Abstract
Following viral infection, cells rapidly present peptides from newly synthesized viral proteins on MHC class I molecules, likely from rapidly degraded forms of nascent proteins. The nature of these defective ribosomal products (DRiPs) remains largely undefined. Using inhibitors of RNA polymerase II that block influenza A virus neuraminidase (NA) mRNA export from the nucleus and inhibit cytoplasmic NA translation, we demonstrate a surprising disconnect between levels of NA translation and generation of SIINFEKL peptide genetically inserted into the NA stalk. A 33-fold reduction in NA expression is accompanied by only a 5-fold reduction in Kb-SIINFEKL complex cell-surface expression, resulting in a net 6-fold increase in the overall efficiency of Ag presentation. Although the proteasome inhibitor MG132 completely blocked Kb-SIINFEKL complex generation, we were unable to biochemically detect a MG132-dependent cohort of NA DRiPs relevant for Ag processing, suggesting that a minute population of DRiPs is a highly efficient source of antigenic peptides. These data support the idea that Ag processing uses compartmentalized translation, perhaps even in the nucleus itself, to increase the efficiency of the generation of class I peptide ligands.
By recognizing foreign and abnormal self-peptides bound to MHC class I molecules, cytotoxic CD8+ T cells play a critical role in immunosurveillance of intracellular pathogens and neoplasias. Although the basic outlines of Ag processing, the process of degrading proteins into peptides and the loading of peptides onto MHC class I molecules, are delineated, many questions of importance to immunology and basic cell biology remain unanswered. A central question is the nature of the substrates that give rise to class I peptide ligands. There is mounting evidence that a large fraction of peptides derive from defective ribosomal products (DRiPs): a subset of newly synthesized proteins rapidly degraded by the cell (1). Defining the biochemical nature of DRiPs will likely provide novel insights into protein translation and cell quality-control decisions and is practically important for rationally manipulating CD8+ T cell activation (vaccines) and deactivation (autoimmunity).
DRiPs have been widely studied in the context of viral infections, where the transient and synchronous nature of viral gene expression simplifies kinetic analysis and clearly links viral protein synthesis tightly to peptide generation (2–4). Although most viral proteins studied are extremely stable metabolically (half-lives on the order of days), the rate of peptide generation closely tracks the rate of source protein synthesis, strongly implicating DRiPs as the major source of peptides. Using influenza A virus (IAV) modified to express the SIINFEKL reporter peptide in the stalk of the neuraminidase (NA) (5), in conjunction with the 25-D1.16 mAb specific for Kb-SIINFEKL complexes (6), we showed that Ag presentation occurred in complete lockstep with NA protein synthesis (7). Precise kinetic measurements indicated that the substrate for NA antigenic peptides has a half-life on the order of 5 min. We were unable to detect a biochemical cohort as a possible source of SIINFEKL; although MG132 blockade of proteasomes completely prevented detection of Kb-SIINFEKL complexes, it did not increase any species of NA that we could detect using a polyclonal Ab raised to the C-terminal region that recognizes denatured NA with high sensitivity.
Amorim et al. (8) reported that the RNA polymerase (RNAP) II inhibitor 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) blocks late IAV gene expression by preventing export of the corresponding IAV mRNAs. Recognizing this as a potential tool for separating viral protein expression from viral peptide generation, we examined the effect of DRB on NA-SIINFEKL biogenesis and Kb-SIINFEKL generation. In this article, we report a clear dissociation between the two processes.
Materials and Methods
Cells, Abs, and viruses
The mouse cell line L-Kb and the human cell line HeLa-Kb were cultured in DMEM containing 7.5% FCS in a 9% CO2 incubator. In both cell lines, Kb is expressed from a transfected gene under the control of the CMV immediate early promoter. The IAV strain A/Puerto Rico/8/34 (PR8) and modified PR8 virus expressing SIINFEKL in the NA protein (insOVA) (5) were grown in 10-d old embryonated chicken eggs, and infectious allantoic fluid were titered on MDCK cells. mAbs 25-D1.16 (anti–Kb-SIINFEKL) and NA2-1C1 (anti-NA) were labeled with Alexa Fluor 647 or Pacific Blue, respectively, using protein labeling kits from Molecular Probes (Eugene, OR), following the manufacturer’s recommended protocol. The anti-NA and anti-nucleoprotein (NP) rabbit polyclonal Ab was described previously (2, 7). Anti-p97 mouse mAb was from Fitzgerald (Concord, MA). The anti-hemagglutinin (HA) mAb CM1 was described previously (9). Donkey anti-rabbit secondary Abs labeled with the infrared (IR) dye 680CW and donkey anti-mouse secondary Abs labeled with the IR dye 800CW were from LI-COR (Lincoln, NE). PE anti-mouse H-2Kb (clone #AF6-88.5) was from BD Biosciences (San Jose, CA).
Viral infections
L-Kb cells and HeLa-Kb cells were resuspended to 2 × 106 cell/ml in autoclavable minimal essential media buffered to pH 6.6. IAV was added at a multiplicity of infection of 10 TCID50, and cells were infected for 30 min in a 37°C water bath with occasional agitation. Cells were washed and cultured at 1 × 106 cell/ml in tissue culture media for the indicated times. In some experiments, DRB (150 μM), brefeldin A (BFA; 10 μg/ml), cyclo-heximide (CHX; 25 μg/ml), actinomycin D (AMD; 1.6 μM), and lep-tomycin B (LMB; 20 nM) were added to infected cells at the indicated times.
Ag-presentation assays and Western blots
Following viral infection, cells (~105) were harvested at the indicated time and maintained at 4°C until the completion of the experiment. Cells were then labeled with Alexa Fluor 647-coupled 25-D1.16 mAbs and Pacific Blue-coupled anti-NA mAb NA2-1C1. Following a 30-min Ab labeling at 4°C, cells were washed in excess PBS and analyzed by flow cytometry using a BD Biosciences LSR II flow cytometer. For H-2Kb stability and recovery assays, cells were harvested at indicated times and kept at 4°C until the completion of the experiment. Cells were then labeled with anti-mouse H-2Kb coupled to PE. In MHC class I-recovery experiments, cells were prechilled on ice for 10 min before being washed in citric acid buffer (pH 3) for 2 min on ice and then washed in excess media. Cells were then cultured in the presence of the inhibitors and stained as above. For Western blots, cells were harvested, washed in PBS, and resuspended to 107 cells/ml in hot SDS-PAGE sample buffer containing MG132 and a protease inhibitor mixture (Roche, Mannheim, Germany). Cells were immediately incubated at 95°C for 20 min with occasional, vigorous vortexing. This “flash” lysis minimizes proteolysis by rapidly inactivating cellular pro-teases. An equal volume of water, containing 50 mM DTT unless otherwise noted, was added to samples, which were briefly boiled and then resolved on a 4–12% PAGE gel, blotted onto nitrocellulose, and exposed to rabbit polyclonal serum against NA, mouse polyclonal serum against HA, or rabbit polyclonal serum against NP, followed by anti-rabbit or anti-mouse secondary Abs coupled to the IR dyes 680CW or 800CW, respectively. Membranes were analyzed using an Odyssey infrared imager (LI-COR).
Isolation of mRNA and real-time PCR
IAV-infected cells were treated 1 h postinfection (p.i.) with DRB or were left untreated and cultured for 4 more hours, at which time 5 × 106 cells were harvested and washed in PBS. Cells were lysed in 50 mM Tris (pH 7.5), 150 mM NaCl, and 0.5% Nonidet P-40 for 5 min on ice. The lysate was centrifuged at 300 × g at 4°C for 5 min, and the supernatant containing the cytosolic fraction and the pellet containing the nuclei were subjected to mRNA isolation using the RNeasy kit (Qiagen, Valencia, CA) and reverse transcribed using SuperScript II (Invitrogen, Carlsbad, CA) with an oligo dT primer, following the manufacturer’s protocol. Real-time PCR was performed with the SYBR Green PCR kit (Applied Biosystems, Foster City, CA) using the following primers: NA 5′-TGTGGCAATGACTGATTGGT-3′ and 5′-TTCTTTAGG-TCGTCCCCTGA-3′ (for NA) and 5′-GGTGAAGGTCGGTGTGAACG-GATTT-3′ and 5′-AATGCCAAAGTTGTCATGGATGACC-3′ (for GAP-DH). PCR reactions and data analysis were performed with the Eppendorf Realplex system; the relative copy number of NA mRNA molecules in each sample was determined by dividing the copy number of NA by the corresponding copy number of GAPDH.
Statistical analysis
Student t test analysis was performed using Prism software (GraphPad, San Diego, CA).
Results
RNAP II inhibitors dissociate NA translation from peptide generation
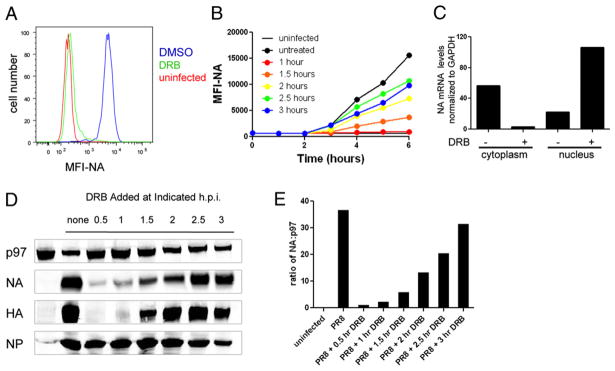
We first extended the findings of Amorim et al. (8) to NA, which was not examined in the original study. We infected L-Kb cells with PR8, added DRB 1 h p.i., and measured cell-surface NA 5 h later by flow cytometry using a NA-specific mAb (Fig. 1A). DRB treatment reduced NA levels >95% compared with untreated cells. We also found that the extent of DRB inhibition of NA expression was inversely related to the time of DRB addition p.i. (Fig. 1B). DRB diminished levels of NA mRNA in the cytoplasm of treated cells and increased NA mRNA in the nucleus, consistent with the reported ability of DRB to retain other late influenza mRNAs in the nucleus (8) (Fig. 1C).
FIGURE 1.
Influenza NA expression is inhibited by DRB treatment. A, L-Kb cells were infected with insOVA for 30 min and then cultured for 1 h before the addition of DRB. After five additional hours in culture, cells were analyzed for cell-surface NA expression by flow cytometry. B, Cells were infected as in A, and DRB was added to infected cells at the indicated times. Cells were collected every hour and stored at 4°C until the completion of the experiment and analyzed for cell surface NA. The mean fluorescence intensity (MFI) of the cell population is reported on the y-axis. C, mRNA from cytosolic and nuclear cell fractions was subjected to reverse transcription, followed by real-time PCR. Levels of NA mRNA in each sample were normalized to GAPDH mRNA levels to control for differences in sample recovery. D, Cells were infected and treated with DRB at the indicated times. Six hours p.i., total-cell lysates were prepared and analyzed by SDS-PAGE and Western blot analysis for NA, HA, NP, and cellular p97. E, The relative amount of NA present in cells is plotted for each DRB treatment.
To establish that DRB inhibition was due to reduced NA synthesis and not interference with NA intracellular trafficking, we measured NA in total-cell extracts prepared by “flash” lysing cells in SDS containing-buffer at 95°C and immunoblotting with rabbit polyclonal Abs raised to a synthetic peptide corresponding to the 15 C-terminal residues of NA (7). This confirmed that NA expression is inhibited by DRB in a manner dependent on its time of addition to cells (Fig 1D). As reported by Amorim et al., HA expression is also severely reduced by DRB, whereas NP expression is affected minimally, if at all, as a result of the rapid expression of NP mRNA. Quantitation of the immunoblots (using the stable cellular protein p97 to normalize data for differences in cell extraction/protein transfer) demonstrated that DRB added within the first hour p.i. reduced NA expression by ≥97% relative to untreated PR8-infected cells (Fig. 1E).
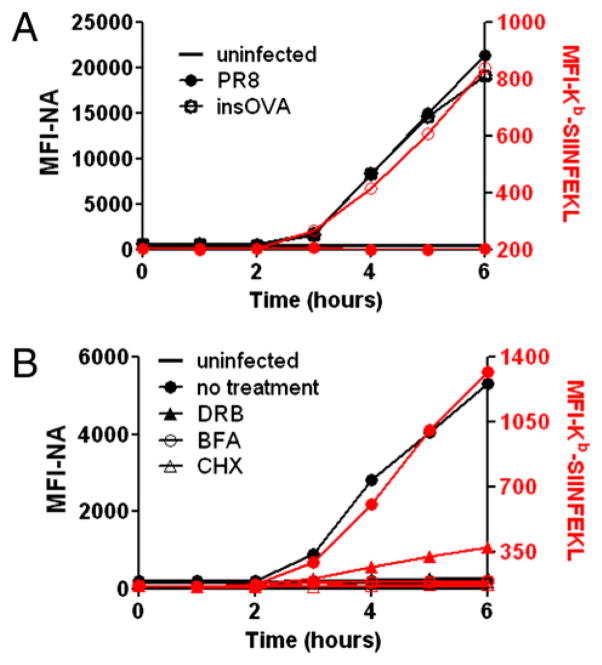
To examine the effect of DRB on Kb-SIINFEKL generation, we infected L-Kb cells with insOVA, a PR8 virus with SIINFEKL inserted into the NA stalk at amino acid position 43 (5, 7). SIINFEKL insertion does not detectably affect NA biogenesis (7). PR8-infected cells served as a specificity control for 25-D1.16 staining (Fig. 2A). As previously reported, Kb-SIINFEKL complexes were detected with nearly identical kinetics to NA itself, pointing to the predominance of DRiPs as a source of SIINFEKL (7). Remarkably, there was still considerable expression of Kb-SIINFEKL in the presence of DRB, despite a near complete blockade of NA surface expression (Fig. 2B). Importantly, DRB-treated cells reached levels of Kb-SIINFEKL expression by 5 h p.i. that equaled levels achieved at 3 h in untreated cells, despite the complete absence of staining of DRB-treated cells by the NA-specific mAb, demonstrating the disconnect between surface expression of NA versus Kb-SIINFEKL resulting from DRB treat-ment. Cells treated with CHX or BFA served as a standard for complete abrogation of Kb-SIINFEKL generation.
FIGURE 2.
MHC class I Ag presentation is partially diminished by DRB treatment of infected cells. A, L-Kb cells were infected with PR8 or insOVA for 30 min and cultured for 6 h; cells were collected each hour for analysis of cell-surface NA (black, left axis) and Kb-SIINFEKL (red trace, right axis). B, L-Kb cells infected with insOVA were treated with DRB, CHX, or BFA 1 h p.i and cultured for 6 h. Cells were collected at indicated times for analysis, as in A.
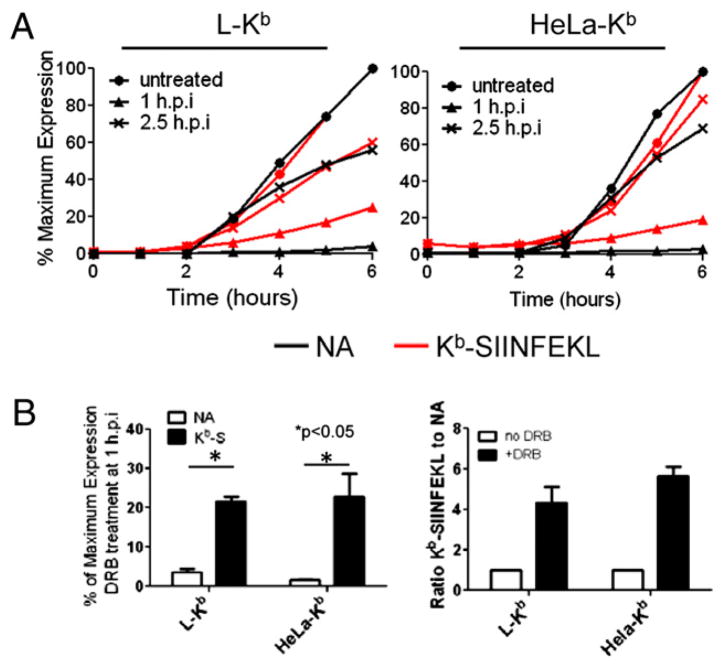
We then compared Kb-SIINFEKL generation in cells treated with DRB 1 and 2.5 h p.i (Fig. 3). In L-Kb and HeLa-Kb cells, addition of DRB at 2.5 h p.i resulted in a parallel decrease in NA and Kb-SIINFEKL, as clearly seen by plotting the percentage of maximal expression at each time point in the experiment. By contrast, early addition of DRB disproportionately inhibited cell-surface expression of NA (20- and 33-fold for L-Kb and HeLa Kb cells, respectively) relative to Kb-SIINFEKL (4- and 5-fold for L-Kb and HeLa Kb cells, respectively). The relative ratio of Kb-SIINFEKL to NA (defined as 1 when no DRB was added) increased 5-fold (L-Kb cells) or 6-fold (HeLa Kb cells) (Fig. 3B). Thus, DRB treatment greatly increases the efficiency of Ag presentation, defined as complexes generated per protein synthesized.
FIGURE 3.
DRB-insensitive Ag presentation is dependent on the timing of treatment. A, L-Kb and HeLa-Kb cells were infected and cultured as in Fig. 1. DRB was added 1 or 2.5 h p.i. At indicated times, cells were collected and analyzed by flow cytometry for cell-surface NA (black trace) and Kb-SIINFEKL (red trace), and the relative amounts of each cell-surface molecule was plotted as a percentage of maximum expression (obtained from untreated cells at 6 h p.i). B, At 6 h p.i., the amount of cell-surface NA and Kb-SIINFEKL (Kb-S) on infected cells treated with DRB 1 h p.i was determined on L-Kb and HeLa-Kb cells and plotted as the percentage of the maximum level obtained in untreated cells. The relative ratio of Kb-SIINFEKL/NA for DRB-treated cells was compared with untreated cells and plotted for each cell type.
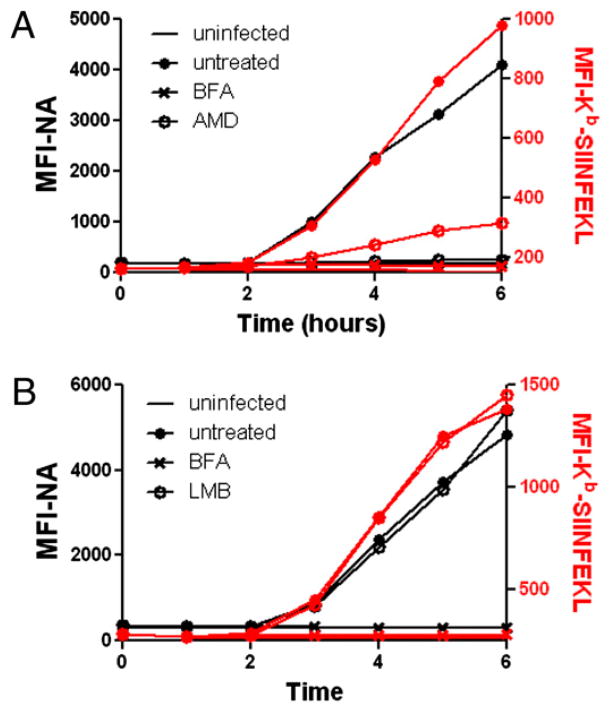
Amorim et al. (8) showed that, like DRB, AMD blocks export of late IAV mRNAs from the nucleus. Because AMD is distinct chemically from DRB, it is unlikely to share off-target effects. AMD behaved nearly identically to DRB in disproportionately blocking expression of NA relative to Kb-SIINFEKL. By contrast, leptomycin B, which blocks nuclear export of viral ribonucleo-protein complexes (but not viral mRNA), had no effect on NA expression or Kb-SIINFEKL generation (Fig. 4).
FIGURE 4.
AMD, but not LMB, prevents NA synthesis. A, L-Kb cells were infected with insOVA for 30 min and cultured for 6 h. At 1 h p.i, cells were left untreated or were treated with AMD. Cell-surface NA (black trace, left axis) and Kb-SIINFEKL (red trace, right axis) were determined by flow cytometry, as in Fig. 2. B, Same as A, except cells were treated with LMB instead of AMD.
Together, these data indicate that although NA expression is highly dependent on nuclear export of NA mRNA, Kb-SIINFEKL is generated at a rate disproportionate to the rate of NA synthesis.
Effect of RNAP II inhibitors is not due to interference with NA biogenesis
There are two general explanations for the effect of RNAP II inhibitors on NA Ag processing. First, it may truly demonstrate that antigenic peptides are synthesized by specialized translation machinery distinct from standard translation. Alternatively, it may be based on disruption of normal NA biogenesis, resulting in a higher DRiP fraction.
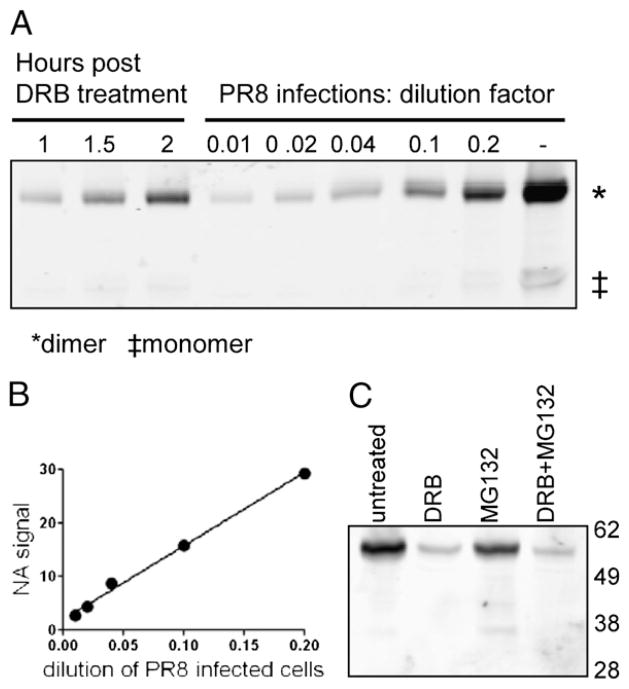
To distinguish these possibilities, we examined the effect of DRB on NA biogenesis. During oligomerization, NA subunits become disulfide cross-linked, an event easily monitored by running SDS-PAGE under nonreducing conditions, where mature NA migrates in dimeric form. Immunoblotting PR8-infected cells treated with DRB at 1, 1.5, or 2 h p.i. revealed that DRB did not alter the dimer/monomer ratio assessed at 6 h p.i. (Fig. 5A). In this experiment, we quantitated the amount of NA expressed in DRB-treated cells by graded addition of extracts from PR8-infected cells into a constant amount of uninfected cell lysates (Fig. 5A). From the standard curve (Fig. 5B), we calculated that addition of DRB 1 h p.i. resulted in a 97% reduction in the amount of NA expressed.
FIGURE 5.
DRB treatment does not prevent NA maturation or degradation. A, Western blot samples were prepared from DRB-treated, PR8-infected cells in the absence of DTT, along with lysates of untreated PR8-infected cells diluted as indicated. NA dimer (~98 kDa) and low levels of NA monomer (~50 kDa) were detected by Western blot. B, The NA signal from diluted PR8 samples was plotted, and the line of best fit was calculated and used to accurately measure levels of NA in DRB-treated cells. C, L-Kb cells were infected with PR8 and cultured for 6 h. After 1 h, cells were left untreated, or DRB was added. At 2 h p.i, cells were treated with MG132. Total-cell lysates were prepared and analyzed for NA expression by Western blot analysis, as in Fig. 1. Approximate molecular mass (in kDa) are shown.
We also examined whether DRB treatment (added 1 h p.i.) induced synthesis of defective NA that could be rescued by adding the proteasome inhibitor MG132 2 h p.i and incubating for an additional 3 h. MG132 failed to detectably increase the amount of full-length or truncated NA recovered in total-cell lysates, as determined by immunoblotting (Fig. 5C).
These data demonstrate that the residual Ag presentation in DRB-treated cells derives from a pool of DRiPs that is too meager to detect by immunoblotting and is not created by the low efficiency of NA biogenesis due to limited levels of NA expression.
RNAP II inhibitors do not limit cellular peptide generation
Could the heightened efficiency of Kb-SIINFEKL generation in the presence of RNAP II inhibitors be due to diminished competition from cellular peptides? RNAP II inhibitors block transcription and rapidly deplete the number of nascent RNA molecules that direct pioneer translation as part of the nonsense-mediated decay (NMD) RNA quality control pathway (10). If pioneer translation products are an important source of class I peptide ligands, as speculated (11), and competition from cellular peptides limits SIINFEKL presentation, then DRB and AMD could reduce cellular peptide supply and increase the efficiency of Kb-SIINFEKL generation from NA.
We addressed this question by treating L-Kb cells with RNAP II inhibitors and measuring overall cell-surface Kb expression by flow cytometry, using directly conjugated AF6-88.5 mAb, which is specific for folded Kb complexed with mouse β2m (12). Although treating cells with CHX to prevent new Ag synthesis, BFA to prevent exocytosis of newly formed peptide–MHC complexes, or MG132 to prevent Ag degradation each resulted in a dramatic decrease in Kb expression, DRB and AMD had less dramatic effects on Kb expression on the surface of infected or uninfected cells (Fig. 6A).
FIGURE 6.
Cell-surface Kb levels are impacted minimally by DRB and AMD treatment. A, L-Kb cells were mock infected or infected with insOVA IAV, then treated with the indicated drugs, and analyzed for surface Kb by flow cytometry. B, Uninfected L-Kb cells or cells infected with insOVA were washed in a cold citric acid buffer to remove existing peptide–MHC complexes from the cell surface and then were cultured with or without indicated inhibitors for specified times. Kb levels on the cell surface were determined by flow cytometry. The mean fluorescence intensity (MFI) of the population analyzed is plotted on the y-axis.
To increase our ability to detect minor effects of the RNAP II inhibitors on class I biogenesis, we repeated this experiment after treating cells with acid to denature cell-surface Kb (Fig. 6B). Following acid stripping, Kb levels quickly recovered on L-Kb cells. As expected, treatment with BFA completely blocked Kb recovery, whereas CHX and MG132 greatly reduced Kb re-expression after the first hour of recovery (during which time pre-formed complexes are delivered to the cell surface), due to their abrogation of DRiP and peptide generation, respectively; the slight difference in the effect of CHX versus MG132 likely reflects residual proteasome-mediated generation of peptides from DRiPs present at the time of CHX addition. Treatment with DRB or AMD reduced the rate of Kb recovery in uninfected and PR8-infected cells by 30–40%. Importantly, inhibition clearly decreased between 3 and 5 h in infected cells, indicating that first, the early effect cannot facilely be attributed to diminished levels of Kb mRNA, a conclusion supported by the minor and constant diminishment of steady-state Kb observed in Fig. 6A.
These data indicate that RNAP II inhibitors retard class I biogenesis in a manner consistent with a potential role for NMD in Ag presentation. However, the magnitude of this effect is not likely sufficient to account for the 5–6-fold increased efficiency in generating Kb-SIINFEKL complexes per NA molecule synthesized, particularly because Kb-SIINFEKL generation in DRB-treated infected L-Kb cell accelerates at a time (3–5 h p.i.) when the effect of DRB on overall peptide generation diminishes (compare Figs. 3A, 6B).
Discussion
The DRiP hypothesis of Ag presentation was formulated largely based upon the observation that antigenic peptides from newly synthesized viral proteins can be presented by MHC class I molecules within 1 h of viral infection (1). It must be stressed that a DRiP is any translation product that does not achieve a stable conformation. Potential DRiPs include miscoded proteins, premature translation-termination products, proteins produced from alternative or downstream initiation codons, proteins that misfold, and “intrinsically disordered proteins” (i.e., properly synthesized proteins are inherently rapidly degraded because of their loose structure in the absence of interacting partners) (13, 14). The contribution of each of these possible sources toward Ag presentation remains to be established. Although progress has been lagging in biochemically characterizing DRiPs, there are clear examples of DRiPs arising from frame shifting (15), downstream initiation (2), and premature termination (16, 17).
In this report, we showed that inhibiting NA mRNA export from the nucleus severely limited NA-SIINFEKL expression but had a more proscribed effect on Kb-SIINFEKL generation. Although peptide generation is diminished by RNAP II inhibitors, a selected source of DRiPs seems to be available for Ag presentation, providing ~20% of SIINFEKL for Ag presentation available in untreated infected cells when inhibitors are added 1 h p.i. (i.e., the ratio of Kb-SIINFEKL complexes generated per source NA-SIINFEKL synthesized increased up to 6-fold relative to untreated cells). How do we explain this remarkable efficiency increase?
We previously proposed that to enhance the sensitivity and speed of immunosurveillance, a subset of ribosomes (immunoribosomes) translate DRiPs that are processed at high efficiency into antigenic peptides (11, 18, 19). Our recent description of noncompetition between peptides binding the same class I allomorph but derived from cytosolic peptide versus standard DRiPs supports this possibility (20). The present results provide further support for this model, if we assume that immunoribosome translation is less affected than is standard translation. These findings also provide a clue to the nature of this translation.
Given the effect of the RNAP II inhibitors on nuclear export, a direct, if iconoclastic, explanation of our findings is that the peptides are translated in the nucleus itself. Nuclear translation was initially described >60 y ago (21) and has come in and out of favor in the intervening years. It remains a highly contentious topic (22, 23). Even opponents of nuclear translation would concede that we cannot eliminate the possibility that a small percentage of protein synthesis occurs in the nucleus.
Indeed, an ever-increasing number of components of protein synthesis have been detected in the nucleus, including ribosomes, initiation factors (24–28), aminoacyl synthetases (29, 30), and aminoacylated tRNA (31). We also note that TAP is present in the inner leaflet of the nuclear membrane (32), as well as in ER elements that tunnel into the nucleus (33). Nuclei are also rich in proteasomes, and we have provided evidence that nuclear pro-teasomes can degrade viral proteins that potentially provide peptides for Ag presentation (34). Thus, many of the relevant participants in translation-linked peptide generation are present in the nucleus. Although our evidence is highly indirect, the effect of RNAP II inhibitors on Kb recovery is consistent with a contribution of NMD to peptide generation. The site of pioneer translation is highly contentious, because it is entwined in the debate regarding nuclear translation itself. Still, our findings support a possible role for nuclear translation in Ag processing and raise the possibility that some immunoribosomes, the hypothetical subset of ribosomes that provide a high-efficiency source of antigenic peptides, are located in the nucleus.
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases.
We thank Glennys Reynoso for providing outstanding technical assistance and Richard Webby (St Jude Research Hospital, Memphis, TN) for generously providing recombinant viruses.
Abbreviations used in this paper
- AMD
actinomycin D
- BFA
brefeldin A
- CHX
cycloheximide
- DRB
5,6-dichloro-1-β-D-ribofuranosylbenzimidazole
- DRiP
defective ribosomal product
- HA
hemagglutinin
- IAV
influenza A virus
- insOVA
IAV strain A/Puerto Rico/8/34 modified to express SIINFEKL peptide inserted in NA
- IR
infrared
- LMB
leptomycin B
- MFI
mean fluorescence intensity
- NA
neuraminidase
- NMD
nonsense-mediated decay
- NP
nucleoprotein
- p.i
postinfection
- PR8
A/Puerto Rico/8/34
- RNAP
RNA polymerase
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Yewdell JW, Antón LC, Bennink JR. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J Immunol. 1996;157:1823–1826. [PubMed] [Google Scholar]
- 2.Berglund P, Finzi D, Bennink JR, Yewdell JW. Viral alteration of cellular translational machinery increases defective ribosomal products. J Virol. 2007;81:7220–7229. doi: 10.1128/JVI.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian SB, Princiotta MF, Bennink JR, Yewdell JW. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J Biol Chem. 2006;281:392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- 4.Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins MR, Webby R, Doherty PC, Turner SJ. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodo-minance hierarchies when antigen is limiting. J Immunol. 2006;177:2917–2925. doi: 10.4049/jimmunol.177.5.2917. [DOI] [PubMed] [Google Scholar]
- 6.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 7.Dolan BP, Li L, Takeda K, Bennink JR, Yewdell JW. Defective ribosomal products are the major source of antigenic peptides endogenously generated from influenza A virus neuraminidase. J Immunol. 2010;184:1419–1424. doi: 10.4049/jimmunol.0901907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amorim MJ, Read EK, Dalton RM, Medcalf L, Digard P. Nuclear export of influenza A virus mRNAs requires ongoing RNA polymerase II activity. Traffic. 2007;8:1–11. doi: 10.1111/j.1600-0854.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- 9.Eisenlohr LC, Gerhard W, Hackett CJ. Acid-induced conformational modification of the hemagglutinin molecule alters interaction of in-fluenza virus with antigen-presenting cells. J Immunol. 1988;141:1870–1876. [PubMed] [Google Scholar]
- 10.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 11.Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Kuhns ST, Pease LR. A region of conformational variability outside the peptide-binding site of a class I MHC molecule. J Immunol. 1998;161:6745–6750. [PubMed] [Google Scholar]
- 13.Wright PE, Dyson HJ. Intrinsically unstructured proteins: reassessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 14.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 15.Fetten JV, Roy N, Gilboa E. A frameshift mutation at the NH2 terminus of the nucleoprotein gene does not affect generation of cytotoxic T lymphocyte epitopes. J Immunol. 1991;147:2697–2705. [PubMed] [Google Scholar]
- 16.Cardinaud S, Starck SR, Chandra P, Shastri N. The synthesis of truncated polypeptides for immune surveillance and viral evasion. PLoS ONE. 2010;5:e8692. doi: 10.1371/journal.pone.0008692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu W, Cochrane M, Leggatt GR, Payne E, Choyce A, Zhou F, Tindle R, McMillan NA. Both treated and untreated tumors are eliminated by short hairpin RNA-based induction of target-specific immune responses. Proc Natl Acad Sci USA. 2009;106:8314–8319. doi: 10.1073/pnas.0812085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yewdell J. To DRiP or not to DRiP: generating peptide ligands for MHC class I molecules from biosynthesized proteins. Mol Immunol. 2002;39:139–146. doi: 10.1016/s0161-5890(02)00097-4. [DOI] [PubMed] [Google Scholar]
- 19.Yewdell JW. The seven dirty little secrets of major histocompatibility complex class I antigen processing. Immunol Rev. 2005;207:8–18. doi: 10.1111/j.0105-2896.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 20.Lev A, Princiotta MF, Zanker D, Takeda K, Gibbs JS, Kumagai C, Waffarn E, Dolan BP, Burgevin A, Van Endert P, et al. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc Natl Acad Sci USA. 2010;107:6964–6469. doi: 10.1073/pnas.0910997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allfrey VG, Mirsky AE, Osawa S. Protein synthesis in isolated cell nuclei. J Gen Physiol. 1957;40:451–490. doi: 10.1085/jgp.40.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlberg JE, Lund E, Goodwin EB. Nuclear translation: what is the evidence? RNA. 2003;9:1–8. doi: 10.1261/rna.2121703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iborra FJ, Jackson DA, Cook PR. The case for nuclear translation. J Cell Sci. 2004;117:5713–5720. doi: 10.1242/jcs.01538. [DOI] [PubMed] [Google Scholar]
- 24.Dostie J, Lejbkowicz F, Sonenberg N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J Cell Biol. 2000;148:239–247. doi: 10.1083/jcb.148.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci USA. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iborra FJ, Jackson DA, Cook PR. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293:1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 27.Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, Sonenberg N. A fraction of the mRNA 59 cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci USA. 1992;89:9612–9616. doi: 10.1073/pnas.89.20.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunasekera N, Lee SW, Kim S, Musier-Forsyth K, Arriaga E. Nuclear localization of aminoacyl-tRNA synthetases using single-cell capillary electrophoresis laser-induced fluorescence analysis. Anal Chem. 2004;76:4741–4746. doi: 10.1021/ac049567e. [DOI] [PubMed] [Google Scholar]
- 30.Nathanson L, Deutscher MP. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J Biol Chem. 2000;275:31559–31562. doi: 10.1074/jbc.C000385200. [DOI] [PubMed] [Google Scholar]
- 31.Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 32.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 33.Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antón LC, Schubert U, Bacík I, Princiotta MF, Wearsch PA, Gibbs J, Day PM, Realini C, Rechsteiner MC, Bennink JR, Yewdell JW. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol. 1999;146:113–124. doi: 10.1083/jcb.146.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]