical, and the bipolar spindles were eventually assembled within 10 min after NEBD (Figure 4A; see Supplemental Figure 3 online). We found that this morphological change in the spindles was generally unperturbed after Aug3 knockdown (Figure 4A). However, during early prometaphase (1 to 3 min), MT intensity clearly increased in control cells but not in the Aug3 knockdown cells (Figures 4A and 4B). Furthermore, the bipolar spindles were 50% longer (Table 2), a finding that might be explained by a reduction in the number of MT nucleation sites in the spindle (Goshima and Kimura, 2010). During prometaphase, misaligned chromosomes were more frequently observed after Aug3 RNAi (after 8 min of NEBD, zero/five cells and six/seven cells had misaligned chromosomes in control and Aug3 RNAi cells, respectively) (arrowheads, Figure 4A).
Figure 4.
Reduced MT Intensity after Augmin or γ-TuRC Knockdown in Pre-Anaphase.
(A) Time-lapse imaging of a caulonemal cell expressing GFP-tubulin (green) and histone-RFP (red) by spinning-disk confocal microscopy. Ten control cells, nine Aug3 RNAi cells, and 11 GCP4 RNAi cells were observed and representative cells are displayed. Images were acquired every 1 min, each with 13 z-sections (separated by 1 µm), and are displayed after maximum projection. Arrowheads indicate misaligned chromosomes. Bar = 10 µm. Also see Supplemental Movie 3 online.
(B) Reduced MT intensity after Aug3 or GCP4 RNAi (n = 18 to 32). GFP-tubulin intensities were measured at the two points, each 2.7 µm away from the chromosome masses.
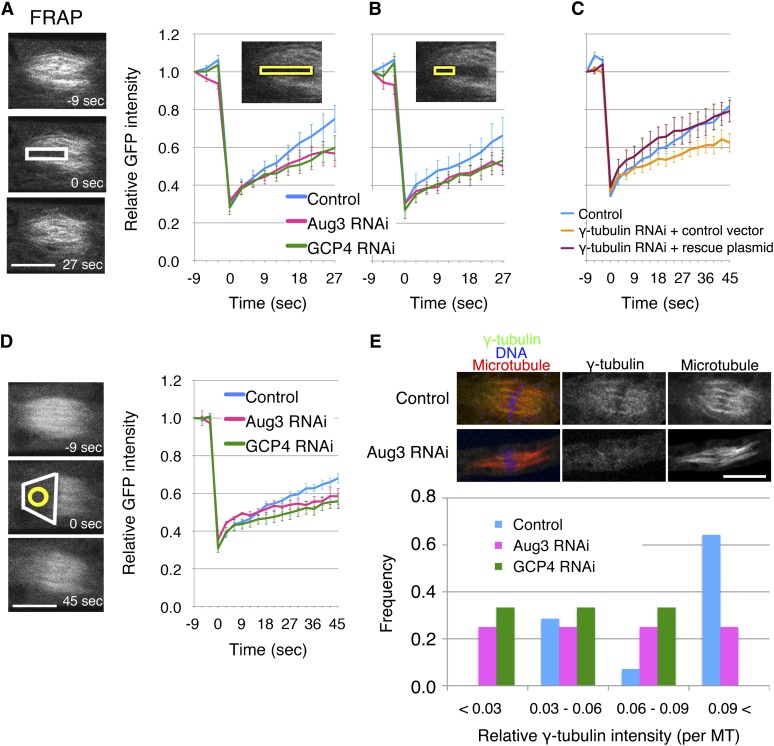
To further test the idea that augmin is responsible for MT generation in pre-anaphase, we performed fluorescence recovery after photobleaching (FRAP) analysis in caulonemal cells that express GFP-tubulin (Figures 5A to 5D; see Supplemental Movie 4 online). In this experiment, GFP signals could be recovered through new spindle MT generation, kinetochore MT polymerization, and poleward transport of the unbleached MTs. To minimize the contribution of the latter two mechanisms, we bleached the area covering the entire length of the half spindle (from chromosomes to poles) and followed the GFP signal recovery in the same area (Figure 5A) or regions far apart from the kinetochore (Figure 5B). In both cases, GFP recovery was slower after RNAi of Aug3. To further verify this result, we bleached the entire half spindle, followed by signal recovery measurement at the area apart from the kinetochore region after RNAi (Figure 5D). We similarly observed slower GFP-tubulin recovery, consistent with the data presented in Figures 5A and 5B. These results suggest that the recovery was due to augmin-dependent de novo MT generation within the spindle. Finally, we stained the MT nucleator γ-tubulin and found that spindle MT localization was diminished after Aug3 RNAi induction (Figure 5E). These results collectively indicate the importance of augmin in mitotic spindle MT generation during pre-anaphase.
Figure 5.
MT Generation Defects after Augmin or γ-TuRC Knockdown in Pre-Anaphase.
(A) and (B) FRAP analysis showed that efficient MT generation in the spindle requires GCP4 and Aug3 (±se, n = 8 each) (white, bleached area; yellow, areas of recovery measurement). Photobleaching was conducted at 0 s. Also see Supplemental Movie 4 online.
(C) Slower recovery after γ-tubulin RNAi (n = 7) was rescued by expressing the RNAi-insensitive γ-tubulin-b gene (n = 6; control n = 6).
(D) Photobleaching of the entire half spindle was conducted at 0 s (white), and fluorescence recovery was monitored at the region apart from the spindle equator (yellow) after RNAi of GCP4 (n = 7) and Aug3 (n = 5; control, n = 11).
(E) Attenuation of spindle γ-tubulin localization after Aug3 RNAi as revealed by immunostaining. Signal intensities of γ-tubulin and GFP-Tubulin on kinetochore Bars = 10 µm. MT bundles were quantified, and the relative values were plotted (n = 6 to 14). Bars = 10 µm.
An Augmin-Dependent Mechanism Predominantly Contributes to Phragmoplast MT Generation
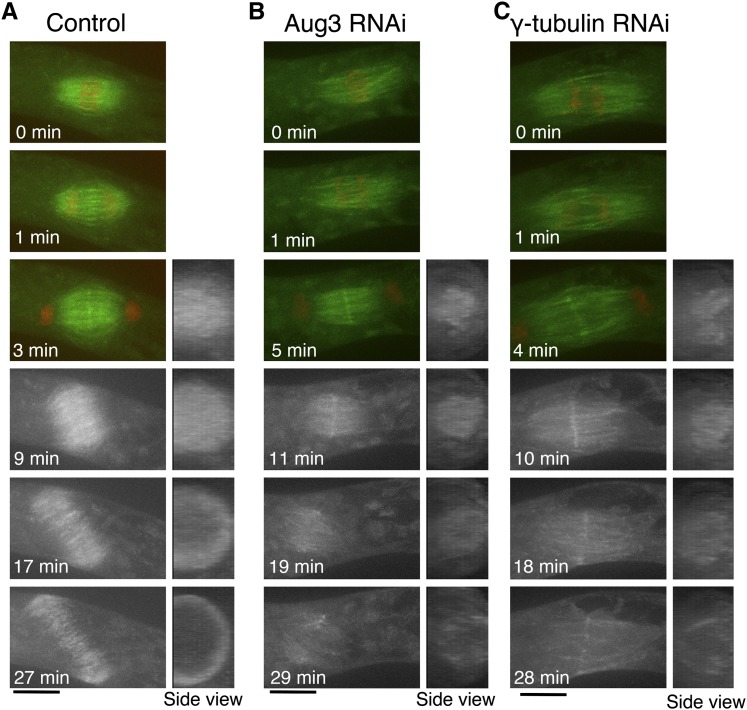
In control cells, sister chromatid separation occurred 10 min after NEBD, after which MT intensity decreased due to kinetochore MT depolymerization (0 to 2 min; Figure 6A; see Supplemental Movie 5 online). However, phragmoplast MTs soon emerged from the middle of the segregating chromosomes, and the ring structure expanded for the next ∼30 min, accompanying cell plate formation. By sharp contrast, the Aug3 RNAi-treated cells that entered anaphase showed reduced MT intensities throughout the cytokinesis phase, except for the midzone in the early phase (5 to 11 min; Figure 6B; see Supplemental Movie 5 online). The fewer postanaphase MTs observed after RNAi may be partly because of the decreased number of MTs before anaphase. However, considering that MT intensity in the control cells increased during the phragmoplast formation process (1 to 3 min; Figure 6A), it is likely that de novo MT generation occurs during this period and that augmin is primarily responsible for the process. Consistent with this interpretation, phragmoplast MT signals precociously diminished before reaching the parental cell wall (19 to 29 min; see Supplemental Movies 5 and 6 online), and the MTs eventually disappeared (see Supplemental Movie 2 online; data not shown). The decrease in phragmoplast MTs after Aug3 knockdown was detected in eight of the observed 12 cells that entered anaphase with bipolar spindles. Assuming that the cells that enter anaphase might have more residual augmin proteins after RNAi than those that collapse their spindles before anaphase, the data suggested that phragmoplast MT formation was highly sensitive to augmin concentration. We concluded that augmin plays a pivotal role in generating MTs during cytokinesis.
Figure 6.
Phragmoplast MT Formation Is Severely Suppressed by Augmin or γ-Tubulin RNAi.
Time-lapse imaging of a caulonemal cell expressing GFP-tubulin (green) and histone-RFP (red) by spinning-disk confocal microscopy. Images were acquired every 1 min, each with 13 z-sections (separated by 1 µm), and are displayed after maximum projection. A hypomorphic line (#24) was used for γ-tubulin in this experiment, which entered anaphase more frequently (five of eight anaphase cells showed this phenotype). Note that a cell was not entirely covered by 13 sections, so that an incomplete ring is seen after projection of the control cell. Also see Supplemental Movies 5 and 6 online. Bars = 10 µm.
γ-TuRC Functions for MT Formation throughout Mitosis
We next characterized the mitotic phenotypes after RNAi of γ-tubulin and GCP4, the components of γ-TuRC (Figure 2A). Long-term live-cell imaging with relatively low resolution showed phenotypes that were indistinguishable from those of augmin RNAi for two γ-tubulin RNAi lines and three independent GCP4 RNAi lines (Figures 3B and 3C, Table 2; see Supplemental Movie 2 online). These defects in the γ-tubulin line were not observed in the absence of β-estradiol or were largely rescued by the expression of an RNAi-insensitive γ-tubulin-b gene, indicating that the phenotype was derived from γ-tubulin knockdown (Figures 3D and 3E, Table 2). We concluded that γ-TuRC is essential for proper spindle and phragmoplast formation, a finding that is consistent with those in other systems (e.g., Binarová et al., 2006; Pastuglia et al., 2006; Kong et al., 2010). Furthermore, higher resolution imaging and quantitative image analyses demonstrated that MT generation is impaired during mitosis after γ-TuRC RNAi, similar to Aug3 (Figures 4 to 6; see Supplemental Movie 4 online). For example, slower recovery after photobleaching of GFP-tubulin was observed for a γ-tubulin RNAi line and was rescued by ectopic expression of an RNAi-insensitive γ-tubulin gene (Figure 5C). In contrast with the Aug3 RNAi, however, we occasionally found prophase cells with fewer MT signals around the NE, indicating that γ-TuRC is also responsible for nucleating cytoplasmic MTs during interphase and prophase, perhaps independent of augmin (Figure 4A; see Supplemental Movie 3 online).
Spindle and Phragmoplast Localization of Endogenous Augmin and γ-TuRC
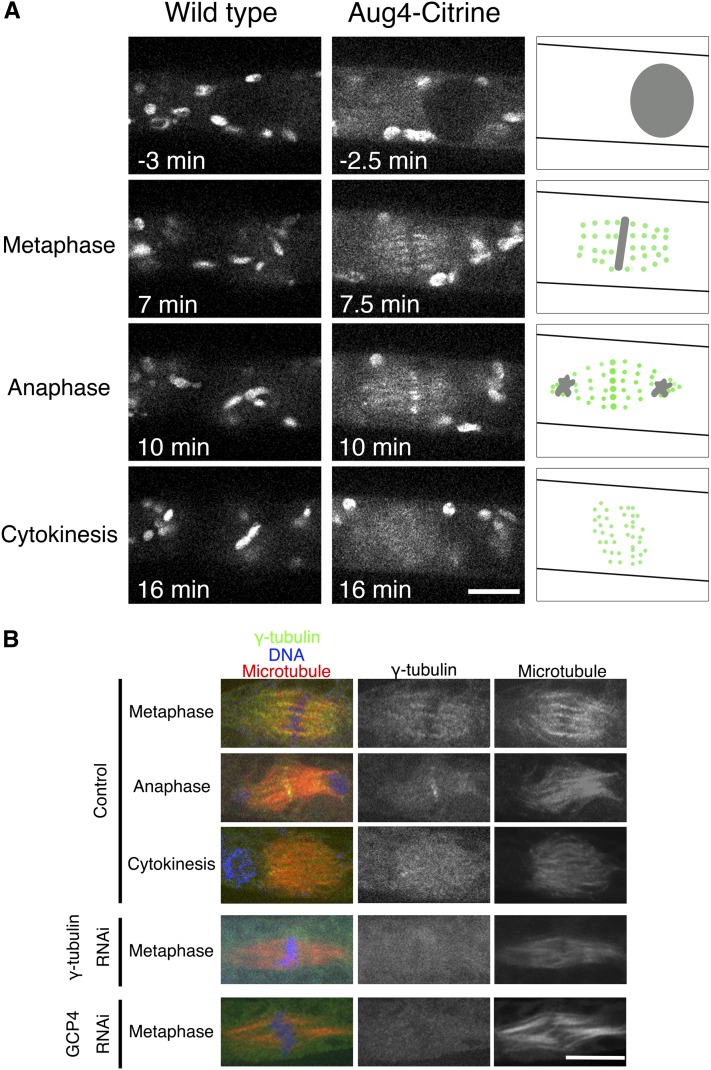
Taking advantage of the unusually high efficiency of homologous recombination in P. patens (Cove et al., 2006), intracellular localization of augmin in living cells was next determined by fusing Citrine, a yellow fluorescent protein variant (Griesbeck et al., 2001), to the endogenous augmin subunit genes (Aug4/C14orf94 and Aug2/CEP27) (see Supplemental Figures 4A and 4B online). The obtained lines grew and executed mitosis in a manner similar to wild-type moss, indicating that Citrine tagging does not impair protein function. In addition to the chloroplast signals that were derived from the autofluorescence of this organelle, many punctate signals were found throughout the spindle and phragmoplasts for the augmin-Citrine lines (Figure 7A; see Supplemental Movie 7 online). A similar localization pattern was observed for γ-tubulin by immunostaining (Figure 7B) and by tagging endogenous γ-tubulin-b (TubG2) with Citrine (see Supplemental Figures 4C and 4D online). Augmin and γ-tubulin were both transiently concentrated at the midzone in anaphase; however, the physiological meaning of this localization is unclear. These data support the idea that augmin and γ-tubulin complexes work together from prometaphase through the completion of cytokinesis to generate mitotic MTs within the spindle and phragmoplast.
Figure 7.
Uniform Spindle and Phragmoplast Localization of Endogenous Augmin and γ-Tubulin.
(A) Spindle and phragmoplast localization of endogenous Aug4 tagged with Citrine. A control wild-type cell without Citrine expression was also imaged under the identical conditions, and fluorescent signals were similarly detected on the chloroplasts but not on the spindle/phragmoplast (left). This indicates that the Aug4-Citrine signals detected on the spindle/phragmoplast represent the localization of these proteins (illustrated on the right). NEBD took place at time 0. Also see Supplemental Movie 7 online.
(B) Spindle and phragmoplast localization of endogenous γ-tubulin revealed by immunostaining. The signals were not detected when γ-tubulin or GCP4 was knocked down. Note that chloroplast signals disappeared during cell fixation.
Bars = 10 µm.
DISCUSSION
In this study, we developed a conditional RNAi system that enables us to combine knockdown of essential mitotic genes with high-resolution live microscopy in P. patens. Our loss-of-function analyses of augmin and γ-TuRC constitute a proof of principle and provide new insight into acentrosomal MT formation during plant mitosis.
Augmin Is a Conserved MT Generator in the Spindle
We found a MT generation defect in pre-anaphase after augmin knockdown, which was consistent with previous results in metazoan cells that possess centrosomes and in an in vitro study using Xenopus laevis egg extracts but had not been shown in plants or other naturally acentrosomal cells (Goshima et al., 2008; Meireles et al., 2009; Uehara et al., 2009; Ho et al., 2011; Petry et al., 2011). The key data that led to this finding were based on three-dimensional time-lapse confocal microscopy, quantitative image analyses, and FRAP after RNAi, which might be more difficult to apply in other acentrosomal cell types that are often embedded in thick tissues. Our data implicate that the importance of augmin-dependent MT generation during spindle assembly may be conserved in many cell types that execute spindle assembly without the use of centrosomes.
However, other mechanisms also appear to play some role in the spindle assembly process in moss; bipolarity was established, and many chromosomes were rapidly aligned in the middle in the presence of reduced augmin levels in caulonemal cells. Although it is not ruled out that more complete depletion of augmin might result in disappearance of spindle MTs, caulonemal cells are assumed to have additional mechanisms to generate spindle and prophase MTs, such as those involving other proteins that activate γ-TuRC. In yeast and animal cells, centrosomin family proteins play this role (Janson et al., 2005; Samejima et al., 2008; Choi et al., 2010); however, no genes homologous to this family can be found in plants, suggesting that unidentified proteins induce γ-TuRC–dependent MT generation in plants. Interestingly, the presence of an augmin- and chromosome/centrosome-independent MT generation pathway has been suggested also in animal cells; in every acentrosomal system so far tested, chromosome-distal MTs are still formed in the absence of augmin (Goshima et al., 2008; Meireles et al., 2009; Petry et al., 2011). Because basic molecular and cell biology tools have become available, it would be interesting to search for new factors responsible for MT nucleation in P. patens in the future.
A possible mechanism that leads to MT generation by augmin during prometaphase would involve localization and activation of γ-TuRC by spindle-associated augmin, as proposed in animal somatic cells (Goshima et al., 2008; Uehara et al., 2009), since moss augmin was localized at spindle MTs and required for γ-tubulin localization onto the spindle. In animals, the initial template MTs for MT amplification are postulated to be centrosomal as well as chromosome-induced MTs that appear after NEBD (Goshima et al., 2008). However, our live imaging in moss suggests that MTs surrounding NE before NEBD could be the major templates. In mammalian cells, augmin is activated by a polo-like kinase in prometaphase (Johmura et al., 2011). It remains to be discovered whether the analogous activation mechanism is present in land plants that have no polo-like kinases in their genomes.
Augmin’s role in spindle pole organization has been also proposed in other systems, in which multipolar or monopolar spindles are frequently observed after augmin inhibition (Goshima et al., 2008; Lawo et al., 2009; Uehara et al., 2009; Ho et al., 2011; Petry et al., 2011). We sometimes observed collapse of the bipolar spindle, suggesting that augmin is needed for spindle pole organization in caulonemal cells as well. However, the collapse event was detected after a long arrest in prometaphase. We therefore prefer the model that the primary role of augmin in P. patens is to prepare sufficient numbers of MTs in the spindle and that the observed defect in pole organization is the consequence of reduced numbers of MTs. In fact, it has been shown that pole disorganization is not merely caused by dysfunctions of MT-cross-linking proteins at the pole but also by deficiency in other processes in the spindle, such as kinetochore-MT attachment (Manning and Compton, 2007) or MT plus-end dynamics (Goshima et al., 2005).
Augmin Is Indispensable for Postanaphase MT Generation
A recent report using FRAP analysis established that phragmoplast MTs are constantly generated in tobacco bright yellow-2 cells (Smertenko et al., 2011). However, the molecular players responsible for postanaphase MT formation have not yet been identified. This study showed that MT generation occurs incessantly during phragmoplast formation and expansion in moss cells and is entirely dependent on augmin and γ-tubulin. Thus, our data strongly suggest that augmin is the dominant player in the activation of γ-TuRC and the generation of phragmoplast MTs in moss. MT leftovers from metaphase (i.e., nonkinetochore MTs) might serve as the templates for new MT generation by augmin/γ-TuRC after anaphase.
The dominant contribution of the augmin-mediated mechanism in postanaphase MT generation has not been previously shown in plants and animals. In Arabidopsis, MT organization defects, but not generation defects, were reported by fixed-cell observation after augmin or γ-TuRC depletion and by live imaging of a hypomorphic γ-tubulin mutant (Binarová et al., 2006; Pastuglia et al., 2006; Kong et al., 2010; Ho et al., 2011). This discrepancy might be due to the differences in species and/or cell types. Alternatively, however, it is possible that the observed organization defects in Arabidopsis are also associated with MT generation defects. In fact, when we observed the phragmoplast structure after Aug3 or γ-tubulin RNAi at a given time point, it indeed looked disorganized, often with longer MT bundles like those observed in the Arabidopsis mutant (e.g., 25 min in Supplemental Movie 5 online). The appearance of abnormally long MTs is consistent with the mathematical model in which a reduction in MT nucleation sites leads to individual MT elongation (Goshima and Kimura, 2010). In animal somatic cells, lack of augmin-dependent MTs during anaphase is compensated for by centrosomal MTs and likely also by acentrosomal MTs from other sources (Goshima et al., 2008; Meireles et al., 2009; Uehara et al., 2009; Wainman et al., 2009; Uehara and Goshima, 2010). However, augmin contribution to central spindle assembly in the absence of centrosomes has not yet been investigated in animals. It is tempting to speculate that loss of centrosomes during plant evolution or animal oocyte development is generally compensated for by augmin machinery reinforcement.
METHODS
Moss Culture and Transformation
The human histone H2B-mRFP fusion gene, directed by the E7113 promoter (Mitsuhara et al., 1996), was integrated into the GFP-tubulin line GTU14 (Hiwatashi et al., 2008). The transgenic line had no detectable growth retardation or morphological defects. BCDAT agar medium was used for regular culturing of protonemata at 25°C under continuous white light (Nishiyama et al., 2000). Imaging of protonemal cells that had been cultured on glass-bottom plates with BCD agar medium at 24 to 25°C was performed. Transformation was performed by the standard polyethylene glycol–mediated method as previously described (Nishiyama et al., 2000). To observe RNAi phenotypes, we cultured protonemata for 3 to 7 d in the presence of 1 µM β-estradiol.
Plasmids
A rough map of the inducible RNAi vector (pGG626) is provided in Supplemental Figure 1A online. All the RNAi constructs were created using the Gateway system (Invitrogen) with pGG626 containing the recombination cassette sequences. Citrine was fused to the endogenous γ-tubulin-b (TubG2), Aug4, and Aug2 genes using the plasmids with ∼1-kb C terminus and 3′-untranslated region sequences of the genes (see Supplemental Table 2 online). The rescue plasmid was constructed by synthesizing a γ-tubulin-b gene whose codon usage was altered.
RNAi Line Selection and Rescue Experiment
Target gene sequences (300 to 1000 bp) were inserted into the pGG626 RNAi vector by the Gateway LR reaction. When available, cDNAs were used as templates in PCR; otherwise, genomic DNA was used instead. Primers are listed in Supplemental Table 3 online. The plasmids were introduced into protoplasts of the GFP-tubulin/histone-RFP line after linearization, and hygromycin-resistant clones were selected. Although the transformed fragment (>10 kb) was longer than those used for Citrine tagging (∼5 kb), the transformation efficiency was comparable; typically, ∼30 lines were obtained after transformation of moss protonemata cultured in 10, 10-cm agar plates. After β-estradiol treatment, histone-RFP images were acquired using a wide-field microscope with ×10 objective lens, followed by automated quantification of the RFP intensity. Although our RNAi plasmids were designed to integrate into the PIG1 locus of the genome (see Supplemental Figure 1A online), a DNA sequence known to be intergenic and nonessential for moss (Okano et al., 2009), it was possible that each line would have different copy numbers of the RNAi cassette. We selected RNAi lines without determining the location of integration; thus, some lines might have had RNAi plasmids at unanticipated locations. However, where applicable, two or more nonoverlapping RNAi were used to validate the unexpected phenotype. The first stable RNAi line was selected ∼2.5 years ago and has been kept at 4°C with passages every ∼6 months. We have not noticed that the phenotype has weakened during this long-term storage. In the rescue experiment, RNAi-insensitive γ-tubulin-b gene or the control vector was transformed into the γ-tubulin RNAi line, and five or seven stable lines were obtained, respectively. These cells were cultured in the presence of β-estradiol, and time-lapse imaging was performed using a wide-field microscope with ×10 objective lens.
Biochemistry
RNAi cell extracts for SDS-PAGE were prepared as follows. Protonemata were first cultured on the BCDAT plate in the absence of β-estradiol, followed by subculturing after passage using glass beads twice every 5 d in the presence or absence of β-estradiol. At days 9 to 11, the protonemata were flash-frozen by liquid nitrogen, and cell extracts were prepared using an SDS-containing buffer. We expected that multiple cycles of homogenizing-subculturing would maintain most of the cells in a vegetative growing phase. We confirmed the appearance of the mitotic phenotype for γ-tubulin and Aug3 RNAi lines that were treated in this way; mitotic duration at days 9 and 10 was 43 ± 50 min (n = 34) and 66 ± 57 min (n = 20), respectively, which were significantly longer than control cells (10 min). Immunoblotting was performed using anti-γ-tubulin (G9, 1:10,000; a gift from Tetsuya Horio [University of Kansas] and Tomohiro Akashi [Nagoya University, Nagoya, Japan] [Horio et al., 1999]), anti-GFP (JL8, which also recognized Citrine; Clontech), affinity-purified anti-XMAP215 (rabbit polyclonal, 1:100), and anti-Mis12 (rabbit polyclonal, 1:100). DNA gel blotting was performed using the AlkPhos Direct kit (GE Healthcare) for genomic DNAs extracted from protonemata. qRT-PCR was performed using Power SYBR Green PCR Master Mix and the 7500 Real-Time PCR system (model 7500; Applied Biosystems) for RNAs extracted from protonemata. The results were normalized with results for TUA1 (α-tubulin). Primers for qRT-PCR are listed in Supplemental Table 4 online.
For mass spectrometry, a large-scale immunoprecipitation was performed using the ectopically expressing Aug4-Citrine and the control untagged lines that were each grown to confluence in 10, 15-cm culture plates. Cells were quickly frozen using liquid nitrogen, crushed using a mortar and pestle, and suspended in 15 mL of HB100 (50 mM HEPES-KOH, pH 7.6, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1% Triton, 1 mM DTT, and protease inhibitor cocktails). After centrifugation at 11,050g for 30 min, the supernatant was subjected to filtration using a 0.20-µm filter. The filtered supernatant was added to 200 μL (bed volume) of anti-GFP–conjugated beads (MBL) and rotated for 4 h at 4°C. After the immunoprecipitants were washed with 15 mL of HB100, they were eluted by 0.2 M Gly, pH 2.0, followed by trichloroacetic acid precipitation.
For gel filtration chromatography, the cell extract was prepared as described above, and a Superdex 200 10/300 column attached to the Biologic system (Bio-Rad) was used. Molecular size markers were used as described previously (Goshima et al., 2008).
Liquid Chromatography Coupled with Tandem Mass Spectrometry
Liquid chromatography–tandem mass spectrometry was performed as described previously (Nozawa et al., 2010). In brief, samples were separated with SDS-PAGE, and each lane was cut into nine pieces. Tandem mass spectra of the trypsinized peptides from each gel section obtained by LXQ (Thermo Finnigan) were obtained using Mascot (Matrix Science) and searched against the National Center for Biotechnology Information nonredundant protein database for Physcomitrella patens. The tandem mass spectrometry-based peptide and protein identifications were validated by Scaffold (Proteome Software).
Microscopy
A wide-field microscope TE2000 (Nikon) attached with a motorized stage and cooled charge-coupled device camera (Micromax; Roper) was used for long-term fluorescent imaging in most cases (×10, 0.30 numerical aperture [NA] objective lens). Sometimes the Ti microscope (Nikon) equipped with a more sensitive electron-multiplying charge-coupled device camera (Evolve; Roper) was used. Images were acquired at multiple sites every 3 min. During the intervals between image acquisitions, cells were illuminated with 500 lx white light. The TE2000 microscope equipped with spinning-disk confocal unit CSU-X (Yokogawa) and an electron-multiplying charge-coupled device camera (ImagEM; Hamamatsu) was used for higher resolution imaging (×100, 1.40-NA lens). The microscope was controlled by the Micromanager software. FRAP of GFP-tubulin was performed with an Olympus FV1000 with a ×60/1.42-NA (half spindle bleach) or ×100/1.40-NA (others) objective lens. Images were acquired every 3 s. Bleaching was conducted after three frames of prebleach imaging. GFP intensity of the bleached area was normalized using the intensity of a nonbleached area of similar size after background subtraction. Immunostaining of γ-tubulin was performed as described (Hiwatashi et al., 2008) with slight modification. In brief, cells expressing GFP-tubulin and histone-RFP were cultured in a glass-bottom dish with a thin layer of BCD agar medium. Cells were fixed with 8% paraformaldehyde, 100 mM PIPES, pH 6.8, 1mM MgCl2, 50 mM EGTA, 0.1% Nonidet P-40, and 1% DMSO for 40 min. Using this method, mitotic caulonemal cells could be found with relative ease, although not all the cells could be stained by antibodies. DNA was stained by 4′,6-diamidino-2-phenylindole.
Automated Quantification of Histone-RFP Signals
We developed a MATLAB code with which histone-RFP signals were automatically detected, and the intensity was calculated. This code comprises three processes: image adjustment, signal detection, and intensity measurement. In the image adjustment process, images acquired by fluorescent microscopy with a ×10 objective lens were subjected to noise reduction by a median filter. In the signal detection process, the adjusted images were binarized, and histone-RFP signals were detected based on signal size and circularity. Finally, the signal intensity was determined after background subtraction. This code is available as Supplemental Data Sets 1 to 4 online.
Accession Numbers
Sequence data from this article can be found in the National Center for Biotechnology Information database under the following accession numbers: TubG1/γ-tubulin-a as XM_001778132, TubG2/γ-tubulin-b as XM_001779791, GCP4 as XM_001758839, Aug2 (partial) as XM_001754622, Aug3 as XM_001753452, Aug4 as XM_001768696, XMAP215 as XM_001764090, Mis12 as XM_001757947, and FtsZ2-1 as XM_001765901.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Development of an Inducible RNAi System.
Supplemental Figure 2. RNAi Confirmation and Identification of the Augmin Complex in P. patens.
Supplemental Figure 3. Asymmetric MT Distribution in Early Mitosis.
Supplemental Figure 4. Construction of the Aug4-Citrine and γ-Tubulin-Citrine Lines.
Supplemental Table 1. Nomenclature of the Augmin and γ-TuRC Subunits Used in This Study.
Supplemental Table 2. Primer Sequences for Citrine Tagging.
Supplemental Table 3. Primer Sequences for RNAi.
Supplemental Table 4. Primer Sequences for qRT-PCR.
Supplemental Movie 1. Protonemal Cell Growth and Division.
Supplemental Movie 2. Mitotic Delay, Abnormal Spindle Shape, and Failure of Cytokinesis after Knockdown of Augmin (Aug3 Subunit) or γ-TuRC (γ-Tubulin and GCP4 Subunits).
Supplemental Movie 3. Time-Lapse Observation of Early Mitosis (NEBD to Anaphase Onset) in Control, Aug3 RNAi, and GCP4 RNAi Cells.
Supplemental Movie 4. FRAP of GFP-Tubulin of Control, GCP4 RNAi, and Aug3 RNAi Spindles.
Supplemental Movie 5. Time-Lapse Observation of Late Mitosis (Anaphase Onset to Late Cytokinesis) in Control, Aug3 RNAi, and γ-Tubulin RNAi Cells.
Supplemental Movie 6. Three-Dimensional Reconstruction of Phragmoplast MT Rings.
Supplemental Movie 7. Localization of Endogenous Aug4 or Aug2 Tagged with Citrine during Mitosis.
Supplemental Data Set 1. Matlab Source Code for Histone-RFP Intensity Measurement 1.
Supplemental Data Set 2. Matlab Source Code for Histone-RFP Intensity Measurement 2.
Supplemental Data Set 3. Matlab Source Code for Histone-RFP Intensity Measurement 3.
Supplemental Data Set 4. Matlab Source Code for Histone-RFP Intensity Measurement 4.
Supplementary Material
Acknowledgments
We thank Mitsuyasu Hasebe for his generous support on this project. We also thank Yoshikatsu Sato for moss lines; Tetsuya Horio and Tomohiro Akashi for antibodies; Jean-Pierre Zrÿd for plasmids; Futamura Chemical Industries for the cellophane; Kyowa Hakko Kogyo for Driselase; Koji Nagao, Sachiko Shibata, and Natsuko Shirai for liquid chromatography–tandem mass spectrometry analyses; Yasunori Machida for qRT-PCR analyses; and Takashi Murata and Ken Kosetsu for their valuable discussion. This work was supported by the Next Generation grant (Japan Society for Promotion of Science), the Inoue Foundation, and Human Frontier Science Program (to G.G.).
AUTHOR CONTRIBUTIONS
Y.N., T.M., Y.H., and G.G. designed research. All the authors performed experiments. R.F. contributed new computational tools. Y.N., T.M., and G.G. analyzed data. G.G. wrote the article.
Glossary
- MT
microtubule
- γ-TuRC
γ-tubulin ring complex
- RNAi
RNA interference
- dsRNA
double-stranded RNA
- RFP
red fluorescent protein
- qRT-PCR
quantitative real-time PCR
- NEBD
nuclear envelope breakdown
- NE
nuclear envelope
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- NA
numerical aperture
References
- Ambrose J.C., Li W., Marcus A., Ma H., Cyr R. (2005). A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 16: 1584–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R.C., Vidali L., Kleinman K.P., Bezanilla M. (2008). Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 54: 863–875 [DOI] [PubMed] [Google Scholar]
- Bannigan A., Scheible W.R., Lukowitz W., Fagerstrom C., Wadsworth P., Somerville C., Baskin T.I. (2007). A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 120: 2819–2827 [DOI] [PubMed] [Google Scholar]
- Bezanilla M., Pan A., Quatrano R.S. (2003). RNA interference in the moss Physcomitrella patens. Plant Physiol. 133: 470–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Perroud P.F., Pan A., Klueh P., Quatrano R.S. (2005). An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol. (Stuttg.) 7: 251–257 [DOI] [PubMed] [Google Scholar]
- Binarová P., Cenklová V., Procházková J., Doskocilová A., Volc J., Vrlík M., Bögre L. (2006). Gamma-tubulin is essential for acentrosomal microtubule nucleation and coordination of late mitotic events in Arabidopsis. Plant Cell 18: 1199–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., Hyman A.A. (2008). XMAP215 is a processive microtubule polymerase. Cell 132: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.K., Liu P., Sze S.K., Dai C., Qi R.Z. (2010). CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. (2005). The moss Physcomitrella patens. Annu. Rev. Genet. 39: 339–358 [DOI] [PubMed] [Google Scholar]
- Cove D., Bezanilla M., Harries P., Quatrano R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57: 497–520 [DOI] [PubMed] [Google Scholar]
- Djondjurov L.P., Yancheva N.Y., Ivanova E.C. (1983). Histones of terminally differentiated cells undergo continuous turnover. Biochemistry 22: 4095–4102 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., Raff J. (2008). A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J.H., Cove D.J., Lloyd C.W. (1985). Immunofluorescence microscopy of microtubules in intact cell lineages of the moss, Physcomitrella patens. I. Normal and CIPC-treated tip cells. J. Cell Sci. 75: 131–147 [DOI] [PubMed] [Google Scholar]
- Goshima G., Kimura A. (2010). New look inside the spindle: microtubule-dependent microtubule generation within the spindle. Curr. Opin. Cell Biol. 22: 44–49 [DOI] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. (2003). Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160: 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R.D. (2008). Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181: 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Nédélec F., Vale R.D. (2005). Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 171: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. (2007). Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger C.L., Cyr R.J. (2000). Microtubule reorganization in tobacco BY-2 cells stably expressing GFP-MBD. Planta 210: 502–509 [DOI] [PubMed] [Google Scholar]
- Griesbeck O., Baird G.S., Campbell R.E., Zacharias D.A., Tsien R.Y. (2001). Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276: 29188–29194 [DOI] [PubMed] [Google Scholar]
- Harries P.A., Pan A., Quatrano R.S. (2005). Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Sano T., Kutsuna N., Kumagai-Sano F., Hasezawa S. (2007). Contribution of anaphase B to chromosome separation in higher plant cells estimated by image processing. Plant Cell Physiol. 48: 1509–1513 [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y., Obara M., Sato Y., Fujita T., Murata T., Hasebe M. (2008). Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell 20: 3094–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.M., Hotta T., Kong Z., Zeng C.J., Sun J., Lee Y.R., Liu B. (2011). Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell 23: 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T., Basaki A., Takeoka A., Yamato M. (1999). Lethal level overexpression of gamma-tubulin in fission yeast causes mitotic arrest. Cell Motil Cytoskeleton 44: 284–295 [DOI] [PubMed] [Google Scholar]
- Inoué S., Sato H. (1967). Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 50(6, suppl.) 259–292 [PMC free article] [PubMed] [Google Scholar]
- Janson M.E., Setty T.G., Paoletti A., Tran P.T. (2005). Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J. Cell Biol. 169: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura Y., Soung N.K., Park J.E., Yu L.R., Zhou M., Bang J.K., Kim B.Y., Veenstra T.D., Erikson R.L., Lee K.S. (2011). Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl. Acad. Sci. USA 108: 11446–11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K., Habermann B., Hyman A.A. (2002). XMAP215: A key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12: 267–273 [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Ohsugi M., Ellenberg J. (2011). Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell 146: 568–581 [DOI] [PubMed] [Google Scholar]
- Kong Z., Hotta T., Lee Y.R., Horio T., Liu B. (2010). The gamma-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell 22: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai F., Yoneda A., Tomida T., Sano T., Nagata T., Hasezawa S. (2001). Fate of nascent microtubules organized at the M/G1 interface, as visualized by synchronized tobacco BY-2 cells stably expressing GFP-tubulin: Time-sequence observations of the reorganization of cortical microtubules in living plant cells. Plant Cell Physiol. 42: 723–732 [DOI] [PubMed] [Google Scholar]
- Kurihara D., Matsunaga S., Uchiyama S., Fukui K. (2008). Live cell imaging reveals plant aurora kinase has dual roles during mitosis. Plant Cell Physiol. 49: 1256–1261 [DOI] [PubMed] [Google Scholar]
- Lambert A.M., Bajer A.S. (1972). Dynamics of spindle fibers and microtubules during anaphase and phragmoplast formation. Chromosoma 39: 101–144 [Google Scholar]
- Lawo S., et al. (2009). HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19: 816–826 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2006). Not so divided: The common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 7: 147–152 [DOI] [PubMed] [Google Scholar]
- Mahoney N.M., Goshima G., Douglass A.D., Vale R.D. (2006). Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16: 564–569 [DOI] [PubMed] [Google Scholar]
- Manning A.L., Compton D.A. (2007). Mechanisms of spindle-pole organization are influenced by kinetochore activity in mammalian cells. Curr. Biol. 17: 260–265 [DOI] [PubMed] [Google Scholar]
- Marshall W.F. (2009). Centriole evolution. Curr. Opin. Cell Biol. 21: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T.L., Kao L.R., Kaufman T.C. (2001). Zygotic development without functional mitotic centrosomes. Curr. Biol. 11: 116–120 [DOI] [PubMed] [Google Scholar]
- Meireles A.M., Fisher K.H., Colombié N., Wakefield J.G., Ohkura H. (2009). Wac: A new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Moritz M., Agard D.A. (2001). Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11: 174–181 [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Murata T., Tanahashi T., Nishiyama T., Yamaguchi K., Hasebe M. (2007). How do plants organize microtubules without a centrosome? J. Integr. Plant Biol. 49: 1154–1163 [Google Scholar]
- Nishiyama T., Hiwatashi Y., Sakakibara I., Kato M., Hasebe M. (2000). Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Nozawa R.S., Nagao K., Masuda H.T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C. (2010). Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12: 719–727 [DOI] [PubMed] [Google Scholar]
- Okano Y., Aono N., Hiwatashi Y., Murata T., Nishiyama T., Ishikawa T., Kubo M., Hasebe M. (2009). A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc. Natl. Acad. Sci. USA 106: 16321–16326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Verbrugghe K.J., Skop A.R. (2005). Midbodies and phragmoplasts: Analogous structures involved in cytokinesis. Trends Cell Biol. 15: 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M., Azimzadeh J., Goussot M., Camilleri C., Belcram K., Evrard J.L., Schmit A.C., Guerche P., Bouchez D. (2006). Gamma-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., Pugieux C., Nédélec F.J., Vale R.D. (2011). Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 108: 14473–14478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.J., Bezanilla M. (2010). Evolutionary crossroads in developmental biology: Physcomitrella patens. Development 137: 3535–3543 [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Samejima I., Miller V.J., Groocock L.M., Sawin K.E. (2008). Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J. Cell Sci. 121: 3971–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel G., Schnepf E. (1980). Polarity and growth of caulonema tip cells of the moss Funaria-Hygrometrica. Planta 147: 405–413 [DOI] [PubMed] [Google Scholar]
- Schuh M., Ellenberg J. (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130: 484–498 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Piette B., Hussey P.J. (2011). The origin of phragmoplast asymmetry. Curr. Biol. 21: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Strepp R., Scholz S., Kruse S., Speth V., Reski R. (1998). Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95: 4368–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.Y., Ngo B., Tapadia A., Hsu P.H., Wu G., Lee W.H. (2011). Aurora-A phosphorylates Augmin complex component Hice1 protein at an N-terminal serine/threonine cluster to modulate its microtubule binding activity during spindle assembly. J. Biol. Chem. 286: 30097–30106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Goshima G. (2010). Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol. 191: 259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R.S., Tomioka A., Petry S., Vale R.D., Obuse C., Goshima G. (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 106: 6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Augustine R.C., Kleinman K.P., Bezanilla M. (2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19: 3705–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Burkart G.M., Augustine R.C., Kerdavid E., Tüzel E., Bezanilla M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009a). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., van Gisbergen P.A., Guérin C., Franco P., Li M., Burkart G.M., Augustine R.C., Blanchoin L., Bezanilla M. (2009b). Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainman A., Buster D.W., Duncan T., Metz J., Ma A., Sharp D., Wakefield J.G. (2009). A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev. 23: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Heald R. (2008). Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 265: 111–158 [DOI] [PubMed] [Google Scholar]
- Whittington A.T., Vugrek O., Wei K.J., Hasenbein N.G., Sugimoto K., Rashbrooke M.C., Wasteneys G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Wu G., Lin Y.T., Wei R., Chen Y., Shan Z., Lee W.H. (2008). Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol. Cell. Biol. 28: 3652–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.