Abstract
Limitations of assay variability, labor costs, and availability of cells can affect the conduct of large population-based studies. The ability to determine optimal conditions for laboratory assessment of immune outcomes, including measurement of cytokines, can reduce the number of peripheral blood mononuclear cells (PBMCs) needed, reduce the labor costs involved, and the variability in secreted cytokine response by pooling cytokines from the same cell culture supernatant. Previously, we used response surface methodology to predict optimal conditions for vaccinia virus-stimulated cytokine responses in recipients of smallpox vaccine. Here we apply the same approach for a measles vaccine study.
PBMCs were collected from vaccinated subjects, and seven cytokines (IFN-γ, IL-2, TNF-α, IL-10, IFN-α, IFN-λ1, and IL-6) involved in measles virus-specific cytokine immune responses were examined. PBMCs were stimulated with differing multiplicity of infection (MOI) and days in culture (incubation time). Response surface methodology was used to select the optimal MOI and incubation time for each secreted cytokine.
Our results demonstrate that each cytokine’s optimal conditions (MOI and incubation time) differ for each virus (measles vs. vaccinia) and each cytokine’s optimal conditions for each virus can be predicted using response surface methodology. These conditions allow for cytokines with overlapping optimal conditions to be pooled from the same supernatant in culture to reduce the number of PBMCs used, the costs involved, and assay variability. Therefore, response surface methodology is an effective technique that can be used to optimize antigen-specific secreted cytokines prior to population-based studies.
Keywords: Response surface methodology, measles virus, vaccinia virus, cytokine, ELISA
Introduction
Cytokines play an important role in the immune response following vaccination via their role in the regulation of innate and adaptive immunity (Ovsyannikova et al., 2003). Specific to measles virus, the cytokine response is primarily driven by CD4+ T cell subsets and inflammatory cytokines (Dhiman et al., 2005). As recent studies show, these cytokines may be effective markers used to quantify cell-mediated responses following vaccination (Ovsyannikova et al., 2003). Understanding both the mechanism and functions of cytokine pathways is important to comprehend the immune response post-vaccination (Ryan et al., 2009).
Although optimal conditions of viral multiplicity of infection (MOI) and incubation time to detect cytokine response have been published for smallpox (vaccinia) vaccine studies (Ryan et al., 2009), no such data are available for measles vaccine studies. Here we describe new findings of optimized cytokine assay conditions in response to measles virus stimulation based on response surface methodology. We compare the results to previous findings based on vaccinia virus stimulation in order to be able to optimize antigen-specific secreted cytokines prior to different population-based studies.
Material and Methods
We used cryopreserved peripheral blood mononuclear cells (PBMCs) from three measles-mumps-rubella (MMR) vaccinated donors who were selected based on their measles virus-stimulated IFNγ secretion values determined by ELISA. Cells were isolated, cryopreserved and subsequently thawed for culture in RPMI-1640 culture media (Gibco, Life Technologies, Grand Island, N.Y.) containing 5% fetal calf serum (Hyclone), as we have previously described (Ryan et al., 2009; Dhiman et al., 2010). Measles virus (Edmonston vaccine strain) was grown and titered on Vero cells, as previously described (Haralambieva et al., 2010). Cytokines were quantified in cultured PBMCs (in 96-well plate format, 2 × 105 cells per well) following in vitro live measles virus stimulation (in triplicate) or in unstimulated cell cultures (also in triplicate), using a modified surface response methodology matrix with combinations of MOIs and incubation times as follows (Table 1): MOIs of 0.05, 0.1, 0.5, and 1 and length of cell culture of 12 h, 24h, 48h (2 days), 72h (3 days), 96h (4 days), and 120h (5 days) (Ryan et al., 2009). Due to limitations in the number of PBMCs, the experiment focused on a subcollection of combinations based on methodological findings from our earlier studies and the literature (Ryan et al., 2009; Dhiman et al., 2010; Haralambieva et al., 2010). Cells stimulated with 5μg/mL PHA (Sigma) were used as a positive control. Cell-free supernatants were harvested and used for cytokine measurements by ELISA following the manufacturer’s recommendations for each kit (R&D Systems, Minneapolis, MN for IFN-λ1; Mabtech, Cincinnati, OH for IFN-α and BD Biosciences Pharmingen, San Diego, CA for the remaining cytokines) (Haralambieva et al., 2010).
Table 1.
Matrix table used on optimization subjects to statistically predict MOI and time.
Time
 MOI 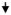
|
12h | 24h | 2d | 3d | 4d | 5d |
|---|---|---|---|---|---|---|
| 0.05 | x | x | x | |||
| 0.1 | x | x | ||||
| 0.5 | x | x | x | |||
| 1.0 | x |
Marked points indicate points statistically determined for optimization experiments to encompass all combinations of incubation times and MOIs
Identification of the optimum time and MOI combinations of selected cytokines (IFN-γ, IL-2, IL-6, IL-10, TNF-α, IFN-α, and IFN-λ1) was carried out using response surface methodology (Ryan et al., 2009; Dhiman et al., 2010). This method possesses all of the desirable characteristics inherent in any regression model, including the ability to interpolate and estimate values of the response variable at unmeasured levels of the experimental variables. For each cytokine of interest, we modeled secretion as a quadratic response function of the (log-transformed) time and MOI variables, resulting in a three-dimensional surface that ideally resembles a convex “hill,” with a corresponding peak occurring at the estimated point of maximum cytokine secretion. Cytokine secretion was modeled as the difference between the mean of the stimulated cell and the mean of the unstimulated cells, divided by a pooled standard error estimate. Our methods are identical to those described previously (Ryan et al., 2009). Briefly, after determining the estimated peak secretion value, and the combination of time and MOI at which this occurred, we calculated the standard error of the response at that point. All such values of time and MOI that yielded an estimated response falling within one standard error of the peak value were considered statistically equivalent to the peak. Specific values of the factor combinations were then selected for each of the cytokines being studied. All analyses were carried out using the SAS software system (SAS Institute, Inc., Cary, N.C.).
Results
Subjects’ PBMCs were stimulated with measles virus and the optimal conditions were separately determined by response surface methodology for each cytokine. The response surface for IL-10 secretion is shown in Figure 1 as a representative example. The peak in the three-dimensional surface of Panel A depicts the maximum cytokine response. The two-dimensional surface in Panel B displays all the points within one standard error of the maximum. Any point contained within the values displayed in Panel B was eligible to be chosen as the “optimal laboratory conditions.” The optimal conditions and ranges for each measles and vaccinia virus-specific secreted cytokine as determined using this methodology are shown in Table 2. Cytokine-specific optimal conditions differ across pathogens, sometimes dramatically (e.g., IL-6 and IL-2). Specific cytokines with overlapping optimal conditions for measles virus were pooled together, which allowed for the same supernatant from cultures to be used to detect different cytokines. This saved time, money, samples, and labor. Cytokines TNF-α and IFN-α were pooled at 24 hours at an MOI of 1.0. Cytokines IL-2 and IL-10 were pooled at 48 hours at an MOI of 0.5. Finally, cytokines IL-6, IFN-γ, and IFN-λ1 were pooled at 72 hours at an MOI of 1.0.
Fig. 1.
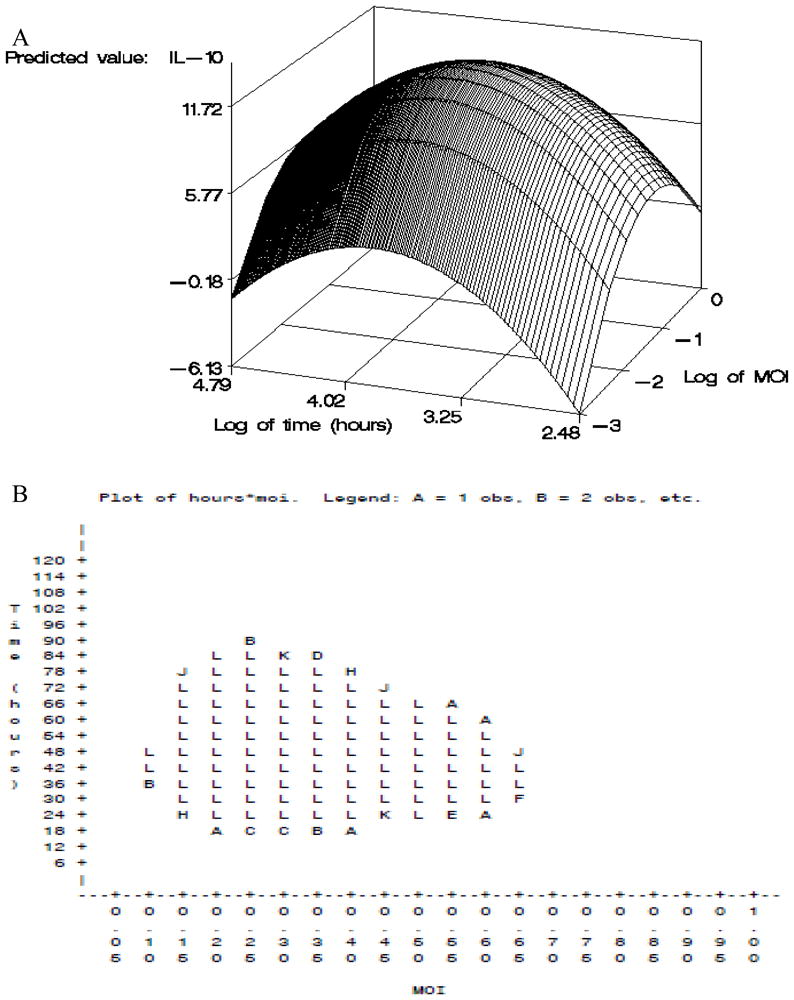
A) Response surface methodology cytokine optimization strategy for IL-10 stimulated with live measles virus. Panel A depicts the three-dimensional surface response plotting log of MOI and time. For IL-10, maximum response occurred at an MOI of 0.5 and 41 hr.
B) Response surface methodology cytokine optimization strategy for IL-10 stimulated with live measles virus. Panel B depicts all times and MOIs that fall within one standard error of the maximum (statistically significant to the maximum). All values are recorded as their original sampling units.
Table 2.
Response surface methodology results including optimal ranges of MOI and time (hours) of culture for detection of measles virus and vaccinia-specific cytokines
| IL-10 | IFN-α | IFN-γ | IL-6 | TNF-α | IL-2 | IFN-λ1 | |
|---|---|---|---|---|---|---|---|
| Measles | |||||||
| Range of MOI | 0.1–0.65 | 0.15–1 | 0.05–1 | 0.25–1 | 0.2–1 | 0.05–1 | 0.45–1 |
| Range of time (hours) | 20–86 | 18–84 | 18–114 | 60–120 | 12–38 | 18–120 | 60–120 |
| Optimal MOI | 0.25 | 0.55 | 0.2 | 1 | 1 | 0.25 | 1 |
| Optimal time (hours) | 41 | 37.5 | 43.5 | 120 | 13 | 64.5 | 120 |
| Smallpox | |||||||
| Range of MOIb | 0.05 | 0.05 | 0.05 | 3–5 | 0.2–1 | 3.5–5 | NAa |
| Range of time (hours)b | 175–192 | 96 | 96 | 192 | 12–65 | 12–36 | NAa |
| Optimal MOIb | 0.05 | 0.05 | 0.05 | 5 | 0.4 | 5 | NAa |
| Optimal time (hours)b | 192 | 96 | 96 | 192 | 25 | 16 | NAa |
NA indicates that virus stimulated cytokines values were not available/tested for comparison
All smallpox data is obtained after stimulation with inactived vaccinia virus data as cited Ryan et al. (Ryan et al., 2009)
Discussion
Using response surface methodology, we were able to identify the optimum incubation time and MOI combinations of selected cytokine assays (IFN-γ, IL-2, IL-6, IL-10, TNF-α, IFN-α, and IFN-λ1). Cytokines from cultures were pooled within a range of 24 to 72 hours and an MOI of 0.5 to 1.0. In a previous study, our group determined optimal conditions for vaccinia-specific secreted cytokines (Ryan et al., 2009). Our current results for measles virus show that optimal secreted cytokine conditions differ for measles and vaccinia viruses. For each viral antigen, this methodology can optimize secreted cytokine responses, ultimately improving the ELISA assay by reducing PMBCs used and assay variability.
Young et al. (Young et al., 2008) suggested the requirement for an affordable and effective way to measure cytokines. Many new techniques such as multiplex, flow cytometry, and mass spectrometry have advantages in measuring cytokines, yet disadvantages lie in their sensitivity, complications, and cost (Young et al., 2008) (Osuchowski et al., 2005) (Listvanova et al., 2003) (Bozza et al., 2007). However, the ELISA assay has many advantages including simplicity, effectiveness, quantification of antigen-specific release of each cytokine, and relatively low cost. Disadvantages include the need for large amounts of PMBCs and limits in detection. These disadvantages can be overcome through response surface methodology-predicted values by pooling cytokines from the same supernatant in culture to reduce PBMCs used and assay variability. This eliminates any variables associated with different blood draws and handling conditions (Osuchowski et al., 2005) (Ryan et al., 2009) (Listvanova et al., 2003) (Ray et al., 2006).
The strength of our study is that the optimization and pooling of supernatants substantially limited the number of PBMCs needed for cytokine quantification and the technicians time and labor costs for assays. Using response surface methodology, incubation times could be predetermined, thereby eliminating the need to test every single time and MOI combination (12.6×106 cells used compared to 4.2×107 cells for each subject). In addition to the optimal levels, optimal ranges were discovered that permitted cytokine pooling, which allowed several cytokines to be assayed from the same supernatant, reducing the number of PBMCs used (4.6×107 cells used compared to 1.8×108 cells for each subject)(Ryan et al., 2009; Ray et al., 2006).
A limitation of the study design was that the optimal conditions for measles virus-stimulated cytokine responses that were originally determined using the same response surface methodology matrix (as previously used for vaccinia virus) did not always reflect measles virus-specific stimulation. While using this same matrix for vaccinia virus, each measles virus-specific cytokine had an optimal MOI of 5.0 and many had optimal incubation times of up to eight days (IL-6, IFN-α, IFN-λ1). It is likely that cytokine release in cell cultures with prolonged incubation periods reflect cell death and not virus stimulation. Excluding an MOI of 5.0 and incubation times longer than five days gave more relevant (antigen stimulated) virus-specific cytokine response.
In summary, using response surface methodology, we optimized MOI and incubation conditions for determing the optimal conditions for measles-specific cytokine secretion in subjects’ PBMCs after measles vaccination. This approach allowed for a reduction in assay variability and number of PBMCs needed, as well as labor costs. Our results indicate the applicability of this methodology to optimize secreted cytokine responses for each specific viral antigen in large population-based studies.
Highlights.
Response surface methodology can predict optimal cytokine response by MOI and incubation time.
The procedure reduces labor and the number of PBMCs used by optimizing and pooling cytokines.
Different results for measles and vaccinia viruses indicate the need to optimize across different experiments.
Acknowledgments
We thank the Mayo Clinic Vaccine Research Group staff and subjects who participated in our studies. We thank V. Shane Pankratz and Caroline L. Vitse for their help with this manuscript. This work was supported by NIH grants AI 33144, AI 48793 (which recently received a MERIT Award), and was made possible by the Rochester Epidemiology Project (Grant Number R01 AG034676 from the National Institute on Aging).
Abbreviations
- PBMCs
Peripheral Blood Mononuclear cells
- ELISA
enzyme linked immunosorbent assay
- MOI
Multiplicity of infection
- ELISPOT
Enzyme-linked immunosorbent spot
- IL
interleukin
Footnotes
CONFLICT OF INTEREST: Dr. Poland is the chair of a safety evaluation committee for novel non-measles vaccines undergoing clinical studies by Merck Research Laboratories. Drs. Ovsyannikova, Haralambieva, and Vierkant declare no potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Bozza FA, Salluh JI, Japiassu AM, Soares M, Assis EF, Gomes RN, Bozza MT, Castro-Faria-Neto HC, Bozza PT. Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit Care. 2007;11:R49. doi: 10.1186/cc5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhiman N, Haralambieva IH, Vierkant RA, Pankratz VS, Ryan E, Jacobson RM, Ovsyannikova IG, Poland GA. Predominant inflammatory cytokine secretion pattern in response to two doses of live rubella vaccine in health vaccinees. Cytokine. 2010;50:24. doi: 10.1016/j.cyto.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhiman N, Ovsyannikova IG, Ryan JE, Jacobson RM, Vierkant RA, Pankratz VS, Jacobsen SJ, Poland GA. Correlations among measles virus-specific antibody, lymphoproliferation and Th1/Th2 cytokine responses following measles-mumps-rubella-II (MMR-II) vaccination. Clin Exp Immunol. 2005;142:498. doi: 10.1111/j.1365-2249.2005.02931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haralambieva IH, Ovsyannikova IG, Dhiman N, Vierkant RA, Jacobson RM, Poland GA. Differential cellular immune responses to wild-type and attenuated edmonston tag measles virus strains are primarily defined by the viral phosphoprotein gene. J Med Virol. 2010;82:1966. doi: 10.1002/jmv.21899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Listvanova S, Temmerman S, Stordeur P, Verscheure V, Place S, Zhou L, Locht C, Mascart F. Optimal kinetics for quantification of antigen-induced cytokines in human peripheral blood mononuclear cells by real-time PCR and by ELISA. J Immunol Methods. 2003;281:27. doi: 10.1016/s0022-1759(03)00267-9. [DOI] [PubMed] [Google Scholar]
- 6.Osuchowski MF, Siddiqui J, Copeland S, Remick DG. Sequential ELISA to profile multiple cytokines from small volumes. J Immunol Methods. 2005;302:172. doi: 10.1016/j.jim.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Ovsyannikova IG, Reid KC, Jacobson RM, Oberg AL, Klee GG, Poland GA. Cytokine production patterns and antibody response to measles vaccine. Vaccine. 2003;21:3946. doi: 10.1016/s0264-410x(03)00272-x. [DOI] [PubMed] [Google Scholar]
- 8.Ray CA, Dumaual C, Willey M, Fill J, O’Brien PJ, Gourley I, Devanarayan V, Konrad RJ. Optimization of analytical and pre-analytical variables associated with an ex vivo cytokine secretion assay. J Pharm Biomed Anal. 2006;41:189. doi: 10.1016/j.jpba.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Ryan JE, Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Poland GA. Response surface methodology to determine optimal cytokine responses in human peripheral blood mononuclear cells after smallpox vaccination. J Immunol Methods. 2009;341:97. doi: 10.1016/j.jim.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young SH, Antonini JM, Roberts JR, Erdely AD, Zeidler-Erdely PC. Performance evaluation of cytometric bead assays for the measurement of lung cytokines in two rodent models. J Immunol Methods. 2008;331:59. doi: 10.1016/j.jim.2007.11.004. [DOI] [PubMed] [Google Scholar]


