Abstract
The functional cycle of the Rac1 GTPase involves a large number of steps, including post-translational processing, cytosolic sequestration by RhoGDIs, translocation to specific subcellular localizations, activation by GDP/GTP exchange, inactivation by GTP hydrolysis, and re-formation of cytosolic Rac1/RhoGDI inhibitory complexes. Here, we summarize the current knowledge about the regulation of those steps. In addition, we discuss a recently described, cytoskeletal-dependent feed-back loop that favors the efficient translocation and activation of Rac subfamily proteins during cell signaling. This route is mediated by a heteromolecular protein complex composed of the cytoskeletal protein coronin1A, the Dbl family member ArhGEF7, the serine/threonine kinase Pak1, and the Rac1/RhoGDI dimer. This route promotes the translocation of Rac1/RhoGDI to F-actin-rich juxtamembrane areas, the Pak1-dependent release of Rac1 from the Rac1/RhoGDI complex, and Rac1 activation. This pathway is important for optimal Rac1 activation during the signaling of the EGF receptor, integrins, and the antigenic T-cell receptor.
Keywords: ArhGEF7, F-actin, Lipid rafts, PAK, Rac1, RhoGDI, coronin, cytoskeleton, β-Pix
Rac1, one of the best characterized members of the Rho/Rac GTPase subfamily, regulates ubiquitous processes such as the formation of membrane ruffles and lamellipodia, cell adhesion, proliferation, intercellular attraction/repulsion, and transcriptomal dynamics. In addition, it modulates cell-type-specific processes such as axon migration/guidance, phagocytosis, or the formation of the immunological synapse.1 To trigger most of those functions, Rac1 has to fulfill two basic conditions. One of them is to be anchored at the plasma membrane to make it possible the subsequent activation of its primary effectors in the correct subcellular localization. The second condition is that it has to be bound to GTP, since this is the only conformational state compatible with the interaction of most downstream effectors.1 These requirements only change in few signaling scenarios, such as the cell cycle-regulated transfer of Rac1 to the nucleus2 or the indistinctive binding of GDP-Rac and GTP-Rac1 to mTOR.3
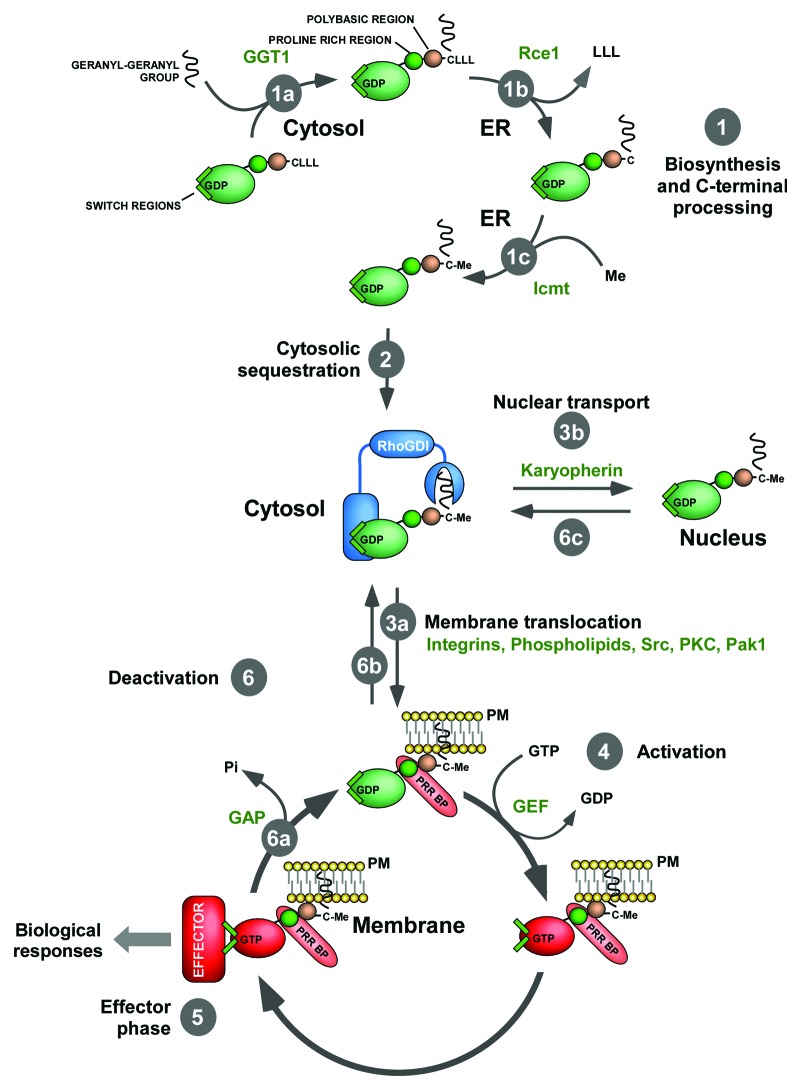
Like the mythical Greek Odysseus, the travel of Rac1 from the cytosol to the plasma membrane is a stepwise mechanism subjected to multiple regulatory challenges (Fig. 1). The first step required to reach its particular Ithaca is the attachment of a geranyl-geranyl group to the most C-terminal cysteine residue of the translated Rac1 protein, a modification catalyzed in the cytosol by type I geranyl-geranyl transferase (GGT1)1,4 (Fig. 1, step 1a). Rac1 then moves to the endoplasmic reticulum, where it is subjected to further processing by the protease Rce1 and the methyltransferase Icmt.1,5 Rcc1 cleaves off the C-terminal Rae1 LLL tripeptide (Fig. 1, step 1b). Icmt incorporates a methyl group at the α-carboxyl group of the C-terminal cysteine that becomes exposed upon the completion of the Rcc1-catalyzed reaction (Fig. 1, step 1c). Based on data gathered using Ras subfamily proteins, it has been assumed that the prenylation, cleavage and methylation steps were conditio sine qua non for the biological activity of Rac1 proteins.1,4,5 Such model is consistent with current experimental evidence,6,7 although some recent reports have unexpectedly shown that Rac1 can be functional in primary GTT1 deficient macrophages8 and in embryonic fibroblasts obtained from Rce1–/– and Icmt–/– mice.9
Figure 1. Depiction of the main steps of the functional cycle of Rac1. Enzymes and other cellular factors collaborating in each of those steps are shown in green. Other symbols are indicated in the figure. Please note that the sequence of the C-terminal LLL tripeptide is different in some Rac1 orthologs, such as those present in horses (TVF), chimpanzees (LQL) or Drosophila (ALL). The status of Rac1 inside the nucleus is still poorly characterized so we have not incorporated the activation/inactivation cycle of Rac1 in that compartment. It is also unclear whether the insertion of Rac1 in membranes is achieved when in the GDP- or GTP-bound state. The latter case has not been contemplated in the scheme for the sake of simplicity. Abbreviations used are: ER, endoplasmic reticulum; GAP, GTPase activating protein; Me, methyl group; Pi, inorganic phosphate; PKC, protein kinase C; PM, plasma membrane; PRR BP, proline-rich region binding protein. Other abbreviations have been described in the main text.
After its transit through the endoplasmic reticulum, Rac1 moves back to the cytosol where it stays in an inactive reservoir until the reception of extracellular signals by the cell. This pool is stabilized by the formation of stoichiometric complexes with RhoGDIs10 (Fig. 1, step 2). RhoGDIs perform both negative and positive actions in this complex. On the negative side, they inhibit the usually high intrinsic GDP/GTP exchange of Rac1, thus favoring the maintenance of the bound GTPase in the inactive conformation in non-stimulated cells. This function is mediated by the direct interaction of the RhoGDI molecule with the Rac1 switch regions.10 In addition, they use a deep hydrophobic cavity to trap the Rac1 prenyl group, a mechanism that keeps the GTPase away from the membrane in a cytosolic, fully soluble pool10 (Fig. 1). On the positive side, RhoGDIs protect the bound Rac1 molecules from proteolytic degradation11 and, in addition, are likely involved in the final transit of Rho/Rac GTPases to the plasma membrane.10
The cytosolic reservoir of Rac1 is rapidly mobilized to the plasma membrane upon cell stimulation1 (Fig. 1, step 3a) or, alternatively, to the nucleus in the G2 phase of the cell cycle2 (Fig. 1, step 3b). The main factors involved in the former mobilization route are the Rac1 GEFs, a group of enzymes containing either Dbl-homology or Dock-homology domains that promote the rapid transition of Rac1 from the inactive (GDP-bound) to the active (GTP-bound) state12 (Fig. 1, step 4). However, alternative physiological activation steps exist, such as the transglutaminase-dependent serotonylation of Rac1 downstream of G-coupled receptors.13 This mobilization step is also facilitated by the C-terminal Rac1 polybasic and proline-rich regions. The former region favors the formation of hydrophilic interactions with the negatively-charged heads of lipids only present in the plasma membrane, because Rac-related proteins lacking this hydrophilic signal are localized in intracellular vesicles14,15 (Fig. 1, step 3a). The Rac1 proline-rich region interacts with the SH3 region of ArhGEF7 (also known as β-Pix), a Dbl-homology family protein that can associate with F-actin rich areas of the cell16 (Fig. 1, step 3a). Recent data indicate that other ancillary signals collaborate in this process, including the post-translational modification of Rac1 by phosphorylation,17 sumoylation18 and ubiquitination.19,20 Rac1 localization can be also regulated positively by extrinsic signals such as the presence of specific phospholipids (PtdIns(4,5)P2, PtdIns(3,4,5)P3)21,22 and lipid rafts23-25 at the plasma membrane (Fig. 1, step 3a). At the end of the effector phase (Fig. 1, step 5), the standard regulatory model holds that Rac1 goes back to the GDP bound state via the action of Rho/Rac GTPase activating proteins12 (Fig. 1, step 6a), re-associates with RhoGDI molecules (Fig. 1, step 6b) and moves back to the inactive cytosolic reservoir until a new stimulation cycle starts.1 In addition to this standard model, recent data demonstrated that Rac1 can undergo activation/inactivation cycles by shuttling between the plasma membrane and endocytic compartments.23,26 The shuttling of Rac1 in and out of the nucleus requires the Rac1 polybasic region and karyopherin α2,27 an importin that transfers cargo molecules inside the nucleus (Fig. 1, step 3b). Whether this step requires GEFs, carrier proteins, or intracellular docking proteins is unknown as yet. The mechanism by which Rac1 is returned to the cytosol remains ill defined (Fig. 1, step 6c).
Whereas the GTPase/RhoGDI complex and the catalytic steps involved in Rac1 GDP/GTP exchange and GTP hydrolysis are well understood in structural terms,10,12 the dynamic aspects that modulate the release of Rac1 from RhoGDIs during the activation process are not well understood. For instance, we do not know whether Rac1 dissociates from the RhoGDI before contacting the upstream GEFs or, alternatively, whether the GEFs promote both the dissociation and activation of the GTPase. In either case, it is unclear the intracellular cues used by either Rac1 or the Rac1/RhoGDI complex to reach the subcellular regions where the stimulated GEFs are localized. Finally, we do not know whether these Rac1 mobilization-related steps are general and/or cell type-specific. Recent inroads in this subject suggest that the release of Rac1 is mediated by the direct phosphorylation of RhoGDI by either upstream (Src, protein kinase C) or downstream (Pak1) kinases.28-31 In any case, it is still difficult to explain how the cytosolic RhoGDI/Rac1 complexes manage to get close to those kinases during cell stimulation.
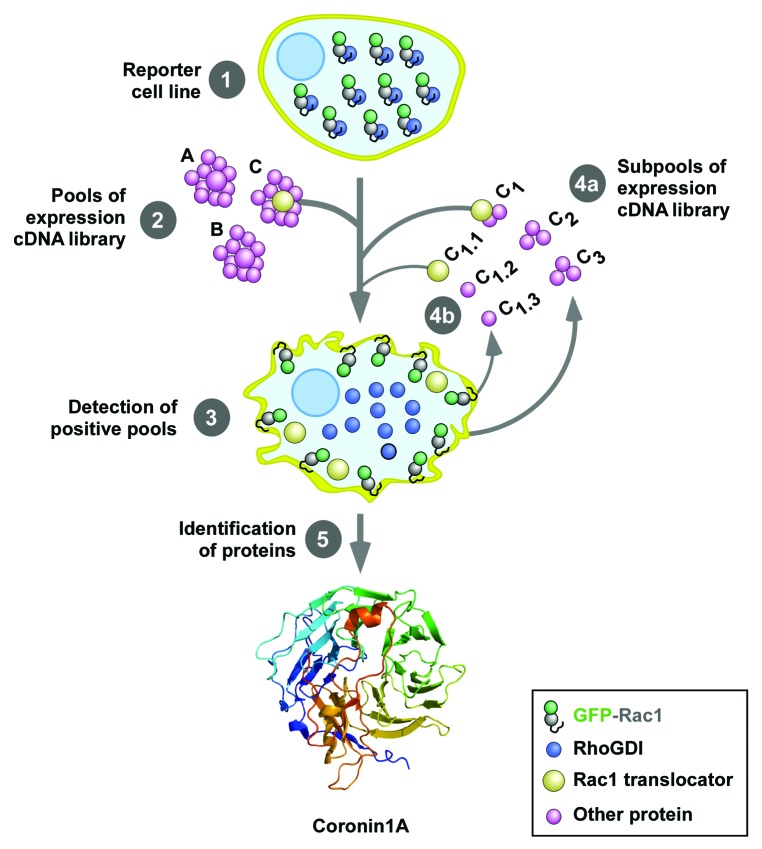
To shed light on this process, we decided to search for proteins involved in the regulation of the translocation of Rac1 to the plasma membrane using a genome-wide functional screen (Fig. 2). This approach led to the isolation of coronin1A (Coro1A) as a protein capable of inducing the translocation and activation of Rac1 during cell signaling (Fig. 2, bottom).32 Coro1A belongs to a large family of cytoskeletal regulators that show a phylogenetic distribution from unicellular eukaryotes to humans.33 These proteins control the bundling of F-actin filaments, the growth and orientation angle of new F-actin branches, and the disassembly of old actin cables.33 Such activities suggested to us that Coro1A could be possibly involved in a cross-talk between the F-actin cytoskeleton and the process of Rac1 translocation and/or activation. Consistent with this idea, we could demonstrate that the overexpression of Coro1A induced the F-actin-dependent translocation and activation of Rac1 in the plasma membrane. Conversely, its inactivation led to ineffective activation of Rac1 and Rac1-downstream routes. This pathway was active in a number of cell types (COS1, 293T, Jurkat cells) and cellular stimulation conditions (EGF signaling, integrin-mediated adhesion, T-cell receptor stimulation).
Figure 2. Scheme of the cellomic screen used in the work reviewed here. The first screening was conducted using an expression library of 135,000 independent cDNA clones obtained from human T cells. To this end, we used a reporter HEK293T stably expressing a cytoplasmic EGFP-Rac1 protein (step 1). To increase the efficiency of the screening, this cell line also expressed anti-apoptotic proteins to avoid the loss of clones due to the presence of pro-apoptotic molecules in the transfected cDNA pools. The screening was conducted by transfecting separate pools of 90 cDNAs in the reporter cell line using the calcium phosphate precipitation method (step 2) and the subsequent score of cells showing plasma membrane localized EGFP-Rac1 using epifluorescence microscopy (step 3). Positive pools were progressively subdivided and transfected in the reporter cell line (steps 4a and 4b) until the isolation of the cDNA clones responsible for the Rac1 translocating activity (step 5). The crystal structure of Coro1A, one of the clones identified in this screening, is shown at the bottom. The structure is composed of WD40 domains arranged in a prototypical β-propeller conformation. See inset at the bottom for the shape and color code use for the indicated proteins used in these experiments.
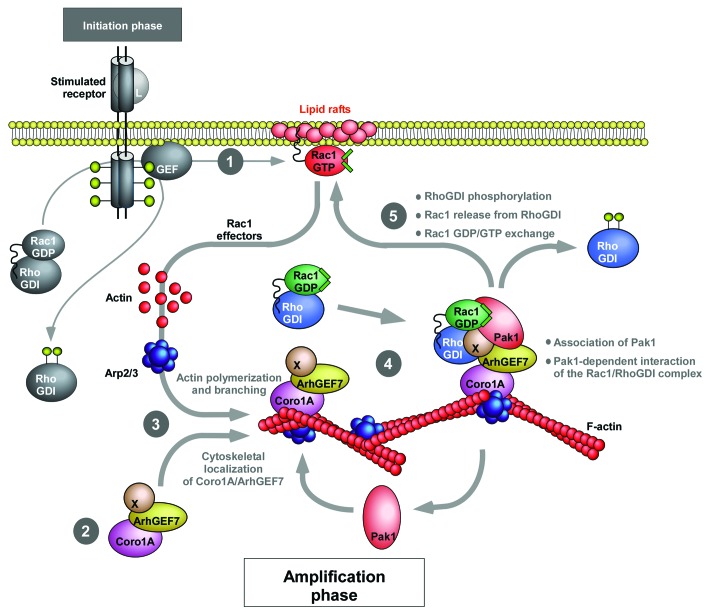
Mechanistic studies revealed that Coro1A promotes the translocation and activation of Rac1 via the formation of a F-actin-dependent, heteromolecular complex with ArhGEF7, Pak1, RhoGDI and Rac1. According to our proposed model (Fig. 3), the formation of this Rac1 translocation complex requires upstream signals such as Rac1 activity, aggregated lipid rafts in the plasma membrane and the presence of F-actin in cells (Fig. 3, stage 1). In the absence of those upstream signals, Coro1A can only associate in a stable manner with ArhGEF7 in the cytoplasm (Fig. 3, stage 2). Upon conditions triggering F-actin polymerization, this inert complex is tethered to membrane ruffles and lamellipodia, a process made it possible by the intrinsic F-actin binding properties of Coro1A (Fig. 3, stage 3). The F-actin bound complex binds then Pak1 and Rac1/RhoGDI (Fig. 3, stage 4), leading to the phosphorylation of RhoGDI by Pak1, the release of Rac1 from the phosphorylated RhoGDI and, finally, to Rac1 activation (Fig. 3, stage 5). Several properties of this new translocation process are important from a signaling point of view. First, the F-actin binding properties of Coro1A offer a rather simple explanation for understanding how the cytosolic Rac1/RhoGDI pool is shuttled toward the plasma membrane during cell stimulation conditions. Second, its dependency on upstream Rac1 signals suggest that the Coro1A route is involved in the generation of secondary waves of Rac1 activation rather than being directly involved in the initial burst of Rac1 activity that takes place upon the reception of the extracellular signal. Thus, according to our proposed regulatory model, cells will have to trigger Rac1 or RhoG activation in order to engage the downstream Coro1A-dependent relay mechanism (Fig. 3). Finally, the need of pre-formed F-actin cytoskeletal structures to engage the Coro1A-dependent translocation route suggests that F-actin can induce a positive feed-back mechanism that will allow the generation of additional waves of Rac1 activation and F-actin polymerization at the plasma membrane (Fig. 3).
Figure 3. Schematic representation of the regulatory model for the proposed Coro1A-mediated translocation and activation of Rac1 reviewed here. The first stimulus triggering the first burst of Rac1 activity via a Coro1A-independent route is shown on the left in gray color and thin lanes. The Coro1A-based relay mechanism involved in the subsequent amplification of Rac1 signals is shown with thicker lanes. L, ligand; X, a putative Rac1 GEF factor associated to ArhGEF7. Phosphorylated residues are shown as yellow lollipops. See further details in the main text of this work.
Despite these advances, the role of Coro1A in the translocation and activation of Rac1 is far from being an open and shut case. Thus, the mechanism that regulates the assembly of Pak1 and the RhoGDI/Rac1 pair onto the Coro1A/ArhGEF7 complex has not been fully elucidated yet. Our data indicate that such assembly requires the presence of ArhGEF7 in the Coro1A complex, a result consistent with the known physical interaction between ArhGEF7 and Pak1.34 However, the requirement of F-actin for the formation of the entire Coro1A/Pak1/RhoGDI complex also indicates that additional ancillary partners and/or signals must also contribute to this process. Whether these extra elements are proteins, membrane lipids, and/or other upstream signals remains to be determined. It is also unclear the Rac1 GEF in charge of activating Rac1 upon its release from the Coro1A-nucleated protein complex (Fig. 3). One option is ArhGEF7 itself, since it is obvious that its presence in the Coro1A complex will ensure its close proximity with the released Rac1 molecules (Fig. 3). However, the catalytic activity of ArhGEF7 is controversial, suggesting that other GEFs could be involved (Fig. 3, protein labeled as X). It is possible therefore that such activation step is at the hands of other GEFs that, due to their localization in F-actin cables or the plasma membrane, could be in close proximity to the Coro1A-nucleated complex. The resolution of all those lingering issues will need further experimental work in the near future. It is also worth noting that recent data obtained in our laboratory suggest that Coro1A can use alternative mechanisms to induce the translocation of Rac1 to the plasma membrane. Hence, we have observed that Coro1A can trigger the membrane localization of Rac1R66E, a mutant protein that cannot bind RhoGDI. This alternative route is mechanistically distinct from that characterized in this reviewed publication, because it cannot be inhibited by blocking Pak1 function (Castro-Castro A, Bustelo XR, unpublished observations). Although these results are probably not physiologically relevant given the lack of significant amounts of RhoGDI-free Rac1 proteins in the cytosol, they are interesting because they reveal a pathway that can further enhance the membrane anchoring of Rac1 upon its liberation from the RhoGDI complexes by the Coro1A/ArhGEF7/Pak1 complex. Although this alternative translocation mechanism remains to be elucidated, we surmise that it could involve the generation of F-actin-dependent “permissive” conditions at the plasma membrane. Consistent with this view, we have observed that the overexpression of Coro1A induces lipid raft aggregation in the plasma membrane. Furthermore, we found that the aggregation of lipid rafts promotes the translocation of the Rac1R66E mutant to the plasma membrane (Castro-Castro A, Bustelo XR, unpublished observations). Finally, it should be pointed out that Coro1A and ArhGEF7 are not ubiquitously expressed, so it is unlikely that the Rac1 translocation/activation mechanism reviewed here will be utilized by all cell types. In this context, it will be interesting to investigate whether other cytoskeletal proteins fulfill Coro1A-like functions in cells that do not express Coro1A. This is a feasible scenario, because our genome-wide functional screen has resulted in the identification of a second WD40 family protein (WDR26) that can also induce the translocation of Rac1 to the plasma membrane (Castro-Castro A, Bustelo XR, unpublished data). It is also possible that the increase in the membrane-localized pool of Rac1 can be achieved by routes alternative to those reported here. For example, our cellomic screen has also identified transmembrane proteins that favor the membrane localization of Rac1 by blocking the internalization of lipid rafts. Unlike the case of Coro1A-dependent route, this alternative pathway cannot be inhibited by either Rac1 dominant negative mutants or Pak1 interference strategies. Taken together, these results suggest that cells can resort to many pathways to regulate the translocation of Rac1 during cell signaling.
Acknowledgments
X.R.B.'s work is supported by grants from the NIH (5R01-CA73735-13), the Spanish Ministry of Science and Innovation (SAF2009-07172, GEN2003-20239-C06-01), the Red Temática de Investigación Cooperativa en Cáncer (RD06/0020/0001), the Castilla y León Autonomous Government (GR97), and the 7th Framework European Union Program (FP7-HEALTH-2007-A-201862). X.R.B.’s lab is a Consolidated Cancer Research Group of the Asociación Española contra el Cáncer.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/19111
References
- 1.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–70. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaelson D, Abidi W, Guardavaccaro D, Zhou M, Ahearn I, Pagano M, et al. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J Cell Biol. 2008;181:485–96. doi: 10.1083/jcb.200801047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saci A, Cantley LC, Carpenter CL. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seabra MC. Membrane association and targeting of prenylated Ras-like GTPases. Cell Signal. 1998;10:167–72. doi: 10.1016/S0898-6568(97)00120-4. [DOI] [PubMed] [Google Scholar]
- 5.Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–12. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 6.Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, et al. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J Biol Chem. 2008;283:25150–63. doi: 10.1074/jbc.M800882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cushman I, Casey PJ. RHO methylation matters: a role for isoprenylcysteine carboxylmethyltransferase in cell migration and adhesion. Cell Adh Migr. 2011;5:11–5. doi: 10.4161/cam.5.1.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan OM, Ibrahim MX, Jonsson IM, Karlsson C, Liu M, Sjogren AK, et al. Geranylgeranyltransferase type I (GGTase-I) deficiency hyperactivates macrophages and induces erosive arthritis in mice. J Clin Invest. 2011;121:628–39. doi: 10.1172/JCI43758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaelson D, Ali W, Chiu VK, Bergo M, Silletti J, Wright L, et al. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell. 2005;16:1606–16. doi: 10.1091/mbc.E04-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–63. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, et al. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol. 2010;12:477–83. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Walther DJ, Peter JU, Winter S, Höltje M, Paulmann N, Grohmann M, et al. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell. 2003;115:851–62. doi: 10.1016/S0092-8674(03)01014-6. [DOI] [PubMed] [Google Scholar]
- 14.Filippi MD, Harris CE, Meller J, Gu Y, Zheng Y, Williams DA. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nat Immunol. 2004;5:744–51. doi: 10.1038/ni1081. [DOI] [PubMed] [Google Scholar]
- 15.Prieto-Sánchez RM, Bustelo XR. Structural basis for the signaling specificity of RhoG and Rac1 GTPases. J Biol Chem. 2003;278:37916–25. doi: 10.1074/jbc.M301437200. [DOI] [PubMed] [Google Scholar]
- 16.ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J Cell Biol. 2006;172:759–69. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon T, Kwon DY, Chun J, Kim JH, Kang SS. Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at serine 71 of Rac1. J Biol Chem. 2000;275:423–8. doi: 10.1074/jbc.275.1.423. [DOI] [PubMed] [Google Scholar]
- 18.Castillo-Lluva S, Tatham MH, Jones RC, Jaffray EG, Edmondson RD, Hay RT, et al. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat Cell Biol. 2010;12:1078–85. doi: 10.1038/ncb2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nethe M, Anthony EC, Fernandez-Borja M, Dee R, Geerts D, Hensbergen PJ, et al. Focal-adhesion targeting links caveolin-1 to a Rac1-degradation pathway. J Cell Sci. 2010;123:1948–58. doi: 10.1242/jcs.062919. [DOI] [PubMed] [Google Scholar]
- 20.de la Vega M, Kelvin AA, Dunican DJ, McFarlane C, Burrows JF, Jaworski J, et al. The deubiquitinating enzyme USP17 is essential for GTPase subcellular localization and cell motility. Nat Commun. 2011;2:259. doi: 10.1038/ncomms1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–61. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–3. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 23.Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat Cell Biol. 2007;9:1381–91. doi: 10.1038/ncb1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RG, Schwartz MA. Integrins regulate Rac targeting by internalization of membrane domains. Science. 2004;303:839–42. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- 25.del Pozo MA, Balasubramanian N, Alderson NB, Kiosses WB, Grande-García A, Anderson RG, et al. Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat Cell Biol. 2005;7:901–8. doi: 10.1038/ncb1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palamidessi A, Frittoli E, Garré M, Faretta M, Mione M, Testa I, et al. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–47. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 27.Sandrock K, Bielek H, Schradi K, Schmidt G, Klugbauer N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic. 2010;11:198–209. doi: 10.1111/j.1600-0854.2009.01015.x. [DOI] [PubMed] [Google Scholar]
- 28.DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol Biol Cell. 2006;17:4760–8. doi: 10.1091/mbc.E06-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol Cell. 2004;15:117–27. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Price LS, Langeslag M, ten Klooster JP, Hordijk PL, Jalink K, Collard JG. Calcium signaling regulates translocation and activation of Rac. J Biol Chem. 2003;278:39413–21. doi: 10.1074/jbc.M302083200. [DOI] [PubMed] [Google Scholar]
- 31.Abramovici H, Mojtabaie P, Parks RJ, Zhong XP, Koretzky GA, Topham MK, et al. Diacylglycerol kinase zeta regulates actin cytoskeleton reorganization through dissociation of Rac1 from RhoGDI. Mol Biol Cell. 2009;20:2049–59. doi: 10.1091/mbc.E07-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lérida I, Muriel O, et al. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 2011;30:3913–27. doi: 10.1038/emboj.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rybakin V, Clemen CS. Coronin proteins as multifunctional regulators of the cytoskeleton and membrane trafficking. Bioessays. 2005;27:625–32. doi: 10.1002/bies.20235. [DOI] [PubMed] [Google Scholar]
- 34.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–92. doi: 10.1016/S1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]