Abstract
The ranges of arctic–alpine species have shifted extensively with Pleistocene climate changes and glaciations. Using sequence data from the trnH-psbA and trnT-trnL chloroplast DNA spacer regions, we investigated the phylogeography of the widespread, ancient (>3 million years) arctic–alpine plant Oxyria digyna (Polygonaceae). We identified 45 haplotypes and six highly divergent major lineages; estimated ages of these lineages (time to most recent common ancestor, TMRCA) ranged from ∼0.5 to 2.5 million years. One lineage is widespread in the arctic, a second is restricted to the southern Rocky Mountains of the western United States, and a third was found only in the Himalayan and Altai regions of Asia. Three other lineages are widespread in western North America, where they overlap extensively. The high genetic diversity and the presence of divergent major cpDNA lineages within Oxyria digyna reflect its age and suggest that it was widespread during much of its history. The distributions of individual lineages indicate repeated spread of Oxyria digyna through North America over multiple glacial cycles. During the Last Glacial Maximum it persisted in multiple refugia in western North America, including Beringia, south of the continental ice, and within the northern limits of the Cordilleran ice sheet. Our data contribute to a growing body of evidence that arctic–alpine species have migrated from different source regions over multiple glacial cycles and that cryptic refugia contributed to persistence through the Last Glacial Maximum.
Keywords: Arctic-alpine plants, cpDNA, phylogeography, Pleistocene glaciations, refugia
Introduction
Changing climates during the last 2 million years have profoundly influenced the distribution and abundance of temperate, boreal, and arctic species. Many species have undergone major range shifts in response to changing sea levels or glacial advances and retreats. During the Last Glacial Maximum (LGM) approximately 18,000 years ago, the Fenno-Scandian Ice Sheet in northern Europe and the Laurentide and Cordilleran Ice Sheets in North America extended over large areas of these continents (Ehlers and Gibbard 2007). Many tundra plants nevertheless have broad present-day distributions in both arctic and alpine environments, a pattern which may reflect repeated cycles of expansion, contraction, and migration over several glacial cycles. Molecular studies in combination with fossil evidence have shown diverse phylogeographic patterns among these widespread species, demonstrating how they came to occupy their present ranges and where refugia occurred (Brochmann et al. 2003; Brochmann and Brysting 2008). Both arctic refugia and southern mountain ranges have been suggested as source regions for arctic plants (Murray 1995; Brochmann and Brysting 2008). However, the extent and timelines of contributions to the arctic flora from these different source areas are still not well understood.
Beringia (Yukon to eastern Siberia) provided a large high-latitude ice-free area during the Pleistocene and has long been proposed as a major refugium for many arctic–alpine plant species during glacial periods (Hultén 1937). This hypothesis is well supported by molecular evidence (e.g., Tremblay and Schoen 1999; Abbott et al. 2000; Abbott and Comes 2003; Alsos et al. 2005; Eidesen et al. 2007; Skrede et al. 2009). Although Hultén (1937) considered Beringia the most important refugial region, he suggested that the broad distributions of many tundra species implied dispersal from additional refugia. Many arctic–alpine species persisted in refugia south of continental ice in both Europe and North America (Abbott and Brochmann 2003; Brochmann et al. 2003; Birks 2008; Brochmann and Brysting 2008); examples are Arabis alpina (Koch et al. 2006) and Dryas octopetala (Skrede et al. 2006). Additional refugia may also have existed along glacial margins, in areas such as coastal eastern Greenland (Funder 1979) and coastal British Columbia (Heusser 1989; Ogilvie 1989; Hebda et al. 1997; King et al. 2009). Recent genetic evidence has indicated survival of tundra plant and animal species in unexpected refugia within the overall margins of continental ice sheets of the Late Pleistocene, both in Europe (Westergaard et al. 2011) and in western North America (Loehr et al. 2006; Marr et al. 2008; Shafer et al. 2010a). For tundra plant species, refugia have been suggested in northwestern Europe (Westergaard et al. 2011), northeastern Canada (Tremblay and Schoen 1999), northwestern Canada (Eriksen and Töpel 2006), and northern interior British Columbia (Marr et al. 2008), but further work is needed to determine their overall importance.
Genetic evidence of rapid spread has been found in some arctic–alpine plants. For example, the ranges of Vaccinium uliginosum (Alsos et al. 2005; Eidesen et al. 2007) and Rubus chamaemorus (Ehrich et al. 2008) expanded rapidly following the LGM. Long-distance dispersal and colonization is supported by molecular evidence for a number of arctic species (Brochmann and Brysting 2008), and is the most plausible explanation for current genetic characteristics of many plants found on the remote archipelago of Svalbard (Alsos et al. 2007). However, the potential for rapid migration does not preclude the possibility of survival in multiple locations, including cryptic refugia previously unknown from fossil or palynological evidence (Stewart and Lister 2001; Provan and Bennett 2008). For example, molecular evidence suggests that some European tree species had a more complex glacial history than previously thought, including survival through the LGM in unexpected locations (Birks and Willis 2008). Tundra plants also had complex responses to past climate change (Schönswetter et al. 2005), and may have spread multiple times from steppe or montane habitats (Tkach et al. 2008). An understanding of how dispersal and refugia shaped their present distributions can provide keys to assessing their potential responses to future climate change.
Oxyria digyna Hill is a widespread arctic–alpine perennial herb (Fig. 1) that occurs in tundra habitats over much of the northern hemisphere (Fig. 2), including nearly all of arctic North America and Eurasia and mountain ranges of southern Russia, the Himalayan region, southern Europe, and western North America (Hultén 1971). Its fossil record extends back to the late Tertiary, with macrofossils dated at ∼3 million years reported from arctic Canada (Ellesmere, Meighen, and Prince Patrick Islands, Beaufort Formation; Mathews and Ovenden 1990). It grows in a variety of mesic tundra environments, including scree with little or no soil development. The flowers of Oxyria digyna are inconspicuous and wind-pollinated. The winged seeds are dispersed moderately well by wind (Tackenberg and Stöcklin 2008) and the species establishes effectively on glacial forelands (Whittaker 1993; Stöcklin and Bäumler 1996). Arctic and alpine populations show ecotypic differences in flower and rhizome production and in growth responses to temperature and day length (Mooney and Billings 1961; Heide 2005). Oxyria digyna is taxonomically distinct, with no closely related taxa, and is generally treated as a single well-defined species. Oxyria digyna is largely diploid (2n= 14), with a few hexaploids (2n= 42) reported from Russia (Fedorov 1974; Goldblatt and Johnson 1979–2010).
Figure 1.

Oxyria digyna, Mt. Lassen, California.
Figure 2.
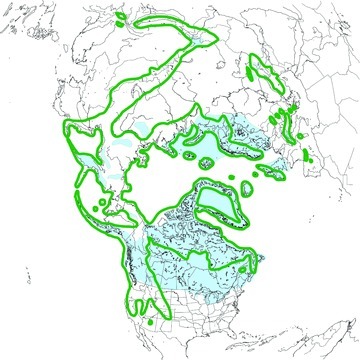
Worldwide geographic distribution of Oxyria digyna (green outline) and maximum extent of ice sheets during the Last Glacial Maximum (blue shading). Range map is redrawn from Hultén (1971) and glacial boundaries from Ehlers and Gibbard (2007).
The history of the tundra floras of the northern hemisphere has been much more extensively investigated in Europe than in North America. In particular, phylogeographic studies of arctic–alpine plants with a focus on western North America are still relatively few (Jorgensen et al. 2003; Bain and Golden 2005), and the contributions of refugia hypothesized for this region (especially cryptic refugia) are not well understood. In a previous study (Marr et al. 2008), we used restriction fragment analysis to identify patterns of genetic diversity in western North American populations of Oxyria digyna in relation to recent glacial history and the locations of refugia. Here we used cpDNA sequence data to reconstruct the broader northern hemisphere phylogeographic history of Oxyria digyna, taking advantage of the much greater power of sequence data to resolve phylogenetic relationships among intraspecific lineages and reveal their early history. We used a combination of analytic approaches together with increased haplotype resolution and greater sampling around the northern hemisphere to address the following questions: (1) what is the geographic structure of genetic variation in Oxyria digyna and how does it compare with other arctic species? (2) How did the species and its major lineages diversify over time and disperse into their present geographic range? (3) What was the contribution of different refugia to the persistence of different lineages in western North America during the LGM?
Methods
Sampling
We sequenced 181 individuals of Oxyria digyna from 140 localities in North America, Europe, and Asia (Table 1). Sample localities were concentrated in western North America, but we obtained samples from much of the geographic range. We usually sampled one plant per population, but included 1–2 additional individuals in some populations from a previous study that used different methods (Marr et al. 2008). We also sampled four outgroup taxa: Oxyria sinensis Hemsl., Rheum palmatum L., Rumex lapponicus Czernov, and Rumex paucifolius Nutt. (Table 1).
Table 1.
Collection locality information for all samples and combined trnH-psbA and trnT-trnL haplotypes for Oxyria digyna. Localities are arranged by continent and geographic region, and within these in order of decreasing latitude.
| Country, locality | Population ID | Latitude | Longitude | Collector | Year | 1Clade and haplotype |
|---|---|---|---|---|---|---|
| Oxyria digyna | ||||||
| Europe and Asia | ||||||
| China, Yunnan, Gaoligong Shan | YN | 25° 13' N | 98° 28' E | P. Fritsch | 2006 | F-42 |
| Greenland, Johannes V. Jensens Land | GN | 83° 24' N | 29° 39' W | D. Mazzuchi | 2006 | A-7 |
| Greenland, Kap K0benhavn | KAP | 82° 24' N | 22° 12' W | D. Mazzuchi | 2006 | A-7 |
| Greenland, Zackenberg Station | ZAC | 74° 28' N | 20° 34' W | K. Westergaard | 2007 | A-7 |
| Greenland, Osterdalen Valley | OST | 69° 15' N | 53° 31' W | K. Westergaard | 2006 | A-14 |
| Greenland, Narsaq | NAR | 60° 57' N | 46° 03' W | K. Westergaard | 2006 | A-14 |
| Iceland, Skalafellsjokull | SKA | 64° 13' N | 15° 44' W | M. Reynolds | 2011 | A-14 |
| Nepal, Annapurna Sanctuary | A | 28° 32' N | 83° 55' E | W. Mackenzie | 2005 | F-41 |
| Nepal, Khumbu Himal, Mt. Everest National Park | NEP | 27° 55' N | 86° 45' E | A. C. Byers | 1984 | F-43 |
| Norway, Endalen Valley, Svalbard | END | 78° 11' N | 15° 46' E | K. Westergaard | 2006 | A-14 |
| Norway, Stranda, Mt. Dalsnibba | NO | 62° 03' N | 7° 15' E | O. Finch | 2003 | A-14 (2) |
| Russia, Altai Republic, Rudnik Mine/Korumduairya Creek | RUD | 50° 20' N | 87° 45' E | K. L. Marr, R. J. Hebda | 2010 | F-45 (2) |
| Russia, Altai Republic, Karakjel Lake/Karasu Creek | KRK | 50° 02' N | 88° 02' E | K. L. Marr, R. J. Hebda | 2010 | F-44 |
| Russia, Altai Republic, Talaya Pass | TAL | 50° 00' N | 89° 16' E | K. L. Marr, R. J. Hebda | 2010 | F-45 |
| Russia, Kamchatka, Chazma River | KM | 58° 01' N | 158° 39' E | W. Merilees | 2004 | A-4 |
| Russia, Magadan Oblast, near Pobieda | POB | 64° 28' N | 144° 55' E | K. L. Marr, R. J. Hebda | 2011 | A-l |
| Russia, Magadan Oblast, divide between Indygirka and Kolyma Rivers | IKP | 63° 27' N | 146° 27' E | K. L. Marr, R. J. Hebda | 2011 | A-l |
| Russia, Magadan Oblast, Burkhalinsky Pass | BUR | 62° 42' N | 148° 48' E | K. L. Marr, R. J. Hebda | 2011 | A-l |
| Russia, Magadan Oblast, Talaya Pass | TAY | 61° 10' N | 152° 00' E | K. L. Marr, R. J. Hebda | 2011 | A-l |
| Russia, Magadan Oblast, Armansky Pass | ARM | 59° 40' N | 150° 37' E | K. L. Marr, R. J. Hebda | 2011 | A-8 |
| Russia, Sakha Republic, near Ust Nera | UST | 64° 29' N | 143° 15' E | K. L. Marr, R. J. Hebda | 2011 | A-l |
| Russia, Wrangel Island | WRA | 71° 15' N | 179° 40' W | A. Benedict | 2009 | A-9 |
| Russia, Yamalo-Nenetsky Autonomous | YA | 65° 27' N | 66° 50' E | I. G. Alsos | 2004 | A-12 (2) |
| Region, Chernaya Mts. | ||||||
| North America | ||||||
| Canada, Nunavut, Alexandra Fjord, Ellesmere | AF | 78° 58' N | 75° 45' W | H. Guest | 2003 | A-6 |
| Canada, Nunavut, Ellef Ringnes Island | RG | 78° 47' N | 103° 32' W | M. Reynolds | 2005 | A-7 |
| Canada, Nunavut, Pond Inlet | PON | 72° 45' N | 77° 35' W | P. Smith | 2011 | A-7 |
| Canada, Nunavut, Roche Bay | RY | 68° 29' N | 82° 39' W | B. Apland | 2006 | A-7 |
| Canada, Nunavut, High Lake Camp | HL | 67° 23' N | 110° 51' W | B. Apland | 2006 | A-7 |
| Canada, Nunavut, Apex, Baffin Island | BN | 63° 44' N | 68° 26' W | L. Gillespie | 2004 | A-7 |
| Canada, NWT, Prince Patrick Island | UL | 76° 14' N | 119° 20' W | M. Reynolds | 2005 | A-7 |
| Canada, NWT, Banks Island, DeSalis Bay | DS | 71° 27' N | 121° 52' W | L. Gillespie | 2004 | A-7 |
| Canada, NWT, Palmer Lake Site 9 | PLK | 64° 32’ N | 129° 34' W | A. Veitch | 2007 | A-6 |
| Canada, NWT, Dechen la' North Canol Road | DEC | 63° 24' N | 129° 36' W | B. Krieckhaus | 2007 | A-5 |
| Canada, YT, Shingle Point | SH | 69° 59' N | 137° 25' W | B. Bennett | 2005 | A-8 |
| Canada, YT, Ivvavik National Park | IV | 69° 37' N | 140° 50' W | B. Bennett | 2005 | A-8 |
| Canada, YT, Upper Fish Creek, Richardson Mts. | UF | 67° 54' N | 136° 33' W | B. Bennett | 2006 | A-8 |
| Canada, YT, Mt. Klotz camp | KLO | 65° 22' N | 140° 11' W | B. Bennett | 2007 | A-6 |
| Canada, YT, Ogilvie Mts. (site 3) | G3 | 64° 47' N | 137° 43' W | H. Guest | 2005 | A-3 (2) |
| Canada, YT, Bonnet Plume, Gillespie Lake | BT | 64° 44' N | 133° 59' W | B. Bennett | 2005 | A-l |
| Canada, YT, Tombstone Territorial Park, North Fork Pass | TB | 64° 35' N | 138° 17' W | K. L. Marr | 2004 | A-6, A-8, |
| Canada, YT, MacMillan Pass | MP | 63° 17' N | 130° 09' W | B. Apland | 2006 | A-3 |
| Canada, YT, Asi Keyi Special Management Area | AK | 61° 34' N | 140° 50' W | B. Bennett | 2004 | A-6 |
| Canada, YT, Outpost Mt. | OM | 60° 59' N | 138° 25' W | H. Guest | 2004 | A-6 |
| Canada, YT, Kotaneelee Range | KO | 60° 13' N | 124° 07' W | B. Bennett | 2004 | A-l |
| Canada, BC, Tagish Highland, TeePee Mt. | TEE | 59° 44' N | 134° 40' W | K. L. Marr, R. J. Hebda | 2008 | A-2 |
| Canada, BC, Liard Plateau, Caribou Mts. | CRM | 59° 43' N | 125° 28' W | K. L. Marr, R. J. Hebda | 2007 | A-l |
| Canada, BC, Ruby Mt. | RB | 59° 42' N | 133° 22' W | K. L. Marr | 2004 | A-6, B-15 |
| Canada, BC, Chuck Creek | CC | 59° 42' N | 136° 36' W | H. Guest | 2004 | A-ll |
| Canada, BC, Horseranch Range | HR | 59° 28' N | 128° 56' W | K. L. Marr, R. J. Hebda | 2004 | A-6 |
| Canada, BC, Cassiar Mts., “Dakota” Lake | DL | 59° 25' N | 130° 13' W | K. L. Marr, R. J. Hebda | 2004 | A-l, A-10, B-15 |
| Canada, BC, Kawdy Plateau, High Tuya Lake | HT | 59° 15' N | 130° 31' W | K. L. Marr, R. J. Hebda | 2004 | A-l, B-15 |
| Canada, BC, Muskwa Ranges, Terminal Range | TER | 59° 02' N | 125° 55' W | K. L. Marr, R. J. Hebda | 2007 | A-l |
| Canada, BC, Nimbus Mt. | NIM | 59° 02' N | 132° 30' W | K. L. Marr, R. J. Hebda | 2008 | B-15 |
| Canada, BC, Muskwa Ranges, Nonda Creek | N | 59° 00' N | 125° 30' W | K. L. Marr | 2003 | A-1, B-18 (2) |
| Canada, BC, Stikine Ranges, Four Mile Creek | 4MC | 58° 51' N | 129° 31' W | K. L. Marr, R. J. Hebda | 2007 | A-6 |
| Canada, BC, French Range, Rath Mt. | RTM | 58°45' N | 130° 25' W | K. L. Marr, R. J. Hebda | 2007 | A-6 |
| Canada, BC, Stikine Ranges, Little Blue Sheep Lake | LBS | 58° 44' N | 128° 15' W | K. L. Marr, R. J. Hebda | 2007 | A-6 |
| Canada, BC, Dome Mt. | DM | 58°31' N | 129° 35' W | K. L. Marr, R. J. Hebda | 2004 | A-6 (2) |
| Canada, BC, Snow Peak | SP | 58° 20' N | 130° 23' W | W. Mackenzie | 2003 | B-15 |
| Canada, BC, Three Sisters Range | SI | 58° 20' N | 129° 45' W | W. Mackenzie | 2003 | A-6 |
| Canada, BC, Stikine Ranges, Shea Mt. | SHM | 58° 19' N | 128° 57' W | K. L. Marr, R. J. Hebda | 2007 | A-6 |
| Canada, BC, mountains south of Pitman River | PIT | 57° 53' N | 127° 54' W | K. L. Marr, R. J. Hebda | 2009 | A-6 |
| Canada, BC, Thudaka Range | THU | 57° 48' N | 126° 37' W | K. L. Marr, R. J. Hebda | 2009 | B-15 |
| Canada, BC, Spatsizi Provincial Park, Airplane Creek | SZ | 57° 41' N | 128° 52' W | R. J. Hebda | 2005 | A-6, B-15 |
| Canada, BC, Mt. Edziza | E | 57° 38' N | 130° 40' W | C. Howers | 2005 | B-15 (2), B-17 |
| Canada, BC, Moosehorn Lake | MOO | 57° 35' N | 127° 13' W | K. L. Marr, R. J. Hebda | 2009 | B-15 |
| Canada, BC, Brothers Lake | BRO | 57° 12' N | 127° 26' W | K. L. Marr, R. J. Hebda | 2009 | E-34 |
| Canada, BC, Kemess North | K | 57° 04' N | 126° 45' W | J. Pojar, W. Mackenzie | 2003 | A-l |
| Canada, BC, Wrede Range | WRE | 56° 38' N | 126° 12' W | K. L. Marr, R. J. Hebda | 2009 | B-15 |
| Canada, BC, Oweegee Range | OW | 56° 34' N | 129° 28' W | K. L. Marr, R. J. Hebda | 2006 | B-17 |
| Canada, BC, Chase Mt. | C | 56° 33' N | 125° 16' W | K. L. Marr, R. J. Hebda | 2003 | B-15 |
| Canada, BC, Mt. Henri | MT | 56° 29' N | 124° 45' W | K. L. Marr, R. J. Hebda | 2003 | E-34 |
| Canada, BC, Ludington Peak | LP | 56° 28' N | 123° 23' W | K. L. Marr, R. J. Hebda | 2003 | B-18 |
| Canada, BC, Needham Creek | NE | 56° 24' N | 123° 31' W | K. L. Marr, R. J. Hebda | 2003 | A-l, B-15, B-18 |
| Canada, BC, Hanna Ridge | HN | 56° 14' N | 129° 26' W | K. L. Marr, R. J. Hebda | 2006 | B-17 |
| Canada, BC, Mt. Tommy Jack | TO | 56°03' N | 127° 46' W | K. L. Marr, R. J. Hebda | 2006 | E-34 |
| Canada, BC, Morphee Mt. | MR | 55°26' N | 123° 02' W | K. L. Marr, R. J. Hebda | 2003 | E-40 |
| Canada, BC, Murray Mt. | M | 55° 24' N | 122° 37' W | K. L. Marr, R. J. Hebda | 2003 | E-39 |
| Canada, BC, Mt. Crum, “Holzworth Meadows” | H | 55° 01' N | 121° 31' W | K. L. Marr, R. J. Hebda | 2003 | E-40 |
| Canada, BC, Babine Mts. | BA | 55° 00' N | 126° 56' W | K. L. Marr, R. J. Hebda | 2002 | E-34 (2) |
| Canada, BC, Nass Ranges, Mt. Couture | CU | 54° 54' N | 128° 42' W | K. L. Marr, R. J. Hebda | 2006 | E-34 |
| Canada, BC, Sleeping Beauty Mt. | SB | 54° 34' N | 128° 52' W | M. Cheney | 2003 | B-17 (2) |
| Canada, BC, Mt. Thornhill | TL | 54° 31' N | 128° 27' W | G. Allen | 2004 | B-17, E-34 |
| Canada, BC, Nadina Mt. | NA | 54° 06' N | 126° 52' W | K. L. Marr, R. J. Hebda | 2002 | E-34 (2) |
| Canada, BC, Tweedsmuir Peak | TW | 53° 39' N | 126° 29' W | K. L. Marr, R. J. Hebda | 2002 | D-32 (2) |
| Canada, BC, Canoe Mt. | CA | 52° 43' N | 119° 11' W | K. L. Marr | 2002 | E-35 (2) |
| Canada, BC, Mt. De La Touche. | LT | 52° 42' N | 132° 02' W | G. W. Douglas | 2003 | B-17 |
| Canada, BC, Itcha Ilgachuz | CH | 52° 42' N | 124° 51' W | R. Coffey | 2003 | E-35 |
| Canada, BC, Mackenzie Mt. | MK | 52° 35' N | 126° 09' W | W. Mackenzie | 2003 | E-35 |
| Canada, BC, Tweedsmuir Park, Crystal Lake trail | CL | 52° 32' N | 125° 49' W | R. Coffey | 2003 | D-32, E-35 |
| Canada, BC, Caput Mt. | CAP | 52° 25' N | 120° 36' W | F. Osorio | 2006 | E-40 |
| Canada, BC, Wells-Gray Provincial Park, Trophy Mt. | TR | 51° 47' N | 119° 55' W | K. L. Marr | 2002 | E-40 |
| Canada, BC, Niut Range | NT | 51° 43' N | 124° 39' W | W. Mackenzie | 2003 | E-35 (2) |
| Canada, BC, Brisco Range, Kindersley Pass | KIN | 50° 42' N | 116° 00' W | K. L. Marr | 2007 | E-34 |
| Canada, BC, Bugaboo Pass | BO | 50°41' N | 116° 45' W | O. McDadi | 2004 | E-34, E-40 |
| Canada, BC, Paradise Mine | PA | 50° 29' N | 116° 18' W | K. L. Marr | 2002 | D-32, E-34 |
| Canada, BC, Vancouver Island, Merry Widow Mt. | MW | 50° 21' N | 127° 18' W | K. L. Marr, R. J. Hebda | 2005 | E-34 |
| Canada, BC, Kootenay Lake | KOO | 50° 18' N | 117° 08' W | W. Mackenzie | 2007 | E-39 |
| Canada, BC, Whistler Mt. | WH | 50° 04' N | 122° 57' W | K. L. Marr | 2003 | E-35 |
| Canada, BC, Manning Park | MA | 49° 6' N | 120° 46' W | K. L. Marr | 2002 | E-40 (2) |
| Canada, BC, Gimli Peak | GI | 49° 46' N | 117° 39' W | H. Guest | 2006 | E-35 |
| Canada, BC, Vancouver Island, above Circlet Lake | FP | 49° 41' N | 125° 22' W | K. L. Marr | 2003 | E-34 |
| Canada, BC, Fernie, Three Sisters trail | 3S | 49° 35' N | 115° 08' W | K. L. Marr | 2002 | E-35 (2) |
| Canada, BC, Vancouver Island, Mt. Arrowsmith | AR | 49° 14' N | 124° 36' W | K. L. Marr | 2004 | D-32 |
| Canada, BC, Silver Tip Mt. | SM | 49° 10' N | 121° 13' W | G. W. Douglas | 2003 | E-35 |
| Canada, BC, Kishinena Range | KR | 49° 02' N | 114° 16' W | W. Mackenzie | 2005 | E-35 |
| Canada, AB, Pipestone R. Trail | PP | 51° 35' N | 116° 09' W | S. White | 2003 | E-35 |
| Canada, AB, Lower Fish Lake | LF | 51° 31' N | ii6° irw | S. White | 2003 | E-35 |
| USA, AK, Beaufort Sea coast, Camden Bay | BFS | 69° 58' N | 144° 46' W | J. Jorgensen | 2007 | A-13 |
| USA, AK, Anaktuvuk Pass, Alaska | ANA | 68° 08' N | 151° 36' W | B. Krieckhaus | 2008 | A-6 |
| USA, AK, Dot Lake | DK | 63°39' N | 144° 37' W | H. Guest | 2005 | A-10 |
| USA, AK, Richardson Highway | RH | 63°21' N | 145° 42' W | H. Guest | 2005 | A-l (2) |
| USA, AK, Caribou Creek | CR | 62° 37' N | 143° 28' W | H. Guest | 2005 | A-10 |
| USA, AK, Chitina Rd. | CM | 61°34' N | 144° 36' W | H. Guest | 2005 | A-3 (2) |
| USA, AK, Crow Pass | CW | 61° 02' N | 149° 07' W | H. Guest | 2005 | A-l |
| USA, AK, Bering Sea, St. Matthew Island | HSM | 60° 33' N | 172° 42' W | CAN 587881 | 1944 | A-l |
| USA, AK, Carbon Mt. | CO | 60° 27' N | 143° 53' W | A Batten | 2005 | B-18, B-19 |
| USA, AK, Kenai Fjords | KF | 59° 27' N | 149° 57' W | M. Carlson | 2005 | B-15 |
| USA, AK, Chichagof Island, Hoonah Ridge | CF | 57° 42' N | 135° 02' W | B. Krieckhaus | 2006 | B-15 (2) |
| USA, AK, Kodiak Island | KD | 57° 16' N | 154° 12' W | C. Parker | 2005 | B-15 |
| USA, AK, Unalaska Island, Pyramid Peak | UNF | 53° 51' N | 166° 32' W | B. Krieckhaus | 2007 | B-16 |
| USA, WA, Winchester Mt. Lookout | WIN | 48° 57' N | 121° 39' W | K. L. Marr | 2007 | E-34 |
| USA, WA, Mt. Baker, Bagley Lakes Trail | BAG | 48° 51' N | 121° 41' W | K. L. Marr | 2007 | E-34 |
| USA, WA, Hart's Pass | HP | 48° 44' N | 120° 40' W | K. L. Marr | 2003 | E-35, E-40 |
| USA, WA, Olympic Mts., Obstruction Point | OP | 47° 55' N | 123° 23' W | K. L. Marr | 2003 | B-21, D-32 |
| USA, WA, Chinook Pass | 1R | 46° 52' N | 121° 31' W | G. Allen, K. L. Marr | 2004 | E-39 |
| USA, MT, Anaconda Pintlar Range, Seymour Lake | SY | 46° 03' N | 113° 58' W | H. Guest | 2006 | E-35 |
| USA, MT, Absaroka Mts. | AB | 45° 40' N | 110° 34' W | P. Lesica | 2004 | B-23, B-25 |
| USA, MT, Hyalite Creek | HY | 45° 26' N | 110° 57' W | K. L. Marr | 2005 | D-30, E-37 |
| USA, ID, Heaven's Gate | HG | 45° 22' N | 116° 30' W | G. Allen, K. L. Marr | 2004 | B-24 |
| USA, ID, Seven Devils Mts. | SD | 45°21' N | 116° 31' W | G. Allen, K. L. Marr | 2004 | E-36 |
| USA, OR, Mt. Hood, Elliott Glacier | EG | 45° 24' N | 121° 40' W | G. Allen, K. L. Marr | 2004 | B-20 (2) |
| USA, OR, Three-Fingered Jack | TJ | 44° 24' N | 121° 48' W | G. Allen, J. Antos | 2004 | E-39 |
| USA, OR, Strawberry Mts. | SW | 44° 18' N | 118° 41' W | G. Allen, K. L. Marr | 2004 | B-22, E-38 |
| USA, OR, Mt. Ashland | AS | 42° 05' N | 122° 43' W | G. Allen, J. Antos | 2004 | D-31 |
| USA, WY, Wind River Range, Haystack Mt. | HM | 42° 44' N | 109° 10' W | J. Dunn | 2006 | C-28 |
| USA, WY, Medicine Bow Mts. | MD | 41° 21’ N | 106° 20’ W | H. Guest | 2006 | C-27 |
| USA, CO, Forest Lakes | FL | 39° 55' N | 105° 40' W | K. L. Marr | 2003 | C-26, C-29 |
| USA, CA, Mt. Lassen | L | 40° 29' N | 121° 31' W | G. Allen, J. Antos | 2004 | D-31 |
| USA, CA, sw of Bishop, Paiute Pass Trail | PU | 37° 14' N | 118° 39' N | H. Guest | 2006 | E-35 |
| Outgroups | ||||||
| Oxyria sinensis | ||||||
| China, Yunnan, Deqing | 57° 59' N | 98° 27' E | L. Q. Jun | 2006 | ||
| Rheum palmatum | ||||||
| Canada, Jardin botanique de Montréal (cultivated) | — | — | G. Allen | 2008 | ||
| Rumex lapponicus | ||||||
| Canada, BC, Anthony Creek | 56° 49' N | 128° 35' E | K. L. Marr, R. J. Hebda | 2005 | ||
| Rumex paucifolius | ||||||
| USA, Wyoming, Wind River Mts., Green River Lake | 43° 17' N | 109° 50' E | G. Allen, J. Antos | 2005 | ||
Parenthese indicate multiple samples from the same locality.
DNA extraction, amplification, and sequencing
DNA was extracted from silica-dried leaves (or in a few cases, pressed specimens) from 1–3 plants per population, using either a modified CTAB method (Doyle and Doyle 1987) or DNeasy Plant Mini Kits (Qiagen, Valencia, CA). We amplified and sequenced two cpDNA noncoding regions, the trnH-psbA and trnT-trnL spacers, in all samples including outgroups. We also sequenced a third region, the ndhJ-trnF spacer, in a subset of 37 Oxyria digyna samples and in the outgroups. For the trnH-psbA region we used the primers psbA (Sang et al. 1997) and trnH (Tate and Simpson 2003). For the trnT-trnL region we used forward primers Tab A (Taberlet et al. 1991) or trnA2 (Cronn et al. 2002) with reverse primers Tab B (Taberlet et al. 1991) or Oxy88 (Marr et al. 2008). For the ndhJ-trnF region we used forward primer ndhJ (Shaw et al. 2007) and reverse primer trnF (Dumolin–Lapegue et al. 1997).
PCR was performed in 50 µL volumes with the following reagents: 5 µL template DNA (1:10 dilutions), 5 µL 10× PCR Buffer, 5 µL 2 mM dNTPs, 0.25 µL Taq DNA polymerase (New England Biolabs, Ipswich, MA), and 2.5 µL of 5 µM of each primer. All PCR reactions were carried out using Techne TC-312 thermal cyclers. PCR parameters for amplification of the trnH-psbA and trnT-trnL regions were 3 min at 94°C, 30 cycles of 94°C for 30 s, 57°C for 1 min, and 72°C for 1 min, with a final 10 min at 72°C. Parameters for the ndhJ-trnF region were 5 min at 80°C, 30 cycles of 95°C for 1 min, 50°C for 1 min, and 65°C for 4 min, with a final 5 min at 65°C. In preparation for sequencing, PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA). All samples were sequenced in both primer directions using the amplifying primers. DNA sequencing was carried out by Macrogen Inc. (Seoul, Korea), using ABI 3700 or ABI 3730XL PRISM capillary sequencers (Applied Biosystems, Foster City, CA). To minimize PCR errors, all rare sequence variants were reamplified and resequenced.
Data analyses
Sequences for each DNA region were aligned using ClustalX (Thompson et al. 1997) and Jalview (Waterhouse et al. 2009), and manually adjusted to minimize the number of length variations. Three of the four outgroup taxa were highly divergent from Oxyria digyna in all sequenced regions and were not used further. Oxyria sinensis aligned satisfactorily with Oxyria digyna for the ndhJ-trnF region only. We performed maximum parsimony (MP) and maximum likelihood (ML) analyses of the ndhJ-trnF region with Oxyria sinensis as outgroup; the resulting trees showed little resolution of relationships among Oxyria digyna haplotypes and are not reported here. All reported analyses are based on the trnH-psbA and trnT-trnL spacer regions.
Haplotype networks for the trnH-psbA and trnT-trnL noncoding regions, separately and in combination, were constructed using the statistical parsimony software TCS v. 1.21 (Clement et al. 2000). An ILD test (Michewich and Farris 1981), as implemented in PAUP 4.0b10, was carried out using 500 random replicates to check for partition heterogeneity of the two regions. Indels were treated as unit characters and included in all analyses. To obtain estimates of support for branches of the combined network, we performed MP and Bayesian phylogenetic analyses on a data set consisting of one sequence for each combined haplotype (with indels coded as 0/1 characters in a separate data partition). MP analysis was performed with PAUP 4.0 version b10 (Swofford 1998), with clade support estimated from 500 bootstrap replicates using a heuristic search with TBR branch swapping. Bayesian analysis was carried out with MrBayes v. 3.1.2 (Huelsenbeck and Ronquist 2001) under the assumption of a strict molecular clock, using a GTR substitution model as determined with jModelTest 0.1.1 (Guindon and Gascuel 2003; Posada 2008). We executed two sets of four Markov chain Monte Carlo (MCMC) runs for 2 million generations, sampling every 100th generation, with burnin of 25%.
We estimated time to most recent common ancestor (TMRCA) for major lineages (with ≥5 samples) identified in the phylogenetic analyses, using BEAST software (Drummond and Rambaut 2007). Analyses were based on the complete data set of 181 sequences and all characters except indels (which were excluded because they are difficult to incorporate into coalescent analyses). Two independent analyses were each run for 108 generations under a GTR substitution model, selected using jModeltest 0.1.1. Analyses were combined and evaluated using Tracer 1.5 (Rambaut and Drummond 2007). In preliminary analyses assuming a relaxed (uncorrelated lognormal) molecular clock or exponential population growth, marginal posterior distributions of (respectively) clock rate variation and population growth rate included zero; we therefore assumed a strict molecular clock and constant population size for all subsequent runs. In order to estimate TMRCA in years, replicate analyses were carried out with calibration based on either fossil evidence or a published mutation rate. For fossil calibration we assigned an age of 3.0–3.4 million years, the estimated age of the oldest known Oxyria digyna macrofossil (Matthews and Ovenden 1990), to the root of the tree (using a normally distributed prior of 3.2 ± 0.14 million years, to give a 95% confidence interval of approximately the above range). For calibration from mutation rate, we used a mean substitution rate of ∼1.52 ± 0.06 × 10–9 substitutions per site per year as estimated for chloroplast noncoding regions by Yamane et al. (2006).
We investigated possible expansion events in the histories of major lineages of Oxyria digyna by performing mismatch analyses using Arlequin v. 3.5 (Excoffier et al. 2005) on all lineages with five or more samples (Clades A, A + C, B, D, E, and F). Mismatch analyses test the hypothesis that a mismatch distribution (the distribution of all possible pairwise differences for a group of sequences) approximates the unimodal shape predicted for a population or lineage that has undergone a demographic or spatial expansion event (Excoffier 2004); the mean number of pairwise differences can be used to infer time since an expansion event.
In western North America, the geographic area for which we had the highest sampling density, we assessed spatial patterns of genetic diversity for five different geographic regions. These were delineated to approximately reflect differences in tundra environment (high arctic to southern alpine) and glacial history (glaciated or not), as follows: (1) Alaska and the Yukon; (2) Northwest Territory and Nunavut; (3) northern British Columbia (latitudes 54.5–60.0°N); (4) southern British Columbia (latitudes 49.0°–54.5°N); and (5) continental western USA (south of 49.0°N). We tabulated overall numbers of haplotypes and numbers of haplotypes unique to a region (across all clades and within major clades) and calculated genetic diversity, Ĥ (the probability that two randomly chosen haplotypes in a sample are different) and molecular diversity, π (the mean number of pairwise differences between haplotypes in a sample), both within clades and overall clades for each region. Both diversity measures are sensitive to numbers and relative abundances of haplotypes; π also reflects divergence among haplotypes. All genetic diversity measures were calculated using Arlequin v. 3.5 (Excoffier et al. 2005).
Results
Sequence variation
Total aligned length of the trnH-psbA region was 315 bp, with 24 variable characters (18 substitutions, five indels of 5 to 8 bp in length, and one single-base T repeat). Total aligned length of the trnT-trnL region was 910 bp, with 68 variable characters used for analyses (41 substitutions and 27 indels of 3 to 19 bp); five additional 1- or 2-bp repeats could not be aligned unambiguously and were omitted from all analyses. There was no evidence for heterogeneity of the two regions (ILD test in PAUP*, P= 0.11) and all inferences are based on the combined two-region data set with an aligned length of 1225 bp. Sequences representing all distinct variants found in this study are deposited in GenBank; accession numbers are JQ080918-JQ080962 for the trnH-psbA region and JQ080963-JQ081007 for the trnT-trnL region.
Phylogenetic relationships among haplotypes
TCS analysis of the combined trnH-psbA and trnT-trnL data set yielded a total of 45 haplotypes (Fig. 3), which formed distinct groups corresponding with strongly supported clades (all with Bayesian posterior probabilities of 0.99–1.00) identified in the phylogenetic analyses (Fig. 3a). These included a widespread clade (A) comprising nearly all arctic samples; four clades (B to E) with variously overlapping geographic distributions in western North America; and a clade (F) found only in the mountains of central and southern Asia. A single, additional haplotype (26) was of indeterminate phylogenetic position but had greatest sequence similarity to Clade C (Fig. 3b). Haplotypes in sampled populations are indicated in Table 1.
Figure 3.
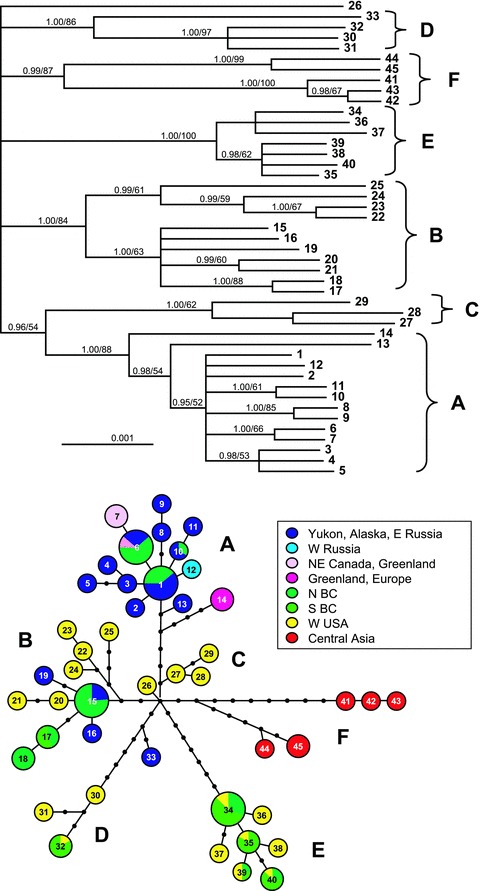
Relationships among Oxyria digyna haplotypes from the combined analysis of trnH - psbA and trnT-trnL cpDNA spacer regions. (A) Bayesian phylogenetic tree, with clade support indicated by Bayesian posterior probabilities and maximum parsimony bootstrap values. (B) TCS haplotype network. Circle diameters are proportional to the number of samples of each haplotype; connecting dots indicate inferred haplotypes not found in the data set. Colors indicate the geographic occurrences of haplotypes.
Although each of the six major clades was strongly supported, relationships among them were mostly unresolved in the Bayesian phylogenetic analysis (Fig. 3a). Clades A and C formed a well-supported higher-level clade, A + C (with posterior probability of 0.96; Fig. 3a).
Timelines and clade histories
Estimated TMRCAs for the analyzed major clades suggest that at least some of these have a very long history spanning much of the Pleistocene (Table 2). Estimated coalescence times were similar whether calibrated with fossil evidence or mutation rate. Clade F was identified as the oldest clade (mean TMRCA 2.58–3.02 × 106 years). The combined Clades A + C (mean TMRCA 1.42–1.66 × 106 years) and Clade D (mean TMRCA 1.27–1.45 × 106 year) appear to be substantially older than the remaining clades (A, B, and E), of which the youngest, Clade E, had a mean TMRCA of 458–516 × 103 years (Table 2). Data for Clade C were insufficient for analysis.
Table 2.
Estimates of time to most recent common ancestor (TMRCA) in years before present for major lineages of Oxyria digyna, based on combined trn H- psb A and trn T-L cpDNA sequence data.
| Lineage | TMRCA (from fossil date)1 Mean ± SD | TMRCA (from substitution rate)2 Mean ± SD |
|---|---|---|
| O. digyna (all extant clades combined)3 | – | 3,705,000 ± 821,000 |
| Clade A | 769,000 ± 279,000 | 893,000 ± 266,000 |
| Clades A + C | 1,415,000 ± 545,000 | 1,656,000 ± 583,000 |
| Clade B (all) | 777,000 ± 294,000 | 911,000 ± 288,000 |
| Clade B (northern subclade) | 475,000 ± 191,000 | 561,000 ± 190,000 |
| Clade D | 1,265,000 ± 518,000 | 1,454,000 ± 507,000 |
| Clade E | 458,000 ± 204,000 | 516,000 ± 199,000 |
| Clade F | 2,576,000 ± 696,000 | 3,024,000 ± 1,188,000 |
based on late Tertiary macrofossils of O. digyna (Matthews and Ovenden 1990).
based on substitution rate of Yamane et al. 2006) for cpDNA noncoding regions.
calculated from substitution rate only.
Mismatch analyses of major clades (A, A + C, B, D, E, and F) suggested spatial expansion events in the histories of all clades (P > 0.05 for all tests of fit to the model). Mismatch distributions were approximately unimodal for all of these clades, but with a second, small peak (suggesting the possibility of multiple expansion events) in Clade A + C and in Clade D. Estimates of τ had very broad 95% confidence limits for all clades and were insufficiently accurate for useful estimates of times since expansion; these are not reported here.
Haplotype diversity and geographic distribution
Haplotype diversity of Oxyria digyna in western North America (Table 3) varied among geographic regions, both overall and within clades. Overall haplotype diversity measures Ĥ and π were high in Regions 1 (Alaska/Yukon), 3 (northern British Columbia), and 5 (continental western USA) and lowest in Region 2 (Nunavut and Northwest Territories). Haplotype diversity was low in Region 4 (southern British Columbia), in marked contrast to Region 3 (northern British Columbia). The high overall values of π in several regions, especially Regions 3 and 5, reflect the presence of haplotypes belonging to divergent major clades in this region (Table 3). Haplotypes from several clades were present in all regions except Region 2. Within individual clades, haplotype diversity varied with latitude across geographic regions, tending to be highest either in the north (Region 1) or in the south (Region 5) (Table 3). These two geographic regions also harbored unique (region-specific) haplotypes for all major clades that were present, and unique haplotypes were concentrated in these regions (Table 3).
Table 3.
Oxyria digyna genetic diversity statistics by geographic region for localities in western North America, based on combined trn H- psb A and trn T-L cpDNA sequence data. Diversity measures for each region are given for all haplotypes combined (in bold) and for haplotypes within each major clade.
| Region | Clade | No. of samples | No. of haplotypes | No. of unique haplotypes | Gene diversity, Ĥ | Molecular diversity, π |
|---|---|---|---|---|---|---|
| Region 1 (Alaska/Yukon) | All | 29 | 11 | 6 | 0.894 | 4.259 |
| A | 23 | 6 | 3 | 0.834 | 1.652 | |
| B | 5 | 4 | 2 | 0.900 | 2.400 | |
| D | 1 | 1 | 1 | 0 | 0 | |
| Region 2 (Nunavut/NWT) | A (All) | 9 | 3 | 2 | 0.556 | 1.667 |
| Region 3 (northern BC) | All | 51 | 11 | 2 | 0.842 | 6.753 |
| A | 21 | 5 | 2 | 0.638 | 0.895 | |
| B | 22 | 3 | 0 | 0.507 | 1.221 | |
| E | 8 | 3 | 0 | 0.607 | 1.643 | |
| Region 4 (southern BC) | All | 39 | 6 | 0 | 0.775 | 4.289 |
| B | 4 | 1 | 0 | 0 | 0 | |
| D | 5 | 1 | 0 | 0 | 0 | |
| E | 30 | 4 | 0 | 0.653 | 1.076 | |
| Region 5 (w USA s of 49°N) | All | 26 | 20 | 15 | 0.979 | 6.200 |
| B | 7 | 6 | 6 | 0.952 | 4.762 | |
| C | 3 | 3 | 3 | 0.909 | 3.000 | |
| D | 4 | 3 | 2 | 0.833 | 3.333 | |
| E | 11 | 7 | 3 | 0.909 | 1.927 | |
| hap. 26 | 1 | 1 | 1 | 0 | 0 |
Three of the major lineages appear to have very disparate geographic distributions, with little overlap (Fig. 4a). The single Eurasian lineage, Clade F, is genetically and geographically remote from all other haplotype groups detected (although it was undoubtedly undersampled in this study). Clade C was found only in a small region of the Rocky Mountains in the western USA. The grouping of this small clade with the northern Clade A in the phylogenetic analysis suggests a formerly more contiguous distribution over the intervening region. The largely arctic Clade A is the most widely distributed lineage (Fig. 4a, b), spanning a continuous longitudinal range of at least 195° (from eastern Greenland across North America to eastern Siberia), and extending to 270° with inclusion of the Yamalo–Nenetsky region of Russia. Clade A comprises a subclade of closely related haplotypes (1 to 13) and a single divergent haplotype (14) of trans-Atlantic distribution (Greenland, Svalbard, and northern Europe). The main subclade is widely distributed, with a concentration of haplotypes in northwestern North America. Within this subclade, the earliest diverging haplotypes 1 and 13 were found from southwestern Alaska to northern British Columbia (Fig. 3b, 4b), with the highest diversity of haplotypes also in this region.
Figure 4.

Geographic distributions of all haplotypes (Fig. 3, Table 1) from the combined analysis of trnH - psbA and trnT-trnL cpDNA spacer regions of 181 samples of Oxyria digyna. (A) Major clades. (B) Individual haplotypes within major clades (haplotype 26 is included on the Clade C map). Haplotypes occurring at multiple localities are indicated by circles and those found at only one locality by triangles. Colors are used only to differentiate haplotypes and do not carry geographic information. Haplotype numbers correspond to those in Fig. 3.
Clades B, D, and E all occur in western North America, mostly south of Alaska and the Yukon, with broad geographical overlap (Table 3, Fig. 4a, b). Their relative abundances and the distribution patterns of haplotypes within each clade suggest different histories. Clade B includes 11 haplotypes divided into two well-marked and geographically distinct subclades (Fig. 3, Fig. 4b). The predominantly northern subclade (haplotypes 15 to 21) occurs primarily from southern coastal Alaska to northern British Columbia, with a disjunct southern lineage (haplotypes 20 and 21). A second, southern subclade (haplotypes 22 to 25) is restricted to cool, damp alpine habitats in the western USA. Clade D is a sparse clade with four divergent haplotypes (30 to 33) distributed over a large geographic region in western North America (Fig. 4b). Haplotype 33 in this clade, identified from a single northern Yukon locality, is sister to a clade of the remaining three haplotypes, found further south. Clade E with seven haplotypes (34–40) is geographically well-defined, occurring from north-central British Columbia to central California. Clades B, D, and E overlap in a large contact zone extending over much of the formerly glaciated region of western North America.
Discussion
An old and complex history
Oxyria digyna is an old and morphologically distinct species (Murray 1995; Brochmann and Brysting 2008). Its diversity of chloroplast haplotypes is consistent with its long fossil history extending back to the late Tertiary (Fredskild and Røen 1982; Mathews and Ovenden 1990; West 2000; Akopian et al. 2008; Bennike et al. 2010). Chloroplast haplotype diversity in Oxyria digyna is greater than in most other widely distributed arctic–alpine species investigated to date, including V. uliginosum (Alsos et al. 2005; Eidesen et al. 2007), A. alpina (Koch et al. 2006), and Juncus biglumis (Schönswetter et al. 2007), and is comparable to genetic diversity of multispecies lineages in genera such as Hordeum (Jakob and Blattner 2006). Alsos et al. (2005) identified three major cpDNA lineages within the arctic shrub V. uliginosum, which also has a Tertiary fossil history, and concluded that the earliest divergence of these lineages probably occurred at the beginning of the Quaternary glaciations (>700,000 years). Our estimated overall TMRCA for Oxyria digyna based on published molecular substitution rates suggests an origin as long ago as the late Tertiary (∼3.7 million years; Table 2), which is consistent with the fossil evidence. Murray (1995) suggested that Oxyria digyna is one of the oldest arctic–alpine species.
Oxyria digyna shows a striking phylogenetic split into at least six deeply divergent cpDNA lineages. Although relationships among these clades were not resolved in the phylogenetic analyses, estimated times to most recent common ancestor (TMRCA) for the different clades indicate that they are of substantially different ages. All of the major clades appear to be old (∼0.5–2.5 × 106 years), and some lineages (clade A + C, clade D, clade F) may have originated in the early Pleistocene. Molecular evidence increasingly indicates (Brochmann and Brysting 2008) that many species in the relatively young (2–3 million year old) arctic tundra ecosystem harbor greater genetic variation than expected. However, among arctic species for which data are available, only Saxifraga oppositifolia appears to have as much divergence among major cpDNA lineages (Abbott et al. 2000) as does Oxyria digyna. The presence of divergent lineages within a species indicates not only a long history, but sufficiently large populations that genetic diversity is maintained over time. Although shifts in climate through the Pleistocene may have led to genetic bottlenecks for many arctic–alpine species, highly cold-adapted species may have maintained very large ranges that simply shifted in response to climate cycles (Brochmann and Brysting 2008). This is a likely scenario for the very widespread and cold-tolerant Oxyria digyna.
Pan-arctic taxa are of diverse origins. Some of the oldest species may have arisen in southern alpine habitats and others in long-persistent high-latitude regions such as Beringia (Abbott and Brochmann 2003; Brochmann and Brysting 2008). Among widely distributed species, multiple sources seem likely. Evidence concerning the geographic origins of Oxyria digyna is equivocal. Its nearest relatives occur in Asia; its only congener, Oxyria sinensis, is restricted to western China and the sister taxon of Oxyria is the wholly Eurasian genus Rheum (Frye and Kron 2003). However, the oldest known fossils of Oxyria digyna are in arctic Canada and in Greenland. Oxyria digyna may have originated in central or eastern Asia but diversified in North America early in its evolution. The high haplotype diversity of major clades in northwestern North America suggests a long history of Oxyria digyna in this region. The complex patterns of lineage distribution and overlap in western North America likely reflect a series of regional extinction and recolonization cycles associated with multiple climatic fluctuations and glacial episodes in the later Pleistocene.
Histories of major lineages
The differing geographic distributions of Oxyria digyna lineages imply different Pleistocene histories. The western North American Clade C and the Eurasian Clade F were found only in southern alpine areas. Oxyria digyna is disjunct in northern and southern Russia (Hultén 1971; Fig. 2) and Clade F may represent a southern lineage, although it is probably more widespread than our sampling indicates. Hultén (1937) and others have suggested that many widely distributed arctic species migrated alternately south and north as glaciers advanced and retreated, sometimes resulting in disjunct geographic ranges with isolated southern populations.
In contrast, the widespread arctic distribution of Clade A suggests a high latitude origin. Clades A and C form a higher-level clade, which could have spread from Asia or possibly originated in western North America with subsequent divergence of Clades A and C to the north and south. The high haplotype diversity of Clade A in northwestern North America and the distribution of its ancestral haplotypes are consistent with a long presence in the Beringian region, with more recent eastward and westward spread through arctic regions. Oxyria digyna is moderately well dispersed by wind (Tackenberg and Stöcklin 2008) and probably was able to spread rapidly into new habitat as continental ice sheets receded and climatic conditions became more favorable. Central and eastern regions of arctic Canada and northern Greenland are occupied almost exclusively by a single derived haplotype, suggesting recent migration. The history of arctic lineages of Oxyria digyna in northern Asia is less clear. Fossils indicate that it occurred in northern Siberia during and preceding the LGM (Kienast et al. 2001; Lopatina and Zanina 2006), and clade A haplotypes in northwestern Siberia are closely related to those from western North America. Broad geographic regions of low haplotype diversity, with higher diversity in refugial areas such as Beringia, have been observed in other arctic plants, including A. alpina (Koch et al. 2006), Ranunculus glacialis (Schönswetter et al. 2003), and Draba species (Skrede et al. 2009). The single amphi-Atlantic haplotype 14 in Clade A may represent a distinct lineage that persisted through the LGM in either European or northeast North American refugia.
Three lineages of Oxyria digyna (Clades B, D, and E) have an unusually broad zone of overlap extending over much of British Columbia and adjacent regions. The sympatry of these divergent cpDNA lineages and the geographic distributions of haplotypes within each lineage indicate a long and complex history of Oxyria digyna in western North America, involving repeated expansion, migration, and local extinction. Mismatch analyses suggested spatial expansion events in the histories of all these clades, though we could not ascertain whether these events coincided with initial clade divergence or occurred subsequently. The differences in estimated age and relative abundance of these clades suggest successive episodes of migration from one or more source regions. Clade D is an old (∼1.2–1.5 × 106 years), sparse clade and probably represents an early-colonizing lineage of which only remnant haplotypes now remain. Clades B and E, in contrast, are of more recent origin (∼0.4–0.9 × 106 years). Clades B and D both show evidence of range fragmentation resulting from glacial advances. Clade B has a particularly complex pattern of north–south disjunctions between related haplotypes, suggesting multiple fragmentation events in its history.
Persistence through the last glacial maximum in western North America
In species with a long history, early distributions of lineages are difficult to infer because of the strong influence of later events. Present-day spatial patterns of genetic variation are often best understood in relation to the most recent glaciation and associated climatic changes. This is true for Oxyria digyna, although the time since the LGM (18,000 years) represents less than 1% of the history of the species (>3 million years) and thus almost all of the observed haplotypes must predate the LGM. Most tundra and boreal species now present in western North America persisted through the last glacial maximum north or south of the continental ice sheets (Jorgensen et al. 2003; Beatty and Provan 2010); some also persisted in refugia between these limits (Loehr et al. 2006; King et al. 2009). Oxyria digyna gives evidence of persistence in all of these areas, with different refugia harboring different combinations of clades.
The ice-free area of Beringia formed a northern refugium extending over most of Alaska and the northern half of the Yukon (Fig. 2) in which Clades A and D almost certainly persisted through the LGM. Clade A is widespread in the Beringian region, which is also where its haplotype diversity is highest. Clade D includes a haplotype (33) that was found only in Beringia; the other haplotypes occur much farther south and likely persisted elsewhere. Clade B may also have persisted in Beringia or nearby cryptic refugia; it is distributed along the southern margin of Alaska including the Aleutian Islands.
Areas south of the continental ice sheets also provided extensive suitable habitat. Two separate lineages within Clade B and all haplotypes of Clade C are presently restricted to cool moist alpine habitats south of the limits of continental ice. These are probably remnants of much more extensive populations that were widespread during the LGM and subsequently became isolated into small, disjunct patches of alpine habitat. Clade E and the southern subclade of group D also occur south of the glacial boundary, but extend north into formerly glaciated regions of British Columbia. These likely persisted, at least in part, south of the continental ice sheets, migrating northward as the ice receded. Clade E occurs from the western USA north over much of British Columbia, with some haplotypes found almost entirely within the limits of the Cordilleran ice sheet but with highest diversity of haplotypes in the south. Northward movement from south of the Cordilleran ice sheet and persistence within its boundaries are both plausible for this clade.
Our evidence suggests persistence of Oxyria digyna within the boundaries of the Cordilleran ice sheet during the LGM. We observed marked differences in haplotype diversity between northern and southern British Columbia (Table 3, Fig. 4b), although these regions, both within the boundaries of the Cordilleran ice sheet, are thought to have had a similar glacial history over the last 25,000–30,000 years. Genetic diversities of Clades A and B were nearly as high in northern British Columbia as in adjacent unglaciated regions further north, and included haplotypes found only within the boundaries of the Cordilleran ice sheet. In contrast, within-clade genetic diversities in the southern part of the ice sheet are much lower, implying continuous ice cover at the LGM. One hypothesis to explain these differences is that the extensive tundra habitat of northern refugia facilitated more extensive postglacial migration of Oxyria digyna from the north. However, haplotype diversity of Oxyria digyna is even higher south of the Cordilleran ice sheet than north of it (Table 3), yet most of this variation is not found in southern British Columbia. Thus persistence of Oxyria digyna in ice-free habitats within the northern Cordilleran ice sheet, rather than differential subsequent migration, appears to be the most likely explanation for the observed pattern of haplotype diversity. Persistence in northern British Columbia during the LGM is also indicated for two vertebrates (Loehr et al. 2006, Shafer et al. 2010a), and may have been possible for other species (Shafer et al. 2010b). The occurrence of refugial habitat within the margins of continental ice sheets in Europe is similarly supported by recent molecular studies (Brochmann et al. 2003; Westergaard et al. 2011).
Conclusions
Oxyria digyna is a very old species, as indicated by both fossil evidence and the cpDNA patterns reported here. It has higher genetic diversity than most arctic–alpine species studied thus far, reflecting its age, but also suggesting a widespread distribution over time with few bottlenecks. Its history rivals that of multispecies complexes, reflecting both ancient and more recent cycles of range contraction and expansion. We show that different lineages within a single species may have very different histories and dispersal patterns spanning hundreds of thousands of years. Although the geographical origin of Oxyria digyna is most likely in Eurasia, our evidence suggests a long and complex history of this species in western North America. Differing ages of lineages and distributions of haplotypes indicate repeated migrations over multiple glacial cycles. Our genetic evidence indicates refugia for Oxyria digyna not only north and south of continental ice sheets but also within the limits of the northern Cordilleran ice sheet in northern British Columbia. Thus our findings support the emerging generalization that cryptic refugia were important for plant survival and recolonization of glaciated regions, as already demonstrated for Europe (Brochmann et al. 2003; Brochmann and Brysting 2008).
Acknowledgments
We are grateful to the many colleagues who provided plant samples (listed in Table 1) and assisted us in other ways. We thank the Canadian Museum of Nature and the University of Colorado for permission to sample tissue from herbarium specimens. Field work in Russia was made possible by generous support from the John and Joan Walton Innovation Fund and the October Hill Foundation. Logistical support and assistance with travel and collecting arrangements in Russia were provided by A. Berkutenko, A. Shmakov and especially M. Olonova, to whom we are very grateful. Lab work was supported by funding from the Royal British Columbia Museum and by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada to G. A. Allen and R. J. Hebda. Finally, we thank J. A. Antos, B. Koop, R. J. Nelson, and several anonymous reviewers for comments on previous versions of the manuscript.
References
- Abbott RJ, Comes HP. Evolution in the Arctic: a phylogeographic analysis of the circumarctic plant, Saxifraga oppositifolia (Purple saxifrage) New Phytol. 2003;161:211–224. [Google Scholar]
- Abbott RJ, Smith LC, Milne RI, Crawford RMM, Wolff K, Balfour J. Molecular analysis of plant migration and refugia in the Arctic. Science. 2000;289:1343–1346. doi: 10.1126/science.289.5483.1343. [DOI] [PubMed] [Google Scholar]
- Abbott RJ, Brochmann C. History and evolution of the arctic flora: in the footsteps of Eric Hultén. Mol. Ecol. 2003;12:299–313. doi: 10.1046/j.1365-294x.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- Akopian J, Gabrielyan I, Freitag H. Fossil fruits of Salsola L. s.l. and Halanthium K.Koch (Chenopodiaceae) from Lower Pleistocene lacustrine sediments in Armenia. Feddes Repertorium. 2008;119:225–236. [Google Scholar]
- Alsos IG, Eidesen PB, Ehrich D, Skrede I, Westergaard K, Jacobsen GH, Landvik JY, Taberlet P, Brochmann C. Frequent long-distance plant colonization in the changing Arctic. Science. 2007;316:1606–1609. doi: 10.1126/science.1139178. [DOI] [PubMed] [Google Scholar]
- Alsos IG, Engelskjøn T, Gielly L, Taberlet P, Brochmann C. Impact of ice ages on circumpolar molecular diversity: insights from an ecological key species. Mol. Ecol. 2005;14:2739–2753. doi: 10.1111/j.1365-294X.2005.02621.x. [DOI] [PubMed] [Google Scholar]
- Bain JF, Golden JL. Chloroplast haplotype diversity patterns in Packera pauciflora (Asteraceae) are affected by geographical isolation, hybridization, and breeding system. Can. J. Bot. 2005;83:1039–1045. [Google Scholar]
- Beatty GE, Provan J. Refugial persistence and postglacial recolonization of North America by the cold-tolerant herbaceous plant Orthilia secunda. Mol. Ecol. 2010;19:5009–5021. doi: 10.1111/j.1365-294X.2010.04859.x. [DOI] [PubMed] [Google Scholar]
- Bennike O, Knudsen KL, Abrahmsen N, Böcher J, Cremer H, Wagner B. Early Pleistocene sediments on Store Koldewey, northeast Greenland. Boreas. 2010;39:603–619. [Google Scholar]
- Birks HH. The late-Quaternary history of arctic and alpine plants. Plant Ecol. Divers. 2008;1:135–146. [Google Scholar]
- Birks HJB, Willis KJ. Alpines, trees, and refugia in Europe. Plant Ecol. Divers. 2008;1:147–160. [Google Scholar]
- Brochmann C, Brysting AK. The Arctic – an evolutionary freezer? Plant Ecol. Divers. 2008;1:181–195. [Google Scholar]
- Brochmann C, Gabrielsen TM, Nordal I, Landvik JY, Elven R. Glacial survival or tabula rasa? The history of North Atlantic biota revisited. Taxon. 2003;52:417–450. [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cronn RC, Small RL, Haselkorn T, Wendel JF. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am. J. Bot. 2002;89:707–725. doi: 10.3732/ajb.89.4.707. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;9:11–15. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214–221. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumolin-Lapegue S, Pemonge M-H, Petit RJ. An enlarged set of consensus primers for the study of organelle DNA in plants. Mol. Ecol. 1997;6:393–397. doi: 10.1046/j.1365-294x.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- Ehlers J, Gibbard PL. The extent and chronology of Cenozoic Global Glaciation. Quat. Int. 2007;164–165:6–20. [Google Scholar]
- Ehrich D, Alsos IG, Brochmann C. Where did the northern peatland species survive the dry glacials: cloudberry (Rubus chamaemorus) as an example. J. Biogeogr. 2008;35:801–814. [Google Scholar]
- Eidesen PB, Alsos IG, Popp M, Stensrud Ø, Suda J, Brochmann C. Nuclear vs. plastid data: complex Pleistocene history of a circumpolar key species. Mol. Ecol. 2007;16:3902–3925. doi: 10.1111/j.1365-294X.2007.03425.x. [DOI] [PubMed] [Google Scholar]
- Eriksen B, Töpel MH. Molecular phylogeography and hybridization in members of the circumpolar Potentilla sect. Niveae (Rosaceae) Am. J. Bot. 2006;93:460–469. doi: 10.3732/ajb.93.3.460. [DOI] [PubMed] [Google Scholar]
- Excoffier L. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol. Ecol. 2004;13:853–864. doi: 10.1046/j.1365-294x.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fedorov AA. Chromosome numbers of flowering plants. Koenigstein, Germany: Koeltz Science Publishers; 1974. [Google Scholar]
- Fredskild B, Røen U. Macrofossils in an interglacial peat deposit at Kap København, North Greenland. Boreas. 1982;11:181–185. [Google Scholar]
- Frye ASL, Kron KA. rbcL phylogeny and character evolution in Polygonaceae. Syst. Bot. 2003;28:326–332. [Google Scholar]
- Funder S. Ice-age plant refugia in East Greenland. Palaeobiol. Palaeoclimateol. Palaeoecol. 1979;28:279–295. [Google Scholar]
- Goldblatt P, Johnson DE, editors. Index to plant chromosome numbers. St. Louis, MO: Missouri Botanical Garden; 1979–2010. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hebda RJ, Howes D, Maxwell B. Brooks Peninsula as an ice age refugium. In: Hebda RJ, Haggarty J, editors. Brooks Peninsula, An Ice Age Refugium. Victoria, British Columbia: Ministry of Environment, Lands and Parks; 1997. pp. 15.1–15.7. B.C. Parks Occasional Paper No. 5. [Google Scholar]
- Heide OM. Ecotypic variation among European arctic and alpine populations of Oxyria digyna. Arct. Antarct. Alp. Res. 2005;37:233–238. [Google Scholar]
- Heusser CJ. North Pacific coastal refugia – the Queen Charlotte Islands in perspective. In: Scudder GGE, Glessler N, editors. The Outer Shores. Skidegate, British Columbia: Queen Charlotte Islands Museum Press; 1989. pp. 91–106. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinform. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hultén E. Outline of the history of arctic and boreal biota during the Quaternary Period. New York: Lehre J. Cramer; 1937. [Google Scholar]
- Hultén E. The circumpolar plants. 2, Dicotyledons. Stockholm: Almqvist & Wiksell; 1971. [Google Scholar]
- Jakob SS, Blattner FR. A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol. Biol. Evol. 2006;23:1602–1612. doi: 10.1093/molbev/msl018. [DOI] [PubMed] [Google Scholar]
- Jorgensen JL, Stehlik I, Brochmann C, Conti E. Implications of ITS sequences and RAPD markers for the taxonomy and biogeography of the Oxytropis campestris and O. arctica (Fabaceae) complexes in Alaska. Am. J. Bot. 2003;90:1470–1480. doi: 10.3732/ajb.90.10.1470. [DOI] [PubMed] [Google Scholar]
- Kienast F, Siegert S, Dereviagin A, Mai DH. Climatic implication of Late Quaternary plant macrofossil assemblages from the Taymyr Peninsula Siberia. Glob. Planet. Change. 2001;31:265–281. [Google Scholar]
- King MG, Horning ME, Roalson EH. Range persistence during the last glacial maximum: Carex macrocephala was not restricted to glacial refugia. Mol. Ecol. 2009;18:4256–4269. doi: 10.1111/j.1365-294X.2009.04280.x. [DOI] [PubMed] [Google Scholar]
- Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae) Mol. Ecol. 2006;15:825–839. doi: 10.1111/j.1365-294X.2005.02848.x. [DOI] [PubMed] [Google Scholar]
- Loehr J, Worley K, Grapputo A, Carey J, Veitch A, Coltman DW. Evidence for cryptic glacial refugia from North American mountain sheep mitochondrial DNA. J. Evol. Biol. 2006;19:419–430. doi: 10.1111/j.1420-9101.2005.01027.x. [DOI] [PubMed] [Google Scholar]
- Lopatina DA, Zanina OG. Paleobotanical analysis of material from fossil gopher burrows and upper Pleistocene host deposits, the Kolyma River lower reaches. Stratigr. Geo. Correl. 2006;14:549–560. [Google Scholar]
- Marr KL, Allen GA, Hebda RJ. Refugia in the Cordilleran ice sheet of western North America: chloroplast DNA diversity in the Arctic-alpine plant Oxyria digyna. J. Biogeogr. 2008;35:1323–1334. [Google Scholar]
- Matthews JV, Jr, Ovenden LE. Late Tertiary plant macrofossils from localities in arctic/subarctic North America: a review of the data. Arctic. 1990;43:364–392. [Google Scholar]
- Michewich MF, Farris SJ. The implications of congruence in Menidia. Syst. Zool. 1981;30:351–370. [Google Scholar]
- Mooney HA, Billings WD. Comparative physiological ecology of arctic and alpine populations of Oxyria digyna. Ecol. Monogr. 1961;31:1–29. [Google Scholar]
- Murray DF. Causes of arctic plant diversity. In: Chapin RS, Korner C, editors. Arctic and Alpine Diversity: Patterns, Causes and Ecosystem Consequences. Heidelberg: Springer; 1995. pp. 21–32. [Google Scholar]
- Ogilvie RT. Disjunct vascular flora of northwestern Vancouver Island in relation to Queen Charlotte Islands’ endemism and coastal Pacific refugia. In: Scudder GGE, Glessler N, editors. The Outer Shores. Skidegate, British Columbia: Queen Charlotte Islands Museum Press; 1989. pp. 127–130. [Google Scholar]
- Posada D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4. 2007. Available at: http://beast.bio.ed.ac.uk/Tracer.
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae) Am. J. Bot. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Schönswetter P, Paun O, Tribsch A, Niklfeld H. Out of the Alps: colonization of Northern Europe by East Alpine populations of the glacier buttercup Ranunculus glacialis L. (Ranunculaceae) Mol. Ecol. 2003;12:3373–3381. doi: 10.1046/j.1365-294x.2003.01984.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Suda J, Popp M, Weiss-Schneeweis H, Brochmann C. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Mol. Phylogenet. Evol. 2007;42:92–103. doi: 10.1016/j.ympev.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol. Ecol. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Shafer ABA, Côté SD, Coltman DW. Hot spots of genetic diversity descended from multiple Pleistocene refugia in an alpine ungulate. Evolution. 2010a;65:125–138. doi: 10.1111/j.1558-5646.2010.01109.x. [DOI] [PubMed] [Google Scholar]
- Shafer ABA, Cullingham CI, Côté SD, Coltman DW. Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol. Ecol. 2010b;19:4589–4621. doi: 10.1111/j.1365-294X.2010.04828.x. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schillng EE, Small RL. Comparison on whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in Angiosperms: the tortoise and the hare III. Am. J. Bot. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Skrede I, Borgen L, Brochmann C. Genetic structuring in three closely related circumpolar plant species: AFLP versus microsatellite markers and high-arctic versus arctic–alpine distributions. Heredity. 2009;102:293–302. doi: 10.1038/hdy.2008.120. [DOI] [PubMed] [Google Scholar]
- Skrede I, Eidesen PB, Portela RP, Brochmann C. Refugia, differentiation and postglacial migration in arctic-alpine Eurasia, exemplified by the mountain avens (Dryas octopetala L.) Mol. Ecol. 2006;15:1827–1840. doi: 10.1111/j.1365-294X.2006.02908.x. [DOI] [PubMed] [Google Scholar]
- Stewart JR, Lister AM. Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 2001;16:608–613. [Google Scholar]
- Stöcklin J, Bäumler E. Seed rain, seedling establishment and clonal growth strategies on a glacier foreland. J. Veg. Sci. 1996;7:45–56. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tackenberg O, Stöcklin J. Wind dispersal of alpine plant species: a comparison with lowland species. J. Veg. Sci. 2008;19:109–118. [Google Scholar]
- Tate JA, Simpson BB. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst. Bot. 2003;28:723–737. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach NV, Hoffmann MH, Röser M, von Hagen KB. Temporal patterns of evolution in the Arctic explored in Artemisia L. (Asteraceae) Plant Ecol. Divers. 2008;1:161–169. [Google Scholar]
- Tremblay NO, Schoen DJ. Molecular phylogeography of Dryas integrifolia: glacial refugia and past recolonization. Mol. Ecol. 1999;8:1187–1198. doi: 10.1046/j.1365-294x.1999.00680.x. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RG. Plant life of the Quaternary cold stages: Evidence from the British Isles. CambridgeUK: Cambridge Univ. Press; 2000. [Google Scholar]
- Westergaard KB, Alsos IG, Popp M, Engelskjøn T, Flatberg KI, Brochmann C. Glacial survival may matter after all: nunatak signatures in the rare European populations of two west-arctic species. Mol. Ecol. 2011;20:376–393. doi: 10.1111/j.1365-294X.2010.04928.x. [DOI] [PubMed] [Google Scholar]
- Whittaker RJ. Plant population patterns in a glacier foreland succession: pioneer herbs and later-colonizing shrubs. Ecography. 1993;16:117–136. [Google Scholar]
- Yamane K, Yano K, Kawahara T. Pattern and rate of indel evolution inferred from whole chloroplast intergenic regions in sugarcane, maize and rice. DNA Res. 2006;13:197–204. doi: 10.1093/dnares/dsl012. [DOI] [PubMed] [Google Scholar]


