This work shows that an E3 ubiquitin ligase interacts with, relocalizes, and negatively impacts the abundance of the symbiosis receptor kinase SYMRK. Our results implicate SINA E3 ligases in the turnover of SYMRK and suggest a clearance mechanism involving uptake from the plasma membrane.
Abstract
The Lotus japonicus SYMBIOSIS RECEPTOR-LIKE KINASE (SYMRK) is required for symbiotic signal transduction upon stimulation of root cells by microbial signaling molecules. Here, we identified members of the SEVEN IN ABSENTIA (SINA) E3 ubiquitin-ligase family as SYMRK interactors and confirmed their predicted ubiquitin-ligase activity. In Nicotiana benthamiana leaves, SYMRK–yellow fluorescent protein was localized at the plasma membrane, and interaction with SINAs, as determined by bimolecular fluorescence complementation, was observed in small punctae at the cytosolic interface of the plasma membrane. Moreover, fluorescence-tagged SINA4 partially colocalized with SYMRK and caused SYMRK relocalization as well as disappearance of SYMRK from the plasma membrane. Neither the localization nor the abundance of Nod-factor receptor1 was altered by the presence of SINA4. SINA4 was transcriptionally upregulated during root symbiosis, and rhizobia inoculated roots ectopically expressing SINA4 showed reduced SYMRK protein levels. In accordance with a negative regulatory role in symbiosis, infection thread development was impaired upon ectopic expression of SINA4. Our results implicate SINA4 E3 ubiquitin ligase in the turnover of SYMRK and provide a conceptual mechanism for its symbiosis-appropriate spatio-temporal containment.
INTRODUCTION
Prominent plant adaptations to nutrient limitation include alterations of root system architecture and symbiotic interactions of roots with bacteria and fungi (Den Herder et al., 2010). Many plant developmental programs, including symbiosis, are controlled by receptor-like kinases (RLKs) (De Smet et al., 2009; Albert et al., 2010). The outcome of RLK signaling not only depends on the availability and distribution of the ligand (Kwak and Schiefelbein, 2008), but also relies on the amount of (active) membrane-associated RLK. However, very little is known about posttranslational RLK regulation to mediate signaling activity.
Ubiquitin-mediated proteasomal protein degradation is an important step in the regulation of protein activities in eukaryotes (Hare et al., 2003; Moon et al., 2004; Smalle and Vierstra, 2004; Groothuis et al., 2006; Zeng et al., 2006). Although data for ubiquitination of RLKs are still limited, posttranslational modifications are likely to be involved in determining receptor fate and localization, for example, by promoting endocytosis, which controls receptor recycling and availability (Robatzek et al., 2006; Geldner et al., 2007), and also by targeting receptors for degradation to alleviate the signaling response (Le Roy and Wrana, 2005; Goff and Ramonell, 2007; Lu et al., 2011). In plants, E3 ligase–mediated regulation has been demonstrated for only few RLKs, such as FLAGELLIN-INSENSITIVE2 (FLS2) and CHITIN-ELICITOR RECEPTOR KINASE1, two receptor kinases that are ubiquitinated by the bacterial E3 ligase AvrPtoB, which overcomes plant defense responses by targeting critical pattern recognition receptors (Göhre et al., 2008; Gimenez-Ibanez et al., 2009). Also, plant E3 ligases, such as PUB12 and PUB13, directly ubiquitinate FLS2 (Lu et al., 2011). Conversely, RLKs might regulate E3 ligases via phosphorylation; for example, the rice (Oryza sativa) E3 ligase XB3 is a substrate of XA21 (Wang et al., 2006). Moreover, PUB12 and PUB13 are phosphorylated by BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase1, a step required for FLS2-mediated degradation (Lu et al., 2011).
The enzymatic reaction of E3 ligases involves the covalent linkage of ubiquitin molecules to a Lys residue of the targets or of ubiquitin itself (Passmore and Barford, 2004; Li et al., 2007). The energy-requiring step is the E1-mediated activation of ubiquitin. Subsequently, E2 ubiquitin-conjugating enzyme binds ubiquitin and delivers it to the substrate, via binding to the E3 ubiquitin ligase that specifically binds the target substrate. This ubiquitination results in the default consequence, namely, proteasomal degradation of the target via the 26S proteasome (Ciechanover, 1998; Hershko and Ciechanover, 1998; Bachmair et al., 2001). The E3 RING finger or HECT domain is responsible for binding to ubiquitinated E2 enzymes and is therefore present in all E3 ligases identified so far (Joazeiro and Weissman, 2000; Pickart, 2001).
The SEVEN IN ABSENTIA (SINA) family of E3 ligases was first identified in Drosophila melanogaster and later found in mammals and plants (Carthew and Rubin, 1990; Della et al., 1993). The amino acid sequence consists of a variable N-terminal region, a conserved C3HC4 RING domain, and a typical SINA domain (Hu and Fearon, 1999), which includes two Zn2+ finger motifs and a substrate binding and dimerization (SBD) domain (Depaux et al., 2006). Mammalian and vertebrate SINA proteins function in a broad range of processes, such as Drosophila photoreceptor development, and responses to hypoxia and stress (Franck et al., 2006; Cooper et al., 2008; House et al., 2009). Their identified substrates include mainly transcription factors and cytosolic proteins, but also membrane proteins (Kim et al., 2004). In the plant kingdom, very little is known about SINA E3 ligase mechanisms and functions. SINA dimers were shown to be the active form in plants (Xie et al., 2002), but in mammalian cells, SINA also participates in an Skp Cullin F-Box-like complex (Santelli et al., 2005). Plant SINA families comprise highly conserved proteins with six members in Arabidopsis thaliana and rice, and 10 in poplar (Populus trichocarpa; Den Herder et al., 2008; Wang et al., 2008). In Arabidopsis, SINAT5 regulates lateral root numbers via specific targeting of the NAC1 transcription factor (Xie et al., 2000, 2002), and SINAT2 interacts with the RAP2.2 transcription factor (Welsch et al., 2007).
Plant roots engage in advantageous mutualistic interactions with microorganisms. For instance, arbuscular mycorrhiza (AM) improves nutrient uptake via a fungal partner (Parniske, 2008; Bonfante and Genre, 2010), and this symbiosis is formed by the majority of land plants. Legumes and actinorhizal plants develop root nodule symbiosis with nitrogen-fixing bacteria. These are accommodated in a specialized organ, the root nodule, and provide nitrogen to the host plant (Oldroyd and Downie, 2008). Perception of a rhizobial signaling molecule, the Nod factor, via Nod Factor receptors in the epidermis (Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Mulder et al., 2006; Radutoiu et al., 2007; Madsen et al., 2010), initiates symbiotic signal transduction processes which result in nodule organogenesis and initiation of bacterial infection. Root hairs curl and entrap bacteria, thus initiating infection thread (IT) formation (Timmers, 2008). Via the IT, rhizobia are guided to the nodule cortex cells, into which they are released from ITs into plant membrane-enclosed compartments and differentiate into nitrogen-fixing bacteroids (Brewin, 2004; Oldroyd and Downie, 2008; Timmers, 2008).
Fine-tuning of symbiosis restricts microbial colonization to appropriate levels and avoids detrimental defense responses (Gibson et al., 2008). Involvement of E3 ligase–mediated posttranslational regulation in symbiotic signaling and development is indicated by the demonstrated role of anaphase-promoting complex (APC)CCS52A in endoreduplication (Vinardell et al., 2003), the isolation of nsRING, which is involved in rhizobial infection (Shimomura et al., 2006), the Lotus japonicus CERBERUS and orthologous Medicago truncatula LIN genes encoding E3 ligase U box proteins, which are required during early stages of IT formation (Kuppusamy et al., 2004; Kiss et al., 2009), and the E3 ligase PUB1, which interacts with the symbiotic LysM RLK LYK3, which is crucial for the initiation of nodule symbiosis. PUB1 is an active E3 ligase in vitro and can be phosphorylated by LYK3 in vitro, but a regulatory effect on the receptor was not observed (Mbengue et al., 2010). In M. truncatula, introduction of the Arabidopsis dominant-negative SINAT5DN form resulted in a symbiotic infection phenotype (Den Herder et al., 2008).
The L. japonicus SYMBIOSIS RECEPTOR-LIKE KINASE (SYMRK) has an intracellular kinase domain (KD) that is active in its autophosphorylated form and is required for root symbiosis (Yoshida and Parniske, 2005). SYMRK is a common SYMBIOSIS (SYM) gene, required to initiate both AM (Endre et al., 2002; Stracke et al., 2002; Demchenko et al., 2004) and nodulation on legumes or the actinorhiza plants Casuarina glauca and Datisca glomerata (Gherbi et al., 2008; Markmann et al., 2008). During nodulation, a role in initiation of infection, as well as in rhizobial internalization and symbiosome formation, was proposed based on RNA interference results obtained with Sesbania rostrata SYMRK or M. truncatula DMI2 (Capoen et al., 2005; Limpens et al., 2005),
Proteins that interact with SYMRK or the Medicago sativa ortholog NODULATION RECEPTOR KINASE have been described. These include 3-Hydroxy-3-Methylglutaryl CoA Reductase1 (Kevei et al., 2007), SYMRK INTERACTING PROTEIN1, which binds the NODULE INCEPTION (NIN) promoter (Zhu et al., 2008), and a remorin protein (Lefebvre et al., 2010). Here, we identified SINA E3 ubiquitin ligases as SYMRK interactors. Since SINA4 overexpression specifically impaired SYMRK protein stability and proper rhizobial infection, we postulate that SINA4 is a negative regulator of SYMRK.
RESULTS
L. japonicus SYMRK Kinase Interacts with SINA E3 Ligases
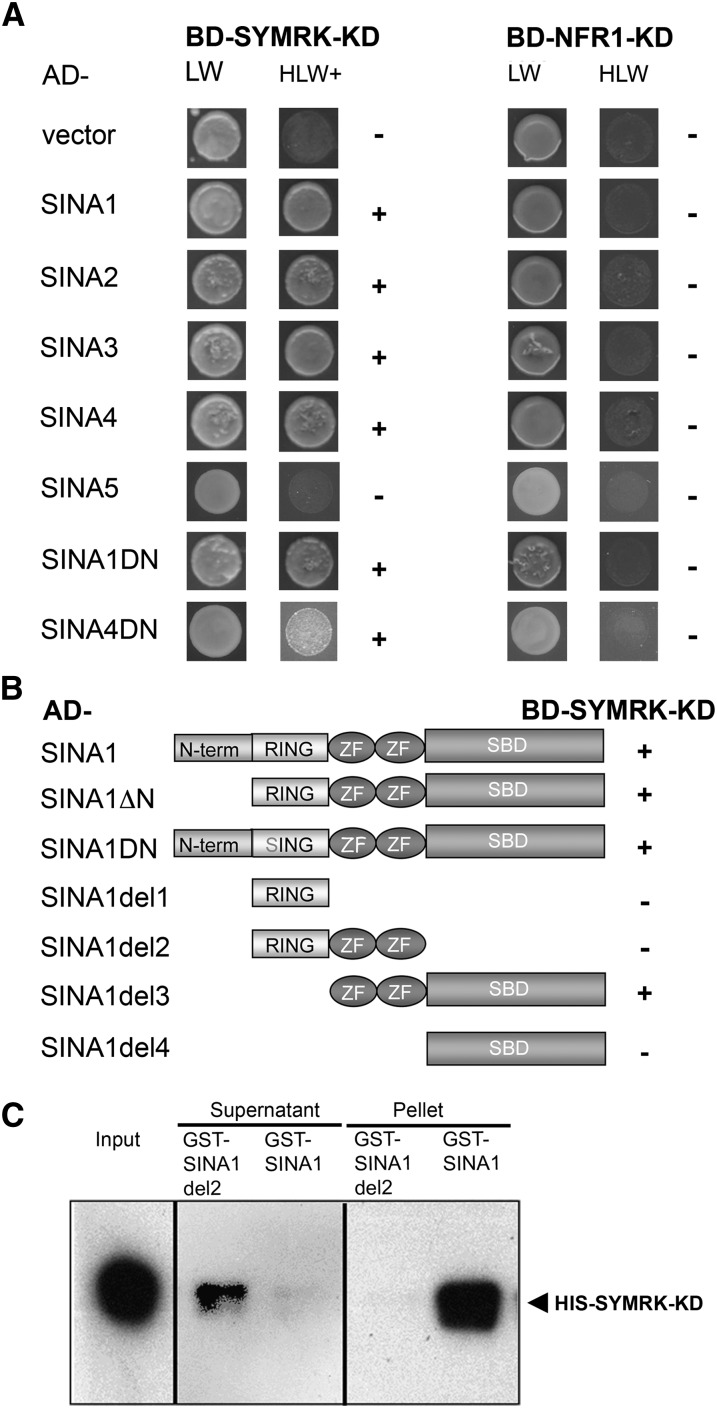
To identify SYMRK-interacting proteins, the SYMRK-KD was used for a Gal4 yeast two-hybrid (Y2H) screen with an L. japonicus cDNA library derived from RNA of roots inoculated with Mesorhizobium loti (Poulsen and Pødenphant, 2002). Six independent clones coding for proteins with sequence similarity to SINA E3 ligases were isolated: SINA1 (two clones), SINA2 (two clones), SINA3 (one clone), and SINA4 (one clone) (Figure 1A). All cDNA clones encoded 5′-proximal truncated versions of the SINA coding regions (see Supplemental Figure 1A online). Although clones for SINA5, a fifth member of the family, were not isolated from the Y2H screen, SINA5 transcripts could be detected in the Y2H cDNA library by PCR. The full coding sequences for SINA1 to 5 were isolated from L. japonicus cDNA. By contrast, SINA6, which shares 94% nucleotide sequence identity with SINA1, has been identified in the L. japonicus genome database (Sato et al., 2007) but could not be isolated from L. japonicus cDNA or from the Y2H library as a transcript. The L. japonicus SINA protein family members are highly conserved with an average 62 to 85% amino acid sequence identity, including conserved Cys and His residues in the RING and Zn2+ finger, required for C3HC4 RING E3 ligase activity and a substantially diverged N-terminal region (see Supplemental Figure 1 and Supplemental Data File 1 online).
Figure 1.
SYMRK-KD Interacts with L. japonicus SINA E3 Ligases in Yeast and in Vitro.
(A) Y2H assay with L. japonicus SINA E3 ligases or dominant-negative (DN) versions of SINA1 and SINA4 fused to the Gal4 activation domain (AD) and the KDs of SYMRK or NFR1 fused to the Gal4 binding domain (BD). Coexpression of BD-SYMRK-KD and AD-SINA1/2/3/4 or AD-SINA1DN/4DN resulted in yeast growth on triple selection medium (HLW+). No growth was detected for BD-SYMRK-KD in combination with AD-SINA5 or coexpression of BD-NFR1-KD with any of the five SINAs.
(B) Y2H assays with (truncated) AD-SINA1 versions and BD-SYMRK-KD. Interaction could only be observed between BD-SYMRK-KD and AD-SINA1 versions containing the SINA domain (zinc finger + SBD).
(C) In vitro pull-down assay with glutathione-coated beads on reactions containing His-tagged SYMRK-KD (His-SYMRK-KD) and (truncated) GST-tagged SINA1 (GST-SINA1/SINA1del2) versions. Reactions were subjected to immunoblot analysis using an antibody against the His-tag (αHis). His-SYMRK-KD could only be coimmunoprecipitated with GST-SINA1.
LW, Leu/Trp-deficient medium; HLW, Leu/Trp/His-deficient medium; HLW+, HLW medium with 3-aminotriazole (5 mM); + or −, yeast growth observed or not; N, variable N-terminal domain; RING, E3 activity domain; SING, inactive RING mutated in the conserved Cys-47 to Ser; ZF, zinc finger motif.
The interaction between SYMRK-KD and SINA1, 2, 3, and 4 was confirmed in yeast, but SINA5 did not interact with SYMRK-KD in yeast (Figure 1A; see Supplemental Table 1 online). To analyze the specificity of the interaction between SINAs and the SYMRK kinase, we tested the KD of the Nod Factor receptors NFR1 and NFR5 (Madsen et al., 2003; Radutoiu et al., 2003) and of a close Lotus homolog of the Arabidopsis FLS2 receptor (FLS2-like KD). Expression of the NFR1 KD in yeast was confirmed (see Supplemental Figure 2A online), but no interaction was observed with any of these RLK domains in combination with SINA1, 2, 3, 4, or 5 (Figure 1A; see Supplemental Table 1 online).
To map the SINA1 domain(s) relevant for the interaction with SYMRK-KD, we analyzed deletion constructs of SINA1 (Figure 1B). As described earlier for homologous proteins from other species (Hu and Fearon, 1999), the SINA domain, consisting of the Zn2+ finger motifs and SBD, was required for interaction. However, the variable N-terminal sequence and the RING domain were not required for interaction (Figure 1B; see Supplemental Figure 1A online). Three out of the six SINA family members, namely, SINA2, SINA3, and SINA5, lacked a C-terminal extension of 14 amino acids (see Supplemental Figure 1A online, red boxed area). Since SINA3 interacted strongly with SYMRK (Figure 1), this extension does not seem critical for the interaction. The functionality of the SYMRK interaction domain of SINAs was confirmed in an in vitro binding assay demonstrating direct interaction between SINA1 and SYMRK-KD (Figure 1C). For this assay, glutathione S-transferase (GST)-SINA1 was pulled down with glutathione beads and immunoblot analysis with α-His antibody was used to detect HIS-SYMRK-KD. HIS-SYMRK-KD was detected in the pellet fraction when GST-SINA1 was used as bait, but not when SINA1 lacking the SBD domain (GST-SINA1-del2) was used as bait (Figures 1B and 1C).
As shown before for plant and mammalian SINAs, the formation of homo- and heterodimers is required for E3 ligase activity (Den Herder et al., 2008; Hu and Fearon, 1999; Xie et al., 2002). Therefore, we tested whether dimerization also occurred with L. japonicus SINAs and observed homo- and heterodimers in several combinations in yeast (see Supplemental Table 1 online) and in planta using bimolecular fluorescence complementation (BiFC; Kerppola, 2008; see Supplemental Table 2 and Supplemental Figure 3A online). We found that four L. japonicus SINA E3 ligase proteins dimerize and specifically interact with the SYMRK-KD in yeast. SINA dimerization was consistently observed in plant cells as small dots in the cytoplasm in close vicinity of the plasma membrane (see Supplemental Figure 3A online).
In Vitro E3 Ligase SINA Activity and Dominant-Negative Inhibition
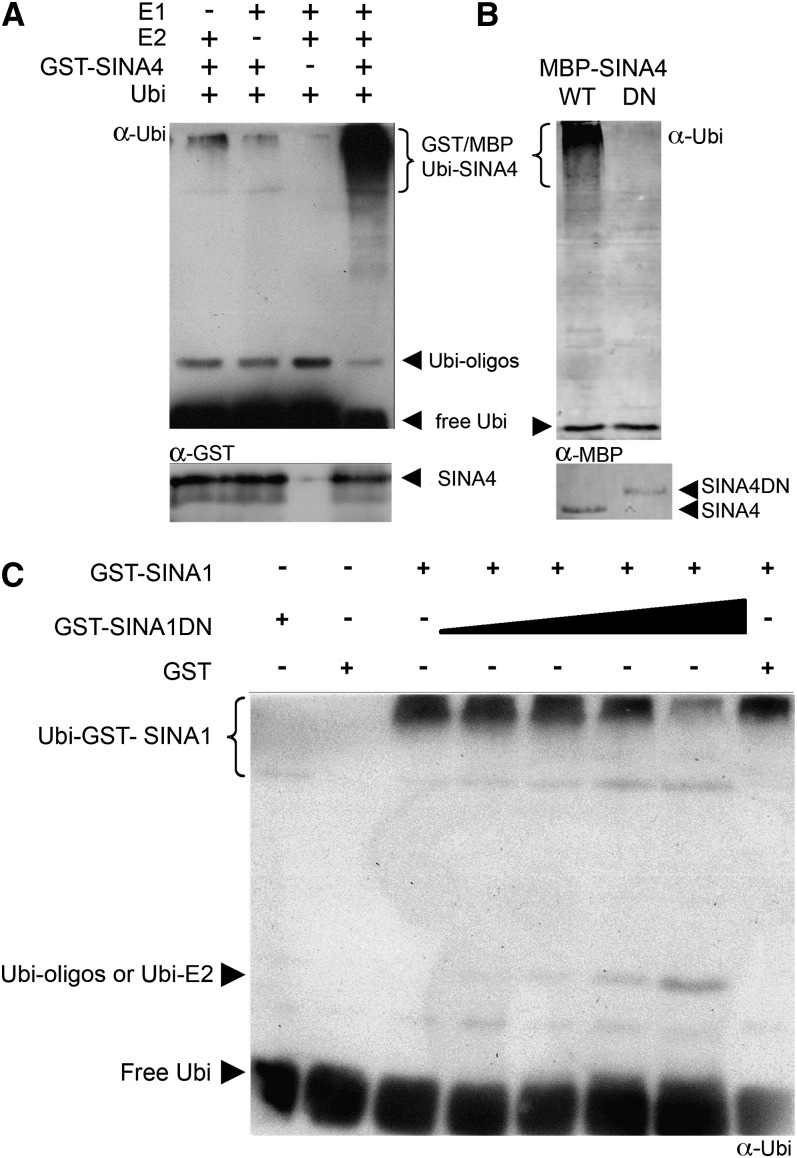
The predicted function of SINAs was analyzed by testing their enzymatic E3 ligase activity via in vitro ubiquitination assays (Figure 2). We confirmed that SINA4 is an active E3 ligase since incubation of SINA4 with E1 ubiquitin-activating and E2 ubiquitin-conjugating enzymes, ubiquitin, and ATP resulted in polyubiquitination in vitro (Figure 2A). Similar in vitro polyubiquitination activity was observed for SINA1 and SINA3 (see Supplemental Figure 4 online). By contrast, SINA2 did not show any in vitro activity (see Supplemental Figure 4 online).
Figure 2.
Dominant-Negative Inhibition of SINA in Vitro E3 Ligase Activity.
(A) In vitro ubiquitination assay with GST-tagged SINA4 (GST-SINA4) in the presence (+) or absence (−) of yeast E1 ubiquitin-activating and human UBC4b E2 ubiquitin-conjugating enzymes, ubiquitin and ATP. Reactions were analyzed for ubiquitination and the presence of the recombinant proteins by immunoblots using antibodies against ubiquitin (α-Ubi; Affiniti) and the GST tag (α-GST). Multiple bands of high molecular weight indicated polyubiquitination activity of GST-SINA4 in the presence of E1 and E2 enzymes, ubiquitin, and ATP.
(B) In vitro ubiquitination assay with MBP-tagged SINA4 (MBP-SINA4WT) and the dominant-negative (DN) SINA4 (MBP-SINA4DN). Reactions were analyzed for ubiquitination and the presence of the recombinant proteins by immunoblot using antibodies against ubiquitin (α-Ubi; Santa Cruz) and the MBP tag (α-MBP). Multiple bands of high molecular weight indicated polyubiquitination activity of MBP-SINA4 but not of the inactive MBP-SINA4DN.
(C) In vitro ubiquitination activity of GST-tagged SINA1 (GST-SINA1) and dominant-negative (DN) SINA1 (GST-SINA1DN) and the effect of GST-SINA1DN on GST-SINA1 polyubiquitination activity. Reactions were analyzed for ubiquitination and the presence of the recombinant proteins by immunoblots using antibodies against ubiquitin (α-Ubi; Affiniti) and the GST tag (α-GST). Multiple bands of high molecular weight indicated polyubiquitination activity of GST-SINA1 but not of GST-SINA1DN. The presence of inactive GST-SINA1DN inhibited GST-SINA1 polyubiquitination activity in vitro.
Active SINA E3 ligases function as dimers to ubiquitinate their targets, or themselves, resulting in polyubiquitinated proteins, which are subsequently degraded in the cytosol (Joazeiro and Weissman, 2000). The predicted regulatory function of the SINA E3 ligases was further confirmed through the generation of stable inactive dominant-negative SINA1 and SINA4 mutant forms (SINA1DN and SINA4DN), in which a conserved Cys is substituted with a Ser in the RING domain (C47S or C66S, respectively; see Supplemental Figure 1A online), thereby abolishing E3 activity, but retaining dimerization and substrate binding capacity (see Supplemental Table 1 online; Xie et al., 2002). The loss of E3 ligase activity of both proteins, SINA4DN (Figure 2B) and SINA1DN (Figure 2C), was confirmed in vitro; increasing amounts of SINA1DN caused a decrease in the amount of polyubiquitinated SINA1 (Figure 2C). These dominant-negative mutant forms were also found to interact with SYMRK-KD, but not with NFR1-KD in yeast (Figure 1A), thus retaining the binding specificity of the wild-type proteins.
SINA Subcellular Localization
We examined the subcellular localization of SINA1 and 4 by transient expression of SINA–fluorescent protein fusions. SINA4:YFP (for yellow fluorescent protein) expressed under the control of the SINA4 promoter in L. japonicus roots resulted in a very weak fluorescent signal, which is probably a reflection of both the weak promoter activity of SINA4 (see below and Supplemental Figure 8 online), and the autocatalytic turnover of SINAs (Figure 2; see Supplemental Figure 3C online). The latter problem could be overcome using the proteasomal inhibitor MG132 (see Supplemental Figures 3C and 3H and Supplemental Table 2 online) or by expression of the inactive SINA4DN:YFP. In L. japonicus roots, a high background fluorescence overlapping the emission spectrum of the YFP fluorescence made it difficult to distinguish the real YFP signal. However, in both, Nicotiana benthamiana leaf epidermal cells and L. japonicus root protoplasts, the SINA1 and SINA4 protein fusions with green fluorescent protein (GFP) and cyan fluorescent protein (CFP), respectively, upon addition of MG132, as well as their dominant-negative counterparts, were consistently localized in the cytosol and/or cytosolic dots of variable size (see Supplemental Figure 3, and Supplemental Table 3, and Supplemental Methods 1 online).
In Planta Interaction between SINAs and SYMRK
In planta interaction between full-length SYMRK and SINA1DN or SINA4DN was tested via BiFC in N. benthamiana. SYMRK-mOrange expressed on its own under the control of the 35S promoter localized to the plasma membrane (Figures 3C and 3D; see Supplemental Figure 5 online). However, upon coexpression of SINA4DN:YFPC and SYMRK:YFPN, the YFP fluorescence, indicative of interaction, localized predominantly, if not always, in dots at the cytosolic interface with the plasma membrane (see Supplemental Figure 2C online; Figure 3E). The YFP signal for BiFC between SINA-YFPN and SINA-YFPC, indicative of self-interaction, was very similar (cf. Figure 3E with Supplemental Figure 3A online). Hence, both interaction signals resemble the localization of the SINA proteins when expressed on their own (see Supplemental Figures 3A to 3C online; Figures 3A, 3B, and 3E).
Figure 3.
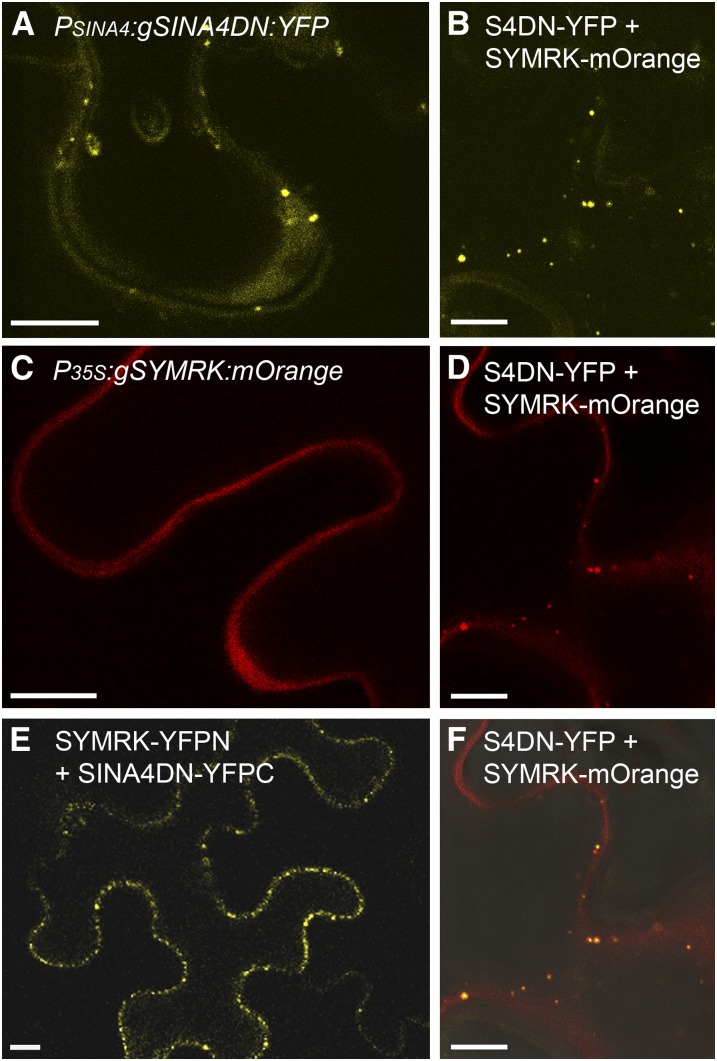
SINA4DN Interacts with, Colocalizes with, and Relocalizes SYMRK.
Confocal laser scanning microscopy images of N. benthamiana epidermal leaf cells 4 DAI with Agrobacterium. Bars = 10 μm.
(A) Expression of ProSINA4:gSINA4DN:YFP, a genomic SINA4 construct converted to the dominant-negative (DN) mutant allele (PSINA4:gSINA4DN:YFP with protein S4DN-YFP). Bright fluorescent dots moving through the cytoplasm were observed.
(C) Expression of Pro35S:gSYMRK:mOrange, a genomic SYMRK construct (P35S:gSYMRK:mOrange with protein SYMRK-mOrange). SYMRK-mOrange fluorescence signal was detected at the plasma membrane.
(B) and (D) Coexpression of ProSINA4:gSINA4DN:YFP (protein S4DN-YFP) and Pro35S:gSYMRK:mOrange (protein SYMRK-mOrange). SYMRK-mOrange partially relocalized to cytosolic aggregating dots (D), which partially colocalized (orange-colored dots) with the S4DN-YFP signal ([B] and [F]).
(F) Merged image showing the colocalization of SYMRK-mOrange and S4DN-YFP signals in (B) and (D).
(E) Coexpression of the BiFC constructs Pro35S:SYMRK:YFPN (SYMRK-YFPN) and Pro35S:SINA4DN:YFPC (S4DN-YFPC). Reconstitution of functional YFP resulted in bright dots along the cytosolic interface with the plasma membrane.
The intensity of the fluorescent signal and the estimated percentage of cells showing fluorescence were taken as an indication of the strength of the interaction (see Supplemental Figure 2C and Supplemental Table 2 online). The BiFC signal observed for SINA1DN:YFPC and SYMRK:YFPN coexpression was rather weak (10 to 30% of total visible epidermal cells [TVECs] showed fluorescent signal), but the signal observed for SINA4DN:YFPC and SYMRK:YFPN coexpression was much brighter and more abundant (31 to 70% of TVEC) (see Supplemental Figure 2C and Supplemental Table 2 online), indicating a stronger interaction between SYMRK and SINA4 in planta. In addition, the interaction of SYMRK with SINA2 and SINA3 resulted in a bright BiFC signal for the inactive SINA2 (31 to 70% of TVEC) and a positive but very low and infrequent YFP signal for SINA3 (10 to 30% of TVEC) (see Supplemental Table 2 online).
SINA4 Relocalizes SYMRK in Planta
We tested whether SINA4 can alter SYMRK localization by expressing fluorescent tagged versions of the two proteins separately and together in N. benthamiana. Transient expression of the genomic construct ProSINA4:SINA4:YFP (followed by MG132 proteasomal inhibitor treatment) and its dominant-negative form ProSINA4:SINA4DN:YFP in N. benthamiana leaves resulted in easily detectable cytosolic localization (Figure 3; see Supplemental Figures 3B and 3C online). Confocal imaging on N. benthamiana epidermal cells expressing the ProSINA4:SINA4DN:YFP- or ProSINA4:SINA4:YFP-derived fusion protein showed bright mobile dots in the cytosol (similar to 35S-driven expression of SINA4; Figures 3A and 3B; see Supplemental Figure 3B online).
Transient coexpression of ProSINA4:SINA4DN:YFP and Pro35S:SYMRK:mOrange resulted in bright cytosolic dots for the SINA4DN-YFP signal as typically observed also when this construct was expressed on its own. However, the otherwise exclusive SYMRK-mOrange localization at the plasma membrane was altered (cf. Figure 3C with Figures 3B, 3D, and 3F) and now partially colocalized with the SINA4DN-YFP dots (Figure 3F). This SINA4-mediated relocalization of SYMRK protein, in combination with the Y2H and BiFC data, strongly supports the hypothesis of a physical interaction between SINA4 and SYMRK in planta.
SINA4 Specifically Interacts with SYMRK Kinase
To determine the specificity of the SINA/SYMRK interaction, we tested the SINA interaction with NFR1. The expression of the SINA-HA-YFPC proteins, as well as the SYMRK-YFPN and NFR1-YFPN proteins, was confirmed via immunoblot analysis (see Supplemental Figure 2B online). However, no interaction was observed for NFR1 with SINAs by BiFC, confirming the Y2H data and validating the positive and specific BiFC interaction of SYMRK with the SINAs (see Supplemental Figure 2C online).
The requirement of an active or specifically phosphorylated kinase for its interaction with E3 ligases and subsequent vice versa ubiquitination has been demonstrated in animals and plants (Göhre et al., 2008; Lovly et al., 2008). We tested whether the kinase activity of SYMRK influenced SINA interaction using the SYMRKK622E inactive mutant form (Yoshida and Parniske, 2005). The Y2H interaction studies using SYMRK-KD or SYMRKK622E-KD showed indistinguishable growth when coexpressed with SINA1, 3, and 4. The interaction of SYMRK-KD with SINA2 was already weaker on quadruple selection media compared with other SINAs, and the interaction of SINA2 with SYMRKK622E-KD could not be detected in the Y2H assays (see Supplemental Table 1 online). However, in N. benthamiana, both SYMRK and SYMRKK622E full-length protein coexpression resulted in a similar BiFC fluorescence pattern and strength with SINA1DN, SINA2, and SINA4DN (see Supplemental Table 2 online). Thus, kinase activity may quantitatively contribute to, but is not essential for, interaction between the SYMRK-KD and SINAs.
SINA4 Specifically Affects SYMRK Stability in N. benthamiana
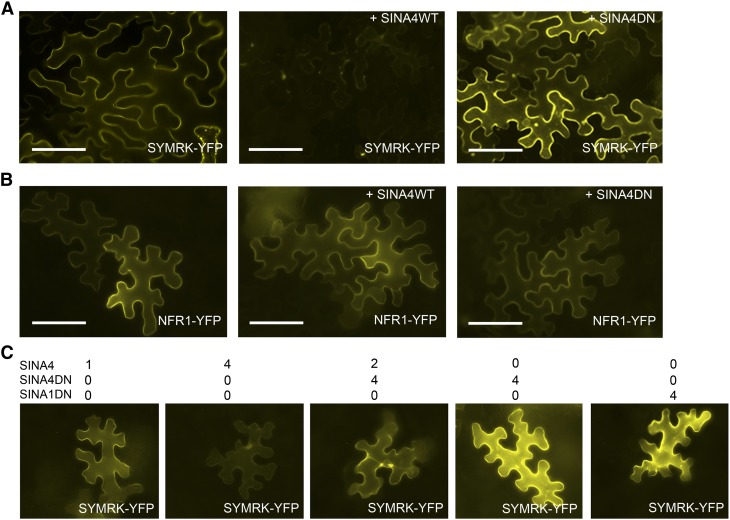
E3 ligases can potentially modulate RLK localization and stability (De Smet et al., 2009; Gimenez-Ibanez et al., 2009; Lu et al., 2011). The induced relocalization of SYMRK by SINA4 might lead to subsequent protein degradation. To test the effect of SINA4 on SYMRK protein stability, we performed coexpression experiments in N. benthamiana epidermal leaf cells. We found that transient coexpression of Pro35S:gSYMRK:YFP and Pro35S:SINA4 reproducibly led to fainter YFP signal, indicating decreased levels of SYMRK-YFP compared with expression of Pro35S:gSYMRK:YFP alone (Figure 4A). By contrast, Pro35S:SINA4DN or Pro35S:SINA1DN coexpressed with Pro35S:gSYMRK:YFP resulted in a bright fluorescent signal, indicating accumulation of SYMRK-YFP protein. We hypothesize that dominant-negative SINAs stabilized SYMRK by binding to it, thus protecting it from endogenous N. benthamiana SINA homologs (Figures 4A and 4C; see Supplemental Figures 6A, 6C, and 6D online).
Figure 4.
SINA4 Coexpression Influences SYMRK but Not NFR1 Protein Abundance and Subcellular Localization.
Epifluorescence microscopy images of N. benthamiana epidermal leaf cells 4 DAI with Agrobacterium. Bars = 100 μm.
(A) Coexpression of Pro35S:gSYMRK:YFP (SYMRK-YFP) and Pro35S:SINA4WT (SINA4WT) or Pro35S:SINA4DN (SINA4DN). The presence of SINA4WT decreased the SYMRK-YFP fluorescence signal, whereas the presence of SINA4DN increased the SYMRK-YFP fluorescence signal.
(B) Coexpression of Pro35S:NFR1:YFP (NFR1-YFP) and Pro35S:SINA4WT (SINA4WT) or Pro35S:SINA4DN (SINA4DN). No SINA4WT- or SINA4DN-dependent changes in the strength of the NFR1-YFP fluorescence signal or the localization of NFR1-YFP were observed.
(C) Coexpression of Pro35S:gSYMRK:YFP (SYMRK-YFP) and Pro35S:SINA4WT (SINA4WT), Pro35S:SINA4DN (SINA4DN), or Pro35S:SINA1DN (SINA1DN). SINA4 destabilized and relocalized SYMRK-YFP in a dosage-dependent manner, whereas SINA4DN or SINA1DN stabilized SYMRK and SINA4DN inhibited the effect of SINA4. Numbers indicate the fold excess of suspension volume of Agrobacteria carrying SINA constructs relative to that of Agrobacteria carrying SYMRK used to infiltrate the leaves.
[See online article for color version of this figure.]
Immunoblot analysis using a SYMRK-specific antibody raised against the extracytoplasmic part of SYMRK (SYMRK-EXT) demonstrated that the amount of SYMRK full-length protein decreased or increased in the presence of SINA4 or SINA4DN, respectively (see Supplemental Figure 6A online). These results indicate that SINA4 targets SYMRK-YFP for degradation. Consistent with the intracellular localization of SINA4, we observed that full length SYMRK protein as well as a truncated cytosolic SYMRK-YFP (tSYMRK-YFP) fragment that was consistently detectable upon expression of the full-length SYMRK-YFP in N. benthamiana, were reduced or increased upon coexpression of SINA4 or SINA4DN, respectively (see Supplemental Figure 6C online). On the other hand, the amount of a SYMRK fragment specifically recognized by the α-SYMRK-EXT antibody (see Supplemental Figure 6A online, fSYMRK) was similar when expressing SYMRK:YFP alone or together with SINA4, indicating that expression of SYMRK:YFP as such was not impaired by the SINA cotransformation (see Supplemental Figure 6A online, cf. lanes 1 and 3).
To test the specificity of SINA4’s interaction and destabilization activity, we coexpressed Pro35S:SINA4DN with Pro35S:NFR1:YFP in N. benthamiana. Plasma membrane–localized NFR1-YFP protein was not obviously affected upon coexpression, demonstrating the target specificity of SINA4 (Figure 4B; see Supplemental Figure 7B online). Finally, we immunoprecipitated SYMRK-YFP from N. benthamiana crude extract, and starting from equal amounts of material expressing SYMRK-YFP, we obtained a different quantity of SYMRK-YFP elution products upon coexpression with different SINAs. The active SINA4 resulted in decreasing SYMRK-YFP amounts, while coexpression of active SINA1 or SINA2 did not decrease SYMRK-YFP amounts (see Supplemental Figure 6D online). Taken together, our data provide strong evidence for a specific and negative effect of SINA4 on SYMRK protein stability.
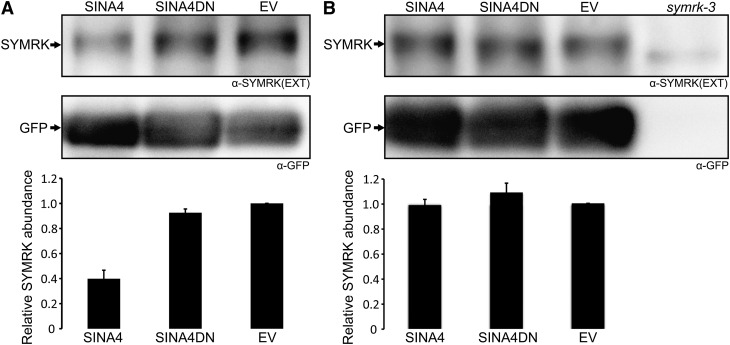
SINA4 Affects SYMRK Protein Stability in L. japonicus Nodulated Roots
To investigate the influence of SINA4 on SYMRK levels during nodulation, we expressed SINA4 or SINA4DN under the control of the lotus ubiquitin promoter (Maekawa et al., 2008) in L. japonicus hairy roots. Expression of SYMRK was analyzed via immunoblot using α-SYMRK-EXT antibody (Figure 5). Upon inoculation with the compatible rhizobial strain M. loti MAFF303099 DsRed, transgenic roots expressing ProUbi:SINA4 contained less SYMRK full-length protein than control roots transformed with the empty vector, whereas noninoculated ProUbi:SINA4 expressing transgenic lotus roots did not show a reduced level of SYMRK protein (Figure 5). From these data we concluded that ectopic expression of SINA4 negatively influenced SYMRK levels during nodulation. No significant increase of SYMRK was observed in SINA4DN-expressing roots (Figure 5), suggesting that additional factors limit SYMRK levels in the root.
Figure 5.
Ectopic Expression of SINA4 Affects SYMRK Abundance in L. japonicus Transgenic Nodulated Roots.
SINA4 variants were expressed in L. japonicus Gifu wild-type hairy roots under the control of the ubiquitin promoter using the vectors ProUbi:SINA4 (SINA4), ProUbi:SINA4DN (SINA4DN), or the empty vector control (EV). At 14 DAI with M. loti (A) or mock treatment (B), transgenic roots were harvested and extracts analyzed by immunoblots for the presence of SYMRK [using α-SYMRK(EXT) antibody, top panels]. The signal from the GFP transformation marker (using α-GFP antibody, bottom panels) was used as internal standard for the quantification of relative abundance of SYMRK. Graphs represent mean values ± sd of two technical replicates. Lane symrk-3, L. japonicus symrk-3 roots (symrk null mutant; negative control).
(A) The relative abundance of SYMRK was reduced in nodulated roots expressing SINA4 compared with the empty vector control. SINA4DN expression did not change SYMRK abundance in nodulated roots.
(B) The relative abundance of SYMRK was not affected in non-nodulated roots expressing either SINA4 or SINA4DN compared the empty vector control.
SINA4 and SYMRK Expression Patterns in Lotus Roots
The transcript abundance of SINA1, 2, 3, and 4 was determined by quantitative RT-PCR on RNA from L. japonicus root tissue inoculated with M. loti rhizobia or an AM fungus. Marker genes for both symbiotic interactions were used as controls for induction. Upon inoculation, NIN (Schauser et al., 1999) and PHOSPHATE TRANSPORTER4 (PT4) (Javot et al., 2007) were induced up to 700- and 7000-fold, respectively (see Supplemental Figures 7A and 7C online). For all four SINA genes, transcripts were detected in noninoculated roots and in nodulated tissue. Also, SINA4 expression increased, up to threefold at 10 d after rhizobial inoculation compared with noninoculated roots, and weak induction could be detected for SINA1 and SINA3 (see Supplemental Figure 7 online).
We also analyzed promoter activity using promoter-β-glucuronidase (GUS) fusions, testing ProSYMRK:GUS and ProSINA4:GUS activity in transgenic hairy roots of L. japonicus. In noninoculated roots, expression of the ProSINA4:GUS construct resulted in weak blue staining in the root zone susceptible for rhizobial infection and more clear staining in the root tip, as well as at lateral root initiation sites (see Supplemental Figures 8A and 8B online). SYMRK promoter activity in noninoculated roots was most pronounced in the epidermis, including root hair cells (see Supplemental Figures 8E and 8F online), while no staining was observed at the root tip or at lateral root initiation sites (data not shown).
After rhizobial inoculation, blue staining indicative of SINA4 promoter activity was observed in the vicinity of M. loti fluorescence and in the vicinity of nodule primordia (see Supplemental Figure 8C online). Also, the SYMRK promoter showed activity in nodule primordia (see Supplemental Figure 8G online), which is consistent with earlier observations in M. truncatula (Bersoult et al., 2005). During further development of the nodules, SINA4 promoter activity became weaker at the nodule base (see Supplemental Figure 8D online) and finally disappeared in fully matured nodules (data not shown), which was also the case for SYMRK promoter activity (see Supplemental Figure 8H online).
In summary, SYMRK and SINA4 have distinct but partially overlapping (such as root hairs and the nodule meristems/primordia) expression patterns during rhizobial infection and early nodule development. Interaction between the two proteins at these locations could affect SYMRK abundance, thereby modulating the tissue susceptibility to symbiotic signaling.
Ectopic Expression of SINA4 in L. japonicus Results in Impaired Rhizobial Infection of Root Hairs and Nodule Cortical Cells
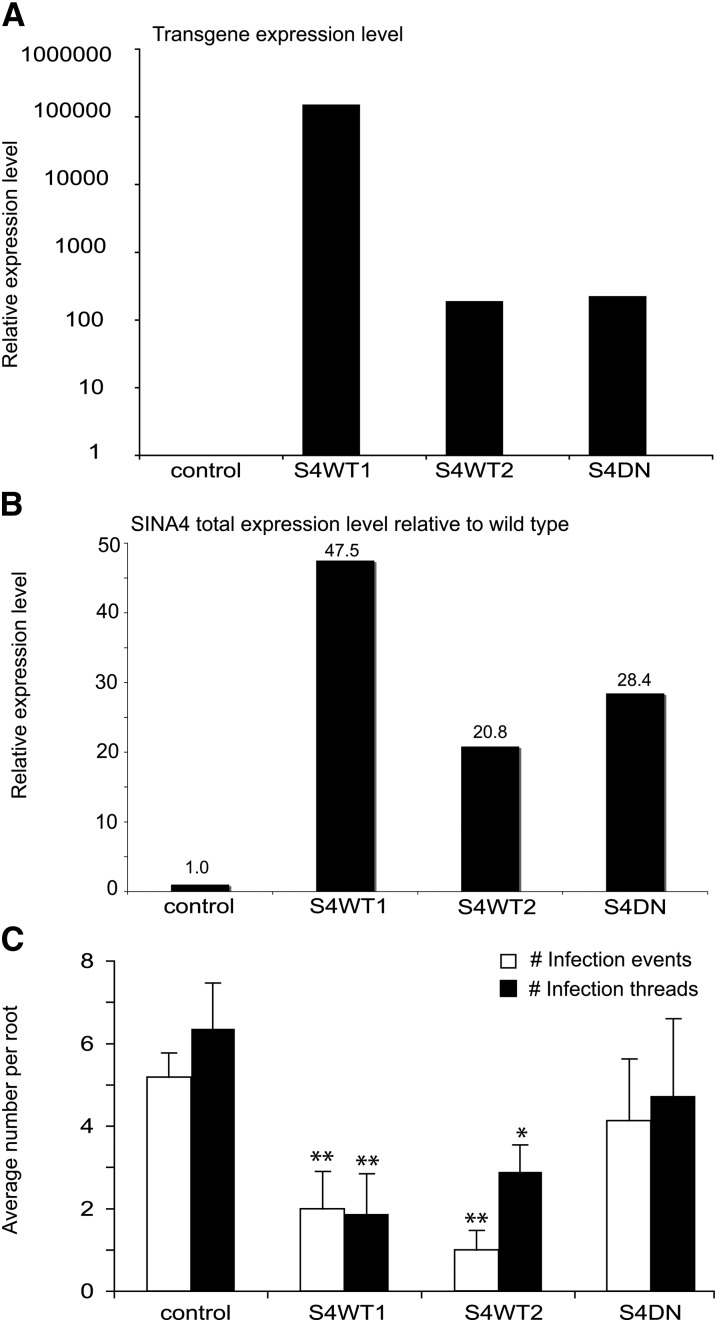
Our results provide strong evidence that SINA4, but not SINA1, negatively regulates SYMRK abundance. We therefore hypothesized that SINA4 should specifically exert a negative effect on nodulation. To test this, we generated stable transgenic L. japonicus plants expressing wild-type or dominant-negative (DN) versions of SINA1 or SINA4. Lines with strong transgene expression were chosen for phenotypic analysis (Figures 6A and 6B). Plants transformed with the pK7WG2D empty vector were used as controls.
Figure 6.
Rhizobial Infection Is Impaired in L. japonicus Transgenics Ectopically Expressing SINA4.
(A) Relative transgenic SINA4 transcript level in L. japonicus stable transgenic lines expressing Pro35S:SINA4WT (S4WT1 and S4WT2) and Pro35S:SINA4DN (S4DN). Quantitative RT-PCR was performed on cDNA of root RNA samples. All lines showed a high transgene transcript accumulation; by contrast, no transgene amplification was detected in the controls which was set as 1 for a Ct value of >40.
(B) Total SINA4 transcript level in L. japonicus stable transgenic lines S4WT1, S4WT2, and S4DN relative to wild-type plants. Quantitative RT-PCR was performed on cDNA of root RNA samples. The numbers indicate the fold overexpression of SINA4.
(C) Effect of ectopic expression of SINA4 variants on rhizobial infection. Plants were grown in plastic containers, and the average numbers of rhizobial infection events (curled root hairs with entrapped bacterial colony, white bars) and infection threads (black bars) were quantified 7 DAI with M. loti. The average numbers of infection events and infection threads were significantly reduced in both Pro35S:SINA4WT lines compared with the control lines (transgenic MG20 plants transformed with the empty vector construct) or Pro35S:SINA4DN lines. Error bars indicate se over a population of n = 10 to 15; *P < 0.05; **P < 0.005 compared with controls, as determined by a two-tailed Student’s t test.
All transgenic lines ectopically expressing SINAs did not display any obvious nonsymbiotic or developmental phenotypes. Consistent with the wild type–like overall growth performance of the transgenic lines, the primary root length was similar for all lines in all conditions tested compared with the empty vector control (see Supplemental Figure 9D online).
Plants of two independent transgenic lines, Pro35S:SINA4WT1 and 2, were significantly impaired during rhizobial infection of root hairs and nodule tissue upon inoculation with M. loti R7A expressing lacZ. The average number of infection events and infection threads per plant incubated at 7 d after inoculation (DAI) was significantly reduced compared with empty vector control plants and to Pro35S:SINA4DN plants (Figure 6C; see Supplemental Figure 9A online). Despite the impairment of the early infection stages, the nodule numbers counted at 26 DAI were not significantly different for Pro35S:SINA4WT compared with the empty vector control and were higher for Pro35S:SINA4DN plants grown under nonoptimal conditions (closed plastic containers; see Methods), which might indicate a slightly better nodulation when SINA4 function is inhibited (see Supplemental Figure 9B online).
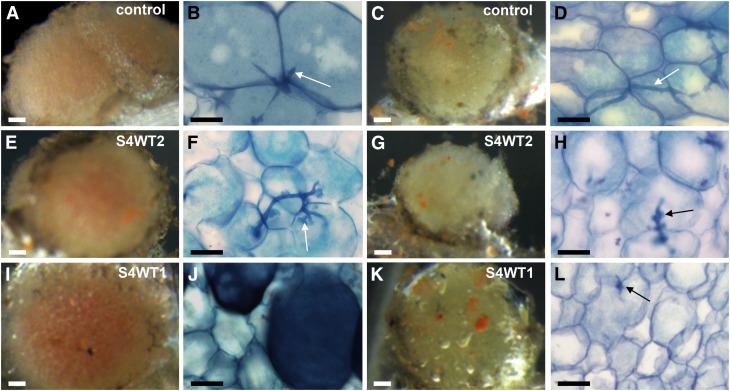
Stereomicroscopy analysis revealed that one-third of the Pro35S:SINA4WT nodules appeared white and small compared with only ∼8% of the nodules on the empty vector control or Pro35S:SINA4DN plants (see Supplemental Figure 9C online). Sectioning followed by toluidine blue staining of both nodule types, white and pink ones, revealed that Pro35S:SINA4WT nodules were not properly infected (Figure 7). Both the pink and the white nodules in the empty vector control (Figures 7A to 7D), the wild type (data not shown), or Pro35S:SINA4DN (data not shown) had thin, normal-looking transcellular infection threads (arrows in Figures 7B and 7D), whereas the infection threads visible in the Pro35S:SINA4WT nodules had a heavily branched and rather thick appearance (arrows in Figures 7F and 7H). Moreover, both the white and pink nodules of the controls and wild types or Pro35S:SINA4DN nodules contained infected cells that were always packed with bacteroids (seen as blue staining inside the cells in white and pink nodules; Figures 7B and 7D), whereas in the Pro35S:SINA4WT2 line, only the pink nodules were largely packed with bacteroids (Figures 7E and 7F), while the Pro35S:SINA4WT2 white nodules were almost devoid of bacteria (Figures 7G and 7H). An even stronger phenotype was observed for the Pro35S:SINA4WT1 plants in which the white nodules were empty (Figures 7K and 7L), and in which the pink nodules contained few infected cells. These Pro35S:SINA4WT1 pink nodules displayed abnormally large, blue cells (Figures 7I and 7J), similar in size to normally infected cells (Figure 7B), but without visible bacteroids inside (Figure 7J).
Figure 7.
L. japonicus Stable Transgenic Plants Ectopically Expressing SINA4 Develop Nonfunctional and Abnormally Infected Nodules.
Plants of two independent L. japonicus stable transgenic lines expressing Pro35S:SINA4WT (S4WT1 and S4WT2) and of a MG20 control line transformed with the empty vector (control) were inoculated with M. loti, and nodules were analyzed 26 d later. Nodule sections were stained with toluidine blue and pink (left panel), and white small senescent nodules (right panels), which made up one-third of the nodules on S4WT plants, were analyzed microscopically (see Supplemental Figure 8C online). (A), (C), (E), (G), (I), and (K) show stereomicroscopy images; (B), (D), (F), (H), (J), and (L) show light microscopy images. Bars = 0.1 mm (stereomicroscopy images) and 25 µm (light microscopy images).
(A) to (D) Nodule development and infection on L. japonicus MG20 control lines transformed with the empty vector. In pink nodules of these control plants, infected cells harboring bacteroids in the cytoplasm were larger than noninfected cells (B). By contrast, infected cells of the white nodules were smaller due to reduced bacterial occupancy (D). Arrows indicate transcellular infection threads visible as thin blue lines ([B] and [D]).
(E) to (L) Nodule development and infection on L. japonicus stable transgenic lines expressing Pro35S:SINA4WT. Abnormal infection centers were observed. S4WT2 plants ([E] to [H]) exhibited an intermediate phenotype featuring pink infected nodules ([E] and [F]) and abnormally highly branched infection threads (arrow; [F]). Abnormal infection threads were also found within the white nodules ([G] and [H]). The infected cells of white nodules also contained fewer bacteroids compared with the control (H). A stronger phenotype was observed for S4WT1 plants. The white nodules ([K] and [L]) were mostly devoid of bacteria with only few remaining infection threads (arrow; [L]). Moreover, huge blue-stained cells were present in the infection center (J) much larger than the uninfected cells in (L).
To determine the specificity of these effects of ectopically expressing SINA4 on nodulation, we analyzed Pro35S:SINA1WT-expressing plants. These Pro35S:SINA1WT lines did not show different infection thread or nodule numbers compared with control plants or the wild type, and they also displayed normally infected nodule tissue (see Supplemental Figure 10 online). However, the ectopic expression of the inactive SINA1DN led to a reduced number of infection threads and nodules compared with the controls.
In conclusion, the reduced bacterial colonization phenotype of Pro35S:SINA4WT appears to be specific to SINA4 and comprises impaired infection thread formation and a strong reduction in bacteroid abundance.
DISCUSSION
SYMRK triggers a program for the intracellular uptake of bacteria (Markmann et al., 2008); however, this program must be contained to restrict infection to appropriate cells and tissues. Here, we identified SINA E3 ligases as novel interaction partners of SYMRK. One family member, SINA4 could be identified as a negative regulator of SYMRK stability and symbiotic infection.
We observed that targeting of SYMRK by the E3 ligase SINA4 caused degradation of SYMRK in planta. In N. benthamiana, we observed a SINA4-mediated decrease of the SYMRK-YFP signal, concomitant with a relocalization to the cytosol and nuclear periphery, which are sites of proteasomal degradation. This regulatory effect of SINA4 observed in N. benthamiana was specific for SYMRK because another symbiotic receptor-like kinase, NFR1, did not disappear or relocalize upon coexpression.
Also, SINA4DN or SINA1DN stabilized SYMRK-YFP, indicating that the inactive dominant-negative SINA proteins bind SYMRK in planta and protect it from degradation. The fact that only the inactive SINA1DN and not the active SINA1 has an effect on SYMRK protein level is consistent with the lack of an obvious nodulation phenotype in Pro35S:SINA1WT. Since SINA1 does not appear to target SYMRK for destruction, the mechanism by which SINA1DN negatively influences symbiosis remains to be elucidated.
To identify the cell types in which such a negative regulation of SYMRK by SINA4 might take place, we compared the expression domains of the corresponding genes in roots by means of promotor:GUS fusions. Both expression domains were partially overlapping in the epidermis, including root hair cells, and, importantly, upon rhizobial inoculation, expression of both SINA4 and SYMRK was observed around rhizobial infection patches and nodule primordia. These overlapping expression patterns of SINA4 and SYMRK thus revealed potential cell types and developmental stages at which posttranslational regulation of SYMRK by SINA4 is possible.
The concept of negative regulation of symbiosis by SINA4 is further supported by the phenotype observed in transgenic L. japonicus lines ectopically expressing the active SINA4 protein. A drastic reduction in rhizobial infection and a high proportion of developmentally impaired nodules was observed. These nodules carried infection threads, but rhizobial occupancy was strongly reduced, rhizobia did not differentiate into bacteroids, and the number of infected cells was reduced. Moreover, the large proportion of small white nodules indicated a developmental delay or premature arrest. Ectopic expression of SINA4 affected infection in the epidermis as well as in the nodule cortex. It appears likely that this effect is mediated through ectopic and inappropriate degradation of SYMRK in cells types that normally do not express SINA4, which is supported by the observation that SYMRK protein levels are reduced in inoculated roots overexpressing SINA4. However, it should be stressed that our data do not exclude the possibility that SINA4 negatively regulates additional symbiosis-relevant proteins.
Higher protein levels of SYMRK were observed when SINA4DN or SINA1DN was coexpressed in N. benthamiana and thereby could protect SYMRK against degradation; however, the L. japonicus lines expressing SINA4DN did not show increased SYMRK protein levels or increased infection compared with controls. Thus, it is likely that multiple layers of regulation determine whether the cells are susceptible to infection. By contrast, the lines expressing SINA1DN did show a difference in nodule development compared with controls, similar to the observations in M. truncatula Pro35S:At SINAT5DN (Den Herder et al., 2008). This observation supports the possibility that the SINA gene family members target additional symbiosis relevant proteins for destruction.
SINA4 showed polyubiquitination activity in vitro and, therefore, potentially ubiquitinates SYMRK upon interaction. Moreover, the observed destabilization of SYMRK by the SINA4 E3 ligase calls for a model involving SYMRK as a SINA4 ubiquitination substrate. Although we could detect SYMRK ubiquitination in N. benthamiana, this also occurred in the absence of SINA4; hence, it was difficult to test SINAs against this background. Moreover, despite several attempts using a range of different commercially available E2 ligases, we could not detect SINA-mediated SYMRK ubiquitination in vitro. One possibiity is that SINA4 needs to be specifically activated in planta by certain proteins or conditions because we do observe a degradation of SYMRK protein upon coexpression with SINA4 E3 ligase in N. benthamiana transiently cotransformed leaves (Figure 4; see Supplemental Figure 6 online), and also in L. japonicus nodulated roots, when SINA4 was expressed from the strong and constitutive ubiquitin promoter (Figure 5).
In some instances, specific phosphorylation of human E3 proteins is required to provide specificity or to (de)activate the proteins (Ichimura et al., 2005; Khurana et al., 2006). Similarly, the rice XA21 receptor kinase phosphorylates XB3 E3 ligase, which in turn stabilizes the kinase (Wang et al., 2006). In addition, Arabidopsis S-domain receptor kinases were found to interact with Arabidopsis PUB-ARM E3 ligases, acting as a phosphorylation substrate and functioning in the abscisic acid response (Samuel et al., 2008). We observed in vitro phosphorylation of all SINA proteins tested by both SYMRK and NFR1 KDs (G. Den Herder, E.B. Madsen, and M. Parniske, unpublished results). So, SINA phosphorylation is not specific in vitro but possibly occurs in planta, although we cannot conclude whether SINA phosphorylation, potentially via SYMRK, would contribute to its E3 activity. Moreover, we cannot conclude whether phosphorylation of SINA4 could lead to ubiquitination of other nodulation-related targets, a mechanism which has been proposed for the LYK3–PUB1 interaction in M. truncatula (Mbengue et al., 2010).
As SYMRK is a common SYM gene, AM symbiosis could also be affected in Pro35S:SINA4 and Pro35S:SINA4DN lines. However, a wild type–like fungal infection and a similar number of arbuscules were observed in these lines. Also, SINA1 expression is slightly upregulated at 3 weeks after inoculation during AM symbiosis, but the Pro35S:SINA1 and Pro35S:SINA1DN transgenic lines developed AM that was indistinguishable from the controls (G. Den Herder and M. Parniske, unpublished results) in our assay system, which involved relatively strong infection pressure from Rhizophagus irregularis supported by chive nurse plants. Perhaps SYMRK does not require such SINA-mediated fine-tuned regulation during AM symbiosis, or a so far unknown AM-expressed E3 ligase targets SYMRK during fungal infection.
In conclusion, we demonstrated that the SINA4 E3 ligase acts as a regulator of SYMRK protein levels and is therefore able to indirectly manipulate rhizobial invasion during root symbiosis. Based on interaction studies, E3 ligase stability assays, and the symbiotic phenotype, we could assign a specific function for SINA4 in targeting SYMRK for degradation. The identification of L. japonicus SINAs, and particularly SINA4, as a key regulator of SYMRK, provides an important step forward in understanding how receptor kinases can be targeted for degradation to mitigate signaling.
METHODS
Sequence and Phylogenetic Analysis
SINA1-5 full coding sequences and genomic sequences were identified using the genomic database of Lotus japonicus (http://www.kazusa.or.jp/lotus/) and isolated via PCR using specific primers (see Supplemental Table 4 online) and L. japonicus Gifu B-129 cDNA. L. japonicus SINA protein sequence alignment was performed using ClustalW (EMBL-European Bioinformatics Institute). Conserved regions were defined and labeled with BioEdit 7.0.9 (Hall, 1999). Phylogenetic analysis of the conserved 654-bp nucleotide sequences, which were guided by the amino acid alignment regions displaying at least 70% identity, was performed as described by Den Herder et al. (2008). Supplemental Figure 1B online presents the result of the neighbor-joining bootstrap analysis with 1000 replicates, for which the support values are labeled on each branch. The human Siah1 sequence (Hu and Fearon, 1999) was used to root the tree.
Y2H Analysis
Y2H analysis was performed according to standard procedures (Stratagene Product Manual 235702; pBD-Gal4 Cam Phagemid vector kit) using the yeast strain AH109 (Clontech). As bait protein, the SYMRK-KD construct was cloned into the pBD-Gal4 vector to perform a screening on the L. japonicus cDNA library derived from RNA of inoculated roots, harvested 5 and 12 DAI (Poulsen and Pødenphant, 2002). Synthetic dropout media (-Trp, -Leu, -His, with [+] or without 5 mM 3-amino-1,2,4-triazole) were used for selection of the protein interactions. Pairwise interactions were tested using the full coding sequence of SINA genes in the pBD-Gal4 and pAD-Gal4 vectors, as well as the deletion constructs used in Figure 1B.
Escherichia coli Expression and Purification of Recombinant Proteins
After cloning of the maltose binding protein (MBP)- or GST-tagged SINA constructs and E. coli expression, protein purification was performed as described by Tirichine et al. (2006) and Yoshida and Parniske (2005), respectively.
In Vitro Ubiquitination Assay
Ubiquitination assays were performed by adding 500 ng SINA E3 ligase or the dominant-negative version to the reaction mixture (2 mM MgCl2, 2 mM ATP, 50 mM Tris, pH 7.5, and 0.5 mM DTT) in the presence or absence of 60 ng E1 (BostonBiochem E-301; from yeast), 300 ng E2 UbcH5b (His6-tagged human recombinant; Biomol UW9060-0100), and 1.2 µg ubiquitin (Biomol UW8795-0005) in a total volume of 30 µL. The reactions were incubated for 3 h at 30°C. The reaction was stopped by adding SDS-PAGE sample buffer and boiling for 5 min. Reactions were analyzed by SDS-PAGE and subsequent immunoblot analysis as described above using the antiubiquitin antibody (1/3000; Santa Cruz Biotechnology P4D1 [sc-8017] or Affiniti [Biomol/Enzo Life Sciences; UG9510]) to detect the ubiquitinated fraction and the α-MBP antibody (1/5000; New England Biolabs, E8030S) or the α-GST antibody to detect the tagged SINA proteins.
Transient Transformation of Nicotiana benthamiana
The subcellular localization and BiFC experiments using fluorescent fusion and split-YFP constructs, respectively, were analyzed in N. benthamiana as described by Yano et al. (2008). The constructs used for subcellular localization are listed in Supplemental Table 3 online, and two different binary vector systems, namely, pSPYCE/pSPYNE (Walter et al., 2004) and pAMPAT-35S GW (Raffaele et al., 2009), were used for BiFC experiments. Using the pSPYNE/pSPYCE system, the best expression signals were observed at 4 to 5 DAI, whereas experiments using the pAMPAT 35S GW split-YFP vectors gave the most reliable result at 2 to 3 DAI. Treatment of the leaf discs with the proteasome inhibitor MG132 (Alexis Biochemical) was performed by incubating leaf discs and gently shaking at room temperature for 20 h in 50 µM MG132. Mock treatment was performed in DMSO/water. For epifluorescence microscopy, the Leica DMI4000B inverse fluorescence microscope was used set as described by Yano et al. (2008), using the appropriate emission filters (dsRED/GFP/YFP and CFP), and confocal laser scanning microscopy was done with the Leica SP5 microscope with fluorescent excitation and emission spectra set as follows: in colocalization experiments, YFPv excitation at 514 nm and emission at 522 to 540 nm; mOrange excitation at 561 nm and emission at 570 to 600 nm (excluding overlapping signals). When mOrange was expressed alone, excitation was at 514 nm and emission was at 540 to 600 nm. When imaged for RFP, excitation was at 561 nm and emission was at 565 to 600 nm. In Supplemental Figures 3D to 3F online, we show that when free RFP (in the cytosol) is imaged at these spectral settings, which are very similar to the mOrange colocalization spectral settings, no YFP dots are visible on the image.
N. benthamiana Protein Extraction and Immunoprecipitation of SYMRK-YFP
Single leaf discs of N. benthamiana leaves transiently expressing the different transformed constructs were excised from the leaf, water infiltrated, and immediately analyzed for fluorescent signal on an inverted epifluorescence microscope (Leica) at different time points (4 to 5 DAI), harvested as quickly as possible, and frozen in liquid nitrogen for protein extraction. Homogenization was done in a Retsch mill (30 s−1; 2 min). Crude extraction was performed by adding 200 μL of extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM DTT, 1% PVPP, and 1% Triton) or extraction buffer used for immunoprecipitation of SYMRK-YFP (50 mM imidazole, pH 7.0, 200 mM NaCl, 1 mM Na-EDTA, 5 mM EGTA, 2% Triton X-100, 1% SDS, 1 mM DTT, PhosStop [Roche], protease inhibitors [P9599; Sigma-Aldrich), 1 mM PMSF, 10% Suc, and 50 µM MG132). After centrifugation for 10 min at 4°C at 10,000g, the supernatant was taken as crude extract. The microscopy and protein expression analysis were performed at least three times for each construct and condition tested, and this method was suitable to deliver reproducible results.
For immunoprecipitation experiments, crude extracts were ultracentrifuged for 20 min at 130,000g to remove insoluble precipitated proteins. The supernatant was taken and added to 800 μL dilution buffer (50 mM imidazole, pH 7.0, 200 mM NaCl, 1 mM EDTA, 2% TritonX-100, 1% SDS, and protease inhibitors [Sigma-Aldrich]). To prepare the preclearance and immunoprecipitate, 2 × 10 μL Protein A sepharose and 15 μL GFP binder resin (Chromotek) per reaction was washed twice in 0.5 mL dilution buffer and spun down at 400g at 4°C for 20 s. The diluted supernatants were added to 10 μL of Protein A sepharose for 30 min on a rotor at 4°C, a preclearance step that was repeated once. The remaining supernatant was used for the immunoprecipitation and added to the prewashed GFP binding matrix, which was incubated in a rotor at 4°C for 30 min. Samples were transferred to a Mobicol tube to start the washing. The resin was spun down at 400g for 60 s at 4°C. The column was washed five times with 500 μL washing buffer (50 mM imidazole, pH 7.0, 600 mM NaCl, 5 mM Na-EDTA, 2% TritonX-100, and 1% SDS). Elution was done with 20 μL 0.1 M Gly, pH 2.4, which was left for 5 min on the column before centrifugation and addition of 2 μL Tris-HCl buffer, pH 8.0. This step was repeated once with another 20 μL 0.1 M Gly, pH 2.4.
SDS-PAGE sample buffer was added, and samples were heated for 10 min at 55°C and loaded on a 10% acrylamide gel for SDS-PAGE (Laemmli, 1970). Immunoblot analysis was performed with the appropriate antibody: Detection of YFP was done with the α-GFP (polyclonal rabbit; Rockland; 1:3000), and BiFC constructs were detected using α-HA conjugated to horseradish peroxidase (Roche; 1/2000) and α-cMyc (Roche, clone 9E10, 1:3000). After immunodetection, membranes were stained with Coomassie Brilliant Blue as a loading control.
L. japonicus Plant Growth and Inoculation with Mesorhizobium loti
All plants used were derived from L. japonicus ecotype B-129 Gifu or MG20 Miyakojima (Kawaguchi et al., 2001), both being referred to as the wild type throughout this study. For rhizobial inoculation, seedlings were grown as described by Tansengco et al. (2003), subsequently incubated in translucent closed plastic containers containing 300 mL Seramis (Mars) and 70 mL liquid Broughton and Dilworth (BD) medium (Broughton and Dilworth, 1971) or in 1-liter Weck jars, containing 350 mL Seramis and 100 mL BD medium, sealed with Micropore tape. M. loti R7A (Sullivan et al., 2002) carrying the pXLGD4 plasmid for expression of the LacZ marker gene was used for inoculation at a final OD600 of 0.05. The bacterial inoculum was prepared after culturing the rhizobia in liquid trypton yeast extract medium for 2 to 3 d at 28°C (220 rpm). Subsequently, rhizobial cultures were spun down and resuspended in BD.
Agrobacterium tumefaciens–Mediated Transformation of L. japonicus Ecotype Gifu B-129 and Miyakojima MG20
L. japonicus transgenic lines were obtained using the Agrobacterium strain AGL1 transformed with the binary vector pK7WG2D-35S:SINA1(WT or DN), pK7WG2D-35S:SINA4(WT or DN), and the empty vector control pK7WG2D (referred to as control plants). Transformation was performed as described by Kato et al. (2005), but using geneticin (pK vectors) as selection marker. Segregation of SINA4 and SINA4DN expressing T1 lines was followed in the T2 plants via in vivo screening of the constitutively expressed endoplasmic reticulum-GFP marker present on the T-DNA. L. japonicus hemizygous T1 parental lines showed a 3:1 Mendelian segregation, indicative for 1 insertion, and T2 homozygous progeny were further used for phenotyping experiments in T3 and T4.
The transgene expression level was determined via quantitative RT-PCR on root material (from a single root system of the T3 and from a pool of five root systems of T4 inoculated plants) using forward primers specific for the SINA1 or SINA4 gene and reverse primers in the three prime terminator sequence, allowing to select for the lines with the highest transgene expression (see Supplemental Table 4 online). Figure 6A and Supplemental Figure 10A online present the relative differences in between the different lines, with the controls given a cycle threshold (Ct) value >40. The total SINA4 expression level was also determined via quantitative real-time RT-PCR using the same samples and methods, but using a forward and reverse primer specific for SINA4, allowing us to determine the fold of overexpression of total SINA4 transcript level compared with the controls (Figure 6B). The Pro35S:SINA1(WT or DN) constructs were transformed into Gifu B-129 background, whereas Pro35S:SINA4(WT or DN) constructs were transformed into MG20 background. Both lines were phenotyped using appropriate control lines (the pK7WG2D empty vector transformed in Gifu and MG20 background, respectively) or the wild type.
Effect of SINA4 Expression on SYMRK Stability in Transgenic Inoculated Roots
Agrobacterium rhizogenes transformation of L. japonicus Gifu wild-type plants was performed using the A. rhizogenes AR1193 strain transformed with pUB-GW-GFP (empty vector control), ProUbi:SINA4 or ProUbi:SINA4DN binary plasmids as described by Charpentier et al. (2008). Plants with transgenic roots were inoculated with 25 mL of M. loti MAFF303099, carrying a dsRED marker, at a final OD600 of 0.05, in Weck jars with 300 mL of sand/vermiculite and 50 mL Fahraeus plant mixture and incubated in the growth chamber (16 h light/8 h dark; 24°C) for 14 d. Transformed roots were harvested and frozen with liquid nitrogen and homogenized in the presence of liquid nitrogen in a mortar. Proteins were extracted by adding 100 μL of extraction buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA, 2 mM DTT, 1% polyvinylpolypyrrolidone, 10% Suc, 1% Triton, 1% SDS, 5 M urea, and 2 M thiourea) per 100 mg tissue. The extract was incubated for one hour at 37°C and subsequently centrifuged for 10 min at 4°C at 10,000g. The supernatant was analyzed SDS-PAGE and immunoblots.
Phenotyping of L. japonicus Roots and Nodules
Plants were grown in sterile conditions and inoculated with M. loti R7A expressing a LacZ marker gene. Staining for β-galactosidase activity of LacZ-expressing rhizobia was done as described by Tansengco et al. (2003) on inoculated roots 5 and 7 DAI (infection events and infection thread quantification) and at 26 DAI (nodule phenotyping). In each experiment, T3 and/or T4 progeny were used for on average n = 10 to 15 plants per time point. A Leica MZFLIII stereomicroscope was used to count the infection events (meaning a root hair curl with entrapped blue-stained bacterial microcolony) and the infection threads on one side of the root, as held on a microscopy slide (the other side was not included to prevent double counting on the same root). The nodule number was determined at 26 DAI, and the percentage of pink (leghemoglobin producing) and white (young) or small, white (early senescent) nodules was defined in both conditions tested. Statistical analysis of the quantified infection events, infection threads, and nodules involved calculation of the se for the mean value of each population tested (which was done by dividing the sd by the square root of n plants), and the differences compared with the wild type were subjected to a two-tailed t test, with a significant outcome (*) of P < 0.05 or (**) of P < 0.005.
For experiments in Figures 6, 7, and Supplemental Figures 9B, 9D, and 10 online, we used closed plastic containers that provided high humidity but represented conditions that were below optimum for rhizobial infection and nodulation. For experiments in Supplemental Figures 9 and 10C online, we used 1-liter glass jars (Weck), which allowed an increased number of symbiotic infection and nodulation events per root system.
A difference in nodulation efficiency was observed in all conditions between the MG20-derived and Gifu-derived transgenic lines. Since the Gifu background produced more infection and nodulation events than MG20, the quantitative values were compared with the control line (empty vector transgenics) derived from the same ecotype.
Microscopy and Sectioning of L. japonicus Nodule Tissue
Nodule tissue (26 DAI) was fixed in 0.1 M Na-cacodylate buffer, pH 7.4, containing 2.5% of glutaraldehyde (Sigma-Aldrich), and nodule samples were dehydrated in a series of 10 to 70% ethanol (each step 1 h at 4°C and 70% overnight at 4°C) and embedded in Technovit 7100 (Kulzer Histo-Technik) according to the manufacturer’s instructions. Semithin sections of 4 μm were taken with a rotary microtome, mounted on Superfrost Plus slides (Roth), and stained with 0.5% toluidine blue before mounting with DEPEX (VWR) and examination using the Leica DMI4000B inverse microscope.
Plasmid Construction
Constructs used for Y2H analysis, protein expression in E. coli, BiFC, transient subcellular localization, and plant transformation were made using Gateway technology (Invitrogen) and Phusion Taq Polymerase (Finnzymes). Entry clones were constructed by BP reactions or TOPO cloning, and the sequence-verified inserts were transferred to the proper destination vectors by Gateway LR reactions. The SINA1-5 coding sequences were isolated from cDNA by conventional PCR using sequence-specific primers (see Supplemental Table 4 online). BP reaction resulted in pDONR207-SINA1 and pDONR201-SINA5, whereas bidirectional TOPO cloning resulted in pENTR-SD-SINA2, 3, and 4. Deletion constructs for SINA1 were constructed by PCR using primers containing an EcoRI-SalI restriction site (see Supplemental Table 4 online), to obtain all constructs in pENTR-SD-TOPO vectors. Introduction of the dominant-negative mutation C47S for SINA1 (SINA1DN) and C66S for SINA4 (SINA4DN) required introduction of a point mutation, which was performed using mutated primers (see Supplemental Table 4 online) and using a PCR-based method as described (Cormack, 1997). The entry clone for the ProSINA4:gSINA4:YFP construct pENTR-TOPO-D-ProSINA4:gSINA4(w/oStop) was made via TOPO cloning of the amplified fragment containing the promoter of SINA4, 2.5 kb upstream of the ATG, and the genomic SINA4 sequence (gSINA4) until stop codon using sequence-specific primers (see Supplemental Table 3 online) on L. japonicus Gifu genomic DNA. The ENTRY clone for the ProSINA4:gSINA4DN:YFP construct pENTR-TOPO-D-ProSINA4:gSINA4DN(w/oStop) was made via whole plasmid amplification on this clone using hot start Phusion polymerase and mutation primers (see Supplemental Table 4 online). The DONR207-ProSINA4 clone was made by BP reaction (Invitrogen) using the AttB-containing amplicon of the SINA4 promoter, 2.5 kb upstream of the ATG, which also includes the untranslated region genomic fragment (see Supplemental Table 4 online).
The SYMRK-KD and SYMRKK622E-KD entry vectors were used as described by Yoshida and Parniske (2005). The ProSYMRK entry clone was made using sequence specific primers (with EagI TOPO site sense sequence; see Supplemental Table 3 online) using Phusion polymerase to amplify the promoter 4979 bp upstream of the start codon from a plasmid containing the genomic sequence of SYMRK (gSYMRK) and TOPO cloning in the pENTR-TOPO-D vector. The genomic SYMRK entry clone pENTR-D-ATGgSYMRK:MYC used for BiFC and all other N. benthamiana experiments contains the following insert between the recombination sites: the gSYMRK sequence (as in Lotus T11E23 [TAC containing Lotus SYMRK, AP004579]) starting from the ATG start codon (as defined in coding sequence of SYMRK, AF492655) until the KpnI restriction site of exon XIII. This site is followed by the remaining coding sequence of exonXIII to exonXV, without stop codon, coupled to an SpeI site and MYC tag (see Supplemental Table 4 online). The NFR1 entry clone contains the coding sequence, which was amplified by PCR with specific primers for TOPO cloning (see Supplemental Table 4 online). The KD of the LjFLS2-like gene was cloned in the entry vector using primers with EcoRI-SalI sites (see Supplemental Table 4 online) and PCR on L. japonicus cDNA after identification of the closest At-FLS2 homolog in the Lotus Tentative Consensus database (http://compbio.dfci.harvard.edu).
The following destination vectors were used: for Y2H, modified pBD-GAL4 Cam (Stratagene) and pGAD424 (Clontech) vectors in which Gateway cassettes were introduced; for bacterial protein expression, N-terminal MBP fusions, pKM596 (Fox and Waugh, 2003); for N-terminal GST fusions, pDEST15 (Invitrogen) and pGEX4T (Amersham); for BiFC, pSPYNE/pSPYCE 35S GW (Walter et al., 2004) and pAMPAT-35S-GW-YFPN/C (kindly provided by Laurent Deslandes, Laboratoire des Interactions Plantes-Microorganismes Toulouse, Institut National de la Recherche Agronomique-Centre National de la Recherche Scientifique; Raffaele et al., 2009); for mOrange fusions, pAMPAT-derived p35S-GW-mOrange-nos (Bayle et al., 2008); for C-terminal and N-terminal CFP, GFP, and YFP fusions, pAMPAT-MCS derivative (GenBank accession number AY436765; see Supplemental Table 3 online); for ProSINA4:gSINA4:YFP, pHYWG2,0 (Karimi et al., 2002; Invitrogen) from which the 35S promoter was cut out; for plant expression, pK7WG2D (35S-driven expression) and pKWGFS7 (promoter-GUS analysis) (Karimi et al., 2002; Invitrogen); for ProUbi:SINA4 or ProUbi:SINA4DN constructs for hairy root transformation, the destination vector pUB-GW-GFP (Maekawa et al., 2008), with the use of the pENTR-SD-SINA4 or pENTR-SD-SINA4DN as donor vectors, respectively.
In Vitro Pull-Down Assay
The recombinant proteins GST-SINA1 and GST-SINA1-del2 (Figure 1B) were purified from E. coli cultures and together with 6xHIS-SYMRK-KD (Yoshida and Parniske, 2005) assayed for direct binding in vitro. The 6xHIS-SYMRK-KD protein was incubated for a maximum of 15 min in binding buffer (50 mM HEPES, 50 mM NaCl, 10 mM MgCl2, 1 mM ZnCl2, 0.25% Nonidet P-40, 25 mM NaF, and proteinase inhibitor cocktail [Sigma-Aldrich]). The incubated protein was then added to GST-sepharose beads and incubated for 2 h at 4°C on a rotor. Separation of the bead-containing pellet by centrifugation precleaned the protein in the supernatant (input), which was subsequently mixed with 1 μg of GST-SINA1 or GST-SINA1-del2 and fresh glutathione sepharose beads and incubated overnight at 4°C on a vertical rotor. Centrifugation was performed to separate nonbound protein in the supernatant and the GST binding fraction in the pellet. The latter was subsequently washed three times with 1 mL binding buffer, and after centrifugation, SDS-PAGE sample buffer was added, and the samples were boiled and analyzed by SDS-PAGE (Laemmli, 1970) and subsequent immunoblot analysis. The separated proteins were electrophoretically transferred onto a Hybond-P polyvinylidenefluoride membrane (GE Healthcare Amersham) by wet blotting. Membranes were blocked in 5% powdered milk in TBS (100 mM Tris-Cl, pH 7.5, and 150 mM NaCl) and washed three times in TBS-T (+0.1% Tween-20), incubated with α-HIS antibody (1/6000; Roche) and α-GST antibody (1/5000; Upstate Biotechnology; Millipore, #06-332) for 1 h in TBS-T to detect the tagged proteins. Subsequently, washing in TBS-T was performed three times, followed by incubation for 1 h with secondary α-rabbit or α-mouse antibody coupled to horseradish peroxidase (Amersham NA934 or Biomol 610-1302, respectively) or to fluorescent dye IR680 (rabbit; Invitrogen A21076) or IR800 (mouse; Biomol 610-132-121). After washing, the protein antibody complexes on the blots were detected either using the ECL chemiluminescence detection kit (GE Healthcare RPN2135) according to the manufacturer’s instructions or using an Odyssee IR-fluorescence scanner (Li-Cor, version 2.1).
L. japonicus Root RNA Isolation and Quantitative RT-PCR Analysis
Total RNA extraction from L. japonicus roots was performed as described by Kistner et al. (2005). First-strand cDNA synthesis reactions were performed using 400 ng of total RNA with the Superscript III First-Strand Synthesis Supermix for quantitative RT-PCR (Invitrogen), according to the manufacturer’s protocol. For quantitative RT-PCR reactions, the Platinum SYBR Green qPCR Supermix-UDG (Invitrogen) was used according to the guidelines of the manual, in a final volume of 10 μL. A reaction using RNA template without RT was used as a control for the presence of genomic DNA contamination. Quantitative PCR amplification was performed with the following program: 94°C for 5 min, 40× (94° for 30 s, 57° for 30 s, and 72° for 30 s), 95°C for 1 min, 55°C for 1 min (iCycler IQ Real Time PCR detection system, software version 3.1; Bio-Rad), and the relative expression was calculated by normalization with the gene expression of the constitutively expressed L. japonicus elongation factor 1-α EF1 and L. japonicus Ubiquitin (see Supplemental Table 3 online) using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Amplification efficiency for each primer pair was calculated using the LinReg software (version 7.5; Ramakers et al., 2003) to obtain comparable results. Reactions were performed in duplicate and averaged. Primers used for specific amplification of the SINA1-4 transcripts (see Supplemental Figure 7 online) and the marker genes NIN and PT4 during symbiotic development were unique in the LjGI index release 6.0 (The Institute for Genomic Research) and are given in Supplemental Table 4 online.
SINA4 and SYMRK Promoter Activity in L. japonicus
A. rhizogenes transformation of L. japonicus Gifu B-129 and MG20 was performed using the A. rhizogenes AR1193 strain transformed with the pKWGFS7-ProSINA4 or pKWGFS7-ProSYMRK binary plasmid as described by Charpentier et al. (2008). Two weeks after transformation, transgenic roots emerged and were inoculated with M. loti MAFF303099, carrying a DsRED marker, in open pots with sand/vermiculite mixture and incubated in the growth chamber (16 h light/8 h dark; 24°C) for the times indicated. Roots were harvested and GUS activity was visualized by X-gluc staining of the roots as described by Groth et al. (2010). The ProSYMRK:GUS transgenic roots were incubated for only 4 h, whereas the ProSINA4:GUS transgenic roots were incubated overnight at 37°C for staining. A stereomicroscope was used for inspection and visualization of blue stain and dsRED localization.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: HE664113, HE664114, HE664115, HE664116, HE664117, and HE664118 (for the L. japonicus SINA1, SINA2, SINA3, SINA4, SINA5, and SINA6 genes, respectively).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. L. japonicus SINA E3 Ligase Family and Phylogenic Relation to Other SINAs.
Supplemental Figure 2. L. japonicus SINAs Interact with SYMRK and Not with NFR1 in Yeast and N. benthamiana.
Supplemental Figure 3. SINA4 Subcellular Localization and SINA Dimerization in Planta.
Supplemental Figure 4. GST-SINA1, -SINA2, and -SINA3 Exhibit E3 Ligase Activity in Vitro.
Supplemental Figure 5. SYMRK Localizes to the Plasma Membrane in N. benthamiana.
Supplemental Figure 6. SINA4 Activity Specifically Affects SYMRK but Not NFR1 Protein Abundance in N. benthamiana.
Supplemental Figure 7. L. japonicus SINA Symbiotic Expression Patterns.
Supplemental Figure 8. SINA4 and SYMRK Promoter Activity during L. japonicus Root and Nodule Development.
Supplemental Figure 9. Phenotypic Analysis of L. japonicus Stable Transgenics Ectopically Expressing Pro35S:SINA4WT (S4WT1/2) and Pro35S:SINA4DN (S4DN1).
Supplemental Figure 10. Phenotypic Analysis of L. japonicus Transgenics Ectopically Expressing Pro35S:SINA1WT and Pro35S:SINA1DN.
Supplemental Table 1. Combinations Tested for Interaction in Gal4 Yeast Two-Hybrid Assays.
Supplemental Table 2. Combinations Tested for Interaction in Bimolecular Fluorescence Complementation Assays in N. benthamiana.
Supplemental Table 3. List of Fluorescent Fusion Constructs Used for N. benthamiana Transient Transformation.
Supplemental Table 4. Primer Sequences Used for Cloning and Quantitative RT-PCR.
Supplemental Data Set 1. Text File of the Alignment Corresponding to the Phylogeny in Supplemental Figure 1 Online.
Supplemental Methods 1. Supplemental Methods for the Supplemental Data.
Supplementary Material
Acknowledgments
We thank Ive De Smet for helping with the writing and for critical discussions. We also thank Carsten Poulsen (Aarhus University) or providing the L. japonicus Y2H cDNA library, Esben Madsen and Jens Stougaard (Aarhus University) for providing the pADGal4- and pBDGal4-NFR1 and NFR5 plasmid clones, Ulrich Rothbauer and Heinrich Leonhardt (University of Munich) for kindly providing GFP binder resin (now available through Chromotek), Gabrielle Büttner and Ute Bergmann (University of Munich) for technical assistance, and Sarah Gardner (The Sainsbury Laboratory, Norwich, UK) for help with the Y2H screens. This work was supported by the German Research Foundation priority program SPP1212 “Microbial Reprogramming of Plant Cell Development.”
AUTHOR CONTRIBUTIONS
G.D.H., S.Y., M.A.-L., and M.P. designed the research. G.D.H., S.Y., M.A.-L., and M.K.R. performed research. G.D.H. and M.P. wrote the article.
Glossary
- RLK
receptor-like kinase
- SBD
substrate binding and dimerization
- AM
arbuscular mycorrhiza
- Y2H
yeast two-hybrid
- KD
kinase domain
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- GUS
β-glucuronidase
- DAI
days after inoculation
- MBP
maltose binding protein
- BandD
Broughton and Dilworth
- TVEC
total visible epidermal cells
References
- Albert M., Jehle A.K., Lipschis M., Mueller K., Zeng Y., Felix G. (2010). Regulation of cell behaviour by plant receptor kinases: Pattern recognition receptors as prototypical models. Eur. J. Cell Biol. 89: 200–207 [DOI] [PubMed] [Google Scholar]
- Arrighi J.F., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A., Novatchkova M., Potuschak T., Eisenhaber F. (2001). Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6: 463–470 [DOI] [PubMed] [Google Scholar]
- Bayle V., Nussaume L., Bhat R.A. (2008). Combination of novel green fluorescent protein mutant TSapphire and DsRed variant mOrange to set up a versatile in planta FRET-FLIM assay. Plant Physiol. 148: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersoult A., Camut S., Perhald A., Kereszt A., Kiss G.B., Cullimore J.V. (2005). Expression of the Medicago truncatula DM12 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol. Plant Microbe Interact. 18: 869–876 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Genre A. (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1: 48 [DOI] [PubMed] [Google Scholar]
- Brewin N.J. (2004). Plant cell wall remodelling in the rhizobium-legume symbiosis. Crit. Rev. Plant Sci. 23: 293–316 [Google Scholar]
- Broughton W.J., Dilworth M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W., Goormachtig S., De Rycke R., Schroeyers K., Holsters M. (2005). SrSymRK, a plant receptor essential for symbiosome formation. Proc. Natl. Acad. Sci. USA 102: 10369–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Rubin G.M. (1990). seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63: 561–577 [DOI] [PubMed] [Google Scholar]
- Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. (1998). The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17: 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S.E., Murawsky C.M., Lowe N., Travers A.A. (2008). Two modes of degradation of the tramtrack transcription factors by Siah homologues. J. Biol. Chem. 283: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B. (1997). Directed mutagenesis using the polymerase chain reaction. Curr. Protoc. Mol. Biol. 37: 8.5.1–8.5.10 [DOI] [PubMed] [Google Scholar]
- Della N.G., Senior P.V., Bowtell D.D. (1993). Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 117: 1333–1343 [DOI] [PubMed] [Google Scholar]
- Demchenko K., Winzer T., Stougaard J., Parniske M., Pawlowski K. (2004). Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol. 163: 381–392 [DOI] [PubMed] [Google Scholar]
- Den Herder G., De Keyser A., De Rycke R., Rombauts S., Van de Velde W., Clemente M.R., Verplancke C., Mergaert P., Kondorosi E., Holsters M., Goormachtig S. (2008). Seven in absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiol. 148: 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G., Van Isterdael G., Beeckman T., De Smet I. (2010). The roots of a new green revolution. Trends Plant Sci. 15: 600–607 [DOI] [PubMed] [Google Scholar]
- Depaux A., Regnier-Ricard F., Germani A., Varin-Blank N. (2006). Dimerization of hSiah proteins regulates their stability. Biochem. Biophys. Res. Commun. 348: 857–863 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voss U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Fox J.D., Waugh D.S. (2003). Maltose-binding protein as a solubility enhancer. Methods Mol. Biol. 205: 99–117 [DOI] [PubMed] [Google Scholar]
- Franck T., Krueger R., Woitalla D., Müller T., Engelender S., Riess O. (2006). Mutation analysis of the seven in absentia homolog 1 (SIAH1) gene in Parkinson’s disease. J. Neural Transm. 113: 1903–1908 [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi H., Markmann K., Svistoonoff S., Estevan J., Autran D., Giczey G., Auguy F., Péret B., Laplaze L., Franche C., Parniske M., Bogusz D. (2008). SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc. Natl. Acad. Sci. USA 105: 4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson K.E., Kobayashi H., Walker G.C. (2008). Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42: 413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D.R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J.P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Goff K.E., Ramonell K.M. (2007). The role and regulation of receptor-like kinases in plant defense. Gene Regul. Syst. Bio. 1: 167–175 [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Groothuis T.A., Dantuma N.P., Neefjes J., Salomons F.A. (2006). Ubiquitin crosstalk connecting cellular processes. Cell Div. 1: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98 [Google Scholar]
- Hare P.D., Seo H.S., Yang J.Y., Chua N.H. (2003). Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr. Opin. Plant Biol. 6: 453–462 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67: 425–479 [DOI] [PubMed] [Google Scholar]
- House C.M., Möller A., Bowtell D.D. (2009). Siah proteins: Novel drug targets in the Ras and hypoxia pathways. Cancer Res. 69: 8835–8838 [DOI] [PubMed] [Google Scholar]
- Hu G., Fearon E.R. (1999). Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 19: 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T., Yamamura H., Sasamoto K., Tominaga Y., Taoka M., Kakiuchi K., Shinkawa T., Takahashi N., Shimada S., Isobe T. (2005). 14-3-3 proteins modulate the expression of epithelial Na+ channels by phosphorylation-dependent interaction with Nedd4-2 ubiquitin ligase. J. Biol. Chem. 280: 13187–13194 [DOI] [PubMed] [Google Scholar]
- Javot H., Pumplin N., Harrison M.J. (2007). Phosphate in the arbuscular mycorrhizal symbiosis: Transport properties and regulatory roles. Plant Cell Environ. 30: 310–322 [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A., Weissman A.M. (2000). RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kato T., Goto Y., Ono K., Hayashi M., Murooka Y. (2005). Expression of a major house dust mite allergen gene from Dermatophagoides farinae in Lotus japonicus accession Miyakojima MG-20. J. Biosci. Bioeng. 99: 165–168 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Motomura T., Imaizumi-Anraku H., Akao S., Kawasaki S. (2001). Providing the basis for genomics in Lotus japonicus: The accessions Miyakojima and Gifu are appropriate crossing partners for genetic analyses. Mol. Genet. Genomics 266: 157–166 [DOI] [PubMed] [Google Scholar]
- Kerppola T.K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37: 465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z., et al. (2007). 3-Hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19: 3974–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana A., Nakayama K., Williams S., Davis R.J., Mustelin T., Ronai Z. (2006). Regulation of the ring finger E3 ligase Siah2 by p38 MAPK. J. Biol. Chem. 281: 35316–35326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Jeong W., Ahn K., Ahn C., Kang S. (2004). Siah-1 interacts with the intracellular region of polycystin-1 and affects its stability via the ubiquitin-proteasome pathway. J. Am. Soc. Nephrol. 15: 2042–2049 [DOI] [PubMed] [Google Scholar]
- Kiss E., Oláh B., Kaló P., Morales M., Heckmann A.B., Borbola A., Lózsa A., Kontár K., Middleton P., Downie J.A., Oldroyd G.E., Endre G. (2009). LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol. 151: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Webb K.J., Szczyglowski K., Parniske M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K.T., Endre G., Prabhu R., Penmetsa R.V., Veereshlingam H., Cook D.R., Dickstein R., Vandenbosch K.A. (2004). LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol. 136: 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2008). A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr. Biol. 18: 1949–1954 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lefebvre B., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy C., Wrana J.L. (2005). Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 6: 112–126 [DOI] [PubMed] [Google Scholar]
- Li W., Tu D., Brunger A.T., Ye Y. (2007). A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446: 333–337 [DOI] [PubMed] [Google Scholar]
- Limpens E., Mirabella R., Fedorova E., Franken C., Franssen H., Bisseling T., Geurts R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lovly C.M., Yan L., Ryan C.E., Takada S., Piwnica-Worms H. (2008). Regulation of Chk2 ubiquitination and signaling through autophosphorylation of serine 379. Mol. Cell. Biol. 28: 5874–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., Janes E.K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Comm. 1: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kusakabe M., Shimoda Y., Sato S., Tabata S., Murooka Y., Hayashi M. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21: 375–382 [DOI] [PubMed] [Google Scholar]
- Markmann K., Giczey G., Parniske M. (2008). Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 6: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., Moreau S., Rivas S., Timmers T., Hervé C., Cullimore J., Lefebvre B. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L., Lefebvre B., Cullimore J., Imberty A. (2006). LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 16: 801–809 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Passmore L.A., Barford D. (2004). Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 379: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Poulsen C., Pødenphant L. (2002). Expressed sequence tags from roots and nodule primordia of Lotus japonicus infected with Mesorhizobium loti. Mol. Plant Microbe Interact. 15: 376–379 [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M., Albrektsen A.S., James E.K., Thirup S., Stougaard J. (2007). LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J.M., Deprez R.H., Moorman A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Mudgil Y., Salt J.N., Delmas F., Ramachandran S., Chilelli A., Goring D.R. (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147: 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelli E., Leone M., Li C., Fukushima T., Preece N.E., Olson A.J., Ely K.R., Reed J.C., Pellecchia M., Liddington R.C., Matsuzawa S. (2005). Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J. Biol. Chem. 280: 34278–34287 [DOI] [PubMed] [Google Scholar]
- Sato S., Nakamura Y., Asamizu E., Isobe S., Tabata S. (2007). Genome sequencing and genome resources in model legumes. Plant Physiol. 144: 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Shimomura K., Nomura M., Tajima S., Kouchi H. (2006). LjnsRING, a novel RING finger protein, is required for symbiotic interactions between Mesorhizobium loti and Lotus japonicus. Plant Cell Physiol. 47: 1572–1581 [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Sullivan J.T., et al. (2002). Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184: 3086–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansengco M.L., Hayashi M., Kawaguchi M., Imaizumi-Anraku H., Murooka Y. (2003). crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol. 131: 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers A.C. (2008). The role of the plant cytoskeleton in the interaction between legumes and rhizobia. J. Microsc. 231: 247–256 [DOI] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Vinardell J.M., Fedorova E., Cebolla A., Kevei Z., Horvath G., Kelemen Z., Tarayre S., Roudier F., Mergaert P., Kondorosi A., Kondorosi E. (2003). Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 15: 2093–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang M., Jin Y., Fu J., Zhu Y., Zheng J., Hu J., Wang G. (2008). Genome-wide analysis of SINA family in plants and their phylogenetic relationships. DNA Seq. 19: 206–216 [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Pi L.Y., Chen X., Chakrabarty P.K., Jiang J., De Leon A.L., Liu G.Z., Li L., Benny U., Oard J., Ronald P.C., Song W.Y. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18: 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R., Maass D., Voegel T., Dellapenna D., Beyer P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N.-H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Guo H.-S., Dallman G., Fang S., Weissman A.M., Chua N.-H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Parniske M. (2005). Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J. Biol. Chem. 280: 9203–9209 [DOI] [PubMed] [Google Scholar]
- Zeng L.R., Vega-Sánchez M.E., Zhu T., Wang G.L. (2006). Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res. 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zhu H., Chen T., Zhu M., Fang Q., Kang H., Hong Z., Zhang Z. (2008). A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol. 148: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.