Abstract
The cytotoxic T lymphocyte antigen-4 (CTLA4) gene is a key negative regulator of the T lymphocyte immune response. It has been found that CTLA4 +49A>G (rs231775), +6230G>A (rs3087243), and 11430G>A (rs11571319) polymorphisms are associated with susceptibility to many autoimmune diseases, and can down-regulate the inhibition of cellular immune response of CTLA4. Three SNPs in CTLA4 were genotyped by using the PCR and DNA sequencing methods in order to reveal the susceptibility and pathology correlation to pulmonary tuberculosis in Southern Han Chinese. We found that the frequency of CTLA4 +49AG genotype in the pulmonary tuberculosis patients (38.42%) was significantly lower than that of the healthy controls (49.77%), (Pcor=0.038, OR 0.653, 95% CI 0.436-0.978). But, no associations were found between the other 2 SNPs (+6230G>A, 11430G>A) and tuberculosis (P>0.05). Haplotype analysis showed that the frequency of haplotype AGG in the healthy controls group (6.9%) was significantly higher than the pulmonary tuberculosis patients group (1.4%), (global P=0.005, Pcor=0.0002, OR 0.183, 95% CI 0.072-0.468). In addition, haplotype GGA was found to be significantly related to tuberculosis with double lung lesion rather than single lung lesion (Pcor=0.042). This study is the first to report that genetic variants in the CTLA4 gene can be associated with pulmonary tuberculosis in Southern Han Chinese, and CTLA4 +49AG genotype as well as haplotype AGG may reduce the risk of being infected with pulmonary tuberculosis. The GGA haplotype was related to tuberculosis with double lung lesion, which provides a new experimental basis to clarify the pathogenesis of pulmonary tuberculosis.
Keywords: Pulmonary Tuberculosis, Single-nucleotide polymorphism, CTLA4 gene, Southern Han Chinese, Lung lesion.
Introduction
Tuberculosis (TB) is the leading cause of death among infectious diseases 1, 2. Although about one third of the world's population is thought to be infected with Mycobacterium tuberculosis (MTB), only 5-15% of people develop clinically active TB during their lifetime 3. This indicates that genetic differences between individuals play an important role in susceptibility to TB. Cumulative evidence suggests that different susceptibility genes can lead to inconsistent incidence rate of TB in ethnic groups 4. Our previous findings also suggest that both genetic and environmental factors are associated with the pathogenesis of TB 5-7.
Cellular immune response and delayed-type hypersensitivity reactions are considered to play a major role in the immunopathogenesis of pulmonary TB 8. T cell activation requires two signals: one signal triggered by MHC-peptide complex and another from CD28 on the T cell's surface binding to the B7 molecules on the antigen presenting cell (APC). The cytotoxic T lymphocyte associated antigen 4 (CTLA4) is a CD28-family receptor, which inhibits T cell proliferation through combination with B7 molecules 9. Human CTLA4 gene is located on chromosome 2q33, and the single-nucleotide polymorphism (SNP) +49A>G (rs231775) is located in the first exon of the CTLA4 gene. A +49A>G base substitution can lead to threonine to alanine amino acid change in the coding region of CTLA4 10. The CTLA4 +6230 (rs3087243) and CTLA4 +49 SNPs have been shown to be in strong linkage disequilibrium, resulting in excellent combinations of haplotype analysis 11, 12. Previous studies have demonstrated that the +49 SNP of the CTLA4 is related to the development of Graves' disease 13, systemic lupus erythematosus 14 and other autoimmunity diseases, while CTLA4 +6230 SNP have been shown to be associated with susceptibility to Hepatitis B virus infection 15 and inflammatory bowel disease 16. Thye et al. 17 found that the CTLA4 +6230G allele contributes to pathology of TB in the African population. However, no previous study has investigated whether polymorphic sites in CTLA4 correlate with the development of pulmonary TB in Southern Han Chinese.
The aim of the present study was to investigate the occurrence of CTLA4 SNPs and their association with susceptibility to pulmonary TB in Southern Han Chinese. The patients were divided into single lung lesion and double lung lesion groups in order to reveal the correlation between CTLA4 SNPs and the severity of lung damage, and to clarify the role of CTLA4 in the pathogenesis of pulmonary TB.
Material and Methods
Patients and Control Subjects
A total of 274 Southern Han Chinese subjects with pulmonary TB, aged 18-70 years (mean age 39.5 ± 16.3 years) were recruited from the Sixth Hospital of Shaoxing (133 subjects) and Hangzhou Red Cross Hospital (141 subjects). The control group comprised 266 healthy subjects, aged 20-62 years (mean age 36.3 ± 10.9 years), unrelated blood donors with no history of TB or other immune diseases. Females constituted 41.2% of the pulmonary TB patients, and 40.6% of healthy controls (Table 1). CTLA4 +49 SNP consisted of 203 TB patients and 221 healthy controls from above, while CTLA4 +6230 and 11430 SNPs consisted of 264 TB patients and 262 healthy controls. This study was approved by the Ethics Committee of the Faculty of Medicine (Zhejiang University, China), and informed consents were obtained from all subjects before blood sampling.
Table 1.
Characteristics of healthy controls and TB patients.
| Patients from Shaoxing Sixth Hospital (N = 133) | Patients from Hangzhou Red Cross Hospital (N = 141) | Control Group (N= 266) |
P | |
|---|---|---|---|---|
| Age, years range (mean ± SD) | 18-70 (41.5 ± 15.8) | 18-67 (37.4 ± 17.4) | 20-62 (36.3 ± 10.9) | 0.573a |
| Gender: female, no. (%) | 51 (38.3) | 62 (44.0) | 108 (40.6) | 0.922b |
| Body mass index (mean ± SD) | 21.5 ± 3.0 | 20.7 ± 3.3 | 23.0 ± 3.9 | 0.103a |
| Tuberculin skin test (>10 mm), no. (%) | 102 (76.69) | 127 (90.07) | ND | / |
| Positive sputum smear, no. (%) | 81 (60.9) | 83 (58.9) | ND | / |
| Presence of TB history of relatives, no. (%) | 13 (9.8) | 13 (9.2) | 19 (7.1) | 0.364b |
| BCG vaccination, no. (%) | 56 (42.1) | 70 (49.6) | 151 (56.8) | 0.155b |
TB: tuberculosis; N: number of subjects; ND: not determined. aP value between total patients and controls, for t test. bP value between total patients and controls, for χ2 test.
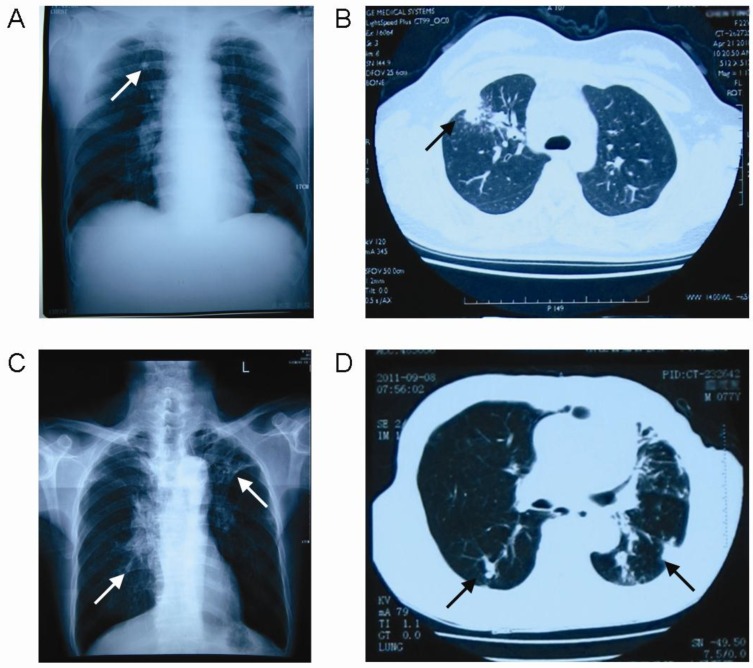
Patients were diagnosed according to the diagnostic criteria for pulmonary TB of Ministry of Health of China 18. All patients meet one of the following pulmonary TB diagnostic criteria: (1) positive sputum examination (smear or culture); (2) negative sputum examination, chest X-ray and CT revealing evidence of typical active TB; (3) pathological diagnosis of TB in lung specimens; (4) suspected of having pulmonary TB after clinical follow-up and X-ray observations, and excluding other lung diseases; (5) clinically ruling out other causes of pleural effusion, and diagnosis of tuberculous pleurisy. In the present study, 59.9% TB patients were diagnosed by the positive sputum smear and 40.1% were diagnosed by chest X-ray and CT revealing evidence of typical active TB. According to the radiographic findings (Figure 1), we divided TB patients into two groups: single lung lesion group with 70 subjects and double lung lesion group with 69 subjects.
Figure 1.
Radiographic findings of the pulmonary tuberculosis patients. Arrow: lesions. A: The chest X-ray of single lung lesion patient; B: The chest CT of single lung lesion patient; C: The chest X-ray of double lung lesion patient; D: The chest CT of double lung lesion patient.
Genotyping
Genomic DNAs were extracted from the peripheral blood leukocytes with DNA extraction kit (QIAamp® DNA Blood Min Kit, Germany). SNPs in the CTLA4 gene were analyzed by polymerase chain reaction (PCR) and direct sequencing. Primers were designed by Prime 5.0. The CTLA4 +49 (rs231775) PCR primers were: forward TTCAAGTGCCTTCTGTGTGTG and reverse AATCACTGCCCTTGACTGCT. The PCR was performed by denaturing at 94°C for 5 min, followed by 35 cycles at 94°C for 50 s, 59°C for 50 s, and 72°C for 35 s, and a final extension at 72°C for 10 min. The CTLA4 +6230 (rs3087243) PCR primers were: forward AGGCAGCAGGTGGCAGAAT and reverse TAAGCAAGAATCACAGAGGGC, which contains 11430 SNP (rs11571319). The PCR was performed by denaturing at 94°C for 5 min, followed by 35 cycles at 94°C for 50 s, 58.5°C for 50 s, and 72°C for 40 s, and a final extension at 72°C for 10 min. The amplified products were purified by PCR cleaning kit (AxyPrep PCR cleaning kit, USA) and then identified by scanning with the ABI 3100 sequencer (Applied Biosystems, Carlsbad, CA, USA).
Statistical Analysis
Hardy-Weinberg equilibrium was assessed by using the chi-square test for each group. The chi-square test was used to compare allele and genotype distribution in the TB patients and healthy controls by the GraphPad Prism version 5.01 software, while n<5, a correction of the two-tailed Fisher exact method was used. Odds ratios (OR) and 95% confidence intervals (CI) were calculated by Miettinen method. A P value of <0.05 was considered to be statistically significant. Haplotype frequencies and associations were calculated by Haploview version 4.2, which uses the expectation-maximization algorithm. Pairwise linkage disequilibrium was estimated by calculating pairwise D' and r2.
Corrections for multiple testing of single variants were made by False Discovery Rate (FDR) correction. The statistical analyses were performed using the R computer package software (version 2.13.0) for global and haplotype-specific P values. The study sample provided 75% power to identify significant genotypic differences between TB cases and controls at a statistical support level of α=0.05 with an OR of 1.4 and a minor allele frequency of 0.1 applying a multiplicative model.
Results
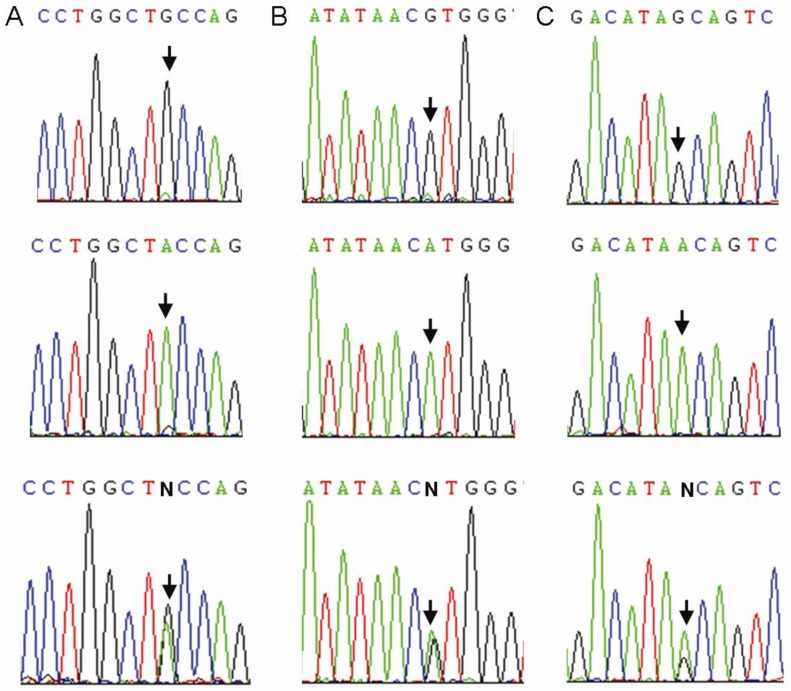
We sequenced all samples of the healthy control group and the TB patients group. Three CTLA4 SNPs: +49A>G (rs231775), +6230G>A (rs3087243), and 11430G>A (rs11571319) were found in Southern Han Chinese (Figure 2). Otherwise, 9 SNPs (rs138279736, rs146541851, rs16840275, rs145950656, rs231721, rs35482731, rs139354616, rs34031880 and rs59387738) with minor allele frequencies (MAF) of less than 1% were removed from the association analysis. All CTLA4 SNPs were in Hardy-Weinberg equilibrium (Table 2).
Figure 2.
Sequence diagrams of three SNPs of CTLA4. Arrow: single-nucleotide polymorphism. A: rs231775; B: rs3087243; C: rs11571319.
Table 2.
CTLA4 allele frequencies in controls (N=266) and TB patients (N=274).
| SNP sites | Allele | Controls N (Freq) | TB patients N (Freq) | P value | OR (95% CI) |
|---|---|---|---|---|---|
| +49 | G | 296 (66.97) | 280 (68.97) | ||
| rs231775 | A | 146 (33.03) | 126 (31.03) | 0.534 | 0.912 (0.683-1.218) |
| HWE(P) | 0.063 | 0.145 | |||
| +6230 | G | 430 (82.06) | 421 (79.73) | ||
| rs3087243 | A | 94 (17.94) | 107 (20.27) | 0.439 | 1.130 (0.829-1.540) |
| HWE(P) | 0.055 | 0.411 | |||
| 11430 | G | 445 (84.92) | 436 (84.17) | ||
| rs11571319 | A | 79 (15.08) | 82 (15.83) | 0.736 | 1.059 (0.757-1.483) |
| HWE(P) | 0.051 | 0.102 |
SNP: single-nucleotide polymorphism; HWE: Hardy-Weinberg equilibrium; N: number of alleles; Freq: frequency; OR: Odds Ratio (TB patients/controls); 95% CI: 95% Confidence Intervals.
There were no significant differences in allele frequencies in 3 SNPs between the healthy control group and the TB patients group (Table 2). The CTLA4 +49 AG genotype frequency was significantly higher in the healthy control group compared to the TB patients group (Pcor=0.038, OR 0.653, 95% CI 0.436-0.978) (Table 3). The frequencies of +6230AG and 11430 AG were higher in the TB patients compared to the healthy controls, but the difference was not significant (P>0.05).
Table 3.
CTLA4 genotype frequencies in controls (N=266) and TB patients (N=274).
| SNP sites | Genotype | Controls N (Freq) | TB patients N (Freq) | P value | Pcor1 | OR (95% CI) |
|---|---|---|---|---|---|---|
| +49 | GG | 93 (42.08) | 101 (49.75) | 1 | ||
| rs231775 | AA | 18 (8.15) | 24 (11.83) | 0.550 | 1.228 (0.626-2.407) | |
| AG | 110 (49.77) | 78 (38.42) | 0.038* | 0.038 | 0.653 (0.436-0.978) | |
| +6230 | GG | 181 (69.08) | 170 (64.39) | 1 | ||
| rs3087243 | AA | 13 (4.96) | 13 (4.93) | 0.877 | 1.065 (0.480-2.362) | |
| AG | 68 (25.96) | 81 (30.68) | 0.225 | 1.268 (0.863-1.863) | ||
| 11430 | GG | 193 (73.66) | 187 (72.20) | 1 | ||
| rs11571319 | AA | 10 (3.82) | 10 (3.86) | 0.945 | 1.032 (0.420-2.537) | |
| AG | 59 (25.52) | 62 (23.94) | 0.697 | 1.085 (0.720-1.633) |
SNP: single-nucleotide polymorphism; N: number of alleles; Freq: frequency; OR: Odds Ratio (TB patients/controls); 95% CI: 95% Confidence Intervals. *P<0.05.
1False Discovery Rate (FDR) corrected P value.
Based on the radiographic findings in TB patients, we divided 273 patients into the single lung lesion group (N=70) and the double lung lesion group (N=69) (Figure 1). There were no significant differences in CTLA4 +49A/G, +6230G/A, and 11430G/A allele frequencies (Table 4) and genotype frequencies (Table 5) between the single lung lesion and double lung lesion groups (P>0.05).
Table 4.
CTLA4 allele frequencies in single lung lesion patients (N=70) and double lung lesion patients (N=69).
| SNP sites | Allele | Single lung lesion N (Freq) | Double lung lesion N (Freq) | P value | OR (95% CI) |
|---|---|---|---|---|---|
| +49 | G | 93 (66.43) | 89 (64.49) | ||
| rs231775 | A | 47 (33.57) | 49 (35.51) | 0.734 | 1.089 (0.664-1.787) |
| HWE(P) | 0.096 | 0.874 | |||
| +6230 | G | 111 (79.29) | 95 (75.40) | ||
| rs3087243 | A | 29 (20.71) | 31 (24.60) | 0.449 | 1.249 (0.702-2.222) |
| HWE(P) | 0.469 | 0.138 | |||
| 11430 | G | 121 (86.43) | 104 (82.54) | ||
| rs11571319 | A | 19 (13.57) | 22 (17.46) | 0.388 | 1.347 (0.691-2.626) |
| HWE(P) | 0.768 | 0.069 |
SNP: single-nucleotide polymorphism; HWE: Hardy-Weinberg equilibrium; N: number of alleles; Freq: frequency; OR: Odds Ratio (double lung lesion/single lung lesion); 95% CI: 95% Confidence Intervals.
Table 5.
CTLA4 genotype frequencies in single lung lesion patients (N=70) and double lung lesion patients (N=69).
| SNP sites | Genotype | Single lung lesion N (Freq) | Double lung lesion N (Freq) | P value | OR (95% CI) |
|---|---|---|---|---|---|
| +49 | GG | 34 (48.57) | 29 (42.03) | 1 | |
| rs231775 | AA | 11 (15.71) | 9 (13.04) | 0.936 | 0.959 (0.349-2.636) |
| AG | 25 (35.72) | 31 (44.93) | 0.310 | 1.454 (0.705-2.997) | |
| +6230 | GG | 45 (64.29) | 38 (60.32) | 1 | |
| rs3087243 | AA | 4 (5.71) | 6 (9.52) | 0.509 | 1.776 (0.467-6.764) |
| AG | 21 (30.00) | 19 (30.16) | 0.858 | 1.071 (0.503-2.282) | |
| 11430 | GG | 52 (74.29) | 45 (71.43) | 1 | |
| rs11571319 | AA | 1 (1.43) | 4 (6.35) | 0.192 | 4.622 (0.498-42.900) |
| AG | 17 (24.28) | 14 (22.22) | 0.905 | 0.952 (0.422-2.144) |
SNP: single-nucleotide polymorphism; N: number of alleles; Freq: frequency; OR: Odds Ratio (double lung lesion/single lung lesion); 95% CI: 95% Confidence Intervals.
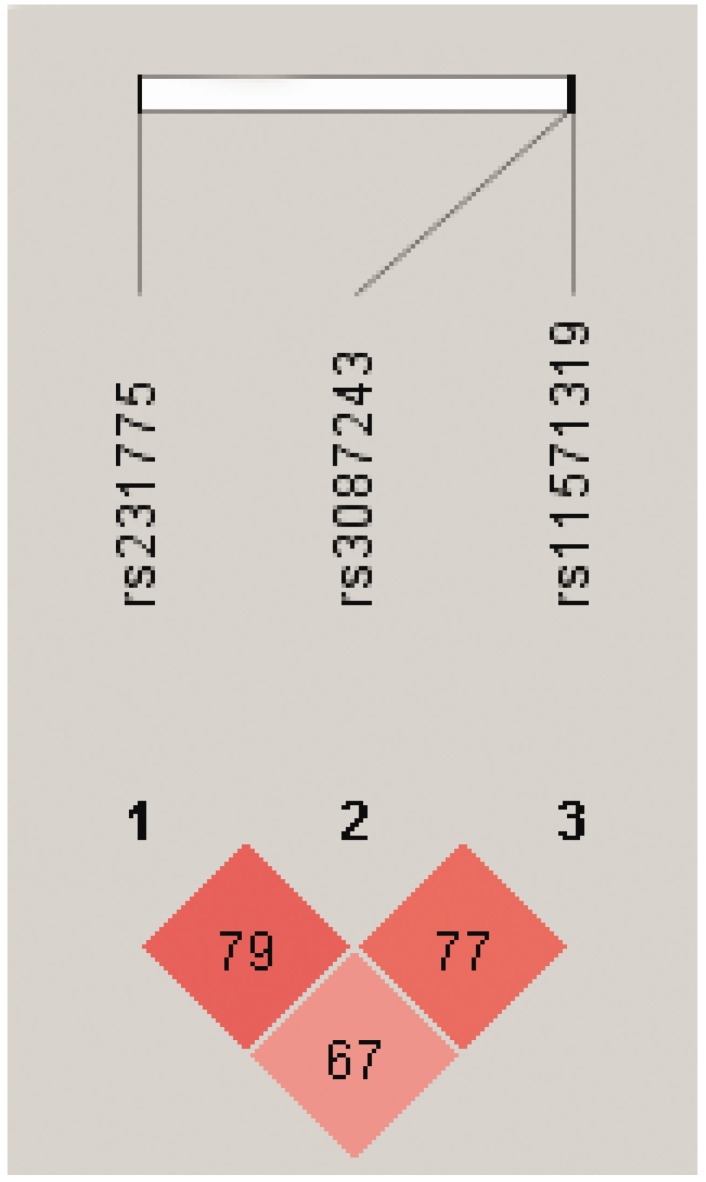
We further performed pairwise linkage disequilibrium analyses (Figure 3) and found that there was a strong linkage disequilibrium between CTLA4 +49 and +6230, +6230 and 11430 (D'>0.75). We also found that the frequency of haplotype AGG was significantly higher in the healthy controls than the TB patients (global P=0.005, Pcor=0.0002, OR 0.183, 95% CI 0.072-0.468) (Table 6). There was no significant difference in the frequency of haplotype AGG between the single lung lesion group and the double lung lesion group. However, the frequency of haplotype GGA in the double lung lesion patients was higher than the single lung lesion group (Pcor=0.042), indicating that this haplotype is significantly related to TB with double lung lesion (Table 7).
Figure 3.
Pairwise linkage disequilibrium based on 3 CTLA4 SNPs using HaploView 4.2 software. Red squares represent high pairwise linkage disequilibrium, coloring down to white squares of low pairwise linkage disequilibrium. The numbers in the individual squares are D' multiplied by 100.
Table 6.
CTLA4 haplotype frequencies in controls and TB patients.
| Haplotype | CTLA4 SNP | Control Freq | TB patients freq | P value2 | Pcor3 | OR (95%CI) | ||
|---|---|---|---|---|---|---|---|---|
| +49 | +6230 | 11430 | ||||||
| Global P=0.0051 | ||||||||
| 1 | G | G | G | 0.608 | 0.630 | 0.632 | 0.768 | 1.072 (0.807-1.424) |
| 2 | A | A | G | 0.155 | 0.164 | 0.666 | 0.603 | 1.086 (0.747-1.578) |
| 3 | A | G | A | 0.106 | 0.132 | 0.252 | 0.359 | 1.285 (0.836-1.976) |
| 4 | A | G | G | 0.069 | 0.014 | <0.0001* | 0.0002 | 0.183 (0.072-0.468) |
| 5 | G | G | A | 0.038 | 0.020 | 0.168 | 0.290 | 0.550 (0.232-1.304) |
| 6 | G | A | G | 0.015 | 0.032 | 0.070 | 0.101 | 2.366 (0.908-6.164) |
Freq: frequency; OR: Odds Ratio (TB patients/controls); 95% CI: 95% Confidence Intervals. *P<0.05.
1P values obtained testing an overall association between haplotypes and the phenotype.
2 Adjusted for age and sex.
3P values of haplotype-specific associations.
Table 7.
CTLA4 haplotype frequencies in single lung lesion patients and double lung lesion patients.
| Haplotype | CTLA4 SNP | Single lung lesion Freq | Double lung lesion freq | P value2 | Pcor3 | OR (95%CI) | ||
|---|---|---|---|---|---|---|---|---|
| +49 | +6230 | 11430 | ||||||
| Global P=0.3741 | ||||||||
| 1 | G | G | G | 0.633 | 0.579 | 0.204 | 0.419 | 0.723 (0.437-1.194) |
| 2 | A | A | G | 0.194 | 0.212 | 0.435 | 0.736 | 1.276 (0.691-2.359) |
| 3 | A | G | A | 0.135 | 0.134 | 0.956 | 0.987 | 1.020 (0.500-2.083) |
| 4 | A | G | G | 0.023 | 0.009 | 0.371 | 0.388 | 0.378 (0.041-3.453) |
| 5 | G | A | G | 0.015 | 0.025 | 0.594 | 0.553 | 1.622 (0.270-9.753) |
| 6 | G | G | A | 0.000 | 0.041 | 0.018* | 0.042* | --- |
Freq: frequency; OR: Odds Ratio (double lung lesion/single lung lesion); 95% CI: 95% Confidence Intervals. *P<0.05.
1P values obtained testing an overall association between haplotypes and the phenotype.
2 Adjusted for age and sex.
3P values of haplotype-specific associations.
Discussion
TB is an infectious disease caused by MTB. The adaptive immune system plays an important role in susceptibility to TB 8. Besen et al. 19 compared the clinical, radiological, and laboratory characteristics of individuals with pulmonary TB co-infected or not with HIV and confirmed the importance of T lymphocyte in the immunopathogenesis of pulmonary TB. Our experiments indicate that CTLA4 SNPs are associated with pathological lung injury and susceptibility to pulmonary TB.
Different gene polymorphisms can lead to different incidence of pulmonary TB in different ethnic groups 4. According to the NCBI database (dbSNP), the frequencies for the CTLA4 +49AG in Asians (36.5%) and Sub-Saharan Africans (47.8%) were lower than the European population (58.3%). Since China, India and Africa have a higher incidence of TB, we speculated that the CTLA4 +49AG is negatively correlated with susceptibility to pulmonary TB. Our results suggest that in the Southern Chinese Han population, the frequency of CTLA4 +49AG in TB patients (38.42%) was significantly lower than the control subjects (49.77%), and there was a negative correlation with susceptibility to TB (Pcor=0.038, OR 0.653, 95% CI 0.436-0.978) (Table 3). These findings confirmed our speculation. CTLA4 +49 A>G caused an amino acid change at position 17 in exon 1 from threonine to alanine, leading to affinity differences between CTLA4 and B7. Ligers et al. 20 and Sun et al. 21 indicated that +49A>G polymorphism causes decreased CTLA4 molecule production resulting in lower surface expression and lower affinity of CTLA4 for B7-1 molecule, which leads to a lower inhibition of T lymphocyte activity and a higher inhibition of the development of many tumors. Walldén et al. 22 found that CTLA4 polymorphisms were associated with increased secretion of cytokines such as, IFN-gamma and IL2, and end up with enhanced immune response. Therefore, we proposed that after the population carrying the CTLA4 49AG genotype is infected with MTB, the CTLA4-mediated inhibition of T lymphocytes is reduced, resulting in the enhancement of T lymphocyte response for the removal of pathogens and protection against pulmonary TB.
There were no significant differences between TB patients and healthy controls in allele or genotype frequencies of CTLA4 +6230 and 11430. However, in a Norwegian case-control study, Zhu et al. 23 found that +49A as well as +6230G polymorphisms are protective factors of chronic obstructive pulmonary disease (COPD). Thye et al. 17 demonstrated that in an African population, +6230G contributed to pathology rather than to protection in pulmonary TB. Furthermore, in our data, there were also no significant differences in all 3 SNPs' allele frequencies and genotype frequencies between single lung lesion group and double lung lesion group. However, a study in an African population 17 showed a significant influence of the +6230G variant on the disease severity as radiographically assessed by the extent of opacities and size and number of pulmonary cavities. The +6230G had been found to be associated with severe pathology in pulmonary TB. These inconsistent results may reflect the heterogeneity of case-control association studies in different ethnic populations. This has been confirmed by Delgado et al. 4 ethnic-specific genetic studies of pulmonary TB and our previous studies 5-7. It is noteworthy that the frequency distribution of these SNPs was not same in all races. The polymorphisms such as, +49A and +6230A were much higher in the European population than the Asian and African populations. This difference, along with gene-gene interactions and environmental and cultural factors, and even the variations in mycobacterium strains, are making the understanding of the observed differences between ethnic groups even more complicated.
In our haplotype analysis, the frequency of AGG haplotype was significantly higher in the healthy controls compared to the TB patients (global P=0.005, Pcor=0.0002, OR 0.183, 95% CI 0.072-0.468) (Table 6). Ueda et al. 24 found that the change in +6230A>G enabled decreased mRNA level of soluble CTLA4, thus affecting its combination with B7 molecules and resulting in the decreased inhibition of T cell activation. Although, this change has been challenged in a recent study 25, but still some studies indicate that +6230GG polymorphism in a population significantly decrease soluble CTLA4 levels in peripheral blood 26. Therefore, we speculate that there is a functional decline in the inhibition of T cell activation in the AGG haplotype group, leading to a stronger T cell immune response.
We found that the frequency of haplotype GGA in the double lung lesion group was higher than the single lung lesion group (Pcor=0.042) (Table 7). Thye et al. 17 suggested that haplotype -1765A/-1722A/-1661A/-443T/+49G/+923C/+6230G is associated with the severity of opacities and cavities in the lung. As the radiological findings of the lung lesions including, opacities and cavities are considered to result from cellular infiltration and necrosis 27, our observations suggest that a reduced activity of CTLA4 causes an increased cellular infiltration of the lung, which may lead to more serious lung lesions. This is in agreement with the concept that pathology of pulmonary TB to some extent is immune mediated. Considered all, together, two kinds of CTLA4 polymorphisms such as, +49G and +6230G can lead to enhanced T cell immune response, but the respective polymorphisms are not associated with the severity of lung lesion. So, we thought that only the presence of +49A>G and +6230A>G polymorphisms together in individuals could significantly enhanced the T cell immune response in pulmonary TB, and cause double lung lesion.
In conclusion, our data suggested that the CTLA4 +49AG is a protective factor to pulmonary TB in Southern Han Chinese. The haplotype AGG was related to healthy controls while, the haplotype GGA was associated with double lung lesion of pulmonary TB. This is the first report that shows an association of CTLA4 gene polymorphisms with TB in the Southern Han Chinese population. It provides a new potential genetic protective factor for pulmonary TB, and shows that +6230 site associated with TB in the African population may not affect the severity of lung damage in the Chinese population.
In the present study, we analyzed alleles, genotypes and haplotypes, and concluded that the +49A>G polymorphism of CTLA4 was associated with pulmonary TB, and may have a protective function. This indicates an important role of T cell activation in the immunopathogenesis of TB.
Acknowledgments
This work was supported by grants from Zhejiang Province Special Sci-Tech Projects (No.2009C03011-3), National Natural Science Foundation of China (No.81072724), and National Special Sci-Tech Projects (No.2012ZX10005001-006, No.2008ZX10005-010).
References
- 1.Vashishtha VM. WHO Global Tuberculosis Control Report 2009: Tuberculosis elimination is a distant dream. Indian Pediatr. 2009;46:401–2. [PubMed] [Google Scholar]
- 2.Dye C. Global epidemiology of tuberculosis. Lancet. 2006;367:938–40. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- 3.Rosman MD O-EA. Clinical presentation and treatment of tuberculosis. In: Fishman AP, editor. Fishman's Pulmonary Diseases and Disorders. New York, USA: McGraw-Hill; 1998. pp. 2483–502. [Google Scholar]
- 4.Delgado JC, Baena A, Thim S. et al. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002;186:1463–8. doi: 10.1086/344891. [DOI] [PubMed] [Google Scholar]
- 5.Xue Y, Jin L, Li AZ. et al. Microsatellite polymorphisms in intron 2 of the toll-like receptor 2 gene and their association with susceptibility to pulmonary tuberculosis in Han Chinese. Clin Chem Lab Med. 2010;48:552–6. doi: 10.1515/CCLM.2010.154. [DOI] [PubMed] [Google Scholar]
- 6.Zhang YX, Xue Y, Liu JY. et al. Association of TIRAP (MAL) gene polymorhisms with susceptibility to tuberculosis in a Chinese population. Genet Mol Res. 2011;10(1):7–15. doi: 10.4238/vol10-1gmr980. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Jiang F, Wei L. et al. Polymorphic Allele of Human MRC1 Confer Protection against Tuberculosis in a Chinese Population. Int J Biol Sci. 2012;8(3):375–82. doi: 10.7150/ijbs.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aktas E, Ciftci F, Bilgic S. et al. Peripheral Immune Response in Pulmonary Tuberculosis. Scand J Immunol. 2009;70(3):300–8. doi: 10.1111/j.1365-3083.2009.02294.x. [DOI] [PubMed] [Google Scholar]
- 9.Anderson DE, Sharpe AH, Hafler DA. The B7 CD28/CTLA-4 costimulatory pathways in autoimmune disease of the central nervous system. Curr Opin Immunol. 1999;11:677–83. doi: 10.1016/s0952-7915(99)00036-9. [DOI] [PubMed] [Google Scholar]
- 10.Harper K, Balzano C, Rouvier E. et al. CTLA-4 and CD28 activated lymphocyte molecules are closely related in both mouse and human as to sequence, message expression, gene structure, and chromosomal location. J Immunol. 1991;147:1037–44. [PubMed] [Google Scholar]
- 11.Amundsen SS, Naluai AT, Ascher H. et al. Genetic analysis of the CD28/CTLA4/ICOS (CELIAC3) region in coeliac disease. Tissue Antigens. 2004;64:593–9. doi: 10.1111/j.1399-0039.2004.00312.x. [DOI] [PubMed] [Google Scholar]
- 12.Munthe-Kaas MC, Carlsen KH, Helms PJ. et al. CTLA-4 polymorphisms in allergy and asthma and the TH1/ TH2 paradigm. J Allergy Clin Immunol. 2004;114:280–7. doi: 10.1016/j.jaci.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 13.Kouki T, Sawm Y, Gardine CA, et a1. CTLA-4 Gene polymorphism at position 49 in exon 1 reduced the inhibitory function of CTLA-4 and contributes to the pathogenesis of Groves'disease. J Immunol. 2000;165:6606–11. doi: 10.4049/jimmunol.165.11.6606. [DOI] [PubMed] [Google Scholar]
- 14.Ulker M, Yazisiz V, Sallakci N. et al. CTLA-4 gene polymorphism of exon 1 (+49 A/G) in Turkish systemic lupus erythematosus patients. Int J Immunogenet. 2009;36(4):245–50. doi: 10.1111/j.1744-313X.2009.00856.x. [DOI] [PubMed] [Google Scholar]
- 15.Chloe L, Timothy L, Richard A. et al. Cytotoxic T-Lymphocyte Antigen 4 Gene and Recovery from Hepatitis B Virus Infection. J Virol. 2004;78(20):11258–62. doi: 10.1128/JVI.78.20.11258-11262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Repnik K, Potocnik U. CTLA4 CT60 single-nucleotide polymorphism is associated with Slovenian inflammatory bowel disease patients and regulates expression of CTLA4 isoforms. DNA Cell Biol. 2010;29(10):603–10. doi: 10.1089/dna.2010.1021. [DOI] [PubMed] [Google Scholar]
- 17.Thye T, Scarisbrick G, Browne ENL. et al. CTLA4 Autoimmunity-Associated Genotype Contributes to Severe Pulmonary Tuberculosis in an African Population. PLoS ONE. 2009;4(7):e6307. doi: 10.1371/journal.pone.0006307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diagnostic criteria for pulmonary tuberculosis. Ministry of Health of China; http://www.moh.gov.cn/publicfiles/business/cmsresources/zwgkzt/cmsrsdocument/doc3242.pdf. [Google Scholar]
- 19.Besen A, Staub GJ, Silva RM. Clinical, radiological, and laboratory characteristics in pulmonary tuberculosis patients: comparative study of HIV-positive and HIV-negative inpatients at a referral hospital. J Bras Pneumol. 2011;37(6):768–75. doi: 10.1590/s1806-37132011000600010. [DOI] [PubMed] [Google Scholar]
- 20.Ligers A, Teleshova N, Masterman T. et al. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun. 2001;2(3):145–52. doi: 10.1038/sj.gene.6363752. [DOI] [PubMed] [Google Scholar]
- 21.Sun T, Zhou YF, Yang M. et al. Functional genetic variations in cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple types of cancer. Cancer Res. 2008;68(17):7025–34. doi: 10.1158/0008-5472.CAN-08-0806. [DOI] [PubMed] [Google Scholar]
- 22.Walldén J, Ilonen J, Roivainen M. et al. Effect of HLA genotype or CTLA-4 polymorphism on cytokine response in healthy children. Scand J Immunol. 2008;68(3):345–50. doi: 10.1111/j.1365-3083.2008.02144.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu G, Agusti A, Gulsvik A. et al. CTLA4 gene polymorphisms are associated with chronic bronchitis. Eur Respir J. 2009;34(3):598–604. doi: 10.1183/09031936.00141808. [DOI] [PubMed] [Google Scholar]
- 24.Ueda H, Howson JM, Esposito L. et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–11. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 25.Purohit S, Podolsky R, Collins C. et al. Lack of correlation between the levels of soluble cytotoxic T-lymphocyte associated antigen-4 (CTLA-4) and the CT-60 genotypes. J Autoimmune Dis. 2005;2:8. doi: 10.1186/1740-2557-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Repnik K, Potocnik U. CTLA4 CT60 single-nucleotide polymorphism is associated with Slovenian inflammatory bowel disease patients and regulates expression of CTLA4 isoforms. DNA Cell Biol. 2010;29(10):603–10. doi: 10.1089/dna.2010.1021. [DOI] [PubMed] [Google Scholar]
- 27.Lee JY, Lee KS, Jung KJ. et al. Pulmonary tuberculosis: CT and pathologic correlation. J Comput Assist Tomogr. 2000;24:691–8. doi: 10.1097/00004728-200009000-00005. [DOI] [PubMed] [Google Scholar]