NTRC functions in maintaining redox homeostasis of chloroplasts and heterotrophic plastids of Arabidopsis. Leaf-specific expression of NTRC was sufficient to restore leaf and root growth, but root-specific expression of NTRC was not. The results emphasize the function of chloroplasts not only as source of carbon and energy but also of signaling molecules for development of heterotrophic organs.
Abstract
Plastids are organelles present in photosynthetic and nonphotosynthetic plant tissues. While it is well known that thioredoxin-dependent redox regulation is essential for leaf chloroplast function, little is known of the redox regulation in plastids of nonphotosynthetic tissues, which cannot use light as a direct source of reducing power. Thus, the question remains whether redox regulation operates in nonphotosynthetic plastid function and how it is integrated with chloroplasts for plant growth. Here, we show that NADPH-thioredoxin reductase C (NTRC), previously reported as exclusive to green tissues, is also expressed in nonphotosynthetic tissues of Arabidopsis thaliana, where it is localized to plastids. Moreover, we show that NTRC is involved in maintaining the redox homeostasis of plastids also in nonphotosynthetic organs. To test the relationship between plastids of photosynthetic and nonphotosynthetic tissues, transgenic plants were obtained with redox homeostasis restituted exclusively in leaves or in roots, through the expression of NTRC under the control of organ-specific promoters in the ntrc mutant. Our results show that fully functional root amyloplasts are not sufficient for root, or leaf, growth, but fully functional chloroplasts are necessary and sufficient to support wild-type rates of root growth and lateral root formation.
INTRODUCTION
Redox regulation based on disulfide-dithiol interchange of key Cys residues of regulatory enzymes is a rapid and reversible mechanism, which allows the control of metabolic fluxes and its adjustment to ever changing environmental constraints. This is a universal type of regulation, present in all types of organisms, from bacteria to plants and animals (Buchanan and Balmer, 2005; Meyer et al., 2009). Thioredoxins (TRXs), small proteins of 12 to 14 kD with a conserved WC(G/P)PC active site, play a central role in redox regulation. In its reduced state, the TRX is able to reduce disulfides of target proteins so that its own active site becomes oxidized to a disulfide. Thus, for a new catalytic cycle, oxidized TRX needs to be reduced in a reaction catalyzed by NADPH-dependent thioredoxin reductase (NTR). Therefore, in all types of organisms the maintenance of the redox status includes a two-component system formed by NTR and TRX, the so-called NADPH-TRX system (NTS), which uses NADPH as the source of reducing power and, in eukaryotic cells, is localized to the cytoplasm and mitochondria (Jacquot et al., 2009).
Although the NTS is universal, plants have several unique features of their redox regulation. In bacteria, yeast, and animals, NTR and TRX are encoded by one to two genes, and the plant genomes so far sequenced reveal the presence of two genes encoding NTR, called NTRA and NTRB, but a large number of up to 11 genes encoding the h-type (h for heterotrophic) TRXs (Gelhaye et al., 2005; Meyer et al., 2005). In plants, as in other eukaryotes, NTS is localized in mitochondria and cytoplasm, NTRA being the major cytosolic isoform, whereas NTRB is more abundant in mitochondria (Laloi et al., 2001; Reichheld et al., 2005). Regarding h-type TRXs, most of them are predicted to be localized to the cytoplasm, but alternative localization to mitochondria (Gelhaye et al., 2004) and nucleus (Serrato et al., 2001; Serrato and Cejudo, 2003) has been reported. Even a double targeting to nucleus and mitochondria has been described for a novel o-type TRX from pea (Pisum sativum; Martí et al., 2009). Moreover, in cereal seed cells that suffer oxidative stress during development and germination, NTR also accumulates in the nucleus and has been proposed to function as an antioxidant based on its ability to reduce 1-Cys peroxiredoxin (PRX) (Pulido et al., 2009).
A remarkable characteristic of redox regulation in plants and algae is the presence of a specific and complex set of TRXs in chloroplasts. These include types f, m, x, and y (Collin et al., 2003) and additional TRXs and TRX-like proteins more recently identified, such as High Chlorophyll Fluorescence164 (Motohashi and Hisabori, 2006), Chloroplastic Drought-induced Stress Protein32 (CDSP32) (Broin et al., 2000), TRX z (Arsova et al., 2010; Chibani et al., 2010), or the family of atypical TRXs called ACHT (for Atypical Cys His-rich Trxs) (Dangoor et al., 2009). The chloroplast also contains a specific system for TRX reduction, which is dependent on ferredoxin (Fd) reduced by the photosynthetic electron transport chain and an Fd-dependent TRX reductase (FTR) (Schürmann and Buchanan, 2008). Therefore, redox regulation in chloroplasts has been considered to rely on reduced Fd, thus being light dependent, in contrast with redox regulation in heterotrophic organisms, and nonphotosynthetic plant tissues, which use NADPH as source of reducing power. In chloroplasts, NADPH is produced during the day as the final product of the photosynthetic electron transport chain in a reaction catalyzed by Fd-NADP+ oxidoreductase (FNR) (Ceccarelli et al., 2004; Lintala et al., 2007) and also during the night by the oxidative pentose phosphate pathway (Neuhaus and Emes, 2000). Therefore, the use of reduced Fd, but not NADPH, for redox regulation in this organelle was considered to be due to the lack of an enzyme able to use NADPH rather than to the lack of NADPH itself. This view of the redox regulation of the chloroplast changed after the discovery of the bimodular enzyme named NADPH-thioredoxin reductase C (NTRC), which is localized in chloroplasts (Serrato et al., 2004). NTRC is composed of NTR and TRX domains and conjugates both activities to efficiently reduce 2-Cys PRXs using NADPH as a source of reducing power (Moon et al., 2006; Pérez-Ruiz et al., 2006; Pérez-Ruiz and Cejudo, 2009). Hence, NTRC allows the use of NADPH to maintain redox homeostasis in the chloroplast (Spínola et al., 2008).
The severe phenotype of an Arabidopsis thaliana NTRC knockout mutant, which is highly dependent on photoperiod and darkness (Pérez-Ruiz et al., 2006; Lepistö et al., 2009), shows that the function of NTRC is very important for plant growth and development. Moreover, the comparison of the ntrc mutant with mutants lacking TRX x and 2-Cys PRX suggested that NTRC is the principal reductant of 2-Cys PRX in the chloroplast (Pulido et al., 2010). Among the redox-regulated processes of the chloroplast, in which NTRC plays a role, some are dependent on its ability to reduce 2-Cys PRX, such as chlorophyll synthesis (Stenbaek et al., 2008; Stenbaek and Jensen, 2010). However, the phenotype of the ntrc mutant is more severe than the phenotype of the 2-Cys PRX double mutant, suggesting that NTRC has additional functions, which are independent of 2-Cys PRX reduction (Pulido et al., 2010). Some of these functions have already been identified and include aromatic amino acid and auxin synthesis (Lepistö et al., 2009) and starch biosynthesis, since NTRC is involved in the redox regulation of ADP-glucose pyrophosphorylase (AGPase) (Michalska et al., 2009).
Interestingly, redox regulation of AGPase was severely affected not only in leaves of the Arabidopsis NTRC knockout mutant, but also in roots. This finding revealed the involvement of NTRC in redox regulation in plant heterotrophic tissues and therefore implied the localization of NTRC in these tissues. Thus, the first objective of this work was to establish the pattern of expression of the NTRC gene and the subcellular localization of the enzyme in Arabidopsis plants. Our results show a pattern of broad expression of NTRC in both photosynthetic and nonphotosynthetic tissues and the localization of the enzyme in any type of plastids. This localization of NTRC led to the hypothesis that the enzyme functions as a general molecular switch able to convert NADPH into redox signals in plastids and thus might serve to integrate redox regulation between photosynthetic and nonphotosynthetic tissues. However, while the function of the chloroplast for plant growth is well known, very little is known about the function of nongreen plastids. To gain insight into the relative function of root amyloplasts in plant growth, compared with chloroplasts, we constructed transgenic Arabidopsis plants expressing NTRC exclusively in leaves or in roots. The phenotypes of these plants show the supreme importance of the chloroplast for the growth of nonphotosynthetic plant tissues, including roots. By contrast, root amyloplasts have a low impact on root growth.
RESULTS
The NTRC Gene Is Expressed in Photosynthetic and Nonphotosynthetic Tissues of Arabidopsis Plants
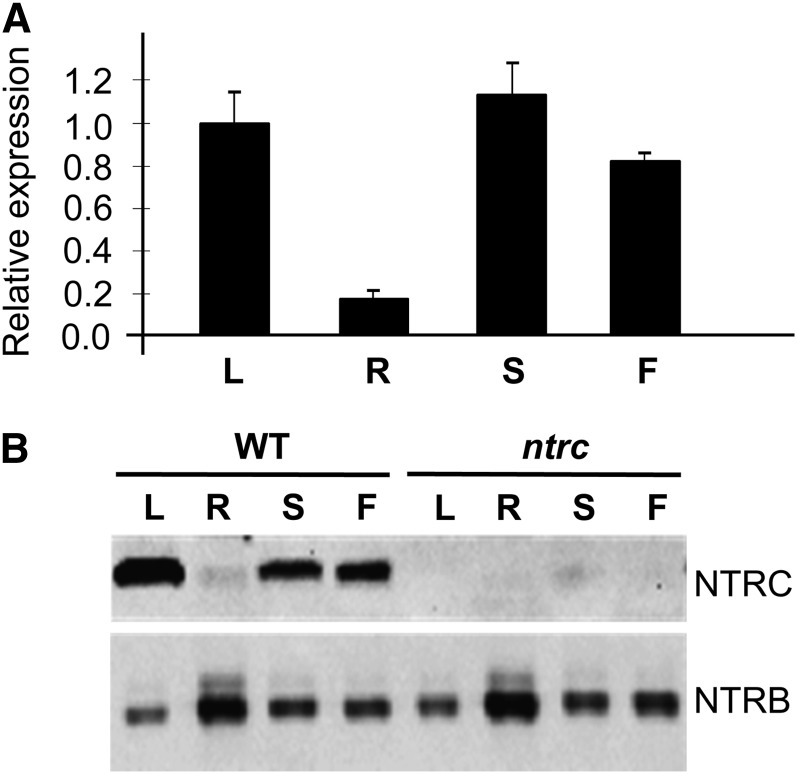
To establish the pattern of expression of the NTRC gene in Arabidopsis, the levels of transcripts in different organs of mature plants was analyzed by quantitative PCR (qPCR). This analysis confirmed the expected high expression of this gene in leaves and also revealed a high content of transcripts in stems and flowers, whereas in roots, the presence of NTRC transcripts was detected at a much lower level (Figure 1A), a pattern that was confirmed with data from Genevestigator (see Supplemental Table 1 online). In agreement with this pattern of expression, immunoblot analysis showed a high amount of NTRC protein in leaves, stems, and flowers and a much lower amount in roots (Figure 1B). Furthermore, this analysis confirmed the absence of NTRC in any of the organs of the ntrc knockout mutant (Figure 1B).
Figure 1.
Expression Pattern of NTRC in Arabidopsis.
(A) qPCR analysis of NTRC transcripts in different organs of Arabidopsis wild-type plants. The amount of transcripts in each organ was represented as arbitrary units relative to the level in leaves, which was set to 1.0. Analysis was performed three times on two independent biological samples, and the mean values ± se are indicated.
(B) The amount of NTRC protein was determined by immunoblot probed with an anti-NTRC polyclonal antibody. Samples (30 μg of protein) were subjected to SDS-PAGE, transferred onto nitrocellulose membranes, and probed with anti-NTRC or anti-NTRB antibodies to test even protein loading, as indicated. Arabidopsis wild-type and ntrc mutant plants were grown under long-day conditions for 48 d. RNA, for qPCR, and protein, for immunoblot analysis, were extracted from leaves (L), roots (R), stems (S), and flowers (F). WT, wild type.
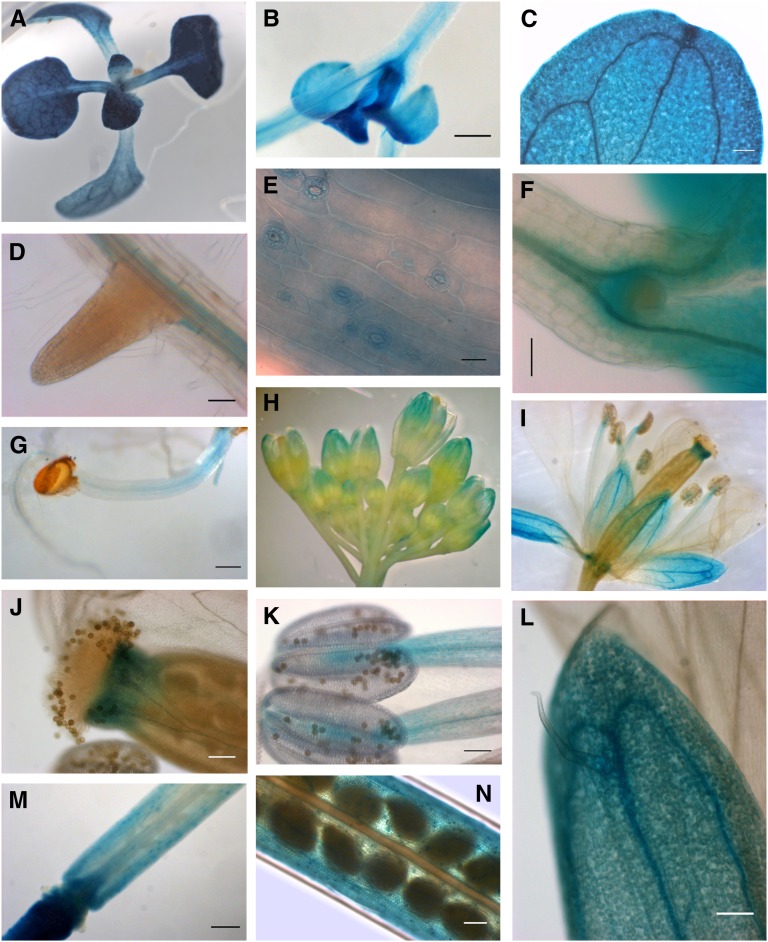
To further analyze the pattern of expression of NTRC in Arabidopsis, a 1.05-kb fragment containing its putative promoter (see Supplemental Figure 1 online) was transcriptionally fused to the β-glucuronidase (GUS) reporter gene and introduced into Arabidopsis wild-type plants. GUS staining of transgenic lines confirmed the high level of expression driven by the NTRC promoter in green tissues, such as cotyledons and first leaves of seedlings grown under long-day (Figure 2A) or short-day conditions (Figure 2B). Expression was high in leaf mesophyll cells in agreement with the high content of NTRC transcripts in leaves, but GUS staining revealed a higher expression associated with the vascular tissue (Figure 2C) as well as in stem guard cells (Figure 2E). NTRC promoter-driven expression was also detected in roots (Figures 2D and 2G) and hypocotyls (Figure 2G), staining being associated with the vascular tissue and the base of hypocotyls diverging to the cotyledons (Figure 2F). GUS staining was also analyzed in reproductive organs of the transgenic plants, which showed the expected high expression of NTRC in green tissues, such as sepals of inflorescences and flowers (Figures 2H, 2I, and 2L). In addition, GUS staining revealed NTRC expression in stigma (Figure 2I and 2J), anthers (Figures 2I and 2K), siliques, and silique petioles (Figures 2M and 2N).
Figure 2.
Histochemical Localization of GUS Expression in Arabidopsis Plants Transformed with the NTRCpro-GUS Reporter Gene.
(A) to (F) GUS staining of 10-d-old seedlings grown under long-day conditions ([A] and [C] to [E]) or under short-day conditions ([B] and [F]).
(G) Etiolated seedlings showing GUS staining in root and hypocotyl.
(H) to (N) GUS staining of inflorescence (H), flower (I), stigma (J), anthers (K), sepal (L), silique petiole (M), and silique (N) of Arabidopsis plants grown for 42 d under long-day conditions.
Bars = 20 μm in (E), 50 μm in (D) and (F), 100 μm in (C), (J), (K), (L), and (N), and 200 μm in (B), (F), and (G).
NTRC Is Localized in Plastids of Photosynthetic and Nonphotosynthetic Tissues in Arabidopsis Plants
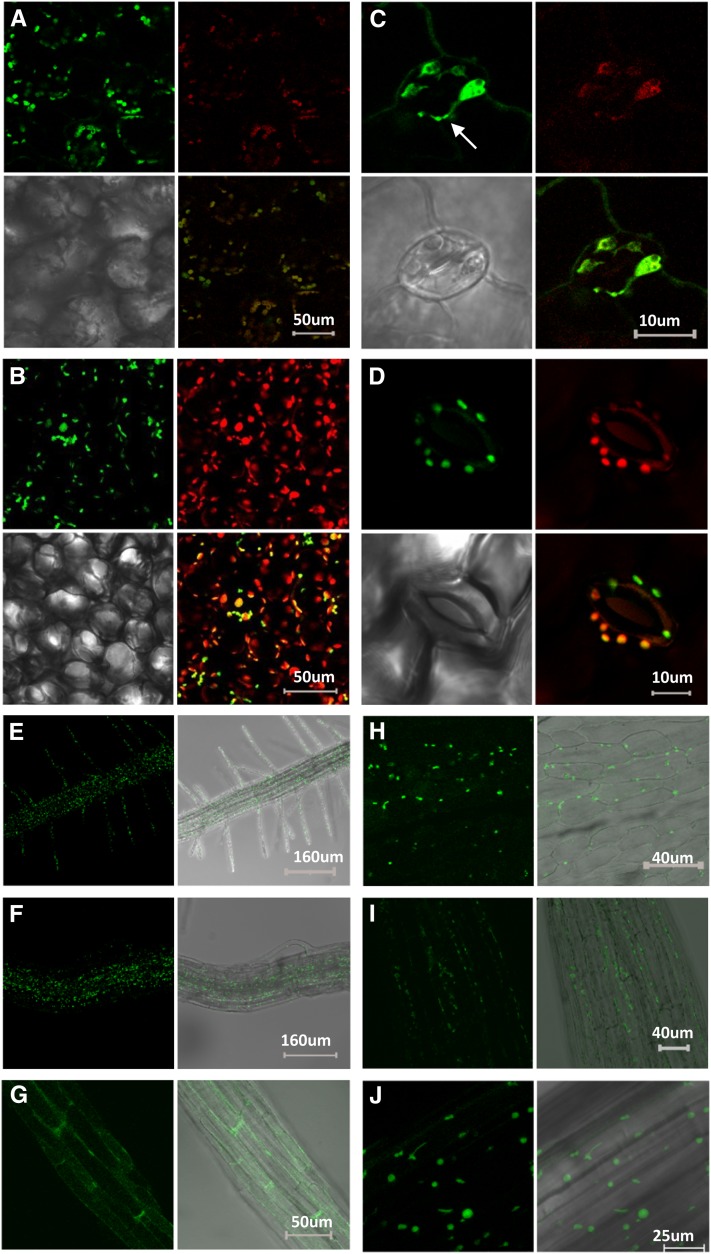
Once we had established the broad distribution of NTRC expression in photosynthetic and nonphotosynthetic tissues of Arabidopsis plants, we designed a set of new transgenic lines to analyze the subcellular localization of the enzyme. To that end, a translational fusion of Arabidopsis NTRC, including its signal peptide, with green fluorescent protein (GFP) was expressed in Arabidopsis wild-type and ntrc mutant plants under the control of the cauliflower mosaic virus (CaMV) 35S or the NTRC promoters. The expression of NTRC-GFP in ntrc mutant plants partially complemented the mutant phenotype (see Supplemental Figure 2 online), thus showing the functionality of the fusion protein. Confocal microscopy of the transgenic plants revealed coincidence of the green fluorescent signal, corresponding to NTRC-GFP, with chlorophyll red fluorescence in leaf mesophyll and guard cells of plants expressing the fusion protein under the CaMV 35S promoter (Figures 3A and 3C) or the NTRC promoter (Figures 3B and 3D), and revealed the presence of the protein in stromules, which were clearly labeled in lines expressing the NTRC-GFP fusion protein under the 35S promoter (Figure 3C, arrow).
Figure 3.
Subcellular Localization of NTRC in Arabidopsis Plants.
(A) to (D) Confocal microscopy micrographs of mesophyll ([A] and [B]) and guard cells ([C] and [D]) of Arabidopsis plants transformed with the NTRC-GFP fusion protein expressed under the CaMV 35S promoter ([A] and [C]) or the NTRC gene promoter ([B] and [D]). Plants were grown for 18 d.
(E) to (J) Subcellular localization of NTRC in nonphotosynthetic tissues of Arabidopsis. Confocal microscopy micrographs showing plastid localization of NTRC in roots of 5-d-old seedlings with the NTRC-GFP fusion protein expressed under the CaMV 35S promoter (E) or the NTRC gene promoter (F) and a root of plants expressing the GFP protein not fused to NTRC (G). Plastid localization of NTRC in petal (H) and anther (I) of 48-d-old plants and hypocotyl of 5-d-old Arabidopsis seedlings grown under darkness (J) and expressing the NTRC-GFP fusion protein under the 35S promoter.
Red, chlorophyll autofluorescence; green, GFP fluorescence. Arrow indicates a chloroplast stromule. Sizes of bars are as indicated.
The analysis of the green fluorescence signal in primary and secondary roots of transgenic plants showed the localization of NTRC in amyloplasts either in plants expressing the fusion protein under the 35S (Figure 3E) or the NTRC promoter (Figure 3F), in contrast with the diffuse signal observed in transgenic plants expressing GFP not fused to NTRC (Figure 3G). The analysis of petals (Figure 3H) and anthers (Figure 3I) showed the localization of NTRC in plastids in these tissues. Finally, hypocotyls of etiolated plants showed the localization of NTRC in etioplasts (Figure 3J), indicating the localization of NTRC in plastids of photosynthetic and nonphotosynthetic tissues of Arabidopsis plants, regardless of growth under light or dark conditions. Therefore, the analysis of plants expressing the GUS reporter gene under the NTRC promoter in conjunction with plants expressing the NTRC-GFP fusion protein under the 35S and NTRC promoters allow the conclusion that NTRC is widely expressed in both photosynthetic and nonphotosynthetic tissues, with the enzyme being localized to plastids.
NTRC Is Involved in Maintaining the Redox Homeostasis of Plastids from Photosynthetic and Nonphotosynthetic Tissues
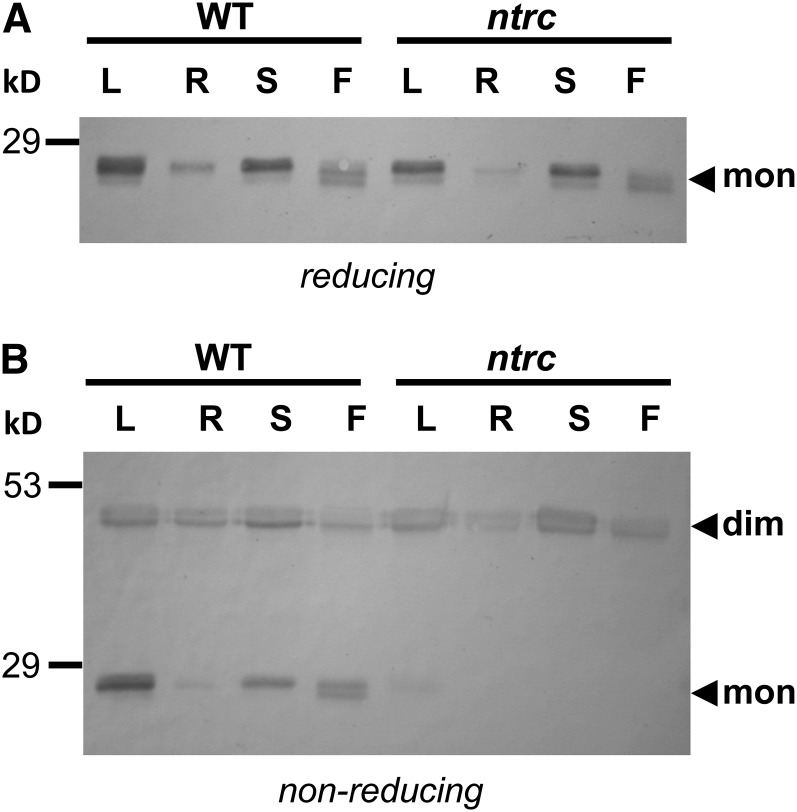
Previous studies performed with the aid of different Arabidopsis mutants showed that the redox status of the chloroplast 2-Cys PRX is essentially controlled by NTRC (Pulido et al., 2010). The finding of the localization of NTRC in plastids of nonphotosynthetic tissues of Arabidopsis plants suggested that this enzyme might also be involved in the maintenance of the redox homeostasis of these plastids. This possibility was tested by the analysis of the redox status of the 2-Cys PRX in different tissues of wild-type and ntrc mutant plants. Immunoblot analysis, under reducing conditions, confirmed the expected expression of 2-Cys PRX in organs with green tissues, such as leaves, stems, and flowers, and revealed the presence of the enzyme, though at lower level, in roots (Figure 4A). These results are in agreement with the content of transcripts of 2-Cys PRXs A and B genes obtained from Genevestigator, which showed lower expression of both genes in roots than in leaves, stems, and flowers (see Supplemental Table 1 online). Gel electrophoresis under nonreducing conditions showed the decreased content of monomeric 2-Cys PRX, indicative of unbalanced redox status, in any of the organs analyzed of the NTRC knockout mutant compared with wild-type plants (Figure 4B). Therefore, these results show that NTRC is involved in the maintenance of the redox homeostasis of chloroplasts and nonphotosynthetic plastids.
Figure 4.
Unbalanced Redox Status of 2-Cys PRXs in Photosynthetic and Nonphotosynthetic Tissues of ntrc Mutant Plants.
Protein extracts from leaves (L), roots (R), stems (S), and flowers (F) of Arabidopsis wild-type (WT) and ntrc mutant plants were subjected to SDS-PAGE under reducing (7.5 μg of protein loaded) (A) or nonreducing conditions (15 μg of protein loaded) (B), as indicated, electrotransferred to nitrocellulose sheets, and probed with anti-2-Cys PRX antibodies. “mon” indicates the monomeric and “dim” the dimeric form of the enzyme. Molecular mass markers (kD) are indicated on the left.
Restitution of Chloroplast, but Not of Amyloplast, Redox Homeostasis Is Sufficient for Wild-Type Root Growth
The finding that NTRC is involved in redox regulation in plastids from photosynthetic and nonphotosynthetic tissues led us to test whether the enzyme has any function integrating redox regulation in both types of plastids and the contribution of each of them to plant growth. To this end, we took advantage of the Arabidopsis NTRC knockout mutant, which was used to generate plants expressing NTRC exclusively in leaves or in roots with the aim of restituting NTRC-dependent redox regulation in either photosynthetic or nonphotosynthetic organs. For leaf-specific expression, the Arabidopsis NTRC cDNA was expressed in the ntrc mutant background under the control of the Rbcs gene promoter (Donald and Cashmore, 1990), whereas the promoters of the phosphate transporter genes Pht1,2 and Pht1,3 (Mudge et al., 2002) were used for root-specific expression. In parallel, Arabidopsis transgenic plants expressing the GUS reporter gene under the control of these promoters confirmed the expected expression exclusively in leaves, for the Rbcs promoter, or in roots, for the Pht1,2 and Pht1,3 promoters (see Supplemental Figure 3 online).
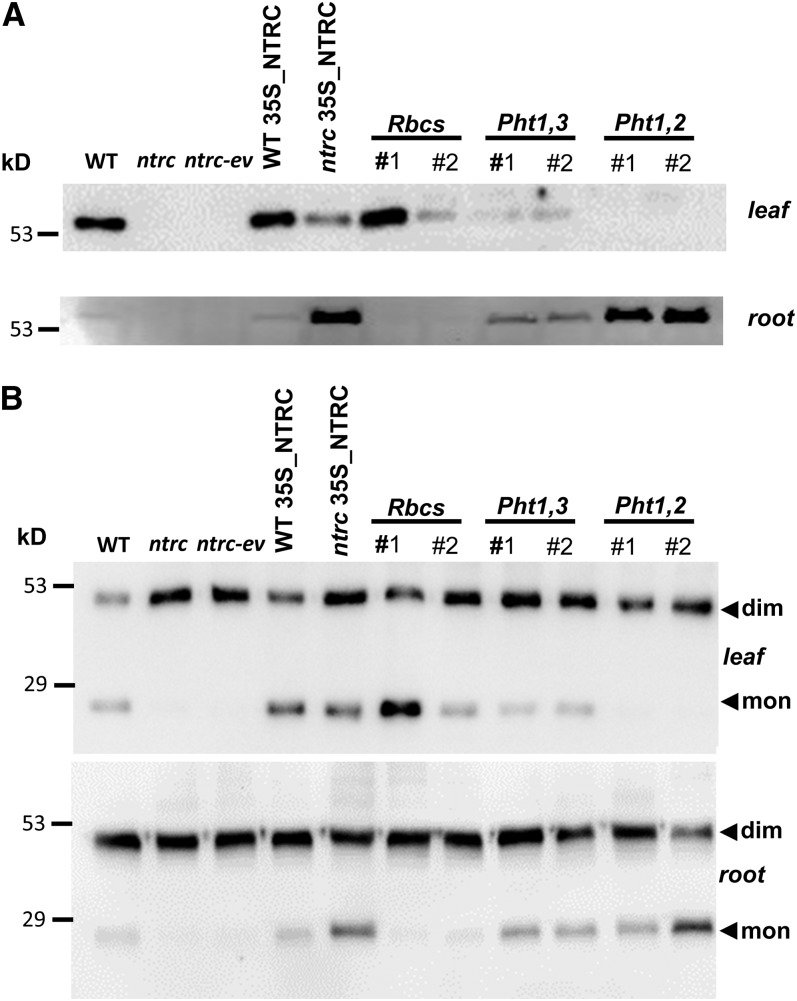
Of the transgenic plants obtained for each construct, two representative lines for each promoter (termed #1 and #2) were chosen for further analysis. In addition, for comparative purposes, transgenic plants expressing NTRC in leaves and roots, under the CaMV 35S promoter, or transformed with the empty vector, were also included. The immunoblot analysis of extracts from leaves and roots shows the expected content of NTRC (high in leaves and low in roots) in wild-type plants, while NTRC was undetectable in either organ of the ntrc mutant or the mutant transformed with the empty vector (Figure 5A). Plants expressing NTRC under the CaMV 35S promoter were obtained either in the wild-type or in the ntrc background. A line in the wild-type background (WT35S_NTRC) showing high expression in leaves and low in roots, and a line in the mutant background (ntrc35S_NTRC) with the opposite pattern, high expression in roots and low in leaves, were chosen (Figure 5A). Finally, among the transgenic plants with organ-specific promoters, those expressing NTRC under the Rbcs promoter accumulated NTRC exclusively in leaves, with line #1 showing higher content than line #2, whereas transgenic plants with the Pht1,2 promoter showed root-specific accumulation of NTRC (Figure 5A). However, the Pht1,3 promoter, which produced the expected high expression of NTRC in roots, was not specific since, although at a low level, the enzyme was also detected in leaves (Figure 5A). To test the functionality of NTRC in the transgenic lines, the redox status of 2-Cys PRX was determined. Figure 5B shows the expected low content of monomeric 2-Cys PRX, indicative of unbalanced redox status, in leaves and roots of the ntrc mutant, compared with the wild-type plants. A high level of monomeric 2-Cys PRX in leaves and roots was recovered in transgenic plants expressing NTRC under the 35S promoter, the level in roots being higher in the ntrc35S_NTRC line in agreement with the higher content of NTRC in roots of these plants. Similarly, transgenic lines expressing NTRC under the Pht1,3 promoter recovered the redox status of 2-Cys PRX in leaves and roots (Figure 5B), in agreement with the presence of NTRC in roots but also in leaves (Figure 5A) of these plants. The redox status of the 2-Cys PRX was restored in leaves of plants expressing NTRC under the Rbcs promoter, but not under the Pht1,2 promoter, in agreement with the presence or absence, respectively, of NTRC in leaves of these plants (Figures 5A and 5B). Similarly, plants showing leaf-specific expression of NTRC, under the control of the Rbcs promoter, showed almost undetectable amounts of monomeric 2-Cys PRX in roots, which was recovered in plants with root-specific expression of NTRC, with the Pht1,2 promoter (Figure 5B). Therefore, these results show that the redox status of the 2-Cys PRXs in leaves and roots is highly dependent on the presence of NTRC in these organs and thus confirm the functionality of NTRC in the transgenic lines under analysis.
Figure 5.
Effect of the Presence of NTRC in Leaves and Roots on the Redox Status of 2-Cys PRX.
(A) Immunoblot analysis of the content of NTRC in leaf (30 μg of protein) and root (50 μg of protein) extracts from the wild type (WT), ntrc mutant, ntrc mutant transformed with the empty vector (ntrc-ev), and transgenic lines expressing NTRC under the CaMV 35S, Rbcs, Pht1,3, and Pht1,2 promoters, as indicated, in the ntrc mutant background. Proteins were subjected to SDS-PAGE under reducing conditions, electrotransferred to nitrocellulose sheets, and probed with anti-NTRC antibodies.
(B) Aliquots (15 μg of protein) of the same protein samples indicated above were subjected to SDS-PAGE under nonreducing conditions, electrotransferred to nitrocellulose sheets, and probed with anti-2-Cys PRX antibody. “mon” indicates the monomeric and “dim” the dimeric form of the enzyme. Molecular mass markers (kD) are indicated on the left.
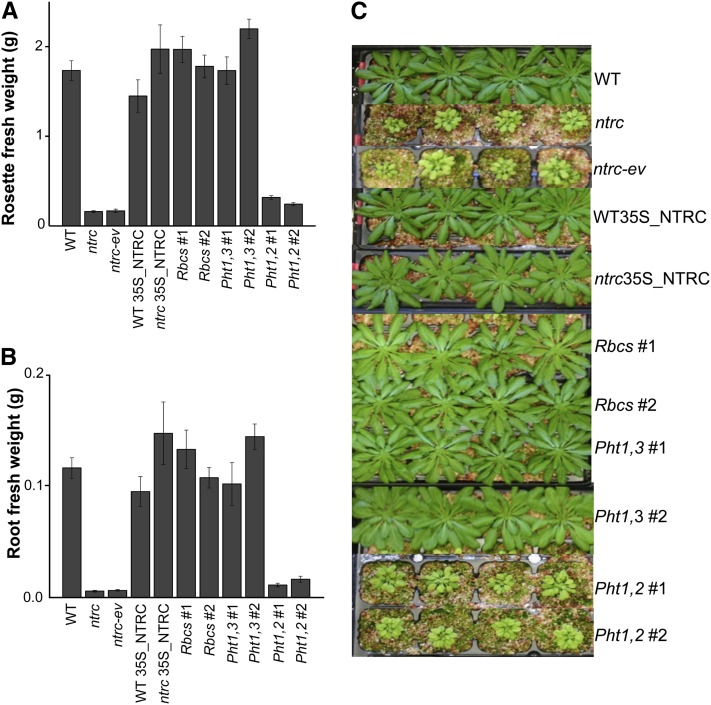
The phenotype of these transgenic plants was thereafter analyzed with the aim of testing the effect of NTRC-dependent redox regulation of plastids from leaves and roots, respectively, on plant growth. Plants were grown under short-day conditions, which were previously described to cause a more severe phenotype on the ntrc mutant (Pérez-Ruiz et al., 2006; Lepistö et al., 2009). Under these conditions, the NTRC knockout plants showed the characteristic inhibition of growth, with the corresponding lower leaf and root fresh weight (Figures 6A to 6C). As expected, the constitutive expression of NTRC, under the 35S promoter, in leaves and roots of the ntrc mutants, recovered the wild-type phenotype in terms of both leaf and root fresh weight (Figures 6A to 6C). In agreement with these results, plants expressing NTRC under the Pht1,3 promoter, which contained NTRC both in leaves and roots, also showed recovery of the wild-type phenotype with respect to both leaf and root growth (Figures 6A to 6C), although these plants displayed a different distribution of NTRC, with a higher content in roots than in leaves, compared with the wild-type plants (Figure 5A). Interestingly, the expression of NTRC exclusively in leaves was sufficient to completely recover the wild-type phenotype in terms of leaf and root fresh weight (Figures 6A to 6C). In sharp contrast, the expression of NTRC exclusively in roots resulted in only a slight increase of leaf and root fresh weight compared with ntrc mutant plants (Figures 6A to 6C).
Figure 6.
Effect of NTRC Expression in Photosynthetic and Nonphotosynthetic Tissues on Plant Growth.
(A) and (B) Wild-type (WT), ntrc mutant, and the different transgenic lines, as indicated, were grown under short-day conditions for 53 d, and rosette leaves (A) or roots (B) from seven plants were dissected and weighed. Mean values ± se are shown. The experiment was repeated at least three times with similar results and a representative one is shown.
(C) Photographs of representative plants of each of the lines under analysis.
Chloroplast Redox Homeostasis Is Essential for Root Growth and Lateral Root Formation
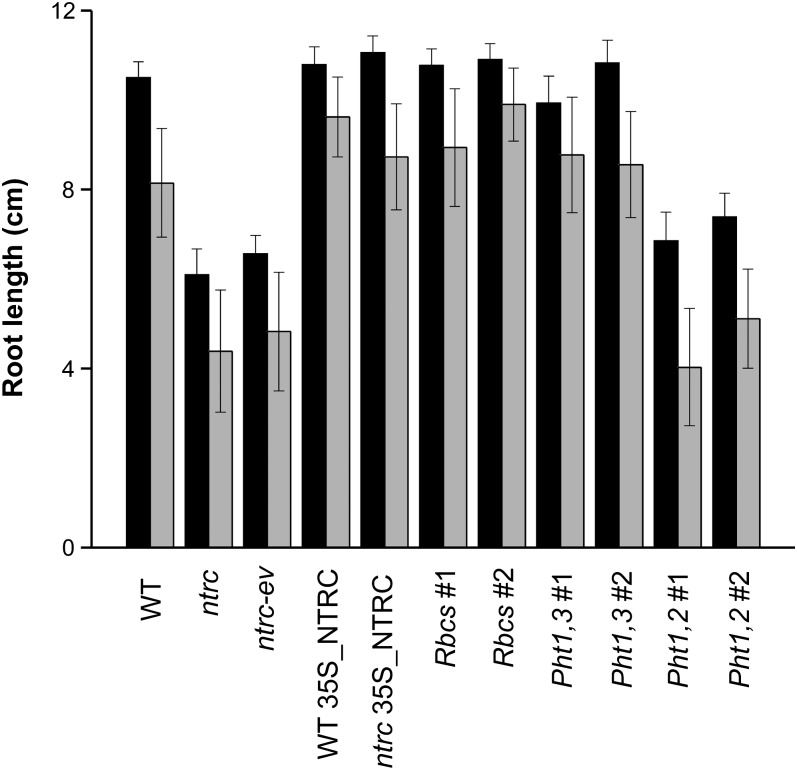
The poor effect of root-specific expression of NTRC on root growth suggests that amyloplast function required fully functional chloroplasts. As a well-recognized function of photosynthesis is to provide Suc as source of carbon and energy for sink organs, we analyzed in more detail the rate of root growth and the effect of the addition of Suc to the growth medium (Figure 7). The rate of root growth was lower in NTRC knockout plants than in wild-type plants, and feeding with Suc had a slightly positive effect on both types of plants. As expected, constitutive expression of NTRC in leaves and roots, under the 35S or the Pht1,3 promoter, recovered wild-type rates of root growth. The expression of NTRC exclusively in leaves was sufficient to recover wild-type root growth despite the fact that root amyloplasts showed signs of redox unbalance (Figure 5B). Notably, expression of NTRC exclusively in roots, under the Pht1,2 promoter, was not sufficient to recover wild-type root growth regardless of the addition of Suc to the medium. Therefore, the reestablishment of the redox homeostasis of root amyloplasts is not sufficient for recovery of root growth, whereas functional chloroplasts are necessary and sufficient to recover root growth independently of the redox status of the root amyloplasts.
Figure 7.
Effect of NTRC Expression in Photosynthetic and Nonphotosynthetic Tissues on Root Growth.
Root length for each of the Arabidopsis lines, as indicated, was determined as follows. Seven-day-old seedlings grown in absence of any added sugar were transferred onto fresh MS plates supplemented with 30 mM, final concentration, Suc (black bars) or mannitol (gray bars) for 14 additional days under short-day conditions on vertical-oriented plates. Assays were repeated three times with at least 21 plants per treatment. Mean values ± se are shown. WT, the wild type.
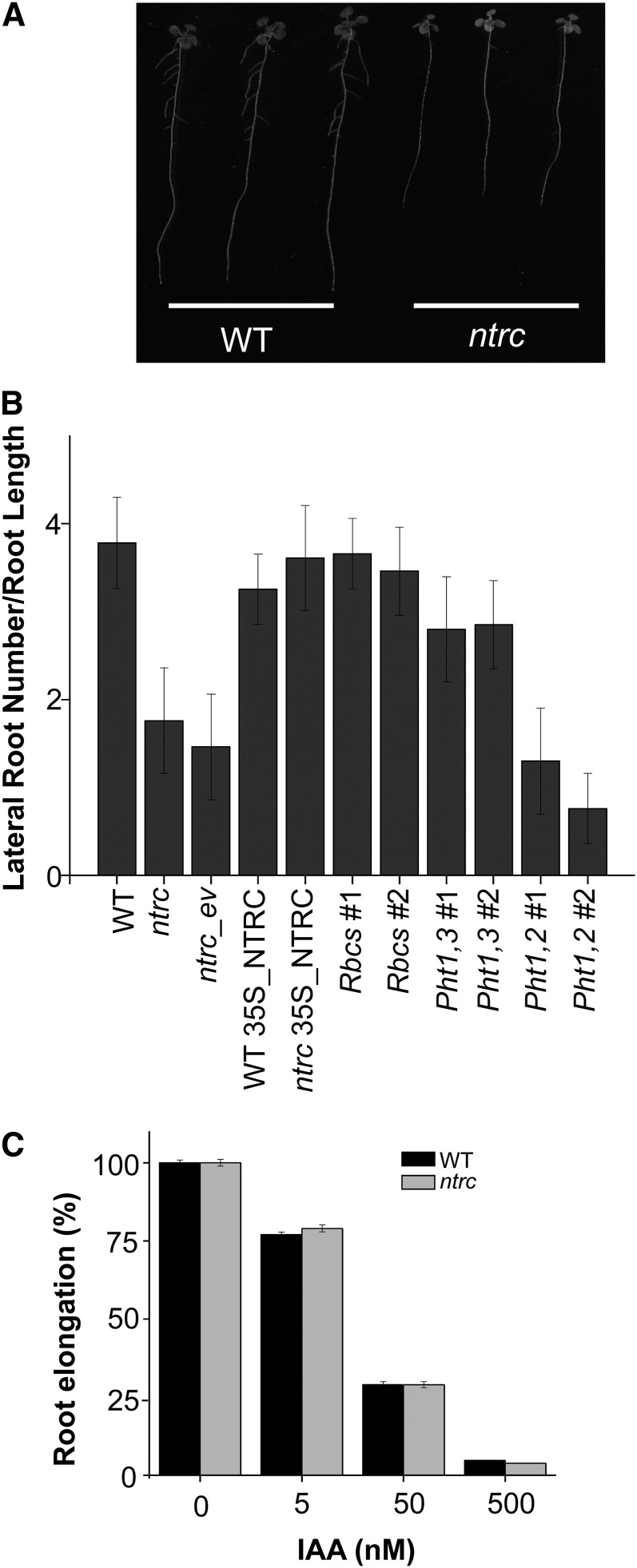
The weak effect of exogenous Suc on root growth in plants expressing NTRC exclusively in roots suggested that chloroplasts provide something other than carbon and energy for root growth. To further analyze this possibility, we studied root phenotypes caused by NTRC deficiency. During early seedling growth, the ntrc mutant shows a slow rate of root growth, but also fewer lateral roots than the wild-type plants (Figures 8A and 8B). As lateral root formation is highly influenced by auxins, this phenotype of the ntrc mutant suggests the involvement of NTRC in auxin signaling. Treatment with exogenous indole-3-acetic acid (IAA) exerted a similar inhibitory effect on root growth in wild-type and ntrc mutant plants (Figure 8C), showing that NTRC deficiency does not affect auxin sensitivity. We then analyzed the numbers of lateral roots in Arabidopsis lines with organ-specific expression of NTRC to determine the effect of chloroplasts and amyloplasts on lateral root formation. As expected, expression of NTRC in leaves and roots, under the 35S or the Pht1,3 promoter, rescued wild-type numbers of lateral root formation (Figure 8B). Interestingly, leaf-specific expression of NTRC, with the Rbcs promoter, was sufficient to recover wild-type numbers of lateral roots, whereas root-specific expression of NTRC, with the Pht1,2 promoter, was not (Figure 8B). Inhibition of root growth in response to exogenous IAA treatment was similar for all transgenic lines under analysis (see Supplemental Figure 4 online), showing no alteration of auxin sensitivity. Therefore, restitution of chloroplast redox homeostasis is sufficient for lateral root formation even in plants with impaired amyloplasts.
Figure 8.
NTRC Is Involved in Lateral Root Formation in Arabidopsis Seedlings.
(A) Images of 11-d-old wild-type (WT) and ntrc mutant seedlings, as indicated, grown under long-day conditions.
(B) Quantification of the number of lateral roots related to root length of seedlings of the different Arabidopsis lines, as indicated, grown for 11 d under long-day conditions.
(C) Five-day-old wild-type and ntrc mutant seedlings grown on MS media were transferred to media supplemented with increasing concentrations of IAA for an additional period of 3 d. Root length was expressed as a percentage of root elongation of untreated seedlings, which was considered 100%. Assays were repeated three times with at least 21 plants per treatment. Mean values ± se are indicated.
An Alternative Pathway for Redox Regulation Is Expressed in Roots
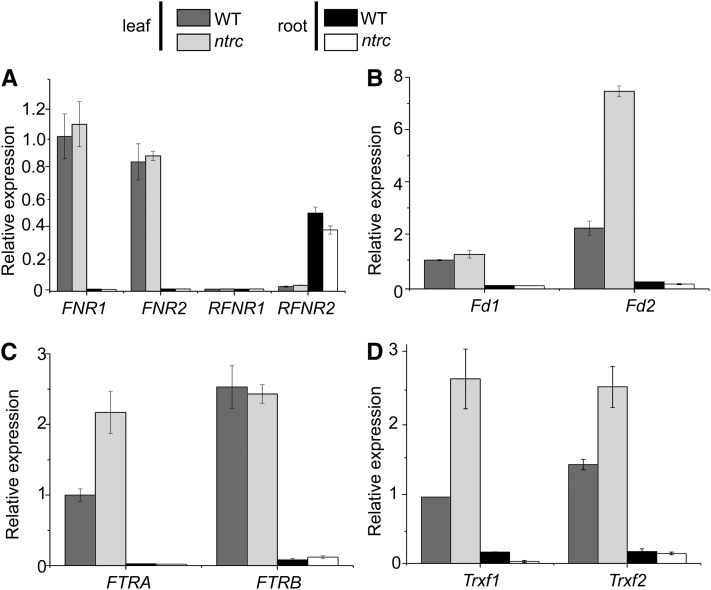
The recovery of the wild-type rate of root growth and lateral root formation in plants expressing NTRC exclusively in leaves indicates that the presence of NTRC in root amyloplasts is not essential for root growth, thus suggesting the presence of an alternative pathway for redox regulation in amyloplasts. Redox regulation by this pathway would require the transfer of electrons from NADPH, generated from sugars by the oxidative pentose phosphate pathway, to amyloplast TRXs with the participation of FNR, Fd, and FTR. To test for the presence of this alternative pathway in roots, the content of the corresponding gene transcripts was analyzed by qPCR in wild-type and ntrc mutant plants. Of the four genes encoding FNR in Arabidopsis, FNR1 and FNR2 showed leaf-specific expression, whereas RFNR2 was expressed at higher level in roots and RFNR1 showed a poor expression in both tissues (Figure 9A). No significant difference of transcript content was observed in wild-type and mutant plants (Figure 9A). In agreement with these results, Genevestigator data (see Supplemental Table 2 online) show higher expression of FNR1 and FNR2 genes in leaves than in roots, whereas RFNR2 is expressed at higher levels in roots. However, the Genevestigator data predict higher expression of RFNR1 than found in our qPCR-based analysis (Figure 9A). Genes encoding Fd (Fd1 and Fd2), the catalytic (FTRB) and regulatory (FTRA) subunits of FTR and the two genes encoding type-f TRXs (Trxf1 and Trxf2), which were chosen as example of TRXs, showed higher expression in leaves but were also detected in roots (Figures 9B to 9D), in agreement with Genevestigator data (see Supplemental Table 2 online). The analysis of expression of these genes in the ntrc mutant revealed higher expression of Fd2, FTRA, and the two genes encoding TRX f in leaves, but not in roots (Figures 9A to 9D). Therefore, genes encoding the alternative pathway for redox regulation are expressed in roots though at lower level than in leaves, like the NTRC gene.
Figure 9.
Expression of FNR, Fd, FTR, and Type-f TRX in Leaves and Roots of Arabidopsis Wild-Type and ntrc Mutant Plants.
qPCR analysis of transcripts of genes encoding FNR (A), Fd (B), FTR (C), and TRX f (D) in leaves and roots of Arabidopsis wild-type (WT) and ntrc mutant plants, which were grown during 14 d under short-day conditions on plates containing MS medium supplemented with Suc. The amount of transcripts is represented as arbitrary units relative to the level of one of the genes for each family in leaves, which was set to 1.0. Analysis was performed three times on two independent biological samples and the mean values ± se are indicated.
DISCUSSION
NTRC Is Important for Redox Homeostasis of Plastids of Photosynthetic and Nonphotosynthetic Tissues
The recent finding that the redox regulation of AGPase was severely altered in roots of the Arabidopsis NTRC knockout mutant (Michalska et al., 2009) implied the presence of NTRC in roots, in contrast with the previously established view of NTRC as an enzyme exclusive to photosynthetic tissues (Serrato et al., 2004; Moon et al., 2006; Lepistö et al., 2009). Thus, the first objective of this work was to establish the pattern of expression of NTRC in mature Arabidopsis plants. Both qPCR and immunoblot analyses, as well as Genevestigator data, confirmed the expression of NTRC in organs with photosynthetic cells and revealed the presence of the enzyme, though at a lower levels, in roots (Figure 1). This broad pattern of expression was confirmed by NTRCpro-GUS transgenic lines, which showed high level of expression driven by the NTRC gene promoter in green tissues but also in root or hypocotyl, in which it is associated to the vascular tissue. Moreover, the analysis of these plants revealed a more unexpected expression in tissues of reproductive organs, such as stigma and anthers. Therefore, although NTRC is expressed at high levels in photosynthetic tissues, in agreement with previous reports (Serrato et al., 2004; Moon et al., 2006; Alkhalfioui et al., 2007; Lepistö et al., 2009), the different approaches performed in this work reveal a broad pattern of expression in either photosynthetic and nonphotosynthetic tissues for this gene.
Different studies based on immunoblot analysis of purified chloroplasts from rice (Oryza sativa) leaves (Serrato et al., 2004), Arabidopsis plants expressing an NTRC-GFP fusion protein (Moon et al., 2006), or immunogold labeling (Pérez-Ruiz et al., 2009) clearly indicated that NTRC is a chloroplast-localized enzyme, in agreement with the high level of expression of the gene in green tissues and the function of the enzyme in chloroplast-localized processes, such as chlorophyll and starch synthesis (Stenbaek et al., 2008; Stenbaek and Jensen, 2010; Michalska et al., 2009). However, the finding of NTRC expression in nonphotosynthetic tissues raised the question of the subcellular localization of the enzyme in these tissues. This question was addressed by the generation of Arabidopsis lines expressing an NTRC-GFP fusion protein, which confirmed the localization of the enzyme in chloroplasts and showed the localization of NTRC in plastids of any of the nonphotosynthetic tissues analyzed, including roots, hypocotyls, anthers, and petals (Figure 3).
The presence of NTRC in plastids of nonphotosynthetic tissues suggested that redox regulation might be an important component for the control of the metabolic pathways in these organelles and that NTRC, which is able to use NADPH for redox regulation, may play a central function in these plastids with no photochemical reactions. Although 2-Cys PRXs were described as chloroplast-localized enzymes (Baier and Dietz, 1997), immunoblot analysis under reducing conditions showed that these enzymes are also present in nonphotosynthetic tissues of Arabidopsis (Figure 4A). Moreover, expression in nonphotosynthetic tissues of the genes encoding 2-Cys PRXs was confirmed by Genevestigator data (see Supplemental Table 1 online), thus indicating that the redox state of these proteins may be taken as a marker of the redox homeostasis of nonphotosynthetic plastids. In chloroplasts, 2-Cys PRXs were proposed to be reduced by CDSP32 (Broin et al., 2002; Rey et al., 2005) and TRX x (Collin et al., 2003), but NTRC appeared to be the main reductant of this enzyme in vivo (Pulido et al., 2010). Indeed, immunoblot analysis under nonreducing conditions showed a lower content of the monomeric form of the 2-Cys PRX in the ntrc mutant not only in leaves but in any of the other tissues analyzed here (Figure 4B), thus indicating the involvement of NTRC in the maintenance of the redox status in plastids from photosynthetic and nonphotosynthetic tissues. These results are in agreement with the previous finding that NTRC is involved in the redox regulation of AGPase in roots (Michalska et al., 2009) and emphasize the notion that redox regulation occurs in plastids of nonphotosynthetic tissues.
NTRC Is a Redox Switch Able to Convert NADPH into Redox Signal
In chloroplasts, redox regulation relies on Fd reduced by the photosynthetic electron transport chain. This is the source of reducing power to the FTR/TRX system, which thus participates in the control of the redox status of the numerous TRX targets so far identified (Buchanan and Balmer, 2005). As a complementary pathway for chloroplast redox regulation, NTRC allows the use of NADPH, produced both by the oxidative pentose phosphate pathway and by photosynthetic electron transport (Spínola et al., 2008). In contrast with the thorough knowledge of redox regulation in chloroplasts, very little is known about the function and redox regulation of plastids in heterotrophic tissues. Plastids from some nonphotosynthetic tissues, such as amyloplasts of cereal endosperm, are specialized in starch synthesis (Tetlow et al., 2008). However, recent proteomic analyses suggest that these plastids perform a complex diversity of metabolic pathways (Balmer et al., 2006a, 2006b; Dupont, 2008). The finding of a complete Fd/FTR/TRX system in amyloplasts isolated from wheat (Triticum aestivum) endosperm and the identification of TRX targets in this organelle suggested that redox regulation may be an important component of the regulation of starch synthesis but also of amino acid and lipid biosynthesis (Balmer et al., 2006a). Moreover, the presence of TRX y in Arabidopsis roots (Collin et al., 2004) and of TRXs f and m in pea roots and flowers (de Dios Barajas-López et al., 2007; Traverso et al., 2008) lend further support to the notion that redox regulation is a relevant aspect of plastid function in nonphotosynthetic tissues. The identification of TRX y targets in Arabidopsis roots suggests that metabolic pathways, including amino acid, lipid, and phenylpropanoid metabolism, protein degradation and folding, and the response to oxidative stress, are redox-regulated processes (Marchand et al., 2010).
The localization of NTRC in plastids of photosynthetic and nonphotosynthetic tissues of Arabidopsis reported here (Figure 3) supports the notion of plastid redox regulation as a general phenomenon in plants. Since heterotrophic plastids lack photochemical reactions, reducing power to support redox regulation in these plastids relies entirely on NADPH produced from sugars by the oxidative pentose phosphate pathway (Kammerer et al., 1998; Neuhaus and Emes, 2000). The biochemical properties of NTRC, including its high affinity for NADPH and the presence of a TRX domain at the C terminus, allow us to propose that NTRC acts as a redox switch able to convert reducing power in the form of NADPH, which might be indicative of the capacity to perform biosynthetic metabolism, into a redox signal through the thiol groups of its TRX domain. Thus, NTRC may constitute a direct pathway for redox homeostasis in heterotrophic plastids (Figure 10). However, transfer of electrons from NADPH to Fd, catalyzed by FNR, is also possible in heterotrophic plastids, as suggested by the high expression of RFNR2 in roots (Figure 9A), in agreement with the previous report of a root-specific form of this enzyme (Oji et al., 1985). Though at a lower level than in leaves, qPCR analyses showed the expression in roots of genes encoding Fd, the catalytic and regulatory subunits of FTR and TRX f1 and TRX f2 (Figures 9B to 9D), a pattern confirmed by the Genevestigator data (see Supplemental Table 2 online). Therefore, the Fd/FTR/TRX pathway might also be operative in nonphotosynthetic plastids (Figure 10), as it is in chloroplasts. The fact that plants that lack NTRC in root amyloplasts, but express the enzyme in chloroplasts, show wild-type levels of root growth suggests that the Fd/FTR/TRX pathway compensates for NTRC deficiency in these organelles, although the roots of these plants will probably have unbalanced metabolite levels. The identification of specific targets of NTRC and the different TRXs of root amyloplasts will help to establish the metabolic processes that depend on NTRC or the Fd/FTR/TRX pathway.
Figure 10.
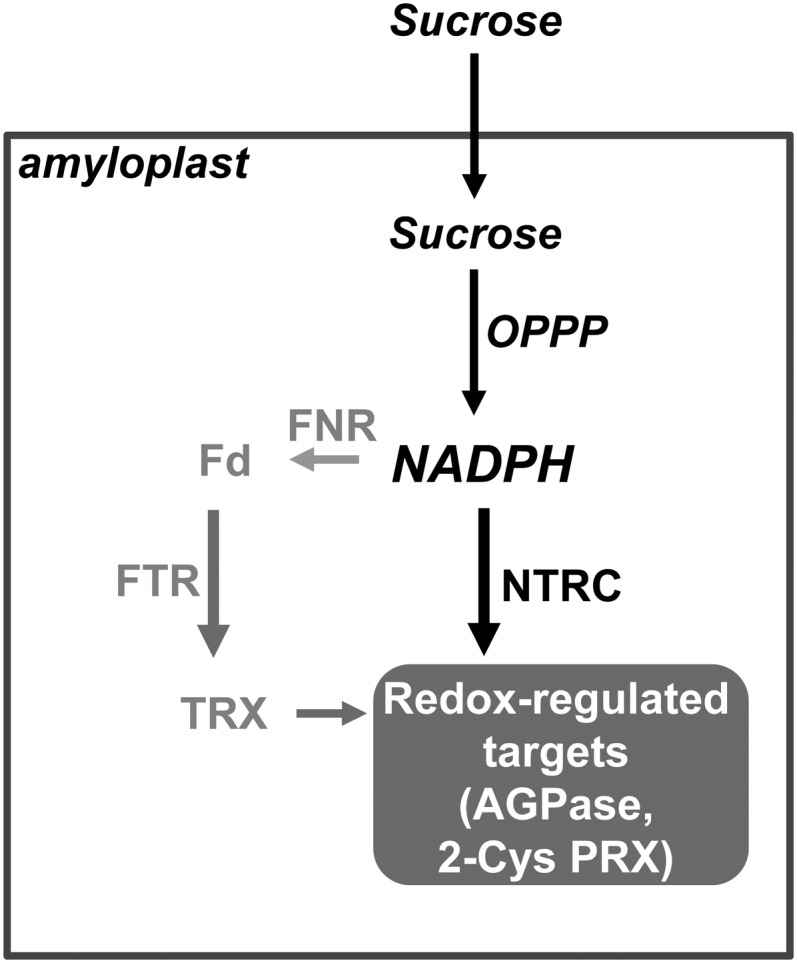
Schematic Representation of Alternative Pathways for Redox Regulation in Root Amyloplasts.
In the absence of photochemical reactions, redox regulation in root amyloplasts depends on NADPH produced from Suc by the oxidative pentose phosphate pathway (OPPP). While NTRC is able to directly use NADPH for redox regulation, the alternative pathway requires the reduction of Fd catalyzed by FNR and the transfer of reducing power to amyloplast TRXs catalyzed by FTR. Evidence has been reported showing NTRC-dependent redox regulation in amyloplasts of AGPase (Michalska et al., 2009) and 2-Cys PRX (this work).
Chloroplast Function Is Sufficient to Support Root Growth in Plants with Impaired Amyloplast Redox Homeostasis
It has long been known that light, as the primary source of energy for plants, is used for the production of photosynthates in chloroplasts of source tissues, which are transported to sink tissues to support their growth. Therefore, growth of heterotrophic tissues is highly dependent on photosynthetic tissues. Moreover, it has been proposed that photosynthates form part of a signaling network integrating both photosynthetic and nonphotosynthetic tissues (Koch, 1996; Paul and Foyer, 2001). Based on the finding of an Fd/TRX system in wheat amyloplasts, Balmer et al. (2006a) proposed that TRX might act integrating redox regulation between both types of organelles, but due to the scarce knowledge of redox regulation in heterotrophic tissues, this proposal has not been tested yet. The presence of NTRC in both photosynthetic and heterotrophic plastids and the unique properties of the enzyme serving as a switch of NADPH into redox signal suggest that NTRC might be important for such integration.
In this work, we took advantage of the severe phenotype of the Arabidopsis NTRC knockout mutant to address whether NTRC functions in coordinating plastid redox regulation between photosynthetic and heterotrophic tissues and the relevance of each type of plastid for plant growth. Leaf-specific expression of NTRC restored redox homeostasis in leaf chloroplasts and recovered wild-type leaf and root phenotypes. By contrast, restitution of redox homeostasis exclusively in roots was insufficient to recover leaf growth and remarkably was also insufficient for recovery of root growth and lateral root formation. Therefore, these results establish that chloroplast function is necessary and sufficient to reach wild-type rates of root growth, thus emphasizing the essential function of chloroplasts for growth of heterotrophic tissues.
The poor growth of roots of transgenic lines expressing NTRC exclusively in roots might be due to deficiency of sucrose supply for amyloplast function since these plants have impaired chloroplast redox homeostasis. However, addition of Suc to the culture medium exerted a poor effect on root growth in these plants, thus showing that leaf chloroplast function is required to provide other metabolites and/or signaling molecules, besides Suc, for root growth. In this regard, it should be mentioned that NTRC deficiency causes decreased auxin content (Lepistö et al., 2009). Moreover, the ntrc mutant shows not only slower root growth but also impairment of lateral root formation (Figure 8), which is a process profoundly affected by shoot-derived auxins at early stages of seedling growth (Bhalerao et al., 2002). The lateral root formation phenotype of the NTRC-deficient mutant was rescued by expression of NTRC exclusively in leaves, but not in roots (Figure 8B), thus showing that the recovery of the redox homeostasis of the chloroplast is sufficient to rescue lateral root formation regardless of amyloplast function. These results point to chloroplasts as source of signaling molecules important for development of heterotrophic organs and are in agreement with the model proposed by Ljung et al. (2005) according to which IAA synthesized in leaves at early stages of seedling development is important for lateral root formation, before roots gain more competence for auxin synthesis. Deficiency of NADP-dependent TRX and glutathione systems, as occurs in the triple mutant ntra ntrb cad2, affects auxin signaling. This triple mutant shows lower auxin content, which causes a severe phenotype, including defects of secondary root growth, vasculature, and pin-like phenotype (Bashandy et al., 2010). By contrast, the auxin-related phenotype of the ntrc mutant is less severe affecting only lateral roots. Because NTRC deficiency affects auxin synthesis (Lepistö et al., 2009) but not auxin sensitivity (Figure 8C), the lateral root formation phenotype of the ntrc mutant might indicate a lower content of auxins, but more work is still needed to test this possibility.
In conclusion, the results presented here show that NTRC is broadly expressed in all plant tissues, localizes to plastids, and is involved in the maintenance of plastid redox homeostasis of photosynthetic and heterotrophic tissues. The biochemical properties of NTRC allowed us to propose its function as a redox switch converting NADPH into redox signal, thus enabling a direct use of NADPH for redox regulation. The finding that restitution of chloroplast redox homeostasis is sufficient to recover root-related phenotypes in plants with impaired amyloplast redox homeostasis emphasizes the essential role of the chloroplast for the growth of heterotrophic tissues. Consequently, green plastids are not only a source of carbon and energy but also of signaling molecules, which may be important in coordinating the growth of photosynthetic and heterotrophic organs of the plant.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana wild-type (ecotype Columbia), ntrc mutant, line SALK_012208 (Serrato et al., 2004), and the transgenic plants generated in this work were grown in soil supplemented with Hoagland medium in culture chambers under long-day (16 h light/8 h dark) or short-day (8 h light/16 h dark) conditions at 22°C during the light and 20°C during dark. The light intensity was set at 140 μmol m−2 s−1. For root growth experiments, seeds were surface sterilized (3 min in 70% [v/v] ethanol followed by 4 min in 30% bleach), placed on plates containing 1× Murashige and Skoog (MS) media solidified with 0.8% agar, and stratified for 2 d at 4°C. Plates were oriented vertically and incubated in a growth chamber under short-day conditions. Seven-day-old seedlings were transferred onto fresh MS plates supplemented with 30 mM Suc or 30 mM mannitol for 14 additional days. For auxin treatments, seedlings grown on MS media for 5 d were transferred to media containing IAA for an additional period of 3 d. Root length and number of lateral roots were measured with the ImageJ software.
qPCR and Immunoblot Analysis
Total RNA (1 µg) was extracted from tissues dissected from mature plants grown for 48 d using Trizol and retrotranscribed by means of the QuantiTect RT kit (Qiagen). Real-time PCR was performed in a total reaction volume of 20 μL containing primers (4 µM each), cDNA (40 ng), and 10 μL of iQ SYBR Green Supermix (Bio-Rad). Results obtained from three independent biological samples (three analytical replicates each) are represented as 2−ΔΔCT (cycle threshold) as described by Livak and Schmittgen (2001). Ubiquitin10 was used as reference gene. Gene-specific primers used are described in Supplemental Table 3 online. Fluorescence of PCR products was determined continuously by the iQ5 cycler (Bio-Rad).
Immunoblot analysis was performed as previously described (Kirchsteiger et al., 2009) using as probes previously described antibodies (anti-NTRC, anti-NTRB, and anti-2Cys Prx). For optimized resolution, SDS-PAGE was performed with 12 to 15% acrylamide/bisacrylamide gels.
Generation of Arabidopsis Transgenic Lines Expressing the NTRCpro:GUS Gene
The NTRCpro:GUS fusion was constructed in the binary vector pGII0229 (Hellens et al., 2000) by a three-step process. First, a 276-bp fragment containing the nopaline synthase gene terminator (NOSter) was amplified by PCR from the pBI121 plasmid using oligonucleotides F-NOSter-NotI and R-NOSter-SacI, introducing NotI and SacI restriction sites, underlined in Supplemental Table 4 online, used to clone the NOSter fragment into the pGII0229 vector. The NTRC gene (At2g41680) is separated from the flanking At2g41690 gene by ∼2.7 kb. Both genes are transcribed in opposite directions so that this 2.7-kb sequence, which shows an even distribution of putative cis-acting elements (Davuluri et al., 2003) (see Supplemental Figure 1 online), may contain the promoters of both genes. Thus, to analyze the putative NTRC promoter, avoiding as much as possible interference from the flanking At2g41690 gene promoter, we selected a 1.05-kb fragment upstream of the NTRC gene coding sequence (see Supplemental Figure 1 online). This fragment was amplified from Arabidopsis (ecotype Columbia) genomic DNA with oligonucleotides FpNtrc-HindIII and RpNtrc-XbaI, introducing HindIII and XbaI restriction sites, underlined in Supplemental Table 4 online, which were used to insert the fragment into the pGII0229-NOSter plasmid. Finally, the UidA gene from Escherichia coli, encoding the GUS reporter enzyme, was amplified with oligonucleotides F-GUS-XbaI and R-GUS-NotI, introducing XbaI and NotI sites, underlined in Supplemental Table 4 online, which were used to transcriptionally fuse the GUS reporter gene to the NTRC promoter. The final construct was sequenced and introduced into Agrobacterium tumefaciens (C58pMP90). Arabidopsis plants were then transformed by the floral dip method (Clough and Bent, 1998), and homozygous plants for the transgene were selected.
Histochemical GUS Staining
Histochemical detection of GUS activity was performed using the substrate 5-bromo-4-chloro-3-indolyl glucuronide (Jefferson et al., 1987) by vacuum-infiltrating seedlings in assay buffer (50 mM sodium phosphate, pH 7.0, 0.2% Triton X-100, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 10 mM EDTA) containing 0.05% 5-bromo-4-chloro-3-indolyl glucuronide. Samples were incubated at 37°C overnight wrapped in aluminum foil to keep them in the dark. Green tissues were cleared with ethanol prior to observation.
Generation of Arabidopsis Transgenic Lines Expressing the NTRC-GFP Fusion Protein
Subcellular localization of NTRC was analyzed in Arabidopsis transgenic plants expressing the NTRC-GFP fusion protein. Given the putative presence of a transit peptide at the N terminus of NTRC, the GFP was translationally fused at the C terminus of the enzyme. To this end, the cDNA encoding GFP was digested from the peGFP plasmid (kindly provided by Thomas Roitsch, Graz University, Austria) by digestion with SmaI and XbaI. These restriction sites were then used to clone this fragment into the pBIBA7 vector (Becker, 1990) to generate the pBIBA7-GFP plasmid. Then, the full-length coding sequence of Arabidopsis NTRC was amplified by PCR from plasmid pUNI_U14278 (The Arabidopsis Information Resource accession sequence 504962595) with oligonucleotides Ntrc-F-KpnI and Ntrc-R-SmaI, introducing KpnI and SmaI restriction sites (underlined in Supplemental Table 4 online), the mutation of the stop codon TGA to TCA (bold), and the insertion of an additional nucleotide (lowercase) to keep the coding frame. The NTRC fragment was then introduced in the pBIBA7-GFP plasmid to generate the pBIBA7-NTRC-GFP construct for expression under the CaMV 35S promoter. For expression of the NTRC-GFP fusion protein under the control of the NTRC gene promoter, the XbaI-NotI fragment containing the UidA gene coding sequence, in the NTRCpro:GUS construct described above, was replaced by an XbaI-NotI fragment containing the coding sequence of the NTRC-GFP fusion protein. All plasmids were checked by sequencing and introduced into Agrobacterium (C58pMP90) to transform Arabidopsis plants by the floral dip method, as indicated above.
Generation of Arabidopsis Transgenic Lines with Leaf- and Root-Specific Expression of NTRC
The Pht1,2 (2000 bp), Pht1,3 (1647 bp), and Rbcs1A (1700 bp) promoters were PCR amplified from wild-type Arabidopsis (ecotype Columbia) genomic DNA using specific primers with Gateway tails. The forward primers contain the AttB1 tail, and the reverse primers contain the AttB2 tail (see Supplemental Table 4 online). Specific sequences for each oligonucleotide were as follows: Pht1,2-Fw, Pht1,2-Rev, Pht1,3-Fw, Pht1,3-Rev, Rbcs-Fw, and Rbcs-Rev (see Supplemental Table 4 online). All PCR products were introduced into the Gateway pDONR207 (Invitrogen) vector using BP clonase, generating promoter entry clones. The promoter fragments were then transferred into the pGWB3 and pGWB1 destination vectors (Nakagawa et al., 2007) using LR Clonase II (Invitrogen). Restriction sites BamHI and KpnI (underlined in Supplemental Table 4 online) were incorporated into the specific sequence primer to facilitate subsequent cloning.
The full-length NTRC cDNA from Arabidopsis (DNA stock number U-14278) was amplified by PCR with oligonucleotides, which added KpnI and SacI restriction sites, underlined in Supplemental Table 4 online, at the 5′ and 3′ ends, respectively. The fragment was cloned into the pGEMt vector (Promega), which was sequenced in both strands. The NTRC cDNA was digested with KpnI and SacI and fused to the KpnI-SacI site of a binary vector pGWB1 to yield plasmids pGWB1-Pht1,2-At-NTRC, pGWB1-Pht1,3-At-NTRC, and pGWB1-Rbcs-At-NTRC.
The constructs were integrated into the Arabidopsis T-DNA insertion mutant SALK_012208 (ntrc) by an Agrobacterium-mediated (C58pMP90) floral dip procedure (Clough and Bent, 1998), and transgenic seedlings were selected on half-strength MS medium containing 20 mg⋅L−1 hygromycin. Several independently transformed plants were obtained with each construct. An empty vector transgenic line, which showed no difference to wild-type plants, was used as control.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At2g41680 (NTRC), At3g11630 (2-Cys-PRX A), At5g06290 (2-Cys PRX B), At1g10960 (Fd1), At1g60950 (Fd2), At5g66190 (FNR1), At1g20020 (FNR2), At4g05390 (RFNR1), At1g30510 (RFNR2), At5g23440 (FTRA), At2g04700 (FTRB), At3g02730 (Trxf1), At5g16400 (Trxf2), At5g43370 (Pht1,2), At5g43360 (Pht1,3) and AT1G67090 (Rbcs1A), and At4g05320 (UBQ10).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Scheme Showing the Position of the NTRC Gene (Atg41680) and the Flanking At2g41690 Gene in Arabidopsis and Distribution of Possible cis-Acting Elements.
Supplemental Figure 2. Characterization of Transgenic Plants Expressing the NTRC-GFP Fusion Protein or GFP under the Control of the CaMV 35S Promoter in Wild-Type and ntrc Mutant Backgrounds.
Supplemental Figure 3. Histochemical Localization of GUS Expression under the Control of Organ-Specific Promoters.
Supplemental Figure 4. Effect of IAA Treatment on Root Growth Inhibition.
Supplemental Table 1. Expression of Genes Encoding NTRC, 2-Cys PRX A, and 2-Cys PRX B in Different Arabidopsis Organs Based on Data Obtained from Genevestigator.
Supplemental Table 2. Expression of Genes Encoding the Alternative Pathway for Redox Regulation in Root Amyloplasts Based on Data Obtained from Genevestigator.
Supplemental Table 3. Gene-Specific Oligonucleotides Used for qPCR Analysis.
Supplemental Table 4. Oligonucleotides Used for Constructs to Generate Transgenic Lines.
Supplementary Material
Acknowledgments
This work was supported by European Regional Development Fund–cofinanced grants from the Ministry of Science and Innovation (BIO2010-15430) and Junta de Andalucía (BIO-182 and CVI-5919). We thank Thomas Roitsch (Graz University, Austria) for kindly providing the plasmid peGFP. We also thank Alicia Orea (Instituto de Bioquímica Vegetal y Fotosíntesis, University of Seville–Consejo Superior de Investigaciones Cientificas) for technical assistance with the confocal microscopy analysis. We thank Anna Lindahl for critical reading of the manuscript.
AUTHOR CONTRIBUTIONS
F.J.C. designed the research. K.K., J.F., M.B.P., and M.G. performed research. All the authors analyzed data. F.J.C. wrote the article.
Glossary
- TRX
thioredoxin
- NTR
NADPH-dependent thioredoxin reductase
- NTS
NADPH-thioredoxin system
- Fd
ferredoxin
- FTR
Fd-dependent TRX reductase
- FNR
Fd-NADP+oxidoreductase
- NTRC
NADPH-thioredoxin reductase C
- AGPase
ADP-glucose pyrophosphorylase
- qPCR
quantitative PCR
- GUS
β-glucuronidase
- GFP
green fluorescent protein
- CaMV
cauliflower mosaic virus
- IAA
indole-3-acetic acid
- MS
Murashige and Skoog
References
- Alkhalfioui F., Renard M., Montrichard F. (2007). Unique properties of NADP-thioredoxin reductase C in legumes. J. Exp. Bot. 58: 969–978 [DOI] [PubMed] [Google Scholar]
- Arsova B., Hoja U., Wimmelbacher M., Greiner E., Ustün S., Melzer M., Petersen K., Lein W., Börnke F. (2010). Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M., Dietz K.-J. (1997). The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: Its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 12: 179–190 [DOI] [PubMed] [Google Scholar]
- Balmer Y., Vensel W.H., Cai N., Manieri W., Schürmann P., Hurkman W.J., Buchanan B.B. (2006a). A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc. Natl. Acad. Sci. USA 103: 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y., Vensel W.H., DuPont F.M., Buchanan B.B., Hurkman W.J. (2006b). Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. J. Exp. Bot. 57: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Bashandy T., Guilleminot J., Vernoux T., Caparros-Ruiz D., Ljung K., Meyer Y., Reichheld J.-P. (2010). Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R.P., Eklöf J., Ljung K., Marchant A., Bennett M., Sandberg G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Becker D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broin M., Cuiné S., Eymery F., Rey P. (2002). The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell 14: 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broin M., Cuiné S., Peltier G., Rey P. (2000). Involvement of CDSP 32, a drought-induced thioredoxin, in the response to oxidative stress in potato plants. FEBS Lett. 467: 245–248 [DOI] [PubMed] [Google Scholar]
- Buchanan B.B., Balmer Y. (2005). Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Ceccarelli E.A., Arakaki A.K., Cortez N., Carrillo N. (2004). Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP(H) reductases. Biochim. Biophys. Acta 1698: 155–165 [DOI] [PubMed] [Google Scholar]
- Chibani K., Tarrago L., Schürmann P., Jacquot J.-P., Rohuier N. (2010). Biochemical properties of poplar thiredoxin z. FEBS Lett. 585: 1077–1081 [DOI] [PubMed] [Google Scholar]
- Collin V., Issakidis-Bourguet E., Marchand C., Hirasawa M., Lancelin J.-M., Knaff D.B., Miginiac-Maslow M. (2003). The Arabidopsis plastidial thioredoxins: New functions and new insights into specificity. J. Biol. Chem. 278: 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V., Lamkemeyer P., Miginiac-Maslow M., Hirasawa M., Knaff D.B., Dietz K.-J., Issakidis-Bourguet E. (2004). Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol. 136: 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dangoor I., Peled-Zehavi H., Levitan A., Pasand O., Danon A. (2009). A small family of chloroplast atypical thioredoxins. Plant Physiol. 149: 1240–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dios Barajas-López J., Serrato A.J., Olmedilla A., Chueca A., Sahrawy M. (2007). Localization in roots and flowers of pea chloroplastic thioredoxin f and thioredoxin m proteins reveals new roles in nonphotosynthetic organs. Plant Physiol. 145: 946–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald R.G.K., Cashmore A.R. (1990). Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 9: 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont F.M. (2008). Metabolic pathways of the wheat (Triticum aestivum) endosperm amyloplast revealed by proteomics. BMC Plant Biol. 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E., et al. (2004). A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc. Natl. Acad. Sci. USA 101: 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E., Rouhier N., Navrot N., Jacquot J.-P. (2005). The plant thioredoxin system. Cell. Mol. Life Sci. 62: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Jacquot J.-P., Eklund H., Rouhier N., Schürmann P. (2009). Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci. 14: 336–343 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B., Fischer K., Hilpert B., Schubert S., Gutensohn M., Weber A., Flügge U.-I. (1998). Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: The glucose 6-phosphate/phosphate antiporter. Plant Cell 10: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchsteiger K., Pulido P., González M.C., Cejudo F.J. (2009). NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Mol. Plant 2: 298–307 [DOI] [PubMed] [Google Scholar]
- Koch K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Laloi C., Rayapuram N., Chartier Y., Grienenberger J.M., Bonnard G., Meyer Y. (2001). Identification and characterization of a mitochondrial thioredoxin system in plants. Proc. Natl. Acad. Sci. USA 98: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A., Kangasjärvi S., Luomala E.M., Brader G., Sipari N., Keränen M., Keinänen M., Rintamäki E. (2009). Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol. 149: 1261–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintala M., Allahverdiyeva Y., Kidron H., Piippo M., Battchikova N., Suorsa M., Rintamäki E., Salminen T.A., Aro E.M., Mulo P. (2007). Structural and functional characterization of ferredoxin-NADP+-oxidoreductase using knock-out mutants of Arabidopsis. Plant J. 49: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C.H., Vanacker H., Collin V., Issakidis-Bourguet E., Maréchal P.L., Decottignies P. (2010). Thioredoxin targets in Arabidopsis roots. Proteomics 10: 2418–2428 [DOI] [PubMed] [Google Scholar]
- Martí M.C., Olmos E., Calvete J.J., Díaz I., Barranco-Medina S., Whelan J., Lázaro J.J., Sevilla F., Jiménez A. (2009). Mitochondrial and nuclear localization of a novel pea thioredoxin: Identification of its mitochondrial target proteins. Plant Physiol. 150: 646–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y., Buchanan B.B., Vignols F., Reichheld J.P. (2009). Thioredoxins and glutaredoxins: Unifying elements in redox biology. Annu. Rev. Genet. 43: 335–367 [DOI] [PubMed] [Google Scholar]
- Meyer Y., Reichheld J.P., Vignols F. (2005). Thioredoxins in Arabidopsis and other plants. Photosynth. Res. 86: 419–433 [DOI] [PubMed] [Google Scholar]
- Michalska J., Zauber H., Buchanan B.B., Cejudo F.J., Geigenberger P. (2009). NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc. Natl. Acad. Sci. USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.C., Jang H.H., Chae H.B., Lee J.R., Lee S.Y., Jung Y.J., Shin M.R., Lim H.S., Chung W.S., Yun D.J., Lee K.O., Lee S.Y. (2006). The C-type Arabidopsis thioredoxin reductase ANTR-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochem. Biophys. Res. Commun. 348: 478–484 [DOI] [PubMed] [Google Scholar]
- Motohashi K., Hisabori T. (2006). HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 281: 35039–35047 [DOI] [PubMed] [Google Scholar]
- Mudge S.R., Rae A.L., Diatloff E., Smith F.W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Neuhaus H.E., Emes M.J. (2000). Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Oji Y., Watanabe M., Waliuchi N., Okamoto S. (1985). Nitrite reduction in barley-root plastids – Dependence on NADPH coupled with glucose-6-phosphate and 6-phosphogluconate dehydrogenases, and possible involvement of an electron carrier and a diaphorase. Planta 165: 85–90 [DOI] [PubMed] [Google Scholar]
- Paul M.J., Foyer C.H. (2001). Sink regulation of photosynthesis. J. Exp. Bot. 52: 1383–1400 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz J.M., Cejudo F.J. (2009). A proposed reaction mechanism for rice NADPH thioredoxin reductase C, an enzyme with protein disulfide reductase activity. FEBS Lett. 583: 1399–1402 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz J.M., González M., Spínola M.C., Sandalio L.M., Cejudo F.J. (2009). The quaternary structure of NADPH thioredoxin reductase C is redox-sensitive. Mol. Plant 2: 457–467 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz J.M., Spínola M.C., Kirchsteiger K., Moreno J., Sahrawy M., Cejudo F.J. (2006). Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P., Cazalis R., Cejudo F.J. (2009). An antioxidant redox system in the nucleus of wheat seed cells suffering oxidative stress. Plant J. 57: 132–145 [DOI] [PubMed] [Google Scholar]
- Pulido P., Spínola M.C., Kirchsteiger K., Guinea M., Pascual M.B., Sahrawy M., Sandalio L.M., Dietz K.J., González M., Cejudo F.J. (2010). Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J. Exp. Bot. 61: 4043–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J.P., Meyer E., Khafif M., Bonnard G., Meyer Y. (2005). AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 579: 337–342 [DOI] [PubMed] [Google Scholar]
- Rey P., Cuiné S., Eymery F., Garin J., Court M., Jacquot J.-P., Rouhier N., Broin M. (2005). Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J. 41: 31–42 [DOI] [PubMed] [Google Scholar]
- Schürmann P., Buchanan B.B. (2008). The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Serrato A.J., Cejudo F.J. (2003). Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta 217: 392–399 [DOI] [PubMed] [Google Scholar]
- Serrato A.J., Crespo J.L., Florencio F.J., Cejudo F.J. (2001). Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol. Biol. 46: 361–371 [DOI] [PubMed] [Google Scholar]
- Serrato A.J., Pérez-Ruiz J.M., Spínola M.C., Cejudo F.J. (2004). A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 279: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Spínola M.C., Pérez-Ruiz J.M., Pulido P., Kirchsteiger K., Guinea M., González M.C., Cejudo F.J. (2008). NTRC new ways of using NADPH in the chloroplast. Physiol. Plant. 133: 516–524 [DOI] [PubMed] [Google Scholar]
- Stenbaek A., Hansson A., Wulff R.P., Hansson M., Dietz K.-J., Jensen P.E. (2008). NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 582: 2773–2778 [DOI] [PubMed] [Google Scholar]
- Stenbaek A., Jensen P.E. (2010). Redox regulation of chlorophyll biosynthesis. Phytochemistry 71: 853–859 [DOI] [PubMed] [Google Scholar]
- Tetlow I.J., Beisel K.G., Cameron S., Makhmoudova A., Liu F., Bresolin N.S., Wait R., Morell M.K., Emes M.J. (2008). Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 146: 1878–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso J.A., Vignols F., Cazalis R., Serrato A.J., Pulido P., Sahrawy M., Meyer Y., Cejudo F.J., Chueca A. (2008). Immunocytochemical localization of Pisum sativum TRXs f and m in non-photosynthetic tissues. J. Exp. Bot. 59: 1267–1277 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.