Abstract
High BAALC expression levels are associated with poor outcome in cytogenetically normal acute myeloid leukemia (CN-AML) patients. Recently, miR-3151 was discovered in intron 1 of BAALC. To evaluate the prognostic significance of miR-3151 expression levels and to gain insight into the biologic and prognostic interplay between miR-3151 and its host, miR-3151 and BAALC expression were measured in pretreatment blood of 179 CN-AML patients. Gene-expression profiling and miRNA-expression profiling were performed using microarrays. High miR-3151 expression was associated with shorter disease-free and overall survival, whereas high BAALC expression predicted failure of complete remission and shorter overall survival. Patients exhibiting high expression of both miR-3151 and BAALC had worse outcome than patients expressing low levels of either gene or both genes. In gene-expression profiling, high miR-3151 expressers showed down-regulation of genes involved in transcriptional regulation, posttranslational modification, and cancer pathways. Two genes, FBXL20 and USP40, were validated as direct miR-3151 targets. The results of the present study show that high expression of miR-3151 is an independent prognosticator for poor outcome in CN-AML and affects different outcome end points than its host gene, BAALC. The combination of both markers identified a patient subset with the poorest outcome. This interplay between an intronic miR and its host may have important biologic implications.
Introduction
Acute myeloid leukemia (AML) is a clinically, cytogenetically, and molecularly heterogeneous disease. Despite recent advances in our understanding of the mechanisms of leukemogenesis and the identification of markers that allow molecular-based stratification to risk-adapted therapies, the majority of patients with AML are not cured.1 The clinical outcome is particularly poor in older (≥ 60 years of age) patients, who have long-term survival rates of only 7%-15%.2
To date, the prognostic impact of molecular genetic markers has been studied most extensively in patients with cytogenetically normal AML (CN-AML).3 In this large cytogenetic subset, several molecular markers have been found to be associated with outcome.3–9 Among them, mutations in the nucleophosmin (nucleolar phosphoprotein B23, numatrin; NPM1) and CCAAT/enhancer binding protein (C/EBP), apha (CEBPA) genes are associated with favorable outcome and have been included as provisional entities in the World Health Organization classification of AML.10 In addition, these 2 mutations and the presence of internal tandem duplications of the fms-related tyrosine kinase 3 gene (FLT3-ITD) that has been associated with unfavorable outcome have been incorporated into a standardized reporting system for genetic abnormalities suggested by an international expert panel on behalf of the European LeukemiaNet (ELN).11
Another strong prognostic factor associated with treatment resistance and poor outcome is high expression of the brain and acute leukemia cytoplasmic (BAALC) gene, which was identified by cDNA-based representational difference analysis in leukemia patients.12 The prognostic impact of BAALC expression has been most extensively studied in CN-AML patients.13–16 Although recent data suggest that BAALC contributes to leukemogenesis by interfering with normal patterns of myeloid differentiation,17 the function of this gene and the mechanism(s) through which its high expression levels affect clinical outcome negatively remain(s) unknown.
Recently, small RNA deep sequencing of melanoma18 and pediatric acute lymphoblastic leukemia samples19 identified a new miR, miR-3151, embedded in intron 1 of the BAALC gene. miRs are short, noncoding RNAs that hybridize to and regulate the expression of targeted mRNAs20 and have been implicated in leukemogenesis.21 Furthermore, expression levels of miRs may be used to refine risk assessment in AML in CN-AML patients.22,23
miR genes are located throughout the genome, but approximately one-third of mammalian miRs reside within the introns of host genes.24,25 Some of these intronic miRs have been found to act in functional synergism with their host genes.26 These findings led us to hypothesize that miR-3151 could be overexpressed in patients with elevated BAALC levels and thus might contribute to the adverse prognostic impact of its host gene, either acting in concert with BAALC or perhaps even being the element predominantly responsible for the adverse outcome observed in BAALC-overexpressing AML patients.
In the present study, we measured miR-3151 and BAALC expression in a cohort of molecularly well-characterized de novo CN-AML patients to assess the impact of miR-3151 expression alone and in combination with the expression of its host BAALC on outcome and also explored some downstream effects of miR-3151 expression.
Methods
Patients, treatment, and cytogenetic studies
One-hundred-seventy-nine patients 60 years of age or older with de novo CN-AML who were treated with intensive cytarabine/daunorubicin-based regimens on Cancer and Leukemia Group B (CALGB) frontline clinical protocols (for details see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), were included in this study. All study protocols received institutional review board approval at the participating institutions. Cytogenetic analyses were performed on pretreatment BM samples by CALGB-approved institutional cytogenetic laboratories as part of CALGB 8461, a prospective cytogenetic companion study.27 The diagnosis of normal cytogenetics was based on the analysis of ≥ 20 BM metaphase cells and confirmed by central karyotype review.28 All patients gave informed consent for the research use of their specimens in accordance with the Declaration of Helsinki.
Molecular analyses
Pretreatment peripheral blood samples were analyzed for miR-3151 and BAALC expression levels by real-time RT-PCR. The TaqMan assays were carried out for each sample in triplicate using TaqMan Primer-Probe sets for BAALC and miR-3151 and the respective housekeeping genes 18S and RNU44 (Life Technologies Corporation/Applied Biosystems) according to protocol instructions (see supplemental Methods).
Additional molecular markers were analyzed centrally in pretreatment BM or peripheral blood samples as described previously and included: FLT3-ITD,29 FLT3 tyrosine kinase domain mutations (FLT3-TKDs),30 partial tandem duplication of the myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) gene (MLL-PTD),31 mutations in the NPM1,32 CEBPA,33 tet methylcytosine dioxygenase 2 (TET2),6 additional sex combs like 1 (Drosophila [ASXL1]),7 DNA (cytosine-5-)-methyltransferase 3 alpha (DNMT3A),8 runt-related transcription factor 1 (RUNX1),9 Wilms tumor 1 (WT1),5 and isocitrate dehydrogenase 1 (NADP+), soluble (IDH1) and isocitrate dehydrogenase 2 (NADP+), mitochondrial (IDH2)4 genes, and expression levels of the v-ets erythroblastosis virus E26 oncogene homolog (avian; ERG)16 and meningioma (disrupted in balanced translocation) 1 (MN1)34 genes.
GEP and MEP
Gene-expression profiling (GEP) of samples was performed using the Affymetrix U133 plus 2.0 array (Affymetrix) and miR-expression profiling (MEP) was performed using The Ohio State University custom miR array (OSU_CCC Version 4.0), as described previously (see supplemental Methods for details).32,35 The miR microarray data have been deposited in ArrayExpress under accession number E-MTAB-1074 and the microarray expression data under accession number E-MTAB-1075.
Validation of FBXL20 and USP40 as direct miR-3151 targets
For stable expression of miR-3151, the miR-3151 stem-loop was cloned into a lentiviral expression vector, as described in supplemental Methods, using lentiviral miR-scramble as the respective control for all experiments. To analyze the effects of forced miR-3151 expression on the predicted target genes, we assessed the expression of FBXL20 and USP40 on mRNA level, as described in the supplemental materials, and compared it with the effects of cells infected with scramble control using real-time RT-PCR. Western Blotting to test the effects of miR-3151 on protein level was performed as described in detail in the supplemental materials. The 3′-untranslated regions of FBXL20 and USP40 were cloned into a luciferase reporter vector (wild-type vs mutated miR-3151–binding sequence, see supplemental Table 3 for details) and luciferase activity was assessed as described in supplemental Methods.
Definition of clinical end points and statistical analysis
The main objective of this study was to evaluate the prognostic impact on clinical outcome in older CN-AML patients of miR-3151 expression alone and in combination with its host gene, BAALC. Median expression levels of miR-3151 and BAALC were used to define low and high miR-3151 and BAALC expressers, respectively, for all analyses (see supplemental Methods for details).
Definitions of clinical end points (ie, complete remission [CR], disease-free [DFS], and overall survival [OS]) and details of statistical analyses, including variable selection for statistical modeling, are provided in supplemental Methods. Associations between patients with low and high expression of miR-3151 for baseline demographic, clinical, and molecular features were compared using the Fisher exact and Wilcoxon rank-sum tests for categorical and continuous variables, respectively. Estimated probabilities of DFS and OS were calculated using the Kaplan-Meier method, and the log-rank test was used to evaluate differences between survival distributions. Multivariable logistical regression models were constructed to analyze factors related to the probability of achieving CR using a limited backward selection procedure. Multivariable proportional hazards models were constructed for DFS and OS to evaluate the impact of miR-3151 expression by adjusting for other variables using a limited backward selection procedure. For achievement of CR, estimated odds ratios (ORs) and for survival end points, hazard ratios (HRs) with their corresponding 95% confidence intervals were obtained for each significant prognostic factor.
For the GEP and MEP, summary measures of gene and miR expression, respectively, were computed, normalized, and filtered (supplemental Methods). The profiles were derived by comparing gene expression between low and high miR-3151 expressers. Univariable significance levels of P = .001 for GEP (P = .005 for MEP) were used to determine the probe sets (probes) that comprised the signatures.
All clinical analyses were performed by the Alliance for Clinical Trials in Oncology Statistics and Data Center.
Results
Associations of miR-3151 expression with clinical and molecular characteristics
Patients with high miR-3151 expression had lower percentages of circulating blasts (P = .02), and were more likely to be NPM1 wild-type (P < .001), belong to the ELN Intermediate-I Genetic Group,11 and harbor mutations of RUNX1 (P < .001) than low expressers (P = .05; Table 1). In addition, high miR-3151 expression was associated with high expression levels of its host gene, BAALC (P < .001), and also of MN1 (P = .05). Approximately two-thirds of the patients had a concordant expresser status for miR-3151 and BAALC expression levels, whereas one-third of the patients exhibited up-regulation of only 1 of the 2 markers.
Table 1.
Clinical and molecular characteristics according to miR-3151 expression status in CN-AML patients 60 years of age or older
| Characteristic | Low miR-3151* (n = 90) | High miR-3151* (n = 89) | P† |
|---|---|---|---|
| Age, y | .99 | ||
| Median | 68 | 68 | |
| Range | 60-79 | 60-81 | |
| Sex, n (%) | .88 | ||
| Male | 48 (53) | 49 (55) | |
| Female | 42 (47) | 40 (45) | |
| Race, n (%) | .10 | ||
| White | 79 (88) | 83 (95) | |
| Nonwhite | 11 (12) | 4 (5) | |
| Hemoglobin, g/dL | .47 | ||
| Median | 9.3 | 9.5 | |
| Range | 6.0-11.7 | 6.5-15.0 | |
| Platelet count, × 109/L | .36 | ||
| Median | 60 | 69 | |
| Range | 4-481 | 11-850 | |
| WBC count, × 109/L | .10 | ||
| Median | 26.2 | 38.7 | |
| Range | 1.4-450.0 | 1.1-434.1 | |
| Percentage of blood blasts | .02 | ||
| Median | 65 | 39 | |
| Range | 0-96 | 0-99 | |
| Percentage of BM blasts | .52 | ||
| Median | 70 | 69 | |
| Range | 11-97 | 4-96 | |
| Extramedullary involvement, n (%) | 21 (24) | 24 (28) | .73 |
| NPM1, n (%) | < .001 | ||
| Mutated | 71 (79) | 43 (49) | |
| Wild-type | 19 (21) | 45 (51) | |
| FLT3-ITD, n (%) | 1.00 | ||
| Present | 33 (37) | 33 (38) | |
| Absent | 57 (63) | 55 (63) | |
| CEBPA, n (%) | .83 | ||
| Mutated | 12 (13) | 13 (15) | |
| Single mutated | 7 | 8 | |
| Double mutated | 5 | 5 | |
| Wild-type | 78 (87) | 76 (85) | |
| ELN Genetic Group, n (%)‡ | .05 | ||
| Favorable | 50 (56) | 35 (40) | |
| Intermediate-I | 40 (44) | 52 (60) | |
| RUNX1, n (%) | < .001 | ||
| Mutated | 4 (5) | 19 (24) | |
| Wild-type | 80 (95) | 59 (76) | |
| FLT3-TKD, n (%) | 1.00 | ||
| Present | 7 (8) | 7 (8) | |
| Absent | 82 (92) | 79 (92) | |
| WT1, n (%) | .08 | ||
| Mutated | 3 (3) | 9 (10) | |
| Wild-type | 87 (97) | 79 (90) | |
| TET2, n (%) | .33 | ||
| Mutated | 24 (27) | 30 (35) | |
| Wild-type | 65 (73) | 56 (65) | |
| MLL-PTD, n (%) | .50 | ||
| Present | 3 (5) | 6 (8) | |
| Absent | 63 (95) | 67 (92) | |
| IDH1, n (%) | 1.00 | ||
| R132 mutated | 12 (13) | 10 (11) | |
| V71I mutated | 0 | 1 (1) | |
| Wild-type | 77 (87) | 76 (87) | |
| IDH2, n (%) | .71 | ||
| IDH2 | 20 (22) | 17 (20) | |
| R140 mutated | 20 | 14 | |
| R172 mutated | 0 | 3 | |
| Wild-type | 69 (78) | 70 (80) | |
| ASXL1, n (%) | .13 | ||
| Mutated | 9 (10) | 16 (19) | |
| Wild-type | 79 (90) | 69 (81) | |
| DNMT3A, n (%) | .74 (mut vs wt) | ||
| Mutated | 27 (31) | 29 (35) | |
| R882 | 15 | 19 | .56 (R882 vs wt) |
| Non-R882 | 12 | 10 | 1.00 (non-R882 vs wt) |
| Wild-type | 59 (69) | 55 (65) | |
| ERG expression group, n (%)* | .40 | ||
| High | 42 (58) | 35 (50) | |
| Low | 31 (42) | 35 (50) | |
| BAALC expression group, n (%)* | < .001 | ||
| High | 34 (39) | 56 (66) | |
| Low | 54 (61) | 29 (34) | |
| MN1 expression group, n (%)* | .05 | ||
| High | 18 (37) | 31 (56) | |
| Low | 31 (63) | 24 (44) |
The median expression value was used as the cutoff point.
P values for categorical variables are from the Fisher exact test; P values for continuous variables are from the Wilcoxon rank-sum test.
The ELN Favorable Genetic Group comprises patients with mutated CEBPA and those with mutated NPM1 without FLT3-ITD; the ELN Intermediate-I Genetic Group includes patients with CEBPA wild-type who are FLT3-ITD–positive and NPM1-mutated, FLT3-ITD–negative and NPM1 wild-type, or FLT3-ITD–positive and NPM1 wild-type.
Prognostic value of miR-3151 expression in CN-AML
Patients with high miR-3151 expression had a lower CR rate (P = .005, 62% vs 81%) compared with low expressers. With a median follow-up time for living patients of 5.1 years (range, 4.1-11.6), high miR-3151 expressers had a shorter DFS (P = .003, HR = 1.76; Figure 1A). At 3 years after CR achievement, only 7% of high miR-3151–expressing patients were disease free compared with 26% of low expressers. High miR-3151 expressers also had a shorter OS (P < .001, HR = 1.86; Figure 1B). Three years after diagnosis, 10% of high miR-3151 expressers were still alive compared with 32% of low expressers.
Figure 1.
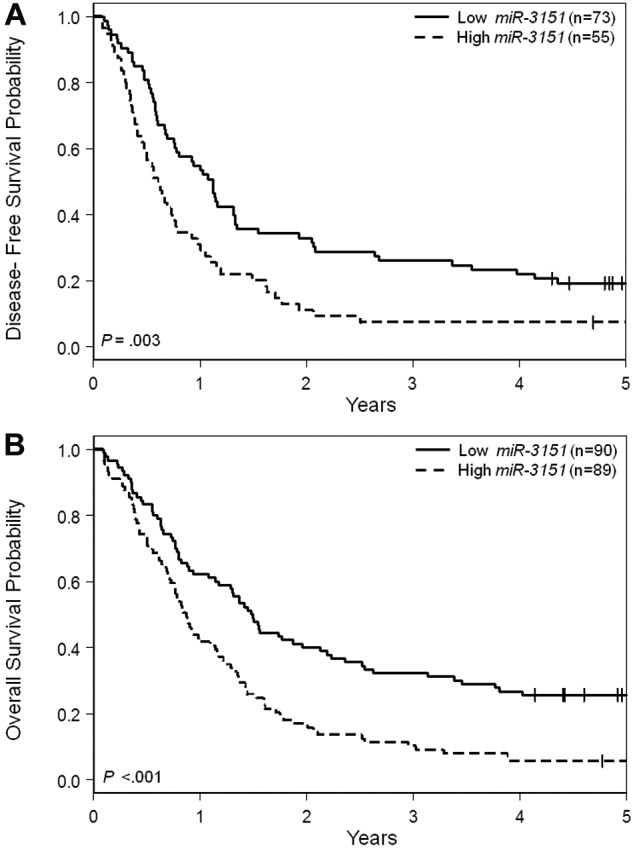
Outcome of CN-AML patients 60 years of age or older with respect to miR-3151 expression. (A) Disease-free survival. (B) Overall survival.
Because miR-3151 expression levels were associated with the expression levels of its host gene, BAALC, we analyzed the impact on outcome end points of both genes using bivariable models. Analyses showed that high expression levels of either marker had significant adverse impact on CR (miR-3151, P < .001, OR = 0.47; BAALC, P < .001, OR = 0.3), DFS (miR-3151, P = .01, HR = 1.68; BAALC, P = .003, HR = 1.82), and OS (miR-3151, P = .002, HR = 1.68; BAALC, P < .001, HR = 2.01). Therefore, miR-3151 and BAALC independently added information for determination of all outcome end points (supplemental Table 1).
Despite the strong association of miR-3151 and BAALC expression levels, approximately one-third of patients were discordant in expresser status of the 2 markers (Table 1). Therefore, we next investigated whether their combination (miR-3151/BAALC, high/high, high/low, low/high, low/low) would reveal differences in impact on outcome end points. Patients who had high expression of both miR-3151 and BAALC demonstrated the lowest CR rates (50%), whereas patients who highly expressed only 1 of the 2 markers had intermediate CR rates (low miR-3151/high BAALC, 71%; high miR-3151/low BAALC, 79%), and low expressers of both markers had the highest CR rate (87%, P < .001). Patients with high expression of both miR-3151 and BAALC had significantly shorter DFS and OS than those expressing both markers at low levels (P < .001 for both DFS and OS; Figure 2A-B) or those who exhibited high expression of only one of the markers (DFS, P = .03, OS, P = .01). Of patients who expressed both the miR and its host gene at high levels, only 1 survived longer than 3 years after diagnosis.
Figure 2.
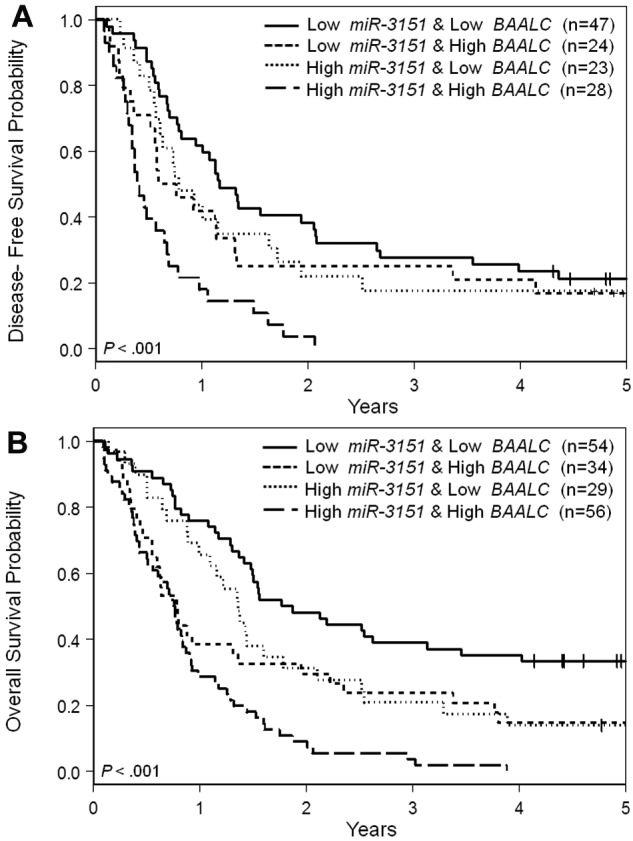
Outcome of CN-AML patients 60 years of age or older with respect to miR-3151 and BAALC expression. (A) Disease-free survival. (B) Overall survival.
In multivariable analyses (Table 2), after adjustment for BAALC expression status and WBC count, patients with high miR-3151 expression had a trend toward a lower CR rate (P = .13, OR = 0.56). High miR-3151 expressers had shorter DFS (P < .001, HR = 2.38), after adjustment for FLT3-TKD and ERG expression status, and shorter OS (P = .009, HR = 1.69) after adjustment for DNMT3A R882 mutations and ERG and BAALC expression status.
Table 2.
Multivariable analysis for outcome according to the miR-3151 expression status in older patients with CN-AML
| End point | Variables in final models | OR/HR | 95% CI | P |
|---|---|---|---|---|
| CR* | miR-3151, high vs low | 0.56 | 0.27-1.19 | .13 |
| BAALC, high vs low | 0.23 | 0.10-0.52 | < .001 | |
| WBC, continuous, 50-unit increase | 0.65 | 0.48-0.88 | .01 | |
| DFS† | miR-3151, high vs low | 2.38 | 1.47-3.85 | < .001 |
| FLT3-TKD, positive vs no TKD | 0.28 | 0.11-0.73 | .009 | |
| ERG, high vs low | 2.41 | 1.50-3.86 | < .001 | |
| OS‡ | miR-3151, high vs low | 1.69 | 1.14-2.50 | .009 |
| ERG, high vs low | 1.71 | 1.16-2.53 | .007 | |
| DNMT3A | ||||
| R882 v wild-type | 1.66 | 1.04-2.66 | .04 | |
| Non-R882 vs wild-type | 1.11 | 0.61-2.01 | .74 | |
| BAALC, high vs low | 1.93 | 1.32-2.82 | < .001 |
CR indicates complete remission; OR, odds ratio; HR, hazard ratio; OS, overall survival; DFS, disease-free survival; OR > 1.0, a higher CR rate for the higher values of the continuous variables and the first category listed for the categorical variables; OR < 1.0, a lower CR rate for the higher values of the continuous variables and the first category listed for the categorical variables; HR > 1.0, a higher risk for an event for the first category listed for the categorical variables; HR < 1.0, a lower risk for an event for the first category listed for the categorical variables; and 95% CI, 95% confidence interval..
Variables considered in the model based on univariable analyses were: miR-3151 expression (high vs low; median cut), BAALC expression (high vs low; median cut), ERG expression (high vs low; median cut), FLT3-ITD (positive vs no ITD), NPM1 (mutated vs wild-type), WT1 (mutated vs wild-type), MLL-PTD (present vs absent), ASXL1 (mutated vs wild-type), RUNX1 (mutated vs wild-type), WBC (continuous, 50-unit increase), platelets (continuous, 50-unit increase), and age (continuous, 10-year increase).
Variables considered in the model based on univariable analyses were: miR-3151 expression (high vs low; median cut), BAALC expression (high vs low; median cut), ERG expression (high vs low; median cut), FLT3-ITD (positive vs no ITD), FLT3-TKD (positive vs no TKD), NPM1 (mutated vs wild-type), DNMT3A (R882 vs Non-R882 vs wild-type), RUNX1 (mutated vs wild-type), WT1 (mutated vs wild-type), and WBC (continuous, 50-unit increase).
Variables considered in the model based on univariable analyses were: miR-3151 expression (high vs low; median cut), BAALC expression (high vs low; median cut), ERG expression (high vs low; median cut), FLT3-ITD (positive vs no ITD), NPM1 (mutated vs wild-type), ASXL1 (mutated vs wild-type), RUNX1 (mutated vs wild-type), WT1 (mutated vs wild-type), DNMT3A (R882 vs non-R882 vs wild-type), platelets (continuous, 50-unit increase), and WBC (continuous, 50-unit increase).
miR-3151 expression in the context of ELN Genetic Groups
The ELN Genetic Groups have been shown to be associated with outcome in older CN-AML patients.36 Moreover, recent studies have demonstrated that the addition of selected new molecular markers can further improve prognostication within the ELN Genetic Groups.6,7,34 Therefore, we investigated whether miR-3151 expression levels could improve outcome prediction within those ELN Genetic Groups that include CN-AML. Within the ELN Favorable Genetic Group, there were no differences in outcome between high and low miR-3151–expressing patients. However, in the ELN Intermediate-I Genetic Group, high miR-3151 expression levels identified patients with particularly poor prognosis for all 3 outcome end points. Only 52% of high miR-3151 expressers in this ELN group achieved a CR, compared with 78% of low miR-3151 expressers (P < .001). Likewise, patients with high miR-3151 expression levels had significantly shorter DFS (< .001; Figure 3A) and OS (P < .001; Figure 3B) compared with low miR-3151 expressers. For all 3 end points, the outcome of the low miR-3151 expressers classified in the ELN Intermediate-I Group was similar to that of both high and low miR-3151–expressing patients in the ELN Favorable Genetic Group (Figure 3A-B and supplemental Table 2).
Figure 3.
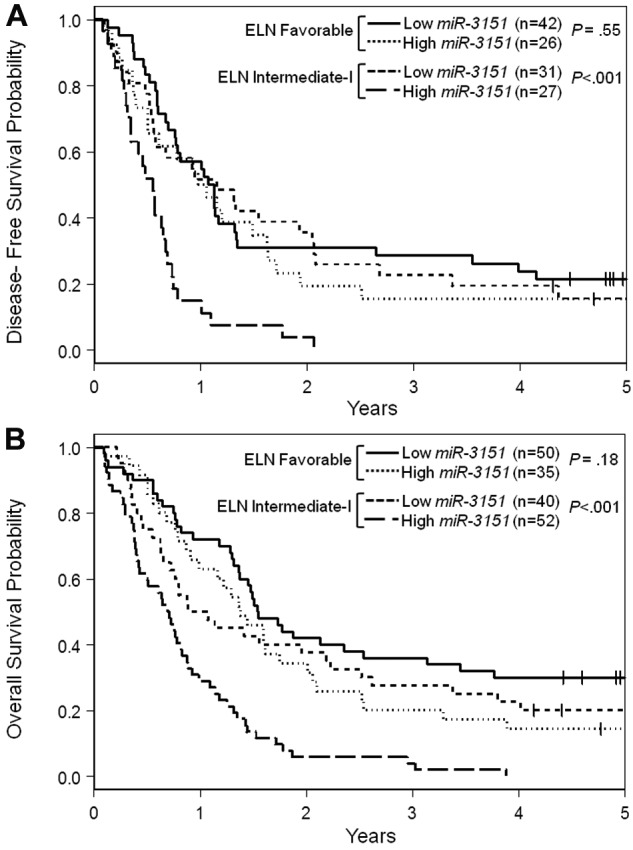
Outcome of CN-AML patients 60 years of age or older with respect to miR-3151 expression and ELN Genetic Group. (A) Disease-free survival. (B) Overall survival.
Biologic insights
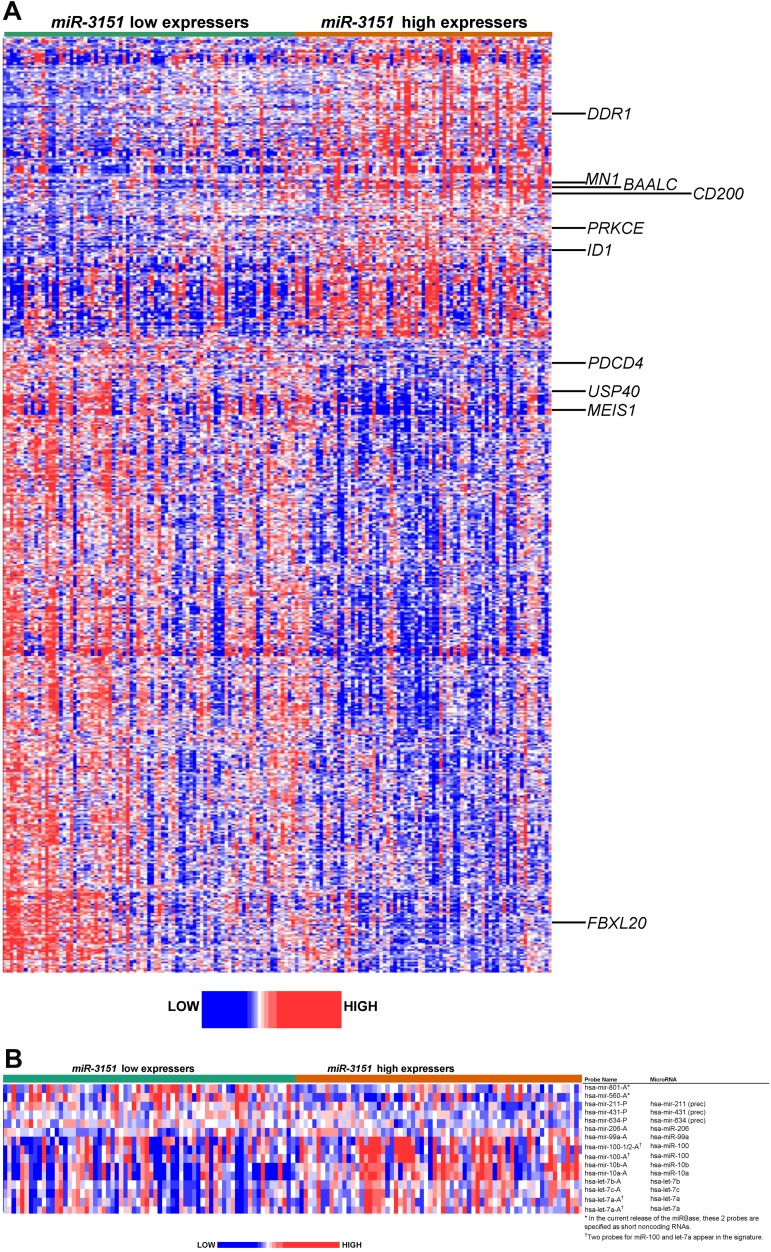
To gain biologic insights into miR-3151–associated leukemia, we derived a gene-expression signature comparing high versus low miR-3151 expressers. High miR-3151 expresser status was associated with the differential expression of 597 probe sets, representing 374 annotated genes. Of these, 192 probe sets (116 annotated genes) were up-regulated and 405 probe sets (258 annotated genes) were down-regulated (Figure 4A and supplemental Tables 4 and 5).
Figure 4.
Heat map of the derived gene- and miR-expression signature associated with miR-3151 expression in CN-AML patients 60 years of age or older. Expression values of the probe sets (probes) are represented by color, with blue indicating expression less than and red indicating expression greater than the median value for the given probe set (probe). (A) GEP: up-regulated and down-regulated genes that are mentioned in the text are indicated along the side. (B) MEP: up-regulated and down-regulated miRs are indicated along the side.
High miR-3151 expressers exhibited up-regulation of genes previously associated with worse outcome in CN-AML, including the transcriptional coregulator MN1,34 the dominant negative helix-loop-helix protein ID1,37 the miR-3151 host BAALC, and the surface marker CD200.38 In addition, we observed up-regulation of genes encoding several kinases, such as DDR1, which has been shown to be important for cell growth and differentiation by activation of NOTCH1,39 and PRKCE,40 which has been shown to be involved in apoptosis and several cellular signaling pathways.
Among the most down-regulated genes in high miR-3151 expressers was the HOX cofactor MEIS1, which is a key regulator in developmental processes and the absence of which is known to cause disturbances in the colony-forming ability of hematopoietic stem cells,41and the tumor suppressor PDCD4, which has been shown to contribute to retinoic acid-induced granulocytic differentiation.42 Furthermore, down-regulation of 23 genes encoding different zinc finger proteins (ZNFs), which are known to be involved in transcriptional regulation, was found in high miR-3151 expressers. Of the 258 down-regulated genes, 73 were in silico predicted targets of miR-3151 (www.microrna.org; supplemental Table 5).
Pathway analysis of the miR-3151–associated expression signature showed an enrichment of genes involved in transcriptional regulation, posttranslational modification, cell-cycle control, cellular development, and cancer pathways, suggesting an impact of miR-3151 on basic regulatory functions (http://ingenuity.com; supplemental Table 6).
To further investigate the networking processes of known miRs in which miR-3151 might be involved, we derived miR-expression signatures associated with miR-3151 expresser status (Figure 4B). We found 15 differentially expressed probes, representing 14 miRs, 5 up-regulated and 9 down-regulated in high compared with low miR-3151 expressers. Among the down-regulated miRs were let-7a, let-7b, and let-7c, which are known to suppress tumorigenesis by participating in many cell-proliferation pathways43 and the down-regulation of which has been previously associated with AML leukemogenesis.22 We also observed down-regulation of miR-10a and miR-10b, which are miRs embedded in HOX gene clusters, and miR-99a and miR-100, the reduced expression of which has been implicated in tumor progression in cervical and prostate cancers.44,45
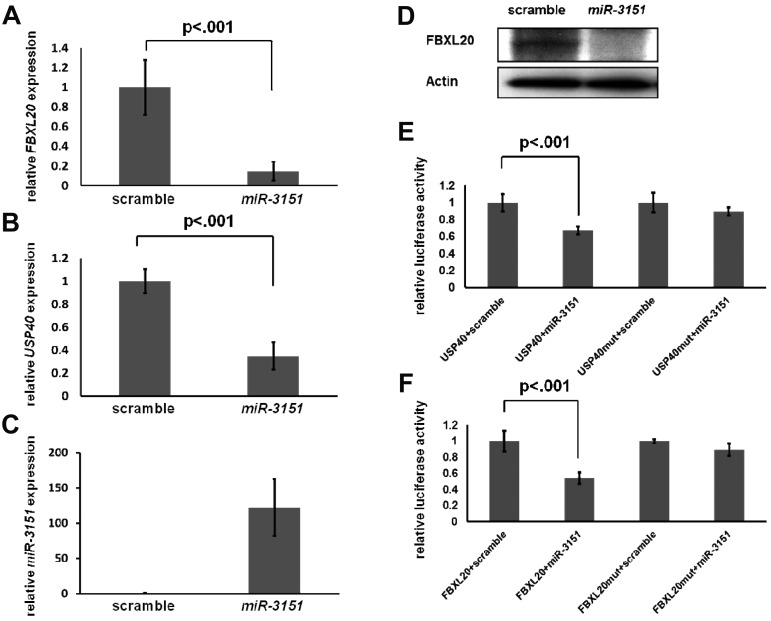
To gain initial insights into the downstream effects of high miR-3151 expression, we sought to validate 1 or 2 of the down-regulated genes in the miR-3151–associated gene expression signature as a direct target of miR-3151. For the identification of potential candidate genes, we searched among the in silico predicted targets for probe sets of annotated genes that showed at least a 25% down-regulation with P < .0001. Only 6 of the 73 genes fulfilled these criteria (see supplemental Table 5 gray highlights). Among these, we selected as potential candidates those with a described function that was associated with the pathways shown to be preferentially involved (supplemental Table 6). Strikingly, 2 of the 6 genes, the F-box and leucine-rich repeat protein 20 (FBXL20) and the ubiquitin-specific protease 40 (USP40), are involved in the ubiquitination pathway, suggesting an impact of miR-3151 on this important posttranslational regulatory process. FBXL20 is a member of the F-box gene family.46 As a part of the SCF (Skp, Cullin, F-box containing) ubiquitin ligase complex, it is responsible for the ubiquitination of proteins, thereby labeling them for consecutive proteasomal degradation. USP40 belongs to the family of cysteine proteases that function as deubiquitinating enzymes.47
To validate FBXL20 and USP40 as direct targets of miR-3151, we stably overexpressed miR-3151 in KG1 cells using a lentiviral system. Forced expression of miR-3151 resulted in significant down-regulation of the FBXL20 and USP40 transcripts (FBXL20, 85% decrease, Figure 5A; USP40, 66% decrease, Figure 5B; both P < .001) compared with scramble control. miR-3151 also down-regulated FBXL20 on the protein level (Figure 5D). However, none of the commercially available USP40 Abs showed a band at the predicted protein size of 140 kDa. To demonstrate that FBXL20 and USP40 are direct targets of miR-3151, the respective 3′-untranslated regions of both genes with the sequence containing the predicted miR-3151–binding sites were cloned into luciferase reporter vectors. The luciferase assays demonstrated a 54% and 33% decrease in luciferase activity for the FBXL20 and USP40 constructs, respectively, after addition of miR-3151 compared with scramble control (Figure 5E-F). The observed down-regulations were abrogated after mutation of the seed sequence of the predicted miR-3151–binding sites (Figure 5E-F). These results demonstrate that FBXL20 and USP40 are direct targets of miR-3151.
Figure 5.
Validation of FBXL20 and USP40 as direct targets of miR-3151. Expression levels of FBXL20 and USP40 as determined by RT-PCR in KG1 cells infected in triplicate with miR-3151 compared relative to scramble control are shown. Forced miR-3151 expression resulted in an 85% decrease of FBXL20 (± SD; A) and a 66% decrease of USP40 expression levels (± SD; B, both P < .001; expression levels are displayed relative to scramble control). (C) Validation of increased miR-3151 expression levels after lentiviral infection. (D) Western blot of FBXL20 expression comparing miR-3151 infected KG1 cells versus scramble control. miR-3151 expression results in elimination of the FBXL20 protein. The effect of miR-3151 on luciferase activity is shown for the FBXL20 (E) and USP40 (F) 3′-UTRs (cloned 3′ of the luciferase gene). The addition of miR-3151 resulted in a decrease of luciferase activity compared with scramble control of 54% for FBXL20 and of 33% for USP40 (± SD; both P < .001). This effect was abrogated after mutation of the respective binding sequences of the predicted miR-3151–binding sites.
Discussion
In the present study, we report an intronic miR as an independent prognostic factor for outcome in older CN-AML patients. Furthermore, our results suggest that miR-3151 may act in concert with its host gene, the established molecular prognostic marker BAALC.
In considering previous findings of a similar nature, the interplay of miR-33 and its SREBP host gene is an illustrative example. It has been shown that the intronic miR-33 targets the ATP-binding cassette transporter A1 (ABCA1), an important regulator of high-density lipoprotein synthesis and reverse cholesterol transport, thereby acting in synergism with SREBP to control cholesterol homeostasis.26 Recently, cooperation between intronic miRs and their host genes has been also demonstrated in prostate cancer48 and hepatocellular carcinoma.49 However, to our knowledge, similar mechanisms have not been reported so far in AML. Even though it was not the aim of our study to determine a functional mechanism of miR-3151 and BAALC interaction, our data suggest that both markers contribute independently to poor outcome in CN-AML patients.
We also found that, like BAALC, higher expression levels of miR-3151 were associated with poor prognosis. However, the hosted miR and the hosting gene have an independent clinical significance and had different effects on specific outcome end points. Whereas miR-3151 expression did not remain an independent predictor in the multivariable model for achievement of CR, BAALC expression remained a strong prognostic factor for CR. This is consistent with the findings of our group and others that CR rate is an outcome end point that is strongly affected by aberrant BAALC expression levels.13,15,16 Conversely, high miR-3151 expression and not BAALC expression was a strong predictor of DFS. Finally, both miR-3151 and BAALC expression levels remained important prognostic factors for OS.
We also found that patients overexpressing both miR-3151 and its host gene BAALC had particularly poor outcome for all outcome end points. In contrast, patients exhibiting up-regulation of only 1 of the 2 markers had a significantly better outcome; their DFS was comparable to that of low miR-3151/low BAALC-expressing patients and their OS was intermediate in comparison with OS of both groups of patients who had concordant miR-3151 and BAALC expresser status. These findings suggest that miR-3151 and BAALC act independently to affect outcome, thereby possibly creating a synergism to support leukemogenesis.
In the present study, by deriving a GEP signature comparing high and low miR-3151–expressing patients, we were able to gain initial insights into the biology and possible downstream effects of miR-3151 in CN-AML patients. We showed that high miR-3151-expressing patients also had up-regulation of genes that are known prognosticators of worse outcome in AML patients, such as MN1, ID1, and CD200 and the miR-3151 host gene BAALC. Among the down-regulated genes associated with high miR-3151 expression, we found several that were implicated in hematopoietic differentiation, including MEIS1, the absence of which has been shown to interfere with the normal development of hematopoietic precursors.42 In addition, we observed a down-regulation of multiple genes encoding ZNF proteins, suggesting a role for miR-3151 in pretranscriptional regulation.
Interestingly, pathway analysis of the miR-3151–associated gene-expression signature suggested an involvement of miR-3151 in general regulatory processes on both the transcriptional and posttranslational levels. Identification of direct target genes of miR-3151 belonging to these pathways may pinpoint a root cause for the downstream effects, including the pathophysiological consequences seen in high miR-3151–expressing CN-AML patients. Indeed, we were able to validate FBXL20 and USP40 as direct targets of miR-3151. Both genes are involved in the ubiquitination pathway, which is known to be important for cell-cycle control, cell growth, and a multitude of transcriptional and posttranscriptional regulatory processes.46,47,50 Even though FBXL20 and USP40 are likely only the first of several important miR-3151 targets, their involvement may initiate and direct future research into our understanding of the function of miR-3151.
The miR-3151–associated GEP signature shared some features with a recently described GEP signature associated with BAALC expression in older CN-AML patients, in which we also reported an up-regulation of MN1 and CD200 and down-regulation of MEIS1.16 However, changes in the expression of other genes were unique to the GEP signature associated with miR-3151 (eg, up-regulation of ID1 and down-regulation of ZNFs). Moreover, key components of the BAALC signature (eg, up-regulation of CD34 and PROM1 and down-regulation of several HOX gene clusters) were not found in our miR-3151–associated signature, suggesting important differences in the biologic activities of miR-3151 and BAALC.
Regarding the derived MEP signature, we found 14 miRs to be differentially expressed between high and low miR-3151 expressers. Among these miRs, a key component was down-regulation of members of the let-7 family, which are known tumor suppressors and the down-regulation of which is found in various types of cancer,43 linking the let-7 family members to the HMGA2 and RAS pathways.
Comparing the derived MEP signature with the signature described to be associated with BAALC expression,16 we observed interesting similarities but also differences. High BAALC-expressing patients exhibited down-regulation of miR-99a, miR-100, and let-7b, whereas down-regulation of let-7a and let-7c seemed to be uniquely associated with high miR-3151 expression levels. Down-regulation of miR-9 was only observed in the signature associated with high BAALC expression levels. No up-regulated miR was shared by both MEP signatures.
The fact that we were able to identify miR-3151 as the second miR after miR-181a21,23 that independently affects outcome of CN-AML patients provides further support to the importance of aberrantly expressed miRs as prognostic factors in CN-AML.
It is noteworthy that miR-3151 is not simply a new molecular marker, but to our knowledge is the first example of how known factors (here BAALC) in AML leukemogenesis may be supported by miRs located in the locus itself. In the case of the BAALC gene, this finding is of special interest because the gene's function and therefore the reasons for its strong prognostic impact in CN-AML patients are unknown.
In conclusion, we have shown in the present study that high expression of miR-3151 is an independent prognostic factor associated with poor outcome in older CN-AML patients. The partly discordant expresser status of miR-3151 and its host gene BAALC, the independent impact of the 2 genes on outcome, and the fact that patients overexpressing both miR-3151 and its host gene have significantly worse outcomes than those exhibiting up-regulation of only 1 of the genes suggest that the 2 genes contribute to the aggressiveness of the disease through different mechanisms. We conclude that determining the expression levels of miR-3151 at diagnosis will help to improve the risk stratification of older CN-AML patients. The development of therapies targeting miR-3151 up-regulation with synthetic inhibitors may provide novel, more effective strategies for personalized treatment of these patients.
Supplementary Material
Acknowledgments
The authors thank Donna Bucci of the CALGB Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center (Columbus, OH) for sample processing and storage services; Lisa J. Sterling and Colin G. Edwards (CALGB Cytogenetics Data Management Center at The Ohio State University Comprehensive Cancer Center) for data management; and The Ohio State University Comprehensive Cancer Center's Nucleic Acid and Microarray Shared Resources for technical support.
This work was supported in part by the National Cancer Institute (grants CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658, and CA129657), the Coleman Leukemia Research Foundation, the Deutsche Krebshilfe–Dr Mildred Scheel Cancer Foundation (H.B.), the Pelotonia Fellowship Program (A.-K.E.), and the Conquer Cancer Foundation (J.H.M.).
Footnotes
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
Presented in part at the 53rd Annual Meeting of the American Society of Hematology, San Diego, CA, December 10, 2011.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.-K.E., G.M., K. Mrózek, S.M.T., A.d.l.C., and C.D.B. designed the study, analyzed the data, and wrote the manuscript; A.-K.E., S.S., H.B., S.P.W., K.H.M., J.H.M., Y.-Z.W., and R.P. performed the laboratory-based research; K. Maharry, M.D.R., D.N., and S.L. performed the statistical analyses; G.M., M.R.B., B.L.P., T.H.C., J.O.M., J.E.K., M.W., M.A.C., R.A.L., and C.D.B. were involved directly or indirectly in the care of patients and/or sample procurement; and all authors read and agreed upon the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
For a complete list of participating Alliance institutions, principal investigators, and cytogeneticists, please see the supplemental Appendix.
Correspondence: Clara D. Bloomfield, MD, The Ohio State University Comprehensive Cancer Center, 1216 James Cancer Hospital, 300 West 10th Ave, Columbus, OH 43210; e-mail: clara.bloomfield@osumc.edu.
References
- 1.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Fröhling S, Schlenk RF, Kayser S, et al. Cytogenetics and age are major determinants of outcome in intensively treated acute myeloid leukemia patients older than 60 years: results from AMLSG trial AML HD98-B. Blood. 2006;108(10):3280–3288. doi: 10.1182/blood-2006-04-014324. [DOI] [PubMed] [Google Scholar]
- 3.Mrózek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109(2):431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcucci G, Maharry K, Wu Y-Z, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(14):2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker H, Marcucci G, Maharry K, et al. Mutations of the Wilms tumor 1 gene (WT1) in older patients with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(5):788–792. doi: 10.1182/blood-2010-01-262543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metzeler KH, Maharry K, Radmacher MD, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2011;29(10):1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30(7):742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendler JH, Maharry K, Radmacher MD, et al. Poor outcome of RUNX1-mutated (RUNX1-mut) patients (pts) with primary, cytogenetically normal acute myeloid leukemia (CN-AML) and associated gene- and micro-RNA expression signatures [abstract]. Blood (ASH Annual Meeting Abstracts) 2011;118(21):1476–1477. Abstract 3454. [Google Scholar]
- 10.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 11.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 12.Tanner SM, Austin JL, Leone G, et al. BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc Natl Acad Sci U S A. 2001;98(24):13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldus CD, Tanner SM, Ruppert AS, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B study. Blood. 2003;102(5):1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 14.Bienz M, Ludwig M, Leibundgut EO, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11(4):1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 15.Eid MA, Attia M, Abdou S, et al. BAALC and ERG expression in acute myeloid leukemia with normal karyotype: impact on prognosis. Int J Lab Hematol. 2010;32(2):197–205. doi: 10.1111/j.1751-553X.2009.01168.x. [DOI] [PubMed] [Google Scholar]
- 16.Schwind S, Marcucci G, Maharry K, et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(25):5660–5669. doi: 10.1182/blood-2010-06-290536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuser M, Berg T, Kuchenbauer F, et al. Functional role of BAALC in leukemogenesis [letter]. Leukemia. 2012;26(3):532–536. doi: 10.1038/leu.2011.228. [DOI] [PubMed] [Google Scholar]
- 18.Stark MS, Tyagi S, Nancarrow DJ, et al. Characterization of the melanoma miRNAome by deep sequencing. PLoS One. 2010;5(3):e9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schotte D, Akbari Moqadam F, Lange-Turenhout EAM, et al. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25(9):1389–1399. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 22.Garzon R, Volinia S, Liu C-G, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwind S, Maharry K, Radmacher MD, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(36):5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10a):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths-Jones S. Annotating noncoding RNA genes. Annu Rev Genomics Hum Genet. 2007;8:279–298. doi: 10.1146/annurev.genom.8.080706.092419. [DOI] [PubMed] [Google Scholar]
- 26.Najafi-Shoushtari SH, Kristo F, Li Y, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328(5985):1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrd JC, Mrózek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100(13):4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 28.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. Int J Oncol. 2008;33(2):239–244. [PMC free article] [PubMed] [Google Scholar]
- 29.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a Cancer and Leukemia Group B study. Cancer Res. 2001;61(19):7233–7239. [PubMed] [Google Scholar]
- 30.Whitman SP, Ruppert AS, Radmacher MD, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111(3):1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caligiuri MA, Strout MP, Schichman SA, et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56(6):1418–1425. [PubMed] [Google Scholar]
- 32.Becker H, Marcucci G, Maharry K, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(4):596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26(31):5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwind S, Marcucci G, Kohlschmidt J, et al. Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood. 2011;118(15):4188–4198. doi: 10.1182/blood-2011-06-357764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mrózek K, Marcucci G, Maharry K, et al. Prognostic utility of the European LeukemiaNet (ELN) genetic-risk classification in adults with de novo acute myeloid leukemia (AML): a study of 1,550 patients (pts) [abstract]. Blood (ASH Annual Meeting Abstracts) 2011;118(21):190–191. Abstract 414. [Google Scholar]
- 37.Tang R, Hirsch P, Fava F, et al. High Id1 expression is associated with poor prognosis in 237 patients with acute myeloid leukemia. Blood. 2009;114(14):2993–3000. doi: 10.1182/blood-2009-05-223115. [DOI] [PubMed] [Google Scholar]
- 38.Tonks A, Hills R, White P, et al. CD200 as a prognostic factor in acute myeloid leukaemia [letter]. Leukemia. 2007;21(3):566–568. doi: 10.1038/sj.leu.2404559. [DOI] [PubMed] [Google Scholar]
- 39.Kim H-G, Hwang S-Y, Aaronson SA, Mandinova A, Lee SW. DDR1 receptor tyrosine kinase promotes prosurvival pathway through Notch1 activation. J Biol Chem. 2011;286(20):17672–17681. doi: 10.1074/jbc.M111.236612. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Knauf JA, Elisei R, Mochly-Rosen A, et al. Involvement of protein kinase Cepsilon (PKCepsilon) in thyroid cell death: a truncated chimeric PKCepsilon cloned from a thyroid cancer cell line protects thyroid cells from apoptosis. J Biol Chem. 1999;274(33):23414–23425. doi: 10.1074/jbc.274.33.23414. [DOI] [PubMed] [Google Scholar]
- 41.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15(10):5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozpolat B, Akar U, Steiner M, et al. Programmed cell death-4 tumor suppressor protein contributes to retinoic acid-induced terminal granulocytic differentiation of human myeloid leukemia cells. Mol Cancer Res. 2007;5(1):95–108. doi: 10.1158/1541-7786.MCR-06-0125. [DOI] [PubMed] [Google Scholar]
- 43.Boyerinas B, Park S-M, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17(1):F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 44.Sun D, Lee YS, Malhotra A, et al. miR-99 family of microRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71(4):1313–1324. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li BH, Zhou JS, Ye F, et al. Reduced miR-100 expression in cervical cancer and precursors and its carcinogenic effect through targeting PLK1 protein. Eur J Cancer. 2011;47(14):2166–2174. doi: 10.1016/j.ejca.2011.04.037. [DOI] [PubMed] [Google Scholar]
- 46.Bai C, Sen P, Hofmann K, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86(2):263–74. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 47.Quesada V, Díaz-Perales A, Gutiérrez-Fernández A, et al. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun. 2004;314(1):54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 48.Poliseno L, Salmena L, Riccardi L, et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3(117):ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y, Lu Y, Zhang Q, et al. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40(10):4615–4625. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21(6):816–24. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.