Abstract
Cohen syndrome (CS) is an autosomal recessive disease caused by mutations in the COH1 gene. It is characterized by intellectual disability, hypotonia, joint hyperlaxity, severe myopia, characteristic facial dysmorphisms and, in some cases, intermittent isolated neutropenia. We investigated an Italian patient with CS together with his family. Genetic analysis disclosed 2 novel mutations: the first is an intronic mutation (c.8697–9A>G) creating a new splice site 8 nucleotides upstream, and the second is a duplication of 1 base (c.10156dupA) generating a premature stop codon. The compound heterozygous mutations explain the proband's phenotype and improved the knowledge of genotype-phenotype correlation.
Key Words: COH1, Cohen syndrome, Mutation, Splicing
Cohen syndrome (CS; OMIM 216550) is a rare autosomal recessive disorder most prevalent in the Finnish and Amish population but also described in several other ethnic groups [Douzgou and Petersen, 2011]. The phenotype spectrum can be very broad, depending, at least in part, on the different type of mutations. Mental retardation, delayed development, clumsiness and muscle weakness, microcephaly, joint hyperlaxity, ophthalmological anomalies, narrow hands and feet, characteristic facial features, and cheerful disposition are the most common findings [Kolehmainen et al., 2004]. The characteristic facial features include high-arched or wave-shaped eyelids, long and thick eyelashes, thick eyebrows, high-arched palate, short philtrum, thick hair and low hairline, prominent nasal root, and prominent upper central incisors. Other signs and symptoms include intermittent isolated granulocytopenia, especially in Finnish patients, truncal obesity in teen years, scoliosis, mitral valve collapse, short stature, and high-pitched voice [Kivitie-Kallio and Norio, 2001; Kolehmainen et al., 2003].
The COH1 gene (VPS13B, OMIM 607817), located on q22.2 locus of chromosome 8, is the only CS-related gene so far identified. It spans approximately 864 kb and is composed of 62 exons. The longest transcript [NM 017890.3, isoform 5, NCBI] is 14.0111 bp long encoding for a 4.022-amino acid protein with putative transmembrane domains and functional motifs specific for intracellular vesicle-mediated protein sorting [Seifert et al., 2011]. COH1 protein is widely expressed in the whole body and, with some differences between isoforms, mostly expressed in the central nervous system, blood, muscles, and heart [Seifert et al., 2009].
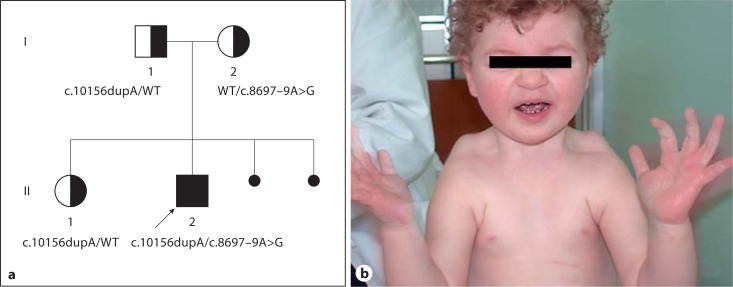
Here we report the identification of 2 novel mutations in COH1 in an Italian CS patient (fig. 1A).
Fig. 1.
a Pedigree of the Italian family and compound heterozygosity of the proband, formed by the c.10156dupA mutation, inherited from the proband's father and also present in his sister, and the c.8697–9A>G mutation, inherited from the mother. In black the 2 mutations are shown and in white the wild type alleles (WT). b The proband at 2.5 years of age.
Case Report
The proband (II:2) is the second male child of nonconsanguineous Italian parents (fig. 1B). The couple has a healthy, first-born female child. Amniocentesis was performed because of maternal age and fetal karyotype resulted 46,XY normal. During pregnancy, the mother reported decreased fetal movements since the eight month. Nevertheless, the boy was born at term with an APGAR score of 8/10 at 1 and 5 min and birth parameters were: weight 2,490 g (<3th percentile), height 48 cm (10th percentile), and OFC 32 cm (<3th percentile). Early hypotonia with poor sucking was evidenced, and it was necessary to implement breast feeding. Psychomotor development was delayed: he was able to sit without help at 9 months and walk at 2 years. At examination at 2 years and 6 months, the patient was not able to speak. Auditory brainstem response revealed a slight augmentation of the IV-V complex, a clear sign of midbrain damage. Ophthalmological evaluation showed exotropia with abnormal fundus and congenital macular atrophy. However, evoked visual potentials and electroretinogram were normal.
Cerebral investigations such as nuclear magnetic resonance and electroencephalogram showed a minor dilatation of subtentorial space, especially at the frontal side, and minor paroxysmal abnormalities in the frontal hemisphere during sleeping, respectively. No abnormalities were detected by abdominal ultrasound. Blood cell count showed a recurrent neutropenia (700 mm3, 400 mm3, 1,500 mm3). At neurological examination, the patient showed diffuse hypotonia and joint laxity. The global intellectual quotient at 2 years and 7 months was 39 and auxological parameters were: weight 13.5 kg (25th–50th percentile), height 91.4 cm (25th–50th percentile), and OFC 44 cm (<3th percentile). Clinical diagnosis revealed dysmorphic facial features and fine curly hair, short philtrum, prominent nasal bridge and bulbous tip, and malar hypoplasia. The hands were small with tapering fingers. Due to the clinical signs and dysmorphological traits, CS was hypothesized and molecular analysis was performed. Written consent was obtained from all family members.
Direct sequencing of all coding regions and exon-intron boundaries of the COH1 gene was conducted on genomic DNA. All variants were confirmed by repeated sequencing of both DNA strands on a second PCR product and tested in 100 healthy Italian controls. Total RNA was isolated directly from freshly extracted blood using the RNA Blood Mini Kit (Qiagen, Germany), and cDNA was synthesized with Transcriptor First Strand cDNA Synthesis Kit (Roche, Germany), according to the manufacturer's instructions. Primer sequences and the PCR protocol are available on request.
Results and Discussion
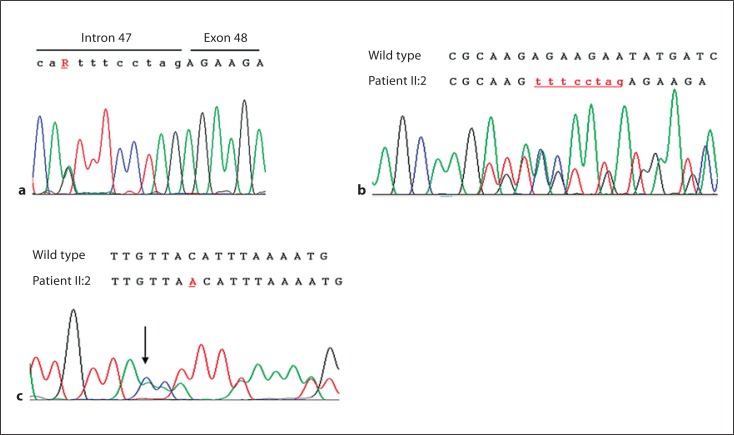
Sequencing of the COH1 gene revealed a nucleotide change (A to G) at position c.8697–9 [NM 017890.3, NCBI] (fig. 2A). This point mutation is present, in heterozygous state, only in the proband (II:2) and his mother (I:2). Bioinformatic analysis using the Human Splicing Finder [Desmet et al., 2009] web tool indicated the activation of a cryptic acceptor splice site in exon 48. Direct sequencing of the cDNA established the generation of an alternative splicing, leading to an extension of 8 nucleotides at RNA level (fig. 2B). This addition of 8 bp is predicted to lead to a frameshift and subsequent truncation of the protein to 2921 residues, p.R2899SfsX24 [NP 060360.3, NCBI] (wild type: 4022 residues) or an unstable mRNA molecule that is rapidly degraded. In the same region, another point mutation has already been reported by Kolehmainen et al. [2004] together with another point mutation, c.8697–2A>G, resulting in a premature stop codon.
Fig. 2.
a Partial genomic sequence of the COH1 gene in the proband carrying the heterozygous mutation c.8697–9A>G (R). b RT-PCR analysis of COH1 exons 47–48 in the proband. Upper cases denote the coding region and lower cases the intron region. c Partial genomic sequence of the COH1 gene in the proband carrying the heterozygous mutation c.10156dupA.
A second mutation was detected in the proband (II:2), his father (I:1), and his sister (II:1). The duplication of base A at position c.10156 [NM 017890.3, NCBI] generates a frameshift in the open reading frame with a premature stop codon at protein level, p.T3361NfsX3 [NP 060360.3, NCBI] (fig. 2C). The novel mutated alleles have not been previously reported in the literature nor detected in a series of 200 normal chromosomes.
Features of our patient present overlapping of Finnish and Irish travelers’ phenotypes. On one hand, the proband shows a friendly behavioral profile in association with severe involvement of mainly the posterior eye segment and neutropenia; on the other hand, as in an Irish traveler's cohort, growth parameters are characterized by a deficiency and a severe language impairment. If we compare the proband with the CS Italian population [Katzaki et al., 2007], we can identify most of the signs previously reported except for the minor dilatation of the subtentorial space evidenced at magnetic resonance.
In conclusion, here we describe 2 additional COH1 mutations leading to truncated or rapidly degraded alleles in agreement with the vast majority of COH1 alterations so far reported [Kolehmainen et al., 2004; Seifert et al., 2009; Parri et al., 2010].
References
- 1.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. (2009);37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douzgou S, Petersen MB. Clinical variability of genetic isolates of Cohen syndrome. Clin Genet. (2011);79:501–506. doi: 10.1111/j.1399-0004.2011.01669.x. [DOI] [PubMed] [Google Scholar]
- 3.Katzaki E, Pescucci C, Uliana V, Papa FT, Ariani F, et al. Clinical and molecular characterization of Italian patients affected by Cohen syndrome. J Hum Genet. (2007);52:1011–1017. doi: 10.1007/s10038-007-0208-4. [DOI] [PubMed] [Google Scholar]
- 4.Kivitie-Kallio S, Norio R. Cohen syndrome: essential features, natural history, and heterogeneity. Am J Med Genet. (2001);102:125–135. doi: 10.1002/1096-8628(20010801)102:2<125::aid-ajmg1439>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Kolehmainen J, Black GC, Saarinen A, Chandler K, Clayton-Smith J, et al. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am J Hum Genet. (2003);72:1359–1369. doi: 10.1086/375454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolehmainen J, Wilkinson R, Lehesjoki AE, Chandler K, Kivitie-Kallio S, et al. Delineation of Cohen syndrome following a large-scale genotype-phenotype screen. Am J Hum Genet. (2004);75:122–127. doi: 10.1086/422197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parri V, Katzaki E, Uliana V, Scionti F, Tita R, et al. High frequency of COH1 intragenic deletions and duplications detected by MLPA in patients with Cohen syndrome. Eur J Hum Genet. (2010);18:1133–1140. doi: 10.1038/ejhg.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seifert W, Holder-Espinasse M, Kühnisch J, Kahrizi K, Tzschach A, et al. Expanded mutational spectrum in Cohen syndrome, tissue expression, and transcript variants of COH1. Hum Mut. (2009);30:E404–E420. doi: 10.1002/humu.20886. [DOI] [PubMed] [Google Scholar]
- 9.Seifert W, Kühnisch J, Maritzen T, Horn D, Haucke V, Hennies HC. Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J Biol Chem. (2011);286:37665–37675. doi: 10.1074/jbc.M111.267971. [DOI] [PMC free article] [PubMed] [Google Scholar]