Abstract
Objective
To investigate the genetic basis and molecular characteristics of the isolated form of ectopia lentis.
Methods
We ascertained a consanguineous Pakistani family with multiple individuals with ectopia lentis. All affected as well as unaffected members with isolated ectopia lentis underwent detailed ophthalmologic and medical examination. Blood samples were collected and DNA was extracted. A genome-wide scan was completed with 382 polymorphic microsatellite markers, and logarithm of odds (LOD) scores were calculated.
Results
Maximum 2-point LOD scores of 5.68 and 2.88 at θ=0 were obtained for markers D8S285 and D8S260, respectively, during the genome-wide scan. Additional microsatellite markers refined the disease locus to a 16.96-cM (14.07-Mb) interval flanked by D8S1737 proximally and D8S1117 distally.
Conclusions
We report on a new locus for nonsyndromic autosomal recessive ectopia lentis on chromosome 8q11.23-q13.2 in a consanguineous Pakistani family.
Clinical Relevance
Identification of genetic loci and genes involved in ectopia lentis will enhance our understanding of the disease at a molecular level, leading to better genetic counseling and family screening and possible future development of better treatment.
The most common cause of ectopia lentis is trauma to the eye, but familial simple ectopia lentis without any systemic abnormality has been described.1,2 Ectopia lentis may occur because of flawed zonule formation.3 Mutations of the fibrillin genes are the bases of Marfan and Beal syndromes, both of which may include ectopia lentis. The most common ocular problems associated with isolated forms of ectopia lentis are myopia, astigmatism, and anisometropia, the severity of which depends on the extent of dislocation of the lens.
The most common cause of ectopia lentis is trauma to the eye, but heritable forms also exist.2 Although rare, familial simple ectopia lentis unassociated with any systemic abnormality has been described. It is most often inherited as an autosomal dominant disorder, but autosomal recessive inheritance has also been reported.4–7 Mutations in the fibrillin 1 gene (FBN1 [OMIM 134797]) have been associated with ectopia lentis, both in an autosomal dominant as well as autosomal recessive fashion.8–12 Recently, mutations in thrombospondin repeat–containing 1 (AD-AMTSL4 [OMIM 610113]) were associated with the simple form of ectopia lentis13; ADAMTSL4 belongs to a large superfamily of proteins, ADAMTS-like protein 4 precursor (thrombospondin repeat–containing protein 1), that are involved in various biological processes, including connective tissue organization, coagulation, and cell migration.14
We report a consanguineous family with 7 affected members ascertained from the Punjab province of Pakistan. All affected family members examined have had dislocated lenses. Clinical physical examination showed no signs of cardiac abnormalities or skeletal features associated with syndromes including ectopia lentis. A genome-wide linkage scan was performed and linkage analysis provided evidence of a new locus for autosomal recessive simple ectopia lentis on chromosome 8q11.23-q13.2.
METHODS
CLINICAL ASCERTAINMENT
A family with nonsyndromic ectopia lentis was recruited to participate in a collaborative study between the National Centre of Excellence in Molecular Biology, Lahore, Pakistan, and the National Eye Institute, Bethesda, Maryland, to identify new disease loci that cause inherited visual diseases. Institutional review board approval was obtained from the National Centre of Excellence in Molecular Biology, and the National Eye Institute for this study. All participating subjects gave informed consent, consistent with the tenets of the Declaration of Helsinki.
A detailed medical history was obtained by interviewing family members. Affected individuals underwent comprehensive ophthalmologic examinations, including visual acuity, slitlamp biomicroscopy, measurement of intraocular pressure, and dilated funduscopy. Blood samples were collected from affected and unaffected family members. DNA was extracted by a nonorganic method as described previously by Kaul et al.15
GENOTYPE ANALYSIS
A genome-wide linkage scan was performed, with 382 highly polymorphic fluorescent microsatellite markers from the ABI PRISM Linkage Mapping Set MD-10 (Applied Biosystems, Foster City, California) having an average spacing of 10 cM. Multiplex polymerase chain reactions (PCRs) were carried out in a 5-μL mixture containing 40 ng of genomic DNA, various combinations of 10-μM dye-labeled primer pairs, 0.5 μL of 10X GeneAmp PCR Buffer (Applied Biosystems), 1mM dNTPs, 2.5mM magnesium chloride, and 0.2 U of Taq DNA polymerase (Applied Biosystems). Amplification was performed in a GeneAmp PCR System 9700 (Applied Biosystems). Initial denaturation was carried out for 5 minutes at 95°C; followed by 10 cycles of 15 seconds at 94°C, 15 seconds at 55°C, and 30 seconds at 72°C; and then 20 cycles of 15 seconds at 89°C, 15 seconds at 55°C, and 30 seconds at 72°C. The final extension was performed for 10 minutes at 72°C, followed by a final hold at 4°C. The PCR products from each DNA sample were pooled and mixed with a loading cocktail containing high-density 400 size standards (Applied Biosystems). The resulting PCR products were separated on an ABI 3100 DNA Analyzer and analyzed by using the GeneMapper software package (Applied Biosystems).
LINKAGE ANALYSIS
Two-point linkage analyses were performed using the FASTLINK version of MLINK from the LINKAGE Program Package.16,17 Maximum logarithm of odds (LOD) scores were calculated using ILINK. Autosomal recessive ectopia lentis was analyzed as a fully penetrant trait with an affected allele frequency of 0.001. The marker order and distances between the markers were obtained from the Marshfield database (http://research.marshfieldclinic.org/) and the National Center for Biotechnology Information chromosome 8 sequence maps (http://www.ncbi.nlm.nih.gov). For the initial genome scan, equal allele frequencies were assumed, while for fine mapping, allele frequencies were estimated from 96 unrelated and unaffected individuals from the Punjab province of Pakistan.
RESULTS
The family (PKCC033) described in this study is from the Punjab province of Pakistan (Figure 1). Displacement of lenses was detected during ophthalmological examinations of the unoperated, affected individuals, though no signs of microspherophakia were present in these patients. Subluxated lens (inferior displacement of the lens) was detected bilaterally in individuals 8 and 9, the right eye of individual 12, and the left eyes of individuals 11, 16, and 19 (Figure 2). Luxated lenses (superior or lateral displacement of the lens) were detected in both eyes of individual 18 and the right eyes of individuals 11, 16, and 19. Displacement of the lens in all of the affected individuals is congenital and unrelated to trauma or accident. Ophthalmological examination (slitlamp biomicroscopy, funduscopy, and tonometry) of family members did not reveal any signs or symptoms of retinal dystrophy during funduscopy or abnormal intraocular pressure. Physical examination revealed no signs of skeletal abnormality, cardiovascular disease, or disability in individuals with ectopia lentis. The members of the family are of normal height, and no individuals showed obesity or mental disability. Additionally, there were no complaints of deafness or other communication disorder in the family. Taken together, these results suggest a simple form of ectopia lentis segregating in the PKCC033 family.
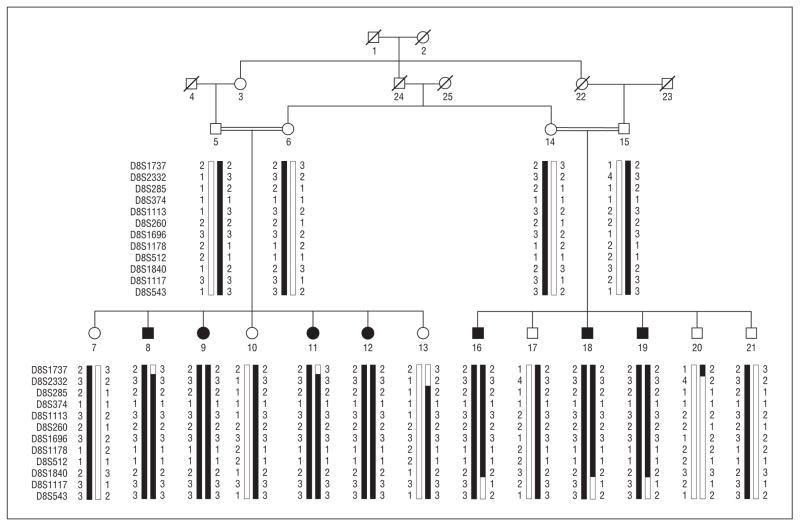
Figure 1.
Pedigree drawing and haplotypes of chromosome 8q markers of family PKCC033. The haplotypes of 12 adjacent chromosome 8q microsatellite markers are shown with alleles forming the risk haplotype shaded black and alleles not cosegregating with disease phenotype shown in white. Double lines between individuals indicate consanguineous mating.
Figure 2.
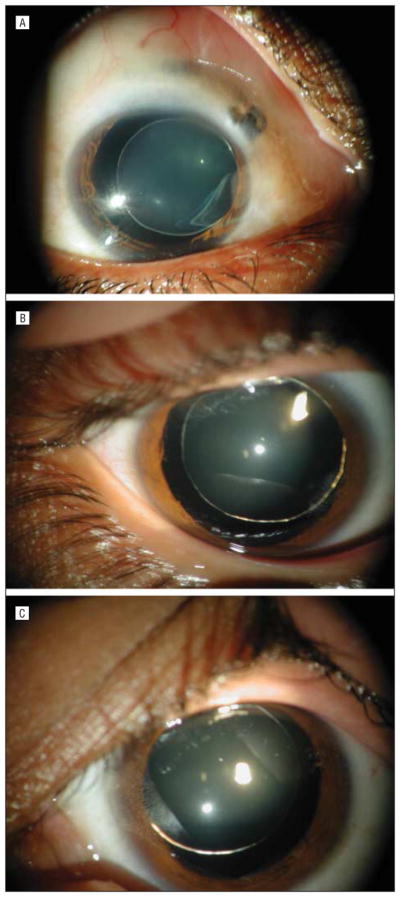
Photographs showing displacement of lenses in family PKCC033. A, Left eye of individual 11. B, Right eye of individual 18. C, Left eye of individual 19.
Initially, linkage to FBN1 and ADAMTSL4 was excluded with primers specific for these loci. A genome-wide linkage scan was performed and scores greater than 1.5 were obtained only for markers D8S285 and D8S260, 2 adjacent markers in the MD-10 mapping set, which yield LOD scores of 5.68 and 2.88, respectively, at θ=0. Fine mapping was carried out by genotyping additional short tandem-repeat markers, including D8S2332, D8S374, D8S1113, D8S1696, D8S1178, D8S512, and D8S1840, which yielded LOD scores of 5.53, 1.11, 5.31, 3.71, 5.26, 2.88, and 5.47, respectively, at θ=0 (Table).
Visual inspection of the haplotypes in family members supports the results of the linkage analysis. As shown in Figure 1, there is a proximal recombination in affected individuals 8 and 11 at D8S1737. Similarly, there are distal recombination events in affected individuals 16, 18, and 19 at D8S1117. Alleles for D8S2332, D8S285, D8S374, D8S1113, D8S260, D8S1696, D8S1178, D8S512, and D8S1840 were homozygous for all affected individuals. This places the disease locus in a 16.96-cM (14.07-Mb) interval flanked by D8S1737 and D8S1117.
COMMENT
Herein, we report a new locus for isolated autosomal recessive ectopia lentis on chromosome 8q11.23-q13.2 in a consanguineous Pakistani family recruited from the Punjab province of Pakistan. Lack of any skeletal abnormalities, cardiovascular diseases, physical disabilities, and/or mental disability in family PKCC033 strongly suggests that the affected individuals have the simple form of ectopia lentis. A maximum LOD score of 5.68 was obtained with D8S285 at θ=0, and the disease locus cosegregates with markers in a 16.96-cM (14.07-Mb) interval flanked by D8S1737 and D8S1117 in family PKCC033. The lack of significant LOD scores other than for markers in the chromosome 8q region during the genome-wide linkage scan, homozygous alleles for all the affected individuals for markers in the linked region of chromosome 8q, along with lack of homozygosity in all the unaffected individuals strongly suggest that the disease locus maps to the 8q11.23-q13.2 region in this consanguineous Pakistani family.
A new locus for the simple form of ectopia lentis extends the genetic heterogeneity of ectopia lentis. Recently, mutations in ADAMTSL4 were identified in affected individuals of Arabian descent associated with the simple form of ectopia lentis.13 In contrast to the affected individuals we described herein, amblyopia, retinal detachment, and cataracts were present in a family described by Ahram and colleagues.13 This suggests that mutations responsible for the disease phenotype in PKCC033 only affect the integrity of the zonular fibers. More than 100 genes reside in the critical interval, but none of them have structural or functional similarity with FBN1 or ADAMTSL4. Among the genes in the critical interval, CPA6 (OMIM 609562) and SDCBP (OMIM 602217) are compelling candidates. CPA6 is a member of a subfamily of carboxypeptidases and is expressed in the eye rectus muscles, though its full role remains unknown. SDCBP encodes a protein that helps in cytoskeletal membrane organization, cell adhesion, protein trafficking, and the activation of transcription factors.
The search for the disease-causing mutation is in progress, and identification of the disease-causing gene will help to improve our understanding of ectopia lentis at the molecular level. Understanding of the pathophysiology and molecular mechanism of ectopia lentis will lead to better treatment and therapy.
Table.
Two-Point LOD Scores of Chromosome 8q Markers for Family PKCC033
| Marker | Genetic Distance, cM | Physical Distance, Mb | Two-Point LOD Score
|
Zmax | θmax | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| θ=0 | θ=0.01 | θ=0.05 | θ=0.09 | θ=0.1 | θ=0.2 | θ=0.3 | |||||
| D8S1737 | 68.27 | 55.78 | −∞ | 0.24 | 1.35 | 1.57 | 1.58 | 1.41 | 0.96 | 1.58 | 0.10 |
| D8S2332 | 69.40 | 56.13 | 5.53 | 5.43 | 5.01 | 4.60 | 4.55 | 3.24 | 2.23 | 5.53 | 0.00 |
| D8S285a | 71.00 | 57.06 | 5.68 | 5.58 | 5.11 | 4.75 | 4.59 | 3.43 | 2.22 | 5.68 | 0.00 |
| D8S374 | 73.82 | 58.32 | 1.11 | 1.09 | 0.97 | 0.86 | 0.83 | 0.54 | 0.27 | 1.11 | 0.00 |
| D8S1113 | 77.89 | 59.70 | 5.31 | 5.22 | 4.80 | 4.354 | 4.25 | 3.14 | 1.99 | 5.31 | 0.00 |
| D8S260a | 79.36 | 61.82 | 2.88 | 2.83 | 2.62 | 2.41 | 2.35 | 1.77 | 1.14 | 2.88 | 0.00 |
| D8S1696 | 79.94 | 63.84 | 3.71 | 3.6 | 3.44 | 3.095 | 3.02 | 2.28 | 1.49 | 3.71 | 0.00 |
| D8S1178 | 79.94 | 64.16 | 5.26 | 5.16 | 4.73 | 4.23 | 4.18 | 3.18 | 1.98 | 5.16 | 0.00 |
| D8S512 | 81.68 | 65.39 | 2.88 | 2.80 | 2.60 | 2.36 | 2.31 | 1.69 | 1.07 | 2.88 | 0.00 |
| D8S1840 | 82.84 | 68.54 | 5.47 | 5.40 | 4.90 | 4.51 | 4.41 | 3.28 | 2.11 | 5.47 | 0.00 |
| D8S1117 | 85.23 | 69.85 | −∞ | −1.59 | −0.1 | 0.39 | 0.46 | 0.66 | 0.5 | 0.66 | 0.20 |
| D8S543 | 87.54 | 70.10 | −∞ | 1.45 | 2.43 | 2.53 | 2.51 | 2.03 | 1.27 | 2.53 | 0.09 |
Abbreviations: LOD, logarithm of odds; Mb, megabase pair; Zmax, maximum LOD score; θmax, value of recombination fractions that gives the maximum LOD score.
Marker included in genome-wide scan.
Acknowledgments
Funding/Support: This study was supported in part by the Higher Education Commission and Ministry of Science and Technology, Islamabad, Pakistan.
Footnotes
Author Contributions: Drs Kaul, S.A. Riazuddin, Hejtmancik, and S. Riazuddin contributed equally to this work.
Financial Disclosure: None reported.
Additional Contributions: The authors are thankful to all of the family members for their participation in this study.
References
- 1.Nelson LB, Maumenee IH. Ectopia lentis. Surv Ophthalmol. 1982;27(3):143–160. doi: 10.1016/0039-6257(82)90069-8. [DOI] [PubMed] [Google Scholar]
- 2.Trivedi HL, Venkatesh R. Ectopia lentis: Weill Marchesani syndrome. Bombay Hosp J. 2009;51(1):49–54. [Google Scholar]
- 3.Ashworth JL, Kielty CM, McLeod D. Fibrillin and the eye. Br J Ophthalmol. 2000;84(11):1312–1317. doi: 10.1136/bjo.84.11.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaureguy BM, Hall JG. Isolated congenital ectopia lentis with autosomal dominant inheritance. Clin Genet. 1979;15(1):97–109. doi: 10.1111/j.1399-0004.1979.tb02033.x. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz C, Rivas F, Villar-Calvo VM, Serrano-Lucas JI, Cantu JM. Familial simple ectopia lentis: a probable autosomal recessive form. Ophthalmic Paediatr Genet. 1986;7(2):81–84. doi: 10.3109/13816818609076113. [DOI] [PubMed] [Google Scholar]
- 6.al-Salem M. Autosomal recessive ectopia lentis in two Arab family pedigrees. Ophthalmic Paediatr Genet. 1990;11(2):123–127. doi: 10.3109/13816819009012957. [DOI] [PubMed] [Google Scholar]
- 7.Edwards MJ, Challinor CJ, Colley PW, et al. Clinical and linkage study of a large family with simple ectopia lentis linked to FBN1. Am J Med Genet. 1994;53 (1):65–71. doi: 10.1002/ajmg.1320530114. [DOI] [PubMed] [Google Scholar]
- 8.Adès LC, Holman KJ, Brett MS, Edwards MJ, Bennetts B. Ectopia lentis phenotypes and the FBN1 gene. Am J Med Genet A. 2004;126A(3):284–289. doi: 10.1002/ajmg.a.20605. [DOI] [PubMed] [Google Scholar]
- 9.Dietz HC, Pyeritz RE. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum Mol Genet. 1995;4(spec No):1799–1809. doi: 10.1093/hmg/4.suppl_1.1799. [DOI] [PubMed] [Google Scholar]
- 10.Lönnqvist L, Child A, Kainulainen K, Davidson R, Puhakka L, Peltonen L. A novel mutation of the fibrillin gene causing ectopia lentis. Genomics. 1994;19(3):573–576. doi: 10.1006/geno.1994.1110. [DOI] [PubMed] [Google Scholar]
- 11.Vanita V, Singh JR, Singh D, Varon R, Robinson PN, Sperling K. A recurrent FBN1 mutation in an autosomal dominant ectopia lentis family of Indian origin. Mol Vis. 2007;13:2035–2040. [PubMed] [Google Scholar]
- 12.Yu R, Lai Z, Zhou W, Ti DD, Zhang XN. Recurrent FBN1 mutation (R62C) in a Chinese family with isolated ectopia lentis. Am J Ophthalmol. 2006;141(6):1136–1138. doi: 10.1016/j.ajo.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 13.Ahram D, Sato TS, Kohilan A, et al. A homozygous mutation in ADAMTSL4 causes autosomal-recessive isolated ectopia lentis. Am J Hum Genet. 2009;84(2):274–278. doi: 10.1016/j.ajhg.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386(pt 1):15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaul H, Riazuddin SA, Shahid M, et al. Autosomal recessive congenital cataract linked to EPHA2 in a consanguineous Pakistani family. Mol Vis. 2010;16:511–517. [PMC free article] [PubMed] [Google Scholar]
- 16.Schäffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44(4):225–237. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 17.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36(2):460–465. [PMC free article] [PubMed] [Google Scholar]