Full text
PDF



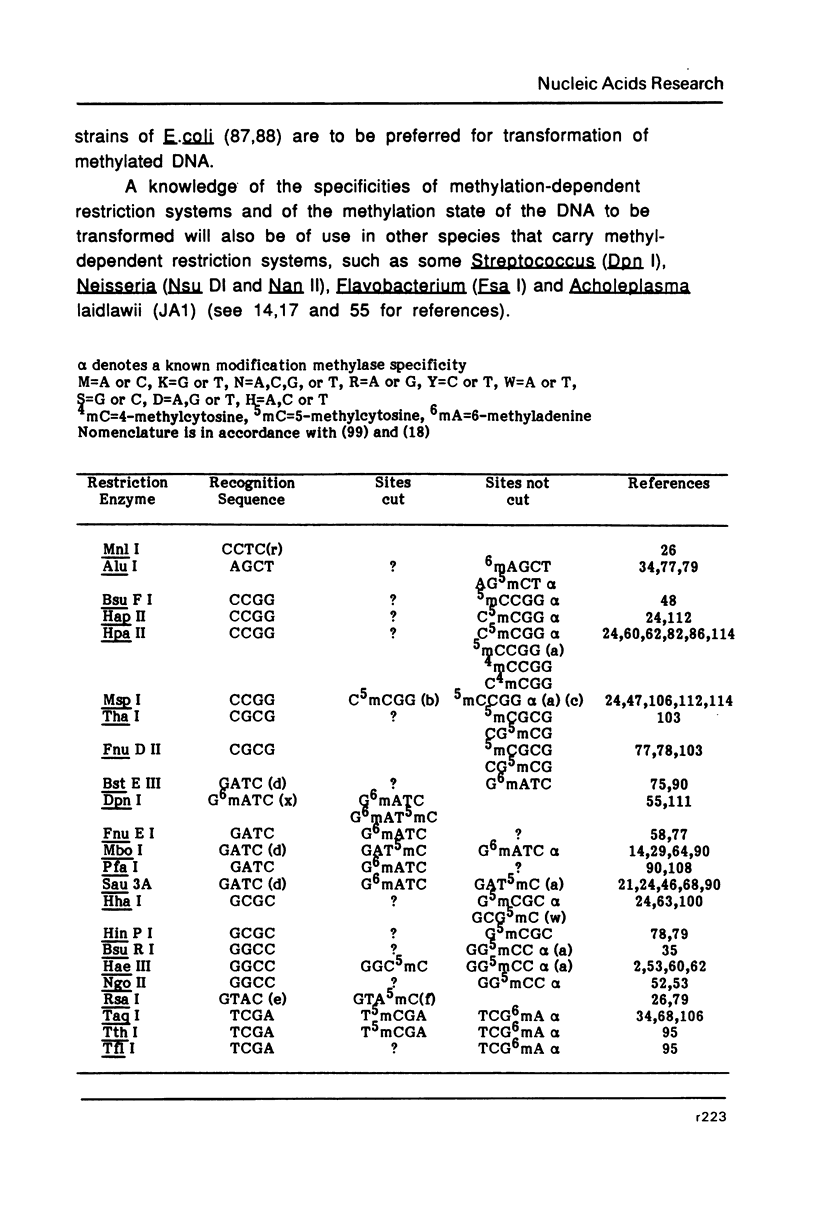
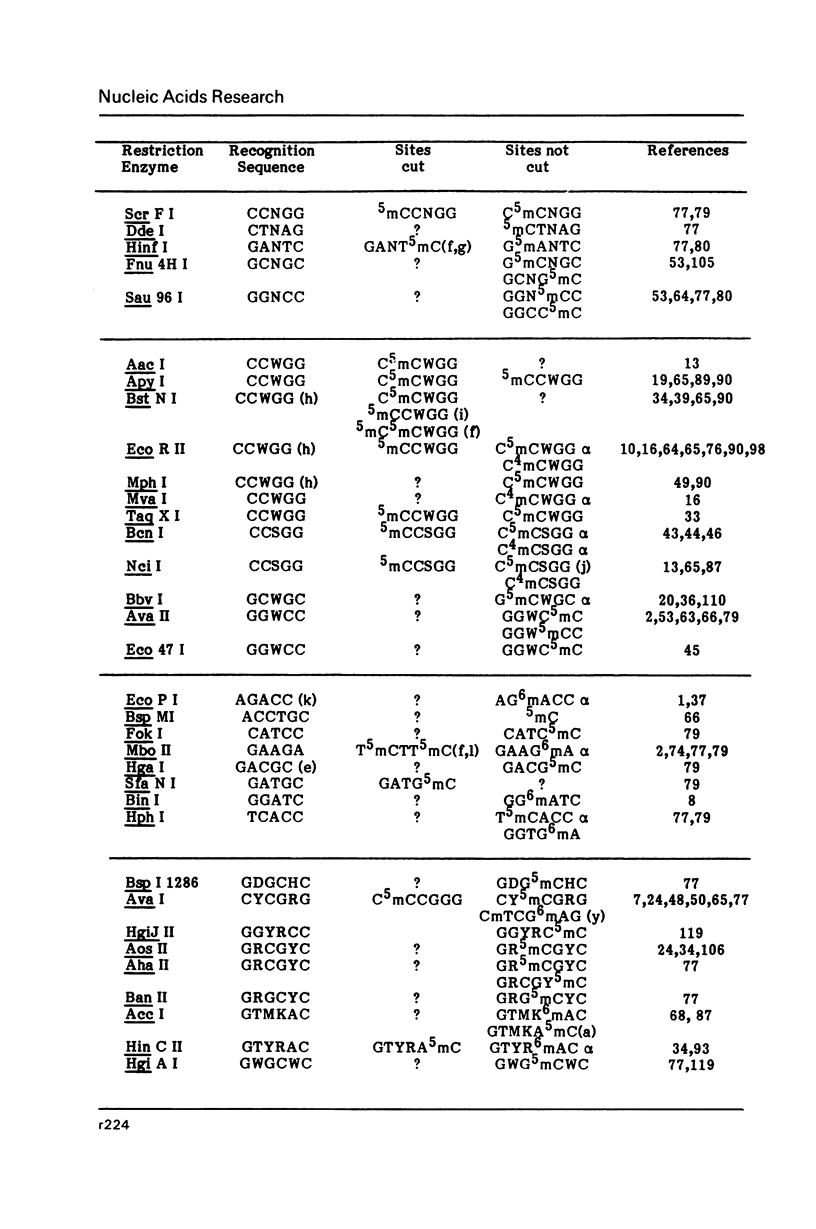
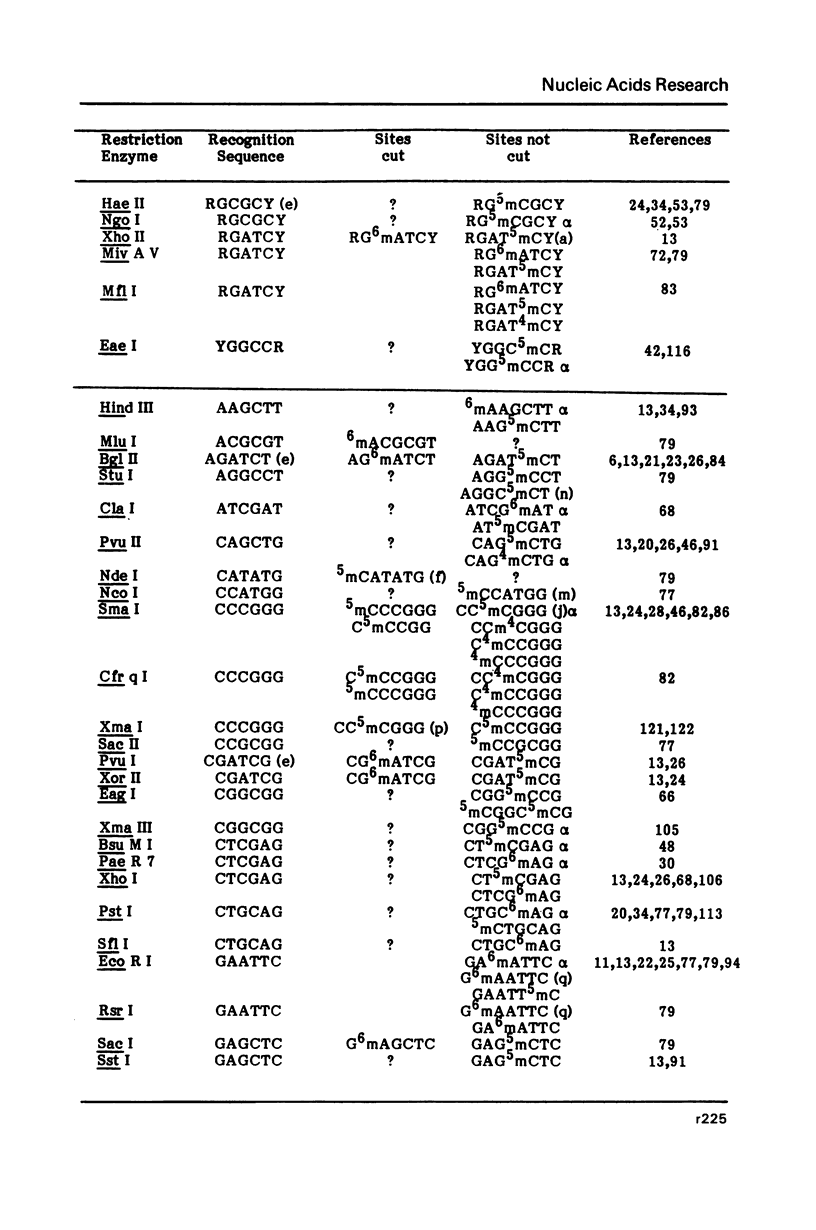
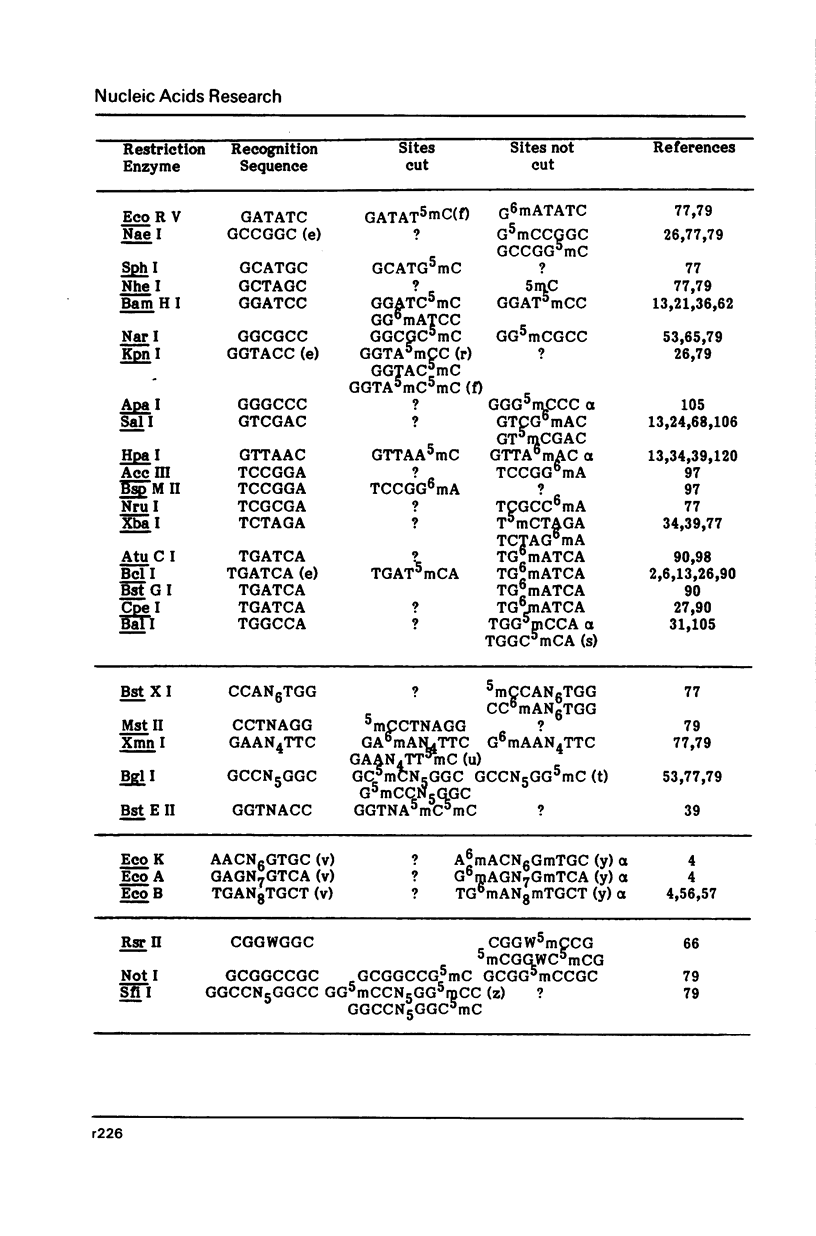




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K. A cautionary note on the use of certain restriction endonucleases with methylated substrates. Gene. 1980 Oct;11(1-2):169–171. doi: 10.1016/0378-1119(80)90097-9. [DOI] [PubMed] [Google Scholar]
- Baumstark B. R., Roberts R. J., RajBhandary U. L. Use of short synthetic DNA duplexes as substrates for the restriction endonucleases Hpa II and Mno I. J Biol Chem. 1979 Sep 25;254(18):8943–8950. [PubMed] [Google Scholar]
- Bingham A. H., Atkinson T., Sciaky D., Roberts R. J. A specific endonuclease from Bacillus caldolyticus. Nucleic Acids Res. 1978 Oct;5(10):3457–3467. doi: 10.1093/nar/5.10.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Smith B. A. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell. 1979 Aug;17(4):889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the methylation by the EcoRI modification methylase of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7279–7286. [PubMed] [Google Scholar]
- Bron S., Murray K., Trautner T. A. Restriction and modification in B. subtilis. Purification and general properties of a restriction endonuclease from strain R. Mol Gen Genet. 1975 Dec 30;143(1):13–23. doi: 10.1007/BF00269416. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Roberts R. J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982 Feb 11;10(3):913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., deBoer E., Wright S., Grosveld F. G., Flavell R. A. The sequence GGCmCGG is resistant to MspI cleavage. Nucleic Acids Res. 1983 Jun 11;11(11):3559–3569. doi: 10.1093/nar/11.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V., Klimasauskas S., Kersulyte D., Vaitkevicius D., Lebionka A., Janulaitis A. Investigation of restriction-modification enzymes from M. varians RFL19 with a new type of specificity toward modification of substrate. Nucleic Acids Res. 1985 Aug 26;13(16):5727–5746. doi: 10.1093/nar/13.16.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bächi B., Reiser J., Pirrotta V. Methylation and cleavage sequences of the EcoP1 restriction-modification enzyme. J Mol Biol. 1979 Feb 25;128(2):143–163. doi: 10.1016/0022-2836(79)90123-2. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985 May 10;13(9):3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. P., Dobritsa S. V. DNA protection with the DNA methylase M . BbvI from Bacillus brevis var. GB against cleavage by the restriction endonucleases PstI and PvuII. Gene. 1980 Jul;10(2):105–112. doi: 10.1016/0378-1119(80)90128-6. [DOI] [PubMed] [Google Scholar]
- Dreiseikelmann B., Eichenlaub R., Wackernagel W. The effect of differential methylation by Escherichia coli of plasmid DNA and phage T7 and lambda DNA on the cleavage by restriction endonuclease MboI from Moraxella bovis. Biochim Biophys Acta. 1979 May 24;562(3):418–428. doi: 10.1016/0005-2787(79)90105-9. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Hedgpeth J., Boyer H. W., Goodman H. M. Physical identity of the SV40 deoxyribonucleic acid sequence recognized by the Eco RI restriction endonuclease and modification methylase. Biochemistry. 1974 Jan 29;13(3):503–512. doi: 10.1021/bi00700a016. [DOI] [PubMed] [Google Scholar]
- Dybvig K., Swinton D., Maniloff J., Hattman S. Cytosine methylation of the sequence GATC in a mycoplasma. J Bacteriol. 1982 Sep;151(3):1420–1424. doi: 10.1128/jb.151.3.1420-1424.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Gelinas R. E., Myers P. A., Roberts R. J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977 Jul;114(1):169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Brooks J. E. Cloned restriction/modification system from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1983 Jan;80(2):402–406. doi: 10.1073/pnas.80.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Storm K., Bald R. Restriction and modification in Bacillus subtilis. Localization of the methylated nucleotide in the BsuRI recognition sequence. Eur J Biochem. 1978 Oct 16;90(3):581–583. doi: 10.1111/j.1432-1033.1978.tb12638.x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S., Keister T., Gottehrer A. Sequence specificity of DNA methylases from Bacillus amyloliquefaciens and Bacillus brevis. J Mol Biol. 1978 Oct 5;124(4):701–711. doi: 10.1016/0022-2836(78)90178-x. [DOI] [PubMed] [Google Scholar]
- Jacobs D., Brown N. L. Isolation and characterization of the M.EaeI modification methylase. Biochem J. 1986 Sep 1;238(2):613–615. doi: 10.1042/bj2380613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulaitis A., Petrusyté M., Butkus V. Three sequence-specific endonucleases from Escherichia coli RFL47. FEBS Lett. 1983 Sep 19;161(2):213–216. doi: 10.1016/0014-5793(83)81010-2. [DOI] [PubMed] [Google Scholar]
- Janulaitis A., Petrusyté M., Butkus V. Three sequence-specific endonucleases from Escherichia coli RFL47. FEBS Lett. 1983 Sep 19;161(2):213–216. doi: 10.1016/0014-5793(83)81010-2. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S. Restriction and modification in Bacillus subtilis: sequence specificities of restriction/modification systems BsuM, BsuE, and BsuF. J Bacteriol. 1983 Nov;156(2):800–808. doi: 10.1128/jb.156.2.800-808.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet E., Cedar H. Effect of CpG methylation on Msp I. Nucleic Acids Res. 1983 Jun 11;11(11):3571–3580. doi: 10.1093/nar/11.11.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch C., Hagblom P., Normark S. Sequence-specific DNA modification in Neisseria gonorrhoeae. J Bacteriol. 1983 Sep;155(3):1324–1332. doi: 10.1128/jb.155.3.1324-1332.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lautenberger J. A., Kan N. C., Lackey D., Linn S., Edgell M. H., Hutchison C. A., 3rd Recognition site of Escherichia coli B restriction enzyme on phi XsB1 and simian virus 40 DNAs: an interrupted sequence. Proc Natl Acad Sci U S A. 1978 May;75(5):2271–2275. doi: 10.1073/pnas.75.5.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. I. Purification, subunit structure, and catalytic properties of the modification methylase. J Biol Chem. 1972 Oct 10;247(19):6176–6182. [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Kessler L. G., Bittner M. Site-specific cleavage of DNA at 8- and 10-base-pair sequences. Proc Natl Acad Sci U S A. 1984 Feb;81(4):983–987. doi: 10.1073/pnas.81.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M., Cantor C. R. Purification of Mbo II methylase (GAAGmA) from Moraxella bovis: site specific cleavage of DNA at nine and ten base pair sequences. Nucleic Acids Res. 1985 Oct 25;13(20):7171–7182. doi: 10.1093/nar/13.20.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M. The effect of site specific methylation on restriction endonuclease digestion. Nucleic Acids Res. 1985;13 (Suppl):r201–r207. doi: 10.1093/nar/13.suppl.r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Purification and characterization of two new modification methylases: MClaI from Caryophanon latum L and MTaqI from Thermus aquaticus YTI. Nucleic Acids Res. 1981 Dec 21;9(24):6795–6804. doi: 10.1093/nar/9.24.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The effect of sequence specific DNA methylation on restriction endonuclease cleavage. Nucleic Acids Res. 1981 Nov 25;9(22):5859–5866. doi: 10.1093/nar/9.22.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The effect of site specific methylation on restriction endonuclease cleavage (update). Nucleic Acids Res. 1983 Jan 11;11(1):r169–r173. doi: 10.1093/nar/11.1.235-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The frequency and distribution of methylatable DNA sequences in leguminous plant protein coding genes. J Mol Evol. 1983;19(5):346–354. doi: 10.1007/BF02101638. [DOI] [PubMed] [Google Scholar]
- Nathans D., Smith H. O. Restriction endonucleases in the analysis and restructuring of dna molecules. Annu Rev Biochem. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- Nelson M., Christ C., Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984 Jul 11;12(13):5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Ueda T. Synthesis of decadeoxyribonucleotides containing N6-methyladenine, N4-methylcytosine, and 5-methylcytosine: recognition and cleavage by restriction endonucleases (nucleosides and nucleotides part 74). Nucleic Acids Res. 1987 Jan 12;15(1):219–232. doi: 10.1093/nar/15.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Streeck R. E., Zachau H. G. Patchwork structure of a bovine satellite DNA. Cell. 1979 Nov;18(3):883–893. doi: 10.1016/0092-8674(79)90140-5. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prère M. F., Fayet O. Susceptibility of Neisseria gonorrhoeae DNA to cleavage by restriction endonuclease KpnI. Ann Inst Pasteur Microbiol. 1985 May-Jun;136A(3):329–338. doi: 10.1016/s0769-2609(85)80095-8. [DOI] [PubMed] [Google Scholar]
- Quint A., Cedar H. In vitro methylation of DNA with Hpa II methylase. Nucleic Acids Res. 1981 Feb 11;9(3):633–646. doi: 10.1093/nar/9.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Urieli S., Pollack Y., Gruenbaum Y., Glaser G. Studies on the biological role of dna methylation; IV. Mode of methylation of DNA in E. coli cells. Nucleic Acids Res. 1980 Apr 25;8(8):1783–1792. doi: 10.1093/nar/8.8.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1984;12 (Suppl):r167–r204. doi: 10.1093/nar/12.suppl.r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizès G., Patillon M., Kovoor A. A restriction endonuclease from Agrobacterium tumefaciens. FEBS Lett. 1977 Oct 1;82(1):69–70. doi: 10.1016/0014-5793(77)80887-9. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. II. Partial recognition site base sequences. J Mol Biol. 1973 Dec 25;81(4):445–459. doi: 10.1016/0022-2836(73)90516-0. [DOI] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977 Oct 25;252(20):7265–7272. [PubMed] [Google Scholar]
- Sato S., Nakazawa K., Shinomiya T. A DNA methylase from Thermus thermophilus HB8. J Biochem. 1980 Sep;88(3):737–747. doi: 10.1093/oxfordjournals.jbchem.a133026. [DOI] [PubMed] [Google Scholar]
- Schlagman S., Hattman S., May M. S., Berger L. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R. Eco RII). J Bacteriol. 1976 May;126(2):990–996. doi: 10.1128/jb.126.2.990-996.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Smith H. O. Nucleotide sequence specificity of restriction endonucleases. Science. 1979 Aug 3;205(4405):455–462. doi: 10.1126/science.377492. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J Bacteriol. 1985 Oct;164(1):490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck R. E. Single-strand and double-strand cleavage at half-modified and fully modified recognition sites for the restriction nucleases Sau3a and Taqi. Gene. 1980 Dec;12(3-4):267–275. doi: 10.1016/0378-1119(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Strobl J. S., Thompson E. B. Methylation of either cytosine in the recognition sequence CGCG inhibits ThaI cleavage of DNA. Nucleic Acids Res. 1984 Nov 12;12(21):8073–8083. doi: 10.1093/nar/12.21.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Dobritsa A. P. On the nature of the cytosine-methylated sequence in DNA of Bacillus brevis var. G.-B. Biochim Biophys Acta. 1975 Sep 12;407(1):61–72. doi: 10.1016/0005-2787(75)90023-4. [DOI] [PubMed] [Google Scholar]
- Vovis G. F., Lacks S. Complementary action of restriction enzymes endo R-DpnI and Endo R-DpnII on bacteriophage f1 DNA. J Mol Biol. 1977 Sep 25;115(3):525–538. doi: 10.1016/0022-2836(77)90169-3. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Y., Langtimm C. J., Chatterjee R., Walder J. A. Cloning of the MspI modification enzyme. The site of modification and its effects on cleavage by MspI and HpaII. J Biol Chem. 1983 Jan 25;258(2):1235–1241. [PubMed] [Google Scholar]
- Wang R. Y., Zhang X. Y., Khan R., Zhou Y. W., Huang L. H., Ehrlich M. Methylated DNA-binding protein from human placenta recognizes specific methylated sites on several prokaryotic DNAs. Nucleic Acids Res. 1986 Dec 22;14(24):9843–9860. doi: 10.1093/nar/14.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead P. R., Brown N. L. EaeI: a restriction endonuclease from Enterobacter aerogenes. FEBS Lett. 1983 May 2;155(1):97–101. doi: 10.1016/0014-5793(83)80217-8. [DOI] [PubMed] [Google Scholar]
- Whitehead P. R., Jacobs D., Brown N. L. Restriction endonucleases from Herpetosiphon giganteus: an example of the evolution of DNA recognition specificity? Nucleic Acids Res. 1986 Sep 11;14(17):7031–7045. doi: 10.1093/nar/14.17.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiatr C. L., Witmer H. J. Selective protection of 5' ... GGCC ... 3' and 5' ... GCNGC ... 3' sequences by the hypermodified oxopyrimidine in Bacillus subtilis bacteriophage SP10 DNA. J Virol. 1984 Oct;52(1):47–54. doi: 10.1128/jvi.52.1.47-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Hammer S. M., Hirsch M. S., Mulder C. Methylation of the viral genome in an in vitro model of herpes simplex virus latency. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2207–2210. doi: 10.1073/pnas.79.7.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Mulder C. Detection of methylated sequences in eukaryotic DNA with the restriction endonucleases Smai and Xmai. J Mol Biol. 1981 Jul 25;150(1):133–136. doi: 10.1016/0022-2836(81)90328-4. [DOI] [PubMed] [Google Scholar]
- van Ormondt H., Lautenberger J. A., Linn S., de Waard A. Methylated oligonucleotides derived from bacteriophage fd RF-DNA modified in vitro by E. coli B modification methylase. FEBS Lett. 1973 Jul 1;33(2):177–180. doi: 10.1016/0014-5793(73)80186-3. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]



