Abstract
Biomass estimation of arbuscular mycorrhiza (AM) fungi, widespread plant root symbionts, commonly employs lipid biomarkers, predominantly the fatty acid 16:1ω5. We briefly reviewed the application of this signature fatty acid, followed by a case study comparing biochemical markers with microscopic techniques in an arable soil following a change to AM non-host plants after 27 years of continuous host crops, that is, two successive cropping seasons with wheat followed by amaranth. After switching to the non-host amaranth, spore biomass estimated by the neutral lipid fatty acid (NLFA) 16:1ω5 decreased to almost nil, whereas microscopic spore counts decreased by about 50% only. In contrast, AM hyphal biomass assessed by the phospholipid (PLFA) 16:1ω5 was greater under amaranth than wheat. The application of PLFA 16:1ω5 as biomarker was hampered by background level derived from bacteria, and further enhanced by its incorporation from degrading spores used as microbial resource. Meanwhile, biochemical and morphological assessments showed negative correlation for spores and none for hyphal biomass. In conclusion, the NLFA 16:1ω5 appears to be a feasible indicator for AM fungi of the Glomales group in the complex field soils, whereas the use of PLFA 16:1ω5 for hyphae is unsuitable and should be restricted to controlled laboratory studies.
1. Introduction
The chemotaxonomic use of lipids has a long tradition in microbiological research [1–3]. Due to the different enzymatic capabilities in lipid metabolism, fatty acids display a great structural diversity and biological specificity, providing an integrated and quantitative measure of microbial biomass and community structure in different environments [4]. In particularly, PLFAs have been employed in soil ecosystems as biomarkers for bacteria, saprotrophic fungi, and AM fungi; see Zelles [5] and Joergensen and Wichern [6] for detailed reviews. Moreover, as the lipid pattern of faunal consumers reflects the fatty acid composition of their diet, trophic biomarker fatty acids for major food resources in soil decomposers have been assigned [7].
Bacteria and fungi are important drivers of soil processes, predominantly nutrient mineralization and transfer to plants. Among the different mycorrhizal types, arbuscular fungi that form symbiosis with the roots of about 80% of all vascular plants are the dominant fungal symbionts that support plant growth [8, 9]. The AM fungal extraradical mycelium (ERM) spreads beyond the rhizosphere of host plants, providing additional surface area for the acquisition of phosphorus and nitrogen [10–12]. In recent years, global interest in sustainable agricultural practices has led to increase in the production and application of AM fungal inoculum in arable soils, which requires reliable methods for their quantification. AM fungi may occur naturally in arable soils, but their density and diversity may be increased by farm management practices such as fertilization or crop types [13–15].
Despite the importance of AM fungi for soil nutrient cycling, information on their distribution is inadequate due to analytical difficulties that are limiting the ability to study processes at the microscale. Four main approaches for quantifying fungal contribution to soil microbial communities are commonly used in soil ecology: (1) microscopic methods, (2) selective inhibition, (3) specific cell membrane components, and (4) specific cell wall components, with microscopic methods generally recording the lowest values [6]. Classical morphological estimation of AM fungal communities in soil includes membrane filter [16], grid line intersection [17], spore counts [3], and aqueous filtration extraction method and quantification of extraradical mycelium based on morphological criteria [18]. Molecular techniques have also been developed to determine AM fungi [19, 20], but difficulties involved in the sequencing as well as the absence of a sequence for some species constrain the use of this approach. More advanced, and particularly for quantification of AM fungal biomass, is the lipid biomarker technique that is being applied to determine the distribution of AM fungi in soil over the last two decades.
Olsson et al. [21, 22] were the first to use signature fatty acid 16:1ω5 regularly to assess growth and interactions of AM fungi in experimental soil-plant systems (Table 1). They proposed the 16:1ω5 as marker fatty acid, with the PLFA fraction assigning viable fungal hyphal biomass, and the NLFA fraction determining storage lipids such as spores [23]. This biomarker approach was also adopted for mycelial structures within plant roots, that is, the NLFA for energy storage in vesicles and the PLFA for membrane constituents such as intraradical mycelium or arbuscles [24, 25]. Based on this, both the 16:1ω5 PLFA and NLFA are widely used as indicators for AM fungi across soil ecosystems (Table 1). In a recent review, Joergensen and Wichern [6] proposed 345 as conversion factor of nmol PLFA to biomass C of AM fungi. This factor, a weighted mean based on literature data originating from four studies, can be criticized. However, it demonstrates the common use of 16:1ω5 PLFA in AM investigations, even though its biomarker value may be hampered.
Table 1.
Application of the marker fatty acid 16:1ω5 to assess the distribution of arbuscular mycorrhiza fungi in artificial and natural soil systems. NLFA: neutral lipid fatty acid, PLFA: phospholipid fatty acid.
| Authors | Lipid fraction and application (marker for) | Soil system |
|---|---|---|
| Olsson et al. [21] | NLFA-storage lipids | Plant mesocosms with γ radiated or autoclaved soil |
| Olsson et al. [22] | PLFA hyphae; conversion factor 38 for nmol PLFA to fungal hyphal length is given | |
|
| ||
| NLFA-energy storage (vesicles) | ||
| Larsen and Bødker [24] Van Aarleand Olsson [25] |
PLFA-membrane constituents (hyphae, arbuscles) | Mycelia structures in plant roots |
| NLFA/PLFA-storage status of fungi | ||
|
| ||
| Olsson and Wilhelmsson [26] | ||
| Hedlund [27] | ||
| Balser et al. [28] | ||
| Hebel et al. [29] | PLFA-hyphal biomass | Grassland, mixed-wood forest stands, arable land, sand dunes; burned forest soil |
| Huang et al. [30] | ||
| Royer-Tardif et al. [31] | ||
| Marshall et al. [32] | ||
|
| ||
| Olsson and Wilhelmsson [26] Hedlund [27] |
NLFA-storage lipids | Arable land, sand dunes |
| van Groenigen et al. [33] Yao and Wu [34] |
NLFA-fungal biomass | Arable land, grassland |
| Bradley et al. [35] | Total lipids (NLFA + PLFA) fungal biomass | Grassland |
|
| ||
| Aliasgharzad et al. [36] | NLFA/PLFA separation between arbuscular mycorrhizal fungi (high ratio) and bacteria (low ratio) | Seminatural sandy grassland |
|
| ||
| Olsson et al. [37] Schnoor et al. [38] |
NFLA-13C allocation in fungal storage lipids | Pot soil with plants in greenhouse |
Despite the proposed primary origin of 16:1ω5 PLFA, it may additionally be found, although in smaller amounts, in the lipids of other organisms, particularly in soil-inhabiting bacteria [39, 40]. On the other hand, the 16:1ω5 NLFA is not only present in spores but also forms the transport vehicle for carbon between intraradical and extraradical mycelium [41]. As assigned by 13C labelling studies, the fungus converts sugars taken up in the root compartment into lipids [42, 43], which can be used to assess the shift of carbon from roots into associated microbial communities without extraction, purification and identification of fungal mycelium [44, 45]. These stable isotope studies indicated that the NLFA 16:1ω5 is a good tool to assign AM fungal biomass in soil but showed no evidence for the application of the PLFA fraction. Despite these discrepancies several recent studies employed the PLFA marker for AM extraradical mycelium in field soils (Table 1).
This general application of 16:1ω5 as biomarker for AM fungi across different soil ecosystems calls for deeper insight to ensure reliability in the quantification of the fungal hyphae or spore biomass. We therefore performed a case study on the dynamics of AM fungal spores and extraradical mycelium comparing lipid biomarker and morphological approaches to determine the distribution of AM fungi in an arable field soil. A long-term fertilizer experiment was used, with the shift to a nonmycorrhizal host plant after 27 years of host crop cultivation. This unique experimental design offers the possibility to assess whether biochemical and microscopic techniques yield similar results under distinct changes in the mycorrhizal symbiosis in situ, without artificial manipulations (e.g., selective inhibition technique). Investigating the same plots in two successive vegetative periods allowed (i) to assign the diminishing of AM fungi in the absence of a host plant, (ii) to screen the background signal from the remaining vegetation period using both methods, and (iii) to determine correlations between the changes within biochemical marker and morphological assessments.
2. Materials and Methods
2.1. Field Site
The study was conducted at a long-term arable field site established in 1980 at the Institute of Biodynamic Research (IBDF) Darmstadt in Germany. The field site is located at 49°N, 8°E, and 100 m above sea level, with annual mean air temperature of 9.5°C and precipitation of 590 mm. The soil type is haplic cambisol comprising 87% sand, 8% silt, and 5% clay in the topsoil. The experimental setup was a two-factorial design amended with mineral (NPK) and organic (cattle manure + biodynamic preparations) fertilizers applied at low and high amounts. These were implemented in a split block design with four replicate plots of 5 m × 5 m each. Except fertilization, all other farming practices such as irrigation, tillage, and crop rotation were similar across the 27 years since the establishment of the long-term field experiment. Plots amended with mineral fertilizer received N, P, K applied in rates of kg ha−1y−1 as 60, 50, 75 (low), and 140, 100, and 125 (high), respectively. Organic plots received composted cattle manure with the addition of biodynamic preparations spread as solid fertilizer before ploughing and milling of the soil. The application rate was calculated to achieve similar nitrogen input as at the mineral fertilizer plots, which resulted in the variation of phosphorus and potassium amounts depending on the manure properties of a given year. On average, organic plots received less P (−25% for low and −38% for high) and 26% more K than mineral plots. For more details on farm management, see Ngosong et al. [46].
2.2. Sampling
Since the long-term field site was established, there has been 27 years continuous mycorrhizal host crop rotation including lupine angustifolius, winter rye, potato, spring wheat, and clover. The present investigations were conducted during two successive cropping seasons with the cultivation of spring wheat (Triticum aestivum cv. Passat) in 2007 and a shift to amaranth (Amaranthus hypochondriacus) in 2008. The former is a well-known host plant for AM fungi while the latter is recognized as non-host [8, 47]. Soil at the wheat plots was sampled four months after sowing and three weeks before crop harvest, while amaranth plots were sampled two months after sowing and two months before crop harvest. For examination of AM fungal morphological structures (hyphal length, spores) and lipids (PLFAs, NLFAs), one soil sample was taken from each replicate plot (n = 4 per treatment) at 0–5 and 5–10 cm depth using 5 cm diameter soil core. Soil was stored at −20°C prior to analyses. Additionally, random samples of wheat and amaranth roots were collected from the respective plots and analysed for infection by AM fungi.
2.3. Morphological AM Fungal Investigation
Morphological assessment of AM fungal structures comprised the colonization of crop roots, length of extraradical mycelium, and number of spores in the bulk soil. For the assessment of root infection rate, fine roots (0.7–1.0 g) were stained with trypan blue in lactic acid and the colonized roots assessed by modified intersection method with 250–300 intersections counted per sample [17]. Fungal spores were isolated from 80 g air-dried soil by sieving and decanting method, with subsequent sucrose gradient centrifugation [48], and counted using the agar film technique [49].
The hyphal length was estimated from soil using a modified membrane filtration technique. Soil sample (1.0 g) was homogenized with 100 mL deionized water in a laboratory mixer (Waring Commercial; Connecticut, USA) for 60 seconds. The suspension was poured through a 40-micrometer filter and washed carefully with water to eliminate fine soil particles. Remaining material was transferred into a petri dish and stained with a few drops of 0.05% trypan blue in lactic acid. The suspension was transferred into a glass beaker and diluted to 300 mL volume. A subsample of the suspension was filtered on a 0.45 μm mesh width membrane filter (MicronSep; GE Water & Process Technologies, USA) using a bottleneck filtration unit (NALGENE Reusable Bottle Top Filter Unit; Nalge Company, New York, USA). The membrane filter was mounted onto microscopic slides and observed under the microscope at 200x magnification, and the AM fungal hyphal length estimated by a modified gridline intersection method [50, 51].
2.4. Fatty Acid Analysis
Lipids were extracted from 4 g soil (wet weight) using the modified Bligh and Dyer method according to Frostegård et al. [52]. Fractionation into NLFAs, glycolipids, and PLFAs was performed using silica acid columns (HF BOND ELUT–SI, Varian Inc.), and the different fractions were eluded with chloroform, acetone, and methanol, respectively. Lipid methanolysis of PLFA and NLFA fractions was conducted in 0.2 M methanolic KOH, and methyl nonadecanoate (19 : 0) was added as internal standard; for more details see Ngosong et al. [53].
Fatty acid methyl esters (FAMEs) were identified by chromatographic retention time comparison with a standard mixture composed of 37 different FAMEs ranging from C11 to C24 (Sigma-Aldrich, St Louis, MO, USA). Analysis was performed by gas chromatography using a GC-FID Clarus 500 (PerkinElmer Corporation, Norwalk, USA) equipped with HP-5 capillary column (30 m × 0.32 mm i.d., film thickness 0.25 μm). To verify correct identification of FAMEs (chain length and saturation), a range of soil samples were analyzed by mass spectrometry using a 3400/Saturn4 Diontrap GC/MS system (Varian, Darmstadt, Germany), equipped with a HP-5 capillary column (50 m × 0.32 mm i.d., film thickness 0.17 μm). A mass range from 50 to 500 m/z was monitored twice a second in Scan mode; for more details see Ngosong et al. [46]. The signature fatty acid 16:1ω5 was used as biomarker for AM fungi, where the PLFA fraction represents fungal extraradical mycelium and the NLFA spore for biomass [23, 54, 55].
2.5. Statistical Analysis
The effects of crop plant shift on AM fungal fatty acid marker and morphological estimations were tested using STATISTICA 6.0 for Windows [56]. Data were subjected to nonparametric statistics using Kruskal-Wallis. Significant effects (P < 0.05) of the different factors are indicated in figures. Additionally, the Spearman Rank Order Correlations between biochemical and microscopically derived results were performed.
3. Results
The morphological examination of crop roots revealed 32–67% colonization of wheat by AM fungi, whereas the nonmycorrhizal host amaranth was not infected (data not presented). This clearly indicates the absence of an active amaranth-fungal symbiosis. In conformity, in the absence of a host plant, AM fungal spore biomass assessed by the NLFA 16:1ω5 almost disappeared in amaranth soils, with less than 0.1 nmol g−1 DW, compared to 0.9–7.9 nmol g−1 DW for wheat soils across depths (Figure 1(a)). Similarly, the microscopic counted spore numbers decreased, but only by 55%, and ranged between 121 and 205 and 87 and 125 spores g−1 DW soil for wheat and amaranth plots, respectively (Figure 1(b)). The relationship between NLFA and microscopic spore estimates was negatively correlated (r = −70, P < 0.05) across fertilizers, depths, and crop plant. Overall, the estimation of AM fungi in the arable soil using signature fatty acid and microscopic techniques mirrored the same trend, but to a different extent.
Figure 1.
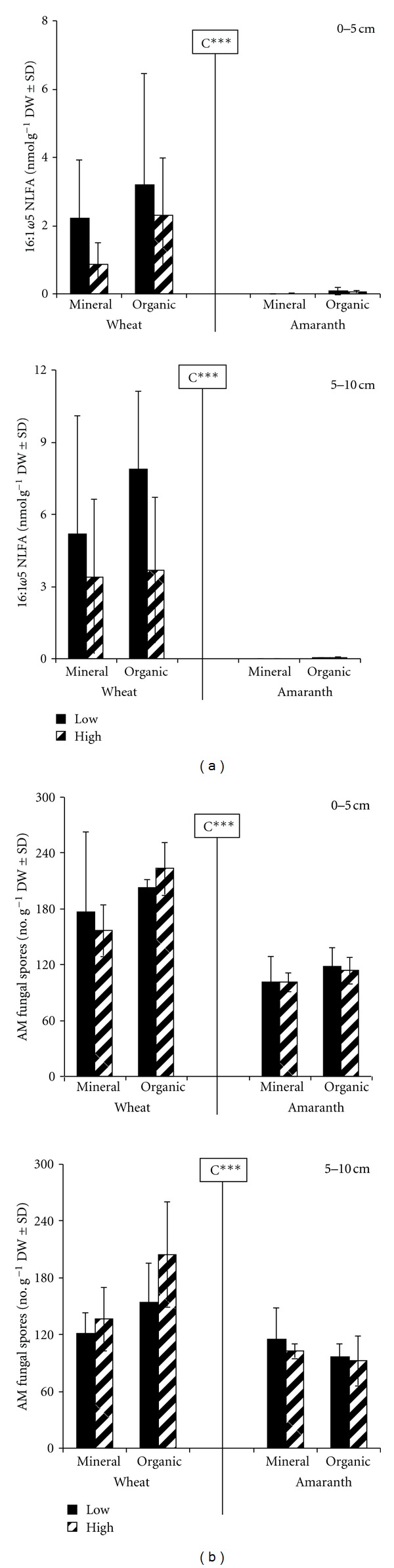
Comparison of microscopic and biochemical estimations of arbuscular mycorrhiza spores in an arable field soil cultivated with wheat or amaranth as crop; 0–5 and 5–10 cm soil depths; amendments with mineral (NPK) and organic (cattle manure + biodynamic preparations) fertilizers applied at low and high amounts; C = crop type; Kruskal-Wallis with *** for P < 0.001. (a) AM spores assessed by neutral lipid fatty acid 16:1ω5 (nmol g−1 DW soil ± SD), (b) Spore density counts (No. g−1 DW soil ± SD).
In contrast to AM fungal spores, hyphal biomass assessed by the marker PLFA 16:1ω5 increased significantly under amaranth with 0.4–1.1 nmol g−1 DW compared to 0.1–0.8 nmol g−1 DW for wheat soil; see Ngosong et al. [46]. This corresponds to an increase by 24–65% in the upper soil and 39–79% in the lower soil layer at amaranth plots. Meanwhile, AM hyphal length under amaranth determined morphologically ranged between 1.5 and 4.0 m g−1 DW and 1.8 and 2.5 m g−1 DW soil at 0–5 and 5–10 cm depth, respectively (Table 2). This contradicts the absence of amaranth root infection by the fungus, and the strong decrease in spore numbers at those plots. Nonetheless, there was no observed correlation (r = −0.13, P > 0.05) between morphological AM hyphal length and biochemical PLFA 16:1ω5 hyphal biomass under amaranth across fertilizer types and soil depth. In addition, there was no correlation (r = −0.10, P > 0.05) between wheat root infection rate and the PLFA 16:1ω5 at wheat plots.
Table 2.
Arbuscular mycorrhizal (AM) extraradical mycelium length and hyphal biomass in amaranth plots estimated morphologically and by phospholipid fatty acid biomarker 16:1ω5 (% DW soil ± SD), respectively, at 0–5 and 5–10 cm soil depths, amended with mineral (NPK) and organic (cattle manure + biodynamic preparations) fertilizers, applied at low and high amounts.
| Mineral | Organic | |||
|---|---|---|---|---|
| Low | High | Low | High | |
| 0–5 cm | ||||
| PLFA 16:1ω5 (nmol g−1 dry soil) | 0.43 ± 0.22 | 0.55 ± 0.22 | 0.63 ± 0.07 | 1.13 ± 0.66 |
| Hyphal length (m g−1 dry soil) | 3.96 ± 2.39 | 1.53 ± 0.70 | 2.08 ± 1.12 | 2.35 ± 1.36 |
|
| ||||
| 5–10 cm | ||||
| PLFA 16:1ω5 (nmol g−1 dry soil) | 0.51 ± 0.18 | 0.53 ± 0.3 | 0.76 ± 0.17 | 0.71 ± 0.14 |
| Hyphal length (m g−1 dry soil) | 2.46 ± 1.37 | 1.78 ± 1.16 | 1.82 ± 1.01 | 1.99 ± 1.71 |
4. Discussion
The present investigation focuses on the correlation of morphological and biochemical estimates of AM fungal dynamics in light of the shift from host to non-host crops. The response of microbial communities, including AM fungi, in relation to fertilizer type and amount as demonstrated by lipid data is discussed in detail elsewhere [46]. When comparing the NLFA signature fatty acid with microscopic estimations, both approaches mirrored the same trend but to a different extent. For the fatty acid, the decline in spore biomass without a host plant was severe with almost nil left, whereas the number of spores remaining was about 50%. Firstly, these differences may be due to low NLFA yield since Olsson [23] suggested that, for efficient extraction of lipids, the spore wall must be broken. On the other hand, Madan et al. [57] reported only small and nonsignificant impact when spores were crushed before analysis. Secondly, Olsson and Johansen [58] demonstrated that AM fungal hyphae contain a significant portion of the NLFA 16:1ω5 used for carbon transport in lipids. Since hyphae are decomposed much faster than spores, this may have contributed to the diminishing of the NLFA signal within one crop cycle. However, as spores form 90% of the external fungal tissue and 20% of spore mass is NLFA [58], the impact of lipids from extraradical hyphae appears rather low. Thirdly, the signature fatty acid 16:1ω5 is common in Glomales, whereas it is rare or lacking in other groups such as Gigaspora [54, 59]. For the latter, several long chain fatty acids such as 20:1ω9 20:2ω6 and 22:1ω9 have been proposed as biomarkers [57, 60]. Hence, the signature fatty acid partially reflects the dynamics of AM in soil, but not of the entire fungal population. This is supported by the observation of larger spores (e.g., Gigaspora) during microscopic examination, which cannot be detected by 16:1ω5. Fourthly, microscopic counts are constrained by the fact that newly formed fungal spores are not distinguishable from those formed earlier in the season [61, 62], resulting in potential leftovers from the previous crop. In sum, the NLFA 16:1ω5 reflected the decline of AM fungal spores after the change to a non-host crop, but it predominantly represented the Glomalen species within the population. Hence, it represents a reliable quantitative estimate of the fungal spore biomass when used in that regard, which is in line with recent studies that applied stable isotopes to assess carbon transfer from roots to AM fungi [44, 45].
In contrast to AM fungal spores, hyphal biomass assessed by the PLFA 16:1ω5 biomarker was higher in the non-host compared to the host crop soils. This is surprising as PLFAs are easily decomposed through enzymatic actions in soil and thus are assumed to reflect the occurrence of living organisms [2]. Meanwhile, the longevity of AM hyphae in soil has rarely been measured although it is assumed to be short. Staddon et al. [63] assigned a high turnover rate with an extraradical hyphal live from 5 to 6 days only. On the other hand, Steinberg and Rillig [64] reported that even under relatively favorable conditions for decomposition (18°C; 15% moisture) about 60% of hyphal length were still present 150 days after being separated from their host. However, there was no correlation of the PLFA estimates to hyphal length in amaranth soil, or to root infection rate at wheat plots, indicating a weak relationship between morphological and biochemical measurements. The significant increase of the PLFA 16:1ω5 at amaranth compared to wheat plots by up to 79% [46] suggests that bacteria used degrading spores as carbon source, thereby assimilating the marker fatty acid. Such trophic transfer of lipids between microorganisms and their substrates was frequently reported [7]. The extraradical mycelium has been assigned as large and rapid mycorrhizal pathway of carbon into other rhizosphere microorganisms [44, 45, 63]. Our results indicate that also fungal spores are attractive resources that form a considerable microbial carbon pool in the bulk soil.
In conclusion, investigating the development of AM fungi in an arable soil following the change of host crop revealed strong analytical discrepancies between biochemical and microscopic techniques. For the application of PLFA 16:1ω5, the background concentration derived from other soil organisms, and particularly bacteria can be too high to correctly quantify mycelium in microbial active soils. Meanwhile, the NLFA 16:1ω5 appears to be a reliable marker for AM fungal storage lipids such as spores, yet it cannot assign other than the Glomales group. Moreover, the occurrence of NLFA 16:1ω5 in extraradical mycelium in soil can superimpose on the overall signal. Clearly, the interpretation that NLFA biomarker arises solely from spores, and PLFA biomarker from mycelia is gross oversimplification. Meanwhile, the approach to combine both the phospholipid and neutral lipid fractions, as marker for AM fungi is no remedy, since it is hampered by the assimilation of signature fatty acid by other decomposers. Overall, these results strongly challenge the use of AM biomarkers, necessitating more comparative in situ based studies to identify their structural and functional origin, in order to effectively assign the dynamics of 16:1ω5 in complex field soils.
Acknowledgments
The authors extend gratitude to the counties Brandenburg, Thüringen, and Leibniz Society for financial support. They are grateful to J. Raupp for the possibility to use the long-term field experiment at the Institute of Biodynamic Research Darmstadt (IBDF) for our studies. They also thank M. Oltmanns (IBDF Darmstadt), M. Jarosch (IBDF Darmstadt), and S. Jeserigk (IGZ Großbeeren) for field and laboratory assistances, respectively.
References
- 1.Lechevalier H, Lechevalier MP. Chemotaxonomic use of lipids-an overview. In: Ratledge C, Wilkinson SG, editors. Microbial Lipids. London, UK: Academic Press; 1988. pp. 869–902. [Google Scholar]
- 2.Tunlid A, White DC. Use of lipid biomarkers in environmental samples. In: Fox A, Morgan SL, Larsson L, Odham G, editors. Analytical Microbiology Methods, Chromatography and Mass Spectrometry. New York, NY, USA: Plenum Press; 1990. [Google Scholar]
- 3.Koske RE, Gemma JN. A modified procedure for staining roots to detect VA mycorrhizas. Mycological Research. 1989;92:486–505. [Google Scholar]
- 4.White DC, Stair JO, Ringelberg DB. Quantitative comparisons of in situ microbial biodiversity by signature biomarker analysis. Journal of Industrial Microbiology. 1996;17(3-4):185–196. [Google Scholar]
- 5.Zelles L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biology and Fertility of Soils. 1999;29(2):111–129. [Google Scholar]
- 6.Joergensen RG, Wichern F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biology and Biochemistry. 2008;40(12):2977–2991. [Google Scholar]
- 7.Ruess L, Chamberlain PM. The fat that matters: soil food web analysis using fatty acids and their carbon stable isotope signature. Soil Biology and Biochemistry. 2010;42(11):1898–1910. [Google Scholar]
- 8.Smith SE, Read DJ. Mycorrhizal Symbioses. 3rd edition. London, UK: Academic Press; 2008. [Google Scholar]
- 9.Veresoglou SD, Menexes G, Rillig MC. Do arbuscular mycorrhizal fungi affect the allometric partition of host plant biomass to shoots and roots? A meta-analysis of studies from 1990 to 2010. Mycorrhiza. 2011;22:227–235. doi: 10.1007/s00572-011-0398-7. [DOI] [PubMed] [Google Scholar]
- 10.Neumann E, George E. Colonisation with the arbuscular mycorrhizal fungus Glomus mosseae (Nicol. & Gerd.) enhanced phosphorus uptake from dry soil in Sorghum bicolor (L.) Plant and Soil. 2004;261(1-2):245–255. [Google Scholar]
- 11.Govindarajulu M, Pfeffer PE, Jin H, et al. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature. 2005;435(7043):819–823. doi: 10.1038/nature03610. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka Y, Yano K. Nitrogen delivery to maize via mycorrhizal hyphae depends on the form of N supplied. Plant, Cell and Environment. 2005;28(10):1247–1254. [Google Scholar]
- 13.Ryan MH, Chilvers GA, Dumaresq DC. Colonisation of wheat by VA-mycorrhizal fungi was found to be higher on a farm managed in an organic manner than on a conventional neighbour. Plant and Soil. 1994;160(1):33–40. [Google Scholar]
- 14.Gavito ME, Miller MH. Changes in mycorrhiza development in maize indeed by crop management practices. Plant and Soil. 1998;198(2):185–192. [Google Scholar]
- 15.Oehl F, Sieverding E, Mäder P, et al. Impact of long-term conventional and organic farming on the diversity of arbuscular mycorrhizal fungi. Oecologia. 2004;138(4):574–583. doi: 10.1007/s00442-003-1458-2. [DOI] [PubMed] [Google Scholar]
- 16.Abbott LK, Robson AD, De Boer G. The effects of phosphorus on the formation of hyphae in soil by the vesicular arbuscular mycorrhizal fungus Glomus fasiculatum . New Phytologist. 1984;97:437–446. [Google Scholar]
- 17.Kormanik P, McGraw AC. Quantification of vesicular-arbuscular mycorrhizae in plant roots. In: Schenck NC, editor. Methods and Principals of Mycorrhizal Research. Minn, USA: The American Phytopathological Society; 1982. pp. 37–45. [Google Scholar]
- 18.Olsson PA, Van Aarle IM, Gavito ME, Bengtson P, Bengtsson G. 13C incorporation into signature fatty acids as an assay for carbon allocation in arbuscular mycorrhiza. Applied and Environmental Microbiology. 2005;71(5):2592–2599. doi: 10.1128/AEM.71.5.2592-2599.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redecker D. Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza. 2000;10(2):73–80. [Google Scholar]
- 20.Schwarzott D, Schüßler A. A simple and reliable method for SSU rRNA gene dna extraction, amplification, and cloning from single AM fungal spores. Mycorrhiza. 2001;10(4):203–207. [Google Scholar]
- 21.Olsson PA, Bååth E, Jakobsen I, Soderstrom B. The use of phospholipid and neutral lipid fatty acids to estimate biomass of arbuscular mycorrhizal fungi in soil. Mycological Research. 1995;99(5):623–629. [Google Scholar]
- 22.Olsson PA, Francis R, Read DJ, Söderström B. Growth of arbuscular mycorrhizal mycelium in calcareous dune sand and its interaction with other soil microorganisms as estimated by measurement of specific fatty acids. Plant and Soil. 1998;201(1):9–16. [Google Scholar]
- 23.Olsson PA. Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiology Ecology. 1999;29(4):303–310. [Google Scholar]
- 24.Larsen J, Bødker L. Interactions between pea root-inhabiting fungi examined using signature fatty acids. New Phytologist. 2001;149(3):487–493. doi: 10.1046/j.1469-8137.2001.00049.x. [DOI] [PubMed] [Google Scholar]
- 25.Van Aarle IM, Olsson PA. Fungal lipid accumulation and development of mycelial structures by two arbuscular mycorrhizal fungi. Applied and Environmental Microbiology. 2003;69(11):6762–6767. doi: 10.1128/AEM.69.11.6762-6767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson PA, Wilhelmsson P. The growth of external AM fungal mycelium in sand dunes and in experimental systems. Plant and Soil. 2000;226(2):161–169. [Google Scholar]
- 27.Hedlund K. Soil microbial community structure in relation to vegetation management on former agricultural land. Soil Biology and Biochemistry. 2002;34(9):1299–1307. [Google Scholar]
- 28.Balser TC, Treseder KK, Ekenler M. Using lipid analysis and hyphal length to quantify AM and saprotrophic fungal abundance along a soil chronosequence. Soil Biology and Biochemistry. 2005;37(3):601–604. [Google Scholar]
- 29.Hebel CL, Smith JE, Cromack K. Invasive plant species and soil microbial response to wildfire burn severity in the Cascade Range of Oregon. Applied Soil Ecology. 2009;42(2):150–159. [Google Scholar]
- 30.Huang Y, Michel K, An S, Zechmeister-Boltenstern S. Changes in microbial-community structure with depth and time in a chronosequence of restored grassland soils on the Loess Plateau in north-west China. Journal of Plant Nutrition Soil Science. 2011;174:765–774. [Google Scholar]
- 31.Royer-Tardif S, Bradley RL, Parsons WFJ. Evidence that plant diversity and site productivity confer stability to forest floor microbial biomass. Soil Biology and Biochemistry. 2010;42(5):813–821. [Google Scholar]
- 32.Marshall CB, McLaren JR, Turkington R. Soil microbial communities resistant to changes in plant functional group composition. Soil Biology and Biochemistry. 2011;43(1):78–85. [Google Scholar]
- 33.van Groenigen KJ, Bloem J, Bååth E, et al. Abundance, production and stabilization of microbial biomass under conventional and reduced tillage. Soil Biology and Biochemistry. 2010;42(1):48–55. [Google Scholar]
- 34.Yao H, Wu F. Soil microbial community structure in cucumber rhizosphere of different resistance cultivars to fusarium wilt. FEMS Microbiology Ecology. 2010;72(3):456–463. doi: 10.1111/j.1574-6941.2010.00859.x. [DOI] [PubMed] [Google Scholar]
- 35.Bradley K, Drijber RA, Knops J. Increased N availability in grassland soils modifies their microbial communities and decreases the abundance of arbuscular mycorrhizal fungi. Soil Biology and Biochemistry. 2006;38(7):1583–1595. [Google Scholar]
- 36.Aliasgharzad N, Mårtensson LM, Olsson PA. Acidification of a sandy grassland favours bacteria and disfavours fungal saprotrophs as estimated by fatty acid profiling. Soil Biology and Biochemistry. 2010;42(7):1058–1064. [Google Scholar]
- 37.Olsson PA, Rahm J, Aliasgharzad N. Carbon dynamics in mycorrhizal symbioses is linked to carbon costs and phosphorus benefits. FEMS Microbiology Ecology. 2010;72(1):125–131. doi: 10.1111/j.1574-6941.2009.00833.x. [DOI] [PubMed] [Google Scholar]
- 38.Schnoor TK, Mårtensson LM, Olsson PA. Soil disturbance alters plant community composition and decreases mycorrhizal carbon allocation in a sandy grassland. Oecologia. 2011;167:809–819. doi: 10.1007/s00442-011-2020-2. [DOI] [PubMed] [Google Scholar]
- 39.Nichols P, Stulp BK, Jones JG, White DC. Comparison of fatty acid content and DNA homology of the filamentous gliding bacteria Vitreoscilla, Flexibacter, Filibacter . Archives of Microbiology. 1986;146(1):1–6. [Google Scholar]
- 40.Zelles L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere. 1997;35(1-2):275–294. doi: 10.1016/s0045-6535(97)00155-0. [DOI] [PubMed] [Google Scholar]
- 41.Bago B, Pfeffer PE, Zipfel W, Lammers P, Shachar-Hill Y. Tracking metabolism and imaging transport in arbuscular mycorrhizal fungi. Plant and Soil. 2002;244(1-2):189–197. [Google Scholar]
- 42.Pfeffer PE, Douds DD, Bécard G, Shachar-Hill Y. Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiology. 1999;120(2):587–598. doi: 10.1104/pp.120.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gavito ME, Olsson PA. Allocation of plant carbon to foraging and storage in arbuscular mycorrhizal fungi. FEMS Microbiology Ecology. 2003;45(2):181–187. doi: 10.1016/S0168-6496(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 44.Olsson PA, Johnson NC. Tracking carbon from the atmosphere to the rhizosphere. Ecology Letters. 2005;8(12):1264–1270. [Google Scholar]
- 45.Drigo B, Pijl AS, Duyts H, et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2 . Proceedings of the National Academy of Sciences of the United States of America. 2010;107(24):10938–10942. doi: 10.1073/pnas.0912421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngosong C, Jarosch M, Raupp J, Neumann E, Ruess L. The impact of farming practice on soil microorganisms and arbuscular mycorrhizal fungi: crop type versus long-term mineral and organic fertilization. Applied Soil Ecology. 2010;46(1):134–142. [Google Scholar]
- 47.Peterson RL, Ashford AE, Allaway WG. Vesicular-arbuscular mycorrhizal associations of vascular plants on Heron Island, a Great Barrier Reef coral cay. Australian Journal of Botany. 1985;33(6):669–676. [Google Scholar]
- 48.Ianson DC, Allen MF. The effect of soil texture on extraction of vesicular-arbuscular mycorrhizal spores from arid soils. Mycologia. 1986;78:168–164. [Google Scholar]
- 49.Thomas A, Nicholas DP, Parkinson D. Modifications of the agar film technique for assaying lengths of mycelium in soil. Nature. 1965;205(4966):p. 105. [Google Scholar]
- 50.Newman EI. A method of estimating the total length of root in a sample. Journal of Applied Ecology. 1966;3:139–145. [Google Scholar]
- 51.Tennant D. A test of a modified line intersect method of estimating root length. Journal of Ecology. 1975;63:995–1001. [Google Scholar]
- 52.Frostegård Å, Tunlid A, Bååth E. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Applied and Environmental Microbiology. 1993;59(11):3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ngosong C, Raupp J, Scheu S, Ruess L. Low importance for a fungal based food web in arable soils under mineral and organic fertilization indicated by Collembola grazers. Soil Biology and Biochemistry. 2009;41(11):2308–2317. [Google Scholar]
- 54.Graham JH, Hodge NC, Morton JB. Fatty acid methyl ester profiles for characterization of glomalean fungi and their endomycorrhizae. Applied and Environmental Microbiology. 1995;61(1):58–64. doi: 10.1128/aem.61.1.58-64.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olsson PA, Larsson L, Bago B, Wallander H, Van Aarle IM. Ergosterol and fatty acids for biomass estimation of mycorrhizal fungi. New Phytologist. 2003;159(1):7–10. doi: 10.1046/j.1469-8137.2003.00810.x. [DOI] [PubMed] [Google Scholar]
- 56. StatSoft, Statistica. Version 6.0 for windows, StatSoft, Tusla, Okla, USA, 2001.
- 57.Madan R, Pankhurst C, Hawke B, Smith S. Use of fatty acids for identification of AM fungi and estimation of the biomass of AM spores in soil. Soil Biology and Biochemistry. 2002;34(1):125–128. [Google Scholar]
- 58.Olsson PA, Johansen A. Lipid and fatty acid composition of hyphae and spores of arbuscular mycorrhizal fungi at different growth stages. Mycological Research. 2000;104(4):429–434. [Google Scholar]
- 59.Bentivenga SP, Morton JB. Stability and heritability of fatty acid methyl ester profiles of glomalean endomycorrhizal fungi. Mycological Research. 1994;98(12):1419–1426. [Google Scholar]
- 60.Sakamoto K, Iijima T, Higuchi R. Use of specific phospholipid fatty acids for identifying and quantifying the external hyphae of the arbuscular mycorrhizal fungus Gigaspora rosea . Soil Biology and Biochemistry. 2004;36(11):1827–1834. [Google Scholar]
- 61.Lee Pau Ju, Koske RE. Gigaspora gigantea: seasonal abundance and ageing of spores in a sand dune. Mycological Research. 1994;98(4):453–457. [Google Scholar]
- 62.Clapp JP, Young JP, Merryweather JW, Fitter AH. Diversity of fungal symbionts in arbuscular mycorrhizas from a natural community. New Phytologist. 1995;130(2):259–265. [Google Scholar]
- 63.Staddon PL, Ramsey CB, Ostle N, Ineson P, Fitter AH. Rapid turnover of hyphae of mycorrhizal fungi determined by AMS microanalysis of 14C. Science. 2003;300(5622):1138–1140. doi: 10.1126/science.1084269. [DOI] [PubMed] [Google Scholar]
- 64.Steinberg PD, Rillig MC. Differential decomposition of arbuscular mycorrhizal fungal hyphae and glomalin. Soil Biology and Biochemistry. 2003;35(1):191–194. [Google Scholar]


