Plastid terminal oxidase (PTOX) is distantly similar to alternative oxidases (AOXs). Both reengineering of AOX1a into the plastid and overexpression of AOX2 are able to rescue the variegation phenotype of the Arabidopsis immutans mutant, which lacks PTOX. We propose that AOXs function in chloroplasts to supplement PTOX activity during early events in chloroplast biogenesis.
Abstract
The immutans (im) variegation mutant of Arabidopsis thaliana is caused by an absence of PTOX, a plastid terminal oxidase bearing similarity to mitochondrial alternative oxidase (AOX). In an activation tagging screen for suppressors of im, we identified one suppression line caused by overexpression of AOX2. AOX2 rescued the im defect by replacing the activity of PTOX in the desaturation steps of carotenogenesis. Similar results were obtained when AOX1a was reengineered to target the plastid. Chloroplast-localized AOX2 formed monomers and dimers, reminiscent of AOX regulation in mitochondria. Both AOX2 and AOX1a were present in higher molecular weight complexes in plastid membranes. The presence of these proteins did not generally affect steady state photosynthesis, aside from causing enhanced nonphotochemical quenching in both lines. Because AOX2 was imported into chloroplasts using its own transpeptide, we propose that AOX2 is able to function in chloroplasts to supplement PTOX activity during early events in chloroplast biogenesis. We conclude that the ability of AOX1a and AOX2 to substitute for PTOX in the correct physiological and developmental contexts is a striking example of the capacity of a mitochondrial protein to replace the function of a chloroplast protein and illustrates the plasticity of the photosynthetic apparatus.
INTRODUCTION
The immutans (im) variegation mutant of Arabidopsis thaliana is a powerful tool to gain insight into mechanisms of chloroplast biogenesis (Aluru et al., 2006; Yu et al., 2007; McDonald et al., 2011). Cells in the green sectors have morphologically normal chloroplasts, whereas cells in the white sectors are heteroplastidic and contain abnormal plastids that lack pigments and organized lamellae, as well as rare, normal-appearing chloroplasts (Wetzel et al., 1994). Early HPLC analyses showed that the white sectors accumulate high levels of phytoene, a colorless C40 carotenoid intermediate, indicating that im is impaired in the activity of phytoene desaturase (PDS), the plastid enzyme that desaturates phytoene in an early step of the carotenoid biosynthetic pathway (Wetzel et al., 1994). All of the steps of carotenogenesis occur in the plastid via nuclear-encoded enzymes that are imported into the organelle posttranslationally (Hirschberg, 2001), and an inhibition of the PDS step would result in lack of accumulation of downstream, colored (photoprotective) carotenoids. Under excess light conditions, this would lead to the generation of white, photooxidized plastids. Normal-appearing chloroplasts, on the other hand, are able to bypass the need for IM gene product activity, perhaps because of intrinsic differences in plastid biochemistry that make some less susceptible to photooxidation during their conversion from proplastids to chloroplasts in the developing leaf primordium or from etioplasts to chloroplasts in greening seedlings.
IM is a distantly related plastid homolog of alternative oxidase (AOX), a mitochondrial inner membrane protein that functions in the alternative (cyanide-resistant) pathway of respiration (Carol et al., 1999; Wu et al., 1999). Central among its physiological functions, AOX is an important sensor of cellular redox balance (Giraud et al., 2008; McDonald, 2008). Similar to AOX, IM has quinol oxidase activity in vivo and in vitro; consequently, it has been designated plastid terminal oxidase (PTOX) (Joët et al., 2002; Josse et al., 2003). PTOX resides at the nexus of a growing number of biochemical pathways in the plastid, including carotenoid biosynthesis, chlororespiration, and photosystem I (PSI) cyclic electron transport (Okegawa et al., 2010; McDonald et al., 2011). It is also a central regulator of photosystem II (PSII) excitation pressure during early chloroplast biogenesis (Rosso et al., 2009) and in photosynthetic marine organisms growing in iron-limited environments (Bailey et al., 2008). Related to its redox surveillance function, PTOX acts as a stress-induced safety valve for the dissipation of excess light energy in some species (Niyogi, 2000; Streb et al., 2005; Rosso et al., 2006, 2009; Shahbazi et al., 2007; Stepien and Johnson, 2009; Savitch et al., 2010; McDonald et al., 2011).
AOX was early recognized by Siedow and colleagues as a diiron carboxylate (DOX) protein (Moore et al., 1995). The relatedness of PTOX and AOX was based on sequence comparisons of a conserved region of AOX termed the DOX domain (∼37% amino acid sequence similarity between PTOX and AOX from diverse sources) (Carol et al., 1999; Wu et al., 1999). This domain contains the active site and is composed of a four-helix bundle that provides six ligands for binding the diiron center (Andersson and Nordlund, 1999; Berthold and Stenmark, 2003; Fu et al., 2005; Moore and Albury, 2008). No crystallographic structures are available for AOX or PTOX, but they have been modeled as interfacial membrane proteins that associate with one leaflet of the bilayer in a manner similar to diiron proteins for which high-resolution x-ray crystal structures have been obtained in nonplant systems (Berthold and Stenmark, 2003). In this model, the four-helix bundle extends from the inner membrane into the matrix (for AOX) or from the thylakoid membrane into the stroma (for PTOX) (Andersson and Nordlund, 1999; Lennon et al., 2003). Whereas PTOX activity in vitro is specific for plastoquinol (PQ; its presumed substrate in plastid membranes) (Josse et al., 2003), it is an open question whether AOX can use substrates other than ubiquinol, which is found in mitochondrial, but not plastid, membranes (McDonald, 2008).
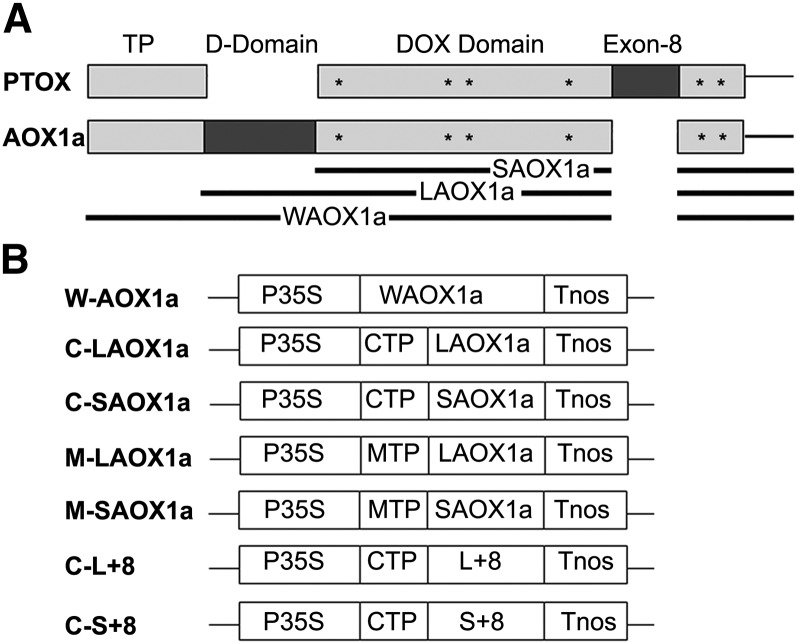
Although AOX and PTOX have conformational elements in common, each contains unique functional domains (Figure 1A). These include, most prominently, an ∼40–amino acid dimerization domain (D-Domain) in AOX that is missing in PTOX. This D-Domain contains a conserved, regulatory Cys that participates in disulfide bond formation between adjacent monomers, giving rise to an inactive dimer; in its reduced state, this Cys binds organic acids, causing enzyme activation (Rhoads et al., 1998; Vanlerberghe et al., 1998; Umbach and Siedow, 2000; Umbach et al., 2006). PTOX, on the other hand, contains a conserved 16–amino acid domain near the active site (Exon 8 Domain) that is missing in AOX (Finnegan et al., 2003; McDonald et al., 2003; Fu et al., 2005). This sequence is required for protein stability (Fu et al., 2005). The lack of a D-Domain in PTOX suggests that it does not dimerize and exists only as a monomer (McDonald et al., 2011). However, this has not been demonstrated experimentally.
Figure 1.
AOX1a Plasmid Constructs.
(A) Structure of PTOX and AOX1a proteins. Both proteins have a DOX domain (gray) with iron binding ligands (asterisks) and an N-terminal targeting sequence (transit peptide [TP]) specific for the plastid (in the case of PTOX) or mitochondrion (in the case of AOX1a). AOX1a has a unique dimerization domain (D-Domain) that contains a regulatory Cys, whereas PTOX has a unique Exon 8 Domain required for protein stability. WAOX1a, the full-length AOX1a amino acid sequence from Arabidopsis; LAOX1a, the AOX1a sequence without its transit peptide; SAOX1a, AOX1a without its TP and D-Domain.
(B) Constructs used in this study. P35S, CaMV 35S promoter; Tnos, terminator of nos (nopaline synthetase); CTP, chloroplast targeting sequence from pea (Pisum sativum) RbcS (Rubisco small subunit); MTP, mitochondrial targeting sequence from tobacco β-ATPase (β-subunit of ATP synthase); WAOX1a, LAOX1a, and SAOX1a, as in (A). IM Exon 8 was inserted into LAOX1a and SAOX1a to generate L+8 and S+8, respectively.
Phylogenetic analyses have revealed that AOX and PTOX are derived from a primitive oxygen reductase that scavenged dioxygen in the highly reducing atmosphere of early life on earth (Gomes et al., 2001). Although horizontal gene transfer events can’t be ruled out, most evidence supports a vertical inheritance pattern whereby the primitive DOX gene radiated into α-proteobacteria (giving rise to the lineage of AOX genes) and cyanobacteria (the PTOX gene lineage), which are the endosymbiotic ancestors of mitochondria and plastids, respectively (Gray et al., 2001; Martin et al., 2002; Finnegan et al., 2003; McDonald et al., 2003, 2011; Gould et al., 2008). It is thought that endosymbiosis was accompanied by transfer of AOX and PTOX from the symbiont to the host genome and that both genes subsequently acquired signals for targeting back to the evolving organelles (Bock and Timmis, 2008). If this scenario is correct, there must have been strong evolutionary pressure for targeting of AOX and PTOX back to their originating compartments (i.e., one protein could not substitute for the other during the churning process that occurred as gene products were functionally tested many times in different cell compartments) (Bogorad, 2008). This suggests that each protein became optimized for function in a given compartment very early in its evolutionary history, predating the events of endosymbiosis.
Despite the striking differences in regulation, substrate specificity, and phylogenetic distance between AOX and PTOX, we wanted to test experimentally whether one protein could functionally substitute for the other. Our strategy was to take advantage of the availability of null im alleles (Wu et al., 1999) to ask whether variegation can be suppressed in transgenic im that express a chloroplast-targeted AOX. We previously used this strategy to define sequences that are required for PTOX activity in planta using techniques of in vitro mutagenesis (Fu et al., 2005, 2009). Arabidopsis has five AOX gene family members (AOX1a-d and AOX2) (Saisho et al., 1997), all of which are targeted to mitochondria, and we chose to focus on AOX1a, the most highly and ubiquitously expressed of these genes (Clifton et al., 2006; Winter et al., 2007). We found that replacement of the mitochondrial transit sequence of AOX1a with the plastid-specific RbcS transit sequence resulted in incorporation of AOX1a into chloroplast membranes of im, where it rescued the phytoene accumulation defect of im and suppressed im variegation. This indicates that AOX1a, despite being optimized for function in mitochondria, is able to substitute for PTOX activity in chloroplast metabolism and, importantly, that it does so in the correct physiological and developmental contexts to permit normal chloroplast biogenesis.
At the time these experiments were in progress, we were engaged in T-DNA activation tagging experiments to identify factors that can modulate im variegation. We discovered, quite unexpectedly, that activation tagging of the gene for AOX2, a seed-specific member of the mitochondrial AOX gene family, gave rise to all-green plants. AOX2 was localized in chloroplast membranes of the tagged lines and, like chloroplast AOX1a, it was able to rescue the phytoene accumulation phenotype of im and substitute for the activity of PTOX in chloroplast biogenesis. Because plastid targeting in the tagged lines was mediated by endogenous AOX2 sequences, our data suggest that AOX2 is capable of functioning as a dual-targeted protein. Due to their complementarity, we describe both the AOX1a and AOX2 experiments in this article.
RESULTS
Targeting of AOX1a to the Plastid Suppresses im Variegation
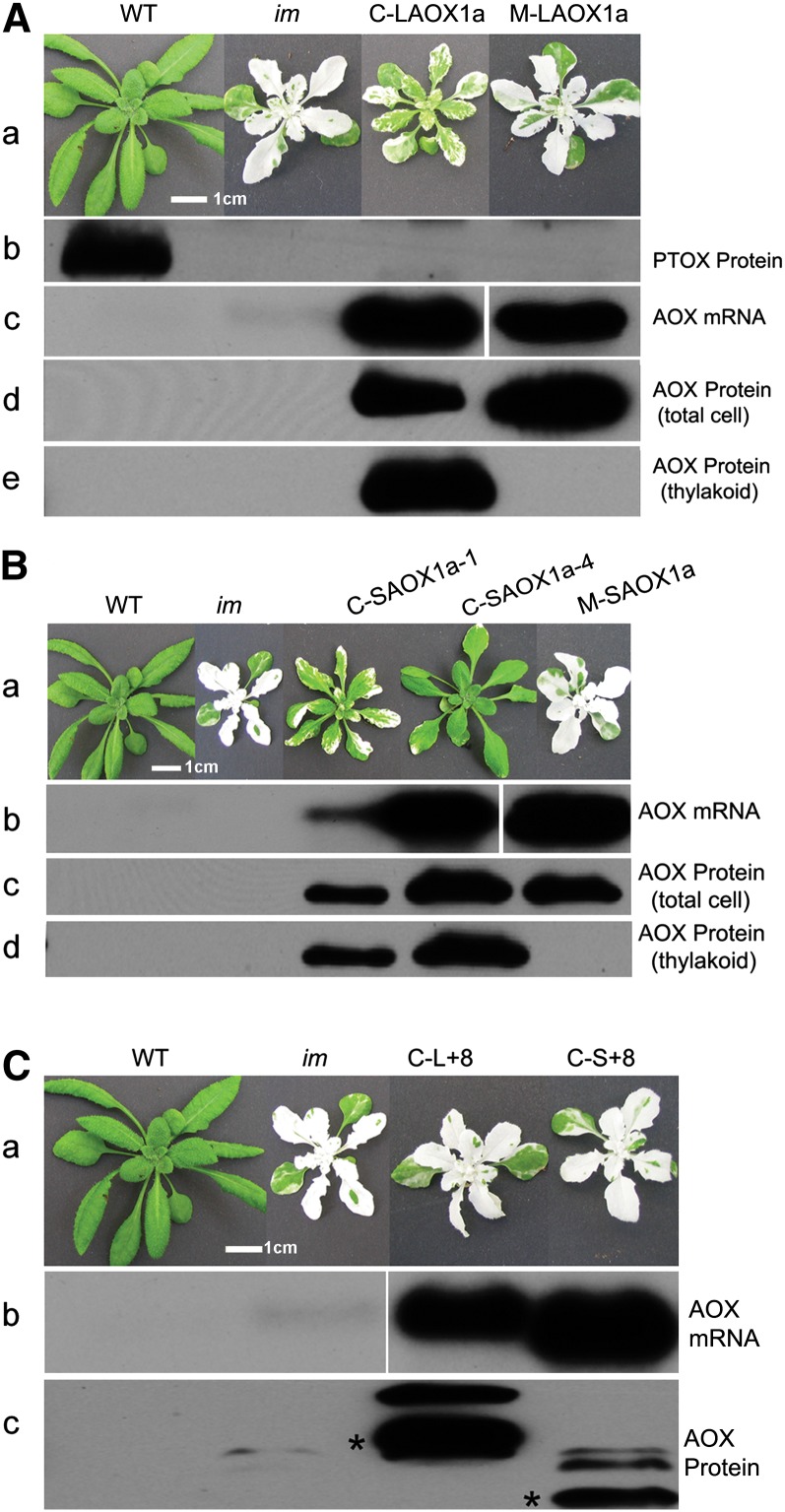
To test whether AOX can functionally substitute for PTOX, we transformed im with a vector containing an AOX1a cDNA whose targeting sequence had been replaced by the chloroplast-specific RbcS targeting peptide (generating C-LAOX1a) (Figure 1B). As a control, the targeting sequence was replaced by the mitochondrial-specific β-ATPase transit peptide (generating M-LAOX1a). Figure 2A shows that transgenic im plants bearing the C-LAOX1a construct have significantly less white sector formation than im, whereas the M-LAOX1a plants closely resemble im. A lack of visible complementation was also observed when im was transformed with a full-length AOX1a (W-AOX1a).
Figure 2.
Expression of AOX1a Constructs in im.
(A) Representative wild type (WT), im, im transformed with C-LAOX1a, and im transformed with M-LAOX1a (panel a). All plants were maintained under continuous illumination (100 μmol m−2 s−1) after an initial 5 d at 15 μmol m−2 s−1. Photographs were taken 4 weeks after germination. Total cell proteins were isolated from 4 mg fresh weight of leaf tissue (“total cell,” panel d) or from pellets obtained by centrifugation of lysed, Percoll gradient–purified chloroplasts (corresponding to 5 μg of chlorophyll) (panels b and e); this fraction is enriched in membranes, primarily thylakoids. The samples were subjected to 12.5% SDS-PAGE, and immunoblot analyses were conducted using PTOX polyclonal antibodies (Fu et al., 2005) or AOX monoclonal antibodies (Elthon et al., 1989; Finnegan et al., 1999). RNA gel blot analyses were performed using total cell RNAs (2 μg) from leaves of the various plants, and these nitrocellulose filters were probed with an AOX1a cDNA (panel c). Bar = 1 cm.
(B) Representative wild type, im, im transformed with C-SAOX1a (lines 1 and 4), and M-SAOX1a (panel a). The plants were maintained as in (A) and photographed 4 weeks after germination. RNA and protein analyses (panels b to d) were conducted as in (A). Bar = 1 cm.
(C) Representative wild type, im, im transformed with C-L+8, and im transformed with C-S+8 (panel a). The plants were maintained as in (A) and photographed 4 weeks after germination. RNA and protein analyses (panels b and c) were conducted as in (A); the proteins were from the pellet fractions of Percoll gradient–purified chloroplasts. In panel c, bands of the predicted sizes of the mature proteins are indicated with asterisks. The origin of the higher molecular weight forms is unknown. Bar = 1 cm.
To confirm these findings, we performed RNA gel blot and immunoblot analyses. For the latter, proteins were isolated from whole leaf extracts or from Percoll gradient–purified chloroplasts and analyzed using either an AOX monoclonal antibody that recognizes all members of the Arabidopsis AOX gene family (Elthon et al., 1989; Finnegan et al., 1999) or a PTOX polyclonal antibody (Fu et al., 2005). As anticipated, PTOX is present in the pellet (membrane) fraction of lysed chloroplasts from wild-type plants (Lennon et al., 2003) but not in those from im or transgenic im plants (Figure 2A, panel b). AOX1a mRNAs and proteins, on the other hand, are plentiful in whole-leaf extracts from the C-LAOX1a and M-LAOX1a lines but are barely detectable in im and the wild type, reflecting the low expression levels of all AOX isoforms in Arabidopsis (Heazlewood and Millar, 2005; Winter et al., 2007) (Figure 2A, panels c and d). AOX proteins are abundant in the chloroplast membrane fractions from C-LAOX1a but not M-LAOX1a (Figure 2A, panel e). Taken together, these data indicate that C-LAOX1a proteins are targeted to chloroplast membranes, where they are able to compensate for the im defect, at least in part.
Do the Exon 8 and D-Domains Affect the Function of Plastid-Localized AOX1a?
To achieve higher levels of complementation, we reasoned that an AOX with greater structural similarity to PTOX than LAOX1a might have enhanced quinol oxidase activity, perhaps better mimicking PTOX conformation in the plastid membrane. To test this hypothesis, we deleted the D-Domain from C-LAOX1a to generate C-SAOX1a; as a control, the D-Domain was deleted from M-LAOX1a to generate M-SAOX1a (Figure 1B). Figure 2B shows two representative C-SAOX1a lines. Both lines are significantly greener than im and the C-LAOX1a plants. The extent of variegation suppression in these lines is positively correlated with transgene copy number (one copy in the C-SAOX1a-1 line versus three copies in the C-SAOX1a-4 line) and also with the amounts of C-SAOX1a mRNAs and proteins in lysed chloroplast pellet fractions (Figure 2B, panels b to d). By contrast, the M-SAOX1a construct does not complement im; although the M-SAOX1a lines have high levels of M-SAOX1a mRNAs and AOX proteins, the proteins are not chloroplast localized. Considered together, the data in Figures 2A and 2B indicate that AOX1a proteins that lack the D-Domain are better able to suppress the im defect than those that contain this domain.
Using techniques of in vitro and in planta mutagenesis, we previously demonstrated that a 16–amino acid sequence near the C terminus is essential for PTOX activity and stability (Exon 8 Domain) (Fu et al., 2005). Interestingly, this domain corresponds precisely to Exon 8 of the IM genomic sequence (Figure 1A). Sequences flanking this domain can be aligned in the Arabidopsis genes for PTOX and AOX, allowing us to pinpoint the site of the Exon 8 insertion in PTOX with respect to AOX. To test whether this sequence influences the activity of chloroplast-localized AOX1a, we inserted it in frame into C-SAOX1a and C-LAOX1a using a multistep in vitro mutagenesis procedure. This gave rise to the C-S+8 and C-L+8 constructs, respectively (Figures 1A and 1B). Figure 2C shows representative transgenics from hundreds of independent transformation events in which the C-S+8 and C-L+8 constructs were introduced into im: None was appreciably greener than im. This apparent lack of complementation is not likely due to a deficiency of transgene expression inasmuch as the transformed lines contain high levels of C-S+8 and C-L+8 mRNAs and chloroplast-localized C-S+8 and C-L+8 proteins (Figure 2C). We speculate that AOX1a is rendered nonfunctional in chloroplasts by introduction of the Exon 8 Domain.
Do Chloroplast AOX1a and PTOX Form Dimers?
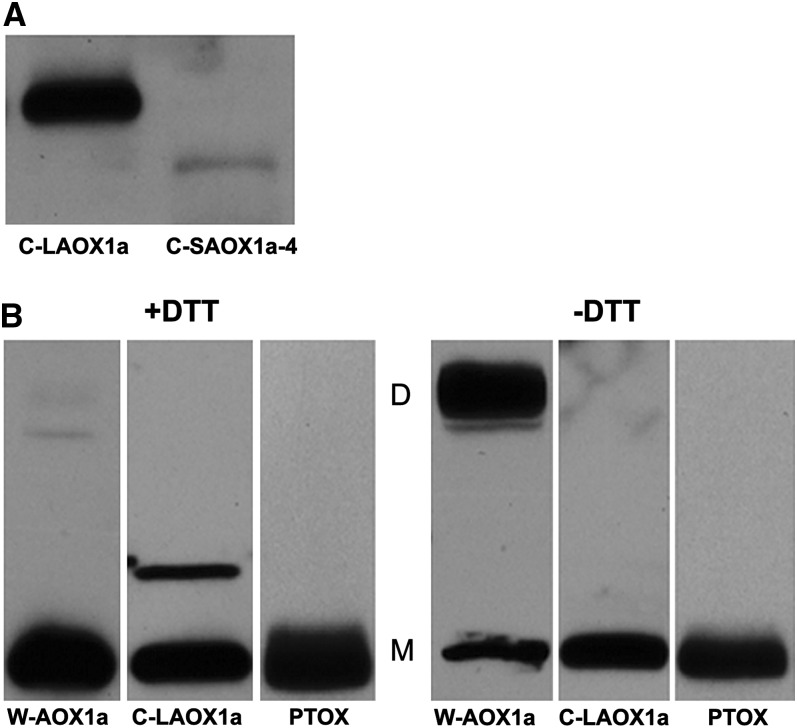
Figure 3A reveals that the C-SAOX1a-4 plants have significantly less chloroplast membrane-associated AOX than the C-LAOX1a plants, despite being considerably more green (Figures 2A and 2B). An attractive hypothesis to explain this observation is that the bulk of chloroplast C-LAOX1a is in an inactive (dimer) form as a consequence of disulfide bond formation between conserved Cys residues in the D-Domains of adjacent monomers, analogous to the situation in mitochondria (Umbach et al., 2006). Presumably, this would not be an option for C-SAOX1a because it lacks a D-Domain and conserved Cys residues (Wu et al., 1999). To test this hypothesis, we performed immunoblot analyses on membrane proteins from gradient-purified chloroplasts of the C-LAOX1a plants; the samples were incubated in the presence or absence of DTT. Mitochondrial AOX (from W-AOX1a plants; Figure 1B) and PTOX (from wild-type plants) served as controls. Figure 3B shows, as anticipated, that AOX forms monomers and dimers in the mitochondrion under nonreducing conditions (−DTT) but only monomers under reducing conditions (+DTT). C-LAOX1a, on the other hand, forms only monomers in the presence or absence of DTT. This suggests that C-LAOX1a is not sequestered in an inactive (dimeric) form in the plastid. Like C-LAOX1a, PTOX forms only monomers in the presence or absence of DTT. This confirms earlier suggestions that PTOX is present as a monomer in thylakoid membranes (Wu et al., 1999).
Figure 3.
Oligomeric State of AOX and PTOX.
(A) Proteins were isolated from the pellet fractions of gradient-purified C-LAOX1a and C-SAOX1a-4 chloroplasts, and immunoblot analyses were conducted as in Figure 2A using monoclonal antibodies to AOX. AOX is smaller in the C-SAOX1a-4 plants because it lacks the D-Domain.
(B) Chloroplast membrane proteins were isolated from W-AOX1a, C-LAOX1a, and wild-type plants and then subjected to SDS-PAGE in the presence or absence of 0.1 mM DTT. Immunoblot analyses were conducted as in Figure 2A: Filters from the W-AOX1a and C-LAOX1a lines were immunoblotted with the AOX antibody, whereas filters from the wild type were immunoblotted with the PTOX antibody. “D” and “M” mark the positions of dimeric (∼64 kD) and monomeric (∼32 kD) forms of AOX, as well as of monomeric PTOX. The origin of the band migrating between the dimeric and monomeric forms in the C-LAOX1a (+DTT) lane is not known.
C-SAOX1a Is More Active in Vitro Than C-LAOX1a
A second hypothesis to explain the finding that the C-SAOX1a-4 plants are greener than the C-LAOX1a plants, despite having considerably less AOX, is that the C-SAOX1a enzyme has a higher activity than the C-LAOX1a enzyme. To test this hypothesis, we used an established in vitro activity assay for AOX that is based on the ability of AOX to confer cyanide-resistant, salicylhydroxamic acid (SHAM)-sensitive oxygen consumption to Escherichia coli membranes (Umbach et al., 2002, 2006). The rationale of this assay is that NADH serves as an electron donor to ubiquinone via a membrane-bound NADH dehydrogenase; electrons are then transferred to molecular oxygen via AOX or the endogenous cytochrome pathway. Flux can be regulated by potassium cyanide (KCN), which inhibits the cytochrome pathway but not AOX activity, or by SHAM, which inhibits AOX but not the cytochrome pathway. In short, O2 consumption occurs by the cytochrome pathway when SHAM is added to the membranes but by AOX when KCN is added. Addition of both inhibitors abolishes O2 consumption.
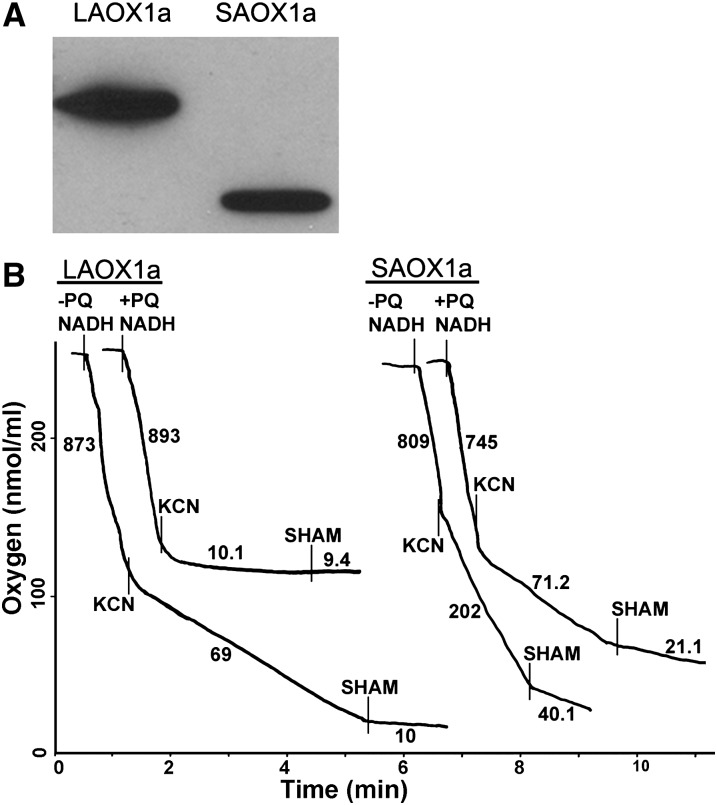
Figure 4 shows the results of in vitro activity assays with LAOX1a and SAOX1a enzymes lacking their N-terminal organelle targeting sequences; both proteins are stably expressed in the bacterial cells (Figure 4A). Figure 4B reveals that SAOX1a-containing E. coli membranes have higher rates of CN-resistant, SHAM-sensitive O2 uptake activity than membranes containing LAOX1a (202 versus 69 nmol O2/min/mg protein, respectively). This indicates that SAOX1a is more active in vitro than LAOX1a. Addition of excess PQ significantly reduces the activity of both enzymes (71.2 nmol O2/min/mg protein for SAOX1a versus 10.1 nmol O2/min/mg protein for LAOX1a), indicative of a preference for endogenous (bacterial) ubiquinol as a substrate. This might not be surprising since AOX has presumably been optimized for ubiquinol as a substrate in mitochondrial membranes.
Figure 4.
In Vitro Activity Assays.
(A) Membranes were isolated from E. coli transformed with LAOX1a or SAOX1a gene constructs lacking their chloroplast targeting sequences. Equal protein amounts were electrophoresed through 12.5% SDS-PAGE gels and then transferred to nitrocellulose membranes. The filters were treated with the AOX antibody.
(B) Oxygen consumption rates (nmol O2/min/mg protein) were determined from the slopes of O2 traces following the addition of 1 mM NADH, 2 mM KCN, and 2 mM SHAM (Umbach et al., 2002). AOX activity is defined as the oxygen consumption rate in the presence of 2 mM KCN minus the oxygen consumption rate in the presence of 2 mM SHAM. E. coli membranes were incubated in the presence or absence of excess decyl-plastoquinone (+/−PQ). Each experiment was repeated four times with similar results.
We previously conducted in vitro activity assays similar to those in Figure 4 to investigate the impact of mutations in active site amino acids that are conserved between AOX and PTOX (Fu et al., 2005, 2009). These studies were supplemented with in planta experiments in which im was transformed with the same mutant constructs, and their ability to complement im was used as a proxy for their relative in vivo activity. We found that the in vitro and in planta studies correlated well with one another (i.e., mutant PTOX enzymes with wild-type in vitro activities were able to complement im completely). Hence, if these in vitro results (Figure 4) can be extrapolated to the in planta situation (Figure 2), our data are consistent with the idea that SAOX1a, despite being present in lower amounts, is more active than LAOX1a in planta and hence better able to complement the defect in im plastids.
Identification of the Suppressor Gene in ATG791
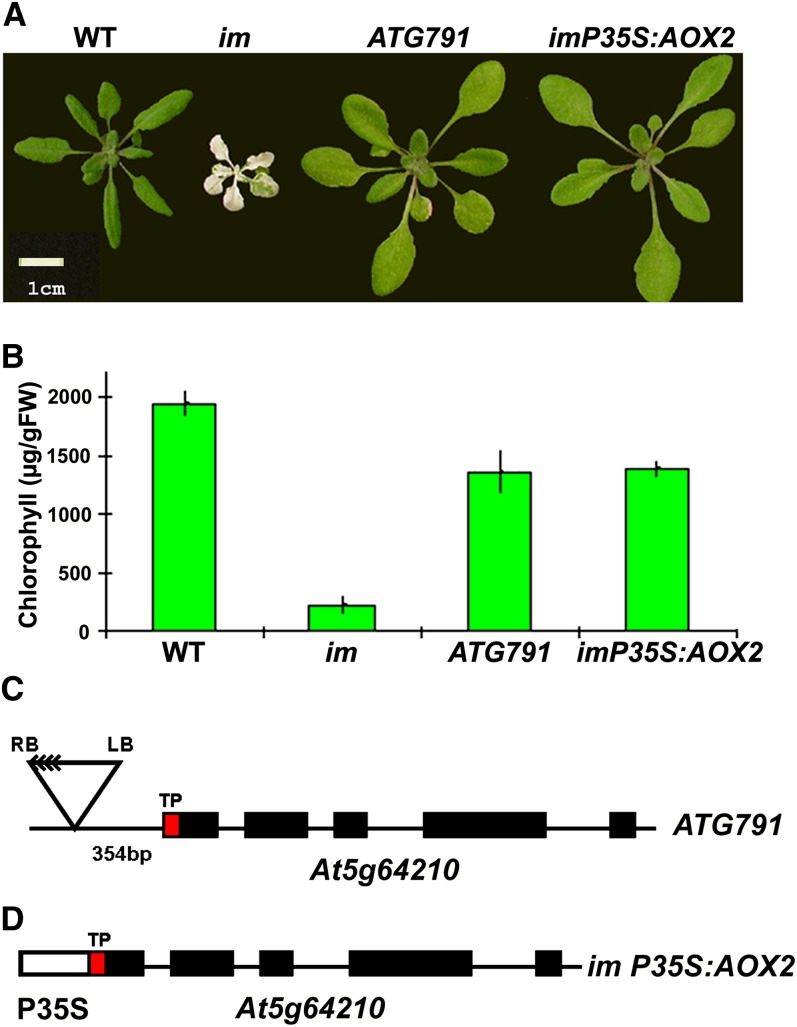
As mentioned earlier, while the studies with plastid-targeted AOX1a were in progress, we were engaged in experiments to isolate activation-tagged mutants of im with nonvariegated phenotypes. One of these lines was chosen for further study (designated ATG791); this line has been stable for a number of generations. The leaves of ATG791 are larger than the wild type and are uniformly pale-green (Figures 5A and 5B). DNA gel blot analyses revealed that ATG791 has two T-DNA inserts (see Supplemental Figure 1A online), both of which cosegregate with the suppression of variegation phenotype. Genomic DNAs flanking these inserts were isolated by plasmid rescue and sequenced, and the inserts were found to reside ∼3.3 Mb apart from one other on chromosome 5 (see Supplemental Figure 1A online). RNA gel blot analyses showed that mRNAs from two of eight genes that flank these inserts are significantly elevated in amount in ATG791, which suggests that they are activation tagged (see Supplemental Figure 1B online). These genes (At5g55180 and At5g64210) reside immediately downstream from each T-DNA insert. At5g64210 has been annotated as AOX2, the nuclear gene for mitochondrial AOX2, whereas At5g55180 is the nuclear gene for glycosyl hydrolase family 17 protein, an enzyme found in the endomembrane system.
Figure 5.
Activation Tagging of im.
(A) Representative 4-week-old wild type (WT), im, ATG791 (the activation-tagged im suppressor line), and AOX2 overexpression plants (im-P35S:AOX2). The plants were grown at 22°C under continuous illumination of ∼15 µmol m−2 s−1 for 7 d and then transferred to ∼100 µmol m−2 s−1 for another 3 weeks. Bar = 1 cm.
(B) Chlorophyll contents were determined on the first two pairs of rosette leaves from 3-week-old plants (on a fresh weight [FW] basis). Each bar represents an average ± sd of three different pooled leaf samples.
(C) Schematic of AOX2 sequences in ATG791. The T-DNA insert (containing four copies of the CaMV 35S enhancer [<]) is located 354 bp upstream of the initiating ATG of the N-terminal plastid targeting sequence of AOX2 (transit peptide [TP] is indicated in red). Exons are filled boxes. LB, left T-DNA border; RB, right T-DNA border.
(D) Schematic of AOX2 sequences in im-P35S:AOX2. P35S is fused to the initiating ATG of the predicted TP of AOX2. Exons are filled boxes.
To determine if either of these is the suppressor gene, transgenic im plants were generated in which full-length cDNAs and genomic DNAs from the two genes were overexpressed using the cauliflower mosaic virus (CaMV) 35S promoter. Controls included the other six genes in the immediate vicinity of the two T-DNAs. We found that im was rescued by overexpression of At5g64210 but not by any of the other constructs. These plants (designated im-P35S:AOX2) closely resemble ATG791 with respect to growth habit, morphology, size, and chlorophyll content (Figures 5A and 5B). Similar results were obtained when genomic DNAs of the eight genes were used rather than cDNAs. Therefore, we conclude that At5g64210 is the suppressor gene in ATG791. Schematic representations of AOX2 gene structure in the tagged line (ATG791) and the overexpression line (im-P35S:AOX2) are illustrated in Figures 5C and 5D.
AOX2 Is Present in Chloroplast Membranes of the im Suppressor Lines
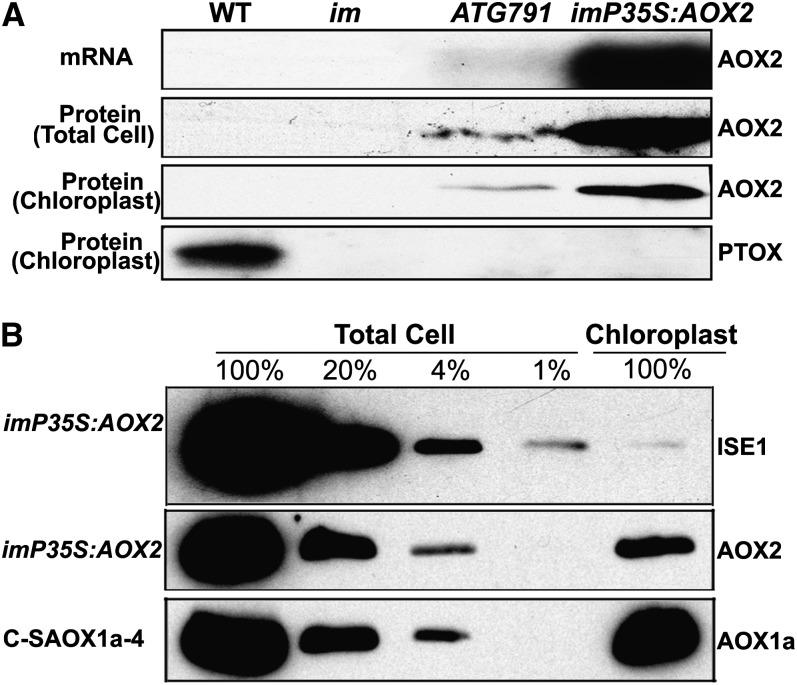
To study the expression of AOX2 in the suppressor (ATG791) and overexpression lines (im-P35S:AOX2), we performed mRNA and protein analyses similar to those for AOX1a (Figure 2). Figure 6A shows that AOX2 mRNAs are not detectable in im and wild-type leaves, confirming the data in Supplemental Figure 1B online. However, their abundance is enhanced in the suppressor line and considerably more so in the overexpression line. Similar expression profiles were observed for AOX2 proteins in samples of total cell protein and in membrane protein fractions from lysed, gradient- purified chloroplasts. As anticipated, PTOX was detectable only in the membrane fraction from isolated wild-type chloroplasts, consistent with its known location in thylakoid membranes (Lennon et al., 2003).
Figure 6.
AOX2 Is Present in Chloroplasts of the Tagged and Overexpression Lines.
(A) RNA gel blot and immunoblot analyses. Total cell RNAs were isolated from 4-week-old plants as in Figure 5A, and RNA gel blots were conducted using an AOX2 gene–specific probe (primers are listed in Supplemental Table 1 online). Rosette leaves from 4-week-old plants also served as the source of total cell proteins and of chloroplast membranes from lysed, Percoll gradient–purified chloroplasts (designated chloroplasts). Equal protein amounts were electrophoresed through 12.5% SDS polyacrylamide gels, and immunoblot analyses were performed using AOX or PTOX antibodies. WT, the wild type.
(B) Total cell proteins or proteins from gradient purified chloroplasts were isolated from the rosette leaves of 4-week-old C-SOX1a-4 and im-P35S:AOX2 plants. Samples containing equal chlorophyll amounts (5 µg chlorophyll) were electrophoresed through 12.5% SDS polyacrylamide gels, and immunoblot analyses were performed using antibodies to AOX and ISE1, a highly expressed mitochondrial-specific protein (Stonebloom et al., 2009). The gels contained a titration series (100, 20, 4, and 1%) of total cell proteins.
Fractions of gradient-purified chloroplasts are often contaminated with small amounts of nonplastid proteins, especially including proteins from mitochondria (Ferro et al., 2003). To examine the extent of mitochondria contamination in our experiments, we performed immunoblot analysis on proteins from purified chloroplast fractions of the AOX2 overexpressors using an antibody to INCREASED SIZE EXCLUSION LIMIT1 (ISE1), a mitochondrial-specific RNA helicase (Stonebloom et al., 2009). Figure 6B shows that ISE1 levels in these fractions are much <1% of those in the total cell sample, compared with ∼20% for AOX2. This suggests that gradient-purified chloroplasts are slightly contaminated with mitochondria. The fact that AOX2 is present at much lower levels in the isolated chloroplast versus total cell fractions further indicates that much of the overexpressed AOX2 in these plants resides elsewhere, likely the mitochondria. Consistent with this interpretation, the levels of AOX1a in the C-SAOX1a-4 plants are similar in the total cell and purified chloroplast fractions. This is as expected since overexpressed AOX1a was targeted to chloroplasts in these transgenics.
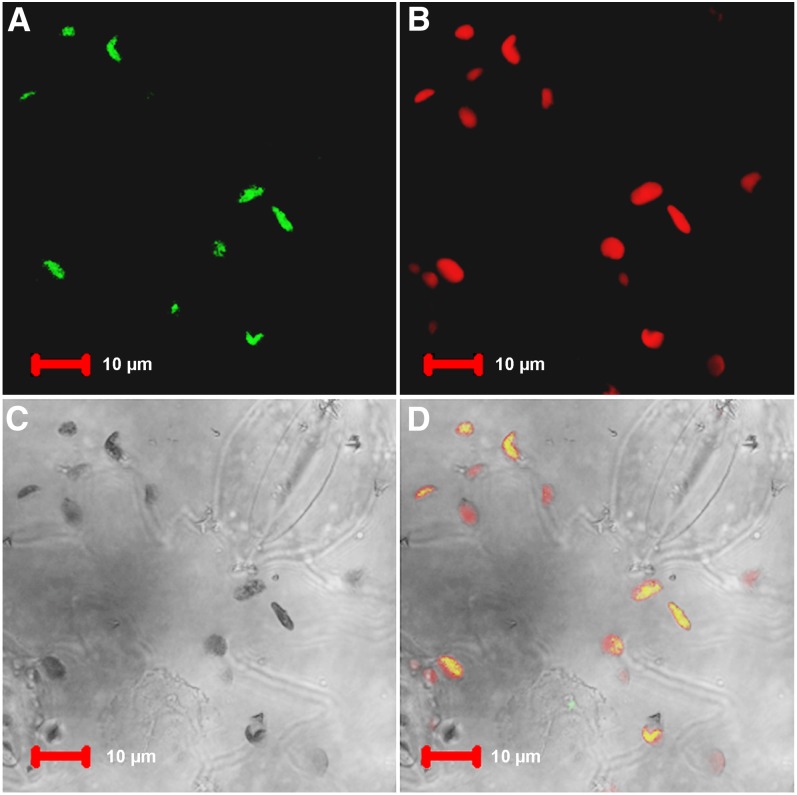
Whereas the above experiments revealed that overexpressed AOX2 can enter chloroplasts, we further assessed whether the protein could be targeted to plastids in a transient expression system. In these studies, we tagged the full-length AOX2 protein with green fluorescent protein (GFP) at its C terminus and transiently expressed the fusion protein in Nicotiana benthamiana. Figure 7 shows that green fluorescence signals from GFP-tagged AOX2 overlaid quite well with the red autofluorescence of chloroplasts. Taken together with the experiments in Figure 6, this indicates that AOX2 can be targeted into plastids using its own localization sequences.
Figure 7.
AOX2-GFP Fusions Are Targeted to Chloroplasts.
An AOX2-GFP fusion construct was transiently expressed in tobacco leaf lower epidermis cells. Sequences in this construct are driven by the CaMV 35S promoter. Bars = 10 μm.
(A) Green fluorescence of AOX2-GFP.
(B) Chlorophyll autofluorescence (red).
(C) Bright field.
(D) A merged image of (A) to (C).
Suborganellar Location and Organization of Chloroplast AOX2
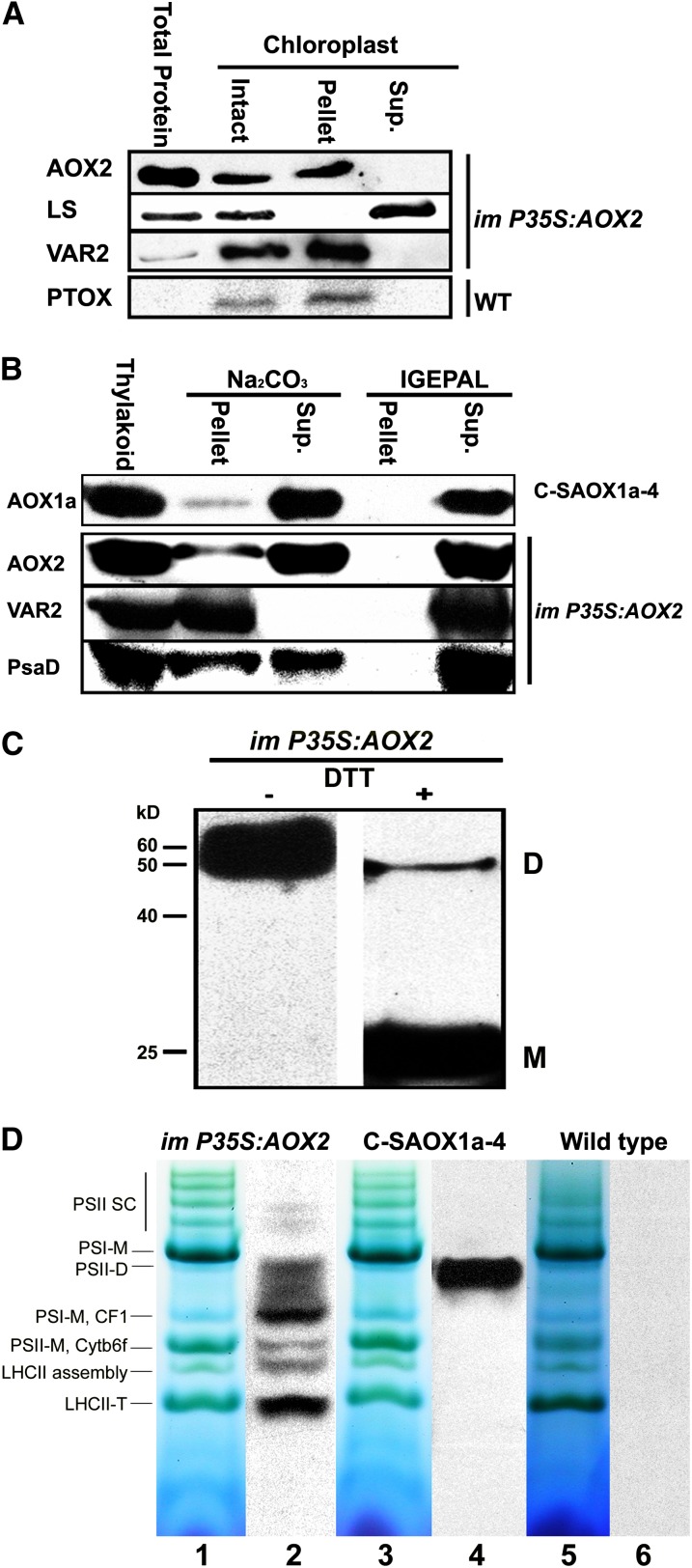
To examine the suborganellar location of chloroplast AOX2, we performed immunoblot analyses on membrane versus soluble proteins from Percoll gradient–purified chloroplasts using im-P35S:AOX2 and wild-type plants. Figure 8A shows that AOX2 is present in the membrane but not soluble fractions. This is similar to VAR2/AtFtsH2, an integral membrane FtsH metalloprotease (Chen et al., 2000), and PTOX, which is a stromal-facing thylakoid membrane protein (Andersson and Nordlund, 1999; Lennon et al., 2003). As anticipated, the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is present in the soluble fraction. We next treated the membrane fractions with 100 mM sodium carbonate (pH 12) or 1% IGEPAL CA-630 to examine whether AOX2 is a peripheral or integral thylakoid membrane protein. In addition to membranes from the AOX2 overexpression line, these experiments included membranes from purified C-SAOX1a-4 chloroplasts (the chloroplast AOX1a overexpression plants; Figure 2). Figure 8B shows that AOX2, AOX1a, VAR2, and PsaD, a peripheral thylakoid membrane protein that binds PSI (Jin et al., 1999), are completely extracted from chloroplast membranes by 1% IGEPAL CA-630. Sodium carbonate, on the other hand, does not extract VAR2, as anticipated for an integral membrane protein, yet partially extracts PsaD, AOX2, and AOX1a. These data support the hypothesis that AOX2 and AOX1a associate with chloroplast membranes, perhaps in a manner similar to PTOX and mitochondrial AOX, both of which are proposed to be interfacial membrane proteins, anchored to one-half of the lipid bilayer (Andersson and Nordlund, 1999; Lennon et al., 2003).
Figure 8.
Localization and Organization of Chloroplast AOX2.
(A) Proteins were isolated from im-P35S:AOX2 and wild-type (WT) whole-leaf extracts (total protein) or from intact, Percoll gradient–purified chloroplasts (intact). In the intact sample, chloroplasts were lysed and separated into supernatant (soluble, stroma) and pellet (membrane, largely thylakoids) fractions. Equal amounts of protein were electrophoresed through 12.5% SDS polyacrylamide gels, and immunoblot analyses were conducted using antibodies to AOX, PTOX, the large subunit of Rubisco (LS), and VAR2/At-FtsH2 (antibodies described in Yu et al., 2011).
(B) Chloroplast membranes (designated “thylakoid”) were isolated from im-P35S:AOX2 and C-SAOX1a-4 seedlings and incubated in either 100 mM sodium carbonate, pH 12, or 1% IGEPAL CA-630. Following incubation, the samples were separated by centrifugation into pellet and supernatant fractions, as described in Methods, and immunoblot analyses were conducted as in (A) using antibodies to AOX, VAR2, and PsaD (Jin et al., 1999).
(C) Proteins from the membrane fractions of isolated chloroplasts from im-P35S:AOX2 rosette leaves were denatured in SDS in the presence or absence of 0.1 M DTT. Samples corresponding to 15 µg chlorophyll were then electrophoresed through 12.5% SDS-PAGE gels, and immunoblot analyses were conducted using the AOX antibody. “D” and “M” mark the positions of dimeric and monomeric forms of AOX, as in Figure 3B.
(D) Percoll gradient–purified chloroplasts were isolated from im-P35S:AOX2, C-SAOX1a-4, and wild-type seedlings, and the membrane fractions were subjected to BN-PAGE on Tris-HCl gradient gels. Immunoblot analyses were then conducted using the monoclonal AOX antibody. The locations of thylakoid complexes are well established in this gel system (Fu et al., 2007). Lanes 1, 3, and 5 are Coomassie blue–stained gels, and lanes 2, 4, and 6 are corresponding blots probed with the AOX antibody. Control chloroplasts from the wild type (lane 6) do not have detectable antibody binding.
Figure 8C shows that chloroplast AOX2 migrates as a dimer in the absence of DTT but primarily as a monomer in its presence. This suggests that chloroplast AOX2 is capable of forming sulfhydryl-linked dimers in thylakoids, reminiscent of the situation in mitochondria. This is in contrast to chloroplast AOX1a, which forms monomers, not dimers, in the absence of DTT (Figure 3B). To determine whether chloroplast AOX2 and AOX1a migrate as higher molecular weight complexes, we performed blue native gel electrophoresis (BN-PAGE) on membrane fractions of lysed, purified chloroplasts from im-P35S:AOX2, C-SAOX1a-4, and wild-type seedlings. The locations of chloroplast complexes and subcomplexes are well established in this gel system (Figure 8D, lanes 1, 3, and 5) (Fu et al., 2007). Figure 8D (lane 2) reveals that AOX2 migrates with subcomplexes of PSI, PSII, CF1, cytochrome b6f, and light-harvesting complex II and that it is significantly enriched in some complexes and less so in others. By contrast, AOX1a migrates at a single position, either coincident with or somewhat smaller than the PSII-D complex (Figure 8D, lane 4). The AOX monoclonal antibody does not react with membrane proteins from wild-type plants (Figure 8D, lane 6), indicating that the binding in lanes 2 and 4 is specific. We conclude that AOX1a and AOX2 are present in higher molecular weight complexes in chloroplast membranes and that these complexes differ for each protein.
Chloroplast AOX2 and AOX1a Replace the Activity of PTOX in Carotenoid Biosynthesis
Although our data indicate that chloroplast-localized AOX1a and AOX2 are able to rescue im, it is worth considering the possibility that these proteins exert their effect on chloroplast biogenesis in an indirect manner, rather than by directly replacing the quinol oxidase activity of PTOX. If a direct effect, it should be possible to demonstrate the involvement of AOX1a and AOX2 in processes that are known to be mediated by PTOX, such as the desaturation steps of carotenogenesis (Wetzel et al., 1994). These steps comprise a redox chain in which electrons are transferred from phytoene and ζ-carotene to the PQ pool via PDS and ζ-carotene desaturase, respectively, and from the PQ pool to molecular oxygen via PTOX (Rosso et al., 2009). The participation of PTOX in this process is especially important during early chloroplast biogenesis when an absence of PTOX, as in im, leads to an overreduced PQ pool (Rosso et al., 2009). This inhibits further electron transfer to this pool and, consequently, phytoene accumulates (Rosso et al., 2009; Foudree et al., 2010).
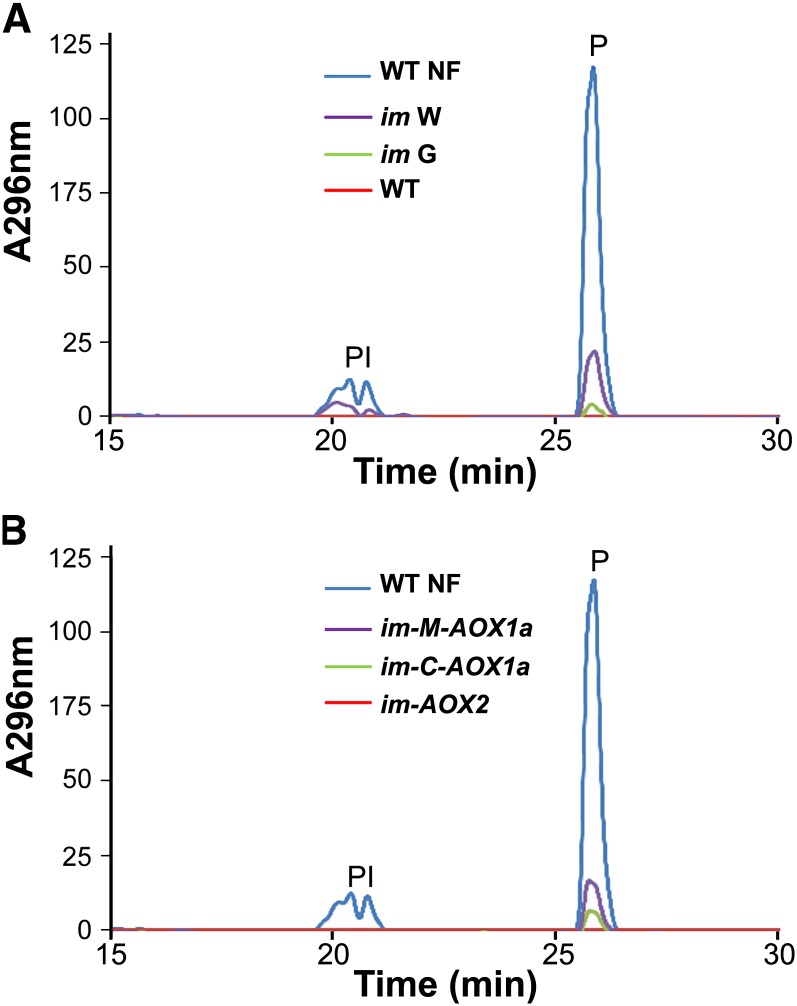
To determine if chloroplast-localized AOX1a and AOX2 are able to substitute for PTOX activity, we tested whether they can rescue the phytoene accumulation defect of im. Figure 9A shows HPLC profiles of phytoene from leaves of the AOX1a and AOX2 overexpression plants, im, and wild-type seedlings treated with norflurazon (NF), a competitive inhibitor for the plastoquinone cofactor binding site of PDS (Breitenbach et al., 2001). Confirming earlier reports (Wetzel et al., 1994), the data show that phytoene is not detectable in wild-type leaves but that it accumulates to high levels in NF-treated leaves and to modest levels in the white sectors of im; small amounts of phytoene are also evident in im green sectors. One reason for the different amounts of phytoene in im versus NF-treated leaves is that NF completely inhibits PDS at the concentration of herbicide used, whereas a lack of PTOX affects PDS activity only indirectly by altering the redox state of the PQ pool; hence, the activity of PDS in im can be modulated by factors such as light intensity and temperature and it is therefore a conditional phenotype (Rosso et al., 2009; Foudree et al., 2010).
Figure 9.
HPLC Analysis of Phytoene Accumulation.
Relative phytoene levels were measured by HPLC analysis of leaf samples from im-P35S:AOX2, C-SAOX1a-4, and M-SAOX1a seedlings. NF-treated wild type (WT) and im (green versus white sectors) served as controls. The absorbance spectra at 296 nm are indicated; for simplicity, only peaks for phytoene (P) and phytoene isomers (PI) are shown.
Figure 9B shows that phytoene does not accumulate in the im-P35S:AOX2 seedlings, but very low levels are present in the C-AOX1a-4 seedlings. The control M-AOX1a seedlings (which overexpress mitochondrial-targeted AOX1a; Figure 1) have levels of phytoene similar to those of im; this is as anticipated since im is not rescued in these seedlings. From these data, we conclude that AOX2 and AOX1a are able to function as quinol oxidases in chloroplast membranes since they are able to replace, at least in part, the quinol oxidase activity of PTOX in carotenoid biosynthesis. The fact that AOX2 completely reverses the phytoene accumulation phenotype of im, in contrast with AOX1a, further suggests that AOX1a is a less effective PQ oxidase than AOX2, at least in the transgenics analyzed. This would be consistent with the less complete suppression of variegation in the C-AOX1a-4 versus im-P35S:AOX2 seedlings (Figures 2B and 5).
Do AOX2 and AOX1a Affect Photosynthetic Function?
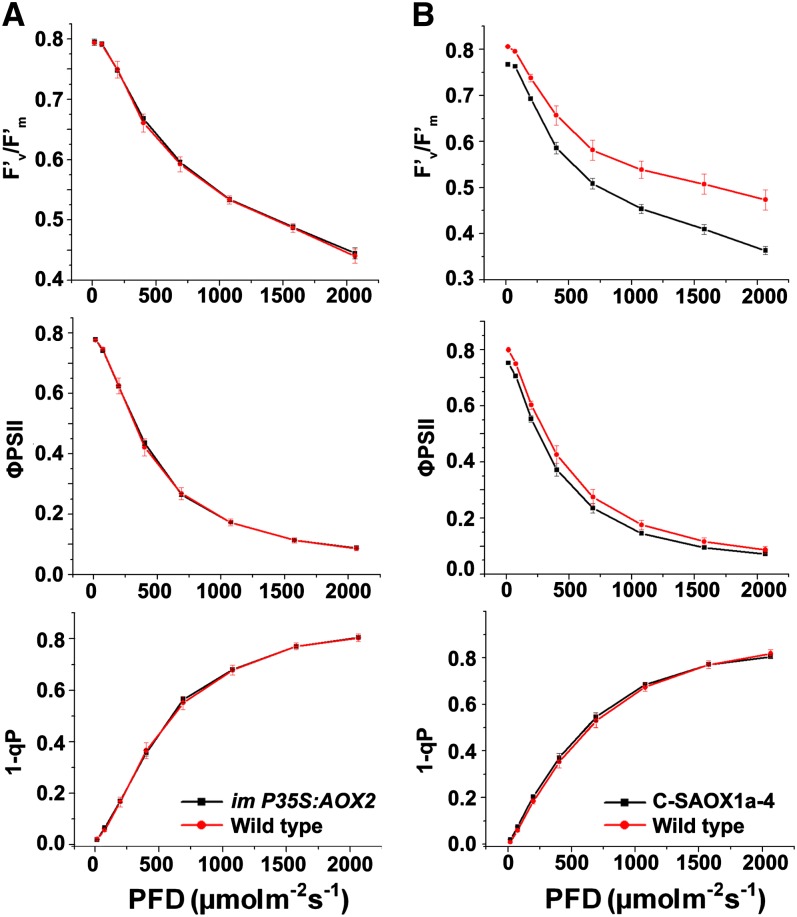
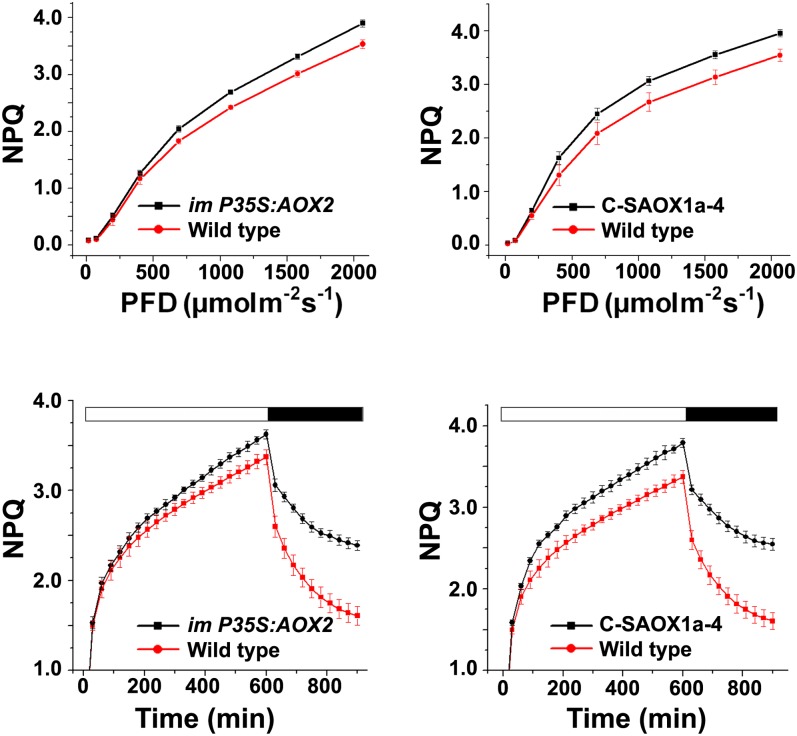
As a first step to determine whether steady state photosynthesis is affected in P35S:AOX2 and C-SAOX1a-4, we measured a number of chlorophyll fluorescence parameters under conditions of changing irradiance. Figures 10A and 10B show that most parameters are strikingly similar in the transgenic versus wild-type plants, suggesting that steady state photosynthesis is not grossly perturbed by the presence of membrane-bound AOX2 and AOX1a. One exception is the lower PSII efficiency in the C-SAOX1a-4 seedlings, which is consistent with the more severe phenotype of these plants versus the im-P35S:AOX2 plants. Another exception is the enhancement of nonphotochemical quenching (NPQ) in both transgenics (Figure 11, top panels); NPQ is a measure of a plant’s ability to dissipate excess light energy as heat (Müller et al., 2001; Li et al., 2009). NPQ rapid induction and dark relaxation kinetics revealed that total NPQ is significantly higher in im-P35S:AOX2 and C-SAOX1a-4 versus the wild type during the induction phase; however, NPQ was not rapidly reversible in either transgenic (Figure 11, bottom panels). This lack of rapid reversibility suggests that C-SAOX1a-4 and im-P35S:AOX2 are primarily defective in photoinhibitory quenching (qI; Müller et al., 2001).
Figure 10.
Chlorophyll a Fluorescence Measurements.
Chlorophyll fluorescence parameters were measured on intact leaves from im-P35S:AOX2 (A) and C-SAOX1a-4 (B) and wild-type seedlings ([A] and [B]) grown for 7 weeks in soil under continuous illumination and temperature (22°C, 100 μmol m−2 s−1). The parameters included the following: F′v/F′m, the maximum efficiency of PSII photochemistry under different photon flux densities (PFD); ΦPSII, the quantum efficiency of PSII photochemistry at different photon flux densities; and 1-qP, the redox state of the QA electron acceptor of PSII (Maxwell and Johnson, 2000; Müller et al., 2001).
Figure 11.
NPQ Analyses.
Steady state light response NPQ (top panels) and rapid induction and dark relaxation NPQ kinetics (bottom panels) were measured on intact leaves from im-P35S:AOX2, C-SAOX1a-4, and wild-type seedlings grown for 7 weeks in soil under continuous illumination and temperature (22°C, 100 μmol m−2 s−1). The data represent the average ± sd of four independent experiments.
DISCUSSION
Chloroplast AOX2 and a Revised Threshold Model of im Variegation
PTOX is expressed in many plastid types (Josse et al., 2000; Aluru et al., 2001; Barr et al., 2004), but the physiological and biochemical processes in which it participates are poorly understood, especially in nongreen plastids. In chloroplasts, PTOX is a versatile alternative electron sink and mediates a number of biochemical pathways that require a terminal quinol oxidase, including carotenoid biosynthesis, chlororespiration, and PSI cyclic electron transport (Wetzel et al., 1994; McDonald et al., 2011; Okegawa et al., 2010). PTOX is also a safety valve under stress conditions, at least in some species (McDonald et al., 2011).
We recently established that developing im thylakoids have higher than normal excitation pressures (a measure of the redox state of the PQ pool) and concluded that PTOX is a central regulator of excitation pressure during early chloroplast biogenesis (Rosso et al., 2009). Its activity at this time is crucial because it provides an electron sink for the harmless dissipation of excess energy when components of the photosynthetic apparatus are synthesized and assembled, prior to the development of fully functional electron transport chains. Our current model of im variegation proposes that developing chloroplasts have heterogeneous excitation pressures because of intrinsic differences in their biochemistry that arise, for example, from gradients in the leaf, sometimes steep, of light and other determinants of light capture and use (Smith et al., 1997). Superimposed on these gradients, individual cells undergo circadian rhythms independently from one another, and outputs from the clock regulate reactive oxygen species scavenging, even under continuous light conditions (Velez-Ramirez et al., 2011). Given the potential for wide variation in excitation pressures, we propose that plastids are above or below thresholds compatible with normal chloroplast differentiation: Plastids with above-threshold values have overreduced thylakoids and are susceptible to photooxidation and/or other reactive oxygen species–mediated damage (giving rise to white plastids), whereas plastids with below-threshold excitation pressures develop into photosynthetically competent chloroplasts (green sectors). Our model further assumes that the pattern of sectoring in im is a reflection of leaf development in dicots, which involves cell and plastid divisions early in differentiation and cell elongation later in development (i.e., the chaotic pattern of sectoring in the mature im leaf is a magnified version of the pattern established in the leaf primordium). The Arabidopsis variegation mutant alx13 seems another newly discovered example to support this hypothesis (Woo et al., 2011).
Although much work has been devoted to understanding the control of excitation pressures during steady state photosynthesis (Rosso et al., 2006; Li et al., 2009), little is known about this topic during chloroplast development. In this article, we focused on activation tagging of im as a tool to gain insight into this question, the rationale being that factors capable of modulating the variegation pattern of im are involved in influencing excitation pressures, either directly or indirectly. We discovered, to our surprise, that activation-tagged AOX2 is able to effectively suppress im variegation and that this occurs by replacement of the quinol oxidase activity of PTOX in the phytoene desaturation step of carotenogenesis. This would restore the ability of the developing chloroplast to quench singlet oxygen and triplet chlorophyll, thus reducing excitation pressures. Rescue of im variegation by chloroplast-targeted AOX1a can be explained in a similar manner.
While our data strongly suggest that chloroplast-localized AOX2 is responsible for the suppression of im variegation, we considered the possibility that this is caused, in part, by a relaxation of excitation pressures in developing im thylakoids due to enhanced AOX2 activities in mitochondria. AOX-mediated affects on chloroplast redox are well documented via such mechanisms as the malate/oxaloacetate shuttle (Scheibe, 2004; Bartoli et al., 2005; Yoshida et al., 2006, 2007; Noguchi and Yoshida, 2008; Foyer and Shigeoka, 2011; Nunes-Nesi et al., 2011). Against this hypothesis is our observation that overexpression of AOX1a in mitochondria does not suppress im variegation (i.e., the W-AOX1a transformants; Figure 1). Thylakoid excitation pressures also are not affected in the Arabidopsis aox1a mutant, which lacks AOX1a (Yoshida et al., 2010). Hence, we feel the preponderance of evidence supports the idea that suppression of im is caused primarily, if not exclusively, by substitution of AOX2 for PTOX activity in the developing plastid.
Subcellular Localization of AOX2
Both observations of overexpressed AOX2 entering purified chloroplasts (Figure 6) and signals from GFP-tagged full-length AOX2 overlying with red autofluorescence of chloroplasts (Figure 7) indicate that AOX2 can be targeted into plastids using its own localization sequences. Also in support of its plastid targeting, AOX2 was found to be induced specifically under treatments affecting chloroplast functions (Clifton et al., 2005).
AOX2, like other AOX family members, has been assumed to reside in mitochondria, even though it lacks a typical mitochondrial targeting signal (Heazlewood and Millar, 2005; Clifton et al., 2006; Kleffmann et al., 2006; Sun et al., 2009). The only direct experimental evidence for the presence of AOX2 in mitochondria comes from experiments in which the putative transit peptide of Arabidopsis AOX2 was fused to GFP, and the construct was transiently expressed in tobacco (Nicotiana tabacum) suspension-cultured Bright Yellow-2 cells (Saisho et al., 2001). It is arguable how well the AOX2 presequence can represent AOX2 itself. This is in seeming contrast with our experiments showing plastid targeting of a full-length AOX2 fused to GFP. However, the two sets of transient expression data are not necessarily in conflict and could have resulted from differences in plastid type, construct, and/or expression system, three elements that commonly complicate interpretations of these sorts of studies (Karniely and Pines, 2005; Carrie et al., 2009a). Nevertheless, considered together with our finding that endogenous AOX2 sequences are able to direct AOX2 into chloroplasts in transgenic plants that overexpress AOX2 (in ATG791 and im-P35S:AOX2), we conclude that AOX2 is targeted to chloroplasts in planta. It does not exclude that possibility that AOX2 also enters mitochondria as well; thus, AOX2 might be a dual-targeted protein.
Although organelle-targeting algorithms predict that hundreds of proteins are dual targeted to mitochondria and plastids (Carrie et al., 2009a), these algorithms do not perform well (Richly and Leister, 2004; Nair and Rost, 2005; Zybailov et al., 2008), and fewer than 100 bona fide dual-targeted proteins have thus far been rigorously identified (Carrie et al., 2009a, 2009b; Yogev and Pines, 2011). Most dual-targeted proteins are involved in biochemical processes that are common to mitochondria and chloroplasts, such as transcription, translation, and DNA recombination surveillance (Millar et al., 2006; Xu et al., 2011). Our proposal that mitochondria and chloroplasts require AOX-like quinol terminal oxidases is consistent with this idea. On the other hand, it is possible that a certain amount of mistargeting might be tolerated by cells as a by-product of the evolutionary development of transit peptides, as suggested in the case of cytochrome c1 of the respiratory chain, which appears to be targeted to chloroplasts as well as mitochondria (Rödiger et al., 2011). The chloroplast targeting of AOX2 and cytochrome c1 suggests that many proteins could have this capacity. One evolutionary advantage of this capacity is that it provides a mechanism for neofunctionalization of the organelle, with control exerted by the nucleus (Rödiger et al., 2011). This might have been especially important during the early stages of endosymbiosis.
Implications from Molecular Characterization of Chloroplast AOX1a and AOX2 Mutants
Although more work will be needed to demonstrate unequivocally that AOX2 is found in plastids under normal physiological conditions, the AOX2 (and AOX1a) overexpression plants are nonetheless promising synthetic systems to explore ways of reengineering photosynthesis and pathways of interorganellar communication (Weber and Osteryoung, 2010; Rochaix, 2011). In particular, they should prove useful in addressing two fundamental questions: How is the photosynthetic apparatus remodeled in response to the presence of non-native redox components? How does AOX function in a foreign redox milieu?
First, it is clear that plastid membranes are able to incorporate large quantities of AOX1a and AOX2 with seemingly few deleterious effects on photosynthesis, physiology, or development. Perhaps the simplest hypothesis is that both proteins are able to be imported into plastids and incorporated into membranes in a compatible conformation. Our data show that AOX2 and AOX1a are active in carotenoid desaturation, but we do not know whether they are present in envelope membranes, along with all other membrane-bound enzymes of carotenogenesis and/or in thylakoids, which contain only PDS (Joyard et al., 2009). Interestingly, PTOX is apparently restricted to the stromal surface of thylakoids and is not found in envelopes (Lennon et al., 2003), which suggests that PDS and PTOX form a functional unit in thylakoids. Because the various plastid membranes have different carotenoid intermediate compositions, as well as different carotenoid enzyme compositions, our data highlight questions about (1) the membrane compartmentation of carotenoid enzymes, especially during chloroplast biogenesis when PTOX activity is required to modulate excitation pressures (Rosso et al., 2009); and (2) the trafficking of carotenoids between the envelope and thylakoid in mature chloroplasts. In this context, it will be of interest to determine the membrane locations of AOX1a and AOX2 and whether they are active in other PTOX-mediated functions, such as chlororespiration and PSI cyclic electron transport, and as a stress safety valve (Okegawa et al., 2010; McDonald et al., 2011).
Second, chloroplast prenlyquinones include plastoquinone 9 (PQ-9), phylloquinone K1, α-tocopherol, and α-tocoquinone, with α-tocopherol predominating in the envelope (Soll et al., 1985) and PQ-9 in thylakoids (Lichtenthaler, 2007). The desaturation steps of carotenoid biosynthesis use PQ-9 (Norris et al., 1995). Although PTOX has a stringent requirement in vitro for PQ and is unable to use ubiquinol, duroquinone, phylloquinone, or benzoquinone (Josse et al., 2003), it has been an open question whether AOX can use substrates other than ubiquinol (McDonald, 2008). One of the striking implications of our studies is that AOX1a and AOX2 are able to use PQ in planta as a substrate for the desaturation reactions of carotenogenesis. This might not be surprising in the case of AOX2, since evolution might have honed the ability of this enzyme to function in both mitochondria and chloroplasts. However, the ability of AOX1a, which is present only in mitochondria, to use PQ suggests that the enzyme displays a certain amount of plasticity in its substrate requirements. Substrate specificity differences between AOX1a, AOX2, and PTOX are likely caused, in part, by differences in active site amino acids, inasmuch as the sequence of a consensus quinol binding site in AOX (Moore and Albury, 2008) is poorly conserved in PTOX (Fu et al., 2009). Aspects of the chloroplast milieu might also influence AOX activities, as illustrated by the observation that removal of the D-Domain enhances chloroplast AOX1a activity in vitro and in planta. We speculate that this favors a conformation or interaction that allows AOX1a to more closely resemble PTOX. On the other hand, addition of the PTOX-specific Exon 8 Domain abolishes AOX1a activity, perhaps by promoting a nonfunctional conformation or unfavorable interactions. Our suite of PTOX (Fu et al., 2005, 2009), AOX1a, and AOX2 mutants should be useful systems to explore quinone biochemistry and mechanisms of AOX and PTOX catalysis.
Third, PTOX and all AOX isoforms have a conserved diiron site in their catalytic domain that is required for activity (Fu et al., 2005). Because both AOX1a and AOX2 are active in chloroplasts, another implication of our data is that correct metallation of AOX occurs (Merchant, 2010). At least for AOX1a, this occurs in an ectopic location.
Fourth, our experiments revealed that AOX2 is present primarily in a dimeric form in mature chloroplast membranes. This suggests that the enzyme is capable of being oxidized, likely at its Cys site, by a redox system that resides in plastids, perhaps a thioredoxin, as demonstrated for mitochondrial AOX (Gelhaye et al., 2004; Balmer et al., 2004). This raises questions about the functional significance of monomeric versus dimeric chloroplast AOX2. If the dimeric form is inactive, similar to dimeric mitochondrial AOX, it is puzzling why all detectable chloroplast AOX2 in the leaf is in this form. One possibility is that dimers represent an artificial interaction produced by overexpression and that only small amounts of the monomer are necessary for activity. This would be analogous to PTOX, which is present at <1% of PSII levels in mature Arabidopsis leaves (Lennon et al., 2003), and consistent with numerous proteomics studies showing that AOX2 is a low abundance protein (Armbruster et al., 2011) (i.e., very small amounts might be necessary for activity). An alternate explanation rests on the assumption that PTOX is not required for steady state photosynthesis in Arabidopsis (Rosso et al., 2006) but only for chloroplast development (Rosso et al., 2009). If the same holds true for chloroplast AOX2, it might not be surprising that most of the enzyme is maintained in an inactive form in mature chloroplasts.
Fifth, our BN-PAGE analyses revealed that AOX1a and AOX2 form high molecular weight membrane complexes, with AOX1a present in what appears to be a single-sized complex and AOX2 in a mixture of complex sizes. These complexes could be homomeric or heteromeric and might include components of the electron transport chain. In this context, it is remarkable that large quantities of AOX1a and AOX2 can be incorporated into chloroplast membranes with little impact on steady state photosynthesis, at least as monitored under the nonstress conditions of our experiments. In fact, an enhanced NPQ capacity is one of the few components that are significantly altered in the AOX1a and AOX2 overexpressors. NPQ is a measure of the amount of feedback deexcitation that occurs when excess absorbed light energy is dissipated as heat (thermal dissipation) (Li et al., 2009), and it has several components: qE, energy-dependent fluorescence quenching; qT, caused by the state transition quenching; and qI, representing the long-term downregulation of PSII (Müller et al., 2001). The fact that both overexpression lines have very slow dark relaxation kinetics suggests that they are primarily defective in qI. A similar phenotype has been reported recently for a chloroplast cyclophilin mutant (cyp38), which defines the gene for a lumen protein necessary for PSII assembly and stabilization (Fu et al., 2007). Although the precise mechanisms of qI are poorly understood, enhanced qI could be caused by alterations in photoprotective mechanisms and/or by photodamage. Our mutants should provide an ideal system for study of this question.
Sixth, a final point worth noting is that the activation-tagged and AOX2 overexpression plants are much larger than normal (Figure 5), indicating that AOX2 is capable of substantially influencing growth. The fact that both transgenic lines have similar growth characteristics, despite widely differing levels of AOX2, further suggests that overexpression can result in enzyme amounts beyond those necessary for optimal growth, at least under the conditions of our experiments. The influence of AOX2 on growth merits further investigation, especially in light of the observation that Arabidopsis growth under nonstress conditions is not markedly affected by overexpression of AOX1a, either in the chloroplast (Figure 2) or mitochondrion (Umbach et al., 2005).
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (Columbia-0) was used as the wild type. Other plants in this study included the spotty allele of im (Wu et al., 1999). Because im has a light-sensitive phenotype, the plants were germinated and grown for 7 d under low light (∼15 µmol m−2 s−1) prior to transfer to high light (∼100 µmol m−2 s−1). All plants in this study were maintained on soil under continuous illumination at 22°C.
Nucleic Acid Manipulations
Genomic DNAs were isolated and DNA gel blot analyses were performed as previously described (Wetzel et al., 1994). Plasmid rescue was conducted using established methods (Weigel et al., 2000). Total cell RNAs were isolated and RNA gel blot analyses were performed also as described (Wetzel et al., 1994). The primers for the DNA and RNA experiments are listed in Supplemental Table 1 online.
Generation of Transgenic Plants
Activation Tagging
The floral dip method (Clough and Bent, 1998) was used for transformation of im with the activation tagging vector pSKI015 (a gift from Joanne Chory at the Salk Institute). T1 plants were germinated on soil, and transgenic plants were selected by spraying with a 1:2000 dilution of Finale (AgrEvo), which contains 5.78% (w/v) ammonium glufosinate (Basta). The T1 plants were self-fertilized, and analyses were performed on T2 and subsequent generation plants.
AOX1a Constructs (Summarized in Figure 1)
An Arabidopsis AOX1a cDNA was obtained by RT-PCR using primers AOX1aF1 and AOX1aR1. For expression in mitochondria, the cDNA was cloned under the control of the CaMV 35S promoter (P35S) of pB003, and the resulting plasmid was named W-AOX1a. For chloroplast expression, a fragment of AOX1a that contains the mature AOX1a protein (i.e., the coding sequence of AOX1a minus the mitochondrial targeting peptide [MTP]) was generated by PCR using primers AOX1aF2 and AOX1aR2. This fragment was cloned behind the pea (Pisum sativum) RbcS chloroplast targeting peptide (CTP) in the binary vector pCB302-1; the resulting plasmid was designated C-LAOX (long AOX). pCB302-1 is designed for targeting proteins into chloroplasts, and transcription in this vector is driven by P35S (Xiang et al., 1999). An AOX1a cDNA lacking its MTP and the immediately flanking, 40–amino acid D-Domain (Umbach and Siedow, 2000) was obtained by PCR using primers AOX1aF3 and AOX1aR3. The amplified sequence was cloned behind the RbcS CTP of pCB302-1; the resulting plasmid was termed C-SAOX (short AOX). The AOX1a fragments in C-LAOX and C-SAOX were also cloned behind the tobacco (Nicotiana tabacum) β-ATPase MTP in pCB302-2; the resulting constructs were designated M-LAOX and M-SAOX, respectively. pCB302-2 is designed for targeting proteins into mitochondria, with transcription being driven by P35S (Xiang et al., 1999).
Compared with AOX, a 16–amino acid sequence is present in PTOX that corresponds precisely to Exon 8 of the genomic sequence (Fu et al., 2005). Sequences flanking this insertion are conserved in PTOX and AOX; thus, the site of insertion can be accurately defined. The Exon 8 sequence was inserted into this position in both LAOX and SAOX using the ExSite PCR-based site directed mutagenesis kit (Stratagene) by four sequential steps (each step adding 12 bp). The sequences were then cloned into pCB302-1 behind the CTP to generate C-L+8 and C-S+8.
Other Overexpression Constructs
Full-length cDNAs (or genomic DNAs) of genes of interest were amplified by PCR or RT-PCR using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen). The DNAs were cloned into the binary vector pB003 behind the CaMV 35 promoter (P35S) (Xiang et al., 1999).
All constructs were sequenced to verify that no errors were introduced by PCR manipulations and then transferred into Agrobacterium tumefaciens by electroporation. The floral dip method was used to transform bolting Arabidopsis plants (Clough and Bent, 1998). BASTA-resistant, T1 transgenic plants were selected on soil by Finale. PCR and DNA gel blotting methods were used to verify that the plants were transformed. Phenotypic analyses were performed on T2 generation plants.
Subcellular Localization of AOX2
To determine the subcellular location of AOX2, an AOX2-GFP fusion construct was created by cloning a full-length AOX2 coding sequence in front of the GFP sequence in pMDC83 (Sokolov et al., 2006); the resulting vector was designated pMDC83-AOX2. To express the fusion protein in tobacco, pMDC83-AOX2 was transformed into Agrobacterium GV3101, which was grown in liquid Luria-Bertani at 28°C, pelleted, and then resuspended in 100 mM MES, 100 mM MgCl2, and 200 µM acetosyringone. The cultures were inoculated at A600 densities of 1.0 and induced by acetosyringone for 2 to 6 h before infiltration. Nicotiana benthamiana was transformed by injecting agrobacteria into leaves with needleless 1-mL syringes. The transfected plants were incubated at 23°C for 3 d before taking images. Bright-field and fluorescent microscopy was performed on a Zeiss 710 Meta UV/VIS confocal microscope.
Protein Analyses
Four-week-old Arabidopsis leaves were homogenized in sample buffer at 4°C then filtered through four layers of Miracloth (Calbiochem) as described previously (Chen et al., 2000). The filtrate was the source of total leaf protein. Intact chloroplasts were isolated from the filtrate using two-step Percoll gradients, also as described (Perry et al., 1991). The chloroplasts were washed twice in SH buffer (0.33 M sorbitol and 50 mM HEPES-KOH, pH 8.0). In some experiments, the intact chloroplasts were lysed in hypotonic buffer (20 mM MOPS and 50 mM EDTA, pH 7.0). The supernatant and pellet fractions were then collected by centrifugation (3000g for 5 min). The pellets were washed twice in SH buffer and then resuspended in SH buffer, while the supernatants were clarified by centrifugation (12,000g for 20 min); the pellets were washed twice in SH buffer and then resuspended in SH buffer. Protein concentrations were measured using the Bio-Rad assay.
To determine the topology of AOX2 in chloroplast membranes, isolated membrane fractions were suspended in either 100 mM sodium carbonate, pH12, or 1% IGEPAL CA-630 and then incubated on ice for 70 min. Following centrifugation (10,000g for 10 min), the pellets were washed twice in SH buffer, while the supernatants were precipitated by acetone. Pellet and acetone extracts were resuspended in the same amount of SDS loading buffer for electrophoresis.
BN-PAGE was performed using established methods (Fu et al., 2007). Briefly, protein complexes from Percoll gradient–purified chloroplasts were solubilized in a buffer containing 1% n-dodecyl-β-d-maltoside. After centrifugation, the supernatant was mixed with Coomassie blue and loaded onto a 4 to 15% Tris-HCl polyacrylamide gradient gel (Bio-Rad). Gel electrophoresis was performed at a constant voltage of 100 V for 6 h at 4°C. The gel was washed three times in transfer buffer with 1% SDS prior to immunoblotting.
Immunoblot analyses were conducted as described (Fu et al., 2005). In brief, protein samples were electrophoresed through 12.5% SDS polyacrylamide gels and then transferred to nitrocellulose membranes. The membranes were incubated with antibodies (described in the text), and protein signals were visualized using the SuperSignal West Pico chemiluminescence kit (Thermo).
In Vitro AOX Activity Assays
AOX activity assays were conducted in vitro using previously described procedures (Fu et al., 2005, 2009). In brief, membranes were isolated from Escherichia coli cells transformed with LAOX1a and SAOX1a (above), and oxygen consumption was monitored with a Clark O2 electrode (Hansatech). Assays were conducted at 25°C with 100 to 200 μg of membrane protein in a 1.5-mL reaction mixture containing 50 mM Tris-maleate, pH 7.5, 10 mM KCl, 5 mM MgCl2, 1 mM EDTA, 5 mM DTT, and 5 mM pyruvate, with or without 0.2 mM decyl-plastoquinone (2,3-methyl-5-decyl-1,4-benzoquinone; Sigma-Aldrich). The reactions were initiated by the addition of NADH (1 mM final concentration), followed by KCN (2 mM final concentration) and SHAM (2 mM final concentration), all available from Sigma-Aldrich.
Chlorophyll and Chlorophyll Fluorescence Measurements
The first two pairs of rosette leaves from 3-week-old seedlings were harvested, weighed, and ground in liquid nitrogen. Chlorophyll was extracted by 95% ethanol in the dark at 4°C, and chlorophyll concentrations were calculated as previously described (Lichtenthaler, 1987).
Chlorophyll fluorescence parameters were measured with an FMS2 fluorometer (Hansatech) on intact leaves from wild-type, im-P35S:AOX2, and C-SAOX1a seedlings grown for 7 weeks in soil under continuous illumination (∼100 µmol m−2 s−1 22°C). F′v/F′m was defined as (F′m − F′o)/F′m; ФPSII was defined as (F′m − Fs)/F′m; NPQ was calculated as (Fm − F′m)/F′m; and 1-qP is equal to (F′o/Fs)(F′m− Fs)/(F′m − F′o). Fm is the maximum fluorescence in the dark-adapted state; F′m is the maximum fluorescence in any light-adapted state; Fs is the steady state fluorescence in the light; F′o is the minimal fluorescence in any light-adapted state; and F′v is variable fluorescence equal to (F′m – F′o) (Maxwell and Johnson, 2000).
For the NPQ rapid induction and dark relaxation kinetics measurements, an actinic light of 2000 µmol m−2 s−1 was provided for 10 min from the dark-adapted state. Relaxation data were acquired by shutting off the actinic source for an additional 5 min. During the entire time period, the fluorescence signal was superimposed with saturating 800-ms pulses of white light every 30s (Müller et al., 2001; Yu et al., 2011).
HPLC Analysis of Carotenoids
Procedures for HPLC analysis of carotenoids have been described (Wetzel et al., 1994). In brief, pigments were extracted from the leaves of 4-week-old wild type, im-P35S:AOX2, C-SAOX1a-4, and M-SAOX1a seedlings. Controls included photobleached leaves from wild-type seedlings treated with NF, an inhibitor of the PDS enzyme. NF-treated tissues accumulate abundant phytoene. Other controls included green and white sectors dissected from im leaves; this procedure yields little contamination of one cell type by the other (∼3%) (Wetzel et al., 1994). Carotenoids were separated by reverse-phase HPLC on an Ascentis C18 column using a 30-min gradient of ethyl acetate (0 to 100%) in acetonitrile-water (9:1) at a flow rate of 1 mL per min. The peaks were monitored at 296 and 440 nm and identified by comparing absorbance spectra and relative retention times with published values (Qin et al., 2007).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries or the Arabidopsis Genome Initiative database under the following accession numbers: At4g22260 (PTOX, IM), At3g22370 (AOX1a), At5g64210 (AOX2), At5g55180 (O-glycosyl hydrolases family 17 protein), At5g55160 (SUM2), At5g55170 (SUM3), At5g55190 (RAN3), At5g64190 (hypothetical protein), At5g64200 (SC35), At5g64220 (calmodulin binding transcription activator2), and CS3639 (im, spotty).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of Activation-Tagged Genes in ATG791.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Kris Niyogi (University of California, Berkeley) for use of instrumentation for the chlorophyll fluorescence measurements and three anonymous reviewers for excellent suggestions on the article. This work was supported by funding to S.R. from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-94ER20147) and a grant to S.L. from the Department of Energy (DE-FG02-12ER16274).
AUTHOR CONTRIBUTIONS
A.F., H.L., and S.R. designed the research. A.F., H.L., F.Y., and S.K. performed the research. A.F., H.L., S.L., and S.R. analyzed the data. A.F. and S.R. wrote the article.
Glossary
- PDS
phytoene desaturase
- PSI
photosystem I
- PSII
photosystem II
- PQ
plastoquinol
- CaMV
cauliflower mosaic virus
- GFP
green fluorescent protein
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- BN
blue native
- NF
norflurazon
- NPQ
nonphotochemical quenching
- PQ-9
plastoquinone 9
- qI
photoinhibitory quenching
- MTP
mitochondrial targeting peptide
- CTP
chloroplast targeting peptide
- SHAM
salicylhydroxamic acid
References
- Aluru M.R., Bae H., Wu D., Rodermel S.R. (2001). The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol. 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru M.R., Yu F., Fu A., Rodermel S. (2006). Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J. Exp. Bot. 57: 1871–1881 [DOI] [PubMed] [Google Scholar]
- Andersson M.E., Nordlund P. (1999). A revised model of the active site of alternative oxidase. FEBS Lett. 449: 17–22 [DOI] [PubMed] [Google Scholar]
- Armbruster U., Pesaresi P., Pribil M., Hertle A., Leister D. (2011). Update on chloroplast research: New tools, new topics, and new trends. Mol. Plant 4: 1–16 [DOI] [PubMed] [Google Scholar]
- Bailey S., Melis A., Mackey K.R., Cardol P., Finazzi G., van Dijken G., Berg G.M., Arrigo K., Shrager J., Grossman A. (2008). Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim. Biophys. Acta 1777: 269–276 [DOI] [PubMed] [Google Scholar]
- Balmer Y., Vensel W.H., Tanaka C.K., Hurkman W.J., Gelhaye E., Rouhier N., Jacquot J.P., Manieri W., Schürmann P., Droux M., Buchanan B.B. (2004). Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. USA 101: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J., White W.S., Chen L., Bae H., Rodermel S. (2004). The GHOST terminal oxidase regulates developmental programming in tomato fruit. Plant Cell Environ. 27: 840–852 [Google Scholar]
- Bartoli C.G., Gomez F., Gergoff G., Guiamét J.J., Puntarulo S. (2005). Up-regulation of the mitochondrial alternative oxidase pathway enhances photosynthetic electron transport under drought conditions. J. Exp. Bot. 56: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Berthold D.A., Stenmark P. (2003). Membrane-bound diiron carboxylate proteins. Annu. Rev. Plant Biol. 54: 497–517 [DOI] [PubMed] [Google Scholar]
- Bock R., Timmis J.N. (2008). Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays 30: 556–566 [DOI] [PubMed] [Google Scholar]
- Bogorad L. (2008). Evolution of early eukaryotic cells: Genomes, proteomes, and compartments. Photosynth. Res. 95: 11–21 [DOI] [PubMed] [Google Scholar]
- Breitenbach J., Zhu C., Sandmann G. (2001). Bleaching herbicide norflurazon inhibits phytoene desaturase by competition with the cofactors. J. Agric. Food Chem. 49: 5270–5272 [DOI] [PubMed] [Google Scholar]
- Carol P., Stevenson D., Bisanz C., Breitenbach J., Sandmann G., Mache R., Coupland G., Kuntz M. (1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C., Giraud E., Whelan J. (2009a). Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 276: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Carrie C., Kühn K., Murcha M.W., Duncan O., Small I.D., O’Toole N., Whelan J. (2009b). Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Chen M., Choi Y., Voytas D.F., Rodermel S. (2000). Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 22: 303–313 [DOI] [PubMed] [Google Scholar]
- Clifton R., Lister R., Parker K.L., Sappl P.G., Elhafez D., Millar A.H., Day D.A., Whelan J. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clifton R., Millar A.H., Whelan J. (2006). Alternative oxidases in Arabidopsis: A comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Elthon T.E., Nickels R.L., McIntosh L. (1989). Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 89: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Finnegan P.M., Umbach A.L., Wilce J.A. (2003). Prokaryotic origins for the mitochondrial alternative oxidase and plastid terminal oxidase nuclear genes. FEBS Lett. 555: 425–430 [DOI] [PubMed] [Google Scholar]
- Finnegan P.M., Wooding A.R., Day D.A. (1999). An alternative oxidase monoclonal antibody recognises a highly conserved sequence among alternative oxidase subunits. FEBS Lett. 447: 21–24 [DOI] [PubMed] [Google Scholar]
- Foudree A., Aluru M., Rodermel S. (2010). PDS activity acts as a rheostat of retrograde signaling during early chloroplast biogenesis. Plant Signal. Behav. 5: 1629–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Shigeoka S. (2011). Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 155: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A., Aluru M., Rodermel S.R. (2009). Conserved active site sequences in Arabidopsis plastid terminal oxidase (PTOX): In vitro and in planta mutagenesis studies. J. Biol. Chem. 284: 22625–22632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A., He Z., Cho H.S., Lima A., Buchanan B.B., Luan S. (2007). A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A., Park S., Rodermel S. (2005). Sequences required for the activity of PTOX (IMMUTANS), a plastid terminal oxidase: In vitro and in planta mutagenesis of iron-binding sites and a conserved sequence that corresponds to Exon 8. J. Biol. Chem. 280: 42489–42496 [DOI] [PubMed] [Google Scholar]
- Gelhaye E., et al. (2004). A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc. Natl. Acad. Sci. USA 101: 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Ho L.H., Clifton R., Carroll A., Estavillo G., Tan Y.F., Howell K.A., Ivanova A., Pogson B.J., Millar A.H., Whelan J. (2008). The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C.M., Le Gall J., Xavier A.V., Teixeira M. (2001). Could a diiron-containing four-helix-bundle protein have been a primitive oxygen reductase? ChemBioChem 2: 583–587 [DOI] [PubMed] [Google Scholar]
- Gould S.B., Waller R.F., McFadden G.I. (2008). Plastid evolution. Annu. Rev. Plant Biol. 59: 491–517 [DOI] [PubMed] [Google Scholar]
- Gray M.W., Burger G., Lang B.F. (2001). The origin and early evolution of mitochondria. Genome Biol. 2: REVIEWS1018. [DOI] [PMC free article] [PubMed]
- Heazlewood J.L., Millar A.H. (2005). AMPDB: The Arabidopsis mitochondrial protein database. Nucleic Acids Res. 33(Database issue): D605–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Jin P., Sun J., Chitnis P.R. (1999). Structural features and assembly of the soluble overexpressed PsaD subunit of photosystem I. Biochim. Biophys. Acta 1410: 7–18 [DOI] [PubMed] [Google Scholar]
- Joët T., Genty B., Josse E.M., Kuntz M., Cournac L., Peltier G. (2002). Involvement of a plastid terminal oxidase in plastoquinone oxidation as evidenced by expression of the Arabidopsis thaliana enzyme in tobacco. J. Biol. Chem. 277: 31623–31630 [DOI] [PubMed] [Google Scholar]
- Josse E.M., Alcaraz J.P., Labouré A.M., Kuntz M. (2003). In vitro characterization of a plastid terminal oxidase (PTOX). Eur. J. Biochem. 270: 3787–3794 [DOI] [PubMed] [Google Scholar]
- Josse E.-M., Simkin A.J., Gaffé J., Labouré A.-M., Kuntz M., Carol P. (2000). A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol. 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2009). Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2: 1154–1180 [DOI] [PubMed] [Google Scholar]
- Karniely S., Pines O. (2005). Single translation—dual destination: Mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 6: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T., Hirsch-Hoffmann M., Gruissem W., Baginsky S. (2006). plprot: A comprehensive proteome database for different plastid types. Plant Cell Physiol. 47: 432–436 [DOI] [PubMed] [Google Scholar]
- Lennon A.M., Prommeenate P., Nixon P.J. (2003). Location, expression and orientation of the putative chlororespiratory enzymes, Ndh and IMMUTANS, in higher-plant plastids. Planta 218: 254–260 [DOI] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler F.W. (1987). Karl Freudenberg, Burckhardt Helferich, Hermann O. L. Fischer: A centennial tribute. Carbohydr. Res. 164: 1–22 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (2007). Biosynthesis, accumulation and emission of carotenoids, alpha-tocopherol, plastoquinone, and isoprene in leaves under high photosynthetic irradiance. Photosynth. Res. 92: 163–179 [DOI] [PubMed] [Google Scholar]
- Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51: 659–668 [DOI] [PubMed] [Google Scholar]
- McDonald A.E. (2008). Alternative oxidase: An inter-kingdom perspective on the function and regulation of this broadly distributed ‘cyanide-resistant' terminal oxidase. Funct. Plant Biol. 35: 535–552 [DOI] [PubMed] [Google Scholar]
- McDonald A.E., Amirsadeghi S., Vanlerberghe G.C. (2003). Prokaryotic orthologues of mitochondrial alternative oxidase and plastid terminal oxidase. Plant Mol. Biol. 53: 865–876 [DOI] [PubMed] [Google Scholar]
- McDonald A.E., Ivanov A.G., Bode R., Maxwell D.P., Rodermel S.R., Hüner N.P.A. (2011). Flexibility in photosynthetic electron transport: the physiological role of plastoquinol terminal oxidase (PTOX). Biochim. Biophys. Acta 1807: 954–967 [DOI] [PubMed] [Google Scholar]
- Merchant S.S. (2010). The elements of plant micronutrients. Plant Physiol. 154: 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Whelan J., Small I. (2006). Recent surprises in protein targeting to mitochondria and plastids. Curr. Opin. Plant Biol. 9: 610–615 [DOI] [PubMed] [Google Scholar]
- Moore A.L., Albury M.S. (2008). Further insights into the structure of the alternative oxidase: From plants to parasites. Biochem. Soc. Trans. 36: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Moore A.L., Umbach A.L., Siedow J.N. (1995). Structure-function relationships of the alternative oxidase of plant mitochondria: A model of the active-site. J. Bioenerg. Biomembr. 27: 367–377 [DOI] [PubMed] [Google Scholar]
- Müller P., Li X.P., Niyogi K.K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R., Rost B. (2005). Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 348: 85–100 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3: 455–460 [DOI] [PubMed] [Google Scholar]
- Noguchi K., Yoshida K. (2008). Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 8: 87–99 [DOI] [PubMed] [Google Scholar]
- Norris S.R., Barrette T.R., DellaPenna D. (1995). Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Araújo W.L., Fernie A.R. (2011). Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 155: 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y., Kobayashi Y., Shikanai T. (2010). Physiological links among alternative electron transport pathways that reduce and oxidize plastoquinone in Arabidopsis. Plant J. 63: 458–468 [DOI] [PubMed] [Google Scholar]
- Perry S.E., Li H.M., Keegstra K. (1991). In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol. 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Qin G., Gu H., Ma L., Peng Y., Deng X.W., Chen Z., Qu L.J. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17: 471–482 [DOI] [PubMed] [Google Scholar]
- Rhoads D.M., Umbach A.L., Sweet C.R., Lennon A.M., Rauch G.S., Siedow J.N. (1998). Regulation of the cyanide-resistant alternative oxidase of plant mitochondria. Identification of the cysteine residue involved in α-keto acid stimulation and intersubunit disulfide bond formation. J. Biol. Chem. 273: 30750–30756 [DOI] [PubMed] [Google Scholar]
- Richly E., Leister D. (2004). An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Rochaix J.-D. (2011). Regulation of photosynthetic electron transport. Biochim. Biophys. Acta 1807: 375–383 [DOI] [PubMed] [Google Scholar]
- Rödiger A., Baudisch B., Langner U., Klösgen R.B. (2011). Dual targeting of a mitochondrial protein: The case study of cytochrome c1. Mol. Plant 4: 679–687 [DOI] [PubMed] [Google Scholar]
- Rosso D., Bode R., Li W., Krol M., Saccon D., Wang S., Schillaci L.A., Rodermel S.R., Maxwell D.P., Hüner N.P. (2009). Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21: 3473–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D., Ivanov A.G., Fu A., Geisler-Lee J., Hendrickson L., Geisler M., Stewart G., Krol M., Hurry V., Rodermel S.R., Maxwell D.P., Hüner N.P. (2006). IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol. 142: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D., Nakazono M., Lee K.-H., Tsutsumi N., Akita S., Hirai A. (2001). The gene for alternative oxidase-2 (AOX2) from Arabidopsis thaliana consists of five exons unlike other AOX genes and is transcribed at an early stage during germination. Genes Genet. Syst. 76: 89–97 [DOI] [PubMed] [Google Scholar]
- Saisho D., Nambara E., Naito S., Tsutsumi N., Hirai A., Nakazono M. (1997). Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol. Biol. 35: 585–596 [DOI] [PubMed] [Google Scholar]
- Savitch L.V., Ivanov A.G., Krol M., Sprott D.P., Öquist G., Huner N.P.A. (2010). Regulation of energy partitioning and alternative electron transport pathways during cold acclimation of lodgepole pine is oxygen dependent. Plant Cell Physiol. 51: 1555–1570 [DOI] [PubMed] [Google Scholar]
- Scheibe R. (2004). Malate valves to balance cellular energy supply. Physiol. Plant. 120: 21–26 [DOI] [PubMed] [Google Scholar]
- Shahbazi M., Gilbert M., Labouré A.M., Kuntz M. (2007). Dual role of the plastid terminal oxidase in tomato. Plant Physiol. 145: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W.K., Vogelmann T.C., DeLucia E.H., Bell D.T., Shepherd K.A. (1997). Leaf form and photosynthesis: Do leaf structure and orientation interact to regulate internal light and carbon dioxide? Bioscience 47: 785–793 [Google Scholar]
- Sokolov L.N., Dominguez-Solis J.R., Allary A.L., Buchanan B.B., Luan S. (2006). A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. USA 103: 9732–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J., Schultz G., Joyard J., Douce R., Block M.A. (1985). Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. Arch. Biochem. Biophys. 238: 290–299 [DOI] [PubMed] [Google Scholar]
- Stepien P., Johnson G.N. (2009). Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte tellungiella. Role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol. 149: 1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S., Burch-Smith T., Kim I., Meinke D., Mindrinos M., Zambryski P. (2009). Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc. Natl. Acad. Sci. USA 106: 17229–17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb P., Josse E.M., Gallouet E., Baptist F., Kuntz M., Cornic G. (2005). Evidence for alternative electron sinks to photosynthetic carbon assimilation in the high mountain plant species Ranunculus glacialis. Plant Cell Environ. 28: 1123–1135 [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D., van Wijk K.J. (2009). PPDB, the plant proteomics database at Cornell. Nucleic Acids Res. 37(Database issue): D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach A.L., Fiorani F., Siedow J.N. (2005). Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach A.L., Gonzàlez-Meler M.A., Sweet C.R., Siedow J.N. (2002). Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochim. Biophys. Acta 1554: 118–128 [DOI] [PubMed] [Google Scholar]
- Umbach A.L., Ng V.S., Siedow J.N. (2006). Regulation of plant alternative oxidase activity: A tale of two cysteines. Biochim. Biophys. Acta 1757: 135–142 [DOI] [PubMed] [Google Scholar]
- Umbach A.L., Siedow J.N. (2000). The cyanide-resistant alternative oxidases from the fungi Pichia stipitis and Neurospora crassa are monomeric and lack regulatory features of the plant enzyme. Arch. Biochem. Biophys. 378: 234–245 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G.C., McIntosh L., Yip J.Y. (1998). Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. Plant Cell 10: 1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez-Ramirez A.I., van Ieperen W., Vreugdenhil D., Millenaar F.F. (2011). Plants under continuous light. Trends Plant Sci. 16: 310–318 [DOI] [PubMed] [Google Scholar]
- Weber A.P.M., Osteryoung K.W. (2010). From endosymbiosis to synthetic photosynthetic life. Plant Physiol. 154: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel C.M., Jiang C.Z., Meehan L.J., Voytas D.F., Rodermel S.R. (1994). Nuclear-organelle interactions: The immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 6: 161–175 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo N.S., Gordon M.J., Graham S.R., Rossel J.B., Badger M.R., Pogson B.J. (2011). A mutation in the purine biosynthetic enzyme ATASE2 impacts high light signalling and acclimation response in green and chlorotic sectors of Arabidopsis leaves. Funct. Plant Biol. 38: 401–419 [DOI] [PubMed] [Google Scholar]
- Wu D., Wright D.A., Wetzel C., Voytas D.F., Rodermel S. (1999). The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Han P., Lutziger I., Wang K., Oliver D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Xu Y.-Z., Arrieta-Montiel M.P., Virdi K.S., de Paula W.B., Widhalm J.R., Basset G.J., Davila J.I., Elthon T.E., Elowsky C.G., Sato S.J., Clemente T.E., Mackenzie S.A. (2011). MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23: 3428–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O., Pines O. (2011). Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta 1808: 1012–1020 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Shibata M., Terashima I., Noguchi K. (2010). Simultaneous determination of in vivo plastoquinone and ubiquinone redox states by HPLC-based analysis. Plant Cell Physiol. 51: 836–841 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Terashima I., Noguchi K. (2006). Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol. 47: 22–31 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Terashima I., Noguchi K. (2007). Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol. 48: 606–614 [DOI] [PubMed] [Google Scholar]
- Yu F., Fu A., Aluru M., Park S., Xu Y., Liu H., Liu X., Foudree A., Nambogga M., Rodermel S. (2007). Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 30: 350–365 [DOI] [PubMed] [Google Scholar]
- Yu F., Park S.S., Liu X., Foudree A., Fu A., Powikrowska M., Khrouchtchova A., Jensen P.E., Kriger J.N., Gray G.R., Rodermel S.R. (2011). SUPPRESSOR OF VARIEGATION4, a new var2 suppressor locus, encodes a pioneer protein that is required for chloroplast biogenesis. Mol. Plant 4: 229–240 [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, n-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.