Abstract
Protein ADP-ribosylation is a reversible posttranslational modification of uncertain significance in cancer. In this study, we evaluated the consequences for cancer susceptibility in the mouse of a genetic deletion of the enzyme responsible for removing mono-ADP–ribose moieties from arginines in cellular proteins. Specifically, we analyzed cancer susceptibility in animals lacking the ADP-ribosylarginine hydrolase (ARH1) that cleaves the ADP ribose–protein bond. ARH1−/− cells or ARH1−/− cells overexpressing an inactive mutant ARH1 protein (ARH1−/− +dm) had higher proliferation rates than either wild-type ARH1+/+ cells or ARH1−/− cells engineered to express the wild-type ARH1 enzyme. More significantly, ARH1−/− and ARH1+/− mice spontaneously developed lymphomas, adenocarcinomas, and metastases more frequently than wild-type ARH1+/+ mice. In ARH1+/− mice, we documented in all arising tumors mutation of the remaining wild-type allele (or loss of heterozygosity), illustrating the strict correlation that existed between tumor formation and absence of ARH1 gene function. Our findings show that proper control of protein ADP-ribosylation levels affected by ARH1 is essential for cancer suppression.
Introduction
Mono-ADP-ribosylation is a posttranslational modification of proteins, in which the ADP-ribose moiety of β-NAD is transferred to specific amino acid residues in target proteins (1). This reaction has been well characterized for bacterial toxins, which thereby alter the activity of critical regulatory proteins in mammalian cells (2, 3). Cholera toxin, for example, ADP ribosylates an arginine in the α-subunit of the stimulatory guanine nucleotide-binding (G) protein of the adenylyl cyclase system, resulting in its activation and an increase in intracellular cAMP (4). Pertussis toxin modifies a cysteine in another family of G proteins, blocking the action of inhibitory agonists on adenylyl cyclase (5). Other toxins use different proteins, and in some instances, different acceptor amino acids (e.g., asparagine) as substrates for ADP ribosylation (3).
Enzymes that catalyze reactions similar to bacterial toxins have been identified in mammalian and avian tissues, including turkey erythrocytes, chicken neutrophils, rat neurons, and mammalian cardiac and skeletal muscle cells (1, 6). These NAD:arginine ADP-ribosyltransferases (ART) vary in cellular localization, some intracellular and others secreted, with a third group linked to the cell surface by glycosylphosphatidylinositol (GPI) anchors (6, 7). These transferases catalyze the stereospecific transfer of ADP-ribose from β-NAD to the guanidino group of arginine (protein), producing the α-ADP–ribosylarginine(protein) (8).
Some natural, substrates and/or effectors of ADP-ribosylation have been defined (9–16). Mono-ARTs in the human airway may have a regulatory role in modulating the innate immune response through modification of the α-defensin human neutrophil peptide-1, HNP-1 (9). ART1 appeared to be responsible for ADP-ribosylation of HNP-1, which reduced its antimicrobial and cytotoxic activities, but did not affect its function as a T-cell chemoattractant or its ability to stimulate of IL-8 release from epithelial cells (9). ADP-ribosylated HNP-1 was found in the airways of patients with asthma and pulmonary fibrosis but not in sputum of patients with cystic fibrosis (10). ADP-ribosylation of HNP-1 by ART1 promoted a novel nucleophilic attack on the guanidine carbon of the ADP-ribosylated arginine with release of ADP-ribose–carbamate and formation of ornithine (11). Replacement of arginine with ornithine reduced the biological activity of HNP-1 and could be a mechanism for irreversible inactivation of the peptide (11). As ADP-ribosylated HNP-1 was isolated from airways of patients with pulmonary disease, it represents an in vivo modification that may have a role in pathogenesis.
Other modifications have been found, after addition of exogenous NAD. Integrin α7 is a major substrate of a GPI-linked ART on the surface of skeletal muscle cells (C2C12) which, based on inhibitor studies, was proposed to be involved in muscle cell differentiation (12, 13). Integrin α7β1, a laminin-binding protein, may be involved in cell adhesion and communication between myoblasts and extracellular matrix (12–14). In murine cytotoxic T cells, a GPl-anchored ART regulated proliferation and cytotoxicity of extracellular β-NAD, possibly via ADP-ribosylation of a regulatory protein, p40, that influences activity of the protein tyrosine kinase p56lck (15, 16).
ADP-ribosylation of arginine may be a reversible modification of proteins. Stereospecific ADP-ribosylarginine hydrolases (ARH1) cleave α-ADP–ribose-arginine (protein), produced in the transferase-catalyzed reaction, to regenerate the arginine-guanidino group (1, 17, 18). Thus, hydrolases complete an ADP-ribosylation cycle that could reversibly regulate the function of substrate proteins (1, 17, 19). In the photosynthetic bacterium Rhodospirillum rubrum (20), an ADP-ribosylation cycle is important in nitrogen fixation, which is controlled by the reversible ADP-ribosylation of dinitrogenase reductase. An ART (termed “DRAT” for dini-trogenase reductase ART) inactivates dinitrogenase reductase by ADP-ribosylation, and DRAG or dinitroreductase ADP-ribosylarginine glycohydrolase regenerates the active form by releasing ADP-ribose from the enzyme (20). This ADP-ribosylation cycle is regulated by environmental signals (e.g., nutrients, light) that determine the requirement for nitrogen fixation and hence the need for active or inactive dinitrogenase reductase (20).
ARHs of ca. 39-kDa were identified in mammalian and avian tissues (19, 21). ARH1 cDNAs from human, rat, and mouse tissues share 82% similarity of deduced amino acid sequences (22, 23), consistent with the cross-reaction of rabbit anti-rat brain ARH1 polyclonal antibodies with ARHs from turkey erythrocytes, and calf, mouse, and human brains (1, 22). Despite their functional similarities, ARH1 molecules from human, rat, and mouse tissues and R. rubrum have only limited regions of similarity in deduced amino acid sequences (22). Replacement of conserved aspartate 60 and/or aspartate 61 with alanine, glutamine, or asparagine in rat brain ARH1 significantly reduced enzyme activity, consistent with requirement for both residues in catalysis (24).
We had reported that effects of cholera toxin, evidenced by both ADP-ribose–arginine content and Gsα modification in murine ARH1−/− cells were greater than those in wild-type cells and were significantly reduced by overexpression of wild-type ARH1 in ARH1−/− cells. Similarly, intestinal loops in the ARH1−/− mouse were more sensitive than their wild-type counterparts to cholera toxin effects on fluid accumulation, Gsα modification, and ADP-ribosylarginine content, based on in vitro and in vivo studies (25). Since the cholera toxin effects result from ADP ribosylation, which is increased in ARH1−/− mice, those data support a role for hydrolase in the intoxication process (25).
ARHs in cells presumably act as components of ADP-ribosylation cycles to regulate levels of ADP-ribosyl(arginine)proteins. To assess their intracellular function(s) and biological effects, ARH1 knockout cells and mice were prepared. We report here our data on the relationship of genotype to behavior of primary embryonic fibroblasts and tumor incidence of ARH1-deficient mice. ARH1−/− cells grown from ARH1 knockout embryos, proliferated faster than ARH1+/+ cells derived from wild-type embryos. They also formed more and larger colonies in soft agar than the ARH1+/+ cells and ARH1−/− cells but not ARH1+/+ cells produced subcutaneous tumors in nude mice. Transfection of wild-type ARH1 gene into the ARH1−/− cells (ARH1−/− +wt) successfully reversed all of those defects, whereas ARH1−/− cells transformed with an inactive double-mutant ARH1 gene construct (ARH1−/− +dm) developed colonies in soft agar and tumors in nude mice. Consistent with cell culture and transplantation results, we observed that ARH1−/− mice spontaneously developed various tumors more frequently than their wild-type littermates. ARH1+/− cells and mice also developed tumors that appeared to result from mutation or loss of the ARH1 allele.
Materials and Methods
Materials
Anti-rabbit and anti-mouse IgG (H+L) antibodies were purchased from Promega; cell proliferation ELISA Kit from Roche; FBS, newborn calf serum, and Dulbecco’s modified Eagle’s medium (DMEM) from Invitrogen; plasmid DNA purification kits from Qiagen; leupeptin, trypsin inhibitor, benzamidine, dithiothreitol, and aprotinin from Sigma; Micro-BCA protein assay reagent kit from Pierce; Dounce all-glass hand-homogenizer from Kontes; Dual Endogenous Enzyme Block, Proteinase K, EnVision System Kit, and rabbit Anti-CD3 antibody from DAKO; rat anti-B cell antibody from BD Pharmingen; rat anti-F4/80 antibody from Serotec; and 3,3′-diaminobenzidine substrate kit from Vector Laboratories.
Supplemental section
Supplemental section contains standard procedures for Western blot analysis, ARH assay, bromodeoxyuridine (BrdU) incorporation assay, MTT assay of viable cells, transfection and clonal selection, immunohistochemistry, MRI scans, inhalation anesthesia of mice for MRI imaging, measurement of cell cycle in ARH1 cells, effect of serum on proliferation of ARH1 cells, and cell culture.
Generation of ARH1 knockout mice
ARH1 knockout mice were generated as reported by Kato and colleagues (25). Animal study protocols (#H-0127 and #H-0172) were approved by the NHLBI Animal Care and Use Committee (ACUC). Heterozygous (ARH1+/−) breeding pairs were used to breed homozygous (ARH1−/−), heterozygous (ARH1+/−), and wild-type (ARH1+/+) mice. ARH1 mice were backcrossed 7 times using C57BL/6J mice (Jackson Laboratory; ref. 25). The genotypes of mice and of primary cultured cells used in the present study were confirmed by PCR and Southern blot analysis as described (25).
Colony formation in soft agar
To assess anchorage-independent growth (26), soft agar clonogenic assays were done. Each well of a 6-well plate contained 2 mL of 0.5% (w/v) Noble agar (Difco) in DMEM with 10% NBCS. Cells, (3 × 103) in 2 mL of 0.375% (w/v) Noble agar in 10% NBCS DMEM were added above the polymerized base solution. All solutions were kept at 40°C before pouring to prevent premature agar polymerization and to ensure cell survival. Plates were incubated (37°C, 5% CO2) under standard conditions for 32 days before colony number and diameter were quantified microscopically (model: MZFL III; Leica).
Tumorigenicity in nude mice
Tumorigenicity of ARH1+/+, ARH1+/−, and ARH1−/− MEFs (passage 5–6) and stably transfected MEFs cells was assessed after subcutaneous injection (1 × 106 cells in 0.15 mL DMEM per site) in 6- to 8-week-old athymic female, Balb/c, nu/nu mice from Charles River(5 mice per group). Tumor volume (mm3, measured 3 times per week using electronic digital caliper from Fowler & NSK) was calculated as length × width2 × 0.5 (27). Mice were euthanized with CO2 when tumors reached a length of 2.0 cm, growth of tumor into the skin was observed, or 60 days after injection, whichever was first. All tumors were excised for histopathologic examination. Tumorigenicity studies, done by Biocon, had been approved according to U.S. law under the Animal Welfare Act (PL89-544 and PL94-279 amended) by the Biocon ACUC.
Statistics
ANOVA methods were used to evaluate the effect of ARH1 knockout on the proliferation of MEFs and their tumorigenic potential. If time and genotype interaction were significant, then time subgroup analyses were explored. To minimize false positive conclusions, all pairwise genotype comparisons were done only if the overall model performance was significant. These comparisons were adjusted for multiple testing. Where the comparisons of genotypes by cell line were not significantly different, then we combined the cell lines of each genotype. A Kruskal–Wallis test was used to compare the distribution of cell type by genotype. A survival analysis by using a log-rank test was used to compare the survival times by genotype. A χ2 test was used to compare the cumulative rate of tumors. Maximum tumor volume was used to compare the genotypes. Finally, a logistic regression was used with binary outcome variables. All statistical tests were considered significant at the 0.05 level (before adjustment for multiple comparisons).
Results
Effect of ARH1 genotype on proliferation of MEFs
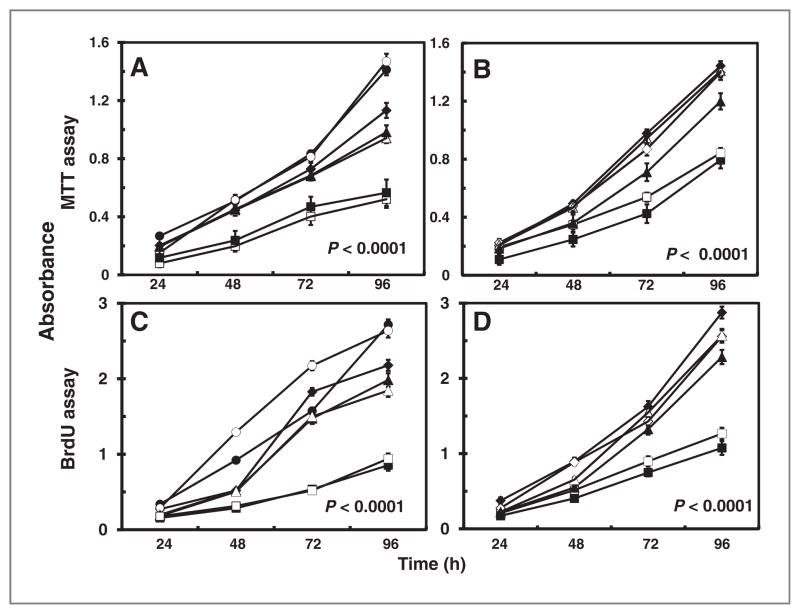
To assess the effects of ARH1 genotype, rates of proliferation of ARH1+/+, ARH+/−, ARH1−/−, ARH1−/− +wt, and ARH1−/− +dm cells were compared. Proliferation of ARH1−/− and ARH1+/− cells was significantly (P < 0.0001) faster than that of ARH1+/+ cells (Fig. 1A and C), and rates for passage 2 and passage 20 cells did not differ significantly. Proliferation of ARH1−/− cells expressing wild-type ARH1, ARH1−/− +wt, was similar to that of ARH1+/+ cells (Fig. 1B and D), whereas those of ARH1−/− +dm, expressing an inactive ARH1 mutant, and ARH1−/− cells were similar. Thus, ARH1 activity affected proliferation.
Figure 1.
Effect of ARH1 genotype in proliferation of MEFs in vitro. A, ARH1+/+ (■, □), ARH1+/− (▲, △), and ARH1−/− (●, ○, ◆) cells (5 × 103) were seeded in 96-well plates and MTT assays were done after growth for the indicated time. B, ARH1−/− mock (◇, ◆), APH1−/− +dm (▲, △), and ARH1−/− +wt (■, □) cells were treated and assayed as in (A). C, ARH1+/+ (■, □), ARH1+/− (▲, △), or ARH1−/− (●, ○, ◆) cells were treated as in (A) before BrdU cell proliferation assays. D, ARH1−/− mock (◇, ◆), APH1−/− +dm (▲, △), or ARH1−/− +wt (■, □) cells were subjected to BrdU assays as in (C). Open and closed symbols represent 2 or 3 different cell lines. Data are means ± SEM of values from 8 assays, 3 experiments. Pairwise comparison showed that all genotypes were significantly different from ARH1+/+ or ARH1−/− +wt (all at P < 0.0001).
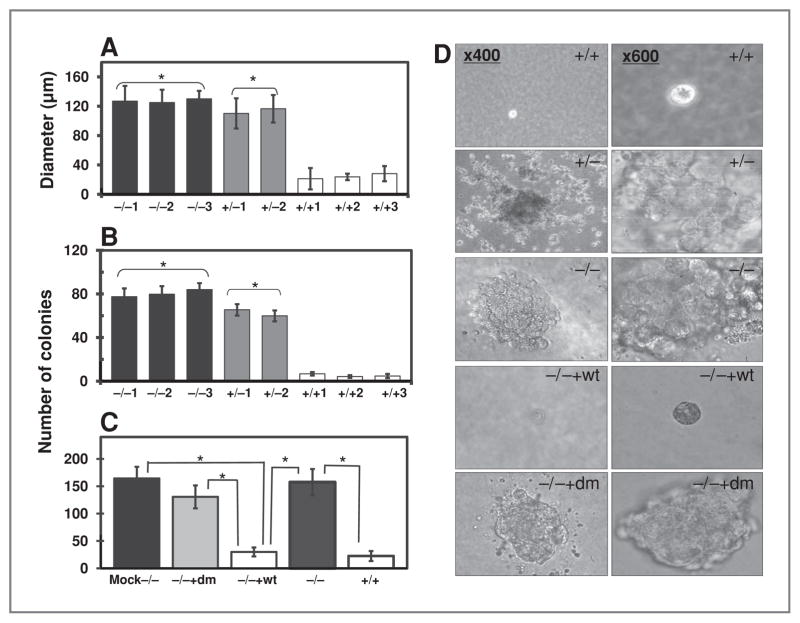
Effect of ARH1 genotype on colony formation in soft agar
To evaluate anchorage-independent growth, we used soft agar clonogenic assays. ARH1−/− and ARH1+/− MEFs formed colonies in soft agar (Fig. 2A, B, and D). ARH1−/− cells expressing ARH1 double-mutant DNA (ARH−/− +dm) and those transfected with control vector (ARH1−/− Mock) formed colonies in soft agar as did ARH−/− cells (Fig. 2C and D). In contrast, behavior of ARH1−/− cells expressing ARH1 wild-type gene (ARH1−/− +wt) in soft agar was similar to that of ARH1+/+ cells (Fig. 2C and D), which rarely formed colonies in soft agar. We concluded that ARH1 genotype and activity can affect anchorage-independent growth and tumorigenesis.
Figure 2.
Effect of ARH1 genotype on growth of MEFs in soft agar. A, diameters of colonies ARH1−/− (black bars), ARH1+/− (gray bars), and ARH1+/+ (open bars) colonies in soft agar after incubation for 32 days (37°C, with 5% CO2). Data are means ± SEM of values from triplicate assays, 3 experiments of each cell line for colony formation in soft agar. Pairwise comparison showed that ARH1−/− and ARH1+/− cells were not different but both were different from ARH1+/+ (*, P < 0.0001). B, numbers of ARH1−/− (black bars), ARH1+/− (gray bars), and ARH1+/+ (open bars) colonies in soft agar after 32 days (37°C, with 5% CO2). Data are means ± SEM of values from triplicate assays, 3 experiments. Pairwise comparison showed that ARH1−/− and ARH1+/− cells were not different but both were different from ARH1+/+ cells (*, P < 0.0001). C, numbers of ARH1−/− mock (black bar), ARH1−/− +dm (gray bar), ARH−/− (dark gray bar), ARH1+/+ (open bar), and ARH1+/++wt (open bar) colonies (all > 100 μm diameter) in soft agar after 32 days. Data are means ± SEM of values from 3 experiments (triplicate assays) for each cell line. Pairwise comparison showed that ARH1−/− mock, ARH1−/−, and ARH1−/− +dm cells were different from ARH1+/++wt or ARH1+/+ (all at *, P < 0.0001). D, appearance (×400 or ×600) of colonies of indicated genotypes in soft agar after 32 days.
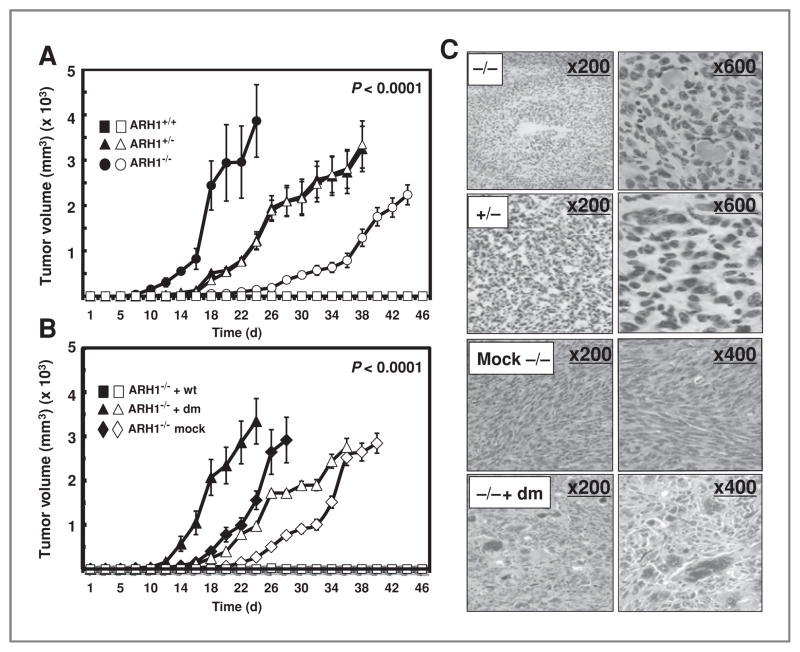
Effect of ARH1 genotype on tumor formation by MEFs in nude mice
Effects of ARH1 genotype and activity on tumor mass in nude mice were quantified 3 times per week. ARH1−/−, ARH1+/−, and ARH1−/− cells expressing an ARH1 double-mutant gene formed tumors in athymic nude mice (Fig. 3A, B, and C). In contrast, ARH1−/− cells expressing an ARH1 wild-type gene and wild-type cells did not form tumors in athymic nude mice (Fig. 3A and B). From these data, ARH1−/− but not ARH1+/+ or ARH1−/− +wt, cells apparently have tumorigenic potential.
Figure 3.
Effect of ARH1 genotype on tumor formation by MEFs in nude mice. A, ARH1+/+ (■, □), ARH1+/− (▲, △), or ARH1−/− (●, ○) cells (1 × 106) were injected subcutaneously in nude mice; width plus length of tumor masses was measured 3 times per week thereafter. Data are means ± SEM of values from 5 mice. These experiments were replicated 3 times. Genotypes were significant predictors of maximum volume (P < 0.0001). Pairwise comparisons indicate that ARH1−/− and ARH1+/− were different from ARH1+/+ (all at P < 0.0001). B, ARH1−/− +wt (■, □), ARH1−/− + dm (▲, △), or ARH1−/− mock (◆, ◇) cells (1 × 106) were injected subcutaneously in nude mice. Width plus length of tumor masses was measured 3 times per week thereafter. Open and closed symbols represent 2 different cell lines. Data are means ± SEM of values from 5 mice of each group in 3 experiments. Groups of ARH1−/− mock and ARH1−/− + dm were significantly different from ARH1−/− +wt (all at P < 0.0001). C, histology of tumors in nude mice injected with ARH1−/−, ARH+/−, ARH1−/− mock, and ARH1−/− +dm cells [hematoxylin and eosin (H&E) stain].
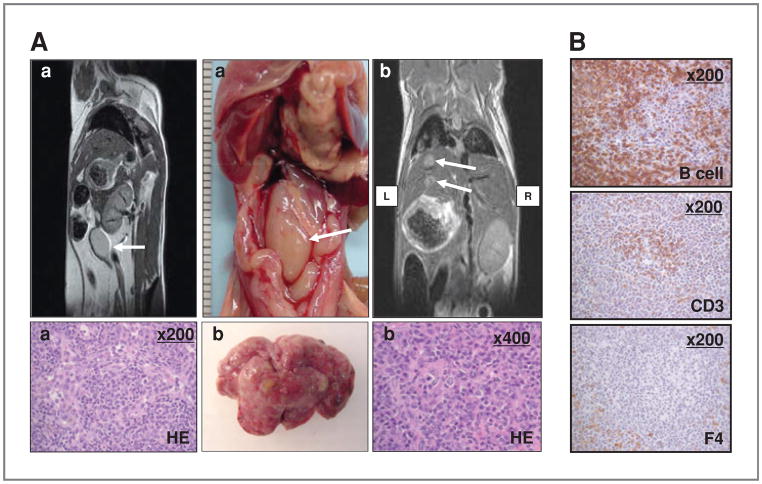
Characterization of tumors in ARH1−/− and ARH1+/− mice
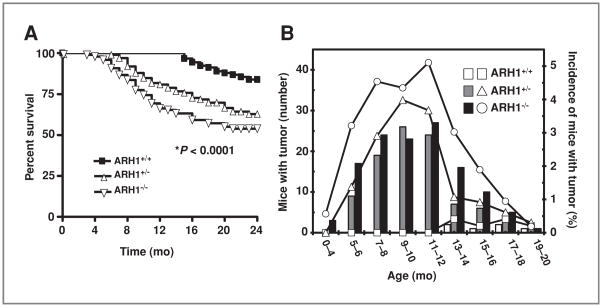
A variety of different spontaneous tumors was seen in ARH1−/− and ARH1+/− mice (Table 1, Fig. 4, Supplementary Figs. S2–5). Tumor development in ARH1 mice (e.g., location, density, and size) was monitored longitudinally by using MRI scans, which enabled comparisons among ARH1−/−, ARH1+/−, and ARH1+/+ mice. By MRI, we determined size of tumor (from 2.5-mm diameter in lung) and metastasis at early stages of tumor growth in ARH1−/−, ARH1+/−, and ARH1+/+ mice (Fig. 4, Supplementary Figs. S2–5). Tumors appeared in ARH1−/− mice as young as 3 months, whereas in ARH1+/− mice, tumors were first detected at 6 months of age (Fig. 5B). ARH1−/− and ARH1+/− mice developed tumors most often between 7 and 12 months of age (Fig. 5B).
Table 1.
Effect of genotype on incidence of tumors in ARH1-deficient mice
| Age | Genotype | Total number of animals | Incidence %
|
Total | ||||
|---|---|---|---|---|---|---|---|---|
| Carcinoma (n)
|
Lymphoma (n) | Sarcoma (n)
|
||||||
| Adeno- | a HC- | Hemangio- | Rhabdomyo- | |||||
| 3–12 months | ARH1+/+ | 315 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 |
| ARH1+/− | 484 | 4.1 (20) | 1.1 (5) | 7.9 (38) | 0.2 (10) | 0.1 (5) | 13.4 | |
| ARH1−/− | 373 | 6.2 (23) | 2.1 (8) | 12.9 (48) | 4.0 (15) | 0.0 (0) | 25.2 | |
| 13–20 months | ARH1+/+ | 175 | 0.0 (0) | 0.0 (0) | 2.9 (5) | 0.6 (1) | 0.0 (0) | 3.5 |
| ARH1+/− | 169 | 3.0 (5) | 0.6 (1) | 7.1 (12) | 0.6 (1) | 0.0 (0) | 11.2 | |
| ARH1−/− | 156 | 5.8 (9) | 0.6 (1) | 8.9 (14) | 5.0 (8) | 0.0 (0) | 20.3 | |
NOTE: P value: Comparing the cumulative number of tumors by genotype. All pairwise comparisons were significant at P < 0.0001 (ARH1−/− vs. ARH1+/−; ARH1−/−, and ARH1+/− vs. ARH1+/+). Genotype was not significantly associated with the type of tumors (P = 0.781). Age was not significantly associated with the type of tumors (P = 0.920).
Hepatocellular (HC).
Figure 4.
Lymphomas in ARH1−/− mice. A (left top), MRI shows tumor mass (lymph node) visible (arrow) in saggital section of 6.5-month ARH1−/− mouse. Below is histology (H&E stain, ×200) of lymphoma in GI lymph node. Findings were similar in 2 MRI experiments. Abdominal lymphoma at necropsy in ARH1−/− mouse (center top), with metastatic lymphoma excised from mouse liver below. At top right, metastatic lymphoma was visible in liver (arrows) on MRI (frontal section) of 6.5-month ARH1−/− mouse. Histology (H&E stain, ×400) of tumor originated in the GI tract of metastatic lymphoma in mouse liver (shown above right). Images in (A-a) were from a different ARH1−/− mouse then images designated (A-b). B, immunoreactivity of lymphoma from ARH1−/− mouse (×200). B-cell, B-cell marker (top); CD3, T-cell marker (center); F4, macrophage marker (bottom). Experiments were repeated with lymphomas from 8 ARH1−/− mice with duplicate analysis in each mouse.
Figure 5.
Effect of genotype on survival and incidence of tumors in ARH1-deficient mice. A, effect of genotype on survival of ARH1-deficient mice. Survival data from 91 ARH1+/+ (■), 99 ARH1+/− (◇), and 98 ARH1−/− mice (▽) followed 24 months. Every mouse developed some form of tumor and died spontaneously or had to be euthanized due to ill health before 24 months. Pairwise comparisons showed that ARH1−/− and ARH1+/− were different from ARH1+/+ (both at *, P < 0.0001). ARH1+/− was not different from ARH1−/− (P = 0.093). B, effect of genotype on incidence of tumor in ARH1-deficient mice. Histogram generated after MRI scanning of all ARH1 genotype mice for tumor detection. 490 ARH1+/+ mice, 653 ARH1+/− mice, and 529 ARH1−/− mice were subjected to MRI scanning and necropsy at 20 months. ARH1+/+ (open bars), ARH1+/− (gray bars), and ARH1−/− (black bars) indicated the number of mice with tumor (left Y axis). ARH1+/+ (□), ARH1+/− (△), and ARH1−/− (○) indicated the percentages of mice with tumor (right Y axis). Comparing the cumulative number of tumors by genotype we obtained at P < 0.0001. All pairwise comparisons were significant at P < 0.0001. ARH1−/− mice did not differ from ARH1+/− mice in terms of the time to first tumor appearance (P = 0.686).
In contrast, tumors developed spontaneously in ARH1+/+ mice only after 12 months of age (3.5% incidence, Table 1, Fig. 5B). Tumor classification was determined via histologic evaluation post-necropsy. Tumors in ARH1−/− mice were mainly adeno- and hepatocellular carcinoma and lymphoma. Spontaneous tumors (e.g., carcinoma, sarcoma, and lymphoma) were more frequent in ARH1−/− (25.2% incidence) and ARH1+/− (13.4% incidence) mice in both 3 to 12 months and 13 to 20 months of age subgroups (Table 1) than in the wild-type mice. Rhabdomyosarcomas (Supplementary Fig. S5) were also seen as well.
The conclusion was that the variable “genotype,” which consists of 3 possible types (ARH1+/+, ARH1+/−, and ARH1−/−), is associated with tumorigenesis. All pairwise comparisons (ARH1−/−, ARH1+/− vs. ARH1+/+, ARH1−/− vs. ARH1+/−) were significant at P < 0.0001. On the basis of the data with all mice in the denominator, genotype is significantly associated with tumor development (P < 0.0001). Age and genotype were not significantly associated with the type of tumors (P value for age is P = 0.781 and for genotype is P = 0.920). ARH1−/− and ARH1+/− mice did not develop any specific type of tumor. However, ARH1−/− mice did not differ from ARH1+/− mice in terms of the time to first tumor appearance (P = 0.686, Fig. 5B).
MRI imaging (Fig. 4A, Supplementary Figs. S2–4) revealed lymphomas in both GI tract and lymph nodes (Fig. 4A), more frequently in ARH1−/− (12.9% incidence) than in ARH1+/− mice (7.9% incidence, Table 1). Immunohistochemistry characterized lymphomas as B cell, rather than T cell (Fig. 4B). ARH1−/− mice developed adenocarcinomas in lung (Supplementary Fig. S2), uterus, and mammary gland (6.2% incidence, Table 1), as well as hepatocellular carcinoma (Supplementary Fig. S2, 2.1% incidence, Table 1). ARH1+/− mice also developed both adenocarcinoma (Supplementary Fig. S3, 4.1% incidence, Table 1) and hepatocellular carcinoma (Supplementary Fig. S4) but at lower frequencies (1.1% incidence, Table 1). Thus, the frequency of tumors in heterozygous mice was less than that in knockout animals, perhaps due to the need (and time required) for inactivation of the single functioning allele. Thus, ARH1 seems to have properties of a tumor-suppressor gene.
Effect of genotype on survival of ARH1-deficient mice
Survival of ARH1−/− (54% survival) and ARH1+/− (64% survival) mice was significantly (P < 0.00001) less than that of ARH1+/+ mice (85% survival) until age 24 months (Fig. 5A). Between ages 24 and 33 months, tumors were found in 31% of ARH1−/− mice, infections (e.g., dermatitis, eye infections) in 14%, and other diseases in 9%, whereas in ARH1+/− mice, tumors were found in 28%, infections (e.g., dermatitis, eye infections) in 22%, and other diseases in 14%. Among ARH1+/+ mice, 16% developed tumors, 48% infections (e.g., dermatitis, eye infections), and 21% other diseases.
Analysis of ARH1 proteins in adenocarcinomas and lymphomas in ARH1+/− mice
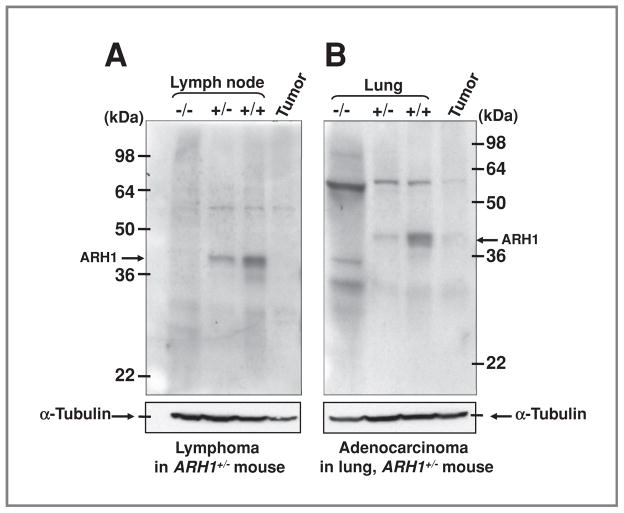
Expression of ARH1 proteins in ARH1−/−, ARH1+/−, ARH1+/+ tissues, and ARH1+/− tumor mass was examined on immunoblots. On the basis of densitometric quantification, ARH1 protein in tissues of ARH1+/− mice without evident tumors was less than that in ARH1+/+ mice. No ARH1 protein was seen on immunoblots of lymphoma from an ARH1+/− female (Fig. 6A). Similarly, ARH1 protein was not detected in adenocarcinoma from ARH1+/− lung, although it was evident in surrounding ARH1+/− nontumorous lung tissue (Fig. 6B). We found 25 adenocarcinomas and 50 lymphomas in ARH1+/− mice. In 16 of 25 adenocarcinoma and 13 of 50 lymphomas of ARH1+/− mice, sufficient tumor tissue was available for analysis. Six of 16 adenocarcinomas, and all 13 lymphomas from ARH1+/− mice had loss of heterozygosity by reverse transcription PCR and absence of ARH1 protein by immunoblotting. Mutations in exon 2 of the ARH1 gene were detected in the remaining 10 of 16 adenocarcinomas by cDNA sequencing (Supplementary Table S4). These mutations were confirmed by sequencing the genomic DNA isolated from adenocarcinomas. It seems that in ARH1 heterozygous mice, the functional allele was either lost or mutated, resulting in tumor development. Sequencing of ARH1 gene from all tumors in heterozygous mice or from nude mice injected with ARH1+/− cells revealed a mutation in exon 2 or exon 3 of the ARH1 gene. Interestingly, these mutations were not found in the ARH1+/− cells, which were injected. It would seem that the cells with an ARH1 mutation had a proliferative advantage.
Figure 6.
ARH1 in tumors from ARH1+/− mice. A, immunoblotting with anti-ARH1 antibodies of proteins in lymph node lysates (50 μg) from ARH1−/−, +/−, +/+ mouse, and lymphoma from lung (50 μg) of ARH1+/− mouse. Arrows indicate 39-kDa ARH1 protein. Below is immunoreactive α-tubulin on the same blot. Arrows indicate 50-kDa α-tubulin. B, immunoblotting as in A of ARH1 in lysates (80 μg) of lung from ARH1−/−, +/3, +/+ mouse, and adenocarcinoma from lung (80 μg) of ARH1+/− mouse. Below is immunoreactive α-tubulin on the same blot.
Supplemental section
Supplemental results section contains the results for effect of ARH1 on cell cycle (Supplementary Fig. S1A), effect of serum on proliferation of ARH1 cells (Supplementary Fig. S1B and C), metastasis in ARH1−/− and ARH1+/− mice (Supplementary Tables S1 and 2 and Supplementary Fig. S3), and multiple tumors in ARH1−/− and ARH1+/− mice (Supplementary Figs. S2 and 4 and Supplementary Table S3).
Discussion
We show here that ARH1 deficiency is associated with elevated frequency and extent of tumorigenesis. The different capacities of ARH1−/−, ARH1−/− +dm, ARH1+/+, and ARH1−/− +wt cells to proliferate evaluated in MTT and BrdU assays paralleled the increased numbers of cells. ARH1−/− cell lines from 14.5-day embryos, and ARH1−/− MEFs stably expressing an inactive, double-mutant ARH1 (ARH1−/− +dm) had higher proliferation rates than did ARH1+/+ or ARH1−/− cells expressing a wild-type ARH1 (ARH1−/− +wt). Consistent with these data, ARH1−/− and ARH1−/− +dm cells formed more and larger colonies in soft agar than did ARH1+/+ cells or ARH1−/− +wt cells. A potential role for ARH1 in tumorigenesis was supported by the finding that ARH1−/− and ARH1−/− +dm cells, but not ARH1+/+ and ARH1−/− +wt cells, produced tumors in nude mice. Thus, ARH1 genotype and enzymatic activity affected cell proliferation, anchorage-independent growth, and tumorigenesis in nude mice. Consistent with an antitumorigenic action of ARH1 was the successful rescue of all of the phenotypes by transfection of the wild-type ARH1 gene, but not the ARH1 double mutant gene, into ARH1−/− cells. These results support the hypothesis that the ARH1 protein plays a key role in cell proliferation. In agreement with the results of soft agar and nude mouse experiments with ARH1−/− cells, tumors developed spontaneously in ARH1−/− and ARH1+/− mice in an age-dependent manner and more frequently than in ARH1+/+ mice. Tumors of diverse types (carcinoma, sarcoma, and lymphoma) occurred in numerous organs/tissues (e.g., lung, liver, spleen, lymph nodes, mammary gland, uterus, vascular, and skeletal muscle).
MRI was used to detect and follow primary and metastatic tumors (size, location, and density). We describe the use of small animal imaging techniques longitudinally to detect, localize, and characterize metastatic lesions in transgenic and nude mice, which enabled us to conclude that MRI is a valuable tool for monitoring tumor dissemination and spread growth from very early to late stages of progression.
In some tumors from ARH1+/− mice or from nude mice after subcutaneous injection of ARH1+/− cells, a clear ARH1 protein band was seen unexpectedly by Western blotting. To determine whether this protein was the product of a mutant gene, ARH1 cDNA from both the tumor and surrounding normal tissues was sequenced. In all instances, mutations in the ARH1 gene were found in the tumor, but not in adjacent nontumor tissue. Notably, no mutation was detected in cDNA from the ARH1+/− cells that had been injected. In all likelihood, an ARH1 mutation in a small population of cells enabled them to proliferate more rapidly than ARH1+/− cells containing a normal allele, thus giving rise to colonies in soft agar and tumors.
We found differences also between ARH1−/− and ARH1+/− mice in frequency of tumors, incidence of metastases, and age at first tumor appearance (Table 1 and Results). As might be expected, frequency of tumors, metastases, and multiplicity of tumors were greater in ARH1−/− than in ARH+/− mice. It is probable that ARH1+/− cells require an additional step, with loss of the active ARH1 allele to enable tumor development. Our findings suggest that loss of ARH1 expression during malignant transformation occurs because of inactivation of a single functional allele, potential mechanisms for which are multiple, including loss of heterozygosity (LOH), and ARH1 gene mutation.
Impairment of cell-cycle regulation involving loss of G1-S control seems to be an important contributing step in tumorigenesis and leads to genomic instability, and inappropriate survival of genetically damaged cells (28, 29). ARH1 mutation seemed to result in loss of G1 control. Moreover, the lack of ARH1 in cells led to more rapid proliferation and tumor formation in nude mice. Loss of ARH1 activity resulted also in spontaneous tumorigenesis. Thus, ARH1 function may influence the G1-S phase of cell cycle and thereby affect growth.
Checkpoint control of S phase prevents the cell from replicating damaged DNA (28). p53-dependent regulation of replication is important in tumorigenesis (28). Cells with DNA damage rapidly increase p53 protein levels by a posttranscriptional mechanism (30, 31), and loss of 1 or 2 p53 alleles has been shown to induce tumors in mice (32, 33). However, no significant difference was observed in p53 protein expression among ARH1−/− mock, ARH1−/− +dm, and ARH1−/− +wt cells (data not shown). Thus, p53 protein expression may not be related directly to the mechanism of tumorigenesis in ARH1-deficient cells.
Posttranslational modifications (e.g., phosphorylation, acetylation) of proteins (e.g., p53, ras, Fox) can regulate cell proliferation and tumor development in mice (34, 35, 36). Posttranslational modifications of p53 by phosphorylation, acetylation, and ubiquitination influence protein stability, and transcriptional activity, with effects on cell proliferation and tumorigenesis (34). Poly (ADP-ribosylation) of proteins by nuclear poly (ADP-ribose) polymerase (PARP-1) contributes to DNA repair, carcinogenesis, chromatin functions, and genomic stability (37). In this manner, posttranslational modifications are involved in disease pathogenesis by altering protein function. In a similar manner, dysregulation of mono-ADP ribosylation of arginines in proteins by ARH1 can affect cell proliferation and tumorigenesis.
Supplementary Material
Acknowledgments
We thank Patricia Zerfas from Diagnostic and Research Service Branch, Division of Veterinary Resources for supporting immunohistochemistry. We also thank Martha Vaughan and Vincent Manganiello for helpful discussions and critical review of the manuscript.
Grant Support
This research was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Williamson KC, Moss J. Mono-ADP-ribosyltransferases and ADP-ribosylarginine hydrolase: a mono-ADP-ribosylation cycle in animal cells. In: Moss J, Vaughan M, editors. ADP-ribosylating toxins and G proteins: insights into signal transduction. Washington, D.C: ASM Press; 1990. pp. 493–510. [Google Scholar]
- 2.Okazaki IJ, Moss J. Characterization of glycosylphosphatidylinositiol-anchored, secreted, and intracellular vertebrate mono-ADP-ribosyl-transferases. Annu Rev Nutr. 1999;19:485–509. doi: 10.1146/annurev.nutr.19.1.485. [DOI] [PubMed] [Google Scholar]
- 3.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–8. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss J, Vaughan M. ADP-ribosylation of guanyl nucleotide-binding regulatory proteins by bacterial toxins. Adv Enzymol Relat Areas Mol Biol. 1988;61:303–79. doi: 10.1002/9780470123072.ch6. [DOI] [PubMed] [Google Scholar]
- 5.Ui M. Pertussis toxin as a valuable probe for G-protein involvement in signal transduction. In: Moss J, Vaughan M, editors. ADP-ribosylating toxins and G proteins: insights into signal transduction. Washington, D.C: ASM Press; 1990. pp. 45–77. [Google Scholar]
- 6.Okazaki IJ, Moss J. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J Biol Chem. 1998;273:23617–20. doi: 10.1074/jbc.273.37.23617. [DOI] [PubMed] [Google Scholar]
- 7.Okazaki IJ, Moss J. Structure and function of eukaryotic mono-ADP-ribosyltransferases. Rev Physiol Biochem Pharmacol. 1996;129:51–104. doi: 10.1007/3-540-61435-4_4. [DOI] [PubMed] [Google Scholar]
- 8.Moss J, Stanley SJ, Oppenheimer NJ. Substrate specificity and partial purification of a stereospecific NAD- and guanidine-dependent ADP-ribosyltransferase from avian erythrocytes. J Biol Chem. 1979;254:8891–4. [PubMed] [Google Scholar]
- 9.Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, et al. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci U S A. 2002;99:8231–5. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paone G, Stevens LA, Levine RL, Bourgeois C, Steagall WK, Gochuico BR, et al. ADP-ribosyltransferase-specific modification of human neutrophil peptide-1. J Biol Chem. 2006;281:17054–60. doi: 10.1074/jbc.M603042200. [DOI] [PubMed] [Google Scholar]
- 11.Stevens LA, Levine RL, Gochuico BR, Moss J. ADP-ribosylation of human defensin HNP-1 results in the replacement of the modified arginine with the noncoded amino acid ornithine. Proc Natl Acad Sci U S A. 2009;106:19796–800. doi: 10.1073/pnas.0910633106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zolkiewska A, Moss J. Processing of ADP-ribosylated integrin alpha 7 in skeletal muscle myotubes. J Biol Chem. 1995;270:9227–33. doi: 10.1074/jbc.270.16.9227. [DOI] [PubMed] [Google Scholar]
- 13.Zolkiewska A, Moss J. Integrin alpha 7 as substrate for a glycosylphosphatidylinositol-anchored ADP-ribosyltransferase on the surface of skeletal muscle cells. J Biol Chem. 1993;268:25273–6. [PubMed] [Google Scholar]
- 14.Zolkiewska A, Nightingale MS, Moss J. Molecular characterization of NAD:arginine ADP-ribosyltransferase from rabbit skeletal muscle. Proc Natl Acad Sci U S A. 1992;89:11352–6. doi: 10.1073/pnas.89.23.11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Nemoto E, Kots AY, Kaslow HR, Dennert G. Regulation of cytotoxic T cells by ecto-nicotinamide adenine dinucleotide (NAD) correlates with cell surface GPI-anchored/arginine ADP-ribosyltransferase. J Immunol. 1994;153:4048–58. [PubMed] [Google Scholar]
- 16.Wang J, Nemoto E, Dennert G. Regulation of CTL by ecto-nictinamide adenine dinucleotide (NAD) involves ADP-ribosylation of a p56lck-associated protein. J Immunol. 1996;156:2819–27. [PubMed] [Google Scholar]
- 17.Moss J, Jacobson MK, Stanley SJ. Reversibility of arginine-specific mono(ADP-ribosyl)ation: identification in erythrocytes of an ADP-ribose-L-arginine cleavage enzyme. Proc Natl Acad Sci U S A. 1985;82:5603–7. doi: 10.1073/pnas.82.17.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss J, Oppenheimer NJ, West RE, Jr, Stanley SJ. Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1986;25:5408–14. doi: 10.1021/bi00367a010. [DOI] [PubMed] [Google Scholar]
- 19.Moss J, Stanley SJ, Nightingale MS, Murtagh JJ, Jr, Monaco L, Mishima K, et al. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J Biol Chem. 1992;267:10481–8. [PubMed] [Google Scholar]
- 20.Lowery RG, Ludden PW. Endogenous ADP-ribosylation in prokaryotes. In: Moss J, Vaughan M, editors. ADP-ribosylating Toxins and G proteins: Insights into Signal Transduction. Washington, D.C: ASM Press; 1990. pp. 459–78. [Google Scholar]
- 21.Moss J, Tsai SC, Adamik R, Chen HC, Stanley SJ. Purification and characterization of ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1988;27:5819–23. doi: 10.1021/bi00415a063. [DOI] [PubMed] [Google Scholar]
- 22.Takada T, Iida K, Moss J. Cloning and site-directed mutagenesis of human ADP-ribosylarginine hydrolase. J Biol Chem. 1993;268:17837–43. [PubMed] [Google Scholar]
- 23.Takada T, Iida K, Moss J. Conservation of a common motif in enzymes catalyzing ADP-ribose transfer. Identification of domains in mammalian transferases. J Biol Chem. 1995;270:541–4. doi: 10.1074/jbc.270.2.541. [DOI] [PubMed] [Google Scholar]
- 24.Konczalik P, Moss J. Identification of critical, conserved vicinal aspartate residues in mammalian and bacterial ADP-ribosylarginine hydrolases. J Biol Chem. 1999;274:16736–40. doi: 10.1074/jbc.274.24.16736. [DOI] [PubMed] [Google Scholar]
- 25.Kato J, Zhu J, Liu C, Moss J. Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice. Mol Cell Biol. 2007;27:5534–43. doi: 10.1128/MCB.00302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato JD, Kan M. Growth of transformed cells in soft agar. In: Bonifacino JS, Dasso M, Harford JB, Schwartz JL, Yamada KM, editors. Current protocols in cell biology. Hoboken, NJ: John Wiley & Sons; 1998. pp. 1.2.1–1.2.15. [Google Scholar]
- 27.Hollingshead M. Intraperitoneal and subcutaneous tumor models for assessing anti-neoplastic agents in rodent. In: Enna SJ, Williams M, Ferkany JW, Frechette R, Kenakin T, Moser P, Ruggeri B, editors. Current protocols in Pharmacology. Hoboken, NJ: John Wiley & Sons; 2002. pp. 5.28.1–5.28.13. [DOI] [PubMed] [Google Scholar]
- 28.Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–28. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 29.Kohn KW, Jackman J, O’Conner PM. Cell cycle control and cancer chemotherapy. J Cell Biochem. 1994;54:440–52. doi: 10.1002/jcb.240540411. [DOI] [PubMed] [Google Scholar]
- 30.Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci U S A. 1992;89:7491–95. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the celluar response to DNA damage. Cancer Res. 1991;51:6304–11. [PubMed] [Google Scholar]
- 32.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 33.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 34.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 35.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 36.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miwa M, Masutani M. PolyADP-ribosylation and cancer. Cancer Sci. 2007;98:1528–35. doi: 10.1111/j.1349-7006.2007.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.