This work identifies the roles of Arabidopsis CDKF;1 and CDKD protein kinases in the regulation of position-specific Ser phosphorylation of RNA polymerase II C-terminal domain (RNAPII CTD) and demonstrates that changes in the deposition of RNAPII CTD phosphoserine marks alter cotranscriptional processing and stability of a set of small RNAs and transcripts involved in their biogenesis.
Abstract
Phosphorylation of conserved Y1S2P3T4S5P6S7 repeats in the C-terminal domain of largest subunit of RNA polymerase II (RNAPII CTD) plays a central role in the regulation of transcription and cotranscriptional RNA processing. Here, we show that Ser phosphorylation of Arabidopsis thaliana RNAPII CTD is governed by CYCLIN-DEPENDENT KINASE F;1 (CDKF;1), a unique plant-specific CTD S7-kinase. CDKF;1 is required for in vivo activation of functionally redundant CYCLIN-DEPENDENT KINASE Ds (CDKDs), which are major CTD S5-kinases that also phosphorylate in vitro the S2 and S7 CTD residues. Inactivation of CDKF;1 causes extreme dwarfism and sterility. Inhibition of CTD S7-phosphorylation in germinating cdkf;1 seedlings is accompanied by 3′-polyadenylation defects of pre-microRNAs and transcripts encoding key regulators of small RNA biogenesis pathways. The cdkf;1 mutation also decreases the levels of both precursor and mature small RNAs without causing global downregulation of the protein-coding transcriptome and enhances the removal of introns that carry pre-microRNA stem-loops. A triple cdkd knockout mutant is not viable, but a combination of null and weak cdkd;3 alleles in a triple cdkd123* mutant permits semidwarf growth. Germinating cdkd123* seedlings show reduced CTD S5-phosphorylation, accumulation of uncapped precursor microRNAs, and a parallel decrease in mature microRNA. During later development of cdkd123* seedlings, however, S7-phosphorylation and unprocessed small RNA levels decline similarly as in the cdkf;1 mutant. Taken together, cotranscriptional processing and stability of a set of small RNAs and transcripts involved in their biogenesis are sensitive to changes in the phosphorylation of RNAPII CTD by CDKF;1 and CDKDs.
INTRODUCTION
The RNA polymerase II C-terminal domain (RNAPII CTD) is composed of Y1S2P3T4S5P6S7 heptapeptide repeats that are highly conserved in eukaryotes (Buratowski, 2009). Position-specific phosphorylation of RNAPII CTD repeats provides a dynamic code for directing and integrating sequential events in transcription, cotranscriptional RNA processing, and chromatin modifications (Sims et al., 2007; Buratowski, 2009; Lenasi and Barboric, 2010). During preinitiation complex assembly, unphosphorylated RNAPII interacts with Mediator, which recruits the general transcription factor TFIIH and associated cyclin H–dependent Kin28/CDK7 kinases that phosphorylate the CTD Ser-5 (S5) and S7 positions (Akhtar et al., 2009; Glover-Cutter et al., 2009; Kim et al., 2009; Boeing et al., 2010; Egloff et al., 2010). In yeast and mammals, deposition of the RNAPII CTD phosphoserine-5 (S5P) mark facilitates promoter clearing and is required for recruitment and activation of capping enzymes that attach a methylguanosine cap to the 5′-ends of nascent transcripts (Ho and Shuman, 1999; Perales and Bentley, 2009; Ghosh et al., 2011). The S7P mark facilitates RNAPII binding of the Integrator complex mediating specific 3′-end processing of small nuclear U1/2 RNAs (Chapman et al., 2007; Egloff et al., 2007, 2010). In addition to Kin28, Bur1 (for Bypass UAS Requirement1) was also identified as a CTD S7-kinase in yeast and proposed to stimulate transcription elongation by its RNAPII association within the coding regions of genes (Tietjen et al., 2010). S2 residues of RNAPII CTD are phosphorylated by the yeast Ctk1 (for Carboxy-Terminal domain Kinase I) and mammalian CDK9 subunits of the P-TEFb positive transcription elongation factor during transcription elongation and termination as well as direct 3′-end processing of transcripts (Ahn et al., 2004; Peterlin and Price, 2006; Egloff and Murphy, 2008; Kim et al., 2010; Mayer et al., 2010; Tietjen et al., 2010). Recently, Hsin et al. (2011) reported that phosphorylation of the CTD Thr-4 residue by CDK9 facilitates the recruitment of 3′-end processing factors to histone genes.
Among the RNAPII CTD kinases, the TFIIH-associated Kin28/CDK7 class plays a distinct role in the coordination of transcription and cell cycle by phosphorylation of CDK-type cell cycle kinases. Cyclin-dependent protein kinases (CDKs) are evolutionarily conserved and represent the main regulatory core of eukaryotic cell cycle. Activation of CDKs requires phosphorylation of a conserved Thr residue in the T-loop by CDK-activating kinases (CAKs) and binding to cyclins (Dewitte and Murray, 2003; De Veylder et al., 2007; Dissmeyer et al., 2007; Harashima et al., 2007; Gutierrez et al., 2009). The Arabidopsis thaliana genome encodes three CAKs (CDKD;1/CAK3, CDKD;2/CAK4, and CDKD;3/CAK2), which are homologous to Kin28 and CDK7 and form different TFIIH-like complexes with cyclin-H;1, TFIIH-2, CAK assembly factor MAT1, and the putative CAK-activating kinase (CAKAK) CDKF;1/CAK1 (Shimotohno et al., 2004, 2006; Van Leene et al., 2010). CDKF;1 appears to be specific to the plant kingdom and phosphorylates the T-loop of CDKD;2 and CDKD;3 in vitro (Shimotohno et al., 2004; Umeda et al., 2005). CDKD;1 and CDKD;3 are localized in nuclei, while CDKF;1 and CDKD;2 show both cytoplasmic and nuclear localization (Shimotohno et al., 2004). Similarly to Kin28/CDK7, the RNAPII CTD is phosphorylated by CDKD;2 and CDKD;3 in vitro (Shimotohno et al., 2004).
In addition to protein coding genes, RNAPII transcribes the precursors of several classes of noncoding RNAs (Voinnet, 2009). So far, little is known about how deposition of specific CTD phosphoserine marks affects the transcription and stability of noncoding RNAs. Two major classes of plant small noncoding RNAs are microRNAs (miRNAs) and short interfering RNAs (siRNAs) that posttranscriptionally regulate a broad range of biological functions (see reviews in Xie et al., 2004, 2010; Voinnet, 2009; Vazquez et al., 2010). The siRNA family is comprised of trans-acting siRNAs (ta-siRNAs), natural-antisense transcript-derived siRNAs (nat-siRNAs), and repeat-associated siRNAs (ra-siRNAs). Precursor microRNAs (pri-miRNAs), ta-siRNAs, and nat-siRNAs are transcribed by RNAPII and cotranscriptionally capped, polyadenylated, and sometimes spliced (Carthew and Sontheimer, 2009; Voinnet, 2009), while ra-siRNA production is mainly dependent on RNAP IV (Djupedal et al., 2005; Kato et al., 2005; Vazquez, 2006; Zheng et al., 2009). Arabidopsis miRNAs located in dsRNA regions of pri-miRNA hairpins are cleaved by DICER-LIKE RIBONUCLEASE III 1 (DCL1) (Kurihara and Watanabe, 2004; Fang and Spector, 2007). Interaction of DCL1 with the dsRNA binding protein HYPONASTIC LEAVES1 (HYL1) and zinc-finger factor SERRATE (SE) is essential for proper trimming and maturation of miRNAs (Kurihara et al., 2006; Machida et al., 2011). In addition, the DCL1-HYL1-SE complex appears to regulate miRNA processing in interaction with the ABH1/CBP80 and CBP20 core subunits of the cap binding complex (CBC) (Laubinger et al., 2008). The nuclear RNA binding protein DAWDLE (DDL) is another DCL1-interacting factor, which is likely implicated in pri-miRNA stabilization (Yu et al., 2008). Whereas the se, cbp80, and cbp20 mutants show enhanced pri-miRNA accumulation and decreased mature miRNA levels, inactivation of DDL causes simultaneous downregulation of unprocessed and mature miRNAs and siRNAs similarly to mutations affecting Mediator (Kim et al., 2008, 2011; Laubinger et al., 2008; Yu et al., 2008). CBC is also required for RNAPII binding of spliceosome components (Görnemann et al., 2005), and Arabidopsis cbp mutants therefore accumulate pre-mRNAs with retained introns (Laubinger et al., 2008). By contrast, association of Drosha/DCL with the spliceosome (Kataoka et al., 2009) interferes with normal spliceosome recruitment causing delayed or alternative splicing of introns located in the vicinity of pre-miRNA loops (Kim and Kim, 2007; Pawlicki and Steitz, 2010).
The CTD of Arabidopsis RNAPII contains 15 consensus and 26 variant heptapeptide repeats (Nawrath et al., 1990), but so far little is known about the specificity and regulation of CTD kinases mediating their position-specific Ser phosphorylation. In this study, we identify the phosphorylation target sites of CDKD and CDKF;1 kinases in RNAPII CTD. Our biochemical and genetic data indicate that CDKF;1 is required for in vivo activation of CDKDs and thus is responsible for the maintenance of phosphoserine marks on the CTD in developing seedlings. Furthermore, we show that mutations of CDKF;1 and CDKDs, altering the Ser phosphorylation pattern of RNAPII CTD, lead to characteristic defects in the transcription and cotranscriptional processing of a set of miRNAs, siRNAs, and some transcripts encoding key components of small RNA biogenesis pathways.
RESULTS
Position-Specific Ser Phosphorylation of RNAPII CTD by the Arabidopsis CDKF;1 and CDKD Kinases
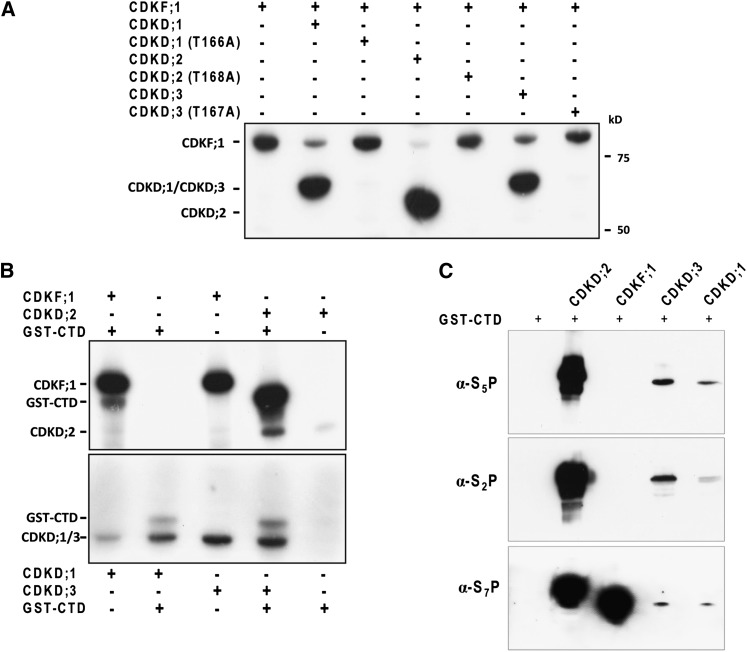
Members of the TFIIH-associated cyclin H–dependent Arabidopsis CDKD kinase family share remarkable sequence conservation (Shimotohno et al., 2003). Nonetheless, previous reports suggest that one of the CDKD family members, CDKD;1, fails to phosphorylate RNAPII CTD and therefore question whether CDKD;1 is a genuine kinase (Shimotohno et al., 2004, 2006). To compare their properties, we purified the catalytic subunits of CDKDs and CDKF;1 in fusion with a N-terminal thioredoxin and C-terminal His6 sandwich tag from Escherichia coli (see Supplemental Figure 1A online) and used them in kinase assays. CDKF;1, which has no known cyclin partner, showed autoactivation by self-phosphorylation and phosphorylated all three CDKDs, including CDKD;1, but not the control thioredoxin protein (Figure 1A; see Supplemental Figure 1B online). Replacement of conserved T-loop Thr residues with Ala inhibited the phosphorylation of mutant CDKD;1 (T166A), CDKD;2 (T168A), and CDKD;3 (T167A) kinases, demonstrating that CDKF;1 phosphorylates the activating T-loops of all three CDKDs (Figure 1A). We also observed that CDKDs undergo low-level autoactivation by self-phosphorylation in the absence of the activating kinase CDKF;1 and associated cyclin H and MAT1 subunits (Figure 1B). All three CDKDs, as well as CDKF;1, phosphorylated a glutathione S-transferase (GST)-CTD substrate, which carried the full-length C-terminal domain of Arabidopsis RNAPII (Figure 1B). Control assays indicated that the observed kinase activities were due to either potential bacterial kinase contamination of purified GST-CTD or phosphorylation of the control GST tag (see Supplemental Figure 1B online). Immunoblotting of GST-CTD kinase reactions with monoclonal antibodies (3E10, 3E8, and 4E12) that specifically recognize the S2P, S5P, and S7P CTD marks (Chapman et al., 2007) indicated that CDKD;1, CDKD;2, and CDKD;3 phosphorylate all three Ser positions of the CTD heptapeptide repeats (Figure 1C). Phosphorylation by CDKD;2 resulted in a higher gel mobility shift of GST-CTD substrate compared with the other kinase reactions, indicating that CDKD;2 had higher CTD kinase activity compared with CDKD;1, and CDKD;3.
Figure 1.
Position-Specific Ser Phosphorylation of RNAPII CTD by CDKD and CDKD-Activating CDKF;1 Kinases.
(A) CDKF;1 phosphorylation of CDKDs is abolished by Ala exchange of conserved T-loop Thr residues indicated in parentheses. kD, molecular mass standard.
(B) CDKF;1 and CDKDs are self-activated by autophosphorylation and phosphorylate the GST-CTD substrate. Kinase-substrate combinations used in the assays are indicated.
(C) Immunoblotting of CDKF;1 and CDKD GST-CTD kinase assays with antibodies recognizing specifically the CTD S2P, S5P, and S7P positions. See also Supplemental Figure 4 online.
Remarkably, CDKF;1, which was previously reported to lack CTD kinase activity (Shimotohno et al., 2004, 2006), also phosphorylated the GST-CTD substrate. Phosphorylation of GST-CTD by CDKF;1 was detectable only with the S7P-specific antibody, indicating that CDKF;1 is a monospecific CTD S7-kinase (Figure 1C). The finding that CDKF;1 is an active RNAPII CTD kinase correlated with the earlier observation that CDKF;1 can complement the fission yeast mcs6 mutation (Umeda et al., 1998; Shimotohno et al., 2004). Mcs6 phosphorylates the RNAPII CTD similarly to its budding yeast Kin28 and human CDK7 homologs (Serizawa et al., 1995; Feaver et al., 1997). Two bootstrap consensus phylogenetic trees created by multiple alignment of yeast, Arabidopsis, and human CAK and CAKAK protein sequences using the COBALT algorithm indicated that CDKF;1 has higher homology to Kin28 and Mcs6 compared with members of the CDKD kinase family (see Supplemental Figures 2, 3A, and 3B online). Comparative analysis of functionally important kinase domains revealed that CDKF;1 contains an unusual insertion of 111 amino acids (Umeda et al., 1998, 2005) and changes in the structures of activation loop and substrate binding pocket (see Supplemental Figure 4 online), suggesting that CDKF;1 has some unique features compared with other CTD kinases.
The results of CTD phosphorylation specificities of CDKD;2 and CDKF;1 were further corroborated by performing in vitro kinase assays with synthetic peptides that carried either consensus or modified CTD heptad dimers fused to C-terminal His6 tags (see Supplemental Figure 1C online). Compared with the wild-type Y1S2P3T4S5P6S7 dimer (peptide 1), CDKD;2 showed very low CTD S7-kinase activity with peptide 2, in which both S2 and S5 positions were replaced with Ala (A). CDKD;2 phosphorylated peptide 4, carrying S2A replacements, with higher efficiency compared with peptide 3, containing S5A exchanges. Thus, CDKD;2 appeared to phosphorylate the S5 positions with preference when S7 residues were present. Compared with wild-type CTD heptads, low-level phosphorylation of mutant peptide variants by CDKD;2 also suggested that initial deposition of CTD S2P or S5P marks could enhance subsequent S7 phosphorylation. Analogous synergy between phosphoserine marks was demonstrated by studies of yeast CTD kinases that prefer partially phosphorylated CTD heptapeptide substrates (Jones et al., 2004; Akhtar et al., 2009). In comparison, CDKF;1 phosphorylated the wild-type and mutant CTD peptides with comparable efficiencies, indicating that S2A and S5A amino acid exchanges in the CTD heptads had no effect on the activity of this monospecific CTD S7-kinase (see Supplemental Figure 1C online). As a further scenario for synergistic phosphorylation of the CTD, this suggested that deposition of the S7P mark by CDKF;1 might facilitate the phosphorylation of CTD S2 or S5 residues by the CDKF;1-activated CDKD kinases.
The results obtained so far indicated that CDKF;1 phosphorylated the T-loops of CDKDs but was not able to phosphorylate the S2 and S5 residues of the CTD, whereas CDKDs phosphorylated all three Ser positions of CTD heptapeptide repeats. This logically led to the question whether CDKF;1 was indeed an upstream CDKD-activating kinase that could stimulate the S2- and S5-kinase activities of CDKDs. Therefore, we performed further GST-CTD kinase assays using a combination of different CDKDs with CDKF;1. In fact, CDKF;1 highly increased both S2- and S5-kinase activities of all three CDKDs, confirming that CDKF;1 is a CDKD/CAKAK (see Supplemental Figures 5A to 5D online).
Developmental Effects of cdkf;1 and cdkd Mutations Suggest Defects in the Small RNA Biogenesis Pathways
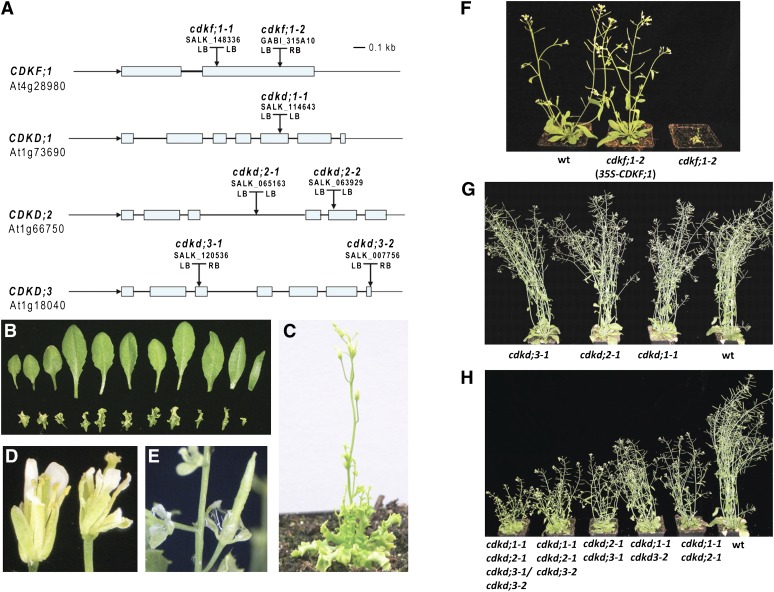
Inactivation of CDKF;1 by T-DNA insertion mutations (cdkf;1-1 and cdkf;1-2; Figure 2A) altered the leaf developmental program of seedlings, leading to the formation of curling serrated leaves (Figures 2B and 2C). As reported previously (Takatsuka et al., 2009), the cdkf;1 mutations also caused cell cycle–related defects, including the inhibition of cell division and elongation leading to arrested root growth and severe dwarfism. Reciprocal crosses between wild-type and hemizygous cdkf;1+/− lines indicated normal female but 30% reduced male transmission of cdkf;1 alleles. Compared with the wild type, cdkf;1 mutants developed much more slowly (Figure 2F) and flowered later during inductive long-day conditions. Their flowers had normal numbers of floral organs but produced no pollen and displayed reduced elongation of stamen filaments, distorted petals, and asymmetric serrated sepals (Figure 2D). Upon pollination with the wild type, cdkf;1 pistils elongated normally and formed fruits (Figure 2E) but produced no viable embryos or seed. All defective traits of cdkf;1 mutants were corrected to the wild type by genetic complementation through transformation of cdkf;1+/− lines with a wild-type CDKF;1 cDNA expression construct (Figure 2F).
Figure 2.
Developmental Defects of the cdkf;1 and cdkd Mutants.
(A) Schematic maps of cdkf;1 and cdkd T-DNA insertion mutant alleles. Exons, blue boxes; introns, black lines; LB and RB, left and right T-DNA border repeats.
(B) Compared with the wild type (top section), the cdkf;1 mutant develops small curly serrated leaves.
(C) cdkf;1 is severely dwarfed and shows retarded development.
(D) cdkf;1 flowers carry serrated sepals, deformed petals, and stamens with short filaments.
(E) Upon pollination with the wild type, cdkf;1 develops normal looking siliques that contain no seed.
(F) The phenotype of cdkf;1-2 is restored to wild type (wt) by genetic complementation with a wild-type cDNA expression construct controlled by the cauliflower mosaic virus 35S promoter.
(G) cdkd single mutants show no change in development compared with the wild type.
(H) cdkd double and d123* triple mutants display different degrees of growth inhibition. See also Supplemental Figure 5 online.
[See online article for color version of this figure.]
In comparison to cdkf;1, knockout mutants of individual CDKD genes did not differ from the wild type (Figure 2G), but double mutants showed reduced growth rates, suggesting a functional redundancy of CDKDs (Figure 2H). Combination of the cdkd;3-2 allele, causing a partial loss of function due to a T-DNA insertion 6 bp upstream of the stop codon, with the cdkd;3-1 null allele (Figure 2A; see Supplemental Figures 6E and 6F online) in a homozygous cdkd;1-1 cdkd;2-1 background led to partial dwarfism and weaker manifestation of curly serrated leaf phenotype compared with cdkf;1 (Figure 2H; see Supplemental Figure 6A online). This hybrid produced 72% aborted ovules, 6% collapsed embryos, 16% homozygous cdkd;1-1 cdkd;2-1 cdkd;3-2, and 6% cdkd;1-1 cdkd;2-1 cdkd;3-1/cdkd;3-2 (named from here on d123*) progeny (see Supplemental Figure 6C online). A single copy of the weak cdkd;3-2 allele, showing lower transcript levels compared with wild-type CDKD;3 (see Supplemental Figure 6F online), was thus sufficient for plant viability in the presence of CDKF;1. On the other hand, the complete lack of triple null cdkd;1-1 cdkd;2-1 cdkd;3-1 mutant offspring indicated that inactivation of all three CDKD genes is lethal.
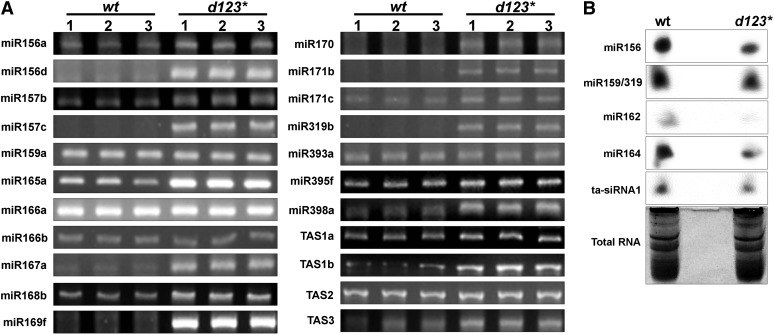
Curling serrated leaves is a typical phenotypic trait suggesting a deficiency of small RNA biogenesis pathways that specify the differentiation of leaf primordia in the se, cbp20, and cbp80 Arabidopsis mutants (Kim et al., 2008; Laubinger et al., 2008). Therefore, we investigated a possible involvement of CDKF;1 and CDKDs in the regulation of small RNAs by RNA gel blot comparison of the levels of several mature miRNAs (miR156, miR165, and miR162) and siRNAs (copia and ta-siRNA1) at 7 d after germination (DAG) in the cdkf;1-2 and cdkd;2-2 mutants compared with the wild type and at 20 DAG in wild-type, cdkf;1-2, and cdkd double and triple mutant seedlings. The levels of all examined small RNAs were reduced in cdkf;1-2 already at 7 DAG (see Supplemental Figure 7 online) and showed a decrease also in the cdkd23 and d123* mutants at 20 DAG (Figure 3) compared with the wild type, suggesting that both classes of RNAPII CTD kinases might be implicated directly or indirectly in the regulation of production of some miRNAs and siRNAs.
Figure 3.
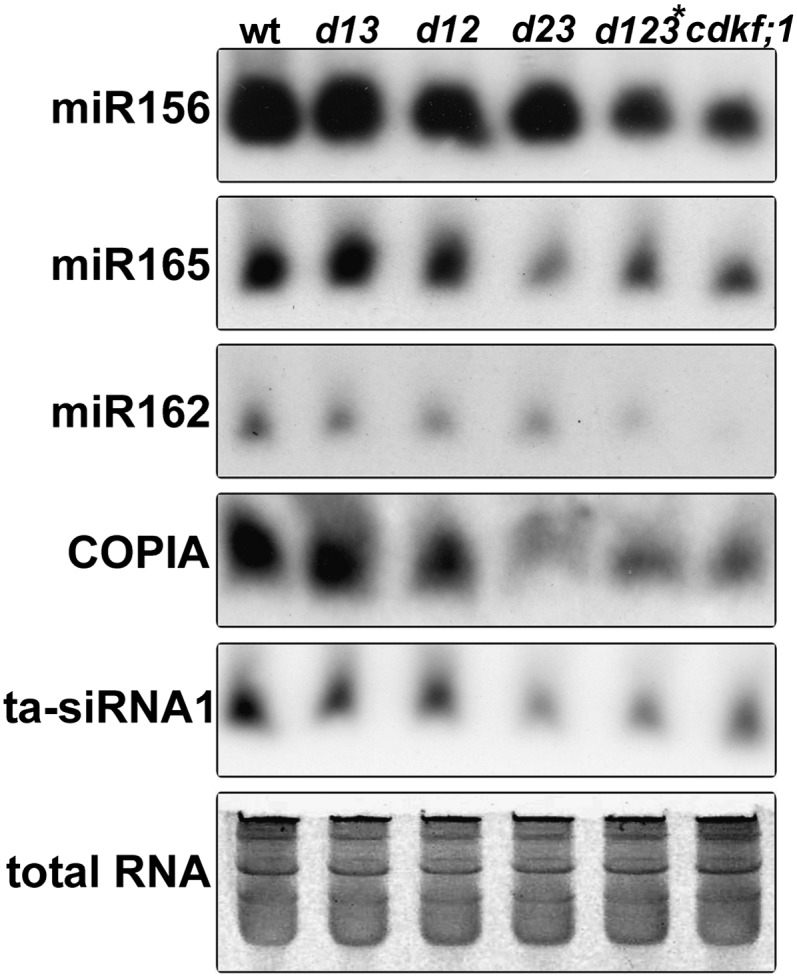
RNA Gel Blot Hybridizations Indicate a Reduction of Mature Small RNA Levels in the cdkf;1, cdkd23, and triple d123* Mutants Compared with the Wild Type.
Total RNA is shown below as a loading control. wt, wild type; d13, cdkd;1-1 cdkd;3-2; d12, cdkd;1-1 cdkd;2-1; d23, cdkd;2-1 cdkd;3-1; d123*, cdkd;1-1 cdkd2-1 (cdkd3-1/cdkd3-2).
Developmental Alterations of RNAPII CTD Phosphorylation in the cdkf;1 and cdkd Mutants
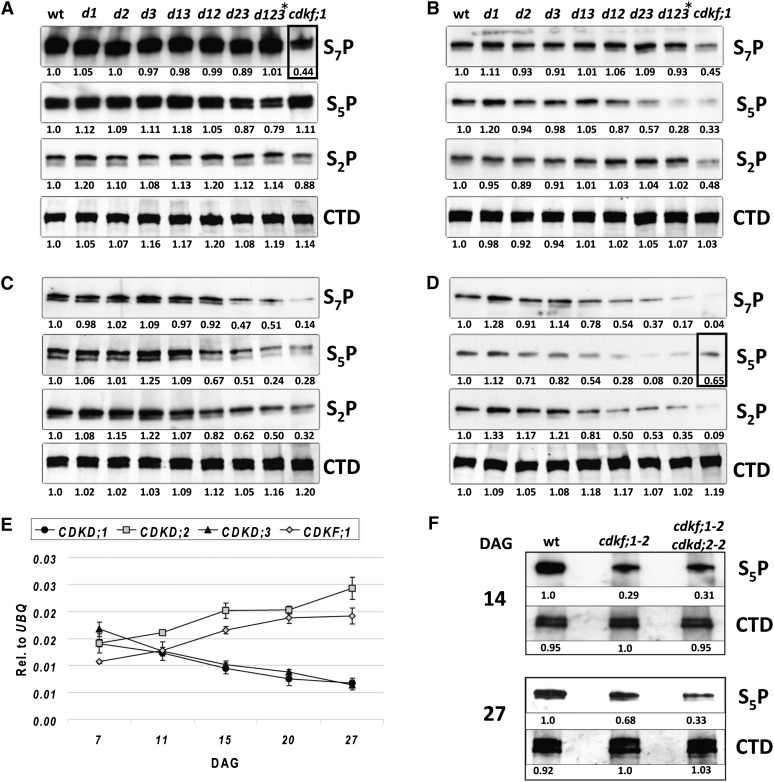
To examine how CDKF;1 and CDKDs contribute to the regulation of the RNAPII CTD phosphoserine code in planta, protein extracts prepared from single cdkf;1-2, double cdkd, and triple d123* mutants at 7, 14, 20, and 27 DAG were immunoblotted with CTD S2P-, S5P-, and S7P-specific monoclonal antibodies following normalization of the samples with the nonphospho-specific 4H8 anti-CTD antibody (Figures 4A to 4D). The level of RNAPII CTD S7P and S5P marks showed over 50 and 20% reduction in the cdkf;1 and d123* mutants at 7 DAG (Figure 4A), which confirmed that in vivo deposition of the CTD S7P and S5P marks is dependent on limiting functions of CDKF;1 and CDKDs, respectively. Comparable levels of CTD S5 mark in wild-type and cdkf;1 seedlings at 7 DAG also indicated that CDKDs, which showed significant autoactivation in vitro, could maintain normal RNAPII CTD S5 phosphorylation in the absence of CDKD-activating CDKF;1 kinase during the first days of germination. Analogously, CDKF;1 together with the activity of the weak cdkd;3-2 allele in the d123* mutant was sufficient to maintain wild-type CTD S7 phosphorylation during early seedling development.
Figure 4.
Developmental Alterations of Position-Specific Ser Phosphorylation of RNAPII CTD in the cdkf;1 and cdkd Single, Double, and Triple Mutants during Seedling Development.
(A) to (D) Quantitative analysis of position-specific CTD Ser phosphorylation with CTD S2P-, S5P-, and S7P-specific antibodies at 7 (A), 14 (B), 20 (C), and 27 (D) DAG. Equal loading of protein samples is indicated by immunoblotting with anti-CTD antibody 4H8 (CTD). Quantitative changes indicated by numerical values were normalized to wild-type control in each lane. Allelic combinations of different cdkd mutants is the same as in Figure 2H, except for d123*, which is cdkd;1-1 cdkd;2-1 (cdkd;3-1/cdkd;3-2). Early reduction of S7P mark in cdkf;1-1 at 7 DAG and compensatory increase of S5P level in cdkf;1-1 at 27 DAG are indicated by frames. wt, wild type.
(E) qRT-PCR comparison of CDKF;1 and CDKD transcript levels relative to UBQ5 standard at different stages of seedling development. The error bars represent sd of the mean of triplicate qRT-PCR measurements using three biological replicates.
(F) Immunoblot analysis with S5P-specific monoclonal antibody indicates that, compared with 14-d-old mutants, enhanced CDKD;2 expression confers a compensatory increase of CTD S5P mark at 27 DAG in cdkf;1-2 seedlings, which is not detectable in the cdkf;1-2 cdkd;2-2 double mutant.
At 14 DAG, cdkf;1 seedlings showed additional reduction of S2P and S5P CTD marks, which indicated a gradual decline of CDKD activities, confirming that CDKF;1 is required for in planta activation of CDKDs (Figure 4B). In comparison, the d123* mutant suffered further reduction of CTD S5P mark at 14 DAG but had still close to wild-type CTD S2P and S7P levels. However, both S2P and S7P CTD marks were reduced by 50% in the d123* mutant at 20 DAG, accompanying an over 70% decline of S5P level (Figure 4C). At 27 DAG, a decreased phosphorylation of all CTD Ser residues was detected also in the cdkd;1-1 cdkd;3-1 (d13), cdkd;1-1 cdkd;2-1 (d12), and cdkd;2-1 cdkd;3-1 (d23) double mutants. At the same time, the CTD S5P level showed a surprising increase in the cdkf;1 mutant due to an age-dependent compensatory mechanism, which reflected differential transcriptional regulation of the CDKD genes. Namely, between 7 and 27 DAG, the transcript levels of CDKD;1 and CDKD;3 gradually declined, while those of CDKD;2 and CDKF;1 increased, resulting in an elevation of S5P levels in the cdkf;1 mutant at 27 DAG (Figures 4D and 4E). This compensatory effect was clearly CDKD;2 dependent as similar increase of CTD S5P level was not observed in the cdkf;1-2 cdkd;2-2 double mutant (Figure 4F).
The CTD S5P Mark Is Required for 5′-Capping of Small RNA Precursors in Arabidopsis
Previous studies demonstrated that 5′-capping of nascent transcripts is dependent on the phosphorylation of RNAPII CTD S5 residues in yeast and mammals (McCracken et al., 1997; Buratowski, 2009; Ghosh et al., 2011). Therefore, using T4 RNA ligase–mediated isolation of uncapped mRNAs (Jiao et al., 2008), we investigated whether the reduction of CTD S5P mark in the d123* mutant at 14 DAG leads to the accumulation of uncapped small RNA precursor transcripts. Compared with the wild type, we in fact detected an enrichment of uncapped transcripts for the majority of examined pri-miRNAs and siRNA precursors in the d123* mutant (Figure 5A). This indicated that proper deposition of the CTD S5P mark is required for 5′-capping of nascent transcripts in Arabidopsis as in yeast and mammals.
Figure 5.
RNAPII CTD S5-Phosphorylation Is Required for 5′-Capping of miRNA and Ta-siRNA Precursors.
For detection and amplification of uncapped miRNA precursors, a T4 RNA ligase–mediated isolation procedure (Jiao et al., 2008) was used with RNA samples of three independent biological replicates.
(A) Accumulation of uncapped small RNA precursors in the d123* mutant at 14 DAG.
(B) Comparison of mature small RNA levels in wild-type and d123* mutant seedlings at 14 DAG. Normalization of total RNAs was used to ensure equal loading.
The CBP20 and CBP80 subunits of the CBC that bind to the 5′-cap structure of nascent transcripts appear to be directly implicated in miRNA processing. Both cbp20 and cbp80/abh1 mutations inhibit the production of mature miRNAs in Arabidopsis without destabilization and depletion of pri-miRNAs (Kim et al., 2008; Laubinger et al., 2008). To examine whether reduction of CTD S5 phosphorylation caused a similar capping defect as the cdp20 and cbp80 mutations, we compared the levels of mature miR156, miR159/319, miR162, miR164, and ta-siRNA1 by RNA gel blotting in wild-type and d123* mutant seedlings at 14 DAG. As expected, the levels of all of examined mature miRNAs and ta-siRNA1 were somewhat lower in the d123* mutant compared with the wild type (Figure 5B). A decrease in mature small RNA levels might either be due to reduced transcription of small RNA precursors (i.e., measured by the total amount of capped and uncapped precursor transcripts) or transcriptional downregulation of genes that are required for processing of small RNA precursors. Therefore, we compared the amounts of 33 miRNA and four ta-siRNA precursors, as well as mRNAs of nine genes implicated in small RNA biogenesis pathways (see Supplemental Table 1 online). Of these, only pri-miR156a, pri-miR165a, and pri-miR171b showed partial reduction accompanying the decline of CTD S5P level in the d123* mutant compared with the wild type at 14 DAG. Taken together, these results showed that deficient deposition of the RNAPII CTD S5P mark negatively influences the processing of small RNA precursors similarly to mutations preventing the recruitment of the CBC complex.
Coregulation of RNAPII CTD S7-Phosphorylation and Transcription of Small Noncoding RNA Precursors and Genes Involved in Their Biogenesis
Specific reduction of S7-phosphorylation in the cdkf;1 mutant at 7 DAG provided a tool to investigate possible regulatory role of the CTD S7P mark in the control of transcription. Using RNA samples from three independent biological replicates of wild-type and cdkf;1-2 seedlings, we performed comparative transcript profiling with Affymetrix ATH1 microarrays and a GeneSpring 11.5 (Agilent) data analysis package. Among a total of 845 genes showing a significant twofold change (P = 0.05) in their mRNA levels, the Gene Ontology (GO) terms in the upregulated group of 478 genes were enriched for responses to stress and hormonal stimuli, while GO terms of the downregulated genes suggested a decline of oxidative stress responses and chloroplast-related functions in the cdkf;1 mutant. As the latter GO terms reflected less robust transcriptional changes, a repeated analysis was performed by lowering the cutoff value (1.5-fold change, P = 0.05). As expected, this resulted in a remarkable enrichment of plastid-related GO terms in the extended downregulated class of 1258 genes, whereas the upregulated class of 1126 transcripts was broadened by GO terms of physiological processes related to stress responses (see Supplemental Tables 2A and 2B online). The gene list was sorted into functionally related classes, including major signaling and metabolic pathways using a combination of GO analysis and publication-based annotation (see Supplemental Data Set 1 online). Subsequent analysis of significant pathways confirmed numerous correlative changes in the expression of known network regulators and their targets. Similarly to mammalian cells, where Ala replacement of the CTD S7 positions had no apparent effect on mRNA levels (Egloff et al., 2007), the reduction of CTD S7 mark in the cdkf;1 mutant did not cause a global decline of transcription, although it altered the abundance of a fraction of mRNAs. Among these, a cluster of transcripts encoding the DCL1, DCL2, ARGONAUTE1 (AGO1), and RNA-DEPENDENT RNA POLYMERASE6 (RDR6) proteins involved in miRNA and ta-siRNA biogenesis (Voinnet, 2009) showed coordinate downregulation in the cdkf;1 mutant (see Supplemental Data Set 1 online). As the cdkf;1 mutation caused a remarkable decrease of mature small RNA levels (Figure 3), transcriptional downregulation of these genes acting in the small RNA biogenesis pathways was notable.
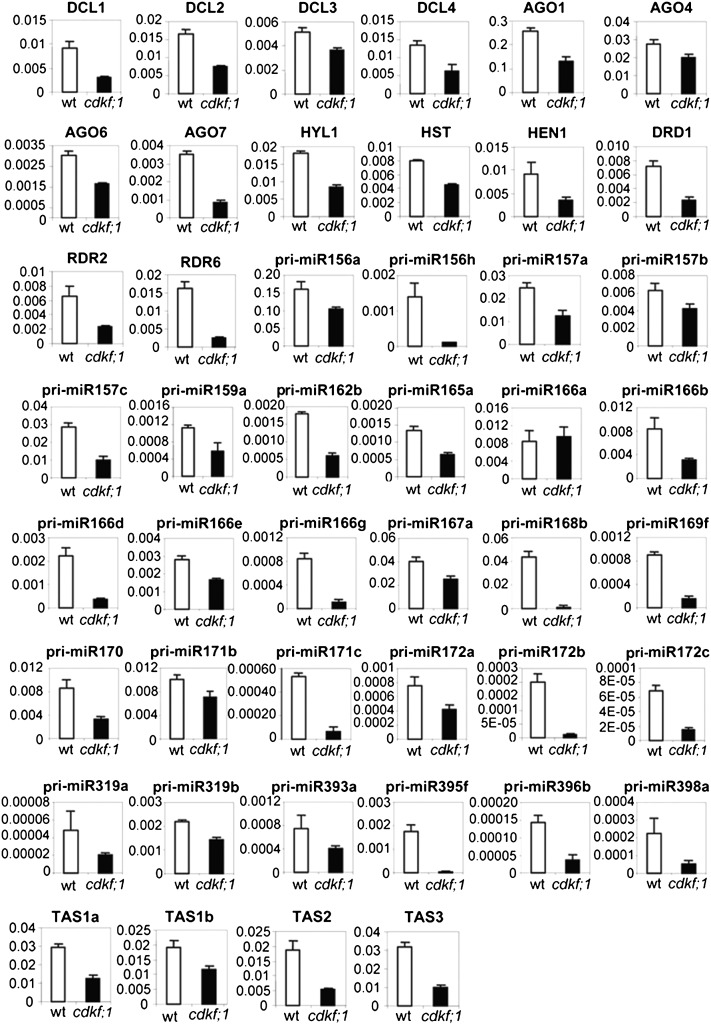
To assess the quality of transcript profiling data, the same 7-DAG wild-type and cdkf;1 RNA templates were used for comparative quantitative real-time PCR (qRT-PCR) measurements of transcript levels of 97 genes chosen arbitrarily from 18 pathway clusters. We detected no conflict in the tendency of observed changes (i.e., up- or downregulation), but the fold change values measured by qRT-PCR for 18 genes were below the cut off limit of Affymetrix data analysis (see Supplemental Data Set 1 online). This discrepancy indicated a higher than expected false discovery rate. Therefore, we next examined whether the observed tendency of parallel downregulation could be validated for other genes in the small RNA biogenesis pathways that were not uncovered in the transcript profiling experiment. qRT-PCR measurements in fact showed that the transcript levels of genes encoding DCL1, 2, 3, and 4; AGO1, 4, 6, and 7; HYL1, HASTY1 (HST1); HUA ENHANCER1 (HEN1); DEFECTIVE IN RNA-DIRECTED DNA METHYLATION1; and RDR2 and RDR6 were significantly lower in the cdkf;1 mutant compared with the wild type (Figure 6). The reduction of the CTD S7-phosphorylation level in the cdkf;1 mutant was thus accompanied by downregulation of genes acting in the biogenesis of miRNAs (i.e., DCL1, HYL1, HST1, and HEN1), miRNA-dependent generation of ta-siRNAs (as for miRNAs and AGO1, RDR6, and AGO7), and formation of nat-siRNAs and some repeat-derived ra-siRNAs (by DCL2, RDR6, DCL1, and HEN1 versus DCL3, RDR2, AGO4, and HEN1, respectively; see review in Vazquez, 2006). At the same time, the qRT-PCR measurements revealed that the levels of 28 tested pri-miRNAs and ta-siRNA source transcripts TAS1, 2, and 3 were also reduced in the cdkf;1 mutant compared with the wild type (Figure 6). Therefore, we concluded that the cdkf;1 mutation affects either the transcription or processing (or both) of these small RNA precursors.
Figure 6.
The cdkf;1-2 Mutation Results in Downregulation of Small RNA Biogenesis Pathways.
qRT-PCR comparison of transcript levels of pri-miRNAs, TAS ta-siRNA precursors, and mRNAs encoding components of small RNA biogenesis pathways in wild-type and cdkf;1-2 seedlings at 7 DAG. The error bars represent the sd of the mean of triplicate qRT-PCR measurements using three biological replicates. wt, wild type.
During seedling development, we observed a comparable reduction of CTD S7P marks in the cdkf;1 and d123* mutants, respectively, at 7 and 20 DAG. Therefore, we argued that the transcription of precursor small RNAs and genes acting in their biogenesis ought to show a similar inhibition in the d123* mutant at 20 DAG as in the cdkf;1 mutant at 7 DAG (Figure 6) if this process was indeed regulated by deposition of the CTD S7P mark. Hence, we investigated the amounts of precursors of 26 miRNAs, ta-siRNA TAS1a/b, 2, 3, and 4, and mRNAs of 15 genes of small RNA biogenesis pathways in the d123* mutant compared with the wild type at 20 DAG. Except pri-miR395f, all small RNA precursors showed at least 1.5-fold lower levels, while nine from the tested genes of small RNA biogenesis pathways were similarly downregulated in the d123* mutant (see Supplemental Table 3 online). As none of these transcripts showed any significant change parallel with the specific 50% reduction of the CTD S5P mark in the d123* mutant at 7 DAG, the results suggested that CTD S7-phophorylation is likely implicated in the regulation of transcription and biogenesis of small RNAs. Nonetheless, we could not exclude that some of the observed transcriptional changes might be due to simultaneous decline of CTD S2P, S5P, and S7P marks in the d123* mutant at 20 DAG, albeit these changes clearly correlated with an exclusive reduction of CTD S7P levels in the cdkf;1 mutant at 7 DAG.
The CDKF;1 Mutation Does Not Impair RNAPII Recruitment to Genes but Causes 3′-End Processing Defects and Enhanced Splicing of Some miRNA Precursors
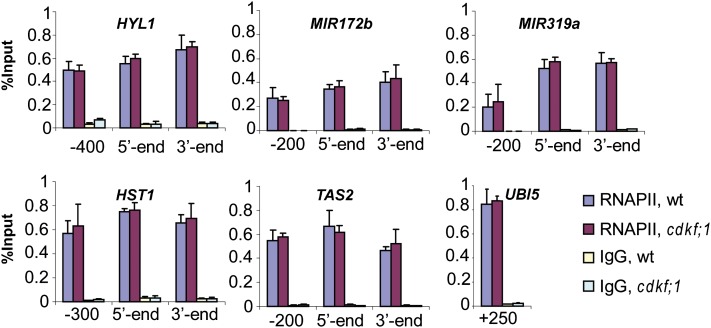
Accompanying the 50% inhibition of CTD S7-phosphorylation at 7 and 20 DAG, the cdkf;1 and d123* mutations led to a similar reduction of precursor miRNA and siRNA levels as the recently described subunit mutations of the RNAPII Mediator complex. The med mutations were shown to inhibit the recruitment of RNAPII to promoter regions of small noncoding RNA genes (Kim et al., 2011). As Mediator is required for RNAPII loading of TFIIH (Boeing et al., 2010), next we asked whether the deficiency of TFIIH-associated CDKF;1 kinase had any effect on the recruitment of RNAPII to protein-coding and small RNA genes. We performed qRT-PCR–based chromatin immunoprecipitation (ChIP) assays with nuclear extracts from wild-type and cdkf;1-2 seedlings at 7 DAG, as well as the RNAPII anti-CTD 4H8 antibody, which detects the RNAPII largest subunit independently of the CTD phosphorylation status (Chapman et al., 2007). The ChIP experiments indicated wild-type RNAPII levels at the promoter regions of HYL1, HST1, MIR172b, MIR319a, and TAS2 genes in the cdkf;1 mutant (Figure 7). Furthermore, the levels of the RNAPII largest subunit detected in association with 3′ and coding regions of these genes were also similar in wild-type and cdkf;1 plants. This suggested that the observed decline in the levels of tested small RNAs and mRNAs was not caused by altered recruitment of RNAPII to genes but rather by possible RNA processing defects.
Figure 7.
ChIP Assays of Representation of RNAPII Largest Subunit on Genes Showing Altered Regulation in the cdkf;1 Mutant.
The qRT-PCR–based ChIP analyses were performed by PCR amplification of promoter, 5′/coding, and 3′-end sequences of HYL1, HST1, TAS2, pri-miRNA172b, and pri-miRNA319a genes, as well as a segment of coding region of the control ubiquitin UBQ5 gene. Error bars represent sd of the mean of three biological replicates. wt, wild type.
[See online article for color version of this figure.]
Phosphorylation of the CTD S7 residue was recently demonstrated to facilitate specific 3′-end processing of small nuclear U RNAs in mammalian cells (Egloff et al., 2007, 2010). This observation prompted us to examine whether the cdkf;1 mutation had any effect on the 3′-polyadenylation status of DCL1, HYL1, HST1, AGO7, MIR156h, MIR157c, MIR168b, MIR169f, MIR172b, MIR319a, TAS2, and TAS3 transcripts that showed downregulation parallel with the decline of the CTD S7P mark at 7 DAG in the mutant seedlings. We performed two independent qRT-PCR comparisons of transcript levels in wild-type and cdkf;1 seedlings using either total [poly(A)− and poly(A)+] RNA or oligo(dT) cellulose-purified poly(A)+ RNA templates isolated from three biological replicates. In the case of total RNA, cDNA templates were synthesized by gene-specific primers designed to the central regions of transcripts, while cDNAs from poly(A)+ RNAs were generated using an oligo(dT) primer. In the two independent sets of experiments, the same gene-specific qRT-PCR primers were used to determine and compare the transcript levels present in either the total or poly(A)+ RNA fractions of wild-type versus cdkf;1 seedlings. Except for MIR168b and TAS3, we found that the relative decrease of all tested transcript levels in the cdkf;1 mutant compared with the wild type was higher in the poly(A)+ than in the total RNA fraction (Table 1). These results thus suggested a potential regulatory link between exclusive reduction of the RNAPII CTD S7P mark and simultaneous deficiency of 3′-polyadenylation of miRNA and ta-siRNA precursors and mRNAs of small RNA biogenesis genes in the cdkf;1 mutant.
Table 1. qRT-PCR Analysis of Small RNA Precursor and Protein Coding Transcripts, Which Show Reduced Levels in the cdkf;1 Mutant Compared to the Wild Type at 7 DAG in the Total RNA and Poly(A+) RNA Fractions.
| Gene | Poly(A)− and Poly(A)+ | Poly(A)+ | |||
|---|---|---|---|---|---|
| Arabidopsis Genome Initiative | Wild Type/cdkf;1 | sd | Wild Type/cdkf;1 | sd | |
| DCL1 | At1G01040 | 1.36 | 0.18 | 1.93 | 0.11 |
| HYL1 | At1G09700 | 1.18 | 0.21 | 1.74 | 0.07 |
| HST1 | At3G05040 | 1.32 | 0.10 | 2.10 | 0.19 |
| AGO7 | At1g69440 | 2.17 | 0.12 | 3.18 | 0.30 |
| MIR156 h | AT5G55835 | 1.70 | 0.08 | 2.85 | 0.17 |
| MIR157c | AT3G18217 | 1.36 | 0.13 | 2.82 | 0.35 |
| MIR168b | AT5G45307 | 1.99 | 0.28 | 2.07 | 0.08 |
| MIR169f | AT3G14385 | 2.51 | 0.33 | 5.46 | 0.29 |
| MIR172b | At5g04275 | 5.47 | 0.63 | 24.68 | 2.34 |
| MIR319a | At4G23713 | 2.58 | 0.31 | 4.00 | 0.20 |
| TAS2 | At2g39681 | 1.02 | 0.09 | 2.12 | 0.21 |
| TAS3 | At3G17185 | 2.68 | 0.18 | 2.4 | 0.31 |
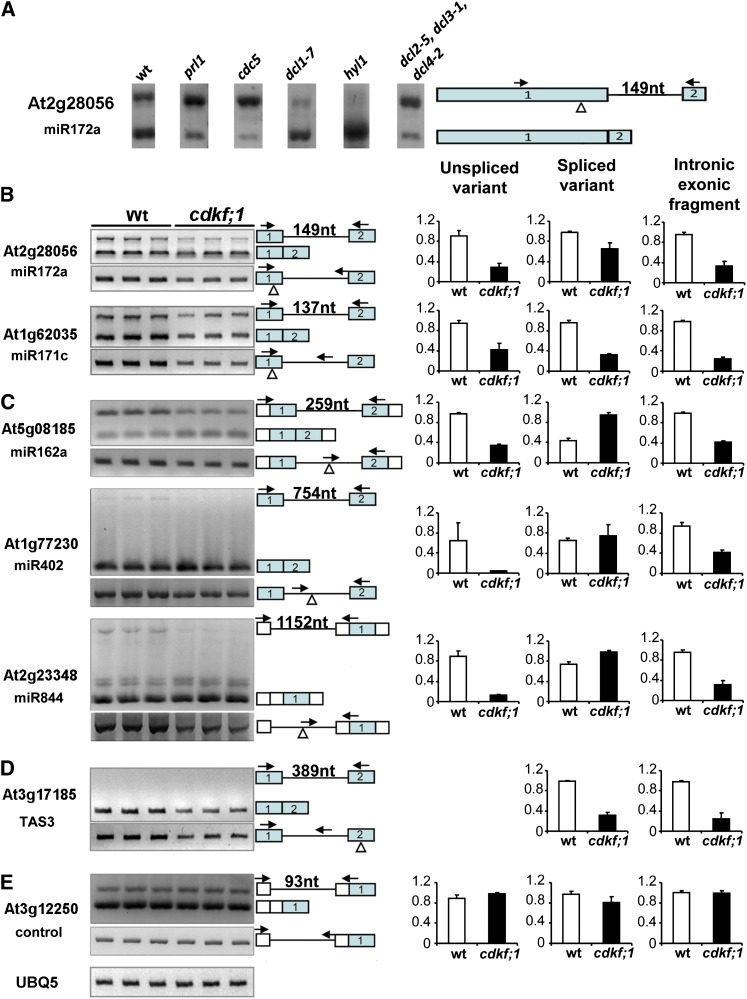
Recently, Kataoka et al. (2009) demonstrated that subunits of the Drosophila melanogaster pre-mRNA microprocessor complex occur in association with the spliceosome and that dicing of pre-miRNAs interferes with the splicing of introns located in their vicinity. As we found that the transcripts of both DCL1 and HYL1 Arabidopsis microprocessor subunits were downregulated by the cdkf;1 mutation, we examined the splicing efficiency of a single intron of pri-miR172a transcript in the dcl1-7 and hyl1 mutants compared with the wild type and a control dcl2-5 dcl3-1 dcl4-1 triple mutant, as well as in lines carrying mutations of the PRL1 and CDC5 subunits of the spliceosome-activating NineTeen Complex (Koncz et al., 2012). Whereas in the latter mutants splicing of partially retained pri-miR172a intron was less efficient compared with the wild type, the removal of intron was dramatically stimulated by the dcl1-7 and hyl1 mutations (Figure 8A). To examine the potential effect of the cdkf;1 mutation on splicing of pre-miRNAs, we performed qRT-PCR measurements of spliced and unspliced isoforms of an extended set of intron-containing pri-microRNAs in the cdkf;1 mutant and the wild type at 7 DAG, including the precursor of TAS3 ta-siRNA and the intronless ubiquitin UBQ5 transcript as normalization standard (Figures 8B to 8E). The predicted intron of TAS3 transcript was not retained either in the wild type or cdkf;1, but the amount of spliced TAS3 RNA was reduced in cdkf;1 as seen in our earlier measurements (Figures 6 and 8D). Similarly, the levels of both unspliced and spliced isoforms of transcripts carrying exonic stem-loops of pre-miRNA172a and 171c were reduced in the cdkf;1 mutant compared with the wild type (Figure 8B). By contrast, the levels of introns that carried internal stem-loops of pre-miRNAs 162a, 402, and 844 were decreased threefold to eightfold in cdkf;1, whereas the amounts of corresponding spliced transcripts were similar or slightly higher compared with the wild type. In conclusion, the cdkf;1 mutation enhanced the splicing of intronic pre-miRNA precursors without destabilizing the resulting spliced transcripts, while it stimulated the destabilization of both spliced and unspliced forms of RNAs that carried either exonic pre-miRNA stem-loops or the TAS3 ta-siRNA.
Figure 8.
The cdkf;1 Mutation Enhances the Splicing of Pre-miRNA–Containing Introns.
(A) Comparison of intron splicing efficiency of pri-miRNA172a in wild-type (wt), prl1, cdc5, dcl1-7, hyl1, and dcl2-5 dcl3-1 dcl4-2 seedlings by RT-PCR. nt, nucleotides.
(B) to (D) RT-PCR assay (left) and qRT-PCR measurements (right).
(B) The levels of introns retained in the vicinity of exonic stem-loops of pre-mRNAs 172a and 171c compared with the level of spliced pri-mRNA isoforms.
(C) Intronic pre-miRNAs 162a, 402, and 844 compared with spliced versions of corresponding transcripts.
(D) Spliced and unspliced forms of TAS3 ta-siRNA precursor.
(E) Normalization of RT- and qRT-PCR assays with the intronless UBI5 (At3g12250) RNA control. Triangles indicate the positions of pre-mRNA stem-loops, arrows mark the positions of exonic and intronic gene-specific primers, and the lengths of introns in nucleotides are indicated. The error bars represent sd of the mean of triplicate qRT-PCR measurements using three biological replicates.
[See online article for color version of this figure.]
Comparison of Transcript Levels of U Small Nuclear RNAs and miRNA Target Genes in the cdkf;1 and d123* Mutants
In mammalian cells, replacement of the CTD S7 residues with Ala leads to a specific decrease in the levels of U1/U2 small nuclear RNAs (snRNAs) (Egloff et al., 2007). By contrast, qRT-PCR analysis of U1a, U1.5, U1.7, U2.2, U2.4, and U2.5 snRNA levels in the cdkf;1-2 mutant during specific reduction of the CTD S7P mark at 7 DAG revealed either significant upregulation or no change compared with the wild type (see Supplemental Figure 8 online). We observed similar alterations in the levels of U snRNAs in the d123* mutant at 14 DAG coinciding with a decline of the CTD S5P mark (see Supplemental Figure 8 online). Consequently, these data indicated that, in contrast with mammalian cells, defective CTD phosphorylation does not lead to destabilization of Arabidopsis U snRNAs.
As several plant miRNAs trigger specific cleavage and subsequent degradation of their target transcripts (Voinnet, 2009), it was also a particular interest to determine whether the observed decline of mature miRNA levels in the cdkf;1 and d123* mutants at 7 and 14 DAG would correlate with an increase in the levels of their target transcripts. The analysis of transcript profiling and confirmatory qRT-PCR data of mRNA levels of 29 target genes of 19 miRNAs revealed significant upregulation of only eight target transcripts in the cdkf;1 mutant (see Supplemental Table 4 online). Analogously, qRT-PCR measurement of transcript levels of 27 miRNA target genes in the d123* mutant identified only three genes showing higher transcript levels compared with the wild type (see Supplemental Table 5 online). As we demonstrated that both cdkf;1 and d123* mutations downregulated numerous key genes in the biogenesis pathways of miRNAs, these results were consistent with the previous reports showing that only very few miRNA target transcripts show significant enrichment in the dcl1, hyl1, and se mutants that are impaired in miRNA biogenesis (Ronemus et al., 2006; Laubinger et al., 2010).
DISCUSSION
Posttranslational modifications of the CTD play a central role in the transcription and cotranscriptional processing of RNAPII-transcribed RNAs (Xiao et al., 2003; Chapman et al., 2007; Egloff et al., 2007; Akhtar et al., 2009; Hsin et al., 2011). Compared with yeast and mammals, so far little is known about the regulation of plant RNAPII CTD codes. Nonetheless, the overall conservation of RNAPII CTD and many of its binding factors suggests that the CTD is involved in similar processes in plants as in other eukaryotes. This is also indicated by the fact that CTD kinases, including the TFIIH-associated CAKs, which represent key regulators of the CTD code during transcription, are evolutionarily conserved. A unique class of CTD kinase that evolved in plants is represented by CDKF;1, which has no known yeast or human counterpart. Nonetheless, CDKF;1 complements the mcs6 and cak1 mutations of fission and budding yeast, respectively (Umeda et al., 1998; Shimotohno et al., 2004). Whereas Mcs6 has CTD and CDK kinase activities, Cak1p has no CTD kinase activity but activates Cdc28p/CDK and Kin28/CAK (Serizawa et al., 1995; Kaldis et al., 1996; Feaver et al., 1997; Espinoza et al., 1998).
CDKDs Are Major CTD S5-Kinases Implicated in 5′-Capping of Small RNA Precursors
Our combined genetic and biochemical studies show that CDKF;1-mediated phosphorylation is required for in vivo activation of functionally redundant CDKD kinases. In the absence of CDKF;1, CDKDs maintain a basic CTD phosphorylation activity, which is sufficient for embryogenesis and seed formation, but declines during development, causing severe defects in the cdkf;1 mutants. Inactivation of all three CDKDs is lethal, likely due to deficiency of CTD S5-phosphorylation, which is required for transcription initiation. In contrast with a previous report (Shimotohno et al., 2004), our data demonstrate that CDKD;1, a third member of the CDKD family, is also an active kinase and, similarly to CDKD;2 and CDKD;3, phosphorylates all Ser residues of Arabidopsis RNAPII CTD. In the cdkd double and d123* triple mutants, reduction of the CTD S2P mark during seedling development showed slower kinetics compared with the depletion of S5P, suggesting a possible involvement of other S2-specific CTD kinases, including candidate CDKC homologs of Ctk1/CDK9 (Fülöp et al., 2005; Cui et al., 2007). Throughout seedling development, the S2P levels were nonetheless lower in cdkf;1 compared with the cdkd mutants, suggesting that CDKF;1 is probably also required for maintenance of the CDKD-independent CTD S2-kinase activity.
In agreement with the observation that 5′-capping of primary transcripts is dependent on the deposition of RNAPII CTD S5P mark in yeast and mammals (McCracken et al., 1997; Ho and Shuman, 1999; Buratowski, 2009; Perales and Bentley, 2009; Ghosh et al., 2011), we showed that CDKD-mediated S5-phosphorylation of the CTD is also required for 5′-capping of miRNAs and ta-siRNAs precursors in Arabidopsis. This implies that the CTD S5P mark is possibly involved in the recruitment and activation of capping enzymes that attach a methylguanosine cap to the 5′-ends of nascent transcripts. CBC, which binds to the 5′-cap structure of RNA precursors including pri-miRNAs, is implicated in miRNA processing (Kim et al., 2008; Laubinger et al., 2008; Gruber et al., 2009). In Arabidopsis and Drosophila, SE/Ars2 is thought to direct components of the DCL1/DROSHA microprocessor to the CBC (Kim et al., 2008; Laubinger et al., 2008). Mutations affecting SE and subunits of the CBC cause a reduction in the levels of mature miRNAs, which is accompanied by parallel accumulation of some unprocessed pri-miRNAs (Kim et al., 2008; Laubinger et al., 2008). We observed a similar decrease of mature miRNAs and siRNAs, as well as accumulation of some of their precursors, accompanying a decline of the CTD S5P mark in the d123* mutant during its early development at 14 DAG. However, accumulation of unprocessed small RNAs clearly occurs at a lower level in d123* seedlings compared with the cbp20 and cbp80 mutants. This is likely due to lower stability of uncapped small RNA transcripts in the d123* mutant. Taken together, these results suggest that CDKD-dependent S5-phosphorylation of RNAPII CTD is involved in the regulation of small RNA processing, most likely through mediating RNAPII recruitment of the CBC during transcription of small RNA precursors in Arabidopsis. Similarly to the cbp mutants, the enrichment of uncapped transcripts during reduction of the CTD S5P mark in the d123* mutant probably represents a general defect that is not limited to miRNA precursors.
CTD S7-Phosphorylation by CDKF;1 and Its Role in the Regulation of Small RNA Pathways
Thus far, the CTD S7P mark is only known to be required for Integrator-mediated 3′-end processing of U1/2 snRNAs in mammals (Egloff et al., 2007, 2010). Ala replacement of CTD S7 positions causes only mild stress-related transcriptional changes in yeast and mammalian cells (Chapman et al., 2007; Egloff et al., 2007). Genome-wide mapping of specific phosphorylated forms of RNAPII CTD in yeast indicates that the CTD code is gene class specific and that the promoter structure and composition of the preinitiation complex might have an influence on the CTD phosphorylation pattern. Among the phosphoserine residues, the CTD S7P pattern is the most divergent as it peaks often discontinuously at either 5′- or 3′-ends of genes, or both. Nonetheless, the CTD S7P mark shows a remarkable ChIP enrichment on introns and noncoding RNA genes (Kim et al., 2010; Tietjen et al., 2010). In this study, we documented that deposition of the CTD S7P mark in Arabidopsis is primarily dependent on CDKF;1, which, to our knowledge, is the first known monospecific CTD S7-kinase. Our data demonstrate that reduction of the CTD S7P mark in the Arabidopsis cdkf;1 mutant is accompanied by 3′-polyadenylation defect of a large set of small RNA precursors and transcripts encoding key factors in their biogenesis pathways. This suggests that deposition of the CTD S7P mark and its recognition might occur in a gene class–specific manner in plants similarly to yeast. Further exploration of this possibility could clarify why pri-miRNAs and genes involved in the small RNA biogenesis pathways are affected simultaneously by reduction of the CTD S7P mark in the cdkf;1 and cdkd mutants.
While the CBP mutations lead to intron splicing defects (Laubinger et al., 2008), the cdkf;1 mutation at 7 DAG enhances the splicing of intronic pre-miRNAs without destabilization of their spliced isoforms. On the other hand, both unprocessed and spliced isoforms of transcripts containing exonic pre-miRNA or ta-siRNA are destabilized in cdkf;1 seedlings. Thus, the effects of the cdkf;1 mutation clearly differ from those of CBC subunit mutations, which lower the mature miRNA levels but lead to the stabilization of uncapped pri-miRNAs (Kim et al., 2008; Laubinger et al., 2008). In addition, our data show that mutations affecting the DCL1 and HYL1 microprocessor subunits result in enhanced splicing of an intron located in the vicinity of exonic pre-miR172a RNA and that spliced and unspliced forms of noncoding transcript carrying the pre-miR172a stem-loop are destabilized in the cdkf;1 mutant. These observations suggests that CDKF;1 might be implicated in a regulatory interplay between the spliceosome and microprocessor complex mediating both processing and quality control of pre-miRNA stem-loop–containing transcripts (Kim and Kim, 2007; Morlando et al., 2008; Kataoka et al., 2009; Pawlicki and Steitz, 2010). In addition, a decline of mature and unprocessed ta-siRNA transcripts in the cdkf;1 mutant predicts a possible role of RNAPII CTD S7-kinase CDKF;1 in the modulation of biogenesis and stability of another class of small RNAs.
Taken together, this study corroborates the previous finding that CDKF;1 is an upstream CDKD-activating kinase and demonstrates that CDKF;1 and CDKDs are major S7- and S5-kinases in Arabidopsis, respectively. Furthermore, our data highlight a possible involvement of CDKF;1 and CDKDs in the regulation of transcription, processing, and stability of small regulatory RNAs. This work is a starting point for in-depth investigations of the CTD code and underlying downstream regulatory mechanisms in Arabidopsis.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used in this study were in the Columbia (Col-0) background. Two different T-DNA insertion mutant alleles of CDKF;1 (At4g28980; SALK_148336, cdkf;1-1; GABI_315A10, cdkf;1-2), CDKD;2 (At1g66750; SALK_065163, cdkd;2-1; SALK_063929, cdkd;2-2), and CDKD;3 (At1g18040; SALK_120536, cdkd;3-1; SALK_007756, cdkd;3-2), and one allele of CDKD;1 (At1g73690; SALK_114643) genes were obtained from the SALK SiGNAL (http://signal.salk.edu; Alonso et al., 2003) and GABI-Kat (http://www.gabi-kat.de; Rosso et al., 2003) collections (Figure 2; see Supplemental Figure 6 and Supplemental Methods 1 online for details of PCR validation of T-DNA insertion mutations and construction of double and triple mutants as well as Agrobacterium tumefaciens–mediated transformation of Arabidopsis). Seedlings were grown in solid Murashige and Skoog Arabidopsis medium containing half concentration of macroelements and 0.5% Suc (Koncz et al., 1994) in a controlled culture room at 22°C with 120 mol/m2 s light intensity and a photoperiod of 8 h light and 16 h dark. Seedlings from in vitro cultures were transferred into soil and grown under 16-h-light/8-h-dark period at 22 to 24°C day and 18°C night temperature and 80 to 120 mol/m2 s light intensity.
Purification of Arabidopsis Proteins Expressed in Escherichia coli
E. coli BL21 (DE3) pLysS cells carrying pGST-CTD (Umeda et al., 1998) were grown at 28°C to OD600 of 0.4 to 0.6. Expression of the GST-CTD protein was induced by 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C. Cells were harvested by centrifugation and then resuspended and sonicated in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM phenylmethylsulfonylfluoride, 0.1 mM benzamidine, 10 µg/mL aprotinin, and 10 µg/mL leupeptin, pH 8.0). The cell lysate was subjected to centrifugation (Sorvall HB-4 rotor; 16,500g for 30 min at 4°C), and the cleared extract was used for affinity purification on glutathione-Sepharose 4B (Amersham Biosciences) as described (Umeda et al., 1998; Bakó et al., 2003). E. coli BL21 (DE3) pLysS cells carrying pET201-Trx-CDKDs-His6, pET201-Trx-CyclinH;1-His6, and pET201-Trx-CDKF;1-His6 (see Supplemental Methods 1 online) were disrupted by sonication in lysis buffer, and upon centrifugation the cleared extracts were affinity purified on nickel-nitriloacetic acid agarose (Qiagen) according to the manufacturer’s instruction. Elution of matrix-bound proteins was performed by increasing the imidazole concentration in stepwise manner from 60 to 250 using 20 mM intervals. Protein profiles of fractions were visualized by SDS-PAGE, and fractions containing apparently homogeneous proteins were pooled. The collected fractions were concentrated using buffer exchange in a 10K Amicon Ultra Centrifugal Filter (Millipore), and the final protein concentrations were adjusted based on Bradford assays (Bio-Rad) using a BSA standard curve.
Protein Kinase Assays
Affinity-purified wild-type CDKF;1 and CDKDs and mutant CDKD proteins were characterized in protein kinase assays. Consensus and altered versions of CTD heptapeptide dimers, carrying Ala exchanges of either S2 or S5 or both positions and C-terminal His6 tags, were synthesized and purified by GenScript. The kinase reactions were performed in kinase buffer (50 mM Tris-HCl, pH 8.0, 15 mM MgCl2, 1 mM DTT, and 5 µCi [γ-32P]ATP) and incubated for 90 min at room temperature. The reactions with GST-CTD and CDKD substrates were separated by 10% SDS-PAGE, whereas the phosphorylated CTD heptapeptide dimers were resolved by 18% SDS-PAGE. Subsequently, the gels were dried and subjected to autoradiography using x-ray films. In all kinase assays, 250 ng CDKF;1/CDKDs and 500 ng substrate were applied, except for the GST-CTD kinase assay shown in Supplemental Figure 5C online, where the concentration of CDKD;2 was reduced to 20 ng.
Phylogenetic Analysis
Multiple alignment of protein sequences was performed using the COBALT software (http://www.ncbi.nlm.nih.gov/tools/cobalt; see Supplemental Data Set 2 online) with default settings (Papadopoulos and Agarwala, 2007) and displayed using the GeneDoc software (http://www.psc.edu/biomed/genedoc/; see Supplemental Figure 2 online). Phylogenetic trees were constructed by MEGA (version MEGA 5.05; http://www.megasoftware.net) with the aligned protein sequences using the default settings of the maximum likelihood (Jones-Taylor-Thornton model of substitution, uniform rates among sites, missing data treatment of use of all sites, Nearest-Neighbor-Interchange among ML Heuristic method, and automatic preparation of initial three) and neighbor-joining algorithm (Poisson correction, uniform rates among sites, homogenous pattern among lineages, and missing data treatment of complete deletion) (see Supplemental Figure 3 Online). Statistical tests of the phylogenies were performed using the bootstrap method with 1000 replicates followed by identification of the consensus tree (Tamura et al., 2011).
Analysis of RNAPII CTD Phosphorylation
For immunoblotting, protein extracts were prepared in extraction buffer (50 mM Tris-HCl, pH 7.5, 10% glycerol, 1 mM EDTA, 150 mM NaCl, 20 mM NaF, and 0.25% Igepal) and, following standardization of protein concentrations, subjected to size separation by SDS-PAGE and electrotransferred onto polyvinylidenfluoride membranes (Millipore). RNAPII CTD levels were normalized using the anti-CTD antibody 4H8 (Abcam; ab5408), Alexa Fluor 680 infrared dye–conjugated rabbit anti-mouse IgG, and an Odyssey Infrared Imaging System (Li-Cor Biosciences). Phosphorylation of S2, S5, and S7 CTD residues was monitored with rat monoclonal antibodies (rmAb) 3E10, rmAb 3E8, and rmAb 4E12 (Chapman et al., 2007). Immunocomplexes were detected by autoradiography of enhanced chemoluminescence generated by horseradish peroxidase–coupled goat anti-rat secondary antibody (Sigma-Aldrich; A 9037). The immunoblot signals were quantified using the AlphaVIEW SA software (Cell Biosciences).
Transcript Profiling with Affymetrix ATH1 Microarrays
Three biological replicates of wild-type and cdkf;1-2 mutant seedlings were grown on solid Murashige and Skoog Arabidopsis medium (Koncz et al., 1994) in Petri dishes at 22°C with 120 mol/m2 s light intensity and an 8-h-light/16-h-dark period. Seedlings were collected at 2 to 3 h after the start of the light period in liquid nitrogen. Total RNA was isolated with RNeasy Mini kit supplied with RNase-Free DNase set (Qiagen) according to the manufacturer's instructions. Biotinylated cRNA was prepared according to a standard Ambion protocol from 1 μg total RNA (MessageAmp II-Biotin Enhanced Kit; Ambion). After amplification and fragmentation, 12.5 μg of cRNA was hybridized for 16 h at 45°C on GeneChip ATH1-121501 Genome Array. GeneChips were washed and stained with Fluidics Script FS450-004 in the Affymetrix Fluidics Station 450 and scanned using a GeneChip Scanner 3000 7G. The data were analyzed with Affymetrix GeneChip Operating Software version 1.4 using Affymetrix default analysis settings and global scaling as normalization method. The primary chip analysis CEL and CHP files and corresponding metadata were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE27687.
Subsequent microarray data analysis was performed using GeneSpring GX10/11.5 (Agilent) software. The data were first summarized with the Robust Multichip Average algorithm using a baseline to the median of all samples followed by quality control with principal component analysis. The raw data were filtered for expression by percentile (upper cutoff 100; lower cutoff 20) and then for error at a coefficient variation <50%. Subsequent statistical analysis with t test for unpaired samples included asymptotic P value computation using 100 permutations and Benjamin-Hochberg multiple testing for false discovery rate. Subsequently, the data were filtered allowing either 1.5-fold or twofold difference between control wild-type and cdkf;1-2 mutant transcript levels and subjected to further statistical analysis of GO terms using a cutoff value <0.1. To assist proper annotation of data to pathways, the Significant Pathway analysis tool with a cutoff P value <0.01 was used from the GeneSpring package, and then the data on pathway allocation of functions were combined with updated information obtained by mining literature references in PubMed.
RT-PCR Splicing Assays
Total RNA was extracted by RNeasy Mini kit supplied with RNase-Free DNase (Qiagen). Two micrograms of total RNA template was reverse transcribed using a Transcriptor First-Strand cDNA synthesis kit (Roche Applied Science) and oligo(dT) primer. The cDNA templates were normalized using UBQ5 (At3g62250) primers (see Supplemental Table 6 online).
qRT-PCR
Total RNA extraction and preparation of the cDNA templates were as described for RT-PCR above using anchored oligo(dT) primers. The reaction mixture was diluted to 100 μL, and 2.5 to 5 μL aliquots were used for real-time PCR assays performed with iQ Supermix (Bio-Rad) in a Bio-Rad iCycler iQ5. All qRT-PCR measurements were performed with triplicates from three independent experiments. Standard curves for all transcripts analyzed were obtained with UBQ5 primers and used for internal sample normalization and quantification as described (User Bulletin; Applied Biosystems).
ChIP
ChIP experiments were performed with chromatin samples prepared from wild-type and cdkf;1-2 seedlings at 7 DAG as described (Searle et al., 2006) using the anti-CTD mouse antibody 4H8 (Abcam) and a control anti-rat IgG (Sigma-Aldrich R3756). Similar amount of untreated sonicated chromatin represented the total input DNA sample. The qRT-PCR measurements with gene-specific oligonucleotides were normalized against a dilution series of the input DNA (1:20, 1:40, 1:80, and 1:160).
Small RNA Detection by RNA Gel Blot Hybridization
Detection of small RNAs was performed by RNA gel blot analysis (Pall and Hamilton, 2008). Total RNA samples were prepared using Trizol extraction (Sigma-Aldrich T9424) according to the manufacturer’s instructions. For comparison of small RNA expression levels in wild-type and mutant seedlings, either 40 or 80 μg of total RNA was size separated in 15% polyacrylamide gels containing 7 M urea under denaturing condition and transferred to Hybond-NX membrane (Amersham Biosciences) by electroblotting. The RNA samples were cross-linked to the membrane using 0.373 g 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide in 10 mL water containing 122.5 μL 1-methylimidazole for 1 h at 60°C. Hybridization was performed at 37°C with oligonucleotides complementary to specific small RNAs (see Supplemental Table 6 online), which were labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs).
Measurement of Uncapped Small RNAs
To determine the levels of uncapped miRNAs/siRNAs, we used the T4 RNA ligase–mediated isolation method described by Jiao et al. (2008) with some modifications. Total RNA samples from three independent biological replicates of wild-type and d123* mutant were extracted using RNeasy mini kit (Qiagen) and subjected to mRNA purification by FastTrack oligo(dT) cellulose kit (Invitrogen). Next, uncapped miRNAs/mRNAs, carrying 5′-monophosphate groups at their 5′-ends (600 ng), were ligated to an RNA adaptor of 45 nucleotides (300 ng). Then, 30 ng was used to prepare cDNA using a Transcriptor First-Strand cDNA synthesis kit (Roche Applied Science) with anchored oligo(dT) primer. The reaction mixtures were diluted to 100 μL, and the cDNA templates were normalized using UBQ5 primers in qRT-PCR assays performed with iQ Supermix (Bio-Rad) in a Bio-Rad iCycler iQ5. Two-microliter aliquots of cDNA templates were mixed in 50 μL final volume with nested primers corresponding to the RNA adaptor and gene-specific reverse primers, and then two 35 cycles of nested PCR were used for amplification of distinct uncapped transcripts (1 μL from the first PCR reaction was used in the second step of nested PCR). Finally, distinct uncapped transcripts were amplified with gene-specific primers using 30 PCR cycles. To evaluate the possible contamination of final PCR products with fragments derived from intact miRNAs with 5′-cap structure, we performed similar control reactions without T4 RNA ligase, which resulted in no amplified products (see Supplemental Figure 9 online).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: CDKD;1 (At1g73690), CDKD;2 (At1g66750), CDKD;3 (At1g18040), and CDKF;1 (At4g28980). Microarray data have been deposited in the Gene Expression Omnibus under accession number GSE27687.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Purification of Recombinant Kinase and Substrate Proteins, Control Kinase Assays, and Phosphorylation of CTD Heptapeptide Dimers by CDKF;1 and CDKD;2 in Vitro.
Supplemental Figure 2. Multiple Sequence Alignment of Yeast, Arabidopsis, and Human CAK and CAKAK Proteins Using COBALT.
Supplemental Figure 3. Phylogenetic Trees of Yeast, Arabidopsis, and Human CAK and CAKAK Proteins.
Supplemental Figure 4. Analysis of Functional Domains of Plant-Specific CDKF;1 Kinase Using NCBI BLAST2.2.19+.
Supplemental Figure 5. CDKF;1 Enhances the Kinase Activities of CDKDs.
Supplemental Figure 6. Characterization of cdkf;1 and cdkd T-DNA Insertion Mutants.
Supplemental Figure 7. Decrease of Mature Small RNA Levels in the cdkf;1 Mutant at 7 DAG.
Supplemental Figure 8. Transcript Levels of Arabidopsis U1/U2 snRNAs in the cdkf;1 and d123* Mutants at 7 and 14 DAG, Respectively, Compared with the Wild Type.
Supplemental Figure 9. Negative Control for T4-RNA Ligase-Mediated Isolation of Uncapped miRNAs.
Supplemental Table 1. Quantitative Analysis of Transcript Levels of a Set of pri-miRNAs, Precursors of TAS ta-siRNAs, and mRNAs Encoding Components of Small RNA Biogenesis Pathways in 14-d-Old Wild-Type and d123* Seedlings.
Supplemental Table 2. Gene Ontology Analysis of Affymetrix Transcript Profiling Data.
Supplemental Table 3. qRT-PCR Comparison of Transcript Levels of a Set of pri-microRNAs, Precursors of TAS ta-siRNAs, and mRNAs Encoding Components of Small RNA Biogenesis Pathways in 20-d-Old Wild-Type and d123* Seedlings.
Supplemental Table 4. Analysis of Transcript Levels of miRNA Target Genes in the cdkf;1 Mutant Compared with the Wild Type at 7 DAG.
Supplemental Table 5. Comparative Analysis of Transcript Levels of a Set of miRNA Target Genes in the d123* Mutant and Wild Type at 14 DAG by qRT-PCR.
Supplemental Table 6. List of Oligonucleotide Primers and Probes.
Supplemental Data Set 1. Pathway-Specific Sorting of Genes Showing Altered Transcript Levels (1.5-Fold Change, P ≤ 0.05) in the cdkf;1-2 Mutant Compared with the Wild Type 7 d after Germination.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 2.
Supplemental Methods 1. Methods for the Supplemental Data.
Supplementary Material
Acknowledgments
We thank M. Umeda for providing cDNA clones, George Coupland and Seth Davis for their critical comments on the manuscript, and Sabine Schaefer for technical assistance. This work was supported by an International Max Planck Research School of the Max Planck Society grant (to M.H.) and SFB635 and Arabidopsis Functional Genomics Network (KO 1438/12-1) grants from the Deutsche Forschungsgemeinschaft (to C.K.).
AUTHOR CONTRIBUTIONS
M.H. and C.K. designed the research. M.H., S.F., B.H., and Z.K. performed the research. M.H. and C.K. analyzed the data. M.H. and C.K. prepared the article.
Glossary
- RNAPII CTD
RNA polymerase II C-terminal domain
- CDK
cyclin-dependent protein kinase
- CAK
CDK-activating kinase
- CAKAK
CAK-activating kinase
- miRNA
microRNA
- siRNA
short interfering RNA
- ta-siRNA
trans-acting siRNA
- nat-siRNA
natural-antisense transcript-derived siRNA
- ra-siRNA
repeat-associated siRNA
- CBC
cap binding complex
- GST
glutathione S-transferase
- DAG
days after germination
- GO
Gene Ontology
- qRT-PCR
quantitative real-time PCR
- ChIP
chromatin immunoprecipitation
References
- Ahn S.H., Kim M., Buratowski S. (2004). Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Akhtar M.S., Heidemann M., Tietjen J.R., Zhang D.W., Chapman R.D., Eick D., Ansari A.Z. (2009). TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bakó L., Umeda M., Tiburcio A.F., Schell J., Koncz C. (2003). The VirD2 pilot protein of Agrobacterium-transferred DNA interacts with the TATA box-binding protein and a nuclear protein kinase in plants. Proc. Natl. Acad. Sci. USA 100: 10108–10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeing S., Rigault C., Heidemann M., Eick D., Meisterernst M. (2010). RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. J. Biol. Chem. 285: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. (2009). Progression through the RNA polymerase II CTD cycle. Mol. Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R.D., Heidemann M., Albert T.K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007). Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Cui X., Fan B., Scholz J., Chen Z. (2007). Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell 19: 1388–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inzé D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Dewitte W., Murray J.A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54: 235–264 [DOI] [PubMed] [Google Scholar]
- Dissmeyer N., Nowack M.K., Pusch S., Stals H., Inzé D., Grini P.E., Schnittger A. (2007). T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I., Portoso M., Spåhr H., Bonilla C., Gustafsson C.M., Allshire R.C., Ekwall K. (2005). RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Murphy S. (2008). Cracking the RNA polymerase II CTD code. Trends Genet. 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Egloff S., O’Reilly D., Chapman R.D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007). Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Szczepaniak S.A., Dienstbier M., Taylor A., Knight S., Murphy S. (2010). The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem. 285: 20564–20569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza F.H., Farrell A., Nourse J.L., Chamberlin H.M., Gileadi O., Morgan D.O. (1998). Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18: 6365–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W.J., Henry N.L., Wang Z., Wu X., Svejstrup J.Q., Bushnell D.A., Friedberg E.C., Kornberg R.D. (1997). Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J. Biol. Chem. 272: 19319–19327 [DOI] [PubMed] [Google Scholar]
- Fülöp K., Pettkó-Szandtner A., Magyar Z., Miskolczi P., Kondorosi E., Dudits D., Bakó L. (2005). The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 42: 810–820 [DOI] [PubMed] [Google Scholar]
- Ghosh A., Shuman S., Lima C.D. (2011). Structural insights to how mammalian capping enzyme reads the CTD code. Mol. Cell 43: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R.P., Bentley D.L. (2009). TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29: 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J., Kotovic K.M., Hujer K., Neugebauer K.M. (2005). Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Gruber J.J., et al. (2009). Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. (2009). The Arabidopsis cell division cycle. The Arabidopsis Book 7: e0120 doi/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H., Shinmyo A., Sekine M. (2007). Phosphorylation of threonine 161 in plant cyclin-dependent kinase A is required for cell division by activation of its associated kinase. Plant J. 52: 435–448 [DOI] [PubMed] [Google Scholar]
- Ho C., Shuman S. (1999). Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Hsin J.P., Sheth A., Manley J.L. (2011). RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 334: 683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Riechmann J.L., Meyerowitz E.M. (2008). Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.C., Phatnani H.P., Haystead T.A., MacDonald J.A., Alam S.M., Greenleaf A.L. (2004). C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 279: 24957–24964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis P., Sutton A., Solomon M.J. (1996). The Cdk-activating kinase (CAK) from budding yeast. Cell 86: 553–564 [DOI] [PubMed] [Google Scholar]
- Kataoka N., Fujita M., Ohno M. (2009). Functional association of the Microprocessor complex with the spliceosome. Mol. Cell. Biol. 29: 3243–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Goto D.B., Martienssen R.A., Urano T., Furukawa K., Murakami Y. (2005). RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Kim H., Erickson B., Luo W., Seward D., Graber J.H., Pollock D.D., Megee P.C., Bentley D.L. (2010). Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Yang J.Y., Xu J., Jang I.C., Prigge M.J., Chua N.H. (2008). Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 49: 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Suh H., Cho E.J., Buratowski S. (2009). Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Kim V.N. (2007). Processing of intronic microRNAs. EMBO J. 26: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., deJong F., Villacorta N., Szakonyi D., Koncz Z. (January 26, 2012). The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3 (online), doi/10.3389/fpls.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Martini N., Szabados L., Hrouda M., Bachmair A., Schell J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, S. Gelvin and B. Schilperoort, eds (Dordrecht-Boston-London: Kluwer Academic Publishers), pp. 1–22 [Google Scholar]
- Kurihara Y., Takashi Y., Watanabe Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Zeller G., Henz S.R., Buechel S., Sachsenberg T., Wang J.W., Rätsch G., Weigel D. (2010). Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proc. Natl. Acad. Sci. USA 107: 17466–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T., Barboric M. (2010). P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 7: 145–150 [DOI] [PubMed] [Google Scholar]
- Machida S., Chen H.Y., Adam Yuan Y. (2011). Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res. 39: 7828–7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Lidschreiber M., Siebert M., Leike K., Söding J., Cramer P. (2010). Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong N., Rosonina E., Yankulov K., Brothers G., Siderovski D., Hessel A., Foster S., Shuman S., Bentley D.L. (1997). 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11: 3306–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J. (2008). Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 15: 902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Schell J., Koncz C. (1990). Homologous domains of the largest subunit of eucaryotic RNA polymerase II are conserved in plants. Mol. Gen. Genet. 223: 65–75 [DOI] [PubMed] [Google Scholar]
- Pall G.S., Hamilton A.J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Papadopoulos J.S., Agarwala R. (2007). COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079 [DOI] [PubMed] [Google Scholar]
- Pawlicki J.M., Steitz J.A. (2010). Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 20: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R., Bentley D. (2009). “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B.M., Price D.H. (2006). Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Ronemus M., Vaughn M.W., Martienssen R.A. (2006). MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18: 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H., Mäkelä T.P., Conaway J.W., Conaway R.C., Weinberg R.A., Young R.A. (1995). Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374: 280–282 [DOI] [PubMed] [Google Scholar]
- Shimotohno A., Matsubayashi S., Yamaguchi M., Uchimiya H., Umeda M. (2003). Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett. 534: 69–74 [DOI] [PubMed] [Google Scholar]
- Shimotohno A., Ohno R., Bisova K., Sakaguchi N., Huang J., Koncz C., Uchimiya H., Umeda M. (2006). Diverse phosphoregulatory mechanisms controlling cyclin-dependent kinase-activating kinases in Arabidopsis. Plant J. 47: 701–710 [DOI] [PubMed] [Google Scholar]
- Shimotohno A., Umeda-Hara C., Bisova K., Uchimiya H., Umeda M. (2004). The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 16: 2954–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R.J., III, Millhouse S., Chen C.F., Lewis B.A., Erdjument-Bromage H., Tempst P., Manley J.L., Reinberg D. (2007). Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka H., Ohno R., Umeda M. (2009). The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. Plant J. 59: 475–487 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen J.R., Zhang D.W., Rodríguez-Molina J.B., White B.E., Akhtar M.S., Heidemann M., Li X., Chapman R.D., Shokat K., Keles S., Eick D., Ansari A.Z. (2010). Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M., Bhalerao R.P., Schell J., Uchimiya H., Koncz C. (1998). A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95: 5021–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M., Shimotohno A., Yamaguchi M. (2005). Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol. 46: 1437–1442 [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F. (2006). Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci. 11: 460–468 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Legrand S., Windels D. (2010). The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 15: 337–345 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Xiao T., Hall H., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D. (2003). Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Khanna K., Ruan S. (2010). Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 21: 790–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C., Chen X. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.