This work shows that NITRIC OXIDE ASSOCIATED1 (NOA1) controlling nitric oxide (NO) biosynthesis is negatively regulated by oleic acid (18:1) via direct binding, leading to protease-dependent NOA1 degradation. Conversely, reductions in 18:1 lead to increased levels of NOA1, inducing NO synthesis and triggering upregulation of NO-responsive nuclear genes, thereby activating disease resistance.
Abstract
The conserved cellular metabolites nitric oxide (NO) and oleic acid (18:1) are well-known regulators of disease physiologies in diverse organism. We show that NO production in plants is regulated via 18:1. Reduction in 18:1 levels, via a genetic mutation in the 18:1-synthesizing gene SUPPRESSOR OF SA INSENSITIVITY OF npr1-5 (SSI2) or exogenous application of glycerol, induced NO accumulation. Furthermore, both NO application and reduction in 18:1 induced the expression of similar sets of nuclear genes. The altered defense signaling in the ssi2 mutant was partially restored by a mutation in NITRIC OXIDE ASSOCIATED1 (NOA1) and completely restored by double mutations in NOA1 and either of the nitrate reductases. Biochemical studies showed that 18:1 physically bound NOA1, in turn leading to its degradation in a protease-dependent manner. In concurrence, overexpression of NOA1 did not promote NO-derived defense signaling in wild-type plants unless 18:1 levels were lowered. Subcellular localization showed that NOA1 and the 18:1 synthesizing SSI2 proteins were present in close proximity within the nucleoids of chloroplasts. Indeed, pathogen-induced or low-18:1-induced accumulation of NO was primarily detected in the chloroplasts and their nucleoids. Together, these data suggest that 18:1 levels regulate NO synthesis, and, thereby, NO-mediated signaling, by regulating NOA1 levels.
INTRODUCTION
Fatty acids (FAs) are essential macromolecules present in all living organisms. FAs not only serve as the major source of reserve energy but also constitute complex lipids that are essential components of cellular membranes. Increasing evidence implicates FAs and their derivatives as signaling molecules, modulating normal and disease-related physiologies in microbes, insects, animals, and plants alike. For example, the T cell response to infection is modulated by eicosapentanoic acid, which induces anti-inflammatory effects (Denys et al., 2001). FAs also serve as alarm molecules to repel phylogenetically related or unrelated species in insects (Rollo et al., 1994). Unsaturated FAs and their derivatives regulate sporulation, sexual structure development, and host seed colonization in mycotoxic Aspergillus spp (Calvo et al., 1999; Wilson et al., 2004). In plants, FAs modulate a variety of responses to both biotic and abiotic stresses (reviewed in Kachroo and Kachroo, 2009; Savchenko et al., 2010). For example, polyunsaturated FA levels in chloroplastic membranes affect membrane lipid fluidity and determine the plants ability to acclimatize to temperature stress (Routaboul et al., 2000; Iba, 2002). Linolenic acid is involved in protein modifications in heat-stressed plants (Yamauchi et al., 2008). FAs also regulate salt, drought, and heavy metal tolerance as well as wounding-induced responses and defense against insect/herbivore feeding in plants (Upchurch, 2008).
De novo FA biosynthesis occurs exclusively in the plastids of all plant cells and leads to the synthesis of palmitic acid and oleic acid (18:1) (Kachroo and Kachroo, 2009). Stearoyl-ACP desaturase (SACPD), which catalyzes the desaturation of stearic acid (18:0) to 18:1, is one of the important soluble chloroplastic enzymes that regulates the generation of monounsaturated FA in plant cells (Shanklin and Cahoon, 1998; Kachroo et al., 2007). The Arabidopsis thaliana genome encodes seven isoforms of SACPD (Kachroo et al., 2007). Yet, a mutation in SUPPRESSOR OF SA INSENSITIVITY OF npr1-5 (SSI2), encoding one of the SACPD isoforms, results in the constitutive activation of defense responses (Chandra-Shekara et al., 2007; P. Kachroo et al., 2001, 2003, 2005; A. Kachroo et al., 2003, 2004, 2007; Venugopal et al., 2009; Xia et al., 2009) and is not compensated for by endogenous expression of the other isoforms. Mutations in genes encoding two other SACPD isoforms do not induce defense signaling, suggesting a specific role for the SSI2 isoform in regulating defense signaling (Kachroo et al., 2007). Detailed characterization has shown that the constitutive defense in ssi2 loss-of-function mutant plants is due to their inability to accumulate chloroplastic 18:1 (P. Kachroo et al., 2001, 2003, 2005; A. Kachroo et al., 2003, 2004, 2007; Chandra-Shekara et al., 2007; Venugopal et al., 2009; Xia et al., 2009), which via an unknown mechanism induces the expression of multiple nuclear-encoded resistance (R) genes (Chandra-Shekara et al., 2007; Venugopal et al., 2009; Xia et al., 2009). Restoration of 18:1 levels via second site mutations in the chloroplast-targeted glycerol-3-phosphate (G3P) acyltransferase (ACT1; A. Kachroo et al., 2003), G3P dehydrogenase (GLY1; Kachroo et al., 2004), or acyl carrier protein 4 (Xia et al., 2009) normalizes R gene expression and, thereby, the altered defense phenotypes of ssi2 plants. In wild-type plants, 18:1 levels can be reduced by the exogenous application of glycerol, which increases ACT1 catalysis and, thereby, 18:1 use (A. Kachroo et al., 2004; P. Kachroo et al., 2005).
Like 18:1, nitric oxide (NO) is a conserved signaling molecule common to plants and animals (Wendehenne et al., 2001; Besson-Bard et al., 2008). In plants, NO is known to participate in several responses, including germination, flowering, stomatal closure, and pathogen defense (Delledonne et al., 1998; Durner et al., 1998; He et al., 2004; Besson-Bard et al., 2008; Wilson et al., 2008). NO biosynthesis in plants is thought to occur via nitrate reductase (NR) and NITRIC OXIDE ASSOCIATED1 (NOA1)–catalyzed reactions (Wendehenne et al., 2001; Desikan et al., 2002; Guo et al., 2003; Crawford, 2006; Besson-Bard et al., 2008). NR is a cytosolic enzyme that catalyzes NAD(P)H-dependent reduction of nitrate to nitrite (Besson-Bard et al., 2008; Moreau et al., 2008). NOA1 was earlier thought to function similar to mammalian NO synthases (Guo et al., 2003) but was recently shown to have GTPase rather than NO synthase activity (Moreau et al., 2008). At present, the relationship between GTPase activity and its role in NO biosynthesis/accumulation or relative contributions of NR and NOA1 pathways to total NO levels in plants remains unclear. Furthermore, the regulation of NO synthesis and how NO exerts its effects in various signaling processes remain largely unclear.
In this study, we evaluated the relationship between low-18:1- and NO-mediated defense signaling pathways. We show that 18:1 synthesized within chloroplast nucleoids regulates the stability of NOA1 and, thereby, NO biosynthesis/accumulation. Reductions in 18:1 levels led to increased levels of NOA1 protein, which in turn increased NO levels. This triggered transcriptional upregulation of NO responsive nuclear genes, thereby activating disease resistance.
RESULTS
ssi2 Loss-of-Function Mutants Accumulate High Levels of Chloroplastic NO
Similar to the ssi2 mutation, application of glycerol induces expression of various nuclear-encoded R genes in wild-type plants in an ACT1-dependent manner (Kachroo et al., 2004; Chandra-Shekara et al., 2007; Venugopal et al., 2009; Xia et al., 2009). These observations suggest that changes in chloroplastic 18:1 levels can induce nuclear gene expression. We hypothesized that 18:1 levels might regulate key molecule(s) that directly or indirectly lead to the induction of R genes. One possibility was that reduction in 18:1 levels induced the formation/accumulation of an intermediate signaling component(s) that directly or indirectly triggered the expression of R genes. To test this hypothesis, we first generated a transcriptional profile of ssi2 plants using Affymetrix arrays and compared this to the transcriptional profiles of wild-type plants exposed to various biotic and abiotic treatments (obtained from the National Center for Biotechnology Information database). Strikingly, the transcription activation profile of ssi2 plants remarkably overlapped with that of NO-treated wild-type plants; of 261 genes reported as induced by 1 mM NO donor, sodium nitroprusside (SNP; Parani et al., 2004), 104 were upregulated in ssi2 plants (see Supplemental Data Set 1 online). Notably, only 81 genes were upregulated when SNP was applied at lower concentrations (0.1 mM; Parani et al., 2004), suggesting that NO modulates gene expression in a concentration-dependent manner. Of the 104 NO-inducible genes upregulated in ssi2 plants, 68 were also induced in the ssi2 sid2 double mutant plants (see Supplemental Data Set 1 online), which exhibit ssi2-like phenotypes due to their low 18:1 levels but contain reduced levels of SA. By contrast, a majority of the NO-responsive genes were expressed at wild-type-like levels in ssi2 act1 double mutants, which are restored in 18:1 levels and exhibit wild-type-like defense responses (A. Kachroo et al., 2003) (see Supplemental Data Set 1 online). Together, these results suggest a correlation between the ssi2 phenotypes and increased expression of NO responsive genes.
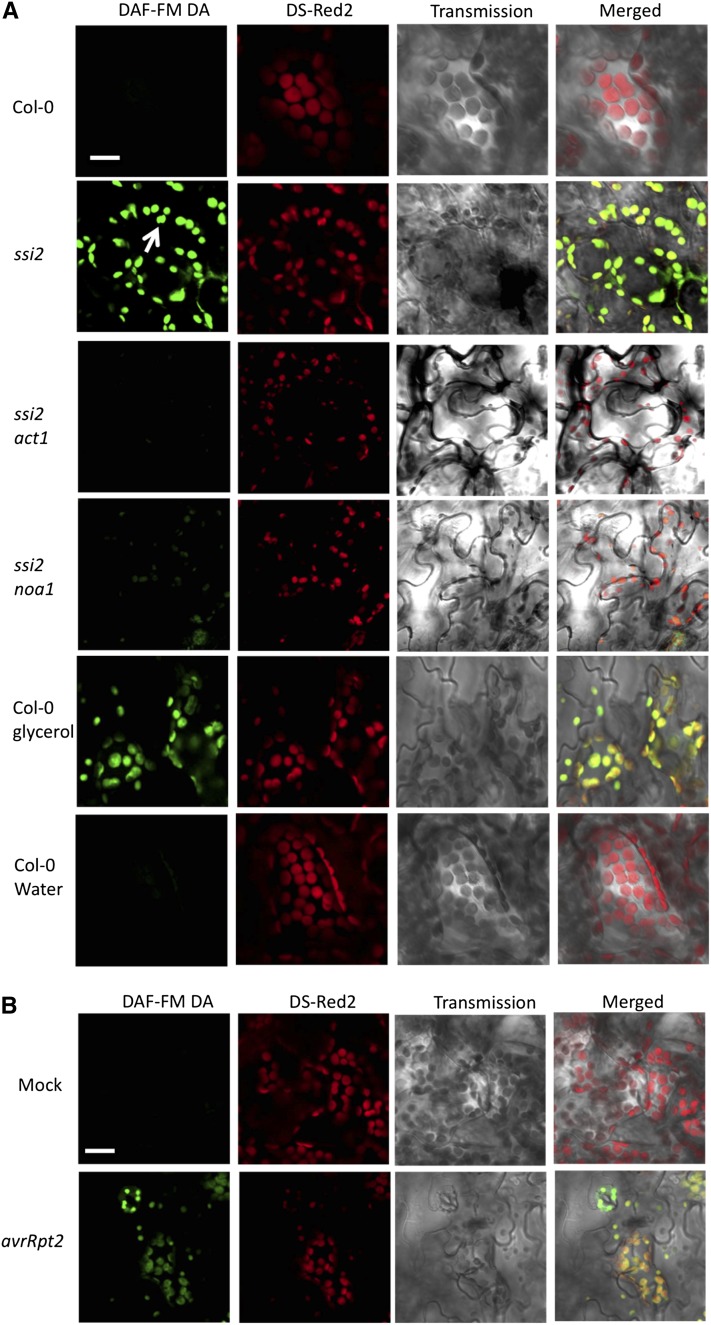
We tested if ssi2 plants accumulated increased NO by staining wild-type and ssi2 plants with the NO-sensitive dye 4-amino-5-methylamino-2,7-difluorofluorescein diacetate (DAF-FM DA; Balcerczyk et al., 2005). The NO donor 2-(N,N-diethylamino)-diazenolate-2-oxide (DEA-NONOate) and nitrous oxide donor sulfo-NONOate were used as positive and negative controls, respectively, to confirm specificity of DAF-FM DA under our conditions (see Supplemental Figure 1A online). Interestingly, confocal microscopy of DAF-FM DA–stained ssi2 leaves showed increased fluorescence (detected as green fluorescence) compared with wild-type plants (Figure 1A), and this correlated with in vitro fluorescence measurements of leaf tissue extracts incubated with DAF-FM DA (see Supplemental Figure 1B online). Consistent with their transcriptional profiles and defense phenotypes, ssi2 sid2 plants showed increased DAF-FM DA fluorescence, but ssi2 act1 plants did not (Figure 1A). The NO levels were also quantified using the Griess reaction assay, which is based on the spontaneous oxidation of NO to nitrite under physiological conditions (Sun et al., 2003) (see Supplemental Figure 1C online). Consistent with the DAF-FM DA–based analysis, Griess assays showed increased levels of nitrite in ssi2 plants (see Supplemental Figure 1C online). Although both methods for NO analysis provided consistent qualitative data, we are unable to make precise quantitative estimates at this time because the method used for in vitro quantification of NO is prone to artifacts (Balcerczyk et al., 2005; Planchet and Kaiser, 2006). To further test the correlation between low 18:1 and high NO, we analyzed NO accumulation in water- and glycerol-treated wild-type Columbia-0 (Col-0) plants. Glycerol application, which lowers 18:1 levels (Kachroo et al., 2004), also induced NO accumulation in the chloroplasts of Col-0 plants (Figure 1A). Unlike glycerol, other osmotic agents that do not lower 18:1 levels, including mannitol or sorbitol did not induce NO accumulation (data shown for mannitol; see Supplemental Figure 1D online). Likewise, glycerol application also induced NO accumulation in Nicotiana benthamiana chloroplasts (see Supplemental Figure 1E online). NO accumulation in response to low 18:1 mimicked pathogen induced accumulation of NO; inoculations with Pseudomonas syringae expressing avrRpt2 resulted in NO accumulation in the chloroplasts within 12 h after inoculation (Figure 1B). Notably, pathogen-induced NO accumulation preceded the increase in salicylic acid (SA) levels (see Supplemental Figure 1F online), which was consistent with the result that exogenous NO induces SA biosynthetic genes and, thereby, SA levels (Durner et al., 1998).
Figure 1.
The ssi2 Plants Accumulate High Levels of Chloroplastic NO.
(A) Confocal micrograph of DAF-FM DA–stained leaves showing subcellular location of NO in wild-type (Col-0) plants, ssi2, ssi2 act1, and ssi2 noa1 mutants, and glycerol-treated Col-0 wild-type plants. Chloroplast autofluorescence (red) was visualized using Ds-Red2 channel. Arrow indicates chloroplast. At least 10 independent leaves were analyzed in four experiments with similar results. Bar = 10 μm.
(B) Confocal micrograph showing pathogen-induced NO accumulation in Col-0 plants at 12 h after inoculation. Plants were inoculated with MgCl2 (mock) or avrRpt2 P. syringae. At least 10 independent leaves were analyzed in four experiments with similar results. Bar = 10 μm.
NOA1-Derived NO Contributes to Defense Phenotypes in ssi2 Plants
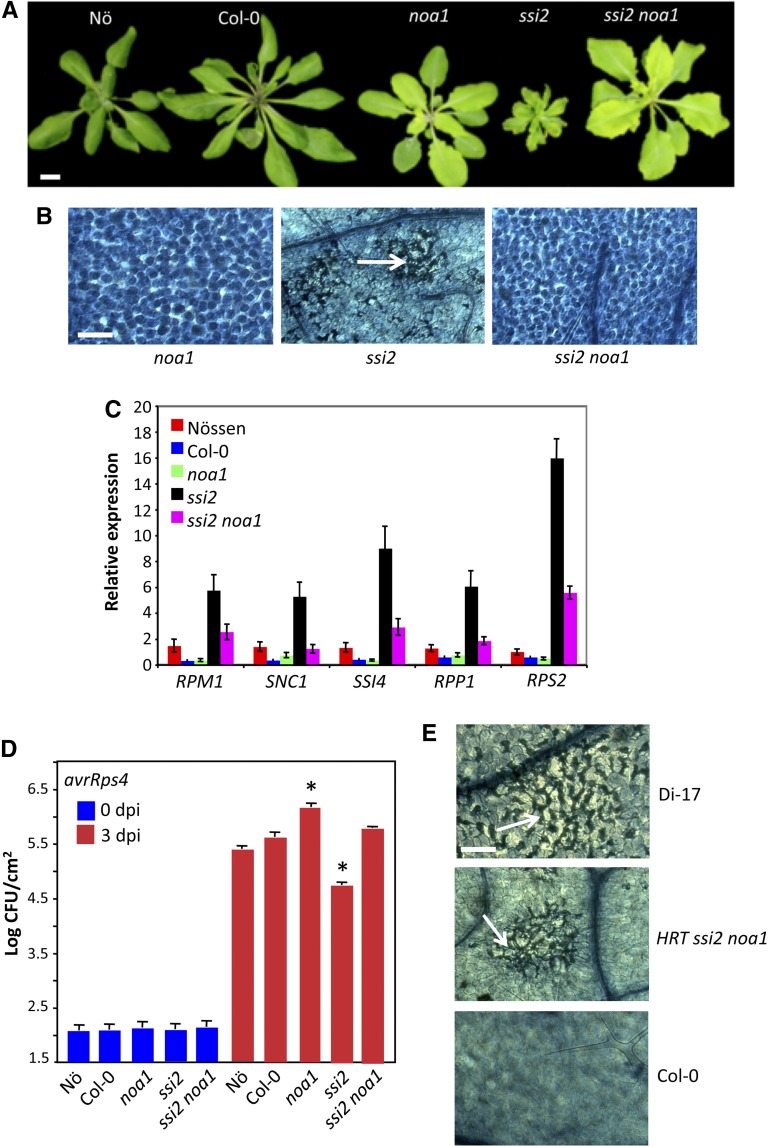
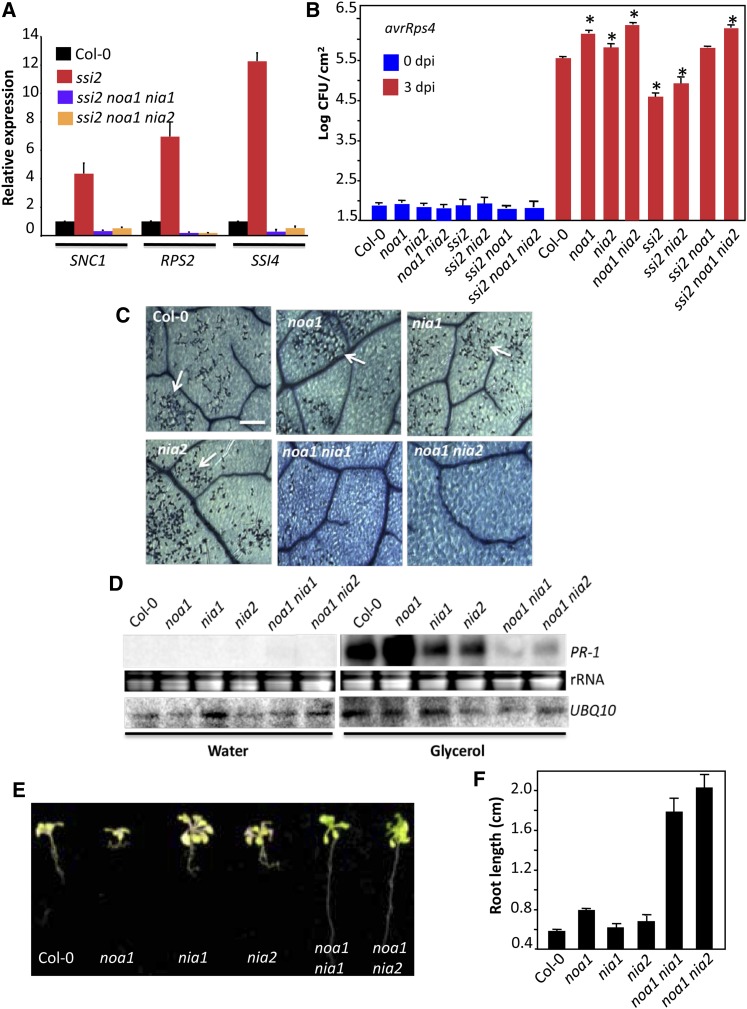
Increased accumulation of NO in the chloroplasts of ssi2 plants and the observation that the chloroplastic NOA1 contributes to elicitor-mediated accumulation of NO (Guo et al., 2003; Zeidler et al., 2004; Gas et al., 2009), prompted us to test the role of NOA1 in ssi2-mediated signaling. We crossed ssi2 mutants with noa1 mutant plants and analyzed F2 progeny for ssi2-like phenotypes. Consistent with digenic segregation, approximately one of 16 F2 plants showed wild-type-like morphology (Figure 2A); 10 of 147 plants contained the ssi2 mutation but showed wild-type-like phenotypes (χ2 = 0.08, P = 0.77). In comparison to ssi2, the ssi2 noa1 plants accumulated much lower levels of NO (Figure 1A; see Supplemental Figures 2A and 2B online) and showed no visible or microscopic cell death (Figures 2A and 2B). To confirm that the restoration of morphological and defense phenotypes in ssi2 noa1 was due to the noa1 mutation, we transformed a wild-type genomic copy of NOA1 into ssi2 noa1 plants and scored phenotypes in T1 and T2 generations (see Supplemental Figures 2C to 2E online). The ssi2 noa1 plants containing the NOA1 transgene showed ssi2-like morphology (see Supplemental Figure 2C online), constitutive cell death (see Supplemental Figure 2D online), and Pathogenesis Related (PR)-1 expression (see Supplemental Figure 2E online), thus confirming a role for NOA1 in ssi2-triggered phenotypes. In contrast with the ssi2 mutation, noa1 did not abolish the constitutive defense phenotypes in another mutant, Constitutive Expressor of PR genes5 (cpr5) (see Supplemental Figure 3 online). Like ssi2, the cpr5 plants are constitutively activated in defense signaling, but this is not due to changes in 18:1 levels (see Supplemental Figure 3E online). Together, these results suggested that NOA1 specifically participates in low 18:1-derived signaling.
Figure 2.
NOA1-Derived NO Contributes to Defense Phenotypes in ssi2 Mutant Plants.
(A) Morphological phenotype of 3-week-old plants. Bar = 0.7 cm.
(B) Microscopy of trypan blue–stained leaves. Arrow indicates dead cells. At least six independent leaves were analyzed in two experiments with similar results. Bar = 270 μm.
(C) Real-time quantitative RT-PCR analysis showing relative levels of indicated R genes. The error bars represent sd (n = 3). The experiment was repeated three times with similar results. FW, fresh weight.
(D) Growth of avrRps4 bacteria on indicated genotypes. The error bars indicate sd (n = 4). Asterisks indicate data statistically significant from the wild type (Col-0, P < 0.05). The experiment was repeated three times with similar results. CFU, colony-forming units.
(E) Trypan blue–stained leaf showing microscopic cell death phenotype on turnip crinkle virus–inoculated leaves (indicated by arrows). This experiment was repeated three times with similar results. Bar = 270 μm.
[See online article for color version of this figure.]
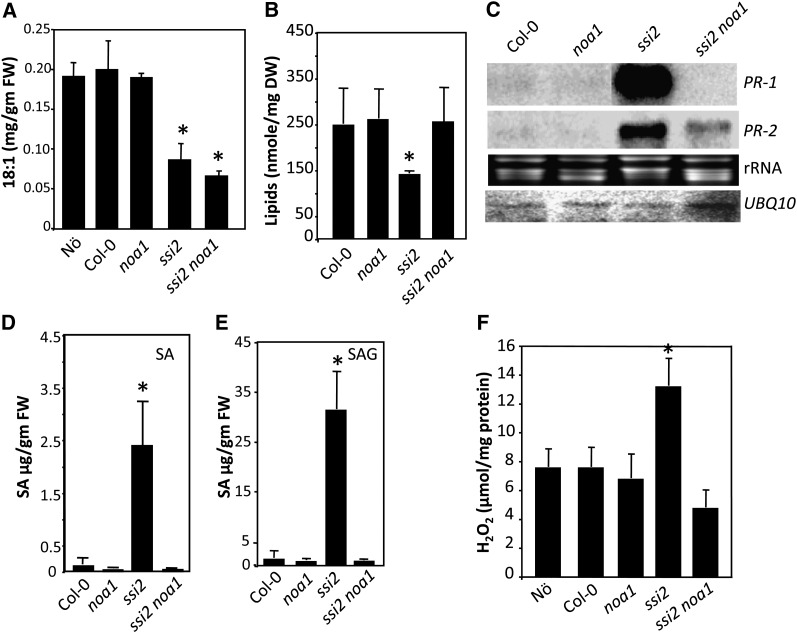
The ssi2 noa1 plants accumulated ssi2-like levels of 18:1 (Figure 3A), suggesting that NOA1 functions downstream of 18:1. Consistent with their wild-type-like morphology, levels of total lipids were significantly higher in ssi2 noa1 compared with ssi2 (Figure 3B), and this correlated with a significant increase in the levels of monogalactosyl diacylglycerol and digalactosyl diacylglycerol lipids in comparison to ssi2 plants (see Supplemental Figure 4 online). The noa1 mutation by itself did not affect the FA or lipid profile in the wild-type background (Figures 3A and 3B; see Supplemental Figure 4 online). We next evaluated the various defense phenotypes in ssi2 noa1 plants to determine if the reduction in NO levels restored ssi2-triggered defense signaling. In comparison to ssi2, the ssi2 noa1 plants showed wild-type-like levels of PR-1 and a significant reduction in PR-2 transcript (Figure 3C) and wild-type-like levels of SA (Figures 3D and 3E) and hydrogen peroxide (H2O2) (Figure 3F). However, ssi2 noa1 plants expressed higher than wild-type levels of R genes, even though these were significantly lower than in ssi2 plants (Figure 2C). Consistent with their R gene expression levels, the resistance of ssi2 noa1 to avrRps4 P. syringae was intermediate to ssi2 and noa1 plants (Figure 2D). Similarly, hypersensitive response formation in response to turnip crinkle virus in ssi2 noa1 plants containing the R gene HRT was intermediate to hypersensitive response on the resistance ecotype Di-17 (HRT/HRT) and susceptible ecotype Col-0 (hrt/hrt) (Figure 2E). Together, these results suggested that ssi2 noa1 plants were not completely restored in R gene expression or pathogen response.
Figure 3.
NOA1 Functions Downstream of 18:1.
(A) Levels of FAs in 4-week-old plants. The error bars represent sd (n = 6). Asterisks denote significant differences with wild-type plants (t test, P < 0.05). FW, fresh weight.
(B) Total lipid levels in indicated genotypes. The error bars represent sd (n = 5). Asterisks denote significant differences with wild-type plants (t test, P < 0.05). The experiment was repeated twice with similar results. DW, dry weight.
(C) RNA gel blot showing transcript levels of PR-1 and PR-2 genes. Ubiquitin mRNA (UBQ10) and ethidium bromide staining of rRNA were used as loading controls. This experiment was repeated four times with similar results.
(D) and (E) SA (D) and SA glucoside (E) levels in indicated genotypes. The error bars represent sd (n = 3). Asterisks denote significant differences with wild-type plants (t test, P < 0.05). The experiment was repeated twice with similar results.
(F) H2O2 levels in indicated genotypes. The error bars represent sd. Asterisks denote a significant difference from the wild type (t test, P < 0.05). H2O2 was quantified from the tissue extracts prepared as described in Methods. The experiment was repeated twice with similar results.
NOA1, NIA1, and NIA2 Contribute Additively to NO Accumulation in ssi2 Plants
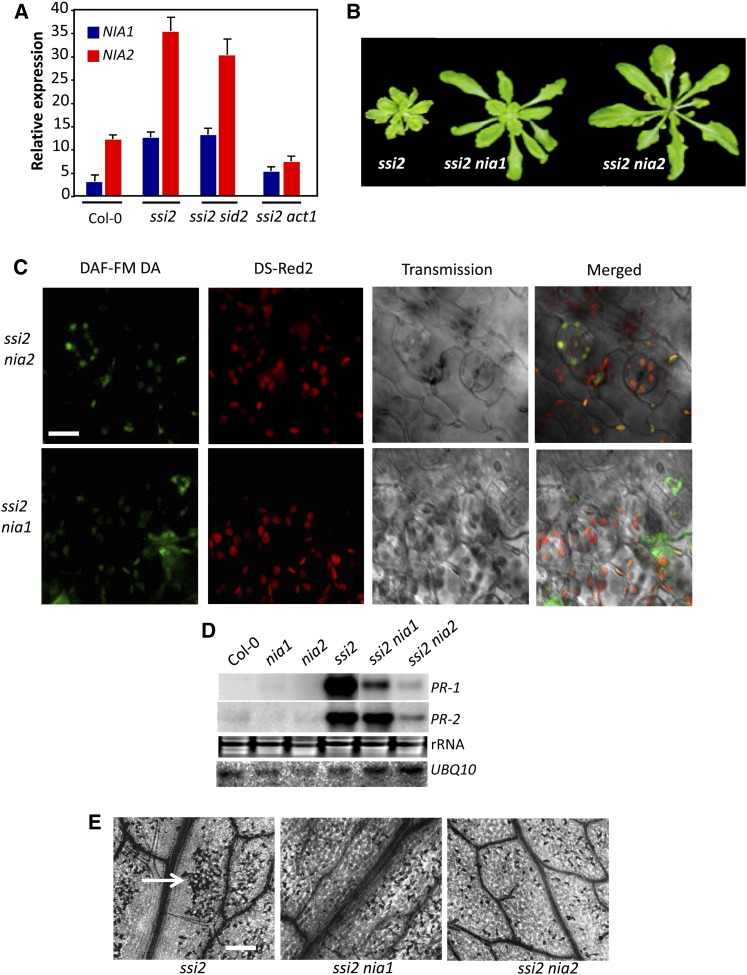
The ssi2 noa1 plants were not completely restored in R gene expression or pathogen response, suggesting that an additional factor(s) contributed to the nominally increased R gene expression in these plants. It was possible that residual NO levels in ssi2 noa1 plants were sufficient to trigger a low level increase in R gene expression. To test this, we assayed R gene expression levels in wild-type plants treated with SNP or DEA-NONOate. Indeed, both SNP and DEA-NONOate were able to induce R gene expression in wild-type plants (see Supplemental Figures 5A and 5B online). This result prompted us to investigate the role of NRs in ssi2 triggered phenotypes, since NO is also generated as a byproduct of the NR (encoded by NIA1 and NIA2 in Arabidopsis) catalyzed reactions (Desikan et al., 2002; Planchet and Kaiser, 2006; Besson-Bard et al., 2008). To determine if NIA1 and/or NIA2 contributed to the accumulation of NO in ssi2 plants, we first evaluated the expression of NIA1 and NIA2 transcripts in wild-type, ssi2, ssi2 sid2, and ssi2 act1 plants. Notably, NIA1 and NIA2 expression correlated with ssi2 phenotypes; the NIA1 and NIA2 transcript levels were elevated in ssi2 and ssi2 sid2 but not in ssi2 act1 plants (Figure 4A; see Supplemental Figure 5C online). Exogenous NO or SA did not induce expression of NIA1 and NIA2 genes (data not shown; see Supplemental Table 1 online), suggesting that their induction was specific to low 18:1 levels. Consistent with this result, expression of NIA1 and NIA2 was also upregulated in ssi2 noa1 plants (see Supplemental Figure 5C online). By contrast, NOA1 expression was not upregulated in the ssi2 plants. Although the ssi2 nia1 plants showed high expression of NIA2, the ssi2 nia2 plants were significantly reduced in expression of both NIA1 and NIA2 genes. We next estimated NR activity in ssi2, ssi2 noa1, ssi2 nia1, and ssi2 nia2 plants. The ssi2 plants show increased NR activity, which correlated with the increased expression of NIA genes in these plants. The NR activity correlated well with the proposed contribution of NIA1 and NIA2 to the total NR activity (Wilkinson and Crawford, 1991); the nia2 and ssi2 nia2 plants showed greatly reduced NR activity (~17% of NR activity detected in wild-type plants), and NR activity in ssi2 nia1 plants was at levels intermediate between wild-type and ssi2 plants. Notably, although noa1 plants showed wild-type-like NR activity, the ssi2 noa1 plants were significantly reduced in their NR activity (see Supplemental Figure 5D online). A likely explanation is that NO generated via NOA1 might regulate NR activity, and this scenario is supported by the observation that exogenous treatment with NO donors increases NR activity (Jin et al., 2009). Together, these results suggested that reduction in 18:1 levels resulted in NO accumulation via the upregulation of the NR activity and the posttranscriptional alteration of NOA1.
Figure 4.
NIA1 and NIA2 Contribute to NO Accumulation in ssi2 Plants.
(A) Real-time quantitative RT-PCR analysis showing relative levels of indicated genes. The experiment was repeated twice with similar results.
(B) Morphological phenotypes of 3-week-old plants.
(C) Confocal micrograph of DAF-FM DA–stained leaves showing subcellular location of NO in ssi2 nia1 and ssi2 nia2 plants (see Supplemental Figure 7B online). Chloroplast autofluorescence (red) was visualized using Ds-Red2 channel. Bar = 10 μm.
(D) RNA gel blot showing transcript levels of PR-1 and PR-2 genes. Ubiquitin mRNA (UBQ10) and ethidium bromide staining of rRNA were used as loading controls. The experiment was repeated three times with similar results.
(E) Microscopy of trypan blue stained leaves. Arrow indicates dead cells. At least six independent leaves were analyzed in two experiments with similar results. Bar = 270 μm.
To determine if the increased expression of NIA1 and NIA2 contributed to the NO-derived phenotypes in ssi2 plants, we generated ssi2 nia1 and ssi2 nia2 plants. Both ssi2 nia1 and ssi2 nia2 plants showed improved morphology (Figure 4B), which correlated with an increase in total lipid and monogalactosyl diacylglycerol levels (see Supplemental Figures 6A and 6B online). The ssi2 nia1 and ssi2 nia2 plants accumulated reduced NO (Figure 4C) or SA (see Supplemental Figure 6C online) and displayed reduced cell death and PR expression (Figures 4D and 4E). However, compared with ssi2 nia1, the ssi2 nia2 plants showed slightly bigger morphology and a pronounced reduction in cell death and PR expression (Figures 4B, 4D, and 4E). A better suppression of ssi2 triggered defense phenotypes in ssi2 nia2 plants correlated with the fact the NIA2 contributes to ~90% of total NR activity (Wilkinson and Crawford, 1991). Intriguingly, even though NIA1 and NIA2 localized to the extrachloroplastic compartment (see Supplemental Figure 7A online), mutations in these lowered chloroplastic NO levels in ssi2 plants (Figures 3C; see Supplemental Figure 7B online). This and the effect of the noa1 mutation on NR activity in ssi2 noa1 plants (see Supplemental Figure 5D online) suggest that NO synthesis and/or accumulation likely involve feedback regulation between NOA1- and NIA1/NIA2-dependent pathways.
To determine if the relative contributions of NOA1 and NIA1/NIA2 resulted in additive effects, we generated and evaluated defense phenotypes in ssi2 noa1 nia1 and ssi2 noa1 nia2 plants. Interestingly, the ssi2 noa1 nia1 and ssi2 noa1 nia2 showed basal level expression of R genes and compromised resistance to avirulent pathogens (Figures 5A and 5B). Consistent with this result, pathogen-treated noa1 nia2 plants showed greater reduction in NO levels compared with single mutant plants (see Supplemental Figures 8A and 8B online). We next assayed glycerol-triggered phenotypes in the noa1, nia1, and nia2 single mutant plants and the noa1 nia1 and noa1 nia2 double mutant plants. As shown earlier, exogenous application of glycerol reduced 18:1 levels in wild-type plants (see Supplemental Figure 9 online), resulting in the induction of cell death and PR-1 expression (Figures 5C and 5D). Glycerol application also lowered 18:1 levels in all mutant genotypes (see Supplemental Figure 9 online). However, glycerol application only induced PR-1 expression and cell death in the noa1, nia1, and nia2 single mutants but not in the noa1 nia1 and noa1 nia2 double mutants (Figures 5C and 5D). Glycerol-mediated depletion of 18:1 also inhibited root growth in wild-type and single mutants but not the double mutant plants (Figures 5E and 5F). Together, these results suggest that the combined loss of NOA1 with NIA1 or NIA2 is essential to completely abolish the increased R expression and altered defense phenotypes under low 18:1 conditions.
Figure 5.
NOA1, NIA1, and NIA2 Contribute Additively to NO Accumulation in ssi2 Plants.
(A) Real-time quantitative RT-PCR analysis showing relative levels of indicated R genes. The error bars indicate sd (n = 3). The experiment was repeated three times with similar results.
(B) Growth of avrRps4 bacteria on indicated genotypes. The error bars indicate sd (n = 4). Asterisks indicate data significantly different from the wild type (Col-0, P < 0.05). The experiment was repeated twice with similar results.
(C) Microscopy of trypan blue–stained leaves. Arrows indicate dead cells. At least six independent leaves were analyzed in two experiments with similar results. Bar = 270 μm.
(D) RNA gel blot showing transcript levels of PR-1 gene in water- and glycerol-treated plants. Ubiquitin mRNA (UBQ10) and ethidium bromide staining of rRNA were used as loading controls. The experiment was repeated three times with similar results.
(E) Morphology and root length of plants grown on Murashige and Skoog medium containing 0.1% glycerol. The experiment was repeated twice with similar results.
(F) Relative root length of plants grown on Murashige and Skoog medium containing 0.1% glycerol. The error bars represent sd (n = 25). The experiment was repeated twice with similar results.
[See online article for color version of this figure.]
NOA1 Localizes to the Chloroplastic Nucleoids
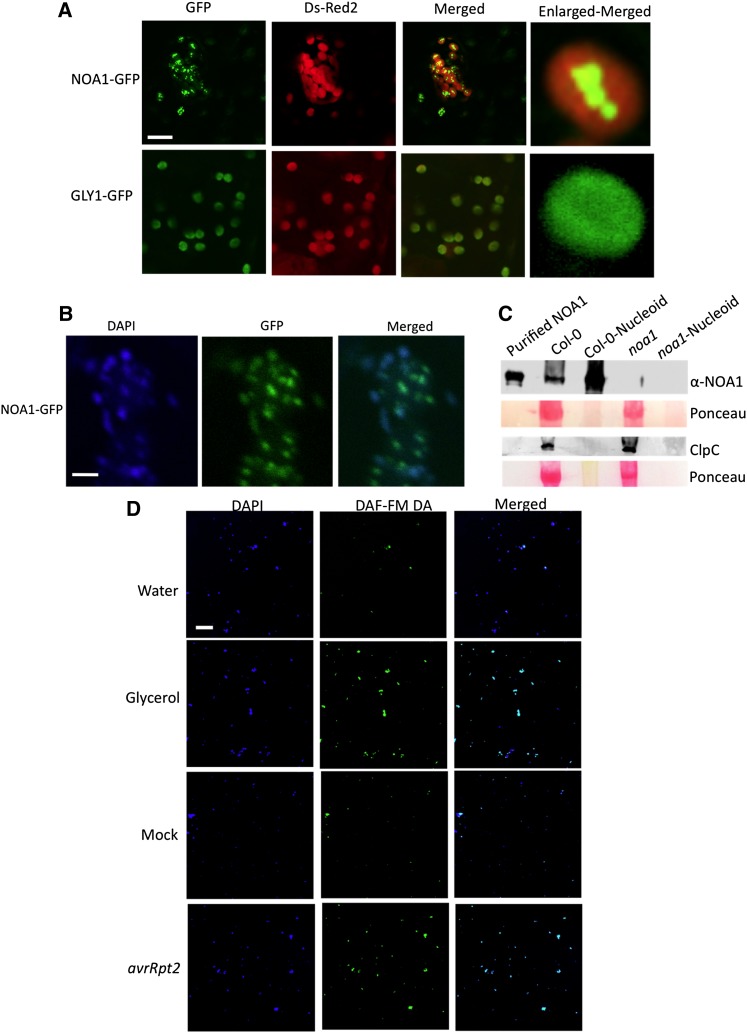
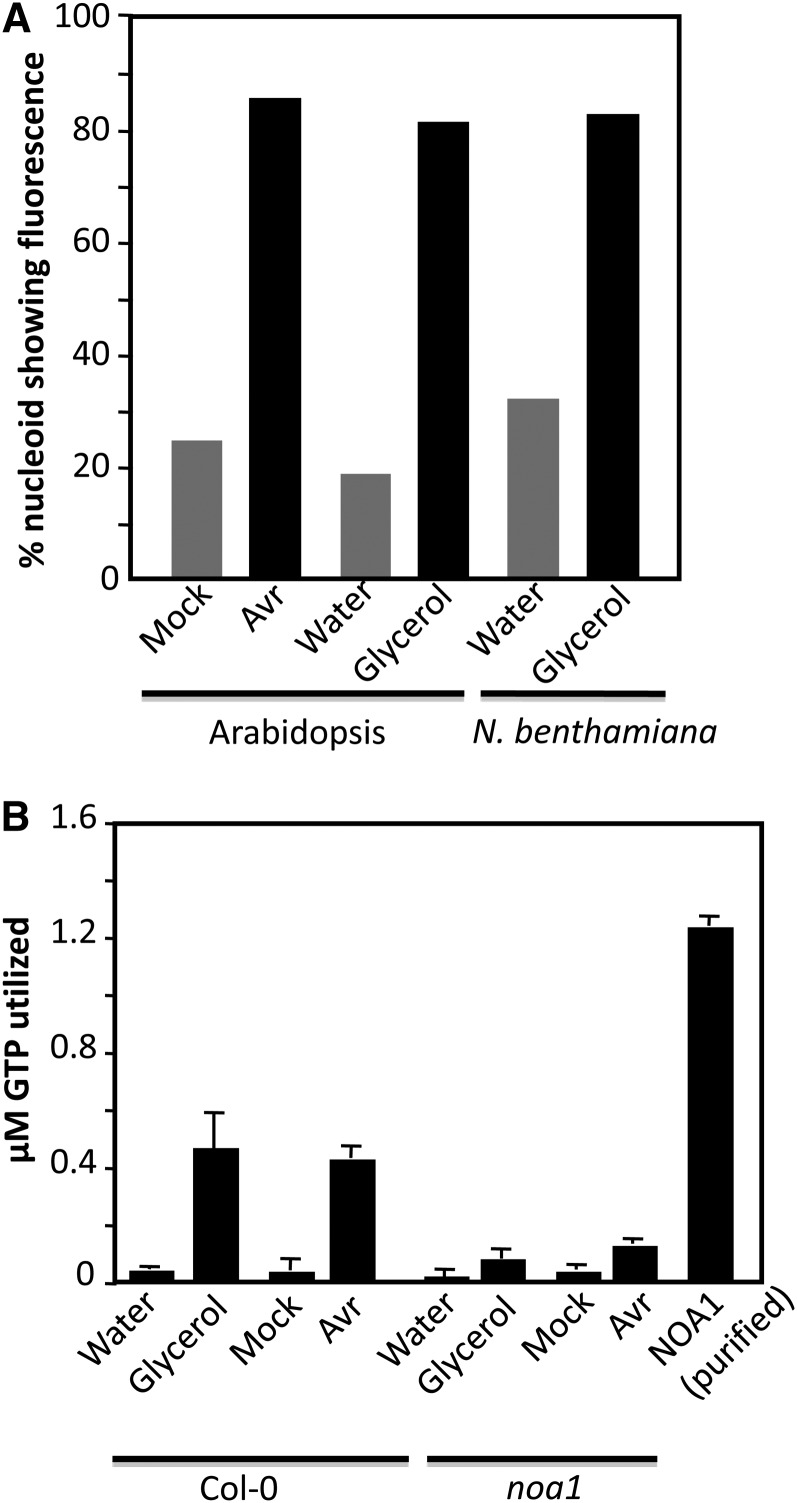
The accumulation of NO in chloroplasts correlated well with the plastidial localization of NOA1-GFP (for green fluorescent protein; Figure 6A). Intriguingly, NOA1-GFP localized in a punctate pattern within the chloroplasts, unlike other chloroplastic proteins like GLY1 (Chanda et al., 2011), which was uniformly distributed in the chloroplast (Figure 6A). Staining with 4′,6-diamidino-2-phenylindole (DAPI) identified the punctate structures as nucleoids, which are nucleus-like bodies that contain genetic material (Figure 6B). The nucleoid-specific localization of NOA1 was further confirmed by protein blot analysis using NOA1-specific antibodies (Figure 6C); NOA1 was enriched in the nucleoid fraction isolated from the wild-type plants compared with total protein extracts. As expected, NOA1 protein was not detected in noa1 mutant plants, which contain a T-DNA insertion in the first exon of the gene (Guo et al., 2003). In contrast with NOA1, stromal protein clpC was detected in total protein extracts but not nucleoid fractions, prepared from wild-type or noa1 plants. The nucleoid-specific localization of NOA1 is consistent with recent proteome analysis of maize (Zea mays) nucleoids (Majeran et al., 2012). These results suggested that perhaps NO synthesis/accumulation was initiated in the chloroplastic nucleoids. Indeed, NO staining did show intensely stained areas within the chloroplasts of ssi2 and pathogen inoculated wild-type plants (see Supplemental Figure 10 online). Furthermore, both pathogen infection and glycerol treatment increased DAF-FM staining of purified nucleoids (Figures 6D and 7A). NOA1 has been shown to possess GTPase activity (Moreau et al., 2008). We determined if increased NO accumulation in the nucleoids also correlated with increased GTPase activity. Interestingly, both pathogen infection and glycerol treatment significantly increased nucleoid associated GTPase activity in wild-type, but not in noa1, plants (Figure 7B). Thus, the increased NO and GTPase activity in the nucleoids also correlated with the localization of NOA1 in these suborganelles.
Figure 6.
NOA1 Localizes to Plastidial Nucleoids.
(A) Confocal micrograph showing localization of NOA1-GFP and GLY1-GFP proteins. Agroinfiltration was used to express proteins in N. benthamiana. Right panel shows enlarged micrographs of individual chloroplasts. The experiment was repeated four times with similar results. Bar = 10 μM.
(B) Confocal micrograph showing nucleoid specific localization of NOA1. Agroinfiltration was used to express NOA1-GFP in N. benthamiana, and the leaves were stained with DAPI prior to microscopy. The experiment was repeated three times with similar results. Bar = 2 μM.
(C) Protein gel blot showing NOA1 levels in the protein extracted from the leaves (total protein extract) or the purified nucleoids. E. coli–purified NOA1 protein was used as a positive control. A replica blot was probed with antibodies against the stromal protein clpC. Ponceau-S staining of the protein gel blot was used as the loading control. The experiment was repeated twice with similar results.
(D) Confocal micrograph showing DAF-FM DA– and DAPI-stained nucleoids. The experiment was repeated twice with similar results. Bar = 20 μm.
Figure 7.
GTPase Activity of the Nucleoid Localizing NOA1 Correlates with NO Accumulation.
(A) Percentage nucleoids showing fluorescence in water-, glycerol-, or pathogen-treated plants. Nucleoids were purified from treated plants and assayed for fluorescence under a confocal microscope. The experiment was repeated twice with similar results.
(B) GTPase activity associated with nucleoids purified from water-, glycerol-, or pathogen-treated plants at 12 h after inoculation. Protein was extracted from 108 nucleoids, and E. coli–purified NOA1 protein was used as a positive control. The experiment was repeated twice with similar results.
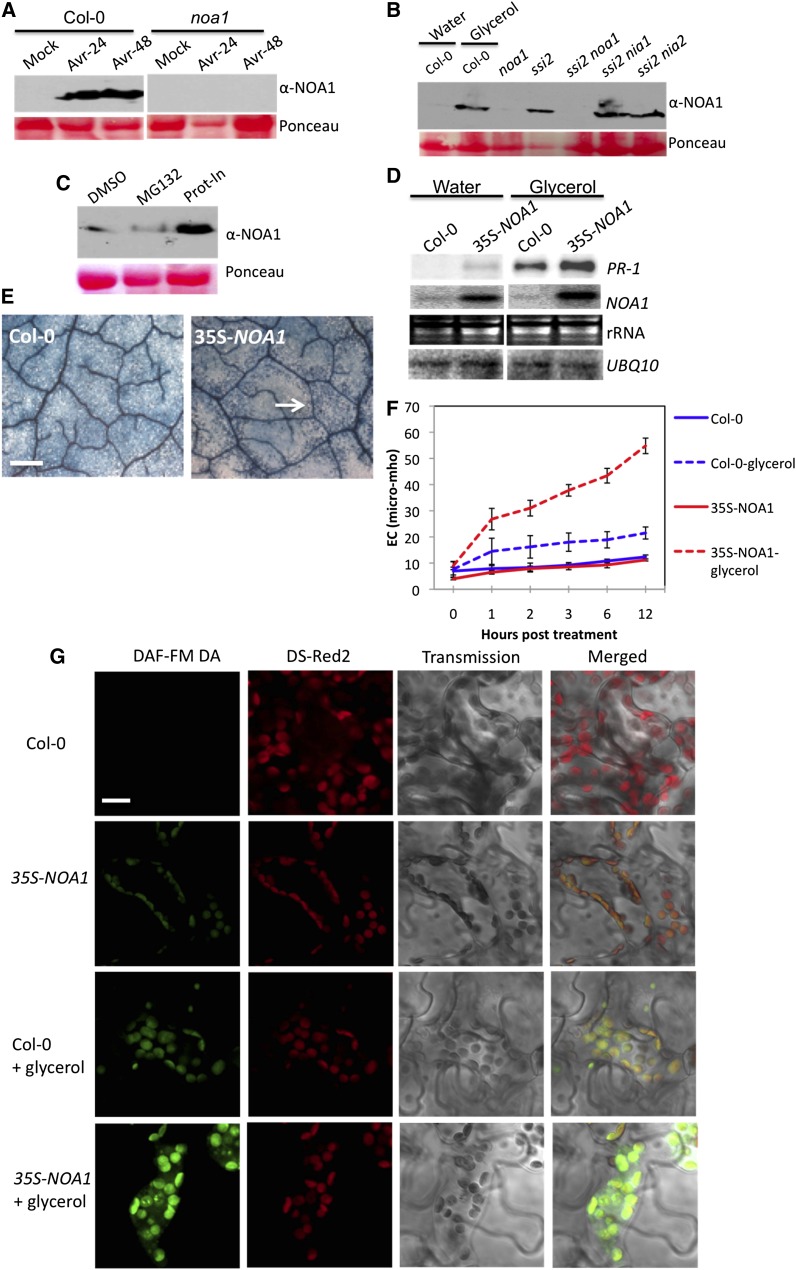
Interestingly, increased GTPase activity in the pathogen-inoculated plants correlated well with an increase in the NOA1 protein levels (Figure 8A), although the NOA1 transcript levels remained unchanged (see Supplemental Table 1 online). Similarly, glycerol treatment also increased NOA1 levels in the wild-type plants (Figure 8B), even though there was no increase in the NOA1 transcript under low 18:1 conditions (glycerol-treated plants; Figure 8D). This suggested that pathogen infection and 18:1 levels regulate the stability of NOA1 at the posttranscriptional level. Consistent with this notion increased levels of NOA1 protein was detected in ssi2, ssi2 nia1, and ssi2 nia2 plants (Figure 8B). A mutation in ssi2 did not increase the levels of three other chloroplastic proteins, suggesting that its effect on NOA1 was a specific phenotype (see Supplemental Figure 11 online). To confirm this further, we examined the effects of the 26S proteasome–specific inhibitor MG132 or protease inhibitor cocktail on NOA1 stability. The Col-0 leaves pretreated with protease inhibitor cocktail accumulated significantly higher levels of NOA1 protein compared with leaves treated with DMSO or MG132 (Figure 8C). Together, these results suggest that normal 18:1 levels promoted the protease-mediated degradation of NOA1.
Figure 8.
Overexpression of NOA1 Potentiates Low-18:1-Triggered Defense Phenotypes.
(A) Protein immunoblot showing NOA1 levels in mock or avrRpt2 bacteria (Avr)–inoculated Col-0 and noa1 plants. Leaves were sampled 24 or 48 h after inoculation. Ponceau-S staining of the immunoblot was used as the loading control. The experiment was repeated twice with similar results.
(B) Protein immunoblot showing NOA1 levels in water- or glycerol-treated Col-0 and untreated 4-week-old ssi2, ssi2 nia1, and ssi2 nia2 plants. Plants were treated with glycerol for 24 h prior to sampling. Ponceau-S staining of the immunoblot was used as the loading control. The experiment was repeated twice with similar results.
(C) Immunoblot showing NOA1 levels in total proteins extracted from Col-0 plants infiltrated with 0.1% DMSO, plant protease inhibitor cocktail (Prot-In), or the 26S proteasome specific inhibitor (MG132) for 24 h.
(D) RNA gel blot showing transcript levels of PR-1 and NOA1 genes in water- and glycerol-treated plants. Ubiquitin mRNA (UBQ10) and ethidium bromide staining of rRNA were used as loading controls. The experiment was repeated twice with similar results. Fold induction of PR-1, normalized with UBQ10, was quantified using Image Quant software.
(E) Microscopy of glycerol-treated leaves stained with trypan blue 24 h after treatment. Arrow indicates dead cells. At least four independent leaves were analyzed in two experiments with similar results. Bars = 270 μm.
(F) Electrolyte leakage in water and glycerol-treated Col-0 and 35S-NOA1 plants. Error bars represent sd (n = 6).
(G) Confocal micrograph of DAF-FM DA–stained leaves showing relative NO levels in water- and glycerol-treated plants. At least four independent leaves were analyzed in two experiments with similar results. Chloroplast autofluorescence (red) was visualized using Ds-Red2 channel. Bar = 10 μm.
We next tested if the overexpression of NOA1 in wild-type plants could relieve the 18:1-mediated repression of NOA1. Notably, 35S-NOA1 plants showed normal phenotype and near basal levels of defense gene expression. However, 35S-NOA1 plants showed increased sensitivity to glycerol; exogenous application of glycerol induced approximately threefold higher levels of PR-1 expression and more pronounced cell death (Figures 8D and 8E). Concurrently, 35S-NOA1 plants showed significantly higher ion leakage (Figure 8F) and elevated NO levels (Figure 8G). These results suggested that, while increased expression of NOA1 in wild-type plants was unable to relieve 18:1-mediated repression, it did potentiate defense phenotypes under low 18:1 conditions.
NOA1 Is an 18:1 Binding Protein
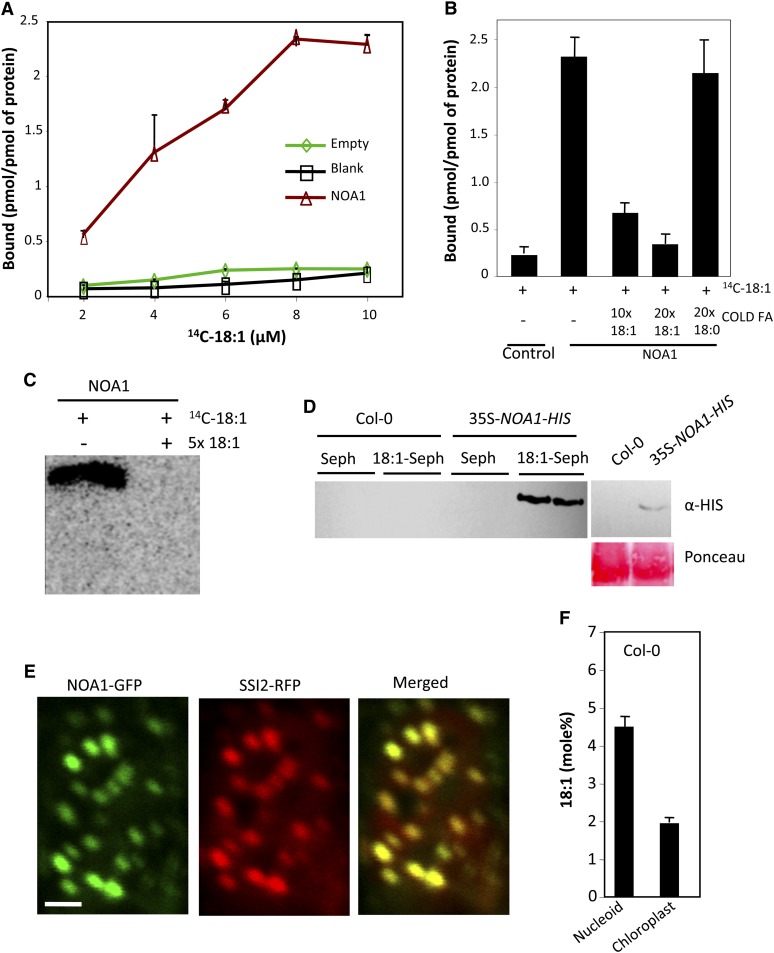
Because exogenous glycerol increased NOA1 levels, we considered the possibility that 18:1 levels regulated the stability of NOA1 by binding to it. Indeed, sequence analysis detected homology to mammalian FA binding domains in the NOA1 protein, and these domains were highly conserved in NOA1-like proteins from other plants (Furuhashi and Hotamisligil, 2008; see Supplemental Figures 12A and 12B and Supplemental References 1 online). To determine if NOA1 bound 18:1, it was important to use a biologically functional form of the protein. Database analysis showed that the transit peptide in NOA1 corresponds to the N-terminal 37 amino acids. However, earlier studies showing GTPase activity associated with NOA1 were performed with the NOA1Δ101 protein lacking the N-terminal 101 amino acids (Moreau et al., 2008). We therefore compared the GTPase activity of Escherichia coli purified NOA1Δ37 with that of NOA1Δ101 (see Supplemental Figure 13A online). Interestingly, NOA1Δ37 showed significantly higher GTPase activity compared with NOA1Δ101 (see Supplemental Figure 13B online), suggesting that the N-terminal 37 to 101 amino acids contributed significantly to the GTPase activity. All binding assays were therefore performed with NOA1Δ37 protein. Six different preparations of NOA1Δ37 bound 18:1 with similar efficiencies (Figure 9A). The binding of NOA1Δ37 to 18:1 saturated at ~20 μM 14C-18:1, and competition assays using cold 18:1 showed a proportionate decrease in the retention of (14C)-18:1, indicating saturable binding between 18:1 and NOA1Δ37 (Figures 9B and 9C). Unlike 18:1, cold 18:0 did not compete with (14C)-18:1 for binding with NOA1Δ37 (Figure 9C). To confirm the 18:1-NOA1 binding, we performed 18:1 affinity chromatography where E. coli purified NOA1 protein was applied to an 18:1 sepharose column. Indeed, NOA1 was specifically retained on the 18:1 sepharose matrix but not on unconjugated sepharose (see Supplemental Figure 13C online). We next generated transgenic plants that overexpressed the NOA1-HIS transgene, and total plant protein extracted from these was applied to 18:1 sepharose column. As with the E. coli–expressed NOA1, the NOA1-HIS protein from plant extracts was also retained on 18:1 sepharose but not on unconjugated sepharose (Figure 9D). While this result confirmed binding between 18:1 and NOA1, it suggested that the 18:1 binding site of NOA1 was not completely saturated with 18:1 in planta. Alternatively, it is possible that 18:1 bound to NOA1 was dissociated during extraction or the prebound 18:1 was exchanged with sepharose-immobilized 18:1, which is known to occur in some FA binding proteins (Smith et al., 1992).
Figure 9.
NOA1 Binds to 18:1 and Colocalizes with SSI2.
(A) 18:1 binding assay performed using 2 μM purified NOA1, 2 μM total protein extracted from pET28a-transformed E. coli (empty), or without any protein (blank). At least four independent binding assays were performed with four different preparations of NOA1 protein.
(B) 18:1 binding assay performed in the presence or absence of 10× and 20× of unlabeled 18:1. NOA1 (2 μM) protein and 8 μM 14C-18:1 were used for the binding assay. The experiment was repeated twice with similar results.
(C) Autoradiograph of NOA1 (2 μM) incubated with 8 μM 14C-18:1 or 14C-18:1 with 5× excess unlabeled 18:1 after electrophoresis on a native PAGE. The experiment was performed twice with similar results.
(D) 18:1 affinity chromatography performed using total protein extracted from 2 g of Col-0 or 35S-NOA1-HIS plants. Left panel shows levels of NOA1 protein in the 35S-NOA1-HIS plants prior to affinity chromatography. The experiment was performed twice with similar results.
(E) Confocal micrograph showing colocalization of NOA1-GFP and SSI2-RFP in N. benthamiana plants. At least four independent leaves were analyzed in two experiments with similar results. Bar = 2 μm.
(F) Relative levels of 18:1 in nucleoid versus whole chloroplasts of Col-0 plants. The error bars indicate sd (n = 6).
[See online article for color version of this figure.]
If low 18:1 was regulating the stability of the nucleoid-localized NOA1, then it might be expected that the 18:1 synthesizing SSI2 was in close proximity to NOA1. Indeed, SSI2 colocalized with NOA1 in the chloroplast nucleoids (Figure 9E) and not exclusively in the stroma as presumed earlier (Shanklin and Somerville, 1991). Unlike SSI2, ACT1, which catalyzes the acylation of 18:1 on G3P (Kunst et al., 1988), was distributed throughout the chloroplasts (see Supplemental Figure 14A online). FA analysis showed that the nucleoids contained higher levels of 18:1 compared with chloroplasts (Figure 9F). Nucleoids also contained other chloroplastic FAs, although their relative levels were different in chloroplast versus nucleoids (see Supplemental Figure 14B online). For instance, palmitic acid was the most abundant FA in the nucleoids as opposed to linolenic acid in the chloroplasts. The nucleoids also contained higher levels of 18:0, which serves as a substrate for the SSI2-catalyzed reaction. As predicted, exogenous application of glycerol lowered 18:1 levels in the nucleoids (see Supplemental Figure 14C online), which is consistent with the low-18:1-mediated increase in NOA1 and subsequent induction of NO levels and defense responses. The close proximity and the same suborganellar localization of SSI2 and NOA1 suggest that in the wild-type plants, NOA1 is present in an 18:1-rich environment within nucleoids, which subjects it to degradation.
DISCUSSION
We show that the 18:1 in wild-type plants regulates the stability of NOA1. Reduction of 18:1, via a genetic mutation in the 18:1-synthesizing SSI2 or exogenous application of glycerol, led to increased accumulation of NOA1 and an increase in chloroplastic NO. Reduction of 18:1 also increased NIA1 and NIA2 gene expression, which in turn contributed to the increased NO in the chloroplasts of ssi2 plants. Notably, both NIA1 and NIA2 are localized outside chloroplasts. This result, together with the observation that ssi2 phenotypes are fully restored in plants lacking NOA1 and one of the NRs (NIA1 or NIA2), suggests that cooperative interaction between NOA1- and NIA1/NIA2-triggered pathways is required for NO accumulation and/or NO-mediated signaling (Figure 10). Previous work has suggested at least two pathways for NO production in isolated soybean (Glycine max) chloroplasts, one of which was dependent on nitrite (Jasid et al., 2006). Thus, it is conceivable that the NIA1/NIA2-catalyzed synthesis of nitrite can regulate NOA1-dependent and/or -independent NO biosynthesis in the chloroplasts.
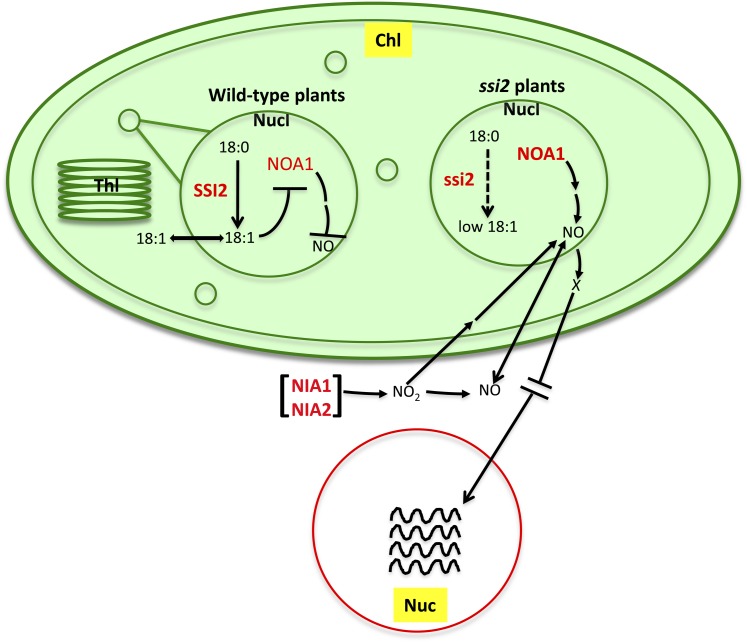
Figure 10.
A Model Illustrating 18:1-Regulated NO Signaling in Plants.
Desaturation of 18:0 to 18:1 is catalyzed by the soluble desaturase SSI2, which is localized in the chloroplastic (chl; shown as an oval) nucleoids (Nucl; shown as small and big circle inside Chl). 18:1 synthesized in the nucleoids is likely exported to stroma, where it participates in glycerolipid biosynthesis, and this reaction is catalyzed by the soluble stromal G3P acyltransferase ACT1. GLY1, a G3P dehydrogenase, which catalyzes biosynthesis of G3P, is also a stromal enzyme (Chanda et al., 2011). 18:1 synthesized in the nucleoids negatively regulates the stability of NOA1, which is also present in the nucleoids. NOA1 levels increase under low 18:1 conditions (due to mutations in SSI2 or after glycerol application) or in response to pathogen inoculation. This in turn initiates NO biosynthesis in the plastids. A reduction in 18:1 also triggers the increased expression of the extrachloroplastic NIA1 and NIA2, which also contribute to plastidial NO biosynthesis/accumulation. Mutations in NIA1/NIA2 affect chloroplastic NO production in response to pathogen infection or low 18:1 levels. This suggests that NIA1/NIA2 either feedback regulate chloroplastic NO biosynthesis or that NO/NO2 made via NIA1/NIA2 enzymes may translocate into chloroplasts and initiate NOA1-dependent/independent NO biosynthesis. At least two pathways for NO production, one of which is triggered in response to nitrite, have been shown to exist in the isolated soybean chloroplasts (Jasid et al., 2006). NO produced in response to pathogen infection or low 18:1 triggers nuclear (Nuc) gene expression (indicated by wavy lines). However, NO was not detected in the nucleus, suggesting that NO-mediated nuclear gene expression occurs possibly via unknown intermediate(s) (indicated by an “X”). Alternatively, NO-triggered nuclear gene expression might involve rapid diffusion of NO to the nucleus. NO-mediated increased gene expression results in SA biosynthesis in the chloroplasts, which further potentiates NO-mediated signaling. Thl, thylakoids. The relative nuclear and chloroplastic sizes are not to scale.
[See online article for color version of this figure.]
NIA2 has been shown to account for ~90% of the total NR activity. However, it is likely that the relative contribution(s) of NIA1 and NIA2 to NO production might depend on specific elicitor-induced pathway. For instance, NIA1 is the major contributor of abscisic acid–induced NO (Neill et al., 2008), whereas NIA2 is required for SA-induced NO production (Hao et al., 2010). Similarly, increased NR activity in response to cytokinin treatment was dependent on NIA1 and correlated with its transcriptional upregulation (Yu et al., 1998). Furthermore, the differential regulation of the NIA1 and NIA2 genes (Cheng et al., 1991; Yu et al., 1998) and proteins (Wang et al., 2010; Park et al., 2011), suggests that these proteins likely play distinct roles in nitrogen and NO metabolism. This is consistent with our result that mutations in either of the NIA genes are sufficient to restore R gene expression in ssi2 noa1 plants. Likewise, mutation in either NIA1 or NIA2 restores glycerol-dependent root length inhibition in noa1 plants.
Interestingly, although NO was primarily detected in the chloroplasts of ssi2 or pathogen/glycerol-treated wild-type plants, it led to the transcriptional upregulation of multiple nuclear genes. Inability to detect NO in the nucleus suggests that nuclear R gene expression is likely mediated via one or more intermediates whose synthesis/activation/localization is dependent on NO levels. However, at this stage, we cannot rule out the possibility that diffusion of low levels of NO and its rapid metabolism in the nucleus results in the altered nuclear gene expression. The fact that tobacco (Nicotiana tabacum) cells treated with the fungal elicitor cryptogein accumulate NO in the chloroplasts as well as nucleus supports the possibility that NO can localize to the nucleus (Foissner et al., 2000). Furthermore, studies in animal systems have suggested that the diffusion of NO through a 4- to 15-μm cellular radius is a rapid process that takes 2 to 30 ms (Lancaster, 1996). Thus, the cellular diffusion of NO, which is thought to be a highly random process, and the ability of NO to react with various cellular components are two key factors that likely govern NO-derived signaling.
In addition to its role in NO synthesis, an allele of NOA1 (RIF1) was recently identified in a screen for mutants affected in the methylerythritol phosphate (MEP) pathway (Flores-Pérez et al., 2008). This raises the possibility that the effect of NOA1 in ssi2-mediated signaling may be via alterations in the MEP pathway. However, posttranscriptional upregulation of MEP pathway enzymes in rif1 cannot be restored by exogenous application of NO, suggesting that the regulation of the MEP pathway by NOA1 is unrelated to its role in NO biosynthesis/accumulation. The MEP pathway functions in the biosynthesis of carotenoids, mono- and diterpenoids, plastoquinones, and the prenyl group of chlorophylls in plant plastids (Rodríguez-Concepción, 2004). Analysis of carotenoids and chlorophyll in ssi2, ssi2 noa1, and noa1 plants showed that the changes in these metabolites do not correlate with the restoration of wild-type-like phenotypes in ssi2 noa1 plants (see Supplemental Figure 15 online). For example, noa1 contained normal levels of chlorophyll, but both ssi2 and ssi2 noa1 contained reduced chlorophyll. Reduced chlorophyll levels in the ssi2 plants correlate well with their altered chloroplast structure (Lightner et al., 1994). By contrast, a reduction in NO levels in ssi2 noa1 plants correlated well with their wild-type-like morphology. Thus, the restoration of a majority of phenotypes in ssi2 noa1 is associated with altered NO levels rather than changes in the MEP pathway.
A recent study suggested that the reduced accumulation of NO in the noa1 plants was due to their inability to accumulate the carbon reserve, Suc (Van Ree et al., 2011). Consistent with this earlier report (Van Ree et al., 2011), our results showed that noa1 accumulated reduced levels of Suc compared with the wild-type plants (see Supplemental Figure 16A online). However, this was also the case for ssi2 and ssi2 noa1 plants. Furthermore, very similar cell death phenotype and NO-specific staining of roots was observed in ssi2 and ssi2 noa1 plants when grown with or without Suc (see Supplemental Figures 16B and 16C online). Together, these results suggest that Suc levels do not contribute to the noa1-mediated restoration of the ssi2-triggered defense phenotypes.
Another possibility is that increased NO levels in ssi2 plants are due to altered nitrogen metabolism in these plants. However, several observations discount this possibility. First, the expression profiles of genes involved in nitrogen metabolism do not correlate with restoration of defense phenotypes in various ssi2 double mutant backgrounds that contain low or high 18:1 levels and show ssi2- or wild-type-like defense phenotypes (see Supplemental Table 2 online). For instance, even though both ssi2 act1 and ssi2 eds1 sid2 show wild-type-like defense phenotypes, only ssi2 act1 showed basal expression of NIA2. Likewise, all ssi2 backgrounds analyzed here showed increased expression of Glu synthase regardless of their 18:1 levels or defense phenotypes. Second, ssi2 plants grown without any external nitrogen source or on soil containing potassium nitrate, ammonium sulfate, or ammonium nitrate showed similar morphological phenotype and contained elevated levels of NO (see Supplemental Figure 17 online). Notably, NO levels were significantly higher in ssi2 plants grown on high-nitrate soil, suggesting that these plants are not impaired in the uptake of nitrate (see Supplemental Figure 17B online).
Increased accumulation of NO under low 18:1 conditions suggests that 18:1 is an important signal that regulates NO synthesis and, thereby, defense signaling. Suborganellar compartmentalization of 18:1 biosynthesis and its use suggest that 18:1 likely shuttles in and out of the nucleoids. Consistent with this notion, pathogen inoculation did not alter 18:1 levels, suggesting that 18:1 flux between stroma and nucleoids or a transient change in 18:1 may play an important role in regulating NOA1. In addition to destabilizing NOA1, binding of 18:1 might also regulate its GTPase activity. Indeed, a marked reduction in GTPase activity in the presence of 18:1 was observed. However, 250 and 500 μM 18:1 was required to inhibit the GTPase activity by 33 and 90%, respectively. These concentrations are higher than the biological levels of 18:1 (~250 μM total 18:1), much of which is conjugated to the membrane lipids. One possibility is that other cellular factors may be required for 18:1-mediated inhibition of GTPase activity at lower concentrations. Interestingly, 18:1 also inhibits NO synthase activity in humans (Davda et al., 1995), suggesting that plants and humans use conserved mechanism to regulate NO levels even though they differ in their biosynthetic processes. The fact that NOA1-like proteins are present in the genomes of all metazoans (Zemojtel et al., 2004) suggests that 18:1-mediated regulation of NOA1-like proteins might contribute to regulation of NO in other nonplant systems. As yet, the link between NOA1-derived NO synthesis and its GTPase activity remains unknown. It is possible that NOA1 serves as an important catalytic component of a larger complex that facilitates NO production and/or accumulation in plants. An alternate possibility is that the GTPase activity of NOA1 regulates the synthesis of enzyme(s) required for NO biosynthesis. The fact that NO-mediated nitration of FAs can transform the FAs into important signaling molecules underscores the overlapping regulatory roles of NO and 18:1 (Baker et al., 2005; Jain et al., 2008). Further work on the compartmentalization of 18:1 and its flux within the chloroplast may provide novel insights into the complex suborganellar regulation of NOA1.
METHODS
Plant Growth Conditions and Genetic Analysis
Plants were grown in MTPS 144 Conviron walk-in chambers at 22°C and 65% relative humidity with 14-h photoperiod. These chambers were equipped with cool white fluorescent bulbs (Sylvania FO96/841/XP/ECO). The PFD of the day period was 106.9 μmol m−2 s−1 (measured using a digital light meter; Phytotronic). Plants were grown on autoclaved Pro-Mix soil (Premier Horticulture). Soil was fertilized once using Scotts Peter’s 20:10:20 peat lite special general fertilizer that contained 8.1% ammoniacal nitrogen and 11.9% nitrate nitrogen. Plants were irrigated using deionized or tap water. For nitrogen treatment, plants were grown on soil containing 5 mM ammonium sulfate, potassium nitrate, or ammonium nitrate. No nitrogen source was added to the control pots.
The ssi2-1 mutant used in this study is an ethyl methanesulfonate–derived mutant described earlier (Kachroo et al., 2001; Shah et al., 2001). The noa1, nia1-5, and nia2-1 plants used in this study are T-DNA–derived knockout mutants described earlier (Guo et al., 2003; Wang et al., 2010). The nia1-2 nia2-5 double mutant seeds were obtained from the ABRC. Crosses were performed by pollinating emasculated flowers of recipient plants with pollen from donor plants. The genotypes were determined by conducting cleaved amplified polymorphic sequence analysis, derived cleaved amplified polymorphic sequence analysis, or PCR. All double and triple mutant plants were confirmed genetically for all loci. The primers used for genotyping are listed in Supplemental Table 3 online.
Complementation and Overexpression
For complementation of ssi2 noa1, a SalI-KpnI genomic fragment spanning the NOA1 promoter, open reading frame, and terminator was amplified from the Col-0 plants and cloned into pCambia binary vector. After confirmation of the DNA sequence, the binary vector was transformed into ssi2 noa1 plants using the floral dip method (Clough and Bent, 1998). The transgenic plants were selected on plates containing hygromycin. The complementation was confirmed by genotype analysis of the T1 plants and by analyzing the segregation of ssi2 and ssi2 noa1 phenotypes in the T2 generation.
For overexpression, a cDNA containing XhoI-XbaI restriction sites was amplified from the Col-0 plants and cloned downstream of 35S-cauliflower mosaic virus promoter in pRTL-β-glucuronidase. After confirmation of the DNA sequence, the HindIII fragment from this recombinant vector was transferred to pBAR1. For NOA1-HIS overexpression, a HIS tag was added at the C-terminal end, and the PCR fragment was cloned into the pSITE vector using Gateway technology (Invitrogen).
GTPase Assay
For the GTPase assay, 10 μM protein was incubated with 50 to 100 μM GTP, 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 150 mM NaCl, 10% glycerol, and 2 mM DTT at 37°C overnight. Samples were boiled for 5 min to stop the reaction and centrifuged, and the supernatant was analyzed by reverse-phase HPLC fitted with C18 5 μm (4.6 × 250 mm) column (Dionex). Nucleotides were separated under isocratic condition at 1 mL/min of 100 mm KH2PO4, pH 6.5, 10 mM tetrabutylammonium bromide, 0.2 mM NaN3, and 7.5% acetonitrile.
Binding Assays
Binding assays were performed as described earlier (Rasmussen et al., 1990). Briefly, 1 to 8 μM 14C 18:1 (specific activity 58.2 Ci/mmol; Perkin-Elmer) was incubated at 37°C with 1 to 20 μM NOA1 protein in a 200-μL reaction volume containing 10 mM potassium phosphate buffer, pH 7.4. After 1 h, the reaction was placed on ice for 15 min, mixed with 400 μL of ice-cold Lipidex-1000, and incubated on ice for 20 min. The reaction was centrifuged at 10,000g for 5 min at 4°C, and the radiolabel in the supernatant was measured using a scintillation counter.
Oleate Agarose Affinity Chromatography
18:1 (Sigma-Aldrich) was coupled to EAH-Sepharose (GE Healthcare) using 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (Fisher Scientific) as described earlier (Peters et al., 1973; Kim et al., 2005). Briefly, 18:1 was coupled by stirring the EAH-Sepharose in 1.5 volumes of 0.1 M sodium oleate at pH 10.0 in the presence of the 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (50 mg/mL of sepharose) for 3 d at 37°C. The matrix was washed extensively at 37°C with 50% (v/v) ethanol followed by washes with 100% ethanol, 0.075 M sodium phosphate (1:1), pH 2.4, and finally with ethanol-0.05 n NaOH (1:1). Unreacted amino groups were blocked by acetylation with acetic anhydride at pH 7.0 at 0°C for 1 h. 18:1 coupling was verified by carrying out binding assays with the 18:1 binding protein BSA. Mock sepharose was prepared from EAH-Sepharose by blocking ligand with 1 M acetic acid.
Chloroplast and Nucleoid Purification
For chloroplast isolation, leaves from wild-type and mutant plants were harvested at the end of the night period. Five grams fresh weight of leaves was homogenized, and the chloroplasts were isolated as described earlier (Aronsson and Jarvis, 2002).
Nucleoid isolation from chloroplasts was performed as described earlier (Jeong et al., 2003). Briefly, intact chloroplasts from 20 g of leaves were pelleted and resuspended in 30 mL of nucleoid extraction buffer containing 17% (w/v) Suc, 20 mM Tris-HCl, 0.5 mM EDTA, 1.2 mM spermidine, 7 mM 2-mercaptoethanol, and protease inhibitor cocktail. A 1/20 volume of 20% (v/v) Nonidet P-40 was added and stirred at 4°C for 30 min. The solution was centrifuged for 10 min at 3000g at 4°C, and the supernatant was recentrifuged at 48,000g for 40 min at 4°C. The pellet was washed twice with nucleoid extraction buffer and resuspended in 100 μL of nucleoid extraction buffer.
NO Staining and Quantification
For NO staining, adaxial side of leaves were infiltrated with 4 μM DAF-FM DA, and, after 5 min of incubation in dark, leaves were observed under Olympus FV1000 confocal laser scanning microscope using a 488-nm laser. For nucleoid staining, nucleoids were incubated in 1 μM DAF-FM DA for 5 min prior to confocal microscopy.
For NO quantification, ~300 mg of leaf tissue was homogenized in 50 mM phosphate buffer, pH 7.0, in dark. The supernatant was incubated with DAF-FM DA for 30 min with constant shaking, and the fluorescence was measured at 495 and 515 nm using a fluorimeter (Molecular Devices). NO quantification using the Greiss method (originally described in Griess, 1879) was performed using reagents assay system from Promega. Briefly, ~300 mg of leaf tissue was homogenized in 50 mM phosphate buffer, pH 7.0, in dark. The supernatant was incubated with sulfanilamide solution for 10 min in dark. To this, 50 μL of the N-(1-naphthyl) ethylendiamine dihydrochloride solution was added followed by another 10 min of incubation in dark. The absorbance was measured at 520 nm using the DTX 880 multimode detector (Beckman Coulter).
H2O2 Quantification
For H2O2 determination, leaves were homogenized in 40 mM Tris-HCl, pH 7.0, and to this 20 μM of 2′,7′-dichlorofluorescein was added. The samples were incubated for 1 h in dark, and the H2O2 levels were measured using a spectrofluorimeter. The concentration of H2O2 was determined as nanograms/milligrams protein by extrapolating from the standard H2O2 curve.
NR Assay
NR activity was measured from leaves of 4-week-old plants as described earlier (Van Ree et al., 2011).
NOA1 Expression and Purification
NOA1 cDNA lacking the N-terminal 37 and 101 amino acids was amplified as a NheI-XhoI fragment from Col-0 and cloned into the pET28a vector. The primers used for PCR are listed in Supplemental Table 2 online. NOA1-HIS protein was purified using a HiTrap Chelating HP column (GE Healthcare) on a fast protein liquid chromatography system. The purified protein was dialyzed using 25 mM Tris-HCl buffer and quantified using Bradford reagent (Bio-Rad).
SA, FA, Suc, and Lipid Analyses
SA and SA glucoside were extracted and measured from ~0.3 g of fresh weight leaf tissue, as described before (Chandra-Shekara et al., 2006).
FA extraction was performed by placing leaf tissue in 2 mL of 3% H2SO4 in methanol. After ~30 min incubation at 80°C, 1 mL of hexane with 0.001% butylated hydroxytoluene was added. The hexane phase was then transferred to vials for gas chromatography (GC). One-microliter samples were analyzed by GC on a Varian fatty acid methyl ester (0.25 mm × 50 m) column and quantified with flame ionization detection. For quantification of FAs, leaves (50 mg) were extracted together with the 17:0 FA internal standard, and the relative levels were calculated based on flame ionization detector peak areas.
For Suc estimation, 10 mg of lyophilized leaf tissues (dry weight) was immersed in 3 mL of 80% ethanol containing 100 μM 2-deoxyglucose as an internal standard. The extract was incubation at 90°C for 5 min and centrifuged at 1000g for 5 min. The supernatant was transferred in another glass vial, and the pellet was washed twice more. The supernatant collected after three extractions was dried under nitrogen gas to ~1 mL and extracted with 1 mL of chloroform. The upper phase was passed through a column containing the mixed-bed resin (TMD-8 hydrogen and hydroxide form; Sigma-Aldrich), dried under nitrogen gas, and derivatized using 100 μL of pyridine and 100 μL of acetic anhydride. The samples were kept at 60°C for 1 h and dried completely under nitrogen gas. The samples were suspended in 500 μL of heptane:toluene (1:1 v/v) and 500 μL of NaHCO3 and vortexed, and the upper phase was concentrated to ~10 μL using nitrogen gas. One-microliter samples were analyzed by GC attached to an electron ionization detector (Hewlett Packard, GCD Systems). The Suc peaks were identified using mass spectrometry.
For lipid extraction, six to eight leaves were incubated at 75°C in isopropanol containing 0.001% butylated hydroxytoluene for ~15 min. To this, 1.5 mL chloroform and 0.6 mL water were added, and the samples were agitated at room temperature for 1 h. The lipids were reextracted in chloroform:methanol (2:1, v/v) until the leaves were bleached. The aqueous content was removed by partitioning with 1 M KCl and water. The lipid extract was dried under a gentle stream of nitrogen gas and redissolved in 0.5 mL of chloroform. Lipid analysis and acyl group identification were performed using the automated electrospray ionization–tandem mass spectrometry facility at Kansas Lipidomics Research Center.
SA, Glycerol, SNP, and NONOate Treatments
SA, glycerol, SNP, and NONOate treatments were performed by spraying 500 μM, 50 mM, 100 to 1000 μM, and 100 μM solutions, respectively.
RNA Extraction, RNA Gel Blot Analyses, and RT-PCR
Small-scale extraction of RNA from two or three leaves (per sample) was performed with the TRIzol reagent (Invitrogen) following the manufacturer’s instructions. RNA gel blot analysis and synthesis of random-primed probes for PR-1 and PR-2 were performed as described previously (Kachroo et al., 2000).
RNA quality and concentration were determined by gel electrophoresis and determination of A260. Reverse transcription and first-strand cDNA synthesis were performed using Superscript II (Invitrogen). Real-time quantitative RT-PCR was performed as described before (Zhang et al., 2009). Each sample was run in triplicate, and ACTIN expression levels were used as internal control for normalization. Cycle threshold values were calculated by SDS 2.3 software. Gene-specific primers used for real-time quantitative RT-PCR analyses are described in Supplemental Table 3 online.
Protein Extraction, Immunoblot Analysis, and Antibody Generation
Proteins were extracted in buffer containing 50 mM Tris-HCl, pH 7.5, 10% glycerol, 150 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 5 mM DTT, and 1× protease inhibitor cocktail (Sigma-Aldrich). Protein concentration was measured with the Bio-Rad protein assay
For Ponceau-S staining, polyvinylidene fluoride membranes were incubated 1 in Ponceau-S solution (40% [v/v] methanol, 15% [v/v] acetic acid, and 0.25% Ponceau-S). The membranes were destained using deionized water. Proteins (30 to 50 μg) were fractionated on a 7 to 10% SDS-PAGE gel and subjected to immunoblot analysis using α-NOA1, α-HIS, or α-GFP (Sigma-Aldrich) antibody. Immunoblots were developed using the ECL detection kit (Roche) or alkaline phosphatase–based color detection. Rabbit anti-NOA1 polyclonal antibodies were generated against NOA1Δ37 protein (Cocalico Biologicals).
Treatment with MG132 and Protease Inhibitors
For 26S proteasome and plant protease inhibitor experiments, 100 μm MG132 (Sigma-Aldrich) or a 1× mixture of plant protease inhibitors (Sigma-Aldrich) was dissolved in 0.1% DMSO and infiltrated into the abaxial surface of Arabidopsis thaliana leaves using a needleless syringe. The protease inhibitor cocktail contained pepstatin A, leupeptin, bestatin, 4-(2-aminoethyl) benzenesulfonyl fluoride, 4-guanidino, and 1,10-phenanthroline.
Transcriptional Profiling
Total RNA was isolated from 4-week-old plants using TRIzol as outlined above. The experiment was performed in triplicate, and a separate group of plants was used for each set. RNA was processed and hybridized to the Affymetrix Arabidopsis ATH1 genome array GeneChip following the manufacturer’s instructions (http://www.affymetrix.com/Auth/support/downloads/manuals/expression_analysis_technical_manual.pdf). All probe sets on the GeneChips were assigned hybridization signal above background using Affymetrix Expression Console Software v1.0 (http://www.affymetrix.com/Auth/support/downloads/manuals/expression_console_userguide.pdf). Data were analyzed by one-way analysis of variance followed by post hoc two-sample t tests for the determination of differential expression reported in Supplemental Data Set 1 online. The P values were calculated individually and in pairwise combination for each probe set.
Pathogen Infection
Inoculations with Pseudomonas syringae DC 3000 were conducted as described before (Chanda et al., 2011). The bacterial cultures were grown overnight in King’s B medium containing rifampicin and/or kanamycin. The cells were washed and suspended in 10 mM MgCl2. The bacterial suspension (106 colony-forming units mL−1) was injected into the abaxial surface of the leaf using a needleless syringe. Three discs from the inoculated leaves were collected and homogenized in 10 mM MgCl2. The extract was diluted and appropriate dilutions were plated on King’s B medium.
Transcripts synthesized in vitro from a cloned cDNA of TCV using T7 RNA polymerase were used for viral infections (Chandra-Shekara et al., 2006). For inoculations, the viral transcript was suspended at a concentration of 0.05 μg/μL in inoculation buffer, and the inoculation was performed as described earlier (Chandra-Shekara et al., 2006). Resistance and susceptibility was scored at 14 to 21 d after inoculation and confirmed by RNA gel blot analysis. Susceptible plants showed stunted growth, crinkling of leaves, and drooping of the bolt.
Confocal Microscopy
For confocal imaging, samples were scanned on an Olympus FV1000 microscope (Olympus America). GFP, cyan fluorescent protein (CFP), and red fluorescent protein (RFP) were excited using 488-, 440-, and 543-nm laser lines, respectively. The various constructs were introduced in Agrobacterium tumefaciens strain LBA4404. Agrobacterium strains carrying various proteins were infiltrated into Nicotiana benthamiana plants expressing RFP- or CFP-tagged nuclear protein H2B or wild-type N. benthamiana plants (Martin et al., 2009). Forty-eight hours later, water-mounted sections of leaf tissue were examined by confocal microscopy using a water immersion PLAPO60XWLSM 2 (numerical aperture of 1.0) objective on an FV1000 point-scanning/point-detection laser scanning confocal 3 microscope (Olympus) equipped with lasers spanning the spectral range of 405 to 633 nm. RFP, CFP, and GFP overlay images (×40 magnification) were acquired at a scan rate of 10 ms/pixel. The GFP channel (488 nm) was used to analyze DAF-FM DA–stained leaves and roots. For nucleoid staining, leaves were infiltrated with 1 mg/mL solution of DAPI ~5 min prior to microscopy. Isolated nucleoids were stained with 0.5 mg/mL of solution of DAPI. The Olympus FLUOVIEW 1.5 was used to control the microscope, image acquisition, and the export of TIFF files.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SSI2 (At2g43710), GLY1 (At2g40690), ACT1 (At1g32200), NOA1 (At3g47450), PR1 (At2g14610), NIA1 (At1g77760), and NIA2 (At1g37130). Microarray data have been deposited at the National Center for Biotechnology Information under accession number GSE36797.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The ssi2 Plants Accumulate High Levels of Chloroplastic NO.
Supplemental Figure 2. Transgenic Expression of NOA1 Restores ssi2-Like Phenotypes in ssi2 noa1 Plants.
Supplemental Figure 3. A Mutation in NOA1 Does Not Restore Constitutive Defense Phenotypes in cpr5 Plants.
Supplemental Figure 4. Profile of Total Lipids Extracted from Wild-Type (Col-0), noa1, ssi2, and ssi2 noa1 Plants.
Supplemental Figure 5. Expression of R and the NIA1/NIA2 Genes Is Induced by NO and Low 18:1 Conditions, Respectively.
Supplemental Figure 6. Mutations in NIA1 and NIA2 Partially Restore ssi2 Phenotypes.
Supplemental Figure 7. NIA1 and NIA2 Are Extrachloroplastic Proteins Required for Chloroplastic NO Accumulation in ssi2 Plants.
Supplemental Figure 8. The noa1 nia Plants Are Compromised in Pathogen-Induced NO Accumulation.
Supplemental Figure 9. The Glycerol-Treated Col-0, noa1, nia1, and nia2 Plants Show Similar Decrease in Their 18:1 Levels.
Supplemental Figure 10. Subcellular Localization of NO in ssi2 and avrRpt2 Inoculated Wild-Type Plants.
Supplemental Figure 11. Levels of clpC and clpP in ssi2 Plants.
Supplemental Figure 12. Fatty Acid Binding Properties of NOA1.
Supplemental Figure 13. N-Terminal 37 to 101 Amino Acids Are Critical for NOA1 GTPase Activity.
Supplemental Figure 14. Localization of ACT1 and Fatty Acid Analysis of Nucleoids.
Supplemental Figure 15. Levels of Chlorophyll and Carotenoids in ssi2 Plants.
Supplemental Figure 16. Suc-Grown ssi2 noa1 Plants Show Wild-Type-Like Phenotypes.
Supplemental Figure 17. ssi2 Plants Grown without Nitrogen Accumulate Elevated NO.
Supplemental Table 1. Transcript Levels of NOA1, NIA1, NIA2, and PR-1 in Response to Pathogen Infections or Exogenous Application of SA.
Supplemental Table 2. Fold Change in Transcript Levels of genes in ssi2, ssi2 sid2, ssi2 act1, and ssi2 eds1 sid2 Plants Compared with Results from Col-0 (Wild-Type) Plants.
Supplemental Table 3. Primer Sequences Used for Genotyping, Real-Time PCR, or Generating Clones for Expression in E. coli and Plants.
Supplemental Data Set 1. Significance of Differential Expression and Fold Change in Transcript Levels of Genes in ssi2, ssi2 sid2, and ssi2 act1 Plants Compared with Results from Col-0 (Wild-type) Plants.
Supplemental References 1. Supplemental References for Supplemental Figure 12.
Supplementary Material
Acknowledgments
We thank Nigel Crawford for noa1 seeds, the ABRC for NIA1, NIA2 Salk lines, and nia1 nia2 double mutant seeds, Zach Adam for clpP and clpC antibodies, Marciel Stadnik for help with NO estimations, Lynnette Dirk for help with NOA1 purification, John Johnson for help with GC, Ludmila Lapchyk for technical help, Amy Crume for managing the plant growth facility, and Daniel Klessig for useful discussions. We thank Barry Pogson and anonymous reviewers for useful suggestions. The lipid analyses were performed at the Kansas Lipidomics Research Center Analytical Laboratory, where instrument acquisition and method development were supported by the National Science Foundation (grant nos. EPS 0236913, MCB 0455318, MCB 0920663, and DBI 0521587), the Kansas Technology Enterprise Corporation, the K-IDeA Networks of Biomedical Research Excellence of the National Institutes of Health (grant no. P20RR16475), and Kansas State University. This work was supported by grants from the National Science Foundation (IOS#051909 and IOS#0749731) to A.K. and P.K.
AUTHOR CONTRIBUTIONS
P.K. and A.K. directed the study. M.K.M. carried out most of the biochemical, molecular, and genetic analysis. A.C.C.-S. isolated nia1 mutant lines and made initial crosses between ssi2 and noa1/nia mutants. R.-D.J. helped with genetic analysis and assayed turnip crinkle virus–induced phenotypes. K.Y. helped with FA profiling. S.Z. developed methods for affinity chromatography and provided affinity resin. B.C. helped with GTPase assays. D.N. carried out SA estimations. The article was prepared by P.K. and A.K. with contributions from all the authors.
Glossary
- FA
fatty acid
- 18:1
oleic acid
- 18:0
stearic acid
- SACPD
stearoyl-ACP desaturase
- G3P
glycerol-3-phosphate
- NO
nitric oxide
- NR
nitrate reductase
- SNP
sodium nitroprusside
- DAF-FM DA
4-amino-5-methylamino-2,7-difluorofluorescein diacetate
- DEA-NONOate
2-(N,N-diethylamino)-diazenolate-2-oxide
- Col-0
Columbia-0
- SA
salicylic acid
- H2O2
hydrogen peroxide
- GFP
green fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
- MEP
methylerythritol phosphate
- GC
gas chromatography
- CFP
cyan fluorescent protein
- RFP
red fluorescent protein
References
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Baker P.R., et al. (2005). Fatty acid transduction of nitric oxide signaling: Multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J. Biol. Chem. 280: 42464–42475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcerczyk A., Soszynski M., Bartosz G. (2005). On the specificity of 4-amino-5-methylamino-2′,7′-difluorofluorescein as a probe for nitric oxide. Free Radic. Biol. Med. 39: 327–335 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A., Pugin A., Wendehenne D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Calvo A.M., Hinze L.L., Gardner H.W., Keller N.P. (1999). Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 65: 3668–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B., Xia Y., Mandal M.K., Yu K., Sekine K.-T., Gao Q.-M., Selote D., Hu Y., Stromberg A., Navarre D., Kachroo A., Kachroo P. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43: 421–427 [DOI] [PubMed] [Google Scholar]
- Chandra-Shekara A.C., Gupte M., Navarre D., Raina S., Raina R., Klessig D., Kachroo P. (2006). Light-dependent hypersensitive response and resistance signaling against Turnip Crinkle Virus in Arabidopsis. Plant J. 45: 320–334 [DOI] [PubMed] [Google Scholar]
- Chandra-Shekara A.C., Venugopal S.C., Barman S.R., Kachroo A., Kachroo P. (2007). Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 7277–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.L., Acedo G.N., Dewdney J., Goodman H.M., Conkling M.A. (1991). Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 96: 275–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford N.M. (2006). Mechanisms for nitric oxide synthesis in plants. J. Exp. Bot. 57: 471–478 [DOI] [PubMed] [Google Scholar]
- Davda R.K., Stepniakowski K.T., Lu G., Ullian M.E., Goodfriend T.L., Egan B.M. (1995). Oleic acid inhibits endothelial nitric oxide synthase by a protein kinase C-independent mechanism. Hypertension 26: 764–770 [DOI] [PubMed] [Google Scholar]
- Delledonne M., Xia Y., Dixon R.A., Lamb C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Denys A., Hichami A., Khan N.A. (2001). Eicosapentaenoic acid and docosahexaenoic acid modulate MAP kinase (ERK1/ERK2) signaling in human T cells. J. Lipid Res. 42: 2015–2020 [PubMed] [Google Scholar]
- Desikan R., Griffiths R., Hancock J., Neill S. (2002). A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99: 16314–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J., Wendehenne D., Klessig D.F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner I., Wendehenne D., Langebartels C., Durner J. (2000). In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. Plant J. 23: 817–824 [DOI] [PubMed] [Google Scholar]
- Furuhashi M., Hotamisligil G.S. (2008). Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 7: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez U., Sauret-Güeto S., Gas E., Jarvis P., Rodríguez-Concepción M. (2008). A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell 20: 1303–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gas E., Flores-Pérez Ú., Sauret-Gueto S., Rodríguez-Concepción M. (2009). Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. Plant Cell 21: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griess P. (1879). Bemerkungen zu der abhandlung der H.H. Weselsky und Benedikt “Ueber einige azoverbindungen.” Chem. Ber. 12: 426–428 [Google Scholar]
- Guo F.Q., Okamoto M., Crawford N.M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Hao F., Zhao S., Dong H., Zhang H., Sun L., Miao C. (2010). Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J. Integr. Plant Biol. 52: 298–307 [DOI] [PubMed] [Google Scholar]
- He Y., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Iba K. (2002). Acclimative response to temperature stress in higher plants: Approaches of gene engineering for temperature tolerance. Annu. Rev. Plant Biol. 53: 225–245 [DOI] [PubMed] [Google Scholar]
- Jain K., Siddam A., Marathi A., Roy U., Falck J.R., Balazy M. (2008). The mechanism of oleic acid nitration by *NO(2). Free Radic. Biol. Med. 45: 269–283 [DOI] [PubMed] [Google Scholar]
- Jasid S., Simontacchi M., Bartoli C.G., Puntarulo S. (2006). Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol. 142: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.Y., Rose A., Meier I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C.W., Du S.T., Zhang Y.S., Lin X.Y., Tang C.X. (2009). Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum). Ann. Bot. (Lond.) 104: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A., Kachroo P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47: 153–176 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15: 2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A., Shanklin J., Whittle E., Lapchyk L., Hildebrand D., Kachroo P. (2007). The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 63: 257–271 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Venugopal S.C., Lapchyk L., Falcone D., Hildebrand D., Kachroo P. (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P., Kachroo A., Lapchyk L., Hildebrand D., Klessig D.F. (2003). Restoration of defective cross talk in ssi2 mutants: Role of salicylic acid, jasmonic acid, and fatty acids in SSI2-mediated signaling. Mol. Plant Microbe Interact. 16: 1022–1029 [DOI] [PubMed] [Google Scholar]
- Kachroo P., Shanklin J., Shah J., Whittle E.J., Klessig D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P., Venugopal S.C., Navarre D.A., Lapchyk L., Kachroo A. (2005). Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol. 139: 1717–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P., Yoshioka K., Shah J., Dooner H.K., Klessig D.F. (2000). Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes, is salicylic acid dependent but NPR-1, ethylene and jasmonate independent. Plant Cell 12: 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Nakatomi R., Akagi T., Hashikawa T., Takahashi R. (2005). Unsaturated fatty acids induce cytotoxic aggregate formation of amyotrophic lateral sclerosis-linked superoxide dismutase 1 mutants. J. Biol. Chem. 280: 21515–21521 [DOI] [PubMed] [Google Scholar]
- Kunst L., Browse J., Somerville C. (1988). Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA 85: 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J.R., Jr (1996). Diffusion of free nitric oxide. Methods Enzymol. 268: 31–50 [DOI] [PubMed] [Google Scholar]
- Lightner J., James D.W., Jr, Dooner H.K., Browse J. (1994). Altered body morphology is cause by increased stearate levels in a mutant of Arabidopsis. Plant J. 6: 401–412 [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Moreau M., Lee G.I., Wang Y., Crane B.R., Klessig D.F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283: 32957–32967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S., Barros R., Bright J., Desikan R., Hancock J., Harrison J., Morris P., Ribeiro D., Wilson I. (2008). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59: 165–176 [DOI] [PubMed] [Google Scholar]
- Parani M., Rudrabhatla S., Myers R., Weirich H., Smith B., Leaman D.W., Goldman S.L. (2004). Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2: 359–366 [DOI] [PubMed] [Google Scholar]
- Park B.S., Song J.T., Seo H.S. (2011). Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. 2: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T., Jr, Taniuchi H., Anfinsen C.B., Jr (1973). Affinity chromatography of serum albumin with fatty acids immobilized on agarose. J. Biol. Chem. 248: 2447–2451 [PubMed] [Google Scholar]
- Planchet E., Kaiser W.M. (2006). Nitric oxide (NO) detection by DAF fluorescence and chemilumisescence: a comparison using abiotic and biotic NO sources. J. Exp. Bot. 12: 3043–3055 [Google Scholar]
- Rasmussen J.T., Börchers T., Knudsen J. (1990). Comparison of the binding affinities of acyl-CoA-binding protein and fatty-acid-binding protein for long-chain acyl-CoA esters. Biochem. J. 265: 849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M. (2004). The MEP pathway: A new target for the development of herbicides, antibiotics and antimalarial drugs. Curr. Pharm. Des. 10: 2391–2400 [DOI] [PubMed] [Google Scholar]
- Rollo C.D., Czyzewska E., Borden J.H. (1994). Fatty acid necromones for cochroaches. Naturwissenschaften 81: 409–410 [Google Scholar]
- Routaboul J.-M., Fischer S.F., Browse J. (2000). Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol. 124: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko T., Walley J.W., Chehab E.W., Xiao Y., Kaspi R., Pye M.F., Mohamed M.E., Lazarus C.M., Bostock R.M., Dehesh K. (2010). Arachidonic acid: An evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 22: 3193–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Kachroo P., Nandi A., Klessig D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25: 563–574 [DOI] [PubMed] [Google Scholar]
- Shanklin J., Cahoon E.B. (1998). Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 611–641 [DOI] [PubMed] [Google Scholar]
- Shanklin J., Somerville C. (1991). Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc. Natl. Acad. Sci. USA 88: 2510–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.F., Tsuchida K., Hanneman E., Suzuki T.C., Wells M.A. (1992). Isolation, characterization, and cDNA sequence of two fatty acid-binding proteins from the midgut of Manduca sexta larvae. J. Biol. Chem. 267: 380–384 [PubMed] [Google Scholar]
- Sun J., Zhang X., Broderick M., Fein H. (2003). Measurement of nitric oxide production in biological systems by using Griess reaction assay. Sensors 3: 276–284 [Google Scholar]
- Upchurch R.G. (2008). Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 30: 967–977 [DOI] [PubMed] [Google Scholar]
- Van Ree K., Gehl B., Chehab E.W., Tsai Y.-C., Braam J. (2011). Nitric oxide accumulation in Arabidopsis is independent of NOA1 in the presence of sucrose. Plant J. 68: 225–233 [DOI] [PubMed] [Google Scholar]
- Venugopal S.C., Jeong R.D., Mandal M.K., Zhu S., Chandra-Shekara A.C., Duroy N., Kachroo A., Kachroo P. (2009). Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 5: e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Du Y., Li Y., Ren D., Song C.-P. (2010). Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22: 2981–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne D., Pugin A., Klessig D.F., Durner J. (2001). Nitric oxide: Comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 6: 177–183 [DOI] [PubMed] [Google Scholar]
- Wilson I.D., Neill S.J., Hancock J.T. (2008). Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 31: 622–631 [DOI] [PubMed] [Google Scholar]
- Wilson R.A., Calvo A.M., Chang P.K., Keller N.P. (2004). Characterization of the Aspergillus parasiticus delta12-desaturase gene: a role for lipid metabolism in the Aspergillus-seed interaction. Microbiology 150: 2881–2888 [DOI] [PubMed] [Google Scholar]
- Wilkinson J.Q., Crawford N.M. (1991). Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell 3: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Gao Q.-M., Yu K., Lapchyk L., Navarre D., Hildebrand D., Kachroo A., Kachroo P. (2009). An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 5: 151–165 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Furutera A., Seki K., Toyoda Y., Tanaka K., Sugimoto Y. (2008). Malondialdehyde generated from peroxidized linolenic acid causes protein modification in heat-stressed plants. Plant Physiol. Biochem. 46: 786–793 [DOI] [PubMed] [Google Scholar]
- Yu X., Sukumaran S., Mrton L. (1998). Differential expression of the Arabidopsis Nia1 and Nia2 genes. cytokinin-induced nitrate reductase activity is correlated with increased nia1 transcription and mrna levels. Plant Physiol. 116: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler D., Zähringer U., Gerber I., Dubery I., Hartung T., Bors W., Hutzler P., Durner J. (2004). Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA 101: 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T., Penzkofer T., Dandekar T., Schultz J. (2004). A novel conserved family of nitric oxide synthase? Trends Biochem. Sci. 29: 224–226 [DOI] [PubMed] [Google Scholar]
- Zhang D.-X., Nagabhyru P., Schardl C.L. (2009). Regulation of a chemical defense against herbivory produced by symbiotic fungi in grass plants. Plant Physiol. 150: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.