Abstract
Bone marrow failure is a nearly universal complication of Fanconi anemia. The proteins encoded by FANC genes are involved in DNA damage responses through the formation of a multisubunit nuclear complex that facilitates the E3 ubiquitin ligase activity of FANCL. However, it is not known whether loss of E3 ubiquitin ligase activity accounts for the hematopoietic stem cell defects characteristic of Fanconi anemia. Here we provide evidence that FANCL increases the activity and expression of β-catenin, a key pluripotency factor in hematopoietic stem cells. We show that FANCL ubiquitinates β-catenin with atypical ubiquitin chain extension known to have nonproteolytic functions. Specifically, β-catenin modified with lysine-11 ubiquitin chain extension efficiently activates a lymphocyte enhancer-binding factor-T cell factor reporter. We also show that FANCL-deficient cells display diminished capacity to activate β-catenin leading to reduced transcription of Wnt-responsive targets c-Myc and Cyclin D1. Suppression of FANCL expression in normal human CD34+ stem and progenitor cells results in fewer β-catenin active cells and inhibits expansion of multilineage progenitors. Together, these results suggest that diminished Wnt/β-catenin signaling may be an underlying molecular defect in FANCL-deficient hematopoietic stem cells leading to their accelerated loss.
Introduction
Fanconi anemia (FA) is the most common inherited bone marrow failure syndrome. The majority of patients develop bone marrow failure within the first decade of life. Patients are also at high risk for the development of acute myeloid leukemia and solid tumors of the head and neck and gastrointestinal tract. The disease is caused by loss-of-function of any 1 of 15 FANC genes.1–6 The nuclear core complex includes FANCA, B, C, E, F, G, L, and M.1 FANCL is a ring type E3 ubiquitin ligase that monoubiquitinates FANCD2 and FANCI, which enhances their function at sites of DNA damage.7–10 The core complex scaffold is essential for facilitating the activity of FANCL because loss of any one of the core proteins results in loss of FANCL E3 ubiquitin ligase function.1
Although FANC proteins are known to execute a normal DNA damage response to crosslinking agents, emerging evidence points to alternative functions for these proteins in hematopoietic stem cells (HSCs), and the loss of these alternative functions may represent a driving force behind the common FA complications of myelodysplasia and acute myeloid leukemia.11–16 Functional defects in FA HSCs exist, including decreased repopulating ability, reduced numbers of HSCs, defective homing capacity, tumor necrosis factor (TNF)–α hypersensitivity, and limited replicative and survival potential compared with normal HSCs.17–26 Collectively these studies support the argument that the FA pathway has a role in maintaining the HSC pool and regulating stem cell fitness.15 However, the molecular mechanisms underlying such HSC defects have not been well characterized.
In light of the well established role of TNF-α in the pathogenesis of marrow failure and leukemia in Fancc-deficient mice,24–26 we first sought mechanistic insights by comparing the transcriptomal response to TNF-α of Fancc-deficient Kit+/Sca1+/Lin− (KSL) marrow cells to the transcriptomal response of wild-type (WT) KSL cells. Ontological analysis of gene expression changes induced by TNF-α revealed that genes encoding proteins of relevance to the Wnt signaling pathway were significantly over-represented (Z = 4.07, supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Although the ontological analysis of such data provides no proof of a particular mechanism directly linking the Wnt pathway to Fancc-deficient cells, the results are compatible with the notion that defective Wnt/β-catenin signaling may underlie the pathogenesis of bone marrow failure in FA.
Given the pivotal position of FANCL as a key E3 ubiquitin ligase whose activity is influenced by at least 7 of the other FA proteins, we hypothesized that FANCL ubiquitinates proteins that control survival and self-renewal of HSCs and sought to identify these additional targets. We focused first on the Wnt/β-catenin pathway because it has an established role in HSC development, cell-fate determination, and maintenance of the stem cell pool27–29 and multiple key proteins in that pathway are regulated by ubiquitination.30 In addition, recent studies demonstrate that polyubiquitination of β-catenin with nonlysine-48 linked ubiquitin chain extension functionally enhances β-catenin activity.31,32 Here we show that FANCL enhances the expression and nuclear activity of β-catenin. This functional interaction is at least in part dependent on the E3 ubiquitin ligase activity of FANCL. Moreover, we demonstrate that this molecular interaction is relevant in loss-of-function studies. We propose that diminished Wnt/β-catenin signaling contributes to the dysfunction of FA HSCs.
Methods
Gene expression profiling of WT versus Fancc-deficient KSL cells with or without TNF-α
See supplemental Methods for details. CEL files and probe set signals, as well as detailed methods and quality control measures, have been deposited in NCBI's Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession no. GSE30318).
Cells and culture conditions
293FT cell line was obtained from Invitrogen and routinely maintained in 10% FBS DMEM supplemented with nonessential amino acids and glutamine. Cord blood units were obtained from the Oregon Cord Blood Program at Oregon Health & Science University (OHSU). Consent forms and Institutional Review Board protocols are established. CD34+ cells were enriched using Miltenyi beads per the manufacturer's protocol. CD34+ enriched cord blood stem and progenitor cells were grown in StemPro (Invitrogen) with 50 ng/mL stem cell factor, 10 ng/mL thrombopoietin, 50 ng/mL Flt3 ligand, and 10 ng/mL IL-6 (Peprotech). Mouse embryonic fibroblasts (MEFs) from WT or Fancc-deficient mice, corrected with Fancc or not corrected,33 were cultured in the same media as 293FT cells. Cells were grown in media containing 0 to 62.5 ng/mL of mitomycin C for 4 days. The BIO [(2'2,3'E)6-bromoindirobian-3'-axime]compound was purchased from Sigma-Aldrich (B1686). Generally, cells were treated with 0.5 to 1μM BIO for 48 hours before experiments.
Constructs
Reporter constructs were generated by incorporating 8X lymphocyte enhancer-binding factor-T cell factor (LEF-TCF) consensus binding sites34 into the pGreenFire1 (pGF1) vector containing eGFP-T2A-lucifersase as the reporter (System Biosciences). Human cDNAs for β-catenin, FANCL, FANCA, FANCC, and FANCG were purchased from Open Biosystems or provided by G.C.B. β-catenin mutants (K19R, K49R, or K19R-K49R) were generated by site-directed mutagenesis with QuickChange II XL (Agilent). Mutations were confirmed by sequencing. cDNAs were cloned into mammalian expression vectors pCDNA6-CMV-V5/His (Invitrogen) or a modified pCDH1-EF1 vector (System Biosciences). We obtained from Addgene the pcDNA3-HA-Ub construct (no. 18712; deposited by Dr Edward Yeh, The University of Texas MD Anderson Cancer Center)35 and the pRK5-HA-Ub mutant constructs (deposited by Drs Ted Dawson, The Johns Hopkins University and Sandra Weller, University of Connecticut Health Center).36,37 Expression of the pmaxGFP construct served as the maxGFP control (Lonza) in some experiments. Glutathione-s-transferase (GST) proteins were generated by cloning cDNAs into pGEX-4T constructs (GE Healthcare Life Sciences). The pLKO.1 FANCL shRNA set generated by The RNAi Consortium was purchased from Open Biosystems.
Quantitative RT-PCR analysis
Total RNA was extracted using RNeasy Kit (QIAGEN) and converted to cDNA using SuperScript VILO reverse transcriptase (Invitrogen). Expression levels were analyzed using Platinum SYBR Green qPRCR Super-mix UDG (Invitrogen) and either the Opticon 2 real-time cycler (MJ Research; Bio-Rad) or the LightCycler 480(Roche). Relative expression was calculated by the equation 2−(ΔΔct) × 100% (see Figure 5A-B; supplemental Figure 2B) and by the Pfaffl methodology (Figure 5E).38 See supplemental Methods for a list of primers.
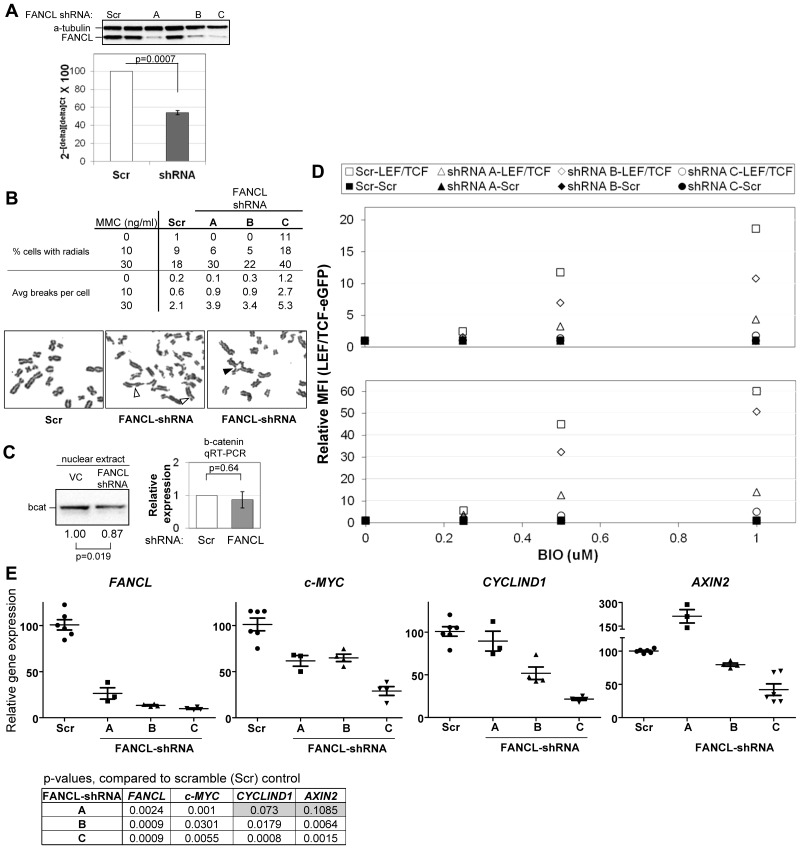
Figure 5.
FANCL-suppression reduces β-catenin expression and activity. (A) pLK0.1 FANCL shRNA constructs and a control shRNA construct (scrambled, scr) were tested for their ability to knockdown exogenous (top panel) and endogenous (bottom panel) expression of FANCL in 293FT cells, by Western blot and qRT-PCR analysis (mean ± SEM), respectively. FANCL shRNA constructs labeled A, B, and C were used for subsequent experiments. (B) Importantly, suppression of FANCL expression reproduces the classic FA phenotype of excessive radial formation (filled arrow head) and chromosome breaks (unfilled arrow head) with mitomycin C treatment compared with scrambled control. The table summarizes the average from 2 separate experiments. (C) Nuclear extracts from cells expressing vector-control (VC) or scrambled shRNA or FANCL shRNA (construct B) were immunoblotted for β-catenin. Shown is a representative blot from 7 experiments (blot). We performed qRT-PCR to determine whether these changes are associated with changes in β-catenin mRNA. Shown are the combined results of FANCL shRNA construct A, B, and C (graph, mean ± SEM). (D) FANCL was suppressed by shRNA expression in 293FT cells stably expressing scramble-eGFP (closed markers) or LEF-TCF-eGFP (open markers) reporters. The cells were assayed for their ability to respond to BIO stimulation using eGFP as the readout. Relative mean fluorescence intensity (MFI) was calculated relative to the no BIO condition. Shown is the average from 2 experiments with multiple independent replicates in each experiment. (E) qRT-PCR analysis of Wnt-responsive targets in the same cells generated from the experiments described in panel D. Horizontal bars indicate mean ± SEM. See “Methods” for details on qRT-PCR data analysis.
Antibodies, Western blot analysis, and immunoprecipitation studies
See supplemental Methods for a list of antibodies. Whole cell lysates were prepared in buffer containing 0.5% Triton X-100, 120mM NaCl, 50mM Tris (pH 8.0), 2mM ethylenediaminetetraacetic acid (EDTA), 1mM Na2-VO4 and 1:300 protease Inhibitor cocktail (P8340; Sigma-Aldrich). Proteins were resolved by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE). ImageJ Version 1.34u software (National Institutes of Health) was used to quantify bands.
Nuclear extracts
Nuclear extracts from 293FT were prepared essentially as described by washing cells in cold phosphate-buffered saline (PBS) and resuspending cells in buffer 1 (25mM HEPES [N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid], 5mM KCl, 0.5mM MgCl2, 1mM dithiothreitol [DTT], 100μM phenylmethylsulfonyl fluoride [PMSF]).39 An equal volume of buffer 2 (buffer 1 with 1% NP-40) was added and then rotated at 4°C for 15 minutes. The supernatant was collected as the cytoplasmic fraction. The pellet was washed in buffer 3 (25mM HEPES, 5mM KCl, 0.5mM MgCl2, 1mM DTT, 100μM PMSF, 0.5% NP-40), and then resuspended in buffer 4 (25mM HEPES, 300mM NaCl, 10% sucrose, 1mM DTT, 1mM PMSF, 0.05% NP-40) and rotated at 4°C for 1 hour. The supernatant was then collected as the nuclear fraction. Purity of nuclear extracts was evaluated by immunoblotting for α-tubulin (a cytoplasmic protein) and Sp1 (a nuclear protein).
LEF-TCF reporter assay
Cells were transfected using Lipofectamine2000 (Invitrogen) following the manufacturer's general guidelines. Both stable cell lines and transient expression of the pGF1 reporter were used for these studies. Cells were trypsinized and diluted in PBS and analyzed using the Guava Easycyte flow cytometer (Millipore). Data were analyzed using FlowJo Version 7.6.1. For the Luciferase assays, cells were lysed in 1× passive lysis buffer and the SteadyGlo reagent was used as the substrate (Promega).
Cell-based ubiquitination assays
293FT cells were transfected with WT FANCL, C307A-FANCL, or a maxGFP control; V5-tagged WT, K19R, K49R, or K19R-K49R β-catenin; and WT or mutant HA-ubiquitin using Lipofectamine2000. Cells were treated with 1uM BIO at the time of media change. Whole cell extracts (approximately 500-750 ug) were incubated with 1 to 2 μg anti-V5 antibody and incubated at 4°C with rocking overnight. Twenty to 40 μL of protein G plus agarose was added and samples were incubated for an additional hour at 4°C with rocking. The beads were then washed 4 times with lysis buffer (0.5% Triton X-100, 120mM NaCl, 50mM Tris [pH 8.0], 2mM EDTA). Proteins were resolved with SDS-PAGE and ubiquitinated β-catenin was detected with an anti-HA antibody.
In vitro ubiquitination assay, measurement of protein stability, and chromosome breakage analysis
See supplemental Methods for details.
CD34+ cord blood stem and progenitor cell studies
CD34+ stem and progenitor cells were transduced with lentiviral particles carrying scrambled control or FANCL shRNA for 48 to 72 hours. Cells were selected in 1 μg/mL puromycin for 48 to 72 hours, and then subjected to experimental assays. For the colony forming assays, cells were plated in MethoCult H4435 (StemCell Technologies) with or without puromycin at 1 μg/mL. For immunofluorescence analysis, CD34+ stem and progenitor cells were spun onto glass slides and fixed with 4% paraformaldehyde (pH 7.4 for 15 minutes. at 37°C). Fixed cells were then permeablized with 0.25% Triton X-100 in PBS for 10 minutes at room temperature. Cells were then blocked (10% mouse and goat serum in PBS) and stained with rabbit anti–β-catenin, 1:50 and FITC-conjugated anti-CD34, 1:11. Alexa Fluor 555 goat anti–rabbit 1:500 was used as the secondary antibody. Cells were viewed and analyzed on a Zeiss LSM710 microscope at the 20× objective. Representative pictures were taken on an Olympus IX71 inverted microscope at the 40× objective. Algorithm-based detection of cytoplasmic and nuclear staining relative to DAPI-labeled nuclei was performed using the multiwavelength scoring module implemented in MetaMorph 7.5 (Molecular Devices). For the reporter assays, cells were also transduced with the LEF-TCF-eGFP reporter and cultured for a total of 7 days (the last 3 days of which included puromycin selection) followed by flow cytometry analysis. Cytokines were replenished once (see “Cells and culture conditions”).
Statistical analyses
Averages are from independent experiments with SD or SEM used to display variability. The Student t test when performed as indicated is with a 2-tailed test.
Results
FANCL overexpression enhances β-catenin activity and protein expression
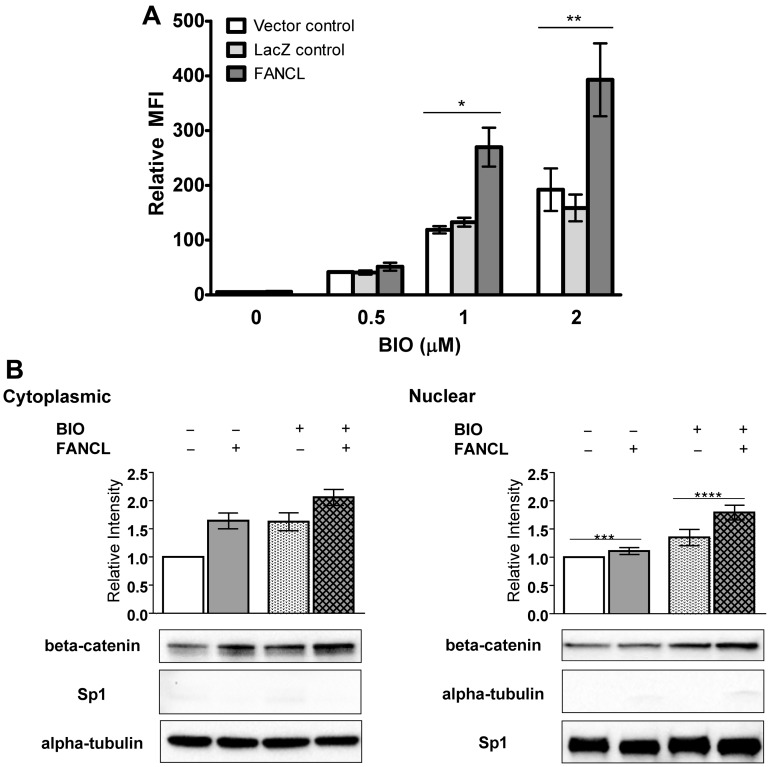
To determine whether FANCL regulates the function of nuclear β-catenin, 293FT cells stably expressing a LEF-TCF-eGFP reporter were transfected with either vector-control, LacZ, or FANCL and treated with BIO [(2′Z,3′E)-6-bromoindirubin-3′-oxime] a potent glycogen synthase kinase-3 (GSK3) inhibitor that activates canonical Wnt pathway (Figure 1A).40 The BIO compound was used to increase β-catenin activity above a detectable threshold. eGFP expression was quantified as a measure of Wnt/β-catenin activity. There was a significant increase in β-catenin activity in cells overexpressing FANCL at the higher concentrations of BIO compared with cells transfected with vector-control or LacZ. There was a dose-responsive effect of FANCL expression when luciferase was used as the readout (supplemental Figure 2A). As expected, the increase in β-catenin activity was associated with a significant increase in cytoplasmic β-catenin with or without BIO (Figure 1B left panel) but only a significant increase in nuclear β-catenin with BIO (Figure 1B right panel) compared with the control condition as shown. There were no significant differences in the loading controls or the purity of the subcellular fractions (Figure 1B, statistical analyses of tubulin and Sp1 expression). Regulation of β-catenin by FANCL is probably occurring at a posttranscriptional level because there was no significant difference in β-catenin mRNA expression (supplemental Figure 2B). We also confirmed that only modest changes in nuclear β-catenin are required for substantial changes in β-catenin activity.41,42 For example, at 1uM BIO there was only a 1.7-fold change in nuclear β-catenin compared with unstimulated cells, but there was a nearly 100-fold increase in LEF-TCF activity (supplemental Figure 2C). Cells were maximally stimulated in the range of 1 to 2μM BIO. There was insufficient inhibition of the highly active GSK3-mediated destruction complex to appreciate the effects of FANCL at 0.5μM BIO (Figure 1A). These results support that FANCL overexpression positively regulates β-catenin activity and protein expression, whereas endogenous mechanisms are active in suppressing this interaction.
Figure 1.
FANCL overexpression enhances β-catenin activity and protein expression. (A) Flow cytometry analysis of LEF-TCF-eGFP reporter assays in 293FT cells transfected with vector-control, LacZ-control, or human FANCL cDNA. Cells were treated with BIO (a GSK3-β inhibitor that activates the Wnt pathway). Data shown are from 4 experiments (mean ± SEM), *P < .01; **P < .05 compared with vector-control and LacZ-control. (B) Immunoblot analysis to quantify β-catenin in cells transfected with FANCL or vector-control. Shown are representative immunoblots for the cytoplasmic (left panel) and nuclear (right panel) fractions and a graph displaying the quantitation from 7 independent blots (mean relative expression to the no BIO and no FANCL control ± SEM), *P = .004; **P = .03; ***not significant; ****P = .008. Loading controls and the purity of the subcellular fractions were evaluated by staining for α-tubulin (n = 6) and Sp1 (n = 5) levels. Shown are representative blots at similar exposure times. We also performed statistical analysis to show that there were no differences in these controls for the pairwise comparisons performed (P values for student t test analysis of tubulin and Sp1 levels were between .77 and .98) and there were no differences globally among the 4 experimental conditions tested (P values for ANOVA analysis of tubulin and Sp1 levels were .36 and .49, respectively).
FANCL and β-catenin interaction may be facilitated by FANCA and FANCG
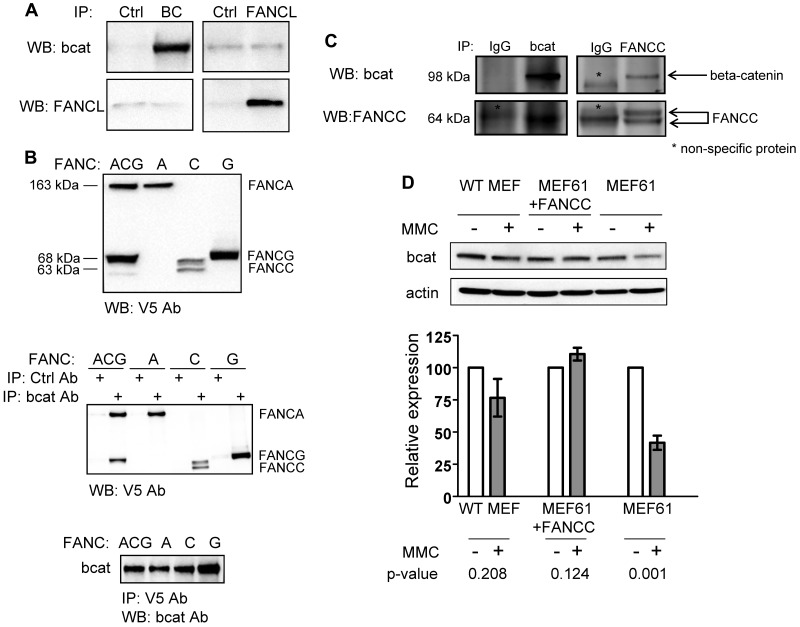
We next sought evidence of a direct interaction between FANCL and β-catenin in coimmunoprecipitation studies using cell extracts from transfected 293FT cells (Figure 2A) and purified GST fusion proteins (supplemental Figure 3A). No stable protein interaction between FANCL and β-catenin was detected above background, suggesting that the interaction of FANCL with β-catenin is transient or unstable, or facilitated by other proteins. Taking into account that the function of FANCL absolutely requires its interaction with other FA proteins in the nuclear core complex, we carried out similar β-catenin binding studies using V5-tagged core complex proteins FANCA, C, and G (Figure 2B top panel). All 3 proteins interacted with β-catenin whether we immunoprecipitated with a β-catenin antibody (Figure 2B middle panel) or a V5 antibody (Figure 2B bottom panel). In addition, using untagged FANCC we confirmed a stable interaction between FANCC and β-catenin (Figure 2C). Interestingly, expression of FANCC was diminished when FANCA and FANCG were overexpressed in the same cells. Quantification by mass spectroscopy confirmed immunoprecipitation of equimolar amounts of FANCA and FANCG using a β-catenin antibody, whereas essentially no FANCC was detected (data not shown). These results are congruent with a published report that FANCA and FANCG form a stable complex.43 However, we could not demonstrate that overexpression of FANCA and FANCG facilitated the interaction of FANCL and β-catenin in immunoprecipitation studies (data not shown). These results suggest that additional proteins, possibly other FA core complex proteins, may be involved in facilitating the interaction between FANCL and β-catenin. We then tested whether Fancc-deficient MEFs have defects in maintaining β-catenin expression under the stress of mitomycin C treatment. Indeed, there was excessive loss of β-catenin expression with mitomycin C treatment of uncorrected Fancc-deficient MEFs compared with WT and Fancc-deficient MEFs corrected by Fancc expression (Figure 2D). Although loss of β-catenin expression may be occurring at a posttranscriptional level, our microarray data indicates that transcriptional changes may also be present (supplemental Table 1).
Figure 2.
FANCL and β-catenin interaction may be facilitated by FANCA and FANCG. (A) Immunoprecipitation studies from cells transfected with cDNAs for human β-catenin and FANCL. (B) Top panel: immunoblot showing expression of C-terminal V5-tagged FANCA, C, and G proteins. Middle panel: immunoprecipitation studies from cells transfected with β-catenin and various V5-tagged FANC proteins as indicated. Bottom panel: reciprocal immunoprecipitation using V5 antibody then probed for β-catenin. At least 3 independent experiments were carried out for these studies. (C) Immunoprecipitation assays were also performed with untagged FANCC with the indicated isotype IgG controls. (D) MEFs from WT mouse and Fancc-deficient MEFs corrected by Fancc cDNA expression serve as control cells. Cells were treated with mitomycin C for 4 days at 62.5 ng/mL and whole cell extracts were analyzed for β-catenin expression. Four independent experiments were performed and shown is a representative result. The data are summarized as mean relative expression to untreated cells (no MMC) for each MEF cell line ± SEM and their corresponding P values. We probed for actin as a loading control for these experiments.
FANCL ubiquitinates β-catenin
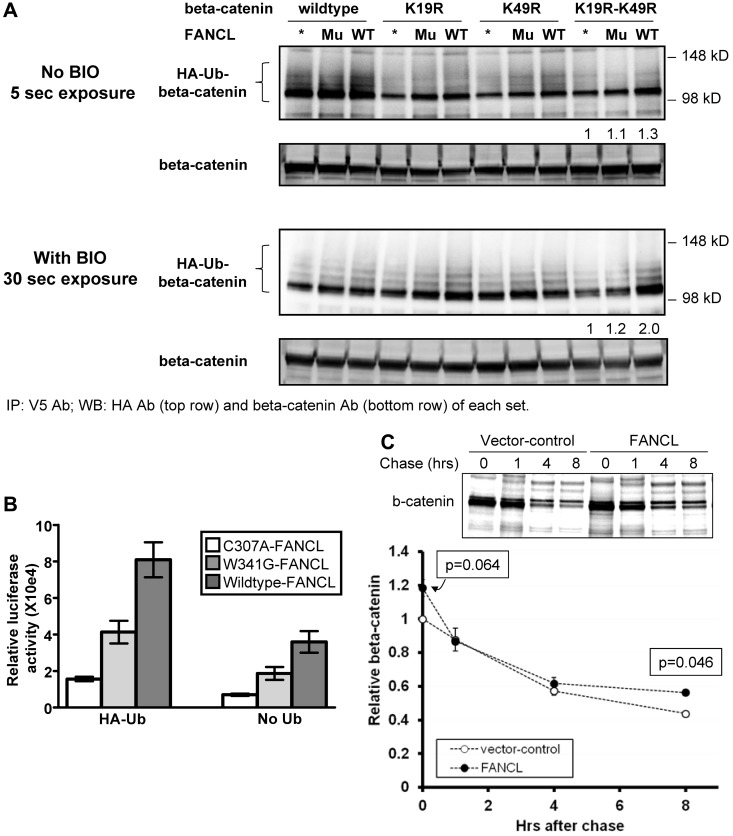
We investigated the possibility that FANCL enhances the expression and nuclear activity of β-catenin by ubiquitination. Recently, studies have shown that Rad6B and E3 identified by differential display (EDD) catalyze polyubiquitination of β-catenin and enhance its stability and nuclear activity.31,32 In both of these reports, ubiquitin chain extension occurred via lysines other than lysine-48. Such atypical ubiquitin chains influence protein function rather than degradation.44 We reduced the contribution of the GSK3-mediated destruction complex targeting β-catenin for ubiquitination by the TrCP-SCF (Skp, Cullin, F-box) E3 ubiquitin ligase complex or by other E3 ubiquitin ligases using K19R and K49R β-catenin mutants.45–49 These lysines are known sites of lysine-48–linked polyubiquitination.46 Ubiquitination of β-catenin was greatest with WT β-catenin and less so with the mutant β-catenin forms, as expected (Figure 3A top panel).46 FANCL ubiquitinated WT β-catenin at a higher level than that observed for the maxGFP and ligase-inactive C307A-FANCL controls. However, the difference was not as obvious with the K19R, K49R, and K19R-K49R β-catenin mutants. Treatment of cells with BIO in similar experiments further diminished the contribution of the GSK3-mediated destruction complex. Consequently, ubiquitinated β-catenin forms are reduced substantially (Figure 3A bottom panel). The blot shown required a much longer exposure time compared with the blot shown in the top panel. Total β-catenin immunoprecipitated was relatively similar among the various conditions despite the differences in ubiquitination. The ability of FANCL to ubiquitinate β-catenin was most obvious in the K19R-K49R β-catenin double mutant (see quantitation in Figure 3A). These studies indicate that FANCL specifically ubiquitinates β-catenin using lysines other than K19 and K49. We also purified GST fusion proteins to test the ability of FANCL to ubiquitinate β-catenin in a cell-free system. There was a significant increase in HA-Ub-GST–β-catenin detected in reactions containing WT GST-FANCL compared with control reactions containing no WT GST-FANCL or containing GST-FANCL-C307A (supplemental Figure 3B). By eliminating one component of the reaction at a time, we also confirmed that the background level of HA-Ub-GST–β-catenin is ATP-dependent and predominately because of the nonspecific, in vitro activity of E1 and E2 (supplemental Figure 3C). There was increased abundance of HA-Ub-GST–β-catenin in the presence of GST-FANCL compared with the background level catalyzed by E1 and E2.
Figure 3.
FANCL ubiquitinates β-catenin. (A) 293FT cells were transfected with a maxGFP control (asterisk), C307A-FANCL mutant (Mu), or WT FANCL (WT); V5-tagged WT, K19R, K49R, or K19R-K49R β-catenin; and hemagglutinin-tagged ubiquitin (HA-Ub). The C307A-FANCL is a ligase-inactive mutant. The β-catenin mutants were used to diminish the contribution of the GSK3-mediated destruction complex targeting β-catenin for polyubiquitination. Whole cell extracts were subjected to immunoprecipitation with V5 antibody and the blots were probed with HA antibody. This experiment was performed 2 independent times for each set. The exposure times for these blots are indicated for comparison of relative signal with or without BIO (GSK3-β inhibitor). We quantified ubiquitinated β-catenin by performing densitometry analysis using ImageJ Version 1.34u between the 98 kDa and 148 kD markers for each blot and expressed the relative signal to the maxGFP control. We also probed the same blots for total β-catenin to show relatively equal immunoprecipitation of total β-catenin. (B) LEF-TCF reporter assays using luciferase as the readout. For this experiment we used 2 ligase-inactive FANCL mutant controls. Raw luciferase values were normalized to FANCL expression relative to C307A-FANCL expression. The results shown are from 3 independent experiments (mean ± SEM); P values are < .01 for the differences noted. (C) Estimation of β-catenin turnover was determined in [35S]-methionine and cysteine pulse-chase labeling experiments to compare the effect of control versus FANCL overexpression. Shown is a representative blot from 3 experiments. The graph summarizes relative β-catenin level to time zero of the “chase,” which begins when the labeling media is replaced with complete unlabeled media (P values are indicated on the graph).
These results suggest that FANCL positively regulates β-catenin by ubiquitination. We then tested whether these ubiquitinated β-catenin forms activate the LEF-TCF-luciferase reporter by expressing WT FANCL or a ligase-inactive FANCL mutant. The C307A-FANCL mutant is defective in FA core complex assembly and binding to E2 ubiquitin-conjugating enzyme, whereas the W341G is only defective in binding to E2.50–52 Coexpression of WT FANCL with ubiquitin led to a substantial activation of the LEF-TCF reporter compared with no ubiquitin coexpression (Figure 3B). This is consistent with our results shown in Figure 1A and supplemental Figure 2A. Without ubiquitin coexpression, the activation of the LEF-TCF reporter by WT FANCL was modest but significantly higher than the ligase-inactive mutant controls. We then quantified β-catenin turnover in cells to test whether FANCL-dependent ubiquitination might enhance β-catenin activity by stabilizing the protein. We transfected cells with vector-control or FANCL and pulse-chase labeled proteins with [35S]-methionine and cysteine (Figure 3C). FANCL expression increased total β-catenin compared with vector-control expression, but there was no significant difference in β-catenin decay as represented by the slopes of the curves. However, because this assay measures total β-catenin turnover and the lysine-48–ubiquitin chain extension on β-catenin is the most abundant species, this was not completely unexpected. These results suggest that a small but active pool of β-catenin may be modified by atypical ubiquitin chains and accounts for the increased β-catenin activity observed in our studies (Figure 1A; supplemental Figure 2A).
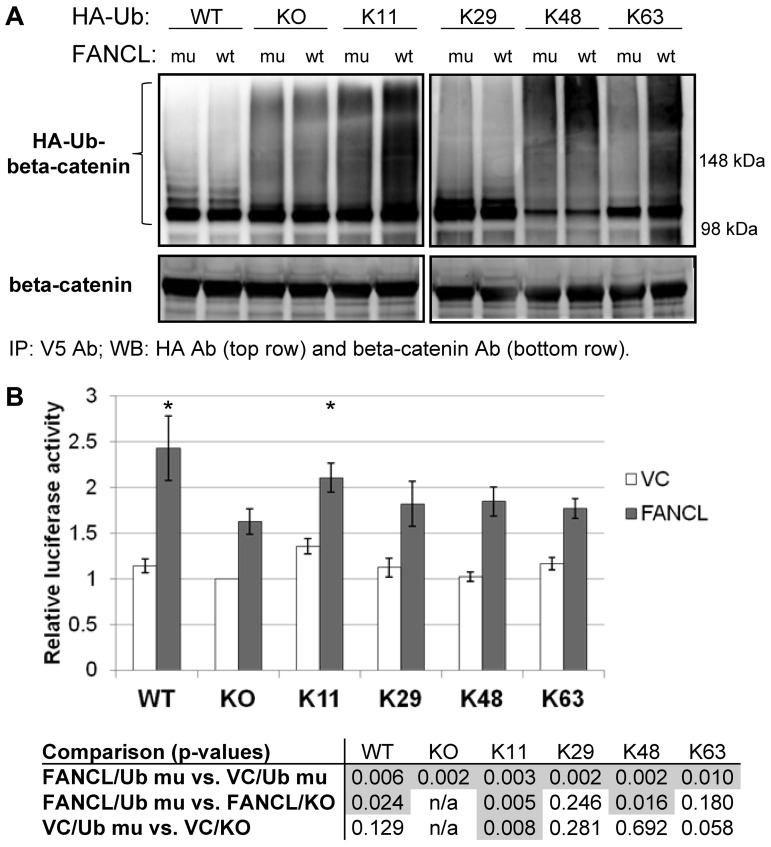
Beta-catenin modified with atypical ubiquitin chains activates the LEF-TCF reporter
Ubiquitination of β-catenin by atypical ubiquitin chain extension has been shown to enhance its function.31,32 To test whether β-catenin is modified by FANCL with atypical ubiquitin chains, we performed additional cell-based ubiquitination assays using HA-Ub mutants that contain only one intact lysine known to be involved in ubiquitin chain extension. The other 6 lysines in any given mutant HA-Ub are mutated from lysine to arginine and cannot participate in ubiquitin chain extension. If HA-ubiquitinated β-catenin forms are detected using a particular Ub mutant, it suggests that FANCL catalyzes ubiquitin chain extension using that particular lysine residue in ubiquitin. We performed these assays in BIO-treated cells expressing the V5-tagged K19R-K49R–β-catenin mutant. The results show that lysine-48–linked polyubiquitination of K19R-K49R–β-catenin in the presence of a GSK3-β inhibitor was minimal as expected (Figure 4A). Compared with C307A-FANCL expression, WT FANCL expression increased polyubiquitinated forms of β-catenin when the lysine-11 and lysine-63 ubiquitin mutants were coexpressed. These results indicate that lysine-11 and lysine-63 atypical ubiquitin chain extension are catalyzed by FANCL. We performed luciferase assays with the LEF-TCF reporter to determine whether β-catenin modified by atypical ubiquitin chains is functionally active. We found that the greatest FANCL-dependent activation of the LEF-TCF reporter occurred with coexpression of the HA-tagged WT ubiquitin, which retains all 7 lysines for ubiquitin chain extension (Figure 4B). The HA-tagged lysine-11 ubiquitin was next best in facilitating the activation of the LEF-TCF reporter indicating that lysine-11 of ubiquitin is used for chain extension. However, because its activity did not equal the WT ubiquitin, these results are most consistent with the idea that FANCL catalyzes polyubiquitination of β-catenin with mixed-chain extension. Increased LEF-TCF activity was observed in all conditions with FANCL overexpression compared with vector-control, regardless of the ubiquitin mutant coexpressed (Figure 4B table, top row). This finding suggests that FANCL uses endogenous ubiquitin to modify β-catenin. Coexpression of the ubiquitin mutants differentially enhanced the effects of FANCL further suggesting preferential use of the WT and lysine-11 for ubiquitin chain extension. We also noted that overexpression of any ubiquitin type with vector-control increased LEF-TCF activity above a no ubiquitin control, suggesting that endogenous E3 ligases actively use ubiquitin to enhance β-catenin activity. Lysine-11 ubiquitin chain extension was most active in this context (Figure 4B table, bottom row).
Figure 4.
β-catenin modified with atypical ubiquitin chains activates the LEF-TCF reporter. (A) Cell-based ubiquitination assays were carried out using HA-Ub mutants that contain only 1 intact lysine known to be involved in ubiquitin chain extension. The other 6 lysines in any given mutant HA-Ub are mutated from lysine to arginine and cannot participate in ubiquitin chain extension. We performed the ubiquitination assays using the V5-tagged K19R-K49R-β-catenin mutant and treated the cells with BIO (GSK3-β inhibitor). The ability of FANCL to catalyze atypical ubiquitin chain extension is compared with the ligase-inactive C307A-FANCL mutant. The experiment was carried out 2 independent times. (B) We performed LEF-TCF reporter assays to determine whether these ubiquitinated β-catenin subtypes are active. Cells were transfected with VC or FANCL, an HA-tagged ubiquitin as shown, and a LEF-TCF luciferase reporter. Luciferase activity was measured and shown are the results from 5 experiments normalized to the VC condition transfected with the knockout (KO) ubiquitin (mean ± SEM). The table below the figure is a compilation of all statistical analyses. Highlighted by shading are P values < .05. We compare FANCL (gray bars) versus vector-control (white bars) for the individual ubiquitin mutants (top row) and all ubiquitin mutants versus its own KO control for FANCL expression (within gray bars, middle row) and for vector-control (within white bars, bottom row). *P < .03 and highlights that the expression of lysine-11 ubiquitin activates the LEF-TCF reporter to the closest approximation of WT ubiquitin expression in FANCL-transfected cells. n/a indicates not applicable.
FANCL-suppression reduces β-catenin expression and activity
To proceed with experiments functionally testing whether endogenous FANCL regulates β-catenin expression, we first screened a set of pLKO.1 Lentiviral FANCL shRNA constructs for their ability to suppress FANCL protein and mRNA expression transiently in 293FT cells (Figure 5A blot and graph, respectively). Suppression of FANCL expression recapitulates the classic FA phenotype of excessive radial formation and chromosome breaks in cells treated with mitomycin C compared with cells expressing a scrambled shRNA (Figure 5B). FANCL shRNA construct C was most effective in suppressing FANCL expression and produced the most severe defects. We next examined whether suppression of FANCL expression results in diminished β-catenin expression. FANCL-deficient cells (construct B) exhibited a significant decrease in nuclear β-catenin compared with cells transfected with vector-control or scrambled shRNA (Figure 5C left panel). We found no significant difference in β-catenin mRNA expression under the same experimental conditions (Figure 5C right panel, combining results of all 3 FANCL shRNA constructs). We performed 2 loss-of-function studies in 293FT cells to investigate whether the regulation of β-catenin by endogenous FANCL is functionally important. As shown in Figure 5D, we suppressed FANCL expression in cells stably expressing the scramble-eGFP or LEF-TCF-eGFP reporter and treated cells with increasing BIO concentrations. Suppression of FANCL expression led to a significant decrease in ability of cells to respond to BIO stimulation for 24 hours and 48 hours. There were no changes in scramble-eGFP-expressing cells indicating that these findings are specific to β-catenin activity. In Figure 5E, we show the results of qRT-PCR analyses with validated primers to quantify changes in Wnt-responsive targets, cyclin D1, c-Myc, and axin2. Loss of β-catenin activity in these cells was indeed associated with loss of mRNA expression of cyclin D1 and c-Myc. The difference in Cyclin D1 expression produced by FANCL shRNA construct A compared with the scrambled shRNA approached statistical significance at 0.073. Loss of Axin2 mRNA expression was only noted for FANCL shRNA constructs B and C. Expression of the FANCL shRNA construct C produced the most severe loss-of-function phenotype in these functional studies compared with constructs A and B. This correlates with the chromosome breakage analysis (Figure 5C). Overall, these results support the conclusion that suppression of FANCL functionally diminishes Wnt/β-catenin signaling and transcription of downstream Wnt-responsive targets.
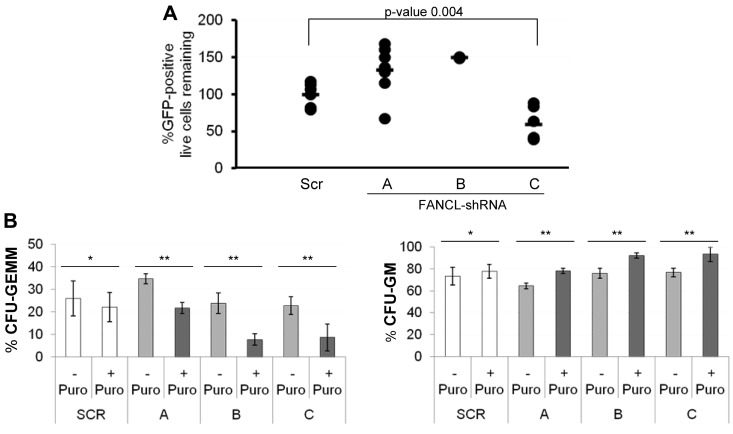
Suppression of FANCL expression in human CD34+ cord blood stem and progenitor cells leads to diminished β-catenin expression and multilineage progenitor expansion
To determine whether our findings are of relevance to human HSCs, we suppressed FANCL expression in human CD34+ hematopoietic stem and progenitor cells using the validated FANCL shRNA constructs. Cells that express the shRNA survived puromycin selection. Because our goal was to evaluate nonmembrane bound β-catenin expression and a low number of CD34+ cells remained in puromycin-selected culture conditions, we initially used immunofluorescence analysis to quantify the effects of scrambled-shRNA versus FANCL-shRNA expression. Shown in supplemental Figure 4A are the results from 1 such experiment. Uniform rules were applied to quantify the results using the MetaMorph software. The data are summarized as the average total intensity of nuclear β-catenin signal relative to the scrambled control in those cells that were defined as CD34+. An example of CD34+ cells scored as positive versus negative for β-catenin expression is shown (supplemental Figure 4B). However, this approach was not sufficiently quantitative especially for the partial FA phenotype produced by FANCL shRNA constructs A and B and β-catenin staining does not necessarily reflect β-catenin activity. To improve the quantitation and interpretation of this experiment, CD34+ cells were transduced with scrambled or FANCL targeted shRNA and the LEF-TCF–eGFP reporter. Cells were maintained in StemPro media with a cytokine cocktail optimal for culturing human hematopoietic stem and progenitor cells while minimizing differentiation in short term cultures. We then evaluated viability and eGFP expression after puromycin selection. The data are shown in Figure 6A. Interestingly, there was an expansion of eGFP-positive cells when either FANCL shRNA construct A or B was expressed compared with the scrambled control. These constructs produce an intermediate severity in radial formation and chromosome breaks with mitomycin C treatment (Figure 5C). In contrast, expression of FANCL shRNA construct C induced a substantial early loss of eGFP-positive CD34+ stem and progenitor cells (P = .004). We also tested the functional consequences of suppressed FANCL expression in CD34+ cells in colony forming assays. There was a significant reduction in multilineage progenitor cells represented by CFU–granulocyte erythrocyte macrophage megakaryocyte (GEMM) in FANCL-deficient CD34+ cells compared with control CD34+ cells (Figure 6B left panel). Yet there was a significant increase in CFU granulocyte-macrophage (GM) colonies, suggesting that suppression of FANCL expression enhances commitment of multipotent cells to more lineage-restricted progenitors (Figure 6B right panel). Together, these results suggest that FANCL regulates β-catenin and pluripotency in hematopoietic stem and progenitor cells, the organ system that most consistently fails in patients with FA.
Figure 6.
Suppression of FANCL expression in human CD34+ cord blood stem and progenitor cells leads to diminished β-catenin activity and multilineage progenitor expansion. (A) Approximately 3000 to 6000 CD34+ cord blood stem and progenitor cells were transduced with scrambled (Scr) or FANCL shRNA and the LEF-TCF-eGFP reporter. Cells were analyzed for eGFP expression after 3 days of puromycin selection (7 days total in culture). Remaining eGFP-positive cells were calculated by multiplying the starting CD34+ cells by the proportion of viable cells and the proportion of eGFP-positive cells. Each data point is expressed relative to the Scr condition. Shown are from 3 independent experiments (except FANCL shRNA construct B) with replicates plotted separately on the graph. The average of the 3 experiments is shown as a horizontal line within the graph. (B) Methylcellulose colony-forming assays were carried out as described. Colony count and type were scored; CFUs, GEMM colonies, and GM colonies. Summarized data are shown for 4 independent experiments (mean ± SEM). There were very few erythroid burst-forming units in these experiments. *Not statistically significant; **P < .04, Student t test. For these studies, transduced CD34+ cells were selected for puromycin resistance. In the colony-forming assays, unselected (no puromycin) cells were also grown to control for any potential effects of viral transduction on cell viability and colony formation that are independent of FANCL suppression.
Discussion
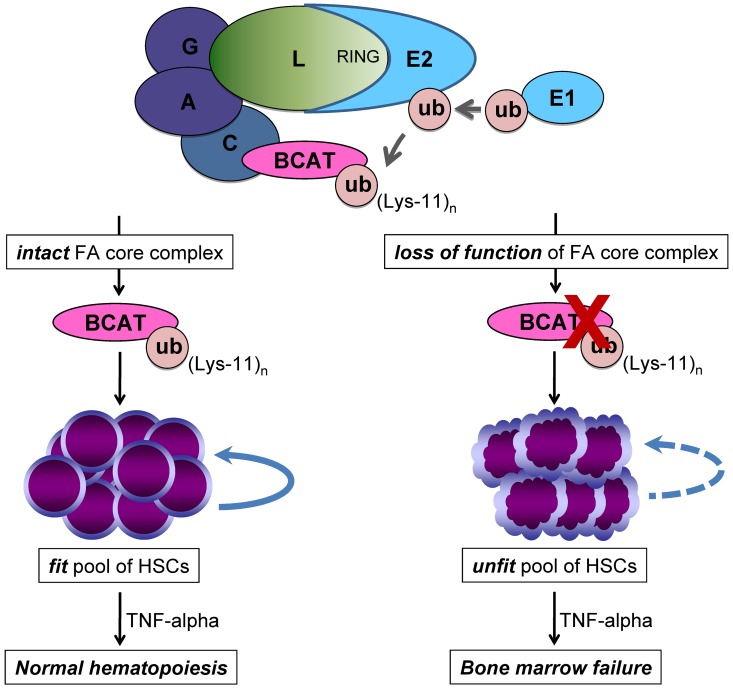
We show here a direct link between FANCL and the function of the Wnt/β-catenin pathway known to be involved in HSC development, cell-fate determination, and maintenance of the stem cell pool.53 We report that FANCL overexpression enhances β-catenin activity and that FANCL-mediates ubiquitination of β-catenin in cell-based assays. Protein modification by ubiquitin is a general mechanism that diversifies both regulation and function of proteins.54 Nonproteolytic functions of ubiquitination have been ascribed to proteins modified by a single ubiquitin molecule (eg, FANCD2) or by polyubiquitination with atypical ubiquitin chains.44 In our studies, we detect atypical ubiquitin chain extension on β-catenin using lysine-11 and lysine-63 ubiquitin residues (Figure 4A). As with Rad6B and EDD, known to positively regulate β-catenin by atypical ubiquitination mechanisms,31,32 we propose that these atypical ubiquitin chains catalyzed by FANCL enhance β-catenin activity in chromatin where it activates Wnt-responsive targets (Figure 7).
Figure 7.
Proposed model based on our finding that FANCL ubiquitinates β-catenin and enhances its nuclear function. Here we show that FANCL ubiquitinates β-catenin with mixed ubiquitin chain extension involving lysine-11 and this small pool of β-catenin is not targeted for ubiquitin-proteosome degradation but has enhanced activity at Wnt-responsive elements. We propose that the loss-of-function of the FA core complex, specifically FANCL, leads to a reduced pool of active β-catenin modified by lysine-11 ubiquitin chain extension. As a result, there is diminished Wnt/β-catenin signaling in FA HSCs. This molecular defect leads to reduced regenerative capacity and self-renewal of FA HSCs and defines an unfit pool of HSCs susceptible to malignant clonal evolution, especially in the presence of a selective pressure such as TNF-α.24,62
Loss-of-function studies confirm the functional relevance of the FANCL/β-catenin interaction. Specifically, suppression of endogenous FANCL markedly inhibited Wnt/β-catenin signaling (Figure 5D). Furthermore, these defects led to diminished gene expression of Wnt-responsive targets, cyclin D1, c-Myc, and axin2 (list of Wnt-responsive targets summarized at http://www.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes). Loss of c-Myc is particularly relevant because it is a key regulator of pluripotency in tissue-specific stem cells including in HSCs.55–57 Accumulating evidence suggests that the Wnt/β-catenin pathway is tightly regulated to maintain HSC function.58 In circumstances of experimental overactivation or suppression, and even transient perturbations of the Wnt/β-catenin pathway, severely impaired HSC repopulation ability has been described.27,28,56 Our work suggests that loss-of-function of FANCL may have such deleterious effects on HSC function.
Indeed, we show that the suppressed function of FANCL in primary human CD34+ hematopoietic stem and progenitor cells caused excessive loss of GEMM colony-forming units (CFUs; Figure 6B). In contrast, there was an expansion of GM colony-forming units. Previous groups have described excessive HSC differentiation and proliferation (loss of quiescence) as a marker of HSC pool depletion.59–61 Our studies also show that there were fewer β-catenin active cells and diminished nuclear β-catenin compared with control (Figure 6A; supplemental Figure 4). These results were most consistent when FANCL suppression was achieved by FANCL shRNA construct C, which produces the most severe FA phenotype in terms of excessive radial formation and chromosome breaks. Partial suppression of FANCL expression (ie, that achieved by FANCL shRNA constructs A and B) in human CD34+ cells led to an expansion of LEF-TCF active cells under these experimental conditions (Figure 6A). This may be a compensatory response to partial suppression of FANCL or reflect mainly the GM progenitor expansion observed in the colony forming assays. Nonetheless, any level of perturbation of FANCL expression results in substantial loss of GEMM CFUs, which is a functional assessment of HSC function. Collectively, these observations suggest that FANCL-deficient CD34+ cells display perturbations in Wnt/β-catenin signaling that may affect HSC function.
We also subscribe to the view that these findings may have therapeutic relevance. Two groups demonstrated that ex vivo exposure of murine Fancc−/− hematopoietic stem and progenitor cells to TNF-α promote the emergence of TNF-α–resistant leukemic stem cells.24,62 These clonal outgrowths display a high frequency of cytogenetic abnormalities and produce acute myeloid leukemia in transplanted recipient mice. These studies support the concept that a combination of intrinsic cellular defects (reducing HSC fitness) and extrinsic pressure (TNF-α selecting somatic mutants with adaptive mutations) drives disease progression in FA (Figure 7).63 Our studies show that Fancc-deficient MEFs display excessive loss of β-catenin expression compared with control MEFs when mitomycin C was selected as the extrinsic pressure (Figure 2D). We suggest that other FA core complex proteins may facilitate FANCL-mediated ubiquitination of β-catenin (Figure 2B). Given that the susceptible pool of HSC may be characterized by deficient Wnt/β-catenin signaling, disease progression might be subverted by manipulating targets of the Wnt/β-catenin pathway. This work provides the basis for additional in vivo confirmatory studies in the hematopoietic system to demonstrate that the interaction of FANCL and β-catenin is functionally important in preventing bone marrow failure and clonal selection.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Drs Jeffrey Tyner, Melissa Wong, and Bill Chang for having provided critical feedback on the paper. They acknowledge Ms Stephanie Willis for sharing her expertise in qRT-PCR analysis and Ms Brieanna Brown and Mr Dorian LaTocha for providing experimental support. They wish to acknowledge Ms Aurelie Snyder from the OHSU Advanced Light Microscopy Core at The Jungers Center (supported by NCRR SIG S10RR023432 and National Institutes of Health [NIH] center grant P30NS061800) for her technical expertise and Dr Larry David for experimental advice and interpretation of data from the Proteomics Shared Resource (supported by the Oregon Opportunity, and NIH center grants 5P30CA069533 and 5P30EY010572).
This work was funded by The OHSU Knight Cancer Institute, The Oregon Medical Research Foundation, and The Aplastic Anemia and Myelodysplasia International Foundation (K.T.D. is the principal investigator), the National Heart, Lung, and Blood Institute (P01 HL048546, G.C.B., A.H.N., S.B.O.), National Cancer Institute (CA138237-01, G.C.B.), and Department of Veterans Affairs (Merit Review, G.C.B.). B.J.D. is supported by the Howard Hughes Medical Institute.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.-H.T.D., M.D.R., C.L.P., S.K., W.D.N., J.E.Y., A. E.H.N., and S.B.O. performed experiments and interpretation of data. The majority of the experiments and the data presented were performed by K.-H.T.D. and M.D.R. K.-H.T.D. supervised the design of the experiments, execution of the experiments, data analyses and presentation, and wrote the majority of the paper. B.J.D. provided equipment and laboratory resources and G.C.B. conducted and analyzed the gene expression microarray experiments and provided selected cDNA constructs. In addition, B.J.D. and G.C.B. provided critical commentary on the hypotheses, experimental design, data analyses, and paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kim-Hien T. Dao, Oregon Health & Science University, Knight Cancer Institute, 3181 SW Sam Jackson Park Rd, UHN73C, Portland, Oregon, 97239-3098; e-mail: daok@ohsu.edu.
References
- 1.Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crossan GP, van der Weyden L, Rosado IV, et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 2011;43(2):147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43(2):142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoepker C, Hain K, Schuster B, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43(2):138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 5.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 6.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 7.Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol. 2010;17(3):294–298. doi: 10.1038/nsmb.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32(6):767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3(2):179–181. [PubMed] [Google Scholar]
- 10.Seki S, Ohzeki M, Uchida A, et al. A requirement of FancL and FancD2 monoubiquitination in DNA repair. Genes Cells. 2007;12(3):299–310. doi: 10.1111/j.1365-2443.2007.01054.x. [DOI] [PubMed] [Google Scholar]
- 11.Pang Q, Keeble W, Christianson TA, Faulkner GR, Bagby GC. FANCC interacts with Hsp70 to protect hematopoietic cells from IFN-gamma/TNF-alpha-mediated cytotoxicity. EMBO J. 2001;20(16):4478–4489. doi: 10.1093/emboj/20.16.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tremblay CS, Huang FF, Habi O, et al. HES1 is a novel interactor of the Fanconi anemia core complex. Blood. 2008;112(5):2062–2070. doi: 10.1182/blood-2008-04-152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanderwerf SM, Svahn J, Olson S, et al. TLR8-dependent TNF-(alpha) overexpression in Fanconi anemia group C cells. Blood. 2009;114(26):5290–5298. doi: 10.1182/blood-2009-05-222414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Li J, Sejas DP, Rathbun KR, Bagby GC, Pang Q. The Fanconi anemia proteins functionally interact with the protein kinase regulated by RNA (PKR). J Biol Chem. 2004;279(42):43910–43919. doi: 10.1074/jbc.M403884200. [DOI] [PubMed] [Google Scholar]
- 15.Bagby GC, Fleischman AG. The stem cell fitness landscape and pathways of molecular leukemogenesis. Front Biosci (Schol Ed) 2011;3:487–500. doi: 10.2741/s167. [DOI] [PubMed] [Google Scholar]
- 16.Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18(12):1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 17.Haneline LS, Gobbett TA, Ramani R, et al. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94(1):1–8. [PubMed] [Google Scholar]
- 18.Kelly PF, Radtke S, von Kalle C, et al. Stem cell collection and gene transfer in Fanconi anemia. Mol Ther. 2007;15(1):211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- 19.Larghero J, Marolleau JP, Soulier J, et al. Hematopoietic progenitor cell harvest and functionality in Fanconi anemia patients. Blood. 2002;100(8):3051. doi: 10.1182/blood-2002-07-2069. [DOI] [PubMed] [Google Scholar]
- 20.Liu JM, Kim S, Read EJ, et al. Engraftment of hematopoietic progenitor cells transduced with the Fanconi anemia group C gene (FANCC). Hum Gene Ther. 1999;10(14):2337–2346. doi: 10.1089/10430349950016988. [DOI] [PubMed] [Google Scholar]
- 21.Raya A, Rodriguez-Piza I, Guenechea G, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460(7251):53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tulpule A, Lensch MW, Miller JD, et al. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115(17):3453–3462. doi: 10.1182/blood-2009-10-246694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Shang X, Guo F, et al. Defective homing is associated with altered Cdc42 activity in cells from patients with Fanconi anemia group A. Blood. 2008;112(5):1683–1686. doi: 10.1182/blood-2008-03-147090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Sejas DP, Zhang X, et al. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117(11):3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sejas DP, Rani R, Qiu Y, et al. Inflammatory reactive oxygen species-mediated hemopoietic suppression in Fancc-deficient mice. J Immunol. 2007;178(8):5277–5287. doi: 10.4049/jimmunol.178.8.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120(Pt 9):1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleming HE, Janzen V, Lo Celso C, et al. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell. 2008;2(3):274–283. doi: 10.1016/j.stem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luis TC, Weerkamp F, Naber BA, et al. Wnt3a deficiency irreversibly impairs hematopoietic stem cell self-renewal and leads to defects in progenitor cell differentiation. Blood. 2009;113(3):546–554. doi: 10.1182/blood-2008-06-163774. [DOI] [PubMed] [Google Scholar]
- 29.Zhao C, Blum J, Chen A, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12(6):528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tauriello DV, Maurice MM. The various roles of ubiquitin in Wnt pathway regulation. Cell Cycle. 2010;9(18):3700–3709. doi: 10.4161/cc.9.18.13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shekhar MP, Gerard B, Pauley RJ, Williams BO, Tait L. Rad6B is a positive regulator of beta-catenin stabilization. Cancer Res. 2008;68(6):1741–1750. doi: 10.1158/0008-5472.CAN-07-2111. [DOI] [PubMed] [Google Scholar]
- 32.Hay-Koren A, Caspi M, Zilberberg A, Rosin-Arbesfeld R. The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol Biol Cell. 2011;22(3):399–411. doi: 10.1091/mbc.E10-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hays LE, Keeble WW, Yates JE, et al. Human FANCC is hypomorphic in murine Fancc-deficient cells. Blood. 2010;116(12):2057–2060. doi: 10.1182/blood-2010-02-266411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13(8):680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 35.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272(45):28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 36.Lim KL, Chew KC, Tan JM, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25(8):2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livingston CM, Ifrim MF, Cowan AE, Weller SK. Virus-Induced Chaperone-Enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 2009;5(10):e1000619. doi: 10.1371/journal.ppat.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gummow BM, Winnay JN, Hammer GD. Convergence of Wnt signaling and steroidogenic factor-1 (SF-1) on transcription of the rat inhibin alpha gene. J Biol Chem. 2003;278(29):26572–26579. doi: 10.1074/jbc.M212677200. [DOI] [PubMed] [Google Scholar]
- 40.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10(1):55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 41.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10(12):1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 42.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16(13):3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Vrugt HJ, Koomen M, Berns MA, et al. Characterization, expression and complex formation of the murine Fanconi anaemia gene product Fancg. Genes Cells. 2002;7(3):333–342. doi: 10.1046/j.1365-2443.2002.00518.x. [DOI] [PubMed] [Google Scholar]
- 44.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9(6):536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marikawa Y, Elinson RP. beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech Dev. 1998;77(1):75–80. doi: 10.1016/s0925-4773(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 46.Winer IS, Bommer GT, Gonik N, Fearon ER. Lysine residues Lys-19 and Lys-49 of beta-catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J Biol Chem. 2006;281(36):26181–26187. doi: 10.1074/jbc.M604217200. [DOI] [PubMed] [Google Scholar]
- 47.Chitalia VC, Foy RL, Bachschmid MM, et al. Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol. 2008;10(10):1208–1216. doi: 10.1038/ncb1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuzawa SI, Reed JC. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell. 2001;7(5):915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- 49.Nastasi T, Bongiovanni A, Campos Y, et al. Ozz-E3, a muscle-specific ubiquitin ligase, regulates beta-catenin degradation during myogenesis. Dev Cell. 2004;6(2):269–282. doi: 10.1016/s1534-5807(04)00020-6. [DOI] [PubMed] [Google Scholar]
- 50.Gurtan AM, Stuckert P, D'Andrea AD. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J Biol Chem. 2006;281(16):10896–10905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- 51.Meetei AR, de Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35(2):165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 52.Hodson C, Cole AR, Lewis LP, Miles JA, Purkiss A, Walden H. Structural analysis of human FANCL, the E3 ligase in the Fanconi anemia pathway. J Biol Chem. 2011;286(37):32628–32637. doi: 10.1074/jbc.M111.244632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 54.Li W, Bengtson MH, Ulbrich A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3(1):e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21(21):3414–3421. doi: 10.1038/sj.onc.1205400. [DOI] [PubMed] [Google Scholar]
- 56.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423(6938):409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 57.Smith K, Dalton S. Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5(6):947–959. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malhotra S, Kincade PW. Wnt-related molecules and signaling pathway equilibrium in hematopoiesis. Cell Stem Cell. 2009;4(1):27–36. doi: 10.1016/j.stem.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barroca V, Mouthon MA, Lewandowski D, et al. Impaired functionality and homing of Fancg-deficient hematopoietic stem cells. Hum Mol Genet. 2012;21(1):121–135. doi: 10.1093/hmg/ddr447. [DOI] [PubMed] [Google Scholar]
- 60.Ito K, Hirao A, Arai F, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431(7011):997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 61.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Li X, Le Beau MM, Ciccone S, et al. Ex vivo culture of Fancc−/− stem/progenitor cells predisposes cells to undergo apoptosis, and surviving stem/progenitor cells display cytogenetic abnormalities and an increased risk of malignancy. Blood. 2005;105(9):3465–3471. doi: 10.1182/blood-2004-06-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bagby GC, Meyers G. Myelodysplasia and acute leukemia as late complications of marrow failure: future prospects for leukemia prevention. Hematol Oncol Clin North Am. 2009;23(2):361–376. doi: 10.1016/j.hoc.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.