Summary
Background
The glutathione S-transferase (GST) enzymes catalyze the conjugation of xenobiotics to glutathione. Based on reports that inherited copy number variations (CNV) modulate some GST gene expression levels, and that the small airway epithelium (SAE) and alveolar macrophages (AM) are involved early in the pathogenesis of smoking-induced lung disease, we asked: do germline CNVs modulate GST expression levels in SAE and AM?
Methods
Microarrays were used to survey GST gene expression in SAE and AM obtained by bronchoscopy from current smokers and nonsmokers, and to determine CNV genotypes.
Results
Twenty six % of subjects were null for both GSTM1 alleles, with reduced GSTM1 mRNA levels seen in both SAE and AM. Thirty % of subjects had homozygous deletions of GSTT1 with reduced mRNA levels in both tissues. Interestingly, GSTT2B, exhibited homozygous deletion in blood in 27% of subjects and was not expressed in SAE in the remainder of subjects but was expressed in AM of heterozygotes and wild type subjects, proportionate to genotype.
Conclusions
These data show a germline CNV-mediated linear relationship of genotype to expression level suggesting minimal compensation of gene expression levels in heterozygotes consistent with GST polymorphisms playing a role in the risk of smoking-associated xenobiotic-induced lung disease.
Introduction
The epithelial surface of cigarette smokers are exposed to large amounts of inhaled compounds in an aerosol of 1014 free radicals per puff, 1010 particulates/mL, and >4000 different compounds including over 60 carcinogens [1–4]. The small airway (bronchi, >6 generations, <2 mm in diameter) epithelium (SAE) takes the brunt of the exposure to the xenobiotics generated by smoking, and is the earliest site of abnormalities in smokers, central to the pathogenesis of chronic obstructive disease (COPD) and adenocarcinoma, the most common smoking-associated lung cancer [5–15]. In addition to the small airways, the alveoli are also exposed to the stress of smoking [9,10,16,17]. The alveoli are protected from xenobiotics, in part, by alveolar macrophages (AM), the lung representative of the mononuclear phenotype systems, equipped with a biologic armamentarium to engulf particulates and render xenobiotics harmless [16,18– 22]. AM play an important role in the pathogenesis of smoking-induced disease in the lower respiratory tract [9,14,18,20,22–28]
As in the other tissues, xenobiotic compounds are enzymatically transformed by airway epithelium and alveolar macrophages to a variety of intermediates by phase I and II enzymes, including the glutathione S-transferases (GSTs) [29–34]. GSTs are an evolutionary-conserved family of dimeric phase II metabolic enzymes that catalyze the conjugation of reduced glutathione with electrophilic compounds, such as xenobiotics present in tobacco smoke, as well as other carcinogens and pesticides, and their isoforms are divided into seven classes: alpha (GSTA), mu (GSTM), pi (GSTP), theta (GSTT), zeta (GSTZ), sigma (GSTS), and omega (GSTO) [29–34]. Though cytosolic GST enzymes are a central part of the lung molecular detoxification arsenal, three of these isoenzyme genes, GSTM1, GSTT1 and GSTT2B (a copy of the GST theta paralog, GSTT2) are located in regions of the genome susceptible to copy number variations (CNV), resulting in gene deletion with different frequencies in different populations [35–38]. Compensatory mechanisms may result in tissue-specific effects of CNV on associated genes expression levels [39–41].
Given this background, and with the knowledge that gene polymorphisms (SNPs) of GSTM1 and GSTT1 are linked to COPD, accelerated decline in lung function, and to lung cancer, the present study was undertaken to determine if the presence of known copy number variable regions in GST isoenzymes results in modifications of GST expression in the SAE and AM [42–46]. To address this issue, SAE and AM were obtained via bronchoscopy from healthy nonsmokers and healthy smokers and, using microarray and TaqMan RT-PCR, assessed for the expression of the GST isoenzymes. Genomic DNA acquired from blood cells of the same individuals was examined by microarray and TaqMan RT-PCR for the presence of copy number variations (CNVs). The data demonstrates that GSTM1 and GSTT1 are significantly expressed in both small airway epithelium and AM, while GSTT2(B) is only expressed in AM. Importantly, the SAE and AM gene expression levels of GSTM1 and GSTT1 correlate with CNV genotype, while the high frequency gene deletion of GSTT2B correlates with expression of GSTT2 in AM. In view of the associations of genetic variants of GSTM1 and GSTT1 with COPD and lung cancer, and that these diseases arise in the SAE and AM principally due to exposure to cigarette smoke with its heavy xenobiotic burden, the observation that the SAE and AM of healthy nonsmokers and smokers exhibits CNV-correlated levels of GST expression suggests that the mechanisms underlying the disease associations with GST isoenzymes include CNV-mediated disturbances in gene expression in lung cells.
Methods
Study Population
In response to advertisements, nonsmokers and smokers were evaluated at the Weill Cornell NIH Clinical and Translational Sciences Center and Department of Genetic Medicine Clinical Research Facility under protocols approved by the Weill Cornell Medical College Institutional Review Board. Written informed consent was obtained from each individual before enrollment. Subjects were deemed to be normal and in good health following standard medical history, physical examination, complete blood count, coagulation profile, serum chemistries and liver function testing, urine studies, chest radiograph, EKG and pulmonary function testing. All were negative for HIV1 and had normal α1-antitrypsin levels (see Supplemental Methods for detailed inclusion/exclusive criteria). For the group of nonsmokers (n=35) and the group of current smokers (n=35), self-reported smoking status was confirmed by urinary tobacco metabolite levels.
Sampling of the Small Airway Epithelium and Collection of Alveolar Macrophages
Small airway epithelium (10th to 12th generation) was collected using flexible bronchoscopy as previously described [47]. Cells were removed from the brush by flicking into 5 ml of ice-cold LHC8 medium (GIBCO, Grand Island, NY), with 4.5 ml immediately processed for RNA extraction, and 0.5 ml to determine the number and types of cells recovered. The expression of genes encoding surfactant and Clara cell secretory proteins confirmed the samples were small airway epithelium [47,48].
Bronchoalveolar lavage fluid was also obtained at the time of bronchoscopy as described previously [49]. Up to a maximum of three sites per individual (right middle lobe, lingula, right lower lobe) were lavaged with a typical volume per site of 100 ml, resulting in a 45–65% return of infused fluid volume. Debris and mucus was removed by filtering the lavage fluid through gauze, after which the fluid was centrifuged at 1,200 rpm for 5 min, at 4°C. Cells were washed twice in RPMI 1640 medium containing 10% fetal bovine serum, 50 U/ml streptomycin, and 2 mM glutamine (Invitrogen, Carlsbad, CA) and seeded overnight in six-well tissue culture plates (2 × 106 in 2 ml/well) at 37°C in a 5% CO2 humidified incubator. The next day, nonadherent cells were gently removed, and cell viability was assessed by Trypan blue exclusion, expressed as a percentage of the total number of recovered cells that were counted on a hemocytometer. Cell differentials were quantified on sedimented cells following cytocentrifugation and the remainder was processed for RNA extraction.
RNA Extraction and Preparation for Microarray
Analyses were performed using the Affymetrix HG-U133 Plus 2.0 microarray. Total RNA was extracted from cells using TRIzol reagent (Invitrogen), and residual DNA was removed by RNeasy MinElute RNA purification kit (Qiagen, Valencia, CA), yielding between 2 and 4 µg RNA per 106 cells. In order to visualize and quantify the degree of RNA integrity, an aliquot of each sample of RNA was analyzed with the Agilent Bioanalyzer. The concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and the samples stored in RNA secure (Ambion, Austin, TX). The GeneChip One-Cycle cDNA Synthesis kit was used to create double stranded complementary DNA from 3 µg of total RNA, followed by a cleanup step using GeneChip Sample Cleanup Module. In vitro transcription was next performed by way of a GeneChip IVT Labeling Kit, followed by additional cleanup and quantification of the biotin-labeled cRNA yield using a spectrophotometer (all reagents from Affymetrix, Santa Clara, CA). In accordance with Affymetrix protocols, the test microarrays were first hybridized and, if quality control was acceptable, hybridization to the gene expression chips was then performed, followed by processing by the Affymetrix GeneChip Fluidics Station 450, and scanning with an Affymetrix GeneChip Scanner 3000 7G (http://affymetrix.com/support/technical/manual/expression_manual.affx). Among the strict quality control measures employed included ensuring (1) RNA quality, assessed by RNA integrity number (RIN) > 7.0; (2) cRNA transcript integrity, assessed by signal intensity ratio of glyceraldehyde-3- phosphate dehydrogenase (GAPDH) 3' to 5' probe sets ≤ 3.0; and (3) multi-chip scaling factor ≤10.0 [50].
Microarray Data analysis and Statistics
The Microarray Suite version 5.0 (MAS 5.0) algorithm from Affymetrix was used to analyze captured images. GeneSpring version 7.3 software (Agilent Technologies) was used to normalize data as follows: (1) per microarray, by dividing raw data by the 50th percentile of all measurements on the individual array; and (2) per gene, by dividing the raw data by the median expression level for all of the genes across all of the arrays in a given dataset. In the current hypothesis-driven study, GSTM1, GSTT1 and GSTT2B were pre-selected for consideration on the basis that they are known to be affected by copy number polymorphisms from a review of the literature, and not on the basis of a global analysis of transcriptomic data or genome-wide genetic variation data. For each of these three pre-specified gene’s, the genes expression levels, as provided by microarray, were compared to the measurement of their specific gene copy number, as assessed by SNP microarray. Correction for false discovery from multiple testing was therefore not performed in the current study. Because equal variance of gene expression among each CNV grouping was not uniformly present, statistical testing for association between copy number and expression levels was done with a Kruskal Wallis test for non-parametric data. Correlations of gene expression level with CNV genotypes were performed using a Kendall tau rank correlation coefficient test. Correlations of gene expression levels for pairs of genes were calculated using Spearman rank correlation. Statistical analyses were carried out using StatView version 5.0 (SAS Institute Inc.). All microarray data has been deposited at the Gene Expression Omnibus (GEO) site (accession number 20250).
Assessment of Copy Number Variations in Genomic DNA
DNA was extracted from blood samples obtained from the study population using the Autogen FX robotic system in accordance with the manufacture’s protocols (Autogen, Holliston, MA). To minimize handling errors, pre-printed bar coded labels were used, and critical steps in the processing of samples were performed with two technicians present. The Affymetrix Human SNP Array 5.0 was used to examine the genomic DNA for CNV regions using Partek Genomics Suite software version 6.4 (Partek Inc., St Louis, MO). Multiple SNP arrays were loaded with data normalized by array to ensure comparable probe intensities relative to a HapMap subject. Search parameters (p<0.00001, >10 probe sets, 2.3<fold-change <1.7, signal/noise ratio ≥0.5) were chosen to allow detection of CNVs in chromosomes harboring cytosolic GST genes. As an additional confirmation of a given CNV call at a given locus, probe intensities of two of the largest CNV probe sets located within the boundaries of the relevant gene and the overlapping reported CNV region according to the Database of Genomic Variants [51] were plotted against each other for the entire study population to examine concordance with Partek calls.
TaqMan RT-PCR Confirmation of Microarray Expression Levels and Copy Number Calls
For gene expression confirmation, cDNA was synthesized from 2 µg RNA in a 100 µl reaction volume using the TaqMan Reverse Transcriptase Reaction k Kit (Applied Biosystems, Foster City, CA) using random hexamers as primers. Triplicate wells were run for each of two dilutions of each sample, 1:50 and 1:100. TaqMan PCR reactions were performed using premade kits (Applied Biosystems) and for each 25 µl reaction volume, 2 µl of cDNA was used. 18S ribosomal RNA served as an endogenous control and relative expression levels were determined using the ΔΔCT method (Applied Biosystems), with the average value for the nonsmokers as the calibrator. The rRNA probe was labeled with VIC dye and the probe for the gene of interest was labeled with FAM (6-carboxy fluorescein), and reactions were ran in an Applied Biosystems 7500 Sequence Detection System.
For copy number call confirmation, genomic DNA from individuals of known CNV genotype (based on Partek microarray analysis), was extracted and purified using a commercial kit and quantified using TaqMan RNase P method (Applied Biosystems). DNA samples were diluted to 5 ng/µl with 1X TE buffer, pH 8.0. Samples labeled with FAM were run against a DNA sample of known copy number for the GST gene of interest, which was used as a calibrator reference (labeled with VIC), together with a No Template Control, to allow detection of contamination and background fluorescence. Ribosomal RNA was used as the internal control (Human Ribosomal RNA Kit, Applied Biosystems). Reactions were carried out using gene-specific TaqMan Copy Number Assays in accordance with associated protocols (Applied Biosytems) and ran in the 7500 Sequence Detection System. CopyCaller™ Software (Applied Biosystems) was used to make the CNV calls.
Results
Study Population and Sampling
Small airway epithelium from healthy nonsmokers (n=35) and healthy smokers (n=35), and alveolar macrophages from healthy nonsmokers (n=22) and healthy smokers (n = 34) were the subject of the analyses. All subjects were deemed to be healthy based on no significant prior medical history, a normal physical examination and unremarkable urine studies, serum chemistries, radiology and pulmonary function studies (Table I; Supplemental Methods). For all subjects, no significant differences were observed between the two groups (nonsmokers vs smokers) with respect to age (p>0.8, pairwise student t test), gender (p>0.1, chi2 test) and ancestry (p>0.2, chi2 test). The combined smokers had an average smoking history of 24 ± 13 pack-yr and their self-reported smoking status was confirmed in all cases by urinary tobacco metabolites levels. SAE samples, of both nonsmokers and smokers contained approximately 6×106 cells of >99% purity, with cell types typical for the small airway epithelium. There was no significant difference in the relative proportions of these airway epithelial subtypes recovered between the two groups of subjects (p>0.05) with the exception of a greater proportion of undifferentiated columnar cells in smokers (p<10−3). Characteristic morphological appearances of both airway epithelium and macrophages were confirmed by microscopy of cytospin preparations from the brushings and cells recovered by lavage, respectively. Approximately twice as many AM were recovered from the lavage samples of smokers vs nonsmokers (31.2 × 106 vs 14.1 × 106 respectively, p <0.005) while the total numbers of other cell types were similar between the two groups (p>0.1).
Table I.
Demographics of the Study Population and Biologic Samples1
| Small airway epithelium | Alveolar macrophages | |||
|---|---|---|---|---|
| Parameter | Healthy nonsmokers |
Healthy smokers |
Healthy nonsmokers |
Healthy Smokers |
| n | 35 | 35 | 22 | 34 |
| Sex (male/female) | 28/7 | 22/13 | 17/5 | 24/10 |
| Age (yr) | 43 ± 11 | 43 ± 6 | 41 ± 8 | 43 ± 7 |
| Race (B/W/O)2 | 17/14/4 | 21/8/6 | 13/6/3 | 20/9/5 |
| Smoking history (pack-yr) | 0 | 24 ± 13 | 0 | 26 ± 17 |
| Urine nicotine (ng/ml) | 0 | 1029 ± 1045 | 0 | 796 ± 878 |
| Urine cotinine (ng/ml) | 0 | 1158 ± 861 | 0 | 1060 ± 717 |
| Blood carboxyhemoglobin (%) |
0.6 ± 0.9 | 2.2 ± 2.0 | 0.6 ± 0.8 | 2.4 ± 2.4 |
| Pulmonary function parameters3 |
||||
| FVC | 108 ± 12 | 109 ± 12 | 106 ± 11 | 109 ± 11 |
| FEV1 | 108 ± 14 | 109 ± 13 | 106 ± 11 | 109 ± 12 |
| FEV1/FVC | 82 ± 7 | 82 ± 4 | 82 ± 5 | 82 ± 4 |
| TLC | 102 ± 13 | 101 ± 12 | 96 ± 8 | 99 ± 12 |
| DLCO | 98 ± 15 | 97 ± 12 | 95 ± 9 | 98 ± 13 |
| Epithelial cells | ||||
| Number recovered | 6.0 × 106 | 6.1 × 106 | ||
| % epithelial cells | 99.5 ± 0.9 | 99.7 ± 0.6 | ||
| % inflammatory cells | 0.5 ± 0.9 | 0.3 ± 0.6 | ||
| % ciliated | 74.4 ± 7.2 | 71.4 ± 7.4 | ||
| % secretory | 7.0 ± 3.7 | 7.4 ± 3.1 | ||
| % basal | 10.7 ± 4.7 | 10.1 ± 3.4 | ||
| % undifferentiated | 7.3 ± 3.2 | 10.8 ± 5.7 | ||
| BAL4 cells | ||||
| Number recovered x106 | 14.1 ± 9.1 | 31.2 ± 20.3 | ||
| % viability | 96.6 ± 1.2 | 96.3 ± 1.1 | ||
| % alveolar macrophages5 | 96.0 ± 1.3 | 95.7 ± 1.7 | ||
| % lymphocytes | 2.7 ± 1.6 | 2.8 ± 1.7 | ||
| % polymorphonuclear cells | 0.9 ± 1.1 | 1.1 ± 1.3 | ||
| % epithelial cells | 0.4 ± 0.7 | 0.4 ± 0.4 | ||
Data is presented as mean ± standard deviation.
B = black, W = white, O = other.
Pulmonary function testing parameters are given as percent of predicted value with the exception of FEV1/FVC, which is reported as % observed; FVC - forced vital capacity, FEV1 - forced expiratoryvolume in 1 sec, TLC - total lung capacity, DLCO - diffusing capacity of the lung for carbon monoxide.
Bronchoalveolar lavage.
Listed is the % AM before purification; after purification the AM were >98% pure.
Expression of Glutathione S-Transferase Genes in Small Airway Epithelium and Alveolar Macrophages
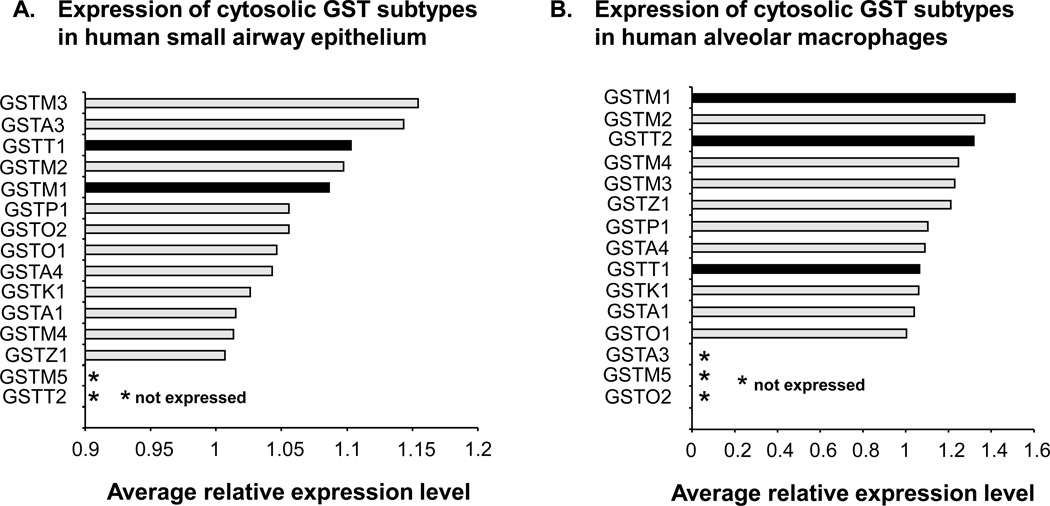
Using an expression criterion of having Affymetrix detection call of “Present” in 50% or more of either nonsmoker or smoker samples, significant expression of all known cytosolic GST genes was observed in SAE of healthy nonsmokers and healthy smokers (all subjects combined) with the exceptions of GSTM5 and GSTT2 (Figure 1A). Similarly, in the case of AM, expression data assessed by microarray, significant expression of all GST genes was seen in nonsmokers and smokers, except for GSTM5, GSTA3 and GSTO2 (Figure 1B). Using the Affymetrix “P” call criteria, of the three GST isoenzyme genes known to be deleted by copy number variation polymorphisms (GSTM1, GSTT1 and GSTT2), two (GSTM1 and GSTT1) were significantly expressed in the small airway epithelium, and all three were significantly expressed in AM. All of the GST genes indicated as being expressed in Figure 1 were expressed when nonsmokers and smokers were considered as two separate groups.
Figure 1.
Expression of cytosolic glutathione S-transferase subtypes in human small airway epithelium and alveolar macrophages for the total study population. A. Relative average gene expression levels of GST isoenzymes in small airway epithelium of healthy nonsmokers (n = 35) and healthy smokers (n = 35). B. Relative average gene expression levels of GST isoenzymes in alveolar macrophages of healthy nonsmokers (n = 22) and healthy smokers (n = 34). For both panels, all cytosolic GSTs are shown on the ordinate and those GST subtypes whose probe sets were called as “Present” by Affymetrix “P” call in fewer than 50% of subjects are referred to as “not expressed” as indicated by an asterisk. GST subtypes known to be affected by common (>5% frequency) heritable copy number variation are highlighted by black bars. The Affymetrix HGU133 Plus 2.0 microarray has no specific probeset for GSTT2B, a duplicate gene of GSTT2, and therefore is not shown.
GSTM1 Copy Number Variation and Correlation with Small Airway Epithelium Gene Expression
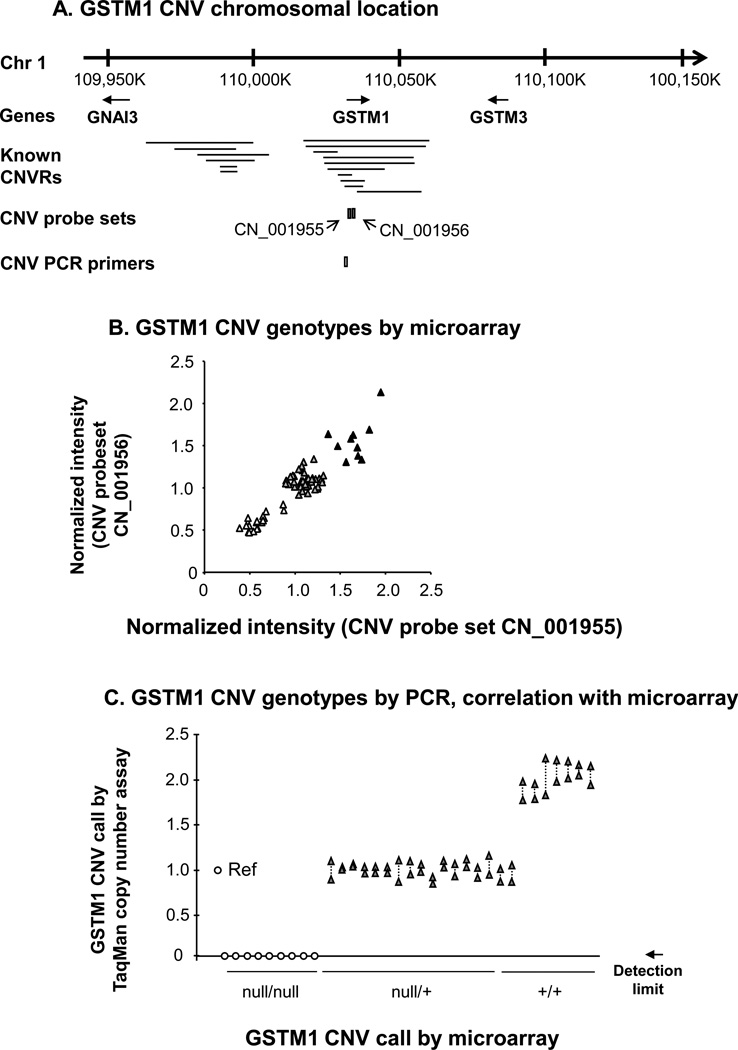
To establish which of the study subject samples had evidence of heritable copy number polymorphisms in the GSTM1 gene, genomic DNA from the entire population of 35 healthy nonsmokers and 35 healthy smokers was hybridized to Affymetrix Human SNP Array 5.0 chips to determine GSTM1 copy number (Figure 2A, B). The data demonstrated that 10 subjects (14%) had the wild type diploid copy number for GSTM1, 42 subjects (60%) were heterozygotes, and 18 subjects (26%) were homozygous for the gene deletion. For a random subset of these subjects, CNV genotypes were confirmed by TaqMan RT-PCR (Figure 2C). For an overview of the frequency of the null allele for GSTM1 and the other GST genes studied in detail, see Table II.
Figure 2.
Identification of heritable GSTM1 copy number variation. A. Region of human chromosome 1 showing the genomic location of the GSTM1 gene in relation to independently reported copy number variable regions (CNVRs) in the Database of Genomic Variants[51]. The relative chromosomal locations of two Affymetrix Human SNP 5.0 CNV probe sets and an Applied Biosystems TaqMan RT-PCR probe for GSTM1 copy number variation are shown below the lines identifying the known CNV. B. GSTM1 CNV genotypes by microarray. Normalized intensity levels from the entire study population (n = 70) of the two CNV probe sets indicated in panel A are plotted on the abscissa and the ordinate. Individuals homozygous null for the GSTM1 CNV (△), heterozygotes (▲) and wild type diploid individuals (▲) are as shown. C. PCR correlation with microarray of GSTM1 genotypes assessed by TaqMan PCR. Represented on the abscissa are the are the TaqMan RT-PCR assays for the GSTM1 CNV using the primers shown in panel A in a random subset of subjects (n = 33) of known GSTM1 genotype [homozygous nulls, heterozygotes (null/+) and diploid wild type, (+/+)] based on microarray results. One reference haploid subject (“Ref”) is shown left most, with TaqMan-derived copy number calls on the ordinate. Null/null individuals had a RT-PCR product below the detection limit as indicated. Shown are duplicate measurements for each subject, linked by dashed lines.
Table II.
Frequency of Cytosolic Glutathione S-Transferase Isoenzyme Copy NumberVariation in the Study Populations
| Frequency of deletion in current study4 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Frequency of deletion in general3 |
Healthy nonsmokers |
Healthy smokers |
|||||||
| GST isoenzyme family 1 |
Chromoso mal location |
Gene subtypes |
No. of CNVs2 |
Eur % |
Afr % |
Eur % |
Afr % |
Eur % |
Afr % |
| alpha (A) | 6p12.1 | GSTA1 | 2 | - | - | - | - | - | - |
| GSTA2 | 2 | - | - | - | - | - | - | ||
| GSTA3 | 0 | - | - | - | - | - | - | ||
| GSTA4 | 1 | - | - | - | - | - | - | ||
| GSTA5 | 0 | - | - | - | - | - | - | ||
| sigma (S) | 4q22.3 | PGDS | 0 | - | - | - | - | - | - |
| theta (T) | 22q11.23 | GSTT1 | 12 | 24[44] | 25[44] | 46 | 47 | 50 | 48 |
| 34[81] | 49[81] | ||||||||
| GSTT2B | 8 | 63[38] | 48[38] | 58 | 46 | 61 | 59 | ||
| mu (M) | 1p13.3 | GSTM1 | 9 | 50 [44] | 30[44] | 61 | 53 | 67 | 50 |
| 71[81] | 51[81] | ||||||||
| GSTM2 | 6 | - | - | - | - | - | - | ||
| GSTM3 | 0 | - | - | - | - | - | - | ||
| GSTM4 | 1 | - | - | - | - | - | - | ||
| GSTM5 | 3 | - | - | - | - | - | - | ||
| pi (P) | 11q13 | GSTP1 | 1 | - | - | - | - | - | - |
| omega (O) | 10q25.1 | GSTO1 | 0 | - | - | - | - | - | - |
| GSTO2 | 0 | - | - | - | - | - | - | ||
| zeta (Z) | 14q24.3 | GSTZ1 | 0 | - | - | - | - | - | - |
GST, glutathione S-transferase. Shown are the cytosolic GST isoenzyme families, each represented by theindicated Greek alphabetical character.
CNV, copy number variation. Shown are the reported number of different CNVs that partially orcompletely overlap the indicated gene, according to the Database of Genomic Variants, accessed onlineon January 20th 2010 at http://projects.tcag.ca/variation/. The term CNV as used here includes genomicamplifications and deletions.
Where the reported copy number variation (CNV) is a gene deletion, the frequency of the null allele ofthe deleted gene in the general population is reported, where known, a dash indicates that no allelefrequency data is available. References are shown in parentheses. Eur = European, Afr = African.
Where a detected CNV results in a deletion of the indicated gene in the present study, the frequency of thenull allele of such a gene is shown.
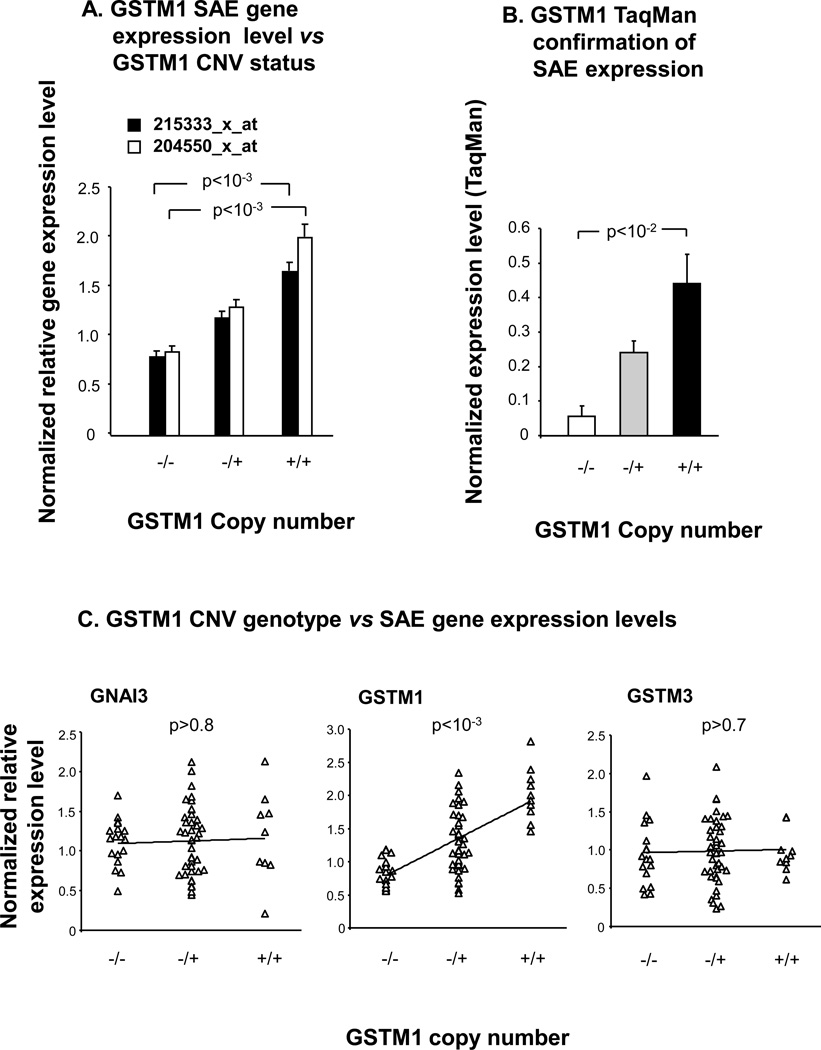
In order to assess the association of GSTM1 CNV genotype with small airway epithelium GSTM1 gene expression, SAE microarray gene expression data from the same 70 individuals who were genotyped, was examined for correlations between genotype and expression level (Figure 3A–C). The association of GSTM1 gene expression with GSTM1 CNV genotype was seen in different GSTM1 probe sets (p<10−3 for both probe sets determined by Kruskal Wallis analysis), and GSTM1 small airway epithelial expression level was verified in a random subset of individuals by TaqMan RT-PCR (Figure 3A,B). To identify potentially spurious associations of genotype with expression, the assessment included two genes flanking GSTM1 but not located within regions known to be copy number variable, GNAI3 and GSTM3. The data shows that, while GSTM1 SAE gene expression level was positively correlated with increasing GSTM1 copy number (p<10−3, tau = 0.404), neither GNAI3 (p>0.8, tau = 0.020) nor GSTM3 (p> 0.7, tau = 0.039) were correlated with GSTM1 CNV genotype (Figure 3C). TaqMan RT-PCR verified the lack of association of SAE expression level of the flanking gene GSTM3 with GSTM1 CNV genotype (p>0.3, data not shown).
Figure 3.
Correlation of small airway epithelium gene expression levels with GSTM1 CNV genotype. A. Microarray. GSTM1 CNV genotype is plotted against normalized average gene expression level for the two indicated Affymetrix GSTM1 expression probe sets (healthy nonsmokers, n=22; healthy smokers, n=34). P values represent Kruskal Wallis tests. B. TaqMan RT-PCR. The normalized average expression level by TaqMan RT-PCR is shown on the ordinate for incremental GSTM1 copy number, for a random subset of individuals (n = 22). P value shown is a Kruskal-Wallis test. C. Comparisons of GSTM1 CNV genotypes vs SAE gene expression levels. The three plots show GSTM1 copy number on the abscissa versus normalized relative gene expression levels on the ordinate, for GNAI3, GSTM1 and GSTM3 respectively, in the total study population (n = 70). Kendall tau rank correlation p values are as shown. For A and B the error bars represent the standard error.
GSTT1 Copy Number Variation and Correlation with Small Airway Epithelium Gene Expression
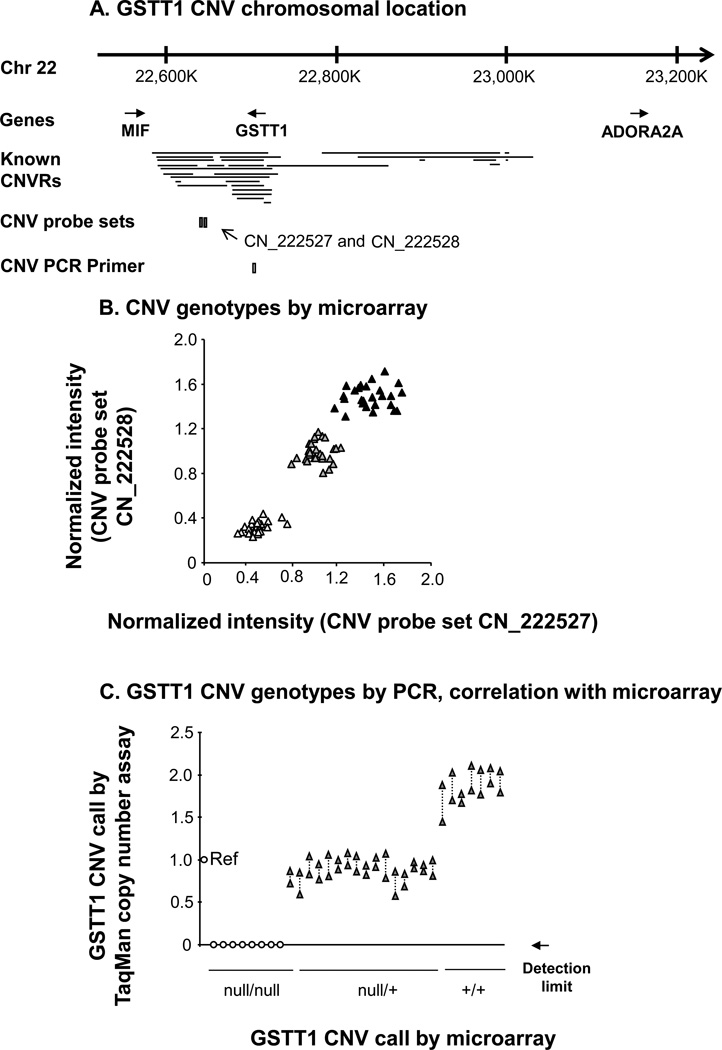
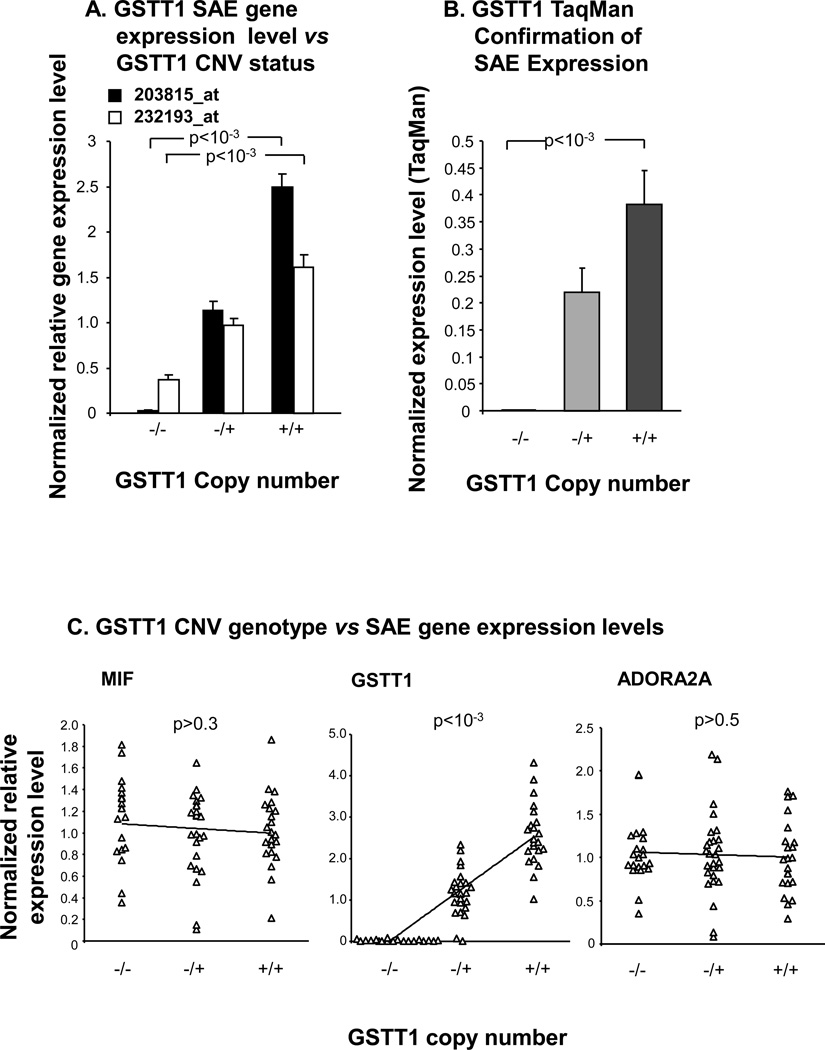
For the copy number variable GST isoenzyme GSTT1, a similar approach to that used for GSTM1 was employed to establish whether or not heritable GSTT1 CNVs were present in the same study population of 35 healthy nonsmokers and 35 healthy smokers (Figure 4). By microarray analysis, the data revealed that 22 subjects (31%) possessed the wild type diploid number of GSTT1 gene copies, 27 subjects (39%) were heterozygotes and 21 individuals (30%) had no copy of GSTT1 in their blood genome (Figure 4A, B). The CNV genotypes identified by microarray were confirmed in a random subset of subjects using TaqMan RT-PCR, with 100% concordance (Figure 4C).
Figure 4.
Identification of heritable GSTT1 copy number variation. A. Chromosome 22 location of the GSTT1 gene in relation to known CNV. The relative locations of two Affymetrix Human SNP 5.0 CNV probe sets and an Applied Biosystems TaqMan RT-PCR primer for GSTT1 copy number variation are also represented below. B. CNV genotypes by microarray. Normalized intensity levels from the study population (n = 70) of the two CNV probe sets indicated in panel A are plotted on the abscissa and the ordinate. Subjects homozygous null for the GSTT1 CNV (△), heterozygotes (▲) and homozygous wild type individuals (▲) are as shown. C. Correlation of PCR assessed GSTT1 CNV genotypes to microarray assessment. TaqMan RT-PCR assays for the GSTT1 CNV using the probe shown in panel A in a random subset of subjects (n = 31) of microarray-defined GSTT1 genotype [homozygous nulls, heterozygotes (null/+) and diploid subjects (+/+)] are represented on the abscissa. The reference haploid subject (“Ref”) is shown on the left, with copy number calls by RT-PCR on the ordinate. Null/null individuals had a RT-PCR product below the detection limit as indicated. Duplicate measurements are shown for each subject, linked by dashed lines.
To investigate the association of GSTT1 CNV genotype with the GSTT1 expression level in SAE, gene expression microarray probe sets specific to GSTT1 were correlated with the identified GSTT1 CNV genotype in the total population of 70 individuals. The nearby flanking genes, MIF and ADORA2A were also examined for potential correlation with GSTT1 CNV genotype, as they are not known to be located within the copy number variable region surrounding GSTT1 (Figure 4A). The data showed that GSTT1 small airway epithelium expression levels were directly proportional to the copy number of GSTT1, with highest expression levels in the wild type subjects, intermediate levels in the heterozygotes and lowest levels of GSTT1 in the homozygous null individuals (Figure 5A;p<10−3 and p<10−3 respectively for two different probe sets based on Kruskal Wallis testing). The correlation of GSTT1 gene expression and copy number was confirmed in a random subset of individuals by RT-PCR (Figure 5B; p<10−3 comparing homozygous nulls with wild types). No correlation was seen between gene expression levels of flanking genes and GSTT1 CNV genotype (Figure 5C, ADORA2A p>0.5, tau = −0.044; MIF p>0.3, tau = −0.082), however there was a strong correlation of GSTT1 expression level in SAE with GSTT1 CNV genotype (Figure 5C,p<10−3, tau = 0.631). TaqMan RT-PCR confirmed no association of MIF expression levels with GSTT1 CNV genotypes (p>0.5, data not shown).
Figure 5.
GSTT1 CNV genotype versus small airway epithelium gene expression. A. For each of the two indicated Affymetrix GSTT1 gene expression probe sets, GSTT1 CNV genotype is plotted against normalized average gene expression level on the ordinate. P values represent Kruskal Wallis test. B. The normalized average expression level by TaqMan RT-PCR is shown on the ordinate versus GSTT1 copy number, for a random subset of individuals (n = 24). P value shown is a Kruskal Wallis test. C. For each of the three indicated genes, GSTT1 copy number is plotted on the abscissa versus each gene’s normalized relative expression levels on the ordinate, in the total study population (n = 70). Kendall tau rank correlation p values are shown. Error bars in panels A, B represent standard error.
GSTM1 and GSTT1 Copy Number Variation and Correlation with Alveolar Macrophage Gene Expression
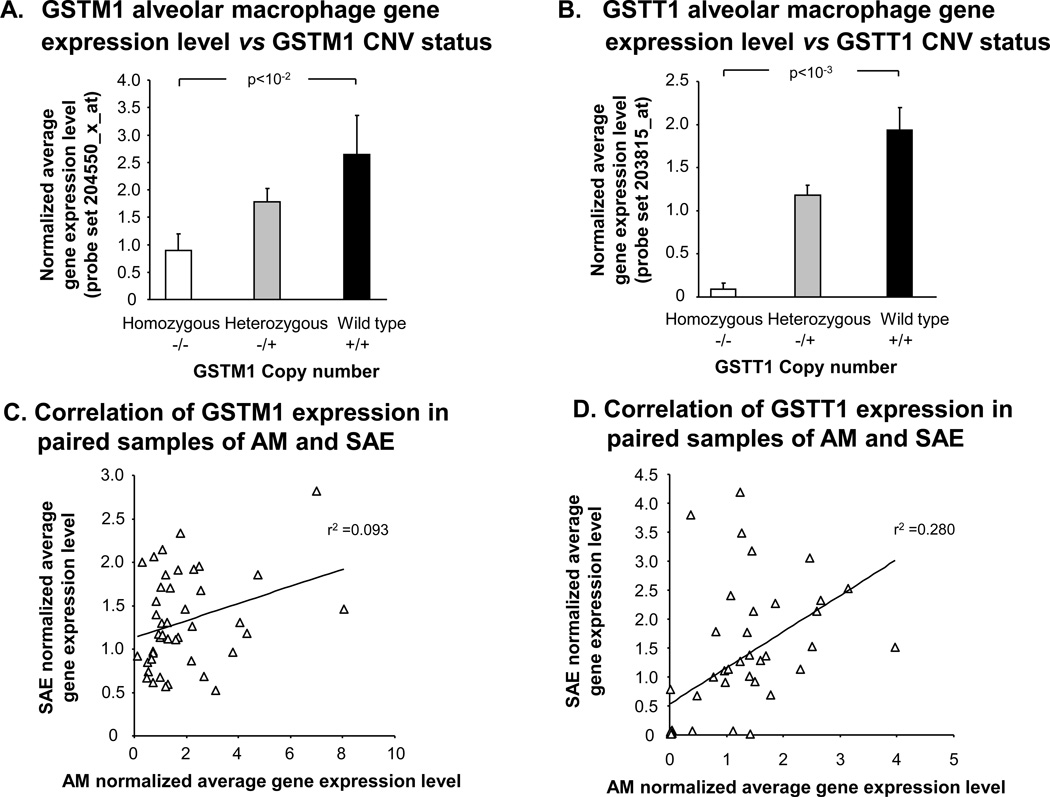
For the purpose of investigating potential correlations of GSTM1 and GSTT1 CNV genotypes with the GSTM1 and GSTT1 expression levels in another lung cell type, microarray gene expression data of AM (n = 22 healthy nonsmokers, n = 34 healthy smokers), were assessed as for the SAE. Similarly to what was observed in the SAE, GSTM1 AM gene expression levels were positively correlated with GSTM1 copy number (Figure 6A,p<10−2 by Kruskal Wallis test). GSTT1 CNV genotypes were also directly proportional to GSTT1 gene expression level in the AM samples (Figure 6B,p<10−3 by Kruskal Wallis test). There was poor correlation of GSTM1 gene expression levels within individuals between SAE and AM samples (r2 = 0.09), and a stronger corresponding, but still weak, correlation (r2 = 0.28) for GSTT1 (Figure 6C, D).
Figure 6.
Correlation of GSTM1 and GSTT1 gene expression levels in alveolar macrophages with CNV genotype. A. GSTM1 gene expression levels vs GSTM1 CNV status. Average normalized gene expression levels in AM for GSTM1 as assessed using microarray are shown on the ordinate vs GSTM1 genotype from healthy nonsmokers (n = 22) and healthy smokers (n = 34). Kruskal Wallis p value is shown. B. GSTT1 gene expression levels vs GSTT1 CNV status. AM average normalized gene expression levels of GSTT1 are shown on the ordinate for each of the indicated GSTT1 CNV genotype groups. P value represents Kruskal Wallis test. C. GSTM1 expression in SAE vs AM. For GSTM1, average normalized microarray gene expression levels in AM is plotted against SAE gene expression for the same individuals (n = 43 ). The Spearman rank correlation is shown. D. GSTT1 expression in SAE vs AM. The correlation of microarray gene expression levels of GSTT1 in the same individuals (n = 43 ) between their AM on the abscissa and SAE on the ordinate. The Spearman rank correlation is shown. Error bars represent the standard error.
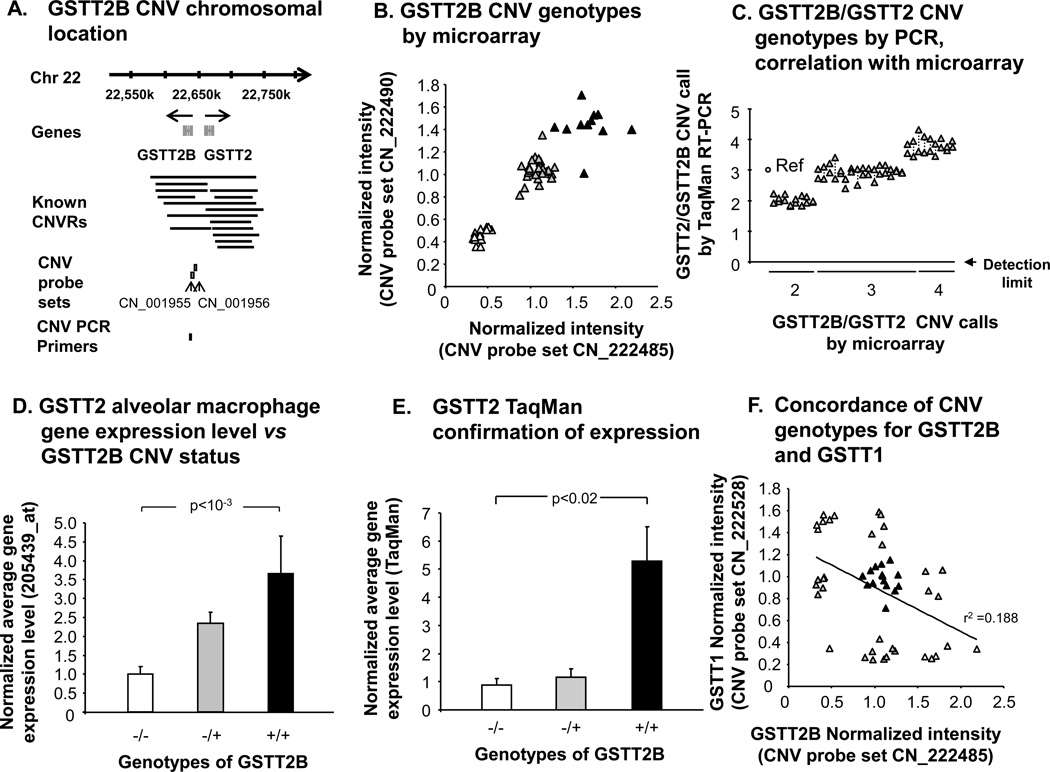
GSTT2B Copy Number Variation and Correlation with GSTT2 Alveolar Macrophage Gene Expression
GSTT2B has also been recently described to manifest a common heritable copy number variation[38]. The Affymetrix probe set for GSTT2 (there is no specific GSTT2B probe set) was expressed in SAE in fewer than 50% of subjects by “P” call, and was therefore not examined further in this cell type. However, the prevalence of GSTT2B CNV in the study population correlated with AM gene expression levels in the study population of 22 healthy nonsmokers and 34 healthy smokers. The data revealed that 12 individuals (21%) were wild types with two copies of GSTT2B, 29 subjects (52%) were heterozygous for the gene deletion, and 15 subjects (27%) were homozygous nulls (Figure 7A,B). The GSTT2B CNV calls of the microarray data was confirmed for a random subset of individuals using TaqMan RT-PCR, with >93 % concordance, using custom-designed primers and probes that detect both GSTT2B and GSTT2 (Figure 7C).
Figure 7.
Identification of genomic GSTT2B CNV and correlation with gene expression in alveolar macrophages. A. Schematic depiction of the chromosomal location of the GSTT2B and GSTT2 genes, showing the known nearby CNV and the locations of two Affymetrix CNV probe sets and TaqMan RT-PCR probe. CNVR, copy number variable region. PCR, polymerase chain reaction. B. GSTT2B CNV genotype assessed by microarray. Normalized intensity levels of the two GSTT2B CNV probe sets shown in panel A are plotted against each other. Those subjects who are homozygous null for the GSTT2B CNV (△), heterozygotes (▲) and individuals with two gene copies (▲) are as shown. C. GSTT1/GSTT2 CNV genotypes. Shown is TaqMan confirmation of the CNV calls made by microarray, in a random subset of individuals (n = 33). A reference haploid subject is shown against whom other individuals of microarray-defined genotype were assessed, with RT-PCR copy number on the ordinate. The custom-designed RT-PCR primers and probe used were specific to both GSTT2 and GSTT2B. Data points for duplicate measurements are shown, linked by dashed lines. D. GSTT2 AM gene expression vs GSTT2B CNV status. Normalized average GSTT2 gene expression level, as assessed by microarray on the ordinate, for healthy nonsmokers (n = 22) and healthy smokers (n = 34) of known GSTT2B CNV genotypes as indicated on the abscissa. P value represents Kruskal Wallis test. E. GSTT2 TaqMan conformation of expression in AM. Shown is TaqMan confirmation of AM GSTT2 gene expression level in a random subset of individuals of known GSTT2B genotype, as shown on the abscissa. The ordinate shows the average normalized GSTT2 expression level. Kruskal Wallis p value is shown. F. Concordance of CNV genotype for GSTT2B and GSTT1. Shown is microarray-defined GSTT1 and GSTT2B CNV probe set intensity levels were plotted against each other. Subjects within each of the nine possible genotype combinations (three by three) are shown in alternating shades for clarity. There is a trend, but only a minor correlation. For panels D, E, error bars indicate mean ± standard error.
The TaqMan RT-PCR expression data using AM mRNA for the flanking gene MIF, which lies outside of the GSTT2B CNV region, demonstrated no correlation with GSTT2B CNV genotype (data not shown). However, in the case of both the gene expression microarray probe set for GSTT2 and a TaqMan RT-PCR assay, AM GSTT2 gene expression levels correlated positively with GSTT2B copy number (Figure 7D, E,p<10−3, p<0.02 respectively, by Kruskal Wallis test). Despite the proximity of GSTT1 and GSTT2B to one another (within 64 kb) within the boundaries of known CNVs on chromosome 22, there was a poor correlation between CNV genotypes of GSTT1 and genotypes of GSTT2B, with only one subject being homozygous null for both deletions, and no subject having both wild type genes (r2=0.19; Figure 7F).
For all of the observed associations of GST isoenzyme copy number with gene expression levels in the present study, there was no significant effect of potential confounding factors, including when parsed by smoking status, with the exception of a higher pack-yr smoking history in the individuals that were heterozygous for the GSTM1 gene deletion when only the AM subject subpopulation was examined (p=0.03; Table III).
Table III.
Effect of Potential Confounding Factors on Associations of GSTM1, GSTT1 and GSTT2 Gene Expression Levels in Small Airway Epithelium and Alveolar Macrophageswith Regional Copy Number Variations1
| p value2 |
|||||
|---|---|---|---|---|---|
| Genetic ancestry |
Smoking status |
Pack yr |
Gender | Age | |
| GSTM1 | |||||
| SAE3 | 0.48 | 0.61 | 0.73 | 0.99 | 0.65 |
| AM | 0.61 | 0.61 | 0.034 | 0.50 | 0.89 |
| GSTT1 | |||||
| SAE | 0.26 | 0.68 | 0.23 | 0.46 | 0.25 |
| AM | 0.53 | 0.75 | 0.88 | 0.39 | 0.28 |
| GSTT2 | |||||
| AM | 0.10 | 0.34 | 0.19 | 0.99 | 0.90 |
Significance by p value of potential confounding variables on observed associations of CNV genotypewith GST isoenzyme gene expression is shown. GSTT2 was not significantly expressed in smallairway epithelium by Affymetrix “P-call”.
p values represent chi-squared tests except for pack-year and age, each of which was tested for interaction with CNV genotype by two factor ANOVA.
SAE, small airway epithelium; AM = alveolar macrophage.
Pack-yr was significantly higher among heterozygotes (haploid) for GSTM1 CNV than amonghomozygous nulls and wild types (diploid). There was a trend towards lower alveolar macrophageGSTM1 gene expression levels in smokers with greater than the median pack-yr smoking history, p=0.07.
Discussion
Smoking places a tremendous xenobiotic burden on the small airway epithelium and alveolar macrophages [1–28]. As with other cell types that express the glutathione S-transferase, these lung cells attempt to biotransform such compounds to innocuous chemicals [29–34]. Based on the knowledge that some members of the glutathione S-transferase family have a high frequency of gene deletion mediated by germline CNV polymorphisms [35–38], the present study asked the question: do germline CNVs influence small airway epithelium and alveolar macrophage gene expression levels of GST subtypes? Through the use of microarray analyses, the data demonstrates that GSTM1 and GSTT1 are expressed in both SAE and AM, while GSTT2 is expressed only in AM. The expression levels of these GST genes are modulated by the CNV genotype, with increasing gene copy number resulting in increased gene expression levels, as evidenced by different gene expression probe sets for each gene and/or confirmed by TaqMan RT-PCR. There was no strong evidence of any compensation at a mRNA level for the reduced gene dosage seen in subjects heterozygous for GST gene deletions; in all 3 cases, gene expression levels generally appeared to fall in a linear manner with falling gene copy number. None of these observations was attributable to effects of age, gender, smoking status, pack-yr or genetic ancestry. Together, these observations suggest that in cells that are confronted by the sustained xenobiotic insult of cigarette smoke, glutathione S-transferase genes known to play a key role in xenobiotic biotransformation are negatively regulated by highly prevalent heritable copy number variation polymorphisms, with a generally linear relationship between gene dose, and gene expression, carrying implications for smoke-induced lung disease pathogenesis.
Copy Number Variations and Gene Expression
Copy number variation polymorphisms, operationally defined as genomic gains and losses of 1 kb or larger, cover as much as 12% of the human genome and will probably turn out to be even more widespread within the genome, as the resolution of the platforms used to identify CNVs improves [40,52]. The most data available on CNV polymorphisms are in the mouse and rat, in which transcripts are over-represented in differentially expressed genes compared to ubiquitously expressed “housekeeping” genes [39,41,53]. Overall, a weak positive correlation was observed in these animal studies between relative gene expression level and the gene copy number, driven by strong correlations in less than a third of these CNV-associated genes.. For approximately two-thirds of CNV-associated genes, the number of gene copies had no effect on relative expression levels in any of several tissues examined. Further, the expression of some genes correlated with gene dosage in some tissues but not in others, implying gene dosage compensation and tissue specific responses to CNV [39,41,53]. Dosage compensation mechanisms have been observed for many genes, and postulated mechanisms proposed include inverse dosage effects and incomplete inclusion of regulatory elements in the gene deletion event [39,53–55]. Examples of tissue-specific gene dosage effects of CNV observed in mice include Rshl2a/b and Sirbp1 [39,40]. Another source of added complexity is the increasing evidence of common somatic mosaicism for copy number variation in different organs and tissues from the same individual [56–58].
For these reasons, the present study was carried out to establish the gene dosage effect of common germline CNVs for glutathione S-transferase genes, genes associated with smoke-induced lung diseases such as COPD and lung adenocarcinoma, in cells that are relevant to the smoke-induced lung diseases, small airway epithelium and alveolar macrophages [5–28]. Perhaps surprisingly, given the importance of these GST isoenzymes and the above observations, we found no convincing evidence of dosage compensation for these common CNVs at the mRNA level. This finding argues more strongly towards the relevance of these gene deletions to xenobiotic-associated lung disease, where no compensatory mechanism against allelic loss of gene expression appears to exist. The three distinct tiers of gene expression levels arising from these common biallelic polymorphisms in SAE and AM helps explain why historically, the results of association studies of polymorphic GST isoenzymes with lung disease are inconsistent, as usually the contributions of all these possible highly prevalent CNV genotypes in a given individual have not been fully addressed in such studies [38,44].
Small Airway Epithelium, Alveolar Macrophages and Xenobiotic Biotransforming Genes
Accumulating evidence has defined the importance of the SAE as the initial site of pathology in smoke-induced lung diseases including COPD and lung adenocarcinoma [5–15]. In addition, AM have long been postulated to play a major role in the development of emphysema [9,14,18,20,22–28]. Both the SAE and AM are important sources of xenobiotic-transforming enzymes such as cytochrome P450 enzymes and glutathione S-transferases which constitute the hosts defenses against attack from the myriad of compounds including many carcinogens present in cigarette smoke [29–34,48]. The present study demonstrates that the gene expression levels in SAE and AM of glutathione S-transferase isoenzymes GSTM1, GSTT1, and in the case of AM only, GSTT2, are reduced in healthy nonsmokers and smokers proportionate to the genes copy number in that individual. This gene dosage effect of CNV was seen with different probe sets for these genes and confirmed by TaqMan RT-PCR. The apparent expression of GSTM1 in homozygous nulls by microarray analysis in the present study is very likely a consequence of background noise from non-specific probe set hybridization, because TaqMan RT-PCR shows absent expression in such individuals. In the case of GSTT2B, a duplicate gene of GSTT2, CNV-mediated GSTT2B deletion reduces gene expression of GSTT2 in the AM, an effect that may be due to the inclusion within the deleted region of an enhancer element for GSTT2 as well as GSTT2B [38]. In the present study, significant expression in SAE and AM was documented for of all the cytosolic GST isoenzymes except for GSTM5 and GSTT2 in SAE and GSTM5, GSTA3 and GSTO2 in AM. Heretofore, GSTT1 was not known to be significantly expressed in SAE, nor is there literature regarding the expression of GSTT2 in AM. Many previous studies of the diversity of GST expression in the lung have focused on whole lung homogenates or proximal, large airway specimens rather than purified samples of small airway epithelium as in the present study, and often do not discriminate between isoenzymes within each of the seven GST classes [29,30,33,34,59].
The fact that the CNV-modulated GST genes are unaffected by smoke exposure is somewhat surprising. Many antioxidant and detoxification genes are significantly up-regulated in airway epithelium by chronic cigarette smoke exposure, based on studies in mice and humans[60–62]. Of note, no study has ever shown smoke-inducibility of GSTM1, GSTT1 or GSTT2 in airway epithelium. In fact, the only human airway epithelium gene expression data showing up-regulation of a GST isoform in response to cigarette smoke implicates GSTA2, which is not a subject of the current study [60]. Microarray studies in mice have shown that chronic cigarette smoke exposure upregulates GSTM2 and GSTO1, which are not affected by common CNV polymorphisms [62].
It is reasonable to suggest that in the setting of the expression of various isoforms of a GST class in a given tissue, redundancy of enzymatic activity would be created making the CNV-mediated alteration in gene dosage clinically irrelevant. A number of groups have previously documented the expression of GST mu isoforms GSTM1–4 in human lymphocytes, which is similar to the GST mu isoform expression profile seen in human small airway epithelium and alveolar macrophages in the present study [63–67]. It has been demonstrated in lymphocytes, that (in the context of the known expression of other GST mu isoforms) the selective activity of GSTM1 towards the substrate trans-stilbene oxide (TSO) correlates with the CNV-mediated deletion mutation of GSTM1 [63,68]. Furthermore, and of relevance to the molecular pathogenesis of smoke-induced lung disease [69], it has also been shown that DNA adduct levels in lymphocytes of smokers are inversely correlated with GSTM1 enzymatic activity towards TSO and positively correlated with daily cigarette consumption [65]. These observations support the concept that expression of various isoforms of a GST class within a given tissue is not sufficient to prevent the development of a clinically relevant CNV-mediated deficit in GST enzymatic activity in the face of chronic cigarette smoke exposure.
Glutathione S-transferase Isoenzyme Polymorphisms and Lung Disease
Gene deletion polymorphisms of GSTM1 and GSTT1 have been well documented [29–38,70–73]. The GSTM1 null allele is thought to have arisen from homologous unequal crossing over between two highly identical 4.2 kb repeated sequences flanking the GSTM1 gene, resulting in a 15 kb deletion including the entire GSTM1 gene [37,72,73]. A similar mechanism involving homologous recombination of two 403 bp flanking repeats has been reported to give rise to the GSTT1 null allele, resulting in a 54 kb deletion that includes the GSTT1 gene in its entirety [73,74]. The GSTT2B CNV has only been more recently identified, and is a 38 kb long deletion of the entire GSTT2B gene located within a 61 kb DNA inverted repeat [38]. Deletion of GSTT2B was shown to result in very low mRNA expression of the nearby duplicate gene GSTT2 in various cell lines, suggesting involvement of a common enhancer element centromeric to GSTT2 and within the CNV region.
All three of these GST gene deletions arise commonly in the population with differing frequencies depending on the ancestral group in question. For example, the GSTM1 CNV-mediated homozygous gene deletion has been reported to have a frequency between 38% and 67% in individuals of European ancestry versus 28% to 35% in individuals of African ancestry [75]. While the CNV-mediated GSTT1 and GSTT2B polymorphisms appear to be biallelic, GSTM1 has evidence of a multiallelic CNV with reports of an uncommon amplification genotype in up to 3% of Saudi Arabians [76]. The present study however, showed no evidence of other than biallelic copy number polymorphisms in GST subtypes.
A number of genomic association studies have linked GSTM1 and GSTT1 CNV-mediated gene deletions to the smoke-induced lung diseases COPD and lung cancer [42–46], although some studies have failed to reproduce these disease associations [77–80]. A number of potential explanations have been put forward for this variability, including inadequately powered studies, effects of population stratification given the known ancestral differences in frequency of the deletions, and the fact that many studies did not discriminate methodologically between wild type individuals and those with a single copy of the gene. However, many of the GST isoenzymes, including GSTM1 and GSTT2B are located within segmental duplications, known to be CNV-enriched throughout the genome, suggesting that there are other, yet to be characterized null alleles of other GST genes or modifying genes that may impact the results of such disease association studies, and may be uncovered in the future as CNV-detection methods are improved [38]. Another potential source of added complexity that the present study does not address, is the increasing evidence of common somatic mosaicism for copy number variation in different organs and tissues from the same individual [56–58].
In conclusion, the SAE and AM, front line cells exposed to the xenobiotics within cigarette smoke and implicated in smoke-induced lung disease, are significant sources of many glutathione S-transferase subtypes including class mu and theta. The data shows that, highly prevalent germline CNV-mediated deletions of GSTM1 and GSTT1 cause a progressive loss of mRNA in SAE of healthy nonsmokers and healthy smokers with no evidence of a compensatory mechanism for the reduced gene dosage at this crucial disease site. The presence of a highly prevalent recently described gene deletion affecting GSTT2B is also confirmed in the present study, with an associated reduction in total GSTT2 gene expression in AM. These data support the concept that the mechanism for associations of CNV-mediated GST gene deletions with smoking-induced lung disease involves an uncompensated loss of gene dosage in SAE and AM with likely resultant loss of some xenobiotic detoxifying capability. Future association studies of GST genes with lung disease should ensure capture of the many varied genotypes brought about by deletion and duplication events in different individuals to clarify the role of this important family of enzymes in complex smoking-induced lung disorders.
Supplementary Material
Acknowledgments
We thank DT Dang, M Teater, L Phipps, M Hicks, JP Murray and T Fukui for technical assistance; and N Mohamed and T Virgin-Bryant for help in preparing this manuscript. These studies were supported, in part, by R01 HL074326; P50 HL084936 and UL1-RR024996.
References
- 1.Pryor WA, Prier DG, Church DF. Electron-spin resonance study of mainstream and sidestream cigarette smoke: nature of the free radicals in gas-phase smoke and in cigarette tar. Environ Health Perspect. 1983;47:345–355. doi: 10.1289/ehp.8347345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 3.Macnee W, Rahman I. Oxidants and antioxidants as therapeutic targets in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1999;160:S58–S65. doi: 10.1164/ajrccm.160.supplement_1.15. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 5.Hogg JC, Macklem PT, Thurlbeck WM. Site and nature of airway obstruction in chronic obstructive lung disease. N Engl J Med. 1968;278:1355–1360. doi: 10.1056/NEJM196806202782501. [DOI] [PubMed] [Google Scholar]
- 6.Niewoehner DE, Kleinerman J, Rice DB. Pathologic changes in the peripheral airways of young cigarette smokers. N Engl J Med. 1974;291:755–758. doi: 10.1056/NEJM197410102911503. [DOI] [PubMed] [Google Scholar]
- 7.Yanai M, Sekizawa K, Ohrui T, Sasaki H, Takishima T. Site of airway obstruction in pulmonary disease: direct measurement of intrabronchial pressure. J Appl Physiol. 1992;72:1016–1023. doi: 10.1152/jappl.1992.72.3.1016. [DOI] [PubMed] [Google Scholar]
- 8.Rosado-de-Christenson ML, Templeton PA, Moran CA. Bronchogenic carcinoma: radiologic-pathologic correlation. Radiographics. 1994;14:429–446. doi: 10.1148/radiographics.14.2.8190965. [DOI] [PubMed] [Google Scholar]
- 9.Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur Respir J. 2003;22:672–688. doi: 10.1183/09031936.03.00040703. [DOI] [PubMed] [Google Scholar]
- 10.Celli BR, Macnee W. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J. 2004;23:932–946. doi: 10.1183/09031936.04.00014304. [DOI] [PubMed] [Google Scholar]
- 11.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, Cherniack RM, Rogers RM, Sciurba FC, Coxson HO, Pare PD. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 12.Petty RD, Nicolson MC, Kerr KM, Collie-Duguid E, Murray GI. Gene expression profiling in non-small cell lung cancer: from molecular mechanisms to clinical application. Clin Cancer Res. 2004;10:3237–3248. doi: 10.1158/1078-0432.CCR-03-0503. [DOI] [PubMed] [Google Scholar]
- 13.Wistuba II. Genetics of preneoplasia: lessons from lung cancer. Curr Mol Med. 2007;7:3–14. doi: 10.2174/156652407779940468. [DOI] [PubMed] [Google Scholar]
- 14.Kim V, Rogers TJ, Criner GJ. New concepts in the pathobiology of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2008;5:478–485. doi: 10.1513/pats.200802-014ET. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturton G, Persson C, Barnes PJ. Small airways: an important but neglected target in the treatment of obstructive airway diseases. Trends Pharmacol Sci. 2008;29:340–345. doi: 10.1016/j.tips.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Tuder RM, Yoshida T, Arap W, Pasqualini R, Petrache I. State of the art. Cellular and molecular mechanisms of alveolar destruction in emphysema: an evolutionary perspective. Proc Am Thorac Soc. 2006;3:503–510. doi: 10.1513/pats.200603-054MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharafkhaneh A, Hanania NA, Kim V. Pathogenesis of emphysema: from the bench to the bedside. Proc Am Thorac Soc. 2008;5:475–477. doi: 10.1513/pats.200708-126ET. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann-Matthes ML, Steinmuller C, Franke-Ullmann G. Pulmonary macrophages. Eur Respir J. 1994;7:1678–1689. [PubMed] [Google Scholar]
- 19.Willey JC, Coy E, Brolly C, Utell MJ, Frampton MW, Hammersley J, Thilly WG, Olson D, Cairns K. Xenobiotic metabolism enzyme gene expression in human bronchial epithelial and alveolar macrophage cells. Am J Respir Cell Mol Biol. 1996;14:262–271. doi: 10.1165/ajrcmb.14.3.8845177. [DOI] [PubMed] [Google Scholar]
- 20.Bezdicek P, Crystal RG. Pulmonary macrophages. In: Crystal RG, West JB, Weibel ER, Barnes PJ, editors. The lung: scientfic foundations. Philadelphia: Lippincott-Raven; 1997. pp. 859–876. [Google Scholar]
- 21.Hukkanen J, Pelkonen O, Hakkola J, Raunio H. Expression and regulation of xenobiotic-metabolizing cytochrome P450 (CYP) enzymes in human lung. Crit Rev Toxicol. 2002;32:391–411. doi: 10.1080/20024091064273. [DOI] [PubMed] [Google Scholar]
- 22.Tetley TD. Macrophages and the pathogenesis of COPD. Chest. 2002;121:156S–159S. doi: 10.1378/chest.121.5_suppl.156s. [DOI] [PubMed] [Google Scholar]
- 23.Chow CK. Cigarette smoking and oxidative damage in the lung. Ann N Y Acad Sci. 1993;686:289–298. doi: 10.1111/j.1749-6632.1993.tb39189.x. [DOI] [PubMed] [Google Scholar]
- 24.Heguy A, O'Connor TP, Luettich K, Worgall S, Cieciuch A, Harvey BG, Hackett NR, Crystal RG. Gene expression profiling of human alveolar macrophages of phenotypically normal smokers and nonsmokers reveals a previously unrecognized subset of genes modulated by cigarette smoking. J Mol Med. 2006;84:318–328. doi: 10.1007/s00109-005-0008-2. [DOI] [PubMed] [Google Scholar]
- 25.Birrell MA, Wong S, Catley MC, Belvisi MG. Impact of tobacco-smoke on key signaling pathways in the innate immune response in lung macrophages. J Cell Physiol. 2008;214:27–37. doi: 10.1002/jcp.21158. [DOI] [PubMed] [Google Scholar]
- 26.Wallace AM, Sandford AJ, English JC, Burkett KM, Li H, Finley RJ, Muller NL, Coxson HO, Pare PD, Abboud RT. Matrix metalloproteinase expression by human alveolar macrophages in relation to emphysema. COPD. 2008;5:13–23. doi: 10.1080/15412550701817789. [DOI] [PubMed] [Google Scholar]
- 27.Winkler AR, Nocka KH, Sulahian TH, Kobzik L, Williams CM. In vitro modeling of human alveolar macrophage smoke exposure: enhanced inflammation and impaired function. Exp Lung Res. 2008;34:599–629. doi: 10.1080/01902140802366261. [DOI] [PubMed] [Google Scholar]
- 28.Shaykhiev R, Krause A, Salit J, Strulovici-Barel Y, Harvey BG, O'Connor TP, Crystal RG. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol. 2009;183:2867–2883. doi: 10.4049/jimmunol.0900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singhal SS, Saxena M, Ahmad H, Awasthi S, Haque AK, Awasthi YC. Glutathione S-transferases of human lung: characterization and evaluation of the protective role of the alpha-class isozymes against lipid peroxidation. Arch Biochem Biophys. 1992;299:232–241. doi: 10.1016/0003-9861(92)90269-3. [DOI] [PubMed] [Google Scholar]
- 30.Cantlay AM, Smith CA, Wallace WA, Yap PL, Lamb D, Harrison DJ. Heterogeneous expression and polymorphic genotype of glutathione S-transferases in human lung. Thorax. 1994;49:1010–1014. doi: 10.1136/thx.49.10.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seidegard J, Ekstrom G. The role of human glutathione transferases and epoxide hydrolases in the metabolism of xenobiotics. Environ Health Perspect. 1997;105(Suppl 4):791–799. doi: 10.1289/ehp.105-1470052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 33.Zhang JY, Wang Y, Prakash C. Xenobiotic-metabolizing enzymes in human lung. Curr Drug Metab. 2006;7:939–948. doi: 10.2174/138920006779010575. [DOI] [PubMed] [Google Scholar]
- 34.Harju T, Mazur W, Merikallio H, Soini Y, Kinnula VL. Glutathione-S-transferases in lung and sputum specimens, effects of smoking and COPD severity. Respir Res. 2008;9:80. doi: 10.1186/1465-9921-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CL, Liu Q, Relling MV. Simultaneous characterization of glutathione S-transferase M1 and T1 polymorphisms by polymerase chain reaction in American whites and blacks. Pharmacogenetics. 1996;6:187–191. doi: 10.1097/00008571-199604000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Gresner P, Gromadzinska J, Wasowicz W. Polymorphism of selected enzymes involved in detoxification and biotransformation in relation to lung cancer. Lung Cancer. 2007;57:1–25. doi: 10.1016/j.lungcan.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Norskov MS, Frikke-Schmidt R, Loft S, Tybjaerg-Hansen A. High-throughput genotyping of copy number variation in glutathione S-transferases M1 and T1 using real-time PCR in 20,687 individuals. Clin Biochem. 2009;42:201–209. doi: 10.1016/j.clinbiochem.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Marotta M, Eichler EE, Eng C, Tanaka H. Linkage disequilibrium between two high-frequency deletion polymorphisms: implications for association studies involving the glutathione-S transferase (GST) genes. PLoS Genet. 2009;5:e1000472. doi: 10.1371/journal.pgen.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 40.Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, Schutz F, Ruedi M, Kaessmann H, Reymond A. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 41.She X, Rohl CA, Castle JC, Kulkarni AV, Johnson JM, Chen R. Definition, conservation and epigenetics of housekeeping and tissue-enriched genes. BMC Genomics. 2009;10:269. doi: 10.1186/1471-2164-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He JQ, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am J Respir Crit Care Med. 2002;166:323–328. doi: 10.1164/rccm.2111059. [DOI] [PubMed] [Google Scholar]
- 43.Cheng SL, Yu CJ, Chen CJ, Yang PC. Genetic polymorphism of epoxide hydrolase andglutathione S-transferase in COPD. Eur Respir J. 2004;23:818–824. doi: 10.1183/09031936.04.00104904. [DOI] [PubMed] [Google Scholar]
- 44.Ye Z, Song H, Higgins JP, Pharoah P, Danesh J. Five glutathione s-transferase genevariants in 23,452 cases of lung cancer and 30,397 controls: meta-analysis of 130 studies. PLoS Med. 2006;3:e91. doi: 10.1371/journal.pmed.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imboden M, Downs SH, Senn O, Matyas G, Brandli O, Russi EW, Schindler C, ckermann-Liebrich U, Berger W, Probst-Hensch NM. Glutathione S-transferase genotypes modifylung function decline in the general population: SAPALDIA cohort study. Respir Res. 2007;8:2. doi: 10.1186/1465-9921-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vineis P, Anttila S, Benhamou S, Spinola M, Hirvonen A, Kiyohara C, Garte SJ, Puntoni R, Rannug A, Strange RC, Taioli E. Evidence of gene gene interactions in lung carcinogenesis in a large pooled analysis. Carcinogenesis. 2007;28:1902–1905. doi: 10.1093/carcin/bgm039. [DOI] [PubMed] [Google Scholar]
- 47.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 48.Plopper CG, Hyde DM, Buckpitt AR. Clara cells. In: Crystal RG, West JB, Weibel ER, Barnes PJ, editors. The lung: scientific foundations. Philadephia: Lippincott-Raven; 1997. pp. 517–534. [Google Scholar]
- 49.Russi TJ, Crystal RG. Use of bronchoalveolar lavage and airway brushingto investigate the human lung. In: Crystal RG, editor. The Lung: Scientific Foundations. Philadelphia: Lippencott-Raven, Inc; 1997. pp. 371–371. [Google Scholar]
- 50.Raman T, O'Connor TP, Hackett NR, Wang W, Harvey BG, Attiyeh MA, Dang DT, Teater M, Crystal RG. Quality control in microarray assessment of gene expression in human airway epithelium. BMC Genomics. 2009;10:493. doi: 10.1186/1471-2164-10-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 52.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, Macdonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- 54.Lee JA, Madrid RE, Sperle K, Ritterson CM, Hobson GM, Garbern J, Lupski JR, Inoue K. Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol. 2006;59:398–403. doi: 10.1002/ana.20732. [DOI] [PubMed] [Google Scholar]
- 55.Montavon T, Le Garrec JF, Kerszberg M, Duboule D. Modeling Hox gene regulation in digits: reverse collinearity and the molecular origin of thumbness. Genes Dev. 2008;22:346–359. doi: 10.1101/gad.1631708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, de Stahl TD, Menzel U, Sandgren J, von TD, Poplawski A, Crowley M, Crasto C, Partridge EC, Tiwari H, Allison DB, Komorowski J, van Ommen GJ, Boomsma DI, Pedersen NL, den Dunnen JT, Wirdefeldt K, Dumanski JP. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piotrowski A, Bruder CE, Andersson R, de Stahl TD, Menzel U, Sandgren J, Poplawski A, von TD, Crasto C, Bogdan A, Bartoszewski R, Bebok Z, Krzyzanowski M, Jankowski Z, Partridge EC, Komorowski J, Dumanski JP. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 58.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pacifici GM, Franchi M, Bencini C, Repetti F, Di LN, Muraro GB. Tissue distribution of drug-metabolizing enzymes in humans. Xenobiotica. 1988;18:849–856. doi: 10.3109/00498258809041723. [DOI] [PubMed] [Google Scholar]
- 60.Hackett NR, Heguy A, Harvey BG, O'Connor TP, Luettich K, Flieder DB, Kaplan R, Crystal RG. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29:331–343. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 61.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adair-Kirk TL, Atkinson JJ, Griffin GL, Watson MA, Kelley DG, DeMello D, Senior RM, Betsuyaku T. Distal airways in mice exposed to cigarette smoke: Nrf2-regulated genes are increased in Clara cells. Am J Respir Cell Mol Biol. 2008;39:400–411. doi: 10.1165/rcmb.2007-0295OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seidegard J, Pero RW, Stille B. Identification of the trans-stilbene oxide-active glutathione transferase in human mononuclear leukocytes and in liver as GST1. Biochem Genet. 1989;27:253–261. doi: 10.1007/BF02401805. [DOI] [PubMed] [Google Scholar]
- 64.Comstock KE, Johnson KJ, Rifenbery D, Henner WD. Isolation and analysis of the gene and cDNA for a human Mu class glutathione S-transferase, GSTM4. J Biol Chem. 1993;268:16958–16965. [PubMed] [Google Scholar]
- 65.Soni M, Madurantakan M, Krishnaswamy K. Glutathione S-transferase Mu (GST Mu) deficiency and DNA adducts in lymphocytes of smokers. Toxicology. 1998;126:155–162. doi: 10.1016/s0300-483x(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 66.Kearns PR, Chrzanowska-Lightowlers ZM, Pieters R, Veerman A, Hall AG. Mu class glutathione S-transferase mRNA isoform expression in acute lymphoblastic leukaemia. Br J Haematol. 2003;120:80–88. doi: 10.1046/j.1365-2141.2003.04039.x. [DOI] [PubMed] [Google Scholar]
- 67.Hofmann T, Kuhnert A, Schubert A, Gill C, Rowland IR, Pool-Zobel BL, Glei M. Modulation of detoxification enzymes by watercress: in vitro and in vivo investigations in human peripheral blood cells. Eur J Nutr. 2009;48:483–491. doi: 10.1007/s00394-009-0039-5. [DOI] [PubMed] [Google Scholar]
- 68.Brockmoller J, Gross D, Kerb R, Drakoulis N, Roots I. Correlation between trans-stilbene oxide-glutathione conjugation activity and the deletion mutation in the glutathione S-transferase class mu gene detected by polymerase chain reaction. Biochem Pharmacol. 1992;43:647–650. doi: 10.1016/0006-2952(92)90591-6. [DOI] [PubMed] [Google Scholar]
- 69.Phillips DH. DNA adducts as markers of exposure and risk. Mutat Res. 2005;577:284–292. doi: 10.1016/j.mrfmmm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 70.Seidegard J, Vorachek WR, Pero RW, Pearson WR. Hereditary differences in the expression of the human glutathione transferase active on trans-stilbene oxide are due to a gene deletion. Proc Natl Acad Sci U S A. 1988;85:7293–7297. doi: 10.1073/pnas.85.19.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300(Pt 1):271–276. doi: 10.1042/bj3000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu S, Wang Y, Roe B, Pearson WR. Characterization of the human class Mu glutathione S-transferase gene cluster and the GSTM1 deletion. J Biol Chem. 1998;273:3517–3527. doi: 10.1074/jbc.273.6.3517. [DOI] [PubMed] [Google Scholar]
- 73.Bolt HM, Thier R. Relevance of the deletion polymorphisms of the glutathione S-transferases GSTT1 and GSTM1 in pharmacology and toxicology. Curr Drug Metab. 2006;7:613–628. doi: 10.2174/138920006778017786. [DOI] [PubMed] [Google Scholar]
- 74.Sprenger R, Schlagenhaufer R, Kerb R, Bruhn C, Brockmoller J, Roots I, Brinkmann U. Characterization of the glutathione S-transferase GSTT1 deletion: discrimination of all genotypes by polymerase chain reaction indicates a trimodular genotype-phenotype correlation. Pharmacogenetics. 2000;10:557–565. doi: 10.1097/00008571-200008000-00009. [DOI] [PubMed] [Google Scholar]
- 75.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997;6:733–743. [PubMed] [Google Scholar]
- 76.McLellan RA, Oscarson M, Alexandrie AK, Seidegard J, Evans DA, Rannug A, Ingelman-Sundberg M. Characterization of a human glutathione S-transferase mu cluster containing a duplicated GSTM1 gene that causes ultrarapid enzyme activity. Mol Pharmacol. 1997;52:958–965. doi: 10.1124/mol.52.6.958. [DOI] [PubMed] [Google Scholar]
- 77.Liu G, Miller DP, Zhou W, Thurston SW, Fan R, Xu LL, Lynch TJ, Wain JC, Su L, Christiani DC. Differential association of the codon 72 p53 and GSTM1 polymorphisms on histological subtype of non-small cell lung carcinoma. Cancer Res. 2001;61:8718–8722. [PubMed] [Google Scholar]
- 78.Miller DP, Liu G, de VI, Lynch TJ, Wain JC, Su L, Christiani DC. Combinations of the variant genotypes of GSTP1, GSTM1, and p53 are associated with an increased lung cancer risk. Cancer Res. 2002;62:2819–2823. [PubMed] [Google Scholar]
- 79.Wang BG, Chen SD, Zhou WP, Zeng M, Li ZB, Cai XL, Wang DQ. [A case control study on the impact of CYP450 MSPI and GST-M1 polymorphisms on the risk of lung cancer] Zhonghua Zhong Liu Za Zhi. 2004;26:93–97. [PubMed] [Google Scholar]
- 80.Calikoglu M, Tamer L, Ates AN, Karakas S, Ercan B. The association between polymorphic genotypes of glutathione S-transferases and COPD in the Turkish population. Biochem Genet. 2006;44:307–319. doi: 10.1007/s10528-006-9031-4. [DOI] [PubMed] [Google Scholar]
- 81.Moyer AM, Salavaggione OE, Hebbring SJ, Moon I, Hildebrandt MA, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Glutathione S-transferase T1 and M1: gene sequence variation and functional genomics. Clin Cancer Res. 2007;13:7207–7216. doi: 10.1158/1078-0432.CCR-07-0635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.