This work identifies the pathogenesis-related protein10 (PR10) as an interacting partner of the leucine-rich repeat protein1 (LRR1). It shows that LRR1 expression enhances PR10-mediated cell death that is dependent on the cytoplasmic localization of the PR10-LRR1 complex.
Abstract
Plants recruit innate immune receptors such as leucine-rich repeat (LRR) proteins to recognize pathogen attack and activate defense genes. Here, we identified the pepper (Capsicum annuum) pathogenesis-related protein10 (PR10) as a leucine-rich repeat protein1 (LRR1)–interacting partner. Bimolecular fluorescence complementation and coimmunoprecipitation assays confirmed the specific interaction between LRR1 and PR10 in planta. Avirulent Xanthomonas campestris pv vesicatoria infection induces PR10 expression associated with the hypersensitive cell death response. Transient expression of PR10 triggers hypersensitive cell death in pepper and Nicotiana benthamiana leaves, which is amplified by LRR1 coexpression as a positive regulator. LRR1 promotes the ribonuclease activity and phosphorylation of PR10, leading to enhanced cell death signaling. The LRR1-PR10 complex is formed in the cytoplasm, resulting in its secretion into the apoplastic space. Engineered nuclear confinement of both proteins revealed that the cytoplasmic localization of the PR10-LRR1 complex is essential for cell death–mediated defense signaling. PR10/LRR1 silencing in pepper compromises resistance to avirulent X. campestris pv vesicatoria infection. By contrast, PR10/LRR1 overexpression in Arabidopsis thaliana confers enhanced resistance to Pseudomonas syringae pv tomato and Hyaloperonospora arabidopsidis. Together, these results suggest that the cytosolic LRR-PR10 complex is responsible for cell death–mediated defense signaling.
INTRODUCTION
Plants have evolved resistance mechanisms to overcome microbial pathogen attack. Disease resistance (R) proteins mediate the recognition of pathogen invasion and elicit downstream signaling responses, leading to plant immunity (Moffett et al., 2002). Programmed cell death (PCD), often the ultimate endpoint of extreme defense, limits the spread of invasive pathogens. In the compatible plant–pathogen interactions, pathogens repress plant immunity using effectors that cause disease symptoms. However, plant recognition of these effectors triggers the hypersensitive response (HR) in the incompatible interactions. This is called effector-triggered immunity and is a stronger immune response than pathogen-associated molecular pattern–triggered immunity.
The leucine-rich repeat (LRR) domain is a conserved feature of many R proteins, including nucleotide binding (NB) and LRR proteins (Dangl and Jones, 2001). Plant NB-LRRs perceive their effector ligands using their N terminus (Gutierrez et al., 2010). LRR-containing receptor-like kinases (RLKs), including FLAGELLIN-SENSITIVE2 and EF-TU RECEPTOR, recognize bacterial flagellin and translational elongation factor EF-Tu, respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). Another LRR-RLK, BRASSINOSTEROID INSENSITIVE1, perceives plant steroid hormone brassinosteroids (Wang et al., 2001). The pepper (Capsicum annuum) LRR1 contains a single LRR domain with five tandem repeats of a 24–amino acid LRR motif (Jung et al., 2004). LRR1, which is lacking a kinase domain, exhibits sequence similarity to RLKs and has been implicated in the plant defense response (Jung et al., 2004; Jung and Hwang, 2007). LRR1 interacts with the pepper HYPERSENSITIVE-INDUCED REACTION PROTEIN1 (HIR1) that triggers HR during avirulent Xanthomonas campestris pv vesicatoria (Xcv) infection. This ultimately leads to compromised HIR1-induced HR (Jung and Hwang, 2007). More recently, LRR1 and HIR1 have been demonstrated to act as cell death regulators associated with plant immunity and disease, respectively (Choi et al., 2011).
Convincing evidence indicates that the pathogenesis-related10 (PR10) family, one of the pathogenesis-related groups, has distinct functions in developmental processes, secondary metabolism, and antimicrobial activity (McGee et al., 2001; Zhou et al., 2002; Hashimoto et al., 2004; Park et al., 2004; Liu and Ekramoddoullah, 2006). Sequence analysis of the PR10 family indicates similarity with a major birch (Betula alba) pollen allergen Bet v 1 (Breiteneder et al., 1989). More recently, a positive transcription factor, WRKYb, was demonstrated to bind the PR10 promoter and activate the defense signaling pathway in pepper (Lim et al., 2011). Several PR10 family genes were isolated from various plant species, including apple (Malus domestica), asparagus (Asparagus officinalis), parsley (Petroselinum crispum), bean (Phaseolus vulgaris), pea (Pisum sativum), sorghum (Sorghum bicolor), potato (Solanum tuberosum), pepper, and rice (Oryza sativa) (Somssich et al., 1986; Matton and Brisson, 1989; Walter et al., 1990; Warner et al., 1992; Lo et al., 1999; Pühringer et al., 2000; Park et al., 2004; Hashimoto et al., 2004). The PR10 genes are induced by microbial attack (Somssich et al., 1986), fungal elicitors (Somssich et al., 1986; Walter et al., 1990), or wounding stress (Warner et al., 1992). In general, PR10 genes are specifically induced in response to abiotic and biotic stress. However, the specific functions of PR10 in plant immunity and cell death signaling remain to be clarified.
Cell death localized at the infection site is the strongest strategy to restrict pathogen growth and development. This cell death response often includes the induction of R gene expression, ion fluxes, and the accumulation of reactive oxygen species (ROS) and defense-related hormones, such as salicylic acid (SA) (Li et al., 2010; Melech-Bonfil and Sessa, 2010). The constitutive activation of defense by genetic modification leads to spontaneous HR-like cell death lesions in the absence of pathogens (Lorrain et al., 2003). These so-called lesion mimic mutants often exhibit dwarf-like phenotypes due to constitutive defense activation (Dangl et al., 1996; Mosher et al., 2010). For example, rice blast lesion mimic mutants, which show a spontaneous cell death phenotype, significantly induce high levels of PR10s (Os-PR10a and Os-PR10b) (Jung et al., 2005, 2006).
Some plant resistance proteins have been convincingly demonstrated to localize and act in specific cellular compartments to trigger innate immunity signaling. Arabidopsis thaliana Toll/Interleukin-1–type NB-LRR receptor RESISTANCE TO PSEUDOMONAS SYRINGAE4 (RPS4) accumulates in the nucleus to trigger ENHANCED DISEASE SUSCEPTIBILITY1–dependent defense signaling (Gassmann et al., 1999; Wirthmueller et al., 2007). Tobacco (Nicotiana tabacum) N protein, which confers resistance against Tobacco mosaic virus, functions in the nucleus to trigger complex downstream defense events (Burch-Smith et al., 2007). In a previous study, the nuclear localization of pepper ABSCISIC ACID-RESPONSIVE1 (ABR1) was shown to be essential as a hypersensitive cell death regulator, despite the lack of a discernible nuclear localization signal (NLS) in ABR1 (Choi and Hwang, 2011). By contrast, some R proteins act in the cytoplasm. Potato NB-LRR receptor RX1, which confers a high resistance to Potato virus X, is located in both the cytoplasm and the nucleus (Slootweg et al., 2010). However, Rx1 is activated in the cytoplasm but not in the nucleus. The other NB-LRR protein, RPM1 (for Resistance to Pseudomonas syringae pv maculicola1), is activated at and functions on the plasma membrane (Gao et al., 2011).
Here, we report the identification of the pepper PR10 as a pepper LRR1-interacting partner using a yeast two-hybrid screen. Bimolecular fluorescence complementation (BiFC) and coimmunoprecipitation (co-IP) assays revealed that the intracellular protein PR10 specifically interacts with LRR1 in planta. Agrobacterium tumefaciens–mediated transient coexpression of PR10 and LRR1 prompted PR10-triggered hypersensitive cell death in pepper and Nicotiana benthamiana leaves. Upon fusing a nonfunctional NLS sequence (nls) to LRR1 and PR10, the cytoplasmic localization of these proteins was shown to be essential for cell death induction. Virus-induced gene silencing (VIGS) of PR10 in pepper compromised the resistance responses against avirulent Xcv infection. By contrast, heterologous PR10 overexpression in Arabidopsis conferred enhanced resistance to Pseudomonas syringae pv tomato and Hyaloperonospora arabidopsidis infection. Taken together, these results demonstrate that cytoplasmic PR10 functions in HR-like cell death and defense signaling. Furthermore, this role is strengthened by interaction with LRR1.
RESULTS
Expression of LRR1 and PR10 in Pepper
RNA and immunoblot analyses were used to characterize further the differential transcriptional and posttranscriptional patterns of PR10 expression in pepper plants (see Supplemental Figure 1 online). Infection with avirulent (incompatible) Xcv strain Bv5-4a triggered the strong and early induction of PR10 in pepper leaves. Although infection with the virulent (compatible) strain Ds1 resulted in strong transcriptional expression of PR10 (see Supplemental Figure 1A online), immunoblot analysis using PR10-specific antibody revealed that the PR10 induction was specific to the pepper leaves infected with the avirulent (incompatible) Xcv strain Bv5-4a (see Supplemental Figure 1B online). Infection with avirulent Xcv strain Bv5-4a caused a rapid induction of the hypersensitive cell death response (HR) in pepper leaves within 24 h of infiltration with a high titer (108 colony-forming units [cfu] mL−1).
PR10 Interacts with LRR1
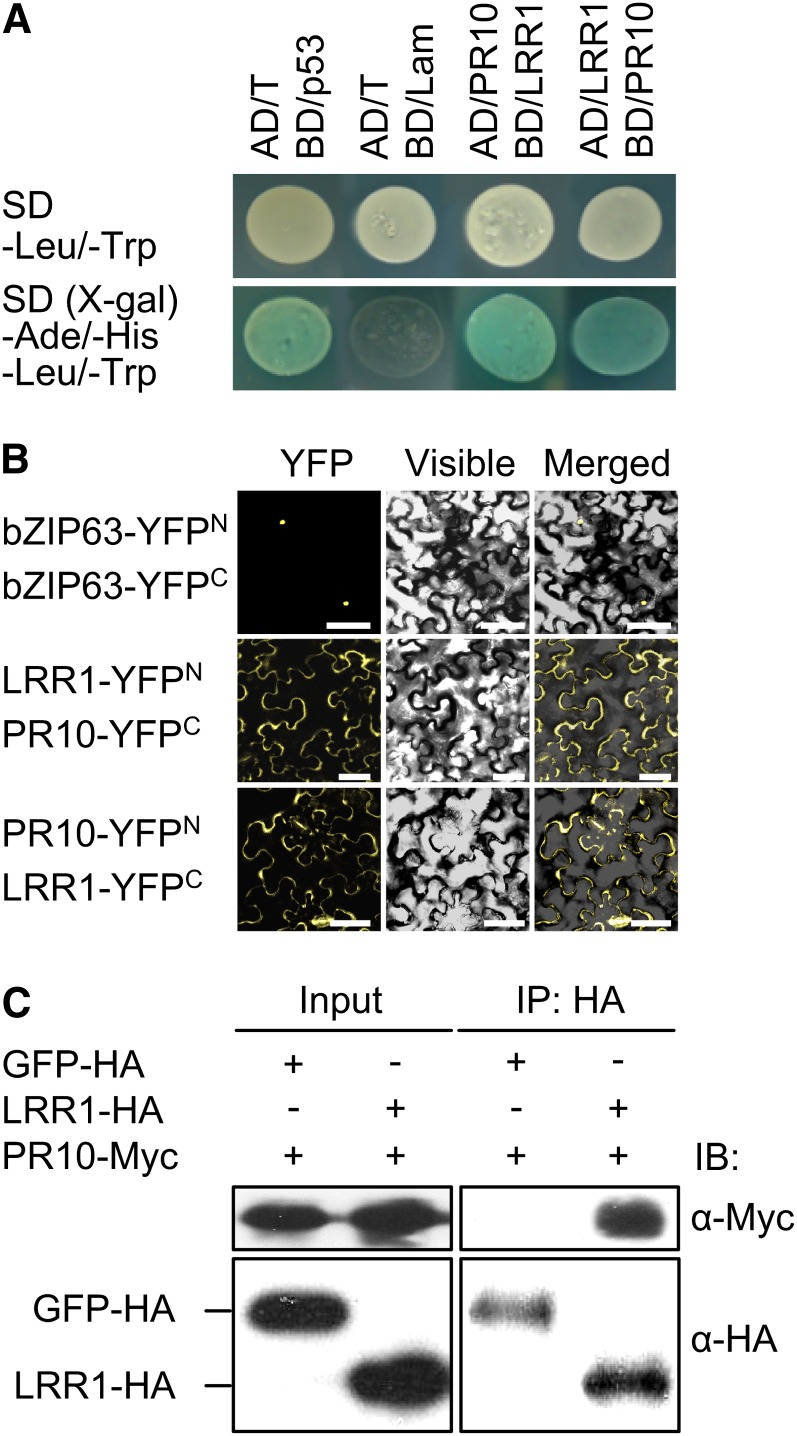
The pepper LRR1 gene was previously isolated from pepper leaves infected with Xcv (Jung et al., 2004). To isolate defense-related proteins in pepper, a pepper cDNA library generated from avirulent Xcv-infected leaves was screened for LRR1-interacting proteins using a GAL4-based yeast two-hybrid system. Among the clones identified from the screening, pepper PR10 was selected for further characterization as an interacting partner of LRR1 (Figure 1A).
Figure 1.
Interaction between LRR1 and PR10 in Yeast and N. benthamiana.
(A) LRR1 interacts with PR10 in a GAL4-based yeast two-hybrid system. SD, synthetic dropout. The plasmids encoding the fusions to the GAL4 AD and the DNA BD are denoted by AD and BD, respectively. Combinations of the Simian Vacuolating Virus 40 large T antigen (AD/T) with the murine p53 (BD/p53) fusion constructs and human lamic C (BD/Lam) were included as positive and negative controls, respectively.
(B) BiFC visualization of the LRR10/PR10 interaction in leaves infiltrated with Agrobacterium. Yellow fluorescence, visible light, and merged images were taken from the epidermal cells. The bZIP63-YFPN and bZIP63-YFPC constructs were used as positive controls. Bars = 50 μm.
(C) co-IP and immunoblotting (IB) of GFP-HA or LRR1-HA and PR10-Myc proteins coexpressed in leaves. GFP-HA was used as a negative control.
The LRR1-PR10 interaction in planta was examined using a BiFC assay (Walter et al., 2004). N- and C-terminal portions of yellow fluorescent protein (YFP) were fused to LRR1 and PR10 to yield LRR1-YFPN and PR10-YFPC, respectively. The reciprocal constructs were also generated (LRR1-YFPC and PR10-YFPN). Interactions between the fusion proteins were visualized in N. benthamiana leaves following Agrobacterium-mediated transient expression. Confocal images of BiFC signals were detected in the cytoplasmic region (Figure 1B), indicating that PR10 binds to LRR1 in the cytoplasm of plant cells. The nuclear localization of bZIP63-YFP fusion proteins was used as a positive control.
Additionally, the LRR1 and PR10 interaction in planta was confirmed by co-IP using a transient coexpression system in N. benthamiana (Figure 1C). Proteins extracted from transiently expressing N. benthamiana leaves were incubated with α-hemagglutinin (HA) antibody to immunoprecipitate LRR1. Potential LRR1 and PR10 complexes were separated by SDS-PAGE. Immunoblotting using α-Myc antibody detected PR10.The co-IP assay revealed that PR10 physically interacts with LRR. By contrast, green fluorescent protein (GFP), included as a negative control, did not exhibit any interaction.
Transient Coexpression of LRR1 and PR10 Induces Cell Death and Defense Responses
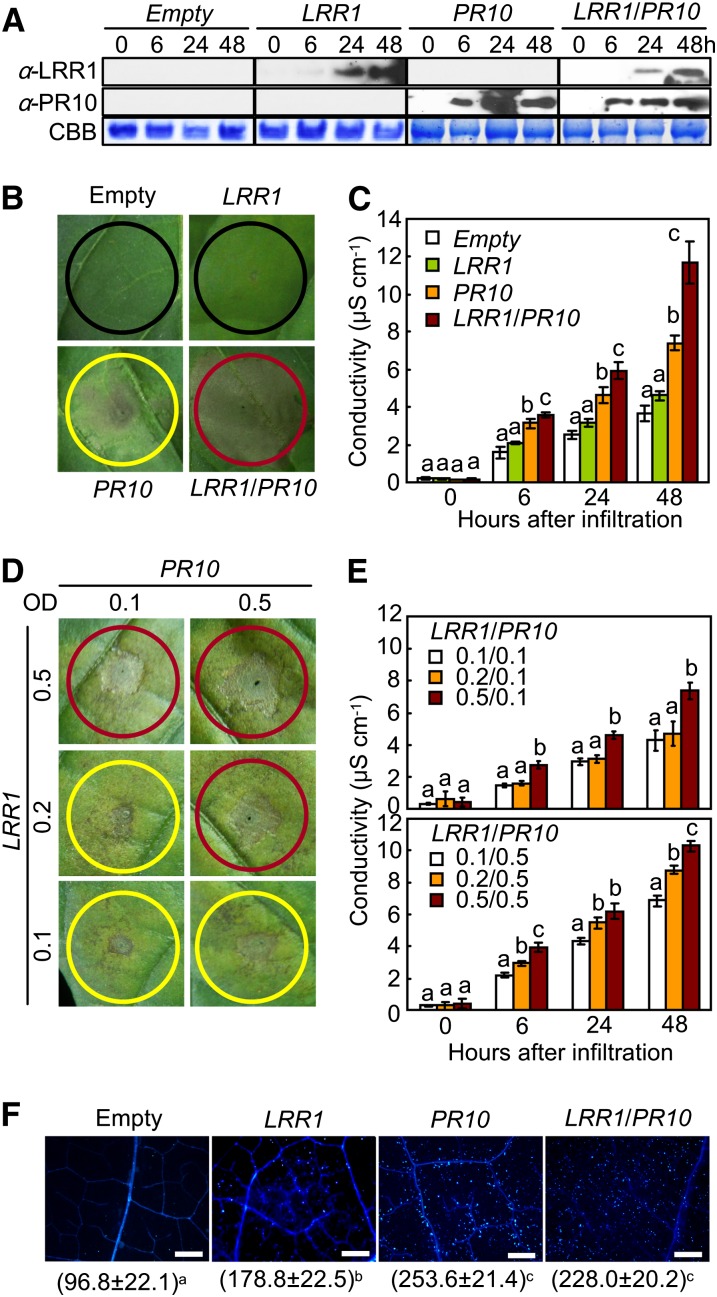
An Agrobacterium-mediated transient expression assay was used to determine the effect of LRR1 and PR10 expression on the induction of the cell death response in pepper leaves (Figure 2). The simultaneous expression of both LRR1 and PR10 in the agroinfiltrated leaves was confirmed using immunoblot analysis (Figure 2A). PR10 was more rapidly and more strongly expressed than LRR1. Plants transiently expressing the empty vector or LRR1 did not trigger any cell death response in pepper leaves (Figure 2B). By contrast, PR10 transient expression induced partial necrotic cell death. However, transient coexpression of LRR1 and PR10 was much more effective in triggering the full hypersensitive cell death response than was transient expression of PR10 alone (Figure 2B). The synergistic effect of LRR1 as a positive regulator of cell death induction by PR10 transient expression was supported by electrolyte leakage data (Figure 2C). Transient coexpression of LRR1 and PR10 resulted in enhanced electrolyte leakage from the pepper leaves, the magnitude of which was greater than that caused by the transient expression of PR10 alone. Infiltration with greater concentrations of Agrobacterium carrying LRR1 resulted in enhanced cell death induction by PR10 transient expression in pepper leaves (Figure 2D). Transient coexpression of LRR1/PR10 by infiltration with higher inoculum ratios of Agrobacterium carrying LRR1 stimulated higher electron leakage from pepper leaf tissues (Figure 2E). The leaves infiltrated with Agrobacterium carrying either PR10 or LRR1/PR10 exhibited significantly higher levels of cell death–associated callose deposition than did the leaves infiltrated with Agrobacterium carrying either the empty vector or LRR1 (Figure 2F).
Figure 2.
Transient Expression of LRR1, PR10, and LRR1/PR10 in Pepper Leaves.
(A) Immunoblot analysis of the transient expression of LRR1, PR10, and LRR1/PR10. CBB, Coomassie blue staining of the gel to show equal loading.
(B) and (C) Cell death phenotypes (B) and electrolyte leakage (C) in leaves infiltrated with Agrobacterium strain GV3101 carrying 35S:00 (empty), 35S:LRR1, 35S:PR10, or 35S:LRR1/35S:PR10. Red, yellow, and black circles indicate full, partial, and no cell death, respectively.
(D) and (E) Cell death phenotypes (D) and electrolyte leakage (E) in leaves infiltrated with Agrobacterium at different inoculum ratios.
(F) Callose deposition (bright blue dots) in leaves infiltrated with Agrobacterium. Number of callose deposits mm−2 represents the mean ± sd from three leaf discs. Bars = 500 μm.
(C), (E), and (F) Data represent the means ± sd from three independent experiments. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher's protected LSD test.
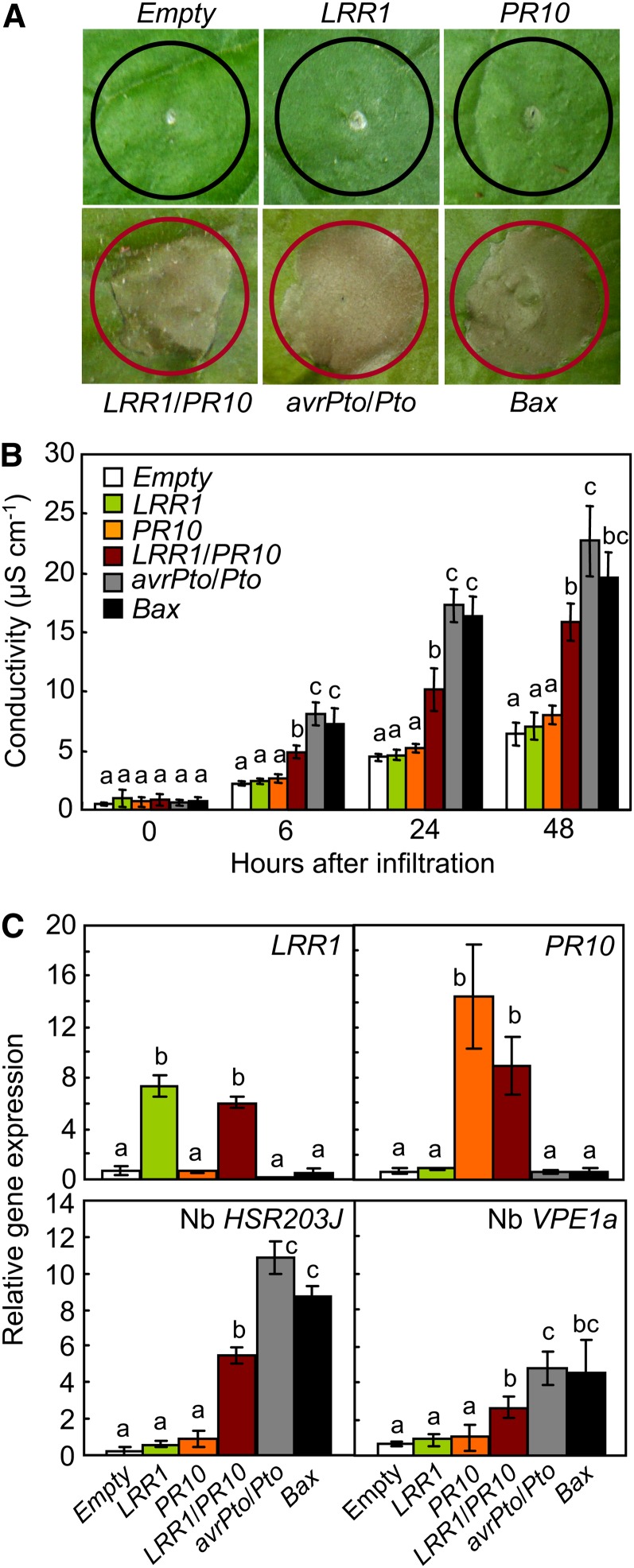
To compare the cell death induction by LRR1/PR10 coexpression with that induced by Bcl2-ASSOCIATED X PROTEIN (BAX) or avrPto/Pto expression, Bax and avrPto/Pto were used as positive inducers of cell death in N. benthamiana leaves (Figure 3). Mammalian Bax is capable of inducing cell death in plants (Lacomme and Santa Cruz, 1999), and the AvrPto interaction with Pto triggers HR in effector-triggered immunity (Tang et al., 1996). In contrast with the results of the pepper transient expression assay, neither LRR1 nor PR10 transient expression was able to trigger the cell death response in N. benthamiana leaves, despite agroinfiltration with an inoculum greater than OD600 = 1.0 (Figure 3A). However, coexpression of LRR1 and PR10 induced a cell death response similar to that triggered by Bax or avrPto/Pto expression. A quantitative analysis using an ion conductivity assay confirmed the cell death induction by LRR1/PR10 transient coexpression (Figure 3B). Electrolyte leakage from leaves transiently expressing LRR1/PR10 was significantly higher than from leaves transformed with empty vector, LRR1 or PR10. However, electrolyte leakage due to LRR1/PR10 expression was somewhat less than the levels induced by Bax or avrPto/Pto expression. Real-time RT-PCR analysis revealed an increase in the expression of LRR1 and PR10 24 h after agroinfiltration (Figure 3C). The coexpression effects of LRR1 and PR10 on the cell death response in leaves were assessed by monitoring the well-known cell death marker genes HYPERSENSITIVE-RELATED203J (HSR203J) (Pontier et al., 1994) and VACUOLAR PROCESSING ENZYME1a (VPE1a) (Zhang et al., 2010). In leaves coexpressing LRR1 and PR10, HSR203J induction was significantly greater than that observed following the transient expression of LRR1 or PR10, though slightly lower than those observed following avrPto or Bax expression (Figure 3C). VPE1a, involved in elicitor-triggered immunity in N. benthamiana (Zhang et al., 2010), was also significantly induced by LRR1 and PR10 coexpression (Figure 3C). However, transient expression of LRR1 or PR10 individually did not trigger HSR203J or VPE1a expression in N. benthamiana leaves (Figure 3C).
Figure 3.
Transient Expression of LRR1, PR10, LRR1/PR10, avrPto/Pto, and Bax in N. benthamiana Leaves.
(A) Cell death phenotypes in leaves infiltrated with Agrobacterium strain GV3101 carrying different constructs. Red and black circles indicate full and no cell death, respectively.
(B) Quantification of electrolyte leakage as ion conductivity to assess the cell death response in leaf discs.
(C) Quantitative real-time RT-PCR analysis of the expression of LRR1, PR10, HSR203J, and VPR1a in N. benthamiana leaves 24 h after agroinfiltration.
(B) and (C) Data represent the means ± sd from three independent experiments. Different letters above the bars indicate significantly different means (P < 0.05), as analyzed by Fisher's protected LSD test.
LRR1 Promotes the Ribonuclease Activity and Phosphorylation of PR10
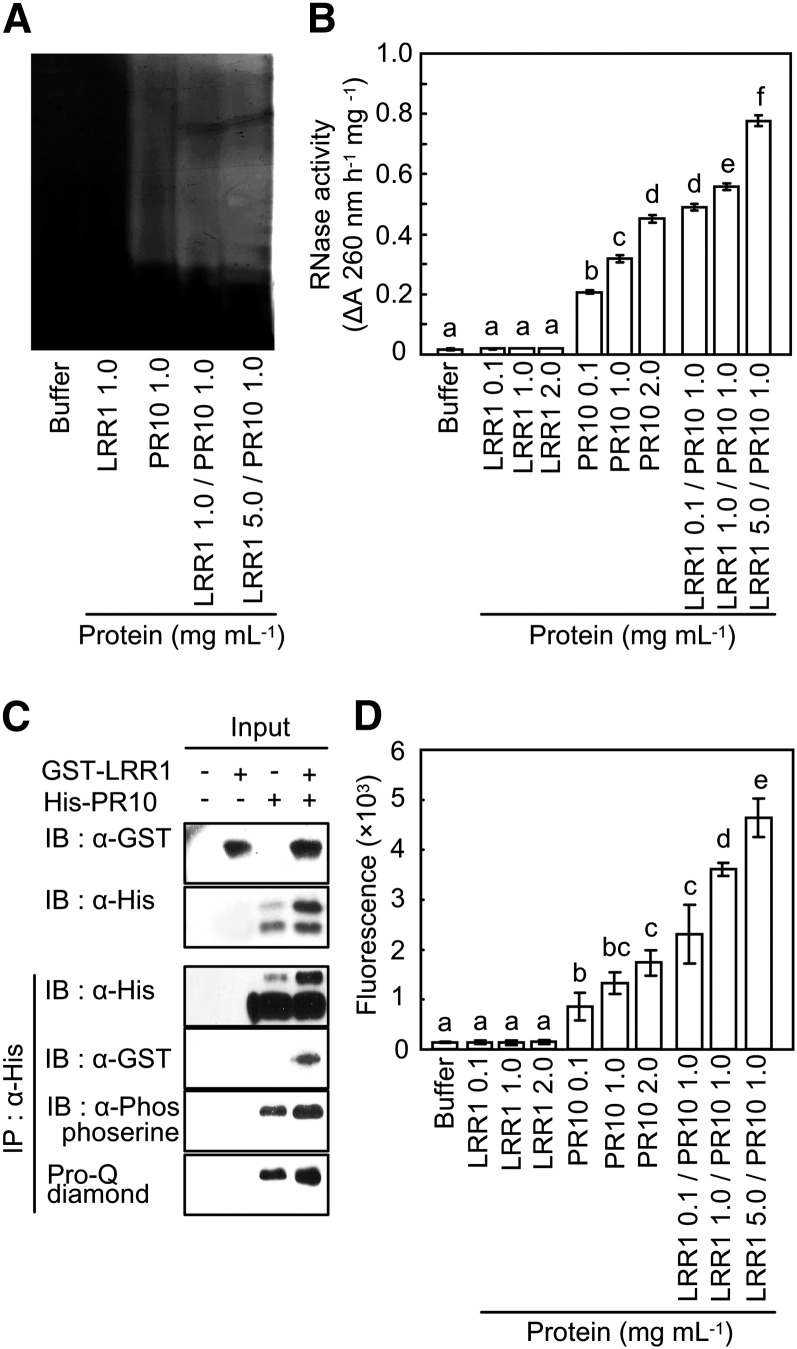
In-gel RNase assay (Bufe et al., 1996) was used to investigate whether LRR1 affects the RNase activity of PR10. The recombinant PR10, but not LRR1, distinctly degraded torula yeast (Candida utiliz) RNA (Figure 4A), indicating that PR10 possesses RNase activity. For the RNA degradation assay, the recombinant proteins (LRR1, PR10, and LRR1/PR10) and elution buffer were incubated with torula yeast RNA. As shown in Figure 4B, recombinant LRR1 with no RNase activity significantly enhanced PR10-mediated RNA degradation in a dose-dependent manner.
Figure 4.
Synergistic Effect of LRR1 on RNase Activity and Phosphorylation of PR10.
(A) Detection of RNase activity of PR10 on 15% acrylamide gel containing yeast RNA. After electrophoresis of recombinant PR10 and LRR1 proteins mixed at different concentrations, the gel was stained with toluidine blue O.
(B) RNase activity assay of PR10 in the presence of LRR1 using yeast RNA.
(C) Pro-Q diamond staining and immunoblot (IB) analysis using a phosphoserine antibody for the detection of PR10 phosphorylation.
(D) Quantification of phosphorylation by the detection of Pro-Q diamond fluorescence using ProXPRESS. Phosphorylation reactions were done using different concentrations of PR10 and LRR1 (0.1 to ~5.0 μg).
(B) and (D) Data represent the means ± sd from three independent experiments. Different letters indicate significantly different means, as analyzed by Fisher's protected LSD test (P < 0.05).
To phosphorylate PR10 by certain protein kinases, we used crude protein extracts from pepper leaves infected with avirulent Xcv Bv5-4a. The phosphorylated PR10 was detectable by Pro-Q diamond gel staining (Figure 4C). Immunoblot analysis using the antiphosphoserine antibody confirmed the phosphorylation of PR10 by protein extracts of pepper leaves (Figure 4C). In parallel, immunoblot analysis using anti-His and anti-glutathione S-transferase (GST) antibodies detected recombinant PR10 and LRR1 in the phosphorylation reactions, respectively. As expected, increases in the amount of LRR1 added to the reactions significantly enhanced the PR10 phosphorylation. Quantification of the Pro-Q diamond staining data by fluorescence measurements revealed that PR10 bound to LRR1 resulted in significantly higher levels of phosphorylation than did PR10 alone (Figure 4D). Collectively, these results indicate that LRR1 positively regulates the phosphorylation and RNase activity of PR10.
Cytoplasmic Localization of the LRR1-PR10 Complex Is Essential for Cell Death Induction
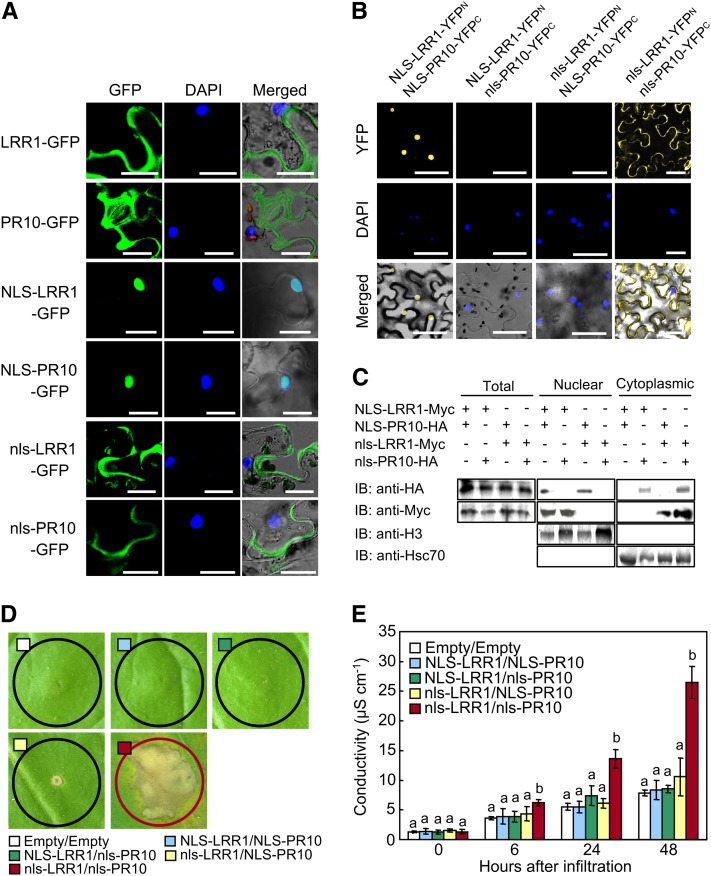
The subcellular locations of GFP-fused LRR1 and PR10 in N. benthamiana leaves were visualized using a confocal microscope. Both proteins were detected in the cytoplasm but not in the nuclei (Figure 5A). To determine the localization sites of LRR1 and PR10 in planta, a time course of BiFC assay signals was analyzed (see Supplemental Figure 2 online). Yellow fluorescent LRR1-PR10 complexes were detected in the cytoplasm 24 h after agroinfiltration. At 48 h after agroinfiltration, the LRR1-PR10 complex initiated an efflux into the extracellular region. To examine whether the protein complex moves from the cytoplasm to the extracellular regions, N. benthamiana leaves were infiltrated with brefeldin A (BFA), an inhibitor of secretion (Staehelin and Driouich, 1997), following the coexpression of LRR1 and PR10. In this localization experiment, BFA treatment resulted in the aggregation of YFP within the cells (see Supplemental Figure 3 online). By contrast, the BiFC signals of the LRR1-PR10 interaction were observed in the cytoplasm and the extracellular space in the absence of BFA. These results indicate that the LRR1-PR10 complex may interact in the cytoplasm to cause secretion into the apoplast via the plasma membrane. Varients of LRR1 and PR10 that would localize to the nucleus were generated by fusion with a well-known Simian Vacuolating Virus 40 NLS (PKKKRKV) (Hodel et al., 2001; Slootweg et al., 2010). Confocal microscopy images demonstrated that the NLS fusion was sufficient to target the NLS-LRR1-GFP or NLS-PR10-GFP proteins to the nuclei (Figure 5A). The nonfunctional NLS mutant (nls) was used to express nls-fused PR10 and LRR1 in the cytoplasm. The NLS or nls fusion proteins were further introduced into the BiFC vectors, pSPYNE and pSPYCE, to investigate which location of the LRR1-PR10 complex is essential for cell death induction. Transient coexpression of NLS-LRR1-YFPN and NLS-PR10-YFPC was detected as yellow fluorescence signals in the nuclei (Figure 5B). This indicates that both fusion proteins localized to the nuclei and interact with each other. Similarly, coexpression of nls-LRR1-YFPN and nls-PR10-YFPC produced BiFC signals in the cytoplasm, indicating a physical interaction between both proteins. However, no yellow fluorescence signals were detected in N. benthamiana leaf cells upon the coexpression of NLS-fused LRR1 and nls-fused PR10, and vice versa. When analyzed by immunoblotting, NLS-fused LRR1 or PR10 proteins were detected in the nuclear fractions. However, nls-fused proteins were observed in the nucleus-depleted fractions (Figure 5C). Histone 3 (H3) and heat shock complex 70 were detected in the nuclear and cytoplasmic fractions, respectively (Figure 5C).
Figure 5.
Cytoplasmic Localization of the LRR1/PR10 Complex Is Essential for Cell Death Induction.
(A) Subcellular localization of NLS- or nls-fused LRR1 and PR10 in N. benthamiana leaves. DAPI images indicate nuclear staining. All images were taken by confocal microscopy 24 h after agroinfiltration. Bars = 20 μm.
(B) BiFC images of NLS- or nls-fused LRR10/PR10 combinations in leaves infiltrated with Agrobacterium. Bars = 50 μm.
(C) Immunoblot (IB) analysis of LRR1-Myc and PR10-HA in nuclear and cytoplasmic fractions of leaves transiently expressing NLS- or nls-fused LRR1 or PR10. Histone H3 and Hsc70 were included as fractionation markers for the nucleus and the cytoplasm, respectively.
(D) Induction of the cell death response by transient expression of nls-fused LRR10/PR10 combinations 3 d after agroinfiltration.
(E) Quantification of electrolyte leakage as ion conductivity to assess the cell death response. Data represent the means ± sd from three independent experiments. Different letters indicate significantly different means, as analyzed by Fisher’s protected LSD test (P < 0.05).
Nuclear or nucleocytoplasmic coexpression of NLS- or nls-fused LRR1 and PR10 did not trigger the cell death response in N. benthamiana leaves (Figure 5D). By contrast, the coexpression of nls-LRR1 and nls-PR10 induced the hypersensitive cell death response 3 d after agroinfiltration and also significantly increased electrolyte leakage from N. benthamiana leaves (Figure 5E). Together, these results suggest that the cytoplasmic coexpression of the LRR1-PR10 complex is required for cell death induction in N. benthamiana leaves.
Silencing of PR10 Attenuates Disease Resistance and Compromises the Hypersensitive Cell Death Response
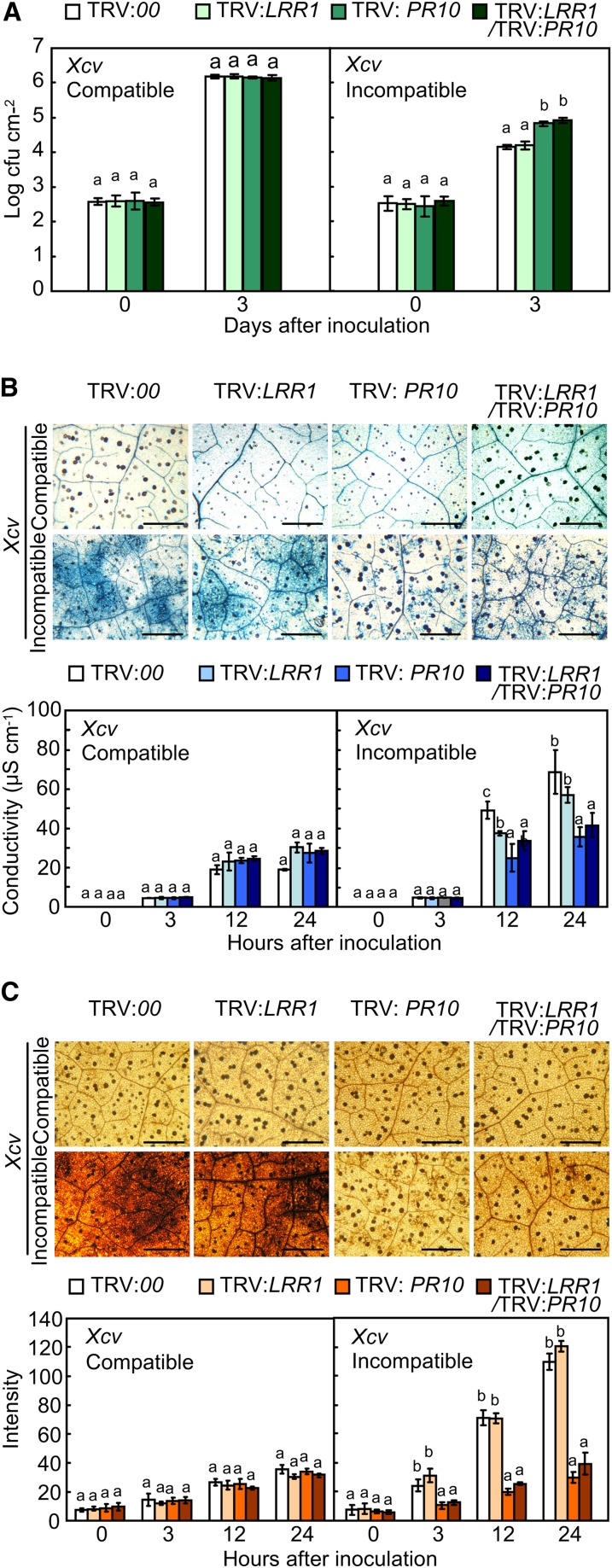
To study the loss of function of PR10 and/or LRR1, we generated LRR1- and/or PR10-silenced pepper plants using the VIGS technique. Quantitative real-time RT-PCR analysis showed that LRR1, PR10, or LRR1/PR10 was effectively silenced in plants infected with Xcv (see Supplemental Figure 4 online). Avirulent Xcv growth in PR10- or LRR1/PR10-silenced leaves increased to significantly higher levels than that in the empty vector controls (Figure 6A). However, the silencing of LRR1 did not enhance avirulent bacterial growth. Notably, the silencing of both genes did not affect the proliferation of Xcv virulent (compatible) Ds1 in pepper leaves. Trypan blue staining of leaves demonstrated that PR10 silencing compromises the HR in the incompatible interaction (Figure 6B, top panel). The visual scores of the cell death phenotype were substantiated by an electrolyte leakage assay (Figure 6B, bottom panel). Inhibition of electrolyte leakage from PR10- or LRR1/PR10-silenced leaves was significantly greater than that in unsilenced or LRR1-silenced leaves during the avirulent Xcv infection. The trypan blue staining and ion conductivity data further indicate that PR10, but not LRR1, triggers pathogen-induced hypersensitive cell death response in pepper. To determine if the silencing of LRR1 and PR10 inhibits ROS accumulation, Xcv-infected leaves were stained with 3,3′-diaminobenzidine tetrahydrochloride (DAB), a histochemical reagent for hydrogen peroxide (H2O2). As expected, PR10- and LRR1/PR10-silenced leaves also accumulated significantly lower levels of H2O2 than the unsilenced and LRR1-silenced leaves at the early stage of Xcv infection (Figure 6C).
Figure 6.
Distinct Responses of LRR1-, PR10-, and LRR1/PR10-Silenced Pepper Plants to Xcv Infection.
(A) Bacterial growth in leaves infected with Xcv (5 × 104 cfu mL−1).
(B) Trypan blue staining (top) with leaves 24 h after inoculation with Xcv (107 cfu mL−1). Quantification of electrolyte leakage (bottom) from leaves infected with Xcv (107 cfu mL−1). Bars = 500 μm.
(C) DAB staining (top) to detect H2O2 production in leaves infected with Xcv (107 cfu mL−1). Quantification of H2O2 production in leaf tissues (bottom), as determined by ImageJ software. Bars = 500 μm.
Data represent the means ± sd from three independent experiments. Different letters indicate significant differences, as determined by Fisher’s protected LSD test (P < 0.05).
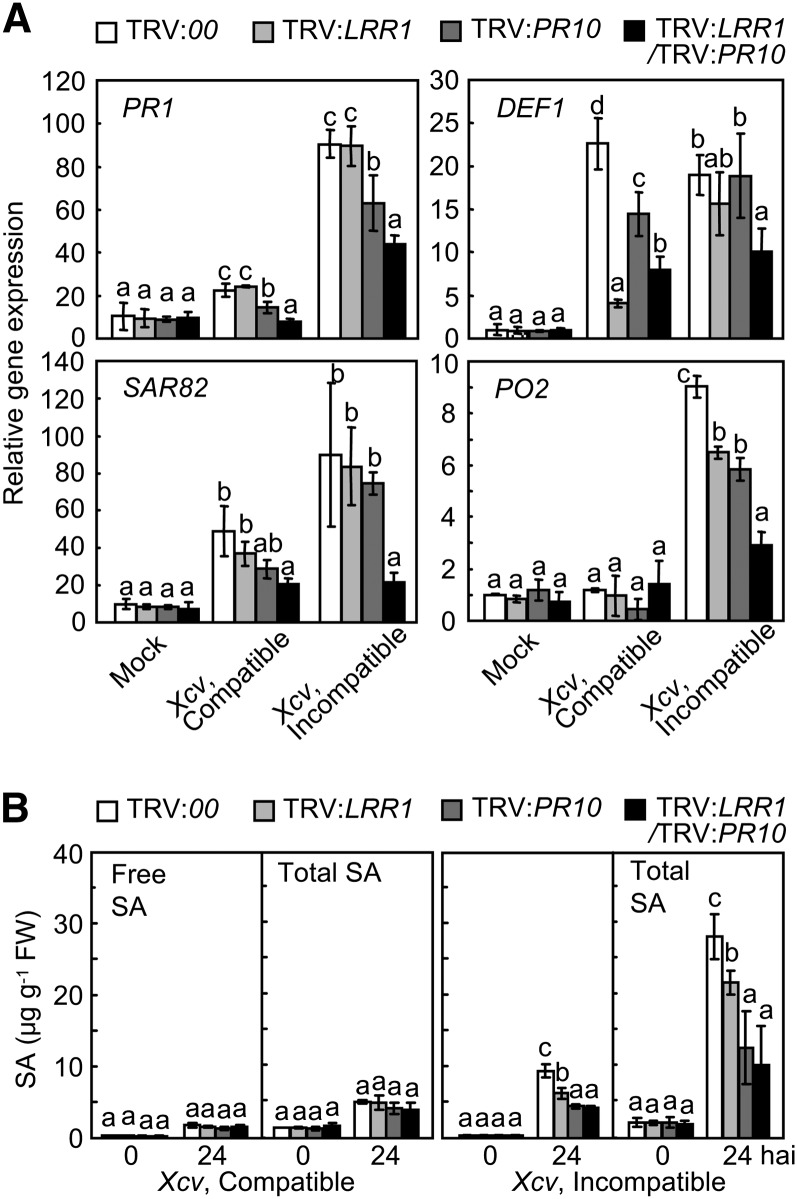
Quantitative RT-PCR analysis was used to investigate whether LRR1 and PR10 silencing regulates defense response genes in pepper leaves during Xcv infection (Figure 7A). PATHOGENESIS-RELATED PROTEIN1 (PR1; Kim and Hwang, 2000), Defensin1 (DEF1; Do et al., 2004), SYSTEMIC ACQUIRED RESISTANCE8.2 (SAR82; Lee and Hwang, 2003), and PEROXIDASE2 (PO2; Choi et al., 2007) were significantly downregulated in pepper leaves that had been silenced for LRR1/PR10 during Xcv infection, especially in the incompatible interactions (Figure 7A). Silencing of LRR1, PR10, or LRR1/PR10 did not significantly compromise the induction of free and total SA by the Xcv virulent (compatible) infection (Figure 7B). By contrast, the accumulated free and total SA levels were significantly lower in all LRR1- and/or PR10-silenced leaves in comparison to the empty vector control leaves 24 h after inoculation with the Xcv avirulent (incompatible) Bv5-4a strain (Figure 7B). Notably, the silencing of PR10 or LRR1/PR10 distinctly suppressed the accumulation of SA and its glycoside (SAG) when compared with LRR1 silencing in pepper leaves during the Xcv virulent (compatible) infection. Together, these results indicate that PR10 is required for the hypersensitive cell death and defense responses, including HR induction, ROS burst, defense-related gene induction, and SA accumulation.
Figure 7.
Silencing of LRR1/PR10 Compromises Defense Gene Expression and SA Accumulation in Pepper Leaves Infected by Xcv.
(A) Quantitative real-time RT-PCR analysis of the expression of defense response genes in the leaves 24 h after Xcv inoculation. Expression values are normalized by the expression level of Ca ACTIN.
(B) Levels of free SA and total SA (free SA plus its conjugate) in the leaves infected by Xcv. FW, fresh weight.
Data represent the means ± sd from three independent experiments. Different letters indicate significant differences, as statistically analyzed by Fisher’s protected LSD test (P < 0.05).
Enhanced Resistance of PR10- and LRR1/PR10-Overexpressing Transgenic Arabidopsis to Bacterial and Oomycete Infection
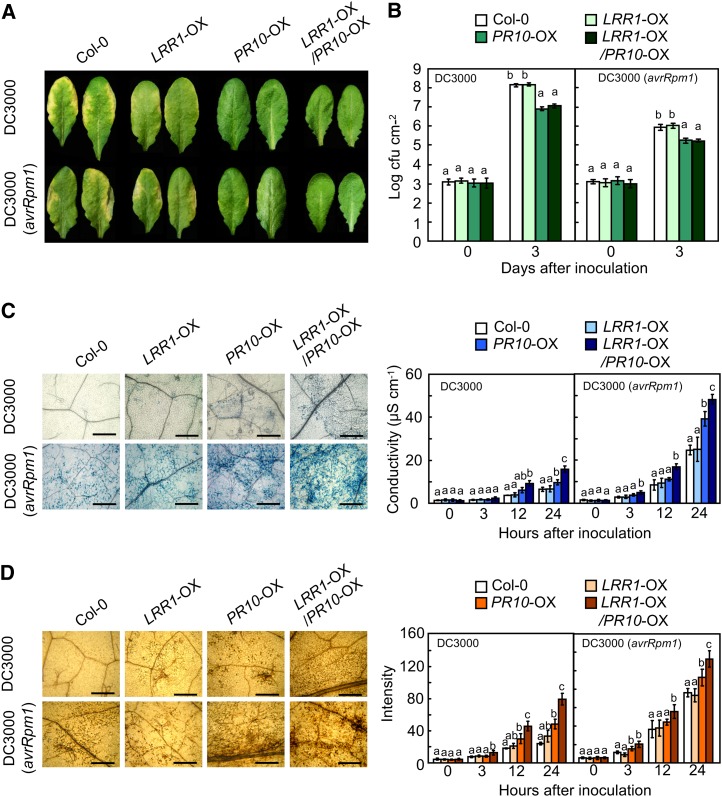
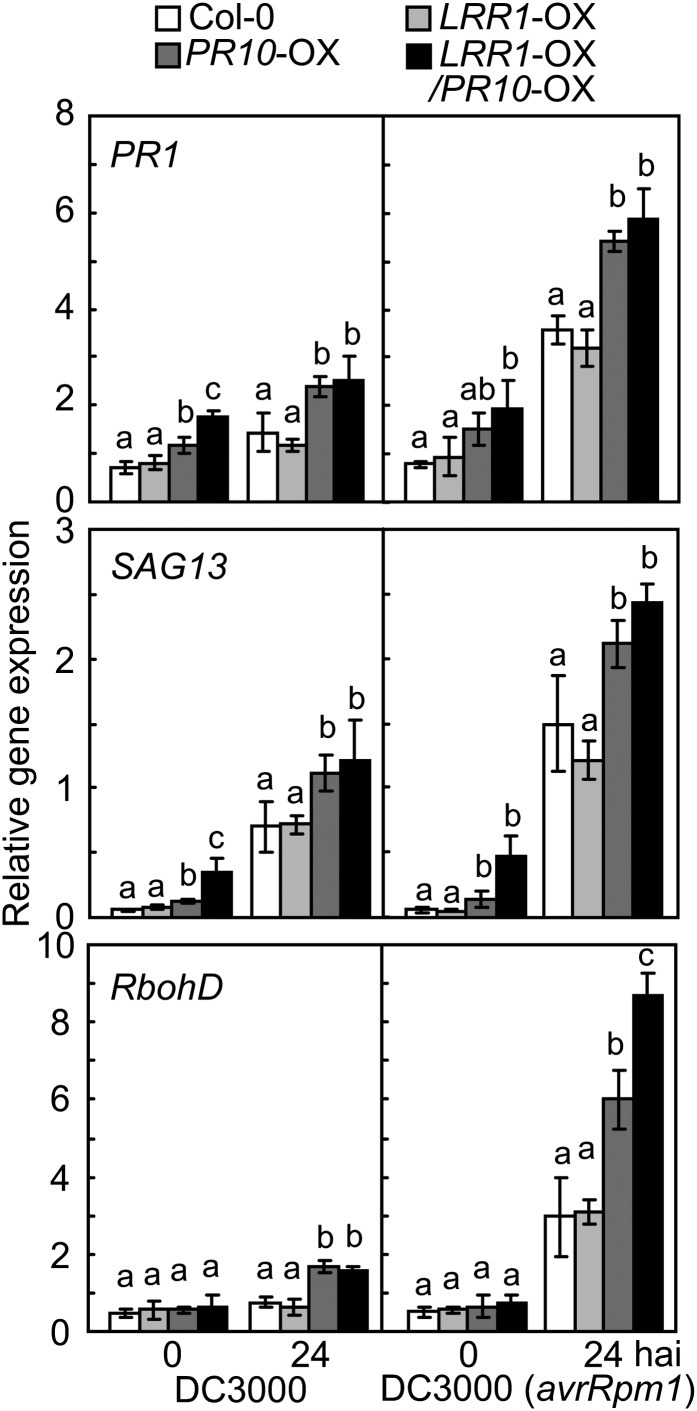
Resistance to P. syringae pv tomato (Pst) infection was investigated using Arabidopsis plants co-overexpressing LRR1, PR10, and LRR1/PR10 (Figure 8). Notably, PR10 overexpression (OX) reduced chlorotic or necrotic disease symptoms in Arabidopsis leaves infected with Pst DC3000 or DC3000 (avrRpm1) (Figure 8A). Bacterial growth in the leaves of wild-type plants was similar to that in LRR1-OX plants. By contrast, PR10 and LRR1/PR10-OX plants exhibited significantly less growth of Pst DC3000 and DC3000 (avrRpm1) than did wild-type and LRR1-OX plants (Figure 8B). Trypan blue–stained micrographs show that cell death induction by Pst infection was significantly accelerated in PR10- and LRR1/PR10-OX plants (Figure 8C). Pst-infected leaves were next stained with DAB, and the brownish color intensity was quantified using ImageJ. As expected, infection with Pst DC3000 and DC3000 (avrRpm1) drastically increased H2O2 accumulation in the leaves of PR10- and LRR1/PR10-OX plants in comparison to wild-type and LRR1-OX plants (Figure 8D). These data suggest that LRR1 positively regulates cell death and the ROS burst caused by PR10 overexpression. The representative defense marker gene PR1 was significantly induced in PR10- and LRR1/PR10-OX Arabidopsis leaves infected by Pst DC3000, particularly in the interaction with Pst DC3000 (avrRpm1) (Figure 9). Expression of SENESCENCE-ASSOCIATED GENE13 (SAG13), a gene encoding a short-chain alcohol dehydrogenase, is known to be required for PCD (Brodersen et al., 2002). The SAG13 transcript levels in PR10- and LRR1/PR10-OX Arabidopsis leaves were significantly greater than those in wild-type Columbia-0 (Col-0) and LRR1-OX leaves during Pst DC3000 infection (Figure 9). NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG D (RbohD) was not constitutively induced in any of the transgenic plants. However, RbohD expression was drastically heightened in PR10- and LRR1/PR10-OX leaves following Pst infection (Figure 9). Details of growth phenotypes and enhanced resistance of PR10 transgenic Arabidopsis to Pst and H. arabidopsidis infection are shown in Supplemental Data Set 1 and Supplemental Figures 5 to 7 online.
Figure 8.
Enhanced Resistance of PR10- and LRR1/PR10-OX Transgenic Arabidopsis Plants to Pst Infection.
(A) Disease symptoms on leaves 72 h after inoculation.
(B) Bacterial growth in the leaves of wild-type and transgenic plants.
(C) Micrographs of the leaves stained with trypan blue (left) 24 h after infiltration and quantification of electrolyte leakage (right) from leaf discs. Bars = 500 μm.
(D) Micrographs of the leaves stained with DAB (left) 24 h after inoculation and intensity of the reddish color from DAB images (right) to assess H2O2 production. Bars = 500 μm.
(B) to (D) Data represent the means ± sd from three independent experiments. Different letters indicate significant differences, as statistically analyzed by Fisher’s protected LSD test (P < 0.05).
Figure 9.
Real-Time RT-PCR Analysis of Defense Marker Gene Expression in LRR1-, PR10-, and LRR1/PR10-OX Transgenic Arabidopsis Plants.
Expression values are normalized by the expression level of ACTIN2. Data represent the means ± sd from three independent experiments. Different letters indicate significant differences, as analyzed by Fisher's protected LSD test (P < 0.05). hai, hours after inoculation with Pst DC3000.
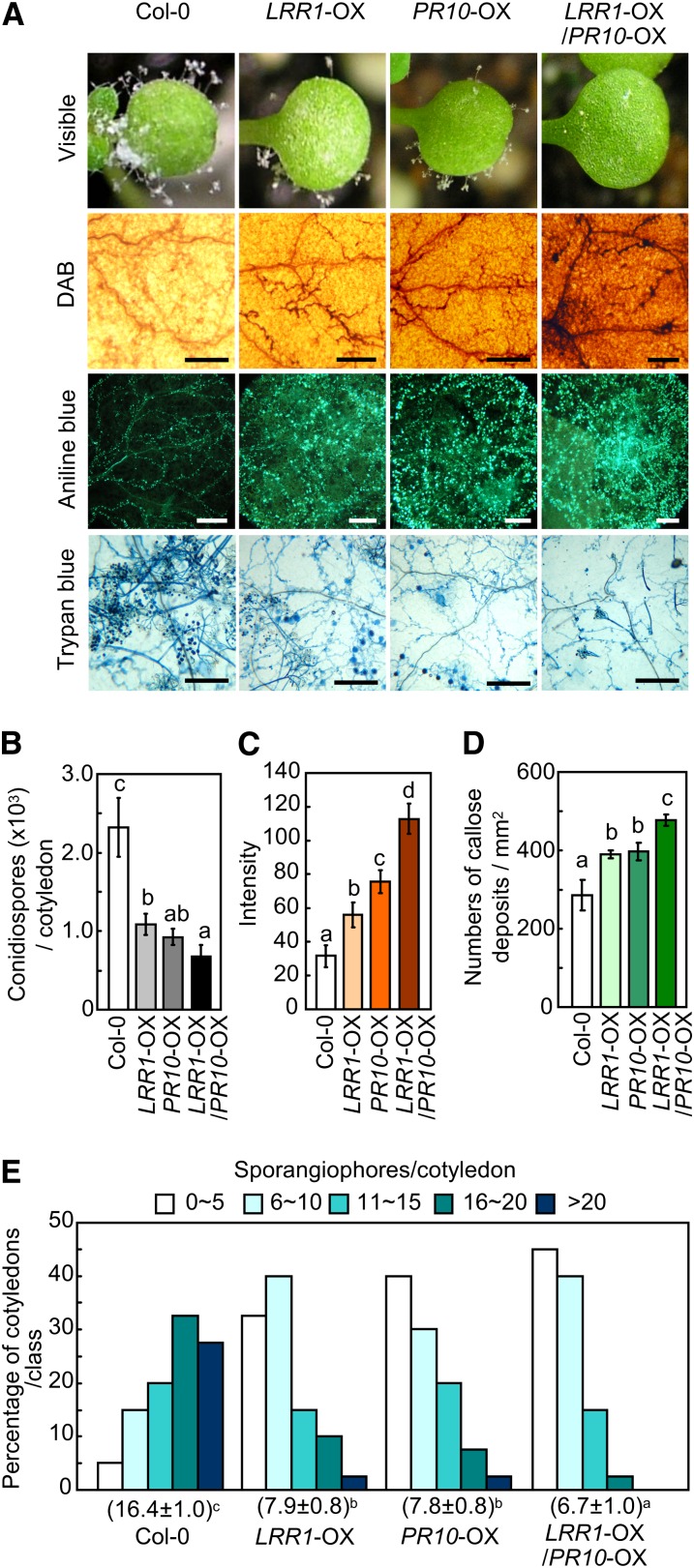
Wild-type Arabidopsis (Col-0) cotyledons displayed a high level of mycelial growth, sporulation, and sporangiophore formation of the oomycete pathogen H. arabidopsidis isolate Noco2 (Figure 10; see Supplemental Figure 8 online). By contrast, transgenic expression of LRR1, PR10, and LRR1/PR10 significantly decreased the growth and development of H. arabidopsidis, ultimately resulting in reduced disease development. Overexpression of these transgenes also conferred enhanced H2O2 production and callose accumulation around the infection sites, preventing further hyphal development (Figures 10A and 10B). Reduced sporulation and conidiopspore production was distinctly noticeable in the PR10- and LRR1/PR10-OX transgenic plants (Figures 10C and 10D). Notably, the double OX transgenic plants of LRR1 and PR10 were more resistant to this oomycete pathogen than were the wild-type and other single OX mutant plants.
Figure 10.
Enhanced Resistance of LRR1-, PR10-, and LRR1/PR10-OX Transgenic Arabidopsis Plants to H. arabidopsidis Infection.
(A) Disease symptoms and cell responses in the cotyledons inoculated with conidiospores of H. arabidopsidis isolate Noco2. Bars = 200 μm.
(B) Quantification of conidiospores on 20 cotyledons.
(C) ROS intensities of leaf tissues, as measured by a color densitometer using ImageJ software after DAB staining.
(D) Quantification of callose deposition by aniline blue staining.
(E) Quantification of asexual sporangiophores on cotyledons. The numbers below each line represent the means ± sd.
(B) to (E) Data represent the means ± sd from three independent experiments. Different letters indicate significant differences, as analyzed by Fisher’s protected LSD test (P < 0.05).
DISCUSSION
In this article, we demonstrate that the pepper pathogenesis-related protein PR10, a member of the Bet v 1 allergen family, is crucial for defense and cell death responses against bacterial pathogen attack. To date, little is known concerning the cellular functions of PR10, although it is known to have ribonuclease and antimicrobial activity (Zhou et al., 2002; Park et al., 2004). In previous proteomics work, PR10 was isolated from pepper leaves infected with an avirulent Xcv strain (Choi and Hwang, 2011). As demonstrated with RNA gel blot and immunoblot analyses, PR10 is strongly induced by avirulent Xcv infection. The convincing data prompted an investigation into the role of PR10 in the hypersensitive cell death and defense responses in plants.
PR10 Interacts with LRR1 to Activate Cell Death and Defense Responses
Up until now, there has been no experimental evidence indicating physical interaction between PR10 and other plant proteins. In this study, we found physical and functional interactions between the PR10 and LRR1 proteins to activate cell death and defense responses in plants. In previous studies, LRR1 was shown to suppress HIR1-triggered cell death as a negative regulator in Arabidopsis and tobacco (Jung and Hwang, 2007; Choi et al., 2011). In contrast with these previous findings, LRR1 physically interacts with PR10 and enhances PR10-triggered cell death and defense responses as a positive regulator. Interestingly, the transient coexpression of LRR1 with PR10 intensifies PR10-triggered cell death, suggesting that the cell death induction by PR10 may require complex formation with LRR1. The conflicting evidence that LRR1 with HIR1 or PR10 acts as a negative or positive regulator, respectively, may reflect the distinct functions of both HIR1 and PR10 for disease-associated and HR-associated cell death, respectively. This suggestion is supported by the experimental finding that silencing of HIR1 in pepper significantly compromised HR and disease-associated cell deaths (Choi et al., 2011). Unlike pepper, transient expression of PR10 alone does not induce a typical cell death response in N. benthamiana leaves. Moreover, immunoblot detection of the transient expression indicated that neither PR10 nor LRR1 induces each other in pepper leaves (Figure 2A). These results suggest that PR10 requires specific pepper components for cell death induction. The Bet v I fold domain present in PR10 has been demonstrated to be responsible for its interaction with several ligands, such as brassinosteroids, cytokinin, and flavonoid (Fujimoto et al., 1998; Mogensen et al., 2002; Marković-Housley et al., 2003; Koistinen et al., 2005), suggesting that it might be involved in PR10 binding to LRR1. However, further studies using domain mutants will be required to elucidate the binding site of PR10 for LRR1.
LRR1 Promotes the Ribonuclease Activity and Phosphorylation of PR10
It has been reported that PR10 is phosphorylated (Ziadi et al., 2001) and that its phosphorylation is required for the defense response to microbial pathogen attack (Park et al., 2004). In this study, we also show that recombinant PR10 has RNase activity and that it is phosphorylated by crude protein extracts from pepper leaves infected by avirulent Xcv Bv5-4a. The plant protein extracts were used as an alternative kinase pool because there is no information about which kinase is involved in PR10 phosphorylation. Notably, LRR1 potentiates the RNase activity of PR10. RNase activity is thought to be essential for the resistance response to microbial pathogens (Barna et al., 1989; Lusso and Kuc, 1995; Galiana et al., 1997; Shivakumar et al., 2000; Park et al., 2004). In this study, LRR1 is also shown to be a potential stimulator of PR10 phosphorylation, which could lead to the activation of defense and HR-like cell death. Together, these results suggest that the phosphorylation and RNase activity of pepper PR10, which is increased by its interaction with LRR1, may be required for cell death and defense induction.
Cytoplasmic Localization of the LRR1-PR10 Complex Is Essential for the Induction of the Cell Death Response
BiFC analysis enables the direct visualization of protein–protein interactions to determine the subcellular site of interaction (Walter et al., 2004). YFP signals of the LRR1-PR10 complex were detected in the cytoplasmic region of N. benthamiana leaves at early time point after agroinfiltration but in both the cytoplasmic and apoplastic regions at late time point. This suggests the export of the LRR1-PR10 complex into the apoplastic space during the cell death response. The localization of the LRR1-PR10 complex in the apoplast region may not be responsible for cell death, but rather a consequence caused by cell wall degradation.
Certain plant disease resistance proteins act in specific subcellular locations to trigger downstream signaling and defense pathways. Some NB-LRR–type immune receptors enter and function in the nucleus (Sheen and He, 2007; Shen and Schulze-Lefert, 2007). Barley (Hordeum vulgare) mildew A 10 (Shen et al., 2007), Arabidopsis RPS4 (Wirthmueller et al., 2007), and tobacco N protein (Burch-Smith et al., 2007) are functional in the nucleus. By contrast, potato Rx1 activates an antiviral mechanism in the cytoplasm but not in the nucleus (Slootweg et al., 2010). The NB-LRR–type R protein, RECOGNITION OF PERONOSPORA PARASITICA1A, is targeted to the plasma membrane and associates with the endoplasmic reticulum and the Golgi apparatus (Michael Weaver et al., 2006). In this study, PR10 interacts with LRR1 in the cytoplasm to trigger defense signaling. The hypersensitive-induced reaction protein, HIR1, was demonstrated to physically interact with LRR1, which possesses four LRR domains for their association (Jung and Hwang, 2007). Interestingly, extracellular LRR1 interacts with membrane-associated HIR1 to form membrane microdomains (Choi et al., 2011). Since LRR1 has a putative signal peptide sequence in N terminus (Jung et al., 2004), this protein is predicted to be involved in the cellular secretion. Choi et al. (2011) showed that LRR1 was localized in small patches at the plasma membrane when it was expressed in onion (Allium cepa) epidermal cells and also that it was enriched in the extracellular protein extract from pepper leaves infected with Xcv. In our experiments, GFP-fused LRR1 under the 35S promoter was predominantly visualized in the cytoplasmic region along with the plasma membrane in epidermal cells of N. benthamiana leaves (Figure 5). Therefore, the relatively small LRR1 protein may flexibly interact with both cytosolic and membrane-anchored proteins during transport from the intracellular region to the apoplastic space.
We attempted to engineer the LRR1-PR10 interaction site to determine whether the cytoplasmic localization of the LRR1-PR10 complex is essential for the cell death induction. The attachment of an NLS (Hodel et al., 2001) recruited both LRR1 and PR10 into the nucleus, where they were able to form a protein–protein association (Figure 5B). However, nuclear pools of the LRR1 and PR10 complex were not able to trigger the HR phenotype. When fused with nls, the LRR1-PR10 complexes were sequestered in the cytoplasm. The coexpression of nls-LRR1 and nls-PR10 induced the hypersensitive cell death response in N. benthamiana leaves (Figure 5E). These results suggest a requirement for the downstream signaling of this complex in the cytoplasm.
LRR1 and PR10 Play a Role in Cell Death and Defense Responses in Pepper and Arabidopsis
PR10 proteins have been identified as essential for the primary detectable defense response of resistant and susceptible plants to biotic attack. The PR10 protein level is significantly higher in Fusarium graminearum–infected resistant maize (Zea mays) inbred CO441 than in F. graminearum–susceptible inbred B73 (Mohammadi et al., 2011). In our study, pepper PR10 is rapidly and strongly induced at transcriptional and protein levels in the incompatible response during Xcv infection. This finding strongly supports a critical role of pepper PR10 in disease resistance. By contrast, silencing of PR10-like proteins in Medicago truncatula results in an antagonistic induction of other pathogenesis-related proteins and in an increased tolerance upon infection with the necrotrophic oomycete Aphanomyces euteiches (Colditz et al., 2007).
In this study, PR10 silencing significantly compromises bacterial growth restriction, cell death response, and ROS accumulation in pepper leaves during infection with incompatible Xcv Bv5-4a. LRR1 positively regulates the PR10-triggered cell death that is accompanied by increased ROS levels. To support these early defense events, the silencing of PR10 or LRR1/PR10 significantly compromised the induction of pepper defense marker genes, such as PR1 (Kim and Hwang, 2000), DEF1 (Do et al., 2004), SAR82 (Lee and Hwang, 2003), and PO2 (Choi et al., 2007), as well as the accumulation of SA, especially by avirulent Xcv infection. In general, the SA-dependent pathway positively regulates defense response against biotrophic pathogen infection (Spoel and Dong, 2008). These findings support the possibility that the LRR1-PR10 complex may regulate the cell death and defense response by affecting downstream defense genes and SA signaling pathways.
Overexpression of both PR10 and LRR1 suppressed Pst DC3000 growth at a level similar to that observed in the single PR10-OX plant. However, the enhanced cell death noted in the PR10/LRR1 double mutant plants supports the potential ability of these genes to function in plant immunity against pathogen invasion. This enhanced cell death effect was also ascertained by a downy mildew infection assay. Co-overexpression of PR10/LRR1 confers enhanced resistance to infection by H. arabidopsidis Noco2, and this resistance is accompanied by an increase in ROS and callose production around the infection sites. These findings suggest that the PR10 and LRR1 complex is effective in triggering the defense response to infection by the biotrophic oomycete.
Proposed Model for the Role of the PR10 and LRR1 Complex in Cell Death–Mediated Defense Signaling
Combining the data presented here, we propose a working model for the role of the PR10 and LRR1 complex in cell death–mediating defense signaling in plants (see Supplemental Figure 9 online). During incompatible Xcv infection, PR10, along with the positive regulator LRR1, induces early defense responses, including callose accumulation, SA and ROS burst, and defense-related gene expression. In addition, LRR1 enhances the phosphorylation and RNase activity of PR10, which may be required for cell death and defense induction. These ultimately lead to HR-like cell death and the basal defense response. In a previous study, transient expression assays showed that LRR1 negatively regulates HIR1-mediated cell death associated with disease (Choi et al., 2011). It seems likely that HIR1 is primarily involved in disease-associated cell death, whereas PR10 may trigger HR cell death and defense. This suggests that LRR1 has a dual function in cell death regulation. There is convincing evidence to indicate that host-controlled PCD is intimately linked with the onset of disease-associated cell death and symptom development (Greenberg and Yao, 2004). During virulent Xcv infection, Xcv effectors suppress pathogen-associated molecular pattern–triggered immunity, where HIR1 signaling induces disease-associated cell death (Choi et al., 2011). It is not determined whether PR10 competes with HIR1 for the interaction with LRR1; however, LRR1 induction may promote PR10-mediated hypersensitive cell death and resistance responses against avirulent Xcv infection. The LRR1-PR10 interaction also activates the cell death pathway in pepper, N. benthamiana, and Arabidopsis. LRR1 physically interacts with PR10 and acts as a positive regulator to enhance PR10-triggered cell death and defense signaling. However, how LRR1 activates PR10-dependent resistance remains to be clarified. More detailed studies of the molecular mechanisms underlying the disease resistance-associated specific interaction between PR10 and LRR1 are needed.
METHODS
Plant Materials and Pathogen Inoculation
Pepper (Capsicum annuum cv Nockwang) and Nicotiana benthamiana plants were raised in a growth room at 26°C with a 16-h-light/8-h-dark cycle. Fully expanded pepper leaves were infiltrated with Xanthomonas campestris pv vesicatoria strains Ds1 (virulent) and Bv5-4a (avirulent).
Arabidopsis thaliana (ecotype Col-0) plants were grown in soil under controlled environmental conditions (16-h-light/8-h-dark cycle, 24°C, and 100 μmol photons m−2 s−1). Pseudomonas syringae pv tomato DC3000 and DC3000 (avrRpm1) were infiltrated into Arabidopsis leaves. Hyaloperonospora arabidopsidis isolate Noco2 was maintained on Arabidopsis (Col-0) seedlings. Ten-day-old Arabidopsis seedlings were spray-inoculated with a suspension of 5 × 104 conidiosporangia mL−1. The disease rating was conducted as previously described (Lee et al., 2008).
RNA Gel Blot and Quantitative Real-Time RT-PCR Analyses
Total RNA was isolated using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. RNA gel blot analysis was conducted following standard procedures. Total RNA was transferred to a nylon membrane and probed with 32P-labeled LRR1 and PR10 open reading frames, which were generated from the plasmid by EcoRI digestion. RT reactions were performed using 1 μg total RNA, 500 ng oligo(dT)18 primer, and Moloney Murine Leukemia Virus Reverse Transcriptase (Enzynomics) at 42°C for 1 h. RT reaction products were used for quantitative real-time RT-PCR analysis. To normalize the transcript levels, β-tubulin and ACTIN2 expression was monitored in N. benthamiana and Arabidopsis, respectively. The gene-specific primer sets used for the quantitative real-time RT-PCR analysis are listed in Supplemental Table 1 online.
Yeast Two-Hybrid Assay
The GAL4 system was used for the yeast two-hybrid assay according to the manufacturer’s instructions (Invitrogen). The LRR1 and PR10 coding regions were PCR amplified and cloned into pCR2.1-TOPO (Invitrogen) using the primers listed in Supplemental Table 2 online. The constructs were recombined into the pGADT7 and pGBKT7 destination vectors, including the GAL4 DNA activation domain (AD) and binding domain (BD), respectively, to create AD/PR10, BD/LRR1, AD/LRR1, and BD/PR10. All of the constructs were transformed into yeast strain AH109. The positive clones were selected by growing on SD/-Leu/-Trp medium, followed by growth on SD/-Leu/-Trp/-Ade/-His medium. The final clones were arrayed on SD/-Leu/-Trp/-Ade/-His medium containing 40 mg L−1 X-Gal and 20 g L−1 Gal.
BiFC Analysis
For the BiFC constructs, LRR1 and PR10 were PCR amplified using the primers listed in Supplemental Table 2 online. The fragments were cloned into pCR2.1/TOPO and then recombined into pSPYNE or pSPYCE, resulting in pSPYNE:LRR1, pSPYCE:PR10, or vice versa, as described (Walter et al., 2004). Agrobacterium tumefaciens strain GV3101 harboring the BiFC constructs was coinfiltrated into N. benthamiana leaves. One day after agroinfiltration, the leaves were visualized using an LSM 5 Exciter confocal laser scanning microscope (Carl Zeiss) with excitation at 514 nm and emission at 525 to 600 nm. When indicated, BFA (10 μg mL−1) was infiltrated into N. benthamiana leaves 24 h after agroinfiltration of the BiFC or GFP constructs.
co-IP
For co-IP constructs, LRR1 and PR10 were PCR amplified using the primers listed in Supplemental Table 2 online. The fragments were cloned into pCR2.1/TOPO and recombined into p35S:6HA or p35S:8Myc. Agrobacterium strain GV3101 harboring the constructs was coinfiltrated into N. benthamiana leaves. Proteins were extracted from leaf samples using extraction buffer (10% glycerol, 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM DTT, 1× complete protease inhibitor cocktail [Roche], and 2% [w/v] polyvinyl polypyrrolidone). The protein extracts were incubated with monoclonal anti-HA or anti-Myc agarose (Sigma-Aldrich) overnight. Beads were collected and washed with immunoprecipitation buffer (10% glycerol, 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.15% Nonidet P-40, and 2 mM DTT). Eluted proteins were analyzed by immunoblotting using monoclonal anti-HA-peroxidase antibody (Sigma-Aldrich) or polyclonal anti-c-Myc-peroxidase antibody (Sigma-Aldrich). Immunodetection was performed using the WEST-ZOL plus protein gel blot detection system (iNtRON Biotechnology).
Antibody Production and Immunoblot Analysis
The polyclonal rabbit antibodies were raised against recombinant PR10 (Young In Frontier). The full-length PR10 and LRR1 coding regions were PCR amplified from pepper cDNA using the primers listed in Supplemental Table 2 online. The fragments of PR10 and LRR1 were integrated into the BamHI and SacI sites of the pET28a expression vector (Invitrogen) expressing an N-terminal His-tag and into the pGEX-5X expression vector (GE Healthcare) expressing an N-terminal GST tag, respectively. His-PR10 and GST-LRR1 fusion proteins were expressed in Escherichia coli BL21, purified, and injected into rabbits. The rabbit antisera were purified using a column conjugated with protein A resin (Sigma-Aldrich).
Total proteins were extracted from the leaves, as described previously (Choi et al., 2008). For immunoblot analysis, equal amounts of protein were separated by SDS-PAGE and blotted to polyvinylidene fluoride membranes. The membranes were probed with LRR1- and PR10-specific antibodies at 1:5000 and 1:10,000 dilution, respectively. Horseradish peroxidase–conjugated goat anti-rabbit IgG (Sigma-Aldrich) was used as a secondary antibody.
RNase Activity Assay
An in-gel RNase activity assay of recombinant PR10 and LRR1 was performed using the torula yeast (Candida utiliz) RNA (Sigma-Aldrich), as described (Yen and Green, 1991). Recombinant PR10 and LRR1 were purified from E. coli transformed with pET28a and pGEX-5X using Ni-NTA resin (Invitrogen) and Glutathione Sepharose 4B (GE Healthcare), respectively. The recombinant or control proteins were mixed with a protein loading dye and loaded onto 15% acrylamide gel containing 2.4 mg mL−1 of yeast RNA. The electrophoresis for protein separation was performed in a Mini PROTEAN 3 cell (Bio-Rad).
RNase activity was determined according to the method of Barna et al. (1989) with minor modifications. Yeast RNA (200 μg) mixed with PR10 and LRR1 (0.1 to ~5.0 μg) was incubated in 400 μL of 100 mM MES, pH 6.0, at 56°C for 30 min. One unit of RNase activity was defined as an increase in absorbance of 1.0 at OD260 after incubation for 30 min.
Phosphorylation Assay
Phosphorylation assay was performed as described by Park et al. (2004), with minor modifications. The pepper leaves inoculated with avirulent Xcv strain Bv5-4a (108 cfu mL−1) were homogenized in extraction buffer (40 mM Tris-HCl, pH 7.8, 1 mM DTT, 10% glycerol, 0.2 mM sodium fluoride, 0.1 mM sodium pyrophosphate, and 1× protease inhibitor cocktail). The phosphorylation reaction was done at 30°C for 20 min after addition of recombinant PR10 and LRR into the buffer (20 μg proteins in the leaf extract, 5 mM MgCl2, 100 μM ATP, and 25 mM KCl). His or GST fusion protein purification was performed using the standard protocol of Ni-NTA purification (Invitrogen) and a GST fusion protein purification system (GE Healthcare), respectively. His or GST fusion protein was immunoprecipitated using a His or GST antibody (Sigma-Aldrich). Immunodetection was performed using anti-His (Santa Cruz), anti-GST (Santa Cruz), and anti-phosphoserine (Sigma-Aldrich) at 1:10,000 dilution.
For detection of phosphoprotein, Pro-Q diamond phosphoprotein staining (Martin et al., 2003; Wang et al., 2005) was used as described by the manufacturer (Invitrogen). After one-dimensional electrophoresis, the gel was stained with Pro-Q diamond in the dark for 60 min and then destained with 100 mL of 20% acetonitrile and 50 mM sodium acetate, pH 4.0. Gel image capture and phosphoprotein quantification were performed using ProXPRESS (Perkin-Elmer LifeScience) with 540/25-nm excitation and 590/30-nm emission filters.
Agrobacterium-Mediated Transient Expression and Subcellular Localization Assays
For the generation of pBIN35S-GFP constructs, LRR1 and PR10 were PCR amplified using the primers listed in Supplemental Table 2 online. The fragments were cloned into pCR2.1/TOPO and recombined into pBIN35S-GFP. Agrobacterium strain GV3101 carrying the constructs was grown overnight in Luria-Bertani liquid medium containing 50 μg mL−1 kanamycin and 50 μg mL−1 rifampicin. Bacterial cells were infiltrated into pepper or N. benthamiana leaves. For comparison, Bax and avrPto/Pto was transiently expressed in N. benthamiana leaves (Choi et al., 2011).
GFP was visualized using a LSM 5 Exciter confocal laser scanning microscope (Carl-Zeiss) with excitation at 48 8 nm and emission at 505 to 530 nm. For visualization of the nuclei, detached leaves were immersed in staining buffer (0.1% 4′,6-diamidino-2-phenylindole [DAPI] in 5% DMSO). The DAPI fluorescence images were obtained using excitation at 405 nm and emission at 435 to 480 nm.
Engineering of Nuclear Expression
For nuclear localization, the NLS was fused to the N terminus of LRR1 or PR10. The nls sequence contains a Lys residue substituted with an Asn residue. The nls, which fails to target to the nuclei, was included as a negative control. The sequences used were NLS (5′-GGCCCTAAAAAGAAGCGTAAGGTT-3′) and nls (5′- GGCCCTAAAAACAAGCGTAAGGTT-3′) (Hodel et al., 2001).
Nuclear fractionation was conducted as previously described (Tameling et al., 2010). Leaf tissues were homogenized in Honda buffer (2.5% Ficoll 400, 5% dextran T40, 0.4 M Suc, 25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 mM phenylmethanesulfonylfluoride, and a complete protease inhibitor cocktail). Triton X-100 was added to a final concentration of 0.5%. The harvested supernatant represented the soluble fraction, and the pellet was purified as a nuclear fraction. The two phase fractions were subjected to immunoblot analysis. Anti-H3 (Abcam) and anti-heat shock complex 70 (Abcam) were used as nuclear and cytosolic protein markers, respectively.
VIGS
To investigate the loss of function of LRR1 and PR10, tobacco rattle virus (TRV)-based VIGS assays were performed as described previously (Liu et al., 2002; Hwang et al., 2011). LRR1- and PR10-specific 5′-untranslated region was PCR amplified and cloned into the pTRV2 vector using the primer sets listed in Supplemental Table 2 online. Agrobacterium GV3101 carrying TRV-derived plasmids (TRV1, TRV2-LRR1, and TRV2-PR10) were cultured in Luria-Bertani liquid medium containing 50 μg mL−1 kanamycin and 50 μg mL−1 rifampicin. The cultures were centrifuged and resuspended in infiltration medium (10 mM MES, pH 5.6, 10 mM MgCl2, and 200 μM acetosyringone). Agrobacterium suspensions containing TRV1 or TRV2 constructs were mixed at a 1:1 ratio and coinfiltrated into the cotyledons of 2-week-old pepper seedlings.
Arabidopsis Transformation
The PR10 coding region was PCR amplified using specific primer sets (see Supplemental Table 2 online) and cloned into the XbaI and BamHI sites of the binary vector pBIN35S (Choi and Hwang, 2011). The resulting binary plasmid was transformed into Agrobacterium strain GV3101 by electroporation. The Agrobacterium-mediated transformation was performed using the floral dipping method (Clough and Bent, 1998). Transgenic plants were selected by germinating seeds on Murashige and Skoog agar medium containing 50 μg mL−1 kanamycin. LRR1-OX Arabidopsis seeds were obtained from the laboratory seed stocks (Jung and Hwang, 2007). Transgenic Arabidopsis plants overexpressing both LRR1 and PR10 were generated by crossing the two OX mutant lines.
Measurement of Ion Conductivity
Ion leakage was measured as previously described (Hwang and Hwang, 2011; Lee et al., 2011). The leaf discs (8 mm in diameter) were washed and incubated in 10 mL of double distilled water for 30 min at room temperature. The ion conductivity was recorded using a sensION7 conductivity meter (Hach).
SA Quantification
SA and its glucoside (SAG) were extracted from pepper leaves and analyzed using HPLC, as described previously (Choi and Hwang, 2011; Kim and Hwang, 2011). HPLC separation was achieved using a 5-μm Xbridge C18 reverse-phase column (4.6 mm × 250 mm; Waters) fitted with an Xbridge C18 guard column (5 mm × 4.6320 mm; Waters). SA levels were measured using a fluorescence detector (Photo Diode Array Detector; Waters) with excitation at 305 nm and emission at 405 nm. 3-Hydroxybenzoic acid (Sigma-Aldrich) was included as an internal standard.
Histochemical Assay
DAB was used to stain H2O2-producing leaves. The leaves were incubated in 1% DAB solution in the dark overnight and destained with lactic acid/glycerol/ethanol (1:1:2). ImageJ software (National Institutes of Health) was used for quantification of H2O2 accumulation from the DAB images. To visualize the cell death response, the infiltrated leaves were stained with trypan blue and destained with saturated chloral hydrate solution (2.5 g mL−1). Epifluorescence microscopy was used to detect callose deposits after staining with aniline blue. Infiltrated leaves were cleared in alcoholic lactophenol (95% ethanol:lactophenol = 2:1; lactophenol, phenol:glycerol:lactic acid:water = 1:1:1:1). Cleared leaves were stained with 0.01% aniline blue in 0.15 M phosphate buffer, pH 9.5. Callose deposition was visualized with a fluorescence microscope under UV light.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: pepper LRR1 (AY237117), pepper PR10 (AF244121), pepper PR1 (AF053343), pepper DEF1 (AF442388), pepper SAR82A (AF313766), pepper PO2 (DQ489711), pepper ACT (GQ339766), N. benthamiana VPE1a (AB075947), N. benthamiana HSR203J (AB091430), N. benthamiana β-TUB2 (U91563), Arabidopsis PR1 (AT2G14610), Arabidopsis SAG13 (AT2G29350), Arabidopsis RbohD (AT5G47910), and Arabidopsis ACT2 (AT3G18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Patterns of PR10 in Pepper Leaves Infected with Xcv.
Supplemental Figure 2. LRR1 and PR10 Interact in the Cytoplasm and Are Released into the Apoplast.
Supplemental Figure 3. Localization of the LRR1 and PR10 Complex in N. benthamiana Leaves Treated with BFA.
Supplemental Figure 4. Quantitative Real-Time RT-PCR Analysis of the Expression of LRR1 and/or PR10 in VIGS Pepper Leaves Infected with Xcv.
Supplemental Figure 5. Enhanced Resistance of PR10-OX Arabidopsis Lines to Pst Infection.
Supplemental Figure 6. PR10 Contributes to Basal Resistance by Callose Deposition.
Supplemental Figure 7. Transgenic Arabidopsis Plants Overexpressing LRR1 and/or PR10.
Supplemental Figure 8. Enhanced Resistance of PR10-OX Arabidopsis Lines to Hpa Infection.
Supplemental Figure 9. Proposed Model for the Role of the PR10 and LRR1 Complex in Cell Death–Mediating Defense Signaling in Plants.
Supplemental Table 1. Gene-Specific Primers for qRT-PCR Used in This Study.
Supplemental Table 2. Primers for Generation of Various Gene Constructs Used in This Study.
Supplemental Data Set 1. Text File of Details of Growth Phenotypes and Enhanced Disease Resistance of PR10 Transgenic Arabidopsis Shown in Supplemental Figures 5 to 8.
Supplementary Material
Acknowledgments
This work was supported by the Next Generation BioGreen21 Program (Plant Molecular Breeding Center; No. PJ008027), Rural Development Administration, Republic of Korea.
AUTHOR CONTRIBUTIONS
D.S.C. and B.K.H. designed the research and wrote the article. D.S.C. and I.S.H. performed the experiments and analyzed the data.
Glossary
- PCD
programmed cell death
- HR
hypersensitive response
- LRR
leucine-rich repeat
- NB
nucleotide binding domain
- RLK
receptor-like kinase
- Xcv
Xanthomonas campestris pv vesicatoria
- ROS
reactive oxygen species
- SA
salicylic acid
- NLS
nuclear localization signal
- BiFC
bimolecular fluorescence complementation
- VIGS
virus-induced gene silencing
- YFP
yellow fluorescent protein
- co-IP
coimmunoprecipitation
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- BFA
brefeldin A
- DAB
3,3′-diaminobenzidine tetrahydrochloride
- H2O2
hydrogen peroxide
- Col-0
Columbia-0
- OX
overexpression
- Pst
P. syringae pv tomato
- AD
activation domain
- BD
binding domain
- DAPI
4′,6-diamidino-2-phenylindole
- TRV
tobacco rattle virus
- cfu
colony-forming units
References
- Barna B., Ibenthal W.D., Heitefuss R. (1989). Extracellular RNase activity in healthy and rust-infected wheat leaves. Physiol. Mol. Plant Pathol. 35: 151–160 [Google Scholar]
- Breiteneder H., Pettenburger K., Bito A., Valenta R., Kraft D., Rumpold H., Scheiner O., Breitenbach M. (1989). The gene coding for the major birch pollen allergen Betv1, is highly homologous to a pea disease resistance response gene. EMBO J. 8: 1935–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H.M., Olszak B., Skov S., Ødum N., Jørgensen L.B., Brown R.E., Mundy J. (2002). Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe A., Spangfort M.D., Kahlert H., Schlaak M., Becker W.-M. (1996). The major birch pollen allergen, Bet v 1, shows ribonuclease activity. Planta 199: 413–415 [DOI] [PubMed] [Google Scholar]
- Burch-Smith T.M., Schiff M., Caplan J.L., Tsao J., Czymmek K., Dinesh-Kumar S.P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D.S., Hwang B.K. (2011). Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23: 823–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.W., Lee B.G., Kim N.H., Park Y., Lim C.W., Song H.K., Hwang B.K. (2008). A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol. 148: 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.W., Kim Y.J., Hwang B.K. (2011). The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant Microbe Interact. 24: 68–78 [DOI] [PubMed] [Google Scholar]
- Choi H.W., Kim Y.J., Lee S.C., Hong J.K., Hwang B.K. (2007). Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 145: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colditz F., Niehaus K., Krajinski F. (2007). Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta 226: 57–71 [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Dietrich R.A., Richberg M.H. (1996). Death don’t have no mercy: Cell death programs in plant-microbe interactions. Plant Cell 8: 1793–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Do H.M., Lee S.C., Jung H.W., Sohn K.H., Hwang B.K. (2004). Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 166: 1297–1305 [Google Scholar]
- Fujimoto Y., Nagata R., Fukasawa H., Yano K., Azuma M., Iida A., Sugimoto S., Shudo K., Hashimoto Y. (1998). Purification and cDNA cloning of cytokinin-specific binding protein from mung bean (Vigna radiata). Eur. J. Biochem. 258: 794–802 [DOI] [PubMed] [Google Scholar]
- Galiana E., Bonnet P., Conrod S., Keller H., Panabières F., Ponchet M., Poupet A., Ricci P. (1997). RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiol. 115: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Chung E.-H., Eitas T.K., Dangl J.L. (2011). Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. USA 108: 7619–7624 Erratum. Proc. Natl. Acad. Sci. USA 108: 8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W., Hinsch M.E., Staskawicz B.J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20: 265–277 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagelin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Greenberg J.T., Yao N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6: 201–211 [DOI] [PubMed] [Google Scholar]
- Gutierrez J.R., Balmuth A.L., Ntoukakis V., Mucyn T.S., Gimenez-Ibanez S., Jones A.M.E., Rathjen J.P. (2010). Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 61: 507–518 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Kisseleva L., Sawa S., Furukawa T., Komatsu S., Koshiba T. (2004). A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 45: 550–559 [DOI] [PubMed] [Google Scholar]
- Hodel M.R., Corbett A.H., Hodel A.E. (2001). Dissection of a nuclear localization signal. J. Biol. Chem. 276: 1317–1325 [DOI] [PubMed] [Google Scholar]
- Hwang I.S., An S.H., Hwang B.K. (2011). Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 67: 749–762 [DOI] [PubMed] [Google Scholar]
- Hwang I.S., Hwang B.K. (2011). The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 155: 447–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E.H., Jung H.W., Lee S.C., Han S.W., Heu S., Hwang B.K. (2004). Identification of a novel pathogen-induced gene encoding a leucine-rich repeat (LRR) protein expressed in phloem cells of Capsicum annuum. Biochim. Biophys. Acta 1676: 211–222 [DOI] [PubMed] [Google Scholar]
- Jung H.W., Hwang B.K. (2007). The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol. Plant Pathol. 8: 503–514 [DOI] [PubMed] [Google Scholar]
- Jung Y.-H., Lee J.-H., Agrawal G.K., Rakwal R., Kim J.-A., Shim J.-K., Lee S.-K., Jeon J.-S., Koh H.-J., Lee Y.-H., Iwahashi H., Jwa N.-S. (2005). The rice (Oryza sativa) blast lesion mimic mutant, blm, may confer resistance to blast pathogens by triggering multiple defense-associated signaling pathways. Plant Physiol. Biochem. 43: 397–406 [DOI] [PubMed] [Google Scholar]
- Jung Y.H., et al. (2006). Differential expression of defense/stress-related marker proteins in leaves of a unique rice blast lesion mimic mutant (blm). J. Proteome Res. 5: 2586–2598 [DOI] [PubMed] [Google Scholar]
- Kim D.S., Hwang B.K. (2011). The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell-death responses. Plant J. 66: 642–655 [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Hwang B.K. (2000). Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant. 108: 51–60 [Google Scholar]
- Koistinen K.M., Soininen P., Venäläinen T.A., Häyrinen J., Laatikainen R., Peräkylä M., Tervahauta A.I., Kärenlampi S.O. (2005). Birch PR-10c interacts with several biologically important ligands. Phytochemistry 66: 2524–2533 [DOI] [PubMed] [Google Scholar]
- Lacomme C., Santa Cruz S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 96: 7956–7961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Choi H.W., Hwang B.K. (2011). The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol. 156: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Hwang B.K. (2003). Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta 216: 387–396 [DOI] [PubMed] [Google Scholar]
- Lee S.C., Hwang I.S., Choi H.W., Hwang B.K. (2008). Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance. Plant Physiol. 148: 1004–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tessaro M.J., Li X., Zhang Y. (2010). Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 153: 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.H., Park C.-J., Huh S.U., Choi L.M., Lee G.J., Kim Y.J., Paek K.-H. (2011). Capsicum annuum WRKYb transcription factor that binds to the CaPR-10 promoter functions as a positive regulator in innate immunity upon TMV infection. Biochem. Biophys. Res. Commun. 411: 613–619 [DOI] [PubMed] [Google Scholar]
- Liu J.J., Ekramoddoullah A.K.M. (2006). The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 68: 3–13 [Google Scholar]
- Liu Y.L., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Lo S.-C.C., Hipskind J.D., Nicholson R.L. (1999). cDNA cloning of a sorghum pathogenesis-related protein (PR-10) and differential expression of defense-related genes following inoculation with Cochliobolus heterostrophus or Colletotrichum sublineolum. Mol. Plant Microbe Interact. 12: 479–489 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Vailleau F., Balagué C., Roby D. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Lusso M., Kuc J. (1995). Increased activities of ribonuclease and protease after challenge in tobacco plants with induced systemic resistance. Physiol. Mol. Plant Pathol. 47: 419–428 [Google Scholar]
- Marković-Housley Z., Degano M., Lamba D., von Roepenack-Lahaye E., Clemens S., Susani M., Ferreira F., Scheiner O., Breiteneder H. (2003). Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J. Mol. Biol. 325: 123–133 [DOI] [PubMed] [Google Scholar]
- Martin K., Steinberg T.H., Cooley L.A., Gee K.R., Beechem J.M., Patton W.F. (2003). Quantitative analysis of protein phosphorylation status and protein kinase activity on microarrays using a novel fluorescent phosphorylation sensor dye. Proteomics 3: 1244–1255 [DOI] [PubMed] [Google Scholar]
- Matton D.P., Brisson N. (1989). Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol. Plant Microbe Interact. 2: 325–331 [DOI] [PubMed] [Google Scholar]
- McGee J.D., Hamer J.E., Hodges T.K. (2001). Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol. Plant Microbe Interact. 14: 877–886 [DOI] [PubMed] [Google Scholar]
- Melech-Bonfil S., Sessa G. (2010). Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 64: 379–391 [DOI] [PubMed] [Google Scholar]
- Michael Weaver L., Swiderski M.R., Li Y., Jones J.D.G. (2006). The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant J. 47: 829–840 [DOI] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen J.E., Wimmer R., Larsen J.N., Spangfort M.D., Otzen D.E. (2002). The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 277: 23684–23692 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., Anoop V., Gleddie S., Harris L.J. (2011). Proteomic profiling of two maize inbreds during early gibberella ear rot infection. Proteomics 11: 3675–3684 [DOI] [PubMed] [Google Scholar]
- Mosher S., Moeder W., Nishimura N., Jikumaru Y., Joo S.H., Urquhart W., Klessig D.F., Kim S.K., Nambara E., Yoshioka K. (2010). The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol. 152: 1901–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.J., Kim K.J., Shin R., Park J.M., Shin Y.C., Paek K.H. (2004). Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 37: 186–198 [DOI] [PubMed] [Google Scholar]
- Pontier D., Godiard L., Marco Y., Roby D. (1994). hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J. 5: 507–521 [DOI] [PubMed] [Google Scholar]
- Pühringer H., Moll D., Hoffmann-Spmmergruber K., Watillon B., Katinger H., Laimer da Camara Machado M. (2000). The promoter of apple Ypr 10 gene, encoding the major allerhen Mal d 1, is stress- and pathogen-inducible. Plant Sci. 152: 35–50 [Google Scholar]
- Sheen J., He P. (2007). Nuclear actions in innate immune signaling. Cell 128: 821–823 [DOI] [PubMed] [Google Scholar]
- Shen Q.-H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ülker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shen Q.-H., Schulze-Lefert P. (2007). Rumble in the nuclear jungle: Compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 26: 4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar P.D., Vasanthi N.S., Shetty H.S., Smedegaard-Petersen V. (2000). Ribonucleases in the seedlings of pearl millet and their involvement in resistance against downy mildew disease. Eur. J. Plant Pathol. 106: 825–836 [Google Scholar]
- Slootweg E., et al. (2010). Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich I.E., Schmelzer E., Bollmann J., Hahlbrock K. (1986). Rapid activation by fungal elicitor of genes encoding “pathogenesis-related” proteins in cultured parsley cells. Proc. Natl. Acad. Sci. USA 83: 2427–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Staehelin L.A., Driouich A. (1997). Brefeldin A effects in plants (Are different Golgi responses caused by different sites of action?). Plant Physiol. 114: 401–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling W.I.L., Nooijen C., Ludwig N., Boter M., Slootweg E., Goverse A., Shirasu K., Joosten M.H.A.J. (2010). RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Frederick R.D., Zhou J., Halterman D.A., Jia Y., Martin G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Walter M.H., Liu J.W., Grand C., Lamb C.J., Hess D. (1990). Bean pathogenesis-related (PR) proteins deduced from elicitor-induced transcripts are members of a ubiquitous new class of conserved PR proteins including pollen allergens. Mol. Gen. Genet. 222: 353–360 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Warner S.A.J., Scott R., Draper J. (1992). Characterisation of a wound-induced transcript from the monocot asparagus that shares similarity with a class of intracellular pathogenesis-related (PR) proteins. Plant Mol. Biol. 19: 555–561 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D.G., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Yen Y., Green P.J. (1991). Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiol. 97: 1487–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Dong S., Wang M., Wang W., Song W., Dou X., Zheng X., Zhang Z. (2010). The role of vacuolar processing enzyme (VPE) from Nicotiana benthamiana in the elicitor-triggered hypersensitive response and stomatal closure. J. Exp. Bot. 61: 3799–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.-J., Lu S., Xu Y.-H., Wang J.-W., Chen X.-Y. (2002). A cotton cDNA (GaPR-10) encoding a pathogenesis-related 10 protein with in vitro ribonuclease activity. Plant Sci. 162: 629–636 [Google Scholar]
- Ziadi S., Poupard P., Brisset M.-n., Paulin J.P., Simoneau P. (2001). Characterization in apple leaves of two subclasses of PR-10 transcripts inducible by acibenzolar-S-methyl, a functional analogue of salicylic acid. Physiol. Mol. Plant Pathol. 59: 33–43 [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.