Abstract
Release of genetically modified (GM) plants has sparked off intensive debates worldwide partly because of concerns about potential adverse unintended effects of GM plants to the agro system and the safety of foods. In this study, with the aim of revealing the molecular basis for unintended effects of a single site insertion GM Kemingdao (KMD) rice transformed with a synthetic cry1Ab gene, and bridging unintended effects of KMD rice through clues of differentially expressed genes, comparative transcriptome analyses were performed for GM KMD rice and its parent rice of Xiushui11 (XS11). The results showed that 680 differentially expressed transcripts were identified from 30-day old seedlings of GM KMD rice. The absolute majority of these changed expression transcripts dispersed and located over all rice chromosomes, and existed physical distance on chromosome from the insertion site, while only two transcripts were found to be differentially expressed within the 21 genes located within 100 kb up and down-stream of the insertion site. Pathway and biology function analyses further revealed that differentially expressed transcripts of KMD rice were involved in certain biological processes, and mainly implicated in two types of pathways. One type was pathways implicated in plant stress/defense responses, which were considerably in coordination with the reported unintended effects of KMD rice, which were more susceptible to rice diseases compared to its parent rice XS11; the other type was pathways associated with amino acids metabolism. With this clue, new unintended effects for changes in amino acids synthesis of KMD rice leaves were successfully revealed. Such that an actual case was firstly provided for identification of unintended effects in GM plants by comparative transciptome analysis.
Keywords: GM KMD rice, comparative transcriptome analysis, differentially expressed genes, changed pathways, unintended effects, changes in amino acid synthesis.
Introduction
The objective for development of genetically modified (GM) plants is aimed at introduction of specific target traits to plants by insertion of defined exogenous genes. These target traits are designed either to be beneficial to plants or to fit the requirements of human beings. These are normally referred to as the intended effects of GM plants 1. However, unintended effects can be resulted from the random insertion of exogenous DNA fragments into the plant genome because the insertion event may alter the expression of certain plant intrinsic genes by disruption, silencing, activation, or modification. Although these unintended effects may be deleterious, beneficial, or neutral with respect to plant health or food safety, the public has shown considerable concern regarding potential hazardous effects of GM plants to agro system and human consumer 2, 3. The large-scale commercial release of GM crops has sparked intense debates worldwide 3, 4.
As a major producer and consumer of rice, China has made great progress in the development of GM rice and has carried out extensive field tests during the past decade 5-8. Quite a lot of attention has been paid on the development of insect-resistant GM rice containing insecticidal delta-endotoxin genes from Bacillus thuringiensis (Bt) 7. Lepidopteran insects frequently cause severe yield losses in rice production 9, and several Bt rice lines targeting Lepidopteran pests have been successfully developed in China 5.
GM KMD rice, derived from the commercial japonica rice variety Xiushui 11 (XS11) by Agrobacterium transformation with a synthetic cry1Ab gene, was reported to be highly resistant to eight lepidopteran rice pest species 7, 10. Molecular characterization via Southern blot and side sequence analysis for exogenous insertion showed that KMD rice is homozygous for the transformed gene and has one single insertion of the gene construct 11-14. Previous studies have showed a few kinds of unintended effects in KMD rice, such as specifically increased susceptibility to rice brown spot mimic lesion disease and rice sheath blight disease 15, 16. Therefore, GM KMD rice could act as model material for development of new approaches and technologies to assess unintended effects caused by foreign gene insertion to plants.
Inserted exogenous genes are initially likely to arouse changes in intrinsic plant genes at the expression level. And then may cause alterations in biological processes and pathways at the physiological and metabolic levels. Finally may lead to occur visible unintended changes to certain plant traits. It is expected that genes with altered expression levels could be detected through transcriptomic analysis, and unintended effects could be identified by moving from the identities of differentially expressed genes to the biological processes and pathways that may link to the ultimate plant traits. For this reason, a number of comparative transcriptome analysis have been performed between GM and comparable non-GM plants, including the model Arabidopsis species for expression of various markers 17, genes related to herbicide resistance 18 and drought tolerance 19, rice plants expressing AFP antifungal protein 20, CsFv antibodies 4 and anthranilate synthase α subunit 21, glyphosate-tolerant soybeans 22, wheat plants expressing phytase 23 and a glutelin subunit 24, maize expressing insect-resistant protein 25, 26, and barley plants expressing 1,3-1,4-β-glucanase and endochitinase 27. Almost all these studies reported that only a few differentially expressed genes were detected in GM plants, and these tiny differentially expressed genes were not enough to cause obvious unintended effects in GM plants, except for a recent report stating that half the transcriptional difference could be associated to the transgene in GM rice for expression of AFP antifungal protein 20. At present, no case of any obvious unintended effects of GM plants uncovered by comparative transcriptome analysis has been reported.
A comparative transcriptome analysis of GM KMD rice and its parent line, XS11, to uncover the molecular basis behind any unintended effects in KMD rice through identification of differentially expressed genes was reported in this study.
Materials and Methods
Plant materials
Seeds of transgenic rice line Kemingdao 1 (KMD) and its corresponding parent, the non-modified isogenic line, japonica rice variety Xiushui 11 (XS11) were used for experiments.
KMD rice expressed a synthetically modified cry1Ab gene derived from Bacillus thuringiensis (Bt) for resistance to Lepidoptera. cry1Ab expression is driven by a maize ubiquitin promoter, the regulatory elements P35S from cauliflower mosaic virus, Tnos from Agrobacterium tumefaciens, and the selection marker genes nptII (neomycin phosphotransferase gene) and hpt (hygromycin phosphotransferase gene) linked in tandem 10, 28. Molecular characterization analysis by Southern blotting indicated that KMD rice was homozygous for the transgene and had single insertion of the gene construct 12, 13. Side sequence analysis for exogenous insert further revealed that the GM KMD was a single insertion transformed case 11, 14.
To confirm that the experimental KMD was a transgenic Bt rice line, the cry1Ab protein expression in KMD seedlings was examined by an immunoaffinity chromatography assay with the BT-cry1Ab/1Ac rapid test strip (Supplementary Material: Figure S1).
Rice sample preparation
Rice seeds were surface-disinfected with 70% (vol/vol) ethanol for 1-2 min and then with a solution of 1% sodium hypochloride for 30 min. After thorough washing with distilled sterile water, seeds were soaked in distilled sterile water for 2 days at 28°C for germination. The rice seedlings were grown in 15 cm diameter soil-filled pots. These smaller pots for planting KMD rice and its corresponding parent XS11 were arranged in parallel rows in a larger pots with 50 x 60 cm, fertilized with a half-strength dose of basal macro- and micro-salt nutrition components of Murashige and Skoog medium 29 through the larger pots. Pots were placed in a climate-controlled chamber on a 16-h-light (30°C)/8-h-dark (26°C) cycle. 30-day old seedling leaves were frozen in liquid nitrogen and kept at -80°C until RNA extraction. More than 20 whole seedling leaves were prepared for each sample, and three samples (replicates) were collected for each rice line.
RNA extraction and quantification
Total RNA was extracted from plant samples using a protocol based on TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, U.S.) according to the manufacturer's instructions. The integrity of extracted RNA was checked by gel electrophoresis and UV spectrometry. Then RNA samples were kept in 70% (vol/vol) ethanol in liquid nitrogen frozen ice and then sent to CapitalBio Corporation (an Affymetrix platform service facility at Beijing) for further quality and quantity examination.
Microarray hybridization
Each RNA sample prepared for hybridization was examined as follows: One microgram of total RNA was reverse-transcribed using a T7 oligo (dT) primer in a first-strand cDNA synthesis reaction. Following RNase H and DNA polymerase mediated second-strand cDNA synthesis, the double-stranded cDNA was purified and used as a template in the subsequent in vitro transcription reaction, which was carried out in the presence of T7 RNA polymerase and the biotinylated nucleotide analog and ribonucleotide mix, in order to perform complementary RNA (cRNA) amplification and biotin labeling. The biotinylated cRNA were then cleaned up, fragmented, and hybridized to the Affymetrix GeneChip® Rice Genome Arrays, which were designed based on the annotation of TIGR (The Institute of Genome Research) version 2.0. The array contained 55,515 probe sets and were used to query 48,564 transcripts of rice japonica subspecies and 1,260 transcripts of rice indica subspecies.
Microarray hybridization was performed at 45°C with rotation lasting for 16h using an Affymetrix GeneChip Hybridization Oven 640. Immediately following hybridization, the arrays were washed and stained (streptavidin-phycoerythrin) at Affymetrix GeneChip Fluidics Station 450 and then scanned with Affymetrix GeneChip® Scanner 3000 7G.
Data analysis
The scanned images obtained were first assessed by visual inspection and then analyzed using Affymetrix GeneChip® Command Console™ (AGCC) software (http://www.affymetrix.com/browse/products.jsp?productId=131429&navMode=34000&navAction=jump&aId=productsNav). The expression flags (indicators of expressed genes) were also determined using Affymetrix® Expression Console™ software (http://www.affymetrix.com/browse/level_seven_software_products_only.jsp?productId=131414&categoryId=35623) application MAS 5.0 algorithm as “present,” “marginal,” and “absent” calls. Then normalization and expression analysis was performed with .CEL files and .mas5.CHP files by DNA-chip analyzer (dChip). All these data have been deposited in the NCBI GEO database under accession number GSE33203. Differentially expressed probe sets were selected using Significance Analysis of Microarrays software (SAM version 3.02, Stanford University, Stanford, CA, U.S. http://www-stat.stanford.edu/~tibs/SAM/index.html) with the two class unpaired method with q value ≤ 5% and fold change ≥ 2.0 (up-regulated) or ≤ 0.5 (down-regulated) relative to the control sample.
The genes expression abundance detected in the microarray experiment was further validated with the quantitative real-time RT-PCR (Q-RT-PCR). The methods and the results were provided in Supplementary Material: Table S1.
Changed pathway assay and functional annotation of differentially expressed genes
Changed pathways and functional annotation of differentially expressed genes of KMD rice were analyzed by the Plant MetGenMAP system 30. This system is freely available at http://bioinfo.bti.cornell.edu/tool/MetGenMAP. All changed pathways were selected by the raw P value at a threshold of 0.05. Significantly changed pathways were selected by the false discovery rate method (FDR) corrected P value at a threshold of 0.05 31. GO term functional classification for differentially expressed genes was performed with three principle GO categories: biological processes, molecular function, and cellular components. Identification of enriched GO terms of changed expression genes focused on biological processes with the FDR multiple test correction methods implemented based on the GO::TermFinder perl module 32.
Amino acid analysis by iTRAQ®-LC-MS/MS
Frozen rice leaves were ground to fine powder in liquid N2. For total amino acid analysis, about 50 mg of fine powder was hydrolyzed in 2 mL of 6 mol/L HCl at 100°C for 24 h in sealed tubes filled with evaporated N2. The powder was diluted 50 fold with deionized water 33. Amino acid analysis by iTRAQ®-LC-MS/MS was performed as described previously 34. Briefly, after samples were diluted and mixed carefully, 40 μL of diluted solution samples were pipetted into Eppendorf tubes and 10 μL of 10% sulfosalicylic acid was added to precipitate proteins. After mixing for 30 min, the samples were centrifuged for 2 min at 10,000 × g. 10 μL of supernatant was mixed with 40 μL of labeling buffer. 10 μL of the diluted supernatant was mixed with 5 μL of iTRAQ® reagent 115 solution and incubated at room temperature for 30 min. Then 5 μL of 1.2% hydroxylamine solution was added. Samples were allowed to evaporate overnight and were reconstituted with 32 μL of iTRAQ® reagent 114-labeled standard mix. Chromatographic separation of isobaric amino acids was achieved at 50°C using an Agilent 1100 HPLC system and an Applied Biosystems C18 5 μm column, 4.6 i.d. × 150 mm. The mobile phase consisted of 0.1% formic acid and 0.01% heptafluorobutyric acid in either water (solvent A) or acetonitrile (solvent B). The column was equilibrated in 98% A and the gradient was 98-72% A for 10 min, 72-0% A after 10 min, and held at 100% B for 16 min. Then the gradient was reestablished with 100-2% B for 16 min, and held at 100% A for 25 min. A flow rate of 800 μL min−1 was used. The injection volume was 2 μL. Tandem mass spectrometry was performed on an API 3200 (Applied Biosystems) with turbo ion spray in positive mode using the following parameters: Ion spray voltage 1500 V; auxiliary gas temperature 700°C; curtain gas, nebulizer gas, and auxiliary gas 20, 70, and 70 arbitrary units, respectively; collision gas medium; entrance potential 10 V; declustering potential 20 V; collision energy 30 V; collision cell exit potential 5 V. Quantitative determination was performed by multiple reaction-monitoring using the internal standard and one transition each for the analyte, according to the manufacturer's instructions. Chromatograms were processed with a beta version of Cliquid® software. The Student's t-tests analysis was used to assess differences in amino acid concentration of rice leaves in KMD versus XS11 using a 95% confidence level. Means were represented with s.e.m.
Results
Overview of differentially expressed transcripts of GM KMD rice compared to its parent of XS11
In order to determine the influences of a single-site insertion of a synthetic cry1Ab gene on expression of rice genes, we compared the transcripts of 30-day old GM KMD rice seedlings leaves to those of XS11. Using Affymetrix microarray analysis, we detected 19,420 rice probes in the two rice lines, accounting for 34.98% of all rice probes on Affymetrix GeneChip® Rice Genome Array. Of these, 718 differentially expressed probes (with fold change ≥ 2.0 or ≤ 0.5) were selected from GM KMD rice. The 718 probes represented 680 differentially expressed rice transcripts (listed in Supplementary Material: Table S2), accounting for 1.36% of all analyzed rice transcripts. Among the 680 differentially expressed transcripts of GM rice, 205 transcripts were up-regulated and 475 transcripts were down-regulated.
Probes of RPTR-Os-A00196-1_s_at, RPTR-Os-K01193-1_at and RPTR-Os-A06498-1_at, which respectively represented 3 transformed elements-beta glucuronidase gene, hygromycin B phosphotransferase gene, and artificial sequence of nopaline synthase gene showed extremely high expression levels in KMD rice relative to XS11 rice (Table 1. ★). This, coupled with the immunoaffinity detection results of Bt cry1Ab protein (Supplementary Material: Figure S1), confirmed that the experimental KMD rice was GM Bt rice. Because high expression levels of transformed elements are expected effects of KMD rice, these three transcripts were discarded and not selected for further analysis.
Table 1.
Expression features of genes located near the insertion site in KMD rice.
| Gene Symbol | Location | Probe Set ID | Fold Change |
|---|---|---|---|
| Os02g0618700 □ | 24614568-24616359 (+) | Os.53063.1.S1_at | Absent calls |
| Os02g0619600 | 24664901-24670392 (-) | Os.18434.1.S1_at | 0.83 |
| Os02g0620100 | 24686644-24690185 (+) | Os.51164.1.S1_at | 1.13 |
| Os02g0620200 | 24690654-24691175 (+) | Os.6623.1.S1_at | 0.99 |
| Os02g0620400 | 24695859-24696717 (+) | Os.37865.1.S1_at | 0.64 |
| Os02g0620500 □ | 24703102-24704598 (+) | OsAffx.12433.1.S1_at | Absent calls |
| insertion site | 24707922-24707956 | RPTR-Os-A00196-1_s_at | 101.87 ★ |
| RPTR-Os-K01193-1_at | 208.51 ★ | ||
| RPTR-Os-A06498-1_at | 334.96 ★ | ||
| Os02g0620600 | 24710277-24712129 (+) | Os.9328.1.S1_at | 0.59 |
| Os02g0620800 □ | 24716249-24717841 (-) | OsAffx.12435.1.S1_at | Absent calls |
| Os02g0621100 ▲ | 24727467-24736677 (+) | Os.53850.1.S1_at | 2.62 |
| Os02g0621300 | 24737440-24743509 (-) | Os.8816.1.S1_at | 1.33 |
| Os02g0621400 □ | 24744227-24744876 (+) | Os.55508.1.S1_at | Absent calls |
| Os02g0621500 | 24747786-24752506 (-) | Os.50341.1.S1_at | 0.87 |
| Os02g0621600 | 24756371-24758680 (-) | Os.17730.1.S1_at | 1.75 |
| Os.17730.1.S1_x_at | 1.72 | ||
| Os02g0621700 | 24761204-24767342 (+) | Os.9758.1.S1_at | 1.02 |
| Os02g0621800 ▼ | 24768844-24774153 (-) | Os.53830.1.S1_at | 0.33 |
| Os02g0622100 | 24786202-24792143 (+) | Os.27664.1.S1_at | 0.89 |
| Os02g0622200 | 24793122-24794433 (-) | Os.16625.1.S1_at | 0.91 |
| Os02g0622300 | 24797477-24798953 (-) | OsAffx.2932.1.S1_s_at | 0.98 |
| Os02g0622400 | 24799323-24802110 (+) | Os.7522.1.S1_at | 1.18 |
| Os02g0622500 | 24802594-24805935 (-) | Os.24611.1.S1_at | 0.99 |
□ Genes expressed in neither KMD nor XS11 rice;▲ Genes up-regulated in KMD rice;▼ Genes down-regulated in KMD rice; ★ Within inserted region of KMD rice, RPTR-Os-A00196-1_s_at, RPTR-Os-K01193-1_at, and RPTR-Os-A06498-1_at, representing beta glucuronidase gene, hygromycin B phosphotransferase gene, and an artificial sequence of nopaline synthase gene, respectively. These showed extremely high expression levels.
Differentially expressed transcripts near the insertion site
In order to determine the expression features of genes near the insertion site, we investigated genes located no more than 100kb up- and down-stream of the insertion site. According to reports, two transition sequences were located at the 3′ and 5′ ends of KMD exogenous insertion 11, 14. The insertion site for exogenous inserts of KMD rice was located in chromosome 2, at the site from 24707922 to 24707956 aligned with the reference sequence of NC-008395.1 (Oryza sativa genome project-ID 9521). There were 20 genes located within 100 kb of the insertion site (from chr2:24607922 to chr2:24807956). The expression features of these 20 genes were as follows: 2 genes were differentially expressed and 4 genes (Os02g0618700, Os02g0620500, Os02g0620800, and Os02g0621400) were found to be unexpressed in both KMD and XS11 rice (Table 1. □), the other 14 genes were not significantly changed in expression levels (fold changes between 0.5 and 2.0, Table 1). Among the two differentially expressed genes, Os02g0621100, encoding a protein containing a Pre-SET motif (a zinc-binding motif), was up-regulated (Table 1. ▲) and Os02g0621800, encoding a long-chain fatty alcohol dehydrogenase family protein, was down-regulated (Table 1. ▼).
Differentially expressed genes distributed over rice chromosomes
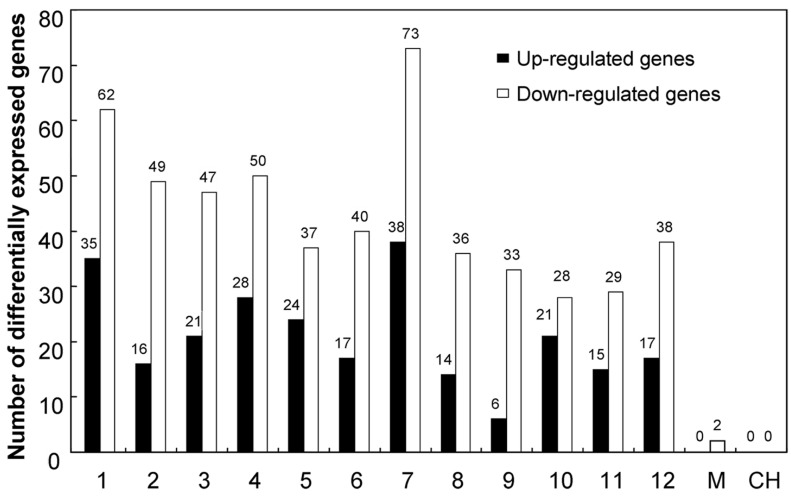
Genes differentially expressed in KMD rice were distributed throughout the rice chromosomes (Figure 1 and Supplementary Material: Table S3). Sixty-five differentially expressed genes (16 up and 49 down) were located in chr2, in which the exogenous cry1Ab was inserted. More differentially expressed genes were found in chr1 (35 up + 62 down = 97); chr3 (21 up + 47 down = 68), and chr7 (38 up + 73 down = 112). Differentially expressed genes were also found in all other rice chromosomes. Interestingly, the two genes located on the mitochondrial chromosome were both down-regulated in KMD rice. Some differentially expressed genes were shown to be two-copy or multi-copy in the rice genome, so a total of 778 differential transcripts were generated from 680 differentially expressed rice transcripts and subjected to chromosome distribution analysis.
Figure 1.
Differentially expressed genes of KMD rice distributed over all rice chromosomes. 1-12: rice chromosome; M: mitochondral chromosome; CH: chloroplast chromosome.
Differentially expressed genes of KMD rice implicated in certain biological processes
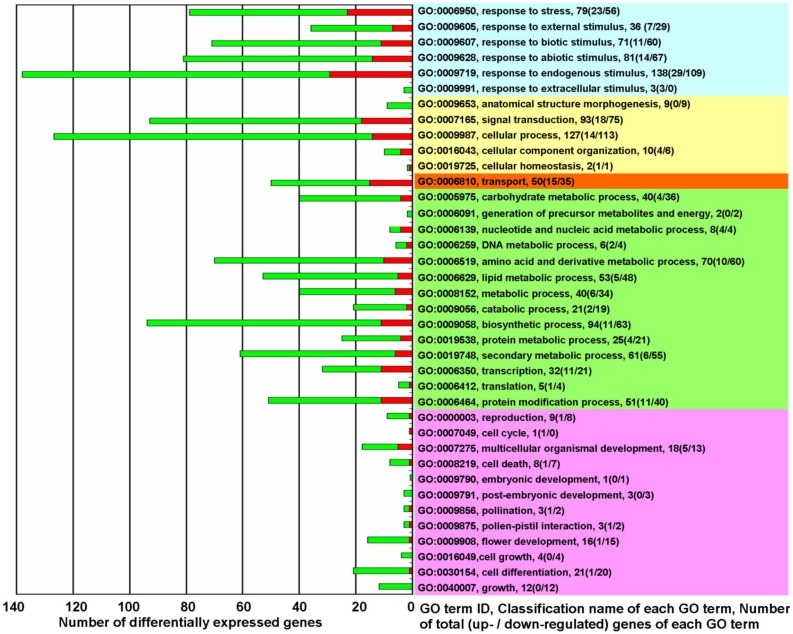
GO term classification analysis for biological processes was performed to determine which differentially expressed genes belonged to which GO terms. Except for 372 unclassified genes, the differentially expressed genes were mainly implicated in the following 5 types of biological processes (Figure 2 and Supplementary Material: Table S4): The first type was the processes of response to various sorts of stresses and stimuli (Figure 2, light blue). The second type was processes involved in plant physiological regulation and maintenance (Figure 2, light yellow). The third type was the processes involved in the directed movement of substances (Figure 2, light red). The fourth type was processes involved in metabolism (Figure 2, light green). The processes of transcription (GO:0006350), translation (GO:0006412), and protein modification (GO:0006464) are classified as metabolic process, but these three processes, coupled with the signal transduction process (GO:0007165), differ from other metabolic processes in that they play essential roles in the initiation of all changed processes. The fifth type was processes associated with plant growth and development (Figure 2, light purple).
Figure 2.
The biological functions of differentially expressed genes in KMD rice★. Red square: Up-regulated genes (fold change ≥ 2.0); Green square: down-regulated genes (fold change ≤ 0.5). Right side - Light Blue: processes for response to various of stress/stimulus; Yellow: processes involved in plant physiological regulation and maintenance; Orange: processes for the directed movement of substances; Green: processes involved in metabolism; Pink: processes associated with plant growth and development. ★ please note that of the 19420 detected probes, accounting for 34.98% of all rice probes on Affymetrix GeneChip® Rice Genome Array, 718 differentially expressed probes were selected with a cut-off q-value ≤ 0.05 for 3 biological replicates and with fold change ≥ 2.0 or ≤ 0.5. There were 34 genes that were represented by 2 probes and 2 genes that were represented by 3 probes. Thus, the 718 probes represented 680 differentially expressed genes. In which 372 unclassified differentially expressed genes (144 up-regulated and 228 down-regulated) were excluded from this figure. GO term classification analysis for biological processes, based on fold change of differentially expressed genes, was performed with FDR multiple-test correction (P value cutoff = 0.05).
Changes in pathways implicated in plant stress and defense responses
123 changed pathways were found when the 680 changed expression genes were analyzed with the Plant MetGenMAP system (Supplementary Material: Table S5). When selected using the false discovery rate (FDR) corrected P value at a threshold of 0.05, most of the changed pathways were cut off. Only 17 pathways changed significantly (Table 2). Among those significantly changed pathways, 8 types of pathway were directly implicated in plant stress and defense responses. This was found to be a prominent feature of gene expression in KMD seedlings. These significantly changed pathways, such as the jasmonic acid biosynthesis pathway; cellulose biosynthesis 35; betanidin degradation 36; phenylpropanoid biosynthesis 37; salicylate biosynthesis 38; 13-LOX and 13-HPL pathway 39; divinyl ether biosynthesis II (13-LOX) 40; and suberin biosynthesis 41, have been previously reported to be involved in stress and defense responses.
Table 2.
Two types of significantly changed pathways in KMD rice.
| Type of pathways | Pathway name | P value (corrected with FDR method) |
|---|---|---|
| Pathways implicated in plant stress/defense responses | jasmonic acid biosynthesis | 0.0007 |
| cellulose biosynthesis | 0.0007 | |
| betanidin degradation | 0.0017 | |
| phenylpropanoid biosynthesis, initial reactions | 0.01408 | |
| salicylate biosynthesis | 0.01408 | |
| 13-LOX and 13-HPL pathway | 0.0243 | |
| divinyl ether biosynthesis II (13-LOX) | 0.0243 | |
| suberin biosynthesis | 0.04152 | |
| Pathways associated with amino acid metabolism | S-adenosylmethionine biosynthesis | 1.45E-05 |
| S-adenosyl-L-methionine cycle | 2.47E-05 | |
| methionine biosynthesis I | 9.46E-05 | |
| methionine biosynthesis II | 0.00296 | |
| superpathway of lysine, threonine and methionine biosynthesis II | 0.01284 | |
| ethylene biosynthesis from methionine | 0.04077 | |
| methionine degradation I (to homocysteine) | 0.04077 | |
| methionine salvage pathway | 0.04152 | |
| histidine biosynthesis I | 0.01408 |
Changes in pathways associated with amino acid metabolism
In addition to the significantly changed pathways implicated in plant stress/defense response, the other 9 kinds of significantly changed pathway were all directly associated with plant amino acid metabolism. Those included the S-adenosylmethionine biosynthesis pathway; the S-adenosyl-L-methionine cycle; methionine biosynthesis I; methionine biosynthesis II; the superpathway of lysine, threonine, and methionine biosynthesis II; ethylene biosynthesis from methionine; methionine degradation I (to homocysteine); methionine salvage pathway; and histidine biosynthesis I. Methionine is a nutritionally essential amino acid 42, 43 and plays important roles in protein construction, the initiation of mRNA translation and indirect regulation of a variety of cellular processes 44. Histidine has been found to play an important role in regulation of biosynthesis of other unrelated amino acids in chelation and transport of metal ions and in plant reproduction and growth 45. Significantly changed in amino acid metabolic pathways are another prominent gene expression feature in the KMD rice seedlings.
Changes in amino acid metabolism
As mentioned above, several biological pathways associated with the metabolism of a few kinds of amino acids were significantly changed in GM KMD rice. Following this indications, the amino acid composition of KMD rice leaves were analyzed using iTRAQ®-LC-MS/MS. The results showed that of the 24 kinds of detectable amino acids, 10 kinds of protein amino acids, specifically L-phenylalanine, L-methionine, L-glutamine, L-leucine, L-valine, L-aspartic acid, L-proline, L-alanine, L-isoleucine, and L-histidine, were significantly changed in 30-day seedling leaves of KMD rice (Table 3). Interestingly, γ-amino-n-butyric acid, a 4-carbon non-protein amino acid, reported to be a typical stress response amino acid in plants 45, 46, was also found to be significantly changed in KMD rice relative to XS11 (Table 3).
Table 3.
Changes in total amino acid composition of KMD rice.
| Amino acid | Concentration (μmol/g FW) | P value | |
|---|---|---|---|
| KMD (means ± s.e.m) | XS11 (means ± s.e.m) | ||
| L-phenylalanine | 33.7 ± 1.57 | 24.61 ± 1.66 | 3.93E-05 |
| L-methionine | 5.02 ± 0.38 | 2.24 ± 0.57 | 1.78E-03 |
| L-glutamine | 19.18 ± 1.5 | 14.92 ± 2.00 | 4.80E-03 |
| L-leucine | 57.73 ± 3.15 | 39.66 ± 0.89 | 5.34E-03 |
| L-valine | 57.82 ± 1.45 | 42.07 ± 3.39 | 6.25E-03 |
| γ-amino-n-butyric acid | 5.39 ± 0.81 | 3.18 ± 0.36 | 1.37E-02 |
| L-aspartic acid | 60.26 ± 3.08 | 40.06 ± 5.56 | 0.020 |
| L-proline | 44.71 ± 4.51 | 34.75 ± 2.04 | 0.024 |
| L-alanine | 85.19 ± 8.46 | 64.70 ± 3.31 | 0.032 |
| L-isoleucine | 36.43 ± 2.87 | 26.92 ± 2.15 | 0.033 |
| L-histidine | 21.27 ± 2.99 | 14.37 ± 0.37 | 0.048 |
| L-glutamic acid | 50.97 ± 12.13 | 35.17 ± 5.17 | 0.067 |
| L-lysine | 53.34 ± 6.8 | 38.14 ± 4.78 | 0.072 |
| L-tryptophan | 9.55 ± 1.62 | 4.89 ± 0.78 | 0.069 |
| L-serine | 109.00 ± 21.67 | 55.39 ± 10.37 | 0.085 |
| glycine | 106.23 ± 17.88 | 69.36 ± 6.40 | 0.107 |
| L-threonine | 35.76 ± 3.89 | 31.40 ± 2.81 | 0.122 |
| L-Ornithine | 25.02 ± 11.89 | 8.10 ± 1.55 | 0.128 |
| L-tyrosine | 20.02 ± 3.62 | 13.18 ± 1.82 | 0.161 |
| ethanolamine (EtN) | 2.85 ± 0.47 | 2.23 ± 0.06 | 0.166 |
| L-arginine (Arg) | 29.00 ± 2.02 | 23.36 ± 2.79 | 0.166 |
| L-citrulline (Cit) | 2.70 ± 0.72 | 3.38 ± 0.22 | 0.283 |
| D,L-β-amino-isobutyric acid (bAib) | 1.50 ± 0.72 | 1.24 ± 0.22 | 0.478 |
| β-alanine (bAla) | 5.63 ± 0.76 | 5.38 ± 0.47 | 0.563 |
Student's t-tests were performed between 30-day seedling leaves of KMD rice and XS11 rice for three biological replicates. Results are shown as mean ± s.e.m. Bold text indicates significant differences between KMD and XS11 rice with P ≥ 0.05.
Discussion
Identification of unintended effects occurring in GM plants and evaluation of their negative influences on the agro-system and on food safety are an important aspect in risk assessment of GM plants 2. Traditionally, multiple approaches were used for GM plants safety assessment, which were mainly performed through comparative analysis in revealing the possibly alteration in GM plants at agronomic traits, environmental adaptability, chemical composition, and other desirable characteristics 47. Although these methods have been used extensively for the identification of unintended effects in GM plants and they play important roles in safety assessment, their methodological limitations for these targeted approach method will never be out of debates. The main cause of these discussions is the fact that only a restricted and biased selection of traits of GM plants can be analyzed, while many other traits changed by genetic modification may be omitted, possibly leaving unintended effects unrevealed 2, 48.
“Profiling” and “omics” technologies, such as transcriptomics, proteomics and metabolomics, are objective, comprehensive and non-selective analytical methods, which allow unbiased, non-targeted profiling of possible changes in GM plants at the transcription and metabolic levels 48. All these changes are the molecular basis for any possible unintended effects that may emerge in GM plants. With the development of these profiling and omics technologies, coupled with traditionally targeted analytical methods, an objective and impartial analytical system for the safety assessment of GM plants can be established. Only then the safety reports made by trained GM plant inspectors are likely to be accepted by most of the public.
GM KMD rice has been reported to show obvious unintended effects, specifically decreased disease resistance 15, 16. However, the molecular basis behind these unintended effects in KMD rice is still unknown. KMD rice may act as a suitable GM plant material to confirm that the new profiling and omics technologies are effective with respect to identification of unintended effects in GM plants.
In this study, with the aim of revealing the molecular basis of unintended effects in KMD rice and possible connecting unintended effects of KMD rice through identification of differentially expressed genes, comparative transcriptome analysis was performed for GM KMD rice and its parent line, XS11. The results showed that 8 kinds of pathways directly implicated in plant stress/defense responses were significantly changed in KMD rice (Table 2). Changes in plant stress/defense response pathways were considerable and appeared that may link to the reported unintended effects of KMD rice, such as increased susceptibility to rice brown spot mimic lesion disease and sheath blight disease 15, 16.
The results also showed that 9 kinds of pathways associated with a few kinds of amino acid metabolism were significantly changed in GM KMD rice (Table 2). The histidine biosynthesis pathway may affect the biosynthesis of other unrelated amino acids. The histidine biosynthesis pathway has been reported to play an important role in regulation of biosynthesis of quite a few unrelated amino acids, such as alanine, aspartate, glutamate, phenylalanine, proline, threonine, tryptophan, lysine, tyrosine, and valine 45, 49. All these indications suggest that any differences in the amino acid composition of GM KMD rice and its non-GM comparable rice line of XS11 merit evaluation. Sensitive amino acid analysis, performed with iTRAQ®-LC-MS/MS, showed that 10 kinds of protein amino acids, such as L-phenylalanine, L-methionine, L-glutamine, L-leucine (Leu), L-histidine, L-glutamine, L-valine, L-aspartic acid, L-proline, L-alanine, L-isoleucine were significantly changed in 30-day seedling leaves of KMD rice relative to XS11 (Table 3). Interestingly, the level a typical plant stress response amino acid, γ-amino-n-butyric acid (GABA), was also found significantly changed in 30-day seedling leaves of KMD rice (Table 3) 46.
As mentioned above, the results of comparative transcriptome analysis not only revealed the molecular basis, especially at biological pathway levels, of the reported unintended effects of KMD rice but also provided direct molecular indications toward other unintended effects in KMD rice. We suggest that comparative transcriptome analysis can serve as an unbiased, comprehensive, and effective technique for the discovery of unintended effects in GM plants through the processes of collecting clues from differentially expressed genes, using these genes to identify potentially altered biology processes and changed pathways, and finally to identify unintended effects in GM plants.
The reasons why certain genes are differentially expressed in KMD rice are still not clear. The first reason may be related to the high-level expression of redundant marker genes, such as the beta glucuronidase gene, selective gene of hygromycin B phosphotransferase gene, and transformed regulation elements gene of artificial sequences for nopaline synthase gene (Table 1). The high-level expression of these redundant genes in GM KMD rice may act as a physiological stressor, altering expression of stress response genes, and bringing on changes in the expression of genes involved in plant physiological regulation and maintenance, metabolism, and substance transport. This interpretation is likely to be supported by the results of biological processes GO term classification analysis of differentially expressed genes in KMD rice (Figure 2 and Supplementary Material: Table S4) and also by the discovery that a typical stress response amino acid, γ-amino-n-butyric acid (GABA), was synthesized at a significantly higher level in 30-day seedling leaves of KMD rice comparing than in XS11 rice (Table 3). Second, near the insertion site in KMD rice, the Os02g0621100 gene, encoding a protein containing a Pre-SET motif (a zinc binding motif), was found to be up-regulated (Table 1. ▲). This pre-SET motif protein contained 9 conserved cysteines that coordinate three zinc ions and plays a role in stabilizing SET domains, while SET domain proteins are lysine methyltransferase enzymes, play fundamental roles in epigenetic regulation of gene activation and silencing in all eukaryotes 50. Thus, changes in the expression of Pre-SET motif protein may influence the activity of SET domain protein, subsequently influencing the expression of genes dependent on SET domain protein. We still cannot be sure if there are variations in KMD rice. If any somatic variations occur in KMD rice during genetic manipulation through tissue culture procedures, these somatic variations may also alter the expression of some genes in KMD rice.
There are many reasons to worry about unintended effects in GM plants. The random insertion of exogenous genes may disrupt or disturb the expression of intrinsic plant genes at or near insertion sites 51. High-level expression of these exogenous genes, such as target genes, reporter genes, selectable marker genes, and other transformed, regulated elements, may place extra physiological stress on the plant's normal metabolism 52. Genetically transformed manipulation through tissue culture procedures may also cause somatic variation in plant cells 53. Various transcriptomic comparison studies have reported that only a few differentially expressed genes were identified between GM and non-GM comparable plants, and these changed expression genes were insufficient to cause any prominent unintended effects in GM plants 4, 17, 25-27, 41, 54. All of these results may reflect the actuality that most GM plants with various unintended effects are discarded during the strict selection procedures. Only healthy GM plant lines with suitable insertion sites and non-genetic variations are kept and used for further study and application.
These limited influence results were likely misinterpreted that no unintended effects could occur in GM plants. However the results of this study provide an actual case for the identification of unintended effects in GM plants by comparative transciptome analysis. Unintended effects are the other side of the development of GM plants. There is no reason to ignore the fact that some unintended effects may have adverse influence on agro systems and on food safety. Especially at present, the technologies for limitation of unintended effects, such as targeted insertion of exogenous genes, concise regulation the expression of exogenous genes, omission of redundant marker genes or transformed regulation elements, and transformed manipulation independent of tissue culture procedures, are still underdeveloped 55. On the other hand, strict safety assessment, both in supervision technologies and policies for GM plants, will promote the improvement of plant genetic transformation technologies in the respects mentioned above and serve to guarantee reasonably safe progress.
Conclusion
The presented results showed that differentially expressed transcripts of GM KMD rice line were distributed over rice chromosomes and implicated in certain biological processes, which related to the unintended effects of GM KMD rice line. So we further proved that differentially expressed transcripts were the molecular basis for cause of unintended effects of GM plants, we also prospected that the genome transcripts profiling method might be a basis and useful way for identification of unintended effects of GM plants and will play an important role in safety assessment of GM plants.
Results in this case mentioned that unintended effects are the other side of the development of GM plants, which may be caused by insert event, high-level expression of extra foreign genes, and even somatic variations occurrence during genetic manipulation through tissue culture procedures. Strict safety assessment, both in supervision technologies and policies for GM plants, will promote the improvement of plant genetic transformation technologies in respects mentioned above and serve to guarantee for promoting the development of safety GM plants with new obtained traits beneficial to plants or fitting requirements of human beings.
Supplementary Material
Fig.S1 and Table S1-S5.
Acknowledgments
We are grateful to Professor Ye GY for kindly providing KMD and Xiushui rice seeds. This work was supported by the National Genetically Modified Organisms Breeding Major Project (2011ZX08011-001) and Special Funding of State Key Laboratory of BPDIP, China (SKL2010SR01).
Abbreviation
- Bt
Bacillus thuringiensis
- cRNA
complementary RNA
- GO
gene ontology
- GM
genetically modified
- KMD
Kemingdao
- Q-RT-PCR
quantitative real-time-PCR
- XS11
Xiushui11.
References
- 1.Kotchoni OS, Gachomo EW, Mwangi M. Commercial production of genetically modified crops: a prognosis towards global acceptance. Int J Agri Biol. 2005;7:681–688. [Google Scholar]
- 2.FAO/WHO. Food derived from modern biotechnology, 2nd ed. ISBN 978-92-5-105914-2. 2009.
- 3.Sorochinskii BV, Burlaka OM, Naumenko VD. et al. Unintended effects of genetic modifications and methods of their analysis in plants. Cytol Genet. 2011;45:324–332. [PubMed] [Google Scholar]
- 4.Batista R, Saibo N, Lourenco T. et al. Microarray analyses reveal that plant mutagenesis may induce more transcriptomic changes than transgene insertion. Proc Natl Acad Sci USA. 2008;105:3640–3645. doi: 10.1073/pnas.0707881105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M, Shelton A, Ye GY. Insect-resistant genetically modified rice in China: from research to commercialization. Annu Rev Entomol. 2011;56:81–101. doi: 10.1146/annurev-ento-120709-144810. [DOI] [PubMed] [Google Scholar]
- 6.Ye GY, Shu QY, Yao HW. et al. Field evaluation of resistance of transgenic rice containing a synthetic cry1Ab gene from Bacillus thuringiensis Berliner to two stem borers. J Econ Entomol. 2001;94:271–276. doi: 10.1603/0022-0493-94.1.271. [DOI] [PubMed] [Google Scholar]
- 7.Shu QY, Ye GY, Cui HR. et al. Transgenic rice plants with a synthetic cry1Ab gene from Bacillus thuringiensis were highly resistant to eight lepidopteran rice pest species. Mol Breed. 2000;6:433–439. [Google Scholar]
- 8.Tu J, Zhang G, Datta K. et al. Field performance of transgenic elite commercial hybrid rice expressing Bacillus thuringiensis-endotoxin. Nat Biotechnol. 2000;18:1101–1104. doi: 10.1038/80310. [DOI] [PubMed] [Google Scholar]
- 9.High SM, Cohen MB, Shu QY. et al. Achieving successful deployment of Bt rice. Trends Plant Sci. 2004;9:286–292. doi: 10.1016/j.tplants.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Cheng XY, Sardana R, Kaplan H. et al. Agrobacterium-transformed rice plants expressing synthetic cry1A(b) and cry1A(c) genes are highly toxic to striped stem borer and yellow stem borer. Proc Natl Acad Sci USA. 1998;95:2767–2772. doi: 10.1073/pnas.95.6.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babekova R, Funk T, Pecoraro S. et al. Development of an event-specific Real-time PCR detection method for the transgenic Bt rice line KMD1. Eur Food Res Technol. 2009;228:707–716. [Google Scholar]
- 12.Wang ZG, Shu QY, Ye GY. et al. Genetic analysis of resistance of Bt rice to stripe stem borer (Chilo suppressalis) Euphytica. 2002;123:379–386. [Google Scholar]
- 13.Wu G, Cui H, Ye G. et al. Inheritance and expression of the cry1Ab gene in Bt (Bacillus thuringiensis) transgenic rice. Theor Appl Gene. 2002;104:727–734. doi: 10.1007/s001220100689. [DOI] [PubMed] [Google Scholar]
- 14.Xie JJ, Wang XF, Peng YF. Side sequence of exogenous insert of transgene paddy strain Kemingdao 1. Patent CN 101240278A. 2007.
- 15.Tang J, Ye G, Yang B. et al. Preliminary study on rice brown spot mimic lesion virulence on Bt gene transformed rice Kemingdao. Chinese J Rice Sci. 2001;15:317–319. [Google Scholar]
- 16.Yuan XP, Zhao XH, Tang J. et al. Resistance evaluation of cry1Ab gene transgenic rice Kemingdao to rice bacterial leaf blight and sheath blight disease. Plant Protection. 2003;29:27–28. [Google Scholar]
- 17.El Ouakfaoui S, Miki B. The stability of the Arabidopsis transcriptome in transgenic plants expressing the marker genes nptII and uidA. Plant J. 2005;41:791–800. doi: 10.1111/j.1365-313X.2005.02350.x. [DOI] [PubMed] [Google Scholar]
- 18.Abdeen A, Miki B. The pleiotropic effects of the bar gene and glufosinate on the Arabidopsis transcriptome. Plant Biotech J. 2009;7:266–282. doi: 10.1111/j.1467-7652.2009.00398.x. [DOI] [PubMed] [Google Scholar]
- 19.Abdeen A, Schnell J, Miki B. Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genomics. 2010;11:69.. doi: 10.1186/1471-2164-11-69. doi:10.1186/1471-2164-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montero M, Coll A, Nadal A. et al. Only half the transcriptomic differences between resistant genetically modified and conventional rice are associated with the transgene. Plant Biotechnol J. 2011;9:693–702. doi: 10.1111/j.1467-7652.2010.00572.x. [DOI] [PubMed] [Google Scholar]
- 21.Dubouzet JG, Ishihara A, Matsuda F. et al. Integrated metabolomic and transcriptomic analyses of high-tryptophan rice expressing a mutant anthranilate synthase alpha subunit. J Exp Bot. 2007;58:3309–3321. doi: 10.1093/jxb/erm179. [DOI] [PubMed] [Google Scholar]
- 22.Cheng KC, Beaulieu J, Iquira E. et al. Effect of transgenes on global gene expression in soybean is within the natural range of variation of conventional cultivars. J Agric Food Chem. 2008;56:3057–3067. doi: 10.1021/jf073505i. [DOI] [PubMed] [Google Scholar]
- 23.Gregersen PL, Brinch-Pedersen H, Holm PB. A microarray-based comparative analysis of gene expression profiles during grain development in transgenic and wild type wheat. Transgenic Res. 2005;14:887–905. doi: 10.1007/s11248-005-1526-y. [DOI] [PubMed] [Google Scholar]
- 24.Baudo MM, Powers SJ, Mitchell RAC. et al. Establishing Substantial Equivalence: Transcriptomics. Methods Mol Biol. 2009;478:247–272. doi: 10.1007/978-1-59745-379-0_15. [DOI] [PubMed] [Google Scholar]
- 25.Coll A, Nadal A, Collado R. et al. Natural variation explains most transcriptomic changes among maize plants of MON810 and comparable non-GM varieties subjected to two N-fertilization farming practices. Plant Mol Biol. 2010;73:349–362. doi: 10.1007/s11103-010-9624-5. [DOI] [PubMed] [Google Scholar]
- 26.Coll A, Nadal A, Palaudelmas M. et al. Lack of repeatable differential expression patterns between MON810 and comparable commercial varieties of maize. Plant Mol Biol. 2008;68:105–117. doi: 10.1007/s11103-008-9355-z. [DOI] [PubMed] [Google Scholar]
- 27.Kogel KH, Voll LM, Schaefer P. et al. Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances. Proc Natl Acad Sci USA. 2010;107:6198–6203. doi: 10.1073/pnas.1001945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu Q, Ye G, Cui H. et al. Development of transgenic Bacillus thuriengiensis rice resistant to rice stem borers and leaf folders. J Zhejiang Agri Uni. 1998;24:579–580. [Google Scholar]
- 29.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 30.Joung JG, Corbett AM, Fellman SM. et al. Plant MetGenMAP: An Integrative Analysis System for Plant Systems Biology. Plant Physiol. 2009;151:1758–1768. doi: 10.1104/pp.109.145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc B: Methodol. 1995;57:289–300. [Google Scholar]
- 32.Boyle EI, Weng SA, Gollub J. et al. GO::TermFinder -open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayer RR, Cherry JH, Rhodes D. Effects of heat shock on amino acid metabolism of cowpea cells. Plant Physiol. 1990;94:796–810. doi: 10.1104/pp.94.2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaspar H, Dettmer K, Chan Q. et al. Urinary amino acid analysis: A comparison of iTRAQ®-C-MS/MS, GC-MS, and amino acid analyzer. J Chromatogr B. 2009;877:1838–1846. doi: 10.1016/j.jchromb.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasternack C. Jasmonates, an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sepulveda-Jimenez G, Rueda-Benitez P, Porta H. et al. A red beet (Beta vulgaris) UDP-glucosyltransferase gene induced by wounding, bacterial infiltration and oxidative stress. J Exp Bot. 2005;56:605–611. doi: 10.1093/jxb/eri036. [DOI] [PubMed] [Google Scholar]
- 37.Humphreys JM, Chapple C. Rewriting the lignin road map. Curr Opin Plant Biol. 2002;5:224–229. doi: 10.1016/s1369-5266(02)00257-1. [DOI] [PubMed] [Google Scholar]
- 38.Shah J. The salicylic acid loop in plant defense. Curr Opin Plant Biol. 2003;6:365–371. doi: 10.1016/s1369-5266(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 39.Noordermeer MA, Veldink GA, Vliegenthart JFG. Fatty acid hydroperoxide lyase: A plant cytochrome P450 enzyme involved in wound healing and pest resistance. Chembiochem. 2001;2:494–504. doi: 10.1002/1439-7633(20010803)2:7/8<494::AID-CBIC494>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 40.Fammartino A, Cardinale F, Goebel C. et al. Characterization of a divinyl ether biosynthetic pathway specifically associated with pathogenesis in tobacco. Plant Physiol. 2007;143:378–388. doi: 10.1104/pp.106.087304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiber L. Transport barriers made of cutin, suberin and associated waxes. Trends in Plant Sci. 2010;15:546–553. doi: 10.1016/j.tplants.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Amir R. Towards improving methionine content in plants for enhanced nutritional quality. Funct Plant Sci Biotechnol. 2008;2:36–46. [Google Scholar]
- 43.Galili G, Amir R, Hoefgen R. et al. Improving the levels of essential amino acids and sulfur metabolites in plants. Biol Chem. 2005;386:817–831. doi: 10.1515/BC.2005.097. [DOI] [PubMed] [Google Scholar]
- 44.Amir R. Current understanding of the factors regulating methionine content in vegetative tissues of higher plants. Amino Acids. 2010;39:917–931. doi: 10.1007/s00726-010-0482-x. [DOI] [PubMed] [Google Scholar]
- 45.Stepansky A, Leustek T. Histidine biosynthesis in plants. Amino Acids. 2006;30:127–142. doi: 10.1007/s00726-005-0247-0. [DOI] [PubMed] [Google Scholar]
- 46.Allan WL, Simpson JP, Clark SM, Shelp BJ. γ-Hydroxybutyrate accumulation in Arabidopsis and tobacco plants is a general response to abiotic stress: putative regulation by redox balance and glyoxylate reductase isoforms. J Exp Bot. 2008;59:2555–2564. doi: 10.1093/jxb/ern122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cellini F, Chesson A, Colquhoun I. et al. Unintended effects and their detection in genetically modified crops. Food and Chemical Toxicology. 2004;42:1089–1125. doi: 10.1016/j.fct.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Kuiper HA, Kok EJ, Engel KH. Exploitation of molecular profiling techniques for GM food safety assessment. Curr Opin Biotechnol. 2003;14:238–243. doi: 10.1016/s0958-1669(03)00021-1. [DOI] [PubMed] [Google Scholar]
- 49.Guyer D, Patton D, Ward E. Evidence for cross-expression in cross-pathway regulation of metabolic gene-expression in plants. Proc Natl Acad Sci USA. 1995;92:4997–5000. doi: 10.1073/pnas.92.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forsbach A, Schubert D, Lechtenberg B. et al. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol Biol. 2003;52:161–176. doi: 10.1023/a:1023929630687. [DOI] [PubMed] [Google Scholar]
- 52.Miki B, Abdeen A, Manabe Y. et al. Selectable marker genes and unintended changes to the plant transcriptome. Plant Biotechnol J. 2009;7:211–218. doi: 10.1111/j.1467-7652.2009.00400.x. [DOI] [PubMed] [Google Scholar]
- 53.Latham JR, Wilson AK, Steinbrecher RA. The mutational consequences of plant transformation. J Biomed Biotechnol. 2006. DOI 10.1155/JBB/2006/25376. [DOI] [PMC free article] [PubMed]
- 54.Barros E, Lezar S, Anttonen MJ. et al. Comparison of two GM maize varieties with a near-isogenic non-GM variety using transcriptomics, proteomics and metabolomics. Plant Biotechnol J. 2010;8:436–451. doi: 10.1111/j.1467-7652.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 55.Tuteja N, Verma S, Sahoo RK. et al. Recent advances in development of marker-free transgenic plants: Regulation and biosafety concern. J Biosciences. 2012;37:167–197. doi: 10.1007/s12038-012-9187-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.S1 and Table S1-S5.