This article presents biochemical, kinetic, and structural characterizations of a bifunctional enzyme that catalyzes both the antepenultimate and the penultimate steps of biotin (vitamin B8) synthesis in Arabidopsis. This work shows that the intermediate substrate of the overall reaction is channeled between both distant active sites, probably through a polar trench at the surface of the enzyme.
Abstract
Diaminopelargonic acid aminotransferase (DAPA-AT) and dethiobiotin synthetase (DTBS) catalyze the antepenultimate and the penultimate steps, respectively, of biotin synthesis. Whereas DAPA-AT and DTBS are encoded by distinct genes in bacteria, in biotin-synthesizing eukaryotes (plants and most fungi), both activities are carried out by a single enzyme encoded by a bifunctional gene originating from the fusion of prokaryotic monofunctional ancestor genes. In few angiosperms, including Arabidopsis thaliana, this chimeric gene (named BIO3-BIO1) also produces a bicistronic transcript potentially encoding separate monofunctional proteins that can be produced following an alternative splicing mechanism. The functional significance of the occurrence of a bifunctional enzyme in biotin synthesis pathway in eukaryotes and the relative implication of each of the potential enzyme forms (bifunctional versus monofunctional) in the plant biotin pathway are unknown. In this study, we demonstrate that the BIO3-BIO1 fusion protein is the sole protein form produced by the BIO3-BIO1 locus in Arabidopsis. The enzyme catalyzes both DAPA-AT and DTBS reactions in vitro and is targeted to mitochondria in vivo. Our biochemical and kinetic characterizations of the pure recombinant enzyme show that in the course of the reaction, the DAPA intermediate is directly transferred from the DAPA-AT active site to the DTBS active site. Analysis of several structures of the enzyme crystallized in complex with and without its ligands reveals key structural elements involved for acquisition of bifunctionality and brings, together with mutagenesis experiments, additional evidences for substrate channeling.
INTRODUCTION
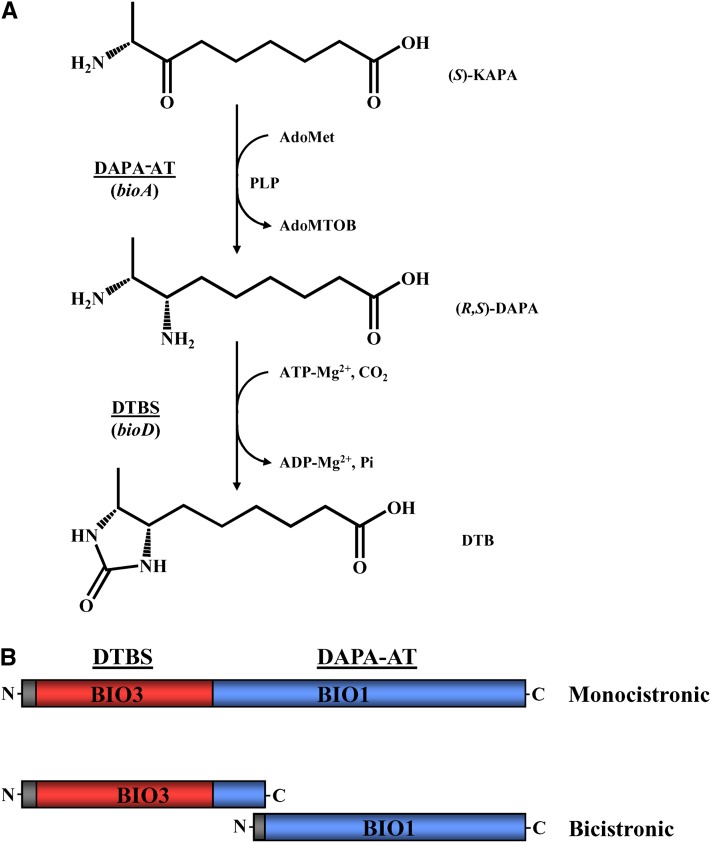
Biotin (vitamin B8) is an essential covalently linked cofactor to some carboxylases, decarboxylases, and transcarboxylases that deal with crucial metabolic processes, such as fatty acid and carbohydrate metabolism (Knowles, 1989; Alban et al., 2000). In mammals, biotin is also known to regulate gene expression through different mechanisms, including histone biotinylation (Zempleni, 2005; Beckett, 2007). Despite its essential functions, de novo synthesis of this vitamin is restricted to bacteria, a few fungi, and plants. Thus, inhibition of the biotin synthesis pathway key enzymes is an attractive approach for antibiotic, fungicide, and herbicide development (Alban et al., 2000; Marquet, 2010). All animals, including humans, cannot synthesize biotin as part of their normal metabolism and therefore rely on the supply of biotin from exogenous sources. The biotin biosynthetic pathway has been extensively studied in bacteria, especially Escherichia coli (reviewed in Streit and Entcheva, 2003, among others). In all known microbes, biotin is synthesized from pimeloyl-CoA through four enzymatic steps comprising 7-keto-8-aminopelargonic acid (KAPA) synthase, 7,8-diaminopelargonic acid aminotransferase (DAPA-AT), dethiobiotin synthetase (DTBS), and biotin synthase encoded by the bioF, bioA, bioD, and bioB genes, respectively. Enzymes encoded by these genes in E. coli, Bacillus sphaericus, and more recently in Bacillus subtilis or Mycobacterium tuberculosis have been characterized biochemically, and/or structurally, and their reaction mechanisms elucidated (Schneider and Lindqvist, 2001; Berkovitch et al., 2004; Dey et al., 2010). In plants, the biosynthetic pathway is similar to the one described for bacteria (Schneider et al., 1989; Shellhammer and Meinke, 1990; Baldet et al., 1993; Patton et al., 1996, 1998). However, if the biotin biosynthetic genes have been identified and genetically characterized in the model plant Arabidopsis thaliana, to date only the first and the last enzymes of the plant pathway, namely, KAPA synthase and biotin synthase, have been characterized at the biochemical level (Picciocchi et al., 2001, 2003; Pinon et al., 2005). The intimate biochemistry of plant DAPA-AT and DTBS reactions remains unknown.
In bacteria, DAPA-AT is a homodimeric pyridoxal 5′-phosphate (PLP)–dependent aminotransferase that catalyzes the conversion of KAPA to DAPA (Figure 1A). In most bacteria, the enzyme uses S-adenosyl-l-Met (AdoMet) as amino group donor, an unusual feature among aminotransferases (Izumi et al., 1975; Breen et al., 2003; Mann and Ploux, 2006). DTBS is a homodimeric enzyme that catalyzes the formation of the ureido ring of dethiobiotin (DTB) from DAPA and CO2 in the presence of ATP and divalent metal ions (Figure 1A) (Alexeev et al., 1995; Huang et al., 1995). In Arabidopsis, genetic studies revealed that DAPA-AT (BIO1; BioA ortholog) and DTBS (BIO3; BioD ortholog) are encoded by adjacent genes, defining a single genetic locus, and are expressed in both single and chimeric BIO3-BIO1 transcripts through alternative splicing events (Muralla et al., 2008). One of the fused transcripts is monocistronic and encodes a bifunctional fusion protein capable of complementing the orthologous auxotrophs of E. coli (bioD and bioA). The second transcript includes 10 additional nucleotides that introduce a premature stop codon. As a consequence, this splice variant is bicistronic, with distinct but overlapping reading frames. This bicistronic transcript is potentially capable of producing separate BIO3 and BIO1 proteins (Figure 1B). Other plants and most fungi also present a chimeric BIO3-BIO1 homolog gene (Hall and Dietrich, 2007; Muralla et al., 2008; Magliano et al., 2011).
Figure 1.
The Two-Step Synthesis of DTB from KAPA.
(A) In bacteria, DTB is synthesized from KAPA by two distinct enzymes encoded by bioA and bioD genes, respectively, catalyzing successively DAPA-AT and DTBS reactions. AdoMTOB, S-adenosyl-2-oxo-4-methylthiobutyric acid.
(B) Representation of BIO3 (BioD ortholog) and BIO1 (BioA ortholog) domains of BIO3-BIO1 fusion protein and of separate BIO3 and BIO1 proteins, putatively encoded by Arabidopsis monocistronic and bicistronic BIO3-BIO1 transcripts, respectively.
[See online article for color version of this figure.]
In this study, we present evidence that the BIO3-BIO1 fusion protein is the major, if not the exclusive, protein form produced by the BIO3-BIO1 locus in Arabidopsis. The enzyme catalyzes both DAPA-AT (BIO1) and DTBS (BIO3) reactions and is targeted to mitochondria. Biochemical and kinetic properties of the pure recombinant mature enzyme were investigated. Our data are consistent with the hypothesis that DAPA, the product of DAPA-AT domain, is not released in the bulk solvent in the course of the reaction but is directly transferred to the DTBS domain for its subsequent conversion to DTB. Finally, the three-dimensional structure of the enzyme was solved to 2.5-Å resolution, in its holoform (i.e., in the presence of bound-PLP) as well as in complex with various substrates and/or products of the enzyme-catalyzed reactions. The structural analyses show key elements involved in acquisition of bifunctionality. These structures and accompanying kinetic data combined with mutagenesis experiments form the basis for mechanistic considerations, in particular with respect to DAPA intermediate transit between both catalytic sites of the bifunctional enzyme.
RESULTS
Expression of the BIO3-BIO1 Locus in Arabidopsis at the Protein Level and Subcellular Distribution
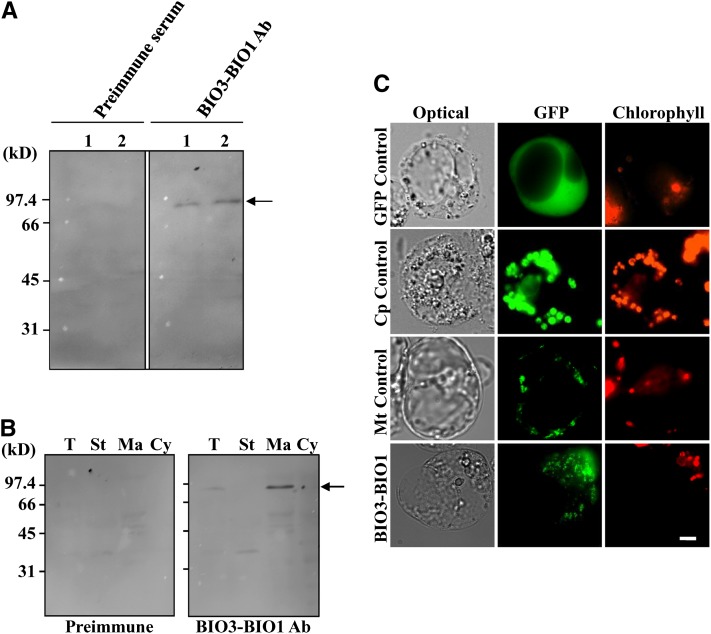
DAPA-AT and DTBS activities in plants are catalyzed by a bifunctional fusion protein, including both reaction domains, BIO1 and BIO3. In addition, the Arabidopsis BIO3-BIO1 gene fusion also produces a bicistronic mRNA transcript with the potential to encode separate proteins (Muralla et al., 2008): a long BIO3 form exhibiting a C-terminal extension and an N-terminal truncated BIO1 form lacking conserved amino acid residues essential for activity in bacteria (Figure 1B). To clarify the situation in the plant cell at the protein level, we performed an immunological characterization of BIO3-BIO1 gene products in Arabidopsis cell extracts using polyclonal antibodies raised against the full-length recombinant BIO3-BIO1 protein overproduced in E. coli (see Supplemental Figure 1 online). Immunoblot analysis of protein samples from whole plants (aboveground organs from 35-d-old plants) or cultured cells identified a single polypeptide of ∼90 kD (i.e., the expected size for the BIO3-BIO1 fusion protein) (Figure 2A). No band of lower molecular mass was specifically detected using BIO3-BIO1 antibodies (the faint signal bands of low molecular mass are nonspecific background from the preimmune).
Figure 2.
Analysis of the Expression of the Arabidopsis BIO3-BIO1 Locus at the Protein Level and Subcellular Distribution.
(A) Immunological detection of BIO3-BIO1 gene products in Arabidopsis. Soluble proteins (40 µg) from aboveground organs from Arabidopsis plants (lane 1) and Arabidopsis cultured cells (lane 2) were separated by SDS-PAGE and analyzed by immunoblotting using affinity-purified antibodies raised against recombinant BIO3-BIO1 (BIO3-BIO1 Ab) or preimmune serum (for negative control).
(B) Immunolocalization of BIO3-BIO1 in Arabidopsis. Soluble proteins (50 µg) from total plant extracts (T), chloroplast stroma (St), mitochondrial matrix (Ma), and cytosolic enriched fraction (Cy) were separated by SDS-PAGE and analyzed by immunoblotting as in (A). Arrows in (A) and (B) point to the BIO3-BIO1 polypeptide band.
(C) Expression of BIO3-BIO1 fused to GFP in Arabidopsis protoplasts. Constructs encoding GFP alone (GFP Control), GFP fused to the C terminus of the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase transit peptide (a chloroplastic marker) from Arabidopsis (Cp Control), GFP fused to the C terminus of the dihydropterin pyrophosphokinase-dihydropteroate synthase transit peptide (a mitochondrial marker) from pea (Pisum sativum; Mt Control), and GFP fused to the C terminus of full-length BIO3-BIO1 (BIO3-BIO1) were introduced into Arabidopsis protoplasts. Images are optical photomicrographs (Optical), GFP fluorescence (GFP; green pseudocolor), and chlorophyll fluorescence (Chlorophyll; red pseudocolor). Bar = 10 µm.
This contrasts with the situation at the mRNA level, where the bicistronic splice variant is somewhat more abundant than the monocistronic version in most parts of the plant (Muralla et al., 2008) (see Supplemental Figure 2 online). To investigate the distribution of the BIO3-BIO1 protein in plant cells, intact chloroplasts and mitochondria from Arabidopsis leaves were purified on Percoll density gradients, thus providing organelles devoid of contamination from the other compartments. Also, a cytosolic-enriched fraction was prepared. Soluble proteins from purified chloroplasts (stroma), mitochondria (matrix), and the cytosolic-enriched fraction were then analyzed by immunoblots. Purified antibody detected the BIO3-BIO1 protein exclusively in the mitochondrial matrix fraction (Figure 2B). As a complementary approach, subcellular localization of BIO3-BIO1 was also investigated by fusing its full-length sequence upstream to the green fluorescent protein (GFP) marker. As shown in Figure 2C, the transient expression of BIO3-BIO1-GFP in Arabidopsis protoplasts resulted in a punctuate pattern of green fluorescence, similar to the one observed with GFP fused to the transit peptide of an authentic mitochondrial protein. Collectively, these results strongly suggest that in Arabidopsis, the BIO3-BIO1 locus produces a unique BIO3-BIO1 fusion protein form that takes place in mitochondria.
Production of the Recombinant Mature BIO3-BIO1 Protein (mBIO3-BIO1)
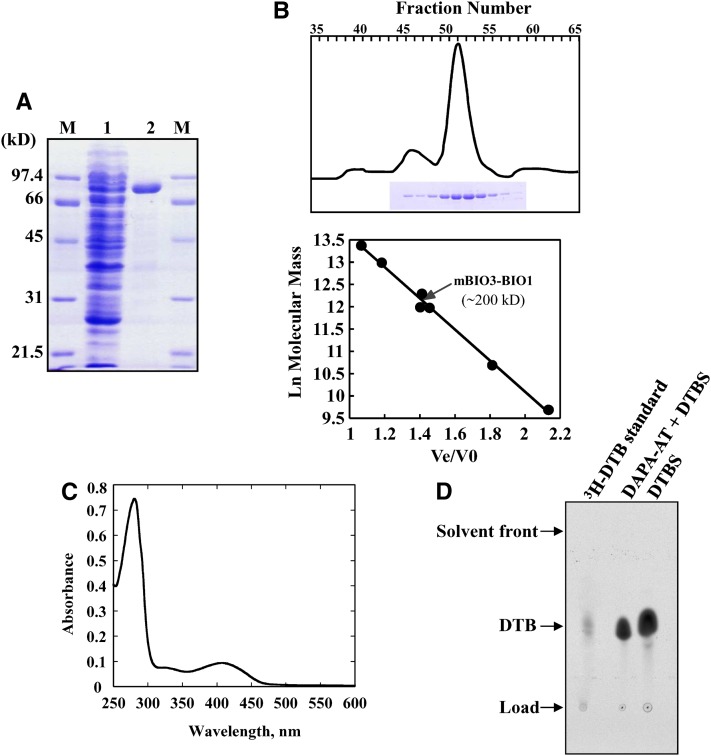
To further characterize the plant DAPA-AT/DTBS reactions, BIO3-BIO1 was overproduced in E. coli as a truncated protein, hereafter designated as mBIO3-BIO1 (for mature BIO3-BIO1) with an N-terminal 22-residue deletion. This sequence was predicted to correspond to the mitochondrial targeting peptide by all the major prediction programs available on the Web and gathered on the Arabidopsis subcellular database (Heazlewood et al., 2007). Since the cloning procedure added an N-terminal hexa-His tag, the recombinant mature protein was purified to near homogeneity by standard nickel-affinity chromatography (Figure 3A). The apparent molecular mass of the native protein, as estimated by gel filtration chromatography on a Superdex S200 column, was found to be ∼200 kD, consistent with a homodimeric structure of the recombinant enzyme in solution (Figure 3B). Typically, 10 to 15 mg of pure enzyme was obtained per liter of cell culture using this two-step purification procedure. UV-visible spectroscopy analysis of the recombinant enzyme revealed an absorption spectrum characteristic of the internal aldimine of PLP-dependent enzymes with an absorbance maximum (band) at 415 nm and a shoulder at 335 nm (Figure 3C).
Figure 3.
Physicochemical and Biochemical Properties of Recombinant Arabidopsis mBIO3-BIO1.
(A) Documentation of mBIO3-BIO1 purification on nickel-nitrilotriacetic acid-agarose resin. Polypeptides were separated by SDS-PAGE and stained with Coomassie blue. Lane 1, soluble proteins (25 µg) from E. coli Rosetta cells producing mBIO3-BIO1; lane 2, proteins eluted from the column (10 µg); lanes M, molecular mass markers.
(B) Purification and molecular mass estimation of native mBIO3-BIO1 by gel filtration. Purified protein was resolved by chromatography onto a Superdex 200 HiLoad column (2.6 × 60 cm; GE Healthcare). Eluted fractions (3 mL) were analyzed by SDS-PAGE (top panel). Standards proteins for column calibration (bottom panel) were as follows: thyroglobulin (669 kD), ferritin (443 kD), catalase (232 kD), γ-globulin (158 kD), aldolase (158 kD), ovalbumin (43 kD), and myoglobulin (17 kD). Ve, elution volume; V0, void volume.
(C) Spectroscopy analysis of purified recombinant mBIO3-BIO1. Absorption spectrum was recorded at 30°C in 100 mM HEPES-KOH, pH 7.5, in the presence of 20 µM pure enzyme.
(D) HP TLC detection of DTB produced by recombinant mBIO3-BIO1. The overall (DAPA-AT + DTBS) reaction and DTBS reaction were performed by measuring the formation of acid-stable [14C]-DTB from acid-labile H14CO3 as described in Methods. Assays contained 0.2 nmol (DAPA-AT + DTBS reaction) or 0.1 nmol (DTBS reaction) enzyme, and reaction mixtures were incubated for 120 or 20 min, respectively. [14C]-DTB produced was separated by TLC and detected by phosphor imaging analysis (Picciocchi et al., 2001). Migration was compared with that of authentic [3H]-DTB standard (Rf = 0.53).
[See online article for color version of this figure.]
Biochemical Characterization of DAPA-AT and DTBS Reactions
To investigate the overall DAPA-AT + DTBS or DTBS alone activities catalyzed by mBIO3-BIO1 in vitro, we developed a radiochemical assay that monitors the formation of acid stable [14C]-DTB from acid-labile 14CO2 in the presence of appropriate substrates and cofactors. This assay, adapted from a previously published procedure designed for measuring E. coli DTBS activity (Gibson et al., 1995), was sensitive and more reliable in our hands than published bioassays (paper disc plate method) and spectrophotometric assays for bacterial DTBS (Krell and Eisenberg, 1970; Gibson et al., 1995; Yang et al., 1997). Identification of [14C]-DTB as a reaction product of the in vitro assays was confirmed by high-performance thin layer chromatography (HP TLC) analysis on a silica gel plate, by comparison of its retention time value with that of authentic [3H]-DTB sample (Figure 3D). Plant DAPA-AT activity was measured using a discontinuous assay in which DAPA synthesized by mBIO3-BIO1 was subsequently titrated using the E. coli monofunctional DTBS (see Methods).
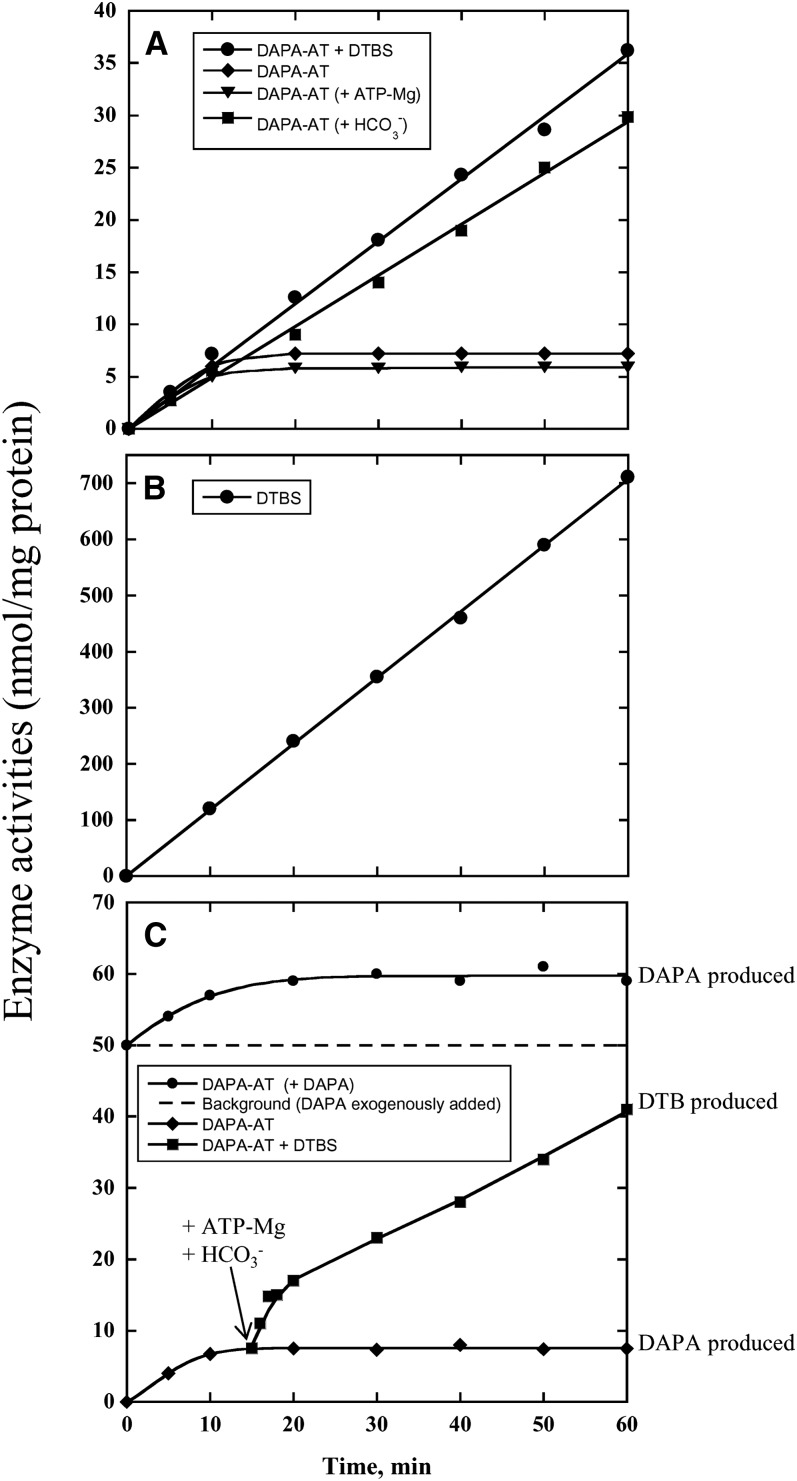
When DAPA-AT + DTBS activity was assayed under optimized conditions, pure recombinant Arabidopsis enzyme exhibited a specific activity of 25 to 35 nmol H14CO3− incorporated h−1 mg−1 protein, confirming that mBIO3-BIO1 is actually a bifunctional enzyme catalyzing both DAPA-AT and DTBS reactions (Figure 4A). Recorded reaction rates were low but consistent with estimated in vivo rates of biotin production and requirement in living organisms (Alexeev et al., 1998; Alban et al., 2000; Farrar et al., 2010; Lin et al., 2010). The reaction was strictly dependent upon the presence of (S)-KAPA, AdoMet, ATP-Mg, and NaHCO3 in the reaction medium. In most bacteria, DAPA-AT uses AdoMet as an amino group donor. B. subtilis uses l-Lys, another unusual amino donor for the reaction (Van Arsdell et al., 2005). Besides AdoMet, however, no other substrate was found to act as an amino donor for Arabidopsis mBIO3-BIO1, among a variety of potential substrates, including l-Asp, l-Glu, l-Met, l-Lys, S-adenosylhomocysteine, and 5′-deoxy-5′-methylthioadenosine. Interestingly, the overall DAPA-AT + DTBS activity catalyzed by mBIO3-BIO1 was linear for at least 60 min under optimized conditions, and no lag in DTB production was observed (Figure 4A). This suggested a direct transfer of the DAPA intermediate between the DAPA-AT and the DTBS domains (Figure 4A).
Figure 4.
Kinetic Study of Arabidopsis mBIO3-BIO1 Catalyzed Reactions.
Activities were assayed under optimized assay conditions as described in Methods. Data are from a representative experiment repeated at least three times.
(A) Time-course formation of DAPA (DAPA-AT reaction; diamonds, triangles, and squares) or DTB (DAPA-AT + DTBS reaction; circles). DAPA-AT was assayed in a reaction medium containing 20 µM (S)-KAPA, 0.2 mM PLP, 3 mM AdoMet, and 0.4 µM enzyme, in the absence (standard assay medium; diamonds) or in the presence of 0.3 mM ATP + 5 mM MgCl2 (triangles) or 10 mM NaHCO3 (squares). DAPA-AT + DTBS was assayed in a reaction medium containing 20 µM (S)-KAPA, 0.2 mM PLP, 3 mM AdoMet, 0.3 mM ATP, 5 mM MgCl2, 10 mM NaHCO3, and 0.4 µM enzyme.
(B) Time-course formation of DTB (DTBS reaction). DTBS was assayed in a reaction medium containing 200 µM DAPA, 0.3 mM ATP, 5 mM MgCl2, 10 mM NaHCO3, and 1 µM enzyme.
(C) Effect of exogenous DAPA on DAPA-AT reaction. DAPA-AT was assayed in a reaction medium containing 20 µM (S)-KAPA, 0.2 mM PLP, 3 mM AdoMet, and 0.4 µM enzyme, in the absence (standard assay medium; diamonds) or in the presence of 2 µM DAPA (circles). After 15 min incubation of the enzyme in the standard DAPA-AT reaction medium, 0.3 mM ATP, 5 mM MgCl2 and 10 mM NaHCO3 were added and the time-course formation of DTB was followed (squares).
When Arabidopsis mBIO3-BIO1 was assayed for DTBS reaction, a specific activity of 700 to 1200 nmol [14C]-DTB formed h−1 mg−1 protein was measured, a value roughly 25 to 30 times higher than the one recorded for the overall DAPA-AT + DTBS reaction (Figure 4B). This suggested that the overall reaction is rate limited by the DAPA-AT activity. Interestingly, it also demonstrated that in vitro the enzyme can use external DAPA for the synthesis of DTB (i.e., that the BIO3 active site is accessible to the outside).
When DAPA-AT was assayed alone, the initial velocity of the reaction was roughly similar to that of the overall reaction, but surprisingly, DAPA formation rapidly reached a plateau (Figure 4A). At best, ∼0.7 to 0.8 nmol DAPA could be synthesized per nmol enzyme monomer in the reaction mixture. A similar kinetic behavior was observed when ATP-Mg, one of the DTBS substrates, was added to the standard DAPA-AT assay. Noteworthy, in the presence of NaHCO3 in lieu of ATP-Mg in the reaction medium, DAPA-AT activity was found to be linear with time, with a resulting specific activity close to that for the overall DAPA-AT + DTBS reaction (Figure 4B). Thus, in the absence of NaHCO3, one of the substrates of the DTBS domain, the DAPA-AT reaction was (sub)stoichiometric, suggesting that the DAPA-AT domain is blocked in a noncatalytically competent form. Interestingly, preincubation of the enzyme in the presence of exogenous DAPA did not prevent de novo DAPA production nor did it preclude appearance of the plateau, showing that the DAPA-AT active site was not accessible to external DAPA and that production of the stable enzyme-DAPA complex was dependent on catalysis (Figure 4C). Also, addition of ATP-Mg and NaHCO3 15 min after the start of the DAPA-AT reaction (i.e., when most of the enzyme is bound up in a noncatalytic enzyme-product complex) resulted in a rapid synthesis of DTB from already produced DAPA, followed by a slower but linear production of DTB, presumably from newly synthesized DAPA (Figure 4C). Collectively, our results suggest that DAPA produced by the DAPA-AT domain is somehow trapped in the active site. Upon addition of the DTBS substrates, DAPA would be rapidly transferred to the DTBS active site for DTB synthesis, without previous release in solution.
To learn more about Arabidopsis DAPA-AT and DTBS reactions, we determined the kinetic parameters for both the overall (DAPA-AT + DTBS) and DTBS activities (Table 1). The most striking feature of these analyses is that the enzyme exhibited a kinetic cooperativity with respect to all tested substrates and for both reactions (Hill coefficient values calculated from Hill plots varied from 1.3 to 2.4 according to the substrate considered). This kinetic behavior has not been described for bacterial monofunctional enzyme counterparts studied so far. It is also interesting to note the ∼30 times lower apparent k0.5 value for ATP measured when the overall DAPA-AT + DTBS reaction was assayed compared with that determined for the DTBS reaction alone, without affecting the catalytic efficiency (unaltered kcat/k0.5 value, since kcat for DTBS reaction is 25-fold higher than for the overall reaction) (Table 1). This probably reflected conformational changes of the ATP binding site upon binding of DAPA-AT substrates or products and suggested interdependence of both DAPA-AT and DTBS domains.
Table 1. Kinetic Parameters for the Arabidopsis Wild Type (mBIO3-BIO1) and F326Y Mutant (mBIO3-BIO1 F326Y) DTBS–DAPA-AT: Comparison with Those from Bacterial Monofunctional DAPA-AT (bioA) and DTBS (bioD).
| Enzyme | Reaction | K0.5 AdoMet (nH) (mM) | K0.5 KAPA (nH) (µM) | K0.5 ATP (nH) (µM) | K0.5 DAPA (nH) (µM) | kcat (min−1) | kcat/K0.5 (min−1 M−1) |
|---|---|---|---|---|---|---|---|
| mBIO3-BIO1a | DAPA-AT + DTBS | 0.63 ± 0.06 (1.34) | 4.78 ± 0.6 (1.29) | 0.56 ± 0.14 (2.4) | 0.072 ± 0.009 | 1.28 × 105 (ATP) | |
| DTBS | 18.8 ± 1.4 (1.47) | 18.7 ± 1.5 (1.75) | 1.85 ± 0.05 | 0.98 × 105 (ATP) | |||
| mBIO3-BIO1 F326Ya | DAPA-AT + DTBS | 1.65 ± 0.19 (1.23) | 4.6 ± 0.6 (1.36) | 0.80 ± 0.21 (2.1) | 0.062 ± 0.005 | 0.77 × 105 (ATP) | |
| DTBS | 29.5 ± 2.7 (1.35) | 28.5 ± 3.8 (1.74) | 2.16 ± 0.11 | 0.73 × 105 (ATP) | |||
| Organism | |||||||
| bioAb | DAPA-AT | 0.15c | <2c | 0.78 | E. coli | ||
| bioAd | DAPA-AT | 0.78c | 3.8c | 1 | M. tuberculosis | ||
| bioDe | DTBS | 5–19c | 1.3–3c | 2–2.9 | E. coli | ||
| bioDf | DTBS | 29c | 2c | – | M. tuberculosis |
The results are means ± sd from three separate determinations, and parameters were estimated by direct fitting of experimental data to the Hill equation by nonlinear regression analysis. Hill coefficient values (nH) are given in parentheses. Empty cell, not applicable; –, not available.
This work.
Km values.
Nevertheless, kinetic parameters (k0.5 and kcat values for the plant bifunctional enzyme) compared well with those (Km and kcat values) of bacterial monofunctional enzymes (Krell and Eisenberg, 1970; Gibson et al., 1995; Yang et al., 1997; Eliot et al., 2002; Mann and Ploux, 2006; Dey et al., 2010) (Table 1). A noticeable exception concerned the kcat value for plant overall reaction (which reflects DAPA-AT activity) that was found to be one order of magnitude lower than that for bacterial DAPA-AT (Eliot et al., 2002; Mann and Ploux, 2006). Many bacterial DAPA-AT enzymes contain a conserved Tyr residue at the N terminus (Tyr-17 in E. coli) that plays an essential role in binding of AdoMet or KAPA substrates and for catalytic efficiency (Sandmark et al., 2004; Dey et al., 2010). A Phe residue at this position (Phe-326 in Arabidopsis) is present in all available plant of fungal sequences (see Supplemental Figure 3 online). This difference could possibly account for the low DAPA-AT activity measured with the Arabidopsis enzyme compared with the bacterial enzymes. To test this hypothesis, we constructed a F326Y BIO3-BIO1 mutant. However, in Arabidopsis, Phe-to-Tyr substitution did not impact importantly the enzyme kinetic parameters. In particular, kcat values were poorly affected (Table 1).
Taken together, our biochemical and kinetic analyses are consistent with a mechanism enabling the enzyme to directly transfer the DAPA intermediate from the DAPA-AT domain active site to the DTBS domain active site without diffusion to the aqueous environment. To address this question and to gain insight into the mechanism of the overall DAPA-AT + DTBS reaction, we undertook a structural characterization of the enzyme holoform (with bound PLP) and the complexes with various substrates and/or products of the enzyme-catalyzed reactions.
Overall Unliganded Structures of mBIO3-BIO1 from Arabidopsis
The native and the selenomethionine ([SeMet]) enzyme have been crystallized without ligand in two different space groups. In both cases, the structures show a dimer made of two monomers related by a noncrystallographic twofold axis. In the space group C2, one dimer of [SeMet]-mBIO3-BIO1 was present in the asymmetric unit. The Cα atoms of the monomers from the [SeMet]-mBIO3-BIO1 can be superimposed with a 0.23 Å root mean square deviation (rmsd) value. In the native form crystallized in the space group P1, two dimers were present in the asymmetric unit and the monomers in each dimer well superimposed (0.21 and 0.19 Å rmsd values for the monomer A/B and C/D, respectively). The two dimers of native mBIO3-BIO1 superimposed with a 1.0 Å rmsd value. [SeMet]-mBIO3-BIO1 can be superimposed with 0.48 and 1.04 Å rmsd values with the dimer A/B and C/D of native mBIO3-BIO1, respectively. These results show that the monomer folds in the dimers are roughly identical.
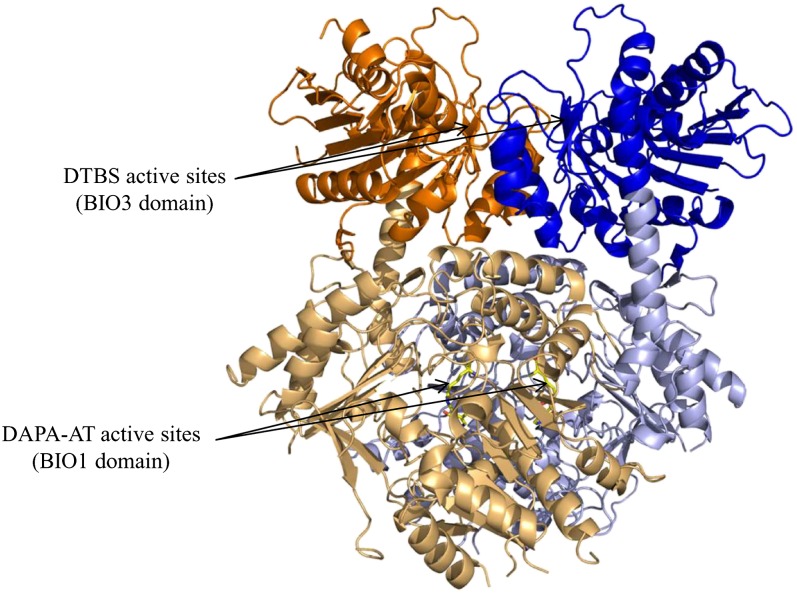
Each monomer can be divided into two domains: the DTBS (BIO3) domain from Ser-6 to Thr-299 (mature enzyme form numbering) and the DAPA-AT (BIO1) domain from Met-300 to Thr-811. A long α-helix Ile283-Val321 (53.3 Å in length) linked the two domains (Figure 5). The catalytic sites of BIO1 and BIO3 are ∼48 Å apart. The monomers interact by involving several zones of both domains. In the space group P1, the superimposition of the dimers (A/B onto C/D) showed large differences in the position of the DAPA-AT domain. These differences were also observed when dimer C/D was superimposed onto [SeMet]-mBIO3-BIO1, mBIO3-BIO1/KAPA, or mBIO3-BIO1/DTB. The average interface area of mBIO3-BIO1 calculated with Protein Interfaces Surfaces Assemblies is 7300 Å2 (22.4% of the accessible surface) for the most complete dimer (A/B) and 5420 Å2 (18.4%) for the second dimer (C/D) (Krissinel and Henrick, 2007). The interface area is 7040 Å2 (22.6%) in [SeMet]-BIO3-BIO1. Because the number of residues observed in the electron density map was different between the dimers in the space group P1 (1525 amino acids in A/B versus 1390 in C/D), the dimer C/D was rebuilt by superimposition with the monomers A and B. In this putative dimer, the interface area observed was 6680 Å2 (20.6%). Differences in buried surfaces result from flexibility of monomers as also revealed by the differences of rebuilt residues in the structures (see Methods). Observation of the molecular surface at the interface revealed the presence of a small discontinuous groove going from the catalytic site of DAPA-AT to the catalytic site of DTBS. This groove is larger in the dimer C/D.
Figure 5.
View of the Overall Fold of the Dimer of mBIO3-BIO1 from Arabidopsis.
The protein is represented as a ribbon, and the DAPA-AT (BIO1) and DTBS (BIO3) domains from each monomer are colored in light blue/light orange or dark blue/orange. PLP is displayed as a yellow stick at the active sites.
Structures of the DTBS and DAPA-AT Domains of the Holoform
The DTBS domain is composed of a nine-stranded β-sheet, with β5 being antiparallel to the others β-strands, surrounded by three α-helices on both sides of the β-sheet. Residues His-9 to Pro-11 form the β-strand β1 involved in the interaction with the DAPA-AT domain through an antiparallel four-stranded β-sheet (see Supplemental Figure 4 online). The two catalytic sites of DTBS are fully accessible to the solvent and are composed of residues from both monomers. The Walker-A motif (Walker et al., 1982) that interacts with phosphates of nucleotide is in the loop between β2 and α1 (Ala22-Thr29). The Walker-B motif (Walker et al., 1982) (Lys177-Glu188) is in the loop connecting α5 to β7 and in β7. Lys-177, Ser-178, Glu-179, Lys-180, and Ser-181 were not observed in the electron density whatever the crystal form was. The Walker-A motif stabilized two sulfate ions in one monomer and only one bound to a magnesium ion in the second monomer in mBIO3-BIO1. The magnesium ion was also stabilized by the Thr-29 hydroxyl group from the Walker-A motif, the Asp-66 and Glu-188 carboxylate groups, and one water molecule. Two other sulfate ions were observed in the dimer. They were stabilized by the Ser-195 hydroxyl group of one monomer and the dipole of the α-helix α7 of the second monomer as well as hydrogen bonds with the Gly-223, Gly-224, Ile-225, and Ser-226 NH groups belonging to the α-helix α7.
The DAPA-AT domain can be divided into three subdomains. The first subdomain (Val339-Asp364) is composed of a three-stranded antiparallel β-sheet, which forms a four-stranded β-sheet with β1 from the DTBS domain. This subdomain is connected to the core of the DAPA-AT domain by two α-helices. The core (Arg424-Ser665) is formed by a seven-stranded β-sheet (β14-β23-β22-β21-β20-β15-β16) with the β-strand β23 being antiparallel to the six others and by an antiparallel three-stranded β-sheet that connects β16 to β20 involving the α-helix α14. The C-terminal part of the DAPA-AT domain (Val732-Thr811) is connected to the core by two α-helices and is formed by an antiparallel three-stranded β-sheet and two α-helices. In the DAPA-AT domain, Lys-644 linked to PLP and Val-583 are in disallowed part of the Ramachandran plot but are well defined in the electron density. A large pocket corresponding to the AdoMet and KAPA binding site is observed in the DAPA-AT domain, going from the solvent to the PLP. PLP from one monomer is stabilized by interacting with Asn-430, Gly-431, Ser-432, Tyr-473, His-474, Gly-475, Glu-581, Asp-615, Val-617, Phe-618, Ala-643, Leu-646, and Thr-647 and with His-679, Ser-680, and Tyr-681 of the second monomer.
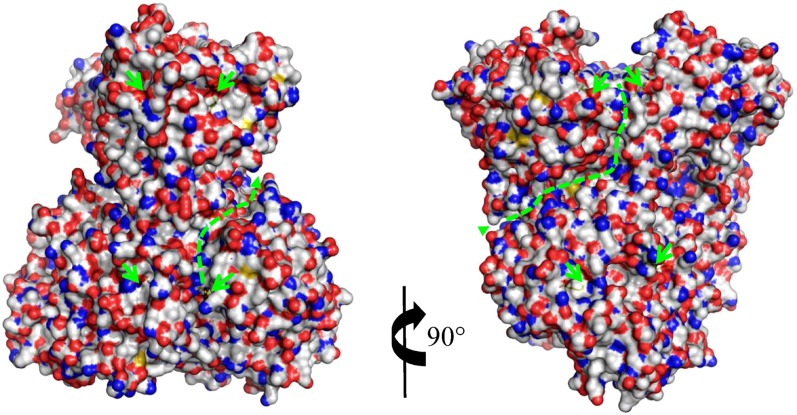
A large polar crevice of ∼12 to 14 Å width at the surface of the enzyme is observed going from the DAPA-AT catalytic binding site to the bottom of the DTBS catalytic site (Figure 6; see Supplemental Figure 5 online).
Figure 6.
Views of the Crevice at the Surface of mBIO3-BIO1 Going from DAPA-AT (BIO1) to DTBS (BIO3).
The two structures of mBIO3-BIO1 are shown after a rotation of 90° following a vertical axis. KAPA observed in the DAPA-AT and DTBS catalytic sites is drawn as a stick and marked by green arrows. The crevice is highlighted with a dashed line.
Structure of mBIO3-BIO1 Bound to KAPA
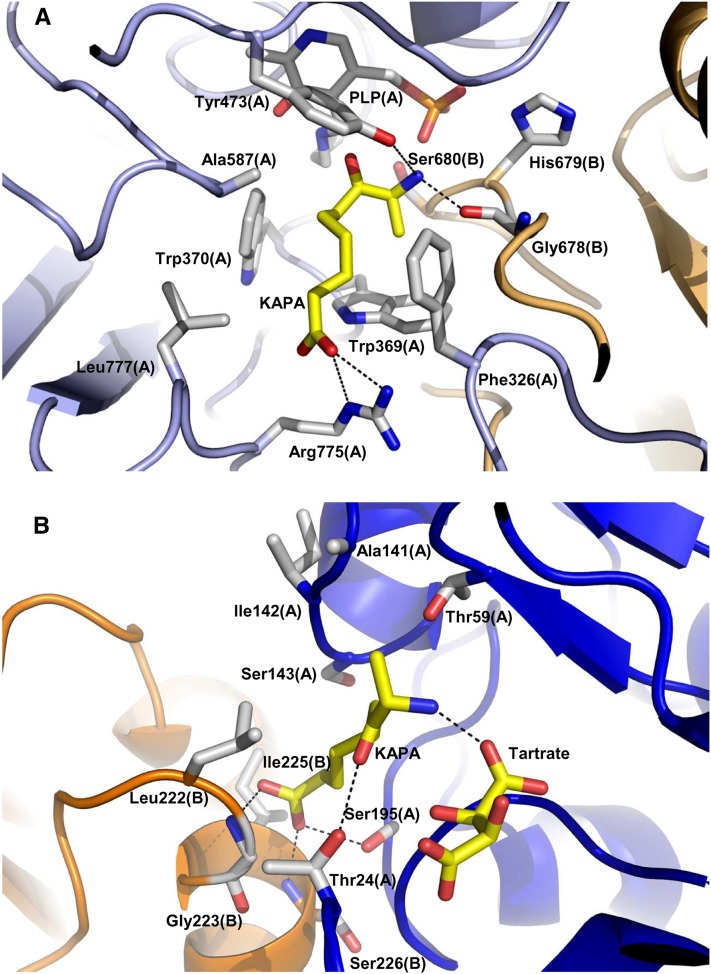
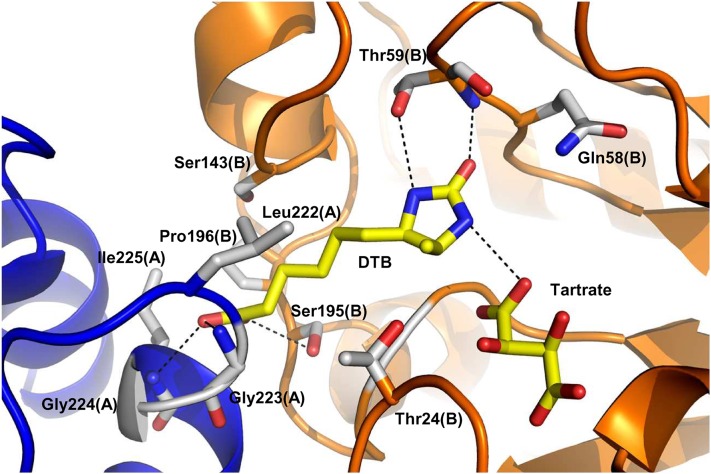
Four KAPA molecules were observed in the electron density map of mBIO3-BIO1/KAPA: Two are bound into the DAPA-AT catalytic site and two were found within the catalytic site of DTBS (Figure 7; see Supplemental Figure 6 online). The presence of KAPA into the DTB binding site likely results from its structural similarity with DAPA and the high KAPA concentration (5 mM) used for cocrystallization experiments. Two tartrate ions from the crystallization solution are also observed in the DTBS domain active site. In the DTBS active site, KAPA interacts with Thr-24, Thr-59, Ala-141, Ile-142, Ser-143, Ser-195, and a tartrate ion of one monomer, and Gly-223, Leu-222, Ile-225, and Ser-226 of the second monomer. The tartrate molecule is hydrogen bonded to Thr-24 and Lys-28 from the Walker-A motif and to the Asp-66 carboxylate group, Lys-55 ammonium group, and Gly-191 nitrogen atom.
Figure 7.
Views of the DAPA-AT (BIO1) and the DTBS (BIO3) Catalytic Sites Bound to KAPA.
(A) The DAPA-AT catalytic site. Each monomer is colored in light blue or light orange. The residues interacting with KAPA and PLP are shown as sticks.
(B) The DTBS catalytic site. Each monomer is colored in dark blue or orange. The tartrate ion and the residues interacting with KAPA are shown as sticks.
Hydrogen bonds were calculated by COOT with a distance cutoff of 3.2 Å. They are drawn with dashed lines.
In the DAPA-AT active site, KAPA is stabilized by several interactions with Phe-326, Trp-369, Trp-370, Tyr-473, Ala-587, Arg-775, Leu-777, and PLP from one monomer and with Gly-678, His-679, and Ser-680 from the other monomer. Phe-326 is located in a loop connected to the α-helix α9, which links BIO3 to BIO1.
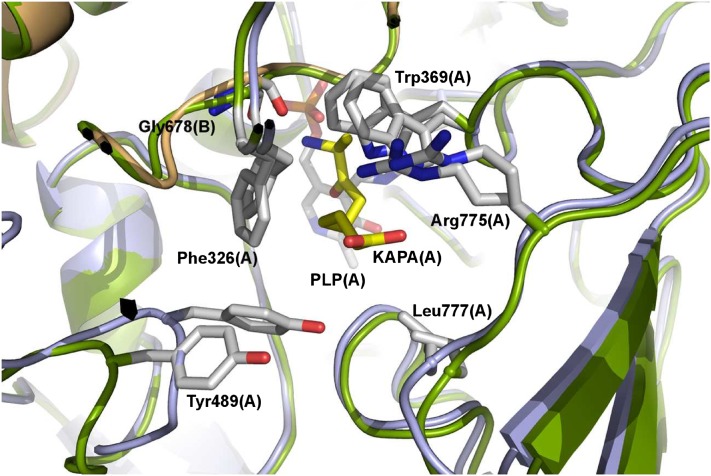
mBIO3-BIO1/KAPA superimposed onto mBIO3-BIO1 with a 0.62 Å rmsd value calculated on Cα. This comparison showed that binding of KAPA leads only to several local conformational changes in the DAPA-AT domain: The Trp-369 side chain rotates to interact with KAPA and the loop connecting α13 to β16 moves toward the binding site and decreases its solvent accessibility (Figure 8). In the DTBS domain, only weak displacements of the main chain are observed (Figure 7B).
Figure 8.
Conformational Changes in the DAPA-AT (BIO1) Catalytic Site upon KAPA Binding.
The unliganded protein is colored in green, and the DAPA-AT bound to KAPA is colored in light blue or light orange. PLP and residues in the active site are drawn as sticks.
Structure of mBIO3-BIO1 Bound to DTB
Two DTB molecules and two tartrate ions were observed bound to the DTBS domains (Figure 9; see Supplemental Figure 6 online). DTB interacts with Thr-24, Gln-58, Thr-59, Ser-143, Gly-191, Ser-195, Pro-196, and tartrate from one monomer and Leu-222, Gly-223, Gly-224, and Ile-225 from the second monomer. The rmsd values with mBIO3-BIO1 and mBIO3-BIO1/KAPA are 0.48 and 0.55 Å, respectively, when calculated on Cα. Conformational changes of the Trp-369 side chain are observed in the DAPA-AT domain as compared with the holoenzyme, although no ligand molecule is bound in the DAPA-AT catalytic site. This observation strongly suggests close communication between both enzyme catalytic sites. Albeit present into the crystallization solution, no ADP was observed in the protein structure resulting from the presence of 0.2 M ammonium tartrate into the crystallization solution that occupied the ATP binding site.
Figure 9.
View of the DTBS (BIO3) Catalytic Site Bound to DTB.
Each monomer is colored in dark blue or orange. The tartrate ion, DTB, and the residues interacting with DTB are shown as sticks. Hydrogen bonds were calculated by COOT with a distance cutoff of 3.2 Å. They are drawn with dashed lines.
Structure Comparisons with Monofunctional Enzymes
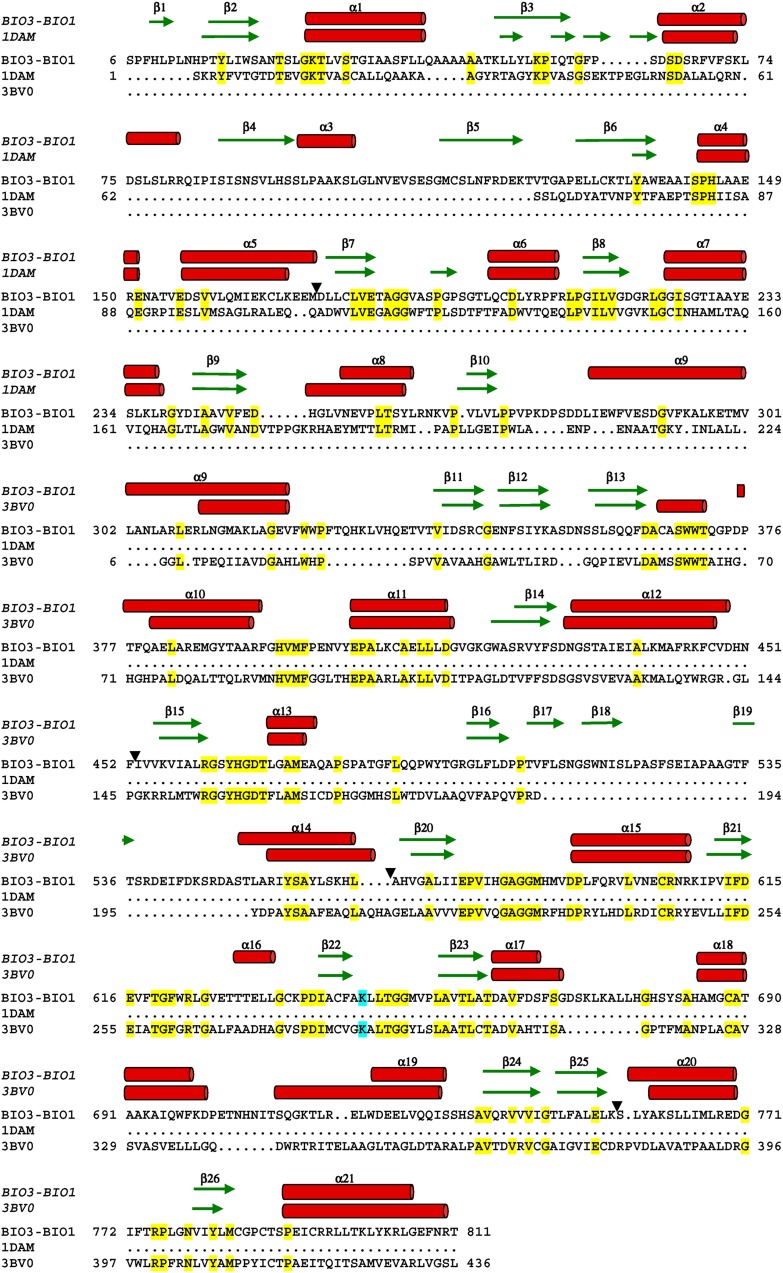
The Arabidopsis DAPA-AT domain from one monomer superimposed with rmsd values in a range 1.78 to 2.01 Å, calculated on Cα, with DAPA-AT from M. tuberculosis (PDB entry: 3BV0) (Dey et al., 2010) and E. coli (PDB entry: 1QJ5) (Käck et al., 1999), respectively. The main structural difference is the additional three β-stranded β-sheet β17-β18-β19 in the DAPA-AT domain of the bifunctional enzyme (Figure 10). The Arabidopsis DTBS domain superimposed with DTBS of known structures with rmsd values in a range from 1.78 Å (PDB entry: 3OF5; DTBS of Francisella tularensis) to 2.47 Å (PDB entry: 3QXX; DTBS of Helicobacter pylori). The Arabidopsis DTBS domain has three more β-strands, β1, β4, and β5, and the α-helix α3 (Figure 10). These low rmsd values reflect the conservation of the BIO3 and BIO1 folds. The dimers of the monofunctional DTBS and DAPA-AT from bacteria superimposed onto Arabidopsis mBIO3-BIO1, and only a few residues are then missing to form the long α-helix α9 connecting BIO3 to BIO1 in mBIO3-BIO1. However, a marked difference between the Arabidopsis and prokaryotic structures is the absence of the crevice at the surface of bacterial DAPA-AT and DTBS proteins.
Figure 10.
Structure-Based Sequence Alignment between mBIO3-BIO1 and DAPA-AT (bioA) from M. tuberculosis (3BV0) (Dey et al., 2010) and DTBS (bioD) from E. coli (1DAM) (Käck et al., 1998).
Only residues observed in the density map are reported in the alignment (gaps are indicated by arrowheads). The conserved residues are highlighted in yellow. The Lys covalently linked to PLP is shaded in cyan. The α-helices and the β-strands are represented by red cylinders and green arrows, respectively.
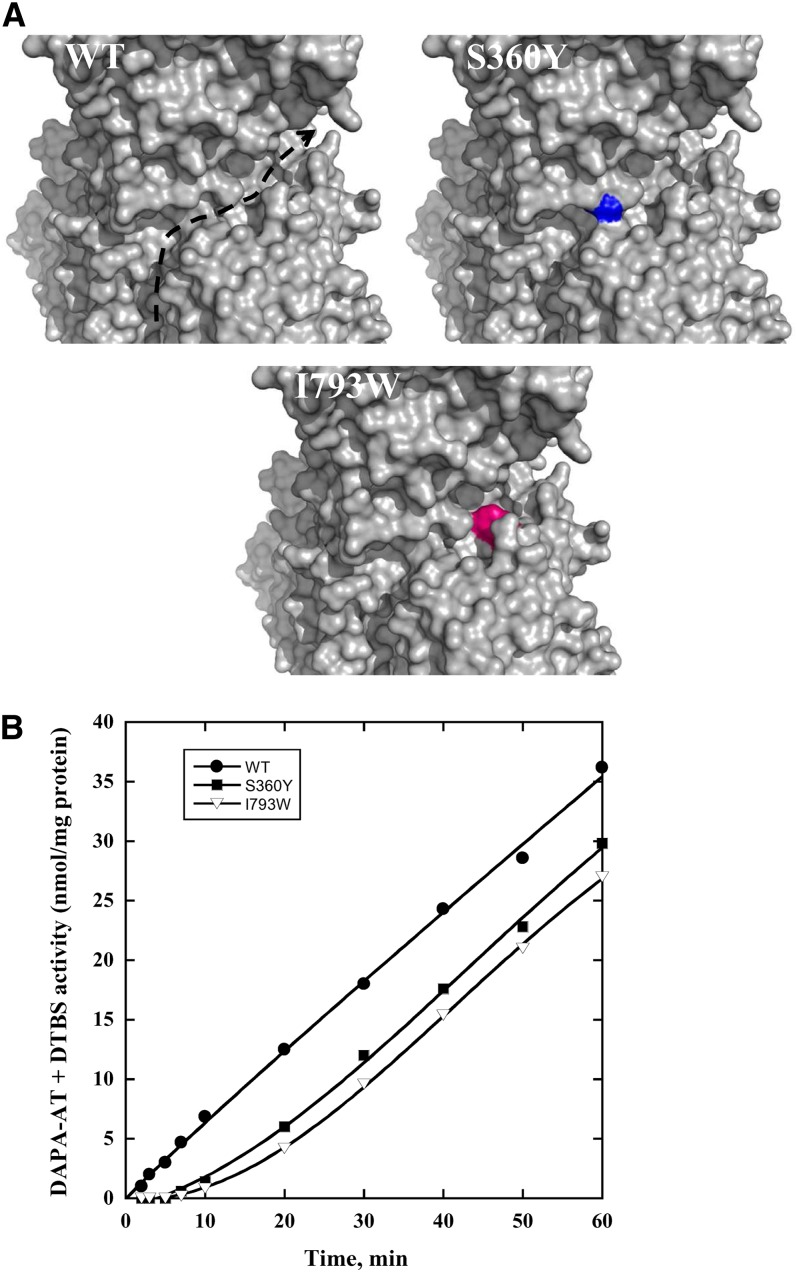
Mutations Obstructing the External Crevice Affect the Channeling
To test the possibility that the external crevice of the Arabidopsis enzyme could be the route for the DAPA intermediate between both plant enzyme actives sites, S360Y and I793W mutants (mature enzyme numbering) were produced and assayed for the overall DAPA-AT + DTBS activity. These amino acid substitutions are predicted to obstruct the observed external crevice (Figure 11A). In contrast with the wild-type enzyme, mutant variants exhibited progress curves with obvious lag phases with extrapolated transient times of ∼10 to 12 min, respectively (Figure 11B), as expected for nonchanneling controls.
Figure 11.
Single Amino Acid Substitutions within the External Crevice of Arabidopsis mBIO3-BIO1 Affect Substrate Channeling.
(A) Close-up view of the crevice of the wild-type (WT) and mutant enzymes. The crevice is shown with an arrow. Tyr-360 and Trp-793 are colored in blue and pink, respectively.
(B) Kinetic study of mBIO3-BIO1 (wild-type, S360Y, and I793W variants) overall catalyzed reaction. DAPA-AT + DTBS activity (time-course formation of DTB) was assayed as in Figure 4A. Data are from a representative experiment repeated three times.
DISCUSSION
The molecular organization of DAPA-AT and DTBS functional domains as a fusion protein, based on homology searches among eukaryotic and prokaryotic genomes, appears to be a common feature to most biotin prototrophic eukaryotes (flowering plants, mosses, green and red algae, and most ascomycete and basidiomycete fungi with sequenced genomes), whereas in prokaryotes, the presence of separate monofunctional BIO1 and BIO3 orthologs seems to be the rule (Rodionov et al., 2002; Hall and Dietrich, 2007; Muralla et al., 2008; Magliano et al., 2011). This suggests that a fusion event between prokaryotic bioD and bioA ancestor genes occurred early in the evolution of modern-day eukaryotes. The ability to produce a bicistronic transcript through differential splicing is probably a more recent event because it seems limited to few angiosperms, including Arabidopsis, Brassica, and rice (Oryza sativa; Muralla et al., 2008). Such read-through transcription of a neighboring tail-to-head tandem genes phenomenon giving rise to mono- and/or bicistronic chimeric mRNA is not uncommon in eukaryotes. For example, computational and experimental analyses estimated that at least 4 to 5% of the tandem gene pairs in the human genome can be eventually transcribed into a single RNA sequence encoding a putative chimeric protein (Parra et al., 2006). Also, up to 155 tandem genes in Arabidopsis and rice plants (110 in Arabidopsis and 45 in rice) could be transcribed into chimeric mRNA potentially encoding chimeric or separate proteins (Shahmuradov et al., 2010). While the functional significance of the production of both mono- and bicistronic chimeric transcripts remains to be established, it may be a means to increase protein variety in eukaryotic genomes.
Here, we demonstrate through immunodetection using antibodies produced with pure recombinant BIO3-BIO1 protein, that in Arabidopsis, the bifunctional fusion protein is the unique enzyme form found in the plant cells. We cannot exclude the possibility that monofunctional BIO3 and BIO1 protein forms are actually translated from the bicistronic transcript, but if so, could be in amounts too low to be accurately detected by an immunodetection approach. Alternatively, separate BIO3 and BIO1 proteins could be produced but rapidly degraded in the plant cell due to instability. Indeed, owing to the overlapping nature of its reading frames, the bicistronic BIO3-BIO1 transcript is predicted to encode a BIO3 protein form presenting a C-terminal extension corresponding to the N terminus of BIO1 domain and a putative N terminus truncated BIO1 domain form lacking regions conserved among all known DAPA-AT and essential for catalysis, which would be catalytically inactive, if actually produced. Interestingly, our initial attempts to overproduce BIO1 protein alone failed. On the other hand, separate long BIO3 protein could be overproduced in an active form in E. coli but proved to be highly susceptible to proteolytic degradation upon purification (see Supplemental Figure 1 online). Another possibility is that bicistronic BIO3-BIO1 transcript could have a function as a large noncoding RNA. Recent evidence points to a widespread role of these molecules in eukaryotic cells (Costa, 2007).
Production of a bifunctional fusion protein through the nonstop transcription of adjacent tandem genes imply that the destination compartment of the protein expressed by the second tail-to-head gene is determined by transit signals of the first gene. Our data are consistent with Arabidopsis BIO3-BIO1 protein being specifically targeted to mitochondria. This underscores the central role played by plant mitochondria in biotin biosynthesis. Indeed, with the noticeable exception of KAPA synthase, the first committed enzyme of biotin synthesis pathway, all the biotin biosynthetic enzymes are localized within mitochondria in Arabidopsis (Baldet et al., 1997; Picciocchi et al., 2001, 2003; Pinon et al., 2005; Arnal et al., 2006). This also emphasizes the complex compartmentalization of the whole biotin network since biotin-utilizing enzyme are scattered among multiple cell compartments (Alban et al., 2000). With few exceptions, such a situation is also prevalent in many plant vitamin pathways, such as those of folates, ascorbate, pantothenate, niacin, or thiamine (Lunn, 2007; Rébeillé et al., 2007).
In this study, we report the biochemical and structural characterizations of the Arabidopsis BIO3-BIO1 protein. The Arabidopsis BIO3-BIO1 protein catalyzes both DAPA-AT and DTBS activities. Kinetics of the DAPA-AT reaction alone suggests that under these assay conditions, DAPA produced is sequestered within the DAPA-AT active site, forming a stable enzyme-product complex preventing further catalytic activity. In the presence of CO2 (a substrate of the DTBS domain), the activity is catalytic, suggesting that DAPA can have access to the DTBS active site, leaving free the DAPA-AT domain for additional turnovers. Finally, the bifunctional enzyme exhibits kinetic cooperativity with respect to all substrates, suggesting conformational changes upon fixation on active sites, consistent with allosteric communication between the two sites. Such conformational changes could favor the transit of DAPA intermediate between both active sites. These changes are particularly well illustrated by the observed kinetics changes relative to ATP when the overall reaction is measured compared with the DTBS reaction alone and by the singular behavior of DAPA intermediate according to the reaction medium conditions. Because DTBS is about 30 times more active than DAPA-AT, it seems likely that the fusion protein arrangement would convey a kinetic improvement owing to proximity effects (reduced soluble phase diffusion) or substrate channeling (direct transfer between catalytic sites).
Several lines of evidence from kinetic and structural studies show that in the course of the overall reaction, DAPA could be directly delivered from the DAPA-AT active site where it is produced to the DTBS active site, without prior diffusion into the bulk solvent. Indeed, when the overall (DAPA-AT + DTBS) reaction is measured in vitro, no delay in the production of DTB, the end product of the whole reaction, is observed (Figure 4). This result contrasts with the expectations for noninteracting DAPA-AT and DTBS activities. In such a situation, the progress curve is predicted to exhibit a lag phase with a transient time (τ) equal to the ratio of Km to Vmax for the second enzyme activity (Ovádi et al., 1989). The value of τ estimated from the kinetic constants in Table 1 is ∼10 min. The absence of such a lag phase in the experimental progress curve for the overall reaction indicates that direct transfer (i.e., channeling) occurs in BIO3-BIO1. Substrate channeling is also supported by the structure analysis of mBIO3-BIO1. The active site for the DAPA-AT domain is a pocket formed by domains of both BIO1 monomers. The active site of each DTBS domain is an open cleft at the interface of both monomers, with more solvent exposed than the DAPA-AT active sites. Both catalytic domains are distant and too far apart (∼48 Å) to allow direct transfer of the DAPA intermediate simply by proximity of the two active sites. DAPA-AT and DTBS active sites are connected by a small groove at the interface of the BIO3-BIO1 monomers. However, in all the structures solved, this groove is interrupted and not wide enough to allow the transfer of a DAPA molecule between both active sites. Thus, this route for the intermediate transfer is hardly predictable unless large protein conformational rearrangements.
Because the three-dimensional structure and the dimer interface are similar to those from bacterial enzymes and no channel or putative channel is observed, the surface was also analyzed. Indeed, surface channeling has been proposed for the bifunctional enzyme dihydrofolate reductase–thymidylate synthase (Knighton et al., 1994). The main difference between the bifunctional and the monofunctional enzymes is an ∼95 Å long and 12 Å average width crevice at the enzyme surface going from the DAPA-AT catalytic site to that of DTBS in each monomer of mBIO3-BIO1. Inspection of the crevice shows that it consisted of polar amino acids corresponding to nonconserved residues belonging to common secondary structures. This polar crevice could be a candidate for the DAPA transfer from DAPA-AT to DTBS. The fact that mutations blocking the trench conduce to the recording of overall DAPA-AT + DTBS activity progress curves exhibiting lag phases with extrapolated transient times close to the theoretical value of τ for noninteracting DAPA-AT and DTBS activities supports this hypothesis.
Interestingly, analysis of the crystal structures also shows that the additional β-strand β-1 and the α-helix α-9 are the key structural elements for acquisition of the bifunctionality. Indeed, α-helix α-9 covalently connects DAPA-AT to DTBS, and the β-strand β-1 of DTBS interacts with the β-strand β-13 of DAPA-AT to stabilize the interaction between the two domains through an antiparallel four-stranded β-sheet together with β-11 and β-12.
Finally, our work raises the question of the biological significance of intermediate channeling, if any, in biotin metabolism. Substrate channeling has many advantages over the free diffusion within the bulk solvent. First, the transit time from one active site to the next is reduced; therefore, unfavorable equilibria, competing enzymatic transformations, or unwanted side reactions can thus be circumvented (Ushiroyama et al., 1992; Herrmann, 1995). Chemically labile intermediates can be protected from breakdown by the aqueous external environment during the transfer between distant active sites (Rudolph and Stubbe, 1995). Sequestration also provides a means of preventing a potentially toxic intermediate product from being released to the solvent (Manjasetty et al., 2003). Last, substrate channeling can trap metabolites with high diffusivity, thus preventing their loss in the surrounding medium or across the cell membranes (Chittur et al., 2001; Barends et al., 2008). DAPA is not known to be particularly labile or toxic and its only fate is biotin synthesis; thus, there is no apparent practical requirement for the sequestration of this intermediate. Because aminotransferases catalyze reversible reactions and because DAPA-AT is much less active than DTBS in Arabidopsis bifunctional enzyme, DAPA trapping in the BIO1 active site and direct transfer to the BIO3 domain upon binding of the BIO3 substrates could be a means of avoiding having the DAPA-AT reaction proceed in the reverse direction, hence increasing the efficiency of the overall reaction. This could be especially critical owing to the low abundance of biotin intermediates in the plant cells (Alban et al., 2000).
In conclusion, we provide an exhaustive biochemical, kinetic, and structural characterization of a bifunctional enzyme of the biotin synthesis pathway. Because this enzyme form is specific to biotin-synthesizing eukaryotes (Hall and Dietrich, 2007; Muralla et al., 2008; Magliano et al., 2011), this characterization of the Arabidopsis enzyme provides a unique working model for ongoing mechanistic studies critical for a better understanding of biotin synthesis and regulation in eukaryotes. Also, elucidation of the structure of a plant bifunctional DTBS–DAPA-AT creates a rich molecular platform that will be useful for the design of new inhibitor families having herbicidal and/or fungal activities.
METHODS
Plant Materials
Arabidopsis thaliana (ecotype Columbia) seeds were grown on soil under greenhouse conditions (23°C with a 16-h photoperiod and a light intensity of 100 µE m−2 s−1) until harvested for analysis. Arabidopsis (ecotype Columbia) cell suspension cultures were grown under continuous white light (40 µE m−2 s−1) at 23°C with rotary agitation at 125 rpm in Gamborg’s B5 medium supplemented with 1 µM 2-naphtalene acetic acid and 1.5% (w/v) Suc.
Chemicals
Rac-KAPA and DAPA were obtained as described previously (Baldet et al., 1993). (S)-KAPA was a kind gift from Olivier Ploux and Stéphane Mann (Mann et al., 2009). Isopropylthio-β-d-galactoside was from Bioprobe Systems. NaH14CO3 (52 mCi/mmol) was from Perkin-Elmer. His binding resin was from Qiagen. All other biochemicals were obtained from Sigma-Aldrich and were the purest grade available.
Protein Determination, SDS-PAGE, and Immunoblot Analysis
Proteins were measured either by the Bradford method (Bradford, 1976) using Bio-Rad protein assay reagent, with γ-globulin as a standard, or for pure proteins by measuring the absorbance at 205 nm (Scopes, 1974). Total soluble proteins from Arabidopsis tissues were extracted by grinding powdered samples in 20 mM MOPS-KOH, pH 7.5, 5% (w/v) glycerol, 1 mM DTT, 1 mM EDTA, 1 mM benzamidine-HCl, 5 mM ε-amino caproic acid, and the complete protease inhibitor cocktail from Roche Applied Science. Samples were centrifuged at 130,000g for 20 min at 4°C, and the supernatant was used as a source of soluble proteins. Arabidopsis cell subfractions were purified as described previously (Pinon et al., 2005). Briefly, chloroplast and mitochondria were isolated from Arabidopsis leaves and purified on Percoll gradients. A cytosolic-enriched fraction was prepared from Arabidopsis cultured cells by gentle rupture of isolated protoplasts followed by differential centrifugation steps. Purity of cytosol, chloroplast, and mitochondrial preparations was assessed by measurement of specific subcellular markers and was found to be higher that 95%, in each case (Pinon et al., 2005). Proteins from Arabidopsis cell subfractions were resolved by SDS-PAGE and electroblotted to nitrocellulose membrane. The blots were probed using affinity-purified BIO3-BIO1 antibodies produced in guinea pigs (Charles River Laboratories, France) and horseradish peroxidase–conjugated anti-guinea pig IgGs, and detection was achieved by chemifluorescence using the ECL Plus Western Blotting detection reagents and a Typhoon 9400 scanner (GE Healthcare Europe).
Cloning, Protein Expression, and Purification
Sequences of primers used in this study are listed in Supplemental Table 1 online. Full-length monocistronic and bicistronic BIO3-BIO1 cDNAs and cDNA encoding mature BIO3-BIO1 protein (mBIO3-BIO1; residues 23 to 833) were obtained by PCR amplification of an RT reaction using total RNAs isolated from Arabidopsis seedlings (RNeasy plant mini kit from Qiagen). Primers were designed on the basis of the predicted sequence of BIO3-BIO1 locus and available cDNA sequences (Muralla et al., 2008) and generated appropriate restriction sites for the cloning of cDNA into expression vectors. The PCR fragments were first cloned into pPCR-Script (Stratagene) for maintenance and sequenced in both strands (Beckman Coulter Genomics). For overexpression, the monocistronic BIO3-BIO1 sequences (full length or devoid of the transit peptide sequence) were inserted between the NdeI and SacI sites of pET-28b (Novagen). This procedure added an N-terminal hexa-His tag. The F326Y, S360Y, and I793W variants of mBIO3-BIO1 were constructed using the Quikchange II site-directed mutagenesis kit (Stratagene) and the pET-mBIO3-BIO1 plasmid as a matrix. The resulting pET-BIO3-BIO1 constructs were introduced first into Escherichia coli DH5α cells and then into the E. coli overexpression host Rosetta 2 (DE3) (Stratagene).
The transformed cell cultures were grown at 37°C in Luria-Bertani medium supplemented with 10 µM pyridoxine, 15 µM thiamin, and the appropriate antibiotics until A600 was 0.6, at which point isopropylthio-β-d-galactoside was added to a final concentration of 0.4 mM. Incubations were continued for 40 h at 20°C. Pelleted cells from 2.5-liter cultures were resuspended in 30 mL of buffer A (20 mM Tris-HCl, pH 8, 500 mM NaCl, 100 µM PLP, and EDTA-free complete protease inhibitor cocktail from Roche Applied Science) supplemented with 10 mM imidazole and disrupted by sonication with a Vibra-Cell disrupter (Branson Ultrasonics) The soluble protein extracts were separated from the cell debris by centrifugation at 18,000g for 30 min and applied onto nickel-nitrilotriacetic acid-agarose columns (1 × 3 cm) previously equilibrated with the same buffer. The columns were washed with buffer A supplemented with 50 mM imidazole. The recombinant proteins were then eluted with buffer A supplemented with 250 mM imidazole. Fractions containing proteins were pooled and dialyzed against Tris-buffered saline. Purifications were achieved by size exclusion chromatography (Superdex 200 HiLoad, 26/60; GE Healthcare) in buffer B (20 mM Tris-HCl, pH 8, 150 mM NaCl, 10 µM PLP, and 10% [v/v] glycerol). Purified enzymes were concentrated by filtration (Macrosep, 30-kD cutoff; Pall Filtron), aliquoted, and stored at –80°C until use.
The seleno-Met F326Y mutant enzyme ([SeMet]-mBIO3-BIO1) was produced and purified similarly except that the expression was performed in the E. coli B834 (DE3) pLys S host strain, and growth was conducted in 1 liter of a minimal medium containing 40 mg of seleno-Met.
The E. coli bioD JW0761 strain carrying the pCA24N-bioD plasmid encoding E. coli DTBS headed by an N-terminal hexa-His tag was obtained from the National BioResource Project: E. coli (National Institute of Genetics). BioD protein was produced and purified in the same way as BIO3-BIO1 proteins, except for the omission of PLP in purification buffers.
GFP Fusion Targeting Analyses
Primers were designed to amplify the sequence encoding for the complete BIO3-BIO1 protein (see Supplemental Table 1 online) and allowed its cloning in frame to the 5′ extremity of the GFP-coding region in the pTH2 vector, at SalI and NcoI sites. The GFP reporter plasmid pTH2 expressed an engineered version of GFP under the control of the cauliflower mosaic virus 35S promoter (Chiu et al., 1996). The transit peptide sequences of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase from Arabidopsis (55 residues, ATS1A gene; GenBank accession number X13611) and dihydropterin pyrophosphokinase-dihydropteroate synthase from pea (Pisum sativum; 28 residues; (Rébeillé et al., 1997)) were used as controls for the targeting of GFP to plastids and mitochondria, respectively. Transient transformation of Arabidopsis protoplasts prepared from a 4-d-old cell suspension culture was achieved using 40 µg of plasmid construct by the polyethylene glycol method, essentially as described (Pinon et al., 2005). Transformed cells were incubated at 23°C for 36 h and analyzed by epifluorescence microscopy using a Zeiss Axioplan2 fluorescence microscope, and images were captured with a digital charge-coupled device camera (Hamamatsu). The filter sets used were Zeiss filter set 13, 488013-0000 (exciter BP 470/20, beamsplitter FT 493, emitter BP 505-530) and Zeiss filter set 15, 488015-0000 (exciter BP 546/12, beamsplitter FT 580, emitter LP 590) for GFP and chlorophyll fluorescence, respectively.
Activity Measurements
The overall (DAPA-AT + DTBS) reaction was assayed by measuring the formation of acid-stable [14C]-DTB from acid-labile H14CO3. The standard assay consisted of 100 mM EPPS, pH 8.6, 1 mM DTT, 0.1 M NaCl, 0.2 mM PLP, 3 mM AdoMet, 20 µM (S)-KAPA, 0.3 mM ATP, 5 mM MgCl2, 10 mM NaH14CO3 (1.6 mCi/mmol), and 0.4 nmol/mL pure mBIO3-BIO1. The assays were initiated by the addition of the enzyme. After 2 to 120 min incubation at 37°C, 500 μL aliquots of the reaction mixture were mixed with 250 μL of 20% (v/v) acetic acid to stop the reaction. The solution was taken to dryness, and the acid-stable radioactivity was quantified in a liquid scintillation counter. Duplicate assays without KAPA were run as controls.
DAPA-AT activity was assayed by measuring the amount of [14C]-DTB formed in a discontinuous coupled reaction using as the coupling enzyme monofunctional DTBS (bioD) from E. coli, which catalyzes the synthesis of DTB from DAPA and CO2 in the presence of ATP-Mg. The standard assay consisted of 100 mM EPPS, pH 8.6, 1 mM DTT, 0.1 M NaCl, 0.2 mM PLP, 3 mM AdoMet, 20 µM (S)-KAPA, and 0.2 to 2 nmol/mL pure mBIO3-BIO1. The reaction was performed at 37°C and arrested by heating 500-μL aliquots of the mixture for 2 min at 80°C and filtration through a 30-kD cutoff membrane (Nanosep; Pall Filtron) to remove the proteins. Duplicate assays without KAPA or AdoMet were run as controls. The amount of DAPA formed was determined by radiochemical assay after complete conversion to DTB, by incubating the filtrate for another 2 h in the presence of 0.3 mM ATP, 5 mM MgCl2, 10 mM NaH14CO3 (1.6 mCi/mmol), and 4 nmol pure bioD. The reaction was stopped by acetic acid treatment and acid-stable radioactivity quantified as described above.
DTBS activity was assayed as described above for the overall (DAPA-AT + DTBS) reaction with the following modifications: The standard assay consisted of 100 mM HEPES, pH 8, 1 mM DTT, 0.1 M NaCl, 0.3 mM ATP, 5 mM MgCl2, 200 µM DAPA, 10 mM NaH14CO3 (1.6 mCi/mmol), and 1 nmol/mL pure enzyme. The reaction was stopped by mixing 100-μL aliquots of the reaction mixture with 250 μL of 10% (v/v) acetic acid. Duplicate assays without DAPA were run as controls.
Kinetics of product formation were linear and rates varied linearly with enzyme concentration, demonstrating adherence to steady state conditions. Kinetic data were fitted to the appropriate rate equations by nonlinear regression analyses using the KaleidaGraph program (Abelbeck Software). Production of [14C]-DTB as the end product of the reactions was controlled by HP TLC analysis and phosphor imaging detection according to the procedure described by Picciocchi et al. (2001).
Crystallization and Data Collection
Searches for crystallization conditions of native mBIO3-BIO1 were performed using the TECAN robot at the Institut de Biologie Structurale in Grenoble, France. One microliter of protein (10 mg/mL) was mixed with 1 μL crystallization solution. The mutation F326Y (mature protein numbering) was found to improve the recombinant enzyme solubility and stability, with no significant impact in Arabidopsis on the enzyme kinetic parameters (Table 1). Then it was subsequently used for crystallization of [SeMet]-mBIO3-BIO1. The native protein (mBIO3-BIO1) was crystallized in 0.1 M BisTris, pH 5.9, 0.2 M LiSO4, and 15% (w/v) polyethylene glycol 3350. [SeMet]-mBIO3-BIO1 F326Y (7.5 mg/mL) was crystallized in the same conditions with the addition of DTT in a 2 to 10 mM range. Extensive searches of crystallization conditions of mBIO3-BIO1/complexes with either 5 mM DAPA, KAPA, DTB, AdoMet, ATP, AMP-PNP, or CO2, or various combinations of these ligands, were performed using the high-throughput crystallization facilities of EMBL. Good diffracting crystals were obtained for mBIO3-BIO1/KAPA and mBIO3-BIO1/ADP/DTB using 0.2 M ammonium tartrate, pH 6.2, and 15% (w/v) polyethylene glycol 3350. Data for the native protein, [SeMet]-mBIO3-BIO1, and mBIO3-BIO1/KAPA were collected on the FIP-BM30A beamline (Roth et al., 2002) at the European Synchrotron Radiation Facility (Grenoble, France) using an ADSC Q315r charge-coupled device detector. Data for mBIO3-BIO1/ADP/DTB were collected on ID14-1 at the European Synchrotron Radiation Facility using an ADSC Q210 charge-coupled device detector. All the data were processed and scaled using XDS (Kabsch, 1993) (see Supplemental Table 2 online).
Single-Wavelength Anomalous Diffraction Phasing
The phase problem could not be solved using molecular replacement and atomic coordinates of the bacterial enzymes. A three-wavelength multiwavelength anomalous diffraction data set was collected on FIP-BM30A at 2.7-Å resolution, at the peak, inflection, and remote high energy of the Se absorption edge. A total of 32 selenium atoms were found using Phenix (Adams et al., 2002) and the data collected at the peak of the Se absorption edge. The resulting phases were further improved by solvent flattening and density averaging using Phenix allowing for partial automated chain tracing (see Supplemental Table 2 online).
Model Building and Refinement
The polyalanine model of [SeMet]-mBIO3-BIO1 was constructed at 2.7-Å resolution, and side chains were added using COOT (Emsley and Cowtan, 2004). The model was refined by restrained refinement and molecular dynamic using Phenix and water molecules were added, also using Phenix. A simulated-annealing 2Fo-Fc omit electron density map was calculated to check the model.
The phase problem for mBIO3-BIO1 was solved by molecular replacement using PHASER (McCoy et al., 2007) and the [SeMet]-mBIO3-BIO1 F326Y atomic coordinates as template. The phase problem for mBIO3-BIO1/ADP/DTB and mBIO3-BIO1/KAPA was solved by molecular replacement using PHASER and the mBIO3-BIO1 atomic coordinates as template. All the structures were refined by restrained refinement and molecular dynamic with Phenix and using noncrystallographic symmetry restrains. In the case of mBIO3-BIO1, the noncrystallographic symmetry was applied only in the dimers. Water molecules were added using Phenix. At the end of the refinement, the occupancies of the ligands (Mg2+, SO42−, DTB, KAPA, and tartrate ion) were also refined. The refinement statistics are given in Supplemental Table 3 online.
In all the models, several residues and side chains were not observed into the electron density maps. Also, the electron density for the six His tag and the preceding five residues at the N terminus was not visible. The structure comparisons with the monofunctional enzymes were performed using the most complete dimer (dimer A/B) of mBIO3-BIO1 (crystals obtained in space group P1). This dimer was also chosen to describe the Arabidopsis mBIO3-BIO1 structure. The structure superimposition was performed using PDBeFold (Krissinel and Henrick, 2004) at European Bioinformatics Institute. All the models were checked using PROCHECK (Laskowski et al., 1993) and WHATCHECK (Hooft et al., 1996). All the figures were prepared using Pymol (Delano, 2002).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL or Arabidopsis Genome Initiative databases under the following accession numbers: BIO3-BIO1 (At5g57590; BOF481); monocistronic BIO3-BIO1 (HQ857557); bicistronic BIO3-BIO1 (HQ857558); E. coli BioD (1DAM; NP_415299.1); and Mycobacterium tuberculosis BioA (3BV0; AAK45886.1). The [SeMet]-mBIO3-BIO1 F326Y, mBIO3-BIO1, mBIO3-BIO1/KAPA, and mBIO3-BIO1/ADP/DTB atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession code numbers 4A0F, 4A0G, 4A0H, and 4A0R, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of BIO3-BIO1 and BIO3 Production in E. coli Using Polyclonal Antibodies Raised against Recombinant Arabidopsis BIO3-BIO1 Protein.
Supplemental Figure 2. Expression Analysis of Arabidopsis Bicistronic and Monocistronic BIO3-BIO1 mRNAs by Real-Time PCR.
Supplemental Figure 3. Part of an Amino Acid Alignment of DAPA Aminotransferases from a Selection of Bacteria, Plants, and Fungi, Highlighting Residue Tyr-17 (E. coli Numbering).
Supplemental Figure 4. Topology of mBIO3-BIO1.
Supplemental Figure 5. View of the Crevice at the Surface of mBIO3-BIO1.
Supplemental Figure 6. Simulated Annealing Omit Maps for the mBIO3-BIO1 Ligands.
Supplemental Table 1. Synthetic Oligonucleotides Used in This Study.
Supplemental Table 2. Data Collection.
Supplemental Table 3. Refinement Statistics.
Supplementary Material
Acknowledgments
We thank Stéphane Mann and Olivier Ploux for supplying (S)-KAPA substrate and Adeline Y. Robin for her help in the preliminary crystallogenesis experiments. We also thank the National BioResource Project (National Institute of Genetics, Japan) for the provision of the E. coli bioD JW0761 strain. We thank Stéphane Ravanel, Gilles Curien, and Michel Matringe for critical reading and helpful comments on the article.
AUTHOR CONTRIBUTIONS
D.C., R.D., and C.A. designed the research. D.C., R.D., V.P., and C.A. performed the research. D.C., R.D., C.M., J.-L.F., and C.A. analyzed data. D.C. and C.A. wrote the article with contribution from R.D.
Glossary
- KAPA
7-keto-8-aminopelargonic acid
- DAPA-AT
7,8-diaminopelargonic acid aminotransferase
- DTBS
dethiobiotin synthetase
- PLP
pyridoxal 5′-phosphate
- AdoMet
S-adenosyl-l-Met
- GFP
green fluorescent protein
- HP TLC
high-performance thin layer chromatography
- DTB
dethiobiotin
- [SeMet]
selenomethionine
- rmsd
root mean square deviation
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. (2002). PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Alban C., Job D., Douce R. (2000). Biotin metabolism in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 17–47 [DOI] [PubMed] [Google Scholar]
- Alexeev D., Baxter R.L., Campopiano D.J., McAlpine R.S., McIver L., Sawyer L. (1998). Rational design of an inhibitor of dethiobiotin synthetase; Interaction of 6-hydroxypyrimidin-4(3H)-one with the adenine base binding site. Tetrahedron 54: 15891–15898 [Google Scholar]
- Alexeev D., Baxter R.L., Smekal O., Sawyer L. (1995). Substrate binding and carboxylation by dethiobiotin synthetase—A kinetic and X-ray study. Structure 3: 1207–1215 [DOI] [PubMed] [Google Scholar]
- Arnal N., Alban C., Quadrado M., Grandjean O., Mireau H. (2006). The Arabidopsis Bio2 protein requires mitochondrial targeting for activity. Plant Mol. Biol. 62: 471–479 [DOI] [PubMed] [Google Scholar]
- Baldet P., Alban C., Douce R. (1997). Biotin synthesis in higher plants: Purification and characterization of bioB gene product equivalent from Arabidopsis thaliana overexpressed in Escherichia coli and its subcellular localization in pea leaf cells. FEBS Lett. 419: 206–210 [DOI] [PubMed] [Google Scholar]
- Baldet P., Gerbling H., Axiotis S., Douce R. (1993). Biotin biosynthesis in higher plant cells. Identification of intermediates. Eur. J. Biochem. 217: 479–485 [DOI] [PubMed] [Google Scholar]
- Barends T.R., Dunn M.F., Schlichting I. (2008). Tryptophan synthase, an allosteric molecular factory. Curr. Opin. Chem. Biol. 12: 593–600 [DOI] [PubMed] [Google Scholar]
- Beckett D. (2007). Biotin sensing: Universal influence of biotin status on transcription. Annu. Rev. Genet. 41: 443–464 [DOI] [PubMed] [Google Scholar]
- Berkovitch F., Nicolet Y., Wan J.T., Jarrett J.T., Drennan C.L. (2004). Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303: 76–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breen R.S., Campopiano D.J., Webster S., Brunton M., Watt R., Baxter R.L. (2003). The mechanism of 7,8-diaminopelargonate synthase; the role of S-adenosylmethionine as the amino donor. Org. Biomol. Chem. 1: 3498–3499 [DOI] [PubMed] [Google Scholar]
- Chittur S.V., Klem T.J., Shafer C.M., Davisson V.J. (2001). Mechanism for acivicin inactivation of triad glutamine amidotransferases. Biochemistry 40: 876–887 [DOI] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Costa F.F. (2007). Non-coding RNAs: Lost in translation? Gene 386: 1–10 [DOI] [PubMed] [Google Scholar]
- Delano W.L. (2002). The PyMOL Molecular Graphics System. (Palo Alto, CA: DeLano Scientific)
- Dey S., Lane J.M., Lee R.E., Rubin E.J., Sacchettini J.C. (2010). Structural characterization of the Mycobacterium tuberculosis biotin biosynthesis enzymes 7,8-diaminopelargonic acid synthase and dethiobiotin synthetase. Biochemistry 49: 6746–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliot A.C., Sandmark J., Schneider G., Kirsch J.F. (2002). The dual-specific active site of 7,8-diaminopelargonic acid synthase and the effect of the R391A mutation. Biochemistry 41: 12582–12589 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Farrar C.E., Siu K.K., Howell P.L., Jarrett J.T. (2010). Biotin synthase exhibits burst kinetics and multiple turnovers in the absence of inhibition by products and product-related biomolecules. Biochemistry 49: 9985–9996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson K.J., et al. (1995). Dethiobiotin synthetase: the carbonylation of 7,8-diaminonanoic acid proceeds regiospecifically via the N7-carbamate. Biochemistry 34: 10976–10984 [DOI] [PubMed] [Google Scholar]
- Hall C., Dietrich F.S. (2007). The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 177: 2293–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 35(Database issue): D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K.M. (1995). The shikimate pathway: Early steps in the biosynthesis of aromatic compounds. Plant Cell 7: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft R.W., Vriend G., Sander C., Abola E.E. (1996). Errors in protein structures. Nature 381: 272. [DOI] [PubMed] [Google Scholar]
- Huang W., Jia J., Gibson K.J., Taylor W.S., Rendina A.R., Schneider G., Lindqvist Y. (1995). Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate. Biochemistry 34: 10985–10995 [DOI] [PubMed] [Google Scholar]
- Izumi Y., Sato K., Tani Y., Ogata K. (1975). 7,8-Diaminopelargonic acid aminotransferase, an enzyme involved in biotin biosynthesis by microorganisms. Agric. Biol. Chem. 39: 175–181 [Google Scholar]
- Kabsch W. (1993). Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26: 795–800 [Google Scholar]
- Käck H., Sandmark J., Gibson K., Schneider G., Lindqvist Y. (1999). Crystal structure of diaminopelargonic acid synthase: Evolutionary relationships between pyridoxal-5′-phosphate-dependent enzymes. J. Mol. Biol. 291: 857–876 [DOI] [PubMed] [Google Scholar]
- Käck H., Sandmark J., Gibson K.J., Schneider G., Lindqvist Y. (1998). Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis. Protein Sci. 7: 2560–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton D.R., Kan C.C., Howland E., Janson C.A., Hostomska Z., Welsh K.M., Matthews D.A. (1994). Structure of and kinetic channelling in bifunctional dihydrofolate reductase-thymidylate synthase. Nat. Struct. Biol. 1: 186–194 [DOI] [PubMed] [Google Scholar]
- Knowles J.R. (1989). The mechanism of biotin-dependent enzymes. Annu. Rev. Biochem. 58: 195–221 [DOI] [PubMed] [Google Scholar]
- Krell K., Eisenberg M.A. (1970). The purification and properties of dethiobiotin synthetase. J. Biol. Chem. 245: 6558–6566 [PubMed] [Google Scholar]
- Krissinel E., Henrick K. (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. (1993). Procheck - A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291 [Google Scholar]
- Lin S., Hanson R.E., Cronan J.E. (2010). Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat. Chem. Biol. 6: 682–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J.E. (2007). Compartmentation in plant metabolism. J. Exp. Bot. 58: 35–47 [DOI] [PubMed] [Google Scholar]
- Magliano P., Flipphi M., Sanglard D., Poirier Y. (2011). Characterization of the Aspergillus nidulans biotin biosynthetic gene cluster and use of the bioDA gene as a new transformation marker. Fungal Genet. Biol. 48: 208–215 [DOI] [PubMed] [Google Scholar]
- Manjasetty B.A., Powlowski J., Vrielink A. (2003). Crystal structure of a bifunctional aldolase-dehydrogenase: Sequestering a reactive and volatile intermediate. Proc. Natl. Acad. Sci. USA 100: 6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S., Colliandre L., Labesse G., Ploux O. (2009). Inhibition of 7,8-diaminopelargonic acid aminotransferase from Mycobacterium tuberculosis by chiral and achiral anologs of its substrate: Biological implications. Biochimie 91: 826–834 [DOI] [PubMed] [Google Scholar]
- Mann S., Ploux O. (2006). 7,8-Diaminoperlargonic acid aminotransferase from Mycobacterium tuberculosis, a potential therapeutic target. Characterization and inhibition studies. FEBS J. 273: 4778–4789 [DOI] [PubMed] [Google Scholar]
- Marquet A. (2010). Biosynthesis of biotin. In Comprehensive Natural Products II, L. Mander and H.-W. Liu, eds (Oxford, UK: Elsevier), pp. 161–180. [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. (2007). Phaser crystallographic software. J. Appl. Cryst. 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R., Chen E., Sweeney C., Gray J.A., Dickerman A., Nikolau B.J., Meinke D. (2008). A bifunctional locus (BIO3-BIO1) required for biotin biosynthesis in Arabidopsis. Plant Physiol. 146: 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovádi J., Tompa P., Vértessy B., Orosz F., Keleti T., Welch G.R. (1989). Transient-time analysis of substrate-channelling in interacting enzyme systems. Biochem. J. 257: 187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Reymond A., Dabbouseh N., Dermitzakis E.T., Castelo R., Thomson T.M., Antonarakis S.E., Guigó R. (2006). Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 16: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton D.A., Schetter A.L., Franzmann L.H., Nelson K., Ward E.R., Meinke D.W. (1998). An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 116: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton D.A., Volrath S., Ward E.R. (1996). Complementation of an Arabidopsis thaliana biotin auxotroph with an Escherichia coli biotin biosynthetic gene. Mol. Gen. Genet. 251: 261–266 [DOI] [PubMed] [Google Scholar]
- Picciocchi A., Douce R., Alban C. (2001). Biochemical characterization of the Arabidopsis biotin synthase reaction. The importance of mitochondria in biotin synthesis. Plant Physiol. 127: 1224–1233 [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A., Douce R., Alban C. (2003). The plant biotin synthase reaction. Identification and characterization of essential mitochondrial accessory protein components. J. Biol. Chem. 278: 24966–24975 [DOI] [PubMed] [Google Scholar]
- Pinon V., Ravanel S., Douce R., Alban C. (2005). Biotin synthesis in plants. The first committed step of the pathway is catalyzed by a cytosolic 7-keto-8-aminopelargonic acid synthase. Plant Physiol. 139: 1666–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F., Alban C., Bourguignon J., Ravanel S., Douce R. (2007). The role of plant mitochondria in the biosynthesis of coenzymes. Photosynth. Res. 92: 149–162 [DOI] [PubMed] [Google Scholar]
- Rébeillé F., Macherel D., Mouillon J.M., Garin J., Douce R. (1997). Folate biosynthesis in higher plants: Purification and molecular cloning of a bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/7,8-dihydropteroate synthase localized in mitochondria. EMBO J. 16: 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D.A., Mironov A.A., Gelfand M.S. (2002). Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 12: 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M., et al. (2002). FIP: A highly automated beamline for multiwavelength anomalous diffraction experiments. Acta Crystallogr. D Biol. Crystallogr. 58: 805–814 [DOI] [PubMed] [Google Scholar]
- Rudolph J., Stubbe J. (1995). Investigation of the mechanism of phosphoribosylamine transfer from glutamine phosphoribosylpyrophosphate amidotransferase to glycinamide ribonucleotide synthetase. Biochemistry 34: 2241–2250 [DOI] [PubMed] [Google Scholar]
- Sandmark J., Eliot A.C., Famm K., Schneider G., Kirsch J.F. (2004). Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis. Biochemistry 43: 1213–1222 [DOI] [PubMed] [Google Scholar]
- Schneider G., Lindqvist Y. (2001). Structural enzymology of biotin biosynthesis. FEBS Lett. 495: 7–11 [DOI] [PubMed] [Google Scholar]
- Schneider T., Dinkins R., Robinson K., Shellhammer J., Meinke D.W. (1989). An embryo-lethal mutant of Arabidopsis thaliana is a biotin auxotroph. Dev. Biol. 131: 161–167 [DOI] [PubMed] [Google Scholar]
- Scopes R.K. (1974). Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 59: 277–282 [DOI] [PubMed] [Google Scholar]
- Shahmuradov I.A., Abdulazimova A.U., Solovyev V.V., Qamar R., Chohan S.N., Aliyev J.A. (2010). Mono- and bi-cistronic chimeric mRNAs in Arabidopsis and rice genomes. Applied and Computational Mathematics 9: 66–81 [Google Scholar]
- Shellhammer J., Meinke D. (1990). Arrested embryos from the bio1 auxotroph of Arabidopsis thaliana contain reduced levels of biotin. Plant Physiol. 93: 1162–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit W.R., Entcheva P. (2003). Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 61: 21–31 [DOI] [PubMed] [Google Scholar]
- Ushiroyama T., Fukushima T., Styre J.D., Spivey H.O. (1992). Substrate channeling of NADH in mitochondrial redox processes. Curr. Top. Cell. Regul. 33: 291–307 [DOI] [PubMed] [Google Scholar]
- Van Arsdell S.W., Perkins J.B., Yocum R.R., Luan L., Howitt C.L., Chatterjee N.P., Pero J.G. (2005). Removing a bottleneck in the Bacillus subtilis biotin pathway: bioA utilizes lysine rather than S-adenosylmethionine as the amino donor in the KAPA-to-DAPA reaction. Biotechnol. Bioeng. 91: 75–83 [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste M., Runswick M.J., Gay N.J. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Sandalova T., Lohman K., Lindqvist Y., Rendina A.R. (1997). Active site mutants of Escherichia coli dethiobiotin synthetase: effects of mutations on enzyme catalytic and structural properties. Biochemistry 36: 4751–4760 [DOI] [PubMed] [Google Scholar]
- Zempleni J. (2005). Uptake, localization, and noncarboxylase roles of biotin. Annu. Rev. Nutr. 25: 175–196 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.