Abstract
Scaffolded DNA origami has recently emerged as a versatile, programmable method to fold DNA into arbitrarily shaped nanostructures that are spatially addressable, with sub-10 nm resolution. Toward functional DNA nanotechnology, one of the key challenges is to integrate the bottom up self-assembly of DNA origami with the top-down lithographic methods used to generate surface patterning. In this report we demonstrate that fixed length DNA origami nanotubes, modified with multiple thiol groups near both ends, can be used to connect surface patterned gold islands (tens of nanometers in diameter) fabricated by electron beam lithography (EBL). Atomic force microscopic imaging verified that the DNA origami nanotubes can be efficiently aligned between gold islands with various inter-island distances and relative locations. This development represents progress toward the goal of bridging bottom up and top down assembly approaches.
In recent years, structural DNA nanotechnology has advanced to the point that fully addressable, self-assembled, DNA nanostructures can be rationally designed and easily constructed1-3. Among these developments, scaffolded DNA origami is especially remarkable for designing and constructing various 2- and 3-D nanostructures with control over structural dimensions and spatial addressability4-11. In this method4 a single stranded, genomic DNA scaffold is folded by approximately 200 short staple strands into a variety of predesigned shapes. Due to the unique sequences of individual staple strands, DNA origami structures possess addressable binding sites with ~ 6 nm resolution and have been utilized as templates to direct the assembly of metal nanoparticles, carbon nanotubes and biological materials12-19. It has been envisioned that, as a fully addressable molecular pegboard, DNA origami could be used as an excellent template to build functional nanoelectronic circuits in a self-assembling system. However, a significant barrier to this is the lack of precise control of the position and orientation of the assembled, solution phase structures on solid substrates, thus limiting the function of the DNA origami directed nanomaterials. While routine, top-down lithographic methods exhibit control of the position and orientation of fabricated structures on a relatively large scale, it is difficult to reach the sub-10 nm regime without the state-of-the-art (and typically expensive) lithographic tools. One unique property of self-assembled DNA origami is that the overall size can be as large as several hundred nanometers, while nanometer precision is maintained through independent, addressable binding sites, making it an ideal candidate for bridging structures formed using bottom-up and top-down nanofabrication methods.
With efforts directed at meeting the above challenges, progress has recently been reported with the site specific attachment of DNA origami structures onto lithographically generated surface patterns. In one example, Soh and co-workers formulated a chemical strategy to attract and immobilize rectangular shaped DNA origami structures onto gold surfaces through charge-charge interaction using a carboxylic acid-terminated self-assembled monolayer20. In another case, triangular shaped DNA origami structures formed in solution were successfully assembled onto hydrophilic oxide surface patterns with matching triangular shape and size, but not to the surrounding hydrophobic substrate21. This method has been further utilized to pattern 5-nm gold particles into ordered, large-area, two-dimensional arrays22, demonstrating the potential of integrating DNA directed self-assembly with lithography to create functional structures. In the above methods, the interactions responsible for the attachment of DNA origami structures to substrates are non-covalent associations such as electrostatic forces. To achieve better chemical selectivity between the lithographically patterned substrate and the DNA nanostructures and to create more complex nanoarchitectures an alternative approach must be employed. The site-specific attachment of DNA origami to lithographically patterned surfaces can be realized through covalent chemical bonds. While electrostatic interactions have the advantage of allowing the target DNA origami structures to dynamically adsorb/desorb and orient/reorient themselves onto the substrate to reach equilibrium, covalent binding may provide a relatively stronger association force and higher selectivity, making the DNA nanostructure registration on the substrate more stable.
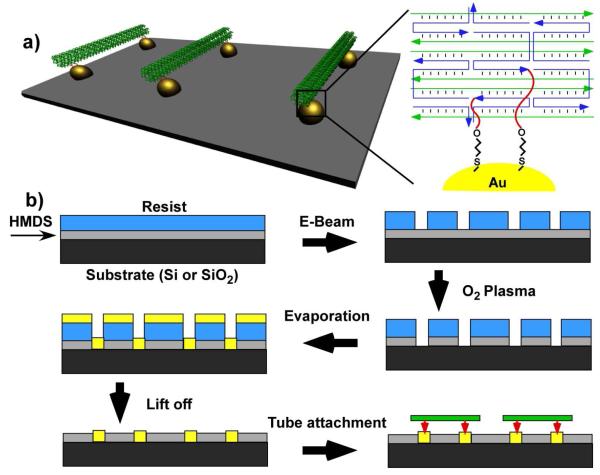
Here we demonstrate a covalent approach to achieve interconnection of EBL generated gold islands of several different patterns using fixed length DNA origami nanotubes. Our strategy is to use thiolated DNA strands extending from both ends of the DNA nanotubes and directly attach the tubes to discrete gold islands on the substrate through well-known, dative covalent, thiol-Au bonds23-24. When the inter-island distance matches the length of the thiolated-tube structure, bridging of the gold islands can be achieved (Figure 1a).
Figure 1.
a) Schematic drawing of gold islands connected by DNA origami tubes on the substrate surface. Thiolated DNA strands (shown in red) are extended from each end of DNA origami tube and the thiol-gold bonds align the tubes along the gold islands. The distance between the thiolated groups is designed so the tube will only connect gold islands that match its length. b) The fabrication of gold islands and binding of DNA origami tubes. An HMDS and PMMA resist layer were spin coated on a clean Si or SiO2 substrate. E-Beam exposure was performed with a designed pattern. After O2 plasma treatment was used to eliminate the exposed HMDS, 2 nm Cr and 7 nm Au were deposited through evaporation. After lift-off with acetone, patterns of gold surface islands with the background covered by HMDS, were fabricated on the substrate. Gold patterned substrate was cut into ~ 5 × 20 mm sections and incubated with freshly purified and reduced DNA origami tube solution to facilitate the specific attachment of the DNA tubes to the gold islands.
The overall fabrication process is illustrated in Figure 1b. First, a clean Si or SiO2 substrate is spin-coated with a monolayer of Hexamethyldisilazane (HMDS) and poly (methyl methacrylate) (PMMA) resist (~ 80 nm). Next, to create well-defined holes, the PMMA resist layer is exposed to an E-beam using a predesigned pattern. After subsequent O2 plasma treatment, 2 nm thick Cr and 7 nm thick Au are deposited consecutively through evaporation. The O2 plasma treatment step cleans the surface to facilitate strong adhesion of the metal islands to the substrate. Finally, acetone lift-off is performed to remove the Au on PMMA and PMMA layers, leaving patterns of gold islands adhered to the substrate with the surrounding surfaces covered by HMDS. The HMDS layer is hydrophobic and prevents the hydrophilic DNA origami structures from non-specifically binding to the substrate in areas without gold islands.
The DNA origami nanotube composed of six DNA helices bundled with a hexagonal cross section was designed and synthesized in a similar fashion as described previously25-26. The fully extended, end-to-end length of the DNA origami nanotube is ~380 nm. The distance between the thiolated groups located near each end is approximately 320 ± 20 nm, considering all possible orientations of these strands. Therefore, the distances between neighboring gold islands on the patterned surface was designed to be within, or far beyond this range to maximize or minimize the probability of bridging two islands by a single DNA nanotube.
A select number of helper strands (4, 2 or 1) located close to both ends of the tube were modified with single stranded regions (33 nucleotide long) extending out from the tube, each carrying a thiol group at the 3′ end (see supporting information for details of the design and DNA sequences). The thiol groups direct the specific and efficient attachment of the DNA nanotubes to the Au islands in the surface assembly stage and the multi-valency provides a way to tune the strength of the DNA nanotube attachment. The DNA origami nanotubes were formed by annealing the M13 scaffold (10 nM) with thiolated strands at a 1:1 ratio and with the rest of the helper strands at a 1:10 ratio. The assembled DNA origami nanotubes were purified using agarose gel electrophoresis to remove any excess helper strands and incorrectly formed tubes; the correctly formed tubes were extracted from the agarose gel with a freeze-squeeze column. The thiol groups in the origami tube were reduced by 20 mM tris (2-carboxyethyl) phosphine (TCEP) for 1 hour before use. Extra TCEP and other small molecules were removed using a Microcon centrifuge device (MWCO 100kD).
The gold island, patterned substrate was cut into 5 mm × 20 mm sections and individual pieces were submersed into a sample tube containing a freshly reduced DNA origami nanotube solution (100 μl and 2 nM) and incubated for 20 hours at room temperature with gentle shaking. The tubes were attached to the gold islands through dative covalent, thiol-Au interactions during the incubation process. Then the substrate was thoroughly rinsed with water, dried under a stream of nitrogen gas, and imaged in air in tapping mode with an atomic force microscope (TM-AFM) (see more details in the supporting information).
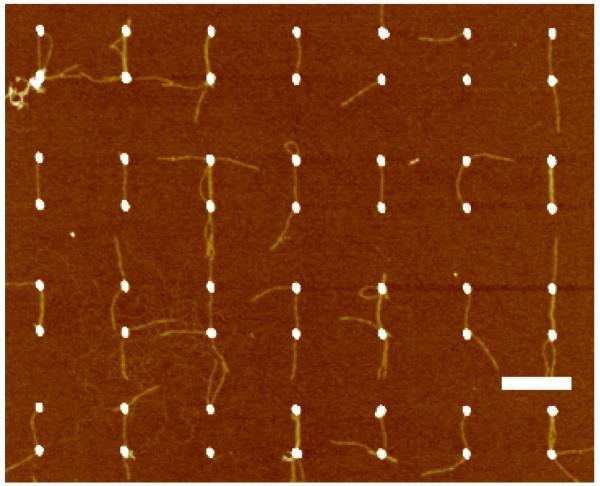
Figure 2 shows a typical AFM image of a gold island patterned substrate section after incubation with 2 nM DNA origami nanotubes for 20 hours. Pairs of gold islands were created on the substrate: each island is 60 nm in diameter with a 300 nm center-to-center distance between the pair; the distance between adjacent island pairs is 600 nm, a distance too large for the 380 nm DNA tube to bridge. The DNA origami tubes contained 4 thiolated groups at both ends providing a tetra-valent interaction with each gold island. Analysis of the AFM images show that ~96% of the gold islands have one or more DNA tubes attached, with the yield of gold islands connected by a single DNA tube ~86%. Occasionally, more than one DNA nanotube was found between the same island pair. For this gold island lattice pattern, all the DNA nanotubes bridging the island pairs were aligned in a parallel fashion due to designed spacing between particles. However, some DNA nanotubes attached to the gold islands at a single end and were orientated in the wrong direction, preventing the other end from reaching an adjacent island.
Figure 2.
AFM images of gold islands connected by DNA origami tubes. The gold islands in the images are 60 nm in diameter. For each pair, the distance between the gold islands is 300 nm, while the distance between neighboring pairs of gold islands is 600 nm. Interconnection can be observed for most gold island pairs. DNA origami tubes were aligned along the pairs of the gold islands with either 0° or 180° orientation. Scale bar is 500 nm.
Several experimental factors were found to affect the efficiency of DNA nanotube attachment to the patterned gold islands: 1) the incubation time, 2) the number of thiol groups at each end of the DNA nanotube, 3) the diameter of the gold islands, and 4) the DNA nanotube concentration used in the incubation step.
First, increasing the incubation time improved the number of correct tube attachments to the gold islands. When the incubation time was increased from 20 to 40 hours while keeping the other conditions the same, the average number of tube interconnections (calculated by dividing the total number of DNA tubes connected between a pair of gold islands by the number of gold pairs in the same image area) increased from 1.33 (276/208) to 1.5 (226/152). However, increasing the incubation time further (up to 70 hours) revealed an increase in nonspecific binding of the DNA origami tubes to the substrate background. The measured surface water contact angle decreased from ≥ 40° to ≤ 20° after 70 hours, indicating partial removal of the HMDS layer during the extended incubation, increasing the surface hydrophilicity, and thus decreasing the specificity of the DNA nanotube attachment to the gold islands.
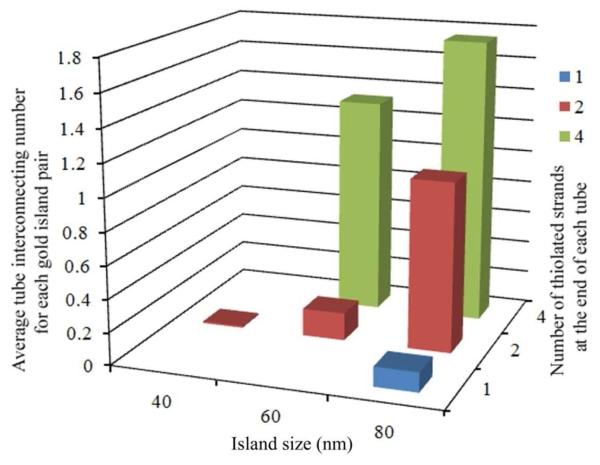
Second, the number of thiol-Au bonds per linkage had a significant effect on the efficiency of attachment. As the number of thiolated groups at each end of origami tube was reduced from 4 to 2, the average number of interconnected islands decreased from 1.33 to 0.17 (19/110), with all other conditions kept the same. Using a single thiol group per binding site resulted in almost no gold island connections, except for the largest island size (80 nm) (Fig 3). This indicates that at least two thiol-Au associations per site are required to achieve efficient binding for this type of DNA origami tube nanostructure. A single DNA origami structure contains ~ 7200 base pairs with a molecular weight of more than 4 Mega Dalton. As the size of a tethered particle increases, the inertia increases and the force required to overcome the random Brownian motion and hold the particle in place also increases. Apparently, a single thiol-Au linkage is not strong enough to hold the whole structure in place.
Figure 3.
Graph showing how the binding efficiency depends on both the number of thiol groups at each end of the DNA tube and the diameter of the gold islands. The average interconnection number was calculated by dividing the total number of observed connections by the total number of gold pairs in the image area. The incubation time for all the samples represented in the graph was 20 hours; DNA tube concentration was 2 nM. Note the flat square corresponding to the 40 nm island indicates zero because we observed no DNA nanotube binding in this case.
The size of the gold islands also plays an important role in the DNA tube attachment efficiency. For a given gold island pattern, when the island size was increased from 60 to 80 nm (for a 2 nM origami tube sample with 2 thiolated DNA strands at each end), the average number of tubes connecting each pair increased from 0.17 to 1.04 (157/151). When origami tubes with 4 thiolated DNA strands at each end were used, the average number of tubes connecting 80 nm gold island pairs was 1.75 (312/178), compared to 1.33 for 60 nm islands (Fig. 3) (see additional AFM images in SI). In contrast, when the island size was reduced to 40 nm, no connected gold island pairs were observed even with 4 thiols per site. It is possible that as the size of the islands decrease, the probability that thiol groups at one end of a tube can attach to the same gold island is significantly reduced, thus the rate of the specific surface adsorption decreases. It was also observed that for substrates with smaller gold islands, many gold islands were missing, maybe due to weaker adhesion of the gold islands to the substrate.
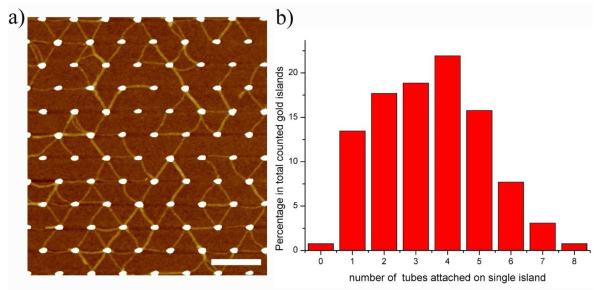
In addition to patterns with gold island pairs, we also fabricated a hexagonal lattice pattern in which each 60 nm diameter gold island is surrounded by 6 islands with equal, 300 nm center-to-center distances. After 20 hours of incubation with 2 nM DNA origami tubes containing 4 thiol groups at each end, interconnection between neighboring gold islands was observed (shown in Fig. 4a). The most common patterns that were observed are triangle or rhombus structures made by origami tubes at the sides and gold islands at the vertices. Fig. 4b contains a histogram showing the distribution of the number of origami tubes that attached to the hexagonal gold island pattern. Statistically, the number of DNA tubes attached to a single gold island exhibited a broad distribution ranging from 0 to 8, with an average of 3.5 (899/259). In order for the hexagonal lattice pattern to be fully interconnected, at least 6 tubes should be attached to each island, forming connections with all the neighboring islands. However, only 11.5% of islands have ≥ 6 tubes attached, with full interconnection not achieved for this condition. To improve the efficiency of attachment we increased the concentration of purified DNA origami tubes 5 fold to 10 nM, concentrating several 2 nM samples using 100 kD Microcon centrifugal filter devices. The average number of tubes attached to a single gold island increased to 4.61 (1730/375), indicating that a higher DNA origami tube concentration can indeed enhance the binding efficiency (see additional AFM images are shown in SI). However, 100% interconnection between all the gold islands was not yet achieved.
Figure 4.
a) AFM image of a hexagonal gold island lattice with by DNA origami tube connections. Scale bar is 500 nm. b) Corresponding histogram of the number of DNA nanotubes attached to each gold island, with a wide distribution from 0 to 8, with 4 tubes the most frequent number observed (22%). Only a small fraction of the gold islands (~11.5%) have 6 or more tubes attached.
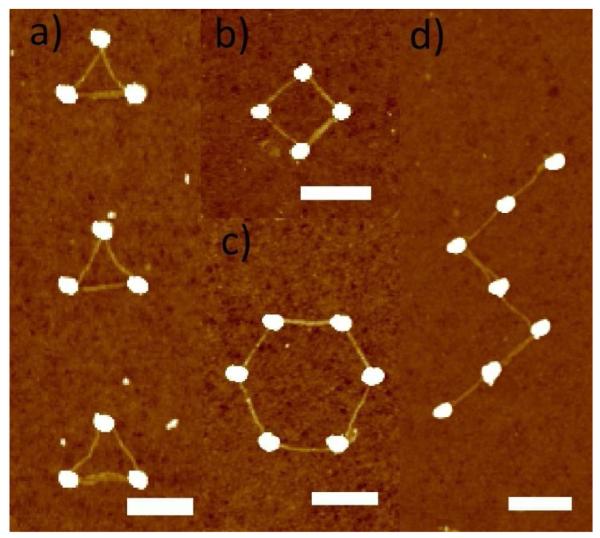
Using the optimized incubation conditions and modified gold island patterns, we further constructed more complex DNA tube patterns as demonstrated in Fig. 5. Clusters of 80 nm gold islands were designed to form the corners of triangle, square, hexagon or opened “z” pattern, with inter-island distances ~ 300 nm within a single group, but > 600 nm between different groups. After 60 hour incubation with 1 nM DNA origami tubes containing 4 thiols at each end, triangle, square, hexagon and “z” shaped inter-connected gold island structures were observed. This result shows that the orientation and position of DNA origami tubes on a substrate can be precisely controlled by gold patterns fabricated using top-down methods.
Figure 5.
Various structures formed by connecting gold islands with DNA origami tubes. a) triangle, b) hexagon, c) square, d) “z” shape. All scale bars are 300 nm.
In summary, we have described a new strategy to connect lithographically patterned gold islands to DNA origami tubes using covalent bond associations. We have found that incubation time, number of thiolated strands, island size and concentration of DNA origami in solution are the major factors that affect the binding efficiency. This interconnection strategy was used to construct complex networks of DNA origami nanotubes, with the orientation of origami tubes precisely controlled by the distance and pattern of the gold islands. These DNA origami nanotubes may be further modified to carry metal nanoparticles or quantum dots, exhibiting enhanced functionality at desired locations along the tubes. In the future, lateral, nucleated reduction could facilitate the formation of fixed length metallic nanowires between gold islands at desired locations on substrate. In addition, other types of chemical bonding strategies for silicon27-28or oxide surfaces29 could be used to improve the selectivity and complexity of the method. The merger of DNA directed self-assembly with top-down approaches may lead to novel methods to fabricate electronic devices. This demonstration of guided DNA self-assembly highlights the possibility of creating fully addressable and massively parallel nanopatterns in the sub-10 nm regime.
Supplementary Material
Acknowledgements
We thank funding supports from ONR, NSF, NIH, ARO, DOE and Sloan Research Fellowship to H.Yan and supports from NSF to H. Yu; H.Yan and Y. Liu are supported as part of the Center for Bio-Inspired Solar Fuel Production, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC0001016. We thank Jeanette Nangreave for help proofreading the manuscript.
Footnotes
Supporting Information Available: DNA sequences, experimental methods, additional atomic force microscopy images. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Seeman NC. Annu. Rev. Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin C, Liu Y, Yan H. Biochemistry. 2009;48:1663–1674. doi: 10.1021/bi802324w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothemund PWK. Nature. 2006;440(7082):297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 5.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 6.Ke Y, Sharma J, Liu M, Jahn K, Liu Y, Yan H. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 7.Douglas S, Dietz H, Liedl T, Högberg B, Graf F, Shih W. Nature. 2009;459(7245):414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzuya A, Komiyama M. Chem. Commun. 2009:4182–4184. doi: 10.1039/b907800b. [DOI] [PubMed] [Google Scholar]
- 9.Dietz Hendrik, Douglas Shawn M., Shih William M. Science. 2009;325(5941):725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke Y, Douglas SM, Liu M, Sharma J, Cheng A, Leung A, Liu Y, Shih WM, Yan H. J. Am. Chem. Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liedl T, Högberg B, Tytell J, Ingber DE, Shih WM. Nature Nanotechnology. 2010;5:520–524. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Science. 2008;319(5860):180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 13.Maune HT, Han SP, Barish RD, Bockrath M, Goddard WA, Rothemund PWK, Winfree E. Nat. Nanotechnol. 2010;5:61–65. doi: 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- 14.Ding BQ, Deng ZT, Yan H, Cabrini S, Zuckermann RN, Bokor J. J. Am. Chem. Soc. 2010;132:3248–3249. doi: 10.1021/ja9101198. [DOI] [PubMed] [Google Scholar]
- 15.Stephanopoulos N, Liu M, Tong GJ, Li Z, Liu Y, Yan H, Francis MB. Nano Lett. 2010;10(7):2714–2720. doi: 10.1021/nl1018468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pal S, Deng Z, Ding B, Yan H, Liu Y. Angew. Chem. Int. Ed. 2010;49:2700–2704. doi: 10.1002/anie.201000330. [DOI] [PubMed] [Google Scholar]
- 17.Gu H, Chao J, Xiao SJ, Seeman NC. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund K, Manzo AJ, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, Taylor S, Pei R, Stojanovic MN, Walter NG, Winfree E, Yan H. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voigt NV, Tørring T, Rotaru A, Jacobsen MF, Ravnsbæk JB, Subramani R, Mamdouh W, Kjems J, Mokhir A, Besenbacher F, Gothelf KV. Nature Nanotechnology. 2010;5:200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 20.Gerdon AE, Oh SS, Hsieh K, Ke Y, Yan H, Soh HT. Small. 2009;5:1942–1946. doi: 10.1002/smll.200900442. [DOI] [PubMed] [Google Scholar]
- 21.Kershner RJ, Bozano LD, Micheel CM, Hung AM, Fornof AR, Cha JN, Rettner CT, Bersani M, Frommer J, Rothemund PWK, Wallraff GM. Nature Nanotechnology. 2009;4:557–561. doi: 10.1038/nnano.2009.220. [DOI] [PubMed] [Google Scholar]
- 22.Hung AM, Micheel CM, Bozano LD, Osterbur LW, Wallraff GM, Cha JN. Nat. Nanotechnol. 2010;5:121–126. doi: 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- 23.Ulman A. Chem. Rev. 1996;96:1533–1554. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 24.Yu HB, Luo Y, Beverly K, Stoddart JF, Tseng HR, Heath JR. Angew. Chem. Int. Ed. 2003;42:5706–5711. doi: 10.1002/anie.200352352. [DOI] [PubMed] [Google Scholar]
- 25.Stearns LA, Chhabra R, Sharma J, Liu Y, Petuskey WT, Yan H, Chaput JC. Angew. Chem. Int. Ed. 2009;45:8494–8496. doi: 10.1002/anie.200903319. [DOI] [PubMed] [Google Scholar]
- 26.Douglas SM, Chou JJ, Shih WM. Proc. Natl. Acad. Sci. USA. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu HB, Webb LJ, Ries RS, Solares SD, Goddard WA, Heath JR, Lewis NS. J. Phys. Chem. B. 2005;109:671–674. doi: 10.1021/jp047672m. [DOI] [PubMed] [Google Scholar]
- 28.Yu HB, Webb LJ, Solares SD, Cao PG, Goddard WA, Heath JR, Lewis NS. J. Phys. Chem. B. 2006;110:23898. doi: 10.1021/jp063655g. [DOI] [PubMed] [Google Scholar]
- 29.Flink S, van Veggel FCJM, Reinhoudt DN. Adv. Mater. 2000;12:1315–1328. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.