Abstract
Eucalyptus trees, mostly native to Australia, are widely planted in the tropics and Southern Hemisphere for the production of wood and pulp. Worldwide surveys of diseases on these trees have yielded a large collection of Ceratocystis isolates from dying trees or from wounds on their stems. The aim of this study was to characterise these isolates and to consider their relatedness to each other. Culture appearance, morphological features and a distinctive fruity odour in all cultures were typical of species in the Ceratocystis fimbriata sensu lato (s. lat.) complex. Phylogenetic analyses of sequences for the combined ITS, βt-1 and TEF1-α gene regions revealed a genetically diverse group of isolates residing in a single large clade, that were distinct from all other species in the C. fimbriata s. lat. complex. Based on morphology and phylogenetic inference, the Eucalyptus isolates are recognised as closely related. The South African isolates are described here as a new species, C. eucalypticola.
Keywords: canker stain diseases, Microascales, tree pathogens, wounds
INTRODUCTION
Eucalyptus species are mostly native to Australia, but have been widely planted in the tropics and Southern Hemisphere. This is because they are adapted to a wide range of different environments and are typically fast growing. It has further been suggested that the success of these trees as non–natives is due to the separation from their natural enemies (Wingfield et al. 2008, Roux & Wingfield 2009). The potential threat of pests and pathogens to the sustainability of eucalypt plantations in areas where they are not native is consequently great and of substantial concern to forestry industries globally (Old et al. 2003, Wingfield et al. 2008).
In order to understand and manage the threat of pests and pathogens to Eucalyptus species grown as non-natives and in plantations, tree health surveys are undertaken regularly. Amongst the pathogens that have been found on these trees, a Ceratocystis sp. in the C. fimbriata s. lat. complex causes serious disease problems in Brazil, the Republic of Congo, Uganda, and Uruguay (Laia et al. 1999, Roux et al. 2000, 2001, 2004, Barnes et al. 2003a). Various other Ceratocystis species in the C. fimbriata s. lat. complex have also been found on naturally occurring or artificially induced wounds on the stems of trees, in various parts of the world. Some of these have been shown to be cryptic taxa that have been provided with names (van Wyk et al. 2007, 2008, 2010a, Rodas et al. 2007, Heath et al. 2009, Kamgan Nkuekam et al. 2012). Several species are thought to be pathogens, while the role of others in tree health is not known.
The genus Ceratocystis comprises a diverse group of fungi, including saprophytes causing blue-stain of lumber and serious pathogens that cause mortality (Kile 1993). The genus is typified by C. fimbriata s. str. that is a pathogen restricted to root crops, specifically sweet potato (Engelbrecht & Harrington 2005). Ceratocystis fimbriata s. lat. represents a diverse assemblage of isolates, some of which have been treated as distinct taxa defined based on phylogenetic inference, morphological differences, and mating behaviour (Barnes et al. 2001, Engelbrecht & Harrington 2005, Johnson et al. 2005, van Wyk et al. 2007, 2008, Heath et al. 2009). However, Ferreira et al. (2010) treated some isolates of the C. fimbriata s. lat. complex from Brazil as representing a particular population of C. fimbriata s. str., rather than as discrete taxa.
Global surveys of the health of Eucalyptus species in plantations have yielded a large collection of isolates that can loosely be accommodated in the C. fimbriata s. lat. complex. The aim of this study was to characterise these isolates and to consider patterns in their distribution on Eucalyptus species worldwide.
MATERIALS AND METHODS
Isolates
Isolates used in this study were obtained from: (1) artificially induced wounds on the stems of Eucalyptus trees in South Africa, Thailand, and Indonesia (Table 1). The isolates were obtained by directly transferring spore masses from the apices of ascomata produced on the wounded inner bark and wood to agar plates. When sporulating structures were absent, the wood samples were placed in moist chambers to enhance sporulation. Spore masses were transferred to 2 % Malt Extract Agar (MEA) in Petri dishes and incubated at room temperature. Additionally, the carrot baiting technique was used to obtain isolates (Moller & DeVay 1968). (2) cultures were sourced from the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI) at the University of Pretoria, South Africa. These isolates had previously been identified as representing the C. fimbriata s. lat. complex and were from diseased Eucalyptus trees in various parts of the world including Brazil, Uganda, Congo, and Uruguay (Table 1).
Table 1. Isolates of Ceratocystis fimbriata s. lat. spp. used in this study.
| Species | Isolate no. | GenBank accession no. | Host | Area |
|---|---|---|---|---|
| C. albifundus | CMW4068 | DQ520638, EF070429, EF070400 | Acacia mearnsii | South Africa |
| C. albifundus | CMW5329 | AF388947, DQ371649, EF070401 | Acacia mearnsii | Uganda |
| C. atrox | CMW19383, CBS120517 | EF070414, EF070430, EF070402 | Eucalyptus grandis | Australia |
| C. atrox | CMW19385, CBS120518 | EF070415, EF070431, EF070403 | Eucalyptus grandis | Australia |
| C. cacaofunesta | CMW15051, CBS152.62 | DQ520636, EF070427, EF070398 | Theobroma cacao | Costa Rica |
| C. cacaofunesta | CMW14809, CBS115169 | DQ520637, EF070428, EF070399 | Theobroma cacao | Ecuador |
| C. caraye | CMW14793, CBS114716 | EF070424, EF070439, EF070412 | Carya cordiformis | USA |
| C. caraye | CMW14808, CBS115168 | EF070423, EF070440, EF070411 | Carya ovata | USA |
| C. colombiana | CMW9565, CBS121790 | AY233864, AY233870, EU241487 | Soil | Colombia |
| C. colombiana | CMW5751, CBS121792 | AY177233, AY177225, EU241493 | Coffea arabica | Colombia |
| C. colombiana | CMW9572 | AY233863, AY233871, EU241488 | Mandarin | Colombia |
| C. eucalypticola | CMW9998, CBS124017 | FJ236721, FJ236781, FJ236751 | Eucalyptus sp. | South Africa |
| C. eucalypticola | CMW10000, CBS124019 | FJ236722, FJ236782, FJ236752 | Eucalyptus sp. | South Africa |
| C. eucalypticola | CMW11536, CBS124016 | FJ236723, FJ236783, FJ236753 | Eucalyptus sp. | South Africa |
| C. eucalypticola | CMW12663 | FJ236724, FJ236784, FJ236754 | Eucalyptus sp. | South Africa |
| C. eucalypticola | CMW15054, CBS124018 | FJ236725, FJ236785, FJ236755 | Eucalyptus sp. | South Africa |
| C. fimbriata s. str. | CMW15049, CBS141.37 | DQ520629, EF070442, EF070394 | Ipomaea batatas | USA |
| C. fimbriata s. str. | CMW1547 | AF264904, EF070443, EF070395 | Ipomaea batatas | Papua New Guinea |
| C. fimbriatomima | CMW24174, CBS121786 | EF190963, EF190951, EF190957 | Eucalyptus sp. | Venezuela |
| C. fimbriatomima | CMW24176, CBS121787 | EF190964, EF190952, EF190958 | Eucalyptus sp. | Venezuela |
| C. larium | CMW25434, CBS122512 | EU881906, EU881894, EU881900 | Styrax benzoin | Indonesia |
| C. larium | CMW25435, CBS122606 | EU881907, EU881895, EU881901 | Styrax benzoin | Indonesia |
| C. manginecans | CMW13851, CBS121659 | AY953383, EF433308, EF433317 | Mangifera indica | Oman |
| C. manginecans | CMW13852, CBS121660 | AY953384, EF433309, EF433318 | Hypocryphalus mangifera | Oman |
| C. neglecta | CMW17808, CBS121789 | EF127990, EU881898, EU881904 | Eucalyptus sp. | Colombia |
| C. neglecta | CMW18194, CBS121017 | EF127991, EU881899, EU881905 | Eucalyptus sp. | Colombia |
| C. obpyriformis | CMW23807, CBS122608 | EU245004, EU244976, EU244936 | Acacia mearnsii | South Africa |
| C. obpyriformis | CMW23808, CBS122511 | EU245003, EU244975, EU244935 | Acacia mearnsii | South Africa |
| C. papillata | CMW8857 | AY233868, AY233878, EU241483 | Annona muricata | Colombia |
| C. papillata | CMW8856, CBS121793 | AY233867, AY233874, EU241484 | Citrus lemon | Colombia |
| C. papillata | CMW10844 | AY177238, AY177229, EU241481 | Coffea arabica | Colombia |
| C. pirilliformis | CMW6569 | AF427104, DQ371652, AY528982 | Eucalyptus nitens | Australia |
| C. pirilliformis | CMW6579, CBS118128 | AF427105, DQ371653, AY528983 | Eucalyptus nitens | Australia |
| C. platani | CMW14802, CBS115162 | DQ520630, EF070425, EF070396 | Platanus occidentalis | USA |
| C. platani | CMW23918 | EF070426, EF070397, EU426554 | Platanus sp. | Greece |
| C. polychroma | CMW11424, CBS115778 | AY528970, AY528966, AY528978 | Syzygium aromaticum | Indonesia |
| C. polychroma | CMW11436, CBS115777 | AY528971, AY528967, AY528979 | Syzygium aromaticum | Indonesia |
| C. polyconidia | CMW23809, CBS122289 | EU245006, EU244978, EU244938 | Acacia mearnsii | South Africa |
| C. polyconidia | CMW23818, CBS122290 | EU245007, EU244979, EU244939 | Acacia mearnsii | South Africa |
| C. populicola | CMW14789, CBS119.78 | EF070418, EF070434, EF070406 | Populus sp. | Poland |
| C. populicola | CMW14819, CBS114725 | EF070419, EF070435, EF070407 | Populus sp. | USA |
| C. smalleyi | CMW14800, CBS114724 | EF070420, EF070436, EF070408 | Carya cordiformis | USA |
| C. smalleyi | CMW26383, CBS114724 | EU426553, EU426555, EU426556 | Carya cordiformis | USA |
| C. tanganyicensis | CMW15991, CBS122295 | EU244997, EU244969, EU244929 | Acacia mearnsii | Tanzania |
| C. tanganyicensis | CMW15999, CBS122294 | EU244998, EU244970, EU244939 | Acacia mearnsii | Tanzania |
| C. tsitsikammensis | CMW14276, CBS121018 | EF408555, EF408569, EF408576 | Rapanea melanophloeos | South Africa |
| C. tsitsikammensis | CMW14278, CBS121019 | EF408556, EF408570, EF408577 | Rapanea melanophloeos | South Africa |
| C. variospora | CMW20935, CBS114715 | EF070421, EF070437, EF070409 | Quercus alba | USA |
| C. variospora | CMW20936, CBS114714 | EF070422, EF070438, EF070410 | Quercus robur | USA |
| C. virescens | CMW11164 | DQ520639, EF070441, EF070413 | Fagus americanum | USA |
| C. virescens | CMW3276 | AY528984, AY528990, AY529011 | Quercus robur | USA |
| C. zombamontana | CMW15235 | EU245002, EU244974, EU244934 | Eucalyptus sp. | Malawi |
| C. zombamontana | CMW15236 | EU245000, EU244972, EU244932 | Eucalyptus sp. | Malawi |
| Ceratocystis sp. | CMW4797 | FJ236733, FJ236793, FJ236763 | Eucalyptus sp. | Congo |
| Ceratocystis sp. | CMW4799 | FJ236734, FJ236794, FJ236764 | Eucalyptus sp. | Congo |
| Ceratocystis sp. | CMW4902 | FJ236715, FJ236775, FJ236745 | Eucalyptus sp. | Brazil |
| Ceratocystis sp. | CMW5312 | FJ236731, FJ236791, FJ236761 | Eucalyptus sp. | Uganda |
| Ceratocystis sp. | CMW5313 | FJ236732, FJ236792, FJ236762 | Eucalyptus sp. | Uganda |
| Ceratocystis sp. | CMW7764 | FJ236726, FJ236786, FJ236756 | Eucalyptus sp. | Uruguay |
| Ceratocystis sp. | CMW7765 | FJ236727, FJ236787, FJ236757 | Eucalyptus sp. | Uruguay |
| Ceratocystis sp. | CMW7766 | FJ236728, FJ236788, FJ236758 | Eucalyptus sp. | Uruguay |
| Ceratocystis sp. | CMW7767 | FJ236729, FJ236789, FJ236759 | Eucalyptus sp. | Uruguay |
| Ceratocystis sp. | CMW7768 | FJ236730, FJ236790, FJ236760 | Eucalyptus sp. | Uruguay |
| Ceratocystis sp. | CMW14631 | FJ236744, FJ236804, FJ236774 | Eucalyptus sp. | Indonesia |
| Ceratocystis sp. | CMW14632 | FJ236743, FJ236803, FJ236773 | Eucalyptus sp. | Indonesia |
| Ceratocystis sp. | CMW16008 | FJ236735, FJ236795, FJ236765 | Eucalyptus sp. | Thailand |
| Ceratocystis sp. | CMW16009 | FJ236736, FJ236796, FJ236766 | Eucalyptus sp. | Thailand |
| Ceratocystis sp. | CMW16010 | FJ236737, FJ236797, FJ236767 | Eucalyptus sp. | Thailand |
| Ceratocystis sp. | CMW16034 | FJ236739, FJ236799, FJ236769 | Eucalyptus sp. | Thailand |
| Ceratocystis sp. | CMW16035 | FJ236738, FJ236798, FJ236768 | Eucalyptus sp. | Thailand |
| Ceratocystis sp. | CMW18572 | FJ236740, FJ236800, FJ236770 | Eucalyptus sp. | Indonesia |
| Ceratocystis sp. | CMW18577 | FJ236742, FJ236802, FJ236772 | Eucalyptus sp. | Indonesia |
| Ceratocystis sp. | CMW18591 | FJ236741, FJ236801, FJ236771 | Eucalyptus sp. | Indonesia |
PCR and sequencing reactions
DNA was extracted from all isolates as described by van Wyk et al. (2006a). Three gene regions were selected for PCR amplification, including ITS1 and ITS2, including the 5.8S rDNA operon, part of the beta-tubulin (βt-1) gene, and part of the Transcription Elongation Factor-1 alpha (TEF1-α) gene region. The reactions and programme for amplification were as described by van Wyk et al. (2006b). The primers utilized were ITS1 and ITS4 (White et al. 1990), βt1a and βt1b (Glass & Donaldson 1995), and EF1F and EF1R (Jacobs et al. 2004).
Sequencing reactions were set up and run as described by van Wyk et al. (2006a). Sequences of the isolates from Eucalyptus were analysed with Chromas Lite 2.01 (http://www.technelysium.com.au). These sequences as well as those for all species in the C. fimbriata s. lat. species complex (Table 1) were aligned using MAFFT (http://timpani.genome.ad.jp/%7emafft/server/) (Katoh et al. 2002). All sequences derived from this study have been deposited in GenBank (Table 1).
Combined gene tree for all described species in the C. fimbriata s. lat. complex
Representative isolates of all described species in the C. fimbriata s. lat. complex were included in this dataset, including those obtained for this study from CMW. The sequences of three gene regions (ITS, βt-1 and TEF1-α) were combined and a partition homogeneity test (PHT) was used to determine if the data from the three regions could be combined, using the software programme PAUP v. 4.0b10 (Swofford 2002). Settings in PAUP were as described in van Wyk et al. (2010a). Ceratocystis virescens was selected as the outgroup taxon.
MrModeltest2 (Nylander 2004) was used to determine the most appropriate model of nucleotide substitution for each of the three gene regions, respectively. These models were then included in the Bayesian analyses using MrBayes (Ronquist & Huelsenbeck 2003). The Bayesian analyses were run as described in van Wyk et al. (2010a).
Combined and separate gene trees of un-named Ceratocystis fimbriata s. lat. isolates obtained from Eucalyptus
This dataset consisted only of Ceratocystis fimbriata s. lat. isolates from Eucalyptus trees and that have not yet been described as separate species. A closely related and previously described species, C. colombiana, also obtained from Eucalyptus, was included as an outgroup. This was done to determine whether these isolates represent one group with no separate grouping or whether geographical grouping exists, as has been documented in C. fimbriata s. lat. (Engelbrecht & Harrington 2005, Ferreira et al. 2010).
Models were obtained for each of the ITS, βt-1 and TEF1-α gene regions with the use of MrModeltest2 (Nylander 2004). Consistent with both the first datasets, these models were incorporated into MrBayes (Ronquist & Huelsenbeck 2003) in order to run Bayesian analyses.
Utilising the C. fimbriata s. lat. isolates from Eucalyptus trees obtained from the CMW culture collection, the Molecular Evolutionary Genetics Analysis software (MEGA) 4 (Tamura et al. 2007) was used to determine the amount of variation for each gene region. The three gene regions were inspected to determine the number of fixed alleles between them. Allele trees were drawn using the software TCS (Clement et al. 2000) from the combined dataset for the Eucalyptus isolates, including the closely related species C. colombiana, known only from Eucalyptus.
Culture characteristics and morphology
Two isolates of Ceratocystis fimbriata s. lat. from Eucalyptus were selected from each country, other than Brazil, for which only one Eucalyptus isolate was available. These were used to describe morphological characteristics. Isolates were transferred to each of five 2 % Malt Extract Agar (MEA) plates and incubated in the dark. The isolates were incubated at 30 °C for 7 d, after which the growth was assessed.
Microscopic examinations were made of isolates from Indonesia, Uruguay, Thailand, and South Africa. Isolates from other countries were excluded because the cultures did not produce ascomata. All taxonomically informative structures were measured from 10 d old cultures on 2 % MEA, mounted in lactic acid. Ten measurements were made for each of the two isolates from Indonesia, Uruguay, Thailand, and South Africa.
A preliminary study of isolates representing the larger collection of C. fimbriata s. lat. isolates from Eucalyptus, and nested together in the same phylogenetic clade, showed that they are morphologically very similar. Consequently, four isolates (CMW 9998, CMW 15054, CMW 10000 and CMW 11536) from Eucalyptus in South Africa were selected for more detailed study. These South African isolates were transferred to five 2 % MEA plates each and incubated at seven different temperatures. These temperatures included 4 °C and six temperatures between 10 °C and 35 °C at 5 °C intervals. Growth was assessed after 7 d of incubation in the dark. Colony colour was assessed for the same isolates used as in the growth studies, grown on 2 % MEA for seven to 10 d at room temperature (25 °C). The colour charts of Rayner (1970) were used for descriptions of colony colour.
Fifty measurements were made of all taxonomically informative characters for isolate CMW 11536 from Eucalyptus in South Africa. An additional ten measurements were made of these structures for isolates CMW 9998 and CMW 10000 and CMW 15054. The minimum, maximum, average and standard deviation (stdv) was calculated for the measurements of each structure and these are presented in this study as; (minimum-) stdv minus the mean – stdv plus the mean (-maximum).
RESULTS
Isolates
Twenty-five isolates obtained from CMW that had been isolated from Eucalyptus trees were included in this study (Table 1). Fifteen of these originated from natural or artificially induced wounds on trees in three countries, South Africa, Thailand, and Indonesia. In addition, ten of the isolates were from trees that are believed to have been killed by the fungus. The latter isolates were from Brazil, Congo, Uganda, and Uruguay.
PCR and sequencing reactions
Results were obtained for three separate datasets. The first provided a broad phylogenetic placement (i.e. Latin American or North American, Asian, and African clade) of the C. fimbriata s. lat. isolates from Eucalyptus. A more focussed analysis determined whether these isolates could be linked to any of the previously described species in the C. fimbriata s. lat. complex that were obtained from Eucalyptus. Thereafter, the isolates from Eucalyptus apparently representing undescribed species were considered in combined as well as single gene trees generated from the sequence data for these isolates. This was to determine whether they could be grouped based on geographical origin.
Combined gene tree for all described species in the Ceratocystis fimbriata s. lat. complex
Amplicons for the three gene regions were on average 500 bp for the ITS and βt-1 gene regions and 800 bp for the TEF1-α region (Table 1). The PHT for the data set including all described species in the C. fimbriata s. lat. complex, had a low value (P=0.01), but could be combined (Cunningham 1997).
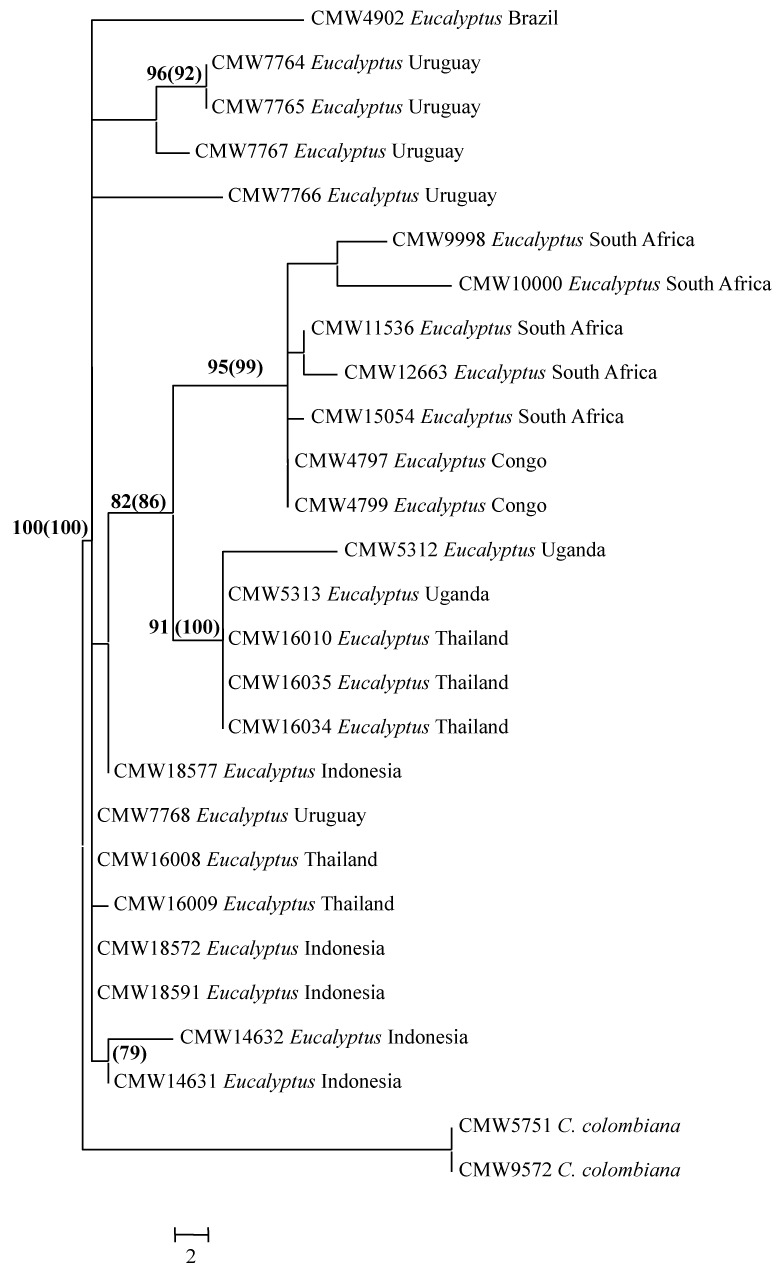
Of the 1 989 characters in this dataset, 1 102 were constant, 45 were parsimony uninformative while 842 were parsimony informative. One hundred and forty two most parsimonious trees were obtained, of which one was selected for presentation (Fig. 1). The tree topology was as follows: Tree length (TL) = 2054 steps, Consistency Index (CI) = 0.7, Retention Index (RI) = 0.9 and Rescaled Consistency (RC) = 0.6. Phylogenetic analyses revealed a clade specific for the isolates from Eucalyptus (Fig. 1). Isolates in this large clade had high bootstrap (88 %) and Bayesian (88 %) support and included some substructure (Fig. 1). The substructure in the large clade for the isolates from Eucalyptus was not strongly supported and these isolates were treated as reflecting a single group of genetically related, but not identical isolates. The closest phylogenetic relative of the isolates in the Eucalyptus clade was C. colombiana (van Wyk et al. 2010a).
Fig. 1.

Phylogenetic tree based on the combined sequences of the ITS, βt and TEF1-α gene regions for isolates from Eucalyptus including those provided the name C. eucalypticola and other described species in the C. fimbriata s. lat. complex. Ceratocystis virescens represents the out-group taxon. Bootstrap values are indicated at the branch nodes and Bayesian values in brackets.
The models obtained using MrModeltest2 were the HKY+I+G model for both the ITS and the TEF1-α genes and the GTR+G model for the βt-1 gene region. Including these models in the Bayesian analyses resulted in a burnin of 7000. These 7000 trees were discarded from the final analyses. The posterior probabilities obtained with the Bayesian analyses supported the bootstrap values obtained in PAUP (Fig. 1).
Combined and separate gene trees for undescribed Ceratocystis fimbriata s. lat. isolates from Eucalyptus
In the dataset for the combined gene regions, there were 1 765 characters of which 1 680 were constant, 31 were parsimony uninformative while 54 were parsimony informative. Twenty-four most parsimonious trees were obtained, one of which was selected for presentation (Fig. 2). The tree topology was as follows: TL = 107 steps, CI = 0.8, RI = 0.9 and RC = 0.7. One well-supported clade (100 % bootstrap, 100 % Bayesian) was observed with high variation. Three clades that were supported within this large clade were also observed (Fig. 2). The models obtained for this dataset were the HKY model for the ITS gene, the F81 model for the βt-1 gene region and the HKY+I model for the TEF1-α gene region. A burn-in of 1000 was obtained and these 1000 trees were discarded from the final analyses. The posterior probabilities obtained with the Bayesian analyses supported the bootstrap values obtained with PAUP (Fig. 2).
Fig. 2.
A phylogenetic tree for the combined sequences of the ITS, βt and EF1-α gene regions, including only the undescribed C. fimbriata s. lat. isolates with Eucalyptus as their host. The closely related species, C. colombiana, is included as outgroup. Bootstrap support is indicated at the branch nodes while Bayesian support is indicated in brackets.
Three well-supported clades were observed; the first included Asian (Indonesia and Thailand) and South American (Brazil and Uruguay) isolates; the second clade included African (Republic of Congo and South Africa) isolates while the third clade included African (Uganda) and Asian (Thailand) isolates. The previously described species, C. colombiana, grouped apart from these three clades (Fig. 2).
Where the data were treated separately, the trees for the ITS, βT and TEF1-α gene regions had a different topology when compared with those for the combined gene regions (Fig. 3). For the ITS gene tree, the same three clades emerged as in the combined dataset and included those for Asian and South American isolates, the African isolates and the African together with Asian isolates. However, only the African and Asian clade had strong support (97 %), the other two clades, African (63 %) and the Asian/ South American (55 %) clades had weak support (Fig. 3). In the case of the βt-1 gene tree, there was no support and all branches collapsed (Fig. 3). For the TEF1-α gene tree, there were two small clades encompassing the South African isolates that had high and medium support (85 % and 65 % respectively), while the rest of the isolates grouped in a single clade with strong (85 %) support (Fig. 3).
Fig. 3.

Three phylograms each representing a single gene region (ITS, βt and TEF-1α, top to bottom) for the undescribed isolates from Eucalyptus representing C. fimbriata s. lat. showing low variation in the three separate gene regions as well as no support for the sub-clades observed in the combined gene trees. No outgroup was assigned to this dataset.
Where data for the C. fimbriata s. lat. isolates were analysed in MEGA, the results showed that in the ITS gene region, the C. fimbriata s. lat. isolates obtained from Eucalyptus were separated from C. colombiana by an average of 23 nucleotide differences (Table 2). Where isolates from different countries were compared, there was also variation in the ITS with a maximum of 13 bp and average of 5 bp differences (Table 2).
Table 2. The number of differences observed between the sequences of the isolates from Eucalyptus (C. fimbriata s. lat.) from Brazil, South Africa, Uruguay, Uganda, Congo, Thailand, Indonesia, and C. colombiana.
| Country | Brazil | South Africa | Uruguay | Uganda | Congo | Thailand | Indonesia | C. colombiana |
|---|---|---|---|---|---|---|---|---|
| Gene region | ||||||||
| ITS | ||||||||
| Brazil | – | 9 | 0 | 6 | 13 | 0 | 0 | 23 |
| South Africa | 9 | 8 | 6 | 6 | 0 | 4 | 9 | 21 |
| Uruguay | 0 | 6 | 4 | 7 | 9 | 0 | 0 | 21 |
| Uganda | 6 | 6 | 7 | 0 | 9 | 0 | 7 | 28 |
| Congo | 13 | 0 | 9 | 9 | 0 | 6 | 11 | 25 |
| Thailand | 0 | 4 | 0 | 0 | 6 | 7 | 0 | 20 |
| Indonesia | 0 | 9 | 0 | 7 | 11 | 0 | 1 | 22 |
| C. colombiana | 23 | 21 | 21 | 28 | 25 | 20 | 22 | 1 |
| βt | ||||||||
| Brazil | – | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| South Africa | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 |
| Uruguay | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Uganda | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Congo | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Thailand | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Indonesia | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| C. colombiana | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 0 |
| TEF | ||||||||
| Brazil | – | 13 | 9 | 12 | 12 | 12 | 12 | 21 |
| South Africa | 13 | 7 | 0 | 0 | 0 | 0 | 0 | 8 |
| Uruguay | 9 | 9 | 9 | 0 | 0 | 0 | 0 | 7 |
| Uganda | 12 | 0 | 0 | 7 | 0 | 0 | 0 | 6 |
| Congo | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| Thailand | 12 | 0 | 0 | 0 | 0 | 1 | 0 | 8 |
| Indonesia | 12 | 0 | 0 | 0 | 0 | 0 | 5 | 8 |
| C. colombiana | 21 | 8 | 7 | 6 | 8 | 8 | 8 | 0 |
Where isolates of C. fimbriata s. lat. from Eucalyptus were compared with C. colombiana in the βt-1 gene region, there were only 3bp differences between them (Table 2). Within the clade representing the C. fimbriata s. lat. group from Eucalyptus, there was only one base pair difference observed in the South African group and no differences between isolates from different countries (Table 2).
For the TEF1-α gene region, there were 21bp differences between the isolate from Brazil and C. colombiana and an average of 8 bp differences between C. colombiana and the other isolates from Eucalyptus. Only the single isolate from Brazil differed from the other isolates while no differences were observed between the isolates from the other countries. The allele networks drawn from the combined gene regions (ITS, βt-1 and TEF1-α) for the C. fimbriata s. lat. obtained from Eucalyptus revealed a single tree with high variation (Fig. 4). There was no obvious geographic structure with regards to the origin of the eucalypt isolates. The previously described species, C. colombiana, formed a separate allele tree (Fig. 4).
Fig. 4.
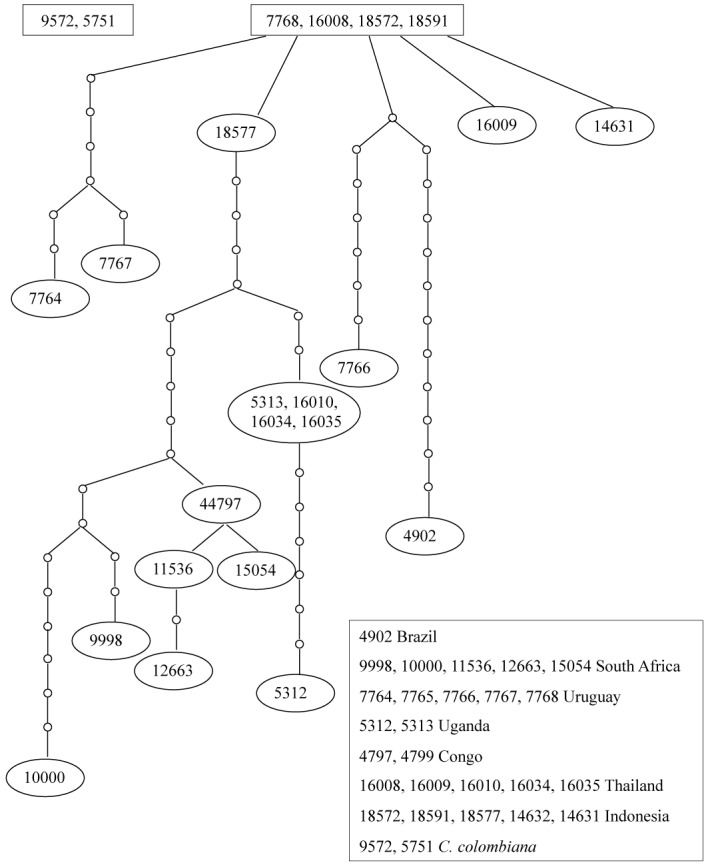
Allele networks obtained from the three combined gene regions (ITS, βt and TEF1-α for all isolates from Eucalyptus as well as C. colombiana. The species C. colombiana is represented as highly different to the Eucalyptus isolates due to the fact that it formed a separate allele tree. The C. fimbriata s.lat. isolates from Eucalyptus all formed one allele tree with high variation observed within the tree.
Culture characteristics and morphology
All isolates from Eucalyptus had a similar greenish olivaceous (33′′′f) (Rayner 1970) colony colour. The cultures had a banana odour similar to that of many Ceratocystis species.
The cultures all grew optimally at 30 °C. No clear morphological differences could be observed between isolates from different countries (Table 3).
Table 3. Morphological comparison of two representative isolates from Indonesia, South Africa, Thailand, and Uruguay. Ten measurements were taken of each structure and the (minimum-) average minus standard deviation – average plus standard deviation and (-maximum) given below.
| Characteristic / Country | Indonesia | South Africa | Thailand | Uruguay |
|---|---|---|---|---|
| Ascomatal bases | ||||
| Shape | Globose | Globose | Globose | Globose |
| Length | (125−)162–199(−200) | (120−)142–190(−202) | (188−)190–197(−200) | (144−)170–197(−200) |
| Width | (143−)173–193(−200) | (132−)143–193(−216) | (154−)177–199(−212) | (141−)164–184(−197) |
| Ascomatal necks | ||||
| Length | (390−)400–450(−470) | (372−)392–460(−486) | (354−)370–400(−424) | (354−)368–386(−409) |
| Width (bases) | (24−)25–35(−40) | (24−)25–35(−42) | (24−)25–35(−39) | (23−)26–32(−38) |
| Width (apices) | (15−)16–18(−20) | (15−)16–20(−22) | (16−)17–19(−20) | (15−)16–22(−25) |
| Ostiolar hyphae | ||||
| Shape | Divergent | Divergent | Divergent | Divergent |
| Length | (36−)43–53(−63) | (39−)40–52(−62) | (33−)35–39(−41) | (38−)41–51(−53) |
| Ascospores | ||||
| Length | 3–5 | 3–5 | 3–4 | 3–4 |
| Width (excluding sheath) | 4–6 | 4–6 | 4–6 | 4–6 |
| Width (including sheath) | 5–8 | 5–7(−8) | 5–7 | 6–7 |
| Primary phialides | ||||
| Length | (69−)70–100(−134) | (73−)76–114(−131) | (67−)76–96(−100) | (73−)75–83(−88) |
| Width (bases) | 4–6 | 4–6 | 4–6 | 2–4 |
| Width (broadest point) | 4–6 | 4–6 | 6–8 | 4–5 |
| Width (apices) | 3–5 | 3–5 | 3–5 | 3–4 |
| Secondary phialides | ||||
| Length | (60−)70–100(−143) | (64−)69–109(−143) | (63−)68–77(−99) | (69−)72–96(−109) |
| Width (bases) | 3–6 | 3–6 | 5–6 | 3–6 |
| Width (apices) | 5–7 | 5–7 | 4–8 | 6–8 |
| Primary conidia | ||||
| Length | (13−)19–20(−24) | (15−)18–24(−25) | (10−)13–17(−18) | (10−)11–15(−18) |
| Width | 4–5 | 4–5 | 3–4 | 2–3 |
| Secondary conidia | ||||
| Length | 6–8 | 6–8 | 6–8 | (7−)9–11 |
| Width | 5–8 | 5–7 | 5–8 | 6–8 |
| Chlamydospores | ||||
| Shape | Globose/Subglobose | Globose/Subglobose | Globose/Subglobose | Globose/Subglobose |
| Length | 10–15 | 10–13 | 12–15 | (6−)7–11(−13) |
| Width | 8–13 | 8–10 | 10–13 | (5−)7–11(−12) |
Isolate CMW 11536 from Eucalyptus in South Africa was chosen to represent the global collection of isolates obtained from Eucalyptus. Three additional isolates (CMW 9998, CMW 10000 and CMW 15054), also from South Africa, were chosen as additional specimens for description. Cultures of these isolates were grown on 2 % MEA, dried down and have been deposited with the National Collection of Fungi (PREM), Pretoria, South Africa. Living cultures are maintained in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI) at the University of Pretoria, South Africa and the Centraalbureau voor Schimmelcultures (CBS) in Utrecht, The Netherlands.
Where growth in culture was characterised based on the average colony diameter (from the five inoculated plates) for the four selected Eucalyptus isolates from South Africa, after 7 d, limited growth was observed at 4 °C (8 mm), 10 °C (7 mm), 15 °C (19 mm) and 35 °C (10 mm). Intermediate growth was observed after 7 d at 20 °C (34 mm) and 25 °C (35 mm), while the optimum temperature for growth in culture was 30 °C at which isolates reached an average of 39 mm diam after 7 d.
TAXONOMY
Isolates of the Ceratocystis from Eucalyptus, originating from many different countries, were phylogenetically distinct from all other Ceratocystis species residing in the C. fimbriata s. lat. clade. They also formed distinct phylogenetic groups based on geographic origin and might be found to represent distinct taxa in the future. For the present, those isolates from South Africa, which also had a morphology different to all described species from Eucalyptus (Table 4) are described as representing a novel taxon.
Table 4. Morphological comparison of previously described species in the C. fimbriata s. lat. species complex obtained from Eucalyptus trees compared to C. eucalypticola.
| Character / Species | C. atrox | C. eucalypticola | C. fimbriatomima | C. neglecta | C. colombiana | C. pirilliformis |
|---|---|---|---|---|---|---|
| Ascomatal bases | ||||||
| Shape | Globose | Globose | Globose | Globose | Globose | Obpyriform |
| Length | (120−)140–180 (−222) | (105−)140–186 (−222) | (142−)173–215 (−234) | (173−)202–244 (−281) | (140−)177–237 (−294) | 145–216(−279) |
| Width | (120−)150–178 (−200) | (118−)146–184 (−216) | (145−)178–225 (−255) | (153−)178–228 (−250) | (140−)177–237 (−294) | 115–186(−206) |
| Ascomatal necks | ||||||
| Length | (270−)310–400 (−460) | (274−)376–464 (−499) | (446−)660–890 (−1070) | (691−)745–840 (−889) | (375−)448–560 (−676) | 372–683(−778) |
| Width (bases) | (21−)26–34(−40) | (19−)25–33(−42) | (28−)32–42(−47) | (27−)31–39(−46) | (24−)27–35(−43) | 18–33(−40) |
| Width (apices) | (13−)14–16(−19) | (14−)16–20(−22) | (16−)18–24(−28) | (14−)16–20(−22) | (12−)14–18(−19) | 12–21(−25) |
| Ostiolar hyphae | ||||||
| Shape | Divergent | Divergent | Divergent | Divergent | Divergent | Convergent |
| Length | (18−)20–26(−28) | (39−)45–59(−66) | (40−)49–61(−68) | (35−)41–49(−54) | (28−)38–46(−52) | N/A |
| Ascospores | ||||||
| Length | 3–4 | 3–5 | 2–4 | 3–6 | 3–4 | 4–6 |
| Width (excluding sheath) | 3–4 | 4–6 | 4–6 | 4–7 | (3−)4–6(−7) | 3–5 |
| Width (including sheath) | 4–6 | 5–7(−)8 | 5–7 | 5–8 | 6–8(−11) | 3–5 |
| Primary phialides | ||||||
| Length | (78−)87–151(−218) | (58−)77–113(−131) | (49−)60–94(−122) | (75−)80–114(−152) | (58−)65–83(−106) | 62–147(−216) |
| Width (bases) | 5–7(−13) | (3−)4–6(−7) | 4–7 | (4−)5–7(−8) | 4–6(−8) | N/A |
| Width (broadest point) | 4–7 | 4–6(−7) | 5–9 | 5–9 | (3−)6–8(−9) | N/A |
| Width (apices) | 4–9 | 3–5 | 3–5 | (3−)4–6(−7) | 3–5(−6) | N/A |
| Secondary phialides | ||||||
| Length | (39−)43–57(−66) | (43−)60–100(−143) | Absent | (38−)48–76(−89) | (42−)49–71(−85) | N/A |
| Width (bases) | 5–7(−9) | (3−)4–6(−7) | Absent | (3−)5–7(−8) | (4−)5–7 | N/A |
| Width (apices) | 4–6)–7) | (4−)5–7(−8) | Absent | (3−)5–7(−8) | (5−)6–8 | N/A |
| Primary conidia | ||||||
| Length | (9−)11–15(−17) | (14−)16–22(−25) | (14−)20–28(−31) | (11−)15–27(−30) | (12−)16–24(−29) | 12–25(−33) |
| Width | 3–5 | 3–5 | 3–5 | (3−)5–6 | 4–6 | 2–5 |
| Secondary conidia | ||||||
| Length | (7−)8–12(−14) | (6−)7–9(−12) | Absent | (6−)10–11 | 9–14 | 4–6 |
| Width | (5−)6–8(−9) | 4–6(−7) | Absent | (4−)5–7(−9) | 6–8(−11) | 3–5 |
| Chlamydospores | ||||||
| Shape | Absent | Globose/Subglobose | Subglobose | Globose | Globose | Oval |
| Length | Absent | (10−)11–13(−15) | (6−)10–14(−15) | (8−)10–12(−13) | 11–14 | 8–12(−13) |
| Width | Absent | 8–10(−11) | (6−)7–11(−12) | (9−)10–14(−16) | 11–15(−17) | 5–8(−10) |
| Reference | Van Wyk et. al. 2007 | This study | Van Wyk et. al. 2008 | Rodas et. al. 2008 | Van Wyk et. al. 2010a | Barnes et. al. 2003 |
Ceratocystis eucalypticola M. van Wyk & M.J. Wingf., sp. nov.
MycoBank MB512397
(Fig. 5)
Fig. 5.
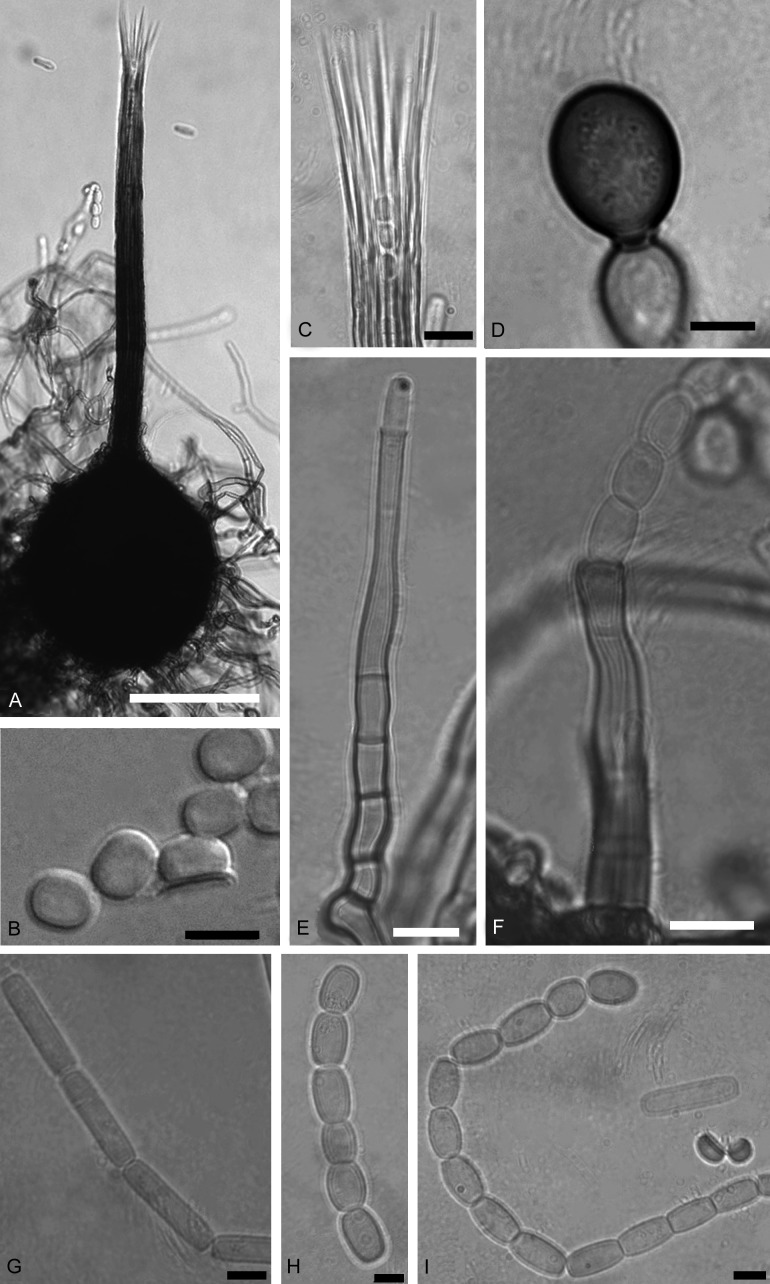
Morphological characteristics of Ceratocystis eucalypticola. a. Ascomata with globose base. b. Hat-shaped (in side view) and cucullate (in top view) ascospores. c. Divergent ostiolar hyphae d. Dark, globose to sub-globose chlamydospore. e. Primary conidiophore, flask-shaped phialide, producing cylindrical conidia. f. Tubular shaped secondary conidiophore, producing a chain of barrel-shaped conidia. g. Chain of cylindrical conidia. h. Chain of barrel-shaped conidia. i. A chain of barrel-shaped conidia, two hat-shaped ascospores and a cylindrical conidium. Bars: a. = 100 μm, b, f–i = 5 μm, c–e = 10 μm.
Etymology: The name refers to Eucalyptus on which the fungus occurs.
All species of Ceratocystis from Eucalyptus are phylogenetically distinct. Colonies of C. eucalypticola are typically green colonies, relatively slow growing, and have a fruity banana odour.
Type: South Africa: Kwa-Zulu Natal: KwaMbonambi, isolated from artificially wounded Eucalyptus, 15 Dec. 2002, M. van Wyk & J. Roux (PREM 60168 – holotype; cultures ex-holotype CMW 11536 = CBS 124016)
Description: Ascomatal bases dark brown to black, globose, un-ornamented (105−)140–186(−222) μm wide, (118−)146–184(−216) μm high. Ascomatal necks dark brown to black at bases becoming lighter towards the apices, (274−)376–464(−499) μm long, apices (14−)16–20(−22) μm wide, bases (19−)25–33(−42) μm wide. Ostiolar hyphae divergent, (39−)45–59(−66) μm long. Ascospores hyaline, hat-shaped in side view, invested in sheath, 3–5 μm long, 4–6 μm wide without sheath, 5–7(−8) μm wide including sheath. Anamorph thielaviopsis-like, conidiophores of two types: Primary conidiophores phialidic, flask-shaped, (58−)77–113(−131) μm long, (3−)4–6 μm wide at the bases, 4–6(−7) μm wide at broadest points and 3–5 μm wide at apices. Secondary conidiophores flaring or wide mouthed, (43−)60–100(−143) μm long, (3−)4–6(−7) μm wide at bases and (4−)5–7(−8) μm wide at apices. Primary conidia cylindrical in shape (14−)16–22(−25) μm long, 3–5 μm wide. Secondary conidia, barrel-shaped, abundant, (6−)7–9(−12) μm long, 4–6(−7) μm wide. Chlamydospores, scarce, hair brown (17′′′′i), globose to sub-globose (10−)11–13(−15) μm long, 8–10(−11) μm wide.
Habitat: Wounded and diseased Eucalyptus.
Known distribution: South Africa.
Other material examined: South Africa: Mpumalanga, Sabie, isolated from artificially wounded Eucalyptus trees, 14 July 2002, M. van Wyk & J. Roux (PREM 60169; living cultures CMW 9998 = CBS 124017); loc. cit., isolated from artificially wounded Eucalyptus trees, 14 July 2002, M. van Wyk & J. Roux (PREM 60170; living cultures CMW 10000 = CBS 124019).
DISCUSSION
Isolates of Ceratocystis fimbriata s. lat. collected from Eucalyptus in Brazil, Indonesia, Republic of Congo, South Africa, Thailand, Uganda, and Uruguay were shown to be phylogenetically related. These included isolates taken from wounds on trees and also those that were associated with trees dying as result of infection by the fungus. Although all isolates from Eucalyptus resided in a single large clade, there was a high degree of diversity among them. It is thus possible that they represent a number of different cryptic species that cannot be resolved. For the present, those isolates from South Africa are provided with the name C. eucalypticola here. Future studies should seek to include additional isolates from Eucalyptus as well as to include sequences for gene regions not considered in this study, and that might discriminate more clearly between species in the C. fimbriata s. lat. complex. Currently, the group is unified based on a specific host and relatively strong phylogenetic similarity. In this respect, it also provides the foundation for further studies including a suite of isolates that would be difficult to obtain.
The species of Ceratocystis most closely related to C. eucalypticola is C. colombiana. Ceratocystis colombiana is a pathogen of coffee trees (Marin et al. 2003) as well as numerous other hosts including indigenous crops in Colombia. Although the two species are phylogenetically related, they are ecologically distinct and are not likely to be confused.
Ceratocystis eucalypticola is one of a number of species in the C. fimbriata s. lat. complex to be described from Eucalyptus trees. Other species from this host include; C. atrox (van Wyk et al. 2007) and C. corymbiicola (Kamgan Nkuekam et al. 2012) from Australia, C. pirilliformis (Barnes et al. 2003b) from Australia and South Africa, C. neglecta (Rodas et al. 2007) from Colombia, C. fimbriatomima (van Wyk et al. 2008) from Venezuela, and C. zombamontana (Heath et al. 2009) from Malawi. All of these species from Eucalyptus can be distinguished from each other based on phylogenetic inference and they have some morphological features that can be used to recognise them.
Morphologically, the specimens of C. eucalypticola cited here resemble species in the C. fimbriata s. lat. complex. The fungus has the typical green colony colour, is relatively slow growing, and has a fruity banana odour. Ceratocystis eucalypticola can be distinguished from other species in the C. fimbriata s. lat. complex in that they occur on Eucalyptus and based on differences in size of some diagnostic characters for this group of fungi.
Ceratocystis eucalypticola includes isolates only from wounds on trees in South Africa in the absence of disease, but is very closely related to isolates that originated from dying trees and that have been shown to be pathogenic (Laia et al. 1999; Roux et al. 2000, 2001, 2004). The species is also closely related to isolates that were collected from wounds on trees in countries other than South Africa where a Ceratocystis disease on Eucalyptus has not been seen. Eucalyptus death associated with C. eucalypticola has never been found in South Africa although trees dying of unknown causes are thought to have died due to infection by this fungus, which can be difficult to isolate. The fungus collected from wounds on trees has also been shown to be pathogenic in greenhouse inoculation trials (Roux et al. 2004, van Wyk et al. 2010b).
Isolates of C. eucalypticola from South Africa represent a clonal population (van Wyk et al. 2006b) and it was most likely introduced into the country. It is thus intriguing that Eucalyptus death associated with this fungus has not been seen. This might be due to planting stock susceptible to C. eucalypticola not having occurred in the country, or that conditions for infection were not suitable. Alternatively, it is possible that trees dying of unexplained causes might have been killed by C. eucalypticola, even though the fungus was not isolated from them. This is a question that is currently being pursued, particularly linked to unexplained Eucalyptus death in South Africa and where Ceratocystis cultures emerge from isolations.
Acknowledgments
We thank the National Research Foundation (NRF), members of the Tree Protection Co-operative Programme (TPCP), the THRIP initiative of the Department of Trade and Industry and the Department of Science and Technology (DST)/NRF Centre of Excellence in Tree Health Biotechnology (CTHB) for funding.
REFERENCES
- Barnes I, Gaur A, Burgess T, Roux J, Wingfield BD, Wingfield MJ. (2001) Microsatellite markers reflect intra-specific relationships between isolates of the vascular wilt pathogen, Ceratocystis fimbriata. Molecular Plant Pathology 2: 319–325 DOI.org/10.1046/j.1464-6722.2001.00080.x [DOI] [PubMed] [Google Scholar]
- Barnes I, Roux J, Wingfield BD, O’Neil M, Wingfield MJ. (2003a) Ceratocystis fimbriata infecting Eucalyptus grandis in Uruguay. Australasian Plant Pathology 32: 361–355 DOI.org/10.1071/AP03032 [Google Scholar]
- Barnes I, Roux J, Wingfield MJ, Old KM, Dudzinski M. (2003b) Ceratocystis pirilliformis, a new species from Eucalyptus nitens in Australia. Mycologia 95: 865–871 DOI org/10.2307/3762015 [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K. (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1660 DOI.org/10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Cunningham CW. (1997) Can three incongruence tests predict when data should be combined? Molecular Biology and Evolution 14: 733-740 [DOI] [PubMed] [Google Scholar]
- Engelbrecht CJB, Harrington TC. (2005) Intersterility, morphology and taxonomy of Ceratocystis fimbriata on sweet potato, cacao and sycamore. Mycologia 97: 57–69 DOI.org/10.3852/mycologia.97.1.57 [DOI] [PubMed] [Google Scholar]
- Feirreira EM, Harrington TC, Thorpe DJ, Alfenas AC. (2010) Genetic diversity and interfertility among highly differentiated populations of Ceratocystis fimbriata in Brazil. Plant Pathology 59: 721–735 DOI.org/10.1111/j.1365-3059.2010.02275.x [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RN, Wingfield BD, Wingfield MJ, Meke G, Mbaga A, Roux J. (2009) Ceratocystis species on Acacia mearnsii and Eucalyptus spp. in eastern and southern Africa including six new species. Fungal Diversity 34: 41–67 [Google Scholar]
- Jacobs K, Bergdahl DR, Wingfield MJ, Halik S, Seifert KA, Bright DE, Wingfield BD. (2004) Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycological Research 108: 411–418 DOI.org/10.1017/S0953756204009748 [DOI] [PubMed] [Google Scholar]
- Johnson JA, Harrington TC, Engelbrecht CJB. (2005) Phylogeny and taxonomy of the North American clade of the Ceratocystis fimbriata complex. Mycologia 97: 1067–1092 DOI.org/10.3852/mycologia.97.5.1067 [DOI] [PubMed] [Google Scholar]
- Kamgan Nkuekam G, Wingfield MJ, Mohammed C, Carnegie AJ, Pegg GS, Roux J. (2012) Ceratocystis species, including two new species associated with nitidulid beetles on Eucalyptus in Australia. Antonie van Leeuwenhoek 101: 217–241 DOI 10.1007/s10482-011-9625-7. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066 DOI.org/10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile GA. (1993) Plant diseases caused by species of Ceratocystis sensu stricto and Chalara. In: Ceratocystis and Ophiostoma: taxonomy, ecology and pathogenicity (Wingfield MJ, Seifert KA, Webber JA, eds): 173–183 St Paul, MN: American Phytopathological Society Press; [Google Scholar]
- Laia ML, Alfenas AC, Harrington TC. (1999) Isolation, detection in soil, and inoculation of Ceratocystis fimbriata, causal agent of wilting, dieback and canker in Eucalyptus. In: Proceedings of the 12th Biennial Conference of the Australasian Plant Pathology Society, 27–30 September (Morin L, ed): 77 Canberra: Australasian Plant Pathology Society; [Google Scholar]
- Marin M, Castro B, Gaitan A, Preisig O, Wingfield BD, Wingfield MJ. (2003) Relationships of Ceratocystis fimbriata isolates from Colombian coffee-growing regions based on molecular data and pathogenicity. Journal of Phytopathology 151: 395–400 DOI org/10.1046/j.1439-0434.2003.00738.x [Google Scholar]
- Moller WJ, DeVay JE. (1968) Carrot as a species-selective isolation medium for Ceratocystis fimbriata. Phytopathology 58: 123–124 [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. [Program distributed by the author.] Uppsala: Evolutionary Biology Centre, Uppsala University; [Google Scholar]
- Old KM, Wingfield MJ, Yuan ZQ. (2003) A manual of diseases of Eucalypts in South-East Asia. Jakarta: Centre for International Forestry Research; [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. Kew: Commonwealth Mycological Institute; [Google Scholar]
- Rodas C, Roux J, van Wyk M, Wingfield BD, Wingfield MJ. (2007) Ceratocystis neglecta sp. nov., infecting Eucalyptus trees in Colombia. Fungal Diversity 28: 73–84 [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 DOI.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Roux J, Coutinho TA, Munjuni Byabashaija D, Wingfield MJ. (2001) Diseases of plantation Eucalyptus in Uganda. South African Journal of Science 97: 16–18 [Google Scholar]
- Roux J, van Wyk M, Hatting H, Wingfield MJ. (2004) Ceratocystis species infecting stem wounds on Eucalyptus grandis in South Africa. Plant Pathology 53: 414–421 DOI.org/10.1111/j.0032-0862.2004.01014.x [Google Scholar]
- Roux J, Wingfield MJ. (2009) Ceratocystis species: emerging pathogens of non-native plantation Eucalyptus and Acacia species. Southern Forests 71: 115–120 DOI org/10.2989/SF.2009.71.2.5.820 [Google Scholar]
- Roux J, Wingfield MJ, Wingfield BD, Bouillett JP, Alfenas AC. (2000) A serious new disease of Eucalyptus caused by Ceratocystis fimbriata in Central Africa. Forest Pathology 30: 175–184 DOI.org/10.1046/j.1439-0329.2000.00202.x [Google Scholar]
- Swofford DL. (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA:Sinauer Associates; [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596–1599 DOI.org/10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- van Wyk M, Pegg G, Lawson S, Wingfield MJ. (2007) Ceratocystis atrox sp. nov. associated with Phoracanthta acanthocera infestations on Eucalyptus in Australia. Australian Journal of Plant Pathology 36: 407–414 DOI.org/10.1071/AP07042 [Google Scholar]
- van Wyk M, Roux J, Barnes I, Wingfield BD, Wingfield MJ. (2006a) Molecular phylogeny of the Ceratocystis moniliformis complex and description of C. tribilliformis sp. nov. Fungal Diversity 21: 181–201 [Google Scholar]
- van Wyk M, Van der Merwe NA, Roux J, Wingfield BD, Kamgan GN, Wingfield MJ. (2006b) Population biology of Ceratocystis fimbriata from Eucalyptus trees in South Africa. South African Journal of Science 102: 259–263 [Google Scholar]
- van Wyk M, Wingfield BD, Marin M, Wingfield MJ. (2010a) Two new Ceratocystis species infecting coffee, cacao, citrus and native trees in Colombia. Fungal Diversity 40: 103–117 DOI.org/10.1007/s13225-009-0005-9 [Google Scholar]
- van Wyk M, Wingfield BD, Wingfield MJ. (2010b) Comparison of procedures to evaluate the virulence of Ceratocystis fimbriata sensu lato isolates from Eucalyptus in South Africa. Southern Forests 72(2): 57–62 DOI.org/10.2989/20702620.2010.507011 [Google Scholar]
- van Wyk M, Wingfield BD, Mohali S, Wingfield MJ. (2008) Ceratocystis fimbriatomima, a new species in the C. fimbriata sensu lato complex isolated from Eucalyptus trees in Venezuela. Fungal Diversity 34: 173–183 [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a sequencing guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds): 315–322 San Diego: Academic Press; [Google Scholar]
- Wingfield MJ, Slippers B, Hurley BP, Coutinho TA, Wingfield BD, Roux J. (2008) Eucalypt pests and diseases: growing threats to plantation productivity. Southern Forests 70: 139–144 DOI.org/10.2989/SOUTH.FOR.2008.70.2.9.537 [Google Scholar]