Abstract
Mai Po Nature Reserve is the largest mangrove ecosystem and the most polluted coastal water body in Hong Kong. Plasmids screening of 100 Vibrio isolates randomly showed 45 % of them contained 1–3 plasmids. These plasmid(s)-bearing isolates could be divided into 12 groups based on their plasmid profiles. Phylogenetic analysis of the partial 16S rRNA gene sequences confirmed that all plasmid(s)-bearing isolates belonged to Vibrio cholerae. Full DNA sequences of the plasmids in Groups I (pVCG1.1 and pVCG1.2), II (pVCG2.1), III (pVCG3.2) and IV (pVCG4.1) have been determined and the results showed that pVCG1.1, pVCG2.1 and pVCG3.2 were almost identical. Plasmids pVCG1.1, pVCG1.2 and pVCG4.1 are comprised of 4,439, 2,357 and 2,163 bp with the overall G+C content of 45.57, 53.54 and 43.09 %, respectively. pVCG1.1 is a novel plasmid, and plasmids pVCG1.2 and pVCG4.1 showed homology of replication initiation proteins to that of the theta type replicons. Attempts to cure the plasmids from their hosts were unsuccessful. These data suggest that plasmids of Vibrio spp. are a significant gene reservoir in the marine ecosystem.
Keywords: Vibrio cholerae, Plasmid, Diversity, Distribution, Mai Po Nature Reserve, Replication
Introduction
Vibrio species are natural inhabitants of aquatic environments, though many of the studies are primarily carried out on clinical strains, the environmental isolates may serve as a reservoir for the wide spread of antibiotic resistance or virulence genes due to horizontal gene transfer (Faruque et al. 1998; Chiang and Mekalanos 1999; Hazen et al. 2010). Plasmids and other mobile genetic elements such as prophages, integrons, genomic islands and transposons play vital and essential roles in the gene transfer processes. The high incidence of plasmid in bacteria of marine sediment suggested that marine environment is an important source for discovery of novel plasmids (Sobecky 1999, 2002; Hazen et al. 2010), but attributing specific traits and functions to the environmental plasmids have been proven difficult.
Of 440 isolates of presumptive marine Vibrios collected from the Gulf of Mexico, 31 % of them harbor plasmids, the plasmid incidence was 1.5-fold higher in oil field than in the control site located 8 km from the impacted sampling sites (Hada and Sizemore 1981). Although attempts have been made to assign phenotypic traits such as hydrocarbon utilization and heavy metal resistance to the plasmid(s)-bearing isolates, no correlation could be established between plasmid content and the resistance determinants. Davidson and Oliver (1986) examined 42 clinical and environmental isolates of Vibrio vulnificus, 5 (12 %) harbored plasmids, with various sizes, attempt to demonstrate a correlation between the presence of plasmids and a variety of phenotypic traits was unsuccessful, only the correlation between the presence of a 6.5-megadalton plasmid and the resistance to vibriostatic agent O/129 was observed. In another study, a relationship between high molecular weight plasmids and eel virulence was suggested by Biosca et al. (1997), who found isolates bearing high molecular weight plasmids showed lower LD50 in eels than those that are plasmid-free. Hoi et al. (1998) observed that 93 of 97 strains isolated from five outbreaks of V. vulnificus in Danish eel farms harbored from 1 to 3 high molecular weight plasmids, supporting the hypothesis of a possible participation of high molecular weight plasmids in virulence of eels.
In Vibrio cholerae, the presence of the conjugative P plasmid, which is not widely disseminated among clinical isolates, significantly attenuates pathogenicity due to plasmid-induced loss of virulence (Sinha and Srivastava 1978). Cook et al. (1984) investigated plasmid profiles of many clinical isolates of classical biotype V. cholerae strains, and found that classical strains possess two plasmids (the smaller one was about 4.7 kb). Bartowsky and Manning (1988) reported three plasmids in a V. cholerae classical strain V58: the P plasmid, a large cryptic plasmid (lcp, 34 kb), and a small cryptic plasmid (scp, 4.7 kb). However, little is known about these cryptic plasmids. Rubin et al. (1998) sequenced a 4.7 kb toxin-linked plasmid, which is identical to the smaller plasmid in the above-mentioned two studies. This plasmid has been designated as pTLC (toxin-linked cryptic). pTLC can exist as both extra-chromosomal double-stranded circular plasmid and tandem duplicated DNA, inserted on the chromosome at a position only 842 bp upstream of the CTX prophage. This plasmid may play a crucial role in the acquisition and replication of CTX prophage.
Systematic studies on plasmid distribution and diversity in marine ecosystems, particularly at the molecular level, should provide new insights and understanding of plasmid function. Mai Po Nature Reserve is the largest mangrove ecosystem and most polluted water body in Hong Kong. This nature reserve plays a very important role in supporting a wide range of wildlife including migratory birds, e.g., the endangered Black-faced Spoonbills with a global population of about 2,000 (Tsim and Lock 2002), of which more than 20 % have been recorded here. This protected area is also an important refueling station for winter birds on their flyway from Arctic and northern China to Australia. The Nature Reserve is threatened by increasing pollutions largely due to the economic development in the adjacent Shenzhen Special Economic Zone of the People’s Republic of China (Cao et al. 2011a, b; Li et al. 2011a, b, c; Shen et al. 2012; Zhao et al. 2012a, b). We have isolated and characterized two novel plasmids from Vibrio species in Mai Po Nature Reserve previously (Zhang et al. 2007; Zhang and Gu 2009; Pan et al. 2010). In this study, a systematic investigation on plasmids diversity and distribution of Vibrio species community was conducted.
Materials and methods
Sampling sites, Vibrio strain isolating, plasmid screening and grouping
Water samples were collected from several previously determined sampling sites (W1, W2, W3, W4 and G12) at Mai Po Nature Reserve (22°29′N to 22°31′N and 113°59′E to 114°03′E) of Hong Kong SAR, PR China (Fig. 1). Surface water was taken to fill 1 L sterile plastic bottles when the tidal level was at approximately 1.5 m at this site. All samples were kept in coolers and transported immediately back to the laboratory after sampling for further processing.
Fig. 1.
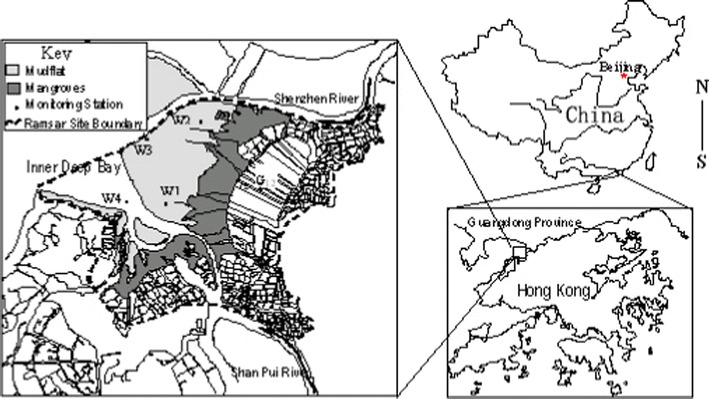
A map of the Mai Po Nature Reserve, Hong Kong showing the area with different habitats and sampling locations (W1–4, G12)
Samples for this study were collected in July 2005. At the time of sampling, in situ temperature, pH, dissolved oxygen, salinity, turbidity and suspended solids were 31.2–32.9 °C, 6.51–7.59, 6.82–13.09 mg/L, 2.2–4.6 ‰, 25.9–84.3 mg/L and 45–158 NTU, respectively. Chemical analysis of the water samples showed that NH4 +-N, NO3 −-N, NO2 −-N, total Kjeldahl N, ortho-P, total P and BOD5 was 3.28–5.52, 0.05–0.27, 0.18–0.33, 1.77–3.24, 0.45–0.67, 0.51–0.82, and 6.12–53.2 mg/L, respectively.
Water samples (0.1 mL) were directly spread onto TCBS agar plates and incubated at 30 °C for about 12 h. Colonies on agar plates were picked and streaked on new TCBS agar plates several times to obtain pure cultures of the isolates. One hundred purified isolates were randomly selected for screening of plasmids. Plasmid screening was carried out using alkaline lysis technique as described by Sambrook et al. (1989). Plasmids for further research were prepared with Qiaprep spin miniprep kit (Qiagen Inc, Valencia, California, USA) for plasmids less than 10 kb, or with alkaline lysis technique for plasmids greater than 10 kb. Each plasmid was separately purified through gel extraction, followed by column purification or ethanol precipitation. Plasmid(s)-bearing isolates were grouped based on their plasmid profiles in terms of the number of plasmid, plasmid size and their RFLP analyses using the enzymes of EcoRI and KpnI.
Bacterial strains, plasmids and molecular genetic techniques
All Vibrio isolates were obtained from the water samples of Mai Po Nature Reserve, Hong Kong. E. coli JM109 cells and pUC19 cloning vector were from laboratory storage. Restriction analysis, DNA ligation and transformation were performed according to standard protocols (Sambrook et al. 1989).
Plasmids sequencing and analyses
Plasmids pVCG1.1, pVCG1.2, pVCG2.1 and pVCG3.2 have a single EcoRI site and pVCG4.1 has a BamHI site, which were used to clone them into EcoRI or BamHI digested pUC19 vector, recombinant plasmids carrying the linear plasmids fragments were designated as pUVG1.1, pUVG1.2, pUVG2.1, pUVG3.2 and pUVG4.1, respectively. The inserted linear plasmids sequences of pUVG1.1, pUVG1.2, pUVG2.1, pUVG3.2 and pUVG4.1 were obtained using pUC19 vector specific primers, and then the internal primers derived from the sequence information through the previous sequencing cycle. Sequences of both strands were determined at Department of Zoology, The University of Hong Kong.
DNA sequence analysis were performed using Bioedit (Hall 1999). Database searches were carried out using BLASTn and BLASTp (Altschul et al. 1997), putative ORFs were determined using both the online ORF finder software on the NCBI website and GeneMark software (http://opal.biology.gatech.edu/GeneMark/).
Phylogenetic analyses
16S rRNA genes were amplified using the universal primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′; E. coli bases 8–27) and reverse primer 1492R (5′-TACCTTGTTACGACTT-3′; E. coli bases 1,507–1,492) (Wilson et al. 1990). Alignment of the 16S rRNA genes and Rep protein sequences were accomplished using Clustal X (Thompson et al. 1997). Phylogenetic analyses were performed using PHYLIP (version 3.6; distributed by J. Felsenstein, University of Washington, Seattle, USA). The trees were constructed by the neighbor-joining method. Bootstrap analyses were performed with 100 re-sampled data sets.
Plasmid curing
Elevated incubation temperature (up to 42 °C), ethidium bromide (500 μg/mL), sodium dodecylsulfate (SDS, 10 %) and acridine orange (500 μg/mL) were employed to eliminate the plasmids from their hosts.
Comparisons of antibiotics and heavy metal ions resistance
Disc diffusion susceptibility testing method was used to determine antibiotics resistance profiles of the plasmid(s)-bearing isolates as described previously (Zhang et al. 2006). The 21 antibiotics (Ampicillin, Carbenicillin, Cephalothin, Chloramphenicol, Clindamycin, Colistin Sulphate, Cotrimoxazole, Erythromycin, Fusidic Acid, Gentamicin, Methicillin, Nalidixic Acid, Nitrofurantoin, Novobiocin, Penicillin G, Streptomycin, Sulphatriad, Sulphamethizole, Sulphamethoxzole, Tetracycline and Trimethoprim) were used in tests. Effects of heavy metal ions on the growth of plasmid(s)-bearing isolates were investigated using the method described previously (Zhang et al. 2006), the selected metals included Cd2+, Zn2+, Cu2+, Mn2+, Pb2+ (test concentrations were 0, 10, 25 and 50 μM in the growth medium), Hg2+ (test concentrations were 0, 10 and 25 μM) and Cr6+ (test concentrations were 0, 50, 100 and 150 μM).
Nucleotide sequences accession numbers
The 16S rRNA gene sequences of the 45 plasmid(s)-bearing V. cholerae isolates were deposited in the GenBank under accession numbers from DQ440932 to DQ440976, The complete nucleotide sequences of pVCG1.1, pVCG1.2 and pVCG4.1 were deposited in GenBank database under accession No. DQ787203, DQ787204 and DQ787205, respectively.
Results
Plasmids screening and grouping
The 45 plasmid(s)-bearing isolates harbored 1–3 plasmids. To determine their plasmid profiles, every single plasmid in the 45 plasmid(s)-bearing isolates was separately purified and their RFLP patterns were obtained using the enzymes of EcoRI and KpnI (data not shown). Based on their plasmid profiles, these plasmid(s)-bearing isolates were divided into 12 groups, each group displayed its own unique plasmid profile (Fig. 2). Every plasmid(s)-bearing isolate was designated with the corresponding group number followed by the isolate number in this group, for example, isolate G1.1 is the first isolate in group I. Each plasmid was also named with the group number and the plasmid number (from large to small), for example, plasmid pVCG1.1 is the largest plasmid in group I and plasmid pVCG1.2 is the second largest plasmid in group I. Group I was the most abundant with 22 isolates, almost half of the 45 plasmid(s)-bearing isolates, this group harbored two plasmids with the size of about 2.4 and 4.4 kb (according to the supercoiled DNA markers). Groups II and IV had one plasmid with size of 4.4 and 2.2 kb, respectively, each group had four isolates. Groups V, VI and VII contained three isolates each. Other groups had one isolate only. 16S rRNA gene alignments showed all the plasmid(s)-bearing isolates belong to Vibrio cholerae (data not shown), a natural inhabitants of aquatic environments.
Fig. 2.
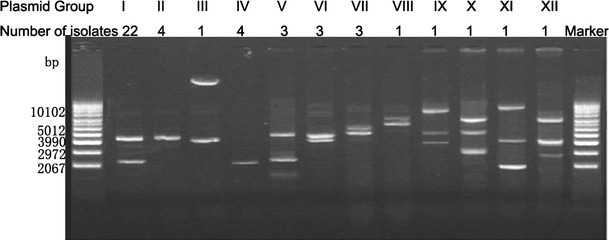
Plasmid profiles of the twelve groups of Vibrio isolates from Mai Po Nature Reserve, Hong Kong. Lanes 1 and 14, Supercoiled DNA ladder marker; plasmid profiles of Group I to Group XII and numbers of isolate in each group are indicated
Sequence analyses
Complete sequences of the plasmids in groups I to IV (pVCG1.1, pVCG1.2, pVCG2.1, pVCG3.2 and pVCG4.1 but not the large plasmid pVCG3.1) were obtained, results showed pVCG1.1 and pVCG2.1 are identical, in pVCG3.2, only a 222 bp fragment (Fig. 3, position 94–316 in pVCG1.1) is lacked compared with its counterpart in groups I and II. Detailed sequence analysis revealed that there are two direct repeats (5′-CCCCTTAAC-3′, position 94–102 and 316–324) flanked this region, so one explanation is the 222 bp fragment was deleted through homologous recombination. However, this cannot exclude other mechanisms. The plasmid physical maps of pVCG1.1, pVCG1.2 and pVCG4.1 are shown in Fig. 3.
Fig. 3.
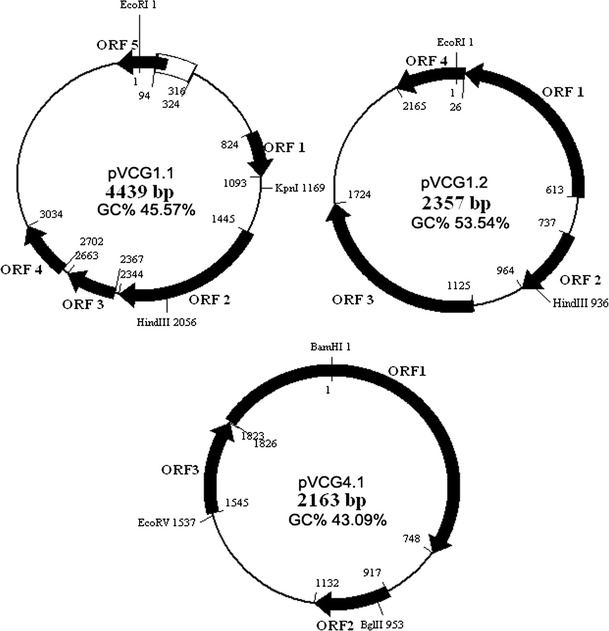
Physical maps of plasmid pVCG1.1, pVCG1.2 and pVCG4.1. Gene position and direction of transcription are indicated by arrows. The fragment (position 94–315) of plasmid pVCG1.1, which lacked in plasmid pVCG3.2 is indicated by open box
The circular plasmids pVCG1.1, pVCG1.2 and pVCG4.1 comprised of 4,439, 2,357 and 2,163 bp with the overall G+C content of 45.57, 53.54 and 43.09 %, respectively. The G+C contents of pVCG1.1 and pVCG4.1 are similar to, but that of pVCG1.2 is much higher than that of the two chromosomes (46.9 and 47.7 %, respectively) of V. cholerae (Heidelberg et al. 2000).
Plasmid pVCG1.1 has five ORFs, encoded the putative proteins of 89, 299, 98, 110 and 97 amino acids, respectively (Fig. 3), but they show no homologies to any known proteins. Most well studied plasmids encoded a replication initiation protein (Rep) which usually determine its replication type (Novick 1987). None of the potential proteins encoded by plasmid pVCG1.1 show any similarity to the known Rep proteins, suggesting that its replication mechanism may be different from that of the known plasmids.
Plasmid pVCG1.2 has four ORFs, encoded the putative proteins of 198, 75, 199 and 72 amino acids, respectively (Fig. 3). Blast searching revealed the predicted protein of ORF1 shows 39 % identity and 59 % similarity over 152 amino acids to the replication initiation protein (Proteins: AAG23805) of Pseudomonas fluorescens plasmid pAM10.6 (Peters et al. 2001). Proteins encoded by other ORFs showed no homology to any known proteins, and their functions are unknown.
Plasmid pVCG4.1 has three ORFs, encoding the putative proteins of 361, 71 and 92 amino acids, respectively (Fig. 3). The predicted protein of ORF1 has 37 % identity and 53 % similarity over 286 amino acids to the replication initiation protein CRI (Proteins: CAA32521) of Enterobacteria phage I2-2 (Stassen et al. 1992). Proteins encoded by the other two ORFs showed no homology to any known proteins.
Since both pVCG1.2 and pVCG4.1 encoded a putative protein similar to the known Rep proteins of theta type replicons. They may belong to the theta type plasmids. The phylogenetic tree of their Rep proteins and those of other theta type replicons is shown in Fig. 4. Plasmids pVCG1.1 and pVCG1.2 co-exist in the same cell, according to incompatibility theory (Novick 1987), they should belong to different incompatibility groups, which in most cases are classified based on replication initiation proteins.
Fig. 4.
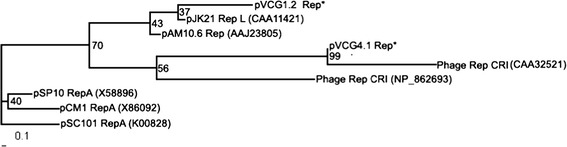
Phylogenetic relationship of plasmid replication initiation proteins of plasmids pVCG1.2 and pVCG4.1 and other referenced protein sequences. The tree was constructed by the neighbor-joining method with Rep protein sequence of plasmid pSC101 as root. Asterisks indicate the proteins in this study, and others are references
So far, including the three plasmids reported here, there are totally five completely sequenced V. cholerae plasmids in NCBI database of plasmid project under V. cholerae genome project (http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=30307). Previously we also identified and sequenced a V. cholerae plasmid, pVC, from marine environment (Zhang et al. 2007). Their main properties were compared in Table 1. Among these plasmids, only pTLC was isolated from a clinical strain, all others were from the marine environmental isolates. Plasmid pVCG1.2 has a much higher GC content, all other sequenced plasmids have a similar GC content to that of the V. cholerae chromosomes.
Table 1.
Vibrio cholerae plasmids comparison
| Plasmids | Accession no. | Size (bp) | GC content (%) | ORFs | Clinical/environmental |
|---|---|---|---|---|---|
| pTLC | AF052650 | 4,719 | 46.6 | 5 | Clinical |
| pSIO1 | AY876057 | 4,906 | 48.0 | 3 | Environmental |
| pVC | AY423429 | 3,806 | 43.3 | 4 | Environmental |
| pVCG1.1 | DQ787203 | 4,439 | 45.6 | 5 | Environmental |
| pVCG1.2 | DQ787204 | 2,357 | 53.5 | 4 | Environmental |
| pVCG4.1 | DQ787205 | 2,163 | 43.1 | 3 | Environmental |
Plasmids curing
To investigate their functions, different approaches (as described in “Materials and methods”) were used in an attempt to cure the plasmid(s) from their hosts in the 12 groups, but none succeeded.
Comparisons of antibiotics and heavy metal ions resistance
Since isolates G1.1, G2.1 and G3.1 share a same plasmid, G3.1 also has another large plasmid pVCG3.1, their antibiotics and heavy metal ions resistance profiles were compared. Results showed isolates G1.1, G2.1 and G3.1 have the same performance to the 21 kinds of tested antibiotics (data not shown), within the tested concentrations, Cd2+, Zn2+, Cu2+, Mn2+, Pb2+ and Cr6+ have the same effect on the growth of isolates G1.1, G2.1 and G3.1 (data not shown), but for Hg2+, only isolate G3.1 showed tolerance to 10 μM Hg2+ (Table 2).
Table 2.
Effects of Hg2+ concentrations on maximum biomass yield of isolates G1.1, G2.1 and G3.1
| Isolates | Hg2+ concentrations (μM) | ||
|---|---|---|---|
| 0 | 10 | 15 | |
| G1.1 | 0.767 ± 0.010 | 0.007 ± 0.001 | 0.007 ± 0.001 |
| G2.1 | 0.631 ± 0.009 | 0.009 ± 0.002 | 0.011 ± 0.002 |
| G3.1 | 0.692 ± 0.024 | 0.297 ± 0.136 | 0.011 ± 0.001 |
SD standard deviation of triplicate
Discussion
In this study, we investigated the plasmids incidence and diversity in Vibrio community of Mai Po Nature Reserve, the incidence of plasmid was relatively high in the environmental Vibrio isolates, plasmid(s)-bearing isolates accounted for 45 % of the Vibrio isolates, and most (82.2 %) of them contained multiple plasmids, isolates containing 2 and 3 plasmids accounted for 66.7 and 15.6 %, respectively. The most abundant plasmid pVCG1.1 showed no homology to the known Rep proteins, and probably represents a novel plasmid, further studies will determine the rep gene and replication mechanism of this plasmid. This phenomenon is in agreement with several other reports (Dahlberg et al. 1997; Sobecky et al. 1997, 1998; Bidinost et al. 1999; Pan et al. 2010), which have showed that plasmids from marine bacteria have no detectable homology with plasmids from clinical isolates, indicating that bacterial isolates obtained from marine environment are a source of novel plasmids.
All attempts to cure the plasmids from their hosts were failed, probably due to the relatively high copy number of the plasmids similar to earlier work (Zhang et al. 2006; 2007). So far, the majority of bacteria adapted to survive and proliferate in the presence of mercury have been shown to reduce Hg2+ to its volatile form Hg° via the well-characterized mer operons (Rochelle et al. 1991; Silver and Walderhaug 1992; Jeffrey et al. 1996; Liebert et al. 1997). Mercury resistant genes were reported to be located on large plasmids (Summers and Silver 1972; Rani and Mahadevan 1994; Silver 1996). In addition, other mercury resistance mechanism such as efflux system in marine bacteria was reported to be plasmid-encoded (Reyes et al. 1999). Isolate G3.1 has a relatively large plasmid pVCG3.1, a relationship of tolerance to 10 μM Hg2+ of G3.1 and presence of plasmid pVCG3.1 may exist but need further direct evidence. Amplification of the possible mer operons in plasmid pVCG3.1 and G3.1 genomic DNA using the primers based on the reported mer operons did not confirm the results (data not shown).
In conclusion, V. cholerae community of Mai Po Nature Reserve, Hong Kong showed high incidence and diversity of plasmids.
Acknowledgments
We would like to thank Jessie Lai for water sample and laboratory support and AFCD of Hong Kong SAR Government for partial financial support and field sampling logistics. Any opinions, findings, conclusions or recommendations expressed in this publication do not reflect the view of the Hong Kong Special Administrative Region Government.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Contributor Information
Ruifu Zhang, Email: rfzhang@njau.edu.cn.
Ji-Dong Gu, Email: jdgu@hkucc.hku.hk.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartowsky EJ, Manning PA. Molecular cloning of the plasmids of Vibrio cholerae O1 and the incidence of related plasmids in clinical isolates and other Vibrio species. FEMS Microbiol Lett. 1988;50:183–190. doi: 10.1111/j.1574-6968.1988.tb02935.x. [DOI] [Google Scholar]
- Bidinost C, Wilderman PJ, Dorsey CW, Actis LA. Analysis of the replication elements of the pMJ101 plasmid from the fish pathogen Vibrio ordalii. Plasmid. 1999;42:20–30. doi: 10.1006/plas.1999.1406. [DOI] [PubMed] [Google Scholar]
- Biosca EG, Amaro C, Larsen JL, Pedersen K. Phenotypic and genotypic characterization of Vibrio vulnificus: proposal for the substitution of the subspecific taxon biotype for serovar. Appl Environ Microbiol. 1997;63:1460–1466. doi: 10.1128/aem.63.4.1460-1466.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Li M, Dang H, Gu J-D. Responses of aerobic and anaerobic ammonia/ammonium-oxidizing microorganisms to anthropogenic pollution in coastal marine environments. Methods Enzymol. 2011;496:35–62. doi: 10.1016/B978-0-12-386489-5.00002-6. [DOI] [PubMed] [Google Scholar]
- Cao H, Li M, Hong Y-G, Gu J-D. Diversity and abundance of ammonia-oxidizing archaea and bacteria in polluted mangrove sediment. Syst Appl Microbiol. 2011;34:513–523. doi: 10.1016/j.syapm.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Chiang SL, Mekalanos JJ. Horizontal gene transfer in the emergence of virulent Vibrio cholerae. In: Rosenberg E, editor. Microbial ecology and infectious disease. Washington DC: ASM Press; 1999. pp. 156–169. [Google Scholar]
- Cook WL, Wachsmuth K, Johnson SR, Birkness KA, Samadi AR. Persistence of plasmids, cholera toxin genes, and prophage DNA in classical Vibrio cholerae O1. Infect Immun. 1984;45:222–226. doi: 10.1128/iai.45.1.222-226.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg C, Linberg C, Torsvik VL, Hermansson M. Conjugative plasmids isolated from bacteria in marine environments show various degrees of homology to each other and are not closely related to well-characterized plasmids. Appl Environ Microbiol. 1997;63:4692–4697. doi: 10.1128/aem.63.12.4692-4697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LS, Oliver JD. Plasmid carriage in Vibrio vulnificus and other lactose-fermenting marine vibrios. Appl Environ Microbiol. 1986;52:211–213. doi: 10.1128/aem.52.1.211-213.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hada HS, Sizemore RK. Incidence of plasmids in Marine Vibrio spp. Isolated from an oil field in the Northwestern Gulf of Mexico. Appl Environ Microbiol. 1981;41:199–202. doi: 10.1128/aem.41.1.199-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hazen TH, Pan L, Gu J-D, Sobecky PA. The contribution of mobile genetic elements to the evolution and ecology of Vibrios. FEMS Microbiol Ecol. 2010;74:485–499. doi: 10.1111/j.1574-6941.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi L, Dalsgaard I, DePaola A, Siebeling RJ, Dalsgaard A. Heterogeneity among isolates of Vibrio vulnificus recovered from eels (Anguilla anguilla) in Denmark. Appl Environ Microbiol. 1998;64:4676–4682. doi: 10.1128/aem.64.12.4676-4682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey WH, Nazaret S, Barkay T. Detection of the merA gene and its expression in the environment. Microb Ecol. 1996;32:293–303. doi: 10.1007/BF00183064. [DOI] [PubMed] [Google Scholar]
- Li M, Cao H-L, Hong Y-G, Gu J-D. Spatial distribution and abundance of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol. 2011;89:1243–1254. doi: 10.1007/s00253-010-2929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Cao H-L, Hong Y-G, Gu J-D. Seasonal dynamics of anammox bacteria in estuarial sediments of Mai Po Nature Reserve revealed by analyzing 16S rRNA and hydrazine oxidoreductase (HZO) genes. Microbes Environ. 2011;26:15–22. doi: 10.1264/jsme2.ME10131. [DOI] [PubMed] [Google Scholar]
- Li M, Hong Y-G, Cao H-L, Gu J-D. Mangrove trees affect the community structure and distribution of anammox bacteria at an anthropogenic-polluted mangrove in the Pearl River Delta reflected by 16S rRNA and hydrazine oxidoreductase (HZO) encoding gene analyses. Ecotoxicology. 2011;20:1780–1790. doi: 10.1007/s10646-011-0711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebert CA, Wireman J, Smith T, Summers AO. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl Environ Microbiol. 1997;63:1066–1076. doi: 10.1128/aem.63.3.1066-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Leung PC, Gu J-D. A new ColE1-like plasmid group revealed by comparative analysis of the replication proficient fragments of Vibrionaceae plasmids. J Microbiol Biotechnol. 2010;20:1163–1178. doi: 10.4014/jmb.1003.03007. [DOI] [PubMed] [Google Scholar]
- Peters M, Jogi E, Suitso I, Punnisk T, Nurk A. Features of the replicon of plasmid pAM10.6 of Pseudomonas fluorescens. Plasmid. 2001;46:25–36. doi: 10.1006/plas.2001.1524. [DOI] [PubMed] [Google Scholar]
- Rani DB, Mahadevan A. Cloning and expression of the mercury resistance genes of marine Pseudomonas sp. strain MR1 plasmid pMR1 in Escherichia coli. Res Microbiol. 1994;145:121–127. doi: 10.1016/0923-2508(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Reyes NS, Frischer ME, Sobecky PA. Characterization of mercury resistance mechanisms in marine sediment microbial communities. FEMS Microbiol Ecol. 1999;30:273–284. doi: 10.1111/j.1574-6941.1999.tb00655.x. [DOI] [PubMed] [Google Scholar]
- Rochelle PA, Wetherbee MK, Olson BH. Distribution of DNA sequences encoding narrow- and broad-spectrum mercury resistance. Appl Environ Microbiol. 1991;57:1581–1589. doi: 10.1128/aem.57.6.1581-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin EJ, Lin W, Mekalanos JJ, Waldor MK. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol Microbiol. 1998;28:1247–1254. doi: 10.1046/j.1365-2958.1998.00889.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shen P-P, Zhou H, Zhao Z, Yu X-Z, Gu J-D (2012) Evaluation of sampling sizes on the intertidal macroinfauna assessment in a subtropical mudflat of Hong Kong. Ecotoxicology. doi:10.1007/s10646-012-0944-x [DOI] [PubMed]
- Silver S. Bacterial resistances to toxic metal ions—a review. Gene. 1996;179:9–19. doi: 10.1016/S0378-1119(96)00323-X. [DOI] [PubMed] [Google Scholar]
- Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha VB, Srivastava BS. Plasmid-induced loss of virulence in Vibrio cholerae. Nature. 1978;276:708–709. doi: 10.1038/276708a0. [DOI] [PubMed] [Google Scholar]
- Sobecky PA. Plasmid ecology of marine sediment microbial communities. Hydrobiologia. 1999;401:9–18. doi: 10.1023/A:1003726024628. [DOI] [Google Scholar]
- Sobecky PA. Approaches to investigating the ecology of plasmids in marine bacterial communities. Plasmid. 2002;48:213–221. doi: 10.1016/S0147-619X(02)00110-5. [DOI] [PubMed] [Google Scholar]
- Sobecky PA, Mincer TJ, Chang MC, Helinski DR. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol. 1997;63:888–895. doi: 10.1128/aem.63.3.888-895.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobecky PA, Mincer TJ, Chang MC, Toukdarian A, Helinski DR. Isolation of broad-host-range replicons from marine sediment bacteria. Appl Environ Microbiol. 1998;64:2822–2830. doi: 10.1128/aem.64.8.2822-2830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen AP, Schoenmakers EF, Yu M, Schoenmakers JG, Konings RN. Nucleotide sequence of the genome of the filamentous bacteriophage I2–2: module evolution of the filamentous phage genome. J Mol Evol. 1992;34:141–152. doi: 10.1007/BF00182391. [DOI] [PubMed] [Google Scholar]
- Summers AO, Silver S. Mercury resistance in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1972;112:1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsim ST, Lock FNY. Knowing Ramsar Wetland. Hong Kong: Cosmos Book Ltd; 2002. [Google Scholar]
- Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Gu J-D. Complete sequence of plasmid pMP1 from the marine environmental Vibrio vulnificus and location of its replication origin. Mar Biotechnol. 2009;11:456–462. doi: 10.1007/s10126-008-9160-3. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang Y, Gu J-D. Identification of environmental plasmid-bearing Vibrio species isolated from polluted and pristine marine reserves of Hong Kong, and resistance to antibiotics and mercury. Antonie van Leeuwenhoek. 2006;89:307–315. doi: 10.1007/s10482-005-9032-z. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang Y, Leung PC, Gu J-D. pVC, a small cryptic plasmid from the environmental isolate of Vibrio cholerae MP-1. J Microbiol. 2007;45:193–198. [PubMed] [Google Scholar]
- Zhao Z-Y, Chu Y-L, Gu J-D (2012) Distribution and sources of polycyclic aromatic hydrocarbons in sediments of the Mai Po Inner Deep Bay Ramsar Site in Hong Kong. Ecotoxicology. doi:10.1007/s10646-012-0948-6 [DOI] [PubMed]
- Zhao Z, Zhuang Y-X, Gu J-D (2012) Abundance, composition and vertical distribution of polycyclic aromatic hydrocarbons in sediments of the Mai Po Inner Deep Bay of Hong Kong. Ecotoxicology. doi:10.1007/s10646-012-0951-y [DOI] [PubMed]


