Abstract
Purpose
The clinical use of BRAF inhibitors is being hampered by the acquisition of drug resistance. This study demonstrates the potential therapeutic utility of the HSP90 inhibitor (XL888) in 6 different models of vemurafenib resistance.
Experimental design
The ability of XL888 to inhibit growth and to induce apoptosis and tumor regression of vemurafenib-resistant melanoma cell lines was demonstrated in vitro and in vivo. A novel mass spectrometry-based pharmacodynamic assay was developed to measure intratumoral HSP70 levels following HSP90 inhibition in melanoma cell lines, xenografts and melanoma biopsies. Mechanistic studies were performed to determine the mechanism of XL888-induced apoptosis.
Results
XL888 potently inhibited cell growth, induced apoptosis and prevented the growth of vemurafenib resistant melanoma cell lines in 3D cell culture, long-term colony formation assays and human melanoma mouse xenografts. The reversal of the resistance phenotype was associated with the degradation of PDGFRβ, COT, IGFR1, CRAF, ARAF, S6, cyclin D1 and AKT, which in turn led to the nuclear accumulation of FOXO3a, an increase in BIM expression and the downregulation of Mcl-1. In most resistance models, XL888 treatment increased BIM expression, decreased Mcl-1 expression, and induced apoptosis more effectively than dual MEK/PI3K inhibition.
Conclusions
HSP90 inhibition may be a highly effective strategy at managing the diverse array of resistance mechanisms being reported to BRAF inhibitors and appears to be more effective at restoring BIM expression and downregulating Mcl-1 expression than combined MEK/PI3K inhibitor therapy.
Keywords: melanoma, BRAF, resistance, therapy, HSP90, vemurafenib
Introduction
Despite the recent clinical success of BRAF inhibitors like vemurafenib (PLX4032) and dabrafenib (GSK2118436) in BRAF mutant melanoma, most of the responses observed are transient, with relapse and resistance occurring in most cases (1, 2). The emerging data suggests that BRAF inhibitor resistance is complex, multi-factorial and results from intrinsic and acquired mechanisms. To date, the loss/inactivation of PTEN function, deletion of the retinoblastoma protein (RB), expression of the MAP kinase family member COT and amplification of cyclin D1 have each been shown to mediate intrinsic resistance by either diminishing the apoptotic response or allowing for cell cycle entry when oncogenic BRAF is inhibited (3–6). Unlike the acquired drug resistance to imatinib seen in chronic myeloid leukemia and to EGFR inhibitors in non-small cell lung cancer, resistance of melanoma cells to BRAF inhibitors does not result from secondary “gate-keeper” mutations in the BRAF kinase (7). Instead, acquired resistance is mediated through constitutive signaling by receptor tyrosine kinases (RTKs) (IGF1R and PDGFR-β), mutations in NRAS or MEK1, the increased expression of COT and as the result of BRAF truncations (4, 8–10). The apparent diversity of resistance mechanisms, and the likelihood that others exist is expected to complicate the design of future clinical trials to prevent or treat resistance to BRAF inhibitors. These observations led us to hypothesize that BRAF inhibitor resistance may be best managed through broadly targeted strategies that inhibit multiple pathways simultaneously.
The heat shock protein (HSP)-90 family of chaperones maintains the malignant potential of cancer cells by regulating the conformation, stability and function of many RTKs and kinases required for oncogenic transformation (11, 12). Many proteins required for melanoma initiation and progression, including mutated BRAF, CRAF, IGF1R, cyclin D1, CDK4 and AKT are known to be clients of HSP90 (13, 14). The role of HSP90 in the stabilization of so many cancer-related proteins has made it an attractive target for therapeutic intervention. At this current time, over 13 small molecule inhibitors of HSP90 are at various stages of pre-clinical and clinical development (12).
Although HSP90 inhibitors have shown only limited single-agent activity, more promising clinical efficacy has been demonstrated when HSP90 inhibitors are combined with other agents. There is now good evidence that HSP90 inhibitors overcome trastuzumab resistance in breast cancer and potentiate the effects of bortezomib in treatment refractory myeloma (15–18). In the current study, we demonstrate that all of the signaling proteins implicated thus far in the escape from BRAF inhibitor therapy are clients of HSP-90, and that the pharmacological inhibition of HSP90 abrogated both acquired and intrinsic vemurafenib resistance by restoring the apoptotic response. These studies support the use of HSP90 inhibitors in overcoming BRAF inhibitor resistance.
Materials and methods
Cell culture and generation of BRAF inhibitor resistance
The parental 1205Lu, WM39 and WM164 melanoma cells lines were a gift from Dr. Meenhard Herlyn (The Wistar Institute, Philadelphia, PA) and were genotyped as beingBRAF V600E mutantin (19). The M229, M229R, M249 and M249R were described in (8). The RPMI7951 melanoma cell line was purchased from ATCC. The identities of all cell lines were confirmed by Biosynthesis Inc (Lewisville, Tx) through STR validation analysis. Naïve and intrinsically resistant lines were cultured in 5% FBS, RPMI. For all studies, all acquired resistant cell lines were maintained in 5% media with the addition of vemurafenib at the following concentrations: 1µM for M229R and M249R, 2µM for WM164R and 3µM for 1205LuR.
Growth inhibition
Cells were plated at a density of 2.5 × 104 cells per ml and left to grow overnight before being treated with increasing concentrations of vemurafenib or XL888as described in (19). Data show the mean of at least three independent experiments ± the S.E. mean.
Western blotting
Proteins were extracted and blotted for as described in (19). After analysis, Western blots were stripped once and re-probed for β-actin or GAPDH to demonstrate even protein loading. The antibodies to IGF1R, PDGFRβ, ARAF, CRAF, phospho-AKT Ser473, total AKT, phospho-ERK, total ERK, cyclin D1, phospho-S6, total S6, phospho-BIM (Ser69), total BIM, HSP70 and MCL-1 were from Cell Signaling Technology (Beverly, MA). Anti-26S was purchased from Abcam (Cambridge, MA) while the antibody against COT was from Santa Cruz Biotechnology (Santa Cruz, CA). For mouse xenograft studies, tumor samples were harvested and immediately placed into RNAlater solution (Invitrogen) prior to protein extraction.
Flow cytometry
Cells were plated into 6 well tissue culture plates at 60% confluency and left to grow overnight before being treated with either 300nM XL888, 3µM AZD6244, 3µM GDC-0941 (Selleck) or the combination of 3µM AZD6244 and 3µM GDC-0941 for 72 or 144 hr. In some studies, RPMI7951, WM793 and 1205Lu cells were treated with 300nM XL888 in the presence or absence of 3µM vemurafenib and harvested after 48 hours. Annexin V and TMRM staining was performed as described in (3).
3D spheroid assays
Melanoma spheroids were prepared using the liquid overlay method (16). Spheroids were either treated for 144 hours with vehicle or 1µM XL888 or for 48 hours with vehicle, 1µM XL888, 3µM vemurafenib or a combination of the two drugs before being washed and analyzed as described in (16).
RNA interference
M229R and 1205LuR were plated at 1×105 and left to grow overnight in RPMI complete media. Complete media was replaced with Opti-MEM (Invitrogen) and Mcl-1 or BIM (both 25nM, Cell Signaling Technologies) siRNA’s in complex with Lipofectamine 2000 (Invitrogen) were added. In addition, scrambled siRNA’s were added as non-targeting controls. A final concentration of 5% FBS in complete RPMI was added the next day. In the BIM studies, cells were transfected for a total of 48 hours prior to a 48hr treatment with 300nM XL888. In the Mcl-1 studies, cells were treated for 96 hr.
Immunofluorescent staining
M229R and 1205LuR cells were seeded at 50% confluency before being treated with 300nM XL888, 3µM AZD6244, 3µM GDC-0941 or AZD6244 + GDC-0941in combination as previously described (25). Cells were stained with antibodies against BIM and FOXO3a followed by staining with secondary anti-rabbit AF488 and imaged with a Leica confocal microscope.
Proteomics Sample Preparation
Proteins were extracted as described for Western Blotting and processed as described in (20).
Liquid chromatography, multiple reaction monitoring mass spectrometry (LC-MRM) analysis
LC-MRM was performed as described in (20). Protein expression was determined using the ratio of peak area of the native peptide to corresponding internal standard; normalization of tissue results was performed using GAPDH to control for cellularity (see Supplemental Table 1). Data were then normalized to the pretreatment (cell lines) or vehicle controls (tissue) and plotted to show the changes in expression after drug treatment.
Human specimen procurement
Patients scheduled to undergo surgical resection for metastatic melanoma were prospectively consented and accrued to an existing melanoma tissue procurement protocol approved by the Moffitt Cancer Center Scientific Review Committee and The University of South Florida Institutional Review Board. Following excision of the specimen in the operating room, fine needle tumor aspirates were taken using a 22-gauge needle for proteomic analysis of the resulting tumor homogenate.
MCL-1 inducible cell line
WM793TR MCL-1 cells were a kind gift from Dr. Andrew Aplin (Kimmel Cancer Center, Philadelphia, PA) (21). Mcl-1 expression was induced by the addition of 100ng/mL doxycycline for 72 hours prior to treatment with 300nM XL888 for an additional 72 hours.
Quantitative real-time PCR
Cells treated for 48 hours with 300nM XL888, 3µM AZD6244, 3µM GDC-0941 or AZD6244 and GDC-0941 in combination prior to RNA isolation. Total RNA was isolated using Qiagen’s RNeasy mini kit. The following TaqMan® Gene Expression Assays primer/probes were used: Hs00197982_m1 (BIM), Hs01050896_m1 (MCL-1), Hs00372937_m1 (BMF), P/N 4319413E (18S) and Hs99999905_m1 (GAPDH). The 18S + GAPDH data were used for normalizing BIM. Q-RT-PCR reactions were performed as previously described (3).
Colony formation
Cells (1×104 per ml) were grown overnight before being treated with vehicle (DMSO) or XL888 (300 nM) for 4 weeks as described in (22), and relative colony density was determined by solubilizing the crystal violet dye in 10% acetic acid followed by measurement of absorbance at 450nm.
Xenograft experiments
BALB SCID mice (The Jackson Laboratory, Bar Harbor, ME) were subcutaneously injected with 2.5 ×106 cells per mouse and grown to approximately 100 mm3 prior to dosing. Mice were treated with either 100mg XL888/kg (n=5) or an equivalent volume of vehicle (10mM HCl), 3x per week by oral gavage. Mouse weights and tumor volumes (L × W2/2) were measured 3X per week. Upon completion of the experiment, vehicle and drug treated tumor biopsies were processed for LC-MRM analysis (as above). Detection of apoptosis by TUNEL staining (Millipore) was carried out according to the manufacturer’s instructions.
Statistical analysis
Data show the mean of at least three independent experiments ± the S.E. mean, unless stated otherwise. Statistically significant results were considered where P≤0.05. Additional statistical analyses are described in the supplemental material.
Results
Inhibition of HSP90 overcomes resistance to vemurafenib resistance mediated through diverse mechanisms
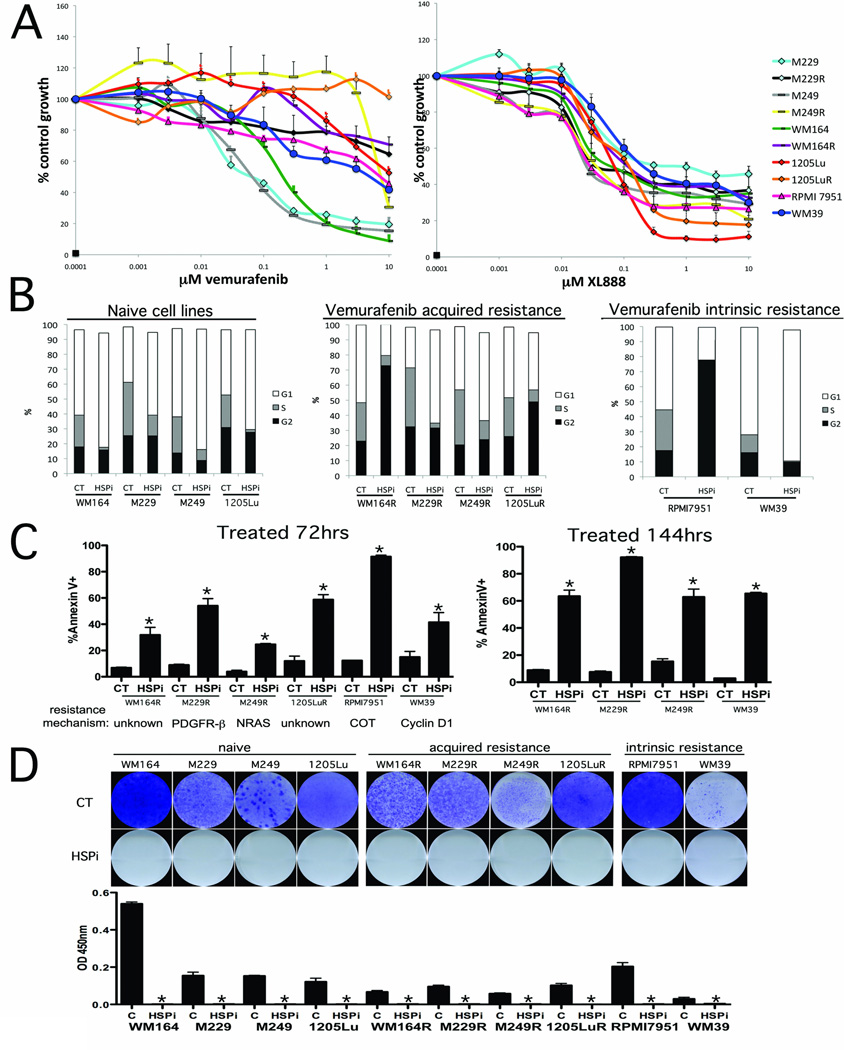
We first assembled a panel of BRAF V600E mutant melanoma cell lines with different mechanisms of intrinsic resistance and acquired vemurafenib resistance (see Supplemental Table 2). Treatment of matched BRAF inhibitor naïve and resistant melanoma cell lines with vemurafenib showed a statistically significant difference in the extent of growth inhibition (P=0.02; t = −.4.38. Figure 1A, Supplemental Figure 1) when resistance was mediated through increased PDGFRβ expression (M229R), and an acquired NRAS mutation (M249R), as well as two lines with uncharacterized mechanisms of resistance (WM164R and 1205LuR) (Figure 1A). Cell lines with amplification of cyclin D1 (WM39) and overexpression of COT (RPMI 7951) showed signs of intrinsic resistance to vemurafenib (IC50> 3µM). By contrast, treatment with the HSP90 inhibitor XL888 led to dose dependent decreases in the growth of all the cell lines with no significant difference in IC50 values observed between the naïve and resistance pairs of cell lines (t = 0.25, p = 0.82)(Figure 1A). The growth inhibitory effects of XL888 were associated with induction of either a G1-phase cell cycle arrest (WM164, M229, M229R, M249, M249R, 1205Lu, WM39) or a G2/M phase cell cycle arrest (WM164R, 1205LuR, RPMI 7951) (Figure 1B). Treatment of all of the vemurafenib resistant melanoma cell lines with XL888 (300 nM) induced high levels (>66%) of apoptosis as shown by Annexin-V binding, caspase-3 cleavage and loss of mitochondrial membrane potential (TMRM) in every cell line tested (Figure 1C and Supplemental Figure 2). The cytotoxic effects of XL888 were durable with no signs of colony formation observed in any of the cell lines (up to 4 weeks: Figure 1D and Supplemental Figure 3).
Figure 1. The HSP90 inhibitor XL888 blocks the growth and survival of melanoma cell lines with diverse mechanisms of vemurafenib resistance.
A: Growth assay showing the response of matched pairs of vemurafenib naïve and resistant melanoma cell lines and melanoma cell lines with intrinsic resistance. (Left) Cells were treated with increasing concentrations of vemurafenib (1nM – 10 µM: 72 hrs) before being subject to the MTT assay. (Right) Cell growth assay showing the response of the cell line panel from (A) to the HSP90 inhibitor (1nM-10 µM: 72 hrs). B: Cell cycle effects of XL888 (300 nM: 24 hrs) upon vemurafenib sensitive and naïve cell lines. Cells were fixed, stained with propidium iodide and distributions analyzed by flow cytometry. C: XL888 induces apoptosis in every model of acquired vemurafenib resistance tested. Cells were treated for either 72 or 144 hrs with XL888 (300 nM) followed by Annexin-V. Apoptosis was measured by flow cytometry. D: (Upper) Colony formation assay showing the long-term effectiveness of XL888. Cell lines were treated with 300nM XL888 for 4 weeks before being fixed and stained with crystal violet. (Lower) Quantification of absorbance after 4 weeks of drug treatment.
Inhibition of HSP90 degrades all of the proteins identified as being critical for vemurafenib resistance
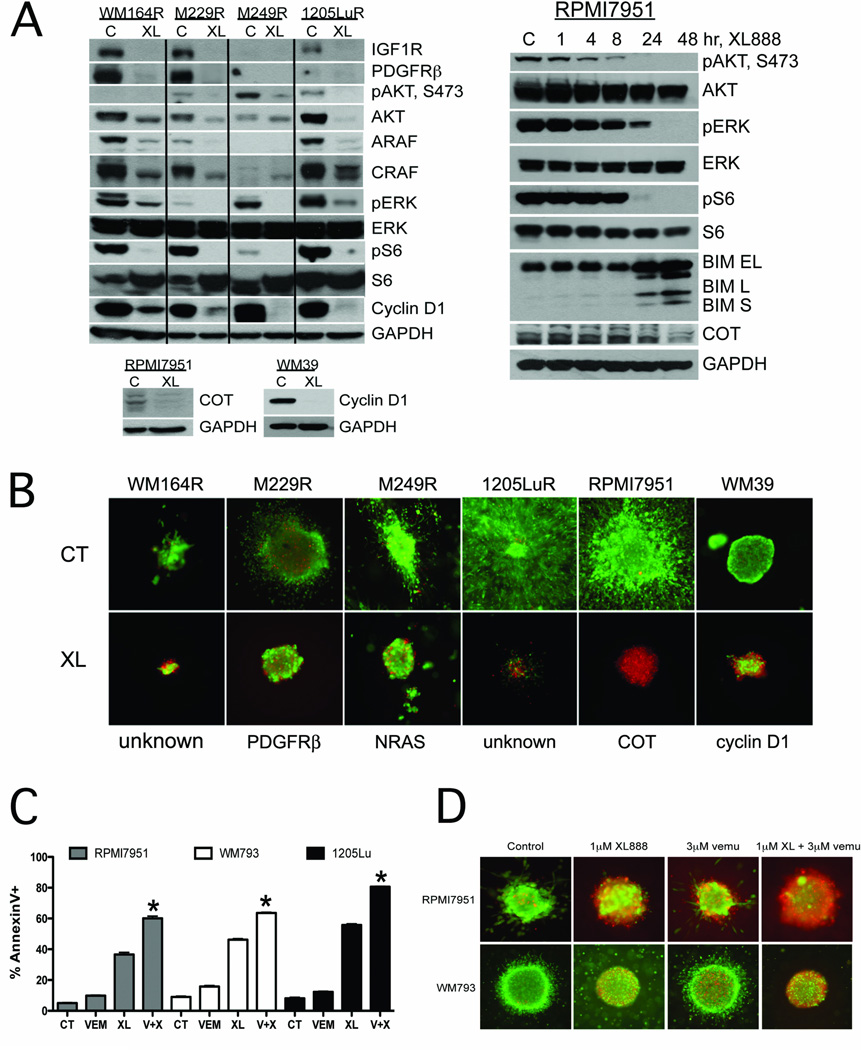
We next asked whether XL888 treatment induced the degradation of all the signaling mediators implicated in acquired and intrinsic resistance (Supplemental Figure 4 summarizes melanoma-relevant HSP90 clients). XL888 treatment (300nM, 48 hrs) led to the degradation of IGF1R, PDGFRβ, ARAF, CRAF and cyclin D1 and the inhibition of AKT, ERK and S6 signaling in all of the cell lines with acquired BRAF inhibitor resistance (Figure 2A). These effects were found to be time-dependent with some sensitive proteins, such as pAKT being downregulated at <8 hrs (Figure 2A). In the intrinsically vemurafenib-resistant melanoma cell lines RPMI7951 and WM39, XL888 treatment was found to degrade both COT and cyclin D1, respectively (Figure 2A). Because the microenvironment modulates the response of melanoma cells to targeted therapies (23), we next grew the panel of vemurafenib-resistant cell lines as collagen implanted 3D spheroids and noted that XL888 was effective at inducing cell death (Figure 2B). In line with the observation that COT mediates resistance to vemurafenib (4), the combination of XL888 with vemurafenib significantly enhanced the level of apoptosis/cytotoxicity in 3D culture in RPMI7951 cells, compared to XL888 alone (Figures 2C,D). A similar enhancement was noted when the vemurafenib + XL888 combination was applied to two melanoma cell lines in which the primary resistance was mediated through PTEN loss (WM793 and 1205Lu) (Figures 2C,D).
Figure 2. XL888 degrades proteins involved in BRAF inhibitor resistance leading to apoptosis induction.
A: (Left) XL888 degrades IGF1R, PDGFRβ, ARAF, CRAF and cyclin D1 and inhibits pAKT, pERK and pS6 signaling in 4 melanoma cell lines with acquired BRAF inhibitor resistance. XL888 degrades the expression of COT and cyclin D1 in melanoma cell lines with intrinsic resistance to vemurafenib. (Right) Time-dependency of the XL888 mediated effects upon pAKT, pERK, pS6, COT and BIM. RPMI7951 cells were treated with XL888 for 0–48 hrs. B: XL888 (1 µM, 144 hrs) is effective at blocking the growth and survival of vemurafenib resistant melanoma cell lines grown as 3D collagen implanted spheroids. Staining shows cell viability, where green corresponds to live cells and red: dead cells. Magnification X4. C: Combining XL888 with vemurafenib leads to enhanced levels of apoptosis in melanoma cell lines with COT overexpression (RPMI7951), or loss of PTEN expression (WM793 and 1205Lu). Cells were treated with vemurafenib (3 µM), XL888 (300nM), or the two in combination for 48 hr. Apoptosis was measured by Annexin-V staining and flow cytometry. D: XL888 in combination with vemurafenib reduces the survival of intrinsically resistant melanoma cell lines grown as 3D collagen-implanted spheroids. RPMI7951 cells (COT amplified) and WM793 cells (PTEN deficient) were treated with either XL888 (1 µM), vemurafenib (3 µM), or a combination of the two for 48 hrs. Viability was measured as described above.
Development of a quantitative pharmacodynamic assay of HSP90 inhibition
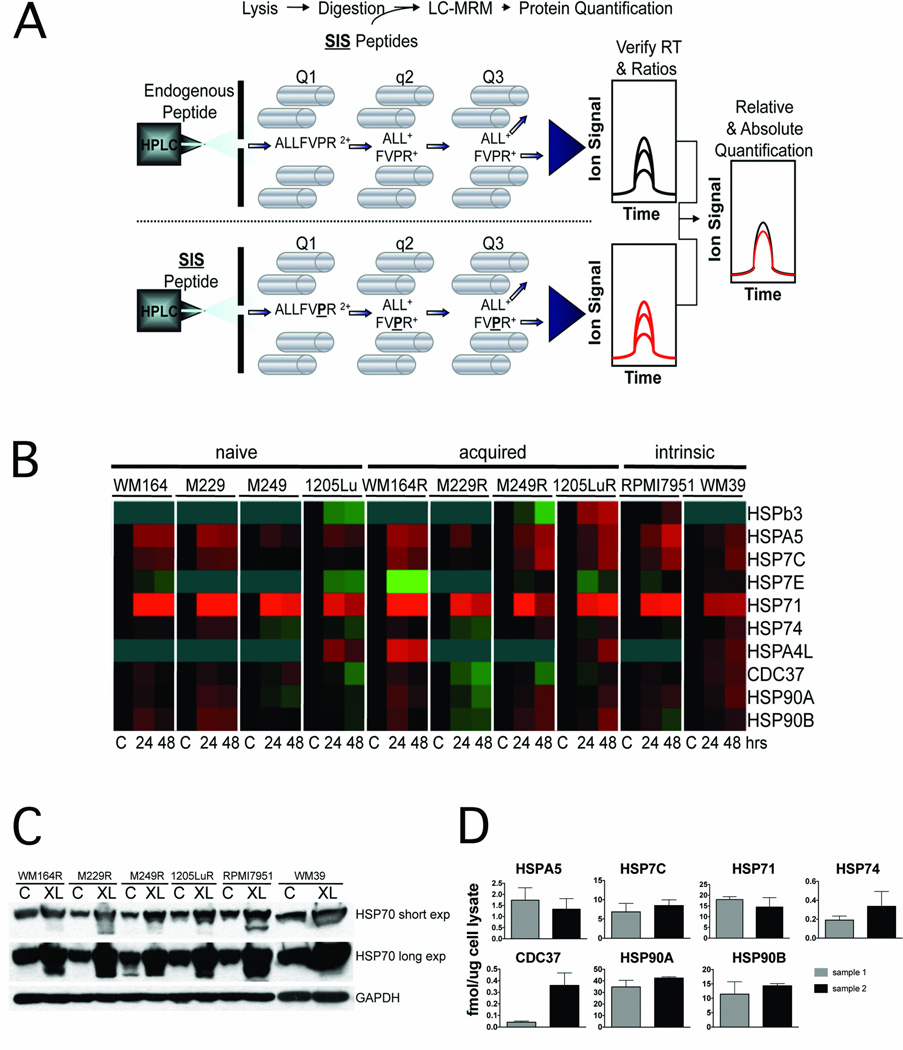
The clinical development of HSP90 inhibitors has been hampered by the lack of a good pharmacodynamic assay for quantifying target inhibition within the tumor (12). As inhibition of HSP90 typically leads to the increased expression of other HSP family members which can be used as a surrogate for HSP90 inhibition, we developed a highly sensitive quantitative LC-MRM assay for the quantification of 11 HSP family members (12) (Figure 3A). Treatment of cell lines that were naïve, intrinsically resistant and with acquired vemurafenib resistance with XL888 (300nM) led to robust time-dependent increases in the expression of HSP70 isoform 1 (HSP71) (Fig. 3B). Western blot experiments confirmed the XL888-dependent increases in HSP70 expression in every cell line evaluated (Figure 3C). The potential clinical relevance of the LC-MRM assay was demonstrated by the successful quantification of HSP70 and other chaperone proteins from fine needle aspirates (~2000 cells) taken from two melanoma specimens (Figure 3D).
Figure 3. Development of a quantitative pharmacodynamic assay for HSP90 inhibition.
A: Work flow of the LC-MRM experiment to measure HSP chaperone levels. After reversed-phase HPLC separation, peptides are selected by their mass-to-charge ratio and dissociated by collisions with background gas before the fragment ions are mass selected to enable specific detection and quantification of individual peptides in complex mixtures. B: Heatmap showing XL888-induced (0–48 hrs, 300 nM) HSP70 expression in all of the melanoma cell lines irrespective of vemurafenib resistance mechanism. C: Western Blot confirmation of HSP70 upregulation following XL888 treatment (300 nM, 48 hr). D: Quantification of absolute (fmol/µg) expression of the HSP chaperone protein expression in fine needle aspirates from two melanoma specimens.
XL888 treatment causes the regression of vemurafenib-resistant xenografts in vivo associated with increased intratumoral HSP70 expression
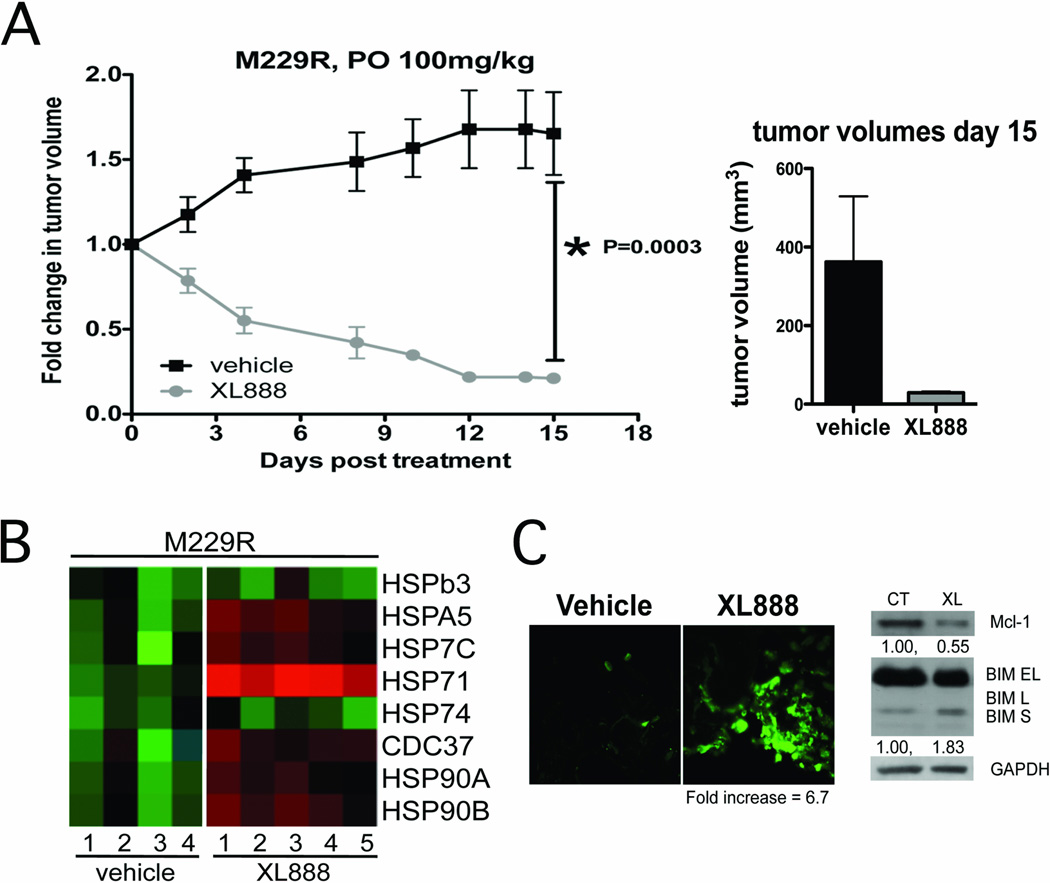
The relevance of HSP90 inhibition as a strategy to overcome BRAF inhibitor resistance in vivo was demonstrated by the ability of XL888 (100 mg/kg, PO, 3X week) to significantly induce the regression of, or growth inhibition of established M229R and 1205LuR xenografts in SCID mice (Figure 4A and Supplemental Figure 5). It was noted that the XL888 was well tolerated by the mice, with no significant alterations in body weigh observed over the study period (Supplemental Figure 6A). XL888 was also noted to be tumor specific in in vitro studies, with minimal growth inhibitory effects observed upon two primary human skin fibroblast cell lines (Supplemental Figure 6B,C).LC-MRM mediated analysis of xenograft samples following 15-days of XL888 treatment showed a robust (8.6-fold) increase in intratumoral HSP70 expression compared to controls (Figure 4B). XL888 treatmentwas noted to be pro-apoptotic in vivo and led to increased TUNEL staining in M229R xenografts associated with increased expression of BIM and decreased expression of Mcl-1 (Figure 4C).
Figure 4. XL888 induces the regression of established M229R xenografts and is associated with increased intratumoral HSP70 expression.
A: XL888 leads to regression of M229R melanoma xenografts. M229R cells were grown until a palpable tumor had formed before being treated with XL888 thrice per week (100 mg/kg) by oral gavage. Data shows growth curves normalized to starting volumes, bar graph shows mean tumor volumes after 15 days of XL888 treatment. XL888 treatment led to significant levels of tumor regression (P=0.003). B: Heatmap showing the increase in HSP70 isoform 1 (HSP71) expression in XL888 treated (15 days, 100mg per kg) xenograft samples compared to vehicle controls. C: XL888 treatment (10 days) led to the induction of apoptosis in established M229R xenografts as measured by increased TUNEL staining (green), and was associated with the induction of BIM expression and the suppression of Mcl-1 expression.
HSP90 inhibition restores nuclear localization of FOXO3a, upregulates BIM expression and inhibits Mcl-1 expression in vemurafenib-resistant cell lines
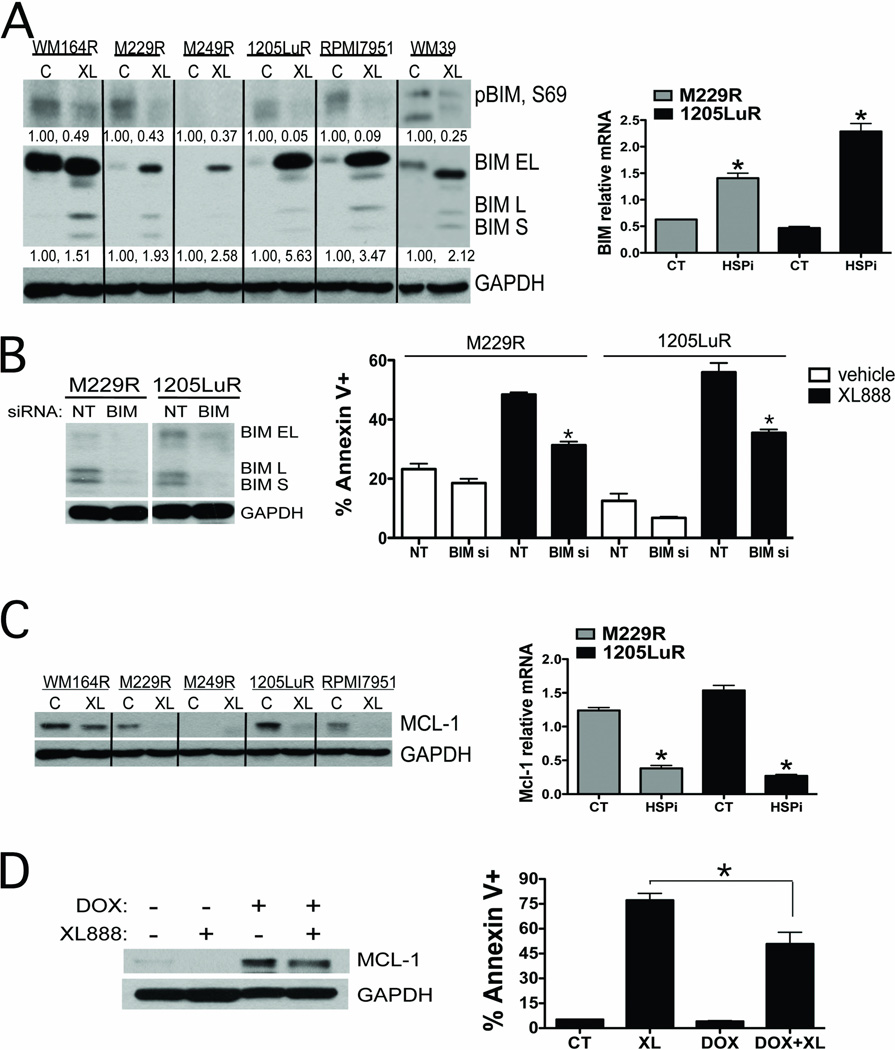
To determine the mechanism of XL888-induced apoptosis in the vemurafenib-resistant melanoma cell lines, we first focused upon BIM. Whereas vemurafenib treatment increased expression of BIM in melanoma cell lines that were drug naïve (3), the resistant cell lines suppressed their expression of BIM even in the continuous presence of vemurafenib (Figure 5A). XL888 treatment reversed this and increased BIM expression, irrespective of resistance mechanism (Figure 5A). It was noted that XL888 treatment increased the expression of BIM-EL, BIM-L and BIM-S expression in the M229R, 1205LuR, RPMI7951 and WM39 cell lines, induced expression of BIM-L and BIM-S in the WM164R cell line and BIM-EL in the M249R cell line (Figure 5A). These effects were mediated in part through increased BIM protein stability as noted by decreased BIM phosphorylation at Ser69 in all of the cell lines tested apart from M249R (Figure 5A). We next asked whether HSP90 inhibition also affected BIM expression at the mRNA level. In vemurafenib naïve cells, inhibition of BRAF leads to the nuclear accumulation of the transcription factor FOXO3a and increased BIM expression (3). In contrast, cell lines with acquired resistance to vemurafenib excluded FOXO3a from the nucleus and suppressed BIM protein and mRNA expression even in the continuous presence of vemurafenib (Figures 5A and Supplemental Figure 7). XL888 treatment reversed these effects and led to the nuclear accumulation of FOXO3a and an increase in BIM mRNA and protein expression (Figures 5A, Supplemental Figure 7). An increase in nuclear size following XL888 treatment was also noted. The importance of BIM expression in the XL888-mediated cell death response was demonstrated by the significant inhibition of apoptosis observed when BIM expression was knocked down by siRNA (Figure 5B).
Figure 5. HSP90 inhibition increased BIM, decreases Mcl-1 and restores apoptosis in vemurafenib-resistant melanoma cell lines.
A: (Left) Western blot showing that XL888 (48 hrs, 300nM) decreases BIM phosphorylation (Ser69) and increases BIM expression. (Right) Quantitative RT-PCR experiment showing that treatment with XL888 (300 nM, 48 hrs) increases the expression of BIM at the mRNA level. B: siRNA knockdown of BIM significantly decreases XL888 (300 nM, 48hrs) mediated apoptosis in two vemurafenib resistant melanoma cell lines (M229R and 1205LuR). C: (Left) Western blot of Mcl-1 expression in vemurafenib resistant melanoma cell lines treated with XL888 (300 nM) for 48 hrs. (Right) Quantitative RT-PCR showing that XL888 (300 nM, 48hrs) treatment downregulates Mcl-1 expression at the mRNA level. D: Induction of Mcl-1 reduces the magnitude of XL888 induced apoptosis. Western blot shows the induction of Mcl-1 following doxycycline treatment. Induction of Mcl-1 (DOX+XL) significantly reduces the level of XL888-induced apoptosis compared to XL888 (XL: 300nM, 72 hrs) alone. *P<0.05
Mcl-1 is pro-survival BH3 family protein member that antagonizes the activity of BIM (21). Treatment of melanoma cell lines in which vemurafenib resistance was mediated through PDGFRβ, COT overexpression and two melanoma cell lines with unknown resistance mechanisms with XL888 (300nM, 48 hrs) led to a marked decrease in the expression of Mcl-1 (Figure 5C). Quantitative RT-PCR experiments showed that XL888 treatment also blocked Mcl-1 expression at the mRNA level (Figure 5C). The importance of Mcl-1 expression for the survival of vemurafenib-resistant melanoma cell lines was confirmed by the significant induction of apoptosis observed following siRNA knockdown of Mcl-1 expression (Supplemental Figure 8). Further evidence for the role of Mcl-1 expression in the drug resistance phenotype came from overexpression studies in which induction of Mcl-1 expression following doxycycline treatment led to a significant reduction in the magnitude of XL888-induced apoptotic response (Figure 5D).
HSP90 inhibition is more effective at inducing BIM expression and apoptosis than combined MEK+PI3K inhibition
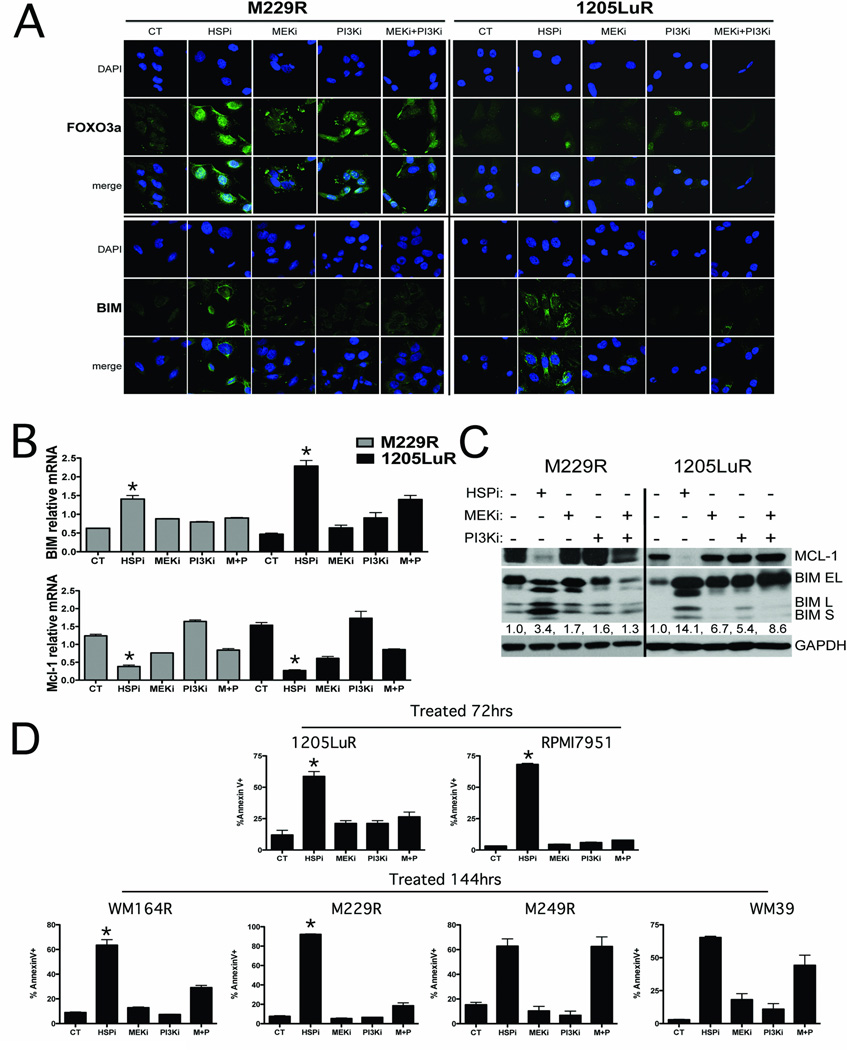
The simultaneous targeting of MEK/ERK and PI3K/AKT signaling is being explored as a strategy for overcoming vemurafenib resistance. We next asked whether HSP90 inhibition was more effective than the MEK+PI3K inhibitor combination at restoring apoptosis in vemurafenib-resistant melanoma cells. Although both XL888 and the PI3K inhibitor GDC-0941 were highly efficient at increasing nuclear accumulation of FOXO3a (Figure 6A), XL888 treatment led to a greater induction of BIM expression at both the protein and mRNA levels and significantly restored the apoptotic response (Figure 6B,C). Similarly, XL888 treatment was also more effective than the MEK or PI3K inhibitor, alone or in combination, at downregulating the expression of Mcl-1 at both the mRNA and protein levels (Figures 6B,C). This was in marked contrast to the responses observed in the parental M229 and 1205Lu cell lines, where the MEK+PI3K inhibitor combination was equally effective as XL888 at inducing BIM expression (Supplemental Figure 9). Although there is evidence that the BH3 protein family member BMF plays a role in the apoptotic response to BRAF inhibition (24), XL888 treatment only weakly induced BMF mRNA expression (Supplemental Figure 10). In contrast, treatment of two vemurafenib-resistant cell lines with either the MEK inhibitor (M229R) or the MEK+PI3K inhibitor (1205LuR) led to a robust induction of BMF expression but induced less apoptosis than following XL888 treatment (Figure 6D, Supplemental Figure 10). As the phosphorylation of BIM by MEK/ERK leads to its proteasomal degradation and the 26S proteasome is an HSP90 client protein, we next determined the contribution of proteasome inhibition to the cytotoxic effects of XL888. Although XL888 treatment was observed to partly degrade the 26S proteasome, HSP90 inhibition had a considerably weaker effect upon proteasomal activity than either the MEK+PI3K inhibitor combination or the proteasome inhibitor (MG-132) (Supplemental Figure 11). In agreement with the marked effects of HSP90 inhibition on BIM and Mcl-1 expression compared to the MEK, PI3K and MEK+PI3K inhibitor combination, XL888 was observed to induce significantly higher levels of apoptosis than each of the other drug combinations in cell lines where resistance was mediated through amplification of COT, PDGFRβ overexpression and in two other models where the resistance mechanism is as yet unknown (Figure 6D). The level of apoptosis induced by the MEK+PI3K inhibitor combination was equivalent to that of the HSP90 inhibitor when resistance was mediated through NRAS mutation or cyclin D1 amplification (Figure 6D).
Figure 6. HSP90 inhibition is more effective at restoring the apoptotic response than combined MEK+PI3K inhibition.
A: Immunofluorescence staining of 1205LuR and M229R cells for BIM and FOXO3a following treatment with either vehicle, XL888 (300nM), AZD6244 (3µM), GDC-0941 (3µM) and the combination of AZD6244 + GDC-0941 (each at 3µM). B: XL888 is more effective than MEK+PI3K inhibitors at increasing BIM and decreasing Mcl-1 mRNA expression in 1205LuR and M229R cell lines. Cells were treated with vehicle, XL888, AZD6244, GDC-0941 and the combination of AZD6244 + GDC-0941 (as above) and Quantitative RT-PCR was performed on BIM and Mcl-1. C: XL888 is more effective than MEK+PI3K inhibitors at increasing BIM and decreasing Mcl-1 protein expression in 1205LuR and M229R cell lines. Cells were treated with vehicle, XL888, AZD6244, GDC-0941 and the combination of AZD6244 + GDC-0941 as above and Western blotting was performed for BIM and Mcl-1. D: XL888 is more effective at inducing apoptosis of melanoma cell lines where resistance is mediated through COT and PDGFRβ expression and in 2 models where the resistance mechanism is unknown. Cells were treated with XL888, AZD6244, GDC-0941 and AZD6244 + GDC-0941 as described above for 72 or 144 hrs. Apoptosis was measured by Annexin-V staining and flow cytometry.
Discussion
The current study addressed whether targeting multiple signaling pathways through the inhibition of HSP90 is sufficient to overcome intrinsic and acquired resistance to the BRAF inhibitor vemurafenib (PLX4032). XL888 is a novel, orally-available HSP90 inhibitor with high selectivity for HSP90α and HSP90β (IC50 - 22nM, 44nM, respectively) and little activity against a panel of 29 other diverse kinases (IC50 all >3600 nM) (25). XL888 inhibited the growth of, and promoted apoptosis in, melanoma cell lines where vemurafenib resistance was mediated through NRAS mutations, PDGFRβ overexpression, COT overexpression and cyclin D1 amplification. It was also pro-apoptotic in two melanoma cell lines with acquired vemurafenib resistance mediated through as yet unknown means. In all of the vemurafenib sensitive cell lines, XL888 induced a G1-phase cell cycle arrest and reduced the percentage of cells in S-phase. In some of the resistance models, XL888 treatment instead induced cell cycle arrest in G2/M, perhaps suggesting an altered signaling dependency following the acquisition of drug resistance. In all cases, the responses to XL888 were highly durable with no resistant colonies emerging following 4 weeks of continuous drug treatment. The prolonged growth inhibition with XL888 was in marked contrast to that observed by our group and reported by others following vemurafenib (or PLX4720) treatment where resistant colonies emerged in every case (8, 22, 26). As the lack of colony formation suggested that all the BRAF inhibitor resistance mechanisms present in our models were abrogated following HSP90 inhibition, we next confirmed that XL888 decreased the expression of the proteins implicated thus far in acquired and intrinsic vemurafenib resistance (IGF1R, cyclin D1, PDGFR-β, AKT, COT, ARAF and CRAF (4, 5, 8, 9, 27)). In each case, not only did XL888 reduce the levels of the RTK/kinases implicated in BRAF inhibitor resistance, it also blocked the signaling activity of the pathways (MEK, AKT, mTOR/S6) involved in therapeutic escape (9, 22, 27, 28). Although IGF1R, COT, AKT, ARAF, MEK and CRAF have previously been reported to be HSP90 clients and subject to proteasome-mediated degradation following HSP inhibition, this is the first report to potentially identify PDGFR-β as a client of HSP90 (a current list of HSP90 clients is maintained at http://www.picard.ch/downloads/Hsp90interactors.pdf).
The potential utility of HSP90 inhibitors in overcoming vemurafenib resistance was illustrated by the ability of XL888 to inhibit multiple, non-overlapping resistance pathways in the same cell line model, e.g., the inhibition of PDGFRβ, IGF1R and COT in the COT amplified cell line and PDGFRβ, IGF1R and COT in the PDGFRβ overexpressing cell line. The fact that melanomas express multiple RTKs and can flexibly switch between multiple signaling pathways suggests that individual melanoma cells may have a number of escape mechanisms at their disposal. Furthermore, effective strategies to manage resistance will need to concurrently target multiple oncogenic pathways (9). There is already evidence from other cancers that HSP90 inhibitors can overcome multiple drug resistance mechanisms. In preclinical studies of breast cancer, inhibitors of HSP90 abrogate diverse trastuzumab resistance mechanisms, including those mediated by PI3K mutations, truncation mutants of p95-HER2 and the upregulation of membrane associated mucin-4 (29–32). In non-small cell lung cancer, inhibition of HSP90 prevents drug resistance associated with the oncogenic switch from EGFR to c-MET (32). HSP90 inhibitors have also proved effective at managing drug resistance in the clinic, with activity being reported against trastuzumab-resistant HER2+ breast cancer and bortezomib-resistant multiple myeloma (16, 17, 33, 34).
The measurement of HSP90 inhibition in vivo has proven to be challenging. Although it is known that HSP90 inhibition is well correlated with the increased expression of the co-chaperone HSP70, which can be quantified in peripheral blood mononuclear cells (PBMCs), this does not correlate well with either intratumoral HSP90 inhibition or clinical activity (12). The high abundance of heat shock chaperone proteins makes them amenable to direct quantification by mass spectrometry with minimal processing (20). As patients with advanced melanoma typically present with accessible cutaneous lesions that can be biopsied or undergo fine needle aspiration, we developed a novel quantitative pharmacodynamic mass spectrometry-based assay for the quantification of HSP90 and its co-chaperones. In agreement with previously published studies on other HSP90 inhibitors, XL888 treatment led to the consistent upregulation in the expression of HSP70 isoform 1 in every vemurafenib sensitive and naïve cell line tested (20, 33, 35). Although there is evidence that increased HSP70 expression limits apoptosis in leukemic cells, the therapeutic relevance of this observation in melanoma is still under investigation (36). The in vivo utility of the LC-MRM technique was demonstrated by the robust increases in HSP70 expression observed in xenografts following XL888 treatment and the ability to quantify levels of HSP90 and its key co-chaperones in small needle biopsies (fine needle aspirates) taken from fresh melanoma specimens. These results demonstrate the utility of LC-MRM based pharmacodynamic assays for measuring intratumoral HSP90 inhibition that can be incorporated into future clinical trials of these drugs.
Inhibition of BRAF, either by siRNA knockdown or small molecule inhibitors of BRAF or MEK, induces apoptosis in BRAF V600E mutant melanoma cells through the pro-apoptotic proteins BIM, BMF and BAD (24, 37–39). BIM (Bcl-2 interacting mediator of cell death) is a BH3 family protein member that plays a key role in the induction of cell death by binding to and antagonizing the pro-survival proteins Bcl-2, Bcl-w, Bcl-XL and Mcl-1 (40, 41). Vemurafenib resistance (both intrinsic and acquired) is characterized by a diminished apoptotic response and impaired BIM expression in the continuous presence of drug. The observation that BIM is regulated both transcriptionally and post-transcriptionally, through many pathways including ERK, AKT, JNK and p38 MAPK, led us to hypothesize that XL888 may overcome vemurafenib resistance by upregulating BIM expression at both the mRNA and protein levels through the simultaneous targeting of multiple signaling pathways (42, 43). Regulation of BIM mRNA is mediated by the transcription factor FOXO3a, which is inactivated following its phosphorylation by AKT at T32, S253 and S315 leading to its nuclear exclusion and localization to the cytoplasm (3, 44). BIM levels are controlled post-translationally through phosphorylation of the protein at a number of sites (including S69) by MEK/ERK signaling, with the phosphorylation of BIM leading to its poly-ubiquitination and proteasomal degradation (39). Our previous studies demonstrated that vemurafenib increased nuclear FOXO3a localization and BIM expression in drug naïve cells leading to increased apoptosis (3). Here we noted that vemurafenib resistance was associated with suppression of nuclear FOXO3a and BIM expression in the continued presence of drug that was reversed upon addition of XL888. Interestingly, XL888 treatment was more effective at restoring the expression of BIM at the mRNA and protein levels and inducing apoptosis than dual inhibition of MEK and PI3K, perhaps suggesting the involvement of other (as yet unidentified) pathways that are also HSP90 clients. Although expression of BIM is regulated both through 26S ubiquitin-dependent and 20S polyubiquitin independent proteasomal mechanisms and the 26S proteasome is a known HSP90 client, we were unable to demonstrate a role for downregulation of the 26S proteasome in the recovery of BIM expression following HSP90 inhibition (45, 46). A number of recent studies have suggested a role for increased BMF (Bcl-2 modifying factor) expression in mediating the apoptotic response of melanoma cells treated with inhibitors of BRAF and MEK (24, 38). Here, we observed that XL888 treatment was a relatively weak inducer of BMF expression in the vemurafenib-resistant melanoma cell lines compared to that seen following MEK or PI3K + MEK inhibition, suggesting that BMF is relatively dispensable in overcoming BRAF inhibitor resistance in our models.
The decision between survival and apoptosis is regulated through the balance of pro and anti-apoptotic Bcl-2 family proteins. Survival of melanoma cells is controlled in part by the anti-apoptotic protein, Mcl-1, whose stability is regulated by the BRAF/MEK/ERK pathway (21). A potential role for Mcl-1 in the tolerance of BRAF inhibition was suggested by the studies showing that acquired vemurafenib resistance led to the recovery of MAPK signaling while resistant cells maintained their Mcl-1 expression in the presence of vemurafenib and that the forced overexpression of Mcl-1 decreased the vemurafenib-induced apoptotic response (21, 22). Inhibition of HSP90 led to the degradation of Mcl-1 protein and reduced Mcl-1 expression at the mRNA level. XL888 was more effective at reducing Mcl-1 mRNA levels than inhibitors of MEK, PI3K and the MEK+PI3K inhibitor combination. It therefore seems likely that the induction of BIM in concert with Mcl-1 downregulation plays a key role in the induction of XL888 mediated apoptosis.
Current preclinical and clinical strategies for managing vemurafenib resistance in melanoma are centered upon combining vemurafenib with inhibitors of the MEK and PI3K/AKT/mTOR pathways (9, 28). Although our study supports use of the MEK+PI3K inhibitor combination when resistance is mediated through NRAS mutations or cyclin D1 amplification, it appears suboptimal when resistance is mediated by increased COT expression, PDGFRβ overexpression and in 2 other cell lines models with undetermined resistance mechanisms. These findings suggest either that other pathways (that also happen to be HSP90 clients) are required for therapeutic escape or that vertical inhibition of the same pathway at multiple points (e.g. ARAF/CRAF/MEK or IGF1R/AKT/S6) simultaneously may be a more effective way of shutting down a signal transduction pathway.
In summary, we have shown for the very first time that all of the signaling proteins implicated thus far in intrinsic and acquired BRAF inhibitor resistance are clients of HSP90 and that inhibition of HSP90 can restore sensitivity to vemurafenib mediated cell death by upregulating expression of BIM and inhibiting expression of Mcl-1. These studies provide the rationale for the dual targeting of HSP90 and BRAF in BRAF mutant melanoma as a strategy to limit the therapeutic escape seen with single agent vemurafenib therapy.
Supplementary Material
Statement of translational relevance.
The impressive clinical response of melanoma patients to the BRAF inhibitor vemurafenib is limited by the onset of resistance. Resistance can be intrinsic or acquired; it is mediated through an array of mechanisms including acquired mutations in NRAS and MEK1, truncated BRAF, overexpression of COT, CRAF, PDGFR-β, cyclin D1 and IGF1R. This apparent diversity of resistance mechanisms, coupled with the phenotypic and cell signaling plasticity of melanoma cells, represents a considerable clinical challenge for which no management strategies currently exist. Here we demonstrate that all of the signaling proteins implicated thus far in the escape from vemurafenib therapy are clients of heat shock protein (HSP)-90. Inhibition of HSP90 using XL888 overcomes both acquired and intrinsic vemurafenib resistance by restoring the apoptotic response, which suggests that the combination of vemurafenib and an HSP90 inhibitor may be a strategy to delay and/or overcome BRAF inhibitor resistance.
Acknowledgements
We would like to thank Dana Aftab (Exelixis) for providing the XL888 and Gideon Bollag (Plexxikon) for providing vemurafenib for these studies. We also thank Laura Hall (Moffitt Molecular Biology Core) for assistance with the qRT-PCR experiments.
Grant support:
K.S.M. Smalley was supported by the NIH/National Cancer Institute (R01 CA161107-01), the Harry Lloyd Trust and the Bankhead-Coley Research Program of the State of Florida (09BN-14). The Moffitt Proteomics Facility is supported by the US Army Medical Research and Materiel Command under Award No. DAMD17-02-2-0051 for a National Functional Genomics Center, the National Cancer Institute under Award No. P30-CA076292 as a Cancer Center Support Grant, and the Moffitt Foundation. The Bankhead-Coley Cancer Research program of the Florida Dept. of Health provided funding for the purchase of the LC-MRM instrumentation (09-BE04-SIG).
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N Engl J Med. 2011 doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smalley KS, Sondak VK. Melanoma--an unlikely poster child for personalized cancer therapy. N Engl J Med. 2010;363:876–878. doi: 10.1056/NEJMe1005370. [DOI] [PubMed] [Google Scholar]
- 3.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010 doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smalley KS, Lioni M, Palma MD, Xiao M, Desai B, Egyhazi S, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther. 2008;7:2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2011 doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittaker S, Kirk R, Hayward R, Zambon A, Viros A, Cantarino N, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Science translational medicine. 2010;2 doi: 10.1126/scitranslmed.3000758. 35ra41. [DOI] [PubMed] [Google Scholar]
- 8.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nature reviews Molecular cell biology. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 12.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nature reviews cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grbovic OM, Basso AD, Sawai A, Ye Q, Friedlander P, Solit D, et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci U S A. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.da Rocha Dias S, Friedlos F, Light Y, Springer C, Workman P, Marais R. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65:10686–10691. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- 15.Banerji U, Affolter A, Judson I, Marais R, Workman P. BRAF and NRAS mutations in melanoma: potential relationships to clinical response to HSP90 inhibitors. Mol Cancer Ther. 2008;7:737–739. doi: 10.1158/1535-7163.MCT-08-0145. [DOI] [PubMed] [Google Scholar]
- 16.Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, et al. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:8302–8307. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, et al. HSP90 Inhibition Is Effective in Breast Cancer: A Phase II Trial of Tanespimycin (17-AAG) Plus Trastuzumab in Patients with HER2-Positive Metastatic Breast Cancer Progressing on Trastuzumab. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5132–5139. doi: 10.1158/1078-0432.CCR-11-0072. [DOI] [PubMed] [Google Scholar]
- 18.Richardson PG, Badros AZ, Jagannath S, Tarantolo S, Wolf JL, Albitar M, et al. Tanespimycin with bortezomib: activity in relapsed/refractory patients with multiple myeloma. British journal of haematology. 2010;150:428–437. doi: 10.1111/j.1365-2141.2010.08264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smalley KS, Contractor R, Haass NK, Lee JT, Nathanson KL, Medina CA, et al. Ki67 expression levels are a better marker of reduced melanoma growth following MEK inhibitor treatment than phospho-ERK levels. Br J cancer. 2007;96:445–449. doi: 10.1038/sj.bjc.6603596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remily-Wood ER, Liu RZ, Xiang Y, Chen Y, Thomas CE, Rajyaguru N, et al. A database of reaction monitoring mass spectrometry assays for elucidating therapeutic response in cancer. Proteomics Clinical applications. 2011;5:383–396. doi: 10.1002/prca.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boisvert-Adamo K, Longmate W, Abel EV, Aplin AE. Mcl-1 is required for melanoma cell resistance to anoikis. Mol Cancer Res. 2009;7:549–556. doi: 10.1158/1541-7786.MCR-08-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br J cancer. 2010;102:1724–1730. doi: 10.1038/sj.bjc.6605714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 24.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70:6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyman SK, Crawley SC, Gong R, Adamkewicz JI, McGrath G, Chew JY, et al. High-content, high-throughput analysis of cell cycle perturbations induced by the HSP90 inhibitor XL888. PLoS ONE. 2011;6:e17692. doi: 10.1371/journal.pone.0017692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang CC, Lai F, Thorne RF, Yang F, Liu H, Hersey P, et al. MEK-Independent Survival of B-RAFV600E Melanoma Cells Selected for Resistance to Apoptosis Induced by the RAF Inhibitor PLX4720. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-2225. [DOI] [PubMed] [Google Scholar]
- 27.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H, Kong X, Ribas A, Lo RS. Combinatorial Treatments That Overcome PDGFR{beta}-Driven Resistance of Melanoma Cells to V600EB-RAF Inhibition. Cancer Research. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandarlapaty S, Scaltriti M, Angelini P, Ye Q, Guzman M, Hudis CA, et al. Inhibitors of HSP90 block p95-HER2 signaling in Trastuzumab-resistant tumors and suppress their growth. Oncogene. 2010;29:325–334. doi: 10.1038/onc.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price-Schiavi SA, Jepson S, Li P, Arango M, Rudland PS, Yee L, et al. Rat Muc4 (sialomucin complex) reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a potential mechanism for herceptin resistance. International journal of cancer Journal international du cancer. 2002;99:783–791. doi: 10.1002/ijc.10410. [DOI] [PubMed] [Google Scholar]
- 31.Nahta R, Esteva FJ. HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast cancer research : BCR. 2006;8:215. doi: 10.1186/bcr1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Research. 2008;68:5827–5838. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson PG, Chanan-Khan AA, Lonial S, Krishnan AY, Carroll MP, Alsina M, et al. Tanespimycin and bortezomib combination treatment in patients with relapsed or relapsed and refractory multiple myeloma: results of a phase 1/2 study. British journal of haematology. 2011;153:729–740. doi: 10.1111/j.1365-2141.2011.08664.x. [DOI] [PubMed] [Google Scholar]
- 34.Arteaga CL. Why Is This Effective HSP90 Inhibitor Not Being Developed in HER2+ Breast Cancer? Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4919–4921. doi: 10.1158/1078-0432.CCR-11-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modi S, Stopeck AT, Gordon MS, Mendelson D, Solit DB, Bagatell R, et al. Combination of trastuzumab and tanespimycin (17-AAG, KOS-953) is safe and active in trastuzumab-refractory HER-2 overexpressing breast cancer: a phase I dose-escalation study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:5410–5417. doi: 10.1200/JCO.2007.11.7960. [DOI] [PubMed] [Google Scholar]
- 36.Beere HM. "The stress of dying": the role of heat shock proteins in the regulation of apoptosis. Journal of Cell Science. 2004;117:2641–2651. doi: 10.1242/jcs.01284. [DOI] [PubMed] [Google Scholar]
- 37.Boisvert-Adamo K, Aplin AE. Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene. 2008;27:3301–3312. doi: 10.1038/sj.onc.1211003. [DOI] [PubMed] [Google Scholar]
- 38.VanBrocklin MW, Verhaegen M, Soengas MS, Holmen SL. Mitogen-activated protein kinase inhibition induces translocation of Bmf to promote apoptosis in melanoma. Cancer Res. 2009;69:1985–1994. doi: 10.1158/0008-5472.CAN-08-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ, et al. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res. 2008;21:534–544. doi: 10.1111/j.1755-148X.2008.00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu SY, Lin P, Hsueh AJ. BOD (Bcl-2-related ovarian death gene) is an ovarian BH3 domain-containing proapoptotic Bcl-2 protein capable of dimerization with diverse antiapoptotic Bcl-2 members. Molecular endocrinology (Baltimore, Md. 1998;12:1432–1440. doi: 10.1210/mend.12.9.0166. [DOI] [PubMed] [Google Scholar]
- 42.Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 43.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Molecular cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 44.Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- 45.Wiggins CM, Tsvetkov P, Johnson M, Joyce CL, Lamb CA, Bryant NJ, et al. BIM(EL), an intrinsically disordered protein, is degraded by 20S proteasomes in the absence of poly-ubiquitylation. Journal of Cell Science. 2011;124:969–977. doi: 10.1242/jcs.058438. [DOI] [PubMed] [Google Scholar]
- 46.Cheung CH, Chen HH, Cheng LT, Lyu KW, Kanwar JR, Chang JY. Targeting Hsp90 with small molecule inhibitors induces the over-expression of the anti-apoptotic molecule, survivin, in human A549, HONE-1 and HT-29 cancer cells. Molecular cancer. 2010;9:77. doi: 10.1186/1476-4598-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.