Abstract
Synthetic duplex DNAs of repeating sequence, such as poly d(TTC).poly d(GAA), were separated into their individual single strands. The various single strands complexed not only, as expected, with their complementary strands, but also with other non-complementary strands. Characterization of such complexes with respect to stoichiometry, Tm values and the dependence of Tm on NaCl concentration showed that a variety of unusual structures could be inferred at physiological salt concentrations. These included extrahelical thymines, G.T oppositions, A.C oppositions and T.C oppositions.
Full text
PDF

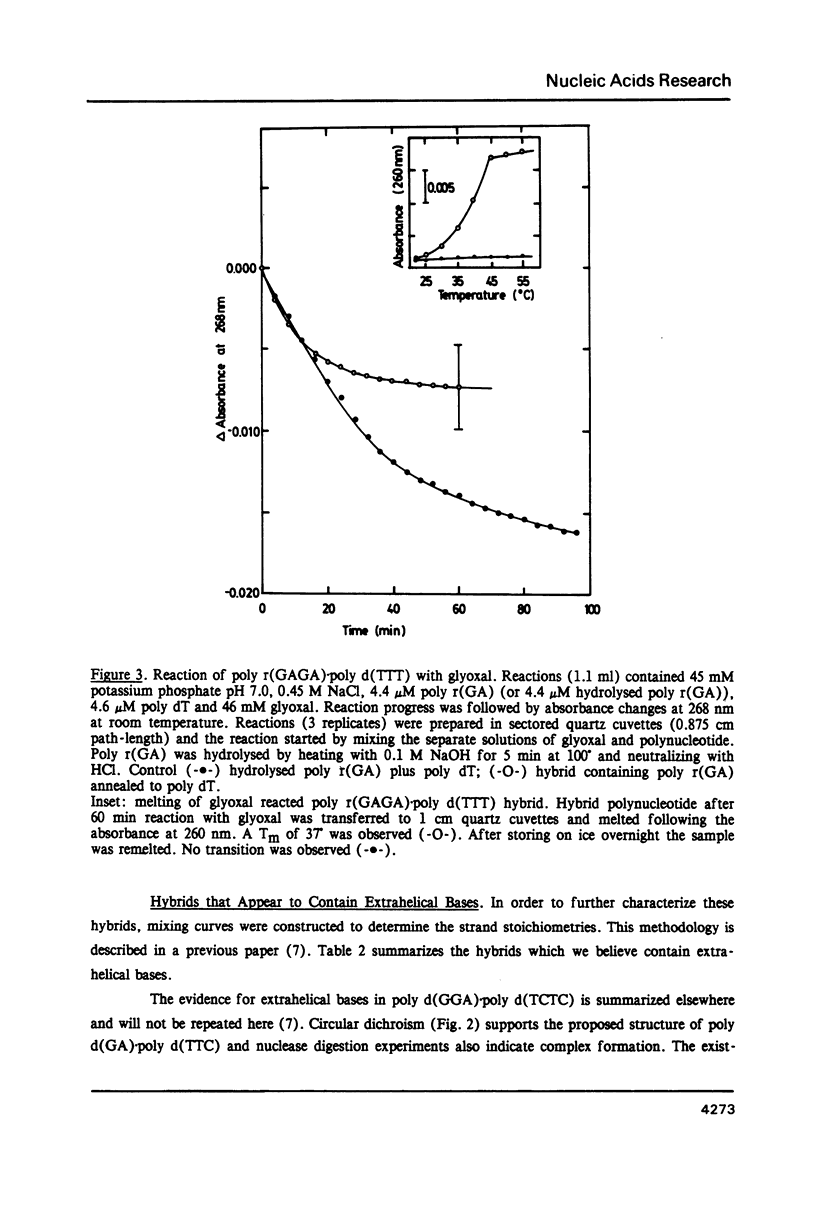
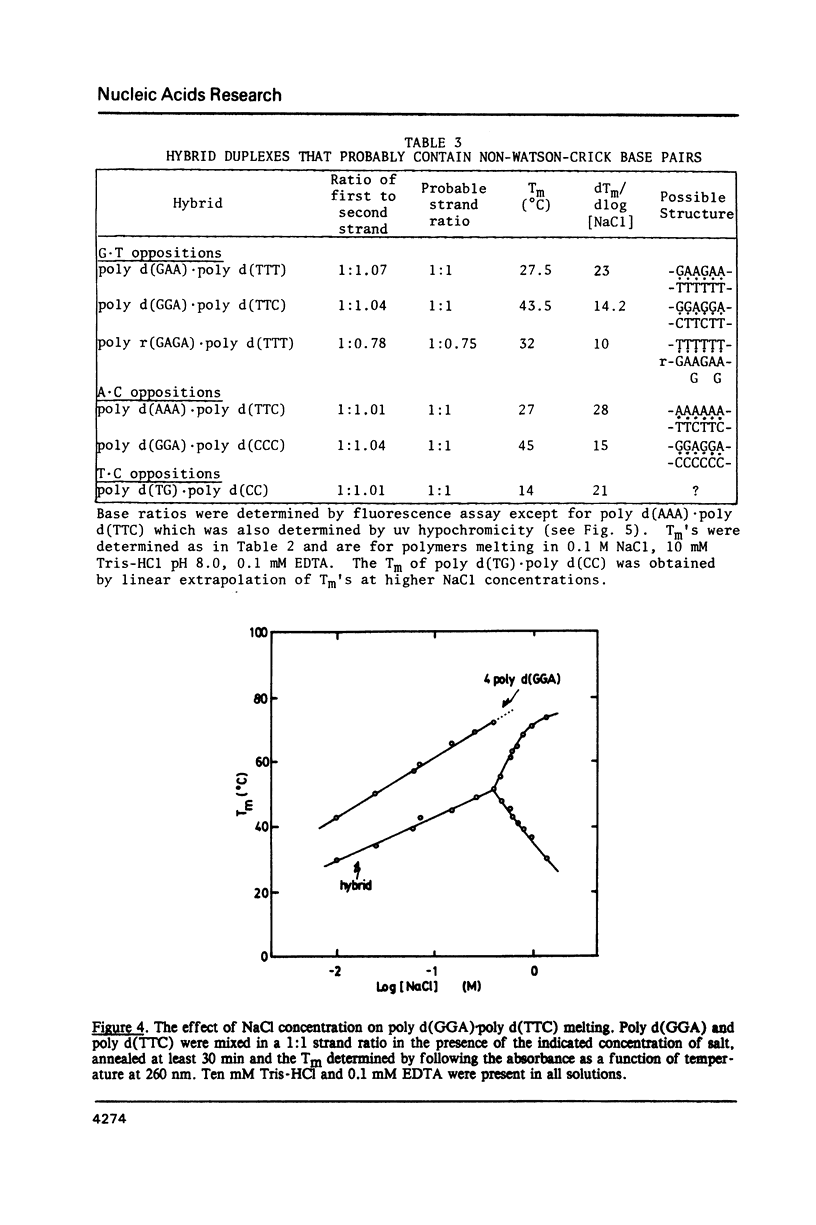
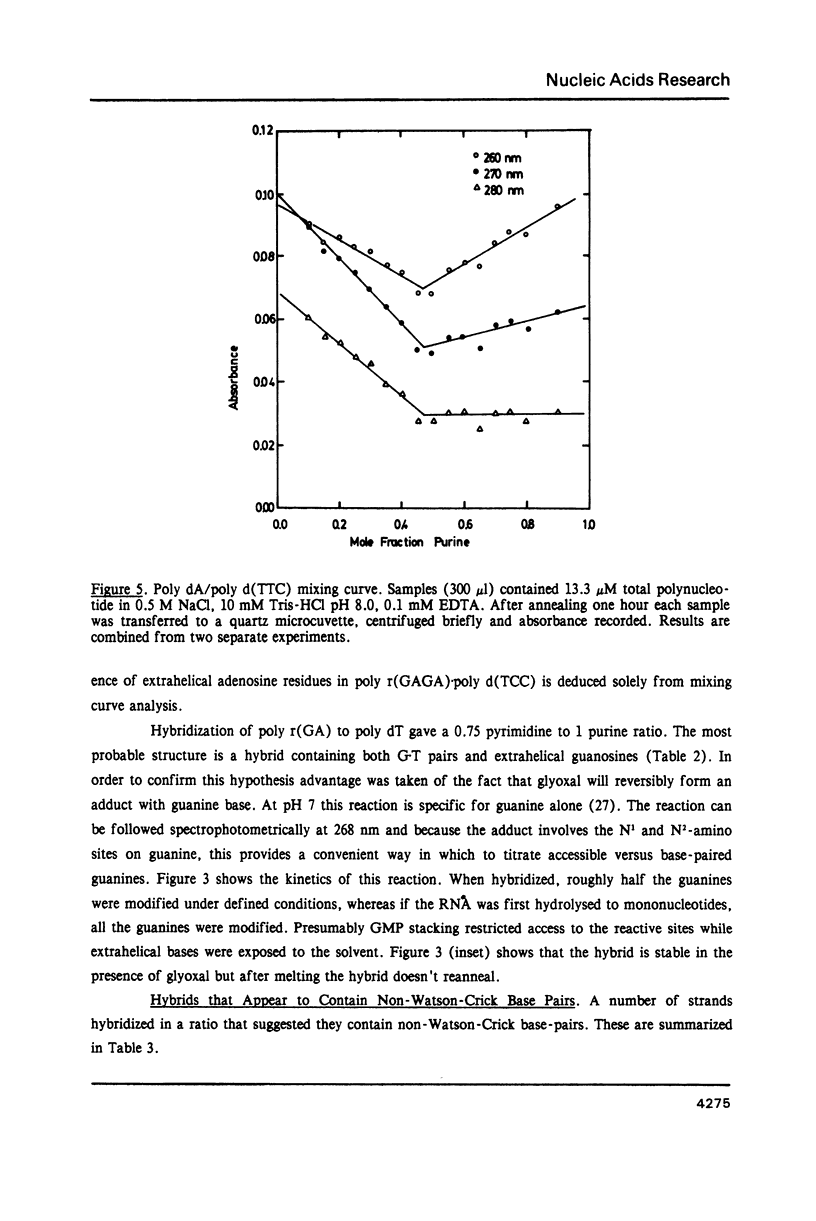

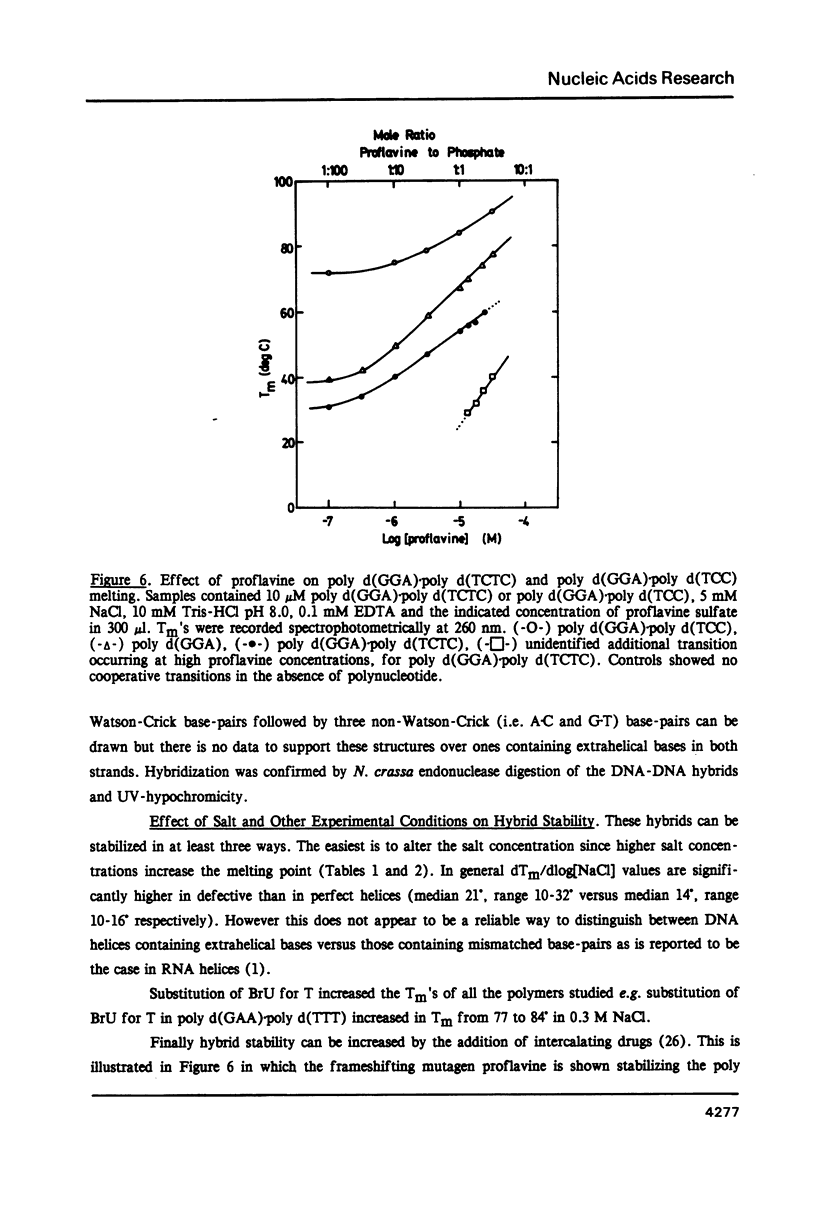



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baroudy B. M., Venkatesan S., Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982 Feb;28(2):315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- Broude N. E., Budowsky E. I. The reaction of glyoxal with nucleic acid components. 3. Kinetics of the reaction with monomers. Biochim Biophys Acta. 1971 Dec 30;254(3):380–388. doi: 10.1016/0005-2787(71)90868-9. [DOI] [PubMed] [Google Scholar]
- Brown T., Kennard O., Kneale G., Rabinovich D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature. 1985 Jun 13;315(6020):604–606. doi: 10.1038/315604a0. [DOI] [PubMed] [Google Scholar]
- Cullis P. M., Wolfenden R. Affinities of nucleic acid bases for solvent water. Biochemistry. 1981 May 26;20(11):3024–3028. doi: 10.1021/bi00514a006. [DOI] [PubMed] [Google Scholar]
- Dodgson J. B., Wells R. D. Synthesis and thermal melting behavior of oligomer-polymer complexes containing defined lengths of mismatched dA-dG and dG-dG nucleotides. Biochemistry. 1977 May 31;16(11):2367–2374. doi: 10.1021/bi00630a009. [DOI] [PubMed] [Google Scholar]
- Drake J. W., Baltz R. H. The biochemistry of mutagenesis. Annu Rev Biochem. 1976;45:11–37. doi: 10.1146/annurev.bi.45.070176.000303. [DOI] [PubMed] [Google Scholar]
- Early T. A., Olmsted J., 3rd, Kearns D. R., Lezius A. G. Base pairing structure in the poly d(G-T) double helix: wobble base pairs. Nucleic Acids Res. 1978 Jun;5(6):1955–1970. doi: 10.1093/nar/5.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. H., Lee J. S., Morgan A. R., Olsen R. K. A method for the specific inhibition of poly[d(A-T)] synthesis using the A-T specific quinoxaline antibiotic TANDEM. Can J Biochem. 1982 Feb;60(2):131–136. doi: 10.1139/o82-018. [DOI] [PubMed] [Google Scholar]
- Evans D. H., Morgan A. R. Extrahelical bases in duplex DNA. J Mol Biol. 1982 Sep;160(1):117–122. doi: 10.1016/0022-2836(82)90134-6. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism evidence for G-U and G-T base pairing in poly[r(G-U)] and poly[d(G-T)]. Biopolymers. 1977 Jun;16(6):1331–1342. doi: 10.1002/bip.1977.360160613. [DOI] [PubMed] [Google Scholar]
- Harwood S. J., Wells R. D. Micrococcus luteus deoxyribonucleic acid polymerase. Studies on the initiation of deoxyribonucleic acid synthesis in vitro. J Biol Chem. 1970 Nov 10;245(21):5625–5634. [PubMed] [Google Scholar]
- Helfgott D. C., Kallenbach N. R. Increased binding of ethidium bromide to polynucleotide duplexes containing mismatched bases. Nucleic Acids Res. 1979 Oct 25;7(4):1011–1017. doi: 10.1093/nar/7.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S., Okazaki R., Tamanoi F. Mechanism of DNA chain growth. XI. Structure of RNA-linked DNA fragments of Escherichia coli. J Mol Biol. 1973 Jul 15;77(4):501–517. doi: 10.1016/0022-2836(73)90219-2. [DOI] [PubMed] [Google Scholar]
- Ho P. S., Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. G.T wobble base-pairing in Z-DNA at 1.0 A atomic resolution: the crystal structure of d(CGCGTG). EMBO J. 1985 Dec 16;4(13A):3617–3623. doi: 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INMAN R. B. TRANSITIONS OF DNA HOMOPOLYMERS. J Mol Biol. 1964 Sep;9:624–637. doi: 10.1016/s0022-2836(64)80171-6. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., Englund P. T., Bertsch L. L. Enzymatic synthesis of deoxyribonucleic acid. XXVI. Physical and chemical studies of a homogeneous deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):2996–3008. [PubMed] [Google Scholar]
- Kallenbach N. R., Litwin S. Statistical analysis of spectral mixing curve data. J Mol Biol. 1971 Dec 28;62(3):608–611. doi: 10.1016/0022-2836(71)90159-8. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U. Unorthodox codon reading and the evolution of the genetic code. Cell. 1981 Feb;23(2):305–306. doi: 10.1016/0092-8674(81)90124-0. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Tinoco I., Jr Mutagen--oligonucleotide complexes with a bulged base as models for frameshift mutation. Nature. 1978 Aug 10;274(5671):609–610. doi: 10.1038/274609a0. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Evans D. H., Morgan A. R. Polypurine DNAs and RNAs form secondary structures which may be tetra-stranded. Nucleic Acids Res. 1980 Sep 25;8(18):4305–4320. doi: 10.1093/nar/8.18.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski R. J., Gray H. B., Jr, Robberson D. L. A sensitive endonuclease probe for lesions in deoxyribonucleic acid helix structure produced by carcinogenic or mutagenic agents. J Biol Chem. 1977 Dec 10;252(23):8740–8746. [PubMed] [Google Scholar]
- Lomant A. J., Fresco J. R. Structural and energetic consequences of noncomplementary base oppositions in nucleic acid helices. Prog Nucleic Acid Res Mol Biol. 1975;15(0):185–218. doi: 10.1016/s0079-6603(08)60120-8. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Uhlenbeck O. C., Doty P. Self-complementary oligoribonucleotides: adenylic acid-uridylic acid block copolymers. J Mol Biol. 1971 Apr 28;57(2):201–215. doi: 10.1016/0022-2836(71)90341-x. [DOI] [PubMed] [Google Scholar]
- Miller J. H. Carcinogens induce targeted mutations in Escherichia coli. Cell. 1982 Nov;31(1):5–7. doi: 10.1016/0092-8674(82)90398-1. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Coulter M. B., Flintoff W. F., Paetkau V. H. Enzymatic synthesis of deoxyribonucleic acids with repeating sequences. A new repeating trinucleotide deoxyribonucleic acid, d(T-C-C)n-d(G-G-A)n. Biochemistry. 1974 Apr 9;13(8):1596–1603. doi: 10.1021/bi00705a007. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Evans D. H., Lee J. S., Pulleyblank D. E. Review: ethidium fluorescence assay. Part II. Enzymatic studies and DNA-protein interactions. Nucleic Acids Res. 1979 Oct 10;7(3):571–594. doi: 10.1093/nar/7.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. R., Lee J. S., Pulleyblank D. E., Murray N. L., Evans D. H. Review: ethidium fluorescence assays. Part 1. Physicochemical studies. Nucleic Acids Res. 1979 Oct 10;7(3):547–569. doi: 10.1093/nar/7.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. R. Studies on polynucleotides. XCIV. Transcription of DNA's with repeating nucleotide sequences. J Mol Biol. 1970 Sep 28;52(3):441–466. doi: 10.1016/0022-2836(70)90412-2. [DOI] [PubMed] [Google Scholar]
- Murray N. L., Morgan A. R. Enzymatic and physical studies on the triplex dTn.dAn.rUn. Can J Biochem. 1973 Apr;51(4):436–449. doi: 10.1139/o73-051. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984 Aug;43(11):2663–2670. [PubMed] [Google Scholar]
- Patel D. J., Pardi A., Itakura K. DNA conformation, dynamics, and interactions in solution. Science. 1982 May 7;216(4546):581–590. doi: 10.1126/science.6280281. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Romaniuk P. J., Hughes D. W., Grégoire R. J., Bell R. A., Neilson T. Effects of internal nonbonded bases and a G.U base pair on the stability of a short ribonucleic acid helix. Biochemistry. 1979 Nov 13;18(23):5109–5116. doi: 10.1021/bi00590a013. [DOI] [PubMed] [Google Scholar]
- Solie T. N., Schellman J. A. The interaction of nucleosides in aqueous solution. J Mol Biol. 1968 Apr 14;33(1):61–77. doi: 10.1016/0022-2836(68)90281-7. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Tazawa I., Tazawa S., Ts'o P. O. Studies on oligonucleotides: thermodynamic and optical properties of the oligoinosinate-polycytidylate complexes. J Mol Biol. 1972 Apr 28;66(1):115–130. doi: 10.1016/s0022-2836(72)80010-x. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]


