Abstract
This paper describes an approach to providing computer-interpretable logical definitions for the terms of the Human Phenotype Ontology (HPO) using PATO, the ontology of phenotypic qualities, to link terms of the HPO to the anatomic and other entities that are affected by abnormal phenotypic qualities. This approach will allow improved computerized reasoning as well as a facility to compare phenotypes between different species. The PATO mapping will also provide direct links from phenotypic abnormalities and underlying anatomic structures encoded using the Foundational Model of Anatomy, which will be a valuable resource for computational investigations of the links between anatomical components and concepts representing diseases with abnormal phenotypes and associated genes.
Phenotypic analysis plays a fundamental role in human genetics diagnostics and research. Phenotypic descriptions in publications describing new disease genes or genotype-phenotype correlations in known syndromes have generally relied on free text descriptions of phenotypic features. In recent years, computational analysis of the spectrum of phenotypic features found in human disease, the so-called ”phenome” [8], has been taken up by a number of groups for a number of goals including prioritizing candidate disease genes [7] and investigating modularity of the genetic disease-phenotype network [6]. In order to fully realize the potential of computational phenome analysis and to optimally use phenotypic information for clinical purposes, there is a clear need for a standardized way of communicating phenotypic information in medical reports, publications, and databases. It is also desirable to be able to compare phenotypic data accross species. In this report, we describe the application of the entity/quality decomposition methodology to the Human Phenotype Ontology in the realm of musculoskeletal phenotypic abnormalities.
I. The Human Phenotype Ontology
Currently, the Online Mendelian Inheritance in Man (OMIM) database [1] is the most important source of information about Mendelian genetic diseases in humans. However, the fact that OMIM does not employ a controlled vocabulary has made computational analysis of this data difficult. The Human Phenotype Ontology (HPO) was initially constructed to comprise terms corresponding to all descriptions used two or more times in OMIM as well as many descriptions used only once. In the process of constructing the HPO, synonyms were merged and domain knowledge was used to create links between the terms. In contrast to the shallow hierarchical structure of the OMIM clinical synopsis section, the leaf nodes of the HPO are typically 5 to 10 levels deep and are organized as a directed acyclic graph (DAG) in which terms may have multiple parent terms. Terms are related to parent terms by is a relationships. The HPO currently provides roughly 8000 terms, each of which describes a single human phenotypic abnormality. Approximately 50,000 annotations to nearly 5,000 mainly Mendelian diseases listed in OMIM are provided, and annotations to other classes of human disease such as chromosomal disorders are currently in preparation (Fig. 1) [9].
Fig. 1.
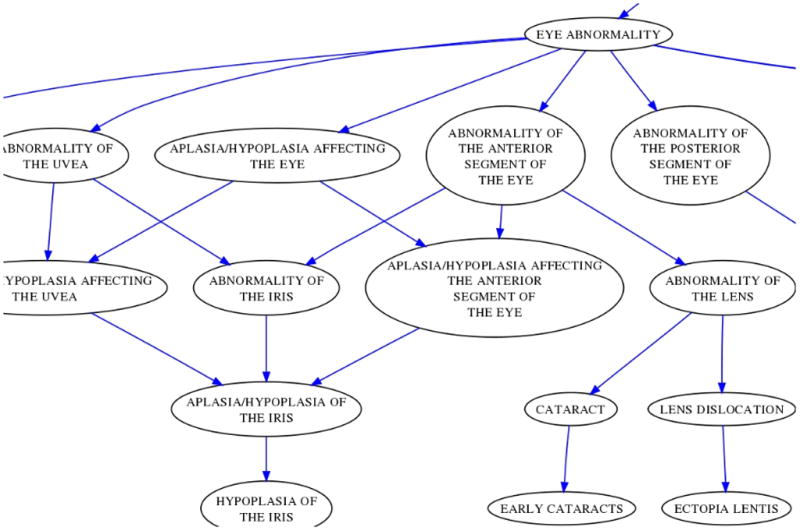
An excerpt of the HPO subgraph of terms used to annotate Marfan syndrome.
Terms in the HPO all possess names that correspond to current medical usage. The syntax of terms in the HPO follow two basic patterns. The first type contains words or terms reflecting the qualities of an entity, for example Mitral valve prolapse (HP:0001634). This term can be broken down into the anatomical entity term Mitral valve and the quality term prolapsed.
The second type of term in HPO contains intrinsic anatomical predicates as well as qualities, often pre-composed into a single canonical term. For instance, the term Arachnodactyly (HP:0001166) consists of a single word which is familiar to most physicians rather than naming a physical entity that has an abnormal quality. However, looking to the definition of the term, we find that Arachnodactyly refers to abnormally long and slender fingers (the word was coined from Greek words meaning spider fingers), that is, there is an abnormal quality (long and slender) of an entity (fingers).
II. PATO: An ontology of phenotypic qualities
PATO is an ontology of phenotypic qualities, intended for use in a number of applications, primarily phenotype annotation. PATO consists of a single hierarchy of qualities and currently offers 2014 terms. PATO is designed to be used in conjunction with ontologies of ”quality-bearing entities”, prominently including ontologies of anatomical entities such as the Foundational Model of Anatomy.(FMA) ontology [11], GO [3], or the cell type ontology [4]. We can use PATO in combination with one of these ontologies to create composite terms. For instance, to describe a ”red-eye” phenotype in Drosophila, we can combine the PATO term red with the Drosophila gross anatomy (FBbt) term ”eye”.
We say that we are ”composing” (or ”coordinating”) the description by using existing terms as elements of the description. Sometimes the composition is done at the time of annotation, in which case it is referred to as ”post-composition”.
There are a number of advantages to associating ontology terms with EQ descriptions, as we shall see in the following sections.
III. Decomposing the Human Skeletal Phenome
There are a number of advantages to composing phenotype descriptions including the fact that it is easier to use inference algorithms if the ”meanings” of the terms of an ontology are broken down into components that allow computerized reasoning as well as the ability to compare phenotypes between different species if a mapping between the respective anatomy ontologies is available. However, a serious disadvantage of this approach is the fact that a post-composed terminology does not always reflect the vocabulary of physicians and scientists involved in medical research, as was mentioned above. Therefore, the HPO has adopted a policy of retaining the medical terminology in common use for the term names, while also providing equivalence mappings to EQ-based compositional descriptions of the terms that can be used for inference or cross-species comparisons. Because of the difficulties in providing exact definitions for medical terms and defining pathophysiologically relevant semantic relationships between them, such a decomposition of HPO terms is a difficult but intellectually rewarding task that requires manual expert curation. The HPO and PATO teams are collaborating on a decomposition of the musculoskeletal subontology of the HPO as a pilot project to identify the curation and annotation strategies that will best work for decomposing the entire human phenome, which will be an ongoing project. In the following, we provide some example decompositions to illustrate our approach.
The majority of terms in the HPO describe abnormalities of anatomic structures. For these HPO terms, therefore, terms from the FMA human anatomy ontology are used to identify the affected entity (bearer of the abnormal quality). An appropriate PATO term is then chosen to describe the abnormal quality that the anatomic structure possesses, which can be described either in qualitative or quantitative terms, and are said to inhere in the bearer. For instance, for the HPO term Reduced bone mineral density (HP:0004349), we combine the FMA term for bone with the PATO term for decreased density. This combination is expressed in obo format notation1 as follows:
[Term] id: HP:0004349 ! Reduced bone mineral density intersection_of: PATO:0001790 ! decreased density intersection_of: inheres_in FMA:30317 ! bone
This states that the phenotype denoted by HP:0004349 is equivalent to all instances of decreased density occurring in bones.
Another example is the decomposition of the HPO term Amyotrophy (HP:0003202) (which is a medical term meaning ”muscular atrophy” or wasting of muscles):
[Term] id: HP:0003202 ! Amyotrophy intersection_of: PATO:0001623 ! atrophied intersection_of: inheres_in FMA:30316 ! muscle
The annotation model can additionally include further qualifiers concerning the developmental stage at which a phenotype holds or qualifiers indicating the expressivity of the phenotype with respect to some baseline (e.g., ”abnormal”). For instance, the HPO term Osteosclerosis refers to an abnormal increase in bone density. The intersection of the FMA term for bone with the PATO term increased density does not faithfully reflect the meaning of the HPO term Ostersclerosis, because not all increased density is abnormal. We therefore add a further intersection with the PATO term pathological:
[Term]
id: HP:0002796 ! Osteosclerosis
intersection_of: PATO:0001788 ! increased density
intersection_of: has_quality PATO:0001869 !
pathological
intersection_of: inheres_in FMA:30317 !
Bone
Once an HPO term has been logically defined in this way, other decompositions can make reference to it. For instance, the HPO term Patchy osteosclerosis refers to irregular areas with increased in bone density such as can be seen in some hereditary disorders such as Hypothyroidism-retardatino-dysmorphism syndrome caused by mutations in the gene encoding tubulin-specific chaperone E [MIM 241410]. Here we represent this as a ”quality aggregate”, using the has part relation to collect the sub-components of the phenotype together:
[Term]
id: HP:0005686 ! Patchy osteosclerosis
intersection_of: PATO:0000001 ! quality
intersection_of: has_part PATO:0001608 ! patchy
intersection_of: has_part HP:0002796 !
Osteosclerosis
There are many cases in which medical terminology does not reflect modern notions about pathophysiology or etiology of disease, meaning that any computational inference techniques that rely on natural language processing techniques to infer the meaning of a term would be doomed to failure. One example is the HPO term Spinal muscular atrophy (HP:0007269), which, despite the name, does not refer to atrophy affecting specifically the muscles of the spine. Rather, spinal muscular atrophy refers to muscular weakness and atrophy related to loss of the motor neurons of the spinal cord and brainstem. This has been decomposed using a relational quality using the FMA terms Motor neuron and Spinal cord. In some cases, this part of the ”phenotype” will be observed by neuropathological analysis [2], although it may be merely inferred in other patients based on clinical findings. The inevitable result of a reduction of the number of motor neurons is amyotrophy of the innervated muscles, and this is encoded by the further intersection of the HPO term Amyotrophy. The full decomposition is:
[Term] id: HP:0007269 ! Spinal muscular atrophy intersection_of: PATO:0002001 ! has fewer parts of type intersection_of: towards FMA:83617 ! motor neuron intersection_of: inheres_in FMA:13478 ! Vertebral column intersection_of: results_in HP:0003202 ! Amyotrophy
IV. Cross-Species Comparisons
The ability to manipulate the mouse genome has rendered the mouse one of the most important model organisms for studying human disease. In the gene driven approach to model discovery targeted mutants are made in specific genes associated with disease in man. In the phenotype driven approach the phenotypes of known or unknown mutations are screened for similarities with human diseases thereby gaining insight into their pathogenesis and genetic aetiology. In both cases establishment of the accurate relationship of mouse phenotypes to human diseases is essential. Although many mouse models display phenotypes that are reminiscent of the phenotypes of humans with inherited mutations at the same genes, important differences between human and mouse phenotypes resulting from mutation in homologous genes are frequently observed [10], [12]. Bridging the gap between mouse phenotypes and human diseases is therefore problematical; partly because formal disease nomenclature differs between mouse and man, but more importantly because not all of the aspects of cognate diseases will be manifested in both species. One approach to discovering a disease model is therefore to break down the summative ( precomposed) diagnosis into its component parts and to search for matches within the resulting pool of constituent phenotype elements across both species. The type of decomposition presented in this paper will provide an important impetus in this direction. As an example, consider the MPO term thymus hypoplasia (MP:0001823) [13], which is defined as ”underdevelopment or reduced size, usually due to a reduced cell number, in the thymus”. This can be decomposed using PATO as the cross product of the mouse anatomy ontology term thymus (MA:0000142) and the PATO term hypoplastic (PATO:0000645), which is defined as ”Underdevelopment or incomplete development of a tissue or organ”. Likewise, the HPO term Thymus hypoplasia (HP:0000778) can be decomposed using the FMA term for thymus (FMA:9607) and the PATO term hypoplastic (PATO:0000645). A number of groups are working on mappings between mouse and human anatomy (e.g., [5], [15]), which can be used together with the the PATO E/Q decompositions developed in our project to search for similar phenotypic abnormalities shared by mouse and human on a systematic level.
V. Conclusions
In this report, we have presented our methodology for providing an E/Q decomposition of the HPO. Currently, approximately 1000 HPO terms have been decomposed, and work on the remaining musculoskeletal terms is expected to be finished shortly. This will be an important step towards linking the HPO to well-established ontologies in the anatomy and molecular biology research communities. This will provide a consistent mapping of anatomical components to diseases and abnormal phenotypes, which will enable the use of clinical data for basic and translational computational biology research. In addition to the links to the FMA described above, we are working on linking the appropriate HPO terms to other ontologies. For instance, the HPO term Glucosephosphate isomerase deficiency (HP:0003290) has been decomposed as the intersection of the PATO term decreased (PATO:0001997) and the GO term glucose-6-phosphate isomerase activity (GO:0004347). Similar decompositions are being made using other ontologies, and other links to pathology ontologies [14] are planned. It is hoped that these refinements will make the HPO useful not only for human geneticists and other physicians interested in phenotypic analysis, but also to molecular biologists and bioinformaticians who are interested in incorporating the human phenotype into investigations on cellular networks and related topics. The HPO is freely available at http://www.human-phenotype-ontology.org.
Acknowledgments
The authors gratefully acknowledge the contributions of Denise Horn and Stefan Mundlos towards improving the structure and terms of the musculoskeletal portion of the HPO. This work was supported by the Deutsche Forschungsgemeinschaft (DFG RO 2005/4-1, SFB 760) and the Berlin-Brandenburg Center for Regenerative Therapies (BCRT) (Bundesministerium für Bildung und Forschung, project number 0313911).
This work was supported by the DFG and the BMBF
Footnotes
Contributor Information
Georgios V. Gkoutos, Email: gg295@gen.cam.ac.uk, Department of Genetics, University of Cambridge, Downing Street, Cambridge, CB2 3EH, England
Chris Mungall, Lawrence Berkeley National Laboratory, Berkeley, California, USA.
Sandra Dölken, Institute for Medical Genetics, Charité Universitätsmedizin Berlin, Augustenburger Platz 1, 13353 Berlin Germany.
Michael Ashburner, Department of Genetics, University of Cambridge, Downing Street, Cambridge, CB2 3EH, England.
Suzanna Lewis, Lawrence Berkeley National Laboratory, Berkeley, California, USA.
John Hancock, MRC Mammalian Genetics Unit, Harwell, England.
Paul Schofield, Department of Anatomy, University of Cambridge, Cambridge, CB2 3EH, England.
Sebastian Köhler, Institute for Medical Genetics, Charité Universitätsmedizin Berlin, Augustenburger Platz 1, 13353 Berlin Germany.
Peter N. Robinson, Email: peter.robinson@charite.de, Institute for Medical Genetics, Charité Universitätsmedizin Berlin, Augustenburger Platz 1, 13353 Berlin Germany
References
- 1.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009 Jan;37(Database issue):D793–D796. doi: 10.1093/nar/gkn665. [Online]. Available: http://dx.doi.org/10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki S, Hayashi M, Tamagawa K, Saito M, Kato S, Komori T, Sakakihara Y, Mizutani T, Oda M. Neuropathological analysis in spinal muscular atrophy type II. Acta Neuropathol. 2003 Nov;106(5):441–448. doi: 10.1007/s00401-003-0743-9. [Online]. Available: http://dx.doi.org/10.1007/s00401-003-0743-9. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000 May;25(1):25–29. doi: 10.1038/75556. [Online]. Available: http://dx.doi.org/10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6(2):R21. doi: 10.1186/gb-2005-6-2-r21. [Online]. Available: http://dx.doi.org/10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodenreider O, Hayamizu TF, Ringwald M, Coronado SD, Zhang S. Of mice and men: aligning mouse and human anatomies. AMIA Annu Symp Proc. 2005:61–65. [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, Liu B, Jiang J, Zhao H, Fan M, Zhang J, Fan Z, Jiang T. Modularity in the genetic disease-phenotype network. FEBS Lett. 2008 Jul;582(17):2549–2554. doi: 10.1016/j.febslet.2008.06.023. [Online]. Available: http://dx.doi.org/10.1016/j.febslet.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Köhler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008 Apr;82(4):949–958. doi: 10.1016/j.ajhg.2008.02.013. [Online]. Available: http://dx.doi.org/10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008 Mar;24(3):103–106. doi: 10.1016/j.tig.2007.12.005. [Online]. Available: http://dx.doi.org/10.1016/j.tig.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008 Nov;83(5):610–615. doi: 10.1016/j.ajhg.2008.09.017. [Online]. Available: http://dx.doi.org/10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenthal N, Brown S. The mouse ascending: perspectives for human-disease models. Nat Cell Biol. 2007 Sep;9(9):993–999. doi: 10.1038/ncb437. [Online]. Available: http://dx.doi.org/10.1038/ncb437. [DOI] [PubMed] [Google Scholar]
- 11.Rosse C, Mejino JLV. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003 Dec;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [Online]. Available: http://dx.doi.org/10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Schofield P, Sundberg J. One medicine, one pathology and the one health concept. J Am Vet Med Ass. 2009 doi: 10.2460/javma.234.12.1530. vol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith CL, Goldsmith C-AW, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6(1):R7. doi: 10.1186/gb-2004-6-1-r7. [Online]. Available: http://dx.doi.org/10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundberg JP, Sundberg BA, Schofield P. Integrating mouse anatomy and pathology ontologies into a phenotyping database: tools for data capture and training. Mamm Genome. 2008 Jun;19(6):413–419. doi: 10.1007/s00335-008-9123-z. [Online]. Available: http://dx.doi.org/10.1007/s00335-008-9123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Bodenreider O. Experience in aligning anatomical ontologies. Int J Semant Web Inf Syst. 2007;3(2):1–26. doi: 10.4018/jswis.2007040101. [DOI] [PMC free article] [PubMed] [Google Scholar]


