Abstract
The economic importance of bovine viral diarrhea is increasing with the emergence of seemingly more virulent viruses, as evidenced by outbreaks of hemorrhagic syndrome and severe acute bovine viral diarrhea beginning in the 1980s and 1990s. It appears that evolutionary changes in bovine viral diarrhea virus were responsible for these outbreaks. The genetic properties of the classical bovine viral diarrhea virus that contribute to the basis of current diagnostic tests, vaccines, and our understanding of pathogenic mechanisms are now being reevaluated because of these “new” virus strains. This shift in virulence has confounded both nomenclature and the significance of current bovine viral diarrhea virus categorization.
The purpose of this review is to summarize our current understanding of bovine viral diarrhea virus with a chronological review of prevailing scientific tenets and practices as described in clinical and scientific North American veterinary journals and textbooks. The first part of this review describes how we have arrived at our current understanding of the viruses, the diseases, and their nomenclature. The second part of the review deals with current concepts in virology and how these concepts may both explain and predict bovine viral diarrhea virus pathogenesis. By reviewing how knowledge of bovine viral diarrhea has evolved and the theories of how the virus itself is able to evolve, the interpretation of diagnostic tests are more effectively utilized in the control and treatment of bovine viral diarrhea virus associated disease.
Bovine viral diarrhea (BVD), mucosal disease (MD), persistent infections (PI), biotypes, and genotypes are all terms that have evolved from attempts to organize the complicated pathogenicity of bovine viral diarrhea virus (BVDV). The historical review of BVD from the first accounts in the 1940s to the devastating acute BVD outbreaks in Ontario in the 1990s and a review of what is known about the virus itself, as presented here, is intended to shed light on the rationale behind the terminology and to assist practitioners in understanding the pathogenesis of infections with BVDV and related viruses.
Medline, accessed via PubMed was used to collect the majority of the references that were used in describing experimental work. The Commonwealth Animal Bureaux (CAB) CD (1979 to present), Agricola (1970 to present), Biological & Agricultural Index (1983 to present), and Biological Abstracts (1980 to present) electronic databases were used for the earlier, specific veterinary publications for the historical review, as were older editions of textbooks such as Veterinary Pathology by Jubb, Kennedy, and Palmer. Old textbooks, as well as older review articles, served as resources to preliminary BVDV research articles providing a picture of the current understanding at the time.
Historical evolution of our understanding of clinical and pathological manifestations of bovine viral diarrhea
A disease of unknown origin affecting cattle was first described in the 1940s. In western Canada, “X disease” was described as a disease with 2 forms; subacute and acute (1). It was believed that the subacute form had likely been present in the region for years: 1 to 2 animals in a herd would be affected and die, with further cases not appearing again for several weeks. The acute form was characterized as a 7- to 10-day illness in young animals, ending in death. Mature cattle exhibited even more severe and explosive signs, dying within 3 to 4 d. The clinical signs included pyrexia; watery and bloody diarrhea; dehydration; tenesmus; tachypnea; tachycardia; drooping ears; anorexia; excessive lachrimation; nasal discharge; hypersalivation; and development of ulcers of the nares, muzzle, lips, and oral cavity mucous membranes. The original report also described skin lesions about the inguinal and perineal regions, the inner thighs, and inside the ears.
Postmortem findings were extensive, though it is unclear whether the full range of lesions were typically observed in each affected animal. Erosive lesions were observed in the nares and in the trachea, as well as throughout the upper and lower alimentary system. The forestomachs were mostly unaffected; the abomasum was described as being nearly denuded of all epithelium. Intestinal lesions were more severe distally and characterized by ulceration. Petechial hemorrhage was found in the cortex of the kidneys, on the ureters, and within enlarged lymph nodes. It was also noted that there was an astonishing decrease in blood volume and that clotting time was also reduced.
The same year, clinicians and scientists at Cornell University reported an outbreak of an apparently new, transmissible disease in cattle during the spring and summer of 1946 (2), similar to what had been reported from western Canada (1). This “new” disease was characterized by severe diarrhea, depression, anorexia, and ulceration of oral mucosa. The clinical signs and postmortem findings were consistent with what was being described as X disease in Saskatchewan. However, Olafson et al (2) in New York restricted the use of the term X disease to a similar though chronic disease characterized by emaciation; thick and dry skin; proliferative lesions on the tongue, palate, and esophagus; and thickening of the gall bladder and bile ducts, resulting in very high mortality rates. Most notably, their attempts at transmission of X disease had failed (2). In contrast, their “new” disease was associated with additional signs of respiratory disease, leukopenia, a drop in milk production, and increased abortion rates, and it was reproducible. The infectious nature of the disease was confirmed by “dosing” a cow with a composite fecal sample from the most severely affected animals in the 1st herd affected. The “dosed” cow did not become ill; however, after 3 wk, other cattle in the herd did. Researchers at Cornell University later reproduced the disease by using a splenic emulsion and blood from acutely ill animals (2). As no bacteria could be cultured from the inoculum, it was assumed that the infectious agent was viral. Inoculated calves were 8 to 14 mo of age. Several exposed experimental animals failed to develop clinical signs, while others showed only mild signs. Animals that developed leukopenia did develop clinical signs. In some animals, pyrexia was followed by salivation and nasal discharge. Severely affected animals developed diarrhea and became dehydrated and emaciated. One to 3 d after the appearance of diarrhea (about day 10), mouth ulcers appeared. In nonfatal cases, oral mucosal lesions healed quickly. Postmortem findings in the more severely affected cattle were consistent with those described for X disease. This reproducible disease, with its varying severity, became known as virus diarrhea (VD) of cattle.
In the 1950s, farmers in Iowa began to experience a disease in cattle that they had not observed before. Although veterinarians at Iowa State University saw that it had similarities to VD of cattle, they believed that they were observing a different disease (3), because the nasal discharge was mucopurulent rather than mucoid; the disease usually did not recur on the same farm during succeeding years; and efforts to reproduce the disease in calves by parenteral injections of blood, tissue extracts, and cerebrospinal fluid resulted only in a transient pyrexia. The gross lesions of the new syndrome, which they termed mucosal disease (MD), varied. They were primarily erosions and hemorrhages of the intestinal tract, with minimal inflammatory cell infiltration.
In 1957, researchers isolated and cultured a virus from a case of MD (4), though reproduction of the disease with this virus still remained elusive. The virus was cytopathic (cp) to the cultured cells, causing morphological changes such as vacuolation and cell death. In the same year, researchers at Cornell University propagated a noncytopathic (ncp) virus from cases of typical VD of cattle (5). The relationship, if any, between these 2 isolates was unknown at the time.
Three years later, researchers at Cornell University grew a cp virus from a case of VD from cattle in Oregon. This virus, named Oregon C24V, when inoculated into cattle, resulted in clinical signs that resembled VD and antibodies that neutralized both cp and ncp strains of VD virus (6). In attempts to clarify the nature of these “new” viruses and the relationship between them, researchers undertook what were, at the time, standard viral characterization studies of physical properties (such as ultrafiltration properties, sedimentation coefficient, and buoyant density) and antigenic properties. A critical discovery was that serum from cattle with typical MD or VD would neutralize the cp effect of the cp viruses in cell culture. Not only did characterization of the physical and antigenic properties allow researchers to determine that the MD virus was very similar to VD viruses, but also that these bovine viruses were related to hog cholera virus (now referred to as classical swine fever virus (CSFV)) (7).
In 1961, the cp VD virus strain Oregon (C24V) was passaged (32 times) in bovine cell culture, until its ability to cause seroconversion without clinical signs was confirmed. This attenuated Oregon virus became the first commercial VD virus vaccine (8). The widespread use of this modified live virus vaccine was subsequently associated with numerous complications, including vaccine-induced MD, reproductive failure, and congenital defects (9).
By the end of the 1960s, what was originally described as X disease had mushroomed into a spectrum of viral diseases, and a general consensus about VD of cattle and MD had emerged (10). Although gross lesions of VD and MD were indistinguishable, there were seemingly substantial differences in clinical disease. Virus diarrhea of cattle was considered to be associated with enzootic disease with sporadic outbreaks with high morbidity but low mortality. Conversely, MD was observed to afflict young cattle with low morbidity but mortality of near 100%. There was little evidence that MD was contagious and attempts to transmit the disease experimentally had failed. When researchers tried to reproduce MD with isolates of cp virus, the resultant disease resembled mild VD of cattle. This observation, as well as similarities in the distribution and microscopic appearance of lesions, contributed to the speculation (though without scientific conviction) that MD and VD of cattle were the ‘same disease with minor variations’ (10).
Research into the pathogenesis of the bovine viral diarrhea-mucosal disease complex (BVD-MD), as it became to be known, in the 1960s and 1970s concentrated on experimental infections, particularly in pregnant cattle and neonatal calves. Abortions and various teratogenic disorders were found to be associated with intrauterine bovine viral diarrhea virus (BVDV) infections. Such experiments also led to the understanding that neonatal calves congenitally infected with BVDV were poor doers and usually did not survive for more than a few months, eventually succumbing to what was previously described as chronic MD (11). These poor doers were discovered to be persistently infected (PI) with BVDV. Most importantly, PI calves were found to have an immunological abnormality; specifically, they did not produce detectable antibodies to BVDV (12). The discovery of a healthy bull persistently infected with a ncp BVDV for more than 2 y with a conspicuous absence of viral antibodies further contributed to the eventual elucidation of BVDV persistent infections (13).
Experiments in which BVDV was inoculated into 58- to 125-day old fetuses of seropositive cows and into seronegative cows that were 42- to 114-days pregnant produced clinically normal calves that were PI with the specific isolate of BVDV, yet these calves remained completely seronegative to any BVDV isolate. Even reinoculation of these PI cattle with their homologous isolate did not produce neutralizing antibody. The PI calves were found to be immunocompetent, as they were able to develop neutralizing antibodies to other infectious agents, such as infectious bovine rhinotracheitis and parainfluenza-3 viruses, and agglutinating antibodies to Pasteurella hemolytica (14). The key to the absence of antibody to BVDV in the PI calves was found to be the timing of their infection in utero with the development of their immune systems, inasmuch as the developing immune system mistakenly recognizes BVDV as “self” and antibodies to that particular BVDV are not made. The ability to cause PI seemed to be restricted to ncp strains. Although cp BVDV was found to be able to cross the placenta, it was unable to cause PI. In addition, ncp BVDV appeared to cause abortion more readily than did the cp BVDV (14,15).
Throughout the 1970s, the invariably fatal MD, first described in 1953 (3), was still only tentatively associated with BVDV. Even though the cp virus isolated from cattle with MD was found to be serologically similar to BVDV, attempts to reproduce the disease resulted only in mild illness. Not until 1984 were researchers able to reproduce classical MD with BVDV (16). Mucosal disease was replicated when a cp BVD-MD virus was inoculated into a PI animal. It was subsequently established that MD occurs only in PI animals after superinfection with a strain of cp BVDV that is antigenically similar to the ncp BVDV already persistently infecting the animal. An antigenically similar cp virus can also arise from mutations in the persisting ncp virus (17).
Now, with the ability to reproduce MD, more planned and valid comparisons to BVD could be made. It was found that the combinations of cp and ncp BVDV in PI animals caused a much more severe disease than either the cp or the ncp virus could alone. This experimental disease caused by the dual infections resembled natural MD (18).
What was known and supposed about BVDV was reviewed in the late 1980s (19). Bovine viral diarrhea virus was classified as a pestivirus, together with border disease virus (BDV) and classical swine fever virus (CSFV), belonging to the family Togaviridae. Cases of severe clinical disease, with severe gross and histological lesions caused by BVDV, were arbitrarily designated as MD. In contrast, the mild disease associated with minimal clinical changes from which ncp BVDV was isolated was called BVD. The significance of ncp BVDV was thought to be solely as a reservoir of viruses, that were detrimental only if infecting pregnant cattle. Indeed, secondary transmission among immunocompetent animals was thought unlikely or insignificant.
As stated above, the ability of ncp BVDV to cause significant disease was somewhat disregarded in light of the elucidation of MD pathogenesis. However, in the late 1980s, a severe hemorrhagic syndrome in adult cattle and veal calves associated with ncp BVDV (without an accompanying cp strain) was reported in several countries (20,21,22). This BVD hemorrhagic syndrome was characterized by fever, marked thrombocytopenia resulting in bloody diarrhea, epistaxis, petechial and ecchymotic hemorrhages on mucosal surfaces, and bleeding from injection sites (22,23). One isolate from the outbreak (isolate 890) was chosen as a reference type and subsequently sequenced (24,25).
In 1993, in the Great Lakes regions (26), Quebec (27), and England (28), outbreaks of disease with clinical signs similar to those of MD and, more variably, to the hemorrhagic syndrome were reported to affect dairy herds, beef cattle, and veal calves. The disease affected all age groups. Arguably, the most severely affected region was southern Ontario. Respiratory disease and diarrhea were the most common initial signs. Oral lesions were frequently observed in older cattle, infected dams aborted, and herd mortality rates averaged 25% (26). As with the previously reported hemorrhagic syndrome, this outbreak of what has become known as “severe acute BVD” was attributed to ncp BVDV, rather than the expected cp/ncp combination that causes MD.
These outbreaks signalled a marked departure from the previous concept that BVD and MD could be reliably distinguished by clinical outcomes and lesions. It suggested that there was more variation in virulence of ncp BVDV than had previously been recognized, and that more sophisticated techniques for virus characterization were necessary for the understanding what was responsible for the wide variation in virulence amongst isolates.
Current concepts in the evolutionary relationship among pestiviruses
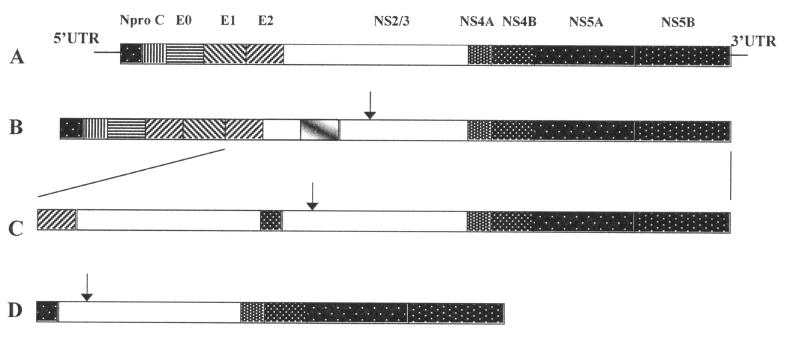
In considering methods for characterizing or evaluating the determinants of BVDV virulence, it is important to review the nature of pestivirus genomes. Pestiviruses are nonsegmented, single, sense stranded (positive polarity (+)) RNA viruses, now classified as a genus of the family Flaviviridae. The flavivirus family has 3 genera: flaviviruses, pestiviruses, and hepatitis C-like viruses. The genomes of BVDV consist of 1 long open reading frame (ORF), flanked by 2 untranslated regions (UTR). The ORF is translated into 1 long polypeptide, which is subsequently cleaved into the individual viral proteins by viral and cellular proteases. With the exception of the first protein, Npro, which is one such viral protease, the BVDV genome is organized with the structural protein genes (the capsid (C), and 3 envelope glycoproteins) at the 5' end of the ORF and the nonstructual protein genes (NS) occupying the last two thirds of the ORF. There are 6 nonstructural protein genes on the noncytopathic BVDV genome (NS2/3, 4A, 4B, 5A, and 5B) (Figure 1A). The nonstructural proteins are either known or assumed to be involved with virus replication. The BVDV gene names and numbers have been designated as such to coincide with the genes of other pestiviruses and the flaviviruses, and the hepatitis C-like viruses, which share a similar genomic arrangement (Figure 1A).
Figure 1. Examples of multiple natural recombination and rearrangements in bovine viral diarrhea virus (BVDV) genomes and deletions of a defective interfering particle thought to be responsible for expression of p80/NS3 and the cytopathic biotype. A) protein genes of a noncytopathic (ncp) BVDV open reading frame (ORF), B) insertion of bovine (ubiquitin) sequence into the NS2/3 gene, C) duplication of BVDV sequence, D) “DI” genomes with large deletions can be packaged into a BVDV vrion using normal virus proteins.
Arrows indicate additional internal protease cleavage site resulting in the cleavage of NS2/3 protein into NS2 and NS3 (p80). Adapted from (29).
The importance of genomic instability among RNA viruses
Phenotypic diversity, such as antigenic variation, infectivity, and replication rates, which can affect viral virulence, can be attributed to genomic reassortments, mutations, or recombinations. Reassortment occurs only in RNA viruses with segmented genomes (such as influenza viruses) and refers to the regrouping of genome segments with segments of other isolates, species, or genera of RNA viruses. Recombination and mutations occur in segmented and nonsegmented RNA viruses and both have been observed in pestiviruses. A mutation can be either a point mutation (change or deletion) of a single nucleotide or the deletion of a section of the genome. Nucleotide point mutations are extremely common in RNA viruses, as viral polymerases are unable to proofread or repair error during replication as DNA coded organisms do. While many point mutations do occur, many will not change either the amino acid or the function of the resultant protein. Nucleotide deletions also occur. Deletions as large as 3102 nucleotides in 1 genome have been observed in a BVDV pair isolated from an animal with MD (29). This deletion mutant was missing most of its structural protein genes (Figure 1D). Such viruses rely on coinfection with other BVDV to provide the missing structural proteins.
Pestiviral recombination events have been well characterized in BVDV. Recombination can occur between host and virus, resulting in host RNA sequences being inserted into the viral genome or by rearrangement, duplication, or both of the viral genome's nucleotide sequence. Either event has been shown to be responsible for the introduction of an additional internal protease cleavage site, which results in the production of the p80 (NS3) protein. This cleaved protein is associated with cytopathogenicity of BVDV in cell culture (29) (Figures 1 and 1C). When this mutation occurs in an animal PI with ncp BVDV, a population of mutated (cp) viruses expands, so that both cp and ncp, but otherwise seemingly identical viruses, exist in the host. It is this particular scenario that results in what is called MD. Some researchers have suggested that differences in the type of genetic alteration (mutation or deletion versus recombination) is specific to whether MD occurs within weeks or months after birth (31). Differences in the pathogenic mechanisms of such “early” or “late onset” MD have yet to be demonstrated in field cases.
Such mechanisms for viral diversity can be advantageous, deleterious, or irrelevant to the evolution and ultimate survival of viruses. Many mutations may simply result in a “nonviable” virus, which, for whatever reason, is unable to replicate. Most mutations will be effectually benign, though accumulation of such benign events may lead to a change in the phenotype of the virus. The most studied recombination or mutational events are those that do change the phenotype of the virus. Most important for the virus are changes that increase or enhance their replication, survival or both. Most important for their hosts are changes to the virulence of the virus or host specificity.
Rates of mutation and thus the generation of diversity are thought to be very rapid among most RNA viruses. Although there are exceptions, the general rate of divergence has been estimated to be 0.03% to 2.0% per nucleotide per year, which is 1 million times that of eukaryotic DNA genomes (32). The rate of RNA virus mutation (or diversification) coupled with the rate of viral replication, thus, could conceivably produce a plethora of genomic variants within a single host. Such variants have been termed quasispecies.
Quasispecies, genotypes, and other subspecies categorizations
Quasispecies have been described in vesicular stomatitis virus, hepatitis C virus (HCV), and BVDV. The study of quasispecies has shown that, despite the stochastic processes of replication and mutation that lead to RNA virus diversity, the population dynamics of RNA virus quasispecies is a nonlinear, deterministic evolutionary behavior (32). All the factors involved in such selection are numerable and unknown, but most certainly include host factors.
Competition among quasispecies is evident in chronic HCV infections. Individuals infected with HCV can be superinfected with different strains, leading to eradication or suppression of the original infecting strain. The ultimate outcome of multiple HCV exposures may be dependent on a single strain establishing its dominance (33). Another HCV quasispecies phenomenon that has been reported is the ability to change quasispecies populations in response to homologous rechallenge. The reincocualtion of chimpanzees that were chronically infected with HCV with the same HCV strain resulted in the emergence of minor quasispecies and a shift in the major quasispecies population (34). More interesting is that this shift in quasispecies dynamics resulted in clinical changes — serum alanine aminotransferase levels, viral load, and antibody titers all rose. Hepatocellular ultrastructural changes that typify HCV infection, which had disappeared prior to reinfection, reappeared.
Environmental factors also affect quasispecies populations. Studies have shown that a minor quasispecies population in humans with chronic HCV infections became the predominant quasispecies during interferon therapy (35). Further research suggests that the presence of specific quasispecies differing in E2 (envelope protein gene) and NS5B (polymerase gene) correlate with the effectiveness of interferon treatment (36).
Quasispecies variation also occurs in pestiviruses (37,38). Although there are no published studies on competition among pestivirus quasispecies, it is likely that the same type of population dynamics exists among quasispecies of pestivirus as among those of HCV. The E2 region in BVDV also contains a hypervariable region or genomic “mutational hot spot” which can be expected to produce quasispecies variation (29). It is this protein that elicits BVDV neutralizing antibodies. However, antibodies to a different, more conserved region of this same protein can be used to determine BVDV genotypes (39). Originally BVDV was segregated into 2 genotypes (BVDV1 and BVDV2), based on the 5' untranslated region (UTR) (40). This region does not code for a protein; however, it is important for the initiation of translation of the RNA ORF and is highly conserved. Subsequently, other regions of the BVDV genome were also found to be highly conserved and, when sequenced, will generally group viruses in much the same way as does the 5'UTR. Based on these conserved regions, phylogenic analysis suggested that BVDV1 and BVDV2 are as different from each other as BVDV1 (for example, reference isolates NADL, SD-1, Oregon, and Osloss ) are from CSFV. This suggests that BVDV1 and BVDV2 genomes are as distinctly different as genomes of BDV are from those of CSFV (40).
As compelling as this seems, such genotypic comparisons are based on arbitrarily chosen genomic regions, which may not code for viral proteins and are of uncertain significance in terms of disease. The initial impetus to subdivide BVDV was an attempt to distinguish what appeared, at the time, to be a newly emerged type of BVDV, which caused the BVDV-associated hemorrhagic syndrome in the 1980s and then later severe acute BVD. Of the BVDV isolates from the severe acute BVD outbreak in the Great Lakes region in 1993 that were genotyped, all were BVDV2. However, genotyping of BVDV isolates in Ontario as far back as 1983 confirmed the existence of BVDV2 long before the appearance of the more severe forms of BVD (26). Type 2 BVDV has also been isolated from other parts of the world where severe acute BVD is rare, or has not been reported (41,42,43,44,45,46).
Furthermore, in the light of quasispecies variability, categorizing BVDV by genotype may be of little consequence, as virulence could very likely be associated with a more variable region, since such regions exist throughout the structural and nonstructural protein coding regions and the 3'UTR.
Of more importance than conserved genomic regions is variability in genomic sequence and its relationship to viral virulence. Direct evidence of variations in BVDV genomic sequences resulting in changes in disease outcomes comes from the mutations and recombinations that lead to the cleavage of NS2/3 to NS2 and NS3 (p80 protein), which is associated with cp effects in cell culture and MD. Cytopathic BDV has also been associated with a mucosal-like disease in sheep (47). Mutations of ncp BVDV, BDV, and CSVF resulting in the switch to the cp phenotype can be generated by extensive in vivo and in vitro passaging (48,49,50,51,52). However, CSFVs are predominantly ncp, yet they are responsible for the most virulent disease caused by any pestivirus. Similarly, ncp BVDV2 can cause severe disease without p80 (25) or evidence of cytopathogenicity in cell culture (26). Thus, other undiscovered virulence factors must exist within the pestivirus genomes.
Diagnostic tests and their interpretation
Virus isolation or detection of virus RNA is the most important test for the determination of BVDV infection. Blood can be used for antemortem diagnosis through either virus isolation or reverse transcriptase-polymerase chain reaction (Rt-PCR). Typically, buffy coat will contain more virus than serum. Isolation of BVDV or Rt-PCR from blood will detect animals that are persistently or acutely infected. Biotype can only be determined by virus isolation. Thus, a diagnosis of MD must involve identification of cp BVDV. Ideally, one would also isolate ncp BVDV and determine that a PI animal was seronegative. However, cp viruses will mask the presence of a ncp virus and only more extensive (nonroutine) tests will detect a coexisting ncp BVDV infection. Seronegativity combined with isolation of cp BVDV is the best confirmation of MD. Seronegativity combined with isolation of ncp BVDV is the best confirmation of a persistent BVDV infection. Unfortunately, this is not always the case, as PI animals can be seropositive (53). This is the result of a BVDV infection with a virus that is antigenically distinct from the persisting virus. As the persistently infecting virus replicates, quasispecies arise that are antigenically different from their parent due to mutations in the E2 region (53). The immune system responds by developing antibodies to these antigenically different quasispecies in the same way that it would to a heterologous infecting BVDV, so these antigenically heterologous viruses are subsequently cleared. Similarly, the identification of PI animals within a herd is confounded by the presence of colostral antibodies, thus, additional tests at a later date are required to confirm whether a young virus-positive animal is or is not PI with BVDV. It is often difficult to persuade a client to agree to the number and complexity of tests required for the identification of PIs within a herd. However, the benefits of identifying and removing PIs from a herd should be stressed, as these animals will inevitably die of MD and will, until then, shed large quantities of virus, perpetuating BVDV-associated reproductive anomalies (abortions, still births, congenital defects), disease, and decreased revenue.
In the case of acute BVDV infections, virus can most often be isolated or RNA can be detected in the blood approximately 4 to 8 d after initial exposure. Inability to detect BVDV in the blood during acute infection is likely due to the production of neutralizing antibodies clearing circulating virus. In this instance, an increase in BVDV antibody titers from acute to convalescent serum samples is most useful. While seroconversion during an acute BVDV infection may prevent virus isolation or a positive Rt-PCR result from blood, virus and viral RNA can be detected postmortem in tissues; BVDV is most often detected in lymph nodes. However, detection of BVDV within peripheral lymph nodes may not be indicative of disease. The detection of BVDV in mesenteric lymph nodes, Peyer's patches, or an aborted fetus, for example, is more easily attributed to disease caused by virulent BVDV.
Genotype is typically determined by Rt-PCR (40); however, genotype-specific antibodies are also available for use with virus isolation (54). As both severe and asymptomatic disease can be caused by either type 1 or type 2 BVDV, genotyping for the diagnosis of BVD is of little relevance. However, it may be useful when evaluating a response to a vaccine. When assessing sera for antibodies following vaccination, the choice of virus used for virus neutralization assays can often be matched to that of the vaccine strain. While the use of the exact strain or, more simply, the particular genotype of the vaccine virus, in such tests is a specific indicator of the immune response to the vaccine; ultimately, the goal is to immunize against heterologous BVDV, both in strain and genotype. Most research suggests that BVDV1-based vaccines do protect against BVDV2 infections (55,56).
Inevitably, the interpretation of BVDV virology and serology must only be considered along with the clinical history and presentation. While the presence of BVDV within a herd will ultimately favor the occurrence of BVDV-associated disease in that herd over a herd that is BVDV free, isolation of BVDV does not mean that BVDV-associated disease will necessarily result. Clinical signs of reproductive anomalies and MD are well described. While severe acute BVD may go unnoticed initially or be misdiagnosed, selected laboratory tests including hematology can greatly aid diagnosis. Lymphopenia, often with neutropenia and with or without thrombocytopenia, is typical of BVDV disease (24,55,57,58).
Summary
Critical review of the milestones that have greatly contributed to the understanding of the pathogenesis of BVDV infections enables scientists to look forward and formulate unanswered questions. The concept that ncp BVDV2 may be of high or low virulence with regard to acute infections is now slowly being accepted. It is not clear if this variation also applies to reproductive failure or congenital defects, or if virulent ncp BVDV can cause persistent infections. It is assumed by many, at least in North America, that highly virulent ncp BVDV1s do not exist. Understanding of virulence determinants, even in cp BVDV, is embryonic. It is known that the presence of cp BVDV is essential to the development of MD, but its exact role is unknown. It appears that the cleavage of the NS2/3 gene product is one such factor, though it is not clear whether this is the only factor or if there are other virulence factors in other parts of the genome. Continued research into the identification of virulence factors of BVDV will be useful in determining the pathogenesis of severe acute BVD and associated reproductive anomalies, as well as providing useful tools and targets for better diagnostic tests and for more effective control of BVDV. Similarly, increased understanding of the nature of BVDV virulence will allow tracking and predictions to be made regarding evolutionary changes, which have and will no doubt continue, to occur.
Footnotes
Acknowledgments
The author thanks Drs. Brian Wilcock and Tony van Dreumel for their insight and their help and advice. CVJ
Dr. Goens' current address is the Department of Virology & Immunology, Southwest Foundation for Biomedical Research, West Loop 410 at Military Drive, San Antonio, Texas, USA 760549.
Address all correspondence and reprint requests to Dr S. Denise Groens; e-mail: dgoens@sfbr.org
References
- 1.Childs, T. X disease in cattle — Saskatchewan. Can J Comp Med 1946;10:316–319. [PMC free article] [PubMed]
- 2.Olafson P, MacCallum AD, Fox A. An apparently new transmissible disease of cattle. Cornell Vet 1946;36:205–213. [PubMed]
- 3.Ramsey FK, Chivers WH. Mucosal disease of cattle. North Am Vet 1953;34:629–634.
- 4.Underdahl NR, Grace OD, Hoerlein AB. Cultivation in tissue culture of cytopathogenic agent from bovine mucosal disease. Proc Soc Biol Med 1957;94:795. [DOI] [PubMed]
- 5.Lee KM, Gillespie JH. Propagation of virus diarrhea virus of cattle in tissue culture. Am J Vet Res 1957;18:952. [PubMed]
- 6.Gillespie JH, Baker JA, McEntee K. A cytopathogenic strain of virus diarrhea virus. Cornell Vet 1960;50:73–79. [PubMed]
- 7.Darbyshire JH. Agar gel diffusion studies with a mucosal disease of cattle. II. A serological relationship between mucosal disease and swine fever. Res Vet Sci 1962;3:125–128.
- 8.Coggins L, Gillespie JH, Robson DS, et al. Attenuation of virus diarrhea virus (Strain Oregon C24V) for vaccine purposes. Cornell Vet 1961;51:539–545. [PubMed]
- 9.Bolin SR. Control of bovine viral diarrhea infection by use of vaccination. Vet Clin North Am Food Anim Pract 1995;11:615–625. [DOI] [PubMed]
- 10.Jubb KVP, Kennedy PC. Pathology of Domestic Animals. 1st ed, vol 2. New York: Academic Pr, 1963;12–21.
- 11.Malmquist WA. Bovine viral diarrhea-mucosal disease: Etiology, pathogenesis, and applied immunity. J Am Vet Med Assoc 1968;763–768.
- 12.Johnson DW, Muscoplat CC. Immunologic abnormalities in calves with chronic bovine viral diarrhea. Am J Vet Res 1973;34:1139–1141. [PubMed]
- 13.Coria MF, McClurkin AW. Specific immune tolerance in an apparently healthy bull persistently infected with bovine viral diarrhea virus. J Am Vet Med Assoc 1978;172:449–451. [PubMed]
- 14.McClurkin AW, Littledike ET, Cutlip RC, Frank GH, Coria MF, Bolin SR. Production of cattle immunotolerant to bovine viral diarrhea virus. Can J Comp Med 1984;48:156–161. [PMC free article] [PubMed]
- 15.Brownlie J, Clarke MC, Howard CJ. Experimental infection of cattle in early pregnancy with a cytopathic strain of bovine virus diarrhoea virus. Res Vet Sci 1989;46:307–311. [PubMed]
- 16.Brownlie J, Clarke MC, Howard CJ. Experimental production of fatal mucosal disease in cattle. Vet Rec 1984;114:535–536. [DOI] [PubMed]
- 17.Sopp P, Hooper LB, Clarke MC, Howard CJ, Brownlie J. Detection of bovine viral diarrhoea virus p80 protein in subpopulations of bovine leukocytes. J Gen Virol 1994;75:1189–1194. [DOI] [PubMed]
- 18.Bolin SR. The pathogenesis of mucosal disease. Vet Clin North Am Food Anim Pract 1995;11:489–500. [DOI] [PubMed]
- 19.Radostits OM, Littlejohns IR. New concepts in the pathogenesis, diagnosis and control of disease caused by the bovine viral diarrhea virus. Can Vet J 1988;29:513–528. [PMC free article] [PubMed]
- 20.Rebhun WC, French TW, Perdrizet JA, Dubovi EJ, Dill SG, Karcher LF. Thrombocytopenia associated with acute bovine virus diarrhea infection in cattle. J Vet Intern Med 1989; 3:42–46. [DOI] [PubMed]
- 21.Perdrizet JA, Rebhun WC, Dubovi EJ, Donis RO. Bovine virus diarrhea — clinical syndromes in dairy herds. Cornell Vet 1987;77:46–74. [PubMed]
- 22.Broes A, Wellemans G, Dheedene J. Syndrome hémorragique chez des bovines infectés par le virus de la diarrhée virale bovine (BVD/MD). Ann Med Vet 1992;137:33–38.
- 23.Corapi WV, Elliott RD, French TW, Arthur DG, Bezek DM, Dubovi EJ. Thrombocytopenia and hemorrhages in veal calves infected with bovine viral diarrhea virus. J Am Vet Med Assoc 1990;196:590–596. [PubMed]
- 24.Bolin SR, Ridpath JF. Differences in virulence between two noncytopathic bovine viral diarrhea viruses in calves. Am J Vet Res 1992;53:2157–2163. [PubMed]
- 25.Ridpath JF, Bolin SR. The genomic sequence of a virulent bovine viral diarrhea virus (BVDV) from the type 2 genotype: detection of a large genomic insertion in a noncytopathic BVDV. Virology 1995;212:39–46. [DOI] [PubMed]
- 26.Carman S, van Dreumel T, Ridpath J, et al. Severe acute bovine viral diarrhea in Ontario, 1993–1995. J Vet Diagn Invest 1998;10:27–35. [DOI] [PubMed]
- 27.Pellerin C, van den Hurk J, Lecomte J, Tijssen P. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology 1994;203:260–268. [DOI] [PubMed]
- 28.David GP, Crawshaw TR, Gunning RF, Hibberd RC, Lloyd GM, Marsh PR. Severe disease in adult dairy cattle in three UK dairy herds associated with BVD virus infection. Vet Rec 1994;134:468–472. [DOI] [PubMed]
- 29.Donis RO. Molecular biology of bovine viral diarrhea virus and its interactions with the host. Vet Clin North Am Food Anim Pract 1995;11:393–423. [DOI] [PMC free article] [PubMed]
- 30.Kupfermann H, Thiel HJ, Dubovi EJ, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol 1996;70:8175–8181. [DOI] [PMC free article] [PubMed]
- 31.Fritzemeier J, Haas L, Liebler E, Moennig V, Greiser-Wilke I. The development of early vs. late onset mucosal disease is a consequence of two different pathogenic mechanisms. Arch Virol 1997;142:1335–1350. [DOI] [PubMed]
- 32.Strauss EG, Strauss JH, Levine AJ. Virus evolution. In: Fields NB, Knipe DM, and Howley PM, eds. Fundamental Virology. Philadelphia: Raven Publ, 1996:141–159.
- 33. Laskus T, Wang LF, Radkowski M, et al. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J Virol 2001;75:2059–2066. [DOI] [PMC free article] [PubMed]
- 34.Wyatt CA, Andrus L, Brotman B, Huang F, Lee DH, Prince AM. Immunity in chimpanzees chronically infected with hepatitis C virus: role of minor quasispecies in reinfection. J Virol 1998;72:1725–1730. [DOI] [PMC free article] [PubMed]
- 35.Sakuma I, Enomoto N, Kurosaki M, Marumo F, Sato C. Selection of hepatitis C virus quasispecies during interferon treatment. Arch Virol 1996;141:1921–1932. [DOI] [PubMed]
- 36.Tao Q, Wei L, Chang J, Wang H, Sun Y. Relationship between interferon therapy and variability in nonstructural gene 5b of hepatitis C virus. J Gastroenterol 1998;33:684–693. [DOI] [PubMed]
- 37. Jones LR, Weber EL. Application of single-strand conformation polymorphism to the study of bovine viral diarrhea virus isolates. J Vet Diagn Invest 2001;13:50–56. [DOI] [PubMed]
- 38.Kiss I, Ros C, Kecskemeti S, Tanyi J, Klingeborn SB, Belak S. Observations on the quasispecies composition of three animal pathogenic RNA viruses. Acta Vet Hung 1999;47:471–480. [DOI] [PubMed]
- 39.Deregt D, van Rijn PA, Wiens TY, van den Hurk J. Monoclonal antibodies to the E2 protein of a new genotype (type 2) of bovine viral diarrhea virus define three antigenic domains involved in neutralization. Virus Res 1998;57:171–181. [DOI] [PubMed]
- 40.Ridpath JF, Bolin SR, Dubovi EJ. Segregation of bovine viral diarrhea virus into genotypes. Virology 1994;205:66–74. [DOI] [PubMed]
- 41.Bolin SR, Ridpath JF. Prevalence of bovine viral diarrhea virus genotypes and antibody against those viral genotypes in fetal bovine serum. J Vet Diagn Invest 1998;10:135–139. [DOI] [PubMed]
- 42.Nagai M, Sato M, Nagano H, et al. Nucleotide sequence homology to bovine viral diarrhea virus 2 (BVDV 2) in the 5' untranslated region of BVDVs from cattle with mucosal disease or persistent infection in Japan. Vet Microbiol 1998;60:271–276. [DOI] [PubMed]
- 43.Becher P, Orlich M, Konig M, Thiel HJ. Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease. J Virol 1999;73:5646–5653. [DOI] [PMC free article] [PubMed]
- 44.Shimazaki T, Sekiguchi H, Nakamura S, Taguchi K, Inoue Y, Satoh M. Segregation of bovine viral diarrhea virus isolated in Japan into genotypes. J Vet Med Sci 1998;60:579–583. [DOI] [PubMed]
- 45.Tessaro SV, Carman PS, Deregt D. Viremia and virus shedding in elk infected with type 1 and virulent type 2 bovine viral diarrhea virus. J Wildl Dis 1999;35:671–677. [DOI] [PubMed]
- 46.Botton SA, da-Silva AM, Brum MC, Weiblen R, Flores EF. Antigenic characterization of Brazilian bovine viral diarrhea virus isolates by monoclonal antibodies and cross-neutralization. Braz J Med Biol Res 1998;31:1429–1438. [DOI] [PubMed]
- 47.Edwards S, Paton D. Antigenic differences among pestiviruses. Vet Clin North Am Food Anim Pract 1995;11:563–577. [DOI] [PubMed]
- 48.Vantsis JT, Barlow RM, Fraser J, Rennie JC, Mould DL. Experiments in Border disease. VIII. Propagation and properties of a cytopathic virus. J Comp Pathol 1976;86:111–120. [DOI] [PubMed]
- 49.Meyers G, Tautz N, Becher P, Thiel HJ, Kummerer BM. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol 1996;70:8606–8613. [DOI] [PMC free article] [PubMed]
- 50.Mittelholzer C, Moser C, Tratschin JD, Hofmann MA. Porcine cells persistently infected with classical swine fever virus protected from pestivirus-induced cytopathic effect. J Gen Virol 1998; 79:2981–2987. [DOI] [PubMed]
- 51.Kosmidou A, Buttner M, Meyers G. Isolation and characterization of cytopathogenic classical swine fever virus (CSFV). Arch Virol 1998;143:1295–1309. [DOI] [PubMed]
- 52.Paton DJ. Pestivirus diversity. J Comp Pathol 1995;112:215–236. [DOI] [PubMed]
- 53.Collins ME, Desport M, Brownlie J. Bovine viral diarrhea virus quasispecies during persistent infection. Virology 1999;259:85–98. [DOI] [PubMed]
- 54.Deregt D, Prins S. A Monoclonal Antibody-Based Immunoperoxidase Monolayer (Micro-Isolation) Assay for Detection of Type 1 and Type 2 Bovine Viral Diarrhea Viruses. Can J Vet Res 1997;62:152–155. [PMC free article] [PubMed]
- 55.Cortese VS, West KH, Hassard LE, Carman S, Ellis JA. Clinical and immunologic responses of vaccinated and unvaccinated calves to infection with a virulent type-II isolate of bovine viral diarrhea virus. J Am Vet Med Assoc 1998;213:1312–1319. [PubMed]
- 56.Dean HJ, Leyh R. Cross-protective efficacy of a bovine viral diarrhea virus (BVDV) type 1 vaccine against BVDV type 2 challenge. Vaccine 1999;17:1117–1124. [DOI] [PubMed]
- 57.Corapi WV, French TW, Dubovi EJ. Severe thrombocytopenia in young calves experimentally infected with noncytopathic bovine viral diarrhea virus. J Virol 1989;63:3934–3943. [DOI] [PMC free article] [PubMed]