Summary
Shigella flexneri is a Gram-negative pathogen that invades the colonic epithelium. While invasion has been thoroughly investigated, it is unknown how Shigella first attaches to the epithelium. Previous literature suggests that Shigella utilizes adhesins that are induced by environmental signals, including bile salts, encountered in the small intestine prior to invasion. We hypothesized that bile would induce adherence factors to facilitate attachment to colonic epithelial cells. To test our hypothesis, S. flexneri strain 2457T was subcultured in media containing bile salts, and the ability of the bacteria to adhere to the apical surface of polarized T84 epithelial cells was measured. We observed a significant increase in adherence, which was absent in a virulence plasmid-cured strain and a type-III secretion system mutant. Microarray expression analysis indicated that the ospE1/ospE2 genes were induced in the presence of bile, and bile-induced adherence was lost in a ΔospE1/ΔospE2 mutant. Further studies demonstrated that the OspE1/OspE2 proteins were localized to the bacterial outer membrane following exposure to bile salts. The data presented are the first demonstration that the OspE1/OspE2 proteins promote initial adherence to the intestinal epithelium. The adhesins required for Shigella attachment to the colonic epithelium may serve as ideal targets for vaccine development.
Keywords: OspE1, OspE2, type-III secretion system, adherence, epithelial cells
Introduction
Shigella flexneri is a Gram-negative, facultative intracellular pathogen that causes approximately 160 million cases of watery diarrhea or bacillary dysentery annually, resulting in one million deaths (Jennison & Verma, 2004). Infection predominately occurs in children under the age of 5 in developing countries, and symptoms of disease include fever, abdominal cramps, and bloody or mucoid stools (Jennison & Verma, 2004). Infection is due to the ability of the pathogen to invade the colonic epithelium, which requires a 220 kilobase virulence plasmid. Encoded on the plasmid are the components of a type-III secretion system (T3SS), the Ipa (invasion plasmid antigen) proteins, and other effector proteins (Schroeder & Hilbi, 2008). The T3SS is a needle-like complex that allows bacterial proteins to be injected directly into the cytoplasm of the host cell. The Ipa proteins are required for invasion of epithelial cells (Schroeder & Hilbi, 2008). During S. flexneri infection, the bacteria transit through M (microfold or membranous) cells, which are specialized antigen-presenting cells of the follicle-associated epithelium (FAE) (Baranov & Hammarstrom, 2004). Transit through M cells allows the bacteria to reach the basolateral pole of the epithelium for efficient invasion (Schroeder & Hilbi, 2008). The FAE is considered the major site of entry for S. flexneri due to the presence of M cells, and studies with the rabbit ligated intestinal loop model have shown that the M cells are also invaded by the bacteria (Sansonetti et al., 1996). However, whether there is a requirement for bacteria to first adhere to the apical surface of the epithelial cells prior to transit through M cells remains unknown.
In order to proficiently colonize the mucosal environment of the gastrointestinal tract, many bacteria, particularly pathogenic Escherichia coli, utilize a range of surface structures such as pili or fimbriae to initiate binding to host cells (Kline et al., 2010). Pili are non-flagellar, proteinaceous, multi-subunit adhesins that include chaperone or usher-assembled pili, such as the Type 1 and Pap (P) pilus of uropathogenic E. coli (Kline et al., 2010). Additional colonization factors, such as fimbrial adhesins, afimbrial adhesins, and bundle forming pili (Torres et al., 2005, Townsend et al., 2001) are utilized by the pathogenic strains of E. coli and Salmonella enterica serotype Typhi. The mechanism by which Shigella first adhere to and colonize the gastrointestinal tract are relatively unknown. Since Shigella evolved from various strains of E. coli (Pupo et al., 2000, Townsend et al., 2001) and given the fact that fimbriae are prevalent among the Enterobacteriaceae (Bloch et al., 1992), we hypothesize that Shigella utilize fimbriae or other adhesins during initial colonization of the colon. Two reports have identified the presence of fimbriae on the surface of Shigella, including type 1 fimbriae (Utsunomiya et al., 1992, Snellings et al., 1997). In addition, comparative genomic studies have identified novel open reading frames in S. flexneri that have been annotated as adhesins (Wei et al., 2003). The data suggest that S. flexneri have the ability to produce adhesins that play a role in efficient adherence to the surface of colonic epithelial cells, yet there is a lack of detailed characterization studies of putative adhesins.
As the bacteria travel through the small intestine, into the colon, and to the site of infection at the FAE, S. flexneri is exposed to various environmental signals, such as bile. The variations in environmental stimuli can lead to changes in gene expression that enhance the virulence of the bacteria. For example, it has been demonstrated that invasion of epithelial cells by S. flexneri is increased in the presence of bile salts (Pope et al., 1995, Olive et al., 2007, Stensrud et al., 2008). Bile salts stimulate the recruitment of the invasins to the tip of the type-III needle complex. Specifically, the bile salt deoxycholate interacts with IpaD, which in turn, interacts with IpaB. This increased recruitment of the invasins leads to more efficient invasion (Olive et al., 2007, Stensrud et al., 2008). Previous studies by Pope, et al demonstrated that bile salts also led to increased adherence of S. flexneri to wepithelial cells (Pope et al., 1995). However, the authors noted that the non-invasive ΔipaB mutant of S. flexneri adhered at the same level as wildtype bacteria to HeLa cells after exposure to deoxycholate; and therefore, concluded that the increase in attachment was independent of invasion. The authors hypothesized that an adhesin secreted through the T3SS is upregulated by bile salts, and that transit through the small intestine is important for the induced expression of this adhesin (Pope et al., 1995).
We sought to exploit the observations of increased adherence of Shigella in the presence of bile to identify the adhesins required for S. flexneri attachment to the colonic epithelium prior to invasion. We observed significant increases in adherence of S. flexneri to the apical surface of polarized T84 cells after exposure to bile salts, and microarray analyses demonstrated that expression of ospE1 and ospE2 was enhanced by bile. A double ΔospE1 plus ΔospE2 mutant did not exhibit a significant increase in adherence following treatment with bile salts, suggesting that these factors represent the bile-induced adhesins. Protein analysis suggested that OspE1 and OspE2 are secreted through the T3SS and are localized to the bacterial outer membrane where they enhance adherence prior to invasion. Identification of the adhesins required for S. flexneri attachment to the colonic epithelium prior to invasion may serve as targets for therapeutics or vaccine development.
Results
Pre-exposure to bile salts increases adherence of Shigella to the apical surface of polarized epithelial cells
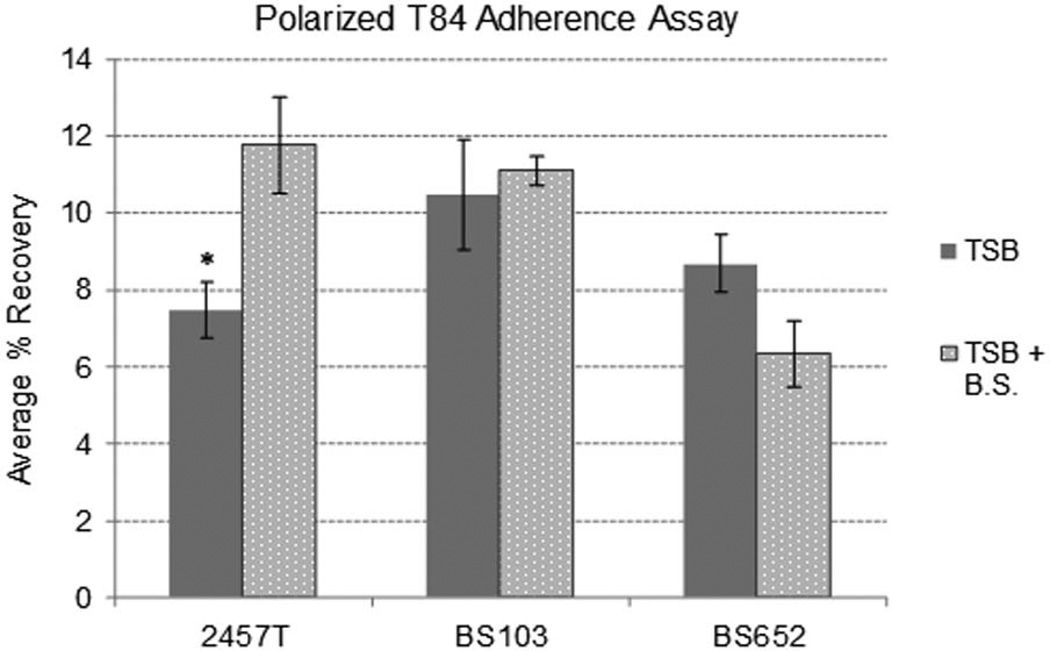
We hypothesized that exposure to bile as Shigella transits through the small intestine serves as an environmental signal to induce the expression of adherence factors. The ability of bile salts to enhance adherence of the bacteria to the apical surface of polarized epithelial cells was assessed by subculturing S. flexneri strain 2457T in tryptic soy broth (TSB) with 0.4% weight-by-volume (w/v) bile salts. Growth curves confirmed that the addition of bile salts did not significantly alter the normal growth kinetics of 2457T (Supplementary Figure 1A). We analyzed adherence to confluent, polarized intestinal T84 cells, which were utilized since the cells are more relevant and mimic the epithelial cells encountered in vivo by S. flexneri (McCormick et al., 1998). 2457T exposed to bile salts during the subculture exhibited a significant and consistent increase in adherence to the apical surface of polarized T84 cells compared to bacteria subcultured in TSB alone (Figure 1). For all adherence assays, the exposure to bile salts occurred for approximately 2.5 hours during subculturing, and no bile salts were added to the tissue culture media during the assay. To determine if the increased recovery of bacteria represented adherence or invasion, gentamicin was added to the polarized cell assay. Only a minimal percentage of infecting bacteria were recovered after gentamicin treatment, ranging from 0.005% to 0.015%, and no significant difference was detected between bacteria pre-treated with or without bile salts. The data support our finding that pre-exposure to bile salts increased adherence at the apical surface, and this increase was not due to invasion. In order to bridge our findings with earlier reports, we also observed an increase in adherence to semi-confluent monolayers of nonpolarized T84 cells (Supplementary Figure 2). These observations agree with previous assays using HeLa cells (Pope et al., 1995).
Figure 1.
Pre-exposure to bile salts increases the adherence of S. flexneri to polarized T84 cells. Bacteria were subcultured with or without 0.4% w/v bile salts and adherence to the apical surface of polarized T84 cells was analyzed. Wildtype S. flexneri strain 2457T consistently had a significant increase in adherence after being exposed to the bile salts during subculture. However, strain BS103 (the virulence plasmid-cured strain of 2457T) and BS652 (the Δspa47 mutant) did not have a significant increase in adherence after exposure to bile salts. Plotted is the average percent (%) recovery of adherent bacteria +/− the standard error for three or more independent, repeated experiments. * = p-value <0.01, as determined by the student’s T-test. The Δspa47 mutant appears to have a decrease in adherence after bile salts exposure, but the p-values were never significant despite extensive analysis. Therefore, we conclude there was no difference in adherence with and without bile salt pre-treatment.
In order to determine if the putative adhesin induced by bile salts was encoded on the chromosome or the virulence plasmid, we analyzed a plasmid-cured strain of 2457T (strain BS103) (Maurelli et al., 1984). Pre-exposure of BS103 to bile salts did not significantly increase adherence to polarized T84 cells when compared to BS103 grown without bile salts (Figure 1). The apparent increase in adherence of BS103 in the absence of bile salts compared to that of 2457T without bile salts is due to the increased growth rate of BS103, which results from a decreased metabolic burden of the plasmid-cured strain (Supplementary Figure 1B). The important observation is that BS103 did not have a significant increase in adherence after exposure to bile salts as was found for 2457T. These data indicated that the putative adhesin induced by bile salts is encoded on the virulence plasmid. We next analyzed a Δspa47 mutant (strain BS652), which lacks only the ATPase that drives the plasmid encoded T3SS. This mutant is unable to invade due to the inability to secrete the T3SS effectors (Venkatesan et al., 1992, Tamano et al., 2000). The Δspa47 mutant also did not manifest a significant increase in adherence after bile salts exposure (Figure 1), indicating that the putative adhesin induced by bile salts is part of or requires a functional T3SS for secretion. Finally, we analyzed a ΔipaD mutant in our polarized T84 adherence assay, since invasion has been shown to be induced through an IpaD interaction with bile salts (Olive et al., 2007, Stensrud et al., 2008). The ΔipaD mutant showed a significant increase in adherence, as was seen for wildtype bacteria (Supplementary Figure 3), demonstrating that the increase in adherence was independent of IpaD.
Microarray analysis identifies candidate adhesin genes
We utilized custom Affymetrix FDA-ECSG arrays to identify candidate adhesin genes induced by bile salts. The microarrays contain probe sets representing 32 E. coli and Shigella strains (Fang et al., 2010). RNA was isolated from 2457T cultures in TSB +/− 0.4% w/v bile salts. Three independent, biological replicates were performed in which 51 induced and 49 repressed genes were significantly altered in the presence of bile salts (Supplementary Table 1). Genes with a greater than 2-fold change and a p-value cutoff of <0.05 were considered significantly altered in the presence of bile salts. Given the fact that bile salts are a stress inducer (Begley et al., 2005, Merritt & Donaldson, 2009), certain results were expected. For example, genes mediating stress responses, such as sodA (superoxide dismutase) and groEL chaperonin (promotes the refolding of proteins under stressful conditions), were induced (Supplementary Table 1). Induction of these genes did not significantly affect the growth rate of the bacteria (Supplementary Figure 1A). Other genes found to be affected by bile salts exposure have not previously been characterized in Shigella. For instance, the entire ccm operon, which includes the genes ccmA through ccmH, was repressed from 2.0 to 3.1-fold (Supplementary Table 1). The roles of these genes in stress responses or other functions remain unknown for Shigella.
With regard to adherence and putative adhesins, some of the results generated from the microarray analysis were surprising. First, previously identified or annotated adhesin genes were not present among the transcriptionally altered genes. Second, the only gene on the virulence plasmid found to be significantly upregulated by bile was ospE1, which was induced 2.8-fold in the presence of bile salts (Supplementary Table 1). Of note, the ospE2 gene, also encoded on the virulence plasmid, is 99% identical to ospE1, and was not present as an independent feature on the array due to this similarity. OspE1 and OspE2 have been previously described as T3SS effector proteins (Miura et al., 2006, Kim et al., 2009, Le Gall et al., 2005); and as a result of this finding, the ospE1 and ospE2 gene products became candidates responsible for the enhanced adherence to T84 cells after bile salts exposure. Additionally, we identified genes in the analysis that were considered potentially relevant since they are genes annotated as putative outer membrane proteins (Supplementary Table 1). For example, yhcN, a putative outer membrane protein and uncharacterized gene encoded on the chromosome, was induced the greatest at 6.3-fold while ybgQ, which encodes another putative outer membrane protein encoded on the chromosome, was decreased 2.1-fold. The induction of yhcN and the repression of ybgQ may also have a role in bile-induced adherence of S. flexneri.
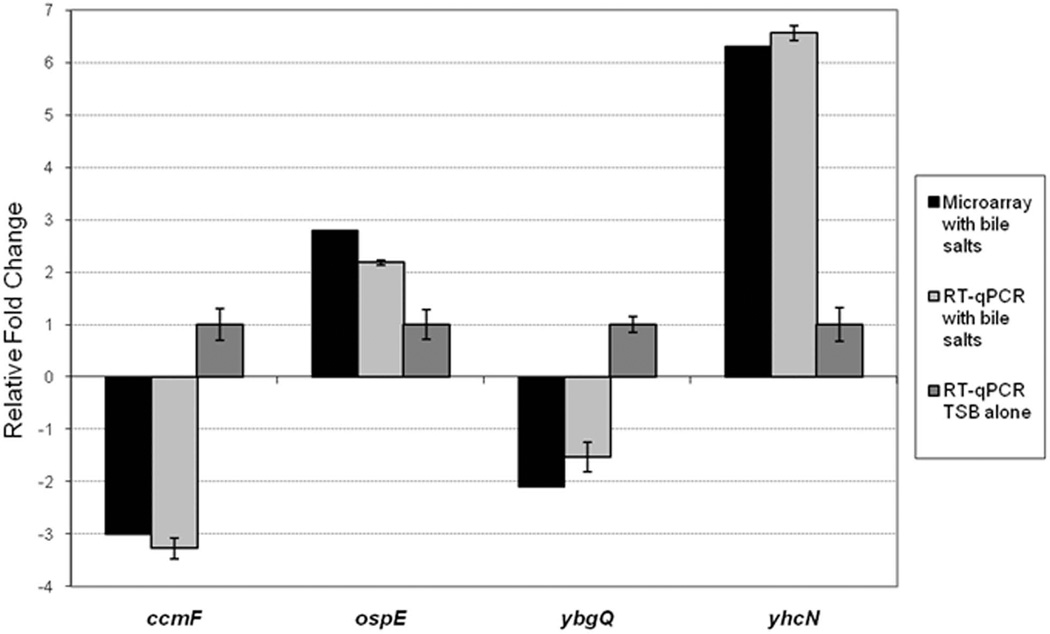
Reverse-transcription quantitative real-time PCR (RT-qPCR) was performed to confirm the results obtained in the microarray analysis (Figure 2). We chose two induced and two repressed genes for the RT-qPCR analysis, and analyzed the RNA used in the three independent microarray experiments. Both ospE1/ospE2 and yhcN were induced in the presence of bile salts while ccmF and ybgQ were repressed in the presence of bile salts. Since the RT-qPCR and microarray results were consistent, we concluded that the relative expression levels determined in the microarray analyses were accurate and significant for ospE1/ospE2, yhcN, ccmF, and ybgQ.
Figure 2.
Reverse transcription quantitative PCR analysis verifies the microarray analysis. RT-qPCR was performed on the 3 independent microarray RNA samples stored at −80°C. The relative fold change +/− the standard error from the ΔCT values is plotted for the four genes. The corresponding changes in expression from the microarray results are plotted in black. “ospE” indicates both ospE1 and ospE2. All p-values were <0.01, except ybgQ, p = 0.065.
OspE1 and OspE2 enhance adherence to polarized T84 cells
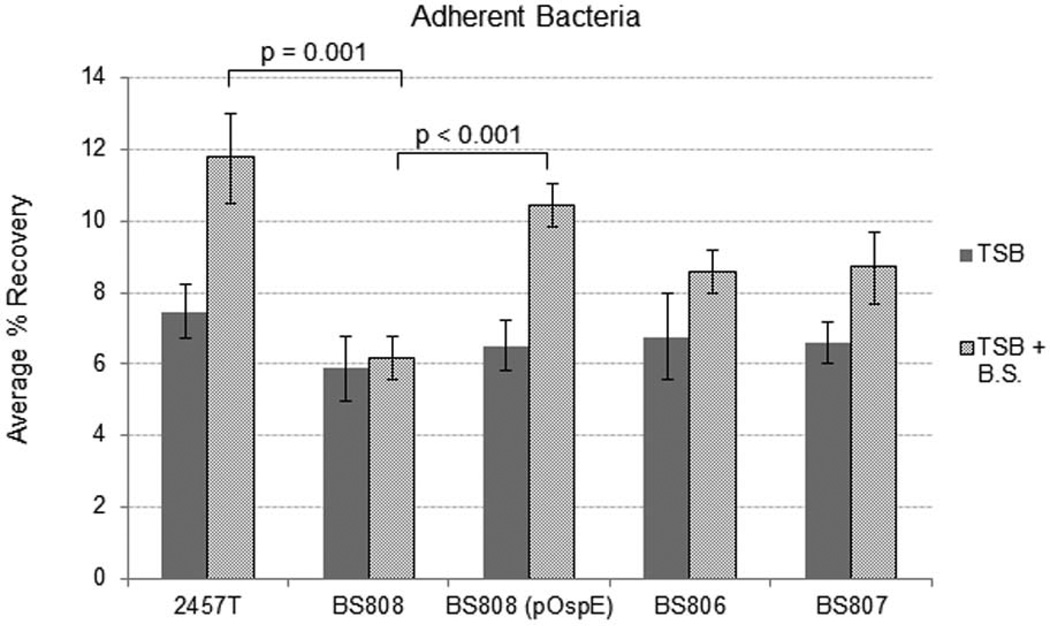
OspE1 and OspE2 were considered to be the most promising candidates for increasing adherence of S. flexneri after exposure to bile salts, given the results obtained from the adherence assays and microarray experiments. We constructed a ΔospE1 and ΔospE2 double mutant (strain BS808) for analysis in the polarized T84 adherence assay. Since ospE1 and ospE2 are 99% identical (only one amino acid difference) and are believed to have resulted from a gene duplication event (Buchrieser et al., 2000), we analyzed the double mutant to ensure there was no redundant function from single gene mutations. As shown in Figure 3, unlike 2457T, the double mutant did not exhibit a significant increase in adherence to the apical surface of polarized T84 after exposure to bile salts. Complementation (BS808 (pOspE)) restored bile-induced adherence to that of wildtype levels (Figure 3). Finally, analysis of the single ΔospE2 mutant (BS806) and the single ΔospE1 mutant (BS807) demonstrated that removal of one of the genes resulted in increased adherence after bile salts exposure, but the increase was not significant when compared to wildtype bacteria (Figure 3). The data confirmed that both OspE1 and OspE2 are involved and required for optimal bile-induced adherence to the apical surface of polarized T84 cells.
Figure 3.
Deletion of ospE1 and ospE2 abolishes the bile-induced increase in adherence. B808, the ΔospE1 + ΔospE2 double mutant, was analyzed for adherence to polarized T84 cells with 2457T as a positive control. 2457T had a significant increase in adherence to the apical surface of T84 cells after being exposed to bile salts during the subculture. However, BS808 did not have a significant increase in adherence after exposure to bile salts. Complementation (BS808 (pOspE)) restored bile-induced adherence. Analysis of the single mutants, BS806 (ΔospE2) and BS807 (ΔospE1), verified that both genes are utilized by S. flexneri to increase adherence following bile salts exposure. The average % recovery for three independent experiments +/- the standard error is plotted. p-values between treatment groups are indicated. There is no significant difference between 2457T and BS808 in the absence of bile salts (p-value = 0.19).
Protein secretion in Shigella is induced after bile salts exposure
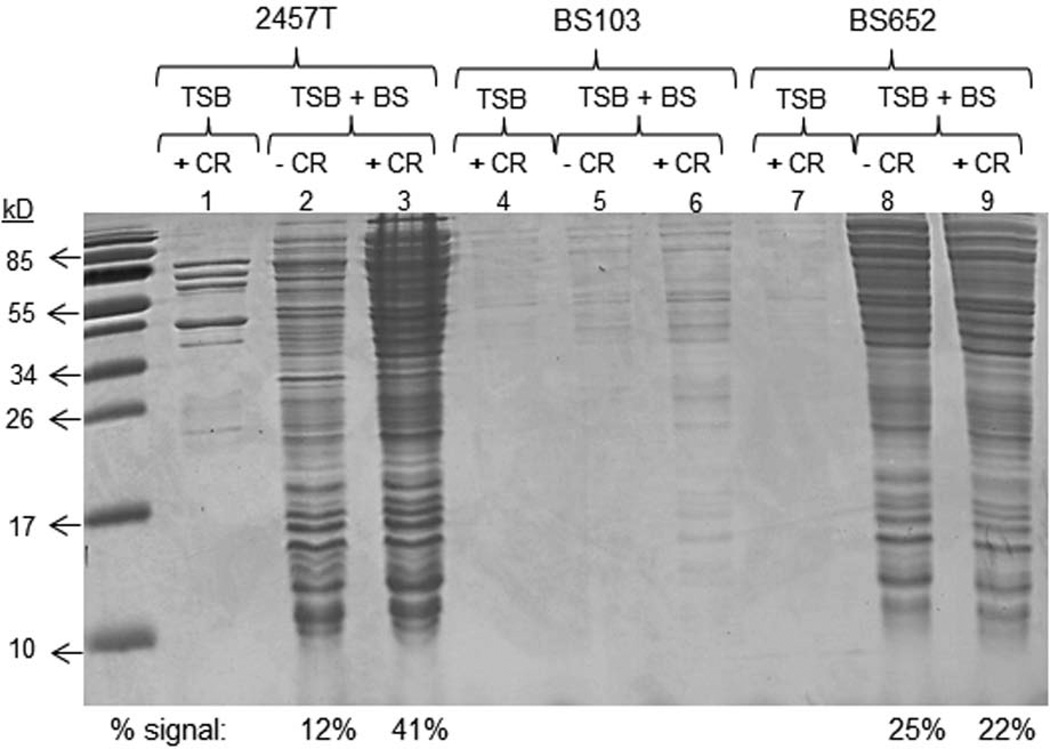
It was previously demonstrated that protein secretion from Shigella is increased following bile salts exposure (Pope et al., 1995). We therefore wanted to determine if the bile-induced protein secretion was dependent on the T3SS. Bacteria were subcultured with or without bile salts as described in the Experimental Procedures. The bacteria were subsequently washed, resuspended in 1X phosphate buffered saline (PBS) with or without Congo red (CR), and incubated at 37°C to measure protein secretion. As shown in Figure 4, wildtype 2457T subcultured in bile salts had a significant increase in the total amount of protein secreted, which was subsequently induced to higher levels by CR treatment. CR is known as an inducer of the T3SS (Bahrani et al., 1997). Secretion was significantly reduced in BS103 compared to 2457T as expected since the strain lacks the virulence plasmid-encoded T3SS; however, some limited secretion was detected in the samples treated with bile salts. The Δspa47 mutant (BS652) had a secretion profile similar to wildtype 2457T after bile salts treatment, but there was no induction with CR as expected since the T3SS is not functional in this mutant. Densitometry analysis of 2457T treated with bile salts and CR induction (lane 3) compared to the Δspa47 mutant under the same conditions (lane 9) revealed that there was an approximate two-fold increase in protein secretion for 2457T (Figure 4). These results demonstrated that the T3SS was induced by bile salts. Both microscopy and the interrogation of cytoplasmic proteins confirmed that the observed induced protein secretion in Figure 4 was not due to bacterial lysis (Supplementary Figures 4 and 5).
Figure 4.
Protein secretion is increased in the presence of bile salts. Supernatants from wildtype S. flexneri strain 2457T, the plasmid-cured strain BS103, and the Δspa47 mutant (BS652) subcultured in tryptic soy broth (TSB) with or without 0.4% w/v bile salts (B.S.) and with or without Congo red (CR) were analyzed on a denaturing protein gel that was subsequently stained with Coomassie blue. For 2457T, more proteins are secreted after exposure to bile salts, which is induced with the addition of CR. BS103 has minimal secretion in all conditions. The Δspa47 mutant had no significant secretion in the absence of bile salts as expected. However, bile salts increased secretion, and comparable levels were obtained irrespective of CR treatment. Densitometry analysis compared the percent of the total signal from the following four conditions: 2457T + B.S. (lane 2), 2457T + B.S. + CR (lane 3), BS652 + B.S. (lane 8), and BS652 + B.S. + CR (lane 9). The percent of the signal for the comparison is displayed below the gel.
OspE1 and OspE2 are secreted through the T3SS but remain associated with the bacterial outer membrane after exposure to bile salts
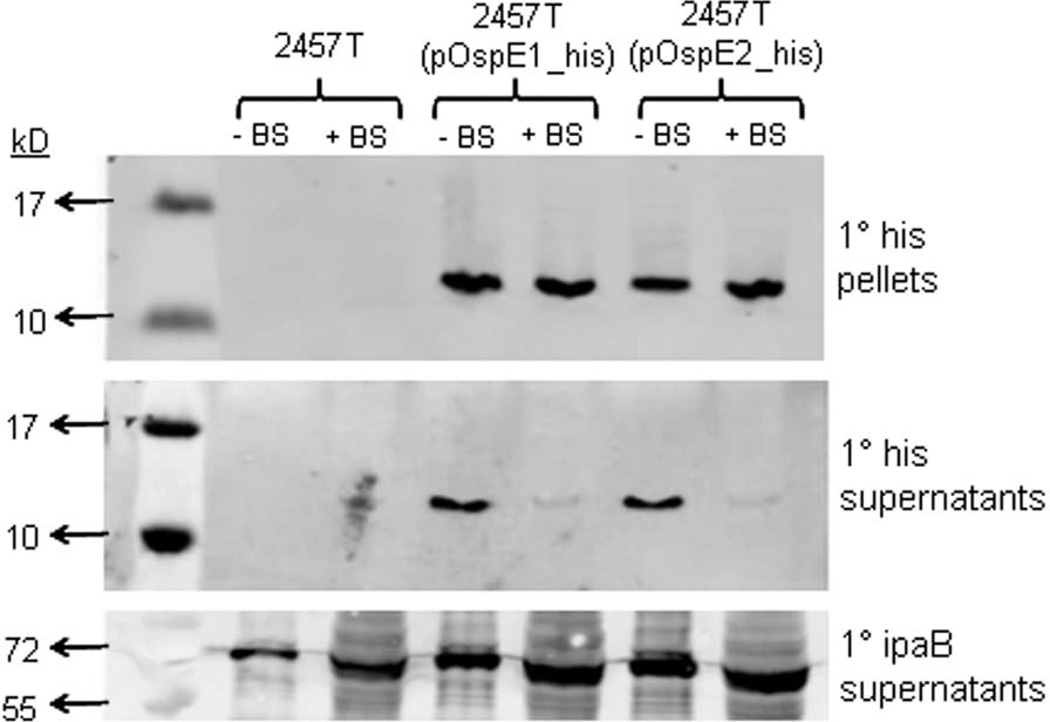
We next wanted to analyze the secretion of OspE1 and OspE2 following bile salts exposure. We constructed C-terminal histidine tagged fusions for OspE1 and OspE2, in which expression was driven by the native promoters (see Experimental Procedures). The plasmids were transformed into 2457T to generate 2457T(pOspE1_his) and 2457T(pOspE2_his). Next, we grew 2457T(pOspE1_his) and 2457T(pOspE2_his) in the presence or absence of bile salts, and subsequently performed the CR secretion assay to analyze T3 secretion. As demonstrated in Figure 5, the his-tagged OspE1 and OspE2 were detected in the pellet fraction in bacteria grown in the presence and absence of bile salts, indicating expression of the tagged proteins from the pGEMT vector in both growth conditions. However, in the supernatants of the CR assay, there was a dramatic difference in the amount of protein detected. After bile salts exposure, only faint bands were detected for OspE1 and OspE2; however, prominent bands were detected in the supernatants when the bacteria were subcultured without bile salts. The data suggested that OspE1 and OspE2 remained associated with the bacteria in the presence of bile salts. Wildtype 2457T (without the tagged constructs) served as a negative control for the primary his antibody to ensure there was no cross-reaction of the antibody with any Shigella proteins. The secretion profile of the invasin IpaB is also shown in Figure 5. Unlike OspE1 and OspE2, IpaB was detected in the supernatants of bacteria grown in both the presence and absence of bile salts. In addition, there was more secretion of IpaB in bacteria exposed to bile salts, verifying that T3 secretion was induced by bile salts.
Figure 5.
OspE1 and OspE2 are not secreted into supernatants after S. flexneri is subcultured in 0.4% w/v bile salts (BS). Bacteria were subcultured with or without bile salts and subsequently incubated in the Congo red (CR) secretion assay. Proteins fractions were processed and analyzed via Western blot analysis using anti-his or anti-IpaB.
Top panel: Pellet fractions from the CR assay demonstrate that the histidine-tagged OspE1 and OspE2 were expressed in 2457T from pOspE1_his or pOspE2_his. No proteins were detected in the negative control 2457T samples.
Middle panel: The supernatants from the CR assay were probed with the anti-his antibody. The tagged versions of OspE1 and OspE2 were secreted (found in the supernatant) when the bacteria were grown in TSB alone. When the bacteria were subcultured with bile salts, no significant secretion into supernatants was detected. No proteins were detected in the negative control 2457T samples.
Bottom panel: The supernatants from the CR assay were probed with the anti-IpaB antibody. IpaB secretion can be detected in all samples. Also, more IpaB was present in the supernatants from bacteria pre-treated with bile salts.
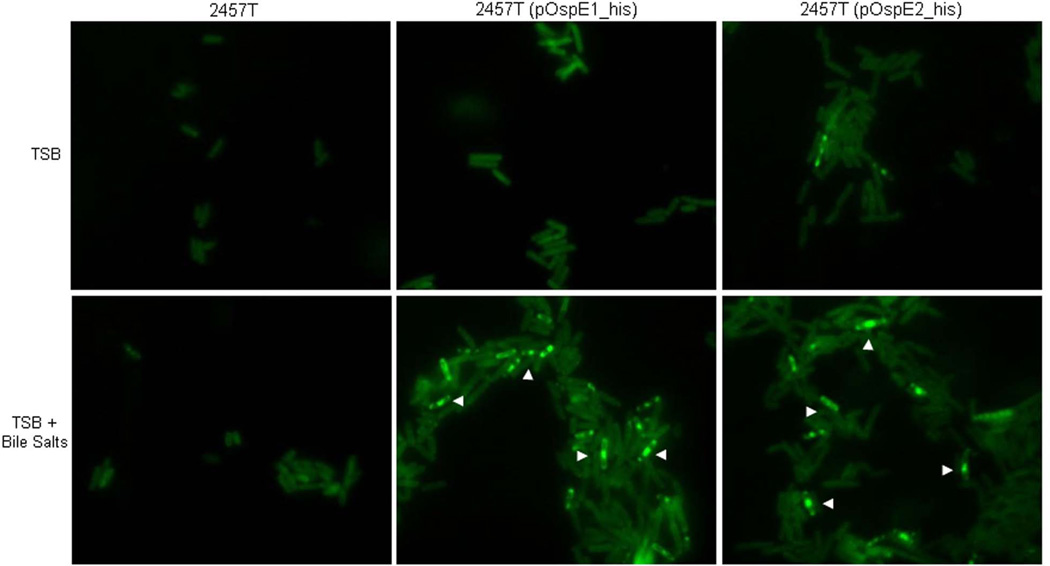
In order to demonstrate that OspE1 and OspE2 associated with the surface of the bacteria after exposure to bile salts, we performed immunofluorescence analysis on the bacteria recovered from the CR assay. To visualize the tagged proteins on the bacterial surface, we chose not to permeabilize the bacteria in the immunofluorescence analysis. Wildtype 2457T served as a negative control and had minimal, background staining (Figure 6). 2457T(pOspE1_his) and 2457T(pOspE2_his) cultured in TSB alone had greater background staining due to the high copy nature of the plasmid expressing the tagged version of OspE1 and OspE2. However, when 2457T(pOspE1_his) and 2457T(pOspE2_his) were subcultured in the presence of bile salts, bright and punctate staining could be visualized on the surface of the bacteria (Figure 6). The data indicated that OspE1 and OspE2 remain associated with the bacterial surface after exposure to bile salts.
Figure 6.
OspE1 and OspE2 are localized to the bacterial surface in the presence of bile salts. Bacteria were prepared as described for the Congo red secretion assay. Afterwards, bacteria were washed, pelleted onto coverslips, fixed, and stained for immunofluorescence analysis (see Experimental Procedures). Top panels are bacteria subcultured in TSB alone, bottom panels are bacteria subcultured in TSB with bile salts. The negative control 2457T had minimal background staining under both conditions. Bacteria harboring pOspE1_his or pOspE2_his had more background than the negative control due to the high copy nature of the plasmid. After subculturing 2457T(pOspE1_his) or 2457T(pOspE2_his) in bile salts, bright and punctate staining above background can be seen on the surface of the bacteria. Arrowheads point to general areas in which the punctate staining can be seen on several bacterial cells.
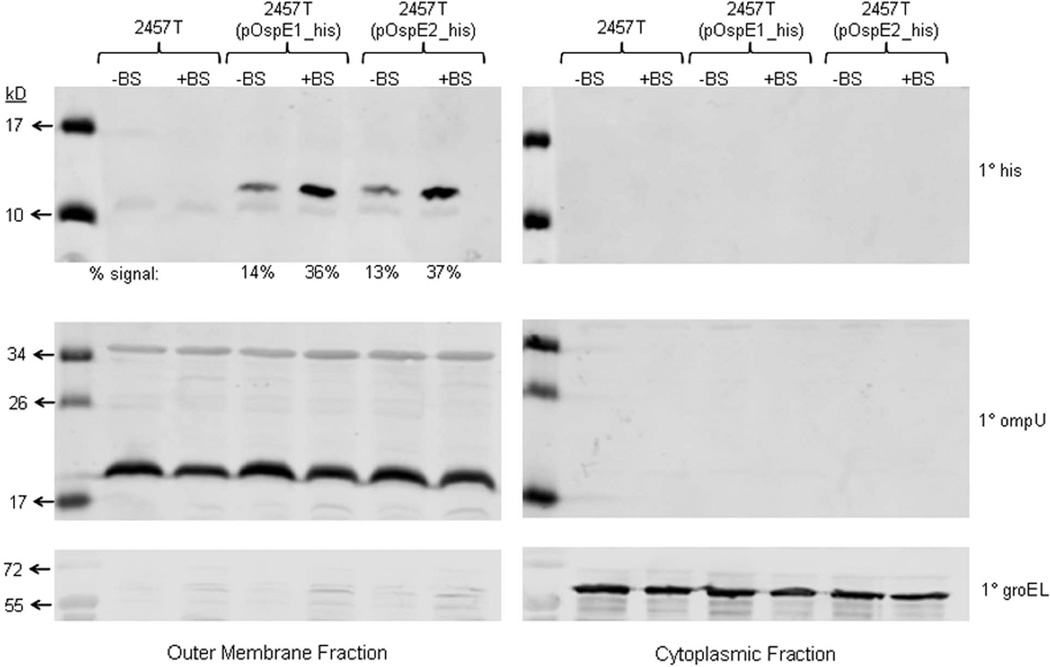
Finally, to demonstrate that OspE1 and OspE2 are localized to the outer membrane of the bacteria following bile salts exposure, we performed fractionation using ultracentrifugation to enrich for the bacterial outer membrane (OM) extracted from the bacterial pellets collected after the CR assay. The Coomassie-stained protein gels for the enriched OM and cytoplasmic fractions are provided in Supplementary Figure 6. In the OM fraction, the banding pattern was similar to the profile obtained from previous OM preparations of S. flexneri (Mukhopadhaya et al., 2006), with predominant bands localized between 34 and 43 kD. Next, we performed Western blot analysis using the anti-his antibody to confirm the OM location of the his-tagged OspE1 and OspE2 in samples pre-exposed to bile salts. As shown in Figure 7, we observed an almost three-fold increase in signal for the tagged proteins in the samples following treatment with bile salts. The densitometry analysis is provided below the blot. The data confirmed that following bile salts exposure, more OspE1 and OspE2 are localized to the bacterial outer membrane. As a control for OM proteins, we probed identical blots with antibodies to V. cholerae OmpU (Figure 7). Blastp analysis of the V. cholerae OmpU protein sequence revealed that S. flexneri has several proteins, ranging in size from 20 kD to 40 kD, that are annotated as outer membrane proteins and have homology to OmpU. The results indicated that we had indeed enriched for outer membrane proteins in the OM fraction. A final control utilized an antibody against the cytoplasmic protein GroEL (60 kD). The signal was localized to the cytoplasmic fraction with no significant signal above background in the OM fraction (Figure 7).
Figure 7.
OspE1 and OspE2 are localized to the bacterial outer membrane after exposure to bile salts. Bacterial pellets were collected after the Congo red secretion assay and the outer membrane (OM) proteins were obtained by ultracentrifugation. The proteins were then analyzed with the anti-his, anti-OmpU, and anti-GroEL antibodies. The OM fraction analysis is on the left while the cytoplasmic fraction analysis is on the right.
Top panel: Analysis of the fractions with the anti-his antibody indicated that more OspE1 and OspE2 were localized to the OM following bile salts exposure. The densitometry analysis is provided below the blot. No signal was detected in the 2457T negative control samples as expected.
Middle panel: Analysis of the OM and cytoplasmic fractions with the anti-OmpU antibody indicated that the OM fractions were enriched for OM proteins in all strains analyzed.
Bottom panel: The anti-GroEL antibody was utilized as a control for the cytoplasmic fractions. A strong signal was obtained for all strains in the cytoplasmic fraction.
Discussion
Previous studies established that bile salt-induced adherence was independent of invasion, but an adherence factor was never identified (Pope et al., 1995). Hence, the goal of this study was to identify the adhesins induced by bile salts. In this study, we demonstrate for the first time that expression of the plasmid-encoded loci ospE1 and ospE2 are induced by bile salts, and that the proteins are localized to the bacterial outer membrane after exposure to bile salts. This localization enhances adherence of S. flexneri to the apical surface of colonic epithelial cells. The 0.4% w/v exposure to bile salts is physiologically relevant since the concentration of bile salts in the small intestine range from 0.2% to 2% w/v depending on diet and the individual (Kristoffersen et al., 2007). The polarized T84 assay was employed in order to utilize an in vitro model that mimics the epithelial cells encountered by the bacteria upon entering the colon. The exposure of bacteria to bile salts prior to the adherence assay was a vital step since the addition of bile salts to the tissue culture media resulted in no significant difference in the recovery of bacteria (data not shown). Therefore, the subculture conditions resulted in the induction of the putative adhesin prior to contact with the epithelial cells. Gentamicin was added to polarized cells to ensure that the difference in adherence was not due to a significant difference in invasion at the apical surface. Apical surface invasion is minimal for S. flexneri since the basolateral surface allows for more efficient invasion of polarized cells (Mounier et al., 1992); however apical binding may be one of the first steps in the invasion process of S. flexneri.
The lack of increased adherence of a plasmid-cured strain (BS103) and a strain with a nonfunctional T3SS (Δspa47) following bile salts exposure allowed the determination that the putative adhesins induced by bile salts were plasmid-encoded. Spa47 is the ATPase that drives the T3SS, and thus a Δspa47 mutant is unable to secrete the T3SS effector proteins (Tamano et al., 2000, Venkatesan et al., 1992). The results of the BS103 and Δspa47 analysis demonstrated that a protein encoded on the virulence plasmid requires the T3SS to be secreted in order to function as an adhesin. We analyzed a ΔipaD mutant since IpaD is a T3SS effector protein encoded on the virulence plasmid that has been shown to bind the bile salt deoxycholate, which increases invasion of the bacteria (Olive et al., 2007, Stensrud et al., 2008). The doubling of adherence of the ΔipaD mutant to the apical surface of T84 cells after bile salts exposure further confirmed that the increase in adherence was independent of the effects of bile salts on IpaD and invasion.
The microarray results identified ospE1 as the only gene on the virulence plasmid induced by bile salts; and therefore, we surmised that the ospE1 and ospE2 genes (having 99% identity) encoded the T3SS effector proteins required for the increase in adherence. Mutant analysis confirmed that both ospE1 and ospE2 were required for the bile-induced adherence. Individual ΔospE2 (BS806) and ΔospE1 (BS807) single mutants had increased adherence following bile salts exposure, but the increase in adherence was less that observed for wildtype bacteria. It is interesting that the adhesive phenotype appears to be additive for the two copies of ospE. More importantly, increased adherence was abolished in the ΔospE1 and ΔospE2 (BS808) double mutant following bile salts exposure. The adherence of the double mutant was comparable to that of BS103 and the Δspa47 mutant. Complementation of the double mutation restored bile-induced adherence to wildtype levels. These data support a role for the plasmid-encoded OspE1 and OspE2 factors in adherence.
In order to function as adhesins, we hypothesized that OspE1 and OspE2 would be present on the outer surface of the bacteria to associate with the apical surface of T84 cells. Previous studies have demonstrated that OspE1 and OspE2 are T3SS effector proteins that play a critical role in the maintenance of epithelial cell integrity once Shigella invades epithelial cells (Kim et al., 2009, Miura et al., 2006, Le Gall et al., 2005). The authors observed that the ΔospE mutants exhibited a cell rounding phenotype that led to detachment of infected cells. The fact that our mutant analyses were performed in polarized assays, in which the cells are confluent and apical surface invasion has not been detected, discounts the possibility that the lack of induced adherence in our experiments could be a result of detachment of rounded, infected cells. The inability of the Δspa47 mutant to demonstrate bile-induced adherence verifies that not only OspE1 and OspE2 are secreted via the T3SS, but also the T3SS is required for surface localization of these bile-induced adherence factors.
To confirm that OspE1 and OspE2 are localized to the bacterial outer membrane after bile salts exposure, we analyzed the secretion and outer membrane localization of the proteins following bile salts exposure. First, in the absence of bile salts, OspE1 and OspE2 were expressed; but instead of remaining associated with the bacterial outer membrane, they were found in the supernatants in the CR assay. The IpaB analysis in the CR assay verified that proteins being secreted through the T3SS were not altered simply due to the exposure to bile salts. Expression of IpaB is not affected by bile salts and it can be secreted similarly into the supernatant after subculturing in both conditions. In addition, the IpaB secretion analysis confirmed that type-III protein secretion is increased after exposure to bile salts. The observed increase in secretion is not due to bacterial lysis (Supplementary Figures 4 and 5). In addition to the induced T3 secretion, it appears that non-T3 secretion is induced as well, and these results agree with previous literature (Pope et al., 1995). Since the non-T3 secretion is not induced in the plasmid-cured strain, we hypothesize that a factor encoded on the virulence plasmid is affecting this protein secretion. While the non-T3 secretion warrants further investigation, our results confirm that bile salts are an inducer of T3 secretion. Secondly, the immunofluorescence experiments confirmed that OspE1/2 were localized to the bacterial surface and were visualized in a pattern that was similar to that found for autotransporters associated with the outer membrane (Jain et al., 2006, Wagner et al., 2009). Finally, the membrane fractionation procedure confirmed that the proteins had induced localization to the bacterial outer membrane following bile salts exposure. Therefore, we conclude that OM localization of OspE1 and OspE2 is due to the induced expression of the genes and likely linked to the induced T3 secretion following bile salts exposure.
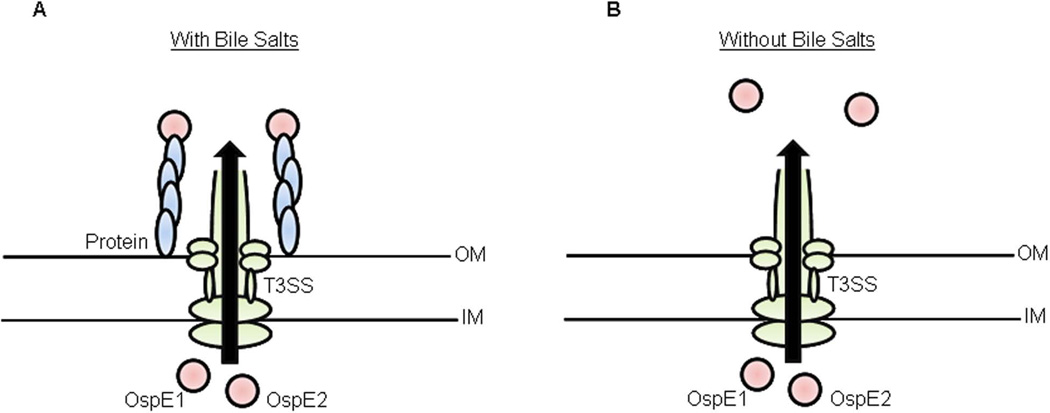
Based on these results, we propose the model presented in Figure 8. As the bacteria transit through the small intestine and into the colon, exposure to bile salts induces ospE1 and ospE2 gene expression. Simultaneously or soon thereafter, T3 secretion is induced to affect secretion of effector proteins and to permit outer membrane localization of OspE1 and OspE2, which enhances bacterial adherence to the apical surface of colonic epithelial cells. While the T3SS is induced by host cell contact (Schroeder & Hilbi, 2008), secretion occurs at a basal level without such contact or other chemical T3 inducers. Several publications have shown secretion of T3 effectors at 37°C in PBS or broth without inducers (Bahrani et al., 1997, Faherty & Maurelli, 2009, Pope et al., 1995). Given these data, T3 secretion most likely occurs in the gut lumen prior to host cell contact. Our data support the hypothesis that T3 effectors are required for pre-invasion virulence.
Figure 8.
Model of the differential functional roles for OspE1 and OspE2 due to bile exposure.
A: As the bacteria transit through the small intestine and into the colon, bile exposure induces the expression and/or secretion of a protein on the outer membrane (OM) of the bacterial cell. OspE1 and OspE2 are subsequently secreted through the T3SS, and bind to the surface of this additional protein. The protein interactions allow the bacteria to bind to the apical surface of human epithelial cells.
B: As the bacteria invade the epithelial cells, exposure to bile is lost. The additional protein is no longer present on the outer membrane of the bacteria. When OspE1 and OspE2 are secreted into the cytoplasm of the host cell, the proteins remain free from the bacterial outer membrane and can bind to integrin-linked kinases to maintain epithelial cell morphology during infection. IM, inner membrane.
As shown in figure 8, we hypothesize that outer membrane localization of OspE1 and OspE2 requires an additional protein that is normally not present (or minimally present) in the absence of bile salts. Another possibility is that bile induces a conformational change in OspE1 and OspE2 that allows more efficient binding to this additional protein to form a protein complex. This adhesin complex subsequently binds to the apical surface of colonic epithelial cells to facilitate adherence. As the bacteria transit through M cells and invade the colonic epithelial cells, the exposure to bile salts is lost. Therefore, the protein that binds OspE1 and OspE2 to the outer membrane is no longer present in sufficient quantities, which allows OspE1 and OspE2 to be secreted via the T3SS into the host cell cytoplasm to maintain epithelial cell integrity during infection (Kim et al., 2009, Miura et al., 2006). Finally, it is important to mention the possibility that bile salts induce a conformational change in OspE1 and OspE2 that could result in an association of the proteins with the lipopolysaccharide (LPS) chains on the OM. We favor the hypothesis that a protein on the OM is binding OspE1 and OspE2 due to the complementation data. We complemented bile-induced adherence to wildtype levels utilizing the high copy pGEMT vector. If bile salts induced a conformational change in OspE1/2 to result in an interaction with LPS, one could speculate that higher levels of adhesion would be observed following complementation with a high copy number plasmid. However, the presence of a greater amount of OspE1/2 did not increase bile-induced adherence above wildtype levels, possibly because an unknown interacting protein remained at limiting quantity. If this unknown protein were induced on a high copy plasmid with OspE1 and OspE2, it is possible that adherence levels would reach above wildtype levels.
Future protein-protein or protein-sugar interaction studies will identify the additional protein or mechanism involved in localizing OspE1 and OspE2 to the bacterial outer membrane after exposure to bile salts. In addition, protein interaction studies with eukaryotic targets will potentially identify the receptor on the apical surface of T84 cells that binds OspE1 and OspE2. The OspE1/2 proteins have already been shown to interact with integrin-linked kinases (Kim et al., 2009), and other eukaryotic proteins are certainly possible. Despite any alternative mechanisms to our protein interaction hypothesis, OspE1 and OspE2 clearly have a role in adherence since the double mutant is unable to exhibit a significant increase in adherence in the T84 assay after exposure to bile salts. Other genes identified in the microarray results, such as yhcN induction or the ccm operon repression, could be important for OspE1 and OspE2 localization to the outer membrane. Future studies will shed light on the interplay of OspE1 and OspE2 with other bacterial proteins or outer membrane structures.
Finally, it is important to note that OspE1 and OspE2 most likely are not the only proteins involved in adherence to epithelial cells. The plasmid-cured strain of Shigella, strain BS103, retained measureable adherence in the polarized T84 assay. In fact, we have yet to identify a mutant that is completely unable to adhere to epithelial cells; and therefore, other adhesins almost certainly play a functional role. Bile salts did not affect the expression of the genes encoding these adhesins, and therefore, were not identified in the microarray analysis. Nevertheless, the data presented here clearly demonstrate that OspE1/2 are induced by bile salts and enable the bacteria to adhere more efficiently to the apical surface of colonic epithelial cells. OspE1 and OspE2 are found in all Shigella species in either one or two copies. In addition, the genes are highly conserved in pathogenic strains of E. coli, Salmonella, and Citrobacter (Miura et al., 2006, Kim et al., 2009). OspE1 and OspE2 could therefore serve as a new set of targets for therapeutics and vaccine candidates. In addition, investigating the role of bile in activating the full virulence potential of S. flexneri could enhance our understanding of this important pathogen, and lead to improved animal models and methods to develop an effective vaccine.
Experimental Procedures
Strains and growth conditions
Table 1 lists the bacterial strains and plasmids used in this study. Bacteria were routinely cultured at 37°C either in tryptic soy broth or Luria-Bertani broth with aeration, or on tryptic soy broth plates with 1.5% agar and 0.025% Congo Red (CR; Sigma). Antibiotics were used at the following concentrations: kanamycin, 50 µg/ml; streptomycin, 50 µg/ml; chloramphenicol, 5 µg/ml; and ampicillin, 100 µg/ml. Bile salts (Sigma) consisted of a mixture of 50% cholate and 50% deoxycholate, and were used at a concentration of 0.4% weight-by-volume (w/v).
Table 1.
Strains and plasmids used in this study.
| Name | Description | Source or reference |
|---|---|---|
| Strains | ||
| 2457T | Wildtype S. flexneri 2a | (Formal et al., 1958) |
| M90T | Wildtype S. flexneri 5 | (Sansonetti et al., 1982) |
| BS103 | Plasmid-cured 2457T | (Maurelli et al., 1984) |
| BS569 | M90T/ipaD2::aphA-3 | (Menard et al., 1993) |
| BS652 | 2457T/spa47::aadA | (Faherty & Maurelli, 2009) |
| BS766 | 2457T transformed with pKM208 | (Faherty & Maurelli, 2009) |
| BS806 | 2457T/ospE2::cat | ATM; this study |
| BS807 | 2457T/ospE1::aphA-3 | ATM; this study |
| BS808 | 2457T/ospE1::aphA-3 + ospE2::cat | ATM; this study |
| BS808 (pOspE) |
BS808 transformed with pOspE+ | This study |
| 2457T (pOspE1_his) |
2457T transform ed with pOspE1_his | This study |
| 2457T (pOspE2_his) |
2457T transformed with pOspE2_his | This study |
| Plasmids | ||
| pKD3 | oriR6K, bla, cat | (Datsenko & Wanner, 2000) |
| pKD4 | oriR6K, bla, aphA-3 | (Datsenko & Wanner, 2000) |
| pKM208 | Temperature-sensitive red-, gam -, lacI- expressing plasmid driven by PTac promoter, bla | (Datsenko & Wanner, 2000) |
| pGEMT | PCR cloning vector, bla, high copy number | Promega |
| pCSF1 | pGEMT::ospE1, wildtype gene cloned into pGEMT | This study |
| pCSF2 | pGEMT::ospE2, wildtype gene cloned into pGEMT | This study |
| pOspE+ | pGEMT::ospE, wildtype gene cloned into pGEMT | This study |
| p(OspE1_his) | pGEMT:: ospE1, ospE1 with C-terminal his tag cloned into pGEMT | This study |
| p(OspE2_his) | pGEMT::ospE2, ospE2 with C-terminal his tag cloned into pGEMT | This study |
Strain construction
The ΔospE1 and ΔospE2 double mutant (strain BS808) was constructed using the λ red linear recombination method as previously described (Faherty & Maurelli, 2009). Since the genes are 99 percent identical, the following modifications were performed to ensure that both genes were deleted. PCR was used to amplify a chloramphenicol resistance cassette gene (cat) from pKD3 (Table 1) which had 5’ and 3’ overhangs identical to the 5’ and 3’ regions of ospE2. Chloramphenicol resistant recombinants were purified and screened via PCR using primers (Supplementary Table 2) that annealed to unique regions up and downstream of ospE2 to detect the size difference due to the insertion of the chloramphenicol cassette. This mutant was used as the donor strain for transduction of 2457T using P1L4, and selection for chloramphenicol resistance generated BS806.
BS806 was retransformed with pKM208 to generate a ΔospE1 mutation. PCR was used to amplify the kanamycin resistance cassette gene (aphA-3) from pKD4 (Table 1) with 5’ and 3’ overhangs identical to the 5’ and 3’ regions of ospE1 internal to the start and stop codons (Supplementary Table 2). These regions of homology were absent in the ospE2 region due to the addition of the chloramphenicol cassette. Kanamycin resistant recombinants were purified and screened via PCR using primers (Supplementary Table 2) that annealed to the unique regions up and downstream of ospE1 to detect the size difference due to the insertion of the kanamycin cassette. This mutant was used as the donor strain for transduction of 2457T using P1L4. Selection for kanamycin yielded BS807 while selection for kanamycin plus chloramphenicol yielded BS808.
Complementation was performed by amplifying the ospE gene and the promoter region by PCR with high fidelity Taq polymerase (Invitrogen) from wildtype 2457T with primers listed in Supplementary Table 2. The procedure was similar to what has been previously described (Kim et al., 2009). The PCR product was purified and ligated into the pGEMT cloning vector (Promega) according to manufacturer’s protocol to generate pOspE+. The ligation reaction was transformed into E. coli DH5α and grown on LB plates with 100 µg/ml ampicillin. Single colonies were restreaked, and Qiagen’s miniprep kit was used to collect the plasmid DNA. The insert was sequenced with primers that anneal outside the multiple cloning site (MCS) for pGEMT and are listed in Supplementary Table 2. After sequence verification, the plasmid was transformed into BS808 and selected on chloramphenicol and ampicillin. Positive colonies were restreaked and PCR verified presence of the insert. The resulting complementation strain was named BS808 (pOspE).
OspE1/OspE2-tag construction
The primers used to construct the C-terminal histidine tags are listed in Supplementary Table 2. First, the genes were amplified by PCR with high fidelity Taq polymerase (Invitrogen) from wildtype 2457T and ligated into the pGEMT cloning vector (Promega) according to manufacturer’s protocol to generate pCSF1 and pCSF2. For this initial cloning step, primers that annealed to unique regions outside of ospE1 and ospE2 were used to ensure the individual genes were cloned as opposed to a mixed population (Supplementary Table 2). pCSF1 and pCSF2 were subsequently used as templates to generate new PCR products for ospE1 and ospE2 in which the 6X his tag was added to the C-termini, followed by a stop codon (Supplementary Table 2). In addition, the native promoter region was present in this construct. The PCR products were again ligated into pGEMT to generate pOspE1_his and pOspE2_his, and both plasmids were subsequently transformed into 2457T. Selection for positive transformants occurred on tryptic soy broth plates with 1.5% agar, 0.025% CR, and 100 µg/ml ampicillin. Sequencing was performed to ensure no mutations were introduced during the cloning process. The primers used for the sequencing annealed outside of the MCS for pGEMT and are listed in Supplementary Table 2. Sequencing was performed, and template DNA was prepared from PCR amplification of the inserts using high fidelity Taq polymerase.
Intestinal T84 cell maintenance and polarization
T84 cells were maintained in DMEM-F12 with 10% fetal bovine serum with penicillin and streptomycin to inhibit bacterial contamination as previously described (Harrington et al., 2005). For polarization of cells, 12-well polycarbonate Transwell filters (Corning) with a 0.4 µM pore were coated with 0.5 mg/ml rat tail collagen (Sigma) and seeded at 1 × 105 cells/filter as previously described (Harrington et al., 2005). Polarization occurred for 10 to 14 days, and polarized cells were used when the transepithelial electrical resistance reached 1000 – 2000 Ω/cm2, which indicates confluency and polarization of the cells. Transepithelial resistance was monitored with an EVOM-G Ohmeter (World Precision Instruments).
Polarized T84 adherence assay
S. flexneri was subcultured in tryptic soy broth (TSB) with or without 0.4% w/v bile salts until the OD600 reached mid-log phase (approximately 0.7). After standardization to an OD600 of 0.35 (which corresponds to approximately 1.0 × 108 CFU/ml), the bacteria were washed with 1X PBS and resuspended in 1 ml tissue culture media (DMEM/F12). The apical media from the polarized T84 cells was removed and 100 µl of the bacteria was applied to the apical surface. The tissue culture plate was then incubated at 37°C with 5% CO2 for 3 hours. Afterwards, 75 µl of the apical media was removed and serial dilutions were made to determine bacterial titers. This portion of bacteria was considered the nonadherent population. Next, the basolateral media was recovered and plated to ensure there was no bacterial contamination or the possibility of invasion at the basolateral side of the cells. The T84 cells were then washed with 1X PBS and lysed with 1% triton-x-100. Serial dilutions were made to determine bacterial titer, and this portion represented adherent bacteria. For all bacterial titers the percent recovery was calculated as [recovered bacterial titer/infecting titer]×100%. Statistical significance was determined by Student’s t-test, and a p-value of ≤ 0.05 was considered significant.
T84 monolayer adherence and invasion assay
In order to analyze adherence or invasion using nonpolarized cells, T84 cells were seeded in a 6-well tissue culture plate to establish a semi-confluent monolayer (approximately 75% confluency was routinely reached). Bacteria were subcultured with or without bile salts, standardized, and washed as described above. The bacteria was resuspended in 1 ml of tissue culture media and placed onto the T84 monolayers (media removed). The tissue culture plates were incubated at 37°C with 5% CO2 for 3 hours. The monolayers were washed with 1X PBS and subsequently lysed with 1% triton-x-100. Serial dilutions were made to determine the amount of cell-associated bacteria, in which the percent recovery was calculated as described above. In order to determine the number of bacteria that invaded the monolayers during the 3-hour incubation, a tissue culture plate was infected and incubated in parallel with the adherence assay. After the 3 hour incubation, the monolayers were washed with 1X PBS and DMEM/F12 media with 50 µg/ml gentamicin was added to the monolayers for a total of 45 minutes (15 minute incubation and wash was followed by 30 minute incubation with fresh media plus gentamicin to ensure removal of all extracellular bacteria). Afterwards, the T84 cells were thoroughly washed and lysed, and serial dilutions were performed to determine the percent of recovered bacteria. Statistical significance was determined by Student’s t-test, and a p-value of ≤ 0.05 was considered significant.
Microarray analysis
In order to identify candidate adhesin genes, microarray analysis was performed on RNA isolated from 2457T grown in TSB +/− 0.4% w/v bile salts. Three independent, biological replicates were performed. Bacterial RNA was isolated using the RNA RiboPure kit (Ambion) according to manufacturer’s protocol, including DNase treatment. Concentrations of total RNA were determined using a NanoDrop ND-1000 spectrophotometer. cDNA was synthesized from total RNA using Superscript III First Strand Synthesis kit (Invitrogen) according to manufacturer’s protocol. Afterwards, template RNA was degraded by incubating the samples at 65°C for 30 minutes with 1N NaOH, which was subsequently neutralized with 1N HCl. Sample cleanup was performed with the MinElute PCR Purification kit and protocol (Qiagen). cDNA probes were prepared by partial digestion of cDNA using the DNA-free DNase kit (Life Sciences, Inc.), optimized for a fragment range of 20–200 bases. The sample ranges were verified by gel electrophoresis on a 4 – 20% polyacrylamide gradient TBE gel (Bio-Rad) and visualized using SYBR gold. Digested cDNA was then labeled with biotin by the addition of biotin-11-ddATP (Perkin-Elmer), 2 µl terminal transferase enzyme (Promega) and appropriate buffers. The reaction was incubated for 2 hours at 37°C and the labeling was quenched by the addition of 0.5 M EDTA. Labeling was verified by mixing the biotin-labeled cDNA with Immunopure NeutrAvidin (Pierce), incubating for 5 minutes at room temperature, and analyzing by gel electrophoresis on a 4 – 20% gradient gel that was followed by visualization with SYBR gold. A clear shift in the size and distribution from unlabeled to labeled DNA was verified prior to hybridization.
The cDNA probes were hybridized to Affymetrix FDA-ECSG custom arrays, which contain probe sets representing 32 E. coli and Shigella strains as well as 46 related plasmid sequences (Fang et al., 2010). Microarrays were processed according to manufacturer’s protocols. Briefly, the Oligo B2 mixture (Affymetrix) was heated to 65°C for 5 minutes and then mixed with the labeled cDNA, DMSO, and the hybridization buffer included in the Affymetrix Hybridization, Wash, and Stain kit. The solution was further denatured at 94°C for 5 minutes, during which time the microarrays were equilibrated with prehybridization buffer and placed in the Affymetrix hybridization oven at 45°C for 10 minutes, rotating at 60 r.p.m. The labeled cDNA was then hybridized to the microarray for 16 hours at 45°C, rotating at 60 r.p.m. The microarrays were washed and stained according to the manufacturer’s specifications using two stain cycles and the prokaryotic washing protocol (http://www.affymetrix.com).
The Affymetrix Gene Chip Operating System (GCOS) was used for data collection, and expression data was normalized and analyzed using the Simpleaffy Bioconductor R package (Gentleman et al., 2004, Wilson & Miller, 2005) and the RMA algorithm (Hubbell et al., 2002, Liu et al., 2002). The intensity value of the three replicates was obtained, combined, and the comparisons were double filtered by fold change (FC≥2) and the t-test p-value (p ≤0.005). The absolute intensity values were transformed into log2 values and subsequent analysis utilized these log transformed normalized values to determine the expression level for the genes on the array (Supplementary Table 1).
Reverse-transcription quantitative PCR analysis
To verify the microarray results, quantitative RT-PCR analysis was performed on the RNA stored at −80°C. The primers for the analysis are listed in Supplementary Table 2. Prior to the analysis, amplification efficiency and specificity were confirmed for each primer. RT-qPCR was performed a two-step reaction in which cDNA was synthesized using SuperScript III first strand synthesis kit (Invitrogen), and quantitative PCR followed using the SYBR green detection method with an ABI 7900HT sequence detection system (Applied Biosystems). Data were collected using the ABI Sequence Detection 1.2 software (Applied Biosystems). All data were normalized to levels of rpoA and analyzed using the comparative cycle threshold (ΔCT) method (Kendall et al., 2010). The expression levels of the target genes under the various conditions were compared using the relative quantification method (Kendall et al., 2010). Real-time data are expressed as the changes in expression levels compared with the WT levels. Statistical significance was determined by Student’s t-test, and a p-value of ≤ 0.05 was considered significant.
Congo red secretion assay
The Congo red (CR) secretion assay was used to identify proteins secreted by the bacteria through the T3SS, and was performed as previously described (Bahrani et al., 1997). Briefly, bacteria were grown to mid-log phase, resuspended in 1X PBS, and CR was added to a final concentration of 30 µg/ml. The bacteria were incubated at 37°C for 1 hour. After incubation, the bacteria were pelleted by centrifugation, and the supernatant was collected and filtered through a 0.22-µm-pore filter and then stored at –20°C. The proteins in the supernatant represent the secreted proteins and were concentrated by trichloroacetic acid precipitation. Trichloroacetic acid pellets were resuspended in 50 µl sodium dodecyl sulfate (SDS) loading buffer for protein analysis and stored at − 20°C. The bacterial pellets, representing the non-secreted proteins, were resuspended in 500 µl of SDS loading buffer and stored at −20°C.
Protein and Western blot analysis
For total protein analysis each sample was resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and Coomassie blue stain was used to visualize total protein. For Western blot analysis proteins were transferred to a PVDF membrane and blocked with 10% dry milk in 1X PBS. His-tagged proteins were detected with anti-his antibody (mouse tetra-his; Qiagen) at a concentration of 1:2,000 in PBS-Tween (PBST) with 10% dry milk overnight at 4°C. IpaB was detected with mouse monoclonal anti-IpaB (1:20,000 dilution) antibody as previously described (Mills et al., 1988). The anti-OmpU antibody was provided by Dr. James Kaper (Sperandio et al., 1996) and diluted 1:3,000, and the anti-GroEL antibody (Sigma) was diluted 1:10,000. After washing, Alexa Fluor 700 goat anti-mouse or anti-rabbit immunoglobulin G (H + L) antibody (Molecular Probes) was added at a 1:3,000 concentration in PBST with 10% dry milk for 1 h. All washes of the PVDF membrane were performed with 1X PBST for 5 minutes at room temperature. Protein gels and Western blots were scanned using the Odyssey infrared detection system (Li-Cor). Densitometry analysis was performed with the software provided by the Odyssey system.
Immunofluorescence analysis
After subculture or the CR assay, bacteria were gently washed and resuspended in 1X PBS. Sterile coverslips were placed in a 24-well sterile tissue culture plate, and approximately 1 × 106 bacteria were placed in the wells. Bacteria were pelleted onto the coverslips by centrifuging the tissue culture plate at 700 × g for 10 minutes. Afterwards, the 1X PBS was removed and the bacteria were fixed to the coverslips with 1X PBS with 3% formaldehyde (36% stock; Sigma) and 0.2% glutaraldehyde (25% stock; Sigma) for at least 30 minutes at 4°C. Samples were subsequently blocked with 10% natural goat serum (NGS; Sigma) for 45 minutes at room temperature. The primary anti-his antibody (mouse tetra-his; Qiagen) was diluted 1:400 in 10% NGS and applied to the bacteria for 1 hour at room temperature. Afterwards, the bacteria were washed with 1X PBS with 0.05% Tween 20. The secondary antibody, Alexa Fluor 488 F(ab')2 fragment of goat anti-mouse IgG (Invitrogen), was diluted 1:200 in 10% NGS and applied to the bacteria for 45 minutes at room temperature. Bacteria were finally washed with 1X PBS with 0.5% Tween 20, and the coverslips were mounted onto glass slides with antifade reagent (Molecular Probes) and stored in the dark. Images were taken using under the 100X objective on a Leica DMRB fluorescence microscope equipped with a DFC300FX digital camera. Exposure time for all images was set to 16,000 milliseconds (ms) to prevent over-exposure of the samples.
Ultracentrifugation
The bacterial pellets obtained after the CR assay were resuspended in a resuspension buffer containing 100 mM NaCl, 10mM MgCl2, 50 mM Tris-Cl, pH 8.0, and 10 µg/ml DNase (Invitrogen). A protease inhibitor cocktail (Sigma) was added according to manufacturer’s protocol. The pellets were then lysed with a microfluidizer (Microfluidics), according to manufacturer’s instructions, by performing a lysis cycle of three passages per strain of bacteria. The lysed cells were centrifuged at 2,000 × g for 10 minutes at 4°C to remove unbroken cells. Next, the supernatants were then centrifuged at 90,000 × g at 4°C for 1 hour. The supernatants containing the cytosolic and periplasmic proteins were removed, and the pellets containing the inner and outer membranes were centrifuged again in the resuspension buffer at 90,000 × g at 4°C for 1 hour to purify the membrane fraction. In order to remove the inner membrane, the pellet fraction was resuspended in an inner membrane solubilzation buffer, which contained 2% w/v Triton x-100, 100 mM NaCl, and 50 mM Tris-Cl, pH8.0. The samples were incubated at 30 minutes at 4°C with gentle shaking. Afterwards, the samples were centrifuged at 90,000 × g at 4°C for 1 hour, and the supernatants containing the solubilized inner membranes were removed. The pellet fractions were resuspended in dH2O and centrifuged at 90,000 × g at 4°C for 1 hour to remove residual Triton x-100. The pellets were collected and resuspended in SDS loading buffer for protein gel analysis.
Supplementary Material
Acknowledgements
We would like to thank Dr. Anthony Maurelli, Uniformed Services University, for the strains, Dr. Edwin Oaks, Walter Reed Army Institute of Research, for the IpaB antibody, and Dr. James Kaper, University of Maryland School of Medicine, for the OmpU antibody used in this study. We would also like to thank Jill Harper, Shweta Singh, Nadia Boisen, and Jane Michalski for technical assistance, and the Center for Vaccine Development for the T32 Vaccinology Fellowship AI07524 awarded to C.S.F. This work was supported by the National Institute of Allergy and Infectious Diseases Grant AI059223 to E.M.B. C.S.F. and J.P.N. were supported in part by NIH U19 AI090873 and grant no. 185872 from the Research Council of Norway’s GLOBVAC program to EntVac. J.C.R. and D.A.R. were supported by funds from the State of Maryland.
References
- Bahrani FK, Sansonetti PJ, Parsot C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immun. 1997;65:4005–4010. doi: 10.1128/iai.65.10.4005-4010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov V, Hammarstrom S. Carcinoembryonic antigen (CEA) and CEA-related cell adhesion molecule 1 (CEACAM1), apically expressed on human colonic M cells, are potential receptors for microbial adhesion. Histochemistry and cell biology. 2004;121:83–89. doi: 10.1007/s00418-003-0613-5. [DOI] [PubMed] [Google Scholar]
- Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Bloch CA, Stocker BA, Orndorff PE. A key role for type 1 pili in enterobacterial communicability. Mol Microbiol. 1992;6:697–701. doi: 10.1111/j.1365-2958.1992.tb01518.x. [DOI] [PubMed] [Google Scholar]
- Buchrieser C, Glaser P, Rusniok C, Nedjari H, D'Hauteville H, Kunst F, Sansonetti P, Parsot C. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faherty CS, Maurelli AT. Spa15 of Shigella flexneri is secreted through the type III secretion system and prevents staurosporine-induced apoptosis. Infect Immun. 2009;77:5281–5290. doi: 10.1128/IAI.00800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Xu J, Ding D, Jackson SA, Patel IR, Frye JG, Zou W, Nayak R, Foley S, Chen J, Su Z, Ye Y, Turner S, Harris S, Zhou G, Cerniglia C, Tong W. An FDA bioinformatics tool for microbial genomics research on molecular characterization of bacterial foodborne pathogens using microarrays. BMC Bioinformatics. 2010;11(Suppl 6):S4. doi: 10.1186/1471-2105-11-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formal SB, Dammin GJ, Labrec EH, Schneider H. Experimenta Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol. 1958;75:604–610. doi: 10.1128/jb.75.5.604-610.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington SM, Strauman MC, Abe CM, Nataro JP. Aggregative adherence fimbriae contribute to the inflammatory response of epithelial cells infected with entero aggregative Escherichia coli. Cell Microbiol. 2005;7:1565–1578. doi: 10.1111/j.1462-5822.2005.00588.x. [DOI] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Jain S, van Ulsen P, Benz I, Schmidt MA, Fernandez R, Tommassen J, Goldberg MB. Polar localization of the autotransporter family of large bacterial virulence proteins. J Bacteriol. 2006;188:4841–4850. doi: 10.1128/JB.00326-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennison AV, Verma NK. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol Rev. 2004;28:43–58. doi: 10.1016/j.femsre.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Kendall MM, Rasko DA, Sperandio V. The LysR-type regulator QseA regulates both characterized and putative virulence genes in enterohaemorrhagic Escherichia coli O157:H7. Molecular Microbiology. 2010;76:1306–1321. doi: 10.1111/j.1365-2958.2010.07174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ogawa M, Fujita Y, Yoshikawa Y, Nagai T, Koyama T, Nagai S, Lange A, Fassler R, Sasakawa C. Bacteria hijack integrin-linked kinase to stabilize focal adhesions and block cell detachment. Nature. 2009;459:578–582. doi: 10.1038/nature07952. [DOI] [PubMed] [Google Scholar]
- Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen SM, Ravnum S, Tourasse NJ, Okstad OA, Kolsto AB, Davies W. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579 [corrected] J Bacteriol. 2007;189:5302–5313. doi: 10.1128/JB.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall T, Mavris M, Martino MC, Bernardini ML, Denamur E, Parsot C. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology. 2005;151:951–962. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, Smeekens SP. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- Maurelli AT, Blackmon B, Curtiss R., 3rd Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun. 1984;43:397–401. doi: 10.1128/iai.43.1.397-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick BA, Siber AM, Maurelli AT. Requirement of the Shigella flexneri Virulence Plasmid in the Ability To Induce Trafficking of Neutrophils across Polarized Monolayers of the Intestinal Epithelium. Infection and Immunity. 1998;66:4237–4243. doi: 10.1128/iai.66.9.4237-4243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt ME, Donaldson JR. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J Med Microbiol. 2009;58:1533–1541. doi: 10.1099/jmm.0.014092-0. [DOI] [PubMed] [Google Scholar]
- Mills JA, Buysse JM, Oaks EV. Shigella flexneri invasion plasmid antigens B and C: epitope location and characterization with monoclonal antibodies. Infect Immun. 1988;56:2933–2941. doi: 10.1128/iai.56.11.2933-2941.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M, Terajima J, Izumiya H, Mitobe J, Komano T, Watanabe H. OspE2 of Shigella sonnei is required for the maintenance of cell architecture of bacterium-infected cells. Infect Immun. 2006;74:2587–2595. doi: 10.1128/IAI.74.5.2587-2595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier J, Vasselon T, Hellio R, Lesourd M, Sansonetti PJ. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect Immun. 1992;60:237–248. doi: 10.1128/iai.60.1.237-248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhaya A, Mahalanabis D, Chakrabarti MK. Role of Shigella flexneri 2a 34 kDa outer membrane protein in induction of protective immune response. Vaccine. 2006;24:6028–6036. doi: 10.1016/j.vaccine.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun. 2007;75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope LM, Reed KE, Payne SM. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect Immun. 1995;63:3642–3648. doi: 10.1128/iai.63.9.3642-3648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Arondel J, Cantey JR, Prevost MC, Huerre M. Infection of rabbit Peyer's patches by Shigella flexneri: effect of adhesive or invasive bacterial phenotypes on follicle-associated epithelium. Infect Immun. 1996;64:2752–2764. doi: 10.1128/iai.64.7.2752-2764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Kopecko DJ, Formal SB. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 2008;21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellings NJ, Tall BD, Venkatesan MM. Characterization of Shigella type 1 fimbriae: expression, FimA sequence, and phase variation. Infect Immun. 1997;65:2462–2467. doi: 10.1128/iai.65.6.2462-2467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Bailey C, Giron JA, DiRita VJ, Silveira WD, Vettore AL, Kaper JB. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect Immun. 1996;64:5406–5409. doi: 10.1128/iai.64.12.5406-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem. 2008;283:18646–18654. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamano K, Aizawa S, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Zhou X, Kaper JB. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect Immun. 2005;73:18–29. doi: 10.1128/IAI.73.1.18-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Dougan G, Baumler AJ. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun. 2001;69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya A, Naito T, Ehara M, Ichinose Y, Hamamoto A. Studies on novel pili from Shigella flexneri. I. Detection of pili and hemagglutination activity. Microbiology and immunology. 1992;36:803–813. doi: 10.1111/j.1348-0421.1992.tb02082.x. [DOI] [PubMed] [Google Scholar]
- Venkatesan MM, Buysse JM, Oaks EV. Surface presentation of Shigella flexneri invasion plasmid antigens requires the products of the spa locus. J Bacteriol. 1992;174:1990–2001. doi: 10.1128/jb.174.6.1990-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Heindl JE, Gray AN, Jain S, Goldberg MB. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J Bacteriol. 2009;191:815–821. doi: 10.1128/JB.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, 3rd, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun. 2003;71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Miller CJ. Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.