This study reports the discovery of the Arabidopsis thaliana augmin complex composed of at least eight subunits, two of which are plant specific, that regulate the function of the γ-tubulin complex during mitosis and cytokinesis.
Abstract
Plant cells assemble the bipolar spindle and phragmoplast microtubule (MT) arrays in the absence of the centrosome structure. Our recent findings in Arabidopsis thaliana indicated that AUGMIN subunit3 (AUG3), a homolog of animal dim γ-tubulin 3, plays a critical role in γ-tubulin–dependent MT nucleation and amplification during mitosis. Here, we report the isolation of the entire plant augmin complex that contains eight subunits. Among them, AUG1 to AUG6 share low sequence similarity with their animal counterparts, but AUG7 and AUG8 share homology only with proteins of plant origin. Genetic analyses indicate that the AUG1, AUG2, AUG4, and AUG5 genes are essential, as stable mutations in these genes could only be transmitted to heterozygous plants. The sterile aug7-1 homozygous mutant in which AUG7 expression is significantly reduced exhibited pleiotropic phenotypes of seriously retarded vegetative and reproductive growth. The aug7-1 mutation caused delocalization of γ-tubulin in the mitotic spindle and phragmoplast. Consequently, spindles were abnormally elongated, and their poles failed to converge, as MTs were splayed to discrete positions rendering deformed arrays. In addition, the mutant phragmoplasts often had disorganized MT bundles with uneven edges. We conclude that assembly of MT arrays during plant mitosis depends on the augmin complex, which includes two plant-specific subunits.
INTRODUCTION
In flowering plants, microtubules (MTs) are nucleated and organized in the absence of a structurally defined MT organizing center like the centrosome. Consequently, the bipolar spindle MT array often exhibits converging but unfocused poles (Palevitz, 1993; Smirnova and Bajer, 1998). Upon the completion of mitosis, the spindle array is replaced by the bipolar phragmoplast in which MTs are oriented with their plus ends facing the division site (Liu et al., 2011b). Within these arrays, MT polymerization takes place continuously to support the rapid reorganization of spindle and phragmoplast (Komaki et al., 2010; Ho et al., 2011a). As the key MT nucleation factor, the γ-tubulin complex is detected along both spindle and phragmoplast MTs with biases toward the MT minus ends facing spindle poles and phragmoplast edges (Liu et al., 1993; Nakamura et al., 2010). The functions of the γ-tubulin complex proteins are essential for MT nucleation and organization during mitosis and cytokinesis in plant cells (Pastuglia et al., 2006; Nakamura and Hashimoto, 2009; Kong et al., 2010).
The association of the γ-tubulin complex with MTs implied a MT-dependent MT nucleation mechanism. In fact, the appearance of the γ-tubulin complex on the MT lattice often precedes new MT nucleation events (Nakamura et al., 2010). Although this γ-tubulin–dependent MT nucleation phenomenon is often observed in the interphase cortical MT array that gives rise to new MT branches at ~40° angles (Murata et al., 2005), it is unclear whether a similar mechanism exists in the spindle and phragmoplast. It is also unknown how the γ-tubulin complex associates with MT lattices prior to initiating MT nucleation. The WD-40 repeat protein NEDD1 (for Neural precursor cell expressed, developmentally down-regulated protein1)/γ-tubulin complex protein-WD has been considered as a targeting factor for the γ-tubulin complex during mitosis in mammalian cells (Lüders et al., 2006). The Xenopus laevis counterpart can cosediment with polymerized MTs, suggesting that it may mediate the interaction between the γ-tubulin complex and MTs (Liu and Wiese, 2008). A homologous protein discovered in plants plays a critical role in MT organization in the spindle and phragmoplast (Zeng et al., 2009). However, it remains unclear how this Arabidopsis thaliana NEDD1 may participate in γ-tubulin–dependent MT nucleation and organization.
It is believed that the γ-tubulin complex is targeted to structurally defined MT organizing center and MT lattices via different anchoring proteins (Kollman et al., 2011). In Drosophila melanogaster, an RNA interference–based screen for spindle defects led to the discovery of dim γ-tubulin genes whose products form the augmin complex that regulates γ-tubulin localization in mitotic spindles, but not at the centrosome (Goshima et al., 2008). A similar complex containing eight HAUS (for homologous to augmin subunits) proteins isolated from mitotic cells regulates spindle assembly and mitotic progression (Lawo et al., 2009; Uehara et al., 2009; Hutchins et al., 2010). A recent study showed that augmin is also required for MT amplification in the central spindle during anaphase (Uehara and Goshima, 2010). Strong interaction between augmin and the γ-tubulin complex can be detected in mitotic but not interphase cells (Teixidó-Travesa et al., 2010). Among augmin subunits, HAUS8/HICE1 is an MT-associated protein (MAP) that directly binds to MTs (Wu et al., 2008). Another subunit, the HAUS6/FAM29A protein, interacts with NEDD1 in mitotic cells (Zhu et al., 2008; Uehara et al., 2009). Collectively, these findings have led to a model of an augmin-NEDD1-γ-tubulin continuum that initiates nascent MT nucleation on existing MTs so that more MTs can be formed within the spindle (Goshima and Kimura, 2010).
Because augmin functions in γ-tubulin localization on spindle MTs but not at the centrosome (Goshima et al., 2008), γ-tubulin–dependent assembly of the acentrosomal spindle in plant cells may employ a similar mechanism. Indeed, the Arabidopsis homolog of HAUS3, AUGMIN subunit 3 (AUG3), exhibits a localization pattern similar to that of γ-tubulin complex proteins (Ho et al., 2011b). AUG3 is an essential protein for gametophyte and sporophyte development, and an aug3 mutation led to impaired mitotic MT arrays that often showed half spindles, elongated spindles, or spindles with unconverged poles (Ho et al., 2011b). In addition, the mutant phragmoplasts frequently had randomly packed MT bundles. Other animal augmin subunits have either low or no sequence similarity to proteins encoded by plant genomes like that of Arabidopsis. If a similar mechanism is shared by animals and plants, plants would be predicted to produce a similar protein complex to fulfill the function.
To examine whether an augmin complex is formed in Arabidopsis, we used a functional AUG3-c-myc fusion protein expressed in transgenic plants for purification and identification of proteins associated with AUG3 in vivo. Reciprocal purifications using later identified AUG subunits uncovered at least eight AUG subunits in the augmin complex. Seven of them are encoded by single genes in Arabidopsis, and their loss-of-function mutations often were sporophyte lethal. Two of the newly identified subunits are homologous only to proteins of plant origin. Based on phenotypic analysis of newly isolated heritable mutations, we report that the function of plant augmin is critical for forming converging spindle poles and organizing MT minus ends at the phragmoplast distal ends. Thus, despite their divergence, the plant and animal augmin complexes have taken on fundamentally similar essential functions in MT nucleation and organization in mitotic MT arrays.
RESULTS
Identification of Six Subunits in the Arabidopsis Augmin Complex
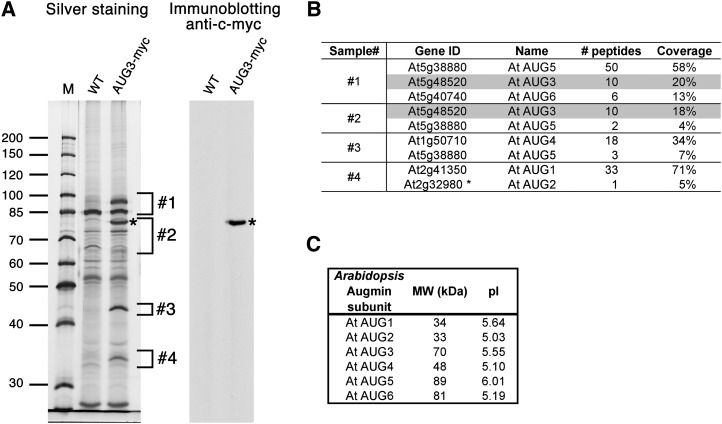
Previously, an AUG3-c-myc fusion protein was expressed in the aug3-1 mutation background and proven to be functional (Ho et al., 2011b). This protein interacted with another putative augmin subunit AUG1 in vivo. We asked whether other proteins could be recovered after the purification scheme was scaled up. Proteins derived from an AUG3-c-myc affinity column gave rise to a number of SDS-PAGE bands that were absent in the wild-type control. Figure 1A also shows the bait detected by an anti-c-myc immunoblotting. To identify these polypeptides copurified with AUG3-c-myc, four gel regions containing distinct bands were excised, as highlighted in Figure 1A, and subjected to trypsin digestion and peptide identification assisted by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Six proteins were detected with 5 to 71% peptide coverage (Figure 1B; see Supplemental Table 1 online), including the previously reported AUG1 and AUG3. Interestingly, all six proteins are acidic with calculated pIs of 5 to 6 (Figure 1C). The four newly identified proteins encoded by At2g32980, At1g50710, At5g38880, and At5g40740 were named AUG2, AUG4, AUG5, and AUG6, respectively. When they were compared with proteins predicted by the genomes of rice (Oryza sativa) and moss (Physcomitrella patens), homologous proteins with high sequence identities were identified (see Supplemental Figures 1 to 4 online). When their sequences were aligned with the corresponding human HAUS proteins, limited amino acid sequence identities of 16 to 19% were found (see Supplemental Figures 1A, 2A, 3A, and 4A online). Notably, the identical amino acid residues between the four Arabidopsis AUGs and the HAUS counterparts were largely conserved in the three examined plant species (see Supplemental Figures 1C, 2B, 3B, and 4C online). Hence, these results further suggest that an augmin complex is formed in Arabidopsis, and at least six of its subunits share amino acid sequence similarity with their human counterparts.
Figure 1.
Purification of the Arabidopsis Augmin Complex and Identification of Six Augmin Subunits.
(A) Proteins copurified with AUG3-c-myc using anti-c-myc antibody were subjected to SDS-PAGE and immunoblotting analysis. Gel was stained with silver nitrate. Asterisks indicate the bait, AUG3-4×c-myc. The numbers next to the gel image indicate the position of the gel regions analyzed in the following LC-MS/MS analysis described in (B). AUG3-myc, transgenic line expressing AUG3-4×c-myc; M, molecular weight markers; WT, wild-type negative control.
(B) MS results obtained from the samples shown in (A). Six augmin subunits, including two previously reported augmin subunits (AUG1 and AUG3), were identified. Gene identification name according to TAIR database (Gene ID), name, number of unique peptides (# peptides), and sequence coverage (Coverage) are listed. The bait AUG3-c-myc is highlighted in gray.
(C) Properties of Arabidopsis augmin subunits. Calculated molecular mass of the proteins (MW [kD]) and theoretical pI are shown.
The AUG1, AUG2, AUG4, and AUG5 Genes Are Essential for Sporophyte Development and Play a Critical Role in Gametogenesis
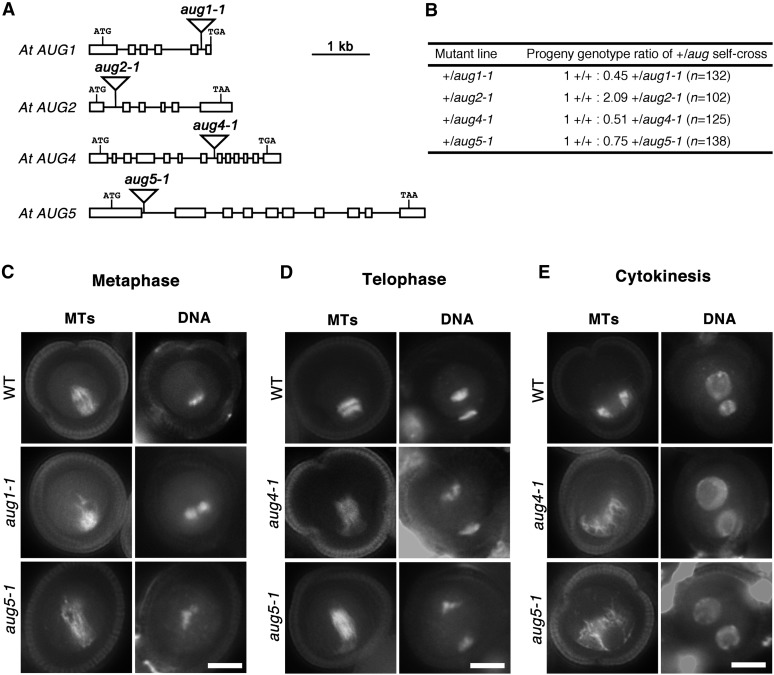
To analyze the functions of the AUG subunits, T-DNA insertional mutations at the AUG1, AUG2, AUG4, and AUG5 loci were isolated (Figure 2A). Unfortunately, no mutations were identified at the AUG6 locus. The aug1-1 mutation had an insertion in the 5th intron, aug2-1 in the 1st intron, aug4-1 in the 7th intron, and aug5-1 in the 1st intron (Figure 2A). Like the aug3-1 mutant (Ho et al., 2011b), the heterozygous mutant plants of aug1-1, aug2-1, aug4-1, and aug5-1 all exhibited distorted genetic segregation patterns when self-pollinated (Figure 2B). Notably, no homozygous progenies were recovered from self-fertilization of the heterozygous plants harboring any one of these four mutations.
Figure 2.
Phenotypic Analysis of Mutations in AUG Genes.
(A) Schematic illustration of the AUG1, AUG2, AUG4, and AUG5 genes and the positions of corresponding T-DNA insertions. Exons and introns are displayed as open boxes and lines, respectively.
(B) Genetic segregation patterns of offspring derived from self-fertilization of the heterozygous aug mutants. All segregation ratios are significantly different from the expected ratio (1 +/+: 2 +/aug: 1 aug/aug) based on the χ2 test (P < 0.5).
(C) to (E) Defects in MT organization caused by aug1-1, aug 4-1, and aug5-1 mutations at metaphase (C), telophase (D), and cytokinesis (E) during pollen mitosis I visualized by immunofluorescence. Bars = 5 μm.
(C) A wild-type (WT) metaphase spindle exhibits a bipolar configuration with converged spindle poles (top panel). In aug1-1 and aug5-1 mutants, metaphase spindles only contain randomly packed MTs in the peripheral side (middle panel) or are elongated and disorganized (bottom panel).
(D) During telophase, compared with the laterally expanding phragmoplast MT array positioned near the cell periphery in the wild type (top panel), elongated MT arrays are observed in aug4-1 and aug5-1 mutants that show no sign of lateral expansion (middle and bottom panels).
(E) During cytokinesis, a curved phragmoplast can be seen in the wild-type cell (top panel). In the aug4-1 and aug5-1 mutants, phragmoplast MTs are disorganized (middle and bottom panels).
The distorted genetic segregation phenotype of the +/aug plants was likely caused by the loss of fertility, since the transmission efficiency of the mutant allele through both the male and female gametes were affected (see Supplemental Table 2 online). In addition, aborted seeds and unfertilized ovules were frequently found in siliques produced by the heterozygous mutants (see Supplemental Figure 5 online), and they could have contributed to the distorted segregation phenotype of the +/aug plants. These results collectively indicate that AUG1, AUG2, AUG4, and AUG5 are essential for both gametophyte and sporophyte development. This is consistent to our earlier finding for AUG3 (Ho et al., 2011b).
Because the developing microgametophytes in Arabidopsis allow us to analyze possible mitotic phenotypes brought about by sporophytically lethal mutations (Liu et al., 2011a), we examined whether the aug1-1, aug2-1, aug4-1, and aug5-1 caused defects in MT organization in mitotic arrays. Defects in the spindle and phragmoplast MT arrays were detected in aug1-1, aug4-1, and aug5-1 mutant cells when compared with cells of the wild-type control (Figures 2C to 2E). In pollen mitosis I, the mitotic apparatus was placed toward the cell periphery as shown by the metaphase spindle with the interior pole pointing at the center of the cell (Figure 2C). The mutant cells often produced two types of spindles: incomplete half spindles anchored to the cell cortex (middle panel in Figure 2C) or elongated spindles across the dividing microspores (bottom panel in Figure 2C). At late anaphase and telophase, the control cells had two reforming nuclei placed peripherally (WT in Figure 2D). By contrast, elongated mutant spindles resulted in the separation of sister chromatids further away from each other (middle and bottom panels in Figure 2D). Compared with the expanding short phragmoplast MT array found in the control cells, mutant cells often produced long MT bundles placed in between two reforming nuclei (Figure 2D). These elongated internuclear MTs did not progress to form a centrifugally expanding phragmoplast array as demonstrated by the control cells (WT in Figure 2E). Instead, MT filaments were randomly packed between two reformed nuclei (Figure 2E). Thus, our results indicate that these augmin subunits play critical roles in MT reorganization during spindle and phragmoplast development.
Augmin Subunits Decorate the Spindle and Phragmoplast MT Arrays
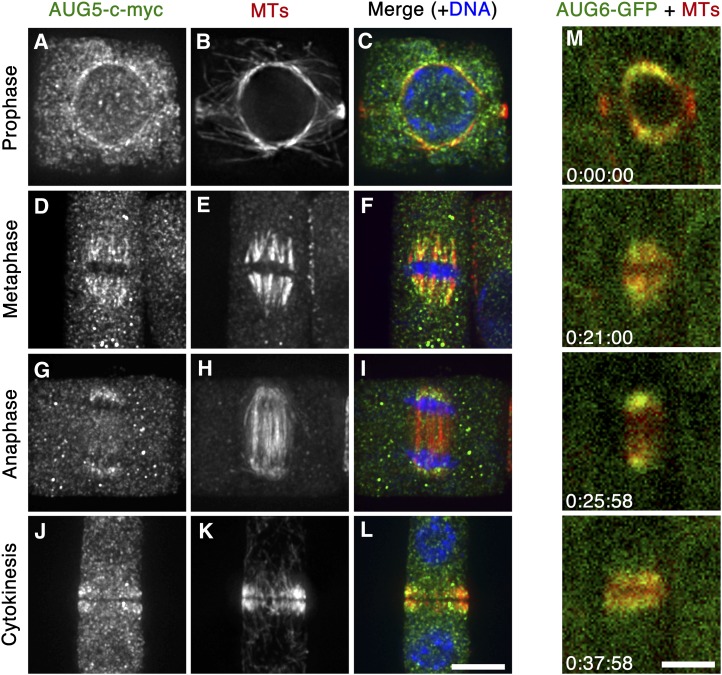
To examine the localization of these newly identified AUG proteins, we generated transgenic plants that expressed AUG4-c-myc or AUG5-c-myc fusion proteins. These proteins were detected by anti-c-myc immunofluorescence in dividing cells and shared a similar if not identical localization pattern (Figure 3; see Supplemental Figure 6 online). For example, the AUG5-c-myc protein was detected on the nuclear envelope decorating the spindle MT array formed at late prophase (Figures 3A to 3C). At metaphase, AUG5-c-myc was found along kinetochore fibers in a punctate manner (Figures 3D to 3F). At late stages of anaphase, the AUG5-c-myc signal was particularly pronounced on the shortening kinetochore fibers near spindle poles (Figures 3G to 3I). Punctate AUG5-c-myc signals also appeared along phragmoplast MTs (Figures 3J to 3L). Thus, the localization pattern of AUG4-c-myc and AUG5-c-myc was similar to that of AUG3 reported previously (Ho et al., 2011b).
Figure 3.
Localization of AUG5 and AUG6 in Mitotic Root Meristematic Cells.
(A) to (L) Immunolocalization of AUG5-c-myc. In merged images, AUG5 is pseudocolored in green, MTs in red, and DNA in blue.
(A) to (C) In a prophase cell, AUG5-c-myc signal is observed along MTs on the nuclear envelope.
(D) to (F) In a metaphase cell, AUG5-c-myc signal appears along kinetochore fiber MTs in a punctate manner.
(G) to (I) During anaphase, AUG5-c-myc particularly decorates the shortening kinetochore fibers at the poles.
(J) to (L) AUG5-c-myc signal decorates phragmoplast MTs.
(M) Live-cell imaging of the AUG6-GFP. Snapshots were extracted from a time-lapse movie of a mitotic root cell expressing AUG6-GFP and mCherry-TUB6 (see Supplemental Movie 1 online). AUG6 and MTs are shown in green and red, respectively. The starting time is set at 0, and snapshot images are taken at times (hour:minutes:seconds) as shown on the bottom left. Like AUG5-c-myc, AUG6-GFP prominently decorates spindle and phragmoplast MT arrays from prophase to cytokinesis.
Bars = 5 μm.
A fusion of the AUG6 genomic sequence including its upstream promoter region and the green fluorescent protein (GFP)-coding sequence was introduced into an Arabidopsis host plant to examine the localization of the AUG6 protein in living cells. In cells expressing both AUG6-GFP and mCherry-TUB6 (for β-tubulin 6), AUG6-GFP was observed along spindle and phragmoplast MTs (Figure 3M; see Supplemental Movie 1 online). Again, the GFP signal was particularly pronounced toward spindle poles during anaphase (snapshot at 25 min; Figure 3M). When fluorescence intensities were scanned across the spindle and phragmoplast, it was obvious that the GFP signal was biased toward the spindle poles (see Supplemental Figure 7 online). While the mCherry-tubulin signal left a very narrow dark line in the phragmoplast midline, the AUG6-GFP signal had a wider dark gap in the phragmoplast midzone (see Supplemental Figure 7 online). This result suggests that AUG6-GFP preferentially localized to MT minus ends in the spindle and phragmoplast MT arrays. Thus, all three proteins showed localization patterns similar to those of AUG1 and AUG3 expressed in either c-myc or GFP fusions (Ho et al., 2011b).
Because the MT minus end–biased localization of the AUG proteins resembled that of the γ-tubulin complex, we tested possible colocalization by double staining of AUG4-myc or AUG5-myc with γ-tubulin. AUG4-myc and AUG5-myc showed colocalization with γ-tubulin in cells of metaphase through telophase (see Supplemental Figure 8 online). This result demonstrates that plant augmin likely interacts with γ-tubulin complex on the spindle and phragmoplast MT arrays.
The colocalization of AUG proteins with γ-tubulin complex prompted us to test further whether the augmin complex interacted with the γ-tubulin complex in vivo. Because such an interaction was detected in mitotic cells in human (Teixidó-Travesa et al., 2010), we used actively dividing tobacco (Nicotiana tabacum) Bright Yellow 2 (BY-2) cells expressing the AUG3-c-myc fusion protein for this purpose. This protein was detected via anti-c-myc immunofluorescence in spindle and phragmoplast MTs in a pattern similar to that seen in Arabidopsis (see Supplemental Figure 9A online). This result suggested that this Arabidopsis-derived AUG3 fusion protein is most likely functional in tobacco cells. Proteins copurified with AUG3-c-myc, detected by the anti-c-myc antibody, were probed with the G9 anti-γ-tubulin antibody. A clear band was detected at the expected ~56-kD position but was not seen in the control experiment using untransformed BY-2 cells (see Supplemental Figure 9B online).
Taken together, these results strongly support the notion that AUG1, AUG3, AUG4, AUG5, and AUG6 are associated in an augmin complex that acts on MTs and interacts with γ-tubulin in spindles and the phragmoplast.
The AUG Complex Contains at Least Two Plant-Unique Subunits
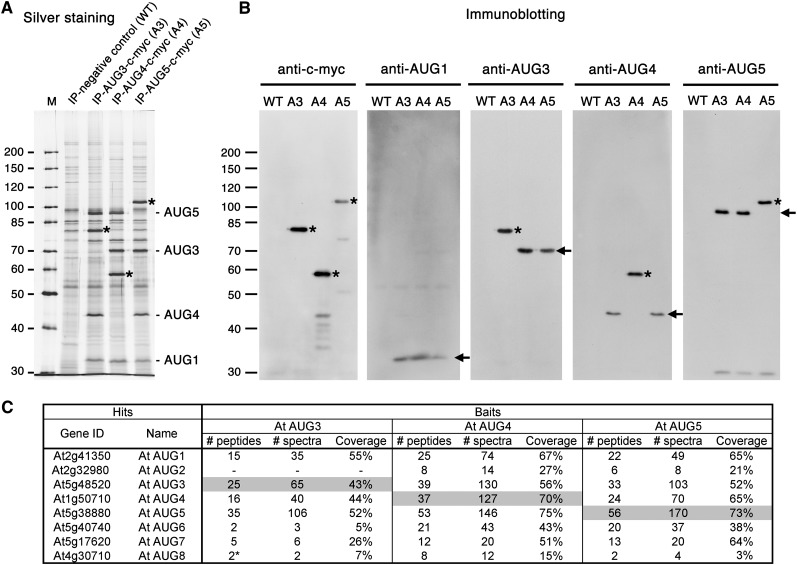
Given that AUG4-myc and AUG5-myc were functional, along with AUG3-c-myc, they were employed as baits for further purification and confirmation of the augmin complex by affinity chromatography. Besides recovering the baits (asterisks, Figure 4A), common bands were revealed by silver staining on SDS-PAGE gels (Figure 4A). These baits were also revealed by immunoblotting with the anti-c-myc antibody (asterisks, Figure 4B). Using antibodies individually raised against AUG1, AUG3, AUG4, and AUG5, the native proteins at 33, 71, 43, and 93 kD, respectively, were detected in the purified protein preparations (arrows, Figure 4B). The anti-AUG3, -AUG4, and -AUG5 antibodies also detected their c-myc–tagged fusion proteins, which migrated behind the native proteins (asterisks, Figure 4B). Interestingly, when a bait was revealed, its corresponding native protein was not detected even though heterozygous plants were used as purification material. In BY-2 cells ectopically expressing AUG3-c-myc, the tobacco AUG1, AUG4, and AUG5 were also copurified with this fusion protein as detected by immunoblotting (see Supplemental Figure 9B online). This result suggested that AUG3-c-myc formed a complex with these endogenous tobacco augmin subunits. Thus, these reciprocal copurifications indisputably support our earlier prediction that these AUG proteins form a stable protein complex in vivo. In addition, the complex likely had only single polypeptides for AUG3, AUG4, and AUG5.
Figure 4.
Reciprocal Purification of Augmin Subunits and Identification of Two Plant-Specific Subunits.
(A) Immunoaffinity purification results using transgenic plants expressing AUG3-c-myc, AUG4-c-myc, or AUG5-c-myc. Purified proteins together with those from a wild-type negative control are revealed in a silver-stained gel. Asterisks indicate the bait for each purification. The positions of AUG1, 3, 4, and 5 are indicated at the right side of the gel image. M, molecular weight markers.
(B) Immunoblotting analysis using anti-c-myc, anti-AUG1, anti-AUG3, anti-AUG4, and anti-AUG5 antibodies. Proteins purified from AUG3-c-myc (A3), AUG4-c-myc (A4), and AUG5-c-myc (A5) include all subunits of the augmin complex. The baits (asterisks) and endogenous augmin subunits (arrows) are revealed by immunoblotting. WT, wild type.
(C) Mass spectrometry results obtained through three independent immunopurifications using AUG3-c-myc, AUG4-c-myc, and AUG5-c-myc as baits. Besides AUG1-6, two additional subunits (AUG7 and AUG8) were identified. Gene ID represents gene identification number by TAIR; # peptides, number of unique peptides; # spectra, number of total spectra; coverage, sequence coverage. The bait of each experiment is highlighted in gray.
Because the animal augmin complexes contain at least eight subunits, we asked whether other proteins were consistently copurified with the three baits used in the purification. To maximize peptide identification, purified proteins were collectively analyzed by a shotgun approach after all purified polypeptides were stacked in an SDS-PAGE resolving gel prior to being subjected to LC-MS/MS analysis. Eight proteins were consistently detected with significant peptide coverage as high as 75% when the three independent baits were used (Figure 4C; see Supplemental Table 3 online). Besides AUG1-6, there were two proteins encoded by the At5g17620 and At4g30710 loci, named AUG7 and AUG8 thereafter, respectively. Again, AUG7 is an acidic protein with pI = 4.82, like the other six augmin subunits identified earlier in Arabidopsis. By contrast, AUG8 is a basic protein with predicted pI of 10.69. Neither AUG7 nor AUG8 shows noticeable amino acid sequence similarity to the human HAUS7 and HAUS8 or other proteins of animal or fungal origin. Thus, they likely are plant-specific augmin subunits.
AUG7 Is Associated with Spindle and Phragmoplast MTs and Plays Essential Roles in Vegetative and Reproductive Growth
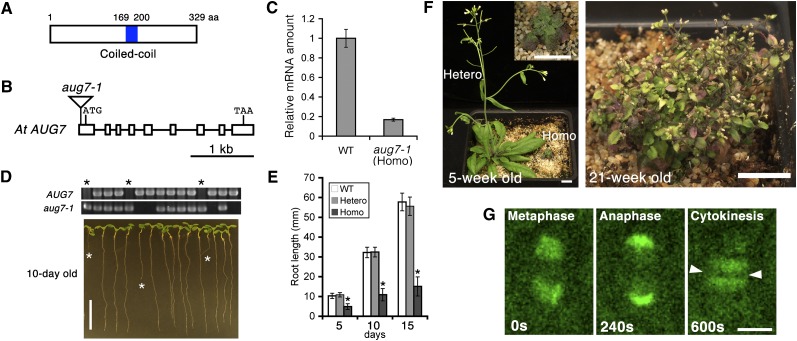
AUG7 contains a predicted central coiled-coil domain (Figure 5A). It shares amino acid sequence identity/similarity of 83.1%/90.9% to the reported tomato (Solanum lycopersicum) NUCLEAR MATRIX PROTEIN1 (NMP1) (Rose et al., 2003) (see Supplemental Figure 10 online). However, the function of NMP1 is unknown.
Figure 5.
Characterization of the AUG7 Gene.
(A) Schematic diagram of the AUG7 protein. A coiled-coil is predicted in the middle of the protein. aa, amino acids.
(B) Schematic representation of the AUG7 gene with exons and introns as open boxes and lines, respectively. The aug7-1 mutation has a T-DNA insertion in the first exon.
(C) Assessment of the expression level of AUG7 by quantitative RT-PCR. The wild-type (WT) expression level was set at 1. The error bars represent sd of four replicates.
(D) Root length test for the offspring derived from self-fertilization of heterozygous aug7-1 mutant. Genotypes of the seedlings were determined based on the detection of the wild-type AUG7 locus (top panel) and/or aug7-1 mutation (middle panel). Among 10-d-old seedlings (bottom panel), the homozygotes have shorter roots (asterisks). Bar = 10 mm.
(E) Comparison of root lengths of wild-type, heterozygous (Hetero), and homozygous (Homo) aug7-1 mutant seedlings at 5, 10, and 15 d after germination. The root lengths of homozygous seedlings are significantly shorter than those of the wild type and heterozygotes (asterisks, t test, P < 10−8). Data are shown as mean ± sd with a minimum of 11 plants.
(F) Pronounced growth retardation caused by the aug7-1 mutation. Five weeks after germination, the homozygous aug7-1 mutant (left panel inset) shows very limited vegetative growth compared with a heterozygous plant, which has already produced an inflorescence (left panel). At 21 weeks after germination, the homozygous aug7-1 plant remains dwarf and produces aborted flowers (right panel). Bars = 10 mm.
(G) Live-cell imaging of AUG7-GFP in a root meristematic cell undergoing mitosis. Snapshots were extracted from Supplemental Movie 2 online. Time is shown in seconds. In metaphase, the AUG7-GFP signal appears in the spindle. The signal is prominent on the shortening kinetochore fiber MTs during anaphase and then seen in the phragmoplast during cytokinesis. Bar = 5 μm.
A mutation was isolated that had a T-DNA insertion in the 5′ untranslated region of the first exon (Figure 5B). This insertion also resulted in a 40-bp deletion 60 bp upstream of the start codon. To determine how the insertion event altered the expression of AUG7, a quantitative RT-PCR experiment was performed: It determined that the homozygous mutant had ~17% of the steady state mRNA level of the wild-type control (Figure 5C). In contrast with the aforementioned +/aug mutants, the self-fertilization progeny of the heterozygous aug7-1 plant showed a normal segregation pattern of +/+:+/aug7-1:aug7-1/aug7-1 = 1:2.05:1.29 (n = 165). However, the aug7-1 homozygous mutant exhibited pleiotropic phenotypes in both vegetative and reproductive growth. At first, we noticed that the homozygous mutant seedlings produced roots with approximately fourfold reduction in length when compared with either the wild-type or heterozygous mutants (Figures 5D and 5E). The homozygous plant also showed seriously retarded growth that became obvious 2 to 3 weeks after germination (Figure 5F). The mutant produced leaves with much reduced sizes that accumulated anthocyanin and appeared purple. Although it remained dwarf, the mutant continued to produce more shoots and eventually inflorescences, and the growth lasted for up to 8 to 10 months after germination. The inflorescences, however, were aborted and the flower buds never opened (Figure 5F). By contrast, the heterozygous plant was indistinguishable from the wild type.
To confirm that the phenotypes described above were caused by the insertional aug7-1 mutation, genetic suppression/complementation experiments were performed using genomic fusions with either the 4xc-myc- or GFP-coding sequence. We recovered plants in the homozygous aug7-1 background that expressed AUG7-c-myc or AUG7-GFP and grew similarly the wild-type control. This supports that the aforementioned phenotypes were indeed caused by the T-DNA insertion and that both fusion proteins were functional.
To test whether AUG7 was associated with other identified AUG subunits in vivo, we performed affinity purification of AUG7-c-myc using an anti-c-myc column. By both protein silver staining and anti-c-myc immunoblotting, the AUG7-c-myc fusion was revealed at 49 kD (see Supplemental Figure 11 online). The purified proteins were also probed with anti-AUG5 antibodies that detected a band at 93 kD, indicating that the AUG5 protein was present. Thus, AUG7 is an integral component of the augmin complex in Arabidopsis.
The tomato AUG7 homolog NMP1 was proposed to be in the nuclear matrix (Rose et al., 2003). However, its localization in dividing cells has not been reported. We observed the AUG7-GFP signal in dividing cells in the root apical meristem by confocal microscopy (Figure 5G; see Supplemental Movie 2 online). The fusion protein decorated the mitotic spindle and appeared pronounced in the spindle poles of the anaphase spindle (Figure 5G, middle panel), a feature shared by both the γ-tubulin complex and other studied AUG proteins. When the cell entered telophase and cytokinesis, AUG7-GFP became associated with the phragmoplast and left a wide dark zone in the middle of the phragmoplast (Figure 5G, arrowheads, right panel). Again, this is similar to the localization pattern of γ-tubulin. We conclude that AUG7 is a bona fide subunit of the Arabidopsis augmin complex.
Reduced AUG7 Expression Compromises the Morphogenesis of the Spindle and Phragmoplast MT Arrays
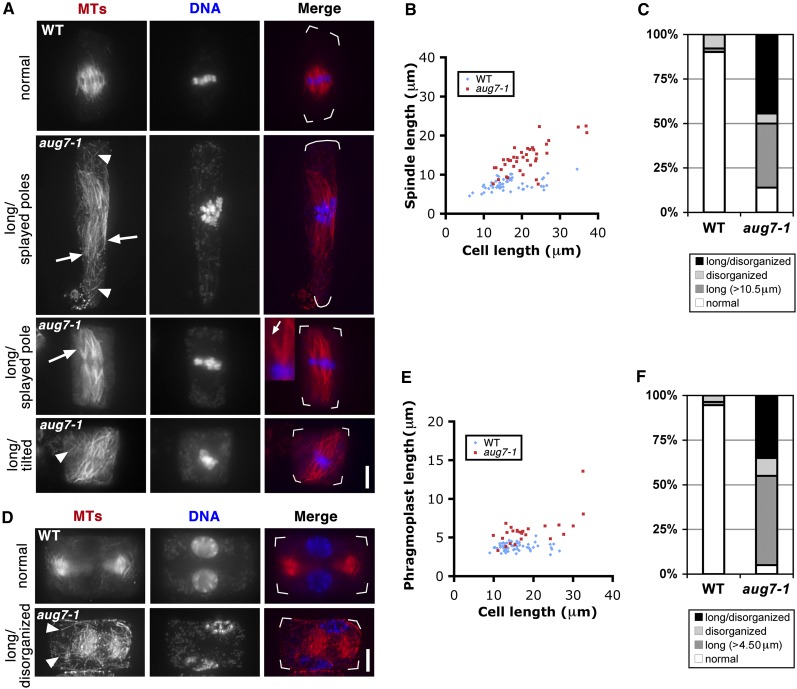
Previously, we were unable to reveal how augmin affected mitosis in somatic cells because the aug3-1 and other aug mutations were sporophyte lethal. Since the homozygous aug7-1 plants had significantly reduced AUG7 expression and showed pronounced growth retardation, we asked whether the phenotype was due to defects in MT organization caused by compromised AUG7 function. Mitotic cells in the root apical meristem were convenient for us to observe spindle and phragmoplast MT arrays by antitubulin immunofluorescence. These cells typically form spindles along the root axis in the wild-type roots. Although the spindles are not perfectly focused, they contain converging poles and are confined in the central region of the cell (WT, Figure 6A). Abnormal spindles were formed in the aug7-1 cells and exhibited distorted morphologies in several aspects (aug7-1, Figure 6A). At first, spindle poles often failed to converge and MTs were splayed more or less toward the cell ends (arrows, Figure 6A), although two half spindles could clearly discerned. Second, spindles often became elongated when compared with the control ones. We compared the spindle lengths and used the cell length as a reference. The wild-type spindles showed consistent lengths of 7.52 ± 1.48 μm no matter how long the cells were (~10 to 30 μm range, 16.5 ± 5.66 μm on average, n = 53). On the other hand, spindles produced by the homozygous aug7-1 cells generally had longer spindles. In addition, longer spindles were found in longer cells (Figure 6B). The lengths of aug7-1 spindles varied from 10 to 25 μm with an average of 14.1 ± 3.81 μm, while the cell lengths ranged from 15 to 40 μm with an average of 21.0 ± 5.92 μm (n = 40) (Figure 6B). Third, spindles inside the aug7-1 mutants often were tilted and oriented diagonally as if they were expanding to the geometric limit (Figure 6A). In addition to these abnormal spindle morphologies, fine and discrete MTs were often detected in the cytoplasm. We quantitatively assessed and compared the spindle morphology in the wild-type and aug7-1 cells by dividing them into four categories: (1) normal spindles oriented along the root axis, (2) long spindles with lengths exceeding 10.5 μm, (3) disorganized spindles with splayed poles or MTs randomly packed near the chromosomes, and (4) disorganized and long spindles. In the wild-type cells, >90% of the spindles were in the “normal” category (n = 51). In aug7-1 cells, abnormal metaphase spindles were found at a very high frequency of 86% (n = 36) (Figure 6C).
Figure 6.
Abnormal MT Organization in the Mitotic Spindle and Phragmoplast of the aug7-1 Mutant.
(A) and (D) Immunofluorescence of MTs in metaphase spindles (A) and phragmoplasts (D) in root meristematic cells of the wild-type (WT) control and aug7-1 mutant. In merged images, MTs are pseudocolored in red and DNA in blue. Cell outline is indicated by white frames in the merged images.
(A) A metaphase spindle of the wild type shows converged spindle poles. The aug7-1 mutant cells often form long, diagonally oriented and/or disorganized spindles with unconverged poles (arrows). Discrete MTs are also detected in the cytoplasm (arrowheads).
(D) Compared with MTs in the wild-type phragmoplast, phragmoplast MTs in aug7-1 tend to be longer and disorganized. Discrete MTs are also detected elsewhere in the cytoplasm (arrowheads). Bar = 5 μm.
(B) and (E) Quantification of the spindle length with the cell length as a reference (B) and the phragmoplast length with the cell length as a reference (E) in the wild-type control and aug7-1 mutant. Generally, longer spindles (B) and phragmoplasts (E) are found in aug7-1 cells compared with those of the wild type.
(C) and (F) Proportions of spindles (C) and phragmoplasts (F) that show normal and abnormal (long, disorganized, and long and disorganized patterns) configurations in the wild type and aug7-1 mutant.
Abnormal phragmoplast MT arrays were also detected in the aug7-1 mutant cells. In the wild type, the control phragmoplasts showed well-organized bipolar MT arrays, and reforming daughter nuclei were positioned vertically on the opposite sides of the phragmoplast (top panels, Figure 6D). The phragmoplasts in the homozygous aug7-1 cells often contained loosely packed fine MTs (bottom panels, Figure 6D). They were often longer than the control phragmoplasts and assumed oblique positions. Concomitant with the oblique orientation of the phragmoplast, the two daughter nuclei were positioned according to the phragmoplast axis (Figure 6D). On average, the mutant phragmoplasts were 5.86 ± 1.91 μm (n = 25) in length, while the wild-type controls were 3.78 ± 0.493 μm (n = 55; Figure 6E). In addition, aug7-1 cells often had fine MTs scattered in the cytoplasm, away from the phragmoplast proper (arrowheads, Figure 6D). Again, when assessed quantitatively, the mutant produced elongated or disorganized phragmoplast MT arrays at a frequency >95% (n = 20), and <6% of the control phragmoplasts were abnormal (Figure 6F). Occasionally, some cells harbored double spindles (see Supplemental Figure 12A online). Other dividing cells of the aug7-1 mutant showed excessive numbers of chromosomes, indicating cells were polyploid (see Supplemental Figures 12B and 12C online).
AUG7 Regulates the Association of γ-Tubulin with the Spindle and Phragmoplast MTs
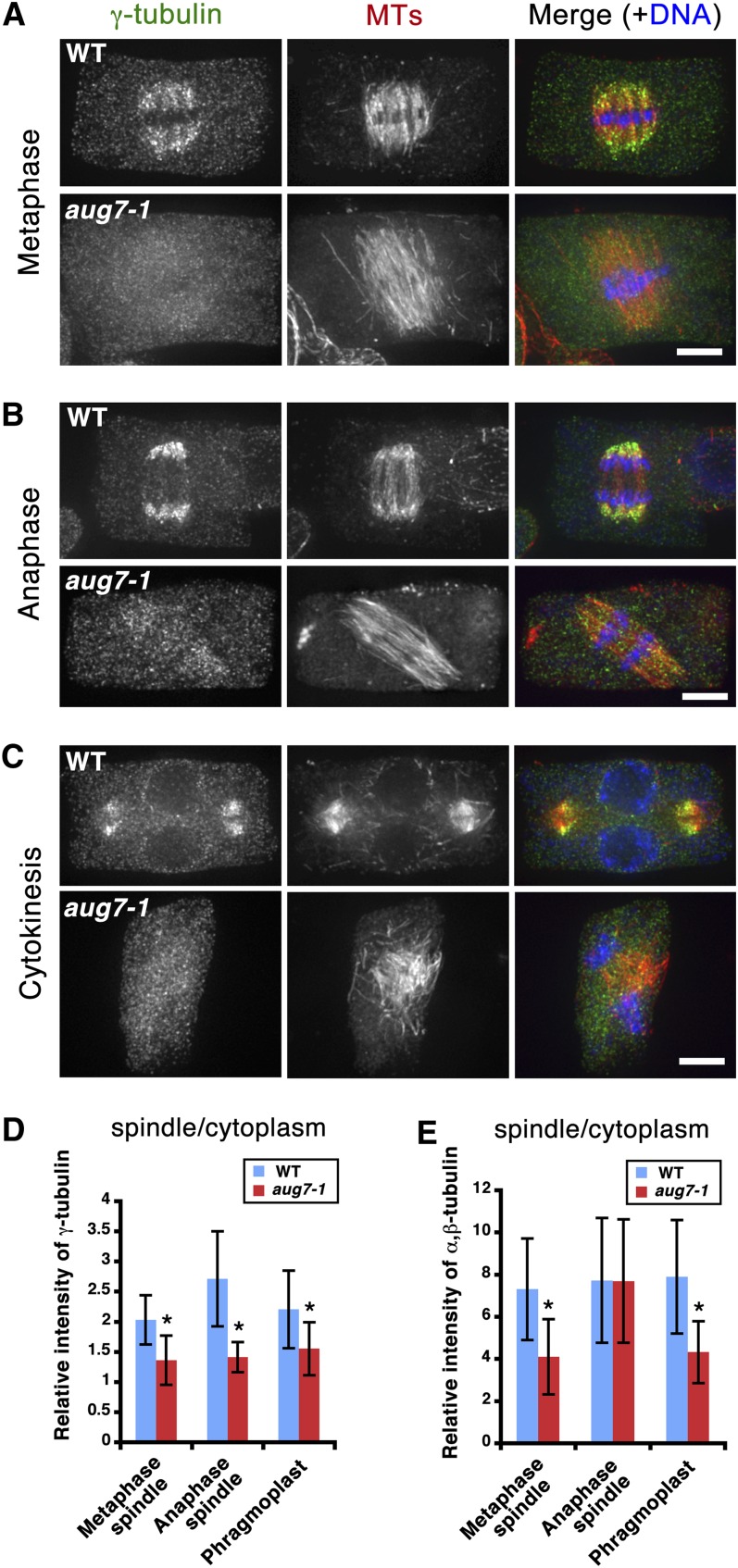
Since the aug7-1 mutation seriously affected the organization of MTs in spindles and phragmoplasts, we asked whether these defects were caused by mislocalization of γ-tubulin inside the spindle and phragmoplast. In the metaphase spindle, γ-tubulin was detected along MTs with biases toward spindle poles where MT minus ends were concentrated (Figure 7A, top panel). In the aug7-1 cells, however, the γ-tubulin signal was hardly detected inside the metaphase spindle and was nearly indistinguishable from the diffuse signal in the cytoplasm (Figure 7A, bottom panel). We quantified the signal intensity of γ-tubulin inside the metaphase spindle and outside the spindle in the cytoplasm. Wild-type metaphase cells had an average ratio of 2.03 ± 0.41 (n = 38), while the homozygous mutant had a ratio of 1.36 ± 0.41 (n = 27), clearly demonstrating drastic reduction of γ-tubulin association with the metaphase spindle in the mutant cells (Figure 7D).
Figure 7.
Delocalization of γ-Tubulin in the Mitotic Spindle and Phragmoplast of aug7-1 Mutant.
(A) to (C) Immunofluorescence of γ-tubulin and MTs in the wild-type and aug7-1 mutant cells. In the merged images, γ-tubulin is pseudocolored in green, MTs in red, and DNA in blue. WT, wild type. Bar = 5 μm.
(A) In a wild-type metaphase cell, γ-tubulin decorates kinetochore fiber MTs with biases toward spindle poles (top panel). Such a pattern is barely detected in the unconverged metaphase spindle of the aug7-1 cell at a similar stage (bottom panel).
(B) During anaphase, γ-tubulin signal is prominently detected along shortening kinetochore fiber MTs in the wild type (top panel), but the signal is greatly diminished in an anaphase aug7-1 cell (bottom panel).
(C) In the wild-type phragmoplast, γ-tubulin localizes on MTs with biases toward minus ends (top panel). In aug7-1, only weak signals are detected on disorganized phragmoplast MTs (bottom panel).
(D) and (E) Quantitative assessment of the fluorescent signal intensities of γ-tubulin (D) and α,β-tubulin (E). The ratios of the spindle- or phragmoplast-localized signals to the cytoplasmic ones are shown as mean ± sd. When marked with asterisks, the decreases in the relative intensity are statistically significant (t test, P < 0.0001).
During anaphase in the wild type, prominent γ-tubulin signals were associated with shortening kinetochore fiber MTs near the spindle poles (Figure 7B, top panel). In the homozygous aug7-1 mutant, again, such a striking localization pattern of γ-tubulin was no longer observed near the spindle poles (Figure 7B, bottom panel). Quantitatively, the γ-tubulin intensity ratio of the polar region and cytoplasm was 2.71 ± 0.79 (n = 9) in wild-type cells but dropped to 1.41 ± 0.25 (n = 5) in the aug7-1 cells (Figure 7D).
A similar γ-tubulin delocalization phenomenon was observed in cells bearing a phragmoplast. Instead of decorating MTs in a manner biased toward their minus ends as in the wild-type phragmoplasts (Figure 7C, top panel), γ-tubulin no longer accumulated on the MTs in the aug7-1 cells undergoing cytokinesis. Instead, the signal became diffuse throughout the cytoplasm (Figure 7C, bottom panel). When the phragmoplast-localized γ-tubulin signal was compared with that in the cytoplasm, a ratio of 2.21 ± 0.64 (n = 43) was found in wild-type cells. This ratio was reduced to 1.55 ± 0.44 (n = 16) in aug7-1 mutant cells (Figure 7D).
We also noticed that the intensities of the MT signals inside the metaphase spindle and the phragmoplast were lower than those in the control cells (Figures 7A and 7C). Quantitatively, the ratios of antitubulin fluorescent intensity in the spindle and phragmoplast to cytosol dropped from 7.31 ± 2.41 (n = 38) and 7.90 ± 2.69 (n = 44) in control cells to 4.11 ± 1.78 (n = 26) and 4.32 ± 1.46 (n = 17) in aug7-1 cells, respectively (Figure 7E). By contrast, the ratios did not show an obvious difference in anaphase spindles (Figure 7E).
Taken together, we conclude that the γ-tubulin localization to spindle and phragmoplast MTs is dependent on AUG7 and likely other augmin subunits. When γ-tubulin is not properly targeted to MTs, MT organization and consequently mitosis can be compromised as demonstrated by the aug7-1 homozygous mutant.
DISCUSSION
In this study, we report the discovery of the Arabidopsis augmin complex composed of at least eight subunits that are associated with the γ-tubulin complex during mitosis and cytokinesis. The function of augmin is required for the acentrosomal mitosis and cytokinesis in plants, as reduced AUG7 expression led to serious defects in MT organization in the spindle and phragmoplast and T-DNA insertional mutants of the AUG1 to AUG5 genes were lethal in both haploid gametophytic and diploid sporophytic cells. Thus, plants and animals deploy an analogous augmin-dependent mechanism, despite variations in the augmin subunits, in regulating the morphogenesis of mitotic and cytokinetic MT arrays.
The Arabidopsis Augmin Complex Contains Both Conserved and Unique Subunits Compared with Its Animal Counterparts
Among the eight augmin subunits reported in the previous and current studies, AUG1 to AUG6 can be aligned with the human HAUS1 to HAUS6 proteins despite very low sequence similarity. This is somewhat surprising because the corresponding subunits in fly and human cells are not absolutely conserved (Lawo et al., 2009; Uehara et al., 2009). Further sequence analysis predicted that proteins like AUG2 and AUG6 share significant structural similarities to their human counterparts. For example, AUG2 contains three predicted coiled-coil domains and the second and third domains have calculated high pI values of 9.99 and 10.00, respectively (see Supplemental Figure 1 online). Similarly, HAUS2 contains coiled-coil domains with predicted pIs for the second and third domains of 10.45 and 9.31, respectively (see Supplemental Figure 1 online). Both AUG6 and HAUS6/FAM29A have four predicted coiled-coils and contain N-terminal basic domains with predicted pIs of 9.18 and 9.14, respectively (see Supplemental Figure 4 online). It remains to be tested whether these deduced features contribute to the assembly of the augmin complex and/or the interaction with the γ-tubulin complex.
Unlike AUG1-6, AUG7, and AUG8 are conserved only within the plant kingdom. Interestingly, Drosophila does not possess a homolog of the human HAUS7/UCHL51P protein nor do humans have a homolog of the Drosophila augmin subunit Dgt8/Wac (Meireles et al., 2009; Uehara et al., 2009). However, some structural features may be conserved among proteins with no obvious sequence conservation. For example, Dgt8/Wac possesses a characteristic central coiled-coil domain through which it interacts with Dgt2 (Meireles et al., 2009). A central coiled-coil domain was also identified in the AUG7 protein reported here. Thus, structural features of coiled-coil domains in AUG7 and other AUG proteins may allow direct interactions as seen between Dgt2 and Dgt8/Wac.
AUG8 is a member of the previously identified QWRF protein family in Arabidopsis, including ENDOSPERM-DEFECTIVE1 (EDE1) and SNOWY COTYLEDON3 (Pignocchi et al., 2009; Albrecht et al., 2010). These proteins may share the MT interaction property, as demonstrated by EDE1 (Pignocchi et al., 2009). The MT binding/bundling subunits of human and Drosophila augmins, Hice1/HAUS8 (410 amino acids) and Dgt4 (188 amino acids), share similarity only in a 49–amino acid stretch despite the drastic difference in protein sizes (Uehara et al., 2009). Based on its cDNA, AUG8 is a 644–amino acid protein with a DUF566 domain of unknown function. Taken together, the AUG8 and its functional counterparts in animals may represent the most variable subunit in the augmin complex from different organisms.
In Drosophila, Msd1/Dgt9 was shown to be a MAP as well (Wainman et al., 2009). This raises a possibility that the augmin complex may contain more than one MAP. Structurally, Msd1/Dgt9 and HAUS2/Cep27 can be paired with each other (Duncan and Wakefield, 2011). Because of its sequence similarity to HAUS2, it would be interesting to test whether AUG2 also interacts with MTs directly.
Except for AUG8, other augmin subunits are encoded by single genes in the Arabidopsis genome. This may explain why loss-of-function mutations in any of the seven genes encoding AUG1-7 would lead to serious defects in mitosis and cytokinesis and likely sporophytic death. A T-DNA insertional mutation in the AUG8 coding region did not cause any noticeable phenotype. This suggests that AUG8 and other QWRF proteins may be functionally redundant in Arabidopsis. It is also plausible that AUG8 and its relatives may be assembled into different complexes. For instance, other QWRF proteins could have been missed in this MS analysis. Collective conclusions can be drawn from genetic analysis of these homologous genes. Nevertheless, an interaction between augmin and MTs may be established through the direct interaction between AUG8 and MTs because AUG8 likely possesses an MT binding property, as demonstrated for EDE1 (Pignocchi et al., 2009).
Unlike other AUG subunits, AUG8 was often detected at low peptide coverage by mass spectrometry (Figure 4C). Two reasons might explain this result. AUG8 may be weakly associated with other AUG subunits that are tightly bound together. Alternatively, the formation of the intact augmin complex may be regulated in a cell cycle–dependent manner. Indeed, recent data suggest that two mitotic kinases, Aurora A and Polo-like kinase 1, phosphorylate Hice1 in human cells (Johmura et al., 2011; Tsai et al., 2011). Whereas phosphorylation at 17 sites by Polo-like kinase 1 was critical for Hice1 to interact with MTs, phosphorylation events by Aurora A at different sites decreases the interaction with MTs and its association with HAUS6/FAM29A. Therefore, cell cycle–dependent phosphorylation may regulate the assembly and function of the augmin complex in Arabidopsis as well.
Although AUG8 was detected with other AUG proteins in our reciprocal immunopurification experiments, it remains to be tested whether it is a functional augmin subunit. Future studies through cell biological and genetic approaches will be necessary to draw conclusions.
Augmin-Dependent Spindle Morphogenesis in Plant Cells
In animal cells, the immediate consequence upon the loss of augmin is the diminished localization of γ-tubulin on spindle MTs, but not at the centrosome (Goshima et al., 2007, 2008; Lawo et al., 2009; Uehara et al., 2009). In plant cells, γ-tubulin and its associated proteins are prominently detected along spindle MTs with a bias toward spindle poles (Liu et al., 1993, 1995; Zeng et al., 2009; Nakamura et al., 2010). Our results indicate that an augmin-dependent mechanism also regulates the localization of γ-tubulin in plant spindles. Together with our report on the aug3 mutant (Ho et al., 2011b), phenotypes exhibited by the aug7-1 cells allow us to gain further insights into the function of MT-localized γ-tubulin in spindle morphogenesis. At first, we found that spindles in the mutant cells have splayed poles. This result suggests that biased localization of γ-tubulin toward the spindle pole and the subsequent efficient nucleation of new MTs are required for the formation of converged spindle poles. This is different from the scenario in the formation of centrosomal spindles in which focused spindle poles can still be maintained when augmin is downregulated (Goshima et al., 2008). This is probably due to the fact that augmin is not required for the localization of γ-tubulin and proteins like NEDD1 at the centrosome (Goshima et al., 2008; Zhu et al., 2008; Lawo et al., 2009). In the absence of augmin, therefore, MTs can still be nucleated from the centrosome during mitosis to establish a bipolar spindle with focused poles. By contrast, augmin is responsible for the maintenance of the converged poles of acentrosomal spindles in plant cells.
Upon augmin downregulation, animal and plant cells share a phenotype of elongated spindles (Goshima et al., 2008; Meireles et al., 2009; Wainman et al., 2009; Ho et al., 2011b). It was postulated that the increase in MT number due to augmin-dependent MT nucleation would shorten individual MTs within the spindle, so that longer spindles would be formed upon the inactivation of MT-dependent MT generation (Goshima and Kimura, 2010). Such a hypothesis would rely on the formation of kinetochore fiber MTs initiated from the centrosome. A similar phenotype of elongated spindles was also found during acentrosomal spindle formation upon the depletion of augmin using X. laevis egg extracts (Petry et al., 2011). However, the meiotic spindle formation under such challenges still relies on the nucleation of MTs from focused/converged spindle poles that are regulated by the cytoplasmic dynein. It was suggested that the lack of sufficient antiparallel MTs near the chromosome would lead to oppositely positioned kinetochore fibers being weakly connected so that spindles are elongated (Petry et al., 2011). In the aug7 mutant cells, spindle poles were splayed so that it would be challenging for the cells to generate centralized MTs toward the chromosomes. It is more likely that MTs generated from the chromosomes may contribute to the assembly of an elongated bipolar spindle in these AUG7-downregulated cells. Earlier observations in Haemanthus and onion (Allium cepa) cells revealed branched MTs along kinetochore fibers, described as microtubular fir trees (MTFTs) (Palevitz, 1988; Smirnova and Bajer, 1992). Unfortunately, MTFTs cannot be easily observed in small Arabidopsis cells. Thus, it awaits examination at high resolution to ascertain whether augmin is required for the formation of MTFTs. If so, augmin would likely contribute to the formation of short MT branches. When its function is compromised, longer MTs would be formed in the spindle.
How would a bipolar spindle be established without the converging poles in the aug7 cells? In the absence of the centrosome, plant cells often form mitotic MT caps on the prophase nuclear envelope and establish bipolar spindles (Lloyd and Chan, 2006). This does not exclude the possibility of a chromosome-based mechanism as observed in animal cells. It has been shown that augmin is not essential for chromosome-mediated MT nucleation for spindle assembly (Goshima et al., 2008; Petry et al., 2011). We suggest that chromosome-based MT nucleation is responsible for the establishment of the bipolar spindle in Arabidopsis cells lacking an efficiently functioning augmin. Although the bipolarity is established, MTs of the mutant spindle would not converge to virtual poles in the absence of poles preestablished by mitotic MT caps at late prophase. By decreases in branching as well as in the degree of pole-oriented convergence, the spindle MT array may become longer. Thus, in addition to augmin, other proteins may be accountable for MT nucleation at polar caps during prophase and on chromosomes during prometaphase.
Similarly, albeit being defective, acentrosomal meiotic spindles were established in a bipolar fashion when augmin was downregulated by RNA interference in frog extracts (Petry et al., 2011). Multiple mechanisms are known to contribute to the assembly of the spindle apparatus in animal cells (Duncan and Wakefield, 2011). Whereas some are dependent on the γ-tubulin complex, others rely on proteins like the Ran GTPase and MAP/motors. Consistent with what has been found in animal cells, augmin is required for localizing the γ-tubulin complex to the spindle in Arabidopsis. The γ-tubulin complex is likely present in cytosolic and MT-localized pools. When it is no longer localized to the spindle apparatus or the phragmoplast, the cytosolic pool may still function in MT nucleation during spindle assembly. Chromosomes would function to anchor MT plus ends so that MT minus ends would be point away from the chromosomes. An alternative solution would rely on proteins like the XMAP215 homolog MOR1/GEM1, which decorates spindle and phragmoplast MTs (Hamada et al., 2004; Kawamura et al., 2006). In addition, the Kinesin-5 motor is another critical player during spindle assembly in plant cells because its loss leads to the collapse of the bipolar array into a monopolar one (Bannigan et al., 2007). These proteins all are essential for mitosis, although phenotypes produced by the loss of their functions differ from one another (Twell et al., 2002; Eleftheriou et al., 2005; Kawamura et al., 2006; Bannigan et al., 2007).
It has been shown that two augmin subunits, HAUS6/FAM29A and HAUS8/Hice1, interact with Ndc80/Hec1, a protein at both the kinetochore and the centrosome, which is required for the assembly of the kinetochore fibers (Bucciarelli et al., 2009; Wu et al., 2009). Surprisingly, downregulation of Hice1 abolishes Hec1 localization only at the centrosome but not at the kinetochores (Wu et al., 2009). The results suggest that Hice1 and Hec1 work together at the centrosome to contribute to spindle assembly initiated at the centrosomes. It would be interesting to test whether augmin and the Ndc80 ortholog in plants work together to contribute to the formation of spindle poles.
We routinely detected diagonally aligned spindles in the aug7 mutant cells. This phenomenon may be a result of prolonged spindle elongation. When spindles continue to elongate in the mutant cells, they would seek extra space in which to expand. Reaching the diagonal position would grant the maximal length for a spindle. Interestingly, the phragmoplasts in mutant cells often were misaligned as well. This suggests that the orientation of the spindle apparatus likely influences that of the phragmoplast.
METHODS
Plant Materials and cDNA Clones
The Arabidopsis thaliana lines used in this study are control Columbia-0 and the SALK_098567 (aug1-1), SALK_059104 (aug2-1), SALK_019542 (aug4-1), SAIL_722_G02 (aug5-1), and SK18263 (aug7-1) mutant lines generated by the Arabidopsis research community (Sessions et al., 2002; Alonso et al., 2003; Robinson et al., 2009), the AUG3-c-myc line (Ho et al., 2011b), the qrt mutant (Preuss et al., 1994), and the mCherry-TUB6 line (Nakamura et al., 2010). Full-length AUG3, AUG4, and AUG5 cDNA clones were generated by colleagues as published earlier (Seki et al., 2002; Castelli et al., 2004). Plant growth conditions and transformation procedures were as described previously (Kong et al., 2010). For the root elongation test, seeds were germinated on vertically placed plates containing 1.2% agar and half-strength strength Murashige and Skoog salts.
Detection/Genotyping of the aug1-1, aug2-1, aug4-1, aug5-1, and aug7-1 Mutations and Phenotypic Characterization
Primers used in this study and their corresponding sequences are listed in Supplemental Table 4 online. The T-DNA insertions at the AUG1, AUG2, AUG4, AUG5, and AUG7 loci were detected by PCR using the following primer sets: aug1-1, 2g1RP3 and LBa1; aug2-1, TH072 and LBb1.3; aug4-1, 1g2RP and LBa1; aug5-1, 5g2RP and GLB3; and aug7-1, sk18263-RP and SK-L1. The wild-type alleles were detected by the following primer sets: AUG1, 2g1RP3 and 2g2LP; AUG2, TH072 and TH071; AUG4, 1g2RP and 1g2LP; AUG5, 5g2RP and 5g2LP; and AUG7, sk18263-RP and sk18263-LP. To identify the T-DNA insertion position in the sk18263 mutant, its genomic DNA was used to amplify the predicted region containing the T-DNA insertion using primers sk18263-LP and sk18263-RP, and the resulting PCR product was then sequenced with primers sk18263-LP and sk18263-RP for both ends.
Backcrosses with wild-type plants were performed at least twice before the characterization, and the observed phenotypes were confirmed to be linked to the T-DNA insertion. The measurement of genetic segregation of self-fertilization and the practice of reciprocal crosses were performed as described previously (Ho et al., 2011b). Defects in male gametogenesis were detected by DNA staining using 4′,6-diamidino-2-phenylindole (aug1-1, aug4-1, and aug5-1) or by dissecting seeds in siliques (aug2-1).
Genetic Suppression/Complementation of the aug4, aug5, and aug7 Mutation and Expression of AUG6-GFP
Genomic fragments of 4.5, 5.9, 6.1, and 3.4 kb for AUG4, AUG5, AUG6, and AUG7, respectively, which contain the promoter regions and the coding sequences, were amplified by PCR using Phusion DNA polymerase (New England Biolabs) and the following primer sets: AUG4, 50710-F and 50710-R; AUG5, 38880-F and 38880-R; AUG6, 40740-F and 40740-R; and AUG7, 17620-F and 17620-R. Each amplified DNA was cloned into the Gateway pENTR/D-TOPO vector (Life Technologies) according to the manufacturer’s instructions. The resulting plasmids were recombined with pGWB16 (for C-terminal 4×c-myc fusions of AUG4, AUG5, and AUG7) or pGWB4 (for C-terminal GFP fusions of AUG6 and AUG7) vectors (Nakagawa et al., 2007) by LR recombination reactions (Life Technologies). Consequently, the pGWB16-AUG4, pGWB16-AUG5, pGWB4-AUG6, pGWB4-AUG7, and pGWB16-AUG7 plasmids were used for Agrobacterium tumefaciens–mediated transformation into the heterozygous mutants +/aug4-1, +/aug5-1, and +/aug7-1 (pGWB16-AUG4, pGWB16-AUG5, pGWB4-AUG7, and pGWB16-AUG7) or Columbia-0 wild-type plants with/without expression of mCherry-TUB6 (pGWB4-AUG6). The resulting fusion proteins were revealed to be functional by their complementation of the corresponding mutant phenotypes. Because of the use of endogenous promoters for all these constructs, the expression level of each fusion protein was expected to be similar to that of the native proteins. Segregation patterns of T-DNA insertions were determined by PCR-based genotyping as described above, using progeny derived from an untransformed heterozygous mutant and two independent transformants.
RNA Extraction and Real-Time Quantitative RT-PCR
RNA samples were prepared from young rosette leaves of 4-week-old aug7-1 lines and wild-type control plants. Protocols for RNA preparation and real-time RT-PCR were adapted from a previous study (Kong et al., 2010), with modifications. In brief, the cDNA samples were diluted to 20 and 5 ng/μL. Triplicate quantitative assays were performed with 1 μL of each cDNA dilution in a 25-μL reaction containing IQ SYBR Green Supermix using an iCycler IQ5 real-time PCR system (Bio-Rad). The relative quantification method delta-delta cycle threshold was used to evaluate quantitative variations between replicates examined. Three biological replicates were performed. The amplification of PP2A, encoding protein phosphatase 2A, was used as a reference to normalize the data (Czechowski et al., 2005). Gene-specific primers for AUG7 were Q-17620_F and Q-17620_R, which span the third and fourth exon and render a 197-bp product from the wild-type locus.
Generation of Anti-AUG3, Anti-AUG4, and Anti-AUG5 Antibodies
The coding regions of AUG3, AUG4, and AUG5 were amplified using Pfx DNA polymerase (Life Technologies) and the BX832241, U50487, and U23033 plasmids as templates with the following primer sets: AUG3, TH121 and TH122; AUG4, U50487_Bam5′ and U50487_Xho3′; and AUG5, Xba23033F and Hind23033R, respectively. The resulting fragments were cloned into the PGEX-KG vector at the EcoRI and SacI sites (AUG3), pGEX-4T-1 vector at the BamHI and XhoI sites (AUG4), or pGEX-KG vector at the XbaI and HindIII sites (AUG5) after the fragments and vectors were digested by each combination of enzymes. The recombinant plasmids coded for the expression of glutathione S-transferase-AUG3 (GST-AUG3), GST-AUG4, or GST-AUG5 fusion protein in bacteria host BL21 (DE3) (Life Technologies). The fusion proteins were purified using immobilized glutathione (Thermo Scientific) according to the manufacturer’s instructions and used as the antigens for immunization of mice (AUG3 and AUG5) or a rat (AUG4). Antibody production and purification were as described previously (Zeng et al., 2009).
Affinity Purification and Mass Spectrometry
Proteins were extracted from control Arabidopsis plants and transformants expressing the 4×c-myc fusions with AUG3, AUG4, AUG5, and AUG7. In brief, 3-d-old etiolated seedlings were frozen in liquid nitrogen and ground to powder using a mortar and pestle. An extraction buffer of 50 mM Tris HCl, pH 8.0, containing 150 mM NaCl and 1% Triton X-100 was added to the powder and incubated at 4 °C for 1 h. The supernatant was collected after centrifugation at 15,000g and filtration through a 0.45-μm Millex HV filter (Millipore). The c-myc fusion proteins were purified with anti-c-myc antibody-conjugated magnetic beads according to the manufacturer’s instructions (Miltenyi Biotec) and subjected to SDS-PAGE with 7.5% polyacrylamide gels. Gels were stained with silver nitrate or transferred to polyvinylidene fluoride membranes (Millipore) for further immunoblotting analysis. The c-myc fusion proteins were detected by the 9E10 anti-c-myc antibody (Developmental Studies Hybridoma Bank at University of Iowa), and AUG1, AUG3, AUG4, and AUG5 were detected by the antibodies described above and previously (Ho et al., 2011b). The anti-AUG3, AUG4, and AUG5 antibodies were diluted at 1:1000.
To identify copurified proteins, the gels were stained with GelCode Blue Safe protein stain (Thermo Scientific) and excised, digested with trypsin, and subjected to LC-MS/MS. To identify peptides, mass spectra were analyzed using X! Tandem (The Global Proteome Machine) with the Arabidopsis IPI protein database (version 3.83) and then loaded into Scaffold 3.0 (Proteome Software). The cutoff setting used is as follows: minimum two unique peptides, protein identification probability >95%, and peptide identification probability >80%. To exclude unrelated proteins, obvious contaminants, such as nonrelated enzymes or proteins, also detected in the negative control were removed. We listed AUG2 in Figure 1B (indicated with an asterisk) despite its detection with single peptide (thus, below our cutoff criteria) since it was validated by another analysis shown in Figure 4C. AUG8 listed in Figure 4C (indicated with an asterisk) was given when peptide identification probability limit was lowered to 50% for AUG3-myc IP.
Protein Purification from Tobacco Suspension-Cultured BY-2 Cells
Tobacco (Nicotiana tabacum) suspension-cultured BY-2 cells were maintained as described elsewhere (Kumagai-Sano et al., 2006). The Arabidopsis AUG3 promoter:AUG3-4xc-myc construct that was used for the complementation experiment for the Arabidopsis aug3-1 mutant in the previous study (Ho et al., 2011b) was transformed into BY-2 cells using an Agrobacterium-mediated method (Kumagai-Sano et al., 2006). Proteins were extracted from evacuolated protoplasts as described (Hamada et al., 2004) and subjected to coimmunoaffinity purification using anti-c-myc antibody as described above.
Immunolocalization and Fluorescence Microscopy
Immunolocalization of tubulins in pollen grains was performed as described previously (Lee et al., 2007). Indirect immunofluorescence of root tip cells or BY-2 cells was performed as described previously (Lee and Liu, 2000). For dual localization study of c-myc fusion proteins and MTs, rabbit anti-c-myc antibodies (Sigma-Aldrich) and fluorescein isothiocyanate–conjugated donkey anti-rabbit IgG (Rockland Immunochemicals) were used. MTs were stained with DM1A anti-α-tubulin (Sigma-Aldrich) followed by Texas Red–conjugated goat anti-mouse IgG (Molecular Probes). For MT staining of the aug7-1 mutant, sheep antitubulin antibodies (Cytoskeleton) were used with Texas Red–conjugated donkey anti-goat IgG (Rockland Immunochemicals). For dual localization with γ-tubulin, G9 mouse monoclonal anti-γ-tubulin antibody (Horio et al., 1999) was used with fluorescein isothiocyanate–conjugated donkey anti-mouse IgG (Rockland Immunochemicals). Images were acquired with a CFI Plan Fluor ×100 objective (numerical aperture of 1.3) under an Eclipse 600 epifluorescence microscope (Nikon) or a UPlan Apo ×100 (numerical aperture of 1.35; Olympus) under a DeltaVision microscope (Applied Precision).
To observe AUG6-GFP, AUG7-GFP, and mCherry-tubulin signals, seeds were germinated on agar medium containing half-strength Murashige and Skoog salt. Seedlings were observed under a Marianas spinning disk confocal microscope (Intelligent Imaging Innovations) using a Plan-Apo ×63 objective (Carl Zeiss) equipped with diode-pumped solid-state lasers with excitation at 488 and 561 nm for GFP and mCherry fusion proteins, respectively. Images were obtained with an electron-multiplying charge-coupled device camera driven by the Slidebook 5.0 software. Figures and movies presented here were assembled in the Metamorph (Molecular Devices) or SoftWoRx (Applied Precision) software packages.
Accession Numbers
The Arabidopsis Information Resource (TAIR) locus identifiers for the genes mentioned in this study are At2g32980 for AUG2, At1g50710 for AUG4, At5g38880 for AUG5, At5g40740 for AUG6, At5g17620 for AUG7, and At4g30710 for AUG8.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Comparison of Plant AUG2 Homologs and Human HAUS2.
Supplemental Figure 2. Sequence Comparison of Plant AUG4 Homologs and Human HAUS4.
Supplemental Figure 3. Sequence Comparison of Plant AUG5 Homologs, Human HAUS5, and Drosophila melanogaster (Fly) Dgt5.
Supplemental Figure 4. Sequence Comparison of Plant AUG6 Homologs, Human HAUS6, and Dm Dgt6.
Supplemental Figure 5. Phenotypes Caused by the aug1-1, aug2-1, aug4-1, and aug5-1 Mutations.
Supplemental Figure 6. Localization of AUG4-c-myc in Mitotic Root Cells.
Supplemental Figure 7. Measurement of the Fluorescence Intensity of AUG6-GFP in the Metaphase Spindle and Phragmoplast Shown in Figure 3M.
Supplemental Figure 8. Dual Localizations of AUG4/AUG5 and γ-Tubulin.
Supplemental Figure 9. Detection of the Interaction between Augmin and γ-Tubulin in Vivo.
Supplemental Figure 10. Sequence Comparison of the AUG7 Homologs.
Supplemental Figure 11. Interaction of AUG7 and AUG5 in Vivo.
Supplemental Figure 12. Dividing aug7-1 Cells with Excessive Numbers of Chromosomes.
Supplemental Table 1. List of the Peptides Identified in the LC-MS/MS Analysis Corresponding to Figure 1.
Supplemental Table 2. Transmission Efficiency of aug Mutant Alleles in Reciprocal Crosses between Heterozygous Mutants (+/aug) and Wild-Type (+/+) Plants.
Supplemental Table 3. List of the Peptides Identified in the LC-MS/MS Analysis Corresponding to Figure 4.
Supplemental Table 4. List of Primers Used in This Work.
Supplemental Movie 1. Time-Lapse Movie of AUG6-GFP in a Dividing Arabidopsis Cell Expressing mCherry-TUB6.
Supplemental Movie 2. Time-Lapse Movie of AUG7-GFP in a Dividing Arabidopsis Cell.
Supplementary Material
Acknowledgments
We thank Takashi Hashimoto at the Nara Institute of Science and Technology in Japan for generously providing the mCherry-TUB6 line, Tsuyoshi Nakagawa at Shimane University in Japan for the pGWB vectors, and Lily Tang and Isabel Parkin at Agriculture and Agri-Food Canada for the SK18263 line. We also thank Savithramma Dinesh-Kumar, Hsou-min Li, and Terry Murphy for their critical comments on the manuscript. T.H. was a Katherine Esau postdoctoral fellow. This report is based on work supported by the National Science Foundation under Grant MCB-0920454. Any opinions, findings, and conclusions or recommendations expressed in this article are those of the authors and do not necessarily reflect the views of the National Science Foundation.
AUTHOR CONTRIBUTIONS
Y.-R.J.L. and B.L. designed the project. T. Hotta, Z.K., Y.-R.J.L., C.-M.K.H., C.J.T.Z., T. Horio, S.F., and T.V. performed specific experiments and analyzed data. T. Hotta, Z.K., Y.-R.J.L., and B.L. wrote the article together. T. Hotta, Y.-R.J.L., and B.L. revised and edited the article.
Glossary
- MT
microtubules
- MAP
microtubule-associated protein
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- GFP
green fluorescent protein
- BY-2
Bright Yellow 2
- MTFT
microtubular fir trees
- TAIR
The Arabidopsis Information Resource
References
- Albrecht V., Simková K., Carrie C., Delannoy E., Giraud E., Whelan J., Small I.D., Apel K., Badger M.R., Pogson B.J. (2010). The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22: 3423–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bannigan A., Scheible W.-R., Lukowitz W., Fagerstrom C., Wadsworth P., Somerville C., Baskin T.I. (2007). A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 120: 2819–2827 [DOI] [PubMed] [Google Scholar]
- Bucciarelli E., Pellacani C., Naim V., Palena A., Gatti M., Somma M.P. (2009). Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and γ-tubulin to promote kinetochore-driven MT formation. Curr. Biol. 19: 1839–1845 [DOI] [PubMed] [Google Scholar]
- Castelli V., et al. (2004). Whole genome sequence comparisons and “full-length” cDNA sequences: a combined approach to evaluate and improve Arabidopsis genome annotation. Genome Res. 14: 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan T., Wakefield J.G. (2011). 50 ways to build a spindle: the complexity of microtubule generation during mitosis. Chromosome Res. 19: 321–333 [DOI] [PubMed] [Google Scholar]
- Eleftheriou E.P., Baskin T.I., Hepler P.K. (2005). Aberrant cell plate formation in the Arabidopsis thaliana microtubule organization 1 mutant. Plant Cell Physiol. 46: 671–675 [DOI] [PubMed] [Google Scholar]
- Goshima G., Kimura A. (2010). New look inside the spindle: Microtubule-dependent microtubule generation within the spindle. Curr. Opin. Cell Biol. 22: 44–49 [DOI] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R.D. (2008). Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181: 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. (2007). Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T., Igarashi H., Itoh T.J., Shimmen T., Sonobe S. (2004). Characterization of a 200 kDa microtubule-associated protein of tobacco BY-2 cells, a member of the XMAP215/MOR1 family. Plant Cell Physiol. 45: 1233–1242 [DOI] [PubMed] [Google Scholar]
- Ho C.M., Hotta T., Guo F., Roberson R.W., Lee Y.R., Liu B. (2011a). Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell 23: 2909–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.M., Hotta T., Kong Z., Zeng C.J., Sun J., Lee Y.R., Liu B. (2011b). Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell 23: 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T., Basaki A., Takeoka A., Yamato M. (1999). Lethal level overexpression of γ-tubulin in fission yeast causes mitotic arrest. Cell Motil. Cytoskeleton 44: 284–295 [DOI] [PubMed] [Google Scholar]
- Hutchins J.R.A., et al. (2010). Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura Y., Soung N.K., Park J.E., Yu L.R., Zhou M., Bang J.K., Kim B.Y., Veenstra T.D., Erikson R.L., Lee K.S. (2011). Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl. Acad. Sci. USA 108: 11446–11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E., Himmelspach R., Rashbrooke M.C., Whittington A.T., Gale K.R., Collings D.A., Wasteneys G.O. (2006). MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140: 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman J.M., Merdes A., Mourey L., Agard D.A. (2011). Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12: 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S., Abe T., Coutuer S., Inzé D., Russinova E., Hashimoto T. (2010). Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in Arabidopsis. J. Cell Sci. 123: 451–459 [DOI] [PubMed] [Google Scholar]
- Kong Z., Hotta T., Lee Y.R., Horio T., Liu B. (2010). The γ-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell 22: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai-Sano F., Hayashi T., Sano T., Hasezawa S. (2006). Cell cycle synchronization of tobacco BY-2 cells. Nat. Protoc. 1: 2621–2627 [DOI] [PubMed] [Google Scholar]
- Lawo S., et al. (2009). HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19: 816–826 [DOI] [PubMed] [Google Scholar]
- Lee Y.R.J., Li Y., Liu B. (2007). Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell 19: 2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.R.J., Liu B. (2000). Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr. Biol. 10: 797–800 [DOI] [PubMed] [Google Scholar]
- Liu B., Ho C.-M.K., Lee Y.-R.J. (2011a). Microtubule reorganization during mitosis and cytokinesis: lessons learned from developing microgametophytes in Arabidopsis thaliana. Front. Plant Sci. 2: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Hotta T., Ho C.-M.K., Lee Y.-R.J. 2011b. Microtubule organization in the phragmoplast. In The Plant Cytoskeleton, B. Liu, ed (New York: Springer), pp. 207–225.
- Liu B., Joshi H.C., Palevitz B.A. (1995). Experimental manipulation of γ-tubulin distribution in Arabidopsis using anti-microtubule drugs. Cell Motil. Cytoskeleton 31: 113–129 [DOI] [PubMed] [Google Scholar]
- Liu B., Marc J., Joshi H.C., Palevitz B.A. (1993). A γ-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Liu L., Wiese C. (2008). Xenopus NEDD1 is required for microtubule organization in Xenopus egg extracts. J. Cell Sci. 121: 578–589 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2006). Not so divided: The common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 7: 147–152 [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U.K., Stearns T. (2006). GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Meireles A.M., Fisher K.H., Colombié N., Wakefield J.G., Ohkura H. (2009). Wac: A new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat. Cell Biol. 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., Hashimoto T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Hashimoto T. (2009). A mutation in the Arabidopsis γ-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122: 2208–2217 [DOI] [PubMed] [Google Scholar]
- Palevitz B.A. (1988). Microtubular fir-trees in mitotic spindles of onion roots. Protoplasma 142: 74–78 [Google Scholar]
- Palevitz B.A. (1993). Morphological plasticity of the mitotic apparatus in plants and its developmental consequences. Plant Cell 5: 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M., Azimzadeh J., Goussot M., Camilleri C., Belcram K., Evrard J.L., Schmit A.C., Guerche P., Bouchez D. (2006). γ-Tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., Pugieux C., Nédélec F.J., Vale R.D. (2011). Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 108: 14473–14478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C., Minns G.E., Nesi N., Koumproglou R., Kitsios G., Benning C., Lloyd C.W., Doonan J.H., Hills M.J. (2009). ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 21: 90–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Robinson S.J., et al. (2009). An archived activation tagged population of Arabidopsis thaliana to facilitate forward genetics approaches. BMC Plant Biol. 9: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Gindullis F., Meier I. (2003). A novel alpha-helical protein, specific to and highly conserved in plants, is associated with the nuclear matrix fraction. J. Exp. Bot. 54: 1133–1141 [DOI] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E.A., Bajer A.S. (1992). Spindle poles in higher plant mitosis. Cell Motil. Cytoskeleton 23: 1–7 [DOI] [PubMed] [Google Scholar]
- Smirnova E.A., Bajer A.S. (1998). Early stages of spindle formation and independence of chromosome and microtubule cycles in Haemanthus endosperm. Cell Motil. Cytoskeleton 40: 22–37 [DOI] [PubMed] [Google Scholar]
- Teixidó-Travesa N., Villén J., Lacasa C., Bertran M.T., Archinti M., Gygi S.P., Caelles C., Roig J., Lüders J. (2010). The gammaTuRC revisited: A comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell 21: 3963–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.Y., Ngo B., Tapadia A., Hsu P.H., Wu G., Lee W.H. (2011). Aurora-A phosphorylates Augmin complex component Hice1 protein at an N-terminal serine/threonine cluster to modulate its microtubule binding activity during spindle assembly. J. Biol. Chem. 286: 30097–30106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D., Park S.K., Hawkins T.J., Schubert D., Schmidt R., Smertenko A., Hussey P.J. (2002). MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat. Cell Biol. 4: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Goshima G. (2010). Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol. 191: 259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R.S., Tomioka A., Petry S., Vale R.D., Obuse C., Goshima G. (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 106: 6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainman A., Buster D.W., Duncan T., Metz J., Ma A., Sharp D., Wakefield J.G. (2009). A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev. 23: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lin Y.T., Wei R., Chen Y., Shan Z., Lee W.H. (2008). Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol. Cell. Biol. 28: 3652–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Wei R., Cheng E., Ngo B., Lee W.H. (2009). Hec1 contributes to mitotic centrosomal microtubule growth for proper spindle assembly through interaction with Hice1. Mol. Biol. Cell 20: 4686–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C.J., Lee Y.R., Liu B. (2009). The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell 21: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Coppinger J.A., Jang C.Y., Yates J.R., III, Fang G. (2008). FAM29A promotes microtubule amplification via recruitment of the NEDD1-γ-tubulin complex to the mitotic spindle. J. Cell Biol. 183: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.