Abstract
Background
The incidence of esophageal adenocarcinoma is rapidly increasing and is now one of the leading causes of cancer death in the western world. MicroRNAs (miRNAs) are small non-coding RNAs which regulate the expression of protein-encoding genes and are involved in the development, progression and prognosis of other malignancies. We hypothesized that global miRNA expression would predict survival and lymph node involvement in a cohort of surgically resected esophagus cancer patients.
Methods
miRNA analysis was performed using a custom Affymetrix microarray with probes for 462 known human, 2102 predicted human, 357 mouse and 238 rat miRNAs. miRNA expression was evaluated in 45 primary tumors, and the association of miRNA expression with patient survival and lymph node metastasis was assessed. The prognostic impact of identified unique miRNAs was verified with quantitative RT-PCR.
Results
Our data indicate that the expression of individual human miRNA species is significantly associated with post-resection patient survival. Using data from five unique miRNAs, we were further able to generate a combined miRNA expression signature that is associated with patient survival (p=0.005; HR = 3.6) independent of node involvement and overall stage. The expression of three miRNAs (miR-99b, miR-199a_3p and _5p) was also associated with the presence of lymph node metastasis.
Conclusions
These results suggest miRNA expression profiling could provide prognostic utility in staging esophagus cancer patients and treatment planning with endoscopic and neoadjuvant therapies. The alterations of specific miRNAs may further elucidate steps in the metastatic pathway and allow for development of targeted therapy.
Keywords: Esophageal cancer, Genetics, genomics, Statistics, survival analysis
INTRODUCTION
Esophageal cancer is composed of two main histologic types: squamous cell carcinoma (SCC) and adenocarcinoma (EAC), with distinct epidemiologic differences between the two diseases (1). Once a rare tumor, incidence data from the Surveillance, Epidemiology and End Results (SEER) program show that EAC has the fastest rate of increase of any tumor type in the United States over the past three decades (2,3). This rapid rise has been attributed at least in part, to the prevalence of obesity, gastroesophageal reflux disease (GERD) and Barrett’s esophagus, a metaplastic change associated with a greater than 30-fold increased risk for EAC (4).
Survival rates from EAC have been historically dismal with almost half of patients presenting with distant disease. With recent advances in endoscopic techniques and with some improvement in response to neoadjuvant therapy, the management of potentially curable patients has been evolving. In experienced centers, patients with early stage, superficial tumors may be offered endoscopic resection or esophagectomy (5,6), while locally advanced tumors will be treated with preoperative chemo- or chemoradiation therapy followed by potentially curative esophagectomy (7–10).
The identification of molecular markers offers potential for improved patient staging and for the development of novel targeted treatments therapy. Recently, we have found that the integration of genomic copy number and gene expression data can accurately predict both survival and lymph node metastasis in esophageal adenocarcinoma (Bandla et al., in submission). We have also shown that microRNAs (miRNA) expression can accurately distinguish between EAC, esophageal SCC, Barrett’s esophagus and normal esophageal epithelium (11). However, there are few reports on the role of miRNA expression and esophageal adenocarcinoma patient outcomes (12,13).
miRNAs are a class of small, non-coding RNAs that are highly conserved and endogenously expressed across many species (14). miRNAs act as part of a multi-protein complex to silence or repress the expression of target genes, and a single miRNA typically influences the expression of many different genes (15). Repression is achieved by the targeting of complementary sequences, primarily on the 3′-untranslated region (UTR), of protein-encoding mRNAs, effecting translational block or degradation of the target mRNA (16,17). Although the full extent of the biological activity of most miRNAs has yet to be identified, they have been suggested to be intrinsic regulators of many cellular processes including cell differentiation, proliferation and apoptosis (18–23). Furthermore, aberrant expression of miRNAs has been linked to development and progression of cancer and has been shown to have prognostic significance in several tumor types, including lung, neuroblastoma, colon, pancreatic and lymphocytic leukemia (24–27). These reports suggest that miRNAs play an influential role in tumorigenic processes, and the identification of miRNAs aberrantly expressed in esophageal adenocarcinomas may lead to a better understanding of disease development and progression.
In order to investigate the role of aberrant miRNA expression in esophageal adenocarcinoma, we developed a custom miRNA microarray platform containing all known human, mouse and rat miRNAs along with over 2,000 novel, predicted human miRNAs. Using this array, we hypothesized that specific miRNAs and miRNA profile signatures could be identified and associated with survival and lymph node involvement of patients with esophageal adenocarcinoma.
MATERIALS AND METHODS
Patient Cohort and Tissue Preparation
Esophageal specimens were obtained from consecutive, eligible patients undergoing esophagectomy for adenocarcinoma during the years 2001 through 2005, at the University of Pittsburgh Medical Center, Pittsburgh, PA. All patients provided informed consent and research was approved by appropriate institutional review boards. No patients received neoadjuvant therapy prior to surgery and clinical characteristics of the cohort are shown in Table 1. Tumors were snap-frozen in liquid nitrogen and subsequently embedded for sectioning on a cryostat. Hematoxylin and eosin stained slides from each specimen underwent pathologic review (by M.W.) to confirm diagnosis and to identify those with >70% tumor cell representation.
Table 1.
Clinical characteristics of esophageal adenocarcinoma patients used in survival analysis.
| Variable | n (%) |
|---|---|
| Gender | |
| Male | 38 (84) |
| Female | 7 (16) |
| N Stage | |
| Node-negative | 24 (53) |
| Node-positive | 21 (47) |
| T Stage | |
| T1 | 20 (44) |
| T2 | 5 (11) |
| T3 | 18 (40) |
| T4 | 2 (5) |
| Overall Stage | |
| I | 14 (31) |
| II | 14 (31) |
| III | 14 (31) |
| IV | 3 (7) |
| Follow-up (months) | |
| Median | 21.8 |
| Range | 1.4 – 55.4 |
miRNA Isolation
RNA was isolated from thirty 5-micron sections of each tumor using the mirVana miRNA Isolation Kit (Ambion, Austin, Texas). Large fraction RNA (containing mRNA and ribosomal RNA) and enriched small RNA (≤200bp) containing miRNA were isolated simultaneously. Integrity of the large fraction RNA was assessed with Agilent Bioanalyzer RNA nano chips and only samples where the RNA integrity number (RIN) was ≥ 6 were used in microarray experiments.
miRNA Array Design
Custom miRNA microarrays (TGmiRV1) were manufactured by Affymetrix (Santa Clara, CA). All miRNA sequences were used in their native form and were tiled 5 times each at different locations across the array. The arrays consist of all human mouse and rat miRNA sequences from the Sanger database (V8.2). In addition, a further 2102 potential mature human miRNA sequences were predicted from pre-mature sequences found in the miRNAmap database (http://mirnamap.mbc.nctu.edu.tw/). Mature sequences were predicted using the miRAlign and ProMir miRNA prediction tools (http://cbit.snu.ac.kr/~ProMiR2/). The array also contained 72 control sequences, along with standard Affymetrix hybridization control probes.
miRNA Labeling
Enriched small RNA was labeled in a final reaction volume of 20μl, containing 4U of poly-A-polymerase (Ambion), 2.5mM MnCl2, 1X E-PAP Buffer (Ambion), 0.33mM UTP (Ambion) and 0.65mM Bio-UTP (Enzo Life Sciences, New York, NY). The reaction mixture was incubated at 37°C for 2 hours. After incubation, unincorporated nucleotides were removed using the MirVana clean-up columns (Ambion) and labeled miRNA was eluted in 30μl of buffer and stored at −20°C until chip hybridization. The labeling reaction mix also contained two synthetic miRNA labeling controls (cel-mir-2 and cel-lin-4), at 0.04fmol (cel-lin-4) and 1.4amol (cel-mir-2).
Array hybridizations
Labeled small RNAs were hybridized on the TGmiRV1 arrays for 16 hours at 45°C in standard Affymetrix MES hybridization buffer. Affymetrix eukaryotic microarray hybridization controls were also included in the hybridization buffer. Arrays were washed and stained on an Affymetrix Fluidics Station and processed arrays were scanned on an Affymetrix G7 scanner.
Data Analysis
miRNA expression estimates were extracted form cel files using Partek Genomic Suite (Partek Inc., St. Louis, MO). Replicate probes were median polished and normalized using Cyclic loess. Normalized data was filtered to include only miRNAs where >50% of samples had a log2 expression value >3.
Statistical Procedures
After normalization and filtering, 199 human miRNAs remained for analysis. Initially, a global test of overall significance, the tail strength statistic (28) was applied, in order to assess of the overall merit of the collection of miRNAs for finding association with overall survival. Individual miRNAs were selected by computing proportional hazards regression coefficients for each miRNA and individual coefficients were tested for significance. To assess the robustness of the survival associations in the absence of an independent patient cohort, p values were recomputed using bootstrap re-sampling (500 samples). In addition, raw p values were adjusted to reflect the expected false discovery rate.
The most significant individual miRNAs were then selected to construct a multi-miRNA signature for association with overall survival. The number of miRNAs for the signature was selected by leave-one-out cross validation by evaluating the predictive ability of multiple miRNA signatures. The top 5 miRNAs were selected and a compound covariate constructed from the individual regression coefficients. This covariate was evaluated in multivariate proportional hazards models to adjust for influence of node status and disease stage. A similar process was followed to find a multi miRNA signature for classifying lymph node status.
miRNA Quantitative RT-PCR
Real time quantitative PCR (qRT-PCR) to verify mature miRNA expression was carried out using the High-Specificity miRNA QRT-PCR Detection Kit (Stratagene Corp, La Jolla, CA) using 100ng of the small RNA fraction. miRNA specific qPCR was performed on an ABI7900HT using a miRNA specific forward primer and universal reverse primer. The expression of individual microRNAs was calculated relative to the expression of an endogenous control (U6 RNA).
RESULTS
Array Sensitivity, Dynamic Range and Specificity
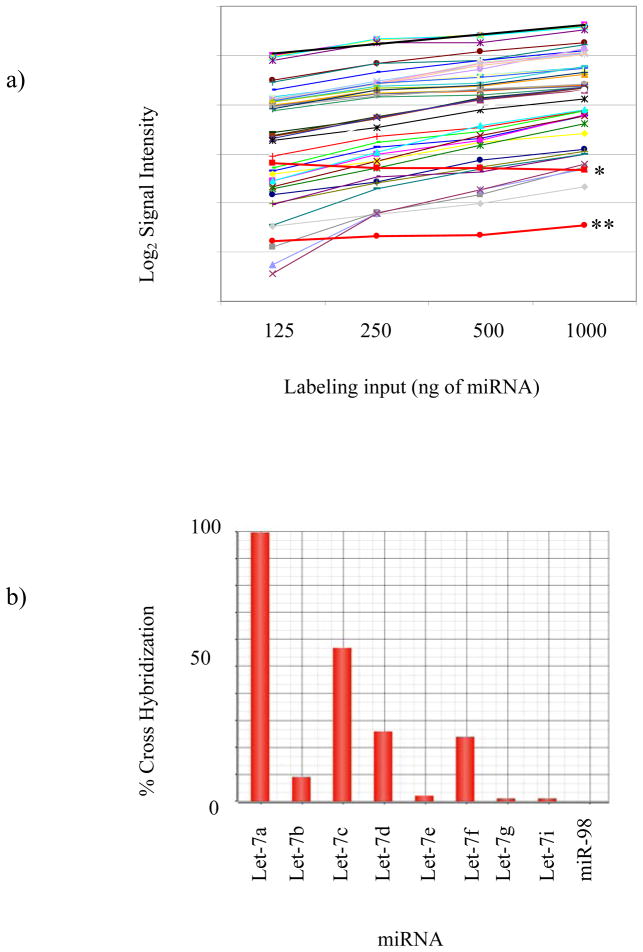
In order to assess the dynamic range, linearity and reproducibility of the arrays, we labeled control miRNA samples at concentrations from 125ng to 1μg and then assessed the expression of 50 randomly selected miRNAs. The average slope for concentration versus signal intensity was 0.96±0.01, consistent with a linear, quantitative response (Figure 1a). In addition, we added 0.04fmol and 1.4amol of two synthetic C. elegans miRNAs, cel-lin-4 and cel-mir-2, in to the labeling mix and the observed intensity for these two species was constant across all arrays, independent of the amount of small RNA labeled. Thus, the detection limit of the system is at least 1.4amol (Figure 1a).
Figure 1.
TGmirV1 Array dynamic range, sensitivity and specificity. a) miRNA was labeled and hybridized at concentrations ranging from 125ng to 1ug. Labeled Cel-mir-2* and Cel-Lin-4** were used as hybridization controls (red lines). The signal to concentration relationship was analyzed for 50 randomly selected miRNAs and the average slope for all miRNAs was 0.96±0.01. b) Array cross hybridization and sequence specificity was assessed using the Let-7 family of miRNAs. Synthetic Let-7a was labeled and hybridized and the percentage of cross-hybridization was assessed for all Let-7 family members.
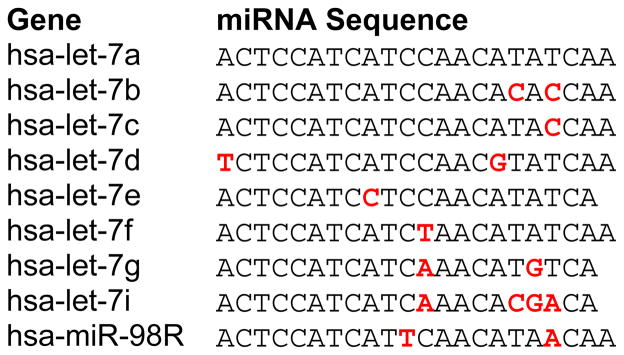
Cross-hybridization is a concern in all microarray experiments, particularly between sequences with high homology. The Let-7 family of miRNAs share on average 90% homology and have been used as the gold standard for assessing cross-hybridization in miRNA microarray and RT-PCR assays (29). Cross-hybridization between Let-7 family members was assessed using a synthetic Let-7a probe labeled and hybridized at amounts equivalent to biological levels. Low cross-hybridization (<10%) was observed between those sequences differing by two or more nucleotides. Of those sequences differing by a single nucleotide, only Let7c showed high (>50%) cross-hybridization with Let7a (Figure 1b; Table 2). The level of cross-hybridization observed on these custom arrays is therefore similar to that reported on other miRNA array platform (29,30).
Table 2.
Let-7 miRNA family sequence conservation, with sequence mismatches shown in red.
|
miRNAs Expressed in Esophageal Adenocarcinoma
A total of 87 of the 462 (19%) known human miRNAs on our array were expressed above background levels in at least 50% of esophageal adenocarcinoma samples. In addition, 112 of the 2101 (5.3%) predicted human miRNAs met the same criteria. By comparison, only 38 of 2563 (1.5%) the human antisense probes were expressed in >50% of samples indicating that non-specific hybridization is unlikely to account for the detection of a large number of the predicted miRNAs. Finally, 9% of miRNAs unique to either rat or mouse were also expressed indicating as yet unidentified human homologs.
Esophageal Adenocarcinoma miRNA Expression and Survival
The expression of 199 verified and predicted human miRNAs was evaluated for association with esophageal adenocarcinoma patient overall survival using the tail-strength statistic (28). We found that the individual p values from Cox regression coefficients for each of the 199 miRNAs produced a tail strength statistic of 0.276, indicating that the p values were lower than would be expected in a random distribution of test statistics. These data indicate that this set of miRNAs contains information relevant to patient survival and justifies exploration of individual miRNAs and miRNA signatures associated with survival.
Association of individual miRNA expression with esophageal adenocarcinoma patient survival was initially explored using a Cox proportional hazard model. A total of ten human miRNA probes were found to correlate with patient survival with bootstrap adjusted p-values <0.05, and eight of these ten also had a false discovery rate of less than 10%. Four of these probes represent miRNAs (miR-143, miR-145, miR-100 and miR-199a_3p) that were already in V8.2 of the Sanger-verified miRNA database. Two others (has-miR-04354a and has-miR-21886) represented potentially novel, predicted miRNA sequences on our array that now match the verified miR-199a_5p. The remaining two probes (hsa-miR-35453, hsa-miR-35463), although predicted as potentially novel miRNAs show very high homology to miR-145 and miR-143 respectively. Thus, these 8 miRNA probes actually identify five unique, verified miRNAs in the current Sanger Database.
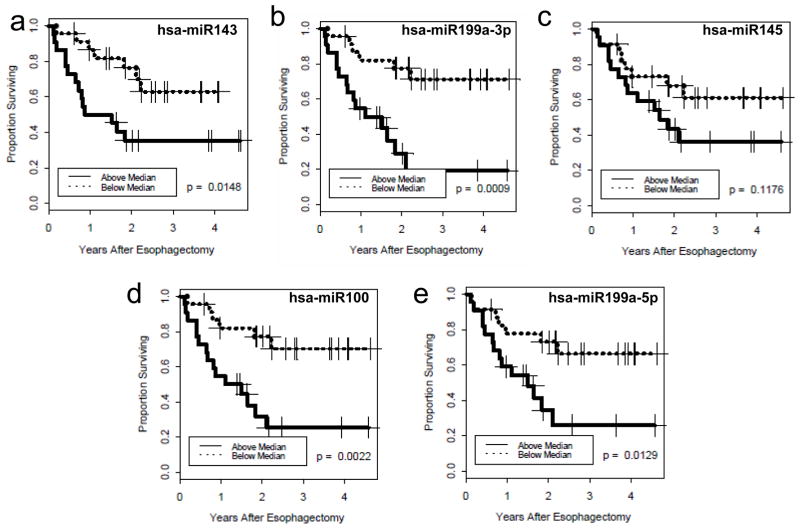
As expected the mouse and rat homologues of human miR-143, miR-145, miR-100 and miR-199a were also shown to associate with patient survival. The predicted, mature miRNA sequences and the probe sequences used for detection on the arrays are shown in Table 3. Kaplan-Meier survival estimates were also generated for individual human miRNAs with patients split into two groups based on the median expression (Figure 2).
Table 3.
Mature miRNA sequences for known and predicted miRNAs involved in esophageal adenocarcinoma survival signature (on custom miRNA array).
| miRNA | Mature Sequence |
|---|---|
| miR-100 | GGAUAUCUAUGUUCGAACA |
| miR-199a | UACAGUAGUCUGCACAUUGGUU |
| miR-143 | UGAGAUGAAGCACUGUAGCUCA |
| miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-04354a | CCCAGUGUUCAGACUACCUGUU |
| miR-21886 | CCCAGUGUUCAGACUACCUGUU |
| miR-35463 | UCCAGUUUUCCCAGGAAUCCCU |
| miR-35453 | UGAGAUGAAGCACUGUAGCUC |
Figure 2.
Kaplan-Meier survival plots for known miRNAs, a) miR-143 (p-value=0.0148), b) miR-199a_3p (p-value=0.0009), c) miR-145 (p-value=0.1176), d) miR-100 (p-value=0.0022) and e) miR-199a_5p (p-value = 0.0129) found to be individually associated with overall survival. Patients are split into high and low expression groups based on the median for each miRNA.
miRNA Expression Survival Signature
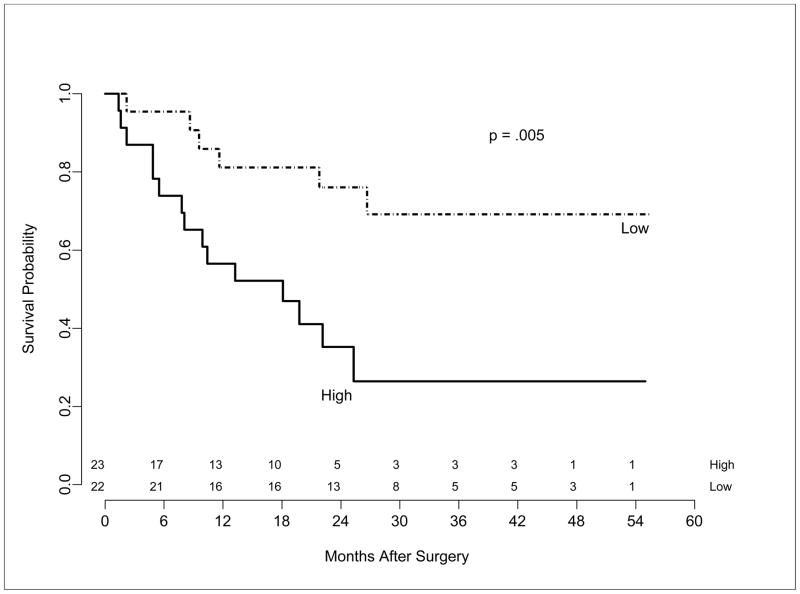
Individually prognostic human miRNAs were then used to develop a miRNA signature to predict esophageal adenocarcinoma patient survival. Based on the combined risk score generated in this model, those patients in the high risk group had a significantly worse survival (p=0.005; HR = 3.6) than those in the low risk group (Figure 3). In addition, the risk score was significantly higher in T3 and T4 tumors compared to T1 and T2 (Wilcoxon test p = .0001). In a multivariate proportional hazards model the miRNA expression signature was also associated with patient survival (p=0.0006; HR=3.25) and this was independent of nodal status (p =0.0099) and overall stage (p=0.0274).
Figure 3.
Kaplan-Meier survival plot for esophageal adenocarcinoma patients based on a miRNA signature. Patients were classified into high or low risk based on a median split of the risk score (log-rank p=0.005; HR =3.6).
miRNA Expression and Lymph Node Metastasis
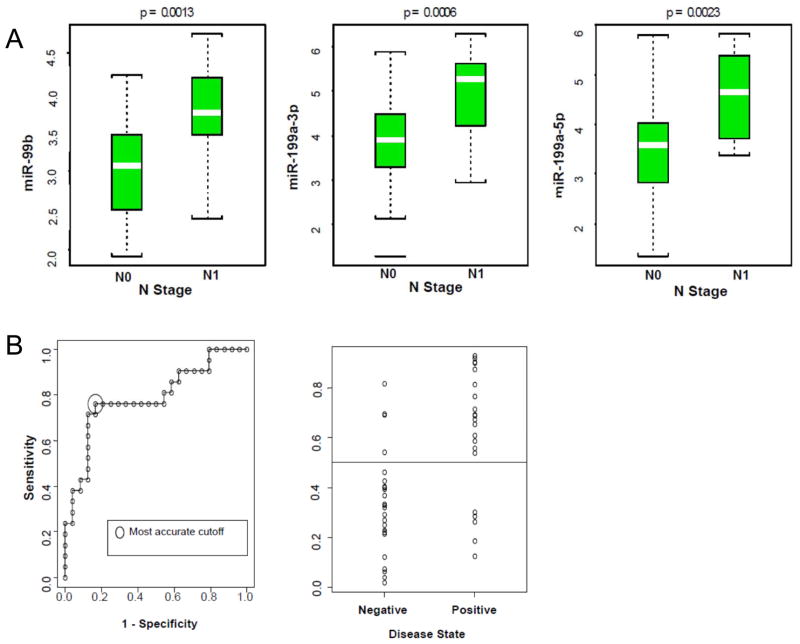
Class comparison analysis identified three miRNAs (mir-99b, mir-199a_3p and mir-199a_5p) that demonstrated a significant association (<10% false discovery rate) with lymph node metastasis in EAC (Figure 4a). Using the three verified miRNAs (mir-99bR and mir-199a_3p and _5p) in a logistic regression model to predict lymph node metastasis, the area under the ROC curve was 0.788 (95% CI; 0.651–0.924) and the cross-validated accuracy was 78% (Figure 4b).
Figure 4.
a) Class comparison analysis, showing the prediction of lymph node metastasis in esophageal adenocarcinoma patients for three miRNAs. b) Using the three verified miRNAs (mir-99bR and mir-199a_3p and _5p) in a logistic regression model to predict lymph node metastasis, the area under the receiver operator curve was 0.788 (95% CI; 0.651–0.924) and the cross-validated accuracy was 78%.
Verification of miRNA Expression by qRT-PCR
Real time qRT-PCR analysis was performed for selected mature miRNAs to verify the prognostic impact of miRNA expression in esophageal adenocarcinoma. cDNA was generated from a subset of esophageal adenocarcinomas from the original set along with 11 samples of normal squamous epithelium.
We sought to confirm the prognostic impact of miR-143 and miR-145 on EAC patient survival. Patient samples were stratified according the median expression of unique miRNAs. Kaplan-Meier survival analysis demonstrated a significant worse survival for patients with high expression of miR-143 (log Rank = 0.037), and miR-145 (Log Rank p=0.023). QRT-PCR confirmed a significantly lower expression of miR-143 (p=0.0049) and miR-145 (p=0.0069) in EAC compared to normal specimens.
Comment
The full extent of the role that miRNAs play in the development and progression of neoplastic disease is still unclear. However, it is apparent that miRNA expression plays a crucial role in regulating gene expression, and their aberrant expression can contribute to the development and progression of cancer (24–27). Although several genomic copy number and gene expression studies have revealed putative oncogenes and tumor suppressor genes that may be involved in esophageal adenocarcinoma, the role of miRNAs and their potential impact on the development of this disease, still remain to be elucidated. Here we used a custom miRNA microarray to explore expression of miRNAs and their association with esophageal adenocarcinoma patient survival and nodal status.
Cox proportional hazard regression analysis of global miRNA expression identified five unique human miRNAs that are associated with EAC patient overall survival. Up-regulation of miRNAs, miR-143, miR-199a_3p, miR-199a_5p, miR-100 and miR-99a predicted a worse survival. Greater numbers of up regulated miRNAs associating with survival is consistent with other studies (24,25). Several of these miRNAs have been previously shown to be deregulated in cancer or associate with cancer patient survival, including miR-143, miR-145 and miR-199a (27).
Several of the miRNAs identified in this study have previously been implicated in cancer development, in particular miR-143 and miR-145. Here, we confirmed their down regulation in EAC compared to normal tissue. This is consistent with other studies showing both miRNA-143 and miR-145 to be under expressed in several human malignancies when compared to their non-cancer counterpart (31–34). Juxtaposed to this, we observe higher expression to associate with worse patient survival, indicating a potential oncogenic function and appearing contrary to their suggested role as tumor suppressors (35). This disparity may be due to the activity of specific miRNAs and specific tissue types or cellular process (36). To further support our findings, a recent study has shown the association of higher miR-143 expression with increasing colon cancer tumor stage and lymph node status, even though its expression is markedly reduced in colon cancer compared to normal (35). These results are analogous to our finding showing higher expression of miR-143 in lymph node positive tumors. This raises the intriguing possibility that loss of expression may be necessary for the initial neoplastic transformation, but their re-expression is necessary for the development of a more aggressive phenotype, and may be involved in the development of metastatic potential.
Our findings suggest that the expression of three related miRNAs miR-199a_3p and _5p and miR-99b were associated with the presence or absence of lymph node involvement. miR-199a was also in the prognostic survival signature. Increased expression of miR-199a recently been shown to associate with a worse survival in several tumors types, including, acute myeloid leukemia, hepatocellular carcinoma and cervical carcinomas (37–39). The tissue specific expression of miRNAs is again highlighted by the association of high miR-99a expression and lymph node metastasis. miR-99a has previously been shown to be down regulated in lung cancer, and suggested to be candidate tumor suppressor in this disease (39). Conversely, miR-99a has been shown to be up-regulated in serous ovarian carcinomas when compared to normal tissue (40) again demonstrating the differential tissue-specific behavior of miRNAs.
Changes in miRNA expression are involved in the pathogenesis of esophageal adenocarcinoma (11–13). The development of a miRNA array containing predicted miRNAs allowed for the discovery of novel miRNAs, many of which are now in the human miRNA database. While more miRNAs are sure to be discovered in the future, our study has identified several known miRNAs that are prognostic in esophageal adenocarcinoma and these may represent new molecular markers for the identification and staging of this disease.
We conclude that miRNA expression profiling may provide prognostic utility in staging patients and treatment planning as the management of esophageal cancer continues to evolve and include endoscopic and neoadjuvant therapies. We acknowledge however that this is preliminary work and needs to be validated in independent patient sets including those receiving neoadjuvant therapy. Finally, the alteration of specific miRNAs may further elucidate steps in the metastatic pathway and allow for development of targeted therapy.
Footnotes
Presented at the Fifty-seventh Annual Meeting of the Southern Thoracic Surgical Association, Orlando, FL, November 3-6, 2010.
Reference List
- 1.Pera M, Manterola C, Vidal O, et al. Epidemiology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:151–159. doi: 10.1002/jso.20357. [DOI] [PubMed] [Google Scholar]
- 2.DeMeester SR. Adenocarcinoma of the esophagus and cardia: a review of the disease and its treatment. Ann Surg Oncol. 2006;13:12–30. doi: 10.1245/ASO.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 3.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 4.Chang JT, Katzka DA. Gastroesophageal reflux disease, Barrett esophagus, and esophageal adenocarcinoma. Arch Intern Med. 2004;164:1482–1488. doi: 10.1001/archinte.164.14.1482. [DOI] [PubMed] [Google Scholar]
- 5.Pech O, Behrens A, May A, et al. Long-term results and risk factor analysis for recurrence after curative endoscopic therapy in 349 patients with high-grade intraepithelial neoplasia and mucosal adenocarcinoma in Barrett’s oesophagus. Gut. 2008;57:1200–1206. doi: 10.1136/gut.2007.142539. [DOI] [PubMed] [Google Scholar]
- 6.Pennathur A, Farkas A, Krasinskas Am, et al. Esophagectomy for T1 esophageal cancer: outcomes in 100 patients and implications for endoscopic therapy. Ann Thor Surg. 2009;87:1048–1054. doi: 10.1016/j.athoracsur.2008.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Portale G, Hagen JA, Peters JH, et al. Modern 5-year survival of resectable esophageal adenocarcinoma: single institution experience with 263 patients. J Am Coll Surg. 2006;202:588–596. doi: 10.1016/j.jamcollsurg.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 8.McKensie S, Mailey B, Artinyan A, et al. Improved Outcomes in the Management of Esophageal Cancer with the Addition of Surgical Resection to Chemoradiation Therapy. Ann Surg Oncol. 2010 Sep; doi: 10.1245/s10434-010-1314-7. epub. [DOI] [PubMed] [Google Scholar]
- 9.Pennathur A, Luketich JD, Landreneau RJ, et al. Long-term results of a phase II trial of neoadjuvant chemotherapy followed by esophagectomy for locally advanced esophageal neoplasm. Ann Thor Surg. 2008;85:1930–1936. doi: 10.1016/j.athoracsur.2008.01.097. [DOI] [PubMed] [Google Scholar]
- 10.Kountourakis P, Correa AM, Hofstetter WL, et al. Combined modality therapy of cT2N0M0 esophageal cancer: the University of Texas M. D. Anderson Cancer Center experience. Cancer. 2010 Oct 19; doi: 10.1002/cncr.25651. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathe EA, Nguyen GH, Bowman, et al. MicroRNA Expression in Squamous Cell Carcinoma and Adenocarcinoma of the Esophagus: Associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Correa AM, Hoque A, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2011;128:132–143. doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14(Spec No 1):R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 15.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 18.Brennecke J, Cohen SM. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 2003;4:228. doi: 10.1186/gb-2003-4-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 20.Jensen RH, Tiirikainen M, You L, et al. Genomic alterations in human mesothelioma including high resolution mapping of common regions of DNA loss in chromosome arm 6q. Anticancer Res. 2003;23:2281–2289. [PubMed] [Google Scholar]
- 21.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 23.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 27.Iorio MV, Visone R, Di LG, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 28.Taylor J, Tibshirani R. A tail strength measure for assessing the overall univariate significance in a dataset. Biostatistics. 2006;7:167–181. doi: 10.1093/biostatistics/kxj009. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Gong J, Zeng H, et al. MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res. 2010;70:2728–2738. doi: 10.1158/0008-5472.CAN-09-3718. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Chen H. The role of microRNAs in colorectal cancer. J Genet Genomics. 2010;37:347–358. doi: 10.1016/S1673-8527(09)60053-9. [DOI] [PubMed] [Google Scholar]
- 34.Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200:154–160. doi: 10.1016/j.cancergencyto.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 36.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer. 2007;46:1000–1010. doi: 10.1002/gcc.20485. [DOI] [PubMed] [Google Scholar]
- 40.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]